Abstract
Selenoproteins contain the amino acid selenocysteine and are found in all domains of life. The functions of many selenoproteins are poorly understood, partly due to difficulties in producing recombinant selenoproteins for cell-biological evaluation. Endogenous mammalian selenoproteins are produced through a noncanonical translation mechanism requiring suppression of the UGA stop codon, and a selenocysteine insertion sequence (SECIS) element in the 3’ untranslated region of the mRNA. Here, recombinant selenoproteins are generated in mammalian cells through genetic code expansion, circumventing the requirement for the SECIS element, and selenium availability. An engineered orthogonal E. coli leucyl-tRNA synthetase/tRNA pair is used to incorporate a photocaged selenocysteine (DMNB-Sec) at the UAG amber stop codon. DMNB-Sec is successfully incorporated into GFP and uncaged by irradiation of living cells. Furthermore, DMNB-Sec is used to generate the native selenoprotein methionine-R-sulfoxide reductase 1 (MsrB1). Importantly, MsrB1 is shown to be catalytically active after uncaging, constituting the first use of genetic code expansion to generate a functional selenoprotein in mammalian systems. The ability to site-specifically introduce selenocysteine directly in mammalian cells, and temporally modulate selenoprotein activity, will aid in the characterization of mammalian selenoprotein function.
Selenoproteins contain the amino acid selenocysteine (Sec), which is a cysteine (Cys) cognate with selenium (Se) in place of sulfur (S). In the majority of characterized selenoproteins, such as glutathione peroxidase and thioredoxin reductases, Sec performs redox-catalytic functions.1 Selenoproteins are found throughout all three domains of life, with 25 selenoproteins expressed in humans.2 However, in some species, Cys-containing homologs nominally perform the same function as selenoproteins. The stringent requirement for Sec in certain organisms has been partially attributed to the reversible nature of Sec oxidation, compared to Cys, thereby conferring resistance to irreversible inactivation under oxidative stress.3 For example, replacement of Sec with Cys in glutathione peroxidase 4 (GPX4) renders neurons susceptible to ferroptotic cell death due to GPX4 inactivation through irreversible Cys oxidation.4
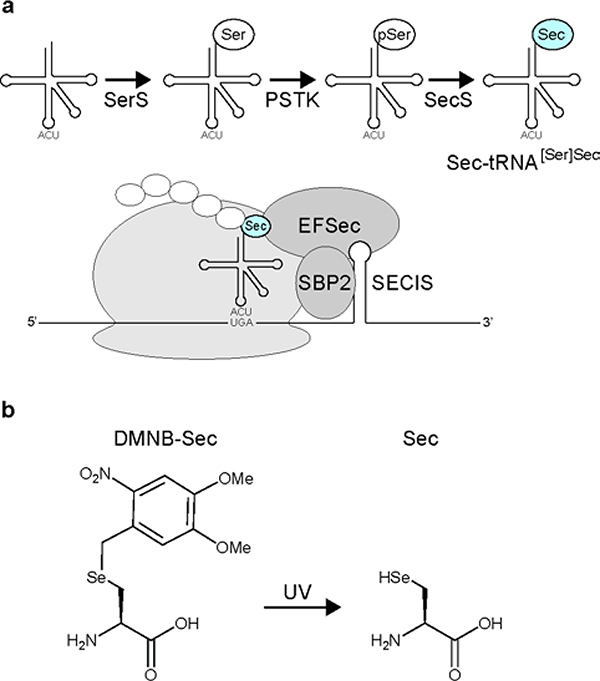
Sec incorporation deviates from canonical protein translation, requiring suppression of the UGA stop codon. In eukaryotes, Sec biosynthesis occurs directly on the suppressor tRNA (tRNA[Ser]Sec). Specifically, tRNA[Ser]Sec is aminoacylated with serine by seryl-tRNA synthetase (SerS), followed by phosphorylation by phosphoseryl-tRNA kinase (PSTK), and subsequent Se incorporation by Sec synthase (SecS), to generate Sec-tRNA[Ser]Sec (Figure 1a).5, 6 Suppression of UGA by Sec-tRNA[Ser]Sec requires a Sec insertion sequence (SECIS) element in the selenoprotein mRNA. The SECIS element appears in the 3’ UTR of the selenoprotein mRNA, and binding of the SECIS binding protein 2 (SBP2) and Sec-specific translation elongation factor (EFSec) are required for successful Sec insertion (Figure 1a).7–9
Figure 1.
