Abstract
In the dataset presented in this article, sixty sugarcane samples were analyzed by eight visible / near infrared spectrometers including seven micro-spectrometers. There is one file per spectrometer with sample name, wavelength, absorbance data [calculated as log10 (1/Reflectance)], and another file for reference data, in order to assess the potential of the micro-spectrometers to predict chemical properties of sugarcane samples and to compare their performance with a LabSpec spectrometer. The Partial Least Square Regression (PLS-R) algorithm was used to build calibration models. This open access dataset could also be used to test new chemometric methods, for training, etc.
Keywords: Micro-spectrometers; Handheld devices; Visible-near infrared spectroscopy; chemometrics, sugarcane
Specifications table
| Subject | VIS-NIR Spectroscopy, micro-spectrometers |
| Specific subject area | Sugarcane characterization |
| Type of data | Table Figure Spectroscopic data |
| How data were acquired | Absorbance spectra data were acquired by measurements on the following spectrometers: LabSpec 4 (ASD i.e. Analytical Spectral Devices, Boulder, CO, USA), NIRscan Nano (Texas instruments Inc., Texas, USA), F750 (Felix Instrument Inc., Camas, WA, USA), MicroNIR1700 (Viavi Solution, Milpitas, CA, USA), MicroNIR2200 (Viavi Solution, Milpitas, CA, USA), NIRONE 2.2 (Spectral Engines, Finland), SCIO (Consumer Physics, Tel Aviv, Israel), and TellSpec (Tellspec Inc., Toronto, Ontario, Canada). Chemical data comes from Cirad Selmet unit database. |
| Data format | Raw Analysed Presented as .csv file formats |
| Parameters for data collection | Dry ground samples from different sugarcane organs were stored under ambient moisture and temperature conditions of the laboratory. Spectra were recorded in reflectance mode in the same small circular cups with quartz glass cover for all spectrometers. Spectralon (99% reflectance) was used as reference background. |
| Description of data collection | The spectra and reference data were recorded in .CSV file formats. For reference data, the first column corresponds to sample identification; the second to the fifth columns correspond to chemical composition: sugar content (TS,% DM), crude protein content (CP,% DM), acid detergent fiber (ADF,% DM), in vitro organic matter digestibility (IVOMD,% of organic matter). For the absorbance data [calculated as log10 (1/Reflectance)], there is one .CSV file for each spectrometer. The file name is the spectrometer name. The first column corresponds to sample identification, and the next ones correspond to absorbance value according to wavelength. The wavelength unit is nanometer, and absorbance ranges depend of each spectrometer. Spectra data were then regressed with TS and CP reference values to generate calibration models. Predicted values and reference values were then compared in repeated cross validation (2 K-fold groups, repeated 50 times) to compare spectrometer performances. These data were recorded in .CSV file formats, one for TS and TP respectively. |
