Abstract
Background
Pandemic COVID-19 by severe acute respiratory syndrome (SARS) coronavirus 2 (SARS-CoV-2) infection is facilitated by the ACE2 receptor and protease TMPRSS2. Modestly sized case series have described clinical factors associated with COVID-19, while ACE2 and TMPRSS2 expression analyses have been described in some cell types. Patients with cancer may have worse outcomes to COVID-19.
Methods
We performed an integrated study of ACE2 and TMPRSS2 gene expression across and within organ systems, by normal versus tumor, across several existing databases (The Cancer Genome Atlas, Census of Immune Single Cell Expression Atlas, The Human Cell Landscape, and more). We correlated gene expression with clinical factors (including but not limited to age, gender, race, body mass index, and smoking history), HLA genotype, immune gene expression patterns, cell subsets, and single-cell sequencing as well as commensal microbiome.
Results
Matched normal tissues generally display higher ACE2 and TMPRSS2 expression compared with cancer, with normal and tumor from digestive organs expressing the highest levels. No clinical factors were consistently identified to be significantly associated with gene expression levels though outlier organ systems were observed for some factors. Similarly, no HLA genotypes were consistently associated with gene expression levels. Strong correlations were observed between ACE2 expression levels and multiple immune gene signatures including interferon-stimulated genes and the T cell-inflamed phenotype as well as inverse associations with angiogenesis and transforming growth factor-β signatures. ACE2 positively correlated with macrophage subsets across tumor types. TMPRSS2 was less associated with immune gene expression but was strongly associated with epithelial cell abundance. Single-cell sequencing analysis across nine independent studies demonstrated little to no ACE2 or TMPRSS2 expression in lymphocytes or macrophages. ACE2 and TMPRSS2 gene expression associated with commensal microbiota in matched normal tissues particularly from colorectal cancers, with distinct bacterial populations showing strong associations.
Conclusions
We performed a large-scale integration of ACE2 and TMPRSS2 gene expression across clinical, genetic, and microbiome domains. We identify novel associations with the microbiota and confirm host immunity associations with gene expression. We suggest caution in interpretation regarding genetic associations with ACE2 expression suggested from smaller case series.
Keywords: translational medical research, gene expression profiling, immunotherapy, tumor microenvironment
Background
Severe acute respiratory syndrome (SARS) coronavirus 2 (SARS-CoV-2), which causes the disease COVID-19, was initially described near the end of 20191 2 and has caused a global pandemic. SARS-CoV-2 is a positive-sense single-strand RNA virus related to the SARS and the Middle East respiratory syndrome (MERS) coronaviruses that have caused previous global health emergencies.3 COVID-19 is characterized predominately by fever, cough, and pneumonia, with some patients presenting with diarrhea and other symptoms.4 5 Mortality rates are described as approximately 10 times higher than seasonal influenza in some clinical subgroups.6
Angiotensin-converting enzyme 2 (ACE2) has been identified as the receptor for the SARS-CoV family,7 and the SARS-CoV-2 spike protein binds ACE2 on host cells with greater affinity than previous SARS-CoV.8 9 Type II transmembrane serine protease TMPRSS2 is the primary human protease that mediates spike protein activation on infected cells, facilitating viral entry via receptor-mediated internalization.9 10 Multiple physiologic roles are known for ACE2 impacting systems such as cardiovascular, nephrology, and immune11 but perhaps most notably related to SARS-CoV-2, pulmonary, where ACE2 has been described to limit severe acute lung injury.12 Analyses of ACE2 protein expression by organ system have suggested high levels in epithelia of the lung and small intestine, consistent with presenting symptoms of patients with COVID-19.13 However, these studies have not integrated analysis of TMPRSS2 and integrated analyses may better inform which organ systems express both genes and may be at greatest infection risk.
Gene expression studies by bulk RNA sequencing and single-cell approaches have attempted to delineate expression patterns of normal airway tract and other tissues.14–16 These studies have suggested high ACE2 expression levels on the epithelia of oral and airway mucosa as well as small intestine. ACE2 has additionally been suggested as an interferon-response gene suggesting a complicated interaction between viral infection and host antiviral response.15 Further, a report has been advanced suggesting that lymphocytes may directly be infected by SARS-CoV-2,17 a finding reported with MERS as well,18 however of unclear clinical significance.
Patients with cancer may be at increased risk for SARS-CoV-2 infection and deleterious outcomes to COVID-19 disease though reports have varied by geography and cancer histology. In a single hospital study from Wuhan, China, patients with cancer made up 1% of the overall prevalence of COVID-19,19 substantially higher than the overall incidence of cancer in the Chinese population at 0.29%.20 Outcomes to COVID-19 appeared to be worse in patients with cancer with increased intensive care unit admission, mechanical ventilation, and mortality, especially those who had recently received chemotherapy or surgery.19 In western populations, risk has appeared to be higher for the cancer population as well. In the UK, a study of 800 patients demonstrated a 28% mortality rate in hospitalized patients with cancer with risk of death associated with age, male sex, and medical comorbidities but not clear interaction between recent treatment with specific anticancer agents.21 In North America, predominately the USA, 30-day all-cause mortality has been described as high for patients with cancer relative to the general population at 13% with regional variation demonstrating higher mortality on the eastern seaboard relative to the middle of the country or Canada.22 In this cohort, similar risk factors including age, male, smoking, increasing medical comorbidities, and performance status were observed.
Increased risk for some patients may also relate to treatment administered though data vary somewhat. In hematologic malignancies, increased mortality after infection has been widely observed23 with some connections to recent chemotherapy.24 In solid tumors, the risk of recent chemotherapy may also be linked to other high risk factors such as age with the impact of treatment being variable.22 Particularly, there has been concern that patients being treated with cancer immunotherapy drugs might be at increased risk given the possible overlapping immune-related toxicities for checkpoint blocking antibodies with the pneumonitis and diarrheal syndromes seen in COVID-19. This remains an area of open investigation with analyses even from the same institution giving conflicting results about the risk of immunotherapy, notably in advanced lung cancer.25 26 Multiple societies, including the Society for Immunotherapy of Cancer, have issued guidance for cancer care during the pandemic as well as the use of immunomodulatory agents such as anti-IL6.27
Several clinical associations have arisen from a smaller series of patients infected with COVID-19. Some include risk factors for poor outcomes such as elevated body mass index (BMI)28 and diabetes29 as well as possible associations with race.30 More broadly, germline genetics and host immune status may have a substantial impact on antiviral host defense,31 similar to what is seen with cancer immunotherapy.32
To better inform considerations surrounding SARS-CoV-2 and COVID-19 in patients with cancer and more broadly in the general population, we performed an integrated analysis of ACE2 and TMPRSS2 gene expression across clinical, genetic, and microbiome domains. We identify novel associations with the commensal microbiota and confirm host immunity associations with gene expression. We suggest caution against overinterpretation of clinical or genetic associations from smaller case series noting that these are not strongly associated with ACE2 or TMPRSS2 gene expression. We hope these data may better inform clinical considerations surrounding risk stratification and prevention approaches.
