Abstract
Background
The novel coronavirus (SARS-CoV-2) shares approximately 80% whole genome sequence identity and 66% spike (S) protein identity with that of SARS-CoV. The cross-neutralization between these viruses is currently not well-defined.
Methods
Here, by using the live SARS-CoV-2 virus infection assay as well as HIV-1 based pseudotyped-virus carrying the spike (S) gene of the SARS-CoV-2 (ppSARS-2) and SARS-CoV (ppSARS), we examined whether infections with SARS-CoV and SARS-CoV-2 can induce cross-neutralizing antibodies.
Findings
We confirmed that SARS-CoV-2 infects cells via angiotensin converting enzyme 2 (ACE2), the functional receptor for SARS-CoV, and we also found that the recombinant receptor binding domain (RBD) of the S protein of SARS-CoV effectively inhibits ppSARS-2 entry in Huh7.5 cells. However, convalescent sera from SARS-CoV and SARS-CoV-2 patients showed high neutralizing activity only against the homologous virus, with no or limited cross-neutralization activity against the other pseudotyped virus. Similar results were also observed in vaccination studies in mice.
Interpretation
Our study demonstrates that although both SARS-CoV and SARS-CoV-2 use ACE2 as a cellular receptor, the neutralization epitopes are not shared by these two closely-related viruses, highlighting challenges towards developing a universal vaccine against SARS-CoV related viruses.
Funding
This work was supported by the National Key Research and Development Program of China, the National Major Project for Control and Prevention of Infectious Disease in China, and the One Belt and One Road Major Project for infectious diseases.
Keywords: COVID-19, SARS-CoV-2, CoV, Cross-neutralization, ACE2
Research in context.
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is a novel coronavirus first reported in Wuhan, China, during December 2019. It is the second known coronavirus to cause Severe Acute Respiratory Syndrome (SARS) in humans, after the “original” Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV). SARS-CoV and SARS-CoV-2 share ∼80% genome similarity overall but only 66% similarity in the Spike (S) protein. It is not known whether antibodies derived from natural infection or vaccination with SARS-CoV can cross-react with SARS-CoV-2, and vice versa.
Evidence before this study
Despite substantial differences at the amino acid level between the S proteins of SARS-CoV and SARS-CoV-2, preliminary studies and prediction models showed that the structures of the S proteins from these viruses remain similar to each other, and hence they likely use the same entry receptor – angiotensin converting enzyme 2 (ACE2). Thus, there is a possibility that antibodies against SARS-CoV S will cross-react with SARS-CoV-2, and vice versa.
Added value of this study
This study shows that SARS-CoV-2 does indeed infect cells via the ACE2, the functional receptor for SARS-CoV. The receptor binding domain (RBD) of the SARS-CoV S protein effectively inhibits SARS-CoV-2 S-pseudotyped virus (ppSARS-2) entry in cells. However, convalescent sera from SARS-CoV and SARS-CoV-2 patients showed high neutralizing activity only against the homologous virus, but no or limited cross-neutralization activity against the heterologous virus. The same trends were also observed in vaccinated mice.
Implications of all the available evidence
Although both SARS-CoV and SARS-CoV-2 were confirmed to use ACE2 as a cellular receptor, neutralization epitopes were not shared by these two viruses. Therefore, other antigens may be necessary for the design and development of a cross-protective vaccine against both SARS-CoV and SARS-CoV-2.
Alt-text: Unlabelled box
1. Introduction
In late December 2019, after several viral pneumonia cases of unknown origin were initially reported in Wuhan, Hubei Province, China, a novel coronavirus was quickly identified as the causative pathogen [1,2]. As of May 23, 2020, over 5100,000 confirmed cases of Coronavirus Disease 2019 (COVID-19) and 330,000 deaths were reported globally [3], with over 200 countries impacted [4], [5], [6], [7]. The causative agent, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), shares approximately 80% genome similarity with that of Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and a lower similarity (50%) with that of Middle East Respiratory Syndrome Coronavirus (MERS-CoV). The latter two viruses were responsible for the outbreaks of severe acute respiratory syndrome in 2002-04, and severe viral pneumonia with renal failure in humans in 2012, respectively [8], [9], [10].
Coronavirus tropism and entry is dependent on the interaction between the S protein and a host cell receptor. It is known that SARS-CoV enters host cells by engaging angiotensin-converting enzyme 2 (ACE2) [11], a type I membrane protein and a metal protease with host distribution including, but not limited to, the lung and gastrointestinal tracts [12]. Studies showed that the viral infection efficiency of SARS-CoV is largely determined by the interaction between S protein and ACE2 [13]. ACE2 has also been shown recently as the cellular receptor for SARS-CoV-2 in virological studies or bioinformatic analyses (PMID: 32015508, 32142651) [9].
