Abstract
RNA N6-methyladenosine (m6A) has emerged as an important epigenetic modification for its role in regulating the stability, structure, processing, and translation of RNA. Instability of m6A homeostasis may result in flaws in stem cell regulation, decrease in fertility, and risk of cancer. To this day, experimental detection and quantification of RNA m6A modification are still time-consuming and labor-intensive. There is only a limited number of epitranscriptome samples in existing databases, and a matched RNA methylation profile is not often available for a biological problem of interests. As gene expression data are usually readily available for most biological problems, it could be appealing if we can estimate the RNA methylation status from gene expression data using in silico methods. In this study, we explored the possibility of computational prediction of RNA methylation status from gene expression data using classification and regression methods based on mouse RNA methylation data collected from 73 experimental conditions. Elastic Net-regularized Logistic Regression (ENLR), Support Vector Machine (SVM), and Random Forests (RF) were constructed for classification. Both SVM and RF achieved the best performance with the mean area under the curve (AUC) = 0.84 across samples; SVM had a narrower AUC spread. Gene Site Enrichment Analysis was conducted on those sites selected by ENLR as predictors to access the biological significance of the model. Three functional annotation terms were found statistically significant: phosphoprotein, SRC Homology 3 (SH3) domain, and endoplasmic reticulum. All 3 terms were found to be closely related to m6A pathway. For regression analysis, Elastic Net was implemented, which yielded a mean Pearson correlation coefficient = 0.68 and a mean Spearman correlation coefficient = 0.64. Our exploratory study suggested that gene expression data could be used to construct predictors for m6A methylation status with adequate accuracy. Our work showed for the first time that RNA methylation status may be predicted from the matched gene expression data. This finding may facilitate RNA modification research in various biological contexts when a matched RNA methylation profile is not available, especially in the very early stage of the study.
Keywords: Epitranscriptomics, next generation sequencing, multiomics, machine learning, supervised learning
Introduction
The structure and function of RNA molecules in cells are regulated with more than 100 chemical modifications, but the specific functions of the majority of those modifications are still enigmatic.1 While most of the RNA nucleotide modifications are believed to be static due to their covalent attachments, m6A is the first type of RNA methylation found to be revisable.2 Even though m6A is the most prevalent internal mRNA decoration found in eukaryotes and RNA of nuclear-replicating viruses, the physiological function and the prevalence of this post-transcriptional RNA modification remain merely partially revealed. It was reported that m6A in nuclear RNA functions as a physiological substrate of fat mass and obesity-associated protein.3 The m6A sites were found on many clock gene transcripts and believed to be related to circadian period elongation and RNA processing delay.4 More recent studies show that m6A is crucial in RNA metabolic processes as it regulates the stability, structure, processing, and translation of RNA. Instability of m6A homeostasis would result in flaws in stem cell regulation, decrease in fertility, and risk of cancer.5
The transcriptome-wide mapping of m6A was available in 2012, due to the invention of methylated RNA immunoprecipitation sequencing method (MeRIP-seq or m6A-seq).6,7 In this method, isolated mRNA is fragmented into ∼100 nucleotides with part of the fragments reserved as untreated input control and the rest precipitated by m6A-specific antibodies. Then, both input control and immunoprecipitated (IP) samples are reverse-transcripted to cDNA. After amplification, the cDNA is subjected to high-throughput sequencing. Next, by comparing signal enrichment of IP samples with input controls, the location of m6A may be identified. The MeRIP-seq plays a pivotal role in the study of m6A. Based on this technique, m6A was found to be a ubiquitous modification of mRNA discovered in the mRNAs of more than 7600 genes. The m6A is enriched around stop codons, within long internal exons and 3′ untranslated regions (3′UTRs). This clustering tenet was consistent in both human and mouse cells, which suggests that m6A may be fundamental in regulating gene expression.6,7 By investigating the whole-transcriptome m6A profiles of 8 types of major human tissues using MeRIP-seq, a positive correlation between m6As and gene expression homeostasis was observed.5 However, the MeRIP-seq process is laborious and time-consuming, and may take up to 9 days to complete.8 Furthermore, the data are still prone to various bias and artifacts.9 Although there are already hundreds of MeRIP-seq experiments collected in existing RNA modification databases such as RMBase10 and MeT-DB,11 they still cover very limited tissue contexts, and a matched RNA methylation profile is usually not available for an arbitrary biological condition.
