Abstract
3D cell culture models have been developed to better mimic the physiological environments that exist in human diseases. As such, these models are advantageous over traditional 2D cultures for screening drug compounds. However, the practicalities of transitioning from 2D to 3D drug treatment studies pose challenges with respect to analysis methods. Patient-derived tumor organoids (PDTOs) possess unique features given their heterogeneity in size, shape, and growth patterns. A detailed assessment of the length scale at which PDTOs should be evaluated (i.e., individual cell or organoid-level analysis) has not been done to our knowledge. Therefore, using dynamic confocal live cell imaging and data analysis methods we examined tumor cell growth rates and drug response behaviors in colorectal cancer (CRC) PDTOs. High-resolution imaging of H2B-GFP-labeled organoids with DRAQ7 vital dye permitted tracking of cellular changes, such as cell birth and death events, in individual organoids. From these same images, we measured morphological features of the 3D objects, including volume, sphericity, and ellipticity. Sphericity and ellipticity were used to evaluate intra- and interpatient tumor organoid heterogeneity. We found a strong correlation between organoid live cell number and volume. Linear growth rate calculations based on volume or live cell counts were used to determine differential responses to therapeutic interventions. We showed that this approach can detect different types of drug effects (cytotoxic vs cytostatic) in PDTO cultures. Overall, our imaging-based quantification workflow results in multiple parameters that can provide patient- and drug-specific information for screening applications.
Keywords: 3D patient-derived tumor organoids, confocal imaging, image analysis, drug screening
Introduction
Diverse in vitro model systems have been developed to study biological phenomena and human disease conditions. Nevertheless, deciding which model system is appropriate to use is a challenging task and often guided by the following criteria: experimental purpose, physiological relevance, reproducibility, and cost. 2D monolayer cell culture has been used for decades because it is easy to maintain and can expand with little cost.1 Most preclinical drug screens rely on this model. However, the current success rate of clinical trials with candidate compounds is extremely low primarily due to toxicity and lack of efficacy.2 One of the main reasons for this poor clinical translation is that 2D models do not mimic in vivo conditions.3
3D cell culture models, such as spheroids and organoids, were developed to better mimic the spatial and microenvironmental information of the in vivo situation. Organoids can be established from embryonic stem cells, adult stem cells, or induced pluripotent stem cells (iPSCs).4–6 Patient-derived tumor organoids (PDTOs), the focus of this work, can be grown directly from patient tissue biopsies or surgically removed tumor tissues. Organoids can form organ-like structures reminiscent of the tissues they originated from and contain both stem and differentiated cell populations resembling patient tissues.7–9 PDTOs can be manipulated in culture to express reporter genes using lentiviral transduction and knock-in or knockout genes of interest with CRISPR-CAS9 techniques.10–12 Organoid cultures can be expanded and stored as a patient-specific biobank and accessed with other patient information from the clinical database.5 These advantages make PDTOs a strong alternative model for drug screening.13,14 Despite the excitement and promise of this model system, significant work remains to establish standard phenotypic analysis methods for interrogation.
Quantitative imaging of organoids provides a window into cellular dynamics within a 3D microenvironment and may offer useful information for drug screening. Taking into consideration the additional challenges with PDTOs over other 3D cultures such as spheroids (i.e., sample thickness, matrix embedding, size heterogeneity, and multiple objects per well), it is critical to understand which features are important to quantify to determine organoid growth and drug response. Several imaging-based studies have measured cell viability and morphology changes with drug treatments in 3D spheroid cultures.15–17 However, to our knowledge, a side-by-side comparison of cell- and organoid-level features from temporal analysis of the same PDTO object has not been done before. To perform such a thorough evaluation, 4D imaging is required with dynamic, multiple z-level scanning, and volumetric reconstruction. Recent advances in high-content imaging systems make this complicated imaging of 3D models possible.13 Our lab, among others, is in the process of developing and standardizing imaging and data analysis pipelines for PDTOs. Here we highlight a multiplexed imaging-based organoid workflow, which draws correlations between multiple parameters and examines PDTO growth dynamics and drug-induced changes that will help build efficient screening workflows for drug discovery in the future.
Materials and Methods
Patient Tissue Processing and Organoid Cultures
Tumor tissue resections from colorectal cancer (CRC) patients were received from the USC Norris Comprehensive Cancer Center following institutional review board (IRB) approval and patient consent. PDTOs were generated following the procedures described previously.18–20 Briefly, tumor tissue was washed with 1× phosphate-buffered saline (PBS), cut into small pieces, and minced using a scalpel before digestion. Tissue was digested in a solution containing 1.5 mg/mL collagenase (Millipore, Burlington, MA; 234155), 20 µg/mL hyaluronidase (MP Biomedicals, Irvine, CA; 100740), and 10 µM Y27632 (Sigma, St. Louis, MO; Y0503) at 37 °C for 30 min. The digested tissue solution was filtered with a 100 µm cell strainer and centrifuged at 900 rpm for 5 min. The cell pellet was washed with DMEM/F12 (Thermo Fisher, Waltham, MA; 11320033) + 10% fetal bovine serum (FBS; Gemini 100-500, Gemini Bio, West Sacramento, CA) three times and resuspended in Cultrex Reduced Growth Factor Basement Membrane Matrix Type 2 (BME; Trevigen, Gaithersburg, MD; 3533-001-02). Sixty microliters of cells and BME mixture was put into each well of a 24-well plate and incubated upside down at 37 °C for 15 min until the mixture solidified as a dome structure.20 Five hundred microliters of organoid growth media (Advanced DMEM/F12 with 10% FBS, 1% penicillin/streptomycin, 1% Glutamax, and 1% HEPES) supplemented with 1× N2 (Sigma Aldrich; 17502048), 1× B-27 (Sigma Aldrich; 17504044), 1 mM N-acetylcysteine (Sigma Aldrich; A7250), 50 ng/mL EGF (Life Technologies, Rockville, MD; PGH 0313), 100 ng/mL Noggin (Tonbo, San Diego, CA; 21-7075-U500), 10 mM nicotinamide (Sigma; N0636), 500 nM A-83-01 (Millipore; 616454-2MG), 10 µM SB202190 (Sigma; 47067), and 0.01 µM PGE2 (Sigma Aldrich; P5640) was added to each well covering the BME dome and incubated at 37 °C/5% CO2. For passaging, organoids/BME domes were scraped and dissociated in 500 µL of gentle cell dissociation reagent (STEMCELL Technology, Cambridge, MA; 07174) by incubating at 4 °C for 15 min on a rocker in 15 mL conical tubes. Organoid suspensions were centrifuged at 900 rpm for 5 min and the cell pellet was resuspended in BME and plated in a 24-well plate following the same procedures as described above.