Methods
Datasets
Sample metadata tables were downloaded from The Cancer Genome Atlas (TCGA) (Genomic Data Commons (GDC) portal: https://portal.gdc.cancer.gov). Out of 11,093 aliquots total, 10,732 were selected to keep one unique aliquot per patient per sample type, as illustrated in online supplementary figure 1. The final cohort consists of 9,657 primary tumors, 367 metastatic samples (all from skin cutaneous melanoma), and 708 normal tissues from 10,038 patients across 34 tumor types (33 primary and one metastatic) (online supplementary tables 1 and 2). Out of 708 normal tissues, 14 tumor types have 15 or more normal samples available and hence were included for statistical comparisons when applicable. The standardized, upper-quartile normalized, batch-corrected, and platform-corrected RNAseq expression of 20,531 genes in RNA-Seq by Expectation Maximization (RSEM)-quantified read count estimates were downloaded from Pan-Cancer Atlas consortium studies (https://gdc.cancer.gov/about-data/publications/pancanatlas) and log2-transformed for further analysis. Fragments Per Kilobase of transcript per Million mapped reads (FPKM) quantification of gene expression and whole-exome sequencing (WES) Binary Alignment Map (BAM) files were downloaded from GDC.33 Demographic and clinical information were retrieved from the TCGA Pan-Cancer Clinical Data Resource.34 BMI and smoking history were retrieved from legacy clinical files. To the authors’ knowledge, only two tumor types had diabetes status information (pancreatic adenocarcinoma (PAAD) and uterine corpus endometrial carcinoma (UCEC)), which was retrieved from clinical XML files on GDC. Eighty-two percent of patients with PAAD and 7% of patients with UCEC have diabetes status recorded. Hence, only PAAD was included in analysis. Commensal microbiota abundance and viral presence of TCGA samples were downloaded from published studies.35 36 Single-cell RNAseq (scRNAseq) gene expression in malignant cells, immune cells, and normal cells were retrieved from nine studies consisting of patients diagnosed with cancer and healthy donors. Links to data files, single-cell cohorts, and bioinformatics software are provided in online supplementary table 3. Data generated in this study are accessible on GitHub repository https://github.com/riyuebao/ACE2_TMPRSS2_multicorrelates.
jitc-2020-001020supp001.pdf (336.7KB, pdf)
jitc-2020-001020supp002.xlsx (1.8MB, xlsx)
ACE2 and TMPRSS2 gene expression correlation and percentile calculation
The gene expression of ACE2 (Entrez Gene ID 59272) and TMPRSS2 (Entrez Gene ID 7113) was retrieved from the RSEM-quantified RNAseq data and used for all analyses described in this study. Spearman’s correlation was calculated between the expression of the two genes in tumor (n=10,024) and normal (n=708) samples across all tumor types and within individual tumor types. The expression percentile was calculated separately within each of the four analysis sets (ACE2 in normal, ACE2 in tumor, TMPRSS2 in normal, TMPRSS2 in tumor) following two steps. First, the median expression of ACE2 or TMPRSS2 was calculated within individual tumor types. Next, tumor types were ranked by the median expression of each gene from higher to lower, and the position of each tumor type in the ranked list was scaled to 0–100, hereafter regarded as “expression percentile” per tumor type, with smaller values indicating top-ranked tumor types. The same process was repeated for each gene in tumor samples (34 tumor types) and normal tissues (14 tumor types).
Analysis of clinical correlates
The expression of ACE2 and TMPRSS2 was compared between designated clinical groups, split by age (younger (<65 years)/older (≥65 years) in tumor or normal), gender (female/male in tumor or normal), race (African American (AA)/Asian/White in tumor or normal), menopause (not post/post in tumor or normal), BMI (level 1 (<25)/level 2 (25–30)/level 3 (30–35)/level 4 (>35)37 in tumor or normal), smoking history (never/light/heavy38 in tumor or normal), tumor stage (I/II/III/IV in tumor), tumor grade (G1G2/G3G4 in tumor). For tumor grade, G1 and G2 were collapsed to indicate low-grade to mid-grade (G1G2), and G3 and G4 were collapsed to indicate high-grade (G3G4). Within each clinical factor, sub-groups of <15 samples were excluded. For each clinical factor, comparisons were performed across all tumors and within individual tumor types. For all tumor types, first, data were fitted into a two-way analysis of variance (ANOVA) model with tumor type and clinical group as variables plus the interaction between the two. Second, if more than two clinical groups exist, Tukey’s honest significance test (HSD) was used with the fitted ANOVA model for pairwise comparisons while controlling for type I errors. Within each tumor type, Tukey’s HSD was used with one-way ANOVA models when more than two groups are present; otherwise Welch Two Sample t-test was used. The list of clinical groups and statistical results are provided in online supplementary tables 4–7.
Analysis of HLA correlates
HLA-A, HLA-B, and HLA-C genotypes were identified for 9,559 patients from TCGA across 34 tumor types using OptiType (version 1.3.2) with WES BAM files. Considering each patient carries two copies of HLA-A, B, or C alleles, both copies were counted toward the total number (19,118) of A, B, or C alleles in the entire cohort. The calculation of HLA prevalence was performed at the allele level. The comparison of ACE2 and TMPRSS2 gene expression between HLA genotypes was performed at patient level, given that gene expression was estimated per sample. First, for each allele, labels 0, 1, or 2 were assigned per patient (0=this patient does not carry this allele; 1=this patient carries one copy of this allele; 2=this patient carries two copies of this allele). Then, gene expression differences were tested using two-way ANOVA models with HLA genotype (0, 1, 2) and tumor type as variables plus the interaction between the two. For each test, at least 15 samples carrying the allele are required. The test was repeated for every HLA-A, B, and C allele with gene expression in tumor or normal samples, followed by Benjamini and Hochberg-False Discovery Rate (BH-FDR) correction for multiple comparisons. Same analysis was repeated with age, gender, and race as covariates in the model.
Analysis of immune gene expression signatures
Five immune responsive and suppressive signatures (interferon-stimulated genes (ISG), T cell-inflamed (Tinfl), myeloid, angiogenesis (angio), and transforming growth factor-β (TGF-β)) (online supplementary table 8) were correlated with ACE2 and TMPRSS2 gene expression in tumor and normal tissues. The expression level of a signature was computed as the average expression of all genes in this signature after centering and scaling using R function scale with parameters scale=TRUE and center=TRUE. Spearman’s correlation was calculated between each signature and ACE2 or TMPRSS2. The full correlation metrics are provided in online supplementary table 9.
Analysis of immune and stromal cell subset correlates
FPKM estimates of RNAseq gene expression was used for quantifying enrichment of 64 tumor and stroma cell types using xCell (version 1.1.0). xCell converts gene expression into rank-based metrics within each sample; hence, normalization and batch correction were not required. To make data comparable across samples, xCell was run once using all samples (n=10,732). Spearman’s correlation was computed between the enrichment score of each cell population and ACE2 or TMPRSS2 expression. The full correlation metrics are provided in online supplementary table 10.
Analysis of viral-associated tumors
The human papillomavirus (HPV), Epstein-Barr virus (EBV), and hepatitis B virus (HBV) were selected for this study as those are the three most prevalent cancer-associated viruses in the cohort. Other viruses detected were excluded. Samples were set to HPV positive/negative by at least 10 normalized reads per million (NRPM) mapped to the HPV genomes, EBV positive/negative by at least 5 NRPM mapped to the EBV genomes, and HBV positive/negative by at least 5 NRPM mapped to the HBV genomes given previously recommended thresholds.36 Stomach adenocarcinoma (STAD), esophageal carcinoma (ESCA), acute myeloid leukemia (LAML), and ovarian serous cystadenocarcinoma (OV) were reset to “negative” for HPV, EBV, and HBV after a manual inspection, which revealed no strong clinical support for viral presence in those tumor types. Within each tumor type, ACE2 and TMPRSS2 gene expression was compared between viral positive and negative tumors using Welch Two Sample t-test. Across all tumor types, two-way ANOVA was used to compare gene expression between viral positive and negative tumors with tumor type and viral group as variables plus the interaction between the two.
Analysis of microbial correlates
The abundance of 1,093 genus-level microbial taxa was quantified from tissue RNAseq data after rigorous quality control, batch correction, and contamination filtering, and normalized to 1 million reads to make data comparable across samples.35 Seven hundred and six normal tissues and 9,801 tumor samples were included in the analysis where data were available. Taxa were filtered to keep bacteria in analysis; viruses and archaea were excluded. Nine hundred and fifty taxa present in at least 20% of samples were kept for statistical testing. Within each tumor type, Spearman’s correlation was computed between each bacteria taxon and ACE2 or TMPRSS2 gene expression in tumor and normal tissues. For each test, at least 15 samples with taxon abundance ≥1 were required. Seventy-five taxa passed FDR-adjusted p<0.05 and Spearman’s ρ>0.5 or <−0.5 in at least one pairwise correlation (online supplementary table 11).