Selenocysteine incorporation and DMNB-Sec structure. (a) Endogenous eukaryotic Sec-incorporation mechanism. (b) Structures of DMNB-Sec and Sec.
Due to the complexity of endogenous selenoprotein production in eukaryotes, recombinant expression has proven challenging. Several methods have therefore been developed to obtain recombinant selenoproteins. In vitro methods include the use of solid-phase selenopeptide synthesis, coupled with native and expressed protein ligation, to produce full-length selenoproteins.10 The recombinant expression of selenoproteins in E. coli has been enhanced by a variety of methods, including the use of reassigned sense codons, engineered tRNAs, and recoded release factor 1 (RF-1)-deficient E. coli.11–15 These prokaryotic expression systems have significantly expanded the ability to produce recombinant mammalian selenoproteins for biochemical characterization. Similar approaches have been taken to increase selenoprotein production yields in eukaryotic cells, including the use of unique SECIS elements from Toxoplasma.16
Genetic code expansion (GCE) technology allows site-specific incorporation of noncanonical amino acids using engineered aminoacyl-tRNA synthetase (RS)/tRNA pairs.17–19 GCE offers an attractive alternative for selenoprotein production that overrides many limitations of the endogenous pathway, including the requirement of the SECIS element and selenium availability. Additionally, GCE enables incorporation of protected analogs of Sec, which can be deprotected and activated on demand. An allyl-protected Sec (ASec) and a photocaged (methyl-(6-nitropiperonyloxymethyl), MeNPOM) Sec (PSc) have been genetically encoded in E. coli using the pyrrolysyl-tRNA synthetase/tRNA pair, and can be uncaged to Sec using palladium (Pd)-catalysis and light, respectively.20, 21 In yeast, a photocaged (4,5-dimethoxy-2-nitrobenzyl, DMNB) Sec (DMNB-Sec) (Figure 1b) was incorporated using the E. coli leucyl-tRNA synthetase/tRNA pair.22 To date, only ASec has been successfully incorporated and uncaged in mammalian cells, therefore additional technologies for mammalian expression of selenoproteins are needed.
Here, we report an alternative platform to generate selenoproteins directly in mammalian cells through incorporation of photocaged DMNB-Sec (Figure 1b). The noncanonical amino acid DMNB-Sec is minimally toxic to mammalian cells at the concentrations required for incorporation. DMNB-Sec was incorporated into the model protein GFP, and the native human selenoprotein MsrB1, in HEK293T cells. Uncaging of the resulting selenoproteins was achieved by minimally invasive 365 nm irradiation of purified protein or living cells expressing the protein.23 Importantly, DMNB-Sec incorporation in MsrB1 was shown to be non-disruptive to protein function, demonstrating the utility of this genetic code expansion platform for studying native selenoprotein function in mammalian cells.
