Abstract
Cerebral edema constitutes an important contributor to secondary injury in acute brain injury. The quantification of cerebral edema in neuroimaging, a well-established biomarker of secondary brain injury, represents a useful intermediate phenotype to study edema formation. Population genetics provides powerful tools to identify novel susceptibility genes, biological pathways and therapeutic targets related to brain edema formation. Here, we provide an overview of the pathogenesis of cerebral edema, introduce relevant genetic methods to study this process, and discuss the ongoing research on the genetic underpinnings of edema formation in acute brain injury. The epsilon 2 and 4 variants within the Apolipoprotein E (APOE) gene are associated with worse outcome after traumatic brain injury and intracerebral hemorrhage, and recent studies link these polymorphisms to inflammatory processes that lead to blood-brain barrier disruption and vasogenic edema. For the Haptoglobin gene (HP), the Hp 2–2 genotype associates with worse outcome after acute brain injury, whereas the haptoglobin Hp 1–1 genotype correlates with increased edema in the early phases of intracerebral hemorrhage. Another important protein in cerebral edema is aquaporin 4, coded by the AQP4 gene. AQP4 mutations contribute to the formation of cytotoxic edema, and further genetic research is necessary to help elucidate the mediating mechanism. Findings supporting the target genes outlined above require replication in larger samples and evaluation in non-white populations. These next steps will be significantly facilitated by the rapid changes observed in the field of population genetics, including large international collaborations, open access to genetic data, and significant reductions in the cost of genotyping technologies.
Keywords: Stroke, Hemorrhagic stroke, Intracerebral hemorrhage, Ischemic stroke, Traumatic brain injury, Brain edema, Stroke genetics
Introduction
Diseases involving acute brain injury, including intracerebral hemorrhage (ICH), ischemic stroke, and traumatic brain injury (TBI), are associated with elevated mortality and morbidity [1]. Stroke is a leading cause of death and adult disability worldwide, while TBI is a contributing factor to a third of all injury-related deaths in the United States [1–4]. For a given patient, the total burden of brain injury corresponds to the sum of the primary and secondary injury components. The primary brain injury is directly related to the mechanisms of the underlying disease and occurs immediately after its onset. The term secondary injury encompasses several biological processes that occur hours to days after the primary insult, either as a direct consequence of the anatomical changes produced by this insult (hydrocephalus and compressive injury) or related to the pathophysiological responses to it (inflammation ad circulatory changes) [5].
The delayed and longer time window of secondary injury provides an appealing opportunity to deploy treatments aimed to reduce the final disability burden related to acute brain injury. One promising final pathway of secondary injury is cerebral edema, a biological process that leads to an increase in brain volume and intracranial pressure, sometimes followed by decreased cerebral perfusion, herniation, and death [1, 5, 6]. Observational studies indicate that cerebral edema is an independent predictor of mortality in TBI and ICH [7] [8]. Understanding the pathophysiology of cerebral edema is key to identifying therapeutic targets of secondary brain injury. Population genetics provides powerful tools to identify novel susceptibility genes, biological pathways and therapeutic targets involved in acute brain injury and edema formation. Thus far, the application of genomic analyses to neuroimaging phenotypes of acute brain injury has been limited by limited sample sizes and incomplete harmonization across the existing data. However, with the creation of large research consortia capable of assembling extremely large sample sizes and the implementation of accurate automated imaging pipelines, the ability to apply genomic analyses to neuroimaging markers has become a reality [3]. In this review, we provide a brief overview of relevant concepts related to population genetics and genomic medicine, review the pathogenesis of cerebral edema, and discuss relevant studies on the genetic underpinnings of edema formation post-acute brain injury.
Population Genetics and Genomic analyses
Population genetics offers tools that help to overcome the limitations of observational studies [9][9]. Examination of inherited genetic variability may shed light on the relationship between genes, different phenotypes, and pathophysiological pathways. Since mutations are randomly distributed during meiosis, the associations between mutation and disease are not influenced by postnatal factors. This model allows for the identification of the genetic and cellular pathways involved in specific conditions. The use of genetic analysis to identify new targets and mechanisms could be a promising strategy for neurocritical care conditions, as most conditions provoke brain edema and are influenced by underlying genetic factors [10–12]. Given that genotyping techniques now capture information from the entire genome, it is possible to exhaustively investigate the cellular processes and pathways involved in brain edema formation [13].
Genetic analyses boost the turnout of the translational research cycle
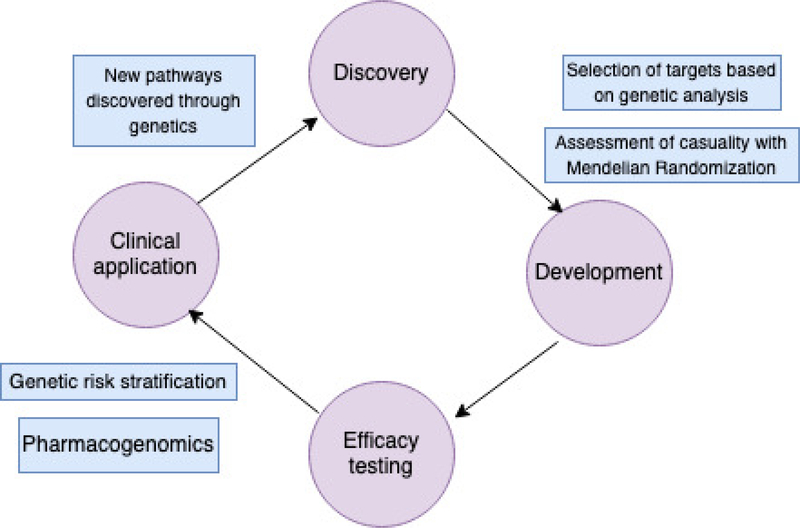
Completing all stages of the translational research cycle (Figure 1), which includes identification of a pathophysiological mechanism, demonstration of its involvement in pathophysiology, and testing agents that act on this pathway, is a demanding task. Moreover, a substantial number of interventions that seemed promising in pre-clinical studies failed to show benefit in expensive and time-consuming clinical trials. Not only can population genetics identify casual relationships between mutations and phenotypes of interest, but it can also help to reduce the proportion of new treatments that do not succeed in clinical trials. A comparative study that aimed to compare clinical interventions with and without genetic support used the Informa Pharmaprojects database along with GWASdb and OMIM, two open-access resources that contain information on genetic risk factors for complex and Mendelian conditions. The authors demonstrated that selection of genetically-supported targets doubles the success of the translational research cycle [14]. New statistical methods, such as Mendelian Randomization, offer even more powerful tools to support these findings [15].