Organoid Labeling with Lentivirus-H2B-GFP and FACS
After removing the culture media, 500 µL of TrypLE (Thermo Fisher; 12605028) solution was added to each well to digest the BME gel by incubating at 37 °C for 10 min. Organoids/BME were scraped into a new 15 mL conical tube and centrifuged at 1400 rpm for 5 min. One milliliter of TrypLE was added to the pellet and incubated at 37 °C for 5 min. After centrifuging at 1400 rpm for 5 min, 2 mL of organoid growth media containing 5 µg/mL polybrene (Sigma; TR-1003-G) was added to resuspend the pellet. Five microliters (40 multiplicity of infection [MOI]) of Lentivirus H2B-GFP (Sigma; 1710229) was added to the organoid suspension and incubated at 37 °C for 30 min. It was then centrifuged at 1400 rpm for 5 min, and both pellet and supernatant were collected. The pellet was resuspended in BME and the organoids/BME were seeded in a 24-well plate. The collected media supernatant was added to the organoids/BME dome to further transduce organoids. The next day, the media was changed with fresh organoid growth media. GFP-positive organoids were confirmed by imaging on an inverted fluorescence microscope (Zeiss, Cambridge, MA; Axio Observer.Z1). GFP-labeled organoids were expanded for fluorescence-activated cell sorting (FACS) analysis. Twenty-four wells of organoids/BME were washed with PBS. Five hundred microliters of TrypLE was added to each well and organoids/BME were harvested by scraping with pipettes. Organoid solution was transferred to a new 15 mL conical tube and incubated at 37 °C for 45 min to dissociate organoids completely to single cells. After digestion, organoid solution was centrifuged at 1400 rpm for 5 min. Pellet was resuspended in 2 mL of PBS. The dissociated organoid solution was filtered with a 100 µm cell strainer. FACS was performed using the ARIA IIu (BD Biosciences, San Diego, CA) to collect GFP-positive single cells. The sorted cells were transferred to a new 15 mL conical tube and centrifuged at 900 rpm for 5 min to remove supernatant. BME was added to the pellet (cell volume/BME volume = 1:100) and the resuspended cells/BME were seeded into a 24-well plate. Organoid growth media was added after the BME was solidified. Growth of GFP-labeled organoids was monitored using an inverted fluorescence microscope.
Drug Treatments
Organoids were dissociated as described in the previous section. Ten microliters of dissociated cells/BME (1000 cells/µL) was seeded in each well of a 96-well plate (Corning Costar, Oneonta, NY; 3904). After 4 days of culturing, organoids were treated with different doses of staurosporine (ST; Sigma-Aldrich; 569396), irinotecan (IR; Sigma-Aldrich; I1406), 5-fluorouracil (5-FU; Selleck Chemicals, Houston, TX; S1209), or SN-38 (Sigma; 7-ethyl-10-hydroxycamtothecin, H0165). DRAQ7 (Abcam, Cambridge, MA; ab109202) was added to each well at 5 µM final concentration 1 h prior to imaging. Drugs and dye were refreshed at 3 days after treatments.
Confocal 3D Live Cell Imaging and Quantitative Image Analysis of Organoids
Time-lapse 3D live cell imaging of H2B-GFP organoids was performed using the Olympus FV3000 laser scanning confocal microscope equipped with a TokaiHit stage-top incubation system to maintain environmental conditions (37 °C and 5% CO2). For static time point imaging, the same organoids were imaged at days 0, 1, 3, and 6 with ST, IR, and FU treatments. SN-38-treated organoids were imaged at days 0, 2, 4, and 7. Tile imaging (2 × 2) with a 10× objective was performed to image the center of each well. GFP (488 nm), DRAQ7 (640 nm), and brightfield (transmitted light) channels were captured with every 5 µm Z step for 800 µm (160 sections). Tile images were stitched to generate a single image file. Olympus image files (.oir) were converted to Imaris file format (.ims) using an Imaris file converter. The same matched area (750 × 750 µm) was cropped and each time point file was added to generate a single time-lapse imaging file. In Imaris, surface rendering was used to detect all organoids and calculate volume based on how much 3D space an organoid object occupies (Organoid surface). Live cell spots were made from H2B-GFP signals to locate individual cell nuclei. Dead cell spots were generated with DRAQ7-stained cells. Colocalization spots were made to calculate the number of true live cells in organoids by subtracting the number of colocalized spots from the GFP-only counts. Organoid surface and spot information was imported to Imaris cell module to track individual organoids with live and dead spot numbers. Organoid 3D volume, surface area, and live and dead cell numbers in tracked organoids were exported from Imaris as Excel data sheets. Measurements of organoid sphericity and two different ellipticities, oblate and prolate, were also exported as morphological features.
Data Processing and Visualization
Data were imported into and processed in the R statistical environment (v3.6.0)21 using the tidyverse package (v1.2.1).22 As part of quality control, organoid track IDs were filtered based on completeness of individual tracking—where organoids with missing time point information were omitted from subsequent analyses. This resulted in minimal loss of data. Second, a size filter was imposed where organoids greater than 2000 cells or less than 50 cells at initial time points per condition were removed from subsequent analyses. This was done in order to maintain reasonable consistency in the overall behavior of the organoids, as organoids that were too big or too small may have differences in their biology (refer to Fig. 2 ). Correlations were assessed using Spearman’s rho (ρ) wherever applicable.
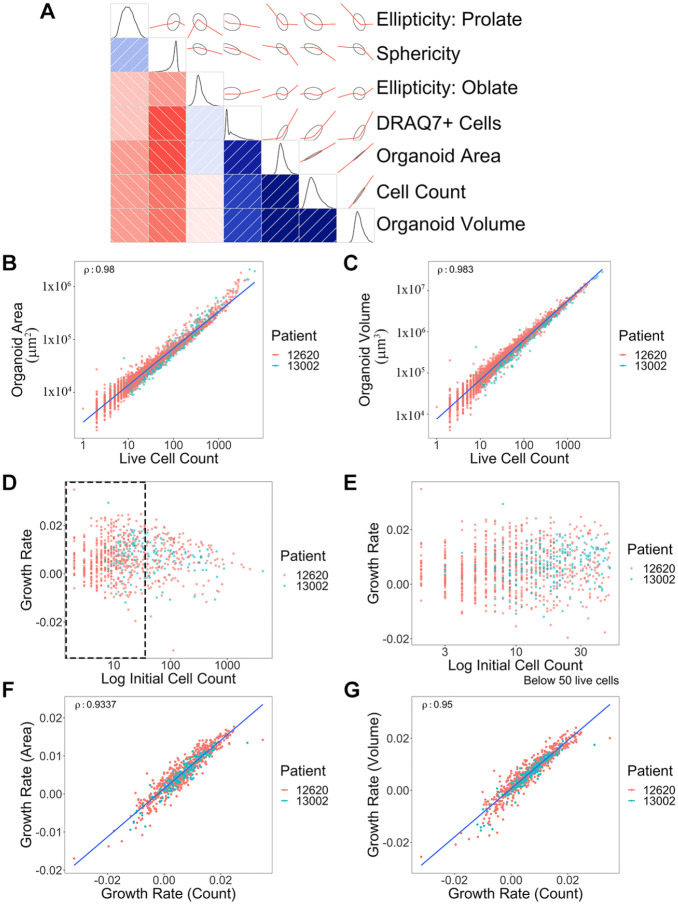
Figure 2.