LASSO regression modeling and variable importance
Least Absolute Shrinkage and Selection Operator (LASSO) regression models of ACE2 or TMPRSS2 gene expression were built in tumor (n=10,024) and normal (n=708) samples separately using 90 features consisting of tissue type as well as clinical (age, gender, race), immune signatures (ISG, T cell-inflamed, myeloid, angiogenesis, TGF-β), immune cell subsets (macrophage M1, macrophage M2, CD8 T cell, CD4 T cell), stroma cell subset (epithelial cell), and 75 microbiota features from microbiota correlation analysis (online supplementary figure 2). Macrophage M1/M2 and T cells were included in the model based on emerging evidence suggesting an important role of macrophage and proinflammatory phenotype39 in COVID-19 disease. Thirty-four tumor types were collapsed into 15 tissue types based on categorizations from The Human Protein Atlas to reduce complexity. Categorical variables were converted to dummy variables using R function dummyVars with parameter fullRank set to TRUE. Data were preprocessed to remove features that have near-zero variance, high correlation (Spearman’s ρ>0.75), or high collinearity. Each feature was scaled and centered. Given the purpose of this analysis was to evaluate the relative importance of features rather than training and validating a predictive model, we did not split samples into training and test sets. Instead, we used all samples with 10-fold cross-validation. Variable importance was reported as raw values and as scaled values to 0–100 (online supplementary table 12). R package caret (version 6.0–84) was used for analysis.
Statistical analysis
In all analyses, a minimal sample size of 15 per group was required for statistical testing. For group-wise comparisons, two-way ANOVA was used with tumor type and group of interest as variables plus the interaction. When more than two groups are present, Tukey’s HSD test was used for pairwise comparisons. Within each tumor type, two-sided Welch Two Sample t-test was used to compare log2-transformed gene expression between groups; for paired samples, two-sided paired t-test was used. Spearman’s correlation was used to determine the relationship between two continuous variables. For multiple comparisons, p values were corrected using BH-FDR method. LASSO regression models were used to evaluate variable importance with 10-fold cross-validation. Those analyses were performed using R (version 3.6.1) and Bioconductor (release 3.10). FDR-adjusted p values less than 0.05 were considered statistically significant.
Results
ACE2 and TMPRSS2 are highly expressed in digestive organs and tumors, however, lower in tumor compared to matched normal
Given the association between COVID-19 disease and various organ-specific symptoms, we investigated the distribution of ACE2 and TMPRSS2 expression in tumor and normal tissues across 34 tumor types consisting of 15 tissue types (online supplementary tables 1–3). None of the LAML samples express TMPRSS2; hence, 33 tumor types were used for the analysis of TMPRSS2.
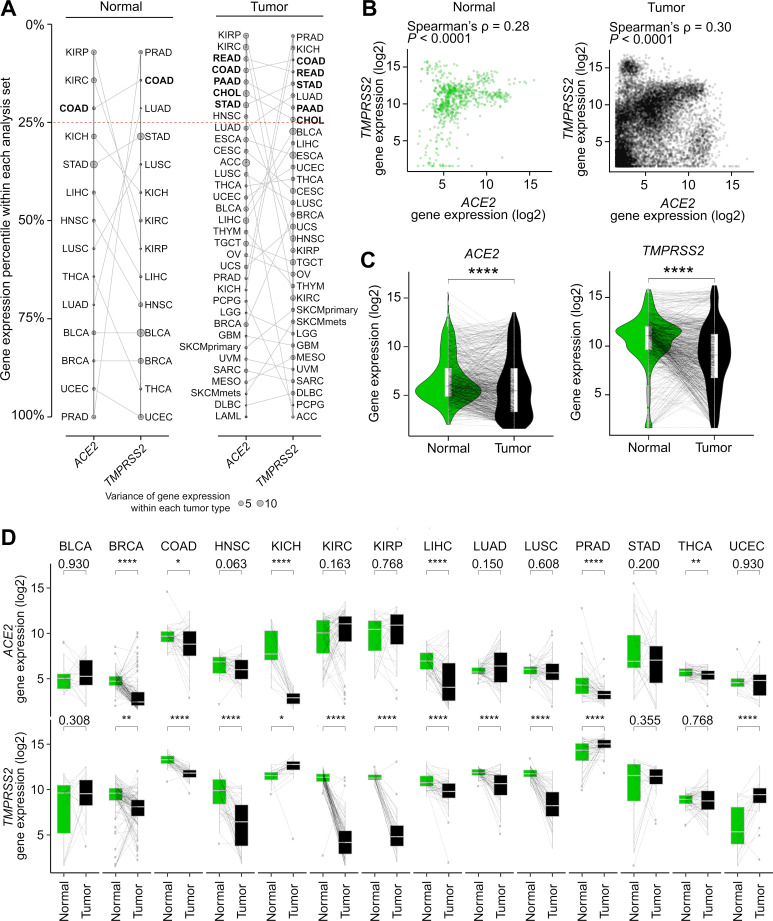
When ranked by expression percentile in each tumor type, cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), PAAD, rectum adenocarcinoma (READ), and STAD are among the top 25% percentile for both genes, all of which are digestive organs (figure 1A). We acknowledge that TMPRSS2 is highly expressed in prostate adenocarcinoma (PRAD) likely due to known TMPRSS2:ERG gene fusion overexpression.40 ACE2 and TMPRSS2 expression showed weak positive correlation in normal (Spearman’s ρ=0.28, FDR-adjusted p<0.0001) and in tumor (ρ=0.30, FDR-adjusted p<0.0001) (figure 1B). Correlation within each tumor type showed a consistent pattern and can be further explored in external data files available on GitHub (https://github.com/riyuebao/ACE2_TMPRSS2_multicorrelates).
Figure 1.
Distribution of ACE2 and TMPRSS2 gene expression in tumor and normal tissues across 34 tumor types. (A) Tumor types ranked by expression percentile in each gene. Five tumor types that show >25% expression percentile within each analysis set in both genes are bolded. Four analysis sets are shown: (left panel) ACE2 in normal (n=14 tumor types), TMPRSS2 in normal (n=14 tumor types); (right panel) ACE2 in tumor (n=34 tumor types), TMPRSS2 in tumor (n=33 tumor types; LAML not shown due to lack of TMPRSS2 expression in this tumor type). (B) Correlation between ACE2 and TMPRSS2 gene expression in normal (n=708 samples) and tumor (n=10,024 samples). (C, D) ACE2 and TMPRSS2 gene expression are higher in normal relative to tumor samples in (C) all tumor types pooled and in (D) individual tumor types. Line connects tumor and matched normal samples from the same patient (n=692 patients). Spearman’s correlation was used in (B). Two-sided paired t-test was used in (C) and (D). P values shown are after FDR correction for multiple comparisons. ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05. For comparisons that do not reach significance level of 0.05, exact p values are shown. A full description of each cancer ID is provided in online supplementary table 1. LAML, acute myeloid leukemia.
In 14 tumor types where 15 or more matched normal tissues are available (online supplementary table 1), we compared gene expression between tumor and matched normal from the same patients. The expression of ACE2 and TMPRSS2 was significantly higher in normal tissues relative to tumors (figure 1C). Within individual tumor types, this pattern was significant for ACE2 in breast invasive carcinoma (BRCA), COAD, chromophobe kidney cancer (KICH), liver hepatocellular carcinoma (LIHC), PRAD, thyroid cancer (THCA), and for TMPRSS2 in eight tumor types (FDR-adjusted p<0.05 and higher in normal). Three tumor types showed elevated levels for both genes in normal tissues compared with tumors (BRCA, COAD, and LIHC) (figure 1D).
