Abstract
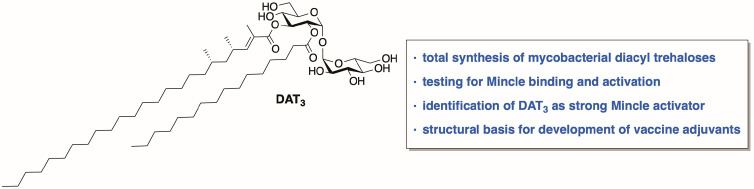
The first asymmetric total synthesis of three structures proposed for mycobacterial diacyl trehaloses, DAT1, DAT2, and DAT3 is reported. The presence of two of these glycolipids, DAT1 and DAT3, within different strains of pathogenic M. tuberculosis was confirmed, and it was shown that their abundance varies significantly. In mass spectrometry, synthetic DAT2 possessed almost identical fragmentation patterns to presumptive DAT2 from Mycobacterium tuberculosis H37Rv, but did not coelute by HPLC, raising questions as the precise relationship of the synthetic and natural materials. The synthetic DATs were examined as agonists for signaling by the C-type lectin, Mincle. The small differences in the chemical structure of the lipidic parts of DAT1, DAT2, and DAT3 led to drastic differences of Mincle binding and activation, with DAT3 showing similar potency as the known Mincle agonist trehalose dimycolate (TDM). In the future, DAT3 could serve as basis for the design of vaccine adjuvants with simplified chemical structure.
Introduction
Mycobacterium tuberculosis (Mtb), which is the causative agent of the disease tuberculosis (Tb), is responsible for the largest number of deaths worldwide by a single pathogen, killing an estimated 1.3 million people annually. The ability of Mtb to survive and persist in the host is estimated to result in billions of latently infected individuals worldwide, with a high incidence of undiagnosed cases.1 After infection of macrophages, Mtb is able to survive and replicate in host phagosomes, while withstanding the hostile acidic environment. The mycobacterial cell envelope is one factor that contributes to the resilience of Mtb within host cells.2 It is a multilayered barrier, composed of many complex lipids, glycolipids, and glycoproteins, many of which are unique to Mtb.3−5 In the last decades, it has been shown that many of these cell wall components have antigenic properties and/or possess immunomodulatory functions. One class of these mycobacterial cell wall components, which consists of diacylated and polyacylated trehaloses, is suggested to be located on the outer part of the mycobacterial cell wall.6 These trehalose-based glycolipids are esterified with palmitic or stearic acid at the 2- and 2′-position, as well as with the Mtb-specific multimethyl-branched acyl residues phthioceranic acid, hydroxyphthioceranic acid, mycosanoic acid, mycolipanolic acid, and mycolipenic acid. Important examples are Ac2SGL,7−9 Sulfolipid-1,10−12 trehalose monomycolate and dimycolate,13,14 diacyl trehaloses (DAT),15−17 and pentaacyl trehaloses (PAT).15,18 Because of the chemical diversity of DAT and the potential for contamination of even small amounts of bioactive molecular variants, testing natural DAT compounds on cells for immune response is not reliable. To establish the molecular structure of these compounds and enable further biological studies, several of the compounds have been the target of total synthesis. In DAT and PAT, both of which have escaped total synthesis until now, the trehalose core is acylated with the methyl-branched fatty acids mycosanoic acid, mycolipanolic acid, and mycolipenic acid (see Figure 1).17,18
Figure 1.
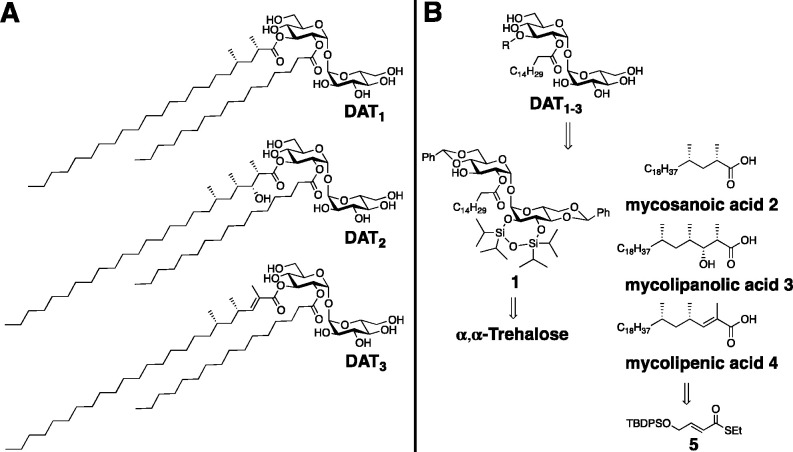
(A) Chemical structures of the mycobacterial diacyl trehaloses DAT1, DAT2, and DAT3. (B) Retrosynthetic analysis.
