Abstract
CD4+ helper T cells (TH) and regulatory T cells (Treg) that respond to common allergens play an important role in driving and dampening airway inflammation in patients with asthma. Until recently, direct, unbiased molecular analysis of allergen-reactive TH and Treg cells has not been possible. To better understand the diversity of these T cell subsets in allergy and asthma, we analyzed the single-cell transcriptome of ~50,000 house dust mite (HDM) allergen-reactive TH cells and Treg cells from asthmatics with HDM allergy and from three control groups: asthmatics without HDM allergy and non-asthmatics with and without HDM allergy. Our analyses show that HDM allergen-reactive TH and Treg cells are highly heterogeneous, and certain subsets are quantitatively and qualitatively different in subjects with HDM-reactive asthma. The number of interleukin (IL)-9 expressing HDM-reactive TH cells is greater in asthmatics with HDM allergy compared to non-asthmatics with HDM allergy, and this IL-9-expressing TH subset displays enhanced pathogenic properties. More HDM-reactive TH and Treg cells expressing the interferon-response signature (THIFNR and TregIFNR) are present in asthmatics without HDM allergy compared with those with HDM allergy. In cells from these subsets (THIFNR and TregIFNR), expression of TNFSF10 was enriched; its product, tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), dampens activation of TH cells. These findings suggest that the THIFNR and TregIFNR subsets may dampen allergic responses, which may help explain why only some people develop TH2 responses to nearly ubiquitous allergens.
One sentence summary
Single-cell transcriptomic analysis of allergen-specific T cells in allergy and asthma reveals new T cell subsets.
Introduction
Asthma is characterized by aberrant type 2 immune responses to common inhaled aeroallergens such as house dust mite (HDM), grass pollen, animal dander, and mold (1–6), leading to ‘asthma attacks’ in sensitized asthmatic subjects in response to inhalation of such allergens (7). The hallmarks of asthma, namely airway narrowing and sputum eosinophilia, have been shown to result from the specific activation of (MHC) class II–restricted CD4+ helper T cells (TH) by challenging asthmatics with synthetic allergen-derived peptides (8–12). Further evidence of the centrality of TH cells in asthma pathology is that their depletion reduces allergic airway inflammation in animal models (13), and that inhibition of TH cell-derived type 2 cytokines (IL-5, IL-13, IL-4) is clinically beneficial in patients with asthma (14–17). However, despite the central role of allergen-reactive TH cells and their products in driving airway inflammation, the full spectrum and function of TH cell subsets that respond to common allergens has yet to be defined. Similarly, though an imbalance between regulatory T cells (Treg) and TH cell responses to allergens is associated with the development of allergy and asthma (18–23), the heterogeneity of allergen-reactive Treg cells remains unstudied.
Previous studies of allergen-reactive T cells have characterized their phenotype based on the expression of cell-surface markers or canonical cytokines (24–26). Due to their relative rarity, analyses of these cells usually require in vitro expansion, which can alter their molecular properties, thus limiting the value of unbiased transcriptomic studies (27–29). Furthermore, transcriptomic studies performed at the whole population level fail to capture cellular heterogeneity and also lack the resolution to detect biological differences associated with asthma or allergy (30). A recent single-cell analysis of TH cells in a mouse models of allergic airway inflammation revealed substantial heterogeneity, and also identified TH subsets that had not been previously described (20).
Characterizing the various subsets of TH and Treg cells in asthmatic subjects and comparing their frequency and properties to those in subjects without asthma is ideally achieved at single-cell resolution. Indeed, single-cell transcriptomic analysis can help define the molecular properties of allergen-reactive TH cells associated with pathology and assess whether these features are the result of an expansion of a pre-existing population of cells or the result of their aberrant differentiation in response to environmental signals (31, 32). To address the latter issue, the subsets of allergen-reactive TH cells must also be defined in subjects without asthma and allergy. Such allergen-reactive TH cells are present even in non-allergic subjects (33–36), although it is not known why or how these cells fail to cause overt allergic responses.
To address these questions in a hypothesis-free manner, we performed single-cell transcriptomic analysis of TH and Treg cells that react to house dust mite allergen (HDM). HDM is one of the most common and ubiquitous allergens, and sensitization is associated with both the onset of allergic asthma and its severity (37–40). The relatively high abundance of HDM-reactive T cells in the blood makes it is possible to isolate sufficient number of cells for high-throughput single-cell transcriptomic analysis. Here, we report on the single-cell transcriptomes of >50,000 HDM-reactive T cells from allergic asthmatic subjects and relevant control groups. Our analysis revealed multiple distinct subsets of TH and Treg cells that are either preferentially expanded or depleted in asthmatic subjects with and without HDM allergy, defined the pathogenic properties of TH subsets associated with allergic asthma, and uncovered a unique HDM-reactive TH subset that is expanded specifically in subjects without HDM allergy.