In this study, we generated pseudotyped viruses bearing the S protein of SARS-CoV (ppSARS) or SARS-CoV-2 (ppSARS2), and examined their usage of ACE2 receptor in SARS-CoV and SARS-CoV-2 infection. We assessed the efficiency of RBD of SARS-CoV spike in blocking SARS-CoV-2 entry and neutralizing activity of sera from mice vaccinated by inactivated SARS-CoV vaccine and/or recombinant SARS-CoV S proteins. We thoroughly evaluated antibody-mediated cross-protection for SARS-CoV-2 and SARS-CoV by determining the cross-neutralizing activity from the sera of convalescent patients infected by either SARS-CoV or SARS-CoV-2.
2. Materials and methods
2.1. Ethics statement
Animal studies were conducted in strict accordance with the Guide for the Care and Use of Laboratory Animals of the People's Republic of China. The study protocol was approved by the Committee on the Ethics of Animal Experiments of the Chinese center for Disease Control and Prevention (China CDC). Consent was provided by the patients for the human antisera to be collected for this work. All experiments with live SARS-CoV-2 was performed in Biosafety Level 3 (BSL-3) containment laboratories at the National Institute for Viral Disease Control and Prevention, China CDC.
2.2. Viruses, recombinant proteins, vaccination and collection of anti-SARS-CoV sera from mice
Live SARS-CoV-2 (Wuhan/IVDC-HB-01/2019) [1] virus was used in this study. Recombinant SARS-CoV RBD proteins were produced from transiently transfected ExpiCHO cell line (Thermo Fisher Scientific) and purified by precise gradient elution Ni-IMAC. Soluble human ACE2 protein was prepared as previously reported [14]. For these experiments, 6–8-week-old female BALB/c mice were purchased from Charles River. Mice were bred in the animal facilities at The National Institute of Occupational Health and Poison Control, Chinese CDC. All mice were housed in pathogen-free animal labs. For vaccination, mice were inoculated via the intramuscular route, with half of the volume injected into each side of the rectus femoris. The blood/sera were collected at 2 weeks after the last injection via the orbital artery after the immunizations. The murine anti-SARS sera were obtained as follows: 1) Group 1 (n = 5) was immunized first with 100 μL of a SARS-CoV inactivated vaccine, then boosted twice with 10 μg recombinant SARS-CoV S protein (3-week intervals), and followed once with 10 μg SARS-CoV S1 (3 weeks after the SARS-CoV S boosts); 2) Group 2 (n = 3) was immunized three times with 100 μL of a SARS-CoV inactivated vaccine with 100 μL Complete Freund's Adjuvant (CFA) at 3-week intervals; 3) Group 3 (n = 3) was immunized three times with 100 μL of SARS-CoV inactivated vaccine with 100 μg alum adjuvant at 3-week intervals; 4) Groups 4 and 5 (n = 5 each group) were immunized with 5 × 107 plaque forming units (PFU) of a recombinant adenovirus serotype 4 vector expressing SARS-CoV S1 (Ad41-SARS-S1) only once (Group 4), or twice (Group 5) at 4 weeks after the first immunization. Control mice (n = 2) were mock vaccinated with 100 μL of Alum adjuvant.
2.3. Preparation of pseudotyped virus
The S gene sequence of SARS-CoV-2 (GISAID, No. EPI_ISL_402119) and SARS-CoV (GenBank No. AY278489.2) were codon-optimized for expression in human cells. HEK 293FT (ATCC), HEK 293T (ATCC), Huh7.5 (ATCC) cell lines were cultured in DMEM medium (Hyclone) with 10% fetal bovine serum (GEMINI) and 1% penicillin-streptomycin (Gibco). Cells were grown at 37 °C, 5% CO2. Pseudotyped viruses were generated as described previously [15]. In brief, the S protein expression plasmid, and plasmids encoding Env-defective, luciferase expressing HIV-1 (pNL4-3. Luc. R-E-) were co-transfected to HEK 293T with X-treme GENE HP DNA Transfection Reagent (Roche), the media was changed at 6 h after transfection, and cell suspensions containing the pseudotyped virus was harvested after 48 h and stored at −70 °C.
