Abstract
Introduction
Head and neck squamous cell carcinomas (HNSCC) arise in the mucosal lining of the upper aerodigestive tract. Risk factors are exogenous carcinogen exposure, human papillomavirus (HPV) infection, and genetic predisposition such as Fanconi anemia (FA). Clinically, tumors are stratified based on stage, site and HPV-status. The majority of HPV-positive and -negative HNSCC is characterized by frequent copy number (CN) changes and an abrogated p53-pathway. A third genetically-defined HPV-negative subclass of HNSCC is emerging: tumors that lack gross chromosomal changes (CN-silent), are mostly TP53-proficient, and have a relatively favorable prognosis.
Methods
A representative panel of HPV-positive, HPV-negative and FA-HNSCC-derived cell lines was genetically characterized.
Results
Despite apparent differences in etiology, FA-HNSCC cell lines show comparable genetic alterations as sporadic non-FA-HNSCC-derived cell lines.
Furthermore, we identified a near diploid CN-silent HPV-negative HNSCC line: VU-SCC-040. Molecular characterization uncovers the absence of TP53 mutations, a functional p53-pathway and a CASP8 mutation. TP53 gene knockout using CRISPR-Cas9 resulted in resistance to MDM2 inhibition. Whereas p53-status is often proposed as a predictive biomarker for treatment response, TP53-knockout did not change sensitivity to cisplatin, Chk1 and Wee1 inhibition. Additionally, 84 CN-silent tumors were identified in the HNSCC PanCancer cohort and shown to be enriched for female gender, HRAS and CASP8 mutations.
Conclusion
HPV-negative and FA-derived HNSCC share comparable CN-profiles and mutation patterns. In contrast, a subclass of CN-silent, HPV-negative and TP53 wild-type HNSCC separates from the majority of HNSCC tumors. We show that VU-SCC-040 is a HNSCC cell model representative of this subclass.
Keywords: Head and neck squamous cell carcinoma, Low-coverage whole genome sequencing, Target-enrichment sequencing, Copy number silent, TP53 wild-type, p53 pathway, Fanconi anemia, Targeted treatment
Introduction
Every year, 500,000 new patients are diagnosed with head and neck squamous cell carcinoma (HNSCC), which develops in the mucosal lining of the oral cavity, pharynx and larynx [1,2]. HNSCC is classified into two molecular subgroups: HPV-positive and HPV-negative tumors [3]. Infection with a high risk human papillomavirus (hrHPV) may result in the development of HPV-positive HNSCC, predominantly in the oropharynx [4], while smoking and concomitant excessive alcohol consumption are the risk factors of HPV-negative HNSCC [5]. Although HNSCC is usually caused by these exogenous factors, patients suffering from the rare genetic predisposition syndrome Fanconi anemia (FA) also carry a high risk for squamous cell carcinomas [6]. FA is caused by germline mutations in one of the 22 Fanconi genes (FANCA-G, FANCI-J, FANCL-W), a pathway essential for interstrand-crosslink removal and DNA repair [7–9]. Cells of FA-patients are characterized by chromosomal breakage due to genetic instability [6,7].
The loss of function of TP53 and CDKN2A occurs early in the oncogenesis of HPV-negative HNSCC [2,10]. These genes are involved in cell cycle regulation and genetic stability, and loss results in considerable genetic aberrations in the majority of HNSCC [11]. This induces a typical pattern of chromosomal gains and losses and aneuploidy that is also observed in HNSCC cell lines [12,13]. Remarkably, a small subset of HPV-negative HNSCC tumors harbors a copy number (CN) silent genetic profile. These tumors are often TP53 wild-type and have a better prognosis than CN-high tumors [5,11,13]. It has therefore been proposed that HPV-negative tumors may be divided into CN-high and CN-silent subclasses [5,13] The latter exceptional subgroup has been recognized recently, and appears to be characterized by HRAS mutations and/or CASP8 inactivation [11], while the TP53 gene is mostly wild-type, although it is unknown whether the p53-pathway is functional in these tumors.
Here, we investigated and characterized a HNSCC cell line panel of the various described etiological origins using genomics, and uncover and characterize a CN-silent TP53 wild-type HNSCC cell line.
Materials and methods
Cell line cultures
A de-identified HNSCC specimen from the oral cavity of an individual diagnosed with FA was obtained through the National Disease Research Interchange (NDRI). The derivative CCH-FAHNSCC-2 cell line was cultured initially on irradiated J2–3T3 feeder cells as described previously for skin keratinocytes [14], and then adapted to growth on plastic in DMEM (Lonza, BE12–707F) substituted with 5% FBS (FBS, Biological Industries, 04–007-1A) and 2 mmol/L L-glutamine (Lonza, BE17–605F).
All cell lines (Table 1) used have been described previously and cultured accordingly [15]. Cell lines were authenticated by visual inspection and on indication by genetic markers including TP53 mutations, HPV-status and were cultured for a maximum of 4 months. Furthermore, all lines were regularly tested for mycoplasma (Mycoalert, Lonza, Verviers, Belgium), and were always negative.
Table 1:
panel of HNSCC cell lines used for low coverage whole-genome sequencing
| Cell line | Gender | Stage | Primary tumor site | HPV status | |
|---|---|---|---|---|---|
| HPV-NEG HNSCC | FaDub | Male | NA | Hypopharynx | Negative |
| UM-SCC-6 | Male | T2N0 | Base of tongue | Negative | |
| UM-SCC-11B‡ | Male | T2N2a | Supraglottic larynx | Negative | |
| UM-SCC-14A | Female | T1N0 | Floor of mouth | Negative | |
| UM-SCC-14Bb ‡ | Female | T1N0 | Floor of mouth | Negative | |
| UM-SCC-14C‡ | Female | T2N0 | Floor of mouth | Negative | |
| UM-SCC-22A | Female | T2N1 | Hypopharynx | Negative | |
| UM-SCC-22B‡ | Female | T2N1 | Hypopharynx | Negative | |
| UM-SCC-35 | Male | T4N1 | Tonsillar fossa | Negative | |
| UM-SCC-38 | Male | T2N2a | Tonsillar pillar | Negative | |
| VU-SCC-040a | Female | T3N0 | Tongue | Negative | |
| VU-SCC-096 | Male | T4N1 | Retromolar trigone | Negative | |
| VU-SCC-120 | Female | T3N1 | Tongue | Negative | |
| VU-SCC-OEb ‡ | Male | Lymph node metastasis | Floor of mouth | Negative | |
| VU-SCC-RO | Male | T3N2b | Oropharynx | Negative | |
| HPV-POS HNSCC | UD-SCC-2 | Male | NA | Hypopharynx | Positive |
| UM-SCC-47 | Male | T3N1 | Tongue | Positive | |
| UM-SCC-104‡ | Male | NA | Floor of mouth | Positive | |
| UT-SCC-45 | Male | T3N1 | Floor of mouth | Positive | |
| VU-SCC-147b | Male | T4N2c | Floor of mouth | Positive | |
| FA-HNSCC | CCH-FAHNSCC-2 | Female | NA | NA | Negative |
| VU-SCC-1131‡ | Female | T4N2b | Floor of mouth | Negative | |
| VU-SCC-1365 | Male | NA | Mouth mucosa | Negative | |
| VU-SCC-1604 | Female | NA | Tongue | Negative | |
TP53 wild-type cell line
Cell lines from a sporadic HNSCC tumor with a de novo Fanconi gene mutation (Stoepker et al., 2015)
Local recurrences of primary tumor
NA - not annotated
Low-coverage whole-genome sequencing (lcWGS)
Procedures of DNA isolation and preparation for lcWGS sequencing on a HiSeq 2500 System (Illumina) have been described previously [10]. Reads were mapped to the human genome (hg19) using BWA-MEM [16], binned in 100 kilobase bins, filtered, and normalized using the QDNAseq package (version 1.14.0) [17]. Segments were determined by DNAcopy [18]. For UM-SCC-35, the CN-profiles were obtained from offtarget reads obtained from exome sequencing, and processed with CopywriteR.For assessment of CDKN2A (p16) losses, an additional analysis using a 10 kb bin size was used, to detect small focal deletions of CDKN2A. A focal loss was characterized as ≤ 3 Mb.
