Abstract
Background:
In December of 2019, coronavirus disease 2019 (Covid-19) was reported in Wuhan, China, and has now rapidly swept around the world. Much research has been carried out since the outbreak, but few studies have focused on the dysfunction of the adaptive immunity.
Methods:
In this retrospective and multi-center study, 373 patients with laboratory-confirmed COVID-19 from Shanghai Public Health Clinical Center and Affiliated Hospital of Putian University were recruited. Demographic, clinical, radiological features, and laboratory data were recorded and analyzed at admission and at discharge. Results of immunological tests were followed up until the patients were discharged.
Results:
Of the 373 patients with COVID-19 pneumonia, 322 were in the non-severe group and 51 were in the severe group. Number of T cells, CD4+ and CD8+ T cells, and total lymphocytes declined remarkably upon admission and elevated when the patients were discharged. At admission, counts of total lymphocytes, T cells, CD4+ and CD8+ T cells, and levels of C3 and C4 in the severe group were lower than those in the non-severe group, whereas the neutrophil to lymphocyte ratio (NLR) was higher in the severe group. Counts of T cells, CD4+ and CD8+ T cells, and total lymphocytes were negatively correlated with lactate dehydrogenase and C-reactive protein.
Conclusion:
COVID-19 might target adaptive immunity and cause a decrease in lymphocytes, especially T cells and subsets. Physicians should pay close attention to the adaptive immunity of patients upon admission. Monitoring NLR, T lymphocytes, and subsets would help physicians with the proper diagnosis and treatment of COVID-19.
The reviews of this paper are available via the supplemental material section.
Keywords: Adaptive immunity, T lymphocytes, T lymphocyte subgroups, COVID-19
Introduction
In December of 2019, a coronavirus 2 (SARS-CoV-2)-associated severe respiratory syndrome novel pneumonia, which has been named as coronavirus disease-2019 (COVID-19), was reported in Wuhan, Hubei Province of China.1 Up until 21 March 2020, data from the World Health Organization (WHO) have shown that more than 292,142 confirmed cases have been identified in 114 countries and regions.2
SARS-CoV-2 belongs to a clade of the sarbecovirus subgenus of the Orthocoronavirinae subfamily.3 Although the pathogenesis of SARS-CoV-2 remains unclear, lymphopenia has been reported to occur in most COVID-19 patients.4,5 The feature has also been found in two other fatal human coronavirus-associated pneumonias: severe acute respiratory syndrome coronavirus (SARS-CoV) and the Middle East respiratory syndrome coronavirus (MERS-CoV).6,7 According to previous studies, T lymphocytes play an important role in the immune response to antiviral infection,8 which indicates that lymphocyte counts could be critical indicators associated with the severity of the pneumonia and the clinical outcome of COVID-19 patients. However, few studies have shed light on these aspects.
Therefore, this study was conducted to investigate the association between immunological indicators including counts of lymphocytes and subgroups and the severity of the pneumonia. We analyzed the immunological indicators of patients with COVID-19 pneumonia with different degrees of severity and compared those indicators between patients at admission and discharge. Correlation of immunological indicators with lactate dehydrogenase (LDH) and C-reactive protein (CRP) was also investigated in the study.
Methods
Study design and participants
In this retrospective study, 373 patients with COVID-19 in the period of 20 January to 29 February 2020 were enrolled at the Shanghai Public Health Clinical Center (SPHCC) and Affiliated Hospital of Putian University, which are both COVID-19 pneumonia-designated hospitals in China. All 373 patients enrolled were diagnosed as having COVID-19 according to the criteria from the Guideline of Diagnosis and Treatment of COVID-19 of National Health Commission of China. Of the 373 patients, 8 patients were classified as mild cases, 314 patients as moderate cases, 33 patients as severe cases, and 18 patients as critical cases. We combined mild and moderate cases into a group named the non-severe group, and combined severe and critical cases into a group named the severe group. The objective of the study and its requirements were explained to the subjects. This case series was approved by the institutional ethics board of the Affiliated Hospital of Putian University, China and the Ethics Committee of SPHCC; oral consent was obtained from patients involved before data were collected retrospectively.
Classification of COVID-19
According to the Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Seventh Version, Released by National Health Commission & State Administration of Traditional Chinese Medicine), severity of COVID-19 infection is classified as follows.
1. Mild cases
The clinical symptoms were mild, and there was no sign of pneumonia on imaging.
2. Moderate cases
Showing fever and respiratory symptoms with radiological findings of pneumonia.
3. Severe cases
Adult cases meeting any of the following criteria:
(a) respiratory distress (⩾30 breaths/ min);
(b) oxygen saturation ⩽93% at rest;
(c) arterial partial pressure of oxygen (PaO2)/ fraction of inspired oxygen (FiO2) ⩽300 mmHg (l mmHg = 0.133 kPa).
4. Critical cases
Cases meeting any of the following criteria:
(a) respiratory failure and requiring mechanical ventilation;
(b) shock;
(c) with other organ failure that requires ICU care.
According to the criteria, the patients were classified as mild cases, moderate cases, severe cases, and critical cases.
Blood routine tests
A total of 2 ml of EDTA anticoagulated peripheral blood of the patients with COVID-19 pneumonia was drawn at admission and discharge, then the blood samples were sent to the clinical labs and analyzed by BC-5800 Automatic Blood Cell Analyzer (Mairui, Shenzhen, China), according to the manufacturer’s instructions. Count and volume distribution of red blood cells and platelets were measured by an electric resistance method. Concentration of hemoglobin was measured by colorimetric method. Counts of white blood cells and subsets were measured by laser flow cytometry. Data can be read from the machine directly, including white blood cells, red blood cells, platelets, lymphocytes, neutrophils, and monocytes. The neutrophil-to-lymphocyte ratio (NLR) was calculated as the ratio of neutrophil count to lymphocyte count.
Flow cytometry of peripheral blood
EDTA anticoagulated peripheral blood samples were collected from COVID-19 patients at admission and another sample from the same patient was collected at discharge. All the blood samples were tested by the clinical labs within 6 h. CD3+/CD4+/CD8+ T cells were measured by multiple-color flow cytometry with human monoclonal anti-CD3-fluorescein isothiocyanate, anti-CD4-phycoerythrin, and anti-CD8-allophycocyanin according to the manufacturer’s instructions as reported in a previous study.9 Lymphocyte subsets detection kits used in this study were purchased from BD company (Becton, Dickinson and Company, Franklin Lakes, USA). The cells were analyzed on a BD FACS Canto II flow cytometry system and analyzed with the BD FACS Diva Software (BD Biosciences, Becton, Dickinson and Company, Franklin Lakes, USA). The percentage and the absolute count of lymphocyte subsets were determined, including T cells (CD3+, CD45+), helper T cells (CD4+, CD3+, CD45+), and suppressor T cells (CD8+, CD3+, CD45+). Each sample was tested in duplicate.