2.4. Pseudotyped virus infection and neutralization experiments
Huh7.5 cells were grown in 96-well plates for infection. Pseudotyped viruses were added to 60–70% confluent cells. After 12 h, infection media was changed to 2% FBS DMEM, and incubated at 37 °C, 5% CO2, for 48 h. The final titer was measured by Bright-Glo firefly luciferase kit (Promega). For neutralization experiments, serum samples were serially diluted in DMEM. Pseudotyped viruses were mixed with the samples and co-incubated for 1 h at 37 °C, followed by incubation with the cells for 12 h, the media was then changed to 2% FBS DMEM, and incubated at 37 °C, 5% CO2 for 48 h. Infection titers were measured by Bright-Glo firefly luciferase kit (Promega). The inhibition ratio was calculated: Inhibition ratio = (Blank RLU – treatment RLU) / Blank RLU *100%. The data were analyzed by one-way or two-way ANOVA with the multiple comparison correction.
2.5. Soluble ACE2 entry inhibition assay
Vero cells were cultured in a 96-well plate and replaced with 2% DMEM before they were added to soluble ACE2 protein at different concentrations [14]. Each concentration was repeated three times in duplicate. SARS-CoV-2 was added to the cells (200 50% tissue culture infective dose, TCID50). Twenty-four hours after infection, the same concentration of diluted ACE2 protein was replenished. Cytopathic effect (CPE) was observed 24 and 48 h after virus infection. The culture supernatant was collected for nucleic acid extraction after 48 h. Real-time fluorescence reverse transcription polymerase chain reaction (RT-PCR) was performed on an ABI Q5 fluorescence quantitative PCR instrument. The Ct value of the measured sample was analyzed with the standard curve to calculate the corresponding TCID50 value of the sample.
2.6. SARS-CoV RBD competition assay
Huh7.5 cells were grown in 96-well plates at one day before infection until 60–70% confluence. Recombinant SARS-CoV RBD protein was serially diluted in DMEM, and then added to cells. After 30 min, pseudotyped viruses were added to cells and incubated at 37 °C, 5% CO2 for 8 h, then the media was changed to 2% FBS DMEM. After 48 h incubation at 37 °C, 5% CO2, the infectious titer was measured by Bright-Glo firefly luciferase kit (Promega). The inhibition ratio was calculated as: Inhibition ratio = (Blank RLU – treatment RLU) / Blank RLU *100%. The data were analyzed by one-way or two-way ANOVA.
2.7. Microneutralization assay
The ability to neutralize SARS-CoV-2 infection of Vero cells was tested by microneutralization assay. Vero cells were maintained in DMEM, supplemented with 10% heat-inactivated fetal bovine sera (FBS, Hyclone) and 2 mM l-glutamine (Invitrogen). The SARS-CoV-2 was grown to stock titers in Vero cells. The microneutralization titer of test antibody was the reciprocal of the highest dilution of test antibody that showed inhibition in all triplicate wells. Microneutralization assay was controlled with back titration, positive control (i.e., serum from a convalescent patient), and negative control (i.e. human sera previously determined to be negative for SARS-CoV-2 specific antibodies).
2.8. Statistical analysis
Statistical analyses were conducted using one-way ANOVA with Bonferroni post-test via the SPSS for Windows software package. Unpaired two-tailed Student's t test was used to compare means between different groups. A value of p < 0.05 indicated statistical significance. The results were expressed as mean ± SD. All figures were generated using the Prism 8 software package (GraphPad Software).
3. Results
3.1. Recombinant RBD proteins from SARS-CoV effectively block viral entry of SARS-CoV-2
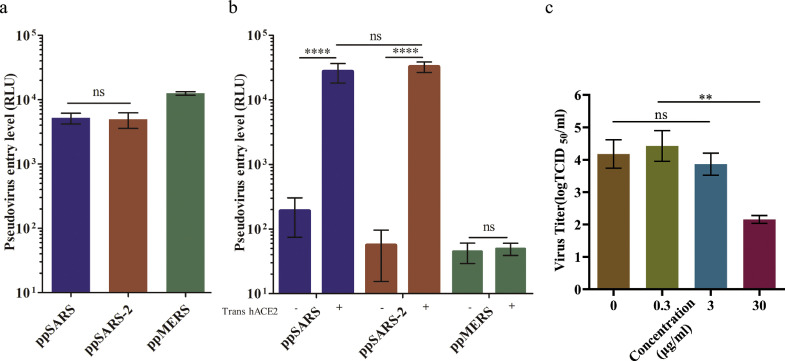
We first assessed the infection efficiency of HIV-1 pseudotyped with S proteins from various coronaviruses including SARS-CoV-2, as well as SARS-CoV and MERS-CoV in the Huh7.5 cell line [16]. Similar levels of viral entry, indicated by luciferase reporter gene expression, were observed for ppSARS and ppSARS-2. Pseudotyped viruses expressing the S proteins from MERS-CoV (ppMERS), which is known to utilize CD26 as an entry receptor [17], also infected Huh 7.5 cells (Fig. 1a).