Correlation of multiple measurements from 3D organoid imaging and growth rate calculations. (A) Correlogram was generated using multiple metrics from 3D organoid image analysis. Organoid morphological and cell features are presented as schematic drawings and overlaid with correlation graphs (red) between the intersection of two parameters. The Pearson correlation of seven metrics used to gauge cell growth and morphology is shown, on the lower triangular half of the diagram—those metrics labeled at the right. The intensity of the red and blue colors represents the strength of negative and positive correlations, respectively, with the directionality and patterns displayed on the opposite side of the diagonal (positive, +40° angle/blue; negative, –40° angle/red), with ellipses surrounding the red lines indicating confidence. Finally, density plots (black) along the diagonal depict the distribution of the data for each metric. (B) Correlation graph of organoid surface area and live cell numbers. Blue line shows a trend. ρ = 0.980. Each dot represents a single organoid. Each patient is distinguished by different color dots (red: 12620; green: 13002). y axis, µm2. (C) Correlation between organoid volume and live cell numbers. ρ = 0.983. y axis, µm3. (D) Distribution of growth rates based on initial organoid sizes. (E) Zoomed-in view of the size distribution graph (black dotted rectangle area in D) based on organoid sizes between 0 to 50 cells. (F) Comparison between area-based growth rate and live cell number-based growth rate. ρ = 0.934. Growth rate was calculated by linear model of log10(live cell or area) ~ time. (G) Comparison between volume-based growth rate and live cell number-based growth rate. ρ = 0.950. Volume growth rate was calculated by linear model of log10(volume) ~ time. Correlations were shown using Spearman’s rho (ρ) value. A total of 826 organoids across two different patients were analyzed.
Growth rates were calculated using three metrics: live cell count, organoid volume, and surface area. In all three instances, a linear model was fit per organoid in the R statistical environment (v3.6.0) using the natural logarithm of one of the three metrics as the response variable, and modeling that as a function of time. The slope of the fitted line was used as the growth rate of the organoid. Data visualization was conducted in the R statistical environment (v3.6.0) using the ggplot2 package (v3.2.1)23 and corrgram package (v1.13; https://CRAN.R-project.org/package=corrgram).24,25
To assess differences in growth rates between drug-treated organoids and control, a one-sided Dunn’s test for multiple comparison using Kruskal–Wallis was employed,26 using the FSA package (v0.8.27).27 The same approach was used to assess differences in live and dead cells between drug-treated groups and control. To assess differences in morphological features, a similar method was used, via a two-sided Dunn’s test. A Mann–Whitney U test was used, where applicable, for pairwise comparisons. All p values were adjusted for multiple testing using a false discovery rate of 5%.28 All the adjusted p values used to claim significant changes are provided as a supplemental Excel file (Suppl. Data S1).
Results
Establishment of Patient-Derived Organoid Imaging and Analysis Workflows
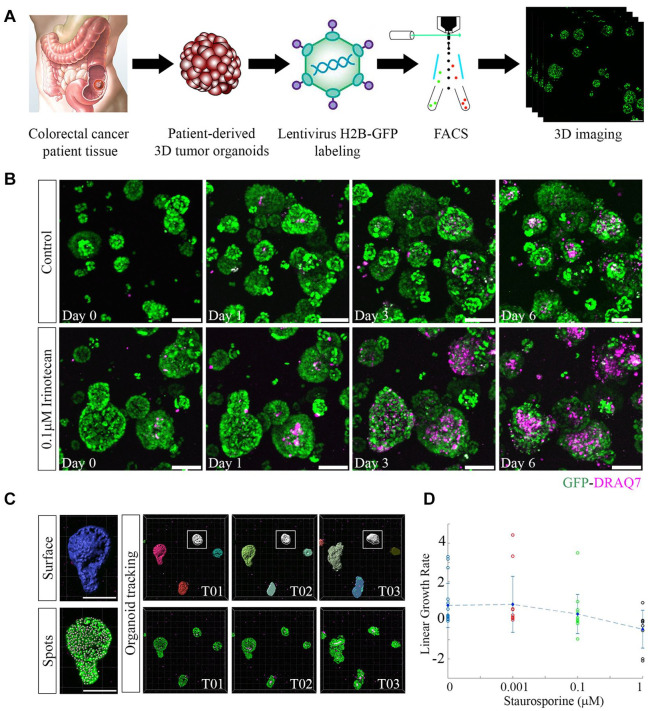
3D PDTOs were established from tumor tissues surgically removed from two different CRC patients (13002: primary colon, stage II-B; 12620: liver metastasis, stage IV-A). Dissociated single cells from the organoids were labeled with H2B-GFP lentivirus and then subjected to FACS to collect pure GFP-labeled cell populations ( Fig. 1A ). Organoids were imaged with multiple Z sections during drug treatments. H2B-GFP-labeled cell nuclei enabled monitoring of cell-level changes such as cell division and migration events (Suppl. Movie S1). DRAQ7 vital dye was added to the organoid cultures to detect dead cell nuclei. For example, drug-treated (0.1 µM IR) organoids showed increased DRAQ7+ dead cells over time compared with untreated control organoids ( Fig. 1B ). Surface and spot rendering visualized organoid- and cell-level regions from the same object, respectively ( Fig. 1C ). This process allows simultaneous measurements of 3D morphological features and cell numbers (live/dead) from individual patient organoids. Multi-time-point 3D confocal imaging data sets were combined as a single time-lapse imaging file to track organoids and cells over time using Imaris software ( Fig. 1C ). Linear growth rate curves based on distinct organoid features were generated and used to examine dose-dependent drug responses with treatment (e.g., ST) ( Fig. 1D ). Automatic spot detection was compared with manual spot identification using GFP fluorescent signal thresholds. A strong correlation was established between the two methods signifying the accuracy of our automated image analysis pipeline (ρ = 0.9801) (Suppl. Fig. S1).
Figure 1.