Confirmation of SARS-CoV-2
Respiratory specimens were collected by the local Centers for Disease Control (CDC) according to the WHO guidance and then delivered to designated authoritative laboratories to detect SARS-CoV-2. Real-time reverse transcription polymerase chain reaction (RT-PCR) assays were performed in accordance with the protocol established by the WHO.10
Data resources
The medical records of patients were analyzed by the research team of the Department of Pulmonary Medicine, Zhongshan Hospital of Fudan University. Epidemiological, clinical, laboratory, radiological characteristics, and the treatment records of the patients were collected from Shanghai Public Health Clinical Center and Affiliated Hospital of Putian University as forms from electronic medical records. Blood was withdrawn and the data were collected at the time when the patients were hospitalized and when they met the discharge criteria: (a) body temperature back to normal for more than 3 days; (b) respiratory symptoms show obvious improvement; (c) pulmonary imaging shows obvious absorption of inflammation; (d) nuclei acid tests negative twice consecutively on respiratory tract samples such as sputum and nasopharyngeal swabs (sampling interval being at least 24 h). The local CDC labs and the Chinese CDC have made a clear definition of COVID-19 infection by throat-swab specimens from the upper respiratory tract. RT-PCR assay was used to confirm the COVID-19 infection.
Outcomes
Data includes symptoms, comorbidity, laboratory, and radiological characteristics of patients at admission, laboratory results of routine blood examinations, biochemical tests, and immunological tests of the last time before the patient was discharged.
Statistical analysis
Continuous variables were presented as mean ± standard deviation. Categorical variables were expressed as frequency and percentage. According to the Chinese guidelines, patients were classified as four groups, namely non-severe (mild and moderate cases) and severe groups (severe and critical cases). Comparison between the two groups was assessed by Student’s t-test or Mann–Whitney rank sum test (for continuous variables) and chi-squared test or Fisher’s exact tests (for categorical variables). To compare the differences of the variables of immunological tests, CRP, and LDH at admission and discharge, paired t-test or Wilcoxon matched-pairs sign-rank test was applied. Regression analysis was used to determine the relationship between LDH and CRP with different variables of immunological examinations, such as T lymphocytes, CD4+ and CD8+ T cells, and total lymphocytes count (TLC) at admission and discharge. Stata version 15 (Stata Corp, LLC, Texas, USA), GraphPad Prism (version 6; GraphPad Software, San Diego, California, USA), and Excel (Version Office 365, Microsoft Corporation, the United States) were used for all statistical analysis and drawing graphs, and the level of significance was set to p < 0.05. The data of significant difference were marked in bold.
Results
Demographics and descriptive
A total of 373 patients with COVID-19 were included in this study, their clinical characteristics are displayed in Table 1. In total 51 of them were in the severe group. The proportion of patients under 39 years old in the severe group was significantly lower than that in the non-severe group (p = 0.0010) and the proportion over 70 years was significantly higher than in the non-severe group (70–79: p = 0.0010; ⩾80: p = 0.0001). Length of hospital stay of patients in the severe group was significantly longer than the non-severe group. A total of 47% of patients [n = 24 (47.06%)] from the severe group had comorbidity, which was significantly higher than the non-severe group [n = 104 (32.30%), p = 0.0390]. Among the comorbidities, prevalence of cardiovascular disease and diabetes in the non-severe group was significantly lower than the severe group (cardiovascular disease: p = 0.0130; diabetes: p = 0.0466). Common self-reported symptoms at onset of illness were fever [n = 184 (48.33%)], cough [n = 127 (34.05%)], and sputum production [n = 78 (20.91%)]. Compared with the non-severe group, a higher proportion of patients in the severe group reported chills/rigors, fatigue, and anorexia (chills and rigors: p < 0.0001; fatigue: p = 0.0160; anorexia: p = 0.0326).
Table 1.
Clinical characteristics of patients with COVID-19.
| Characteristic | Total (n = 373) |
Non-severe (n = 322) |
Severe (n = 51) |
p value |
|---|---|---|---|---|
| Age, n (%) | ||||
| ⩽39 | 128 (34.32) | 121 (37.58) | 7 (13.73) | 0.0010** |
| 40–49 | 62 (16.62) | 55 (17.08) | 7 (13.73) | 0.5500 |
| 50–59 | 67 (17.96) | 60 (18.63) | 7 (13.73) | 0.3960 |
| 60–69 | 77 (20.64) | 63 (19.57) | 14 (27.45) | 0.1960 |
| 70–79 | 32 (8.58) | 21 (6.52) | 11 (21.57) | 0.0010** |
| ⩾80 | 7 (1.88) | 2 (0.62) | 5 (9.80) | 0.0001*** |
| Male | 197 (52.82) | 168 (52.17) | 29 (56.86) | 0.0390* |
| Days from illness onset to admission days | 5.191 (3.651) | 5.249 (3.751) | 4.824 (2.951) | 0.4400 |
| Hospitalization days | 17.48 (8.702) | 16.51 (7.560) | 23.61 (12.33) | <0.0001**** |
| Comorbidity, n (%) | ||||
| Any | 128 (34.32) | 104 (32.30) | 24 (47.06) | 0.0390* |
| Hypertension | 71 (19.03) | 59 (18.32) | 12 (23.53) | 0.3790 |
| Cardiovascular disease | 18 (4.83) | 12 (3.73) | 6 (11.76) | 0.0130* |
| Diabetes | 29 (7.77) | 21 (6.52) | 8 (15.69) | 0.0466* |
| COPD | 4 (1.07) | 2 (0.62) | 2 (3.92) | 0.0920 |
| Chronic liver disease | 14 (3.75) | 10 (3.11) | 4 (7.84) | 0.2086 |
| Others | 29 (7.77) | 24 (7.45) | 5 (9.80) | 0.7634 |
| Symptoms, n (%) | ||||
| Fever | 184 (48.33) | 156 (48.45) | 28 (54.90) | 0.3920 |
| Cough | 127 (34.05) | 106 (32.92) | 21 (41.18) | 0.2480 |
| Headache | 18 (4.83) | 15 (4.66) | 3 (5.88) | 0.9782 |
| Sputum production | 78 (20.91) | 64 (19.88) | 14 (27.45) | 0.2160 |
| Diarrhea | 22 (5.90) | 20 (6.21) | 2 (3.92) | 0.7452 |
| Muscle pain | 34 (9.12) | 31 (9.63) | 3 (5.88) | 0.7990 |
| Chills and rigors | 22 (5.90) | 7 (2.17) | 15 (29.41) | <0.0001**** |
| Fatigue | 54 (14.48) | 41 (12.73) | 13 (25.49) | 0.0160* |
| Anorexia | 18 (4.83) | 12 (3.73) | 6 (11.76) | 0.0326* |
Data was presented as mean (SD).
p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
COVID-19, coronavirus disease 2019; COPD, chronic obstructive pulmonary disease.
Laboratory indices
Laboratory findings upon admission to hospital are summarized in Table 2. All subjects displayed at least one abnormality in imaging, and the most common abnormality was ground-glass opacity [n = 343 (91.96%)]. Unilateral pneumonia was more common in the non-severe group (p = 0.0140), while bilateral pneumonia was more common in the severe group (p = 0.0110). Compared with the non-severe group, levels of aspartate aminotransferase (AST) and total bilirubin (TB) levels in the severe group were significantly higher (AST: p < 0.0001; TB: p = 0.0367).
