Abstract
Gestational exposure to lead (Pb) adversely impacts offspring health through multiple mechanisms, one of which is the alteration of the epigenome including DNA methylation. This study aims to identify differentially methylated CpG sites associated with trimester-specific maternal Pb exposure in umbilical cord blood (UCB) leukocytes. Eighty-nine mother-child dyads from the Early Life Exposure in Mexico to Environmental Toxicants (ELEMENT) longitudinal birth cohorts with available UCB samples were selected for DNA methylation analysis via the Infinium Methylation EPIC BeadChip, which quantifies methylation at >850 000 CpG sites. Maternal blood lead levels (BLLs) during each trimester (T1: 6.56 ± 5.35 µg/dL; T2: 5.93 ± 5.00 µg/dL; T3: 6.09 ± 4.51 µg/dL), bone Pb (patella: 11.8 ± 9.25 µg/g; tibia: 11.8 ± 6.73 µg/g), a measure of cumulative Pb exposure, and UCB Pb (4.86 ± 3.74 µg/dL) were measured. After quality control screening, data from 786 024 CpG sites were used to identify differentially methylated positions (DMPs) and differentially methylated regions (DMRs) by Pb biomarkers using separate linear regression models, controlling for sex and estimated UCB cell-type proportions. We identified 3 DMPs associated with maternal T1 BLL, 2 with T3 BLL, and 2 with tibia bone Pb. We identified one DMR within PDGFRL associated with T1 BLL, one located at chr6:30095136-30095295 with T3 BLL, and one within TRHR with tibia bone Pb (adjusted P-value < .05). Pathway analysis identified 15 overrepresented gene pathways for differential methylation that overlapped among all 3 trimesters with the largest overlap between T1 and T2 (adjusted P-value < .05). Pathways of interest include nodal signaling pathway and neurological system processes. These data provide evidence for differential methylation by prenatal Pb exposure that may be trimester-specific.
Keywords: Developmental exposures, epigenetics, developmental programming, biomarkers, environmental exposure
Background
Lead (Pb) is a ubiquitous environmental pollutant found in air, soil, water, and food. Human exposures mainly occur through ingestion and inhalation, and Pb can be stored long term in teeth and bones. Pb is a potent neurotoxicant that alters brain development resulting in abnormal cognition and behaviors. Early life Pb exposure also increases the risk for developing a variety of adverse health outcomes later in life, including Alzheimer disease, attention-deficit hyperactivity disorder, cardiovascular disease, and intellectual deficits.1-5 The Institute for Health Metrics and Evaluation (IHME) estimated in 2017 that Pb exposure accounted for almost 2% of total all-cause mortality and at least 2.5 million years of healthy life lost (disability-adjusted life years [DALYs]) worldwide due to idiopathic developmental intellectual disability,6 but this may be an underestimate.7
The health effects of prenatal Pb exposure can be framed with the Developmental Origins of Health and Disease (DoHAD) theory,8 which links early-life exposures to the development of disease and adverse health effects later in life. The placenta is not an effective barrier against Pb, and as bone reabsorption increases during pregnancy, Pb is released into the bloodstream from where it can reach the fetus potentially causing abnormalities in development.9-13 The fetus may be exposed to stored Pb in bone from the mother’s previous exposures nearly decades prior as well as to concurrent Pb exposure circulating through the blood.
It has been hypothesized that toxicant exposures, including Pb, may result in epigenetic modifications during sensitive developmental periods, thereby impacting gene regulation and subsequent development and later life health outcomes. Epigenetic modifications, which include DNA methylation, are mitotically heritable and regulate gene expression without altering the underlying DNA sequence. DNA methylation occurs primarily in cytosine-guanine (CpG) dinucleotides, which are enriched in regions called islands, typically located in promoter regions of genes.14,15 Establishment of DNA methylation patterns occurs in early gestation and disturbances during this period may result in DNA methylation variations that are propagated through mitosis to new cells and developing organs thus affecting gene expression in a wide range of tissues associated with various developmental physiological processes.16
We and others have provided evidence for the impact of early-life Pb exposure on the epigenome in animal and human studies. In mice, dams were exposed to 1 of 3 doses of Pb acetate in water prior to mating through weaning, and the offspring were followed through adulthood and compared with unexposed mice. Pb was associated with altered DNA methylation in offspring tail tips immediately following cessation of exposure (3 weeks of age) at 2 metastable epialleles.17 Altered DNA methylation by Pb was also observed in offspring brain (10 months of age) at environmentally responsive retrotransposons.18 In a human birth cohort, we quantified DNA methylation at 4 genes in 247 umbilical cord blood (UCB) leukocyte DNA samples. Biomarkers of prenatal Pb exposure (maternal tibia: 9.82 [SD =9.45] µg/dL, maternal patella: 14.4 [SD = 14.8] µg/dL, UCB: 6.49 [SD = 3.52] µg/dL) were associated with percent DNA methylation in neonates in long interspersed elements 1 (LINE-1), as well as growth-related genes (IGF2 and HSD11B2) providing evidence for maternal cumulative Pb burden influencing the epigenome of a developing fetus.19 Finally, the US prospective pregnancy cohort, Project Viva, conducted an epigenome-wide analysis on 268 UCB samples to evaluate the association between prenatal maternal Pb exposure (mid-to-late gestation Pb in erythrocytes averaged 1.22 [SD = 0.63] µg/dL) and DNA methylation.20 Results identified sex-specific differentially methylated CpG sites by Pb exposure with more found in females (n = 38) than males (n = 2). Mechanistically, metal exposures such as Pb increase oxidative stress and can cause oxidative DNA damage which inhibits the ability of methyltransferases to bind to DNA; this leads to hypomethylation at some loci.21-23 In addition, Pb exposure has been shown to alter expression and function of epigenetic machinery (ie, DNA methyltransferases [DNMTs]). Several human and animal studies suggest DNA methylation changes from developmental Pb exposure are due to altered DNMT expression and activity.21,24-27
Despite mounting evidence for Pb’s effect on the epigenome, no studies have specifically investigated the association between genome-wide DNA methylation and Pb exposure during each of the 3 trimesters of pregnancy, as well as cumulative gestational Pb exposure estimated by maternal patella and tibia measures. We hypothesize that prenatal Pb exposure will be significantly associated with changes in DNA methylation patterns across the genome at birth and that these changes will differ depending on the timing of exposure. A whole genome approach will assist researchers in gaining valuable insight into the effects of prenatal Pb exposure on gene regulation and help to identify potential biomarkers of interest specific to each trimester and cumulative exposure. Here, we performed Pb exposure assessment in multiple biomarkers and DNA methylation profiling in UCB samples from 89 mother-infant pairs enrolled in the Early Life Exposures in Mexico to Environmental Toxicants (ELEMENT) project, a longitudinal birth cohort.