| Data source location | Spectra data, reference data of sugarcane samples were obtained at Cirad, Montepllier, France |
| Data accessibility | Dataset presented as .CSV file formats are available on this article. Dataset can be download in Mendeley repository data: https://data.mendeley.com/datasets/mjttsjfj2s/draft?a = 93aa9093–32f9–43e6–9d42–0c39729caa38 |
Value of the data
-
•
These data can be used to compare the performance of several micro-spectrometers with Labspec spectrometer considered here as reference
-
•
These data can be used to test new chemometric methods
-
•
These data can be used for training
1. Data description
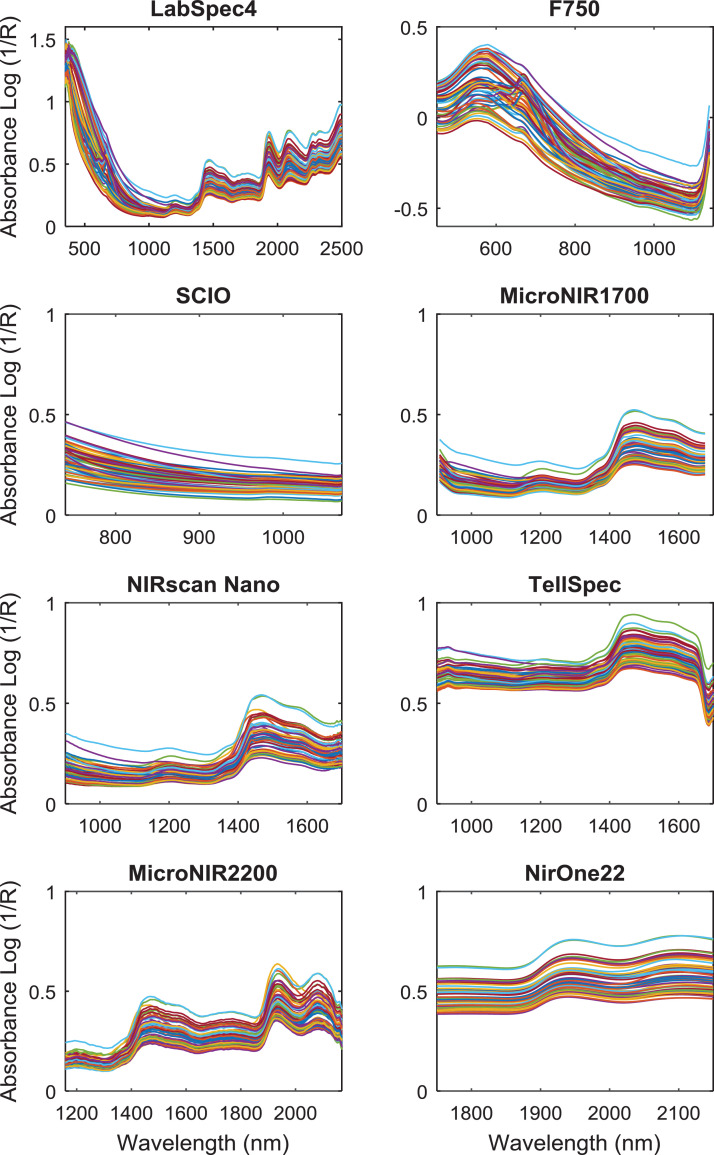
Chemical and Nir spectra measurements were made on 60 sugarcane samples from different plant parts (leaves, stem or whole aerial part). Chemical parameters (total sugar content - TS, crude protein content - CP, acid detergent fiber - ADF, in vitro organic matter digestibility - IVOMD were determined (Table 1). In parallel, reflectance spectra were measured using eight spectrometers (Table 2 and Fig. 1). Absorbance data was calculated as log10 (1/Reflectance). To illustrate, PLS-R results are shown for the total sugar content (Table 3) and crude protein content (Table 4).
Table 1.
Summary statistics of chemical properties of sugarcane samples used for calibration.
| Chemical properties | Unit | Min | Max | Mean | SD |
|---|---|---|---|---|---|
| Total sugar content | % dry matter | 1.1 | 51.0 | 23.4 | 17.3 |
| Crude protein content | 0.9 | 9.6 | 3.1 | 2.1 | |
| Acid detergent fiber fraction | 26.0 | 59.3 | 39.2 | 8.7 | |
| In vitro organic matter digestibility | 13.0 | 66.6 | 41.0 | 15.1 |
Table 2.
Specific characteristics for NIR spectrometers.
| Device and manufacturer | Spectral range (nm) | Resolution (nm) | Technology | Lighting module | Weight |
|---|---|---|---|---|---|
| LabSpec 4 (ASD) |
350–1000 1001–1800 1801–2500 |
3 @ 700 nm 10 @ 1400/2100 nm |
Silicon array InGas photodiode array InGas photodiode array |
1 halogen lamp | 5.44 kg |
| NIRscan Nano (Texas Instrument) | 901–1701 | 10 | 1 photodiode InGaAs | 2 halogen lamps | 85 g |
| F750 (Felix Instrument) | 450–1140 | 8–13 | Diode array | 1 xenon tungsten lamp | 1.05 kg |
| MicroNIR1700 (Viavi) | 908–1676 | 6 | InGas photodiode array | 2 tungsten lamps | <60 g |
| MicroNIR2200 (Viavi) | 1158–2169 | 8 | 1 photodiode InGaAs | 2 tungsten lamps | <60 g |
| NIRONE 2.2 (Spectral Engines) | 1750–2150 | 20–26 | InGaAs photodiode array | 2 tungsten lamps | 15 g |
| SCIO (Consumer Physics) | 740–1070 | Not communicated | 2 photodiodes | 1 LED lamp | 35 g |
| TellSpec (TellSpec) | 900–1700 | 10 | 1 photodiode InGas | 2 halogen lamps | 136 g |
Fig. 1.