Results
Bulk RNA-seq analysis of HDM allergen-reactive T cells does not identify asthma-specific features
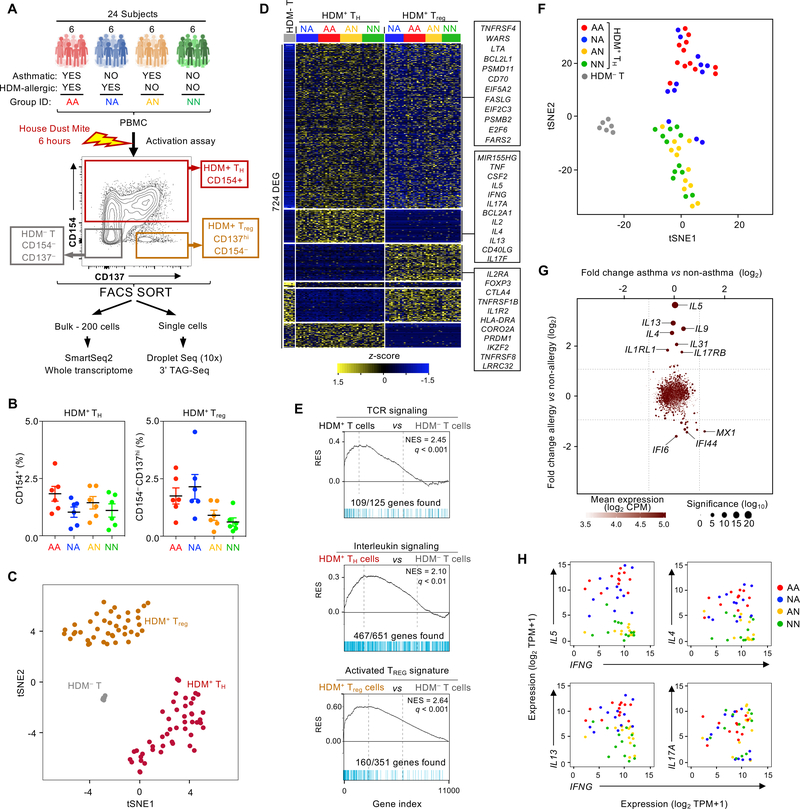
To comprehensively characterize the molecular properties of allergen-reactive TH and Treg cells from patients with asthma, we isolated pure populations of HDM-reactive memory TH and Treg cells ex vivo (see Materials and Methods and fig. S1) from asthmatics with HDM allergy (N = 6) and performed both bulk and single-cell RNA-seq (Fig. 1A). To distinguish the molecular features that are specific to asthma as opposed to HDM allergy, we performed similar assays in HDM-reactive TH and Treg cells isolated from HDM-allergic subjects without asthma (N = 6). Because allergen-reactive T cells are present even in non-allergic subjects (33–36), we also isolated HDM-reactive TH and Treg cells from asthmatic (N = 6) and healthy subjects (N = 6) without HDM allergy to uncover features that may contribute to the lack of HDM allergy i.e., IgE reactivity. In total, we performed 95 bulk RNA-seq and ~50,000 single-cell RNA-seq assays on T cells from a total of 24 subjects (Fig. 1A, and table S1).
Fig. 1.
Bulk RNA-seq analysis of HDM allergen-reactive T cells does not identify asthma-specific features. (A) Schematic representation of the study summarizing the subject groups, activation assay, and sorting and sequencing strategies. (B) Dot plot showing the percentage of CD4 memory HDM-reactive T cells that were CD154+ (left) or CD154– CD137+ (right) for each subject group. Horizontal line mean; error bar, standard error. (C) t-SNE plot of bulk RNA-seq samples: 6 HDM– T cells (from asthma-allergic patients), 24 HDM+ TH (from all patients), 18 HDM+ Treg (from all patients)). (D) Heatmap of row-wise z-score-normalized expression for 724 genes differentially expressed between the 3 groups of cells in Figure 1C. Each column represents data from one subject. (E) Gene set enrichment analysis (GSEA) of grouped bulk RNA-seq datasets presented in Figure 1C. q, false discovery rate; NES, normalized enrichment score; RES, relative enrichment score; list of genes provided in table S4. (F) t-SNE plot of bulk RNA-seq datasets for HDM– T samples (N = 6) and TH samples colored by disease group, where each dot represents one RNA-seq data from one subject (N = 12 per group). (G) Scatter plot shows log2-fold change in expression of significantly differentially expressed genes between asthma (N = 24) versus non-asthma (N = 24) (x axis) and allergic (N = 24) versus non-allergic (N = 24) (y axis) TH samples. (H) Scatter plots show co-expression of the indicated canonical TH1 and TH2 cytokines in TH samples coded by disease group.