Clinical factors and HLA genotypes do not strongly associate with ACE2 and TMPRSS2 gene expression in tumor or normal tissues
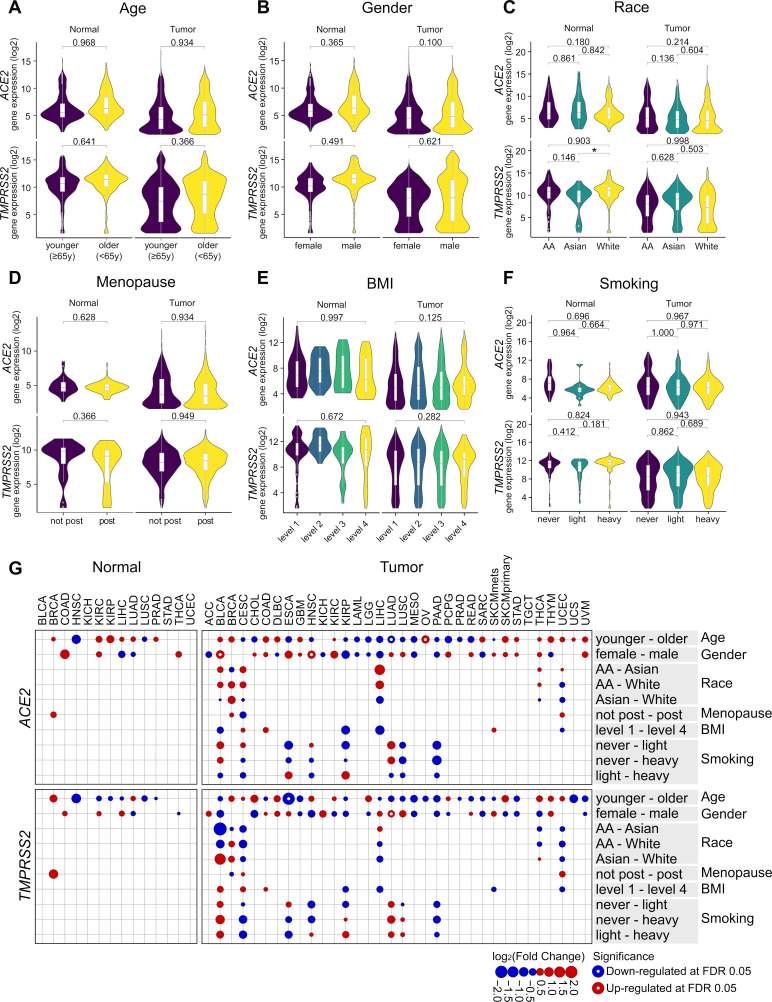
Several clinical observations on COVID-19 indicated clinical factors such as BMI might be associated with severity.5 28 We sought to investigate the association between clinical variables (age, gender, race, tumor stage, tumor grade, menopause, BMI, smoking history) and ACE2 or TMPRSS2 gene expression in tumor or normal tissues from 10,038 patients. For each clinical factor, data were fitted into a two-way ANOVA model with tumor type and clinical factor as variables plus the interaction between the two. Overall, we did not observe significant differences in expression for either gene when comparing designated clinical groups within tumor or normal tissues (figure 2A–F) (online supplementary tables 4 and 5). We observed sporadic clinical groups within individual tumor types that do show significance (eg, age in ESCA for TMPRSS2) (figure 2G) though are of unclear clinical relevance at this time (online supplementary tables 6 and 7). The results suggested those clinical variables are not strongly associated with ACE2 or TMPRSS2 expression. In addition, we investigated the association between the presence of diabetes and gene expression in PAAD, where data were available. Similarly, no significant differences in gene expression were detected in tumor or normal tissues from patients with or without diabetes.
Figure 2.
Clinical correlates of ACE2 and TMPRSS2 expression do not show consistent patterns across individual tumor types. (A)–(F) Gene expression by age (<65 years, ≥65 years), gender (female, male), race (African American, Asian, White), menopause (non-post, post), BMI (levels 1–4), smoking history (never, light, heavy), and by sample type (tumor, normal) when applicable. The criteria for each group definition are described in Methods section. The number of samples in each group is provided in online supplementary tables 4 and 5. In (E), only p values between levels 1 and 4 are shown; the rest is provided in online supplementary table 5. (G) log2 fold change of ACE2 and TMPRSS2 expression between designated clinical groups in each tumor type. Comparisons are shown on the row in the format of “group 1–group 2”, with red and blue circles representing upregulation and downregulation of gene expression in group 1 relative to group 2, respectively. Size of a circle is scaled by the absolute value of log2(fold change). White dot in the center of a circle represents significant comparisons at FDR-adjusted p<0.05 (online supplementary tables 6 and 7). At least 15 samples per group were required for statistical testing; otherwise excluded, hence no circles shown. Two-way ANOVA was used in (A)–(F), with tumor type and clinical group as the variables plus interaction between the two. For clinical factors that have more than two groups (C, E, F), Tukey’s HSD was used with the fitted ANOVA model for pairwise comparisons while controlling for Type I errors. Two-way ANOVA p values after BH-FDR correction are shown in (A), (B), and (D), and Tukey’s HSD p values are shown in (C), (E), and (F). *p<0.05. For comparisons that do not reach significance level of 0.05, exact p values are shown. In (G), Tukey's HSD was used with one-way ANOVA models when more than two groups are present (race, BMI, smoking); otherwise Welch Two Sample t-test was used (age, gender, menopause). ANOVA, analysis of variance; BH, Benjamini and Hochberg; BMI, body mass index; HSD, honest significance test.
Two HLA genotypes (B*46:01, B*54:01) have been reported to be associated with severe clinical outcomes by other groups.41 We investigated the prevalence of the two alleles and identified low prevalence across all tumor types (0.6% and 0.2%, respectively, out of 19,118 HLA-B alleles from 9,559 patients). When looking into individual tumor types, both alleles were found to be significantly enriched in LIHC (FDR-adjusted p<0.0001), which was likely due to the enrichment of Asian populations in this cohort (43%). Comparison of ACE2 or TMPRSS2 gene expression from tumor or normal samples between HLA genotypes showed no significant differences after adjusting for tumor-type specific gene expression, and can be further explored in external data files available on GitHub.
ACE2 correlates with distinct immune gene expression signatures and cell subsets while TMPRSS2 correlates with epithelial cell populations in tumor and normal tissues
To understand potential associations of ACE2 and TMPRSS2 in tissues relevant to patients being treated with cancer immunotherapy, we investigated the correlation between ACE2 or TMPRSS2 and immune gene expression signatures known to be relevant in immuno-oncology (online supplementary tables 8 and 9). ACE2 was positively correlated with ISG signature in tumor samples from 24/34 tumor types (71%, 14 reached FDR-adjusted p<0.05) and normal tissues from 10/14 tumor types (71%, 4 reached FDR-adjusted p<0.05) and negatively correlated with angiogenesis and TGF-β signatures (figure 3A). TMPRSS2 showed a mixed pattern of correlations with those immune responsive or suppressive signatures (figure 3B).
Figure 3.
ACE2 and TMPRSS2 expressions are correlated with distinct immune signatures or cell populations. (A, B) Correlation of five immune gene expression signatures, ISG, Tinfl, myeloid, angio, and TGF-β with (A) ACE2 and (B) TMPRSS2 gene expression. (C, D) Correlation of 64 immune, stroma, and other cell subsets with (C) ACE2 and (D) TMPRSS2 gene expression. n=14 tumor types shown for both genes in normal tissues. n=34 tumor types shown for ACE2 in tumor, and n=33 tumor types shown for TMPRSS2 in tumor. The full correlation statistics are provided as online supplementary tables 8 and 9. Spearman’s correlation was used in (A) to (D). ISG, interferon-stimulated genes; TGF-β, transforming growth factor-β; Tinfl, T cell-inflamed; angio, angiogenesis.