The biological system is a coordinated system. There exists a significant correlation between different types of genetic and epigenetic features that work in harmony to achieve various biological functions. The idea and feasibility of computational predicting omic features have been well documented in previous works,12 eg, Whitaker et al13 predicted the entire human epigenome from DNA motifs via the Epigram pipeline. The possibility of predicting DNA methylome was explored using various approaches.13-20 Recently, Nath et al21 predicted the long non-coding RNA transcriptome with protein-coding genes. In the field of epitranscriptome bioinformatics, although there exists the potential to train the transcriptome-wide prediction model to predict the entire epitranscriptome, most works focused on RNA modification site prediction from a single DNA or RNA sequence.22,23 Please refer to a comprehensive review.24 Recently, Chen et al25 developed the WHISTLE method and constructed so far the most accurate m6A epitranscriptome from sequence and genomic features; however, similar to other approaches, the WHISTLE method provided only a general m6A epitranscriptome without considering its dynamics (or condition-specificity). Machine learning predictors were developed for better characterization of the single-based m6A profile on sub-regions of mRNA, such as the LITHOPHONE26 and WITMSG27 for the prediction of intronic and lncRNA m6A sites. Prediction modeling may also be applied to the functional annotation of m6A28 and other types of internal mRNA modifications such as pseudo-uridine29 and m7G.30 The deep-m6A approach developed by Zhang et al may perform condition-specific quantification of the m6A epitranscriptome at base resolution. However, it requires matched epitranscriptome sequencing data (MeRIP-seq), which may not be available in most biological contexts of interests, thus limiting its usage.31 Compared with the RNA methylation data, gene expression data are more abundant in public databases and are much easier to obtain. Hence, it is highly desirable to develop in silico approaches to predict the condition-specific RNA methylation status from matched gene expression data.
In this article, we sought to computationally predict the RNA methylation status from gene expression data. We first differentiated the methylated and unmethylated RNA sites using classification methods, then estimated the methylation level using regression methods. Our results suggested that gene expression data can be used to construct predictors of RNA methylation status, which provides a new and easier venue for the pilot studies of RNA methylation under various biological contexts.
Materials and Methods
RNA methylation and gene expression data
MeRIP-seq data of 73 IP samples from different types of mouse cells paired with their matched input controls were collected for this study (Supplement S1). Among them, 58 samples were used for training, and the remaining 15 samples served as the testing set. It is worth noting that the input control of MeRIP-seq data is essentially RNA-seq, which corresponds to gene expression data. For the candidate m6A RNA methylation sites, we considered the 102 024 m6A sites reported by base-resolution techniques (m6A-CLIP and miCLIP) that were collected from the WHISTLE project.25 Reads of MeRIP-seq data in IP samples and input samples are both quantified in terms of reads per million reads mapped (RPKM). Furthermore, we used the M-value, ie, log2 ratio of reads in IP to reads in input,32 to determine the status of RNA methylation:
For classification analysis, the methylation status of a site is considered positive when the corresponding M-value is greater than 0; otherwise, it is regarded as negative. For regression analysis, we seek to directly predict the absolute amount of methylation from matched gene expression data after transformation. An independent pair of input and IP data obtained from human cells under different experimental conditions (31 samples, 69 433 sites, see Supplement S2) were also tested to evaluate the generalizability of our predicting scheme on human data.
Furthermore, as a contrast to models based on expression level, we also trained Elastic Net-regularized Logistic Regression (ENLR), Support Vector Machine (SVM), and Random Forests (RF) by incorporating sequence-based features, including the presence of purine, amino group and weak hydrogen bonds, and cumulative frequency of nucleotide with window width 41 bp centered by m6A.25
The 100 sites with the greatest variation in m6A modification level were selected for classification and regression analysis, because these sites were most dynamic and were potentially most responsive to various stimulus and more crucial when studying the context-specificity of m6A RNA methylation. For each target site, the gene expression profile of the corresponding site and its 1000 neighbor sites was selected as our predictive features to construct classifiers and regressors. Because of having a lot more features than samples, the prediction model would result in a typical “large p small n” paradigm33; therefore, both the regression and classification analysis required sparse estimators to avoid overfitting.