Fig. 1.
Cell entry sensitivity test with pseudotyped SARS-CoV, SARS-CoV-2 and MERS-CoV viruses. (a) Huh7.5 cells are sensitive to infection with ppSARS, ppSARS-2 and ppMERS, with similar entry levels between ppSARS and ppSARS-2 (p > 0.1, two-way ANOVA). (b) HEK 293T cells were transiently transfected with the hACE2 expression plasmid. ppSARS and ppSARS-2 were both found to significantly enhance the infection ratio (p < 0.001, two-way ANOVA). Similar levels of entry were observed in hACE2 transfected 293T cells (p > 0.1, two-way ANOVA). (c) VeroE6 cells were infected with live SARS-CoV-2 in the presence of soluble ACE2 at different concentrations, in which 30 µg/mL of soluble ACE2 was found to inhibit virus replication. **P < 0.01, ****P < 0.0001.
We next used 293T cells with or without transfection of human ACE2 (hACE2) to assess the viral infection. Exogenous expression of hACE2 resulted in approximately 200times higher viral entry of both ppSARS and ppSARS-2, confirming that hACE2 expression substantially increasing the infection efficiency (Fig. 1b). To test whether hACE2 is required for SARS-CoV-2 infection, we infected Vero cells with 50 TCID50 of live SARS-CoV-2 virus in the presence of various concentrations of recombinant ACE2, as a soluble form of ACE2-Fc [14]. SARS-CoV-2 amplified over 200 times on Vero cells within 36 h in the absence of any inhibitor, recombinant ACE2-Fc inhibited the infection in a dose-dependent manner, with greater than 60% virus amplification was inhibited at a concentration of 30 µg/mL of ACE2-Fc (Fig. 1c), suggesting ACE2 is required for the SARS-CoV-2 infection in Vero cells.
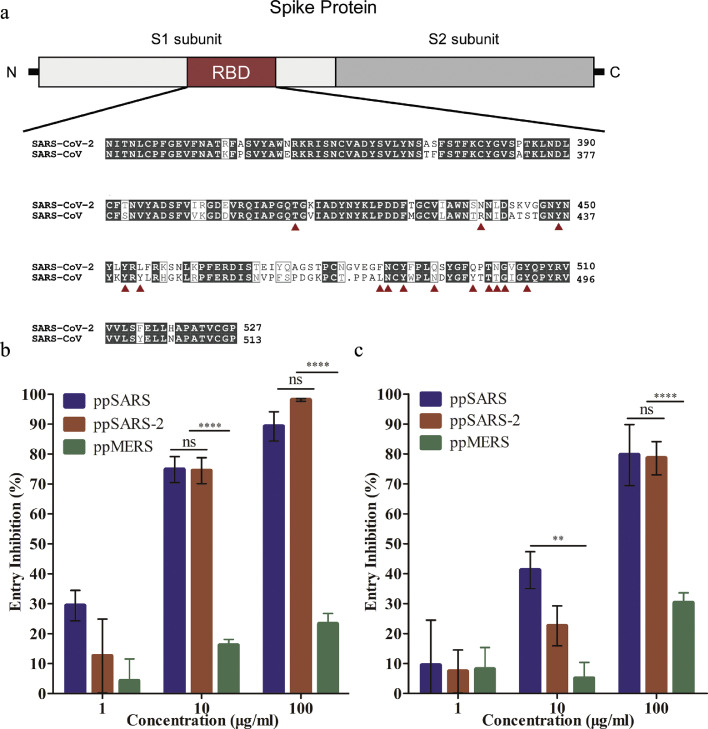
We next examined whether recombinant RBD proteins from SARS-CoV could inhibit SARS-CoV-2 infection. Sequence alignment of RBD of SARS-CoV and SARS-CoV-2 showed relative high conservation of the residues crucial for ACE2 binding (Fig. 2a). Two forms of SARS-CoV RBD recombinant protein were used as entry inhibitor in viral infection assay: 1) recombinant RBD monomer (RBD-monomer); 2) RBD-trimer, in which a T4f motif was fused at the C-terminal of RBD, presumably mimicking a natural form of the RBD in the S trimer. We found that ppSARS and ppSARS-2 can both be blocked by RBD-trimer (Fig. 2b), and to a lesser extent, RBD-monomer (Fig. 2c). RBD-trimer blocked over 70% viral entry of ppSARS and ppSARS-2 at a concentration of 10 µg/mL, and over 85% viral entry at a concentration of 100 µg/mL. 10 µg/mL RBD-monomer blocked 40% ppSARS and 20% ppSARS-2 infection, respectively; while 100 µg/mL RBD-monomer blocked ~80% viral entry of both viruses. As expected, viral infection by ppMERS was not or only slightly affected by the RBDs (Fig. 2b and c). These results are in line with the structural studies that SARS-CoV and SARS-CoV-2 share similar binding site on ACE2 [18].