3D imaging of H2B-GFP-labeled organoids provides high-content information. (A) 3D tumor organoids were generated from patient tissues and transduced with H2B-GFP lentivirus to label individual cell nuclei. Transduced organoids were dissociated and sorted to collect a pure population of fluorescently labeled cells. Regrown H2B-GFP-labeled organoids were imaged with multiple z stacks using a confocal laser scanning microscope. Scale bar, 100 µm. (B) Static time point imaging of control (untreated) and 0.1 µM IR-treated GFP-labeled organoids. Images were captured at multiple time points (days 0, 1, 3, and 6). DRAQ7 vital dye was used to label dead cell nuclei in 3D organoids (purple). DRAQ7+ cells increased over time with the treatment. Scale bar, 100 µm. (C) Organoid surface and individual cell spot detection for each organoid was determined using Imaris software based on GFP intensity. Scale bar, 150 µm. Single organoid tracking occurred over multiple time points (white box, T01-T03). (D) Organoid linear growth rate was calculated using different parameters (live cell count, organoid volume, or organoid surface area) for multiple doses of ST treatments. Each circle denotes an individual organoid and the circle colors indicate different treatment doses. The blue diamond represents the mean growth rate and the vertical lines are the standard deviations.
Multiparametric Analysis to Assess Organoid Growth Rate
Given our 3D imaging and data analysis pipeline, which simultaneously measures multiple features per organoid in a dynamic fashion, we can compare each measurement parameter in parallel. Two PDTOs were treated with three different drugs: ST (0.0001–1 µM), IR (0.01–50 µM), and 5-FU (0.1–100 µM). Organoids were imaged at four different time points before (day 0) and after (days 1, 3, and 6) drug treatments. All quantified features including organoid volume, surface area, live cell numbers, dead cell numbers, sphericity, ellipticity–oblate, and ellipticity–prolate from both untreated control and drug-treated PDTOs were used to make a correlogram ( Fig. 2A ). We found that four parameters, volume, surface area, live cell counts, and dead cell counts, are positively correlated. The first three parameters represent organoid growth. DRAQ7+ dead cells also increase as organoids become large due to an increase in dead cells inside the necrotic region of large organoids ( Fig. 2A ). While prolate and sphericity were reduced as organoids grow and differentiate forming branch-shaped organoids, oblate was increased because organoids become large horizontally.
It is important to determine how best to assess organoid growth and drug response using specific measurements from 3D imaging. To identify the most efficient parameter, organoid volume, organoid surface area, and live cell counts (from individual organoids) were compared and found to have very strong correlations between them ( Fig. 2B,C ). Organoid volume was the parameter most correlated with live cell number (ρ = 0.983), but surface area also showed a high correlation, except toward the high cell number range (ρ = 0.980). This suggests that for large size organoids, volume is the more accurate parameter to measure. These correlations are maintained across control and drug-treated conditions (Suppl. Fig. S2). There was no difference between the two different patient organoid populations (13002 vs 12620).
PDTOs vary in size and shape. To evaluate whether the initial size of organoids impacts growth, we determined growth rates from tracked organoids with different initial starting cell numbers; however, no clear relationship was observed for the 2 PDTOs we tested ( Fig. 2D ). Nevertheless, small size organoids (<50 cells) did show more variation in growth rate values, suggesting that there is a threshold of organoid size that needs to be considered in 3D imaging analysis ( Fig. 2E ). This could be the result of incomplete organoid surface detection in Imaris caused by inadequate GFP signal detection in small organoids. Due to this heterogeneity, we filtered based on size and only used organoids with initial cell numbers ranging from 50 to 2000 to generate organoid growth rate curves. Both volume- and area-based growth rates are highly comparable to live cell number-based growth rate calculations, although the volume-based growth rate had a slightly better correlation (area, ρ = 0.934 vs volume, ρ = 0.950) ( Fig. 2F,G ).
Detecting Drug-Specific Changes in Organoid Models
Anticancer drugs target tumor cells via different mechanisms of action. There are two major drug-induced cell behavior classes, those that stimulate a cytotoxic response leading to cell death and compromised 3D structures versus those that are more cytostatic and inhibit or delay cell cycle progression, resulting in a growth-inhibitory response.29,30 For in vitro drug studies, it is important to be able to distinguish between these two different cellular responses.
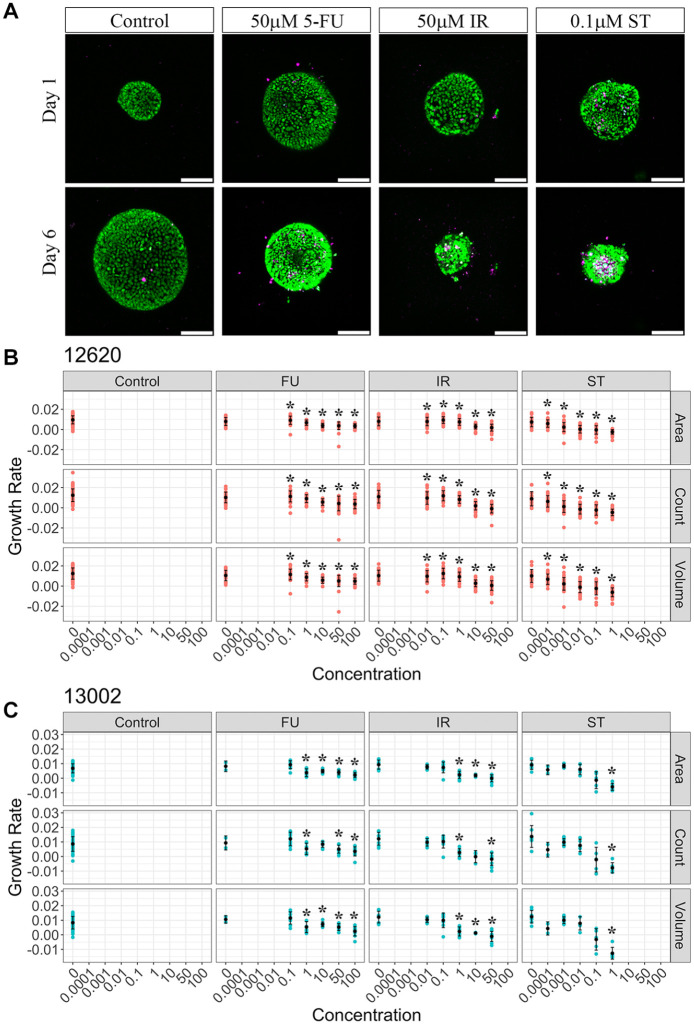