Methods
Study population
The ELEMENT project has used a series of longitudinal birth cohorts to investigate the influence of exposure to Pb and other toxicants—in utero and in childhood—on sensitive periods of development. This project is based on 3 sequentially recruited cohorts comprising 1643 mother-infant pairs, some of whom have been followed for 20+ years.28 This study uses data and biological samples from the second and third birth cohorts, for which 1530 women were originally enrolled and 1012 mother-infant pairs were followed up after birth. Women were recruited from 1997-2000 and 2001-2005, from the Mexican Social Security Institute hospital in Mexico City. Eligibility and exclusion criteria are as previously described.28,29 For all recruited mother-child pairs, data collected include sex, gestational age, socioeconomic status, anthropometry data, and other environmental exposures. Families were followed up at multiple time points from infancy through adolescent years. For this study, we selected 97 ELEMENT participants from the second and third cohorts who had archived UCB samples from which we could isolate DNA. The final study comprised 89 mother-infant dyads whose epigenetic data passed downstream quality control (QC). Characteristics of these 89 participants compared with all women of cohorts 2 and 3 that had least one Pb biomarker measurement (n = 1214) and their children are represented in Supplemental Table 1.
At the time of enrollment, all mothers were informed about the study; those who agreed to participate read and signed a letter of informed consent about the original study. The original research protocol and all amendments to the study protocol were approved by the Ethics Committees of the National Institutes of Public Health of Mexico, participating hospitals, and the Internal Review Board at all participating institutions including the University of Michigan.
Pb exposure assessment and genomic DNA isolation
UCB was collected within 12 hours after birth. Cohort 2 blood lead levels (BLL) from each trimester and UCB were measured using inductively coupled plasma mass-spectrometry (ICP-MS, Thermo Finnigan, Bremen, Germany) at the University of California, Santa Cruz, as described previously.30 Cohort 3 BLLs in maternal venous blood from each trimester and UCB were measured at the Trace Metal Laboratory of the American British Cowdray Hospital using graphite furnace atomic absorption spectrometry (instrument model 3000; PerkinElmer, Norwalk, CT, USA).
Bone Pb was measured in maternal left patella (trabecular bone) and mid-shaft of the left tibia (cortical bone) 1 month post-partum as an indicator of cumulative Pb exposure during pregnancy using a spot-source 109Cd K-shell X-ray fluorescence (K-XRF) instrument. The technical specifications and validation of this instrument are described in detail elsewhere.31 Analysis of means and standard deviations once weekly for QC and calibration measures did not disclose any significant shifts in precision or accuracy. Tibia and patella bone Pb values below the limit of detection (ie, negative values) were dropped from subsequent analyses resulting in 73 patella bone Pb and 46 tibia bone Pb measures.
DNA was isolated from UCB leukocytes using Qiagen kits and standard protocols for blood DNA isolation. Nucleic acid yield and purity were assessed first using a NanoDrop spectrophotometer (ThermoFisher Scientific), and double-stranded DNA was also quantified via a Qubit fluorometer. All DNA samples were stored at −80°C until later use.
Epigenetic analysis
All genomic DNA samples were bisulfite converted (500 ng) using the Zymo EZ DNA Methylation kit (Zymo Research) and the recommended incubation conditions and methods for downstream Infinium analysis. In short, this treatment converts unmethylated cytosines to uracils while leaving methylated cytosines intact. DNA methylation was quantified at 863 904 CpG sites following hybridization to the Infinium MethylationEPIC BeadChip (Illumina).32 Beadchips were processed and scanned on the Illumina iScan at the University of Michigan (UM) Advanced Genomics Core using the Infinium Methylation EPIC kit.
We used a preprocessing pipeline for the resulting raw EPIC data based on functional normalization33 that uses the “minfi” package in R.34 The pipeline included several steps: loading raw image files, linking to phenotypic information, dropping probes with single-nucleotide polymorphisms (SNPs) within query sites, removing poor-quality samples (>5% failed probes) and poorly detected probes (P-value > 1e−6 when comparing to background), normalizing the data to correct for background signal and dye bias, checking and correcting for batch effects (by slide and array), and removing CpG sites on the X and Y chromosomes. After QC and normalization, 786 024 CpG sites and 89 samples passed all QC measures and were subsequently used in statistical analysis.
Estimates of cell-type composition (T lymphocytes [CD4T, CD8T], B cells, NK cells, monocytes, granulocytes, and nucleated red blood cells [nRBCs]) for each sample was performed using an established method based on UCB cell-type-specific differentially methylated regions. Cell-type proportions were adjusted for in downstream statistical analysis to avoid confounding by cell-type composition.35 Beta-values and M-values (log2-transformed beta-values) were then calculated from the processed data and used in statistical analysis.36
Statistical analysis
All statistical analyses were performed in the R Project for Statistical Computing (version 3.4.3). Analyses were performed to identify differentially methylated loci by Pb exposure. Pb biomarkers tested were maternal BLLs at each trimester, UCB Pb levels, and Pb levels in maternal patella and tibia, which were treated as non-transformed continuous variables. Bone and UCB Pb levels were normally distributed. Trimester-specific Pb measures were right-skewed yet all biomarker concentrations were biologically plausible and relevant to this study population. As such the relationships between maternal BLLs and DNA methylation were tested with and without BLL outliers. We performed bivariate analyses relating Pb biomarkers and covariates (eg, cell-type proportions, gestational age, maternal age, maternal education, household income category) to identify potential confounders. We also performed singular value decomposition to identify covariates associated with principal components of the DNA methylation data. We then chose potential confounding variables to include in the final statistical models of site-specific DNA methylation data that were associated with both Pb and DNA methylation. We also retained several variables that had a large effect on DNA methylation but were not associated with Pb (ie, infant sex, cell type proportions). In models with BLLs, offspring sex and estimated cell-type compositions of granulocytes and nRBC35 were included. Models with bone Pb measures as the predictor of interest additionally included maternal age and cohort. Other covariates available in this study population, including gestational age, were not considered as confounders because they were neither associated with prenatal Pb exposure nor DNA methylation in our study sample.
Differentially methylated positions (DMPs) by Pb were identified using the “limma” package.37,38 Briefly, linear regression models were run for each CpG site using M-values, which are logit-transformed beta values representing the proportion of methylation at a given CpG site, with one Pb exposure biomarker as the predictor of interest and the aforementioned covariates included. Limma calculates fold-change in methylation, t-statistics, and P-values by applying empirical Bayes smoothing to the standard errors. To account for multiple testing, we used the false discovery rate (FDR) method39 and consider DMPs with corrected P-value (q-value) < .05 to be statistically significant. For comparison purposes between studies, we also report estimates (β) from the same regression analysis with betas (non-transformed methylation values) as the outcome.
We used the statistical package DMRcate to test associations between each Pb exposure variable and differential methylation at the regional level across the entire genome.40 The Gaussian kernel bandwidth lambda values used was the default of 1000, and 2 CpG sites were considered the minimum consecutive loci to be included in a differentially methylated region (DMR). Differentially methylated regions with FDR < 0.05 were statistically significant.
We used LRpath, a multiple-comparison enrichment testing tool that uses logistic regression to test for gene sets that have higher significance values by raw P-value than expected at random,41 to identify gene pathways enriched for hyper- or hypo-methylation by Pb exposure. It considers CpG sites in known genes, their respective log fold-changes by Pb from linear regression models, and associated P-values.
