Abstract
The presence of a phagocytic peak of photoreceptor outer segments by the retinal pigment epithelium (RPE) one or two hours after the onset of light has been reported for several diurnal and nocturnal species. This peak in phagocytic activity also persists under constant lighting conditions (i.e., constant light or dark) thus demonstrating that the timing of this peak is driven by a circadian clock. The aim of this study was to investigate the change in RPE whole transcriptome at two different circadian times (CT; 1 hour before (CT23) and 1 hour after (CT1) subjective light onset). C57BL/6J male mice were maintained in DD for three days and euthanized under red light (< 1 lux) at CT23 and CT1. RPE was isolated from whole eyes for RNA library preparation and sequencing on an Illumina HiSeq4000 platform. 14,083 mouse RPE transcripts were detected in common between CT23 and CT1. 12,005 were protein coding transcripts and 2078 were non-protein coding transcripts. 2421 protein coding transcripts were significantly upregulated whereas only 3 transcripts were significantly downregulated and 12 non-protein coding transcripts were significantly upregulated and 31 non-protein coding transcripts were significantly downregulated at CT1 when compared to CT23 (p < 0.05, fold change ≥±2.0). Of the protein coding transcripts, most of them were characterized as: enzymes, kinases, and transcriptional regulators with a large majority of activity in the cytoplasm, nucleus, and plasma membrane. Non-protein coding transcripts included biotypes such as long-non coding RNAs and pseudogenes. Gene ontology analysis and ingenuity pathway analysis revealed that differentially expressed transcripts were associated with integrin signaling, oxidative phosphorylation, protein phosphorylation, and actin cytoskeleton remodeling suggesting that these previously identified phagocytic pathways are under circadian control. Our analysis identified new pathways (e.g., increased mitochondrial respiration via increased oxidative phosphorylation) that may be involved in the circadian control of phagocytic activity. In addition, our dataset suggests a possible regulatory role for the identified non-protein coding transcripts in mediating the complex function of RPE phagocytosis. Finally, our results also indicate, as seen in other tissues, about 20 % of the whole RPE transcriptome may be under circadian clock regulation.
Keywords: transcriptome, retinal pigment epithelium, mouse, circadian
1. INTRODUCTION
Circadian rhythms are present in nearly all organisms and function as an anticipatory mechanism to changes in the environment (Takahashi, 2017). These rhythms are driven by circadian clocks located in many organs and tissues (Takahashi, 2017) including the eye (DeVera et al., 2019; Felder-Schmittbuhl et al., 2018). Among the many circadian rhythms present in the eye, the circadian rhythm in phagocytosis of photoreceptor outer segments (POS) by retinal pigment epithelium (RPE) is one of the most well studied (see Ruggiero and Finnemann, 2014 for a recent review). In light:dark (LD) conditions a peak in phagocytic activity occurs 1-2 hours after the light onset and is present in both nocturnal and diurnal species (Bobu and Hicks, 2009; Doyle et al., 2002; Grace et al., 1999; LaVail, 1976; Lewis et al., 2018; Lo and Bernstein, 1981). The rhythm in phagocytic activity also persists when the animals are maintained in constant darkness (DD; Grace et al., 1999; LaVail, 1976) or light (Besharse and Hollyfield, 1979), after the optic nerve has been severed (Teirstein et al., 1980), and once the master circadian clock located in the brain has been ablated (Su Terman et al., 1993). A recent study has also reported that the rhythm in phagocytic activity is likely to be driven by the RPE since the diurnal rhythm of phosphatidylserine exposure on the outer leaflet of the POS was abolished with the loss of functioning integrin receptors (Ruggiero et al., 2012) which are located on the apical surface of RPE cells that face the POS (Nandrot et al., 2004). Finally, it has been demonstrated that the RPE does indeed contain a circadian clock that acts independently from the master circadian clock located in the brain (Baba et al., 2010) and that the RPE clock is entrained to environmental LD cycles by dopamine via dopamine 2 receptors (D2R, Baba et al., 2017).
Many studies have focused on elucidating the molecular mechanisms that drive the RPE peak in phagocytic activity after light onset. These investigations have demonstrated that activation of the integrin signaling pathway is required for the burst in the RPE phagocytic activity observed in the early morning since mice lacking a functioning αvβ5 integrin receptor (Nandrot et al., 2004) or its ligand MFG-E8 (Nandrot et al., 2007) fail to show the morning peak in phagocytic activity. Transcriptomic analysis of the mouse RPE has revealed that phosphoinositide signaling is also implicated in the mechanism leading to the increase in RPE phagocytosis observed after onset of light (Mustafi et al., 2013).
However, it is worth nothing that all these studies have investigated this phenomenon under LD conditions and no study has investigated the molecular drivers under DD lighting conditions. The aim of the present study was to investigate the changes in the mouse RPE whole transcriptome at two different circadian times. Our study revealed that many previously identified pathways associated with RPE phagocytosis such as: integrin signaling, cAMP signaling, focal adhesion, epithelial adherence junction signaling, and protein phosphorylation are indeed under control of the RPE circadian clock. Furthermore, we also identified a role for mitochondrial respiration in association with RPE phagocytic activity which to our knowledge has not been previously associated with RPE phagocytosis in vivo. Finally, our study indicated that about 20% of the total RPE transcriptome may be under the control of the circadian clock.
2. MATERIAL AND METHODS
2.1. Animals
Male C57BL/6J mice (8-10 weeks of age) were purchased from Jackson Laboratory and housed in 12:12 LD with lights on at 7AM and lights off at 7PM. Mice were then placed into DD conditions for three days and sacrificed one hour before subjective light onset (circadian time (CT) 23) and one after subjective light onset (CT1). All experimental procedures were performed in accordance with the NIH Guide on Care and Use of Laboratory Animals and were approved by the Morehouse School of Medicine Animal Care and Use Committee.