To examine drug-specific changes in patient organoids, the organoids were treated with the three different drugs mentioned above. ST is a potent protein kinase C inhibitor that rapidly kills cells31 and was used as a positive control. Two clinically available CRC drugs, IR and 5-FU, were also used to establish dose–response curves in patient organoids. IR is metabolized into SN-38, an inhibitor of topoisomerase I, and blocks DNA replication and transcription.32 5-FU, an antimetabolite drug, inhibits thymidylate synthase and prevents nucleotide synthesis.33 The organoids were imaged and tracked across multiple time points during drug treatments ( Fig. 3A ). Control (i.e., untreated) organoids continue to increase in cell number and size over the duration of the experiments ( Fig. 3A ). The organoids of patient 12620 showed a higher growth rate (mean growth rate ± standard error, 0.0115 ± 0.000263) than those of patient 13002 (mean growth rate ± standard error, 0.00793 ± 0.000407) in the control group, suggesting that the linear growth rates from multiple parameters can visualize interpatient growth dynamic differences. This can also be seen in the live cell counts of individual organoids where larger (i.e., higher live cell counts) organoids were found from patient 12620 compared with patient 13002 ( Fig. 3B,C , Suppl. Fig. S3). It is important to note that the doubling time of cells within 3D organoids is on the order of 3.5–5.25 days (based on average 12620 and 13002 growth rates, respectively) compared with 2D cultures, where cells generally divide at a rapid rate every 1–2 days. Visual examination of 5-FU treatment did not result in a substantial reduction in live cell numbers, yet the organoids did become a more compact spherical shape ( Fig. 3A ). IR-treated organoids showed a dramatic decrease in live cell number with a corresponding increase in the number of DRAQ7-labeled dead cells. Organoid size was also reduced significantly with IR ( Fig. 3A ). Organoids treated with ST displayed similar patterns to IR-treated organoids ( Fig. 3A ). To measure organoid drug responses, organoid surface area, live cell count, and volume-based growth rates were compared for different drug treatment conditions. All three parameters (surface area, live cell count, and volume) showed very similar drug dose–response curves ( Fig. 3B,C ). Organoid linear growth rate was largely decreased with ST and IR treatments in a dose-dependent manner, with a negative growth rate measured at the higher drug concentrations. However, 5-FU showed a less significant reduction in growth rate that, on average, remained a positive value across the drug doses. Given IR is a prodrug, we also tested its active metabolite, SN-38, which showed more DRAQ7+ dead cells and reduced growth rates at lower dose ranges (0.01–10 µM) than IR (0.1–50 µM) (Suppl. Figs. S4 and S5). This suggests that ST, IR, and SN-38 are cytotoxic drugs causing rapid cell death in organoids, while 5-FU appears to be more cytostatic in these specific PDTOs, inhibiting organoid growth but not inducing large amounts of cell death at the concentrations and time duration treated in vitro ( Figs. 3B and C , Suppl. Fig. S3). 5-FU has been shown to have both cytostatic and cytotoxic effects that are dependent on drug concentration and treatment time.34,35 Further exploration is needed to examine whether 5-FU response may vary across PDTOs. Notably, linear growth rate analysis based on quantitative organoid temporal tracking was necessary because measurements of each parameter (area, live cell count, and volume) at a single time point failed to show drug dose responses on organoids (Suppl. Fig. S6). This is most likely a result of intrapatient organoid size and growth rate heterogeneity.
Figure 3.
Measurements of organoid drug response with anticancer drugs. (A) 3D organoid (patient 12620) images showing drug responses at two different time points (days 1 and 6). Representative images from control (no treatment), 50 µM 5-FU-treated, 50 µM IR-treated, and 0.1 µM ST-treated organoid groups. H2B-GFP (green), DRAQ7 (purple); scale bar, 100 µm. (B) Area, live cell count, and volume-based growth rates of patient 12620 organoids in different drug treatment groups (control, 5-FU, IR, and ST). Each red dot represents a single organoid. Mean values and standard deviations were labeled with black dots and lines, respectively. Drug dose was shown as micromolar concentration on the x axis. Significance (*p < 0.05) was indicated with asterisks above each drug concentration group compared with controls (untreated, 0 µM). (C) Area, live cell count, and volume-based growth rates of patient 13002 organoids in different drug treatment groups (control, 5-FU, IR, and ST). Each blue dot represents a single organoid. Mean values and standard deviations were labeled with black dots and lines, respectively. Significance (*p < 0.05) was indicated with asterisks above each drug concentration group compared with controls.
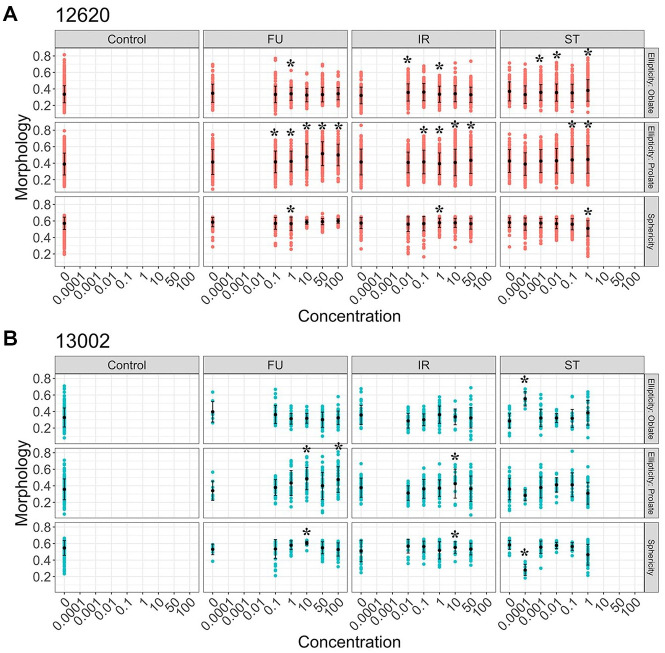
For some drugs such as 5-FU that show marginal growth rate effects, we wanted to explore other parameters, such as 3D morphological changes, that may be early indicators of drug response rather than live and dead cell counts. Our individual organoid tracking pipeline includes the morphological parameters sphericity and ellipticity. When we compared morphology changes within each drug treatment group, 5-FU induced significant changes in organoid prolate ellipticity, suggesting organoid shapes are stretched in the vertical direction with increasing drug concentrations ( Fig. 4 ). The feature mean value of organoid sphericity was also higher in the 5-FU condition compared with the control, although this change was not significant. Both IR- and ST-treated organoids showed similar trends in sphericity to the 5-FU group due to organoid size changes as a result of active cell killing with drugs ( Figs. 3A and 4 ). High-dose (1 µM) ST treatment showed a reversal of the sphericity value because completely dead organoids lose their structure and all the remaining cells spread out, resulting in a less spherical shape ( Figs. 3A and 4 ). PDTOs (12620 and 13002) respond to 5-FU more dynamically by changing morphological readouts rather than live/dead cell counts ( Fig. 4 , Suppl. Fig. S3).