The ACE package [19] was used for ploidy estimations and plotting of absolute CN-profiles (Figure S1). The ACE-models were manually inspected and curated when required.
The frequency plots of CN-aberrations were obtained using the frequencyPlot function of the CGHbase R package [20].
For HPV and EBV viral genome mapping after hg19 alignment, the unmapped (i.e. non-human) reads were subsequently aligned to HPV types 16, 18, 31, 33 and 52 and the EBV genomes obtained from the NIH, and reads were enumerated as a measure of viral presence.
HaloPlex targeted sequencing
HaloPlex targeted sequencing (Agilent, HiSeq 2500 system (Illumina)) of the coding exons of 12 genes frequently mutated in HNSCC has been described previously [10]. In short, sequencing reads were analyzed using Agilent software SureCall version 3.5.1.46 with default Haloplex settings. Duplicates within the sequencing coverage were not removed, per Agilent’s instructions for Haloplex sequencing. Next, all variants were compared to 1000Genomes Phase 3, using a EU_AF 0.01 threshold to discard known SNP variants using Oncotator annotation. Overall, the mean sequencing coverage of the twelve genes in all cell lines was 13,977 (ranging from 10,290 – 16,689). For TP53 specifically, a mean sequencing coverage of 10,757 was obtained (ranging from 117 – 48,606).
CRISPR-Cas9 TP53-knockout
VU-SCC-040 was transduced using polybrene enhancement with the Lenti-Cas9–2A-blast vector with blasticidin S selection marker [21]. After blasticidin S (Sigma, 15205) selection, Edit-R TP53 crRNA (Dharmacon, CR-003329–01) was transiently transfected with tracrRNA (Dharmacon U-002000–05) using Dharmafect 1 transfection reagent (Dharmacon, T-2001). Selection for the TP53-knockout cells was established with 10 μM Nutlin-3a (MedChem, HY-10029) treatment. Mutation status was obtained by Sanger sequencing with BigDye™ Direct Cycle Sequencing Kit (Applied Biosystems, 4458687) according to manufacturer’s protocol. Sequencing primers: Forward TP53 5F M13 TGTAAAACGACGGCCAGTtgttcacttgtgccctgact and Reverse TP53 6R M13 CAGGAAACAGCTATGACCttaacccctcctcccagaga. Sequences were analyzed with TIDE version 2.0.1 [22].
RT-qPCR
TP53 mRNA levels were assessed after RNA isolation (PureLink RNA micro kit, Thermo Fisher, 12183016) and cDNA synthesis (high-capacity cDNA reverse transcription kit, Applied Biosystems, 4368814). RT-qPCR was performed using Taqman Universal PCR Mastermix NoAmpErase UNG (Applied Biosystems, 4324018) with TP53 probe (Applied Biosystems, Hs01034258_g1) and input was corrected with housekeeping gene GUSB (Applied Biosystems, Hs00939627_m1).
Western blot analysis
Protein lysates were obtained using RIPA buffer (Thermo Fisher, 89901) containing HALT protease and phosphatase inhibitor (Thermo Fisher, 78441). Proteins were separated on a Mini-Protean TGX gel (4–20%, Biorad, 456–1094) according to the protocol of the manufacturer. Infrared imaging with Odyssey® CLx Imaging System (Li-COR) was used as readout. Used antibodies: p53 (D-07) (Agilent, M7001), p21 (12D1) (Cell signaling, 2947), β-actin (AC-15) (Sigma, A5441), Cas9 (7A9–3A3) (Cell signaling, 14697), IRDye Odyssey secondary antibodies (Li-COR, 926–68071, 926–32211, 926–68070, 926–32210).
Dose-responses
Responses to Nutlin-3a, Cisplatin (Accord Healthcare, 16729–288-11), Rabusertib (LY2603618, Selleckchem, S2626) and Adavosertib (AZD1775/MK-1775, Biovision, 2373) were analyzed as published previously after 72h treatment and read-out using CellTiter-Blue viability assay (Promega) at GloMAX plate reader (Promega) [23].
Statistical analysis
For statistical analyses R software v.3.4.3, Graphpad Prism v.8 and IBM SPSS v.25 were used.
Results
Low-coverage whole-genome sequencing of HNSCC cell lines
In total, the cell line panel consisted of 24 HNSCC cell lines, of which five were HPV-positive (23%), four were derived from FA-patients (14%) and were HPV-negative, and also the remaining 15 cell lines were HPV-negative (63%). A general overview of the CN-profiles was obtained with low-coverage whole-genome sequencing (Table S1, S2 and Figure 1a–b). Of the FA-HNSCC lines, two were established from a patient with homozygous germline FANCA mutations (CCH-FAHNSCC-2 and VU-SCC-1365), one (VU-SCC-1131) with FANCC and one (VU-SCC-1604) with FANCL mutations [24].