2.2. RPE RNA sampling, library preparation, and high-throughput sequencing
C57BL/6J mice were euthanized at CT23 and CT1 (n=3 mice/time point). RPE cell isolation was performed as previously described (Baba et al., 2010) with a modification that included brief sonication to remove RPE cells from choroid rather than gentle peeling. Briefly, eyes were enucleated under dim red-light conditions (<1 lux) and removal of the anterior segment along with the neural retina was dissected from the posterior segment that contains the RPE, choroid, and sclera. Each dissected posterior segment was placed in 50 μL of RNAlater (Qiagen, 76104). Removal and isolation of RPE cells was done via brief sonication (Fisher, F60) on ice (< 2 secs at 10% max power) and was done in ambient light conditions. Two RPE samples from one mouse were pooled into one tube. The isolated RPE cells were processed for RNA isolation with TRIzol (Ambion, 15596018) following the manufacturer’s instructions. Total RNA samples from the RPE were sent to Omega Bioservices (Norcross, GA) for both library preparation and next-generation sequencing. The QuantSeq™ 3’mRNA-Seq Kit (Lexogen, 015.24) was used for library preparation and was done according to the manufacturer’s instructions. High throughput sequencing was performed on the HiSeq4000 Illumina platform with paired-end 150 bp reads at a sequencing depth of at least 10 million reads.
2.3. RPE RNA-sequence (Seq) analysis
Each FASTA file (i.e., biological sample) was mapped to the University of California–Santa Cruz (UCSC; Santa Cruz, CA, USA) mouse genome assembly and transcript annotation (mm10). Mapping was performed with Bowtie2 (v2.1.0) using default settings. HTSeq-count (PyCharm Community Edition 2016.3.2) was used to generate counts of reads uniquely mapped to annotated genes using the UCSC mouse assembly mm10 gtf file. Settings for HTSeq-count were as follows: mode=union, stranded=no, minimum alignment quality=10, feature type=exon, ID attribute=gene_id, and under advanced options: do not count nonunique or ambiguous counts, ignore secondary and supplementary alignments. A featured count of at least 10 in at least 2 biological samples in the same CT were filtered and subsequently used for data normalization. Featured count data normalization were done with RUVseq (v3.10; Peixoto et al., 2015) using the RUVs function with default settings before manual calculation of CPM (counts per million) for each identified gene. Validation of normalization procedure was done visually with the construction of a principal component analysis graph (PCA) and relative log expression graph (RLE). Additionally, all datasets were filtered for highly expressed genes identified in photoreceptors and the choroid as previously published and made publicly available by Bennis et al. (2015) which allowed for post-hoc RPE tissue enrichment. Differential expression of each transcript common to both time points was defined with a p-value < 0.05 and a fold change ≥±2.0 as calculated by edgeR (v3.24.1) with B-H false positive adjustment. Construction of the heatmaps (ggplot; v3.2.1) and volcano plots (EnhancedVolcano; v3.10) were done in R (v3.5.2). Differentially expressed genes (DEGs; i.e., protein coding transcripts) - which encompass a large majority of identified transcripts - were subjected to gene ontology analysis (Gene ontology consortium, Nucleic acids research, 2019) and ingenuity pathway analysis (IPA, v48207413) for canonical pathway characterization. The RNA-seq data reported in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE143396 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE143396). The complete list of gene ontology terms and canonical pathways identified in IPA are available as supplemental Tables 1 and 2, respectively.
3. RESULTS
3.1. A large number of transcripts were significantly increased at CT1 with respect to CT23
We identified 14,083 transcripts in the mouse RPE. Following data normalization via the use of RUVSeq (v3.10), our normalized RNA-seq dataset demonstrated little z-score variability among the three biological samples for each time point in both the protein coding dataset (Figure 1a) and the non-protein coding dataset (Figure 1c) as depicted with a heatmap. Additionally, validation of the normalization methodology was also established with a visual inspection of correct sampling clustering in a PCA graph (Supplemental figure 1a). An RLE graph was also constructed to visualize the spread of variability of featured count data within each biological sample. As a result of normalization, our dataset correctly centered around zero which indicated little if any sequencing variability within each sample following RUVSeq normalization procedures (Supplemental Figure 1b). As previously reported by Bennis et al. (2015), there are 585 genes highly expressed in the choroid and 1580 genes highly expressed in photoreceptors when compared to the same genes expressed in the RPE. Our post-hoc filtering of our RPE dataset indicated that in less than 2% of the photoreceptor specific genes and less than 0.1% of the choroid specific genes were present in our RPE dataset. Our post-hoc RPE enrichment dataset included 12,005 transcripts that were protein coding (Figure 1b) whereas 2078 transcripts were non-protein coding (Figure 1d). 2421 protein coding transcripts were significantly upregulated whereas only 3 transcripts (Rspo1, Per2, and Hey1) were significantly downregulated at CT1 with respect to CT23 (Figure 1b). 12 non-protein coding transcripts were significantly upregulated and 31 non-protein coding transcripts were significantly downregulated (Figure 1d). Interestingly, our data demonstrated about 20% of the protein coding transcripts showed a significant change in their CPM values at CT1 when compared to CT23.
Figure 1. Visualization of protein and non-protein coding transcripts.