RESULTS AND DISCUSSION
Incorporation of DMNB-Sec into GFP in HEK293T cells
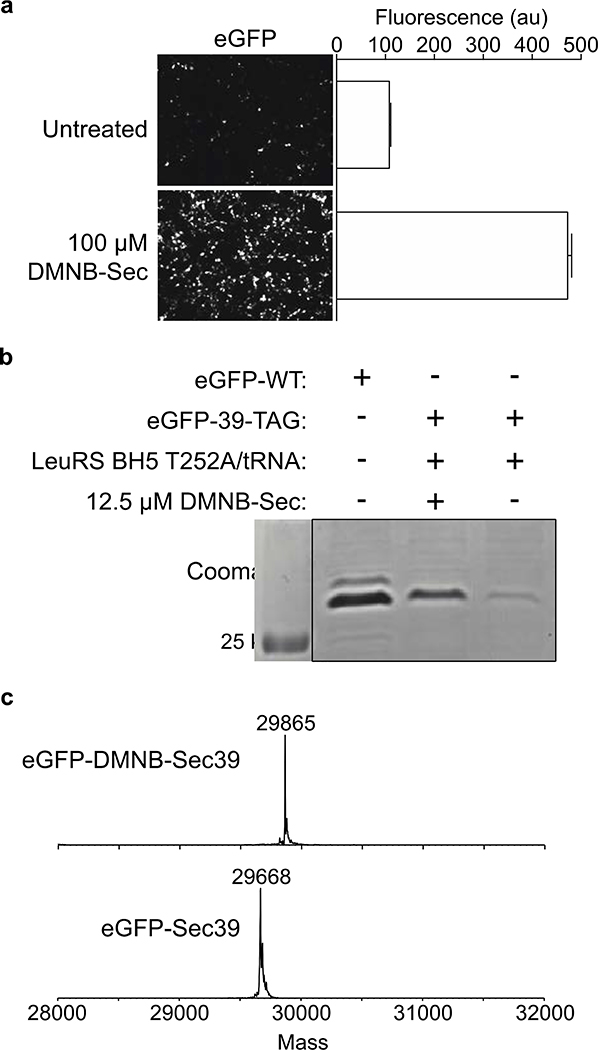
The feasibility of incorporating a photocaged selenocysteine amino acid into proteins in mammalian cells was initially evaluated using GFP as the target protein in HEK293T cells. Photocaged DMNB-Sec (Figure 1b) was synthesized as previously reported.22 Incorporation of DMNB-Sec into GFP in mammalian cells was achieved using the E. coli LeuRS BH5 T252A/tRNA pair, which was originally evolved for photocaged serine (DMNB-Ser) incorporation,24 and subsequently used for DMNB-Sec in yeast.22 LeuRS and 8 copies of the Leu tRNA were cloned into the pAcBac2 vector containing an eGFP-39-TAG reporter (Supplemental Figure 1).25–27 Transient transfection of the pAcBac2 vector into HEK293T cells was followed by growth in the presence or absence of 100 μM DMNB-Sec. Fluorescence of GFP was measured to be 5-fold higher for lysates from cells exposed to DMNB-Sec (Figure 2a), consistent with previously reported data for the yeast LeuRS system with DMNB-Sec and DMNB-Cys.22
Figure 2.
Generation of eGFP-DMNB-Sec39. (a) Fluorescence microscopy and quantification of HEK293T cells transiently transfected with a pAcBac2 plasmid encoding LeuRS BH5 T252A /Leu tRNA/eGFP-39-TAG, in the presence and absence of DMNB-Sec. (b) SDS-PAGE of eGFP-WT and LeuRS BH5 T252A /Leu tRNA/eGFP-39-TAG expressed in the presence and absence of DMNB-Sec (c) ESI-MS analysis of eGFP-DMNB-Sec39 (expected mass: 29865.33) and irradiated eGFP-Sec39 (expected mass: 29669.24)
The toxicity of DMNB-Sec was examined using an MTT assay, and DMNB-Sec was found to be minimally toxic to HEK293T at low micromolar concentrations (EC50 of 207 μM; Supplemental Figure 2). DMNB-Sec was used at concentrations in the range of 12.5–100 μM for all reported experiments. To purify GFP for mass-spectrometry characterization, HEK293T cells were transfected with eGFP-WT or eGFP-39-TAG and grown in the presence or absence of DMNB-Sec. The resulting proteins were purified via a C-terminal 6XHis tag and analyzed by SDS-PAGE (Figure 2b). A measure of purified protein concentration revealed that eGFP-DMNB-Sec39 was expressed and purified with a yield of 45% compared to eGFP-WT. Intact-protein electrospray-ionization mass spectrometry (ESI-MS) confirmed the presence of the expected DMNB-Sec-containing eGFP species (Figures 2c, Supplemental Figure 3). Analysis of eGFP from cells grown in the absence of DMNB-Sec identified a mass consistent with the incorporation of leucine (Leu) or isoleucine (Ile) (Supplemental Figure 4). When eGFP was expressed in the presence of DMNB-Sec, eGFP-Leu/Ile39 was not detectable by ESI-MS (Supplemental Figure 3), suggesting that incorporation of these natural amino acids is suppressed by the presence of DMNB-Sec.