Results
Descriptive statistics of ELEMENT study data
Among the 89 mothers included in this analysis, 41 (46%) had male infants (Table 1). The mean age of women at offspring birth was 26.4 (SD = 4.8) years. The average gestational age at birth was 39.0 (SD = 1.1) weeks. The mean (SD) maternal BLLs averaged over all 3 trimesters was 6.18 (4.51) µg/dL, with the first trimester (T1) mean of 6.56 (5.35) µg/dL, second trimester (T2) mean of 5.93 (5.00) µg/dL, and third trimester (T3) mean of 6.09 (4.51) µg/dL. Maternal BLLs between the trimesters were highly correlated (r > 0.63) according to Spearman rank-order correlation test. Average Pb concentration in patella was 9.42 (10.28) µg/g and in tibia was 7.51 (9.46) µg/g. Umbilical cord blood Pb levels were measured to represent offspring Pb exposure at birth and averaged 4.86 (3.74) µg/dL. Geometric means of BLL among women in this study were more than 4 times higher than women of childbearing ages within a similar time period in the United States according to NHANES (1999-2002 and 2003-2006).42 Sources of Pb exposure in Mexico City during this time included the use of traditional lead-glazed ceramics for cooking, and Pb in gasoline which was not phased out in full until 1997.43 Pregnant women in countries such as Malaysia, India, Iran, Malta, and Bangladesh continue to have exposures similar or up to 2½ times greater on average than that of the women in this study.44-48
Table 1.
Characteristics of ELEMENT mother-infant pairs with UCB DNA methylation data.
| Characteristics | No. | Mean ± SD or N (%) | Range |
|---|---|---|---|
| Mothers | |||
| Age (y) | 89 | 26.4 ± 4.81 | 18.0-37.0 |
| Blood lead (µg/dL) | |||
| First trimester (T1) | 69 | 6.56 ± 5.35 | 0.90-35.8 |
| Second trimester (T2) | 74 | 5.93 ± 5.00 | 0.80-38.2 |
| Third trimester (T3) | 76 | 6.09 ± 4.51 | 0.90-34.0 |
| Average all trimesters | 77 | 6.18 ± 4.51 | 1.17-33.1 |
| Bone lead (µg/g) | |||
| Patella | 73 | 11.8 ± 9.25 | 0.20-42.0 |
| Tibia | 46 | 11.8 ± 6.73 | 0.40-28.8 |
| Household income | 77 | ||
| Lowest | 12 (15.6) | ||
| Low-middle | 31 (40.3) | ||
| Middle | 20 (26.0) | ||
| Middle-high | 9 (11.6) | ||
| Highest | 5 (6.5) | ||
| Maternal education (total years) | 89 | 11.0 ± 2.39 | 3.00-17.0 |
| Children | |||
| UCB lead (µg/dL) | 86 | 4.86 ± 3.74 | 0.00-19.5 |
| Gestational age (wk) | 87 | 39.0 ± 1.09 | 36.0-41.0 |
| Male (%) | 89 | 41 (46.0) | |
Abbreviations: UCB, umbilical cord blood.
We compared participants included in this analysis with all ELEMENT mother-infant pairs from the same cohorts (2 and 3). Demographics and Pb biomarker concentrations were not statistically different between the subset and the entire study (Supplemental Table 1) with the exception of gestational age which was slightly higher in the subset with DNA methylation data.
CpG site-specific analysis and pathways analysis
We found that DNA methylation levels at 3 CpG sites demonstrated significant associations (FDR < 0.05) with T1 BLLs, 1 CpG with T3 BLLs, and 2 with tibia bone Pb levels (Table 2). There were no statistically significant DMPs in analyses with maternal BLLs in T2, UCB Pb, or maternal patella bone Pb. Of the 3 statistically significant DMPs with the maternal BLLs during T1, the sites mapped to RAB5A, EXT1, and a non-genic region (chr8:12615485). The DMPs in RAB5A and EXT1 had less DNA methylation (−0.022% and −0.024%, respectively) per 1 µg/dL increase in maternal T1 BLL. In contrast, the non-genic region (chr8:12615485) was hypermethylated by Pb (0.35% per µg/dL of T1 BLL). The DMP by T3 maternal BLLs was hypomethylated (−0.52% per µg/dL BLL) and in a non-genic region (chr1:160839299). Maternal tibia bone Pb-associated DMPs were hypermethylated in LNFN1 (0.37% per µg/g bone Pb) and hypomethylated in a non-genic region located at chr6:792305 (−0.21% per µg/g bone Pb).
Table 2.
Statistically significant DMPs (q < 0.05) by first-trimester maternal BLL, third-trimester maternal BLL, and maternal tibia Pb.
| Pb biomarker | Probe ID | Gene name | Chr | Pos | β estimate | P-value | Q-value |
|---|---|---|---|---|---|---|---|
| T1 BLL | cg17138393 | RAB5A | chr3 | 19988887 | −0.000221 | 5.46E-08 | 0.0227 |
| cg03390844 | chr8 | 12615485 | 0.00348 | 5.78E-08 | 0.0227 | ||
| cg00984923 | EXT1 | chr8 | 119124069 | −0.000235 | 1.49E-07 | 0.0390 | |
| T3 BLL | cg01328348 | chr1 | 160839299 | −0.00519 | 1.81E-08 | 0.0142 | |
| Tibia Pb | cg00002033 | LRFN1 | chr19 | 39798481 | 0.00367 | 3.57E-08 | 0.0281 |
| cg03463208 | chr6 | 792305 | −0.00205 | 7.23E-08 | 0.0284 |
Abbreviations: BLL, blood lead level; Chr, chromosome; DMP, differentially methylated position; pos, base pair position from hg19.
Results from the analysis are shown for CpG sites associated with Pb biomarker levels below q-value of 0.05 from models adjusted for sex and estimated cell-type proportions (granulocytes and nucleated red blood cells). The P-values and q-values are obtained from the analyses using methylation expressed as M-values as the outcome variable, and reported association estimates (β) are from analysis using methylation beta values as the outcome variable.
LRpath analysis was conducted to test for gene sets enriched among continuous significance values (raw P-values) by each Pb biomarker. After multiple-comparisons correction, there were 109 gene sets enriched with T1 BLLs, 76 with T2 BLLs, 28 with T3 BLLs, and 12 with UCB Pb levels, as well as 15 with patella and 68 with tibia bone measures that met a q-value cut-off of 5% (Supplemental Table 2). Differently methylated genes associated with Pb exposure across all 3 trimesters and at birth were more likely to be enriched in gene pathways involved in detection of chemical stimulus, sensory perception and smell, immunoglobin binding, transmembrane receptor activity, and olfactory receptor activity. Four pathways were statistically significant (q-value < 0.05) among all blood Pb variables, with 35 uniquely overlapping between T1 and T2, 4 uniquely overlapping between T2 and T3, none matching between T1 and T3 only, and 2 matching between T1 and at birth UCB (Figure 1). There were only 4 matching pathways when comparing results from models of tibia and patella bone Pb measures (Supplemental Figure 1).
Figure 1.