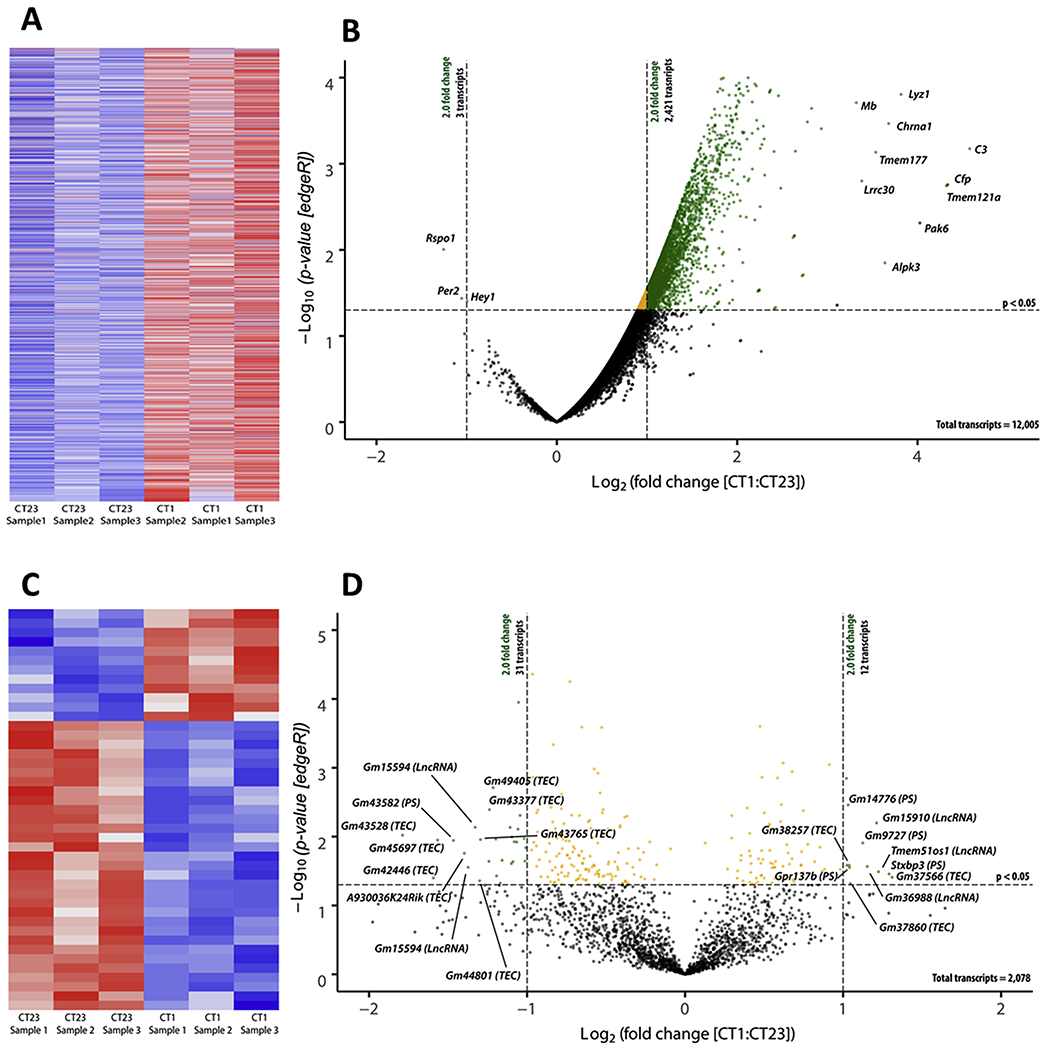
Identified differentially expressed transcripts (CT1 as compared to CT23) as defined by a p-value < 0.05 and a fold change ≥ 2.0 (as calculated by edgeR v3.24.1, B-H adjustment) were plotted in a heatmap and assembled via unsupervised hierarchal clustering for both protein-coding transcripts (a) and non-protein coding transcripts (c). A positive z-score (red) indicated positively regulated transcripts and a negative z-score (blue) indicated negatively regulated transcripts. To visualize the distribution of transcripts with their respective p-value and fold change, a volcano plot was constructed for both protein coding (b) and non-protein coding transcripts (d). Transcripts that were labeled black are not significant, orange transcripts are significantly expressed but do not pass the fold change threshold, and green transcripts are both significantly expressed and pass the fold change threshold. The top 10 upregulated protein coding transcripts and only three downregulated transcripts are plotted on the right and left on the volcano plot, respectively (b). The top 10 up (right) and down (left) expressed non-protein coding transcripts are plotted on the volcano plot with their respective p-value and fold change (d). TEC=To be experimentally confirmed; PS=pseudogene; LncRNA=long non-coding RNA; CT = circadian time; C3 = complement component 3; Cfp = complement factor properdin; Tmem121a = transmembrane protein 151a; Pak6 = p21 (RAC1) activated kinase 6; Lyz1 = lysozyme 1; Chrna1 = cholinergic receptor, nicotinic, alpha polypeptide 1; Alpk3 = alpha-kinase 3; Tmem177 = transmembrane protein 177; Lrrc30 = leucine rich repeat containing 30; Mb = myoglobin; Per2 = period circadian clock 2; Rspo1 = R-spondin 1; Hey1 = hairy/enhancer-of-split related with YRPW motif. Note: definitions of specific biotypes for each transcript can be found on the Ensembl website for further clarification (https://useast.ensembl.org/info/genome/genebuild/biotypes.html).
3.2. Biological characterization of the protein and non-protein coding transcripts
Because we were able to identify over 14,000 transcripts in the mouse RPE, we also sought to characterize the class of each protein coding and non-protein coding transcripts that fell under the definition of differential expression (p < 0.05, fold change ≥ ±2.0). Using the database provided in IPA, a majority of protein coding transcripts were classified as: enzymes, kinases, transcriptional regulators, and transporters (Figure 2a). When the identified protein coding transcripts were categorized based on location of action in a cell, a large majority of the expressed proteins acted in the cytoplasm as well as in the plasma membrane and nucleus (Figure 2b). Using the datamining tool, BioMart (Ensembl release 98) we were able to classify the different classes of non-coding transcripts. Both up and down differentially expressed datasets for non-protein coding transcripts revealed that both sets contained transcripts classified as long-coding RNA (lncRNA) and pseudogenes (PS; Figures 2c and 2d).
Figure 2. Biological Characterization of protein and non-protein transcripts.