Table 2.
Laboratory results of patients with COVID-19.
| Variables | Normal range | Total (n = 373) |
Non-severe (n = 322) |
Severe (n = 51) |
p value |
|---|---|---|---|---|---|
| Abnormalities on imaging examination, n (%) | |||||
| Ground-glass opacity | 343 (91.96) | 294 (91.30) | 49 (96.07) | 0.3747 | |
| Unilateral pneumonia | 78 (20.91) | 74 (22.98) | 4 (7.84) | 0.0140* | |
| Bilateral pneumonia | 266 (71.31) | 222 (68.94) | 44 (86.27) | 0.0110* | |
| White blood cell count, ×109/l | 3.5–9.5 | 5.374 (2.310) | 5.277 (1.963) | 5.978 (3.789) | 0.6029 |
| Hemoglobin, g/l | 115–150 | 135.9 (15.82) | 135.8 (16.05) | 136.7 (14.36) | 0.7063 |
| Platelet count, ×1012/l | 100–300 | 198.0 (72.56) | 200.4 (73.74) | 183.1 (63.25) | 0.0856 |
| Alanine aminotransferase, U/l | 7–40 | 26.96 (19.58) | 26.77 (20.2) | 28.16 (15.27) | 0.0814 |
| Aspartate aminotransferase, U/l | 13–40 | 28.30 (19.94) | 27.23 (20.06) | 34.98 (17.91) | <0.0001**** |
| Total bilirubin, µmol/l | 5.1–19 | 9.588 (4.536) | 9.412 (4.511) | 10.68 (4.583) | 0.0367* |
| Blood urea nitrogen, mmol/l | 2.68–8.2 | 4.626 (2.654) | 4.465 (1.622) | 5.626 (5.814) | 0.2528 |
| Creatinine, µmol/l | 35–84 | 65.87 (25.98) | 64.63 (20) | 73.45 (48.34) | 0.1945 |
Data was presented as mean (SD).
p < 0.05, ****p < 0.0001.
COVID-19, coronavirus disease 2019.
Immunological indicators of different severity
The cell counts of CD3+, CD4+, and CD8+ lymphocytes and TLC in the severe group were significantly lower than those in the non-severe group at admission (CD3+: p < 0.0001; CD4+: p < 0.0001; CD8+: p < 0.0001; TLC: p < 0.0001) and discharge (CD3+: p < 0.0001; CD4+: p < 0.0001; CD8+: p = 0.0086; TLC: p < 0.0001). Although we failed to find any significant difference in the ratio of CD4+ and CD8+ lymphocyte counts (CD4+/CD8+) between groups at admission, CD4+/CD8+ in the severe group was significantly lower at discharge than the non-severe group (p = 0.0464). Neutrophil count was significantly higher in the severe group compared with patients in the non-severe group at admission (p = 0.0436), while no significant difference was observed at discharge. NLR was significantly higher in the severe group compared with patients in the non-severe group both at admission (p < 0.0001) and discharge (p = 0.0010; Tables 3 and 4 and Figure 1)
Table 3.
The levels of LDH, CRP, and immunological indicators of COVID-19 pneumonia patients at admission.
| Variables | Normal range | Total | Non-severe | Severe | p value |
|---|---|---|---|---|---|
| CD3+ lymphocytes, ×106/l | 690–2540 | 857.17 (480.40) | 899.72 (424.50) | 585.74 (690.52) | <0.0001**** |
| CD4+CD3+ lymphocytes, ×106/l | 410–1590 | 503.82 (268.48) | 535.41 (265.67) | 302.28 (187.94) | <0.0001**** |
| CD8+CD3+ lymphocytes, ×106/l | 190–1140 | 321.04 (271.18) | 331.07 (185.65) | 258.26 (564.68) | <0.0001**** |
| CD4+/CD8+ | 0.9–3.6 | 1.89 (1.04) | 1.88 (1.04) | 1.92 (1.07) | 0.8911 |
| C3, g/l | 0.9–1.8 | 1.18 (0.22) | 1.20 (0.22) | 1.07 (0.24) | 0.0004*** |
| C4, g/l | 0.1–0.4 | 0.33 (0.31) | 0.34 (0.33) | 0.28 (0.09) | 0.0323* |
| IgM, g/l | 0.4–2.3 | 1.11 (0.71) | 1.13 (0.74) | 1.00 (0.41) | 0.3313 |
| IgA, g/l | 0.7–4 | 2.62 (1.28) | 2.63 (1.29) | 2.56 (1.24) | 0.4460 |
| IgG, g/l | 7–16 | 12.35 (2.86) | 12.32 (2.67) | 12.54 (3.92) | 0.8905 |
| Monocytes, ×109/l | 0.1–0.6 | 0.47 (0.22) | 0.47 (0.19) | 0.47 (0.36) | 0.9513 |
| NLR | <1.2 | 3.61 (4.45) | 2.93 (1.80) | 7.90 (10.20) | <0.0001**** |
| Neutrophil count, ×109/l | 1.8–6.3 | 3.56 (2.07) | 3.34 (1.43) | 4.96 (4.06) | 0.0436* |
| TLC, ×109/l | 1.1–3.2 | 1.28 (0.61) | 1.34 (0.57) | 0.95 (0.75) | <0.0001**** |
| LDH, IU/l | 109–245 | 251.06 (102.60) | 233.51 (77.26) | 361.18 (159.59) | <0.0001**** |
| CRP, mg/l | 0–10 | 20.17 (25.89) | 16.88 (21.50) | 41.84 (19.14) | <0.0001**** |
Data was presented as mean (SD).
p < 0.05, ***p < 0.001, ****p < 0.0001.
C3, complement 3; C4, complement 4; CD4+/CD8+, the ratio of CD4+ lymphocyte counts to CD8+ lymphocyte counts; COVID-19, coronavirus disease 2019; CRP, C-reactive protein; Ig, immunoglobulin; LDH, lactate dehydrogenase; NLR, neutrophil-to-lymphocyte ratio; TLC, total lymphocyte count.
Table 4.
The levels of LDH, CRP, and leukocyte counts of COVID-19 pneumonia patients at discharge.
| Variables | Normal range | Total | Non-severe | Severe | p value |
|---|---|---|---|---|---|
| CD3+ lymphocytes, ×106/l | 690–2540 | 1279.10 (504.71) | 1342.67 (493.06) | 938.79 (429.62) | <0.0001**** |
| CD4+CD3+ lymphocytes, ×106/l | 410–1590 | 767.93 (329.01) | 810.72 (321.02) | 530.61 (269.47) | <0.0001**** |
| CD8+CD3+ lymphocytes, ×106/l | 190–1140 | 441.36 (200.65) | 455.71 (198.07) | 357.58 (198.21) | 0.0086** |
| CD4+/CD8+ | 0.9–3.6 | 2.00 (1.01) | 2.02 (0.96) | 1.87 (1.27) | 0.0464* |
| NLR | <1.2 | 2.16 (1.84) | 1.99 (1.04) | 3.28 (4.17) | 0.0010** |
| Neutrophil count, ×106/l | 1.8–6.3 | 3.39 (1.39) | 3.35 (1.25) | 3.68 (2.09) | 0.8482 |
| TLC, ×109/l | 1.1–3.2 | 1.78 (0.59) | 1.84 (0.59) | 1.44 (0.48) | <0.0001**** |
| LDH, IU/l | 109–245 | 212.90 (58.50) | 205.84 (49.31) | 260.03 (87.35) | <0.0001**** |
| CRP, mg/l | 0–10 | 1.82 (5.82) | 1.33 (2.57) | 5.02 (14.27) | 0.1671 |
Data was presented as mean (SD).
p < 0.05, **p < 0.01, ****p < 0.0001.