DAT was first isolated in 1989 by Daffé et al. and was initially named SL-IV (Sulfolipid-IV), since the structure was first assigned as 2,3-diacyl-trehalose-2′-sulfate.15,16 The structure of this family of acyl trehaloses was eventually revisited and corrected to be 2,3-diacyl-trehalose, and depending on the nature of the 3-O-acyl group, were termed DAT1, DAT2, or DAT3 (see Figure 1A). In the following reports, these compounds were often referenced as just DAT, presenting a family of mycobacterial glycolipids rather than defined molecular structures.17 Many studies have asserted the antigenic properties of DAT glycolipids by ELISA, but these were tested mainly as mixtures rather than pure compounds. It was demonstrated multiple times that anti-DAT antibodies are present in blood sera of Tb patients but not of healthy controls.17,19−22 This raised great interest in using DAT for the detection and diagnosis of Tb in patients. The reports utilizing ELISA for the detection of anti-DAT antibodies, however, showed a huge variation in sensitivity and specificity, depending on assay design.23
In recent years, research has focused on elucidating the biosynthesis of DAT and unravelling its effect on the immune system.24−27 It was shown that DAT partially inhibits the proliferation of murine T-cells, suggesting a role in immunosuppression and T-cell hyporesponsiveness associated with Tb.28Mtb mutants incapable of synthesizing mycolipenic acid, and therefore deficient in DAT and PAT, show aggregation in liquid culture, because of defects in capsule attachment, indicating that one of the functions of DAT and PAT is anchoring the hydrophilic capsule to the hydrophobic mycolic acid layer of the mycobacterial cell envelope.24,29−31 However, in aerosol infection mouse models using DAT/PAT-deficient mutants, there were no observed differences in growth, compared to wild-type compounds, suggesting that DAT/PAT itself is not necessary for Mtb survival.29
Recently, Mtb cell wall components—such as trehalose dimycolate (TDM, also known as cord factor)—have been identified as high-affinity ligands for macrophage-inducible C-type lectin (Mincle).32−34 The activation of Mincle results in downstream expression of cytokines, chemokines, and growth factors and leads to recruitment of inflammatory cells to the site of activation as a central part of the innate immune response to Mtb.33 Several other Mtb cell wall glycolipids have been identified as Mincle activators,34,35 and there is growing interest in using these Mincle ligands for the development of novel vaccine adjuvants.36
In 2017, it was demonstrated that a DAT-containing extract from Mtb also activated Mincle.35 We realized that, apart from minute amounts of contaminants in natural isolates that can influence the results, the activation of Mincle could very well be dependent on the precise structure of the DAT. Therefore, we sought to synthesize three different DATs with precisely defined molecular structure and stereochemistry to study their Mincle activating properties and to assess the influence of the acyl substituents on Mincle binding. Furthermore, we aimed to confirm the presence of these three DATs in different strains of Mtb, including clinical isolates.
Results and Discussion
Synthesis
DAT1, DAT2, and DAT3 differ in their chiral acyl group esterified with the 3-OH of the trehalose core. Therefore, our synthesis plan involved the preparation of suitably protected 2-palmitoyl trehalose 1 and the three mycobacterial lipids 2, 3, and 4 as key intermediates necessary to construct the target diacyl trehaloses. Trehalose 1 could be obtained starting from α,α-trehalose by a desymmetrization approach previously applied in the synthesis of trehalose-based sulfoglycolipids.37,38 The mycobacterial lipids, on the other hand, can be traced back to the common precursor 5 (Figure 1B). The synthesis of mycolipanolic and mycolipenic acid was previously reported by us and involves copper-catalyzed asymmetric conjugate addition (Cu-cat. ACA) and an Evans’ aldol reaction to introduce the stereocenters.39 We sought to improve the current synthetic procedures to arrive at an efficient, high-yielding total synthesis.
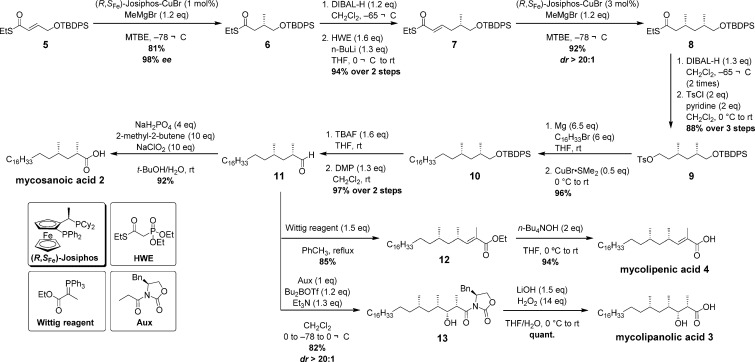
The synthesis of the chiral enantiopure lipids 2, 3, and 4 (see Scheme 1) commenced with Cu-cat. ACA of methylmagnesium bromide to α,β-unsaturated thioester 5 giving 6 in 81% yield and 98% enantiomeric excess (ee). Reduction to the corresponding aldehyde, followed by Horner–Wadsworth–Emmons reaction, produced another α,β-unsaturated thioester 7. The second methyl stereocenter was again introduced by Cu-cat. ACA in excellent yield and diastereomeric ratio (dr). Double DIBAL-H reduction, followed by tosylation, gave 9 in 88% yield over three steps. Tosylate 9 was subjected to a Grignard cross-coupling in the presence copper(I) to install the linear alkyl tail of 10. Removal of the silyl protecting group, followed by Dess-Martin oxidation, gave aldehyde 11 in an excellent yield of 97% over two steps. From 11, all three mycobacterial lipids could be synthesized in a limited number of steps. Mycosanoic acid 2 was obtained in 92% yield after Pinnick oxidation of aldehyde 11. Mycolipenic acid 4 was prepared by first subjecting 11 to a Wittig reaction, followed by alkaline ester hydrolysis. To install the two remaining stereocenters present in 3, an Evans’ aldol reaction was performed, giving 13 in good yield and excellent dr. The aldol product 13 was finally hydrolyzed to give mycolipanolic acid 3. Compared to the previous syntheses of 3 and 4, the yields could be significantly improved by careful optimization of the reactions. For mycosanoic acid, mycolipanolic acid, and mycolipenic acid, excellent overall yields were obtained with 53% over 10 steps, 47% (previously 2%) over 11 steps, and 46% (previously 5%) over 11 steps, respectively, making the synthesis of these chiral lipids highly efficient. In the synthesis of mycolipenic acid, oxidation, Wittig reaction, and ester hydrolysis were significantly improved, whereas in the case of mycolipanolic acid, the Evans’ aldol reaction and the removal of the chiral auxiliary were optimized to give high yields.39
Scheme 1. Asymmetric Synthesis of Mycosanic Acid (2), Mycolipanolic Acid (3), and Mycolipenic Acid (4).