Figure 1.
The cycle of translational research.
Definition of Cerebral Edema
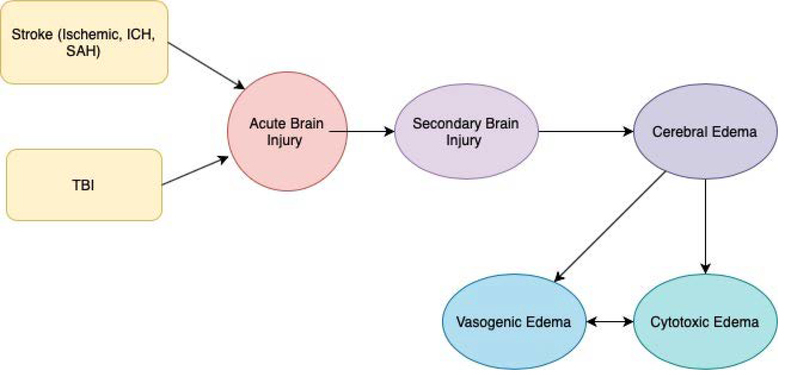
Edema is defined as an increase in brain tissue water that occurs in both the cells and interstitial space [6]. Cerebral edema is mainly characterized as cytotoxic or vasogenic edema [16, 17]. Cytotoxic edema results from failure of homeostatic pumps, leading to an imbalanced ionic gradient. This intracellular osmotic shift causes water to move into the cells, leading to cerebral edema. Vasogenic edema involves inflammatory markers and results from blood brain barrier (BBB) disruption causing an influx of protein rich fluid into the brain [5, 18–21]. Cytotoxic and vasogenic edema closely interact with each other and mutually contribute to edema formation after acute brain injury [22]. The cascade of edema formation is shown in Figure 2.
Figure 2.
Pathophysiological cascade leading to brain edema formation.
ICH – Intracerebral Hemorrhage, SAH – subarachnoid hemorrhage, TBI – traumatic brain injury
Edema formation after intracerebral hemorrhage
The accumulation of brain edema starts a few hours after the onset of an intraparenchymal hemorrhage and peaks between 7–11 days [23, 24]. Edema formation occurs in multiple phases within this time frame. In the first few hours after ICH, clot retraction causes serum proteins to be released, creating an osmotic gradient that pulls water into the interstitial space [22, 24]. In the first few days following the initial ionic edema, neutrophils and macrophages induce inflammatory cytokines to be upregulated. Inflammatory cytokines, such as IL-Iβ, IL-6, and TNF-α, contribute to the BBB breakdown and upregulate matrix metalloproteinases (MMPs), neuropeptide substance P and bradykinins [4, 21, 22, 24, 25]. MMPs, specifically MMP-9, breakdown collagen and other basal lamina proteins integral to the BBB [26, 27]. Substance P has been shown to increase vascular permeability and edema after brain injury [28–30]. Bradykinins are linked to increased edema, vascular permeability, and BBB after ischemia and other forms of brain injury [31, 32]. Additionally, leakage of red blood cells activates a coagulation cascade that produces thrombin, which is a proinflammatory agent at high concentrations [20]. Thrombin further breaks down the BBB and works alongside complement cascade proteins that enter the parenchyma, causing microglia activation, erythrocyte lysis, and release of hemoglobin and iron into brain matter [21, 22, 33, 34]. Toxic products of iron and hemoglobin degradation, such as free radicals, further contribute to secondary injury [4, 5, 22, 24].
Edema formation in traumatic brain injury
Cytotoxic edema is present as early as 2–4 hours after TBI [21]. Impaired oxygen and glucose due to ischemia and hypoxia reduces ATP stores, disrupting the Na+/K+ ATPase function. Sodium cations flow down the electrochemical gradient, accumulating inside the cells, and drawing water into the cellular space. Aquaporins, a class of water-channel proteins located in the basolateral membranes of ependymal cells and astrocytes, colocalize with inward rectifying potassium channels. When injury occurs, water flows through the aquaporin channels and into the cell as a result of the ionic imbalance [1, 4, 6, 21]. Furthermore, the imbalanced ionic and electrochemical gradients cause the release of the excitatory neurotransmitter glutamate. Glutamate triggers pre and post synaptic neurons and leads to water influx in neighboring cells [17]. Calcium regulation is additionally impaired due to the increased cell volume. When calcium is upregulated, inflammatory pathways are triggered, contributing to the further breakdown of the BBB [17]. Vasogenic edema is also facilitated by the mechanical and vascular damage after traumatic injury. Proteins and red blood cells leaked into the parenchyma cause activation of the coagulation cascade. Thrombin acts to break down the BBB and activates microglia, cell lysis, and the release of hemoglobin, as discussed above.
APOE
Genetic variants within APOE may play a role in edema formation in acute brain injury. The apolipoprotein E (APOE) gene codes for the apolipoprotein E protein, which plays a key role in lipid transport and metabolism [35]. The epsilon variants (ε) are the most studied polymorphisms within APOE. The ε variants represent haplotypes built with genotype information from the SNPs rs429358 and rs7412. Using information from these two SNPs, APOE-ε variants are labeled as APOE-ε2, APOE-ε3, and APOE- ε4, with APOE-ε3 being the most common allele found in the most population [36]. APOE is secreted by astrocytes in the central nervous system and has been shown to have BBB stabilization properties [37]. The APOE ε4 is associated with increased risk of cognitive decline, dementia, and Alzheimer’s disease [38, 39]. Current research indicates that the APOE polymorphisms are also involved in many other disease processes, such as arteriosclerosis, traumatic brain injury, and intracerebral hemorrhage [35, 38, 40].
