SUMMARY
LACC1 genetic variants are associated with multiple immune-mediated diseases. However, laccase domain containing-1 (LACC1) functions are incompletely defined. We find that upon stimulation of the pattern-recognition receptor (PRR) NOD2, LACC1 localizes to the endoplasmic reticulum (ER) and forms a complex with ER-stress sensors. All three ER-stress branches, PERK, IRE1α, and ATF6, are required for NOD2-induced signaling, cytokines, and antimicrobial pathways in human macrophages. LACC1, and its localization to the ER, is required for these outcomes. Relative to wild-type (WT) LACC1, transfection of the common Val254 and rare Arg284 immune-mediated disease-risk LACC1 variants into HeLa cells and macrophages, as well as macrophages from LACC1 Val254 carriers, shows reduced NOD2-induced ER stress-associated outcomes; these downstream outcomes are restored by rescuing ER stress. Therefore, we identify ER stress to be essential in PRR-induced outcomes in macrophages, define a critical role for LACC1 in these ER stress-dependent events, and elucidate how LACC1 disease-risk variants mediate these outcomes.
In Brief
Huang et al. show that upon stimulating innate receptors on human macrophages, LACC1 localizes to the endoplasmic reticulum (ER), associates with ER-stress sensors, and increases the unfolded protein response. LACC1-dependent ER stress promotes innate-receptor-induced signaling, cytokines, and antimicrobial pathways. LACC1 disease-risk variants show reduced ER stress-dependent outcomes.
Graphical abstract

INTRODUCTION
Understanding the consequences of genetic variants associated with immune-mediated diseases can often lead to unexpected elucidation of mechanisms and pathways required for maintaining immune homeostasis. LACC1 genetic variants are associated with inflammatory bowel disease (IBD) (Jostins et al., 2012), leprosy (Liu et al., 2015), and juvenile arthritis (Wakil et al., 2015), yet only a limited number of studies has examined the functional consequences of mammalian laccase domain containing-1 (LACC1) protein and its variants (Assadi et al., 2016; Cader et al., 2016; Lahiri et al., 2017; Skon-Hegg et al., 2019). We and others found that the common LACC1 Ile254Val variant (allele frequency 0.226–0.245 in European ancestry individuals per the dbSNP) results in a loss of function as manifested by decreased cytokine secretion, bacterial clearance, ROS production, and fatty-acid oxidation in macrophages (Cader et al., 2016; Lahiri et al., 2017). However, the mechanisms through which LACC1 regulates innate immunity are incompletely defined. Subcellular localization can provide important insight into mechanisms through which proteins mediate their effects, with localization to multiple subcellular compartments allowing for proteins to carry out distinct functions at these respective sites (Yogev and Pines, 2011). Previous studies showed LACC1 localized to the cytosol, mitochondria, and/or peroxisomes in human macrophages (Assadi et al., 2016; Cader et al., 2016; Lahiri et al., 2017). However, our in silico analysis showing LACC1 localization to the ER and a prior study showing that a small percent of LACC1 may also localize to the ER in THP-1 cells under homeostatic conditions (Assadi et al., 2016) set the foundation for additional LACC1 mechanisms. We therefore hypothesized that LACC1 localizes to the ER in human macrophages and regulates key ER functions contributing to pattern-recognition receptor (PRR)-initiated outcomes.
ER stress is a cellular response triggered by an accumulation of misfolded proteins in the ER. ER stress is initiated by three major branches of the unfolded protein response (UPR): IRE1α, PERK, and ATF6 (Grootjans et al., 2016). Dysregulated ER stress is associated with immune-mediated diseases, including IBD (Grootjans et al., 2016; Kaser et al., 2010). Mouse studies showed that deletion of IRE1β, which was primarily expressed in the gastrointestinal epithelia (Bertolotti et al., 2001), or deletion of XBP1 in intestinal epithelial cells (IECs) (Kaser et al., 2008) results in increased susceptibility to intestinal inflammation. Consistently, XBP1 loss-of-function polymorphisms are associated with IBD risk (Kaser et al., 2008). The role of ER stress in IBD pathogenesis has mostly been examined in the context of epithelial cells (Adolph et al., 2013); its role is less well understood in other cell types, including macrophages. A role for ER stress has been implicated in PRR-initiated responses in macrophages; however, the mechanisms and outcomes of ER stress in these cells are incompletely understood. For example, following recognition of microbial products by PRRs on macrophages, how the specific branches of the UPR contribute to PRR-induced outcomes in macrophages is not well defined. IRE1α has generally been found to be activated upon PRR stimulation and to contribute to PRR-induced cytokines in mouse macrophages (Bronner et al., 2015; Martinon et al., 2010). In contrast, the contribution of ATF6 to this outcome has been seen in some (Rao et al., 2014) but not other (Bronner et al., 2015; Martinon et al., 2010) studies. We therefore sought to address the potential roles and mechanisms through which LACC1 contributes to ER stress. We further sought to define how ER stress regulates distinct outcomes in human macrophages, and how LACC1 and immune disease-risk variants in LACC1 might modulate these macrophage outcomes.
In this study using primary human monocyte-derived macrophages (MDMs), we found that NOD2 stimulation induced ER stress with activation of the PERK, IRE1α, and ATF6 pathways. This ER-stress induction required LACC1, which localized to the ER and associated with each of the ER-stress sensors. LACC1 ER localization and the three ER-stress branches were critical for the subsequent PRR-induced signaling, cytokine secretion, and bacterial clearance pathways, including NAPDH oxidase subunit and ROS induction, NOS2 induction, and autophagy pathway induction. Furthermore, compared to wild-type (WT) LACC1, the common LACC1 Val254 and the rare LACC1 Arg284 immune disease-risk variants transfected into MDMs showed reduced NOD2-induced ER stress, signaling, cytokine secretion, and bacterial clearance pathways. Importantly, primary human macrophages from the individuals carrying the LACC1 Val254 risk allele showed decreased PRR-induced outcomes, and restoration of ER stress to the levels observed in MDMs from non-risk LACC1 carriers restored these outcomes. Taken together, we define a critical role for ER stress in PRR-induced signaling, cytokines, and bacterial clearance in human macrophages, and uncover an important role for and mechanisms through which the immune-disease-associated LACC1 gene and its variants modulate these responses.
RESULTS
LACC1 Localizes to the ER
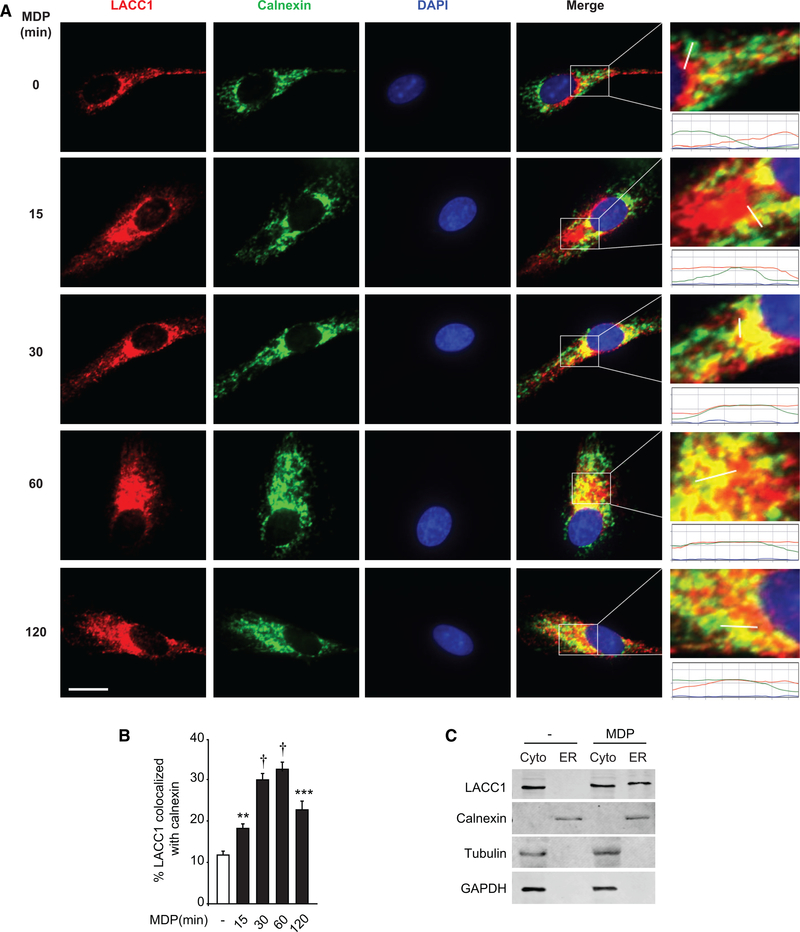
While LACC1 is associated with multiple immune-mediated diseases, only a few recent studies have examined its function in mammalian cells. LACC1 has been shown to colocalize with the mitochondria, cytosol, and peroxisomes (Assadi et al., 2016; Cader et al., 2016; Lahiri et al., 2017). However, our in silico analysis by PSORTII indicated that LACC1 might also localize to the ER, and a THP-1 human macrophage cell line (unstimulated) study showed a low percentage of LACC1 localization to the ER (Assadi et al., 2016). Such localization might provide insight into potentially important undefined LACC1 functions. We therefore asked if LACC1 localized to the ER in primary human MDMs. Using calnexin as an ER marker, a low level of LACC1 localization to the ER was observed at baseline in MDMs through a microscopic approach (Figures 1A and 1B). As LACC1 regulates PRR-induced outcomes (Lahiri et al., 2017), we asked if PRR stimulation regulates LACC1 localization to the ER. We stimulated NOD2, a PRR associated with Crohn’s disease whose ligand is the minimal component of bacterial peptidoglycan, muramyl dipeptide (MDP). LACC1 ER localization increased following NOD2 stimulation with peak colocalization occurring at 30–60 min post-treatment (Figures 1A and 1B). We confirmed specificity of the antibody through a knockdown approach as assessed by both flow cytometry and western blot (Figures S1A and S1B). We further confirmed LACC1 ER localization following NOD2 stimulation through an independent biochemistry approach examining isolated ER fractions (Figure 1C). Therefore, LACC1 partially localizes to the ER following NOD2 stimulation.
Figure 1. NOD2 Stimulation Induces LACC1 Localization to the ER in MDMs.