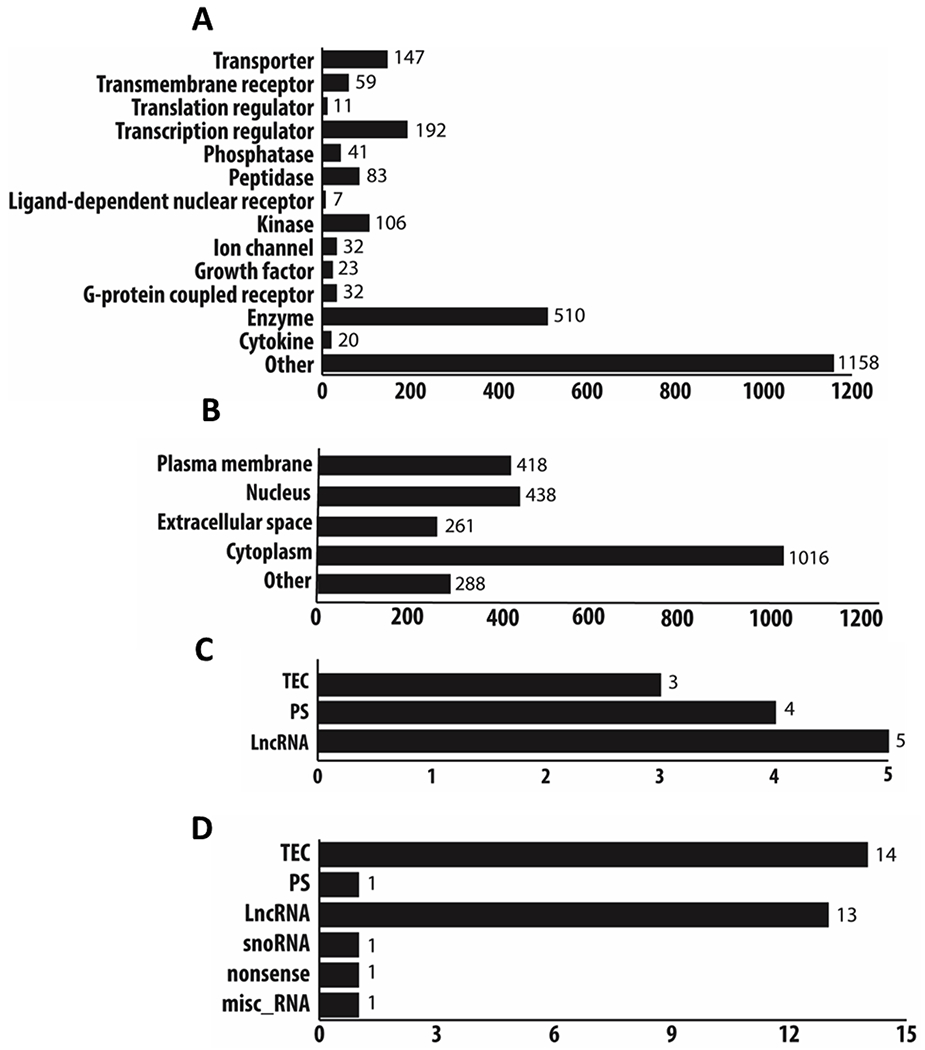
All differentially expressed protein coding transcripts (CT1 as compared to CT23) with a p-value < 0.05 and a fold change ≥ 2.0 (edgeR, v3.24.1, B-H adjustment) were categorized into different class types and location of action in the cell via the ingenuity pathway analysis database (v48207413; a and b). A large majority of protein coding transcripts were characterized into enzymes, kinases, transcriptional regulators, and transporters (a). Cellular localization of the protein-coding transcripts was identified to be expressed in the cytoplasm, nucleus, and plasma membrane (b). Using the datamining tool Biomart (Ensembl release 98), we also classified the non-protein coding differentially expressed transcripts (p < 0.05, fold change ≥±2.0, edgeR, v3.24.1, B-H adjustment) into different categories (c and d). Non-protein coding transcripts that were positively differentially expressed had transcripts that were classified as: TEC, PS, and LncRNA (c). Non-protein coding transcripts that were negatively differentially expressed were classified as: TEC, PS, LncRNA, snoRNA, nonsense, and misc RNA (d). TEC=To be experimentally confirmed; PS=pseudogene; LncRNA=long non-coding RNA. Note: definitions of specific biotypes for each transcript can be found on the Ensembl website for further clarification (https://useast.ensembl.org/info/genome/genebuild/biotypes.html).
3.3. Identification of the transcripts and pathways under circadian control
To gain a better appreciation of the biological processes activated by the circadian clock in the mouse RPE, we subjected all DEGs for gene ontology analysis and characterized them based on known biological processes. As expected, DEGs were associated with gene ontology terms that reflect: cell-cell adhesion, extracellular matrix reorganization, focal adhesion assembly, response to cAMP, and protein phosphorylation were observed (Figure 3). Interestingly, the identification of genes associated with mitochondrial respiration was observed (Figure 3). To determine what biological pathways were activated (i.e., considering the fold change of each DEG), all DEGs were subjected to canonical pathways analysis in IPA. Similar to the gene ontology analysis, canonical pathways that were identified included: actin cytoskeleton signaling, integrin signaling, germ cell-sertoli cell junction signaling, and epithelial adherens junction signaling (Figure 4a). In agreement to the gene ontology analysis, mitochondrial dysfunction which shares many genes with oxidative phosphorylation (Figure 4b) was the greatest associated canonical pathway (Figure 4a). Interestingly, when all the top 10 canonical pathways were plotted for the number of common genes among them, those that displayed more than 20 genes overlapping (dark bold line) were previously identified pathways associated with RPE phagocytosis (Figure 4b). As expected, very little gene overlap was observed in pathways such as colorectal cancer metastasis signaling or regulation of cellular mechanics by calpain protease (light to mid grey line; less than 20 genes) when compared to other phagocytic associated pathways which again had greater gene overlap amongst each other when compared to non-phagocytic biological processes (Figure 4b).
Figure 3. Gene ontology analysis of the protein coding mouse transcriptome.
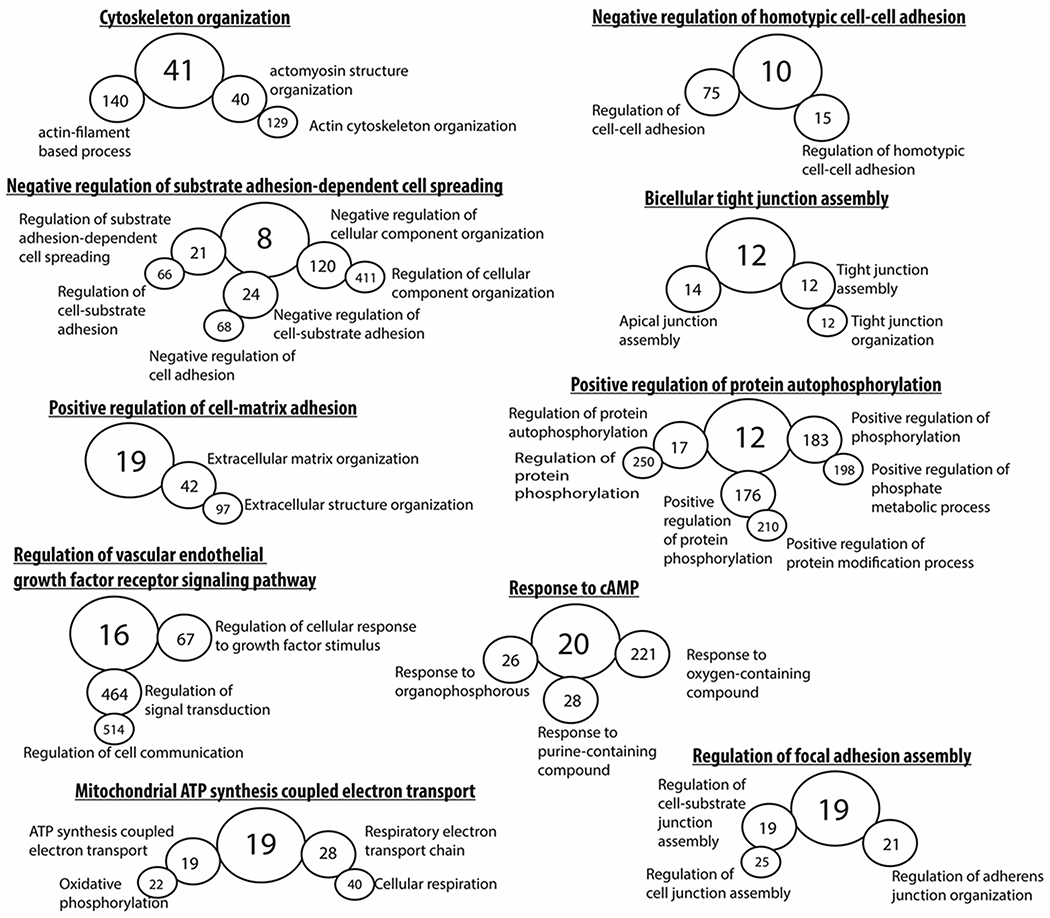
Gene ontology hierarchal analysis of all differentially expressed (CT1 as compared to CT23) protein coding transcripts (p < 0.05, fold change ≥±2.0, edgeR, v3.24.1, B-H adjustment) revealed terms that were consistent to previously known processes involved in RPE phagocytosis such as cell-cell adhesion, tight junction formation, focal adhesion assembly, protein autophosphorylation, and cAMP signaling. Interestingly, the identification of mitochondrial respiration has not been previously demonstrated before in the context of RPE phagocytosis in vivo. Greater circle diameter indicative of parent term in gene ontology hierarchal analysis. The number in each circle represents the exact number of genes that make up that gene ontology term identified in the current study’s dataset.