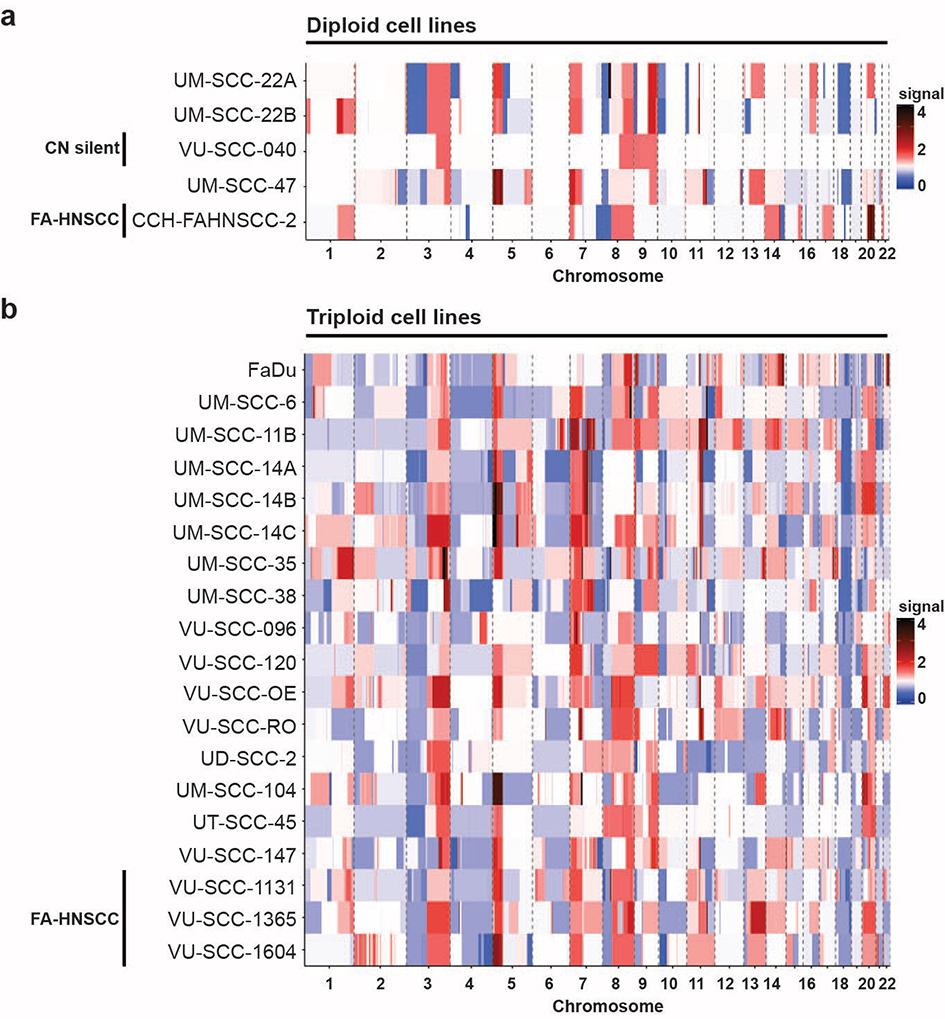
Figure 1. Low-coverage sequencing results of a representative panel of HNSCC cell lines.
Copy number profiles of a panel of HNSCC cell lines were obtained using low coverage whole-genome sequencing. Using the ACE package [19], the absolute copy numbers of all chromosomes per cell line was determined. Red represents a gain of the chromosomal region, blue a loss, white is neutral (no genomic aberration). All HNSCC lines, except for VU-SCC-040, showed large copy number aberrations with a high frequency of gains and losses.
a
ACE analysis revealed that 5 cell lines were near diploid, of which one HPV-positive cell line (UM-SCC-47) and one FA-HNSCC cell line (CCH-FAHNSCC-2). Interestingly, VU-SCC-040, a HPV-negative HNSCC line showed a copy number silent profile with just 3 single gains.
b
The majority of HNSCC lines (n=19) exhibited many copy number gains and losses, and became aneuploid to near 3N.
By using ACE analysis [19] we uncovered that 79% (19 of 24) of the cell lines had a near-triploid aneuploidy (Figure 1b), indicating past events of genomic instability or mitotic errors [2,5], and five lines were near-diploid. Note that UM-SCC-22A and −22B are related as well as cell lines UM-SCC-14A, −14B and −14C [25]. Of the four FA-HNSCC lines, three were near-triploid while CCH-FAHNSCC-2 was the only near-diploid FA-line (Figure 1a), a distribution comparable with the non-FA-lines. Of note, cell line VU-SCC-040 is HPV-negative, near diploid and shows only three copy number gains and a focal loss of CDKN2A.
We further analyzed the number of gains and losses for the HPV-positive, HPV-negative and FA-HNSCC subgroups (Figure S1a–c). Irrespective of tumor subgroup, over 75% of HNSCC lines acquired gains of chromosome arms 3q, 5p and 7p, and losses of 3p, 4p and 18q. HPV-negative HNSCC subsequently gained 8q, 9q and 11p, and lost 7q, 8p, 9p, 11q and 18p. Chromosome arms 9q and chromosome 20 were additionally gained in HPV-positive and all FA-HNSCC cell lines. Furthermore, FA-HNSCC lines lost chromosome arm 10p. Interestingly, chromosome arm 9q is gained in the majority of both HPV-negative and HPV-positive HNSCC, but was gained in only one of four (VU-SCC-1365) FA-HNSCC lines tested (Figure 1b, S2a–c and Table S1).
Target-enrichment sequencing of commonly mutated genes in HNSCC
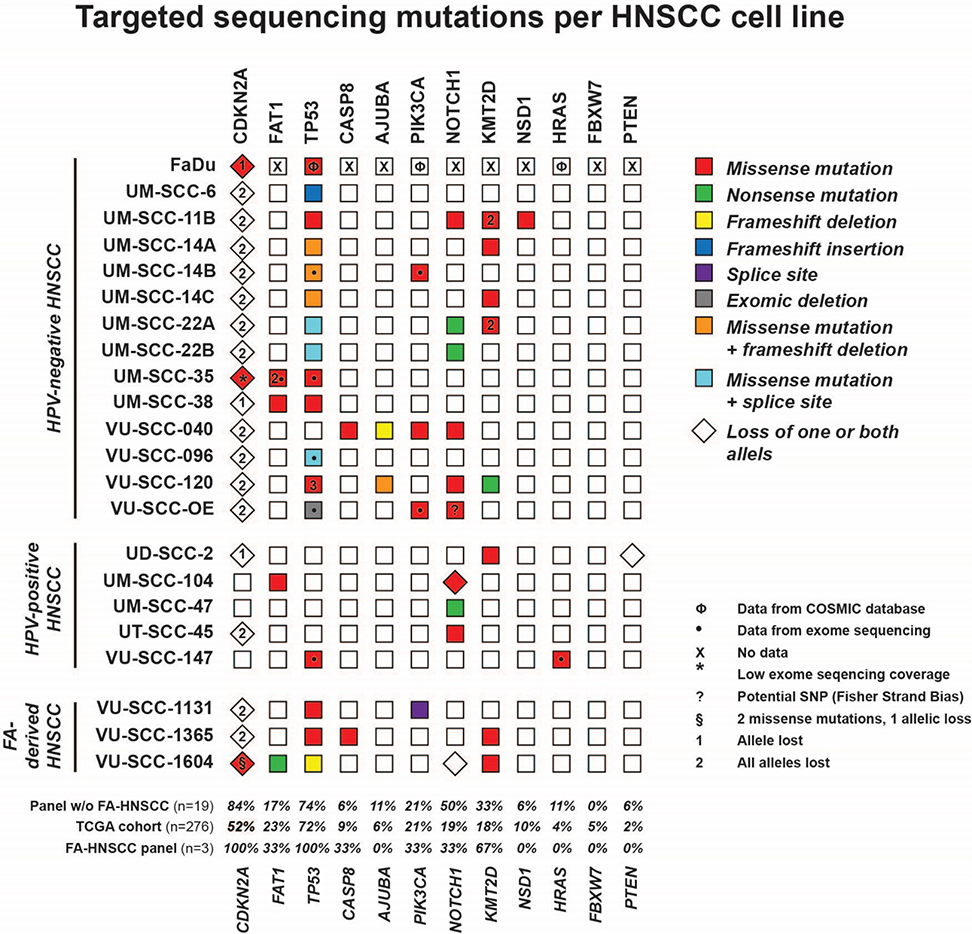
TCGA sequencing efforts revealed genes that are frequently mutated in HNSCC, and may be considered as cancer driver genes [11]. Here, we directly investigated whether the 12 most frequently mutated genes were also altered in the studied HNSCC cell line panel (Figure 2) by using HaloPlex target-enrichment sequencing, or mining of previously obtained exome sequencing data [24]. The mutation status of cell line FaDu was obtained from the COSMIC database.