Classification and regression methods
For classification analysis, ie, to differentiate methylated m6A sites and unmethylated m6A sites, we used ENLR, SVM, and RF for classification, as they were reported to be promising classifiers in previous DNA methylation predicting tasks.13,14,19 The ENLR model minimizes an objective function consisting of negative log-likelihood of logistic regression along with both - and -penalty to obtain a sparse generalized linear model by shrinking the coefficients of less-informative sites into zeros.34 There are 2 parameters to be learned, one is the overall weight of the penalty and the other is the weight between - and -penalty. SVM maps features of interest into a higher dimensional space via kernel functions and generates a decision hypersurface that maximizes the margin between examples from different categories.35 We chose radial basis function as our kernel and tuned the inverse kernel width for the radial basis kernel function as well as the cost regularization parameter which controls the smoothness of the fitted function. RF is an ensemble learning algorithm that generates a specified number of decision trees and cast predictions based on the majority of the votes from an individual tree.36 For RF, we optimized the number of variables randomly sampled as candidates at each split.
For regression analysis, ie, to estimate directly the absolute RNA methylation level of m6A site, we considered the Elastic Net (EN) family with objective function:
where s is the sample size, y is the response, β is the coefficient to be estimated, X is the covariate, is the -norm and is the -norm. Note that when , this model reduces to lasso; when , this model reduces to ridge regression. All analysis was implemented in R. The scripts and processed data used in this project are publicly downloadable on GitHub (https://github.com/xvehao/m6Aprediction).
Results and Discussions
Predict condition-specific m6A sites with classification analysis
For each site, we constructed ENLR, SVM, and RF models as classifiers, ran a 5-fold cross-validation to tune the parameters of each model with the caret R package,37 and then tested their performance on the testing samples. To evaluate the performance of classifiers, we generated the receiver operating characteristic (ROC) curve (true positive rate vs false positive rate under different threshold) and computed the area under the ROC curve (AUC) as the metric. As shown in Figure 1A, positive classification results were achieved from all the 3 classifiers tested with their AUCs substantially higher than the classifiers based on sequence features (highest median AUC = 0.54 by ENLR, see Supplement S3), which suggested that it is indeed possible to predict context-specific m6A RNA methylation sites from matched gene expression data. The medians of AUC of 3 methods across test samples were very close, but the result of SVM had a narrower AUC spread and the highest overall accuracy.
Figure 1.
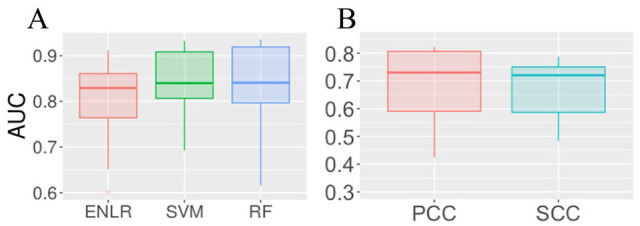
Model performance on the mouse test set: (A) All 3 classifiers have a median AUC over 0.8 in the identification of condition-specific m6A sites. Among them, SVM achieved the best performance as it has the highest AUC and the narrowest AUC spread. (B) Both median PCC and SCC between the predicted value by ENLR and the actual value are above 0.7, which implies a strong linear and monotonic relation between the predicted and actual values. AUC indicates area under ROC; ENLR, Elastic Net-regularized Logistic Regression; PCC, Pearson correlation coefficient; RF, Random Forests; SCC, Spearman correlation coefficient; SVM, Support Vector Machine.
To further investigate the expression level of which kinds of genes were determinant in predicting m6A status, we examined the biological meaning of ENLR model because ENLR can perform feature selection automatically due to its sparse property. We performed gene site enrichment analysis, with DAVID,38 using those sites ever selected as predictors by ENLR as input and all sites as background. Three functional annotation terms were found to be statistically significant with Family Wise Error Rate less than 0.05: phosphoprotein, SRC Homology 3 (SH3) domain, and Endoplasmic Reticulum (ER). This result further attached potential biochemical significance to ENLR model as all 3 terms were found to be closely related to the m6A pathway in literature.