Figure 4.
Drug-specific changes of organoid morphology measurements. Three different morphological measurements, ellipticity–oblate, ellipticity–prolate, and sphericity, are shown for multiple drug treatments at different doses. (A) Morphology changes of 12620 organoids with drug treatments. Each red dot represents a single organoid. All the morphological features were scaled from 0 to 1. Mean value and standard deviation were shown with black dots and lines, respectively. Asterisks indicate significant changes compared with controls (*p < 0.05). (B) Morphology changes in 13002 organoids with drug treatments. Mean value and standard deviation were shown with black dots and lines, respectively. Asterisks show significant changes compared with controls (*p < 0.05).
Discussion
High-content, high-throughput imaging can play an important role in drug discovery. Such imaging-based screens need to be optimized by simplifying unnecessary processes and removing superfluous information. However, it is difficult to determine what is unnecessary without performing pilot experiments to explore various readouts, which was the focus of the work described herein. Moreover, the imaging and data analysis pipelines need to be scalable to enable screens of many chemical compounds. In recent years, significant effort has been placed in instrument and software development to facilitate this throughput.36–38
When incorporating complex multicellular model systems into research investigations, such as 3D spheroids and PDTOs, quantitative multiplexed measurements are key to understanding dynamic growth and drug response readouts. Spheroid cultures are a prevalent in vitro model system used for high-throughput drug screens.39–41 Spheroid size and morphology changes have been measured to determine drug responses previously.42,43 Individual live or dead cells have been analyzed in fixed spheroid cultures using immunostaining techniques for apoptotic markers, such as caspase-3/7 antibodies,15 and in live spheroid cultures using vital dyes, such as EthD-1 and propidium iodide. Although these approaches can be used to either analyze 3D morphological changes or detail cell-level information in spheroids, to our knowledge, there are no comparative studies to identify optimal analysis parameters in 3D PDTO imaging to understand dynamic drug responses. Recently, Karolak et al. measured organoid growth dynamics with surface area and morphology, but this study was purely based on mathematical modeling and in silico simulations.44 We simultaneously compared multiple parameters in PDTOs and found strong correlations among 3D phenotypic measurements of volume, surface area, and live cell counts. While this correlation was maintained for the two patients and drug compounds tested in this study, there may be situations where one parameter could significantly outperform the others. Of note, we found no correlation between organoid initial size (based on cell count) and growth rate. This may be the result of cellular heterogeneity (i.e., stem vs differentiated cell types) between individual organoids but further analysis is required. Additionally, certain research questions may warrant a specific parameter to be measured a priori. For example, if one is interested in tracking the emergence or outgrowth of drug-resistant clones, it will be important to quantify individual cell counts.
Automation of this 3D PDTO imaging pipeline with liquid handling and robotics, as established by other research groups,45,46 will improve the imaging efficiency with multiple patient samples and drug compound libraries. Unlike most spheroid cultures, organoids are grown in 3D extracellular matrices (e.g., BME gel). Automatic dispensing of BME gel together with cells/organoids into multiwell plates is challenging due to temperature control and viscosity. Francies et al. established an automatic process of seeding organoids in 96-well plates to perform subsequent cell viability assays, but the organoids were layered on top of the gel.46 To replicate the 3D environment, it is necessary to optimize the workflow for automatic seeding of organoid/BME gel mixtures to allow for complete embedding of organoids within the ECM. Moreover, implementing machine learning algorithms within our analysis pipeline to detect organoid features will significantly reduce image analysis time.47,48 Patient organoids can behave differently (i.e., growth rates and drug effects) based on their genetic and environmental backgrounds.49 We tested two different patient-derived organoids in this study and observed differential growth rates between them. However, two patients are not enough to draw any conclusions about interpatient heterogeneity. Investigating the biological significance of interpatient organoid heterogeneity will be the topic of future studies.
In vivo patient condition is complex and simple cell culture models cannot recapitulate the real disease situation. Cancer progression and treatment outcomes are often affected by microenvironmental changes including interactions with multiple stromal cell types such as endothelial cells, immune cells, and cancer-associated fibroblasts (CAFs).50–52 In addition to analyzing tumor cell or organoid-only growth rates, it is important to consider other tumor microenvironmental factors to better predict physiological therapeutic outcomes. 3D organoids can be combined with stromal cell cultures such as CAFs to measure tumor–stromal interactions. CAFs secrete a large number of growth factors and cytokines affecting tumor growth and drug resistance.53,54 CellTiter-Glo has been widely used as an assay method for drug screening.55 However, it reads metabolic ATP-level changes from entire cell populations in a single well, and it is difficult to distinguish effects from two different cell types in co-culture conditions. Although CellTiter-Glo can be used to capture patient heterogeneity in organoid models,56 combining our 3D imaging-based multiparametric analysis with microenvironmental perturbations will address drug-specific changes in more physiologically relevant heterocellular conditions.
PDTOs have many advantages compared with other biomimetic model systems.5 If we establish efficient imaging and data analysis workflows, the power of the organoid model system will increase dramatically. We can use 3D PDTO imaging to answer detailed biological questions, predict patient outcomes, and identify effective drug compounds through screening. Toward achieving these goals, we are working on establishing a faster, reliable 3D imaging and analysis process. With improved throughput, multiparametric analysis of 3D PDTOs has a strong potential to be a contender in a new drug discovery solution pipeline.
Supplemental Material
Supplemental material, Supplemental_Data1__for__Comparison_of_cell_and_organoid-level_analysis_of_patient-derived_3D_organoids for Comparison of Cell and Organoid-Level Analysis of Patient-Derived 3D Organoids to Evaluate Tumor Cell Growth Dynamics and Drug Response by Seungil Kim, Sarah Choung, Ren X. Sun, Nolan Ung, Natasha Hashemi, Emma J. Fong, Roy Lau, Erin Spiller, Jordan Gasho, Jasmine Foo and Shannon M. Mumenthaler in SLAS Discovery
Supplemental material, Supplemental_Figures_for_Comparison_of_cell_and_organoid-level_analysis_of_patient-derived_3D_organoids for Comparison of Cell and Organoid-Level Analysis of Patient-Derived 3D Organoids to Evaluate Tumor Cell Growth Dynamics and Drug Response by Seungil Kim, Sarah Choung, Ren X. Sun, Nolan Ung, Natasha Hashemi, Emma J. Fong, Roy Lau, Erin Spiller, Jordan Gasho, Jasmine Foo and Shannon M. Mumenthaler in SLAS Discovery
Acknowledgments
We would like to acknowledge Olympus for the generous instrumentation support and technical guidance as part of the USC–Olympus Innovation Partnership. We gratefully acknowledge Dr. Heinz-Josef Lenz for access to CRC patient tissue samples with USC IRB approval. We thank Chi-Li Chiu for technical advice on Imaris. Colin Flinders helped with FACS of H2B-GFP-labeled patient organoids.