Figure 4. RPE associated phagocytic pathways under circadian control.
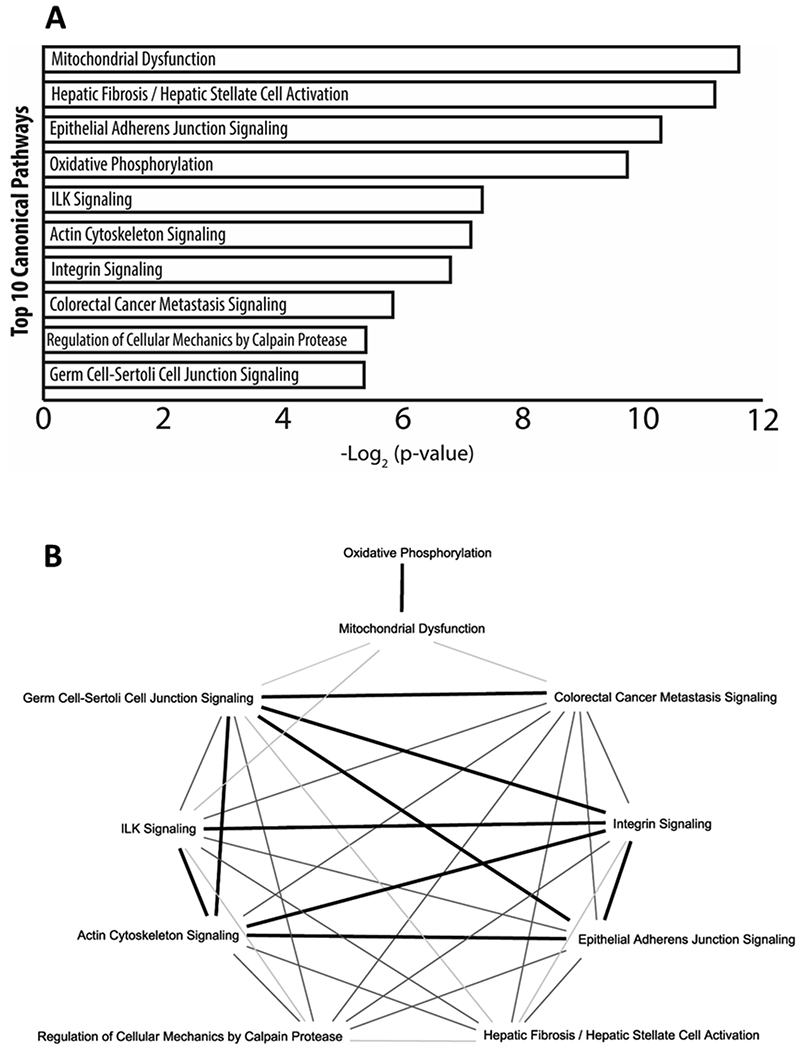
Subjecting all differentially expressed (CT1 as compared to CT23) protein-coding transcripts (edgeR, v3.24.1, B-H adjustment; p < 0.05, fold change ≥ ±2.0) to canonical pathway analysis with IPA (Ingenuity pathway analysis v48207413) revealed previously known biological pathways associated with RPE phagocytosis such as: integrin signaling, actin cytoskeleton signaling, germ cell-sertoli cell junction signaling, and epithelial adherens junction signaling. Much like the gene ontology analysis (Figure 3), the identification of mitochondrial processes was also apparent in the canonical pathway analysis. When the top 10 canonical pathways were plotted for common genes shared among the pathways, it revealed that integrin signaling, ILK signaling, actin cytoskeleton signaling, germ cell-sertoli cell junction signaling, and epithelial adherens junction signaling all had at least 20 genes in common (dark bold line) among them suggesting strong cross talk in these different pathways in mediating the process of RPE phagocytosis (b). While biological processes such as regulation of cellular mechanics by calpain protease and colorectal cancer metastasis signaling had less than 20 genes in common to the other identified pathways (light and medium grey lines, b). Interestingly, while mitochondrial processes were highly expressed in both gene ontology (Figure 3) and in canonical pathway analysis, there is very little overlap (light grey lines) among the other phagocytic associated pathways identified (b).
4. DISCUSSION
The overall aim of this study was to determine the change in the transcriptome of the RPE obtained from mice maintained in DD at two different circadian time points (CT23 and CT1). Our study identified a large number of protein-coding transcripts that were significantly upregulated at CT1 (Figure 1b) and a smaller number of non-coding transcripts that were up- and down- regulated at CT1 (Figure 1d) when compared to CT23. As expected with the large increase in transcriptional activity, a large majority of protein coding transcripts coded for: enzymes, transcriptional regulators, and kinases (Figure 2a) and had locations of action in the cytoplasm, nucleus, and plasma membrane (Figure 2b). Interestingly, the identification of lncRNAs and PS as non-protein coding transcripts revealed another level of complexity mediating the transcriptomic environment of RPE cells (Figures 2c and 2d). As expected, many biological pathways identified by either gene ontology or canonical pathway analysis indicated previously known processes (i.e., integrin signaling, cytoskeleton remodeling and response to cAMP) involved in the regulation of the daily peak of phagocytic activity by the RPE (Figures 3 and 4a). Furthermore, we also identified several transcripts involved in mitochondrial respiration that were upregulated at CT1 when compared to CT23 (Figures 3 and 4a). Finally, our data also indicated that about 20% of the RPE transcriptome may be under the control of a circadian clock.