Animal studies of APOE
Animal models of acute brain injury have pointed to APOE as an important gene in acute brain injury and outcome. In an animal model of subarachnoid hemorrhage (SAH), APOE-deficient mice showed larger edema and greater BBB disruption after SAH than the wild-type mice. Additionally, there was a significant upregulation of inflammatory cytokines, such as Cyclophilin A, IL-Iβ, IL-I6, p-p65, and TNF-α, in the APOE-deficient mice after SAH, as well as increased MMP-9 activity. These results point to APOE as being directly involved in the inflammatory response that disrupts the BBB and contributes to vasogenic edema after acute brain injury [41]. Researchers argue that APOE reduces the inflammatory response in an isoform-specific fashion, with APOE-4 being the least effect of the three isoforms [41–43]. The use of an apolipoprotein peptide mimetic following head injury provides compelling evidence that APOE is involved in inflammation and edema formation after acute brain injury. In one study, researchers used an intravenous injection of a small peptide derived from the APOE receptor binding region in mice following TBI. This peptide has been previously shown to retain all functions of the intact protein [44]. Treatment of mice after TBI with the APOE peptide was associated with better short-term and long-term functional outcome, decreased number of injured hippocampal neurons, and reduction in TNF-α RNA [44]. In a similar study, researchers injected APOE mimetic peptide COG1410 in a subset of mice that underwent a controlled cortical impact injury to model TBI. Treatment with the peptide significantly decreased MMP-9 activity, reduced BBB disruption, reduced volume of the TBI lesion and vasogenic edema, and improved functional outcome [45].
Human studies of APOE
Genetic studies in humans have yielded associations between APOE polymorphisms and multiple diseases and traits. Genetic studies have shown that APOE-ε4 is a predictor of poor functional outcome after head trauma [7, 40, 46–49]. Additionally, several related studies found that the APOE-ε2 allele is associated with larger baseline hematoma volume, increased risk of hematoma expansion, and poor outcome and mortality in hemorrhages located in lobar regions of the brain [50] [51]. In addition, one study reported that the ε4 allele was significantly associated with midline shift and functional outcome in ICH patients [52]. This finding led to the hypothesis that the mechanism linking the APOE phenotype with midline shift and poor outcome was perihematomal edema [52]. Supporting this hypothesis, recent studies found an association between APOE polymorphisms and a number of inflammatory processes involved in the formation of cerebral edema. When the ε4 version of APOE is expressed in neurons, it is susceptible to proteolysis. APOE-ε4 is the least stable isoform and has the greatest neurotoxic effects. Cellular pathways impaired by APO-ε4 include mitochondrial dysfunction, amyloid B production, neurite outgrowth, neuronal apoptosis, and BBB integrity [53]. Taken in consideration with the animal research linking APOE-ε4 to inflammatory cytokines discussed above, it is highly probable that APOE-ε4 facilitates inflammatory processes that contribute to BBB and vasogenic edema after acute brain injury. Further research is needed to establish the genetic relationship between APOE and cerebral edema and the influence of APOE-ε4 at different time points post-acute brain injury.
Haptoglobin
Haptoglobin (Hp) is an acute-phase response protein that plays an antioxidant role by binding and neutralizing hemoglobin (Hb) in the blood [54, 55]. The bound Hp-Hb complex is cleared by CD163 scavenger receptors on macrophages, reducing cytotoxicity, edema, and neuronal damage caused by free Hb [56]. In the human genome, the HP gene at chromosome 16q22 is polymorphic and is represented as Hp-1 and Hp-2. Individuals can either by homozygous for the Hp-1 allele (Hp-1–1), heterozygous (Hp-1–2), or homozygous for the Hp-2 allele (Hp-2–2) [57]. Previous research from both animal and human studies has established that the haptoglobin polymorphism influences Hb clearance, oxidative stress, and is a risk factor for cardiovascular events [55, 58–60].
Animal studies of Haptoglobin
In one rodent study of ICH, rats with reduced blood Hp levels had significantly higher levels of oxidative brain damage and edema 24 hours after ICH induction. Additionally, rats with the Hp gene knocked out had severe neurological outcomes compared to wild types rodents, while rats with Hp2 overexpression had less severe neurological deficits [54]. Additional studies support the finding that Hp overexpression in ICH rodent model decreases oxidative stress, reduces lesion volume, and improves neurological status [59, 61].
Human studies of Haptoglobin
Compared to Hp2–2, Hp1–1 is a superior antioxidant, binds better to hemoglobin, and is removed faster by CD163 [57]. Furthermore, a greater number of cytokines, particularly IL-6 and IL-10, are secreted when the Hp1-Hb complex is bound to CD163 as compared to Hp2-Hb [55]. While hemoglobin cytotoxicity is a deleterious process that contributes to secondary injury after intracerebral hemorrhage, the release of inflammatory cytokines also contributes to the breakdown of BBB and vasogenic edema. One genetic study that characterized patients with ICH by their Hp genotype reported a nonsignificant trend towards increased mortality when comparing persons with Hp-2–1 and Hp-2–2 versus those with Hp1–1. Additionally, participants with the Hp2 allele had significantly worse functional outcomes measured by the modified Rankin scale [62]. Interestingly, a different genetic study found that the Hp1–1 phenotype was associated with increased perihematomal edema (PHE) in the early stages after ICH [56]. The findings that Hp-1–1 acts as a better antioxidant and is associated with improved functional outcome and lower mortality after ICH, while simultaneously being associated with greater PHE, appear to be contradictory to the biological model proposed that PHE is a marker of poor functional outcome after brain injury. In order to explain this contradiction, it is hypothesized that early PHE has protective properties while Hp-Hb binding reduces prolonged inflammation, edema, and subsequently poor outcome [56]. Further genetic studies of haptoglobin and edema formation at different time points following an ICH is necessary. Moreover, it would be beneficial to study the haptoglobin phenotype in the context of TBI to gain a better understanding of its role in secondary injury.
Aquaporins
Aquaporins (AQPs) are a class of water-channel proteins, and AQP4, AQP1, and AQP9 are specifically present in the brain. AQP4, the main water channel found in the brain, is located in end-feet membranes of astrocytes adjacent to capillaries, glial limiting membranes, subependymal astrocytes and ependymal cells. AQP1 is located in the choroid plexus and plays an important role in cerebral spinal fluid production and regulation. AQP9 is located in some astrocytes and ependymal cells and functions to transfer solutes, such as glycerol and urea [63, 64].