Figure 2. Haloplex targeted sequencing of frequently mutated genes in HNSCC.
Schematic overview of the gene mutations per cell line, obtained with targeted Haloplex sequencing. A red box represents a missense mutation, green a nonsense mutation, yellow a frameshift deletion, blue a frameshift insertion, purple a splice site mutation, grey an exomic deletion, orange both a missense mutation and a frameshift deletion, blue both a missense mutations and a splice site mutation. For CDKN2A encoding the p16 protein, the number between brackets indicates a single (1) or homozygous (2) loss of the locus. The mutation annotations per cell line and gene can be found in table S3.
Besides selecting most frequently mutated cancer driver genes, we also mined the TCGA data for mutations in FA genes. In total, 9% of samples sequenced by TCGA had a mono-allelic Fanconi anemia gene mutation according to cBioportal [26]. Hence, mono-allelic FA-mutations are not uncommon in sporadic HNSCC [24,27]. However, a homozygous mutation in an FA-gene is necessary for FA-pathway disruption to cause the FA-associated clinical and cellular phenotypes, except for the FANCB gene that is located on the X-chromosome. For the non-FA cell lines in this cell line panel, heterozygous FANC-mutations have been reported previously for VU-SCC-147, VU-SCC-OE and UM-SCC-14B, and a somatic homozygous FANCM mutation was found in FaDu [24].
All FA-HNSCC lines were confirmed to be HPV-negative by the clinically used HPV DNA PCR [24,28,29] and exhibited genetic losses of CDKN2A and TP53, in line with non-FA HPV-negative HNSCC (Figure 2). The distribution of gene mutations in the total cell line panel is comparable to the TCGA cohort, although we found a larger fraction of CDKN2A alterations. The TCGA CDKN2A percentage shown in Figure 2 represents genomic alterations and protein expression, since focal losses of 9p21 often occur [11]. No genetic alterations in the three sequenced FA-HNSCC cell lines were found in AJUBA, NSD1, HRAS, FBXW7 and PTEN. The occurrence of mutations in FAT1, PIK3CA, NOTCH1 was similar to the frequencies found in the TCGA cohort and the other sequenced HNSCC lines. These results indicate that the genomic driver alterations underlying HNSCC tumorigenesis in FA-patients are largely comparable to those that cause cancer in non-FA-patients.
Of note, we confirmed all previously identified TP53 mutations that were analyzed by classical Sanger sequencing before [30] (Table S3). However, we identified an unreported insertion in exon 4 of TP53 in UM-SCC-6 by Haloplex sequencing that changes the reading frame. The mutation was associated with a loss of 17p and present in all reads while it was absent in any read of other cell lines, now classifying this line as TP53-null. To confirm this insertion, we sequenced exon 4 by Sanger sequencing and could verify the mutation in a stretch of six G nucleotides as identified by Haloplex sequencing (data not shown) [30–32]. Additionally, we therefore analyzed p53 expression by Western blot (see below), and showed that it was indeed negative for p53. We also authenticated the cell lines using the published STR marker profiles and could confirm that the used dell line DNA was derived from UM-SCC-6.
VU-SCC-040 is TP53 wild-type and copy number silent
Within the TCGA PanCancer Atlas, 71% of HNSCC tumors harbored a TP53 alteration, associated with increased mortality (Figure S2a). In HPV-positive tumors, TP53 mutations are typically absent since p53 is abrogated by the viral E6 oncogene. A subgroup of HPV-negative tumors is also p53 wild type and typically characterized by few copy number changes. We noted one HPV-negative cell line, VU-SCC-040, that shows an intact TP53 gene and also very few copy number changes.
The cell line VU-SCC-040 was derived from a T3N0 stage tumor of a female patient who neither smoked nor consumed alcohol [12]. HPV-status was analyzed several times with a clinically used DNA-based PCR for HPV [29] and was always negative. According to the surgery report the tumor was located at the right site of the mobile tongue, crossing the midline and with minimal extension to the floor of mouth. It did not extent into the base of tongue. The tumor was histologically classified as moderately differentiated. These characteristics are in line with clinical data of other CN-silent tumors and do not point to a HPV-related origin [12,13]. The cell line stands out in the tested HNSCC cell line panel for a number of reasons. Low-coverage whole-genome sequencing revealed a CN-silent profile, especially when compared to other HPV-negative HNSCC lines with their respective high-CN (compare Figures 3a with S2b–d). Furthermore, VU-SCC-040 is one of five near-diploid cell lines, with a shallow genetic profile that only comprises a single gain of only 3q, 8q and 9p/q, and a double focal loss of CDNK2A (9p21.3) (Figure 3a). A potential driving Epstein-Barr virus (EBV) infection was also ruled out through alignment of the unmapped low-coverage sequencing reads to the EBV genome, which was in fact negative for all cell lines. Validity of this approach was ascertained using low-coverage sequencing data of EBV-positive gastric cancer as positive controls (data not shown). As an extra control on the negative HPV status, we aligned reads to the HPV-genome of HPV types 16, 18, 31, 33 and 52 and confirmed absence of HPV DNA as previously determined by the clinically used DNA PCR-based HPV testing method [29]. Of note, known HPV-positive cell lines came out as positive and negative as negative, validating the accuracy of this approach (Table S4). Hence a viral etiology seems excluded. Haloplex target-enrichment sequencing of VU-SCC-040 revealed missense mutations in CASP8, PIK3CA, NOTCH1 and a frameshift deletion in AJUBA (Table S3).