The first 2 terms may refer to the formation of a methyltransferases (MTases) complex comprising MTase-like 3 (Mettl3) and MTase-like 14 (Mettl14), called Mettl3-Mettl14 complex. This complex efficiently catalyzes methyl group transfer by using Mettl3 as catalytic core and Mettl14 as an RNA-binding platform. Moreover, Mettl3-Mettl14 complex exhibits a much higher catalytic activity than either Mettl3 or Mettl14 alone in vitro. The Mettl3 MTase domain and the Mettl14 MTase domain are connected by the N-terminal α-helical motif (NHM) and by the C-terminal motif (CTM) with a phosphoserine via a salt bridge. Phosphorylation therefore plays an important regulatory role in MTase binding as it ensures the 2 MTases are connected tightly, and their extensive interaction network is difficult to disrupt.39 SH3 domain is important in salt bridge formation between the conserved acidic residue in the SH3 domain and the favored arginine residue, either N-terminal or C-terminal to the Pro-X-X-Pro motif,40 which is exactly the structure of Mettl3-Mettl14 complex. Moreover, the change in ER also alters the m6A modification. ER stress responses are found to contribute to differential m6A modification,41 and SIALKBH2, an active m6A demethylase, is found to be located in the ER.42 After analyzing the mechanism of ENLR model, we conjecture that the gene expression level of sites regulating phosphoprotein, SH3 domain, and ER might influence the formation of Mettl3-Mettl14 complex and demethylation of m6A in ER, and therefore are associated with the m6A level.
Predict condition-specific m6A level with regression analysis
In the regression analysis, we ran a 5-fold cross-validation to tune the associated parameters with mean squared error (MSE) as the metric. Furthermore, we computed both Pearson correlation coefficient (PCC) and Spearman correlation coefficient (SCC) between predicted methylation value and the MeRIP-seq data to evaluate the performance of our regressor (Figure 1B). While PCC measured the linearity between 2 data sets, SCC accessed the monotonic relationship between them. Both PCC and SCC were within −1 to 1, and a higher absolute value of PCC or SCC implied higher concordance between 2 data sets. The median of both PCC and SCC across different samples was above 0.7, suggesting a strong linear relationship between the predicted value and the actual value, while the median PCC and SCC obtained by model based on sequence-based features were respectively, 0.49 and 0.43 (Supplement S3). We identified 1 outlier in the boxplot with PCC −0.1 and SCC −0.15, which corresponds to Mettl3 knocked-out mouse embryonic stem cell. The abnormal performance of our regressor on this particular sample was likely to be caused by the removal of Mettl3, which was reported to lead to near-complete depletion of m6A on mRNA,43 thus rendering the association between gene expression level and m6A methylation level completely different from that in other types of cells.
Additional investigation on human data
To further investigate whether our predicting scheme could be generalized to different species, we tested our classifiers with independent input and IP data obtained from different types of human cells under heterogeneous experimental conditions (26 samples as training set, 5 as testing). The AUC of each model on test set was shown by Table 1, from which we can see that the gene expression data can serve as covariates for effectively predicting m6A methylation status in human cells as well, with SVM being the best classifier.
Table 1.
Performance of classifiers (AUC) on the human test set.
| Sample | Classification method | ||
|---|---|---|---|
| ENLR | SVM | RF | |
| 1 | 0.90 | 0.91 | 0.91 |
| 2 | 0.82 | 0.86 | 0.89 |
| 3 | 0.82 | 0.88 | 0.87 |
| 4 | 0.85 | 0.91 | 0.76 |
| 5 | 0.77 | 0.80 | 0.71 |
| Average | 0.83 | 0.87 | 0.83 |
Abbreviations: AUC, area under ROC curve, ROC, receiver operating characteristic; ENLR, Elastic Net-regularized Logistic Regression; RF, Random Forests; SVM, Support Vector Machine.
All 3 classifiers have an average AUC above 0.8. SVM achieved the best performance in the identification of condition-specific m6A sites.
The bold values in the table highlight the best performance achieved in each sample.
Conclusions
We implemented both regression and classification to predict RNA m6A methylation from gene expression data. With the positive results, we showed, for the first time, that condition-specific m6A methylation status may be predicted from gene expression data. Gene Site Enrichment Analysis on important sites by ENLR further suggested that those sites related to phosphoprotein, SH3 domain and ER be determinant in m6A methylation status prediction. Therefore, we recommend that features related to those 3 functional terms should be considered in regressors or classifiers in future m6A predicting study. We are optimistic that the accuracy may be further improved with efforts.