Animal studies of Aquaporins
Several animal model studies suggest that AQPs are involved in cytotoxic edema after acute brain injury. In a mouse model of ICH, AQP4 expression was upregulated following the ICH, while AQP4 deficient mice showed worse neurological deficits, and greater edema, BBB disruption, micro-vessel damage, and neuronal death [65]. AQP4 is anchored by the α-syntrophin protein, and deletion of the α-syntrophin protein disturbs the polarization of AQP4 [64, 66]. A-syntrophin-deficient mice resulted in downregulated AQP4 expression in astrocyte perivascular membranes, as well as delayed edema formation [67]. A-syntrophin deletion also resulted in reduced astrocyte swelling after severe hypoosmotic stress and oxygen/glucose deprivation [68]. Additionally, rodent studies that used ethanol treatment after TBI and curcumin treatment after ICH showed reduced AQP4 mRNA and gene expression along with decreased cerebral edema [63, 69]. These findings support the conclusion that AQPs play an important role in cytotoxic edema formation post brain injury. Aquaporins are also implicated in vasogenic edema, as astrocytic end-feet are integral to the BBB, and AQP4 assists in the reabsorption of fluid into extracellular fluid spaces [64, 70].
Human studies of Aquaporins
While there is a growing literature of animal studies supporting AQP4 involvement in cerebral edema, the number of human studies remains limited. One genetic study looked at variations within exome 4 of the AQP4 gene and reported no significant variation among 102 patients with TBI [71]. However, this study was limited to only exon 4 of the AQP-4 gene and did not take into considerations variants that could be present on exons 1–3. Another genetic study of AQP4 that evaluated clinical, neuroimaging, and genetic data from 363 TBI patients identified 7 tag SNPs along the AQP4 gene region and found that two of them, rs3763043 and rs3875089, were associated with clinical outcome as evaluated 6 months after the traumatic event [71].
Sur1 Gene
Sur1, a sulfonylurea receptor and transmembrane protein, is coded by the ABCC8 gene. Sur1 associates with two ion-channels: Kir6.2 and transient receptor potential melastatin 4 (Trpm4). The Sur1-Trpm4 complex is an ATP-sensitive cation channel that is not usually found in the brain, but is upregulated after trauma, ischemia, and hypoxia [72]. Previous work has shown that Sur1-Trpm4 assembles with AQP4 to help modulate astrocyte swelling, a key factor in cytotoxic edema [73]. One study found that Sur1 was overexpressed in many cell types, particularly neurons and endothelial cells, following traumatic brain lesions [74]. Furthermore, Sur1-Trpm4 activity is associated with MMP-9 secretion, a proteinase involved in BBB breakdown and vasogenic edema [26, 27, 72]. To date, there has only been one genetic study of ABCC8 in relation to edema formation. This study used a candidate gene approach to investigate if polymorphisms within this gene region influence edema after TBI. The investigators identified three SNPs, rs2283261, rs3819521, and rs2283258, that were significantly associated with cerebral edema, measured by CT imaging, and increased intracranial pressure. Because all three SNPs are located in non-coding regions of ABCC8, the authors speculate that these SNPs may play a role in the regulation and expression of Sur1 [75].
Hastened pace of discovery in population genetics
The rapid advancements in population genetics research observed in recent years will bring substantial benefits to understanding the genetic factors described above. This transformation is due to many factors, including an evolving research culture that encourages collaboration and public sharing of available data; the creation of large international consortia designed to share ideas, harmonize data, and achieve large sample sizes; and novel analytical methods designed to obtain maximal benefit from Big Data. [76, 77].
Genetic studies are complex, time-consuming and costly. Making genetic data publicly accessible increases the overall impact of these studies by maximizing scientific yield of each dataset. Open access resources can be divided into 4 main categories: repositories that store already generated genetic data; large population studies with attached biobanks; disease-specific studies with genomic data available for public access; and scientific platforms that grant access to data in closed computational environments, allowing investigators to work with data without downloading them to their personal computers and local servers. Table 1 describes some of the numerous open access data resources currently available in population genetics research.
Table 1.
Open access data resources
| Ref. | Name | Website | Resource type | Sample size | Access type |
|---|---|---|---|---|---|
| [81] | UK Biobank | https://www.ukbiobank.ac.uk/ | Observational study | 500,000 | Open |
| [79] | Million Veteran Program | https://www.research.va.gov/mvp/ | Observational study | 1 million | Restricted |
| [80] | All of Us | https://allofus.nih.gov/ | Observational study | 1 million | Open |
| [82] | China Kadoorie Biobank | https://www.ckbiobank.org/ | Observational study | 500,000 | Restricted |
| [78] | dbGAP | https://www.ncbi.nlm.nih.gov/gap/ | Repository | - | Open |
| [83] | EGA | https://ega-archive.org/ | Repository | - | Open |
| [84] | MIMIC-III | https://mimic.physionet.org/ | Database | 61,532 | Open |
| [85] | eICU-CRD | https://eicu-crd.mit.edu/ | Database | 200,000 | Open |
Acronyms: UK = United Kingdom, dbGAP = Database of Genotypes and Phenotypes, EGA = European Genome-phenome Archive, MIMIC-III = Medical Information Mart for Intensive Care-III, eICU-CRD = eICU Collaborative Research Database.
The Database of Genotypes and Phenotypes (dbGaP) sponsored by the NIH [78], the Cerebrovascular Disease Knowledge Portal (CDKP) [79] in the United States, and the European Genome-phenome Archive (EGA) [80] in Europe are three publicly available, large open access repositories. The UK Biobank [81] and the China Kadoorie Biobank [82] are successful examples of large, population-based, prospective studies that include more than 500,000 participants and provide a variety of data, ranging from genetic information to physical measurements. Regarding disease-oriented open access studies, the Ethnic/Racial Variations of Intracerebral Hemorrhage (ERICH) study [83] is a dominant resource in Stroke research and has led to numerous sub-analyses already [84–89]. The ERICH study includes 6,000 ICH cases and matched controls, including individuals of different ethnic background. The American Heart Association Precision Medicine Platform is a scientific platform that offers public access to numerous datasets with genetic and non-genetic information. It’s closed, computational environment uses Jupyter Notebooks to script, as well as Amazon Web Service to offer computational support to researchers [86]. Lastly, the All of Us Research Program, which is planned to start winter of 2020, will enroll at least 1 million participants in the United States from different ethnic backgrounds, providing even greater amounts of data to be used in genetic analyses [80].