We did not find a consistent pattern of ACE2 with cell subsets across all tumor types with the exception of a high positive correlation with macrophage M2 in normal tissues from kidney cancers (kidney renal clear cell carcinoma (KIRC), KICH, kidney renal papillary cell carcinoma (KIRP)) (Spearman’s ρ=0.84, 0.82, 0.79, FDR-adjusted p<0.0001) and STAD (ρ=0.67, FDR-adjusted p<0.001) (figure 3C) (online supplementary table 10). TMPRSS2 was positively correlated with epithelial cell abundance in tumor samples from 29/33 tumor types (88%, 17 reached FDR-adjusted p<0.05) and normal tissues from 14/14 tumor types (100%, 9 reached FDR-adjusted p<0.05) (figure 3D).
With the observation of correlation with specific immune signatures, we sought to investigate whether ACE2 or TMPRSS2 were expressed directly by immune cells or other specific cell types in nine independent single-cell RNAseq studies. This includes tumor and immune cells from patients with cancer diagnosed with glioblastoma or melanoma (Single Cell Portal, Broad Institute), or head and neck cancer.42 In the cohorts of patients with cancer, less than 1% of the malignant cells express ACE2 and/or TMPRSS2, while few to none of the immune cells express either gene. An exploration of three studies focusing on immune cells from healthy donors (13,316 peripheral blood mononuclear cells (Immune Cell Atlas), 39,563 ileum lamina propria immunocytes (Immune Cell Atlas), 594,857 immune cells (Census of Immune Cells, EBI)) further confirmed the lack of ACE2 and TMPRSS2 expression in immune cell populations. Realizing that other studies had reported the genes as highly expressed in lung, heart, brain, and colon, we investigated published large-scale profiling of 702,968 single cells from patients with non-cancer or healthy donors (Human Cell Landscape).43 We found TMPRSS2 was expressed in stomach, colon, kidney, prostate, intestine, jejunum, pancreatic, esophagus, and bladder tissues, while ACE2 was only expressed in jejunum and fetal intestine. Last, we examined a public scRNAseq cohort from patients with COVID-19 infection (https://doi.org/10.5281/zenodo.3747336) and confirmed that ACE2 was not expressed in immune cells, and TMPRSS2 was present in 6 out of 140,956 cells total by one read count. Therefore, we concluded that ACE2 or TMPRSS2 is not expressed in immune cell populations, at least in the cohorts investigated. The expression of both genes in bulk RNAseq data was likely to be derived from non-immune cells, such as epithelial cells in the tissues.
ACE2 and TMPRSS2 gene expression associates with microbiota in normal tissues particularly from colon and stomach adenocarcinoma
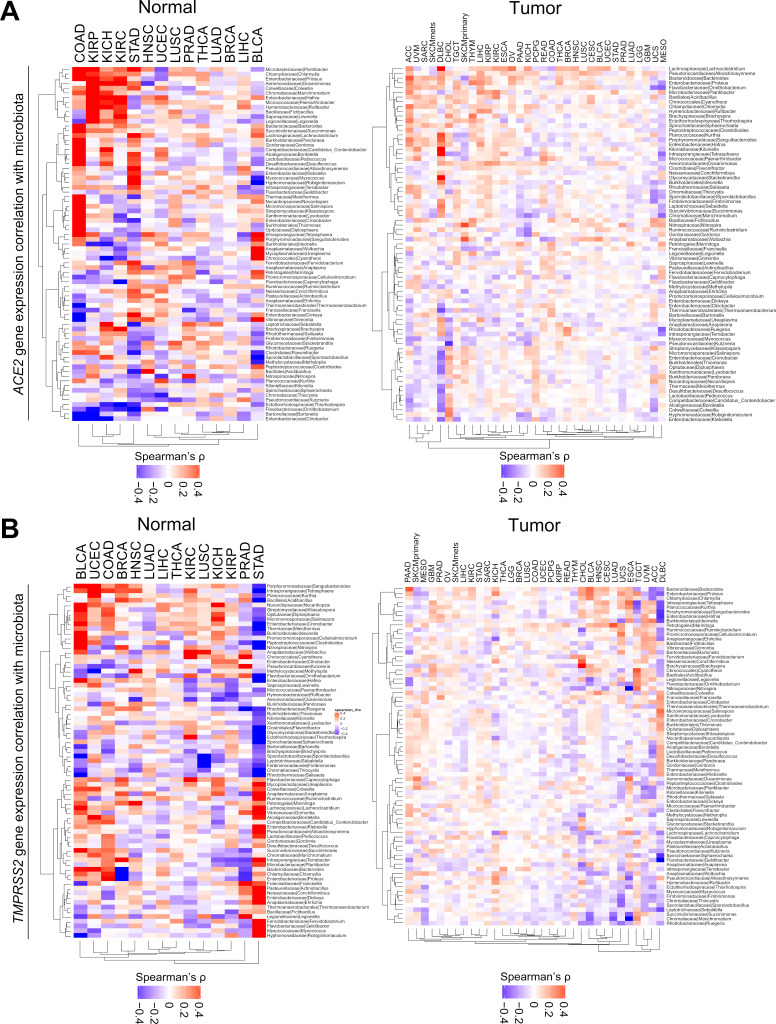
Given associations between strong antitumor immune responses due to the presence of tumor-related virus and particular commensal microbiota, we sought to investigate associations between ACE2 and TMPRSS2 gene expression with the presence of virus or tissue microbiota. Across known viral positive and negative tumor types, we found an inconsistent pattern relative to viral associated versus non-viral associated tumors (HPV, EBV, HBV). We then correlated 1,093 commensal microbiota identified in tumor and normal tissue RNAseq data, as published in Ref. 35 with gene expression. We identified 75 taxa that showed significant and strong positive or negative correlation (Spearman’s ρ>0.5 or <−0.5) with either gene in at least one pairwise correlation (FDR-adjusted p<0.05) (figure 4A and B) (online supplementary table 11). COAD and STAD were the two tumor types demonstrating the strongest and most prevalent positive correlation of ACE2 and TMPRSS2 gene expression with abundance of specific bacteria taxa, respectively. Kidney cancers, including KIRP, KICH, and KIRC, also showed positive correlations between ACE2 and microbiota if not as prevalent as colon or stomach cancers. If ranked by Spearman’s correlation coefficient, Chlamydia was the top microbiota positively correlated with ACE2 in COAD (ρ=0.81, FDR-adjusted p<0.0001) (online supplementary table 11). For both genes, those patterns were less prominent in tumor samples, which could be due to high heterogeneity in tumors. Overall, we observed approximately a 2.6:1 ratio of commensal microbiota for gram-negative to positive groups in the 75 taxa (50 negatives, 19 positives, others undetermined) (online supplementary table 11).
Figure 4.
ACE2 and TMPRSS2 expressions are correlated with distinct microbiota communities. (A, B) Correlation of 75 bacteria taxa with (A) ACE2 and (B) TMPRSS2 gene expression. Within each plot, the left panel shows normal tissues, and the right panel shows tumor samples. Seventy-five taxa were selected by Spearman’s correlation coefficient ρ>0.5 or <−0.5 and FDR-adjusted p<0.05 in at least one pairwise correlation (online supplementary table 11). For both genes, n=14 and n=33 tumor types are shown in normal and tumor samples, respectively. LAML does not have data available for microbiota abundance, hence excluded from analysis for both genes. Spearman’s correlation was used in (A) and (B). LAML, acute myeloid leukemia.