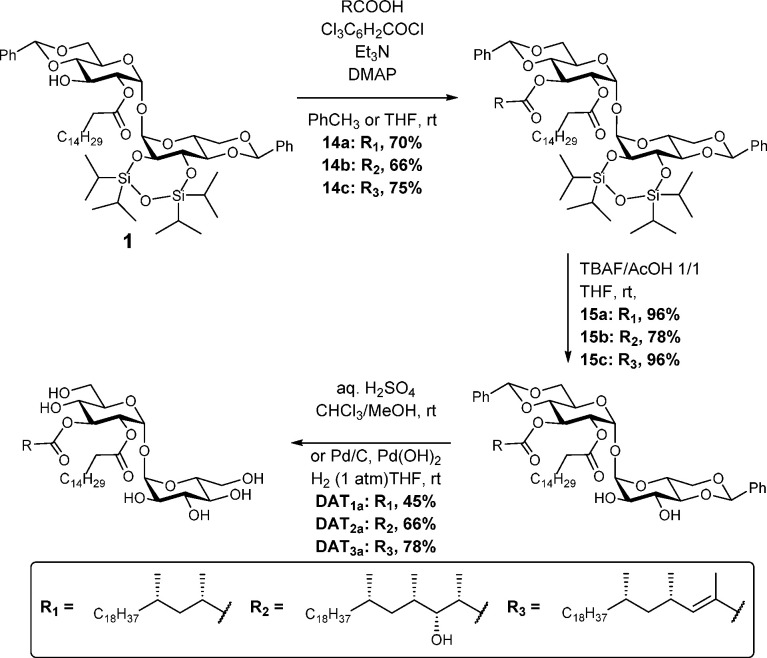
With the enantiopure acids 2–4 in hand, the esterification of palmitoylated trehalose 1 was achieved by following the Yamaguchi procedure. In the case of mycosanoic acid and mycolipenic acid, the corresponding diacylated products were obtained in good yields; however, in order to reach that result for mycolipanolic acid, the esterification procedure needed to be carefully optimized to avoid acyl migration and elimination of the β-hydroxyl of 3. By limiting the number of equivalents of base and keeping the time for acid activation at a minimum, synthesis of 14b could achieved in good yield. Notably, in the case of 14a and 14c, no acyl migration was observed, indicating that the β-hydroxyl in 3 might play a role in acyl migration. Removal of the silyl protecting group of 14a–14c under buffered conditions gave the corresponding diols 15a–15c in good to excellent yields. The final deprotection—the removal of the benzylidene protecting group—was achieved by applying a procedure that was reported by Guiard et al.,37 using aqueous sulfuric acid (DAT1 and DAT3) or by palladium hydrogenolysis (DAT2, to prevent β-hydroxyl elimination) and provided the three di-O-acyl trehaloses DAT1, DAT2, and DAT3 in moderate to good yields. (See Scheme 2.) The spectral data of DAT1 matched the reported NMR data of isolated DAT1a.17 Besra’s report describes the 1H NMR signals of the anomeric protons of DAT1 at 5.25 and 5.05 ppm for the acylated and nonacylated glucose unit, respectively. The spectrum of synthetic DAT1 shows these two anomeric signals at 5.24 and 5.06 ppm, which is in good agreement. Furthermore, H-2 and H-3 (at the positions bearing the acyl moieties) in natural DAT1 appear at 4.83 and 5.40 ppm, respectively, and in synthetic DAT1 at 4.82 and 5.39 ppm, respectively. The 13C signals of the anomeric carbons in natural DAT1 are reported at 95.0 and 92.0 ppm. In the synthetic material, these signals can be found 94.6 and 91.7 ppm, again in good agreement. In addition, the carbonyl carbon signals in synthetic DAT1 resonate at 173.5 and 177.6 ppm and the corresponding signals in natural DAT1 can be found at 173.8 and 177.8 ppm. All in all, these data leave us confident that the structure of synthetic DAT1 is identical to that of natural DAT1, as described by Besra (for more detailed NMR signal comparison, see the Supporting Information). As for synthetic DAT2 and DAT3, the structural identity is beyond reasonable doubt, because the structures of the lipid components have been previously established39 and the nuclear magnetic resonance (1H NMR and 13C NMR) and mass spectra showed patterns very similar to those of synthetic and natural DAT1.