Future directions
The impact of genetics on Neurocritical Care will continue to grow in coming years. Advancements in Neurocritical Care relies on many factors, with collaboration between clinicians, investigators, and research institutions being crucial. An important objective for these collaborations is to integrate genetic analyses with clinical data, such as laboratory results, neuroimaging, and vital signs, provided by Neuroscience Intensive Care Units. While public access to these data will accelerate the pace of discovery in the field, it will inevitably raise challenges related to adequately protecting patients’ privacy. [90].
Conclusions
Brain edema constitutes an important component of secondary brain injury via increased intracranial pressure and dislocation of neuroanatomical structures, sometimes with associated herniation. Given the significant association between cerebral edema with patient outcome after acute brain injury and the extended time frame of edema formation, brain edema represents an appealing target for novel therapies. In this review, we discussed a number of possible genetic loci involved in this process. APOE-ε2 and APOE-ε4 have been shown to be associated with outcome after TBI and ICH, and there is evidence linking these polymorphisms to inflammatory processes that lead to BBB disruption and vasogenic edema. Through its ability to bind and remove hemoglobin, haptoglobin is an important protein with antioxidant properties. The Hp-2 allele has been associated with worse outcome after acute brain injury, while the Hp-1–1 phenotype was associated with increased edema in the early phase after ICH. Future research should be focused on studying the Hp alleles within different time points of edema formation after brain injury. Another important protein in cerebral edema is AQP4, which contributes to the formation of cytotoxic edema. More research is necessary to help elucidate its mechanistic properties. A summary of the genes involved in the pathogenesis of brain edema is shown in Table 2. Ascertaining novel loci related to edema formation and understanding the pathophysiology of the discussed genetic loci is crucial for the identification novel drug targets.
Table 2.
Genes involved in the pathophysiology of brain edema.
| Gene | Principal role | Other diseases related | Potential mechanism of edema formation |
|---|---|---|---|
| APOE | lipid transport and metabolism [46] | cognitive decline, dementia, and Alzheimer’s disease [87, 91], arteriosclerosis, traumatic brain injury, and intracerebral hemorrhage [82, 86, 88] | inflammatory response that disrupts the BBB and contributes to vasogenic edema after acute brain injury [92] |
| Hp | acute-phase response and antioxidant role by binding and neutralizing hemoglobin in the blood [72, 93] | cardiovascular events [58, 60] | oxidative stress [54], the release of inflammatory cytokines, the breakdown of BBB [55] |
| AQP | the main water channel found in the brain, cerebral spinal fluid production and regulation | Epilepsy [94], neurological autoimmune diseases[95], diabetes, artherosclerosis[96], cancer [97], peripheral nerves system damage[98] | cytotoxic and vasogenic edema, BBB disruption, micro-vessel damage, and neuronal death[65] |
| Sur1 | upregulation after trauma, ischemia, and hypoxia [99] | diabetes[99], hypoglycemia[100], autoimmune diseases[101] | modulation of astrocyte swelling, a key factor in cytotoxic edema, BBB breakdown and vasogenic edema[73] |
APOE – the apolipoprotein gene, Hp- haptoglobin gene, BBB – blood-brain barrier, AQP- aquaporin gene, Sur1- a sulfonylurea receptor and transmembrane protein gene
Highlights.
Brain edema is a component of secondary brain injury.
Mechanisms involved include inflammation, vasogenic and cytotoxic edema.
Genes involved brain edema: Apolipoprotein E, Haptoglobin, AQP4, and ABCC8 (Sur1).
Publicly accessible genetic data will help elucidate the genetic factors of edema.
Acknowledgments
Declaration of interests
EK has nothing to disclose. NS has nothing to disclose. GJF is supported by the National Institutes of Health (K76AG059992, R03NS112859 and P30AG021342), the American Heart Association (18IDDG34280056), the Yale Pepper Scholar Award (P30AG021342) and the Neurocritical Care Society Research Fellowship.
References
- 1.Kunz A, Dirnagl U, and Mergenthaler P, Acute pathophysiological processes after ischaemic and traumatic brain injury. 2010. p. 495–509. [DOI] [PubMed]
- 2.Krishnamurthi RV, et al. , The Global Burden of Hemorrhagic Stroke: A Summary of Findings From the GBD 2010 Study. Global Heart, 2014. 9(1): p. 101–106. [DOI] [PubMed] [Google Scholar]
- 3.Bonita R, et al. , The Global Stroke Initiative. The Lancet Neurology, 2004. 3(7): p. 391–393. [DOI] [PubMed] [Google Scholar]
- 4.Winkler EA, Minter D, and Yue JK, Cerebral Edema in Traumatic Brain Injury: Pathophysiology and Prospective Therapeutic Targets. Neurosurgery Clinics of North America, 2016. 27(4): p. 473–488. [DOI] [PubMed] [Google Scholar]
- 5.Leasure A, et al. , Treatment of Edema Associated With Intracerebral Hemorrhage. Current Treatment Options in Neurology, 2016. 18(2): p. 9–9. [DOI] [PubMed] [Google Scholar]
- 6.Jha RM, Kochanek PM, and Simard JM, Pathophysiology and treatment of cerebral edema in traumatic brain injury. Neuropharmacology, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tucker B, et al. , Early Brain Edema is a Predictor of In-Hospital Mortality in Traumatic Brain Injury. Journal of Emergency Medicine, 2017. 53(1): p. 18–29. [DOI] [PubMed] [Google Scholar]
- 8.Z, G., et al. , Perihematomal Edema Expansion Rates and Patient Outcomes in Deep and Lobar Intracerebral Hemorrhage. Neurocritical Care, 2017. 26(2): p. 205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acosta JN, Brown SC, and Falcone GJ, Genetic Variation and Response to Neurocritical Illness: a Powerful Approach to Identify Novel Pathophysiological Mechanisms and Therapeutic Targets. Neurotherapeutics, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bevan S, et al. , Genetic Heritability of Ischemic Stroke and the Contribution of Previously Reported Candidate Gene and Genomewide Associations. Stroke, 2012. 43(12): p. 3161–3167. [DOI] [PubMed] [Google Scholar]