Integration of multidimensional correlates revealed specific contributors shaping ACE2 and TMPRSS2 expression in tumor and normal tissues
To integrate all correlates and evaluate their relative importance in determining the gene expression of ACE2 and TMPRSS2, we built LASSO regression models in tumor and normal tissues separately using features from the clinical, immune, and microbial domains (online supplementary figure 2). Clinical features included were age, gender, and race, while menopause, BMI, and smoking history were excluded because >50% of the samples were missing information. HLA genotype was not included because of many categories and/or levels, which may lead to overfitting. Immune gene expression signatures included ISG, T cell-inflamed, myeloid, angiogenesis, and TGF-β. Immune cell type features included macrophage M1/M2, CD8, and CD4 T cells, and non-immune cell type features included epithelial cells. Microbe features included the 75 bacteria taxa from the analysis above. In addition, we collapsed 34 tumor types into 15 tissue types and included these in the model to account for tissue-specific gene expression variations.
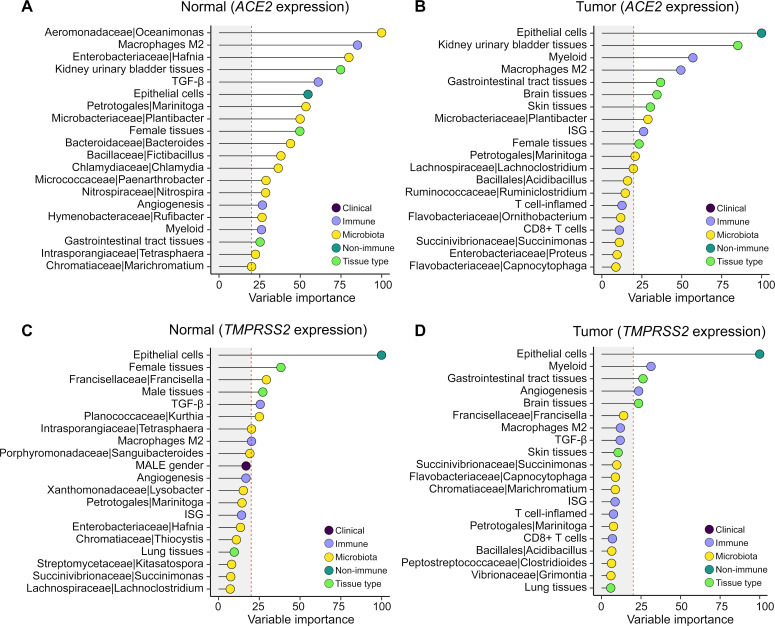
We calculated the importance of each feature in the models with 10-fold cross-validation (online supplementary table 12). After quality control and filtering, among the features kept in each model, immune and epithelial cells were the top-ranked features that predict ACE2 expression in normal tissues and tumors. Microbiota were observed to be important features for ACE2 in normal tissues but not in tumors (figure 5A and B). For TMPRSS2 expression, epithelial cell abundance is the most important predictor in both normal and tumor samples (figure 5C and D). Taken together, these results suggested that immune signatures, epithelial cells, and commensal microbiota were important predictors for ACE2 expression, while TMPRSS2 expression was primarily determined by epithelial cells.
Figure 5.
Variable importance of multidimensional correlates in predicting ACE2 and TMPRSS2 expression. (A, B) Clinical, immune, and microbiota features in association with ACE2 gene expression in (A) normal and (B) tumor samples. (C, D) Clinical, immune, and microbiota features in association with TMPRSS2 gene expression in (C) normal and (D) tumor samples. For each gene, an analysis was performed in normal (n=708) and tumor samples (n=10,024) separately, with workflow illustrated in online supplementary figure 2. Variable importance scaled to 0–100 is shown on the x-axis. Vertical red dashed line labels score=20. Features are shown on the y-axis colored by clinical, immune, non-immune, microbiota, and tissue type. Top 20 features ranked by variable importance higher to lower are shown, and the full list is provided in online supplementary table 12. LASSO regression was used in (A) to (D) with 10-fold cross-validation.
Discussion
We performed a pan-cancer analysis of the receptor that facilitates SARS-CoV-2 infection (ACE2) and the protease that mediates spike protein activation and viral entry (TMPRSS2) by integrating data across six resources including clinical, genetic, transcriptomic and microbiome domains. We found that ACE2 and TMPRSS2 are generally expressed lower in tumors relative to matched normal and that digestive organs (both tumor and normal samples) have the highest expression. Neither clinical factors nor HLA genotypes were consistently associated with gene expression levels. Multiple immune gene expression signatures such as ISG and the T cell-inflamed tumor microenvironment did correlate with ACE2, and inverse correlations were seen with angiogenesis and TGF-β. ACE2 expression correlated with increased macrophage abundance in some tumors, while TMPRSS2 was strongly associated with epithelial cells. Regarding lymphocytes and macrophages, no ACE2 expression was observed in these cells across multiple single-cell sequencing studies. Microbiota contents are clearly associated with ACE2 and TMPRSS2 gene expression levels, possibly suggesting a causal role and the potential to be a modifiable biomarker.
The mortality of COVID-19 disease has been substantially greater than that seen with seasonal influenza and led to the identification of or hypothesis that certain clinical factors may be associated with outcomes. The factors of particular focus included advanced age, BMI, and possibly diabetes or other chronic health conditions such as cardiopulmonary syndromes and immunosuppression or cancer.19 28 29 In addition, certain races or ethnicities have experienced greater morbidity and mortality due to pandemic.30 In our analysis of ACE2/TMPRSS2 gene expression in tumors and matched normal tissues, we observe no consistent association for these factors. Further work would be required to investigate other variables associated with these disease states, such as chronic inflammatory conditions, immunosuppression, and other disparities that may be contributing factors,44 45 and cellular context is important to interpret the complexity of those associations.46
An initial hypothesis when considering the deleterious outcomes for patients with cancer and COVID-19 disease was that cancer tissues themselves might have higher expression of viral entry related genes. We found that gene expression levels did not support this to be the case. Rather cancer tissues broadly have lower expression of ACE2 and TMPRSS2, though the cancers of the digestive tract do have the highest relative level among cancer tissues. This suppressed expression level is consistent with that observed in immuno-oncology gene expression studies,47 in which the T cell-inflamed tumor microenvironment has been observed to be lower in cancer compared with matched normal. ACE2 has been described as a type I interferon-inducible gene.15 Across our analysis, we see strong correlations of ACE2 with type I (ISG) and type II (T cell-inflamed) interferon signatures consistent with this.
Observing higher ACE2 levels in T cell-inflamed tumors may be relevant to the administration of cancer immunotherapy during the COVID-19 pandemic, especially in patients with tumors of the aerodigestive tract such as head and neck, lung, and colorectal/anal tracts. T cell-inflamed gene expression is strongly correlated with treatment response to checkpoint immunotherapy48 and has not been associated with immune-related adverse events (irAE).49 However, if ACE2 and TMPRSS2 levels are high, making viral infection potentially more likely, concomitant treatment with checkpoint blockade may potentially change antiviral host response50 or possibly obscure rapidly delineation of symptoms such as fatigue, dyspnea, diarrhea,51 and complicate irAE management, especially given emerging evidence that corticosteroids may worsen COVID-19 disease.52 To date, it remains unclear whether immune-checkpoint blockade increases risk of SARS-CoV-2 infection or subsequent impact of COVID-19.25 26
Direct infection or dysregulation of immune cell populations is an additional area of concern in patients with cancer and more broadly in infected patients. COVID-19 can manifest with lymphopenia with some autopsy series suggesting lymph node or splenic atrophy.53 Certainly, dysregulated macrophage activity, with the elaboration of IL-6 and other inflammatory cytokines, is a major component of the disease. Studies have raised the possibility that SARS-CoV-2 infects lymphocytes17 or macrophages,53 leading to COVID-19 associate findings. In our study, we investigated the expression of ACE2 and TMPRSS2 across multiple single-cell sequencing databases encompassing nine independent studies. However, we found no evidence of expression in these cells. It must be noted that the possibility exists that type I interferon may induce ACE2 expression, which would not be captured in our analysis. We would note, however, that previous studies have not definitively determined that T cells or macrophages are infected by SARS-CoV-2, and direct viral culture from purified cell populations would be needed to confirm this. Additionally, multiple other known pathologies associated with sepsis and extreme illness could explain these lymph node and splenic findings, and few patients with COVID-19 have been documented to have an extreme viremia consistent with what would be required as a prerequisite to such histologic findings.