HDM-reactive TH cells (0.2–3 % of all memory TH cells) and Treg cells (1–5 % of all memory Treg cells) were detected in all 4 subject groups, including the HDM-allergic and non-allergic subjects (Fig. 1B). Bulk transcriptome analysis showed that HDM-reactive TH and Treg cells clustered separately from one another and from HDM-non-reactive cells (HDM– T cells) (Fig. 1C). 724 transcripts were differentially expressed between both HDM-reactive, activated T cells populations, TH and Treg (following stimulation with HDM peptide/MHC complex from antigen-presenting cells) and HDM– T cells (not stimulated by HDM-allergen derived peptides) (adjusted P-value < 0.01, log2 fold change > 2, Fig. 1D and table S2). As expected, these differentially expressed transcripts were highly enriched for genes in the TCR signaling pathway (Fig. 1E, top panel). Allergen-activated HDM-reactive TH cells expressed greater amounts of several transcripts encoding cytokines (IL-2, −13, −5, - 4, −9, −31 −17F, −22, TNF, IFNG, CSF-2) and chemokines (CCL20, CXCL10) linked to effector functions (Fig. 1E, middle panel). HDM-reactive Treg cells expressed higher levels of genes linked to Treg function, such as IL2RA, FOXP3, CTLA4, IKZF2, TNFRSF8, when compared with HDM– T cells (Fig. 1E, bottom panel, fig. S2, and table S2).
Clustering analysis of HDM-reactive TH cells by disease group showed separation based on HDM allergy status rather than asthma phenotype (Fig. 1F). For example, in HDM-reactive TH cells from HDM-allergic subjects, expression of canonical TH2 cytokines was increased compared with those from HDM-non-allergic subjects (Fig. 1G), whereas no significant differences were observed between the HDM-reactive TH cells from asthmatic versus non-asthmatic subjects with HDM allergy (Fig. 1G). The heterogeneity observed within the HDM-reactive TH population, reflected in the co-expression of transcripts encoding canonical TH1, TH2 and TH17 cytokines (Fig. 1H), is likely to have limited the resolution of bulk transcriptome data to distinguish asthma-specific features.
Single-cell RNA-seq analysis reveals heterogeneity among HDM-reactive TH cells
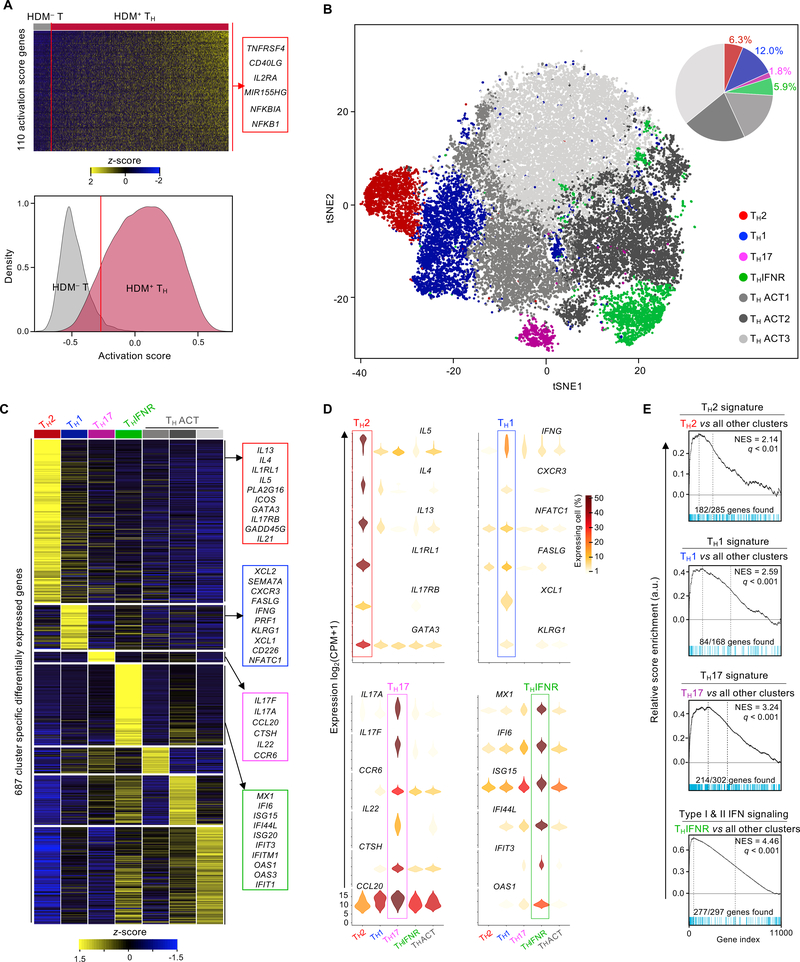
Single cells from all 6 subjects in each disease group were pooled for droplet-based single-cell RNA-seq (10x Genomics platform), and genotype-based deconvolution was employed to obtain subject-specific single-cell transcriptomes and to exclude potential cell doublets (see Materials and Methods, and fig. S3). Our cell isolation strategy, based on the CD154 activation marker, primarily enriches for HDM-reactive TH cells (41–43). Analogous to flow cytometry-based approaches, single-cell transcriptome analysis allows discrimination of activated (true positives) from non-activated (false positives) TH cells. Based on a TH activation signature, derived by comparing HDM-reactive TH and HDM-non-reactive (HDM– T cells) single cells (Fig. 2A and fig. S3), we eliminated potential false positive cells from the HDM-reactive TH cell population (Fig. 2A and fig. S3).