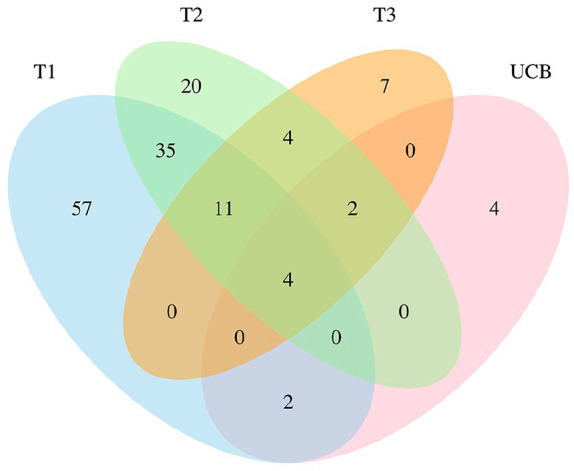
Venn diagram of functional annotations (q-value < 0.05) associated with maternal Pb exposure compared across all 3 trimesters (first trimester, T1; second trimester, T2; third trimester, T3) and umbilical cord blood (UCB).
Significant pathways from Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathways related to neurological development included neurological system processes associated with T1 (q-value = 2.87E10−6) and T2 (q-value = 1.77E10−3) BLLs and tibia bone Pb (q-value = 5.65E10−9). Nodal system pathway was also associated with T1 (q-value = 1.40E10−3) and T2 (q-value = 1.36E10−3) BLLs. Neurotransmitter receptor activity genes were associated with tibia bone Pb (q-value = 7.84E10−3).
Regional analysis
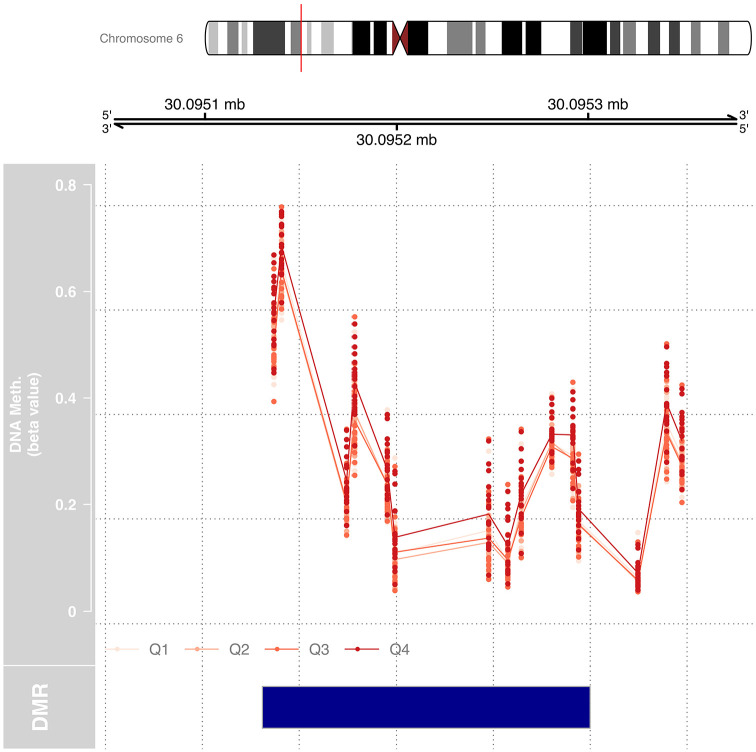
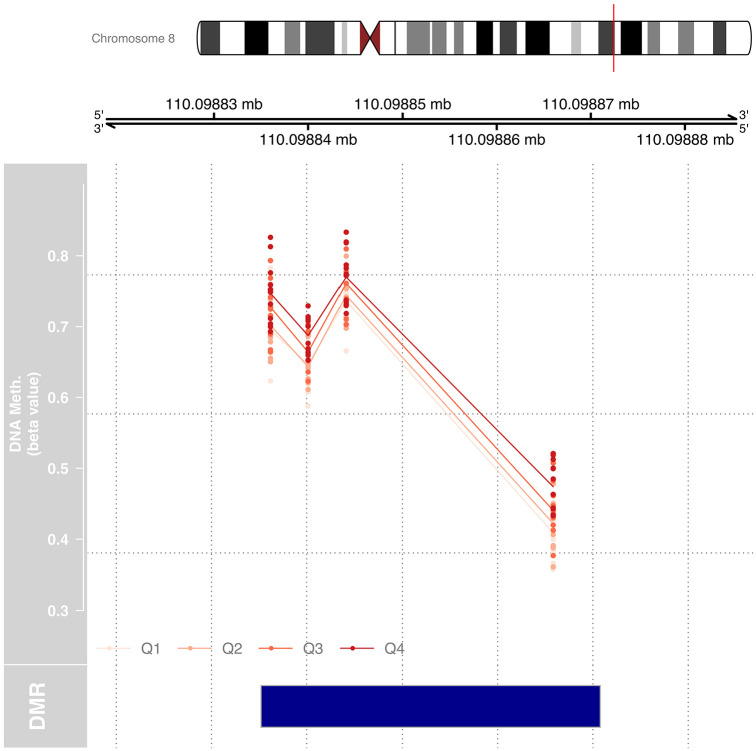
We tested for DMRs by Pb exposure for each trimester using DMRcate, which combines data from at least 2 consecutive CpG sites. DMRcate identified 1 region of differential methylation with T1 BLLs, 1 region with T3 BLLs, and 1 region with tibia bone Pb levels. No regions were identified with T2 BLLs, UCB Pb, or patella bone Pb. The region identified in T1 that was differentially methylated included 4 CpG sites in a region that is located between chr8:17433625-17433761 and is within the promoter of PDGFRL (Figure 2). The region identified in T3 included 12 CpG sites located at chr6:30095136-30095295 (Figure 3). Tibia bone Pb was associated with a region with 4 CpG sites within the promoter of TRHR (Figure 4).
Figure 2.
Differentially methylated region (DMR) by maternal blood Pb concentrations in T1 located within 200 base pairs of the transcription start site of PDGFRL (chr8:17433625-17433761). Linear regression modeling, treating Pb exposure as a continuous variable, adjusted for sex and estimated cell-type proportions. DNA methylation (beta value) is plotted for each sample at 6 CpG sites in the region included 4 sites of the statistically significant DMR. Exposure quartile ranges are represented from light red to dark red with quartile 4 (Q4), representing the top 25% most Pb exposed during T1, being the most methylated at each of the 4 CpG sites within the DMR.
Figure 3.
DMR by maternal blood Pb concentrations in T3 at chr6:30095136-30095295 includes 12 CpG sites within an intergenic region with no known regulatory function. This DMR was selected from a linear regression modeling, treating Pb exposure as a continuous variable, adjusting for sex and estimated cell-type proportions. DNA methylation (beta value) is plotted for each sample at the 12 CpG sites in the DMR. Exposure quartile ranges are represented from light red to dark red with Q4, representing the top 25% most Pb exposed during T3, being the most methylated at each of the 12 CpG sites within the DMR.
Figure 4.