Fig. 2.
Alignment of RBD sequences for SARS-CoV and SARS-CoV-2, and competitive inhibition assays with RBD for pseudotyped SARS-CoV, SARS-CoV-2 and MERS-CoV viruses. (a) SARS-CoV-2 & SARS-CoV receptor binding domain alignment. Amino acid residues known to be important for binding were denoted by red triangles. (b) Competition assay for RBD-trimer or (c) RBD-monomer to the ACE2 receptor to block ppSARS, ppSARS-2 or ppMERS entry. Near-complete inhibition was observed at 100 μg/mL for RBD-trimer, with no significant differences observed between ppSARS and ppSARS-2 (p > 0.1, two-way ANOVA). Approximately 80% inhibition was observed at 100 μg/mL for RBD-monomer, with no significant differences observed between ppSARS and ppSARS-2 (p > 0.1). ** P < 0.01, **** P < 0.0001.
3.2. Mice vaccinated with various SARS-CoV vaccines did not develop notable cross-neutralizing antibodies against SARS-CoV-2
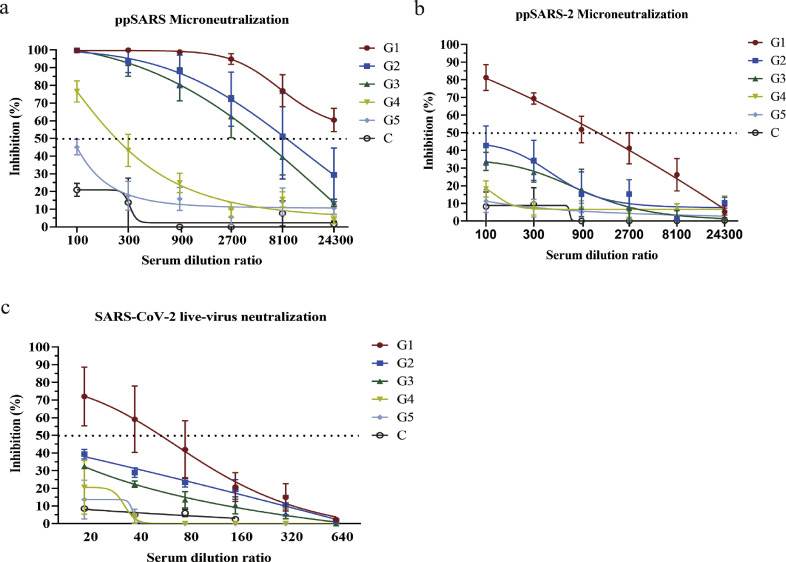
Although SARS-CoV RBD effectively blocked SARS-CoV2 entry, it remained unclear whether mice vaccinated with SARS-CoV vaccines could provide protection against SARS-CoV-2 infection. We assessed neutralizing activity to SARS-CoV and SARS-CoV-2 using the sera from mice vaccinated with several SARS-CoV vaccines (for detailed methodology see Materials and Methods). Sera samples from Groups 1–3, which were first immunized with a SARS-CoV inactivated vaccine, followed by boosting with either recombinant SARS-CoV S protein, S1 protein; or inactivated SARS-CoV vaccine in the presence of Complete Freund's Adjuvant (CFA) or alum adjuvant, were found to completely neutralize ppSARS at dilutions of 1:100, with the ND50 estimated to be over 1:24,300 in Group 1; 1:8780 in Group 2 or 1:5405 in Group 3 (Fig. 3a). Some neutralizing activity against ppSARS was also detected from the sera of Groups 4–5, but at lower serum titers. Importantly, the mouse sera did not show notable neutralizing activity against ppSARS-2, with the exception of Group 1, which exhibited 80% neutralization against ppSARS-2 at a 1:100 dilution (Fig. 3b), with ND50 titers of 1:1351. These results were confirmed by assays using live SARS-CoV-2 infection in Vero cells, in which only antisera from Group 1 showed 70% neutralization at a 1:20 dilution (Fig. 3c), with ND50 titers of 1:56. These results indicated mice vaccinated by SARS-CoV vaccines have limited if any cross-neutralization against SARS-CoV-2.