MDMs were treated with 100 μg/mL MDP for the indicated times. Cells were immunostained for LACC1 (red), calnexin (green), and nucleus (DAPI, blue).
(A) Representative micrographs with scale bar representing 10 μm. Enlarged images with graph of line intensities show pixel densities (y axis) along the indicated line (x axis).
(B) Summary graph indicating percent LACC1 colocalized with calnexin + SEM (25 cells quantified). Representative of three independent experiments.
(C) MDMs were treated with 100 μg/mL MDP for 30 min. ER and cytosolic fractions (cyto) were assessed for LACC1 localization by western blot. Markers for ER (calnexin) and cytosolic (tubulin, GAPDH) fractions are shown. Representative of two independent experiments. **p < 0.01; ***p < 0.001; and †p < 1 × 10—4.
LACC1 Is Required for NOD2-Induced ER Stress in Human MDMs
Given the NOD2-induced LACC1 localization to the ER, we considered ER-associated mechanisms through which LACC1 might be contributing to PRR-induced signaling and cytokine secretion. In particular, as select prior reports found that the ER-stress sensor IRE1α can contribute to PRR-induced cytokines in mouse macrophages (Bronner et al., 2015; Martinon et al., 2010) and physiological responses requiring high levels of protein secretion can activate the UPR (Rutkowski and Hegde, 2010), we considered that LACC1 might be regulating the UPR, which in turn, may regulate PRR-induced outcomes. We will utilize the terminology ER stress in the studies that follow. We first established that in human MDMs MDP treatment activated both the PERK and IRE1α pathways as early as 15 min, as measured by the phosphorylation of PERK and IRE1α, with peak activation at 30 min (Figures 2A and S1C). This was confirmed independently by western blot (Figure 2B). We further measured transcripts of spliced XBP1, occurring downstream of the IRE1α pathway, and CHOP, ERdj4, and BiP, additional ER-stress induced transcripts (Eizirik et al., 2013; Grootjans et al., 2016). Each transcript was maximally induced 4 h after MDP treatment (Figure 2C). We next assessed if LACC1 is required for NOD2-induced ER stress. We silenced LACC1 using a knockdown approach and observed decreased mRNA (Figure S1D) and protein levels by both flow cytometry (Figure S1E) and western blot (Figure S1F). NOD2-induced PERK and IRE1α phosphorylation was significantly diminished following LACC1 knockdown (Figure 2D). Consistently, NOD2-induced ER-stress transcripts were also reduced following LACC1 knockdown in MDMs (Figure 2E). Taken together, LACC1 is required for NOD2-induced ER stress.
Figure 2. ER Stress Pathways Are Required for NOD2-Induced Outcomes and LACC1 Is Required for NOD2-Induced ER Stress.
(A–C) Human MDMs were treated with 100 μg/mL MDP: (A) phospho-PERK and phospho-IRE1α at the indicated times was assessed by flow cytometry with a summary graph of fold change mean fluorescence intensity (MFI) (n = 6); (B) phospho-PERK and phospho-IRE1α at 30 min by western blot; GAPDH and total protein expression served as loading controls; and (C) mRNA expression at the indicated times (n = 4).
(D–J) MDMs were transfected with scrambled or the indicated small interfering RNA (siRNA) and then treated with 100 μg/mL MDP: (D) phospho-PERK and phospho-IRE1α at 30 min with representative histograms showing MFI values and a summary graph with fold MFI change (n = 6) and (E) mRNA expression at 4h (n = 8; similar results in an additional n = 8). (F and G) Fold phospho-proteins at 15 min (n = 6), (H) fold phospho-PERK and phospho-IRE1α at 30 min (n = 6), (I) mRNA expression at 4 h (n = 8; similar results in an additional n = 8), or (J) cytokines at 24 h (n = 4; similar results in an additional n = 4). Significance is compared to (A and C) untreated cells and (D–J) scrambled siRNA-transfected, MDP-treated cells. Comb, combined PERK, IRE1α, and ATF6 siRNA; scr, scrambled; and tx, treatment. Mean + SEM. *p < 0.05; **p < 0.01; ***p < 0.001; †p < 1 × 10—4; and ††p <1 × 10—5.
NOD2 Is Required for MDP-Induced ER Stress
A previous study found that chemical inducers activating the ER stress response in mouse BMDMs did so in a NOD1-and NOD2-dependent manner, but independent of the ligands to these receptors (e.g., C12-iE-DAP and MDP, respectively) (Keestra-Gounder et al., 2016). We therefore assessed if MDP-induced ER stress was NOD2-dependent. Following effective NOD2 knockdown (Figure S1G), MDP-induced PERK and IRE1α activation (Figure S1H), and ER-stress response transcript upregulation (Figure S1I) were dramatically reduced. As a control, we ensured that MDP-induced cytokine secretion was reduced following NOD2 knockdown (Figure S1J). We also established that TriDAP (ligand for NOD1) treatment induced ER-stress pathways and cytokines, and that this was dependent on NOD1 (Figures S1G and S1K–S1M). Moreover, combined NOD1 and NOD2 knockdown resulted in reduced outcomes following either MDP or TriDAP treatment (Figures S1H–S1M). Finally, Keestra-Gounder et al. (2016) found that NOD2 was required for the ER stress induced by the chemical inducers thapsigargin and tunicamycin in mouse BMDMs, but a study in epithelial cell lines found that tunicamycin-induced outcomes were independent of NOD2 (Lopes et al., 2018). We found that thapsigargin-induced ER stress was, in fact, not dependent on NOD2 or NOD1 in human MDMs (Figures S1N–S1P). We ensured that thapsigargin did not reduce cell viability (Figure S1Q). Taken together, MDP treatment induces NOD2-dependent ER stress in human MDMs.
Cytokine Autocrine/Paracrine Loops Regulate NOD2-Induced ER Stress
We next sought to address mechanisms through which LACC1-dependent ER stress was contributing to PRR-induced outcomes. NOD2 stimulation activates MAPK and nuclear factor κB (NF-κB) signaling, which, in turn, is required for multiple downstream outcomes, including cytokines (Hedl and Abraham, 2012; Pauleau and Murray, 2003). To first assess if ER stress regulates NOD2-induced signaling, we knocked down each ER-stress branch and confirmed that knockdown of each resulted in a selective decrease of the targeted transcript (Figure S2A) and protein (Figures S2B and S2C). Cell survival was intact in both untreated and MDP-treated cells upon ER-stress molecule knockdown (Figure S2D). Consistent with the early time at which ER stress is observed (Figure 2A), all three branches of ER stress were required for NOD2-induced MAPK and NF-κB signaling (Figures 2F and 2G). We then sought to confirm that knockdown of each ER-stress branch affected only the activation of the targeted branch. Interestingly, however, NOD2-induced PERK activation was diminished not only following PERK knockdown in MDMs, but also following knockdown of IRE1α and ATF6 (Figure 2H). Similarly, NOD2-induced activation of IRE1α (Figure 2H) and of the ER stress response transcripts (Figure 2I) was dependent on each of the three ER-stress sensor branches. Given the important role of cytokine autocrine/paracrine loops for NOD2-dependent outcomes in MDMs (Hedl and Abraham, 2011a,b, 2014), and that cytokines can induce ER stress (Grootjans et al., 2016), we hypothesized that cytokine autocrine loops might be activating all three branches of ER stress and, therefore, accounting for the cross-regulation observed between the ER-stress pathways in Figures 2H and 2I. We first examined how ER stress affects cytokine secretion following NOD2 stimulation. PERK, IRE1α, and ATF6 were each required for NOD2-induced cytokines (Figure 2J). We then treated MDMs with IL1β, TNF, or IL6 and found that they each activated PERK and IRE1α, and induced ER-stress response transcripts (Figures S2E and S2F). Importantly, blocking each of these autocrine/paracrine loops prior to NOD2 stimulation reduced NOD2-induced ER stress; the combined blockade led to an even greater reduction (Figures S2G and S2H). Taken together, each of the ER-stress branches is required for NOD2-induced signaling and cytokines, and NOD2-induced autocrine/paracrine cytokines contribute to activation of each of the three ER-stress branches.
NOD2-Induced Signaling and Cytokines in LACC1-Deficient Cells Can Be Rescued with Restoration of ER Stress
To define the degree to which LACC1-dependent ER stress contributes to NOD2-induced signaling and cytokines, we asked if restoring ER stress in the absence of LACC1 could rescue NOD2-induced outcomes. To this purpose, we used cyclopiazonic acid (CPA), an inducer of ER stress with low toxicity (Fumagalli et al., 2016), which did not affect MDM viability (Figure S3A). We confirmed that CPA restored NOD2-induced ER stress in LACC1-deficient, MDP-treated MDMs as assessed by PERK and IRE1α activation, as well as ER stress response transcript induction (Figures 3A and 3B). Importantly, CPA treatment restored MAPK and NF-κB signaling and cytokine secretion in NOD2-stimulated, LACC1-deficient MDMs (Figures 3C–3E). We confirmed that complementation of ER stress in LACC1-deficient MDMs could restore NOD2-induced cytokines using two additional ER-stress inducers: tunicamycin (Figures S3B and S3C) and thapsigargin (Figure S3D). Taken together, the decreased NOD2-induced MAPK and NF-κB signaling and cytokines in LACC1-deficient cells can be restored with complementation of ER stress.
Figure 3. Complementing NOD2-Induced ER Stress Restores Signaling and Cytokines in LACC1-Deficient MDMs.
MDMs were transfected with scrambled or LACC1 siRNAs. Cells were then treated with 100 μg/mL MDP ± 10 μM cyclopiazonic acid (CPA): (A) fold phospho-PERK and phospho-IRE1α at 30 min (n = 6), (B) mRNA expression at 6 h (n = 4; similar results in an additional n = 4), (C and D) fold phospho-protein at 15 min (n = 6), or (E) cytokines at 12 h (n = 4; similar results in an additional n = 4). Mean + SEM. Scr, scrambled; Tx, treatment. **p < 0.01; ***p < 0.001; †p< 1 × 10—4; and ††p< 1 × 10—5.