DMR by maternal tibia bone Pb located within 800 base pairs of the transcription start site of TRHR (chr8:110098835-110098870). The DMR was identified by a linear regression model, treating Pb exposure as a continuous variable, and adjusting for sex and estimated cell-type proportions, maternal age, and cohort. DNA methylation (beta value) is plotted for each sample for the 4 CpG sites in the DMR. Exposure quartile ranges are represented from light red to dark red with Q4, representing the top 25% most Pb exposed when cumulative Pb is measured in tibia, being the most methylated at each of the 4 CpG sites within the DMR.
Discussion
We performed an epigenome-wide DNA methylation analysis using DNA extracted from UCB leukocytes and the Illumina MethylationEPIC beadchip in 89 mother-infant pairs to identify differentially methylated genes by trimester-specific and cumulative Pb exposure biomarkers. To our knowledge, this is the first genome-wide DNA methylation study to investigate the association of DNA methylation at birth with prenatal Pb exposure at each of the 3 trimesters of pregnancy, as well as cumulative prenatal Pb exposure in maternal patella and tibia bones. We found that maternal BLLs during T1 and T3, as well as maternal tibia bone Pb levels, were associated with differential methylation at several CpG sites in UCB.
Epidemiological studies of Pb-exposed pregnant women have reported that BLLs below 5 µg/dL can have adverse impacts on neurodevelopment, manifesting for example as decreased scores in the neonatal behavioral neurological assessment test.49 Maternal BLLs under 10 µg/dL have been associated with a 3.5- to 6-point decrease in the Mental Development Index, a measure of IQ using the Bayley Scales of Infant Development, in 24-month-old infants (n = 146).29 Although the inverse association between prenatal Pb exposure and neurodevelopment is known, whether epigenetic perturbation is one of the mechanisms underlying this association is not known.
In the epigenome-wide analysis performed here, the statistically significant DMPs and DMRs associated with biomarkers of prenatal Pb exposure annotated to several genes involved in neurodevelopment or neurologic function. In this study, DNA methylation levels at 3 CpG sites were significantly associated with maternal T1 BLLs, for which one is in a DNase I hypersensitivity site (DHS) within the first exon of RAB5A, a Ras-related protein. DNase I hypersensitivity site are regulatory regions of the genome often located at transcription start sites where the DNA is accessible for transcription factor binding. RAB5A is an important mediator of endocytic pathways due to its role in intracellular membrane trafficking and fusion, and localizing early endosomes.50-53 Evidence from an in vitro study indicates that loss of RAB5A activity may perturb endosomal dynamics and may underlie neuronal dysfunction and degeneration in various motor neuron and neurodegenerative diseases.54 The second CpG site associated with T1 BLLs is located at chr8:12615485, a DHS that is also classified to be within an enhancer region (according to FANTOM5)55 but unclassified to date in terms of the specific gene(s) it may regulate. The third CpG site is in a DHS, a suspected enhancer, and is within 200 base pairs of the transcription start site of EXT1, exostosin glycosyltransferase 1. EXT1 has a primary role in the biosynthesis of heparan sulfate, which is known to regulate various biological processes such as cell proliferation, growth factor signaling, and embryonic development, specifically mammalian neuronal development.56-59
Maternal T1 Pb exposure was also associated with differential methylation in UCB at the regional level, identified by analyzing consecutive probes. We observed a region of 4 CpG sites located at chr8:17433625-17433761 within 200 base pairs of the transcription start site of PDGFRL, platelet-derived growth factor receptor-like protein. This gene encodes a known tumor suppressor that inhibits the growth of colorectal cancer cells in vitro; the function of this gene in development, if any, is not yet known.60
We observed one association between DNA methylation and T3 BLL. The CpG site associated with T3 BLLs is located at chr1:160839299 in an intergenic region within a DHS. In the regional analysis, there was one DMR that was statistically associated with maternal T3 Pb exposure. It annotated to chr6: 30095136-30095295 with 12 methylation sites within an intergenic region with no known regulatory function.
DNA methylation levels at 2 CpG sites were significantly associated with tibia bone Pb, a measure of cumulative Pb exposure throughout and before pregnancy. The first is within a DHS located in the gene LRFN1, Leucine Rich Repeat and Fibronectin Type III Domain Containing 1. LRFN proteins promote neurite outgrowth and synapse formation in hippocampal neurons and have a role in the developing nervous system with involvement in the regulation and conservation of excitatory synapses.61-63 The second is located within a CpG island at chr6:792305 of no known regulatory function to date. We identified one region of differential methylation associated with tibia bone Pb located in the TRHR gene, thyrotropin-releasing hormone receptor, on chr8:110098835-110098870 within 800 base pairs of the transcription start site. This receptor signals synthesis, secretion, and bioactivity of the thyroid stimulating hormone within the pituitary gland in the brain by activating the phosphatidylinositol-calcium-protein kinase C transduction pathway.64,65 We did not identify associations between patella Pb and DNA methylation. Pb in tibia has a residence time of 25 to 30 years66 while patella Pb has a half-life ranging from months to years.67 While both serve as proxies for gestational Pb exposure, differences in half-life and bioavailability (ie, following bone turnover) may influence the relationship between maternal bone Pb and fetal exposure dose throughout pregnancy.
We hypothesized that the greatest number of DMPs by Pb would be identified for T1, yet only a few significant results were obtained for T1, along with T3 and bone Pb. During early gestation, there are 2 major events influencing epigenetic programming: (1) post-fertilization there is a massive wave of demethylation to trigger embryonic developmental; (2) following, there is a subsequent re-methylation that shapes the epigenome in first cell lineages.68,69 These events occur during early T1, and as such environmental perturbation during T1 could lead to epigenetic reprogramming that is propagated across all germ layers and tissues. Differentiation, which is critical for nervous system development, also occurs during T1.70 Throughout the remainder of fetal development, dividing somatic cells are methylated via maintenance epigenetic machinery.16 Thus, while gestational exposures are expected to have the most dramatic effects on the DNA methylome across tissues, subtle changes can still occur in mid- to late-gestation that may be tissue-specific.
Our study is one of a growing body of literature that shows associations between gestational Pb exposure and offspring DNA methylation in rodent models and humans.17-20,71-73 While the DMPs and DMRs identified in this study by T1 and T3 maternal BLL and maternal bone Pb were not the same as those previously reported in other cohorts, collectively these studies show that Pb exposure and DNA methylation vary by timing of exposure, sex, and population. A rural Bangladesh birth cohort study with high environmental Pb exposure performed epigenome-wide DNA methylation profiling using the Infinium HumanMethylation450K BeadChip (450K) on UCB and assessed its association with prenatal Pb exposure measured in maternal urine at 8 weeks’ gestation and in maternal erythrocytes at 14 weeks’ gestation (n = 127).74 This study identified 9 loci associated with urine Pb and 2 loci with erythrocyte Pb. Another study, a US prospective pregnancy cohort, Project Viva, with relatively low levels of Pb exposure (mean maternal erythrocyte Pb, 1.22 ± 0.63 μg/dL) conducted an epigenome-wide analysis using 450K on 268 UCB samples to evaluate the association between prenatal Pb exposure measured on average at 27.9 weeks’ gestation and DNA methylation.20 Their results identified 4 CpG sites in all newborns, as well as sex-specific CpG sites with more found in females (n = 38) than males (n = 2), associated with prenatal Pb. Differences in findings from these cohorts and ELEMENT could be due to differences in exposure levels between the cohorts, participant racial/ethnic background, sex differences, timing of Pb exposure assessment, and statistical power to detect true associations. Even so, our study adds to the previous evidence that prenatal Pb exposure is associated with altered DNA methylation with likely small effect sizes, and this potential mechanism of toxicity or biomarker of response to exposure should be studied in larger cohorts and/or in consortiums.