Nevertheless, our work suffers from the 3 main limitations. First, as the data samples were collected from different public data sets provided by different laboratories under heterogeneous experimental conditions, the systematic error arising in each experiment is unavoidable. Different experiment reagents and protocols (such as polyA selection and ribo-minus in RNA library construction) may lead to a substantial difference between samples. As a consequence, the accuracy of prediction would be undermined. The prediction models could be trained later using the technical independent quantification and the inference methods on MeRIP-Seq. Second, as MeRIP-Seq data are prohibitively difficult to obtain, it is possible that the limited samples in hand may not be representative of all mouse/human cells. Therefore, larger-scale MeRIP-seq data from different tissues and developmental stages are needed for the development of prediction tools with higher accuracy in future studies. Third, while we considered in our prediction only the gene expression data in the form of RNA-seq, it is reasonable to assume that the accuracy may be further improved with other matched omic data, such as proteomic data from LC/MS or DNA methylation data.
Supplemental Material
Supplemental material, sra_meRIP_mouse_xyz3342045810769 for Prediction of RNA Methylation Status From Gene Expression Data Using Classification and Regression Methods by Hao Xue, Zhen Wei, Kunqi Chen, Yujiao Tang, Xiangyu Wu, Jionglong Su and Jia Meng in Evolutionary Bioinformatics
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work has been supported by National Natural Science Foundation of China (31671373); XJTLU Key Program Special Fund (KSF-T-01); Jiangsu Six Talent Peak Program (XYDXX-118).
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: JM and JS initialized the project; ZW, KC, YT, and XW processed the raw data; HX designed and implemented the research plan; HX drafted the article. All authors read, critically revised, and approved the final article.
Availability of Data and Materials: The scripts and the data used in this project are publicly available from GitHub at: https://github.com/xvehao/m6Aprediction.
ORCID iDs: Hao Xue  https://orcid.org/0000-0003-1231-4747
https://orcid.org/0000-0003-1231-4747
Kunqi Chen  https://orcid.org/0000-0002-6025-8957
https://orcid.org/0000-0002-6025-8957
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Boccaletto P, Machnicka MA, Purta E, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303-D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15:293-306. [DOI] [PubMed] [Google Scholar]
- 3. Jia G, Fu Y, Zhao X, et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fustin JM, Doi M, Yamaguchi Y, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793-806. [DOI] [PubMed] [Google Scholar]
- 5. Xiao S, Cao S, Huang Q, et al. The RNA N6-methyladenosine modification landscape of human fetal tissues. Nat Cell Biol. 2019;21:651-661. [DOI] [PubMed] [Google Scholar]
- 6. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201-206. [DOI] [PubMed] [Google Scholar]
- 7. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8:176-189. [DOI] [PubMed] [Google Scholar]
- 9. Zhang T, Zhang SW, Zhang L, Meng J. trumpet: transcriptome-guided quality assessment of m6A-seq data. BMC Bioinform. 2018;19:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xuan J-J, Sun W-J, Lin P-H, et al. RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res. 2018;46:D327-D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu H, Wang H, Wei Z, et al. MeT-DB V2.0: elucidating context-specific functions of N6-methyl-adenosine methyltranscriptome. Nucleic Acids Res. 2018;46:D281-D287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu T, Zheng X, Li B, Jin P, Qin Z, Wu H. A comprehensive review of computational prediction of genome-wide features. Brief Bioinform. 2018:bby110-bby110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Whitaker JW, Chen Z, Wang W. Predicting the human epigenome from DNA motifs. Nat Methods. 2014;12:265-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bhasin M, Zhang H, Reinherz EL, Reche PA. Prediction of methylated CpGs in DNA sequences using a support vector machine. FEBS Lett. 2005;579:4302-4308. [DOI] [PubMed] [Google Scholar]
- 15. Fang F, Fan S, Zhang X, Zhang MQ. Predicting methylation status of CpG islands in the human brain. Bioinformatics. 2006;22:2204-2209. [DOI] [PubMed] [Google Scholar]
- 16. Das R, Dimitrova N, Xuan Z, et al. Computational prediction of methylation status in human genomic sequences. Proc Natl Acad Sci USA. 2006;103:10713-10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zeng H, Gifford DK. Predicting the impact of non-coding variants on DNA methylation. Nucleic Acids Res. 2017;45:e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fan S, Huang K, Ai R, Wang M, Wang W. Predicting CpG methylation levels by integrating Infinium HumanMethylation450 BeadChip array data. Genomics. 2016;107:132-137. [DOI] [PubMed] [Google Scholar]