Scheme 2. Completion of the Total Synthesis of the Mycobacterial Glycolipids DAT1, DAT2, and DAT3.
Detection of DAT1, DAT2, and DAT3 in Mtb Strains
Having completed the total synthesis of DAT1, DAT2, and DAT3 with structures as described in the literature,17 we sought to determine if the synthesized glycolipids match the structures of natural products present in virulent strains of Mtb. Mycobacterial lipid extracts of the reference strain H37Rv and three clinical isolates j257, j011, and j117 were analyzed by means of LC-MS. The extracted-ion chromatograms (Figure 2A) suggest that all three DATs are produced by the laboratory strain H37Rv. Ions consistent with DAT1 and DAT2 were only reliably detected in the H37Rv strain, whereas DAT3 could be detected in all four strains. The corresponding mass spectra of the detected natural DATs matched the calculated m/z of each compound within the expected experimental error of 3–4 ppm. Collision-induced fragmentation (see the data given in the Supporting Information) of the natural and synthetic DATs yielded interpretable cleavages (Figure 2C, H-transfers not shown) that supported the general structures and connectivity. Co-injection (Figure 2B) of synthetic standards and natural lipid mixtures showed a chromatographic match for DAT1 and DAT3. However, synthetic DAT2 eluted more than a minute earlier than the natural compound. Thus, whereas the identity of DAT1 and DAT3 can be considered to have been established beyond a reasonable doubt, we conclude that material with the structure of synthetic DAT2 does not occur in the H37Rv strain. This may mean that an isomer of the proposed structure of DAT2 is present in this strain, or that the structure of natural DAT2 has been incorrectly assigned.15
Figure 2.
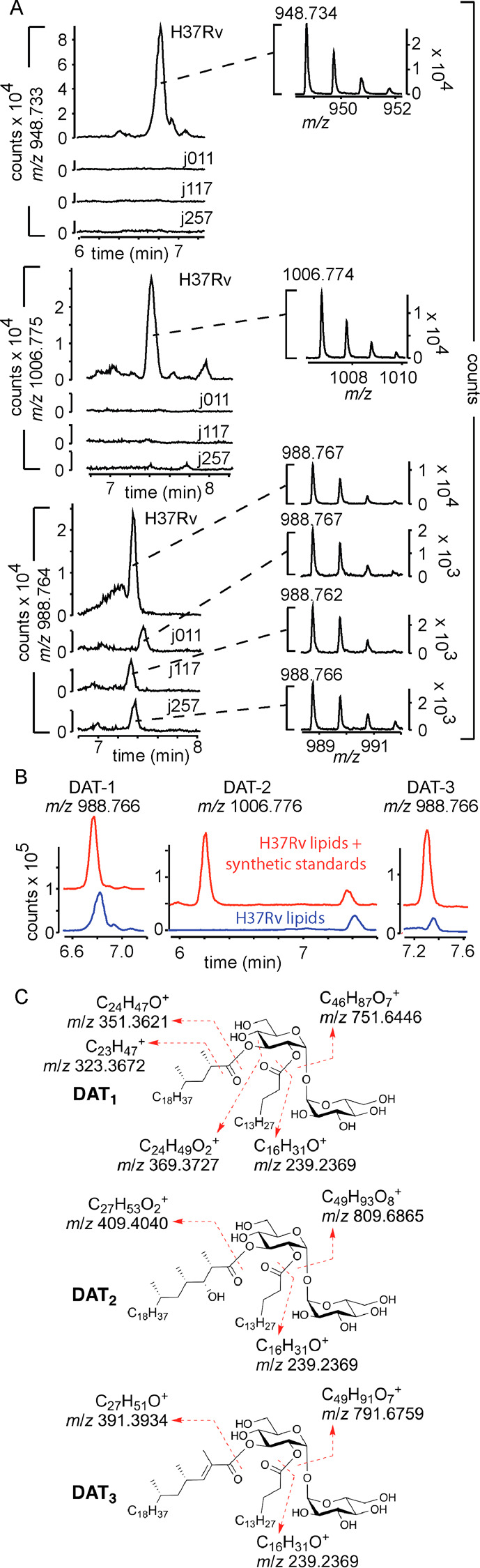
Detection of DAT variants in M. tuberculosis strains. Lipid extracts from four different M. tuberculosis strains were analyzed via high-performance liquid chromatography–mass spectroscopy (HPLC-MS): laboratory strain H37Rv, and three clinical isolates named j257, j011, and j117. Extracted ion chromatograms of ions corresponding with the ammonium adduct of DAT1 (calculated m/z = 948.733), DAT2 (m/z = 1006.775), and DAT3 (m/z = 988.764) showed m/z values consistent with those expected from DATs. (B) Comparison with synthetic standards showed chromatographic coelution for DAT1 and DAT3 but not for DAT2, indicating that synthetic DAT2 is not identical to natural DAT2. (C) CID analysis of the standards and natural compounds (see data given in the Supporting Information) yielded fragmentation patterns diagnostic for the known structures.