Fig. 3.
Microneutralization of antisera from mice immunized with various SARS vaccine candidates with pseudotyped SARS-CoV, pseudotyped and live SARS-CoV-2 viruses. Immunized mice groups were denoted as G1-G5, whereas mock alum-immunized mice were denoted as C. (a) Neutralizing activity can be observed in all five immunized groups. (b) Cross-neutralizing activity to ppSARS-2 can be detected only in Group 1, with no cross-neutralization observed in the other four groups. (c) Reexamination of mice antisera with live SARS-CoV-2 showed that neutralizing activity was only observed in Group 1.
3.3. Convalescent sera from SARS-CoV and SARS-CoV-2 patients exhibit limited or no cross-neutralization against the other virus
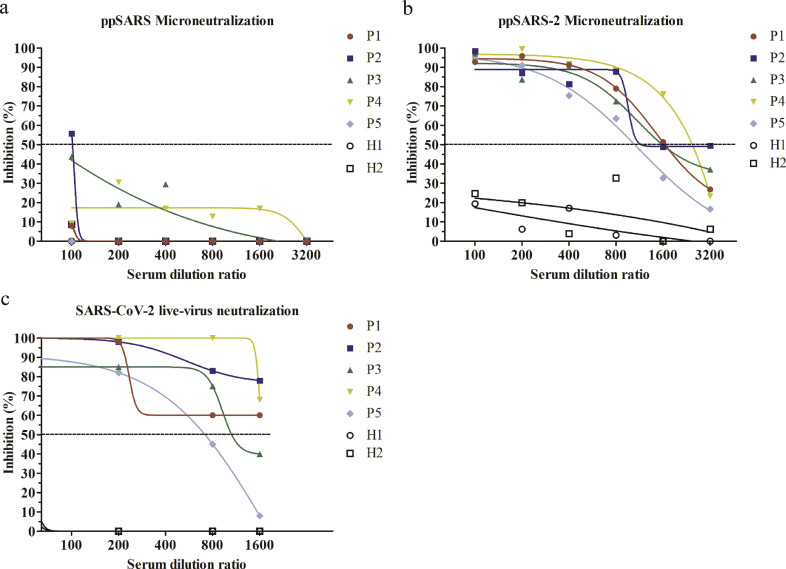
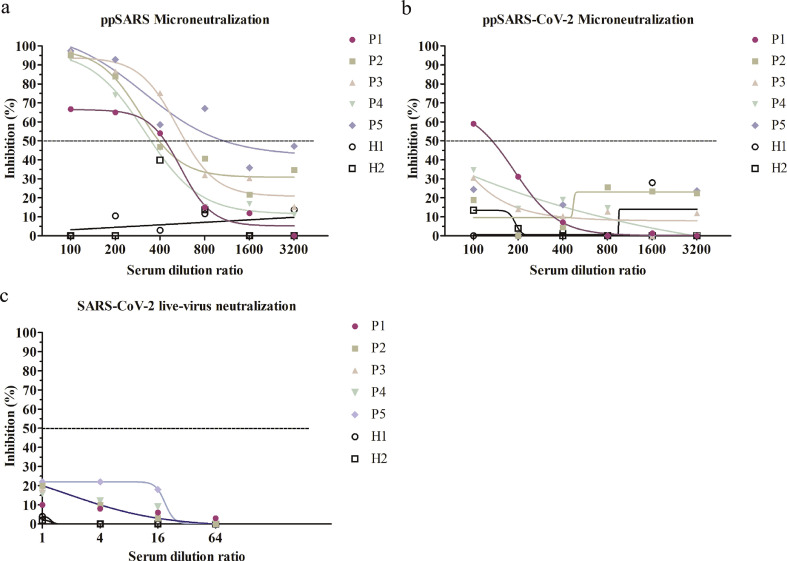
We then examined whether convalescent sera from SARS-CoV and SARS-CoV-2 patients exhibit cross-neutralization activity against the other virus. We first used pseudotyped viruses to evaluate the neutralizing activity of sera collected from convalescent patients of SARS-CoV (n = 5) and SARS-CoV-2 (n = 5). Convalescent sera from SARS-CoV-2 patients effectively neutralized ppSARS-2 infection at serum dilutions of 1:400, while showed no neutralizing activity against ppSARS infection (Fig. 4a and b). The 50% neutralizing dilution (ND50) titer was around 1750 for ppSARS2. In live SARS-CoV-2 microneutralization assay, the sera also showed high neutralizing activity, with an estimated ND50 of 1:1585 (Fig. 4c). Conversely, convalescent sera from SARS-CoV patients blocked ppSARS infection at serum dilutions of 1:200, with an ND50 around 1:630, and showed no neutralizing activity against either ppSARS infection (Fig. 5a and b) or live SARS-CoV-2 infection (Fig. 5c). Collectively, these results show that SARS-CoV and SARS-CoV-2 share limited if any common neutralization epitopes although both viruses use ACE2 as a cellular receptor.