While the mechanism by which Pb perturbs DNA methylation is not entirely known, several studies provide evidence for direct or indirect changes in DNMT expression and activity by Pb. DNA methyltransferases add methyl groups to CpG sites; if DNMTs fail to add methyl marks during cellular replication, DNA can become passively demethylated.75,76 Pb exposure has been shown to increase reactive oxygen species which inhibits the DNMT binding to DNA causing hypomethylation at some but not all loci.21-23,77,78 In addition, several studies have provided evidence for Pb exposure effects on DNMT expression and function which could result in either increases or decreases in methylation.21,24-27
Our study had several strengths including Pb exposure assessment at multiple time periods during gestation, epigenome-wide assessment of DNA methylation, a prospective study design, and rich data on key covariates. At the same time, our study has limitations. First, due to the sample size, we were unable to perform a sex-specific analysis to determine how sex may influence the relationship between DNA methylation and developmental Pb exposure. We were also underpowered to detect all “true” differentially methylated genes by Pb exposure, especially those with small effect sizes. Second, although epigenomic changes in early gestation would be expected to propagate across all tissues, it is still important to consider tissue specificity when conducting differential methylation studies. While brain is the primary tissue of interest in terms of Pb toxicity, blood-based epigenetic measures are typically necessary in longitudinal epidemiological studies. Cross-tissue studies should be conducted to better understand the biological connection between our findings in UCB with DNA methylation in the brain.79 The Toxicant Exposures and Responses by Genomic and Epigenomic Regulators of Transcription (TaRGET II) consortium is currently promoting efforts toward understanding the role of environment in disease susceptibility as a function of epigenomic perturbations across target and surrogate tissue using mouse models (ie, blood and brain), which will provide insights on cross-tissue, inter-individual variation of epigenomic marks.80 Finally, although DNA methylation changes would be expected to alter gene expression, we are unable to test this hypothesis; storage conditions of archived UCB samples in this cohort did not allow for RNA isolation.
In conclusion, we found that T1, T3, and the cumulative measure in tibia of gestational Pb exposure were associated with several statistically significant changes in DNA methylation. In total, 5 of the 6 CpG sites identified within the study were located within DHS which are important functional regions for gene regulation. There was no statistically significant differential methylation by T2 BLL, cord blood Pb, and patella bone Pb. Our findings are of importance because prenatal Pb exposure has been previously associated with adverse neurodevelopmental outcomes, but whether epigenetic mechanisms contribute to these long-term effects is not well characterized. Pb still remains a widespread environmental health problem. Dietary changes and micronutrient supplementation during pregnancy could play an essential role in neutralizing epigenomic perturbations due to environmental Pb exposure,81-83 and the concentration of Pb in circulation can also be reduced by dietary supplementation (ie, to calcium).84 Many public health promotion efforts are centered on primary prevention of Pb exposure in early childhood even though gestation is also a critical developmental period when Pb can affect long-term health. Our results suggest that prenatal Pb exposure may modify DNA methylation profiles at birth, and this should be explored as one potential mechanism underlying Pb’s neurodevelopmental effects.
Supplemental Material
Supplemental material, Supplemental_Figure_and_table for Trimester-Specific Associations of Prenatal Lead Exposure With Infant Cord Blood DNA Methylation at Birth by Christine A Rygiel, Dana C Dolinoy, Wei Perng, Tamara R Jones, Maritsa Solano, Howard Hu, Martha M Téllez-Rojo, Karen E Peterson and Jaclyn M Goodrich in Epigenetics Insights
Acknowledgments
The authors acknowledge the research staff at participating hospitals and the American British Cowdray Hospital in Mexico City for providing research facilities. We thank the mothers and children for participating in the study.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was made possible by U.S. Environmental Protection Agency (US EPA) grants RD834800 and RD83543601 and National Institute for Environmental Health Sciences (NIEHS) grants P20 ES018171, P01 ES02284401, R01 ES007821, R01 ES014930, R01 ES013744, 1U2C ES026553, and P30 ES017885. This work was also supported by University of Michigan (UM) Genome Science Training Grant T32 HG000040 (C.A.R.). This study was also supported and partially funded by the National Institute of Public Health/Ministry of Health of Mexico. The contents of this publication are solely the responsibility of the grantee and do not necessarily represent the official views of the US EPA or the NIH. Furthermore, the US EPA does not endorse the purchase of any commercial products or services mentioned in the publication.
Declaration of conflicts of interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: CAR, DCD, KEP, and JMG contributed to the conception and design of the study. CAR and TRJ performed experiments. CAR performed the statistical analysis, interpretation of data, and wrote the first draft of the manuscript. JMG provided oversight in the analysis, interpretation, and writing processes. DCD, WP, MS, HH, MMT-R, KEP, and JMG were responsible for obtaining data from the cohort and following the cohort. All authors contributed to the manuscript and approved the submission.