- 19. Zhang W, Spector TD, Deloukas P, Bell JT, Engelhardt BE. Predicting genome-wide DNA methylation using methylation marks, genomic position, and DNA regulatory elements. Genome Biol. 2015;16:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Y, Liu T, Xu D, et al. Predicting DNA methylation state of CpG dinucleotide using genome topological features and deep networks. Sci Rep. 2016;6:19598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nath A, Geeleher P, Huang RS. Long non-coding RNA transcriptome of uncharacterized samples can be accurately imputed using protein-coding genes [published online ahead of print January 17, 2019]. Brief Bioinform. doi: 10.1093/bib/bby129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou Y, Zeng P, Li YH, Zhang Z, Cui Q. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016;44:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen W, Xing P, Zou Q. Detecting N6-methyladenosine sites from RNA transcriptomes using ensemble Support Vector Machines. Sci Rep. 2017;7:40242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen X, Sun YZ, Liu H, Zhang L, Li JQ, Meng J. RNA methylation and diseases: experimental results, databases, Web servers and computational models. Brief Bioinform. 2019;20:896-917. [DOI] [PubMed] [Google Scholar]
- 25. Chen K, Wei Z, Zhang Q, et al. WHISTLE: a high-accuracy map of the human N6-methyladenosine (m6A) epitranscriptome predicted using a machine learning approach. Nucleic Acids Res. 2019;47:e41-e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu L, Lei X-J, Fang Z-Q, Tang Y-J, Meng J, Wei Z. LITHOPHONE: improving lncRNA methylation site prediction using an ensemble predictor. Front Genet. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu L, Lei X-J, Meng J, Wei Z. WITMSG: large-scale prediction of human intronic m6A RNA methylation sites from sequence and genomic features. Curr Genom. 2020;21:67-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang S, Xie Y, He Z, et al. m6ASNP: a tool for annotating genetic variants by m6A function. Gigascience. 2018;7:giy035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Song B, Tang Y, Wei Z, et al. PIANO: a web server for pseudouridine site (Ψ) identification and functional annotation. Front Genet. 2020;11:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Song B, Tang Y, Chen K, et al. m7GHub: deciphering the location, regulation and pathogenesis of internal mRNA N7-methylguanosine (m7G) sites in human. Bioinformatics. 2020;3:btaa178. [DOI] [PubMed] [Google Scholar]
- 31. Zhang S-Y, Zhang S-W, Fan X-N, et al. Global analysis of N6-methyladenosine functions and its disease association using deep learning and network-based methods. Plos Comput Biol. 2019;15:e1006663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tang Y, Chen K, Wu X, et al. DRUM: inference of disease-associated m(6)A RNA methylation sites from a multi-layer heterogeneous network. Front Genet. 2019;10:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bernardo J, Bayarri M, Berger J, et al. Bayesian factor regression models in the “large p, small n” paradigm. Bayesian Stat. 2003;7:733-742. [Google Scholar]
- 34. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1-22. [PMC free article] [PubMed] [Google Scholar]
- 35. Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20:273-297. [Google Scholar]
- 36. Tin Kam H. Random decision forests. Paper presented at: Proceedings of 3rd International Conference on Document Analysis and Recognition; August 14-16, 1995, Montreal, QC, Canada. [Google Scholar]
- 37. Kuhn M. Building predictive models in R using the caret package. J Stat Softw. 2008;28:1-26.27774042 [Google Scholar]
- 38. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4: 44-57. [DOI] [PubMed] [Google Scholar]
- 39. Wang X, Feng J, Xue Y, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575-578. [DOI] [PubMed] [Google Scholar]
- 40. Bedford MT, Frankel A, Yaffe MB, Clarke S, Leder P, Richard S. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J Biol Chem. 2000;275:16030-16036. [DOI] [PubMed] [Google Scholar]
- 41. Gokhale NS, McIntyre ABR, Mattocks MD, et al. Altered m6A modification of specific cellular transcripts affects Flaviviridae infection. Mol Cell. 2020;77:542-555.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou L, Tian S, Qin G. RNA methylomes reveal the m(6)A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol. 2019;20:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Geula S, Moshitch-Moshkovitz S, Dominissini D, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002-1006.Citiuri rehenis sande esed utatium, tenis est [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sra_meRIP_mouse_xyz3342045810769 for Prediction of RNA Methylation Status From Gene Expression Data Using Classification and Regression Methods by Hao Xue, Zhen Wei, Kunqi Chen, Yujiao Tang, Xiangyu Wu, Jionglong Su and Jia Meng in Evolutionary Bioinformatics