Sugarcane Absorbance raw spectra collected from the eight devices (for F750 spectra are normalized).
Table 3.
Cross-validation results for total sugar content (% dry matter, SNV: standard normal variate, Der1: Savitzky–Golay derivative 1, Der2: Savitzky–Golay derivative 2, NORM: normalization, RMSECV: Root Mean Square of Standard Error in Cross-Validation).
| Device | Pretreatment | Latent variables | RMSECV | R2cv |
|---|---|---|---|---|
| DLP NIRscan Nano EVM | SNVDer2 | 3 | 6.2 | 0.864 |
| F750 | NORM | 4 | 8.9 | 0.720 |
| LabSpec 4 | SNV | 8 | 2.6 | 0.976 |
| MicroNIR1700 | SNVDer1 | 5 | 3.8 | 0.949 |
| MicroNIR2200 | SNVDer1 | 8 | 2.8 | 0.972 |
| NIRONE 2.2 | SNVDer2 | 7 | 7.7 | 0.791 |
| Scio | SNVDer2 | 4 | 9.4 | 0.687 |
| TellSpec | Der1 | 5 | 9.3 | 0.692 |
Table 4.
Cross-validation results for crude protein content (% dry matter, SNV: standard normal variate, Der1: Savitzky–Golay derivative 1, Der2: Savitzky–Golay derivative 2, NORM: normalization, RMSECV: Root Mean Square of Standard Error in Cross-Validation).
| Device | Pretreatment | Latent variables | RMSECV | R2cv |
|---|---|---|---|---|
| DLP NIRscan Nano EVM | SNVDer1 | 8 | 1.2 | 0.656 |
| F750 | NORM | 3 | 0.7 | 0.874 |
| LabSpec 4 | SNVDer1 | 5 | 0.5 | 0.926 |
| MicroNIR1700 | SNVDer2 | 9 | 0.6 | 0.911 |
| MicroNIR2200 | SNVDer1 | 6 | 0.7 | 0.893 |
| NIRONE 2.2 | SNVDer1 | 8 | 0.6 | 0.900 |
| Scio | – | 5 | 0.7 | 0.874 |
| TellSpec | SNVDer2 | 9 | 1.1 | 0.732 |
2. Experimental design, materials, and methods
2.1. Samples and analyses
Sixty samples of sugarcane from different plant parts (leaves, stem or whole aerial part) were selected from Cirad samples collected in the French West Indies (Guadeloupe) for which the chemical data were available.
Before chemical analysis, the samples were dried for 72 h at 85 °C, milled with a Retsch Ⓡ SM100 mill (Retsch GmBH, Germany) with a 1 mm exit sieve and analyzed in the Cirad Selmet feed laboratory (https://umr-selmet.cirad.fr/en/products-and-services/laboratory-specialized-in-animal-feed-resources) to determine TS content, CP content, ADF, and IVOMD.
CP content was estimated from the total nitrogen content (N) measured by the Kjeldahl method [1], with the relationship CP = N * 6.25. Total mineral content (MM, mineral matter or ash) was determined by ashing at 550 °C. TS content was determined by the modified Luff-Schoorl method [2]. ADF was determined according to the Van Soest method [3]. IVOMD was measured by an enzymatic method (pepsine cellulose) [4].