Fig. 4.
Serum microneutralization of antisera from convalescent SARS-CoV-2 patients with pseudotyped SARS-CoV, pseudotyped and live SARS-CoV-2 viruses. Convalescent SARS-CoV-2 patients were denoted as P1-P5, and healthy controls were denoted as H1-H2. (a) None of the 5 tested patient samples can neutralize ppSARS. (b) All 5 serum samples can effectively neutralize ppSARS-2. (c) All 5 serum samples can effectively neutralize live SARS-CoV-2.
Fig. 5.
Serum microneutralization of antisera from convalescent SARS-CoV patients with pseudotyped SARS-CoV, pseudotyped and live SARS-CoV-2 viruses.Convalescent SARS-CoV patients were denoted as P1-P5, and healthy controls were denoted as H1-H2. (a) Neutralization against ppSARS was observed in all five patients. (b) None of the 5 convalescent serum samples can effectively neutralize ppSARS-2. (c) None of the convalescent samples can effectively neutralize live SARS-CoV-2.
4. Discussion
Vaccines inducing broad and sustained immunity could offer effective protection against infectious diseases such as COVID-19 [19,20]. Unfortunately, there are currently no approved vaccines available against any human coronavirus, despite the huge medical, social and economic impacts imposed by the SARS, MERS, and the COVID-19 pandemics. Developing a vaccine, ideally a universal prophylactic effective against coronaviruses, will be extremely useful for combating the ongoing COVID-19 pandemic as well as future outbreaks caused by SARS-CoV variants and possibly other coronaviruses. SARS-CoV-2 is closely related to SARS-CoV. Although the mechanisms underlying immune protection against SARS-CoV has yet to be fully understood, it is believed that anti-SARS-CoV neutralizing antibodies play a critical role [21,22]. Neutralizing antibodies usually bind to the receptor binding region or the membrane fusion site of the viral entry protein. The S protein of coronavirus is responsible for viral entry, and is the most effective target for inducing neutralizing antibodies. Indeed, antibodies targeting the RBD of the S protein showed the highest neutralizing activity against SARS-CoV infections [23]. Interestingly, although SARS-CoV RBD effectively blocked SARS-CoV-2 infection, our studies revealed that there are limited if any common neutralization epitopes between these two viruses. In our animal studies with established SARS-CoV vaccination regimens [24,25], mice vaccinated with various combinations of inactivated SARS-CoV, recombinant adenovirus expressing SARS-CoV spike protein, or recombinant SARS-CoV S proteins, showed potent neutralizing activity against SARS-CoV but limited or no neutralization activity against SARS-CoV-2 infection. We confirmed this by studying the neutralizing activity of convalescent sera from patients naturally infected by SARS-CoV or SARS-CoV-2. Whether prior SARS-CoV infection can protect a subsequent SARS-CoV-2 infection (or vice versa) remains to be determined, convalescent sera from SARS-CoV and SARS-CoV-2 patients showed high neutralizing activity only against the homologous virus, but with limited or no cross-neutralization activity against the heterologous virus. The results were consistent with other studies which showed the lack of cross-neutralizaiton between these two viruses [26], even though both of them use ACE2 as the receptor to enter the cells [27].
Despite lacking a full immunogenicity profile analysis (including analysis of T-cell responses), as well as animal challenge data from a suitable animal model, our results suggest that current candidate SARS-CoV vaccines may not generate cross-protective immunity against SARS-CoV-2, highlighting challenges towards developing a universal vaccine against SARS-CoV related viruses.