Mincle Activation by DAT1, DAT2, and DAT3
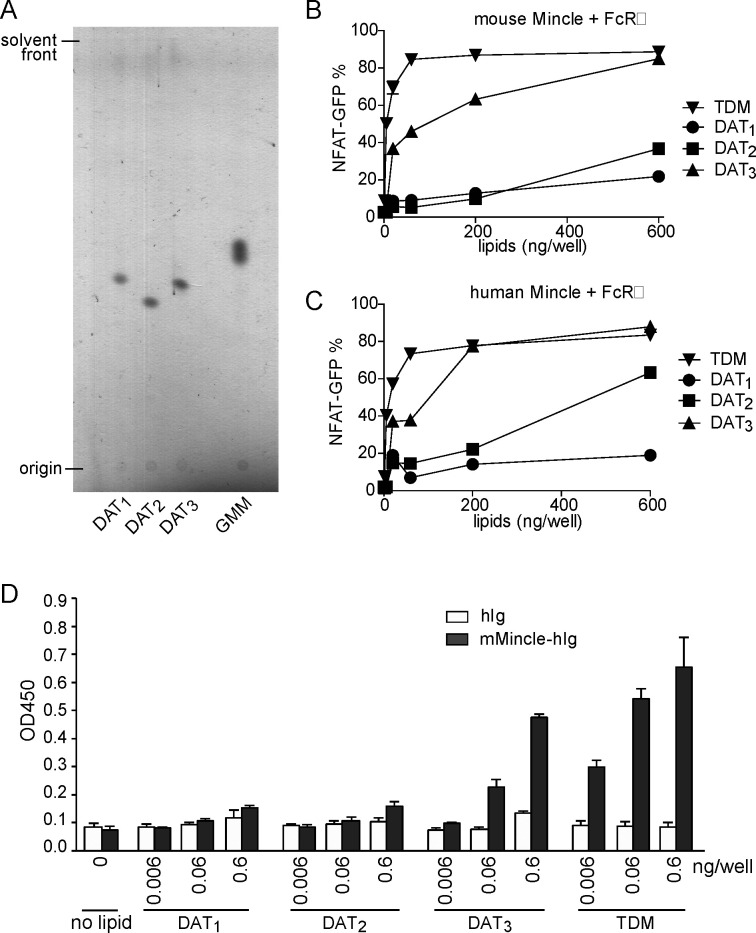
We decided to assess the Mincle activating properties of the synthetic DATs as well (see Figure 3), keeping in mind that our synthetic DAT2 was not present in the studied Mtb strains. Mincle activation was compared to the known Mincle-agonist TDM, which is highly potent. Previous studies have identified various lipidlike trehaloses that activate Mincle, so we expected that all three forms of DAT, which differ in small ways in their alkyl chains, were good candidate activators. Prior to the functional assays, TLC analysis of synthetic DAT1, DAT2, and DAT3 was performed to exclude the presence of glycolipid degradation products and quantification errors (Figure 3A). Mincle activation requires the adaptor protein FcRγ. Therefore, functional Mincle-activation assays were performed by treatment of NFAT-GFP reporter cells expressing murine Mincle and FcRγ (Figure 3B) or human Mincle and FcRγ (Figure 3C) with the synthetic DAT variants and TDM. In both assays, DAT3 was able to activate Mincle. In the case of human Mincle, DAT3 showed similar potency to the highly potent agonist TDM. DAT2 and, remarkably, DAT1 only weakly activated murine Mincle. When using human Mincle-expressing cells, DAT2 showed moderate activation, whereas DAT1 again barely induced Mincle stimulation. In an independent experiment, an ELISA-based technique was applied that was dependent not on cellular activation but only on the detection of physical interaction between DAT and soluble Mincle proteins (Figure 3D). Strong binding of murine Mincle to DAT3 was observed, but only minimal binding to DAT1 and DAT2, thereby confirming the results obtained in the cellular activation assay. These results provide evidence that the chemical structure of the 3-O-acyl substituent (either mycosanoic acid, mycolipanolic acid, or mycolipenic acid) strongly influences Mincle binding and activation.
Figure 3.
Synthetic DAT3 is recognized by human and mouse Mincle. (A) Before functional assays, DAT1, DAT2, and DAT3 were analyzed by thin-layer chromatography for relative quantification and the presence of major breakdown products. (B and C) NFAT-GFP reporter cells expressing mouse Mincle + FcRγ or human Mincle + FcRγ were stimulated with the indicated amount of DAT1, DAT2, DAT3, or TDM. After 24 h, induction of NFAT-GFP was analyzed by flow cytometry. (D) ELISA-based detection of DAT1, DAT2, DAT3, or TDM by mouse Mincle-human Ig Fc (mMincle-hIg) fusion proteins. Bound protein was detected with antihuman Ig-horse radish peroxidase (HRP), followed by the addition of a colorimetric substrate and measurement.
Conclusions
In this study, we have accomplished the first total synthesis of three structurally related mycobacterial DATs. These synthetic DATs were used as a reference in the detection of natural DATs in Mtb by liquid chromatography-mass spectroscopy. This showed that the presence and abundance of DAT1 and DAT3 differs strongly, dependent on the Mtb strain. This has important consequences for the potential use of DATs as markers for Tb infection. In addition, it might explain a posteriori the irreproducibility observed in the many attempts to reliably detect DAT by ELISA. It also showed that the proposed structure of DAT2 does not occur in the studied strains, including the H37Rv strain. An alternative explanation is that the structure of DAT2 has been assigned incorrectly, since, because of the lack of literature NMR data, a comparison with our synthetic material was not possible. This will be further investigated.