C3, complement 3; C4, complement 4; CD4+/CD8+, the ratio of CD4+ lymphocyte counts to CD8+ lymphocyte counts; COVID-19, coronavirus disease 2019; CRP, C-reactive protein; Ig, immunoglobulin; LDH, lactate dehydrogenase; NLR, neutrophil to lymphocyte ratio; TLC, total lymphocyte count.
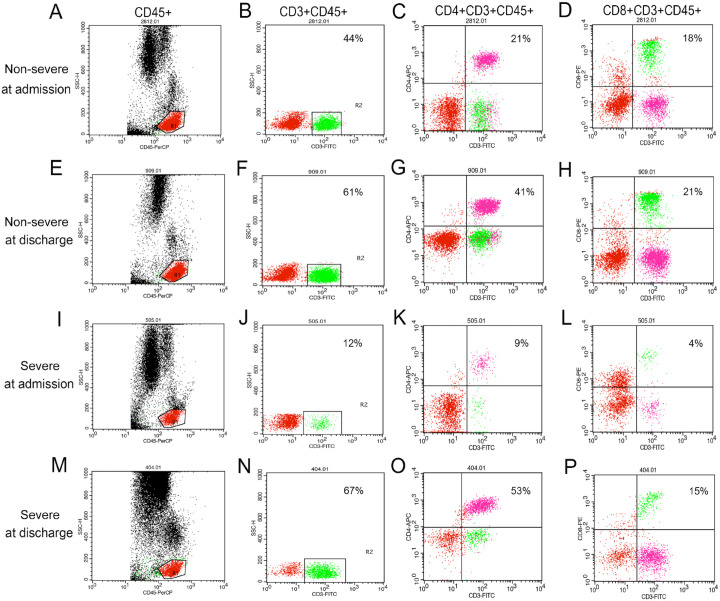
Figure 1.
Flow cytometry scatter plot of leukocytes from COVID-19 patients.
(A–P) Representative flow cytometry scatter plot of CD45+ (A, E, I, M), CD3+CD45+ (B, F, J, N), CD4+CD3+CD45+ (C, G, K, O), CD8+CD3+CD45+ (D, H, L, P) lymphocytes of COVID-19 patients: (A–D) representative flow cytometry scatter plot of one non-severe patient at admission; (E–H) representative flow cytometry scatter plot of the same non-severe patient at discharge; (I–L) representative flow cytometry scatter plot of one severe patient at admission; (M–P) representative flow cytometry scatter plot of the same severe patient at discharge. A–H represent one non-severe patient and the numbers indicate this non-severe patient’s percentages of lymphocytes. I–P represent one severe patient and the numbers indicate this severe patient’s percentages of lymphocyte. COVID-19, coronavirus disease 2019.
At admission, levels of C3 and C4 in the severe group were significantly lower than those in the non-severe group (C3: p = 0.0004; CD4+: p = 0.0323). We failed to find any significant difference in levels of IgM, IgA, or IgG between patients in the severe and non-severe group (Tables 3 and 4).
Leukocytes at admission and discharge
Comparing the immunological indicators of the patients at admission and discharge, we found that CD3+, CD4+, and CD8+ lymphocyte counts and TLC in discharged patients were elevated in both the severe group (CD3+: p < 0.0001; CD4+: p < 0.0001; CD8+: p < 0.0001; TLC: p < 0.0001) and non-severe group (CD3+: p < 0.0001; CD4+: p < 0.0001; CD8+: p < 0.0001; TLC: p < 0.0001) compared with those at admission. CD4+/CD8+ in the non-severe group was elevated at discharge compared with that at admission (p = 0.0005), while no significant difference was found in the severe group. Discharged patients had lower NLR than when they were admitted in both the severe group (p < 0.0001) and non-severe group (p < 0.0001). However, no significant difference in neutrophil counts was found between at admission and discharge (Table 5 and Figure 1).
Table 5.
Comparison of the levels of LDH, CRP, and leukocyte counts of COVID-19 pneumonia patients at admission and discharge.
| Variables | At admission | At discharge | p value |
|---|---|---|---|
| CD3+ lymphocytes, ×106/l | |||
| Total | 857.17 (480.40) | 1279.10 (504.71) | <0.0001**** |
| Non-severe | 899.72 (424.50) | 1342.67 (493.06) | <0.0001**** |
| Severe | 585.74 (690.52) | 938.79 (429.62) | <0.0001**** |
| CD4+CD3+ lymphocytes, ×106/l | |||
| Total | 503.82 (268.48) | 767.93 (329.01) | <0.0001**** |
| Non-severe | 535.41 (265.67) | 810.72 (321.02) | <0.0001**** |
| Severe | 302.28 (187.94) | 530.61 (269.47) | <0.0001**** |
| CD8+CD3+ lymphocytes, ×106/l | |||
| Total | 321.04 (271.18) | 441.36 (200.65) | <0.0001**** |
| non-severe | 331.07 (185.65) | 455.71 (198.07) | <0.0001**** |
| Severe | 258.26 (564.68) | 357.58 (198.21) | <0.0001**** |
| CD4+/CD8+ | |||
| Total | 1.89 (1.04) | 2.00 (1.01) | 0.0024** |
| Non-severe | 1.88 (1.04) | 2.02 (0.96) | 0.0005*** |
| Severe | 1.92 (1.07) | 1.87 (1.27) | 0.7499 |
| NLR | |||
| Total | 3.61 (4.45) | 2.16 (1.84) | <0.0001**** |
| Non-severe | 2.93 (1.80) | 1.99 (1.04) | <0.0001**** |
| Severe | 7.90 (10.20) | 3.28 (4.17) | <0.0001**** |
| Neutrophil count, ×109/l | |||
| Total | 3.56 (2.07) | 3.39 (1.39) | 0.8363 |
| Non-severe | 3.34 (1.43) | 3.35 (1.25) | 0.4558 |
| Severe | 4.96 (4.06) | 3.68 (2.09) | 0.2362 |
| TLC, ×109/l | |||
| Total | 1.28 (0.61) | 1.78 (0.59) | <0.0001**** |
| Non-severe | 1.34 (0.57) | 1.84 (0.59) | <0.0001**** |
| Severe | 0.95 (0.75) | 1.44 (0.48) | <0.0001**** |
| LDH, U/l | |||
| Total | 251.06 (102.60) | 212.90 (58.50) | <0.0001**** |
| Non-severe | 233.51 (77.26) | 205.84 (49.31) | <0.0001**** |
| Severe | 361.18 (159.59) | 260.03 (87.35) | <0.0001**** |
| CRP, mg/l | |||
| Total | 20.17 (25.89) | 1.82 (5.82) | <0.0001**** |
| Non-severe | 16.88 (21.50) | 1.33 (2.57) | <0.0001**** |
| Severe | 41.84 (19.14) | 5.02 (14.27) | <0.0001**** |
Data was presented as mean (SD).
p < 0.01, ***p < 0.001, and ****p < 0.0001.