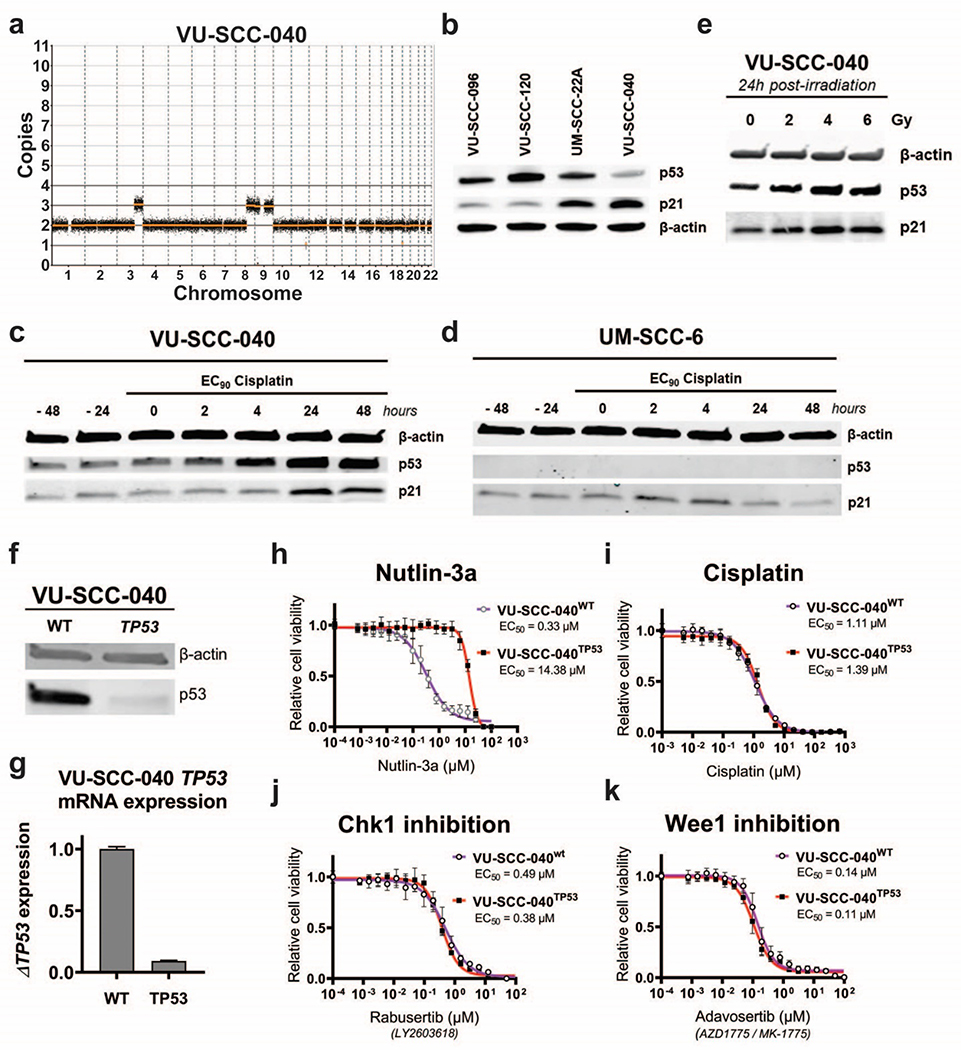
Figure 3. Characterization of HPV-negative, CN-silent and TP53 wild-type cell line VU-SCC-040.
a
Genomic CN-profile of copy number silent cell line VU-SCC-040, which only contains a single gain of 3q, 8q and 9p/q.
b
Western blot analysis of p53 and downstream target p21 in TP53-mutant cell lines VU-SCC-096, VU-SCC-120 and UM-SCC-22A, compared with CN-silent TP53 wild-type lineVU-SCC-040. VU-SCC-040 showed lower levels of p53 compared to the TP53 mutated cell lines, as expected. Furthermore, p21 expression was higher compared to the TP53 mutated cell lines, although UM-SCC-22A showed an intermediated p21 expression level.
c-d
Protein expression of p53 and p21 in TP53 wild-type cell line VU-SCC-040 (c) and TP53-null line UM-SCC-6 (d) was analyzed after treatment of cisplatin with the EC90 concentration for the indicated timespans. Before treatment, levels of p53 and p21 are stable in VU-SCC-040. Upon treatment of cisplatin, p53 expression is increased within 4 hours, and p21 levels increase after 24 hours, strongly suggesting that this part of the pathway is intact. In UM-SCC-6, p53 expression is absent due to a frameshift insertion in exon 1 and consequently, p21 levels do not change upon treatment of cisplatin.
e.
24 hours post-irradiation with 2, 4 and 6 Gy, an increase of p53 was observed for 4 and 6 Gy in VU-SCC-040.
f.
p53 protein expression was investigated in the parental VU-SCC-040wt and CRISPR TP53-knockout line VU-SCC-040TP53. Only a very low p53 expression was observed in the knockout line compared to the wild-type line, in line with the TIDE sequence analysis, and indicating that the majority of cells obtained a TP53 knockout.
g.
mRNA expression of TP53 was significantly reduced in the knockout cell line compared to the parental wild-type VU-SCC-040. Although this is not necessarily observed with a CRISPR-Cas9 approach, it confirms nonsense mediated decay of the RNA levels and the successful knockout of the gene.
h.
VU-SCC-040wt cells are very vulnerable to MDM2 inhibition with Nutlin-3a, as expected for a cell with a functional p53-pathway. For the VU-SCC-040TP53 knockout line, resistance to Nutlin-3a was obtained, in line with the EC50-values of the TP53-mutant HNSCC lines (figure S2f–h).
i.-k.
Dose-response curves of VU-SCC-040wt and VU-SCC-040TP53 showing the relative cell viability after 72h treatment with a dilution range of Cisplatin (i), Chk1 inhibitor Rabusertib (LY2603618) (j) and Wee1 inhibitor Adavosertib (AZD1775 / MK-1775) (k). Surprisingly, no difference in response was found between VU-SCC-040wt and VU-SCC-040TP53, and the EC50 values were within the range of TP53-mutant HNSCC cell lines, and sensitivity much higher than normal cells ([15,37] and van Harten et al., submitted). This indicates that the cell cycle is perturbed in VU-SCC-040, but apparently with an intact p53 pathway.
To broaden the insight in common mutations in CN-silent HNSCC, we selected and analyzed the CN-silent (<10% genomic alterations or an aneuploidy score <10) HPV-negative tumors from the PanCancer Atlas TCGA HNSCC cohort via cBioportal. We identified 84 (19%) CN-silent tumors, of which 37 were TP53 wild-type (Figure S3). When comparing to the CN-high tumors (n=439) in this cohort, the CN-silent tumors were significantly more often found in females (Table 2 and S5). The CN-silent HNSCC subclass (n=84) exhibited significantly less TP53, AJUBA and NSD1 mutations as compared to the overall representation of HNSCC, while an enrichment of HRAS and CASP8 mutations was in line with earlier observations [11]. Furthermore, CDKN2A mutations were more often found in CN-silent TP53-mutated HNSCC, and HRAS, ARID2 and HLA-B mutation were more often found in CN-silent TP53 wild-type HNSCC. Whether these observations point to in fact two molecular subclasses of CN-silent tumors, either TP53 wild type or TP53 mutated, remains to be determined.