Immune responses to cancer and in other settings are increasingly being recognized as influenced by the commensal microbiota.54 We were, therefore, interested in investigating associations of tissue-based microbiota and ACE2 as a surrogate for the risk of SARS-CoV-2 infection. In our analysis, we found strong correlations of specific bacterial flora and high expression of ACE2 in COVID-19 related organs, including colorectal and kidney. Particularly in colorectal, where presentations with diarrhea have been widely described, we note a ratio of at least 2:1 of gram-negative bacteria in the bacteria populations significantly associated with elevated ACE2 expression. A dominance of gram-negative bacteria in the fecal microbiota is assumed at baseline, and yet disequilibrium with an increase of these bacteria is associated with diminished immunological outcomes, especially in immunosuppressed patients.55 In our study, we have analyzed a heterogeneously collected group of tumors and match normal tissues. However, this observation suggests that further investigation of the commensal microbiome in COVID-19 and possibly that bacterial antibiosis related to coronavirus infection might be of relevance in the future.
We note limitations to our report with the acknowledgment that the use of pre-existing data does not fully capture the complexity of active infection by SARS-CoV-2. Rather we sought to investigate factors correlated with viral cell engagement via ACE2 and viral entry via TMPRSS2 as possible associative risk factors that might be entertained on a clinical or translational level when considering risk for patients with cancer and otherwise of COVID-19. Certainly, there may be virus infection-induced changes that are dynamic. However, we believe our analysis to be the most comprehensive catalog of ACE2 and TMPRSS2 correlates to date (34 tumor types from 15 tissue types across 10 038 subjects including both tumor samples and matched normal tissues as well as scRNAseq databases consisting of patients with cancer and healthy donors). We also acknowledge that the microbiota we analyzed were identified from tissue RNAseq data, and the sample collection and preparation of tissue RNAseq was not designed originally to completely rule out potential contamination or confirm the vitality of identified microbes. However, these source data constitute the largest collection of microbiota communities identified from patients with cancer, have previously been used in this manner to build prediction algorithms, and the data were optimized via rigorous methodology to control for noise across the data set.35 We also note that we are unable in this analysis to comment on respiratory or fecal samples from patients infected with COVID-19 and very much look forward to better understanding the functional mechanisms associated with those commensal and pathogenic microbiota related to COVID-19. Last, our work does not determine a causal role of those correlates in driving response or severity of COVID-19 disease and would require further mechanistic studies as well as prospective clinical trials in patients to further develop or investigate interventional approaches.
Conclusion
We have performed a multiomic analysis of ACE2 and TMPRSS2 gene expression related to clinical, genetic, microbiome covariates associated with COVID-19 infection. We have identified novel commensal microbiome associations and further described interferon associated gene expression patterns in normal and tumor tissues related relevant to SARS-CoV-2 infection. These data will hopefully inform sample collection, future analyses, and treatment of patients with cancer and others infected with COVID-19.
Acknowledgments
The authors thank C Reid for technical assistance on retrieving metadata attributes from GDC.
Footnotes
Twitter: @RiyueSunnyBao, @jasonlukemd
Contributors: RB and JJL conceived the study. JJL supervised the project. RB acquired the data, developed the methodology, performed the computations, and analyzed the data. KH parsed the legacy clinical XML files from GDC. KH and LH provided critical feedback on the TCGA data and LASSO regression analysis. RB, KH, LH, and JJL interpreted the results. RB and JJL wrote the manuscript. All authors contributed to the final manuscript.
Funding: JJL acknowledges the Department of Defense Career Development Award (W81XWH-17-1-0265), the Arthur J Schreiner Family Melanoma Research Fund, the J. Edward Mahoney Foundation Research Fund, Brush Family Immunotherapy Research Fund, and Buffet Fund for Cancer Immunotherapy.
Competing interests: JJL declares Data and Safety Monitoring Board: TTC Oncology; Scientific Advisory Board: 7 Hills, Actym, Alphamab Oncology, Arch Oncology, Kanaph, Mavu (now part of AbbVie), Onc.AI, Pyxis, Spring bank, Tempest; Consultancy: Abbvie, Akrevia, Algios, Array, Astellas, Bayer, Bristol-Myers Squibb, Eisai, EMD Serono, Ideaya, Incyte, Janssen, Merck, Mersana, Novartis, PTx, RefleXion, Regeneron, Rubius, Silicon, Tesaro, Vividion; Research Support: (all to institution for clinical trials unless noted) AbbVie, Agios (IIT), Array (IIT), Astellas, Bristol-Myers Squibb, CheckMate (SRA), Compugen, Corvus, EMD Serono, Evelo (SRA), Five Prime, FLX Bio, Genentech, Immatics, Immunocore, Incyte, Leap, MedImmune, Macrogenics, Necktar, Novartis, Palleon (SRA), Merck, Spring bank, Tesaro, Tizona, Xencor; Travel: Akrevia, Bayer, Bristol-Myers Squibb, EMD Serono, Incyte, Janssen, Merck, Mersana, Novartis, Pyxis, RefleXion; Patents: (both provisional) Serial #15/612,657 (Cancer Immunotherapy), PCT/US18/36052 (Microbiome Biomarkers for Anti-PD-1/PD-L1 Responsiveness: Diagnostic, Prognostic and Therapeutic Uses Thereof). RB declares Serial #15/612,657 (Cancer Immunotherapy), PCT/US18/36052 (Microbiome Biomarkers for Anti-PD-1/PD-L1 Responsiveness: Diagnostic, Prognostic and Therapeutic Uses Thereof), PCT/US62/755945 (Compositions for treating infectious, autoimmune, and allergic disease).
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: The original data files were downloaded from Genomic Data Commons (https://portal.gdc.cancer.gov/), Pan-Can Atlas studies (https://gdc.cancer.gov/about-data/publications/pancanatlas), and published studies. Preprocessed data files are provided as supplementary tables. Large-size data objects, as well as additional figures and tables, were deposited to a public, open access GitHub repository (https://github.com/riyuebao/ACE2_TMPRSS2_multicorrelates). Other data will be provided upon request from the corresponding author.