C3, complement 3; C4, complement 4; CD4+/CD8+, the ratio of CD4+ lymphocyte counts to CD8+ lymphocyte counts; COVID-19, coronavirus disease 2019; CRP, C-reactive protein; Ig, immunoglobulin; LDH, lactate dehydrogenase; NLR, neutrophil to lymphocyte ratio; TLC, total lymphocyte count.
Correlation between LDH and Leukocytes
Compared with the non-severe group, LDH levels were significantly higher in patients from the severe group at admission (p < 0.0001) and discharge (p < 0.0001). In addition, LDH levels of discharged patients in both the non-severe group (p < 0.0001) and severe group (p < 0.0001) were significantly lower than those at admission (Tables 3–5). Therefore, we performed a correlation analyses between LDH and immunological indicators (Table 6).
Table 6.
Correlation between the levels of LDH and leukocyte counts in patients with COVID-19 pneumonia.
| Variables | At admission |
At discharge |
||
|---|---|---|---|---|
| r | p | r | p | |
| CD3+ lymphocytes, ×106/l | ||||
| Total | −0.4424 | <0.0001**** | −0.2113 | 0.0020** |
| Non-severe | −0.3543 | <0.0001**** | −0.2108 | 0.0045** |
| Severe | −0.5787 | <0.0001**** | 0.1945 | 0.2861 |
| CD4+CD3+ lymphocytes, ×106/l | ||||
| Total | −0.4121 | <0.0001**** | −0.2139 | 0.0017** |
| Non-severe | −0.3128 | <0.0001**** | −0.2106 | 0.0044** |
| Severe | −0.5624 | <0.0001**** | 0.1943 | 0.2866 |
| CD8+CD3+ lymphocytes, ×106/l | ||||
| Total | −0.3910 | <0.0001**** | −0.2148 | 0.0018** |
| Non-severe | −0.3054 | <0.0001**** | −0.1978 | 0.0079** |
| Severe | −0.5319 | <0.0001**** | −0.1195 | 0.5370 |
| NLR | ||||
| Total | 0.3972 | <0.0001**** | 0.2557 | <0.0001**** |
| Non-severe | 0.2835 | <0.0001**** | 0.2260 | 0.0001*** |
| Severe | 0.6588 | <0.0001**** | 0.2525 | 0.0868 |
| Neutrophil count, ×109/l | ||||
| Total | 0.1673 | 0.0013** | 0.1459 | 0.0057** |
| Non-severe | 0.0741 | 0.1878 | 0.1414 | 0.0125* |
| Severe | 0.5454 | <0.0001**** | 0.2619 | 0.0753 |
| TLC, ×109/l | ||||
| Total | −0.3996 | <0.0001**** | −0.1921 | 0.0003*** |
| Non-severe | −0.3161 | <0.0001**** | −0.1394 | 0.0139* |
| Severe | −0.4688 | <0.0001**** | −0.1496 | 0.3154 |
| CRP, mg/l | ||||
| Total | 0.5966 | <0.0001**** | 0.2669 | <0.0001**** |
| Non-severe | 0.5393 | <0.0001**** | 0.2459 | <0.0001**** |
| Severe | 0.5898 | <0.0001**** | 0.3560 | 0.0223* |
p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
C3, complement 3; C4, complement 4; CD4+/CD8+, the ratio of CD4+ lymphocyte counts to CD8+ lymphocyte counts; COVID-19, coronavirus disease 2019; CRP, C-reactive protein; Ig, immunoglobulin; LDH, lactate dehydrogenase; NLR, neutrophil-to-lymphocyte ratio; r, correlation coefficient; TLC, total lymphocyte count.
At admission, CD3+, CD4+, and CD8+ lymphocyte counts and TLC were negatively correlated with LDH levels in both the non-severe group (CD3+: p < 0.0001; CD4+: p < 0.0001; CD8+: p < 0.0001; TLC: p < 0.0001) and severe group (CD3+: p < 0.0001; CD4+: p < 0.0001; CD8+: p < 0.0001; TLC: p < 0.0001). Neutrophil count in the severe group was positively correlated with LDH level (p < 0.0001), while we failed to find any correlation in the non-severe group. NLR and LDH levels were positively correlated in the non-severe group (p < 0.0001) and severe group (p < 0.0001).
At discharge, CD3+, CD4+, and CD8+ lymphocyte counts and TLC were negatively correlated with LDH level (CD3+: p = 0.0045; CD4+: p = 0.0044; CD8+: p = 0.0079; TLC: p = 0.0139) in the non-severe group, while neutrophil count and NLR were positively correlated with LDH level (neutrophil count: p = 0.0125; NLR: p = 0.0001). However, we failed to find any correlation between leukocyte counts and LDH level in the severe group.
Correlation between CRP and leukocytes
Compared with the non-severe group, CRP levels were significantly higher in patients from the severe group at admission (p < 0.0001). In addition, CRP levels of discharged patients in both the non-severe group (p < 0.0001) and severe group (p < 0.0001) were significantly lower than those at admission (Tables 3–5). Therefore, we performed the correlation analyses between CRP and leukocyte counts (Table 7).
Table 7.
Correlation between the levels of CRP and leukocyte counts in patients with COVID-19 pneumonia.
| Variables | At admission |
At discharge |
||
|---|---|---|---|---|
| r | p | r | p | |
| CD3+ lymphocytes, ×106/l | ||||
| Total | −0.5000 | <0.0001**** | −0.2830 | 0.0002*** |
| Non-severe | −0.4333 | <0.0001**** | −0.2619 | 0.0018** |
| Severe | −0.5844 | 0.0001*** | −0.3043 | 0.1307 |
| CD4+CD3+ lymphocytes, ×106/l | ||||
| Total | −0.4749 | <0.0001**** | −0.2283 | 0.0031** |
| Non-severe | −0.4063 | <0.0001**** | −0.2102 | 0.0127* |
| Severe | −0.5265 | 0.0004*** | −0.2237 | 0.2721 |
| CD8+CD3+ lymphocytes, ×106/l | ||||
| Total | −0.4391 | <0.0001**** | −0.3202 | <0.0001**** |
| Non-severe | −0.3688 | <0.0001**** | −0.2583 | 0.0022** |
| Severe | −0.6187 | <0.0001**** | −0.5290 | 0.0094** |
| NLR | ||||
| Total | 0.4599 | <0.0001**** | 0.2803 | <0.0001**** |
| Non-severe | 0.3627 | <0.0001**** | 0.2487 | <0.0001**** |
| Severe | 0.3778 | <0.0001**** | 0.4482 | <0.0001**** |
| Neutrophil count, ×109/l | ||||
| Total | 0.1800 | 0.0014** | 0.0083 | 0.8848 |
| Non-severe | 0.1256 | 0.0391* | 0.0030 | 0.9612 |
| Severe | 0.3627 | 0.0198* | 0.0816 | 0.6121 |
| TLC, ×109/l | ||||
| Total | −0.4516 | <0.0001**** | −0.3251 | <0.0001**** |
| Non-severe | −0.3840 | <0.0001**** | −0.2777 | <0.0001**** |
| Severe | −0.5448 | 0.0002*** | −0.5748 | 0.0001*** |
p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
C3, complement 3; C4, complement 4; CD4+/CD8+, the ratio of CD4+ lymphocyte counts to CD8+ lymphocyte counts; COVID-19, coronavirus disease 2019; CRP, C-reactive protein; Ig, immunoglobulin; NLR, neutrophil to lymphocyte ratio; r, correlation coefficient; TLC, total lymphocyte count.