To monitor uncaging of the DMNB protecting group, cells expressing eGFP-DMNB-Sec39 were irradiated (365 nm, 10 mins), followed by purification and ESI-MS analysis of the resulting protein. The presence of uncaged eGFP-Sec39 was confirmed by ESI-MS (Figure 2c, Supplemental Figure 5), with no detectable ESI-MS peak observed for the caged eGFP-DMNB-Sec39. In the previously published study that incorporated DMNB-Sec into proteins in yeast, irradiation of yeast lysates resulted in the formation of diselenide dimers as well as dehydroalanine species.22 However, here we report that only the expected monomer was observed in ESI-MS analysis of mammalian-expressed eGFP-Sec39. To further explore the kinetics of uncaging, purified eGFP-DMNB-Sec39 was subjected to irradiation for various time periods (0–20 mins), followed by ESI-MS analysis. Detectable levels of the uncaged eGFP-Sec39 species appears after ~0.5 mins of irradiation at 365 nm, and complete uncaging is observed after ~5 mins of irradiation (Supplemental Figure 6).
Incorporation of DMNB-Sec into human MsrB1 in HEK293T cells
Upon demonstrating the successful incorporation of DMNB-Sec into GFP, a photocaged version of the native mammalian selenoprotein, human methionine-R-sulfoxide reductase 1 (MsrB1), was generated. MsrB1 is localized to the cytoplasm and nucleus, and utilizes an active-site Sec to stereo-selectively reduce methionine-R-sulfoxide.28 Intriguingly, the MsrB1 homologs in humans, MsrA, MsrB2 and MsrB3, utilize a Cys, instead of Sec, to perform similar chemistry. Studies comparing Cys- versus Sec-containing Msr proteins have shown that Sec is advantageous for reductase activity.1, 29, 30 MsrB1 protects proteins from oxidative damage, and enables signaling via dynamic, enzyme-dependent methionine oxidation and reduction.31, 32 The role of MsrB1 has been well characterized for the substrate actin, but regulation of other potential substrates is poorly understood, due in part to the difficulties with overexpression and temporally controlled activation of MsrB1 in mammalian cells. Expression of MsrB1 with a caged active-site Sec residue would facilitate further characterization of the cellular functions of MsrB1.
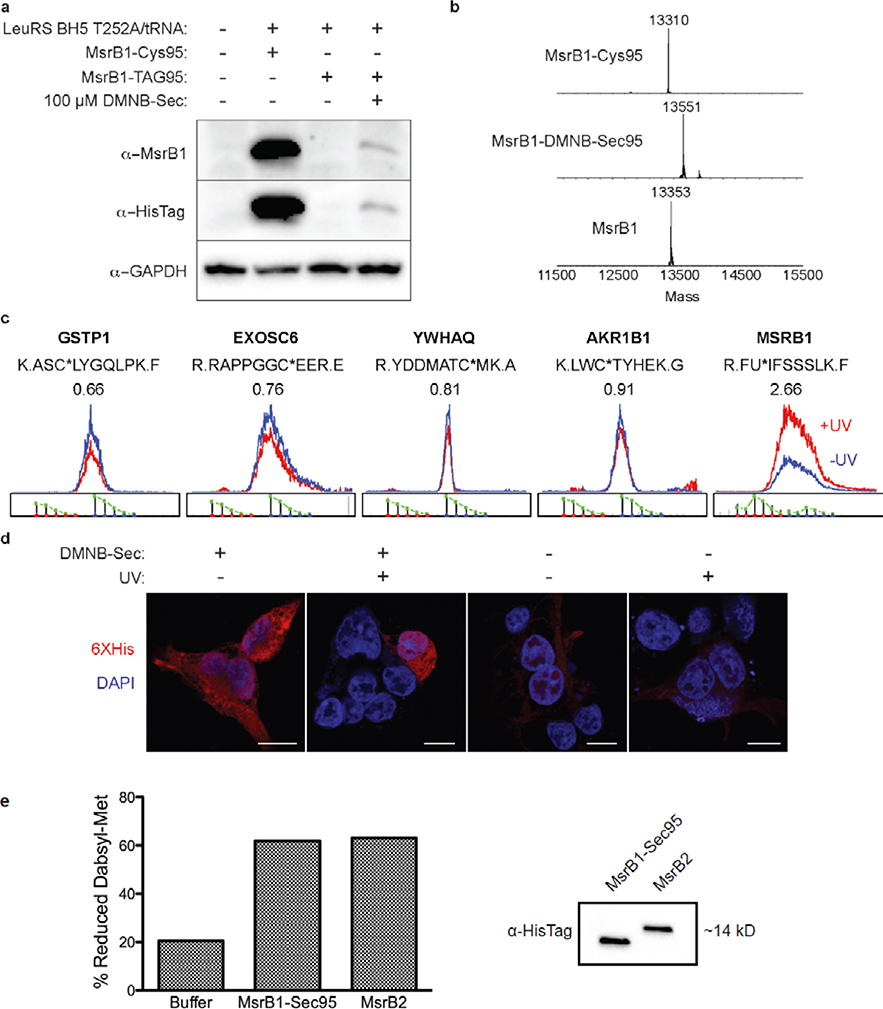