Author contributions
Ren Yang and Jiaming Lan performed data collection, data analysis, data interpretation and writing; Baoying Huang conducted the experiments in BSL-3 and data collection; Ruhan A performed data collection; Mingqing Lu performed the literature search; Wen Wang conducted the animal experiments; Wenling Wang designed the figures; Wenhui Li assisted with data analysis and writing; Yao Deng designed the study and analyzed the data; Gary Wong conducted data analysis, data interpretation and wrote the manuscript; Wenjie Tan conducted study design, data analysis and data interpretation.
Declaration of competing interest
The authors declare no conflict of interests with respect to this study.
Funding sources
The study was supported by the National Key Research and Development Program of China (Nos. 2016YFD0500301) and the National Major Project for Control and Prevention of Infectious Disease in China (Nos. 2016ZX10004001-003) to WT, and the One Belt and One Road Major Project for infectious diseases (2018ZX10101004-003) to GW. GW is supported by a G4 grant from IP, FMX and CAS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Yao Deng, Email: dengyao31@163.com.
Gary Wong, Email: garyckwong@ips.ac.cn.
Wenjie Tan, Email: tanwj28@163.com.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. COVID-19. 2020.
- 4.Hui D.S., I Azhar E., Madani T.A., Ntoumi F., Kock R., Dar O. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health -the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Z., Xiao X., Wei X., Li J., Yang J., Tan H. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J Med Virol. 2020 doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H. First Case of 2019 Novel Coronavirus in the United States. N Engl J Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., et al. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N Engl J Med2020;382(10):970–1. [DOI] [PMC free article] [PubMed]
- 8.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 14.Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24(8):1634–1643. doi: 10.1038/sj.emboj.7600640. 2005-04-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng Y., Lan J., Bao L., Huang B., Ye F., Chen Y. Enhanced protection in mice induced by immunization with inactivated whole viruses compare to spike protein of middle east respiratory syndrome coronavirus. Emerg Microbes Infect. 2018;7(1):60. doi: 10.1038/s41426-018-0056-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao G., Du L., Ma C., Li Y., Li L., Poon V.K. A safe and convenient pseudovirus-based inhibition assay to detect neutralizing antibodies and screen for viral entry inhibitors against the novel human coronavirus MERS-CoV. Virol J. 2013;10:266. doi: 10.1186/1743-422X-10-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu G., Hu Y., Wang Q., Qi J., Gao F., Li Y. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500(7461):227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C., Abiona O. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kardani K., Bolhassani A., Shahbazi S. Prime-boost vaccine strategy against viral infections: mechanisms and benefits. Vaccine. 2016;34(4):413–423. doi: 10.1016/j.vaccine.2015.11.062. [DOI] [PubMed] [Google Scholar]
- 20.Casadevall A. Antibody-based vaccine strategies against intracellular pathogens. Curr Opin Immunol. 2018;53:74–80. doi: 10.1016/j.coi.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burton D.R., Hangartner L. Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu Rev Immunol. 2016;34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Blargan L.A., Goo L., Pierson T.C. Deconstructing the antiviral neutralizing-antibody response: implications for vaccine development and immunity. Microbiol Mol Biol Rev. 2016;80(4):989–1010. doi: 10.1128/MMBR.00024-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niu P., Zhao G., Deng Y., Sun S., Wang W., Zhou Y. A novel human mAb (MERS-GD27) provides prophylactic and postexposure efficacy in MERS-CoV susceptible mice. Sci China Life Sci. 2018;61(10):1280–1282. doi: 10.1007/s11427-018-9343-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spruth M., Kistner O., Savidis-Dacho H., Hitter E., Crowe B., Gerencer M. A double-inactivated whole virus candidate SARS coronavirus vaccine stimulates neutralising and protective antibody responses. Vaccine. 2006;24(5):652–661. doi: 10.1016/j.vaccine.2005.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.See R.H., Zakhartchouk A.N., Petric M., Lawrence D.J., Mok C.P., Hogan R.J. Comparative evaluation of two severe acute respiratory syndrome (SARS) vaccine candidates in mice challenged with SARS coronavirus. J Gen Virol. 2006;87(Pt 3):641–650. doi: 10.1099/vir.0.81579-0. [DOI] [PubMed] [Google Scholar]
- 26.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020 doi: 10.1016/j.cell.2020.03.045. pii:S0092-8674(20)30338-X. [DOI] [PMC free article] [PubMed] [Google Scholar]