We found that small changes in the structure of the branched acyl chain in DAT lead to large differences in recognition by Mincle. It has been shown previously by Decout et al. that one of the molecular requirements for Mincle recognition, besides the trehalose or glucose scaffold, is the presence of two alkyl chains, either as two separate esters or as one fatty acid ester with an alkyl chain branched α to the carbonyl. Moreover, it was previously demonstrated that the lipid chains can be significantly shorter than the C80 lipids present in TDM. For instance, the synthetic Mincle ligand GlcC14C18, a glucose esterified at the 6-position with a C18 alkyl tail with a C14 alkyl branch on the α-position, shows even higher potency than TDM.35 In addition, in a previous report, Mincle activation by β-glucosylceramide, which also contains an unsaturation in the lipid chain, was demonstrated.40 Here, we show that the presence of the α,β-unsaturation in DAT3 enhances Mincle activation drastically, compared to the saturated counterpart DAT1. This leads us to speculate that the double bond either serves as a point of interaction (such as π–π-stacking) with parts of the Mincle binding pocket or induces a specific conformation beneficial for binding. All in all, one might conclude that DAT3 could be an alternative starting point for adjuvant design for TDM, given the higher complexity and lipophilicity of the latter. For future development of Mtb vaccine adjuvants, even simpler DAT analogues could be designed without chiral methyl branches or based on glycose rather than trehalose.
Acknowledgments
We thank T. Tiemersma, R. Sneep, and P. van der Meulen (University of Groningen) for assistance on analyses.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschembio.0c00030.
Synthetic procedures, LC-MS protocols, Mincle functional and binding assay protocols, compound data (1H and 13C NMR, HRMS) (PDF)
Author Contributions
∇ These authors contributed equally.
This work was supported by the Dutch Science Foundation (NWO), the Bill and Melinda Gates Foundation, and AMED (Nos. JP19gm0910010 and JP19ak0101070).
The authors declare no competing financial interest.
Supplementary Material
References
- World Health Organization . (2018) Global Tuberculosis Report 2018 (ISBM No. 978-92-4-156564-6).
- Ehrt S.; Schnappinger D. (2009) Mycobacterial survival strategies in the phagosome: defence against host stresses. Cell. Microbiol. 11 (8), 1170. 10.1111/j.1462-5822.2009.01335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry C. E. III; Lee R. E.; Mdluli K.; Sampson A. E.; Schroeder B. G.; Slayden R. A.; Yuan Y. (1998) Mycolic Acids: Structure, Biosynthesis and Physiological Functions. Prog. Lipid Res. 37 (2-3), 143. 10.1016/S0163-7827(98)00008-3. [DOI] [PubMed] [Google Scholar]
- Karakousis P. C.; Bishai W. R.; Dorman S. E. (2004) Mycobacterium tuberculosiscell envelope lipids and the host immune response. Cell. Microbiol. 6 (2), 105. 10.1046/j.1462-5822.2003.00351.x. [DOI] [PubMed] [Google Scholar]
- Singh P.; Rameshwaram N. R.; Ghosh S.; Mukhopadhyay S. (2018) Cell envelope lipids in the pathophysiology of Mycobacterium tuberculosis. Future Microbiol. 13 (6), 689. 10.2217/fmb-2017-0135. [DOI] [PubMed] [Google Scholar]
- Ortalo-Magné A.; Lemassu A.; Lanéelle M.-A.; Bardou F.; Silve G.; Gounon P.; Marchal G.; Daffé M. (1996) Identification of the Surface-Exposed Lipids on the Cell Envelopes of Mycobacterium tuberculosis and Other Mycobacterial Species. J. Bacteriol. 178, 456. 10.1128/JB.178.2.456-461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougous J. D.; Leavell M. D.; Senaratne R. H.; Leigh C. D.; Williams S. J.; Riley L. W.; Leary J. A.; Bertozzi C. R. (2002) Discovery of sulfated metabolites in mycobacteria with a genetic and mass spectrometric approach. Proc. Natl. Acad. Sci. U. S. A. 99 (26), 17037. 10.1073/pnas.252514899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleron M.; Stenger S.; Mazorra Z.; Wittke F.; Mariotti S.; Böhmer G.; Prandi J.; Mori L.; Puzo G.; De Libero G. (2004) Diacylated Sulfoglycolipids Are Novel Mycobacterial Antigens Stimulating CD1-restricted T Cells during Infection with Mycobacterium tuberculosis. J. Exp. Med. 199 (5), 649. 10.1084/jem.20031097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerdink D.; Horst B. t.; Lepore M.; Mori L.; Puzo G.; Hirsch A. K. H.; Gilleron M.; de Libero G.; Minnaard A. J. (2013) Total synthesis, stereochemical elucidation and biological evaluation of Ac2SGL; a 1,3-methyl branched sulfoglycolipid from Mycobacterium tuberculosis. Chem. Sci. 4 (2), 709. 10.1039/C2SC21620E. [DOI] [Google Scholar]