ORCID iD: Christine A Rygiel  https://orcid.org/0000-0001-7690-8945
https://orcid.org/0000-0001-7690-8945
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Santa Maria MP, Hill BD, Kline J. Lead (Pb) neurotoxicology and cognition. Appl Neuropsychol Child. 2018;8:272-293. [DOI] [PubMed] [Google Scholar]
- 2. Lanphear BP, Hornung R, Khoury J, et al. Low-level environmental lead exposure and children’s intellectual function: an international pooled analysis. Environ Health Perspect. 2005;113:894-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Navas-Acien A, Guallar E, Silbergeld EK, Rothenberg SJ. Lead exposure and cardiovascular disease—a systematic review. Environ Health Perspect. 2007;115:472-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reuben A, Caspi A, Belsky DW, et al. Association of childhood blood lead levels with cognitive function and socioeconomic status at age 38 years and with IQ change and socioeconomic mobility between childhood and adulthood. JAMA. 2017;317:1244-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reuben A. Childhood lead exposure and adult neurodegenerative disease. J Alzheimers Dis. 2018;64:17-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Collaborators GRF. Global, regional, and national comparative risk assessment of 84 behavioural, environmental and occupational, and metabolic risks or clusters of risks for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1923-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shaffer RM, Sellers SP, Baker MG, et al. Improving and expanding estimates of the global burden of disease due to environmental health risk factors. Environ Health Perspect. 2019;127:105001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barker D, Eriksson J, Forsen T, Osmond C. Fetal origins of adult disease: strength of effects and biological basis. Int J Epidemiol. 2002;31:1235-1239. [DOI] [PubMed] [Google Scholar]
- 9. Goyer RA. Lead toxicity: from overt to subclinical to subtle health effects. Environ Health Perspect. 1990;86:177-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Silbergeld EK. Lead in bone: implications for toxicology during pregnancy and lactation. Environ Health Perspect. 1991;91:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chuang HY, Schwartz J, Gonzales-Cossio T, et al. Interrelations of lead levels in bone, venous blood, and umbilical cord blood with exogenous lead exposure through maternal plasma lead in peripartum women. Environ Health Perspect. 2001;109:527-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lafond J, Hamel A, Takser L, Vaillancourt C, Mergler D. Low environmental contamination by lead in pregnant women: effect on calcium transfer in human placental syncytiotrophoblasts. J Toxic Environ Health A. 2004;67:1069-1079. [DOI] [PubMed] [Google Scholar]
- 13. Wani AL, Ara A, Usmani JA. Lead toxicity: a review. Interdiscip Toxic. 2015;8:55-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Illingworth RS, Gruenewald-Schneider U, Webb S, et al. Orphan CpG Islands Identify numerous conserved promoters in the mammalian genome. PLoS Genetics. 2010;6:e1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209-213. [DOI] [PubMed] [Google Scholar]
- 16. Vogel Ciernia A, LaSalle JM. The landscape of DNA methylation amid a perfect storm of autism aetiologies. Nature Reviews Neuroscience. 2016;17:411-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Faulk C, Barks A, Liu K, Goodrich JM, Dolinoy DC. Early-life lead exposure results in dose- and sex-specific effects on weight and epigenetic gene regulation in weanling mice. Epigenomics. 2013;5:487-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Montrose L, Faulk C, Francis J, Dolinoy DC. Perinatal lead (Pb) exposure results in sex and tissue-dependent adult DNA methylation alterations in murine IAP transposons. Environ Mol Mutagen. 2017;58:540-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goodrich JM, Sánchez BN, Dolinoy DC, et al. Quality control and statistical modeling for environmental epigenetics: a study on in Utero lead exposure and DNA methylation at birth. Epigenetics. 2015;10:19-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu S, Hivert MF, Cardenas A, et al. Exposure to low levels of lead in utero and umbilical cord blood DNA methylation in project viva: an epigenome-wide association study. Environ Health Perspect. 2017;125:087019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sanchez OF, Lee J, Yu King Hing N, Kim SE, Freeman JL, Yuan C. Lead (Pb) exposure reduces global DNA methylation level by non-competitive inhibition and alteration of DNMT expression. Metallomics. 2017;9:149-160. [DOI] [PubMed] [Google Scholar]
- 22. Castellani RJ, Lee HG, Perry G, Smith MA. Antioxidant protection and neurodegenerative disease: the role of amyloid-beta and tau. Am J Alzheimers Dis Other Demen. 2006;21:126-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2). Nucleic Acids Res. 2004;32:4100-4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dosunmu R, Alashwal H, Zawia NH. Genome-wide expression and methylation profiling in the aged rodent brain due to early-life Pb exposure and its relevance to aging. Mech Ageing Dev. 2012;133:435-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bihaqi SW, Huang H, Wu J, Zawia NH. Infant exposure to lead (Pb) and epigenetic modifications in the aging primate brain: implications for Alzheimer’s disease. J Alzheimers Dis. 2011;27:819-833. [DOI] [PubMed] [Google Scholar]
- 26. Wright RO, Schwartz J, Wright RJ, et al. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ Health Perspect. 2010;118:790-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu J, Basha MR, Brock B, et al. Alzheimer’s disease (AD)-like pathology in aged monkeys after infantile exposure to environmental metal lead (Pb): evidence for a developmental origin and environmental link for AD. J Neurosci. 2008;28:3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Perng W, Tamayo-Ortiz M, Tang L, et al. Early life exposure in Mexico to environmental toxicants (ELEMENT) project. BMJ Open. 2019;9:e030427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu H, Téllez-Rojo MM, Bellinger D, et al. Fetal lead exposure at each stage of pregnancy as a predictor of infant mental development. Environ Health Perspect. 2006;114:1730-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lamadrid-Figueroa H, Téllez-Rojo MM, Hernández-Cadena L, et al. Biological markers of fetal lead exposure at each stage of pregnancy. J Toxicol Environ Health A. 2006;69:1781-1796. [DOI] [PubMed] [Google Scholar]
- 31. Aro AC, Todd AC, Amarasiriwardena C, Hu H. Improvements in the calibration of 109Cd K x-ray fluorescence systems for measuring bone lead in vivo. Phys Med Biol. 1994;39:2263-2271. [DOI] [PubMed] [Google Scholar]
- 32. Moran S, Arribas C, Esteller M. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics. 2016;8:389-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fortin JP, Triche TJ, Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33:558-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bakulski KM, Feinberg JI, Andrews SV, et al. DNA methylation of cord blood cell types: applications for mixed cell birth studies. Epigenetics. 2016;11:354-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Du P, Zhang X, Huang CC, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mansell G, Gorrie-Stone TJ, Bao Y, et al. Guidance for DNA methylation studies: statistical insights from the Illumina EPIC array. BMC Genomics. 2019;20:366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Benjamini Y, Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J Royal Stat Soc B Methodol. 1995;57:289-300. [Google Scholar]
- 40. Peters TJ, Buckley MJ, Statham AL, et al. De novo identification of differentially methylated regions in the human genome. Epigenet Chromatin. 2015;8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim JH, Karnovsky A, Mahavisno V, et al. LRpath analysis reveals common pathways dysregulated via DNA methylation across cancer types. BMC Genomics. 2012;13:526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ettinger AS, Egan KB, Homa DM, Brown MJ. Blood lead levels in U.S. women of childbearing age, 1976-2016. Environ Health Perspect. 2020;128:17012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pantic I, Tamayo-Ortiz M, Rosa-Parra A, et al. Children’s blood lead concentrations from 1988 to 2015 in Mexico City: the contribution of lead in air and traditional lead-glazed ceramics. Int J Environ Res Public Health. 2018;15:2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hisham HJ, Chuah SY, Syarif HL, Nik Nasri I, Fairulnizam MN. Blood lead levels of pregnant women from the Klang Valley. Med J Malaysia. 1998;53:76-81. [PubMed] [Google Scholar]