NOD2-Induced MAPK and NF-κB Pathways Promote ER Stress Induction
We next examined mechanisms wherein LACC1 regulates ER stress. We hypothesized that one such mechanism may involve proximal LACC1 signaling in the cytoplasm, whereas another may require direct LACC1 localization to the ER. Regarding the first hypothesis, we previously found that following NOD2 stimulation of human MDMs LACC1 associates with the NOD2 signaling complex (containing RIP2, IRAK1, TRAF6, phospho-ERK, phospho-p38, and phospho-IκBα) and contributes to its optimal assembly (Lahiri et al., 2017). A mouse macrophage study found that MAPK and NF-κB pathways were not required for PRR-induced XBP1 splicing (Martinon et al., 2010), yet ERK, p38, and JNK have been shown to activate different branches of ER stress in several cell types (Darling and Cook, 2014). We therefore assessed if RIP2, TRAF6, and MAPK and NF-κB pathways were required for NOD2-induced ER stress and ER-stress-related transcripts in human MDMs and found this to be the case (Figures S3E, S3F, S3H, and S3I). We further confirmed their role in NOD2-induced cytokine secretion (Figures S3G and S3J). Taken together, the proximal NOD2 signaling complex and downstream MAPK and NF-κB signaling pathways are required for NOD2-induced ER stress, and ER stress, in turn, amplifies NOD2-initiated signaling.
LACC1 Localization to the ER Is Required for LACC1 Complex Assembly, ER Stress, Signaling, and Cytokines following NOD2 Stimulation
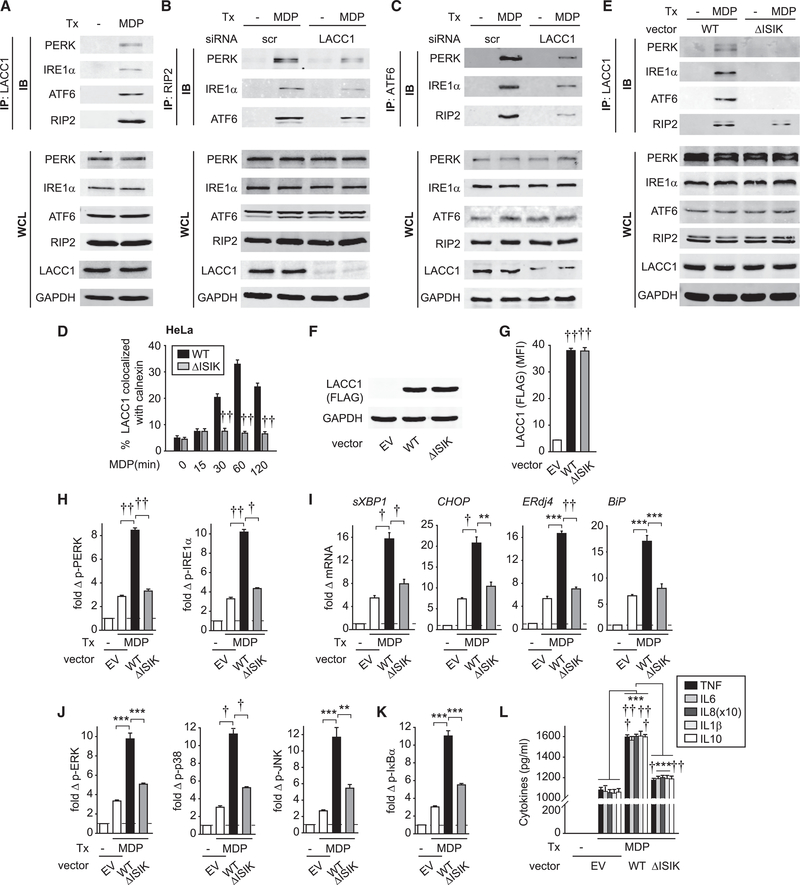
Given NOD2-induced LACC1 localization to the ER, we next asked if LACC1 could form a complex with ER-stress sensors. RIP2, a NOD2 signaling intermediate, was recently reported to localize to the ER upon NOD2 stimulation in a HCT116 colon cancer cell line (Bist et al., 2017). We now also find that RIP2 localizes to the ER in NOD2-stimulated primary human MDMs through both microscopic (Figures S4A and S4B) and cell fractionation (Figure S4C) approaches. We confirmed specificity of the RIP2 antibody through RIP2 knockdown (Figure S4D). As we had previously found that LACC1 associated with RIP2 (Lahiri et al., 2017), this raised the possibility that upon NOD2 stimulation, LACC1 assembles in a complex with RIP2 in the ER along with ER-stress molecules. LACC1 indeed associated with PERK, IRE1α, and ATF6 following NOD2 stimulation (Figure 4A). As expected, LACC1 associated with RIP2 upon NOD2 stimulation (Figure 4A). Using a reciprocal approach through immuno-precipitating RIP2, we established that RIP2 assembled in a complex with PERK, IRE1α, and ATF6 (Figure 4B). Importantly, LACC1 was essential for this assembly, as LACC1 knockdown reduced the NOD2-induced assembly of RIP2 with PERK, IRE1α, and ATF6 (Figure 4B). We confirmed the role of LACC1 for NOD2-induced assembly of ER-stress proteins with RIP2 using an alternative immunoprecipitation approach with ATF6 (Figure 4C). As we previously found that LACC1 did not regulate Dectin-1-induced outcomes (Lahiri et al., 2017), we examined the Dectin-1 pathway as a negative control. Upon either NOD2 or Dectin-1 stimulation, LACC1 did not associate with either Dectin-1 or Syk, whereas Dectin-1 and Syk associated with each other upon Dectin-1 stimulation (Figure S4E).
Figure 4. NOD2-Induced LACC1 Localization to the ER Is Required for Subsequent ER-Stress Molecule Complex Assembly, ER Stress, and Downstream Outcomes.
(A) MDMs were treated with 100 μg/mL MDP for 30 min. LACC1 was immunoprecipitated (IP) and the recruitment of the indicated proteins was assessed by western blot (immunoblot [IB]).
(B and C) MDMs were transfected with scrambled or LACC1 siRNAs, and then treated with 100 μg/mL MDP for 30 min. (B) RIP2 or (C) ATF6 were IP and the recruitment of the indicated proteins assessed by western blot.
(D and E) HeLa cells were transfected with empty vector (EV), WT LACC1 or LACC1-ΔISIK vectors, along with NOD2. (D) Cells were treated with 100 μg/mL MDP for the indicated times and then immunostained for LACC1 and calnexin. Summary graph of percent LACC1 colocalized with calnexin + SEM (25 cells quantified). Data represent one of three independent experiments. (E) Cells were treated with 100 μg/mL MDP for 30 min. LACC1 was IP and the recruitment of the indicated proteins assessed by western blot. (A–C and E) Equivalent expression for the respective proteins is shown in whole-cell lysates (WCLs). GAPDH was used as a loading control.
(F–L) MDMs were transfected with empty vector (EV) or FLAG-LACC1 vectors.
(F) Western blot.
(G) Summary graph of LACC1 MFI as detected by flow cytometry using an anti-FLAG antibody to detect the transfected LACC1 variants (n = 6).
(H–L) Cells were treated with 100 μg/mL MDP: (H) fold phospho-PERK and phospho-IRE1α at 30 min (n = 6, similar results in an additional n = 10), (I) mRNA expression at 4 h (n = 6), (J and K) fold phospho-protein at 15 min (n = 6), or (L) cytokines at 24 h (n = 6). Mean + SEM. Tx, treatment. **p < 0.01; ***p < 0.001; †p< 1 × 10—4; and ††p < 1 × 10—5.
We next asked if the localization of LACC1 to the ER was required for NOD2-induced ER stress and downstream out-comes. Using in silico analysis, we found that LACC1 contained a predicted C-terminal ER-retention motif, ISIK(E). We deleted this motif (Figure S5A), and overexpressed WT LACC1 and the ISIK LACC1 deletion mutant (LACC1-ΔISIK) in HeLa cells. We ensured equivalent expression (Figure 4E). LACC1-ΔISIK failed to localize to the ER upon NOD2 stimulation (Figures S5B and 4D). Consistent with this, LACC1-ΔISIK failed to associate with PERK, IRE1α, and ATF6 following NOD2 stimulation, but still maintained some association with RIP2 (Figure 4E), likely due to complex formation with cytoplasmic RIP2. To assess if LACC1 ER localization was required for the NOD2-induced, ER stress-associated outcomes we had identified in MDMs, we transfected LACC1-ΔISIK into MDMs and ensured expression levels equivalent to that of transfected WT LACC1 (Figures 4F and 4G). Importantly, NOD2-induced ER stress (Figure 4H), ER-stress related transcripts (Figure 4I), signaling (Figures 4J and 4K), and cytokine secretion (Figure 4L) were all decreased in LACC1-ΔISIK compared to WT LACC1-transfected MDMs. Taken together, upon NOD2 stimulation, LACC1 forms a complex with the ER-stress sensors PERK, IRE1α, and ATF6, and LACC1 ER localization is required for the optimal formation of this complex and subsequent NOD2-induced ER stress and ER stress-associated outcomes.
The LACC1 Disease-Risk Variants Val254 and Arg284 Show Decreased NOD2-Induced ER Stress and Downstream Outcomes
We and others found that the Crohn’s disease-and leprosy-associated LACC1 Ile254Val variant results in decreased PRR-induced signaling, cytokines, and bacterial clearance (Cader et al., 2016; Lahiri et al., 2017). Furthermore, a rare variant in LACC1, Cys284Arg, associated with early-onset Crohn’s disease and juvenile arthritis (Patel et al., 2014; Wakil et al., 2015), showed reduced ROS production (Cader et al., 2016). We therefore examined these two variants in the context of NOD2-induced ER stress. We constructed plasmids expressing LACC1 Val254 or Arg284 (variant location as per model in Figure S5A) and transfected them into HeLa cells. NOD2-induced localization of the LACC1 Val254 risk variant to the ER was intact (Figures 5A and S6A), as was its assembly with ER-stress sensors (Figure 5B). However, LACC1 Arg284 showed a decrease in each of these NOD2-induced outcomes (Figures 5A, 5B, and S6A). Importantly, compared to WT LACC1, expression of LACC1 Val254 and particularly LACC1 Arg284 in human MDMs at equivalent levels (Figures 5C and 5D) showed reduced NOD2-induced ER stress (Figure 5E), ER stress-related transcripts (Figure 5F), signaling (Figures 5G and 5H), and cytokine secretion (Figure 5I). Taken together, the common LACC1 Val254 and the rare LACC1 Arg284 disease-associated risk variants demonstrate reduced NOD2-induced ER stress and ER-stress associated outcomes.