3. NIR spectra acquisition
All samples were scanned with following spectrometers: LabSpec 4 (ASD i.e. Analytical Spectral Devices, Boulder, CO, USA), NIRscan Nano (Texas instruments Inc.,Texas, USA), F750 (Felix Instrument (Camas, WA, USA), MicroNIR1700 (Viavi Solution–Milpitas, CA, USA), MicroNIR2200 (Viavi Solution–Milpitas, CA, USA), NIRONE 2.2 (Spectral Engines, Finland), SCIO (Consumer Physics, Tel Aviv, Israel), and TellSpec (Tellspec Inc., Toronto, Ontario, Canada). Because of the different technologies used, spectral range, and resolution are instrument dependent (Table 2).
All samples used in this study were stored before measurement under the ambient moisture and temperature conditions of the laboratory (sample with +/- 8% of moisture content). Spectral measurements of sample were recorded in reflection (Fig. 1) mode in small circular cups (diameter 50 mm) with a quartz glass cover. A background with Spectralon (99% reflectance) was performed before each spectrum sample acquisition. To limit possible device drift, a measurement of this reference was performed before each sample measurement. For the MicroNIR1700 device, the spectral analysis has been done in triplicate but we only show results for one replicate (set 1).
4. Data analysis
All the calculations were run under Matlab (The Mathworks, Natick, MA, USA). Before data processing, spectral resolution was standardized at 2 nm for all spectrometers by filtering. A PLS-R algorithm was used to perform model TS content and CP content [5]. The number of latent variables and the best spectra pretreatment were determined by comparing the performance of two fold-group repeated cross-validation (2 random groups of 30 samples selected at random, repeated 50 times) [6]. Model results for TS content and for CP content (table 3 & 4), for the best spectra pretreatment (among SNV, derivatives, normalization) were evaluated on the basis of the coefficient of determination (R²cv) and the root mean square standard error of cross-validation (RMSECV).
CRediT author statement
Abdallah Zgouz: conceptualization, methodology, investigation, data curation, validation, writing - original draft; Daphné Héran: conceptualization, investigation, methodology; Bernard Barthès: Resources, writing - review & editing; Denis Bastianelli: resources, writing - review & editing; Laurent Bonnal: resources; Vincent Baeten: methodology, validation; writing- reviewing and editing; Sebastien Lurol: resources; Michael Bonin: resources; Jean-Michel Roger: software, writing- reviewing and editing, validation; Ryad Bendoula: funding acquisition, conceptualization, supervision, methodology, validation, writing - review & editing, investigation; Gilles Chaix: project administration, funding acquisition, conceptualization, investigation; methodology, data curation, supervision, validation, writing - original draft, writing - review & editing.
Competing interests
The authors declare that they have no known competing financial interests or personal relationships, which have, or could be perceived to have, influenced the work reported in this article.
Declaration of Competing Interest
None.
Acknowledgments
The authors would like to acknowledge the French near infrared spectroscopy scientific network HelioSPIR for financial support; as well as the Itap unit, and Cirad (Agap unit, Anne Clément Vidal), and Adisseo (Vincent Larat) for financial and/or infrastructure supports.
References
- 1.SMRI Determination of the reducing sugars in raw sugar by the Luff Schoorl method. 1997SMRI Test methods, TM050.
- 2.Commission Regulation (EC) No 152/2009 of 27 January 2009 laying down the methods of sampling and analysis for the official control of feed. Off J Eur Union. Part J. 2009:130. [Google Scholar]
- 3.Van Soest P.J., Robertson J.B., J.B., Lewis B.A., B.A. Methods for dietary fibre, neutral detergent fibre and non starch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991;74:3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2. [DOI] [PubMed] [Google Scholar]
- 4.Aufrère J. Laboratory prediction of forage digestibility by the pepsin-cellulase method. the renewed equations. INRA Prod. Anim. 2007;20(2):129–136. [Google Scholar]
- 5.Wold S., Sjöström M., Eriksson L. Chemometrics and intelligent laboratory systems. Chem. Intell. Lab. Syst. 2001;58:109–130. [Google Scholar]
- 6.Wold S. Cross-validatory estimation of the number of components in factor and principal components models. Techn. 1978;20:397–405. [Google Scholar]