Table 2:
CN-silent versus CN-high HNSCC from PanCancer Atlas TCGA HNSCC cohort
| PanCancer Atlas TCGA HNSCC |
||||||||
|---|---|---|---|---|---|---|---|---|
| CN-silent | CN-high | Chi-sq | ||||||
| CN-silent vs CN-high | ||||||||
| Mean | Count | % | Mean | Count | % | Sig. | ||
| Count | 84 | 439 | ||||||
| Sex | Male | 49 | 58.3% | 334 | 76.0% | |||
| Female | 35 | 41.7% | 105 | 24.0% | < 0.0001* | |||
| Age | 62 | 60 | ||||||
| CDKN2A | Wild-type | 64 | 76.2% | 353 | 80.4% | |||
| Mutation | 20 | 23.8% | 86 | 19.6% | 0.3808 | |||
| FAT1 | Wild-type | 59 | 70.2% | 357 | 81.3% | |||
| Mutation | 25 | 29.8% | 82 | 18.7% | 0.021* | |||
| TP53 | Wild-type | 37 | 44.0% | 129 | 29.4% | |||
| Mutation | 47 | 56.0% | 310 | 70.6% | 0.0085* | |||
| CASP8 | Wild-type | 68 | 81.0% | 413 | 94.1% | |||
| Mutation | 16 | 19.0% | 26 | 5.9% | 0.0001* | |||
| AJUBA | Wild-type | 84 | 100.0% | 416 | 94.8% | |||
| Mutation | 0 | 0.0% | 23 | 5.2% | 0.0328* | |||
| PIK3CA | Wild-type | 66 | 78.6% | 368 | 83.8% | |||
| Mutation | 18 | 21.4% | 71 | 16.2% | 0.2459 | |||
| NOTCH1 | Wild-type | 68 | 81.0% | 371 | 84.5% | |||
| Mutation | 16 | 19.0% | 68 | 15.5% | 0.4239 | |||
| KMT2D | Wild-type | 77 | 91.7% | 370 | 84.3% | |||
| Mutation | 7 | 8.3% | 69 | 15.7% | 0.0780 | |||
| NSD1 | Wild-type | 84 | 100.0% | 380 | 86.6% | |||
| Mutation | 0 | 0.0% | 59 | 13.4% | 0.0004* | |||
| HRAS | Wild-type | 66 | 78.6% | 426 | 97.0% | |||
| Mutation | 18 | 21.4% | 13 | 3.0% | < 0.0001* | |||
| FBXW7 | Wild-type | 77 | 91.7% | 415 | 94.5% | |||
| Mutation | 7 | 8.3% | 24 | 5.5% | 0.3207 | |||
| PTEN | Wild-type | 81 | 96.4% | 429 | 97.7% | |||
| Mutation | 3 | 3.6% | 10 | 2.3% | 0.4856 | |||
| ARID2 | Wild-type | 81 | 96.4% | 423 | 96.4% | |||
| Mutation | 3 | 3.6% | 16 | 3.6% | 0.9820 | |||
| HLA-B | Wild-type | 81 | 96.4% | 424 | 96.6% | |||
| Mutation | 3 | 3.6% | 15 | 3.4% | 0.9266 | |||
Functional analysis of p53-pathway in VU-SCC-040
TP53 is wild-type in VU-SCC-040 and HPV is absent, suggesting a functional p53-pathway. However, its functionality may be abrogated by other mechanisms such as inactivation of p53-activating molecules or downstream targets. We therefore decided to functionally characterize the p53-pathway. Analysis of p53 protein expression revealed that VU-SCC-040 has relatively low expression in comparison with TP53-mutant lines. As most missense mutations cause overexpression of mutant p53, this was expected for cells with a TP53 wild-type signature (Figure 3b). Also the expression of p21, downstream of p53 [33], is relatively high compared to the low expression in the TP53-mutant lines (except for UM-SCC-22A), which also indicates that p53 is functional in VU-SCC-040. What is more, VU-SCC-040 showed p53-pathway activation after treatment with cisplatin (Figure 3c and 3d). Samples were taken before and at the start of EC90 cisplatin treatment to analyze baseline expression (EC90 VU-SCC-040: 40 μM, UM-SCC-6: 4.6 μM). Already after 2h of treatment, an induction of p53 was noted that increased over time. Induction of p21 was also found, but followed later. UM-SCC-6, a TP53-null cell line with a frameshift mutation in exon 4 (Table S3) and 17p loss as described above, lacked p53 and p21 induction. Similar responses of p53 and p21 were obtained 24h after γ-irradiation in VU-SCC-040 (Figure 3e). All these findings suggest that the p53-pathway may indeed be functional in VU-SCC-040.
Next, we generated a VU-SCC-040 TP53-knockout cell line (annotated as VU-SCC-040TP53) using CRISPR-Cas9. The TP53-knockout population was selected with Nutlin-3a treatment, since the relatively low transfection efficiency of VU-SCC-040 prohibited clone selection by limiting dilution. The RNA guides used, target TP53 in exon 5. Sanger sequencing confirmed a TP53 g.13234del (exon 5) in the VU-SCC-040TP53 cell line, as expected (Table S6). TIDE analysis [22] revealed a TP53 alteration rate of 97%. Examination of p53 protein and mRNA expression levels confirmed significant reduction of expression in VU-SCC-040TP53 (Figure 3f–g).
Drug sensitivity in VU-SCC-040 cells with TP53-knockout
Next, we investigated response to MDM2 inhibition by using Nutlin-3a. Nutlin-3a is a specific inhibitor of MDM2, which results in increased expression of p53 [34]. This causes lethality in TP53 wild-type cells, but not in cells with an abrogated p53-pathway. Indeed, VU-SCC-040wt was very sensitive to inhibition with Nutlin-3a (EC50 = 0.33 μM), while the VU-SCC-040TP53 cells showed a drastically reduced sensitivity to Nutlin-3a, (EC50 = 14 μM, 42-times increased as compared to the parental line, Figure 3h). The TP53-mutant HNSCC cell lines UM-SCC-11B, UM-SCC-22A and VU-SCC-120 were all resistant to MDM2 inhibition with EC50-values of 22, 21 and 20 μM, respectively (Figure S2f–h), in line with the EC50 values of VU-SCC-040TP53.
It has been postulated that TP53 mutation status may impact outcome to treatment in HNSCC. For cisplatin, wild-type TP53 has been associated with better responses to treatment [35]. Remarkably, treatment with cisplatin showed no significant difference in the cellular response of TP53 wild-type cells and the VU-SCC-040TP53 cells (Figure 3i, EC50-values of 1.1 and 1.4 μM, respectively). These EC50-values for cisplatin are in the lower range of EC50 values of a larger and previously tested HNSCC cell line panel (0.69 μM - 7.6 μM, median: 1.8 μM) [30].
Chk1 and Wee1 are kinases involved in cell cycle regulation in S- and G2-phase, protecting cells from DNA replication failure and support initiation of DNA repair [36]. It is generally assumed that an altered p53-function enhances therapeutic outcome, which is often used as including criterion for clinical trials with these inhibitors. In our CN-silent TP53 wild-type HNSCC line VU-SCC-040wt, the EC50-value of Chk1 inhibitor Rabusertib (LY2603618) was 0.49 μM (Figure 3j). The range of HNSCC cell lines tested previously in our lab showed an EC50-range of 0.045 – 0.80 μM (mean = 0.50 μM) [37]. Knocking out TP53 in VU-SCC-040 only marginally sensitized the cells to Chk1 inhibition (EC50 = 0.38 μM). Additionally, EC50-values for Wee1 inhibitor Adavosertib (AZD1775/MK-1775) were minimally reduced in VU-SCC-040TP53 (EC50 = 0.14 μM in VU-SCC-040wt and 0.11 μM, in VU-SCC-040TP53) (Figure 3k). The panel of HNSCC cell lines tested with Adavosertib had revealed an EC50-range from 0.043 – 0.39 μM (median: 0.15 μM) (van Harten et al., submitted). These results indicate that loss of TP53 is not by itself responsible for sensitivity to cisplatin, Chk1 and Wee1 inhibition in HNSCC cell lines, at least in a CN-silent background.