At admission, CD3+, CD4+, and CD8+ lymphocyte counts and TLC were negatively correlated with CRP level in both non-severe group (CD3+: p < 0.0001; CD4+: p < 0.0001; CD8+: p < 0.0001; TLC: p < 0.0001) and severe group (CD3+: p = 0.0001; CD4+: p = 0.0004; CD8+: p < 0.0001; TLC: p = 0.0002). Neutrophil count and NLR were positively correlated with CRP level in both the non-severe group (neutrophil count: p = 0.0391; NLR: p < 0.0001) and severe group (neutrophil count: p = 0.0198; NLR: p < 0.0001).
At discharge, CD3+ and CD4+ lymphocyte counts were negatively correlated with CRP level in the non-severe group (CD3+: p = 0.0018; CD4+: p = 0.0127), while we failed to find any correlation between them and CRP level in the severe group. CD8+ lymphocyte count and TLC were negatively correlated with CRP level in both the non-severe group (CD8+: p = 0.0022; TLC: p < 0.0001) and severe group (CD8+: p = 0.0094; TLC: p = 0.0001). NLR was positively correlated with CRP level in both the non-severe group (p < 0.0001) and severe group (p < 0.0001). However, we failed to find any correlation between neutrophil counts and CRP level in the severe group.
LDH, CRP, and complement
Correlation analyses indicated CRP levels were positively correlated with LDH levels in both the non-severe group (p < 0.0001) and severe group (p < 0.0001) at admission, and the positive correlation remained at discharge (non-severe group: p < 0.0001; severe group: p = 0.0223) (Table 6).
Levels of C3 and C4 were positively correlated with LDH level (C3: p = 0.0073; C4: p < 0.0001) in the non-severe group at admission. However, we failed to find any correlation between complement and LDH level in the severe group (Table 8).
Table 8.
Correlation between the levels of CRP, LDH, and complement in patients with COVID-19 pneumonia at admission.
| Variables | LDH |
CRP |
||
|---|---|---|---|---|
| r | p | r | p | |
| C3, g/l | ||||
| Total | 0.0857 | 0.1206 | 0.2177 | 0.0003*** |
| Non-severe | 0.1590 | 0.0073** | 0.2934 | <0.0001**** |
| Severe | 0.0628 | 0.6821 | 0.0919 | 0.5995 |
| C4, g/l | ||||
| Total | 0.2758 | <0.0001**** | 0.2128 | 0.0005*** |
| Non-severe | 0.3703 | <0.0001**** | 0.2705 | <0.0001**** |
| Severe | −0.0258 | 0.8709 | 0.0474 | 0.7932 |
p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
C3, complement 3; C4, complement 4; COVID-19, coronavirus disease 2019; CRP, C-reactive protein; LDH, lactate dehydrogenase; r, correlation coefficient.
In the non-severe group, levels of C3 and C4 were positively correlated with CRP level (C3: p < 0.0001; C4: p < 0.0001) at admission, while we failed to find any correlation between complement and CRP level in the severe group (Table 8).
Discussion
In this retrospective study, 373 patients with laboratory-confirmed COVID-19 from Shanghai and Putian were enrolled, including 8 mild patients, 314 moderate patients, 33 severe patients, and 18 critical patients. Immunological variables and markers for severity of the disease were analyzed, such as CRP and LDH. This is the first study to reveal the dynamic changes of immunological variables during hospitalization and their significant correlation with CRP and LDH at admission and at discharge.
By comparing CRP, LDH, lymphocyte and subtypes, neutrophil count, and NLR at admission with the same variables at discharge, LDH and CRP levels were shown to be significantly higher upon admission to hospital and declined at discharge, in concert with the previous studies.11,12 In contrast, lymphocytes and subtypes were decreased at admission, and increased at discharge. Neutrophils relatively, but not significantly, decreased at discharge compared with neutrophils at admission in severe and critical patients. However, in mild and moderate patients there is no obvious change. NLR, a useful infection biomarker, has been proved to be evidently related to the diagnosis of bacteremia and virus infection.13 In COVID-19 patients of both the severe and non-severe groups, NLR was increased remarkably at admission and declined at discharge. Compared with the non-severe group at admission, neutrophil count, CRP, LDH, and NLR were significantly higher and counts of lymphocytes and subsets were significantly lower in the severe group. At discharge, the differences remained between non-severe and severe groups, except neutrophil count and CRP.
Abnormal changes of CD4+ and CD8+ T cells caused CD4/CD8 ratio disturbance, which shows immune dysfunction in these patients with COVID-19, especially in severe and critical patients. Rapid innate immunity is the first barrier against virus attack, releasing different inflammatory cytokines, and then the adaptive immunity is prepared to respond to the virus appropriately.14 T cells, CD4+, and CD8+ T cells of the adaptive immune system play a critical antiviral role by balancing the fight against pathogens and the risk of causing overwhelming inflammation or autoimmunity.15 CD4+ T cells function by producing virus-specific antibodies via activating T-dependent B cells. CD8+ T cells directly kill viral infected cells. Delayed development of the adaptive system causes prolonged virus clearance, which was common in severe SARS-CoV infection patients.16 Hence, a decrease of lymphocytes and the T cell subgroups could aggravate the patients’ condition of COVID-19, which was consistent with the low number of TLC and lymphocyte subtypes in severe and critical patients.