References
- 1. Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 2020;5:536–44. 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020;395:565–74. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang C, Horby PW, Hayden FG, et al. A novel coronavirus outbreak of global health concern. Lancet 2020;395:470–3. 10.1016/S0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guan WJ, ZY N, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mizumoto K, Chowell G. Estimating risk for death from coronavirus disease, China, January-February 2020. Emerg Infect Dis 2020;26:1251–6. 10.3201/eid2606.200233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li W, Moore MJ, Vasilieva N, et al. Angiotensin-Converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003;426:450–4. 10.1038/nature02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181:271–80. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wrapp D, Wang N, Corbett KS, et al. Cryo-Em structure of the 2019-nCoV spike in the prefusion conformation. Science 2020;367:1260–3. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Walls AC, Park Y-J, Tortorici MA, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020;181:281–92. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bernstein KE, Khan Z, Giani JF, et al. Angiotensin-converting enzyme in innate and adaptive immunity. Nat Rev Nephrol 2018;14:325–36. 10.1038/nrneph.2018.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Imai Y, Kuba K, Rao S, et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 2005;436:112–6. 10.1038/nature03712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hamming I, Timens W, Bulthuis MLC, et al. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol 2004;203:631–7. 10.1002/path.1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zou X, Chen K, Zou J, et al. Single-Cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med 2020;14:185–92. 10.1007/s11684-020-0754-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. CaA Z, Samuel J, Nyquist SK, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is enriched in specific cell subsets across tissues, CELL-D-20-00767 2020. [DOI] [PMC free article] [PubMed]
- 16. Xu H, Zhong L, Deng J, et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci 2020;12:8. 10.1038/s41368-020-0074-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang X, Xu W, Hu G, et al. SARS-CoV-2 infects T lymphocytes through its spike protein-mediated membrane fusion. Cell Mol Immunol 2020. 10.1038/s41423-020-0424-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chu H, Zhou J, Wong BH-Y, et al. Middle East respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J Infect Dis 2016;213:904–14. 10.1093/infdis/jiv380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liang W, Guan W, Chen R, et al. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol 2020;21:335–7. 10.1016/S1470-2045(20)30096-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zheng RS, Sun KX, Zhang SW, et al. Report of cancer epidemiology in China, 2015. Zhonghua Zhong Liu Za Zhi 2019;41:19–28. 10.3760/cma.j.issn.0253-3766.2019.01.005 [DOI] [PubMed] [Google Scholar]
- 21. LYW L, Cazier JB, Team UKCCMP, et al. COVID-19 mortality in patients with cancer on chemotherapy or other anticancer treatments: a prospective cohort study. Lancet 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuderer NM, Choueiri TK, Shah DP, et al. Clinical impact of COVID-19 on patients with cancer (CCC19): a cohort study. Lancet 2020;395:1907–18. 10.1016/S0140-6736(20)31187-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. He W, Chen L, Chen L, et al. COVID-19 in persons with haematological cancers. Leukemia 2020;34:1637–45. 10.1038/s41375-020-0836-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paul S, Rausch CR, Jain N, et al. Treating leukemia in the time of COVID-19. Acta Haematol 2020:1–13. 10.1159/000508199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luo J, Rizvi H, Egger JV, et al. Impact of PD-1 blockade on severity of COVID-19 in patients with lung cancers. Cancer Discov 2020:CD-20-0596. 10.1158/2159-8290.CD-20-0596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robilotti EV, Babady NE, Mead PA, et al. Determinants of severity in cancer patients with COVID-19 illness. medRxiv 2020;2020.2005.2004:20086322. 10.1101/2020.05.04.20086322 [DOI] [Google Scholar]
- 27. Ascierto PA, Fox BA, Urba WJ, et al. Insights from immuno-oncology: the Society for immunotherapy of cancer statement on access to IL-6-targeting therapies for COVID-19. J Immunother Cancer 2020;8:e000878. 10.1136/jitc-2020-000878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lighter J, Phillips M, Hochman S, et al. Obesity in patients younger than 60 years is a risk factor for Covid-19 hospital admission. Clin Infect Dis 2020. 10.1093/cid/ciaa415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guo W, Li M, Dong Y, et al. Diabetes is a risk factor for the progression and prognosis of COVID-19. Diabetes Metab Res Rev 2020:e3319. 10.1002/dmrr.3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Available: https://www.propublica.org/article/early-data-shows-african-americans-have-contracted-and-died-of-coronavirus-at-an-alarming-rate
- 31. Braciale TJ. Hahn Ys: immunity to viruses. Immunological Reviews 2013;255:5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Trujillo JA, Sweis RF, Bao R, et al. T Cell-Inflamed versus non-T Cell-Inflamed tumors: a conceptual framework for cancer immunotherapy drug development and combination therapy selection. Cancer Immunol Res 2018;6:990–1000. 10.1158/2326-6066.CIR-18-0277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang Z, Hernandez K, Savage J, et al. Uniform genomic data analysis in the NCI genomic data commons. bioRxiv 2019;788919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu J, Lichtenberg T, Hoadley KA, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 2018;173:400–16. 10.1016/j.cell.2018.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Poore GD, Kopylova E, Zhu Q, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020;579:567–74. 10.1038/s41586-020-2095-1 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36. Thorsson V, Gibbs DL, Brown SD, et al. The immune landscape of cancer. Immunity 2019;51:411–2. 10.1016/j.immuni.2019.08.004 [DOI] [PubMed] [Google Scholar]
- 37. Simonnet A, Chetboun M, Poissy J, et al. High prevalence of obesity in severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) requiring invasive mechanical ventilation. Obesity 2020;28:1195–9. 10.1002/oby.22831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ghasemi F, Prokopec SD, MacNeil D, et al. Mutational analysis of head and neck squamous cell carcinoma stratified by smoking status. JCI Insight 2019;4. 10.1172/jci.insight.123443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moore JB, June CH. Cytokine release syndrome in severe COVID-19. Science 2020;368:473–4. 10.1126/science.abb8925 [DOI] [PubMed] [Google Scholar]
- 40. Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and Ets transcription factor genes in prostate cancer. Science 2005;310:644–8. 10.1126/science.1117679 [DOI] [PubMed] [Google Scholar]
- 41. Lin M, Tseng H-K, Trejaut JA, et al. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med Genet 2003;4:9. 10.1186/1471-2350-4-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Puram SV, Tirosh I, Parikh AS, et al. Single-Cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 2017;171:1611–24. 10.1016/j.cell.2017.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Han X, Zhou Z, Fei L, et al. Construction of a human cell landscape at single-cell level. Nature 2020;581:303–9. 10.1038/s41586-020-2157-4 [DOI] [PubMed] [Google Scholar]
- 44. Wiebe N, Stenvinkel P, Tonelli M. Associations of chronic inflammation, insulin resistance, and severe obesity with mortality, myocardial infarction, cancer, and chronic pulmonary disease. JAMA Netw Open 2019;2:e1910456. 10.1001/jamanetworkopen.2019.10456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. National Academies of Sciences E, Medicine Communities in action: pathways to health equity. Washington, DC: The National Academies Press, 2017. [PubMed] [Google Scholar]
- 46. Muus C, Luecken MD, Eraslan G, et al. Integrated analyses of single-cell atlases reveal age, gender, and smoking status associations with cell type-specific expression of mediators of SARS-CoV-2 viral entry and highlights inflammatory programs in putative target cells. bioRxiv 2020;2019:049254. [Google Scholar]
- 47. Spranger S, Luke JJ, Bao R, et al. Density of immunogenic antigens does not explain the presence or absence of the T-cell-inflamed tumor microenvironment in melanoma. Proc Natl Acad Sci U S A 2016;113:E7759–68. 10.1073/pnas.1609376113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018;362:eaar3593. 10.1126/science.aar3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Puzanov I, Diab A, Abdallah K, et al. Managing toxicities associated with immune checkpoint inhibitors: consensus recommendations from the Society for immunotherapy of cancer (SITC) toxicity management Working group. J Immunother Cancer 2017;5:95. 10.1186/s40425-017-0300-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Läubli H, Balmelli C, Kaufmann L, et al. Influenza vaccination of cancer patients during PD-1 blockade induces serological protection but may raise the risk for immune-related adverse events. J Immunother Cancer 2018;6:40. 10.1186/s40425-018-0353-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Martins F, Sofiya L, Sykiotis GP, et al. Adverse effects of immune-checkpoint inhibitors: epidemiology, management and surveillance. Nat Rev Clin Oncol 2019;16:563–80. 10.1038/s41571-019-0218-0 [DOI] [PubMed] [Google Scholar]
- 52. Russell B, Moss C, Rigg A, et al. COVID-19 and treatment with NSAIDs and corticosteroids: should we be limiting their use in the clinical setting? Ecancermedicalscience 2020;14:1023. 10.3332/ecancer.2020.1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Feng Z, Wang G, Wang C, et al. The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directly Decimates human spleens and lymph nodes. medRxiv 2020;2020.2003.2027:20045427. [Google Scholar]
- 54. Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018;359:104–8. 10.1126/science.aao3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Taur Y, Pamer EG. The intestinal microbiota and susceptibility to infection in immunocompromised patients. Curr Opin Infect Dis 2013;26:332–7. 10.1097/QCO.0b013e3283630dd3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-001020supp001.pdf (336.7KB, pdf)
jitc-2020-001020supp002.xlsx (1.8MB, xlsx)