Figure 5. LACC1 Val254 and Arg284 Disease-Risk Variants Show Decreased NOD2-Induced ER Stress and Downstream Outcomes.
(A and B) HeLa cells were transfected with empty vector (EV) or WT LACC1, LACC1 Val254, or LACC1 Arg284 vectors and treated with 100 μg/mL MDP for 30 min.
(A) Cells were immunostained for LACC1 and calnexin. Summary graph indicating percent of LACC1 colocalized with calnexin (25 cells quantified). Data represent one of three independent experiments.
(B) (Left) LACC1 was IP and the recruitment of the indicated proteins assessed by western blot (immunoblot [IB]). (Right) Equivalent expression for the respective proteins is shown in whole-cell lysates (WCLs). GAPDH was used as a loading control.
(C–I) MDMs were transfected with empty vector (EV) or FLAG-LACC1 vectors. (C) Western blot and (D) summary graph of flow cytometry using anti-FLAG antibody (MFI) to detect the transfected LACC1 variants (n = 6). Cells were treated with 100 μg/mL MDP: (E) fold phospho-PERK and phospho-IRE1α at 30 min (n = 6, similar results in an additional n = 6), (F) mRNA expression at 4 h (n = 5, similar results in an additional n = 4), (G and H) fold phospho-protein at 15 min (n = 6), or (I) cytokines at 24 h (n = 6). Mean + SEM. NT, no treatment; Tx, treatment. **p < 0.01; ***p < 0.001; †p < 1 × 10—4; and ††p < 1 × 10—5.
LACC1-Dependent ER Stress Is Required for Optimal NOD2-Induced Bacterial Clearance and Bacterial Clearance Mechanisms
Outcomes examined downstream of ER stress have generally focused on protein folding, cytokine secretion, and apoptosis (Grootjans et al., 2016); less is known about how PRR-induced ER stress regulates bacterial clearance, particularly in human MDMs. Impaired microbial clearance in intestinal tissues can increase susceptibility to IBD (Dhillon et al., 2014; Holland, 2010; Muise et al., 2012). The LACC1 Val254 disease-risk variant shows decreased bacterial clearance in human (Lahiri et al., 2017) and mouse (Cader et al., 2016) macrophages. Chronic NOD2 stimulation enhances macrophage-mediated bacterial clearance (Lahiri and Abraham, 2014) and mimics intestinal conditions of ongoing exposure to bacterial products, including to the NOD2 ligand MDP, which is present in the intestinal lamina propria (Feng et al., 2007; Zheng and Abraham, 2013). We therefore focused on chronic NOD2 stimulation for these studies. As the initial step in clearing intracellular bacteria requires bacterial uptake, we first assessed the role of LACC1 in bacterial uptake. Upon stimulating MDMs through NOD2 for 48 h, uptake of Salmonella enterica serovar Typhimurium-GFP dramatically increased (Figures 6A and S6B). The increased bacterial uptake was impaired with LACC1 knockdown (Figure 6A). Optimal bacterial uptake required intact ER-stress sensor pathways (Figure S6C). Importantly, restoring ER stress by CPA in chronic NOD2-stimulated LACC1-deficient MDMs restored bacterial uptake (Figures 6A and S6B). We observed similar results with fluorophore-labeled E. coli bioparticles (Figures 6A and S6B).
Figure 6. LACC1-Mediated ER Stress Is Required for NOD2-Induced Bacterial Clearance and Bacterial Clearance Mechanisms.
(A) MDMs (n = 4; similar results in an additional n = 12) were transfected with scrambled or LACC1 siRNA, then treated with 100 μg/mL MDP for 48 h ± 10 μM CPA, and then co-cultured with S. Typhimurium-GFP or E. coli bioparticles-FITC. Bacterial uptake at 20 min (mean fluorescence intensity [MFI]).
(B) MDMs (n = 6) were transfected with scrambled or the indicated siRNAs alone or in combination (comb.). Cells were co-cultured with E. faecalis, AIEC or S. aureus and assessed for intracellular bacterial clearance. Colony forming units (CFUs). Significance is compared to MDP-treated, scrambled siRNA-transfected cells or as indicated.
(C–G) MDMs were transfected with scrambled or LACC1 siRNAs, and then treated with 100 μg/mL MDP for 48 h ± 10 μM CPA.
(C) Cells were then co-cultured with E. faecalis. CFU (n = 4).
(D–G) Cells were assessed for expression of the indicated measures by flow cytometry (MFI) (n = 6).
(H–Q) MDMs (n = 6) were transfected with empty vector (EV) or the indicated FLAG-LACC1 vectors. Cells were then treated with 100 μg/mL MDP for 48 h and assessed for (H and M) bacterial clearance or (I–L) and (N–Q) expression of the indicated measures by flow cytometry. Mean + SEM. Scr, scrambled; Tx, treatment. *p < 0.05; **p < 0.01; ***p < 0.001; †p < 1 × 10—4; and ††p< 1 × 10—5.
To define the contribution of ER-stress pathways to the clearance of bacteria that are successfully taken up by MDMs, we reduced PERK, ATF6, or IRE1α expression. Upon chronic NOD2-stimulation, each of the PERK, ATF6, and IRE1α pathways alone and somewhat more in combination, were required for the optimal clearance of the intestinal resident bacteria, Enterococcus faecalis; adherent-invasive Escherichia coli (AIEC), which colonizes the ilea of Crohn’s disease patients with increased frequency (Darfeuille-Michaud et al., 2004); and of Staphylococcus aureus (Figure 6B). We confirmed that LACC1 was required for NOD2-enhanced bacterial clearance (Figure 6C). Importantly, restoring ER stress by CPA in chronic NOD2-stimulated LACC1-deficient MDMs restored bacterial clearance (Figure 6C). Therefore, LACC1-dependent ER stress contributes to NOD2-induced bacterial clearance. As such, despite the reduced levels of bacteria due to reduced bacterial uptake in LACC1-deficient MDMs, these MDMs ultimately have higher levels of bacteria. To further assess the LACC1 regulation of intracellular bacteria, we conducted a kinetic study. Although NOD2-stimulated, LACC1-deficient MDMs begin with lower levels of bacteria and a less steep growth line, these cells demonstrated a slower onset of antimicrobial mechanisms and ultimately a less effective reduction in intracellular bacteria (Figure S6D).
We next assessed the contribution of ER stress to select LACC1-dependent anti-microbial pathways mediating the enhanced bacterial clearance in chronic NOD2-stimulated macrophages. Mitochondrial ROS and cellular ROS induction contribute to bacterial clearance (Shiloh et al., 1999). We and others previously found that LACC1 is required for PRR-induced mtROS and ROS (Cader et al., 2016; Lahiri et al., 2017). Importantly, NOD2-induced mtROS and ROS were dependent on each of the ER-stress branches (Figure S6E), and restoration of ER stress in LACC1-deficient MDMs with CPA rescued NOD2-induced mtROS and ROS (Figure 6D). The NADPH oxidase complex contributes to ROS induction, such that we next examined if LACC1 was required for NOD2-induced NADPH oxidase complex expression. Protein expressions of p40phox, p47phox, and p67phox were each induced by NOD2 in a LACC1-dependent manner (Figures 6E and S6F). Complementing ER stress through CPA restored NOD2-induced NADPH oxidase subunit expression in LACC1-deficient MDMs (Figure 6E). Reactive nitrogen species (RNS) and autophagy pathways can also contribute to bacterial clearance (Abraham and Medzhitov, 2011; Shiloh et al., 1999). We established that LACC1 was required for NOD2 induction of the RNS-producing enzyme NOS2 and autophagy pathways as measured by LC3II and ATG5 (Figures 6F, 6G, and S6F). Furthermore, NOS2 and autophagy induction were dependent on ER stress (Figures S6G and S6H), and restoration of ER stress with CPA rescued NOD2-induced NOS2, LC3II, and ATG5 in LACC1-deficient MDMs (Figures 6F and 6G). These LACC1-dependent pathwayswere regulated at the transcriptional level (Figure S6I). Taken together, NOD2-induced bacterial clearance and bacterial clearance mechanisms, which include reactive oxygen and nitrogen species and autophagy pathway induction, are dependent on LACC1-mediated ER stress.
LACC1 ER Localization Is Required for Optimal NOD2-Induced Bacterial Clearance Pathways and LACC1 Disease-Risk Variants Show a Decrease in Bacterial Clearance Mechanisms
We next assessed if LACC1 ER localization was required for the NOD2-induced, LACC1-dependent bacterial clearance mechanisms we had identified. We first transfected WT LACC1 into MDMs and found that this enhanced NOD2-induced bacterial clearance (Figure 6H) and each of the bacterial clearance mechanisms we had defined to be LACC1-dependent (Figures 6I–6L), thereby demonstrating LACC1 contributions through a complementary approach to the knockdown studies above. In contrast, transfection of LACC1-ΔISIK (which fails to localize to the ER and induce ER stress; Figures 4D, 4H, and 4I) in MDMs failed to enhance NOD2-induced bacterial clearance (Figure 6H) and bacterial clearance mechanisms (Figures 6I–6L).
We then assessed how the LACC1 disease-associated variants regulate the NOD2-induced, LACC1-dependent, ER stress-mediated antimicrobial pathways we had defined. Relative to expression of WT LACC1 in human MDMs, expression of LACC1 Val254 showed only a partial increase in NOD2-induced bacterial clearance (Figure 6M) and bacterial clearance mechanisms (Figures 6N–6Q). Expression of the rare LACC1 Arg284 disease-associated variant failed to enhance these outcomes (Figures 6M–6Q). Taken together, LACC1 localization to the ER and LACC1-dependent ER stress contributes to NOD2-induced bacterial clearance and bacterial clearance mechanisms, and the LACC1 disease-associated variants show a decrease in these NOD2-induced outcomes.