- 11.Devan W, et al. , Heritability Estimates Identify a Substantial Genetic Contribution to Risk and Outcome of Intracerebral Hemorrhage. Stroke; a journal of cerebral circulation, 2013. 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korja M, et al. , Genetic Epidemiology of Spontaneous Subarachnoid Hemorrhage. Stroke, 2010. 41(11): p. 2458–2462. [DOI] [PubMed] [Google Scholar]
- 13.Beyene J and Pare G, Statistical genetics with application to population-based study design: a primer for clinicians. European Heart Journal, 2013. 35(8): p. 495–500. [DOI] [PubMed] [Google Scholar]
- 14.Nelson MR, et al. , The support of human genetic evidence for approved drug indications. Nature Genetics, 2015. 47(8): p. 856–860. [DOI] [PubMed] [Google Scholar]
- 15.Davies NM, Holmes MV, and Davey Smith G, Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ, 2018. 362: p. k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klatzo I, Pathophysiological aspects of brain edema. Acta Neuropathologica, 1987. 72(3): p. 236–239. [DOI] [PubMed] [Google Scholar]
- 17.Alluri H, et al. , Blood–brain barrier dysfunction following traumatic brain injury. 2015. p. 1093–1104. [DOI] [PubMed]
- 18.Anttila V, et al. , Analysis of shared heritability in common disorders of the brain. Science, 2018. 360(6395). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes JR, The idiosyncratic aspects of the epilepsy of Fyodor Dostoevsky. Epilepsy and Behavior, 2005. [DOI] [PubMed] [Google Scholar]
- 20.Ziai WC, Hematology and Inflammatory Signaling of Intracerebral Hemorrhage. Stroke, 2013. 44(6 suppl 1): p. S74 LP–S78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chodobski A, Zink BJ, and Szmydynger-Chodobska J, Blood-Brain Barrier Pathophysiology in Traumatic Brain Injury. 2011. p. 492–516. [DOI] [PMC free article] [PubMed]
- 22.Zheng H, et al. , Mechanism and therapy of brain edema after intracerebral hemorrhage. 2016. p. 155–169. [DOI] [PubMed]
- 23.Staykov D, et al. , Natural Course of Perihemorrhagic Edema After Intracerebral Hemorrhage. Stroke, 2011. 42(9): p. 2625–2629. [DOI] [PubMed] [Google Scholar]
- 24.Xi G, Keep RF, and Hoff JT, Mechanisms of brain injury after intracerebral haemorrhage. 2006. p. 53–63. [DOI] [PubMed]
- 25.Lu J, et al. , Systemic inflammatory response following acute traumatic brain injury. Frontiers in bioscience (Landmark edition), 2009. 14: p. 3795–813. [DOI] [PubMed] [Google Scholar]
- 26.Abdul-Muneer PM, et al. , Role of Matrix Metalloproteinases in the Pathogenesis of Traumatic Brain Injury. 2016. p. 6106–6123. [DOI] [PMC free article] [PubMed]
- 27.Feiler S, et al. , Contribution of matrix metalloproteinase-9 to cerebral edema and functional outcome following experimental subarachnoid hemorrhage. Cerebrovascular Diseases, 2011. 32(3): p. 289–295. [DOI] [PubMed] [Google Scholar]
- 28.Vink R, Gabrielian L, and Thornton E, The role of substance P in secondary pathophysiology after traumatic brain injury. 2017. [DOI] [PMC free article] [PubMed]
- 29.Donkin JJ, et al. , Substance P is associated with the development of brain edema and functional deficits after traumatic brain injury. Journal of Cerebral Blood Flow and Metabolism, 2009. 29(8): p. 1388–1398. [DOI] [PubMed] [Google Scholar]
- 30.Turner RJ and Vink R, The role of substance P in ischaemic brain injury. 2013. p. 123–142. [DOI] [PMC free article] [PubMed]
- 31.Wahl M, et al. , Mediators of Vascular and Parenchymal Mechanisms in Secondary Brain Damage. 1993. p. 64–72. [DOI] [PubMed]
- 32.Dobrivojević M, Špiranec K, and Sinđić A, Involvement of bradykinin in brain edema development after ischemic stroke. 2014. p. 201–212. [DOI] [PubMed]
- 33.Lee KR, et al. , Edema from intracerebral hemorrhage: the role of thrombin. Journal of Neurosurgery, 1996. 84(1): p. 91–96. [DOI] [PubMed] [Google Scholar]
- 34.K.R, L., et al. , Mechanisms of edema formation after intracerebral hemorrhage: Effects of thrombin on cerebral blood flow, blood-brain barrier permeability, and cell survival in a rat model. Journal of Neurosurgery, 1997. 86(2): p. 272–278. [DOI] [PubMed] [Google Scholar]
- 35.Waters RJ and Nicoll JAR, Genetic influences on outcome following acute neurological insults. 2005. p. 105–110. [DOI] [PubMed]
- 36.Adler G, et al. , Bosnian study of APOE distribution (BOSAD): a comparison with other European populations. Annals of Human Biology, 2017. 44(6): p. 568–573. [DOI] [PubMed] [Google Scholar]
- 37.Main BS, et al. , Apolipoprotein E4 impairs spontaneous blood brain barrier repair following traumatic brain injury. Molecular Neurodegeneration, 2018. 13(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rasmussen KL, Plasma levels of apolipoprotein E, APOE genotype and risk of dementia and ischemic heart disease: A review. 2016. p. 145–155. [DOI] [PubMed] [Google Scholar]
- 39.Mahoney-Sanchez L, et al. , The Complex Role of Apolipoprotein E in Alzheimer’s Disease: an Overview and Update. Journal of Molecular Neuroscience, 2016. 60(3): p. 325–335. [DOI] [PubMed] [Google Scholar]
- 40.Friedman G, et al. , Apolipoprotein E-epsilon4 genotype predicts a poor outcome in survivors of traumatic brain injury. Neurology, 1999. 52(2): p. 244–8. [DOI] [PubMed] [Google Scholar]
- 41.Pang J, et al. , Potential implications of Apolipoprotein E in early brain injury after experimental subarachnoid hemorrhage: Involvement in the modulation of blood-brain barrier integrity. Oncotarget, 2016. 7(35). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lynch JR, et al. , Apolipoprotein E affects the central nervous system response to injury and the development of cerebral edema. Annals of Neurology, 2002. 51(1): p. 113–117. [DOI] [PubMed] [Google Scholar]