To recombinantly express MsrB1, a pAcBac2 plasmid containing the following elements was generated: (1) LeuRS BH5 T252A; (2) 8 copies of the E. coli Leu tRNA; and, (3) the MsrB1 gene with the amber stop codon (MsrB1-TAG95) or a Cys codon (MsrB1-Cys95) at residue 95 (the site of the endogenous Sec residue), and a C-terminal 6XHis tag. To test for successful incorporation of DMNB-Sec, MsrB1 expression was monitored via immunoblot using antibodies against MsrB1 and the C-terminal 6XHis tag. Importantly, the 6XHis antibody will only detect full-length protein generated via successful amber suppression. Full-length MsrB1 protein was observed for MsrB1-Cys95, and DMNB-Sec treated MsrB1-TAG95 cells (Figure 3a). As expected, no protein was detected in the MsrB1-TAG95 cells in the absence of DMNB-Sec. Comparison of protein yields of MsrB1-Cys95 and MsrB1-DMNB-Sec95 by western blot indicated that expression of the DMNB-Sec-containing MsrB1 was ~10% that of the Cys-containing version.
Figure 3.
Generation of MsrB1-DMNB-Sec95. (a) Immunoblot of MsrB1 expression with a Cys or TAG amber codon at residue 95 in the presence and absence of DMNB-Sec. (b) ESI-MS analysis of MsrB1-Cys95 (expected mass: 13309.97), MsrB1-DMNB-Sec95 (expected mass: 13552.04), and uncaged MsrB1-Sec95 (expected mass: 13356.87). (c) Low pH isoTOP-ABPP analysis showing extracted ion chromatograms for Cys- or Sec-containing peptides from irradiated (red, IA-light) or non-irradiated (blue, IA-heavy) cells expressing MsrB1-DMNB-Sec95. (d) Immunofluorescence of cells transfected with MsrB1-TAG95 in the presence or absence of DMNB-Sec, before and after irradiation. Scale bar represents 10 μm. (e) Methionine sulfoxide reductase activity of buffer alone (negative control), purified uncaged MsrB1-Sec95, and MsrB2 (positive control), measured as the percent reduced dabsyl-Met (dabsyl-MetR) present within the total amount of dabsyl-Met (oxidized and reduced). Immunoblot shows that levels of purified MsrB1-Sec95 is comparable to the amount of MsrB2 used in the activity assay.
Purification and analysis by ESI-MS confirmed the presence of MsrB1-Cys95 and MsrB1-DMNB-Sec95 (Figures 3b, Supplemental Figure 7 and 8). Successful uncaging was confirmed by subjecting purified MsrB1-DMNB-Sec95 to UV irradiation (365 nm, 10 mins), and subsequent ESI-MS analysis (Figure 3b, Supplemental Figure 9). As with eGFP-Sec39, dimer and dehydroalanine formation was not observed by ESI-MS. Additionally, MsrB1-Ile/Leu95 was not detected, confirming that misincorporation of Ile/Leu is negligible in the presence of DMNB-Sec.