Footnotes
Supplemental material is available online with this article.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: the research leading to these results received support from the NCI’s Cancer Systems Biology Consortium U01-CA-232137 grant (Multi-PI: Mumenthaler) and the Stephenson Family Fund.
ORCID iD: Emma J. Fong  https://orcid.org/0000-0001-6806-1959
https://orcid.org/0000-0001-6806-1959
References
- 1. Anton D., Burckel H., Josset E.; et al. Three-Dimensional Cell Culture: A Breakthrough In Vivo. Int. J. Mol. Sci. 2015, 16, 5517–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hay M., Thomas D. W., Craighead J. L.; et al. Clinical Development Success Rates for Investigational Drugs. Nat. Biotechnol. 2014, 32, 40–51. [DOI] [PubMed] [Google Scholar]
- 3. Edmondson R., Broglie J. J., Adcock A. F.; et al. Three-Dimensional Cell Culture Systems and Their Applications in Drug Discovery and Cell-Based Biosensors. Assay Drug Dev. Technol. 2014, 12, 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. DeRose Y. S., Gligorich K. M., Wang G.; et al. Patient-Derived Models of Human Breast Cancer: Protocols for In Vitro and In Vivo Applications in Tumor Biology and Translational Medicine. Curr. Protoc. Pharmacol. 2013, 60, 14.23.1–14.23.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Drost J., Clevers H. Organoids in Cancer Research. Nat. Rev. Cancer 2018, 18, 407–418. [DOI] [PubMed] [Google Scholar]
- 6. Young M., Reed K. R. Organoids as a Model for Colorectal Cancer. Curr. Colorectal Cancer Rep. 2016, 12, 281–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Spence J. R., Mayhew C. N., Rankin S. A.; et al. Directed Differentiation of Human Pluripotent Stem Cells into Intestinal Tissue In Vitro. Nature 2011, 470, 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim M., Mun H., Sung C. O.; et al. Patient-Derived Lung Cancer Organoids as In Vitro Cancer Models for Therapeutic Screening. Nat. Commun. 2019, 10, 3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clevers H. Modeling Development and Disease with Organoids. Cell 2016, 165, 1586–1597. [DOI] [PubMed] [Google Scholar]
- 10. Bolhaqueiro A. C. F., van Jaarsveld R. H., Ponsioen B.; et al. Live Imaging of Cell Division in 3D Stem-Cell Organoid Cultures. Methods Cell Biol. 2018, 145, 91–106. [DOI] [PubMed] [Google Scholar]
- 11. Matano M., Date S., Shimokawa M.; et al. Modeling Colorectal Cancer Using CRISPR-Cas9-Mediated Engineering of Human Intestinal Organoids. Nat. Med. 2015, 21, 256–262. [DOI] [PubMed] [Google Scholar]
- 12. Fujii M., Matano M., Nanki K.; et al. Efficient Genetic Engineering of Human Intestinal Organoids Using Electroporation. Nat. Protoc. 2015, 10, 1474–1485. [DOI] [PubMed] [Google Scholar]
- 13. Li L., LaBarbera D. V. 3D High-Content Screening of Organoids for Drug Discovery. Comp. Med. Chem. III 2017, 1–24. [Google Scholar]
- 14. Miranda C. C., Fernandes T. G., Diogo M. M.; et al. Towards Multi-Organoid Systems for Drug Screening Applications. Bioeng. 2018, 5, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sirenko O., Mitlo T., Hesley J.; et al. High-Content Assays for Characterizing the Viability and Morphology of 3D Cancer Spheroid Cultures. Assay Drug Dev. Technol. 2015, 13, 402–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zanoni M., Piccinini F., Arienti C.; et al. 3D Tumor Spheroid Models for In Vitro Therapeutic Screening: A Systematic Approach to Enhance the Biological Relevance of Data Obtained. Sci. Rep. 2016, 6, 19103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lal-Nag M., McGee L., Titus S. A.; et al. Exploring Drug Dosing Regimens In Vitro Using Real-Time 3D Spheroid Tumor Growth Assays. SLAS Discov. 2017, 22, 537–546. [DOI] [PubMed] [Google Scholar]
- 18. Sato T., Vries R. G., Snippert H. J.; et al. Single Lgr5 Stem Cells Build Crypt-Villus Structures In Vitro without a Mesenchymal Niche. Nature 2009, 459, 262–265. [DOI] [PubMed] [Google Scholar]
- 19. Boj S. F., Vonk A. M., Statia M.; et al. Forskolin-Induced Swelling in Intestinal Organoids: An In Vitro Assay for Assessing Drug Response in Cystic Fibrosis Patients. J. Vis. Exp. 2017, 120, 55159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baker L. A., Tiriac H., Tuveson D. A. Generation and Culture of Human Pancreatic Ductal Adenocarcinoma Organoids from Resected Tumor Specimens. Methods Mol. Biol. 2019, 1882, 97–115. [DOI] [PubMed] [Google Scholar]
- 21. R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. http://www.R-project.org/.