Discussion
Recent efforts to gain insight in mutations and genetic alterations in HNSCC created new opportunities to better understand the disease and its drivers [11,38]. Moreover, the genetic data can also be used to authenticate cell lines. Here, we aimed to investigate the molecular characteristics of the variety of HNSCC subgroups, including FA-associated HNSCC, and we identified and functionally characterized the HPV-negative CN-silent TP53 wild-type HNSCC cell line VU-SCC-040.
FA-patients are rare, with an incidence of around 1 in 100,000 newborns [7] but patients rapidly develop HNSCCs most particularly in the oral cavity and at much younger age. The FA/BRCA-pathway is crucial for the removal of interstrand-crosslinks, adducts that form between DNA strands and disable the formation of DNA replication forks during S-phase [9]. Upon FA/BRCA-pathway activation, the crosslink is removed and DNA is precisely repaired through homologous recombination (HR). The lack of a functional FA/BRCA-pathway renders FA-patients lethally sensitive to DNA cross-linkers such as cisplatin, whilst this is a frequently applied treatment in the management of HNSCC. Investigating the genomic background of the tumors arising in these patients is therefore highly relevant, since treatment of HNSCC in FA-patients meets toxicity limitations and is restricted [6]. Despite the differences in risk factors, either genetic predisposition by DNA repair deficiency or the exposure to exogenous carcinogens, the CN-profiles and mutated genes found seemed very comparable between FA-HNSCC and sporadic non-FA-HNSCC. Hopefully this implies that FA-patients might benefit from the efforts to identify new targeted treatments for HNSCC in general, although increased toxicity in FA-background should always be carefully examined.
Most interestingly, we identified a CN-silent TP53 wild-type cell line, VU-SCC-040. CN-silent HNSCC forms a subgroup of HPV-negative HNSCC, occurring mostly in female patients with no history of smoking and excessive alcohol use [5,13]. This subgroup of tumors exhibits a low number of chromosomal aberrations, are commonly wild-type TP53 and enriched for CASP8 and HRAS mutations. These patients also have a more favorable survival as compared to patients with HNSCC tumors with a CN-high profile [11,13]. The discovery of at least one cell line model confirms the existence of this particular genetic subgroup. The identification of similar cell lines and studies of their specific molecular characteristics such as replication stress, drug responses and radiation sensitivity, would help to further elucidate mechanisms underlying oncogenesis, cell proliferation and drug sensitivities in TP53-proficient tumors. VU-SCC-040 cells are near diploid and exhibit a single gain of chromosomes 3q, 8q and 9 and a focal loss of CDKN2A. Whereas p53 is inactivated by either mutation or HPV-E6 in almost all HNSSC, we showed that the p53-pathway appears to be functional in VU-SCC-040, and likely in this subgroup of tumors.
Surprisingly, the sensitivities to cisplatin, Chk1 and Wee1 inhibition in VU-SCC-040wt overlapped with those found in TP53-mutant HNSCC cell lines tested ([37] and van Harten et al., submitted), while it is generally assumed that treatment with Chk1 and Wee1 inhibitors is only effective in a TP53-mutant background [36]. Furthermore, the introduction of TP53-knockout neither sensitized the cells to Chk1 and Wee1 inhibition, nor made these cells more resistant to cisplatin. Exome sequencing of VU-SCC-040 and preferably more of these CN-silent cell lines, may aid in unraveling the mechanisms of drug response.
In conclusion, we investigated a variety of subgroups of HNSCC by molecular characterization of representative cell lines. FA-HNSCC tumors show large similarities to HPV-negative HNSCC cell lines. Furthermore, we discovered a CN-silent, TP53 wild-type HNSCC cell line with an apparent intact p53 pathway, VU-SCC-040, and target-enrichment sequencing revealed mutations in CASP8, AJUBA, PIK3CA and NOTCH1.
Supplementary Material
Highlights.
Fanconi anemia (FA) derived HNSCC is molecularly very comparable to sporadic non-FA HNSCC.
VU-SCC-040 is a cell model representative for HPV-negative copy-number silent HNSCC.
VU-SCC-040 is TP53 wild-type and the p53-pathway appears to be functional.
VU-SCC-040 adds important information to the published descriptive genetic data.
Acknowledgements
The authors thank Klaas de Lint for his expertise and reagents to generate the TP53-knockout VU-SCC-040 cell line by CRISPR-Cas9 genome editing, and Elizabeth E. Hoskins for establishing the CCH-FAHNSCC-2 cell line.
Financial support: The study was supported the VUmc Cancer Center Amsterdam and Amsterdam UMC.
Footnotes
Conflicts of interest
The authors declared no conflicts of interest.