Zhao et al. suggested that LDH is the potential marker for evaluation of the severity of COVID-19, since LDH is much higher when compared with non-COVID-19 pneumonia.17 Liu et al. found out that CRP and LDH were both highly correlated with disease severity and might be potential predictors.18 Hence, we applied CRP and LDH as markers for severity of COVID-19 in this study. To further study the relationship of immune dysfunction with severity of the disease, we analyzed the correlation of the count of T cells, CD4+T cells, CD8+T cells, neutrophils, and total lymphocytes, as well as CD4/CD8 ratio, NLR with LDH, and CRP at admission and at discharge. Interestingly, we observed a significant negative correlation of lymphocytes and lymphocyte subtypes with LDH and CRP, and a positive correlation of NLR with LDH, and CRP at admission, which has never been proved in other studies. These findings confirmed that the levels of counts of lymphocytes and subtypes and NLR could represent the severity of the pneumonia in addition to the function of adaptive immunity. Lymphopenia is also observed in MERS patients with a lower degree compared with SARS patients.19 In addition, there have been a few studies that proved that the decrease of T lymphocytes is strongly related with the severity of the acute phase of patients affected with SARS, and a T lymphocyte subset analysis would help improve the early diagnosis of the disease.20,21 During the infection, depletion of T cells with antiviral effects may prolong the infection and promote viral survival. Considering lymphopenia and levels of T cell subgroups were much lower in severe and critical patients, with relatively longer hospitalization days, numbers of total lymphocytes, T cells, CD4+, and CD8+ T cells, NLR might suggest a prognosis of COVID-19 patients, which still needs to be further confirmed. However, we failed to find significant correlations of the TLC and the subsets with LDH in the severe group at discharge. As a result, lymphocyte subtypes might be suitable to evaluate the severity of the disease for severe and critical patients at admission but may not be applicable at discharge. This could be the result of a longer recovery time of the adaptive immunity of the severe group compared with the non-severe group.
In addition to the adaptive immune system, we also analyzed immunoglobulins and complement proteins of COVID-19 patients, including C3, C4, IgM, IgA, and IgG. A report shows that the median duration of IgM and IgA detection of COVID-19 was 5 days, and IgG was detected 14 days after symptom onset.22 As a result, there was no difference of immunoglobulins between non-severe and severe groups at admission in our study. The complement system plays an antiviral role in the host immune response to CoV infection by providing a way for the innate immune system to detect and respond to pathogens.23 Levels of C3 and C4 were lower in the severe group compared with the non-severe group at admission, which could explain for the severe and critical patients’ severity. Since there was no significant difference in immunoglobulins between severe and non-severe groups, we only analyzed the correlation of C3 and C4 with LDH and CRP. In our research, there were positive significant correlations between C3 and C4, and LDH and CRP, in mild and moderate patients at admission, but not in the severe group. The results suggest that complement systems might be seriously damaged in severe and critical patients, which caused the lower production of the complement proteins. Hence, there was no significant correlation of C3 and C4 with LDH or CRP.
This study has several limitations. Although this research involves 373 patients, there were only 51 patients in the severe group, which might cause some potential bias. It would be better if more severe and critical COVID-19 patients were included. Despite that, out research demonstrated that adaptive immunity was hampered in COVID-19 patients, especially in severe and critical patients. Levels of lymphocytes, T cells, CD4+, and CD8+ T cells negatively correlate with the severity of this disease marked by LDH and CRP. Level of NLR is positively correlated with LDH and CRP. Monitoring NLR, T lymphocytes, and subset analysis would help identify the severity of pneumonia and assist physicians with proper diagnosis and treatment of COVID-19.
Supplemental Material
Supplemental material, Author_Response_1 for Dysfunction of adaptive immunity is related to severity of COVID-19: a retrospective study by Liang Xie, Qinhan Wu, Qunying Lin, Xuhui Liu, Weihua Lin, Shengyu Hao, Weiping Hu, Guiling Xiang, Hongzhou Lu and Shanqun Li in Therapeutic Advances in Respiratory Disease
Supplemental material, Author_Response_2 for Dysfunction of adaptive immunity is related to severity of COVID-19: a retrospective study by Liang Xie, Qinhan Wu, Qunying Lin, Xuhui Liu, Weihua Lin, Shengyu Hao, Weiping Hu, Guiling Xiang, Hongzhou Lu and Shanqun Li in Therapeutic Advances in Respiratory Disease
Supplemental material, Reviewer_1_v.1 for Dysfunction of adaptive immunity is related to severity of COVID-19: a retrospective study by Liang Xie, Qinhan Wu, Qunying Lin, Xuhui Liu, Weihua Lin, Shengyu Hao, Weiping Hu, Guiling Xiang, Hongzhou Lu and Shanqun Li in Therapeutic Advances in Respiratory Disease
Supplemental material, Reviewer_1_v.2 for Dysfunction of adaptive immunity is related to severity of COVID-19: a retrospective study by Liang Xie, Qinhan Wu, Qunying Lin, Xuhui Liu, Weihua Lin, Shengyu Hao, Weiping Hu, Guiling Xiang, Hongzhou Lu and Shanqun Li in Therapeutic Advances in Respiratory Disease
Supplemental material, Reviewer_2_v.1 for Dysfunction of adaptive immunity is related to severity of COVID-19: a retrospective study by Liang Xie, Qinhan Wu, Qunying Lin, Xuhui Liu, Weihua Lin, Shengyu Hao, Weiping Hu, Guiling Xiang, Hongzhou Lu and Shanqun Li in Therapeutic Advances in Respiratory Disease
Supplemental material, Reviewer_2_v.2 for Dysfunction of adaptive immunity is related to severity of COVID-19: a retrospective study by Liang Xie, Qinhan Wu, Qunying Lin, Xuhui Liu, Weihua Lin, Shengyu Hao, Weiping Hu, Guiling Xiang, Hongzhou Lu and Shanqun Li in Therapeutic Advances in Respiratory Disease
Supplemental material, Supplement_material for Dysfunction of adaptive immunity is related to severity of COVID-19: a retrospective study by Liang Xie, Qinhan Wu, Qunying Lin, Xuhui Liu, Weihua Lin, Shengyu Hao, Weiping Hu, Guiling Xiang, Hongzhou Lu and Shanqun Li in Therapeutic Advances in Respiratory Disease
Footnotes
Author contribution(s): Liang Xie: Formal analysis; Investigation; Software; Writing-original draft.
Qinhan Wu: Formal analysis; Investigation; Writing-original draft.
Qunying Lin: Conceptualization; Data curation; Resources; Writing-original draft.
Xuhui Liu: Conceptualization; Data curation; Resources; Writing-original draft.
Weihua Lin: Conceptualization; Data curation; Writing-original draft.
Shengyu Hao: Methodology; Writing-review & editing.
Weiping Hu: Data curation; Methodology; Writing-review & editing.
Guiling Xiang: Methodology; Writing-review & editing.
Hongzhou Lu: Investigation; Supervision; Validation; Writing-review & editing.
Shanqun Li: Funding acquisition; Investigation; Supervision; Validation; Writing-review & editing.
Conflict of interest statement: The authors declare that there is no conflict of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by grants from the National Key Research and Development Program of China (grant number 2018YFC1313600).
ORCID iDs: Guiling Xiang  https://orcid.org/0000-0001-5293-2908
https://orcid.org/0000-0001-5293-2908
Shanqun Li  https://orcid.org/0000-0002-8096-9780
https://orcid.org/0000-0002-8096-9780
Supplemental material: The reviews of this paper are available via the supplemental material section.
Contributor Information
Liang Xie, Department of Pulmonary Medicine, Zhongshan Hospital, Fudan University, Shanghai, China; Department of Infectious Diseases, Shanghai Public Health Clinical Center, Shanghai, China; Department of Respiratory Medicine, Affiliated Hospital of Putian University, Putian, China.