PRR-Stimulated MDMs from Immune-Mediated Disease-Risk LACC1 Val254 Carriers Show Decreased ER Stress-Dependent Outcomes Relative to Ile254 Carrier Cells, and This Can Be Rescued by Complementing ER Stress
We next assessed if MDMs from individuals carrying the immune-disease risk LACC1 Val254 genotype demonstrate reduced PRR-induced, ER stress-dependent outcomes relative to Ile254 carriers. Consistent with the results examining transfected LACC1 Val254 (Figures 5E and 5F), MDMs from Val254 homozygotes showed less NOD2-induced ER stress and ER-stress-associated transcripts relative to Ile254 homozygotes, with heterozygotes showing an intermediate phenotype (Figures 7A and 7B). We also confirmed that Val254 homozygote cells showed less NOD2-induced MAPK and NF-κB activation (Figure S7A). Furthermore, we confirmed these cells showed less NOD2-induced bacterial clearance (Figure 7G) and less mtROS and ROS (Figure S7B). Importantly, MDMs from LACC1 Val254 homozygote risk carriers demonstrated lower levels of NOD2-induced NADPH oxidase subunit (Figure S7C), NOS2, LC3II, and ATG5 upregulation compared to LACC1 Ile254 carrier MDMs (Figures S7D and S7E). Complementing ER stress by CPA in the immune-disease risk LACC1 Val254 homozygote MDMs restored NOD2-induced ER-stress, signaling, cytokine secretion, bacterial clearance, and bacterial clearance pathways to the levels seen in Ile254 carrier MDMs (Figures 7C–7F, 7H–7L). As myeloid cells in intestinal tissues are exposed to multiple PRR ligands, we confirmed that CPA-mediated ER-stress complementation restored cytokine secretion upon stimulation of additional PRRs (Figure 7F). Taken together, MDMs from LACC1 Val254 immune-disease risk carriers show decreased PRR-induced outcomes relative to Ile254 carrier MDMs and restoring ER stress in these cells can rescue these PRR-induced outcomes.
Figure 7. MDMs from LACC1 Val254 Risk Carriers Demonstrate Reduced NOD2-Induced ER Stress and Downstream Outcomes.
(A and B) MDMs from LACC1 Ile254, Ile/Val254, and Val254 carriers (rs3764147 AA, GA, and GG carriers, respectively) (n = 15/genotype) were left untreated or treated with 100 μg/mL MDP: (A) fold phospho-PERK and phospho-IRE1α at 30 min and (B) mRNA expression at 4 h.
(G) Following 48 h cells were cultured with E. faecalis (CFU).
(C–F and H–L) MDMs from LACC1 Ile254 and Val254 carriers (n = 15/genotype) were left untreated or treated with (C–E and H–L) 100 μg/mL MDP ± 10 μg/mL CPA or (F) 10 μg/mL MDP, 1 μg/mL Pam3Cys, or 0.01 μg/mL lipid A ± 10 μg/mL CPA. (C) Fold phospho-PERK and phospho-IRE1α at 30 min, (D) mRNA expression at 4 h, (E) fold phospho-protein at 15 min, (F) TNF secretion at 24 h, (H) following 48 h intracellular clearance of E. faecalis was assessed (CFU), and (I–L) MFI of mtROS, ROS, or the indicated proteins at 48 h. Mean + SEM. NS, not significant; Tx, treatment. *p < 0.05; **p < 0.01; ***p < 0.001; †p< 1 × 10—4; and ††p< 1 × 10—5.
DISCUSSION
Despite the association of LACC1 genetic variants with IBD, leprosy, and juvenile arthritis, the functions of this protein in mammals have only recently emerged in limited studies and are still incompletely understood. In this primary human MDM study, we found that LACC1 localized to the ER with NOD2 stimulation, associated with ER-stress sensors, and was required for NOD2-induced ER stress, which then mediated a broad range of immune outcomes, including MAPK and NF-κB signaling, and cytokine secretion, as well as bacterial clearance through bacterial uptake, ROS, RNS, and autophagy pathway induction. LACC1 ER localization was required for these outcomes. These NOD2-mediated outcomes required each of the three ER-stress sensor PERK, IRE1α, and ATF6 pathways, and signaling through the ER-stress pathways was in part dependent on upstream MAPK and NF-κB signaling and the early secretion of cytokines, which functioned in an autocrine/paracrine manner. In MDMs transfected with LACC1 Val254 or Arg284 disease-risk variants, NOD2-induced ER stress and each of the ER-stress-dependent outcomes was decreased relative to WT LACC1 transfection. Furthermore, in MDMs from LACC1 Val254 risk carriers, decreased outcomes were restored by ER-stress induction, indicating that LACC1-dependent ER stress is a mechanism regulating the decreased outcomes in cells from these risk individuals. Taken together, our findings define an ER-stress requirement for a broad range of PRR-induced outcomes in human macrophages, including signaling, cytokine secretion, and bacterial clearance. They further identify a critical role for LACC1 in these ER stress-associated outcomes, and for modulation of these outcomes by LACC1 IBD risk variants (Figure S7F).
While dysregulated ER stress has been increasingly implicated in a variety of immune-mediated diseases, such as IBD, studies examining ER stress in IBD have focused predominantly on epithelial cells. The importance of ER-stress outcomes in myeloid cell functions and how PRR stimulation induces specific branches of the UPR has been incompletely understood. Many studies examining ER-stress mechanisms and downstream outcomes utilize chemical ER-stress inducers such as tunicamycin or thapsigargin. One limitation of this approach is the sustained levels of ER stress observed with these chemical inducers. We find that physiological microbial-ligand-induced ER stress is transient and, therefore, can result in distinct cellular outcomes. This is consistent with the roles that UPR pathway activation can play in homeostatic and physiological responses (Rutkowski and Hegde, 2010). Another caveat when utilizing chemical inducers is that different concentrations or durations of chemical-induced ER stress can lead to distinct outcomes in different myeloid cell types, resulting in either the induction (Zhao et al., 2014) or suppression (Komura et al., 2013) of cytokines. The few studies that examined PRR-induced ER stress in myeloid cells showed some discrepant results, in particular regarding which UPR branches are activated following PRR stimulation. For example, in mouse macrophages, stimulation of some but not all TLRs activated the IRE1α pathway, while ATF6 activation was not observed (Bronner et al., 2015; Martinon et al., 2010). This is in contrast with a study where TLR4 stimulation induced ATF6 activation in mouse Kupffer cells (Rao et al., 2014). Furthermore, a mouse macrophage study found that NOD2 regulated ER stress independent of its ligand MDP (Keestra-Gounder et al., 2016). Our study finds that MDP activates all three ER-stress branches in a NOD2-dependent manner in human MDMs. One potential explanation for the distinct ER-stress branches activated upon PRR stimulation in these different studies and the distinct roles they play may be due to differences in the cell types examined and the nature of the autocrine cytokine loops secreted, as we now define that the autocrine/paracrine cytokines can themselves contribute to the ER stress observed with PRR stimulation. Another inconsistency in macrophage studies concerns the role of MAPK and NF-κB signaling in activating ER stress. These pathways were shown to activate ER stress in several studies (reviewed in Darling and Cook, 2014) but not universally (Martinon et al., 2010). We find that MAPK and NF-κB signaling was required for NOD2-induced ER stress, which, in turn, further amplified MAPK and NF-κB signaling in human MDMs. This highlights a PRR-induced feed-forward loop wherein signaling and autocrine/paracrine cytokines result in amplification of ER stress.
The role of ER stress in microbial clearance has been mostly examined in epithelial cells or mouse myeloid cells, with mechanisms mediating this clearance still incompletely defined. Previous studies found that different branches of ER stress can either enhance or inhibit microbial growth depending on the microbe, infection conditions, or the host species (Pillich et al., 2016). We find that in NOD2-stimulated human MDMs, ER stress leads to enhanced bacterial clearance. A prior study showed that IRE1-induced ROS production was required for bacterial clearance in mouse macrophages (Abuaita et al., 2015). Consistently, the NADPH oxidase subunits Nox2 (Li et al., 2010) and p47phox (Galán et al., 2014) were induced by non-PRR-mediated ER stress. We now extend these findings to show that LACC1-dependent ER stress is required for optimal PRR-induced ROS and for the induction of the NADPH oxidase subunits p40phox, p47phox, and p67phox in human MDMs. Furthermore, one study showed that ER stress-induced spliced XBP1 was required for induction of NOS2 in HepG2 cells (Guo et al., 2010), and we find that in human MDMs PRR-induced NOS2 is mediated by LACC1-dependent ER stress. Our findings therefore indicate that PRR-induced, LACC1-dependent ER stress upregulates multiple pathways and mechanisms required for PRR-induced bacterial clearance in macrophages. LACC1 mediates this ER stress through association with the ER stress sensor proteins, but likely through additional mechanisms. While the focus of this study has been human macrophages, it is also likely that LACC1 and the disease-risk Val254 variant regulate ER stress in other cells contributing to immune-mediated diseases. The LACC1 Arg284 disease-risk variant failed to localize to the ER and associate with ER-stress proteins upon NOD2 stimulation. In contrast, the LACC1 Val254 disease-risk variant was not impaired in these functions, despite reduced subsequent ER stress, signaling, cytokines, and bacterial clearance. Fungal and bacterial LACC1 homologs demonstrate polyphenol oxidase activity (Zhu and Williamson, 2004), but human LACC1 has not been identified to demonstrate this same activity (Cader et al., 2016; Lahiri et al., 2017). It is possible that human LACC1 mediates distinct enzymatic activity or other post-translational modifications of the proteins with which it associates and which are then reduced with the LACC1 Val254 variant.
ER-stress dysregulation is observed in multiple immune-mediated diseases and examining disease-associated genes regulating this process may provide insight into mechanisms leading to these diseases. Despite this, few genetic variants directly regulating ER-stress pathways have been described to date, with XBP1 variants being a notable exception (Kaser et al., 2008). We now identify that LACC1, a gene associated with IBD, leprosy, and arthritis, is an important regulator of the physiological and transient ER stress observed with PRR stimulation. Furthermore, whereas increased ER stress is observed in immune-mediated diseases, a failure to properly induce pathways required for the UPR can also confer risk for immune-mediated diseases, as is seen in mice with IRE1β and XBP1 deficiencies (Bertolotti et al., 2001; Kaser et al., 2008). We now identify that loss-of-function immune-mediated disease-risk LACC1 variants result in reduced PRR-induced ER stress. This reduced ER stress, in turn, results in reduced PRR-induced signaling, cytokine secretion, and bacterial clearance. Our study indicates that properly balancing ER stress in macrophages might provide an effective strategy for targeting immune-mediated diseases.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Requests for further information and resources can be directed to the Lead Contact, Clara Abraham (clara.abraham@yale.edu). Unique reagents generated in this study are available with a completed Material Transfer Agreement.