- 22. Wickham H., Averick M., Bryan J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar]
- 23. Wickham H. Ggplot2 Elegant Graphics for Data Analysis. Springer: Berlin, 2016. [Google Scholar]
- 24. Friendly M. Corrgrams: Exploratory Displays for Correlatigon Matrices. Am. Stat. 2002, 56, 316–324. [Google Scholar]
- 25. Write K. Corrgram: Plot a Correlogram. R Package Version 1.13 2018. [Google Scholar]
- 26. Dunn O. J. Multiple Comparisons Using Rank Sums. Technometrics 1964, 6, 241–252. [Google Scholar]
- 27. Ogle D. H., Wheeler P.; Dinno A. FSA: Fisheries Stock Analysis. R Package Version 0.8.27 2020. [Google Scholar]
- 28. Benjamini Y., Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar]
- 29. Shapiro G. I., Harper J. W. Anticancer Drug Targets: Cell Cycle and Checkpoint Control. J. Clin. Invest. 1999, 104, 1645–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kummar S., Gutierrez M., Doroshow J. H.; et al. Drug Development in Oncology: Classical Cytotoxics and Molecularly Targeted Agents. Br. J. Clin. Pharmacol. 2006, 62, 15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gani O. A., Engh R. A. Protein Kinase Inhibition of Clinically Important Staurosporine Analogues. Nat. Prod. Rep. 2010, 27, 489–498. [DOI] [PubMed] [Google Scholar]
- 32. Mullangi R., Ahlawat P., Srinivas N. R. Irinotecan and Its Active Metabolite, SN-38: Review of Bioanalytical Methods and Recent Update from Clinical Pharmacology Perspectives. Biomed. Chromatogr. 2010, 24, 104–123. [DOI] [PubMed] [Google Scholar]
- 33. Longley D. B., Harkin D. P., Johnston P. G. 5-Fluorouracil: Mechanisms of Action and Clinical Strategies. Nat. Rev. Cancer 2003, 3, 330–338. [DOI] [PubMed] [Google Scholar]
- 34. Focaccetti C., Bruno A., Magnani E.; et al. Effects of 5-Fluorouracil on Morphology, Cell Cycle, Proliferation, Apoptosis, Autophagy and Ros Production in Endothelial Cells and Cardiomyocytes. PLoS One 2015, 10, e0115686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Midena E., Lazzarini D., Catania A. G.; et al. Cytostatic and Cytotoxic Effects of 5-Fluorouracil on Human Corneal Epithelial Cells and Keratocytes. Cornea 2013, 32, 338–344. [DOI] [PubMed] [Google Scholar]
- 36. Caicedo J. C., Cooper S., Heigwer F.; et al. Data-Analysis Strategies for Image-Based Cell Profiling. Nat. Methods 2017, 14, 849–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Driscoll M. K., Welf E. S., Jamieson A. R.; et al. Robust and Automated Detection of Subcellular Morphological Motifs in 3D Microscopy Images. Nat. Methods 2019, 16, 1037–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scheeder C., Heigwer F., Boutros M. Machine Learning and Image-Based Profiling in Drug Discovery. Curr. Opin. Syst. Biol. 2018, 10, 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Madoux F., Tanner A., Vessels M.; et al. A 1536-Well 3D Viability Assay to Assess the Cytotoxic Effect of Drugs on Spheroids. SLAS Discov. 2017, 22, 516–524. [DOI] [PubMed] [Google Scholar]
- 40. Hou S., Tiriac H., Sridharan B. P.; et al. Advanced Development of Primary Pancreatic Organoid Tumor Models for High-Throughput Phenotypic Drug Screening. SLAS Discov. 2018, 23, 574–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fang Y., Eglen R. M. Three-Dimensional Cell Cultures in Drug Discovery and Development. SLAS Discov. 2017, 22, 456–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen W., Wong C., Vosburgh E.; et al. High-Throughput Image Analysis of Tumor Spheroids: A User-Friendly Software Application to Measure the Size of Spheroids Automatically and Accurately. J. Vis. Exp. 2014, 89, 51639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Borten M. A., Bajikar S. S., Sasaki N.; et al. Automated Brightfield Morphometry of 3D Organoid Populations by OrganoSeg. Sci. Rep. 2018, 8, 5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Karolak A., Poonja S., Rejniak K. A. Morphophenotypic Classification of Tumor Organoids as an Indicator of Drug Exposure and Penetration Potential. PLoS Comput. Biol. 2019, 15, e1007214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Czerniecki S. M., Cruz N. M., Harder J. L.; et al. High-Throughput Screening Enhances Kidney Organoid Differentiation from Human Pluripotent Stem Cells and Enables Automated Multidimensional Phenotyping. Cell Stem Cell 2018, 22, 929–940.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Francies H. E., Barthorpe A., McLaren-Douglas A.; et al. Drug Sensitivity Assays of Human Cancer Organoid Cultures. Methods Mol. Biol. 2019, 1576, 339–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moen E., Borba E., Miller G.; et al. Accurate Cell Tracking and Lineage Construction in Live-Cell Imaging Experiments with Deep Learning. bioRxiv 2019. 10.1101/803205. [DOI]
- 48. Moen E., Bannon D., Kudo T.; et al. Deep Learning for Cellular Image Analysis. Nat. Methods 2019, 16, 1233–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vlachogiannis G., Hedayat S., Vatsiou A.; et al. Patient-Derived Organoids Model Treatment Response of Metastatic Gastrointestinal Cancers. Science 2018, 359, 920–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shiga K., Hara M., Nagasaki T.; et al. Cancer-Associated Fibroblasts: Their Characteristics and Their Roles in Tumor Growth. Cancers 2015, 7, 2443–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Quail D. F., Joyce J. A. Microenvironmental Regulation of Tumor Progression and Metastasis. Nat. Med. 2013, 19, 1423–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Neal J. T., Li X., Zhu J.; et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell. 2018, 175, 1972–1988.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gascard P., Tlsty T. D. Carcinoma-Associated Fibroblasts: Orchestrating the Composition of Malignancy. Genes Dev. 2016, 30, 1002–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Attieh Y., Vignjevic D. M. The Hallmarks of CAFs in Cancer Invasion. Eur. J. Cell Biol. 2016, 95, 493–502. [DOI] [PubMed] [Google Scholar]
- 55. Hannah R., Beck M., Moravec R.; et al. CellTiter-Glo™ Luminescent Cell Viability Assay: A Sensitive and Rapid Method for Determining Cell Viability. Cell Notes 2001, 2, 11–13. [Google Scholar]
- 56. Kopper O., de Witte C. J., Lõhmussaar K.; et al. An Organoid Platform for Ovarian Cancer Captures Intra- and Interpatient Heterogeneity. Nat. Med. 2019, 25, 838–849. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplemental_Data1__for__Comparison_of_cell_and_organoid-level_analysis_of_patient-derived_3D_organoids for Comparison of Cell and Organoid-Level Analysis of Patient-Derived 3D Organoids to Evaluate Tumor Cell Growth Dynamics and Drug Response by Seungil Kim, Sarah Choung, Ren X. Sun, Nolan Ung, Natasha Hashemi, Emma J. Fong, Roy Lau, Erin Spiller, Jordan Gasho, Jasmine Foo and Shannon M. Mumenthaler in SLAS Discovery
Supplemental material, Supplemental_Figures_for_Comparison_of_cell_and_organoid-level_analysis_of_patient-derived_3D_organoids for Comparison of Cell and Organoid-Level Analysis of Patient-Derived 3D Organoids to Evaluate Tumor Cell Growth Dynamics and Drug Response by Seungil Kim, Sarah Choung, Ren X. Sun, Nolan Ung, Natasha Hashemi, Emma J. Fong, Roy Lau, Erin Spiller, Jordan Gasho, Jasmine Foo and Shannon M. Mumenthaler in SLAS Discovery