References
- [1].Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality - Major patterns in GLOBOCAN 2012, worldwide and Georgia. Bull Georg Natl Acad Sci 2015;9:168–73. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- [2].Leemans CR, Braakhuis BJM, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- [3].Brakenhoff RH, Wagner S, Klussmann JP. Molecular Patterns and Biology of HPV-Associated HNSCC In: Golusiński W, Leemans C, Dietz A (eds) HPV Infection in Head and Neck Cancer. vol. 206 2017. doi: 10.1007/978-3-319-43580-0_4. [DOI] [PubMed] [Google Scholar]
- [4].Castellsagué X, Alemany L, Quer M, Halec G, Quirós B, Tous S, et al. HPV Involvement in Head and Neck Cancers: Comprehensive Assessment of Biomarkers in 3680 Patients. J Natl Cancer Inst 2016;108:djv403. doi: 10.1093/jnci/djv403. [DOI] [PubMed] [Google Scholar]
- [5].Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer 2018;18:269–82. doi: 10.1038/nrc.2018.11. [DOI] [PubMed] [Google Scholar]
- [6].Furquim CP, Pivovar A, Amenábar JM, Bonfim C, Torres-Pereira CC. Oral cancer in Fanconi anemia: Review of 121 cases. Crit Rev Oncol Hematol 2018;125:35–40. doi: 10.1016/j.critrevonc.2018.02.013. [DOI] [PubMed] [Google Scholar]
- [7].Nalepa G, Clapp DW. Fanconi anaemia and cancer: An intricate relationship. Nat Rev Cancer 2018;18:168–85. doi: 10.1038/nrc.2017.116. [DOI] [PubMed] [Google Scholar]
- [8].Mehta PA, Tolar J. Fanconi Anemia. University of Washington, Seattle; 1993. [Google Scholar]
- [9].Patel DR, Weiss RS. A tough row to hoe : when replication forks encounter DNA damage. Biochem Soc Trans 2018;46:1–9. doi: 10.1042/BST20180308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].de Boer DV, Brink A, Buijze M, Stigter-van Walsum M, Hunter KD, Ylstra B, et al. Establishment and Genetic Landscape of Precancer Cell Model Systems from the Head and Neck Mucosal Lining. Mol Cancer Res 2018;17:120–30. doi: 10.1158/1541-7786.MCR-18-0445. [DOI] [PubMed] [Google Scholar]
- [11].Lawrence MS, Sougnez C, Lichtenstein L, Cibulskis K, Lander E, Gabriel SB, et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hermsen MAJA, Joenje H, Arwert F, Welters MJP, Braakhuis BJM, Bagnay M, et al. Centromeric breakage as a major cause of cytogenetic abnormalities in oral squamous cell carcinoma. Genes, Chromosom Cancer 1996;15:1–9. doi:. [DOI] [PubMed] [Google Scholar]
- [13].Smeets SJ, Brakenhoff RH, Ylstra B, Van Wieringen WN, Van De Wiel MA, Leemans CR, et al. Genetic classification of oral and oropharyngeal carcinomas identifies subgroups with a different prognosis. Cell Oncol 2009;31:291–300. doi: 10.3233/CLO-2009-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hoskins EE, Morris TA, Higginbotham JM, Spardy N, Cha E, Kelly P, et al. Fanconi anemia deficiency stimulates HPV-associated hyperplastic growth in organotypic epithelial raft culture. Oncogene 2009;28:674–85. doi: 10.1038/onc.2008.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Nagel R, Martens-de Kemp SR, Buijze M, Jacobs G, Braakhuis BJM, Brakenhoff RH. Treatment response of HPV-positive and HPV-negative head and neck squamous cell carcinoma cell lines. Oral Oncol 2013;49:560–6. doi: 10.1016/j.oraloncology.2013.03.446. [DOI] [PubMed] [Google Scholar]
- [16].Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Scheinin I, Sie D, Bengtsson H, Van De Wiel MA, Olshen AB, Van Thuijl HF, et al. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res 2014;24:2022–32. doi: 10.1101/gr.175141.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics 2007;23:657–63. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- [19].Poell JB, Mendeville M, Sie D, Brink A, Brakenhoff RH, Ylstra B, et al. ACE: absolute copy number estimation from low-coverage whole-genome sequencing data. Bioinformatics 2018. doi: 10.1093/bioinformatics/bty1055. [DOI] [PubMed] [Google Scholar]
- [20].van Wieringen WN, Unger K, Leday GGR, Krijgsman O, de Menezes RX, Ylstra B, et al. Matching of array CGH and gene expression microarray features for the purpose of integrative genomic analyses. BMC Bioinformatics 2012;13:80. doi: 10.1186/1471-2105-13-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 2015;163:1515–26. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- [22].Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 2014;42:e168–e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].de Boer DV, Martens-de Kemp SR, Buijze M, Stigter-Walsum, Van M, Bloemena E, Dietrich R, et al. Targeting PLK1 as a novel chemopreventive approach to eradicate preneoplastic mucosal changes in the head and neck. Oncotarget 2017;8:1–13. doi: 10.18632/oncotarget.17880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Stoepker C, Ameziane N, Van Der Lelij P, Kooi IE, Oostra AB, Rooimans MA, et al. Defects in the Fanconi anemia pathway and chromatid cohesion in head and neck cancer. Cancer Res 2015;75:3543–53. doi: 10.1158/0008-5472.CAN-15-0528. [DOI] [PubMed] [Google Scholar]
- [25].Brenner JC, Graham MP, Kumar B, Saunders LM, Kupfer R, Lyons RH, et al. Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head Neck 2010;32:417–26. doi: 10.1002/hed.21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data: Figure 1. Cancer Discov 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chandrasekharappa SC, Chinn SB, Donovan FX, Chowdhury NI, Kamat A, Adeyemo AA, et al. Assessing the spectrum of germline variation in Fanconi anemia genes among patients with head and neck carcinoma before age 50. Cancer 2017;123:3943–54. doi: 10.1002/cncr.30802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Van Zeeburg HJT, Snijders PJF, Pals G, Hermsen MAJA, Rooimans MA, Bagby G, et al. Generation and molecular characterization of head and neck squamous cell lines of Fanconi anemia patients. Cancer Res 2005;65:1271–6. doi: 10.1158/0008-5472.CAN-04-3665. [DOI] [PubMed] [Google Scholar]
- [29].Nauta IH, Rietbergen MM, van Bokhoven AAJD, Bloemena E, Lissenberg-Witte BI, Heideman DAM, et al. Evaluation of the eighth TNM classification on p16-positive oropharyngeal squamous cell carcinomas in the Netherlands and the importance of additional HPV DNA testing. Ann Oncol 2018;29:1273–9. doi: 10.1093/annonc/mdy060. [DOI] [PubMed] [Google Scholar]
- [30].Martens-de Kemp SR, Dalm SU, Wijnolts FMJ, Brink A, Honeywell RJ, Peters GJ, et al. DNA-Bound Platinum Is the Major Determinant of Cisplatin Sensitivity in Head and Neck Squamous Carcinoma Cells. PLoS One 2013;8. doi: 10.1371/journal.pone.0061555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lee TL, Yang XP, Yan B, Friedman J, Duggal P, Bagain L, et al. A Novel Nuclear Factor-KB Gene Signature Is Differentially Expressed in Head and Neck Squamous Cell Carcinomas in Association with TP53 Status 2007. doi: 10.1158/1078-0432.CCR-07-0670. [DOI] [PubMed] [Google Scholar]
- [32].Sano D, Xie T-X, Ow TJ, Zhao M, Pickering CR, Zhou G, et al. Disruptive TP53 mutation is associated with aggressive disease characteristics in an orthotopic murine model of oral tongue cancer. Clin Cancer Res 2011;17:6658–70. doi: 10.1158/1078-0432.CCR-11-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Deng C, Zhang P, Wade Harper J, Elledge SJ, Leder P. Mice Lacking p21 CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 1995;82:675–84. doi: 10.1016/0092-8674(95)90039-X. [DOI] [PubMed] [Google Scholar]
- [34].Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, Hilton H, et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci U S A 2006;103:1888–93. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhou G, Liu Z, Myers JN. TP53 Mutations in Head and Neck Squamous Cell Carcinoma and Their Impact on Disease Progression and Treatment Response. J Cell Biochem 2016;117:2682–92. doi: 10.1002/jcb.25592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bauman JE, Chung CH. CHK it out! Blocking WEE kinase routs TP53 mutant cancer. Clin Cancer Res 2014;20:4173–5. doi: 10.1158/1078-0432.CCR-14-0720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].van Harten AM, Buijze M, van der Mast R, Rooimans MA, Martens-de Kemp SR, Bachas C, et al. Targeting the cell cycle in head and neck cancer by Chk1 inhibition : a novel concept of bimodal cell death. Oncogenesis 2019;8:38. doi: 10.1038/s41389-019-0147-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cheng H, Yang X, Si H, Saleh AD, Xiao W, Coupar J, et al. Genomic and Transcriptomic Characterization Links Cell Lines with Aggressive Head and Neck Cancers. Cell Rep 2018;25:1332–1345.e5. doi: 10.1016/j.celrep.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.