EXPERIMENTAL MODELS
HeLa cells (ATCC) were used. Primary cells were prepared from healthy blood donors. Studies were approved by the institutional review board at Yale University.
METHOD DETAILS
Primary Myeloid Cell Culture
Human peripheral blood mononuclear cells (PBMCs) were isolated from peripheral blood using Ficoll-Paque (GE Dharmacon, Lafayette, CO). Monocytes were purified from PBMCs by adhesion, and tested for purity (> 98% by CD11c expression) and differentiated as in Hedl et al. (2016) for 7 days with 10ng/ml M-CSF (Shenandoah Biotechnology, Warwick, PA). Genotyping for polymorphisms was performed by Taqman (Applied Biosystems).
MDM Stimulation
Cultured MDMs were treated with MDP (Bachem, Torrance, CA), 10 μM cyclopiazonic acid (CPA, MP Biomedicals, Santa Ana, CA), lipid A (Peptides International, Louisville, KY) or Pam3Cys (MilliporeSigma, Billerica, MA). Supernatants were assayed for TNF (clones MAb1 and MAb11), IL6 (clones MQ2–13A5 and MQ2–39C3), IL8 (clones G265–5 and G265–8), IL10 (clones JES3–19F1 and JES3–12G8) (BD Biosciences, San Jose, CA) or IL1β (clones H1b-27) (Biolegend) by ELISA.
Transfection of Small Interfering RNAs (siRNAs) and DNA Vectors
100nM scrambled or siGENOME or ON-TARGETplus SMARTpool small interfering RNA (siRNA) against LACC1, PERK, IRE1α or ATF6 (Dharmacon, Lafayette, CO) (4 pooled siRNAs for each gene) were transfected into MDMs using Amaxa nucleofector technology (Lonza, Morristown, NJ) for 48h. Studies were then conducted after this 48h interval. For plasmid transfection, 2 μg (for MDMs) or 500 ng (for HeLa cells) empty vector (pcDNA3.0), or C-terminal FLAG-tagged LACC1 WT, Val254, Arg284 or ΔISIK (LACC1 construct with the deletion of the ISIKE motif in the C terminus) vectors, generated through site-directed mutagenesis (Agilent Technologies), were transfected into MDMs by Amaxa nucleofector technology or into HeLa cells by Lipofectamine 2000 (Invitrogen) for 48h. For HeLa cell studies we also transfected 50 ng of NOD2 (pcDNA3.0).
Protein Detection
Proteins were detected in permeabilized (for intracellular expression) cells by flow cytometry with fluorophore-conjugated antibodies to phospho-PERK (Thermo Fisher Scientific), phospho-ERK (197G2), phospho-p38 (28B10), phospho-JNK (G9), phospho-IκBα (14D4), LC3II, FLAG (9A3) (Cell Signaling Technology, Danvers, MA), phospho-IRE1α (Abcam), NOS2 (C-11), p40phox (D-8), p47phox (A-7), p67phox (D-6) (Santa Cruz Biotechnology, Santa Cruz, CA) or ATG5 (EPR1755) (Abcam, Cambridge, MA). Briefly, for intracellular expression cells were fixed with Lyse/Fix Buffer (BD Biosciences) for 10 min, permeabilized for 1h with Perm Buffer III (BD Biosciences), washed and then stained with the indicated antibodies. Isotype controls were included for treated cells.
LACC1 (E-7; Santa Cruz Biotechnology) or RIP2/RICK (25/RIG-G) (BD Biosciences) were immunoprecipitated from MDMs or from transfected HeLa cells using antibody bound to Protein A or Protein G Sepharose (MilliporeSigma, Billerica, MA) and associated proteins were blotted with antibodies to PERK (D11A8), IRE1α (14C10) (Cell Signaling Technology), ATF6 (Proteintech, Rosemont, IL), RIP2, or GAPDH (clone 6C5, MilliporeSigma; clone D16H11, Cell Signaling Technology). LACC1 or the respective protein in whole-cell lysate served as a loading control. In some cases ER fractionation was carried out using a 1.3M sucrose gradient and ultracentrifugation as per Williamson et al. (2015). Calnexin (AF18) and α-tubulin (TU-02) (Santa Cruz Biotechnology) served as fractionation controls. Western blot was performed as per Hedl et al. (2016). Signal was detected by secondary anti-rabbit or anti-mouse IgG Dy-Light 680 or DyLight 800 antibodies (Thermo Fisher Scientific) with the Odyssey Imager (Li-COR Biosciences, Lincoln, NE).
mRNA Expression Analysis
Total RNA was isolated using Trizol reagent (Thermo Fisher Scientific). Quantitative PCR was performed using All-In-One qPCR Mix (Genecopoeia) on the ABI Prism 7000 (Applied Biosystems). Samples were normalized to GAPDH. Primer sequences are shown in Table S1.
Intracellular mtROS and ROS Measurement
Mitochondrial-ROS (mtROS) and ROS were measured by flow cytometry using 5 μM MitoSOX or 10 μM 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) (Invitrogen), respectively, following manufacturer’s instructions.
Intracellular Bacterial Clearance
Macrophages were cultured in triplicate for 1 hour with Enterococcus faecalis, adherent invasive Escherichia coli (AIEC) strain LF82 (a generous gift from Dr. Emiko Mizoguchi) or Staphylococcus aureus at 10:1 MOI, washed with PBS, and incubated in HBSS with 50 μg/ml gentamicin for an additional hour. Cells were washed, lysed with 1% Triton X-100 (MilliporeSigma) and plated on MacConkey or LB agar.
Bacterial Entry/Phagocytosis
FITC-labeled E. coli bioparticles (1.5×106) (Invitrogen) or 5:1 MOI Salmonella enterica serovar Typhimurium-GFP (generously provided by Jorge E. Galan) were co-cultured with MDMs for 20 minutes, cell surface fluorescence was quenched with 0.25mg/ml trypan blue, and cells were then analyzed by flow cytometry.
Microscopy
Cells were fixed in 4% paraformaldehyde or methanol, and then permeabilized with 0.1% Triton X-100 and incubated with anti-LACC1 (MilliporeSigma), calnexin (AF18) (Santa Cruz Biotechnology) or DAPI (Acros Organics) and detected by secondary antibodies labeled with Cy3 or Cy2 (Jackson ImmunoResearch). Fluorescence microscopy was conducted with the Zeiss Axio Observer microscope and pixel density analysis was conducted with ZEN lite (Carl Zeiss Microscopy, Thornwood, NY). Quantification of colocalization was performed using ImageJ.
QUANTIFICATION AND STATISTICAL ANALYSIS
Significance was assessed using two-tailed t test. Bonferroni-Holm correction was used for multiple comparisons where appropriate. The number of samples is indicated in the legends. To keep cytokines on same axis, a multiplier was applied for the higher cytokine levels of IL8 as shown in figure keys. p < 0.05 was considered significant. Lines over multiple bars indicate same significance values for these bars.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal LACC1 | MilliporeSigma | Cat# HPA040150 |
| Rabbit monoclonal LACC1 (E-7) | Santa Cruz Biotechnology | Cat# sc-374553 |
| Mouse monoclonal Calnexin (AF18) | Santa Cruz Biotechnology | Cat# sc-23954 |
| Rabbit monoclonal IRE1α (14C10) | Cell Signaling Technology | Cat# 3294 |
| Rabbit monoclonal PERK (D11A8) | Cell Signaling Technology | Cat# 5683 |
| Rabbit polyclonal ATF6 | Proteintech | Cat# 24169–1-AP |
| Rabbit monoclonal phospho-IRE1 (Ser724) [EPR5253] | Abcam | Cat# ab124945 |
| Rabbit polyclonal phospho-PERK (Thr982) | Thermo Fisher Scientific | Cat# PA5–40294 |
| Mouse monoclonal RIP2/RICK (A-10) | Santa Cruz Biotechnology | Cat# sc-166765 |
| Mouse monoclonal RIP2/RICK (25/RIG-G) | BD Biosciences | Cat# 612349 |
| Annexin V | BD Biosciences | Cat# 556419 |
| Mouse monoclonal a-Tubulin (TU-02) | Santa Cruz Biotechnology | Cat# sc-8035 |
| Mouse monoclonal p40-phox (D-8) | Santa Cruz Biotechnology | Cat# sc-48388 |
| Mouse monoclonal p47-phox (A-7) | Santa Cruz Biotechnology | Cat# sc-17844 |
| Mouse monoclonal p67-phox (D-6) | Santa Cruz Biotechnology | Cat# sc-374510 |
| Mouse monoclonal NOS2 (C-11) | Santa Cruz Biotechnology | Cat# sc-7271 |
| Rabbit polyclonal LC3B | Cell Signaling Technology | Cat# 2775 |