- Goren M. B. (1970) Sulfolipi Di of Mycobacterium Tuberculosis, Strain H37RvII. Structural Studies. Biochim. Biophys. Acta, Lipids Lipid Metab. 210, 127. 10.1016/0005-2760(70)90068-8. [DOI] [PubMed] [Google Scholar]
- Goren M. B.; Brokl O.; Das B. C.; Lederer E. (1971) Sulfolipid I of Mycobacterium tuberculosis, Strain H37Rv. Nature of the Acyl Substituents. Biochemistry 10, 72. 10.1021/bi00777a012. [DOI] [PubMed] [Google Scholar]
- Geerdink D.; Minnaard A. J. (2014) Total synthesis of sulfolipid-1. Chem. Commun. 50 (18), 2286. 10.1039/C3CC48087A. [DOI] [PubMed] [Google Scholar]
- Noll H.; Bloch H.; Asselineau J.; Lederer E. (1956) The chemical structure of the cord factor of Mycobacterium tuberculosis. Biochim. Biophys. Acta 20, 299. 10.1016/0006-3002(56)90289-X. [DOI] [PubMed] [Google Scholar]
- Ryll R.; Kumazawa Y.; Yano I. (2001) Immunological Properties of Trehalose Dimycolate (Cord Factor) and Other Mycolic Acid-Containing Glycolipids – A Review. Microbiol. Immunol. 45 (12), 801. 10.1111/j.1348-0421.2001.tb01319.x. [DOI] [PubMed] [Google Scholar]
- Minnikin D. E.; Dobson G.; Sesardic D.; Ridell M. (1985) Mycolipenates and Mycolipanolates of Trehalose from Mycobacterium tuberculosis. Microbiology 131, 1369. 10.1099/00221287-131-6-1369. [DOI] [PubMed] [Google Scholar]
- Daffé M.; Papa F.; Laszlo A.; David H. L. (1989) GLycolipids of Recent Clinical Isolates of Mycobacterium tuberculosis: Chemical Characterization and Immunoreactivity. Microbiology 135, 2759. 10.1099/00221287-135-10-2759. [DOI] [PubMed] [Google Scholar]
- Besra G. S.; Bolton R. C.; McNeil M. R.; Ridell M.; Simpson K. E.; Glushka J.; van Halbeek H.; Brennan P. J.; Minnikin D. E. (1992) Structural Elucidation of a Novel Family of Acyltrehaloses from Mycobacterium tuberculosis. Biochemistry 31, 9832. 10.1021/bi00155a040. [DOI] [PubMed] [Google Scholar]
- Daffé M.; Lacave C.; Lanéelle M.-A.; Gillois M.; Lanéelle G. (1988) Polyphthienoyl trehalose, glycolipids for virulent strains of the tubercle bacillus. Eur. J. Biochem. 172, 579. 10.1111/j.1432-1033.1988.tb13928.x. [DOI] [PubMed] [Google Scholar]
- Martín-Casabona N.; Gonzalez Fuente T.; Papa F.; Rosselló Urgell J.; Vidal Plá R.; Codina Grau G.; Ruiz Camps I. (1992) Time Course of Anti-SL-IV Immunoglobin G Antibodies in Patients with Tuberculosis-Associated AIDS. J. Clin. Microbiol. 30 (5), 1089. 10.1128/JCM.30.5.1089-1093.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa F.; Luquin M.; David H. L. (1992) DOT-ELISA for detection of phenolic glycolipid PGL-Tb1 and diacyl-trehalose antigens of Mycobacterium tuberculosis. Res. Microbiol. 143, 327. 10.1016/0923-2508(92)90024-I. [DOI] [PubMed] [Google Scholar]
- Tórtola M. T.; Lanéelle M. A.; Martín-Casabona N. (1996) Comparison of Two 2,3-Diacyl Trehalose Antigens from Mycobacterium tuberculosis and Mycobacterium fortuitum for Serology in Tuberculosis Patients. Clin. Diagn. Lab. Immunol. 3 (5), 563. 10.1128/CDLI.3.5.563-566.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz M.; Lanéelle M.-A.; Luquin M.; Torrelles J.; Julián E.; Ausina V.; Daffé M. (1997) Occurrence of an antigenic triacyl trehalose in clinical isolated and reference strains of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 157, 251. 10.1111/j.1574-6968.1997.tb12781.x. [DOI] [PubMed] [Google Scholar]
- Julián E.; Cama M.; Martínez P.; Luquin M. (2001) An ELISA for five glycolipids from the cell wall of Mycobacterium tuberculosis: Tween 20 interference in the assay. J. Immunol. Methods 251, 21. 10.1016/S0022-1759(01)00313-1. [DOI] [PubMed] [Google Scholar]
- Dubey V. S.; Sirakova P. E.; Kolattukudy P. E. (2002) Disruption of msl3 abolishes the synthesis of mycolipanoic and mycolipenic acids required for polyacyltrehalose synthesis in Mycobacterium tuberculosis H37Rv and causes cell aggregation. Mol. Microbiol. 45 (2), 1451. 10.1046/j.1365-2958.2002.03119.x. [DOI] [PubMed] [Google Scholar]