To demonstrate uncaging of MsrB1 directly in cells, we utilized an isoTOP-ABPP method that was modified for selenoprotein detection.33 Typical isoTOP-ABPP monitors Cys reactivity in complex proteomes via the following steps: (1) cell-lysate labeling with an iodoacetamide (IA)-alkyne probe; (2) conjugation of IA-alkyne modified proteins to a cleavable-biotin tag using copper (I)-catalyzed azide-alkyne cycloaddition (CuAAC); (3) enrichment of IA-alkyne modified proteins on streptavidin beads; (4) on-bead trypsin digestion and cleavage of the linker to release IA-alkyne modified peptides for analysis by LC/LC-MS/MS. Incorporation of isotopically light and heavy IA-alkyne probes, or cleavable linkers, enable quantitative comparisons of cysteine reactivity across different biological samples.34–36 Performing the initial IA-alkyne labeling step at low pH (~pH 5.75) was shown to suppress Cys labeling, and thereby enhance the detection of Sec-containing peptides for the analysis of Sec-reactivity changes.33
Low-pH isoTOP-ABPP analysis was used to compare lysates from irradiated (IA-light labeled) and non-irradiated (IA-heavy labeled) HEK cells expressing MsrB1-DMNB-Sec95 (Supplemental Figure 10). The IA-labeled MsrB1 Sec-containing peptide (R.FU*IFSSSLK.F) was identified via LC/LC-MS/MS, and shown to display a light:heavy ratio (R) of 2.66, indicative of ~3-fold increased Sec reactivity in the irradiated sample (Figure 3c). In contrast, identified Cys-containing peptides showed R values of ~1 (Figure 3c), indicating no reactivity change upon irradiation. Importantly, the identified MsrB1 peptide displayed an isotopic envelope characteristic of Se-containing peptide species, confirming Sec incorporation.37 Together, the data from the low pH isoTOP-ABPP analysis confirms the successful incorporation and uncaging of DMNB-Sec in MsrB1 directly in mammalian cells.
Endogenous MsrB1 is known to localize to the cytosol and nucleus of cells.28 To confirm that MsrB1-DMNB-Sec95 shows similar localization patterns, we utilized confocal microscopy to determine the cellular localization of MsrB1-DMNB-Sec95 in irradiated and non-irradiated cells. Regardless of irradiation, the recombinant MsrB1 was shown to display both cytosolic and nuclear localization, consistent with the endogenous protein (Figure 3d). As expected, cells cultured in the absence of DMNB-Sec did not show a significant signal for 6XHis-MsrB1. These imaging studies confirm that photocaged versions of endogenous selenoproteins can be produced and activated in mammalian cells with the expected cellular localization for subsequent biological interrogation.
To confirm that the MsrB1-DMNB-Sec95 expressed in HEK293T cells displays methionine sulfoxide reductase activity, we monitored the ability of the purified uncaged MsrB1 protein to reduce dabsyl-methionine sulfoxide (dabsyl-MetO). Briefly, dabsyl-MetO was incubated with buffer (negative control), uncaged MsrB1-DMNB-Sec95 (MsrB1-Sec95), or human recombinant MsrB2 (positive control). The resulting reaction mixtures were analyzed by LC/MS, whereby levels of oxidized and reduced dabsyl-Met were quantified using absorbance at 466 nm.38 MsrB1-Sec95 reduced dabsyl-MetO at levels comparable to human recombinant MsrB2, producing ~3-fold greater dabsyl-Met relative to the negative control (Figure 3e, Supplemental Figure 11). The observed reductase activity of MsrB1-Sec95 on the dabsyl-MetO substrate confirms that incorporation of DMNB-Sec into MsrB1 does not grossly disrupt protein structure, and subsequent uncaging produces the catalytically active protein product.