| Rabbit monoclonal APG5L/ATG5 [EPR1755(2)] | Abcam | Cat# ab108327 |
| Mouse monoclonal Syk (4D10) | Cell Signaling Technology | Cat# 80460 |
| Rabbit monoclonal Dectin-1 (E1X3Z) | Cell Signaling Technology | Cat# 60128 |
| Mouse monoclonal FLAG Tag (9A3) | Cell Signaling Technology | Cat# 8146 |
| Rabbit monoclonal Calreticulin (D3E6) | Cell Signaling Technology | Cat# 12238 |
| Rabbit monoclonal phospho-p44/42 MAPK (Erk½) (Thr202/Tyr204) (197G2) | Cell Signaling Technology | Cat# 13148 |
| Mouse monoclonal phospho-p38 MAPK (Thr180/Tyr182) (28B10) | Cell Signaling Technology | Cat# 4551 |
| Mouse monoclonal phospho-SAPK/JNK (Thr183/Tyr185) (G9) | Cell Signaling Technology | Cat# 9257 |
| Rabbit monoclonal phospho-IκBα (Ser32) (14D4) | Cell Signaling Technology | Cat# 2859 |
| Mouse monoclonal GAPDH (6C5) | MilliporeSigma | Cat# CB1001 |
| Rabbit monoclonal GAPDH (D16H11) | Cell Signaling Technology | Cat# 5174 |
| Anti-mouse IgG (H+L), F(ab’)2 Fragment (Alexa Fluor 488 Conjugate) | Cell Signaling Technology | Cat# 4408 |
| Anti-rabbit IgG (H+L), F(ab’)2 Fragment (Alexa Fluor 647 Conjugate) | Cell Signaling Technology | Cat# 4414 |
| Cy2 AffiniPure Goat Anti-Mouse IgG (H+L) | Jackson ImmunoResearch | Cat# 115–225-146 |
| Cy2 AffiniPure Goat Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch | Cat# 111–225-144 |
| Cy3 AffiniPure Goat Anti-Mouse IgG (H+L) | Jackson ImmunoResearch | Cat# 115–165-146 |
| Cy3 AffiniPure Goat Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch | Cat# 111–165-144 |
| DyLight 680 Goat Anti-Mouse IgG (H+L) | Thermo Fisher Scientific | Cat# 35519 |
| DyLight 800 Goat Anti-Mouse IgG (H+L) | Thermo Fisher Scientific | Cat# SA510176 |
| DyLight 680 Goat Anti-Rabbit IgG (H+L) | Thermo Fisher Scientific | Cat# 35569 |
| DyLight 800 Goat Anti-Rabbit IgG (H+L) | Thermo Fisher Scientific | Cat# SA510036 |
| Biotin mouse monoclonal TNF (MAb11) | BD Biosciences | Cat# 554511 |
| Mouse monoclonal TNF (MAb1) | BD Biosciences | Cat# 551220 |
| Mouse monoclonal IL-8 (G265–5) | BD Biosciences | Cat# 554716 |
| Biotin mouse monoclonal IL-8 (G265–8) | BD Biosciences | Cat# 554718 |
| Biotin rat monoclonal IL-6 (MQ2–39C3) | BD Biosciences | Cat# 554546 |
| Rat monoclonal IL-6 Antibody (MQ2–13A5) | BD Biosciences | Cat# 554543 |
| Biotin rat monoclonal IL-10 (JES3–12G8) | BD Biosciences | Cat# 554499 |
| Rat monoclonal IL-10 Antibody (JES3–19F1) | BD Biosciences | Cat# 554703 |
| Mouse monoclonal IL-1β Antibody (H1b-27) | BioLegend | Cat# 511602 |
| Goat polyclonal IL-1β Antibody (Poly5174) | BioLegend | Cat# 517403 |
| Bacterial Strains | ||
| Enterococcus faecalis | Yale Microbiology laboratories | N/A |
| Staphylococcus aureus | Yale Microbiology laboratories | N/A |
| Adherent invasive E. coli strain LF82 | Dr. Emiko Mizoguchi; Darfeuille-Michaud et al., 2004 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cyclopiazonic acid (CPA) | MP Biomedicals | Cat# 18172–33-3 |
| Thapsigargin | Enzo Life Sciences | Cat# BML-PE180 |
| Tunicamycin | MP Biomedicals | Cat# 11089–65-9 |
| Ac-muramyl-Ala-D-Glu-NH2 | Bachem | Cat# G-1055 |
| Pam3Cys-Ser-(Lys)4, Hydrochloride | MilliporeSigma | Cat# 506350 |
| Lipid A | Peptide International | Cat# CLP-24005 s |
| TriDAP | InvivoGen | Cat# tlrl-tdap |
| Curdlan | InvivoGen | Cat# tlr-curd |
| Human M-CSF | Shenandoah Biotechnology | Cat# 100–03 |
| Human TNF | PeproTech | Cat# 300–01A |
| Human IL1β | PeproTech | Cat# 200–01B |
| Human IL6 | PeproTech | Cat# 200–06 |
| Human IL8 | PeproTech | Cat# 200–08 |
| Human IL10 | PeproTech | Cat# 200–10 |
| Critical Commercial Assays | ||
| TaqMan Genotyping Master Mix | Applied Biosystems | Cat# 4371355 |
| Pierce 660nm Protein Assay Reagent | Thermo Fisher Scientific | Cat# 22660 |
| Herculase II Fusion DNA polymerase | Agilent Technologies | Cat# 600675 |
| Phusion high fidelity DNA polymerase | NEB | Cat# M0530 |
| All-in-One First strand cDNA synthesis kit | GeneCopoeia | Cat# QP009 |
| All-in-One qPCR Mix | GeneCopoeia | Cat# QP002 |
| Carboxy-H2DCFDA | Invitrogen | Cat# C400 |
| MitoSOX Red mitochondrial superoxide indicator | Invitrogen | Cat# M36008 |
| Lipofectamine 2000 Transfection Reagent | Invitrogen | Cat# 11668019 |
| Experimental Models: Cell Lines | ||
| HeLa cells | ATCC | ATCC CCL-2 |
| Oligonucleotides | ||
| siGENOME SMARTpool siRNA for human LACC1 | Dharmacon | Cat# M-015653–00 |
| siGENOME SMARTpool siRNA for human PERK | Dharmacon | Cat# M-004883–03 |
| siGENOME SMARTpool siRNA for human IRE1a | Dharmacon | Cat# M-004951–02 |
| siGENOME SMARTpool siRNA for human ATF6 | Dharmacon | Cat# M-009917–01 |
| siGENOME SMARTpool siRNA for human TRAF6 | Dharmacon | Cat# M-004712–00 |
| siGENOME SMARTpool siRNA for human RIPK2 | Dharmacon | Cat# M-003602–02 |
| siGENOME SMARTpool siRNA for human NOD1 | Dharmacon | Cat# M-004398–00 |
| siGENOME SMARTpool siRNA for human NOD2 | Dharmacon | Cat# M-003464–01 |
| siGENOME SMARTpool siRNA for human NEMO | Dharmacon | Cat# M-003767–02 |
| siGENOME SMARTpool siRNA for human ERK | Dharmacon | Cat# M-003555–04 |
| siGENOME SMARTpool siRNA for human LACC1 | Dharmacon | Cat# M-015653–00 |
| siGENOME SMARTpool siRNA for human PERK | Dharmacon | Cat# M-004883–03 |
| siGENOME SMARTpool siRNA for human IRE1a | Dharmacon | Cat# M-004951–02 |
| siGENOME SMARTpool siRNA for human ATF6 | Dharmacon | Cat# M-009917–01 |
| siGENOME SMARTpool siRNA for human TRAF6 | Dharmacon | Cat# M-004712–00 |
| siGENOME SMARTpool siRNA for human RIPK2 | Dharmacon | Cat# M-003602–02 |
| siGENOME SMARTpool siRNA for human NOD1 | Dharmacon | Cat# M-004398–00 |
| siGENOME SMARTpool siRNA for human NOD2 | Dharmacon | Cat# M-003464–01 |
| siGENOME SMARTpool siRNA for human NEMO | Dharmacon | Cat# M-003767–02 |
| siGENOME SMARTpool siRNA for human ERK | Dharmacon | Cat# M-003555–04 |
| ON-TARGETplus SMARTpool siRNA for human p38 | Dharmacon | Cat# L-003512–00 |
| siGENOME Non-targeting siRNA Pool | Dharmacon | Cat# D-001206–13 |
| ON-TARGETplus Non-targeting siRNA Pool | Dharmacon | Cat# D-001810–10 |
| See Table S1 for qPCR primers | N/A | N/A |
| Recombinant DNA | ||
| pCDNA3.0-C-FLAG-hLACC1-I254 | Lahiri et al., 2017 | N/A |
| pCDNA3.0-C-FLAG-hLACC1-V254 | Lahiri et al., 2017 | N/A |
| pCDNA3.0-C-FLAG-hLACC1-C284R | This paper | N/A |
| pCDNA3.0-C-FLAG-hLACC1-ΔISIK(E) | This paper | N/A |
| Software | ||
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
| ZEN lite Other |
Carl Zeiss, Germany | https://www.zeiss.com/microscopy/us/products/microscope-software/zen-lite.html |
| Escherichia coli (K-12 strain) BioParticles, Fluorescein | Invitrogen | Cat# E2861 |
| 4’,6’-Diamidino-2-phenylindole dihydrochloride(DAPI) | Acros Organics | Cat# 202710100 |
Highlights.
LACC1 localizes to the ER upon PRR stimulation of human macrophages
LACC1 associates with ER-stress sensors and promotes the UPR upon PRR stimulation
LACC1-dependent ER stress is required for PRR-initiated downstream pathways
Common and rare disease-risk variants in LACC1 show a reduction in ER stress
ACKNOWLEDGMENTS
This work was supported by the Department of Defense (W81XWH-16–1-01–78) and the NIH (R01DK099097, R01DK106593, DK062422, and DKP30–34989).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPORTING CITATIONS
The following reference appears in the Supplemental Information: Wright et al. (2016).
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.11.105.
DATA AND CODE AVAILABILITY
This study did not generate any unique datasets or code.