- 43.Laskowitz DT, et al. , Apolipoprotein E suppresses glial cell secretion of TNFα. Journal of Neuroimmunology, 1997. 76(1–2): p. 70–74. [DOI] [PubMed] [Google Scholar]
- 44.Lynch JR, et al. , A novel therapeutic derived from apolipoprotein E reduces brain inflammation and improves outcome after closed head injury. Experimental Neurology, 2005. 192(1): p. 109–116. [DOI] [PubMed] [Google Scholar]
- 45.Cao FJYWYZJ and Liu JQXCLVMPLFXLSX, Apolipoprotein E-Mimetic COG1410 Reduces Acute Vasogenic Edema following Traumatic Brain Injury. Neurotrama, 2016. 33(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waters RJ and Nicoll JA, Genetic influences on outcome following acute neurological insults. Curr Opin Crit Care, 2005. 11(2): p. 105–10. [DOI] [PubMed] [Google Scholar]
- 47.Niskakangas T, et al. , Association of Apolipoprotein E Polymorphism With Outcome After Aneurysmal Subarachnoid Hemorrhage. Stroke, 2001. 32(5): p. 1181–1184. [DOI] [PubMed] [Google Scholar]
- 48.Nathoo N, et al. , Genetic vulnerability following traumatic brain injury: The role of apolipoprotein E. Molecular pathology : MP, 2003. 56: p. 132–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pruthi N, et al. , Apolipoprotein E polymorphism and outcome after mild to moderate traumatic brain injury: a study of patient population in India. Neurol India, 2010. 58(2): p. 264–9. [DOI] [PubMed] [Google Scholar]
- 50.Brouwers HB, et al. , Apolipoprotein E genotype predicts hematoma expansion in lobar intracerebral hemorrhage. Stroke, 2012. 43(6): p. 1490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Biffi A, et al. , APOE genotype and extent of bleeding and outcome in lobar intracerebral haemorrhage: a genetic association study. Lancet Neurol, 2011. 10(8): p. 702–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.James ML, et al. , Apolipoprotein E modifies neurological outcome by affecting cerebral edema but not hematoma size after intracerebral hemorrhage in humans. J Stroke Cerebrovasc Dis, 2009. 18(2): p. 144–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mahley RW and Huang Y, Apolipoprotein e sets the stage: response to injury triggers neuropathology. Neuron, 2012. 76(5): p. 871–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao X, et al. , Neuroprotective role of haptoglobin after intracerebral hemorrhage. J Neurosci, 2009. 29(50): p. 15819–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Melamed-Frank M, et al. , Structure-function analysis of the antioxidant properties of haptoglobin. Blood, 2001. 98(13): p. 3693–8. [DOI] [PubMed] [Google Scholar]
- 56.Halstead Michael R, et al. , Abstract TP343: The 1–1 Haptoglobin Phenotype is Associated With Perihematoma Edema Progression in Acute Intracranial Hemorrhage. Stroke, 2017. 48(suppl_1): p. ATP343–ATP343. [Google Scholar]
- 57.Guetta J, et al. , Haptoglobin genotype modulates the balance of Th1/Th2 cytokines produced by macrophages exposed to free hemoglobin. Atherosclerosis, 2007. 191(1): p. 48–53. [DOI] [PubMed] [Google Scholar]
- 58.Levy AP, et al. , Haptoglobin genotype is a determinant of iron, lipid peroxidation, and macrophage accumulation in the atherosclerotic plaque. Arterioscler Thromb Vasc Biol, 2007. 27(1): p. 134–40. [DOI] [PubMed] [Google Scholar]
- 59.Leclerc J, et al. , Abstract TP102: Modulation of Neuroinflammation by Haptoglobin Reduces Oxidative Stress and Improves ICH Outcomes. Stroke, 2016. 47(suppl_1): p. ATP102–ATP102. [Google Scholar]
- 60.Marvasti TB, et al. , Haptoglobin 2–2 genotype is associated with presence and progression of MRI depicted atherosclerotic intraplaque hemorrhage. Int J Cardiol Heart Vasc, 2018. 18: p. 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao X, et al. , Cytoprotective role of haptoglobin in brain after experimental intracerebral hemorrhage. Acta Neurochir Suppl, 2011. 111: p. 107–12. [DOI] [PubMed] [Google Scholar]
- 62.Murthy SB, et al. , Presence of Haptoglobin-2 Allele Is Associated with Worse Functional Outcomes After Spontaneous Intracerebral Hemorrhage. World Neurosurgery, 2015. 83(4): p. 583–587. [DOI] [PubMed] [Google Scholar]
- 63.Wang B. f., et al. , Curcumin attenuates brain edema in mice with intracerebral hemorrhage through inhibition of AQP4 and AQP9 expression. Acta Pharmacologica Sinica, 2015. 36(8): p. 939–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Filippidis AS, Carozza RB, and Rekate HL, Aquaporins in Brain Edema and Neuropathological Conditions. Int J Mol Sci, 2016. 18(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang Y, et al. , Effects of Aquaporin-4 on edema formation following intracerebral hemorrhage. Exp Neurol, 2010. 223(2): p. 485–95. [DOI] [PubMed] [Google Scholar]
- 66.Amiry-Moghaddam M, Frydenlund DS, and Ottersen OP, Anchoring of aquaporin-4 in brain: molecular mechanisms and implications for the physiology and pathophysiology of water transport. Neuroscience, 2004. 129(4): p. 999–1010. [DOI] [PubMed] [Google Scholar]
- 67.Amiry-Moghaddam M, et al. , Alpha-syntrophin deletion removes the perivascular but not endothelial pool of aquaporin-4 at the blood-brain barrier and delays the development of brain edema in an experimental model of acute hyponatremia. Faseb j, 2004. 18(3): p. 542–4. [DOI] [PubMed] [Google Scholar]
- 68.Anderova M, et al. , Altered astrocytic swelling in the cortex of alpha-syntrophin-negative GFAP/EGFP mice. PLoS One, 2014. 9(11): p. e113444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang T, et al. , Reduction of brain edema and expression of aquaporins with acute ethanol treatment after traumatic brain injury. J Neurosurg, 2013. 118(2): p. 390–6. [DOI] [PubMed] [Google Scholar]
- 70.Papadopoulos MC, et al. , Aquaporin-4 facilitates reabsorption of excess fluid in vasogenic brain edema. Faseb j, 2004. 18(11): p. 1291–3. [DOI] [PubMed] [Google Scholar]