Qinhan Wu, Department of Pulmonary Medicine, Zhongshan Hospital, Fudan University, Shanghai, China.
Qunying Lin, Department of Respiratory Medicine, Affiliated Hospital of Putian University, Putian, China; The School of Clinical Medicine, Fujian Medical University, Fuzhou, Fujian Province, China.
Xuhui Liu, Department of Infectious Diseases, Shanghai Public Health Clinical Center, Shanghai, China.
Weihua Lin, Department of Respiratory Medicine, Affiliated Hospital of Putian University, Putian, China; The School of Clinical Medicine, Fujian Medical University, Fuzhou, Fujian Province, China.
Shengyu Hao, Department of Pulmonary Medicine, Zhongshan Hospital, Fudan University, Shanghai, China.
Weiping Hu, Department of Pulmonary Medicine, Zhongshan Hospital, Fudan University, Shanghai, China.
Guiling Xiang, Department of Pulmonary Medicine, Zhongshan Hospital, Fudan University, Shanghai, China.
Hongzhou Lu, Department of Infectious Diseases, Shanghai Public Health Clinical Center, 2901 Caolang Road, Shanghai, China, 201508.
Shanqun Li, Department of Pulmonary Medicine, Zhongshan Hospital, Fudan University, 180 Fenglin Rd., Shanghai, 200032, China.
References
- 1. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 2020; 382: 1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization. Coronavirus disease 2019 (COVID-19) situation report – 62, https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200322-sitrep-62-covid-19.pdf?sfvrsn=f7764c46_2 (accessed 21 March 2020).
- 3. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020; 382: 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395: 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 2020; 8: 420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wong RS, Wu A, To KF, et al. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ 2003; 326: 1358–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lau SKP, Lau CCY, Chan KH, et al. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle East respiratory syndrome coronavirus: implications for pathogenesis and treatment. J Gen Virol 2013; 94: 2679–2690. [DOI] [PubMed] [Google Scholar]
- 8. Liu J, Zheng X, Tong Q, et al. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J Med Virol 2020; 92: 491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Falay M, Senes M, Korkmaz S, et al. Biological variation of peripheral blood T-lymphocytes. J Immunol Methods 2019; 470: 1–5. [DOI] [PubMed] [Google Scholar]
- 10. World Health Organization. Coronavirus disease (COVID-19) technical guidance: laboratory testing for 2019-nCoV in humans, https://apps.who.int/iris/bitstream/handle/10665/331501/WHO-COVID-19-laboratory-2020.5-eng.pdf?sequence=1&isAllowed=y
- 11. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020; 395: 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020; 382: 1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Russell CD, Parajuli A, Gale HJ, et al. The utility of peripheral blood leucocyte ratios as biomarkers in infectious diseases: a systematic review and meta-analysis. J Infect 2019; 78: 339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev 2009; 227: 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cecere TE, Todd SM, Leroith T. Regulatory T cells in arterivirus and coronavirus infections: do they protect against disease or enhance it? Viruses 2012; 4: 833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cameron MJ, Bermejo-Martin JF, Danesh A, et al. Human immunopathogenesis of severe acute respiratory syndrome (SARS). Virus Res 2008; 133: 13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao D, Yao F, Wang L, et al. A comparative study on the clinical features of COVID-19 pneumonia to other pneumonias. Clin Infect Dis. Epub ahead of print 12 March 2020. DOI: 10.1093/cid/ciaa247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu Y, Yang Y, Zhang C, et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci 2020; 63: 364–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis 2013; 13: 752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li T, Qiu Z, Zhang L, et al. Significant changes of peripheral T lymphocyte subsets in patients with severe acute respiratory syndrome. J Infect Dis 2004; 189: 648–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li T, Qiu Z, Han Y, et al. Rapid loss of both CD4+ and CD8+ T lymphocyte subsets during the acute phase of severe acute respiratory syndrome. Chin Med J (Engl) 2003; 116: 985–987. [PubMed] [Google Scholar]
- 22. Guo L, Ren L, Yang S, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis. Epub ahead of print 21March 2020. DOI: 10.1093/cid/ciaa310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baker S, Kessler E, Darville-Bowleg L, et al. Different mechanisms of serum complement activation in the plasma of common (Chelydra serpentina) and alligator (Macrochelys temminckii) snapping turtles. PLoS One 2019; 14: e0217626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Author_Response_1 for Dysfunction of adaptive immunity is related to severity of COVID-19: a retrospective study by Liang Xie, Qinhan Wu, Qunying Lin, Xuhui Liu, Weihua Lin, Shengyu Hao, Weiping Hu, Guiling Xiang, Hongzhou Lu and Shanqun Li in Therapeutic Advances in Respiratory Disease
Supplemental material, Author_Response_2 for Dysfunction of adaptive immunity is related to severity of COVID-19: a retrospective study by Liang Xie, Qinhan Wu, Qunying Lin, Xuhui Liu, Weihua Lin, Shengyu Hao, Weiping Hu, Guiling Xiang, Hongzhou Lu and Shanqun Li in Therapeutic Advances in Respiratory Disease
Supplemental material, Reviewer_1_v.1 for Dysfunction of adaptive immunity is related to severity of COVID-19: a retrospective study by Liang Xie, Qinhan Wu, Qunying Lin, Xuhui Liu, Weihua Lin, Shengyu Hao, Weiping Hu, Guiling Xiang, Hongzhou Lu and Shanqun Li in Therapeutic Advances in Respiratory Disease
Supplemental material, Reviewer_1_v.2 for Dysfunction of adaptive immunity is related to severity of COVID-19: a retrospective study by Liang Xie, Qinhan Wu, Qunying Lin, Xuhui Liu, Weihua Lin, Shengyu Hao, Weiping Hu, Guiling Xiang, Hongzhou Lu and Shanqun Li in Therapeutic Advances in Respiratory Disease
Supplemental material, Reviewer_2_v.1 for Dysfunction of adaptive immunity is related to severity of COVID-19: a retrospective study by Liang Xie, Qinhan Wu, Qunying Lin, Xuhui Liu, Weihua Lin, Shengyu Hao, Weiping Hu, Guiling Xiang, Hongzhou Lu and Shanqun Li in Therapeutic Advances in Respiratory Disease
Supplemental material, Reviewer_2_v.2 for Dysfunction of adaptive immunity is related to severity of COVID-19: a retrospective study by Liang Xie, Qinhan Wu, Qunying Lin, Xuhui Liu, Weihua Lin, Shengyu Hao, Weiping Hu, Guiling Xiang, Hongzhou Lu and Shanqun Li in Therapeutic Advances in Respiratory Disease
Supplemental material, Supplement_material for Dysfunction of adaptive immunity is related to severity of COVID-19: a retrospective study by Liang Xie, Qinhan Wu, Qunying Lin, Xuhui Liu, Weihua Lin, Shengyu Hao, Weiping Hu, Guiling Xiang, Hongzhou Lu and Shanqun Li in Therapeutic Advances in Respiratory Disease