As we have previously mentioned integrin signaling plays a key role in the regulation of the daily burst in RPE phagocytic activity (Nandrot et al., 2007, 2004; Ruggiero and Finnemann, 2014). A role for cAMP (cyclic adenosine monophosphate) in the regulation of such a phenomenon is also well established since Besharse, Dunis and Burnside (1982) reported that in a Xenopus eyecup, administration of dibutyryl cyclic adenosine monophosphate reduced the phagosome content of the Xenopus epithelium. Subsequent in vitro studies also reported that increasing the level of intracellular cAMP in RPE cells decreases phagocytic activity (Hall et al., 1993; Kuriyama et al., 1995). In this context it worth mentioning that administration of melatonin - which decreases levels of cAMP in RPE cells (Nash and Osborne, 1995) - has been also implicated in the regulation of the peak in RPE phagocytic activity (Besharse and Dunis, 1983; Laurent et al., 2017). Similarly, it has been shown that the activation of D2R which also decreases intracellular cAMP levels - are in fact involved in the regulation of the daily peak of the phagocytic activity by the RPE (Goyal et al., 2019).
Contrary to what was reported by Mustafi et al., (2013), our data do not directly indicate a role for the phosphoinositide pathway in the control of the circadian peak of RPE phagocytic activity. Such a discrepancy can be explained by the different experimental design of the two studies. In 2013, Mustafi and colleagues investigated the peak under LD conditions and at two different time points (1.5 and 9 hours after light onset) whereas in our study we have compared the changes in the RPE transcriptomic one hour before (CT23) and after (CT1) the expected onset of light under DD conditions. Thus, we believe that such a difference in the collection times makes it difficult to compare the two datasets when it comes to activated biological processes.
A surprising result that was identified by both gene ontology and IPA canonical pathway analysis revealed a role for mitochondrial respiration in contributing to RPE phagocytic activity (Figures 3 and 4a). However, a role for mitochondrial processes have already been demonstrated in professional phagocytes (e.g., macrophages, neutrophils, monocytes, and mast cells) in the immune system (see review: Sancho et al., 2017). Specifically, how metabolic shifts mediated by the mitochondria help promote phagocytic clearance in these professional phagocytes (see review: Langston et al., 2017). Interestingly, a recent publication demonstrated that conditioned media from human umbilical tissue derived cells - which contains MerTK ligands such as protein S and Gas6 (Cao et al., 2016) - restored phagocytic activity and increased mitochondrial processes in cultured RPE tissue from diagnosed age-related macular degeneration patients (Inana et al., 2018). Most recently, a study using electron microcopy to characterize mitochondrial morphology demonstrated that in RPE cells with more distributed mitochondrial networks and greater mitochondrial density had decreased phagosome clearance in zebrafish RPE cells (Toms et al., 2019). As previously highlighted, recent evidence does suggest a role for mitochondrial processes in the context of phagocytic processes and RPE health. This study further suggests the involvement of the mitochondria in the regulation of phagocytic activity.
Several studies in mouse have shown that in many tissues/organs about 10-20 % of the mouse transcriptome is under circadian control (Storch et al., 2002; Zhang et al., 2014). A few studies have also investigated the circadian transcriptome in the eye. Storch et al., (2007) reported that in the retina about 10-15% of the transcriptome is under circadian control and more recently Jiao et al., 2019 have shown that also in the cornea approximately 24% of the genome is rhythmically transcribed under LD conditions. Although our study has only investigated the change at two circadian time points, our results suggest a similar pattern to what has being previously described in many different mouse organs and tissues (Storch et al., 2002; Zhang et al., 2014). Our data suggests that the circadian regulation of RPE functions may be important for the health and viability of this important tissue and in general of the retina.
Although the main scope of the current study was to determine the changes to the RPE transcriptome at two different circadian time points, our study also provided a characterization of the whole mouse RPE transcriptome and expand the dataset first published by Ida et al. (2004) and by Mustafi et al. (2013). Ida and colleagues were able to characterize a little over 2700 genes expressed in RPE/choroid tissue (Ida et al., 2004) via expressed sequence tag (EST) technology. While obviously current technology is much more sensitive to detect transcripts, there was still about 94% of genes identified by EST that were found in our RNA-seq dataset. Interestingly, when similar techniques of RNA detection were used (i.e., RNA-seq) there was about a 91% overlap in the RPE protein-coding transcriptome between the current study’s dataset and the one previously reported by Mustafi et al. (2013) whereas only about 10% of the total identified non-protein coding transcripts overlapped between the two datasets.
Lastly, we were able to further characterize our RNA-seq dataset into different classes for both the protein coding and non-protein coding transcripts (Figure 2). In accordance to previously published reports on RPE phagocytosis, we have identified many of the protein-coding transcripts that when transcribed into proteins are active in the cytoplasm where a vast majority of RPE physiological changes take place (e.g., actin cytoskeletal remodeling and protein phosphorylation). In addition, the identification of the nucleus and the plasma membrane was also not surprising as these two cellular locations are important for facilitating RPE phagocytosis of POS (see reviews: Ruggiero and Finnemann, 2014; Sparrow et al., 2010). Interestingly, many non-protein coding transcripts that were differentially expressed were classed as lncRNAs and PS (Figures 2c and 2d). Recently with the advent of various high-throughput sequencing methodologies, the identification of lncRNAs and PS in biological systems have been associated with regulating the expression of transcripts at various pre- and post-transcriptional levels (see reviews: Tay et al., 2014; Thomson et al., 2016).