REFERENCES
- Abraham C, and Medzhitov R. (2011). Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology 140, 1729–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abuaita BH, Burkholder KM, Boles BR, and O’Riordan MX (2015). The Endoplasmic Reticulum Stress Sensor Inositol-Requiring Enzyme 1α Augments Bacterial Killing through Sustained Oxidant Production. MBio 6, e00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolph TE, Tomczak MF, Niederreiter L, Ko HJ, Bö ck, J., Martinez-Naves, E., Glickman, J.N., Tschurtschenthaler, M., Hartwig, J., Hosomi, S., et al. (2013). Paneth cells as a site of origin for intestinal inflammation. Nature 503, 272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assadi G, Vesterlund L, Bonfiglio F, Mazzurana L, Cordeddu L, Schepis D, Mjösberg J, Ruhrmann S, Fabbri A, Vukojevic V, et al. (2016). Functional Analyses of the Crohn’s Disease Risk Gene LACC1. PLoS ONE 11, e0168276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A, Wang X, Novoa I, Jungreis R, Schlessinger K, Cho JH, West AB, and Ron D. (2001). Increased sensitivity to dextran sodium sulfate colitis in IRE1beta-deficient mice. J. Clin. Invest 107, 585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bist P, Cheong WS, Ng A, Dikshit N, Kim BH, Pulloor NK, Khameneh HJ, Hedl M, Shenoy AR, Balamuralidhar V, et al. (2017). E3 Ubiquitin ligase ZNRF4 negatively regulates NOD2 signalling and induces tolerance to MDP. Nat. Commun 8, 15865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner DN, Abuaita BH, Chen X, Fitzgerald KA, Nuñez G, He Y, Yin XM, and O’Riordan MX (2015). Endoplasmic Reticulum Stress Activates the Inflammasome via NLRP3-and Caspase-2-Driven Mitochondrial Damage. Immunity 43, 451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cader MZ, Boroviak K, Zhang Q, Assadi G, Kempster SL, Sewell GW, Saveljeva S, Ashcroft JW, Clare S, Mukhopadhyay S, et al. (2016). C13orf31 (FAMIN) is a central regulator of immunometabolic function. Nat. Immunol 17, 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, Bringer MA, Swidsinski A, Beaugerie L, and Colombel JF (2004). High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 127, 412–421. [DOI] [PubMed] [Google Scholar]
- Darling NJ, and Cook SJ (2014). The role of MAPK signalling pathways in the response to endoplasmic reticulum stress. Biochim. Biophys. Acta 1843, 2150–2163. [DOI] [PubMed] [Google Scholar]
- Dhillon SS, Fattouh R, Elkadri A, Xu W, Murchie R, Walters T, Guo C, Mack D, Huynh HQ, Baksh S, et al. (2014). Variants in nicotinamide adenine dinucleotide phosphate oxidase complex components determine susceptibility to very early onset inflammatory bowel disease. Gastroenterology 147, 680–689.e2. [DOI] [PubMed] [Google Scholar]
- Eizirik DL, Miani M, and Cardozo AK (2013). Signalling danger: endoplasmic reticulum stress and the unfolded protein response in pancreatic islet inflammation. Diabetologia 56, 234–241. [DOI] [PubMed] [Google Scholar]
- Feng BS, He SH, Zheng PY, Wu L, and Yang PC (2007). Mast cells play a crucial role in Staphylococcus aureus peptidoglycan-induced diarrhea. Am. J. Pathol 171, 537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli F, Noack J, Bergmann TJ, Cebollero E, Pisoni GB, Fasana E, Fregno I, Galli C, Loi M, Soldá T, et al. (2016). Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat. Cell Biol 18, 1173–1184. [DOI] [PubMed] [Google Scholar]
- Galán M, Kassan M, Kadowitz PJ, Trebak M, Belmadani S, and Matrougui K. (2014). Mechanism of endoplasmic reticulum stress-induced vascular endothelial dysfunction. Biochim. Biophys. Acta 1843, 1063–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grootjans J, Kaser A, Kaufman RJ, and Blumberg RS (2016). The unfolded protein response in immunity and inflammation. Nat. Rev. Immunol 16, 469–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Lin EA, Liu P, Lin J, and Liu C. (2010). XBP1U inhibits the XBP1S-mediated upregulation of the iNOS gene expression in mammalian ER stress response. Cell. Signal 22, 1818–1828. [DOI] [PubMed] [Google Scholar]
- Hedl M, and Abraham C. (2011a). Distinct roles for Nod2 protein and autocrine interleukin-1beta in muramyl dipeptide-induced mitogen-activated protein kinase activation and cytokine secretion in human macrophages. J. Biol. Chem 286, 26440–26449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedl M, and Abraham C. (2011b). Secretory mediators regulate Nod2-induced tolerance in human macrophages. Gastroenterology 140, 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedl M, and Abraham C. (2012). Nod2-induced autocrine interleukin-1 alters signaling by ERK and p38 to differentially regulate secretion of inflammatory cytokines. Gastroenterology 143, 1530–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedl M, and Abraham C. (2014). A TNFSF15 disease-risk polymorphism increases pattern-recognition receptor-induced signaling through caspase-8-induced IL-1. Proc. Natl. Acad. Sci. USA 111, 13451–13456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedl M, Yan J, and Abraham C. (2016). IRF5 and IRF5 Disease-Risk Variants Increase Glycolysis and Human M1 Macrophage Polarization by Regulating Proximal Signaling and Akt2 Activation. Cell Rep 16, 2442–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland SM (2010). Chronic granulomatous disease. Clin. Rev. Allergy Immunol 38, 3–10. [DOI] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. ; International IBD Genetics Consortium (IIBDGC) (2012). Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EE, Higgins DE, Schreiber S, Glimcher LH, and Blumberg RS (2008). XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134, 743–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A, Martínez-Naves E, and Blumberg RS (2010). Endoplasmic reticulum stress: implications for inflammatory bowel disease pathogenesis. Curr. Opin. Gastroenterol 26, 318–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keestra-Gounder AM, Byndloss MX, Seyffert N, Young BM, Chávez-Arroyo A, Tsai AY, Cevallos SA, Winter MG, Pham OH, Tiffany CR, et al. (2016). NOD1 and NOD2 signalling links ER stress with inflammation. Nature 532, 394–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komura T, Sakai Y, Honda M, Takamura T, Wada T, and Kaneko S. (2013). ER stress induced impaired TLR signaling and macrophage differentiation of human monocytes. Cell. Immunol 282, 44–52. [DOI] [PubMed] [Google Scholar]
- Lahiri A, and Abraham C. (2014). Activation of pattern recognition receptors up-regulates metallothioneins, thereby increasing intracellular accumulation of zinc, autophagy, and bacterial clearance by macrophages. Gastroenterology 147, 835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri A, Hedl M, Yan J, and Abraham C. (2017). Human LACC1 increases innate receptor-induced responses and a LACC1 disease-risk variant modulates these outcomes. Nat. Commun 8, 15614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Scull C, Ozcan L, and Tabas I. (2010). NADPH oxidase links endoplasmic reticulum stress, oxidative stress, and PKR activation to induce apoptosis. J. Cell Biol 191, 1113–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Irwanto A, Fu X, Yu G, Yu Y, Sun Y, Wang C, Wang Z, Okada Y, Low H, et al. (2015). Discovery of six new susceptibility loci and analysis of pleiotropic effects in leprosy. Nat. Genet 47, 267–271. [DOI] [PubMed] [Google Scholar]
- Lopes F, Keita AV, Saxena A, Reyes JL, Mancini NL, Al Rajabi A, Wang A, Baggio CH, Dicay M, van Dalen R, et al. (2018). ER-stress mobilization of death-associated protein kinase-1-dependent xenophagy counteracts mitochondria stress-induced epithelial barrier dysfunction. J. Biol. Chem 293, 3073–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F, Chen X, Lee AH, and Glimcher LH (2010). TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat. Immunol 11, 411–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muise AM, Snapper SB, and Kugathasan S. (2012). The age of gene discovery in very early onset inflammatory bowel disease. Gastroenterology 143, 285–288. [DOI] [PubMed] [Google Scholar]
- Patel N, El Mouzan MI, Al-Mayouf SM, Adly N, Mohamed JY, Al Mofarreh MA, Ibrahim N, Xiong Y, Zhao Q, Al-Saleem KA, and Alkuraya FS (2014). Study of Mendelian forms of Crohn’s disease in Saudi Arabia reveals novel risk loci and alleles. Gut 63, 1831–1832. [DOI] [PubMed] [Google Scholar]
- Pauleau AL, and Murray PJ (2003). Role of nod2 in the response of macrophages to toll-like receptor agonists. Mol. Cell. Biol 23, 7531–7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillich H, Loose M, Zimmer KP, and Chakraborty T. (2016). Diverse roles of endoplasmic reticulum stress sensors in bacterial infection. Mol. Cell Pediatr 3, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao J, Yue S, Fu Y, Zhu J, Wang X, Busuttil RW, Kupiec-Weglinski JW, Lu L, and Zhai Y. (2014). ATF6 mediates a pro-inflammatory synergy between ER stress and TLR activation in the pathogenesis of liver ischemia-reperfusion injury. Am. J. Transplant 14, 1552–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, and Hegde RS (2010). Regulation of basal cellular physiology by the homeostatic unfolded protein response. J. Cell Biol 189, 783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh MU, MacMicking JD, Nicholson S, Brause JE, Potter S, Marino M, Fang F, Dinauer M, and Nathan C. (1999). Phenotype of mice and macrophages deficient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity 10, 29–38. [DOI] [PubMed] [Google Scholar]
- Skon-Hegg C, Zhang J, Wu X, Sagolla M, Ota N, Wuster A, Tom J, Doran E, Ramamoorthi N, Caplazi P, et al. (2019). LACC1 Regulates TNF and IL-17 in Mouse Models of Arthritis and Inflammation. J. Immunol 202, 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakil SM, Monies DM, Abouelhoda M, Al-Tassan N, Al-Dusery H, Naim EA, Al-Younes B, Shinwari J, Al-Mohanna FA, Meyer BF, and Al-Mayouf S. (2015). Association of a mutation in LACC1 with a monogenic form of systemic juvenile idiopathic arthritis. Arthritis Rheumatol. 67, 288–295. [DOI] [PubMed] [Google Scholar]
- Williamson CD, Wong DS, Bozidis P, Zhang A, and Colberg-Poley AM (2015). Isolation of Endoplasmic Reticulum, Mitochondria, and Mitochondria-Associated Membrane and Detergent Resistant Membrane Fractions from Transfected Cells and from Human Cytomegalovirus-Infected Primary Fibroblasts. Curr. Protoc. Cell Biol 68, 3.27.21–3.27.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PB, McEntegart A, McCarey D, McInnes IB, Siebert S, and Milling SW (2016). Ankylosing spondylitis patients display altered dendritic cell and T cell populations that implicate pathogenic roles for the IL-23 cytokine axis and intestinal inflammation. Rheumatology (Oxford) 55, 120–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev O, and Pines O. (2011). Dual targeting of mitochondrial proteins: mechanism, regulation and function. Biochim. Biophys. Acta 1808, 1012–1020. [DOI] [PubMed] [Google Scholar]
- Zhao C, Pavicic PG Jr., Datta S, Sun D, Novotny M, and Hamilton TA (2014). Cellular stress amplifies TLR¾-induced CXCL½ gene transcription in mononuclear phagocytes via RIPK1. J. Immunol 193, 879–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, and Abraham C. (2013). NF-kB1 inhibits NOD2-induced cytokine secretion through ATF3-dependent mechanisms. Mol. Cell. Biol 33, 4857–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, and Williamson PR (2004). Role of laccase in the biology and virulence of Cryptococcus neoformans. FEMS Yeast Res. 5, 1–10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.