- 71.Dardiotis E, et al. , AQP4 tag single nucleotide polymorphisms in patients with traumatic brain injury. J Neurotrauma, 2014. 31(23): p. 1920–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.King ZA, et al. , Profile of intravenous glyburide for the prevention of cerebral edema following large hemispheric infarction: evidence to date. Drug Des Devel Ther, 2018. 12: p. 2539–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stokum JA, et al. , SUR1-TRPM4 and AQP4 form a heteromultimeric complex that amplifies ion/water osmotic coupling and drives astrocyte swelling. Glia, 2018. 66(1): p. 108–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martinez-Valverde T, et al. , Sulfonylurea Receptor 1 in Humans with Post-Traumatic Brain Contusions. J Neurotrauma, 2015. 32(19): p. 1478–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jha RM, et al. , ABCC8 Single Nucleotide Polymorphisms are Associated with Cerebral Edema in Severe TBI. Neurocrit Care, 2017. 26(2): p. 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eraslan G, et al. , Deep learning: new computational modelling techniques for genomics. Nat Rev Genet, 2019. 20(7): p. 389–403. [DOI] [PubMed] [Google Scholar]
- 77.Zou J, et al. , A primer on deep learning in genomics. Nat Genet, 2019. 51(1): p. 12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tryka KA, et al. , NCBI’s Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Res, 2014. 42(Database issue): p. D975–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gaziano JM, et al. , Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol, 2016. 70: p. 214–23. [DOI] [PubMed] [Google Scholar]
- 80.Sankar PL and Parker LS, The Precision Medicine Initiative’s All of Us Research Program: an agenda for research on its ethical, legal, and social issues. Genet Med, 2017. 19(7): p. 743–750. [DOI] [PubMed] [Google Scholar]
- 81.Bycroft C, et al. , The UK Biobank resource with deep phenotyping and genomic data. Nature, 2018. 562(7726): p. 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen Z, et al. , China Kadoorie Biobank of 0.5 million people: survey methods, baseline characteristics and long-term follow-up. Int J Epidemiol, 2011. 40(6): p. 1652–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lappalainen I, et al. , The European Genome-phenome Archive of human data consented for biomedical research. Nat Genet, 2015. 47(7): p. 692–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Johnson AE, et al. , MIMIC-III, a freely accessible critical care database. Sci Data, 2016. 3: p. 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pollard TJ, et al. , The eICU Collaborative Research Database, a freely available multi-center database for critical care research. Sci Data, 2018. 5: p. 180178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Crawford KM, et al. , Cerebrovascular Disease Knowledge Portal: An Open-Access Data Resource to Accelerate Genomic Discoveries in Stroke. Stroke, 2018. 49(2): p. 470–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Woo D, et al. , The Ethnic/Racial Variations of Intracerebral Hemorrhage (ERICH) study protocol. Stroke, 2013. 44(10): p. e120–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Leasure AC, et al. , Identification and Validation of Hematoma Volume Cutoffs in Spontaneous, Supratentorial Deep Intracerebral Hemorrhage. Stroke, 2019. 50(8): p. 2044–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Walsh KB, et al. , Untreated Hypertension: A Powerful Risk Factor for Lobar and Nonlobar Intracerebral Hemorrhage in Whites, Blacks, and Hispanics. Circulation, 2016. 134(19): p. 1444–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou J and Troyanskaya OG, Predicting effects of noncoding variants with deep learning-based sequence model. Nat Methods, 2015. 12(10): p. 931–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rasmussen KL, Plasma levels of apolipoprotein E, APOE genotype and risk of dementia and ischemic heart disease: A review. Atherosclerosis, 2016. 255: p. 145–155. [DOI] [PubMed] [Google Scholar]
- 92.Marini S, et al. , Comparison of Genetic and Self-Identified Ancestry in Modeling Intracerebral Hemorrhage Risk. Front Neurol, 2018. 9: p. 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim GS, et al. , Critical role of sphingosine-1-phosphate receptor-2 in the disruption of cerebrovascular integrity in experimental stroke. Nat Commun, 2015. 6: p. 7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu H, et al. , Effects of AQP4 and KCNJ10 Gene Polymorphisms on Drug Resistance and Seizure Susceptibility in Chinese Han Patients with Focal Epilepsy. Neuropsychiatr Dis Treat, 2020. 16: p. 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pittock SJ and Lucchinetti CF, Neuromyelitis optica and the evolving spectrum of autoimmune aquaporin-4 channelopathies: a decade later. Ann N Y Acad Sci, 2016. 1366(1): p. 20–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Madonna R, et al. , Co-expression of glycosylated aquaporin-1 and transcription factor NFAT5 contributes to aortic stiffness in diabetic and atherosclerosis-prone mice. J Cell Mol Med, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Magouliotis DE, et al. , Transcriptomic analysis of the aquaporin gene family and associated interactors in rectal cancer. Microrna, 2019. [DOI] [PubMed] [Google Scholar]
- 98.He D, et al. , Autoimmune aquaporin-4 induced damage beyond the central nervous system. Mult Scler Relat Disord, 2017. 18: p. 41–46. [DOI] [PubMed] [Google Scholar]
- 99.Reis AF and Velho G, Sulfonylurea receptor −1 (SUR1): genetic and metabolic evidences for a role in the susceptibility to type 2 diabetes mellitus. Diabetes Metab, 2002. 28(1): p. 14–9. [PubMed] [Google Scholar]
- 100.Jahnavi S, et al. , Novel ABCC8 (SUR1) gene mutations in Asian Indian children with congenital hyperinsulinemic hypoglycemia. Ann Hum Genet, 2014. 78(5): p. 311–9. [DOI] [PubMed] [Google Scholar]
- 101.Makar TK, et al. , Silencing of Abcc8 or inhibition of newly upregulated Sur1-Trpm4 reduce inflammation and disease progression in experimental autoimmune encephalomyelitis. J Neuroinflammation, 2015. 12: p. 210. [DOI] [PMC free article] [PubMed] [Google Scholar]