While the results from this RNA-seq experiment are insightful to RPE biology, there are a few limitations that should be kept in mind. As previously stated, only two circadian time points were considered in assessing the circadian transcriptome of the RPE thereby limiting the l characterization of the RPE circadian transcriptome. Despite these limitations the data obtained in our study are similar circadian transcriptomic results from other tissues previously examined
5. CONCLUSIONS
In conclusion, we believe that the data from this study increases the knowledge of the RPE transcriptome. Additionally, previously identified biological pathways associated with RPE phagocytosis (e.g., integrin signaling, protein phosphorylation, actin cytoskeleton remodeling) seem to be under circadian regulation. Whole transcriptome analysis also revealed various lncRNAs and PS which have been associated with regulating transcription and potentially reveals another level of complexity in the process of RPE phagocytosis. A novel finding from this study also suggests a role for mitochondrial respiration as a contributing factor to the regulation of RPE phagocytic activity.
Supplementary Material
14,083 mouse RPE transcripts were detected in common between CT23 and CT1. 12,005 were protein coding transcripts and 2078 were non-protein coding transcripts.
2421 protein coding transcripts were significantly upregulated whereas only 3 transcripts were significantly downregulated and 12 non-protein coding transcripts were significantly upregulated.
The majority of the upregulated transcripts were classified as: enzymes, kinases, and transcriptional regulators with a large majority of activity in the cytoplasm, nucleus, and plasma membrane.
Our analysis also identified new pathways (e.g., increased mitochondrial respiration via increased oxidative phosphorylation) that may be involved in the circadian control of phagocytic activity.
Finally, our results also indicate, as seen in other tissues, about 20 % of the whole RPE transcriptome may be under circadian clock regulation.
Acknowledgments
FUNDING: This work was supported by the National Institutes of Health [R01EY026291, P30 EY006360, U54NS083932, U54MD007602]
ABBREVIATIONS
- cAMP
cyclic adenosine monophosphate
- CPM
counts per million
- CT
circadian time
- D2R
dopamine 2 receptor
- DD
dark:dark
- DEGs
differentially expressed genes
- Hey1
hairy/enhancer-of-split related with YRPW motif
- LD
light:dark
- lncRNA
long non-coding RNA
- PCA
principal component analysis
- Per2
period circadian clock 2
- POS
photoreceptor outer segment
- PS
pseudogene
- RLE
relative log expression
- RNA-seq
ribonucleic acid-sequencing
- RPE
retinal pigment epithelium
- Rspo1
R-spondin 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Baba K, DeBruyne JP, Tosini G, 2017. Dopamine 2 Receptor Activation Entrains Circadian Clocks in Mouse Retinal Pigment Epithelium. Sci. Rep 7, 1–7. 10.1038/s41598-017-05394-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba K, Sengupta A, Tosini M, Contreras-Alcantara S, Tosini G, 2010. Circadian regulation of the PERIOD 2::LUCIFERASE bioluminescence rhythm in the mouse retinal pigment epithelium-choroid. Mol. Vis 16, 2605–2611. [PMC free article] [PubMed] [Google Scholar]
- Bennis A, Gorgels TGMF, Brink J.B. ten, Spek, van der PJ, Bossers K, Heine VM, Bergen AA, 2015. Comparison of Mouse and Human Retinal Pigment Epithelium Gene Expression Profiles: Potential Implications for Age-Related Macular Degeneration. PLOS ONE 10, 1–23. 10.1371/journal.pone.0141597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besharse JC, Dunis DA, 1983. Methoxyindoles and photoreceptor metabolism: activation of rod shedding. Science. 219, 1341–1343. 10.1126/science.6828862 [DOI] [PubMed] [Google Scholar]
- Besharse JC, Dunis DA, and Burnside B 1982. Effects of cyclic adenosine 3’,5’-monophosphate on photoreceptor disc shedding and retinomotor movement. Inhibition of rod shedding and stimulation of cone elongation. The Journal of General Physiology. 79, 775–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besharse JC, Hollyfield JG, 1979. Turnover of mouse photoreceptor outer segments in constant light and darkness. Invest. Ophthalmol. Vis. Sci 18, 1019–1024. [PubMed] [Google Scholar]
- Bobu C, Hicks D, 2009. Regulation of retinal photoreceptor phagocytosis in a diurnal mammal by circadian clocks and ambient lighting. Invest. Ophthalmol. Vis. Sci 50, 3495–3502. 10.1167/iovs.08-3145 [DOI] [PubMed] [Google Scholar]
- Cao J, Murat C, An W, Yao X, Lee J, Santulli-Marotto S, Harris IR, Inana G, 2016. Human umbilical tissue-derived cells rescue retinal pigment epithelium dysfunction in retinal degeneration. STEM CELLS 34, 367–379. 10.1002/stem.2239 [DOI] [PubMed] [Google Scholar]
- DeVera C, Baba K, Tosini G, 2019. Retinal Circadian Clocks are Major Players in the Modulation of Retinal Functions and Photoreceptor Viability. Yale J. Biol. Med 92, 233–240. [PMC free article] [PubMed] [Google Scholar]
- Doyle SE, Grace MS, McIvor W, Menaker M, 2002. Circadian rhythms of dopamine in mouse retina: the role of melatonin. Vis. Neurosci 19, 593–601. [DOI] [PubMed] [Google Scholar]
- Felder-Schmittbuhl M-P, Buhr ED, Dkhissi-Benyahya O, Hicks D, Peirson N, Ribelayga CP, Sandu C, Spessert R, Tosini G, 2018. Ocular Clocks: Adapting Mechanisms for Eye Functions and Health. Invest Ophthalmol Vis Sci. 59, 4856–4869. 10.1167/iovs.18-24957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal V, DeVera C, Laurent V, Sellers J, Chrenek MA, Hicks D, Baba K, luvone PM, Tosini G, 2019. Dopamine 2 receptor signaling controls the daily burst in phagocytic activity in the mouse retinal pigment epithelium. bioRxiv 789917 10.1101/789917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace MS, Chiba A, Menaker M, 1999. Circadian control of photoreceptor outer segment membrane turnover in mice genetically incapable of melatonin synthesis. Vis. Neurosci 16, 909–918. [DOI] [PubMed] [Google Scholar]
- Hall MO, Abrams TA, Mittag TW, 1993. The phagocytosis of rod outer segments is inhibited by drugs linked to cyclic adenosine monophosphate production. Invest. Ophthalmol. Vis. Sci 34, 2392–2401. [PubMed] [Google Scholar]