- Gonzalo-Asensio J.; Mostowy S.; Harders-Westerveen J.; Huygen K.; Hernandez-Pando R.; Thole J.; Behr M.; Gicquel B.; Martin C. PhoP (2008) A Missing Piece in the Intricate Puzzle of Mycobacterium tuberculosis Virulence. PLoS One 3 (10), e3496 10.1371/journal.pone.0003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzios S. K.; Schelle M. W.; Holsclaw C. M.; Behrens C. R.; Botyanszki Z.; Lin F. L.; Carlson B. L.; Kumar P.; Leary J. A.; Bertozzi C. R. (2009) PapA3 is an Acyltransferase Required for Polyacyltrehalose Biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem. 284 (19), 12745. 10.1074/jbc.M809088200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J. E.; Ramirez A. S.; Salas L. P.; Helguera-Repetto C.; Gonzalez-y-Merchand J.; Soto C. Y.; Hernandez-Pando R. (2013) Transcription of Genes Involved in Sulfolipid and Polyacyltrehalose Biosynthesis of Mycobacterium tuberculosis in Experimental Latent Tuberculosis Infection. PLoS One 8 (3), e58378 10.1371/journal.pone.0058378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra R.; Segura E.; Leyva R.; Esparza L. A.; Lopez-Marin L. M. (2001) Mycobacterial Di-O-Acyl-Trehalose Inhibits Mitogen- and Antigen-Induced Proliferation of Murine T Cells In Vitro. Clin. Diagn. Lab. Immunol. 8 (6), 1081. 10.1128/CDLI.8.6.1-91-1088.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau C.; Neyrolles O.; Bordat Y.; Giroux S.; Sirakova T. D.; Prevost M.-C.; Kolattukudy P. E.; Gicquel B.; Jackson M. (2003) Deficiency in mycolipenate- and mycosanoate-derived acyltrehaloses enhances early interactions of Mycobacterium tuberculosis with host cells. Cell. Microbiol. 5 (6), 405. 10.1046/j.1462-5822.2003.00289.x. [DOI] [PubMed] [Google Scholar]
- Gonzalo Asensio J.; Maia C.; Ferrer N. L.; Barilone N.; Laval F.; Soto C. Y.; Winter N.; Daffe M.; Gicquel B.; Martin C.; Jackson M.; et al. (2006) The Virulence-associated Two-component PhoP-PhoR System Controls the Biosynthesis of Polyketide-derived Lipids in Mycobacterium tuberculosis. J. Biol. Chem. 281 (3), 1313. 10.1074/jbc.C500388200. [DOI] [PubMed] [Google Scholar]
- Walters S. B.; Dubnau E.; Kolesnikova I.; Laval F.; Daffe M.; Smith I. (2006) The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol. Microbiol. 60 (2), 312. 10.1111/j.1365-2958.2006.05102.x. [DOI] [PubMed] [Google Scholar]
- Ishikawa E.; Ishikawa T.; Morita Y. S.; Toyonaga K.; Yamada H.; Takeuchi O.; Kinoshita T.; Akira S.; Yoshikai Y.; Yamasaki S. (2009) Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J. Exp. Med. 206 (13), 2879. 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. J. (2017) Sensing Lipids with Mincle: Structure and Function. Front. Immunol. 8, 1662. 10.3389/fimmu.2017.01662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braganza C. D.; Teunissen T.; Timmer M. S. M.; Stocker B. L. (2018) Identification and Biological Activity of Synthetic Macrophage Inducible C-Type Lectin Ligands. Front. Immunol. 8, 1940. 10.3389/fimmu.2017.01940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decout A.; Silva-Gomes S.; Drocourt D.; Barbe S.; Andre I.; Cueto F. J.; Lioux T.; Sancho D.; Perouzel E.; et al. (2017) Rational design of adjuvants targeting the C-type lectin Mincle. Proc. Natl. Acad. Sci. U. S. A. 114 (10), 2675. 10.1073/pnas.1612421114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenen H.; Bodendorfer B.; Hitchens K.; Manzanero S.; Werninghaus K.; Nimmerjahn F.; Agger E. M.; Stenger S.; Andersen P.; et al. (2010) Cutting edge: Mincle is essential for recognition and adjuvanticity of the mycobacterial cord factor and its synthetic analog trehalose-dibehenate. J. Immunol. 184 (6), 2756. 10.4049/jimmunol.0904013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiard J.; Collmann A.; Gilleron M.; Mori L.; De Libero G.; Prandi J.; Puzo G. (2008) Synthesis of Diacylated Trehalose Sulfates: Candidates for a Tuberculosis Vaccine. Angew. Chem., Int. Ed. 47 (50), 9734. 10.1002/anie.200803835. [DOI] [PubMed] [Google Scholar]
- Wu C.-H.; Wang C.-C. (2014) Strategies for desymmetrising trehalose to synthesise trehalose glycolipids. Org. Biomol. Chem. 12 (30), 5558. 10.1039/C4OB00587B. [DOI] [PubMed] [Google Scholar]
- ter Horst B.; van Wermeskerken J.; Feringa B. L.; Minnaard A. J. (2010) Catalytic Asymmetric Synthesis of Mycolipenic and Mycolipanolic Acid. Eur. J. Org. Chem. 2010, 38. 10.1002/ejoc.200901120. [DOI] [Google Scholar]
- Nagata M.; Izumi Y.; Ishikawa E.; Kiyotake R.; Doi R.; Iwai S.; Omahdi Z.; Yamaji T.; Miyamoto T.; et al. (2017) Intracellular metabolite beta-glucosylceramide is an endogenous Mincle ligand possessing immunostimulatory activity. Proc. Natl. Acad. Sci. U. S. A. 114 (16), E3285 10.1073/pnas.1618133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.