- 45. Srivastava S, Mehrotra PK, Srivastava SP, Tandon I, Siddiqui MK. Blood lead and zinc in pregnant women and their offspring in intrauterine growth retardation cases. J Anal Toxicol. 2001;25:461-465. [DOI] [PubMed] [Google Scholar]
- 46. Vigeh M, Yokoyama K, Mazaheri M, et al. Relationship between increased blood lead and pregnancy hypertension in women without occupational lead exposure in Tehran, Iran. Arch Environ Health. 2004;59:70-75. [DOI] [PubMed] [Google Scholar]
- 47. Magri J, Sammut M, Savona-Ventura C. Lead and other metals in gestational hypertension. Int J Gynaecol Obstet. 2003;83:29-36. [DOI] [PubMed] [Google Scholar]
- 48. Rahman SN, Fatima P, Chowdhury AQ, Rahman MW. Blood level of lead in women with unexplained infertility. Mymensingh Med J. 2013;22:508-512. [PubMed] [Google Scholar]
- 49. Liu Ja, Gao D, Chen Y, Jing J, Hu Q, Chen Y. Lead exposure at each stage of pregnancy and neurobehavioral development of neonates. Neurotoxicology. 2014;44:1-7. [DOI] [PubMed] [Google Scholar]
- 50. Simonsen A, Lippé R, Christoforidis S, et al. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394:494-498. [DOI] [PubMed] [Google Scholar]
- 51. Hoffenberg S, Liu X, Nikolova L, et al. A novel membrane-anchored Rab5 interacting protein required for homotypic endosome fusion. J Biol Chem. 2000;275:24661-24669. [DOI] [PubMed] [Google Scholar]
- 52. Numrich J, Ungermann C. Endocytic Rabs in membrane trafficking and signaling. Biol Chem. 2014;395:327-333. [DOI] [PubMed] [Google Scholar]
- 53. Homma Y, Kinoshita R, Kuchitsu Y, et al. Comprehensive knockout analysis of the Rab family GTPases in epithelial cells. J Cell Biol. 2019;218:2035-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Otomo A, Hadano S, Okada T, et al. ALS2, a novel guanine nucleotide exchange factor for the small GTPase Rab5, is implicated in endosomal dynamics. Hum Mol Genet. 2003;12:1671-1687. [DOI] [PubMed] [Google Scholar]
- 55. Forrest AR, Kawaji H, Rehli M, et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030-1037. [DOI] [PubMed] [Google Scholar]
- 57. Inatani M, Yamaguchi Y. Gene expression of EXT1 and EXT2 during mouse brain development. Brain Res Dev Brain Res. 2003;141:129-136. [DOI] [PubMed] [Google Scholar]
- 58. Yamaguchi Y, Inatani M, Matsumoto Y, Ogawa J, Irie F. Roles of heparan sulfate in mammalian brain development current views based on the findings from Ext1 conditional knockout studies. Prog Mol Biol Transl Sci. 2010;93:133-152. [DOI] [PubMed] [Google Scholar]
- 59. Liu CC, Zhao N, Yamaguchi Y, et al. Neuronal heparan sulfates promote amyloid pathology by modulating brain amyloid-β clearance and aggregation in Alzheimer’s disease. Sci Transl Med. 2016;8:332ra44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Guo FJ, Zhang WJ, Li YL, et al. Expression and functional characterization of platelet-derived growth factor receptor-like gene. World J Gastroenterol. 2010;16:1465-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Morimura N, Inoue T, Katayama K, Aruga J. Comparative analysis of structure, expression and PSD95-binding capacity of Lrfn, a novel family of neuronal transmembrane proteins. Gene. 2006;380:72-83. [DOI] [PubMed] [Google Scholar]
- 62. Mah W, Ko J, Nam J, Han K, Chung WS, Kim E. Selected SALM (synaptic adhesion-like molecule) family proteins regulate synapse formation. J Neurosci. 2010;30:5559-5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang PY, Seabold GK, Wenthold RJ. Synaptic adhesion-like molecules (SALMs) promote neurite outgrowth. Mol Cell Neurosci. 2008;39:83-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hinkle PM, Gehret AU, Jones BW. Desensitization, trafficking, and resensitization of the pituitary thyrotropin-releasing hormone receptor. Front Neurosci. 2012;6:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Weintraub BD, Gesundheit N, Taylor T, Gyves PW. Effect of TRH on TSH glycosylation and biological action. Ann N Y Acad Sci. 1989;553:205-213. [DOI] [PubMed] [Google Scholar]
- 66. Rabinowitz MB, Wetherill GW, Kopple JD. Kinetic analysis of lead metabolism in healthy humans. J Clin Invest. 1976;58:260-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tsaih SW, Korrick S, Schwartz J, et al. Influence of bone resorption on the mobilization of lead from bone among middle-aged and elderly men: the Normative Aging Study. Environ Health Perspect. 2001;109:995-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Reik W, Dean W. DNA methylation and mammalian epigenetics. Electrophoresis. 2001;22:2838-2843. [DOI] [PubMed] [Google Scholar]
- 69. Kundakovic M, Jaric I. The epigenetic link between prenatal adverse environments and neurodevelopmental disorders. Genes (Basel). 2017;8:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Alfano DP, Petit TL. Neonatal lead exposure alters the dendritic development of hippocampal dentate granule cells. Exp Neurol. 1982;75:275-288. [DOI] [PubMed] [Google Scholar]
- 71. Dou JF, Farooqui Z, Faulk CD, et al. Perinatal lead (Pb) exposure and cortical neuron-specific DNA methylation in male mice. Genes. 2019;10:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sen A, Heredia N, Senut MC, et al. Early life lead exposure causes gender-specific changes in the DNA methylation profile of DNA extracted from dried blood spots. Epigenomics. 2015;7:379-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Faulk C, Liu K, Barks A, Goodrich JM, Dolinoy DC. Longitudinal epigenetic drift in mice perinatally exposed to lead. Epigenetics. 2014;9:934-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Engstrom K, Rydbeck F, Kippler M, et al. Prenatal lead exposure is associated with decreased cord blood DNA methylation of the glycoprotein VI gene involved in platelet activation and thrombus formation. Environ Epigenet. 2015;1:dvv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Develop. 2016;30:733-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chen Z-X, Riggs AD. DNA methylation and demethylation in mammals. J Biol Chem. 2011;286:18347-18353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Valko M, Morris H, Cronin M. Metals, toxicity and oxidative stress. Curr Med Chem. 2005;12:1161-1208. [DOI] [PubMed] [Google Scholar]
- 78. Fowler BA, Whittaker MH, Lipsky M, Wang G, Chen XQ. Oxidative stress induced by lead, cadmium and arsenic mixtures: 30-day, 90-day, and 180-day drinking water studies in rats: an overview. Biometals. 2004;17:567-568. [DOI] [PubMed] [Google Scholar]
- 79. Bakulski KM, Halladay A, Hu VW, Mill J, Fallin MD. Epigenetic research in neuropsychiatric disorders: the “tissue issue.” Curr Behav Neurosci Rep. 2016;3:264-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang T, Pehrsson EC, Purushotham D, et al. The NIEHS TaRGET II Consortium and environmental epigenomics. Nature Biotechnology. 2018;36:225-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Cropley JE, Suter CM, Beckman KB, Martin DI. Germ-line epigenetic modification of the murine Avy allele by nutritional supplementation. Proc Natl Acad Sci U S A. 2006;103:17308-17312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ettinger AS, Hu H, Hernandez-Avila M. Dietary calcium supplementation to lower blood lead levels in pregnancy and lactation. J Nutr Biochem. 2007;18:172-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 2007;104:13056-13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hernandez-Avila M, Gonzalez-Cossio T, Hernandez-Avila JE, et al. Dietary calcium supplements to lower blood lead levels in lactating women: a randomized placebo-controlled trial. Epidemiology (Cambridge, Mass). 2003;14:206-212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplemental_Figure_and_table for Trimester-Specific Associations of Prenatal Lead Exposure With Infant Cord Blood DNA Methylation at Birth by Christine A Rygiel, Dana C Dolinoy, Wei Perng, Tamara R Jones, Maritsa Solano, Howard Hu, Martha M Téllez-Rojo, Karen E Peterson and Jaclyn M Goodrich in Epigenetics Insights