- Ida H, Boylan SA, Weigel AL, Smit-McBride Z, Chao A, Gao J, Wistow G, Hjelmeland LM, 2004. EST analysis of mouse retina and RPE/choroid cDNA libraries. Mol. Vis 10, 439–444. [PubMed] [Google Scholar]
- Inana G, Murat C, An W, Yao X, Harris IR, Cao J, 2018. RPE phagocytic function declines in age-related macular degeneration and is rescued by human umbilical tissue derived cells. J. Transl. Med 16, 1–15. 10.1186/s12967-018-1434-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Song R, Regev A, Struhl K, 2015. Many lncRNAs, 5’UTRs, and pseudogenes are translated and some are likely to express functional proteins. eLife. 4, 1–21. 10.7554/eLife.08890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Wu M, Lu D, Gu J, Li Z, 2019. Transcriptional Profiling of Daily Patterns of mRNA Expression in the C57BL/6J Mouse Cornea. Curr. Eye Res 44, 1054–1066. 10.1080/02713683.2019.1625408 [DOI] [PubMed] [Google Scholar]
- Kuriyama S, Abrams TA, Mittag TW, 1995. Isoproterenol Inhibits Rod Outer Segment Phagocytosis by Both cAMP-Dependent and Independent Pathways. Invest Ophthal Vis Sci. 36, 730–736. [PubMed] [Google Scholar]
- Langston PK, Shibata M, Horng T, 2017. Metabolism Supports Macrophage Activation. Front. Immunol 8, 1–7. 10.3389/fimmu.2017.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent V, Sengupta A, Sánchez-Bretaão A, Hicks D, Tosini G, 2017. Melatonin signaling affects the timing in the daily rhythm of phagocytic activity by the retinal pigment epithelium. Exp. Eye Res 165, 90–95. 10.1016/j.exer.2017.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVail MM, 1976. Rod outer segment disk shedding in rat retina: relationship to cyclic lighting. Science 194, 1071–1074. 10.1126/science.982063 [DOI] [PubMed] [Google Scholar]
- Lewis TR, Kundinger SR, Link BA, Insinna C, Besharse JC, 2018. Kif17 phosphorylation regulates photoreceptor outer segment turnover. BMC Cell Biol. 19, 1–16. 10.1186/s12860-018-0177-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W-K, Bernstein MH, 1981. Daily patterns of the retinal pigment epithelium. Microperoxisomes and phagosomes. Exp. Eye Res 32, 1–10. 10.1016/S0014-4835(81)80033-4 [DOI] [PubMed] [Google Scholar]
- Milligan MJ, Lipovich L, 2015. Pseudogene-derived lncRNAs: emerging regulators of gene expression. Front. Genet 5, 1–7. 10.3389/fgene.2014.00476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafi D, Kevany BM, Genoud C, Bai X, Palczewski K, 2013. Photoreceptor phagocytosis is mediated by phosphoinositide signaling. FASEB J. 27, 4585–4595. 10.1096/fj.13-237537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandrot EF, Anand M, Almeida D, Atabai K, Sheppard D, Finnemann SC, 2007. Essential role for MFG-E8 as ligand for αvβ5 integrin in diurnal retinal phagocytosis. Proc. Natl. Acad. Sci 104, 12005–12010. 10.1073/pnas.0704756104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandrot EF, Kim Y, Brodie SE, Huang X, Sheppard D, Finnemann SC, 2004. Loss of Synchronized Retinal Phagocytosis and Age-related Blindness in Mice Lacking αvβ5 Integrin. J. Exp. Med 200, 1539–1545. 10.1084/jem.20041447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash MS, Osborne NN, 1995. Agonist-induced effects on cyclic AMP metabolism are affected in pigment epithelial cells of the Royal College of Surgeons rat. Neurochem. Int 27, 253–262. 10.1016/0197-0186(95)00040-f [DOI] [PubMed] [Google Scholar]
- Peixoto L, Risso D, Poplawski SG, Wimmer ME, Speed TP, Wood MA, Abel T, 2015. How data analysis affects power, reproducibility and biological insight of RNA-seq studies in complex datasets. Nucleic Acids Res. 43, 7664–7674. 10.1093/nar/gkv736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiero L, Connor MP, Chen J, Langen R, Finnemann SC, 2012. Diurnal, localized exposure of phosphatidylserine by rod outer segment tips in wild-type but not Itgb5−/− or Mfge8−/− mouse retina. Proc. Natl. Acad. Sci 109, 8145–8148. 10.1073/pnas.1121101109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiero L, Finnemann SC, 2014. Rhythmicity of the Retinal Pigment Epithelium, in: Tosini G, luvone PM, McMahon DG, Collin SP (Eds.), The Retina and Circadian Rhythms. Springer-Verlag, New York, pp. 95–112. [Google Scholar]
- Sancho D, Enamorado M, Garaude J, 2017. Innate Immune Function of Mitochondrial Metabolism. Front. Immunol 8, 1–9. 10.3389/fimmu.2017.00527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow J, Hicks D, P. Hamel C, 2010. The Retinal Pigment Epithelium in Health and Disease. Curr. Mol. Med 10, 802–823. 10.2174/156652410793937813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch K-F, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ, 2002. Extensive and divergent circadian gene expression in liver and heart. Nature 417, 78–83. 10.1038/nature744 [DOI] [PubMed] [Google Scholar]
- Su Terman J, Reme’ CE, Terman M, 1993. Rod outer segment disk shedding in rats with lesions of the suprachiasmatic nucleus. Brain Res. 605, 256–264. 10.1016/0006-8993(93)91748-H [DOI] [PubMed] [Google Scholar]
- Takahashi JS, 2017. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet 18, 164–179. 10.1038/nrg.2016.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Rinn J, and Pandolfi PP 2014. The multilayered complexity of ceRNA crosstalk and competition. Nature. 505, 344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teirstein PS, Goldman AI, O’Brien PJ, 1980. Evidence for both local and central regulation of rat rod outer segment disc shedding. Invest. Ophthalmol. Vis. Sci 19, 1268–1273. [PubMed] [Google Scholar]
- Toms M, Burgoyne T, Tracey-White D, Richardson R, Dubis AM, Webster AR, Futter C, Moosajee M, 2019. Phagosomal and mitochondrial alterations in RPE may contribute to KCNJ13 retinopathy. Sci. Rep 9, 1–15. 10.1038/s41598-019-40507-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutar L, Özgür A, and Tutar Y 2018. Involvement of miRNAs and Pseudogenes in Cancer. Methods Mol. Biol 1699, 45–66. [DOI] [PubMed] [Google Scholar]
- Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB, 2014. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci 111, 16219–16224. 10.1073/pnas.1408886111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