In summary, studies of selenoprotein function have been hindered by the poor efficiency of recombinant expression of Sec-containing proteins and related mutants, stimulating the development of numerous technologies for the production of selenoproteins in vitro, in E. coli, and in mammalian cells. Of these, GCE methods can introduce protected variants of selenocysteine for controlled activation, and circumvent the requirement for the SECIS element and the dependence on selenium concentrations in the growth media. We adapt the E. coli leucyl-tRNA synthetase/tRNA pair to incorporate the photocaged amino acid DMNB-Sec into GFP and native selenoproteins directly in mammalian cells. Currently, the only available technology for producing protected selenoproteins in mammalian cells, is a GCE platform utilizing ASec, whereby ASec-containing GFP was deprotected directly in mammalian cells using palladium-mediated strategies. ASec was not incorporated into a native selenoprotein, so the effects of palladium-mediated deprotection on selenoprotein function was not investigated. We show that DMNB-Sec incorporation and uncaging by UV irradiation does not disrupt the catalytic activity of MsrB1. Additionally, DMNB-Sec displays low toxicity at the concentrations required for selenoprotein production (12.5–100 μM). In contrast, the required concentrations of ASec (0.2 mM) are toxic to mammalian cells, necessitating supplementation with high concentration of cysteine (3.2 mM) to mitigate toxicity.20 Addition of high cysteine concentrations will perturb cellular redox status, and limit application of ASec for investigating the redox functions of the majority of selenoproteins. Therefore, DMNB-Sec incorporation provides a complementary GCE platform that can be utilized for the study of redox-active selenoproteins in mammalian cells.
The yield of MsrB1-DMNB-Sec95 is significantly lower than that of the Cys-containing MsrB1, necessitating further optimization of the GCE platform. Efforts are underway to improve the efficiency of DMNB-Sec incorporation through optimization of the LeuRS/tRNA pair. Upon optimization, the technology described herein could enable temporally controlled selenoprotein production in mammalian cells for further advancing our understanding of selenoprotein biology. Ultimately, the all-in-one pAcBac2 vector system used to deliver the DMNB-Sec incorporation machinery can be readily packaged into an engineered baculovirus vector that can facilitate highly efficient unnatural amino acid incorporation into a wide variety of mammalian cells and tissues, including challenging cells such as neurons and stem cells.25, 27
METHODS
See the Supporting Information for details.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by NIH grant R01GM117004 and R01GM118431-01A1 to E.W., grants R01GM124319 and R01GM126220 to A.C, and fellowship F32GM131615-01 to J.C.P.
Footnotes
The authors declare no competing financial interests.
Supporting Information
Detailed experimental procedures and supplementary figures and tables. The Supporting Information is available free of charge on the ACS Publications website at DOI:
REFERENCES
- 1.Labunskyy VM, Hatfield DL & Gladyshev VN Selenoproteins: molecular pathways and physiological roles. Physiol Rev 94, 739–777 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kryukov GV et al. Characterization of mammalian selenoproteomes. Science 300, 1439–1443 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Reich HJ & Hondal RJ Why Nature Chose Selenium. ACS Chem Biol 11, 821–841 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Ingold I et al. Selenium Utilization by GPX4 Is Required to Prevent Hydroperoxide-Induced Ferroptosis. Cell 172, 409–422 e421 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Xu XM et al. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol 5, e4 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan J et al. RNA-dependent conversion of phosphoserine forms selenocysteine in eukaryotes and archaea. Proc Natl Acad Sci U S A 103, 18923–18927 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry MJ et al. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3’ untranslated region. Nature 353, 273–276 (1991). [DOI] [PubMed] [Google Scholar]
- 8.Copeland PR, Fletcher JE, Carlson BA, Hatfield DL & Driscoll DM A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J 19, 306–314 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tujebajeva RM et al. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep 1, 158–163 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, Cheng R & Rozovsky S Synthesis and semisynthesis of selenopeptides and selenoproteins. Curr Opin Chem Biol 46, 41–47 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu X, Soll D & Sevostyanova A Challenges of site-specific selenocysteine incorporation into proteins by Escherichia coli. RNA Biol 15, 461–470 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mukai T, Sevostyanova A, Suzuki T, Fu X & Soll D A Facile Method for Producing Selenocysteine-Containing Proteins. Angew Chem Int Ed Engl 57, 7215–7219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thyer R et al. Custom selenoprotein production enabled by laboratory evolution of recoded bacterial strains. Nat Biotechnol 36, 624–631 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brocker MJ, Ho JM, Church GM, Soll D & O’Donoghue P Recoding the genetic code with selenocysteine. Angew Chem Int Ed Engl 53, 319–323 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thyer R, Filipovska A & Rackham O Engineered rRNA enhances the efficiency of selenocysteine incorporation during translation. J Am Chem Soc 135, 2–5 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Novoselov SV et al. A highly efficient form of the selenocysteine insertion sequence element in protozoan parasites and its use in mammalian cells. Proc Natl Acad Sci U S A 104, 7857–7862 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chin JW Expanding and reprogramming the genetic code. Nature 550, 53–60 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Italia JS et al. Expanding the genetic code of mammalian cells. Biochem Soc Trans 45, 555–562 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Young DD & Schultz PG Playing with the Molecules of Life. ACS Chem Biol 13, 854–870 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J et al. Site-Specific Incorporation of Selenocysteine Using an Expanded Genetic Code and Palladium-Mediated Chemical Deprotection. J Am Chem Soc 140, 8807–8816 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welegedara AP, Adams LA, Huber T, Graham B & Otting G Site-Specific Incorporation of Selenocysteine by Genetic Encoding as a Photocaged Unnatural Amino Acid. Bioconjug Chem 29, 2257–2264 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Rakauskaite R et al. Biosynthetic selenoproteins with genetically-encoded photocaged selenocysteines. Chem Commun (Camb) 51, 8245–8248 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Ellis-Davies GC Caged compounds: photorelease technology for control of cellular chemistry and physiology. Nat Methods 4, 619–628 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemke EA, Summerer D, Geierstanger BH, Brittain SM & Schultz PG Control of protein phosphorylation with a genetically encoded photocaged amino acid. Nat Chem Biol 3, 769–772 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Chatterjee A, Xiao H, Bollong M, Ai HW & Schultz PG Efficient viral delivery system for unnatural amino acid mutagenesis in mammalian cells. Proc Natl Acad Sci U S A 110, 11803–11808 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell AL, Addy PS, Chin MA & Chatterjee A A Unique Genetically Encoded FRET Pair in Mammalian Cells. Chembiochem 18, 511–514 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Zheng Y, Lewis TL Jr., Igo P, Polleux F & Chatterjee A Virus-Enabled Optimization and Delivery of the Genetic Machinery for Efficient Unnatural Amino Acid Mutagenesis in Mammalian Cells and Tissues. ACS Synth Biol 6, 13–18 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Kryukov GV, Kumar RA, Koc A, Sun Z & Gladyshev VN Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase. Proc Natl Acad Sci U S A 99, 4245–4250 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HY, Fomenko DE, Yoon YE & Gladyshev VN Catalytic advantages provided by selenocysteine in methionine-S-sulfoxide reductases. Biochemistry 45, 13697–13704 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HY, Zhang Y, Lee BC, Kim JR & Gladyshev VN The selenoproteome of Clostridium sp. OhILAs: characterization of anaerobic bacterial selenoprotein methionine sulfoxide reductase A. Proteins 74, 1008–1017 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao G et al. Methionine sulfoxide reductase B1 (MsrB1) recovers TRPM6 channel activity during oxidative stress. J Biol Chem 285, 26081–26087 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee BC et al. MsrB1 and MICALs regulate actin assembly and macrophage function via reversible stereoselective methionine oxidation. Mol Cell 51, 397–404 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bak DW, Gao J, Wang C & Weerapana E A Quantitative Chemoproteomic Platform to Monitor Selenocysteine Reactivity within a Complex Proteome. Cell Chem Biol 25, 1157–1167 e1154 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weerapana E et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abo M, Li C & Weerapana E Isotopically-Labeled Iodoacetamide-Alkyne Probes for Quantitative Cysteine-Reactivity Profiling. Mol Pharm 15, 743–749 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qian Y et al. An isotopically tagged azobenzene-based cleavable linker for quantitative proteomics. Chembiochem 14, 1410–1414 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Gao J et al. Selenium-Encoded Isotopic Signature Targeted Profiling. ACS Cent Sci 4, 960–970 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarrago L et al. Monitoring of Methionine Sulfoxide Content and Methionine Sulfoxide Reductase Activity In Selenoproteins: Methods and Protocols, Methods in Molecular Biology; Chavatte L, Ed., Humana Press: New York, 2018; vol. 1661; pp 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.