Fig. 2.
Single-cell RNA-seq clustering analysis reveals heterogeneity among HDM-reactive TH cells. (A) Top - heatmap of row-wise z-score-normalized expression for 110 genes used to establish the activation score (see Methods), rows are ordered by hierarchical clustering. Each column represents a single-cell RNA-seq data, ordered by activation score. Right - examples of genes included in the TH activation score. Bottom - histogram shows the density function of HDM– T (N = 3,075, grey) and HDM+ TH (N = 31,105, red) cell activation scores (Methods). The red line indicates the threshold of selection (activation score of −0.27, 5 % of HDM–). (B) t-SNE visualization of Seurat clustering analysis of approximately 28,313 single HDM+ TH cell transcriptomes obtained from all 24 subjects. IFNR, interferon response; ACT, biologically uncharacterized activated T cells (3 groups). Top right - pie chart shows the cell number proportion of each cluster. (C) Heatmap of row-wise z-score-normalized mean expression of cluster-specific differentially expressed genes. Columns represent the average expression for each cluster, ordered based on biological relevance. Right - lists of biologically relevant example genes for each cluster. Equal numbers of cells were sampled from each cluster. (D) Violin plots show log2 (CPM+1) normalized expression in each cluster (3 THACT clusters merged) for 24 cluster-specific signature genes (6 per cell type). Color scale represents the fraction of cells within each cluster expressing the given gene, excluding cells with no expression. (E) GSEA between each cluster and other clusters of single-cell transcriptome datasets presented in Figure 2B. q, false discovery rate; NES, normalized enrichment score; RES, relative enrichment score; list of genes provided in table S4.
Analysis of the single-cell transcriptomes of HDM-reactive TH cells (non-doublet and activation-signature positive) revealed 7 clusters (Materials and Methods, and fig. S4) present at varying frequency among subjects, highlighting the importance of studying cells from multiple subjects (Fig. 2B and fig. S5). To understand the molecular properties unique to each cluster, we performed multiple pair-wise single-cell differential gene expression analyses (Materials and Methods, and table S3). Several hundred genes (N = 687) were especially highly expressed by each cluster, allowing classification into specific TH subsets (Fig. 2C). Cells in cluster 1 were highly enriched for transcripts encoding canonical type 2 cytokine genes (IL5, IL13, IL4), the TH2 master transcription factor GATA3, and receptors (IL1RL1 and IL17RB) for the TH2-polarizing cytokines IL-33 and IL-25, indicating that this cluster represented TH2 cells (Fig. 2D). Notably, the TH2 subset only represented ~6.3 % of the HDM-reactive TH cell population (Fig. 2B). Cluster 2 was enriched for TH1 phenotype- and function-related genes such as IFNG, CXCR3, and PRF1 (44–47) (Fig. 2, C and D). In addition, we found that the expression of genes encoding the chemokines XCL1 and XCL2 was correlating with expression of IFNG (fig. S6), suggesting a potentially important role of these chemokines in the function of TH1 cells (46–49). Cluster 3 was enriched for TH17 phenotype- and function-related genes such as IL17A, IL17F, CCR6, IL22, CTSH, and CCL20 (50–52). The characteristics of cell clusters 1–3 were confirmed by gene set enrichment analysis (GSEA) using curated lists of signature genes (Fig. 2E and table S4). Cells in cluster 4 were very highly enriched for type I and II interferon response genes (IFI6, MX1, ISG20, OAS1, IFIT1, IFI44L) (53, 54), indicating that they represent a previously uncharacterized TH subset, which we termed TH subset expressing the interferon response signature (THIFNR) (Fig. 2, C and D). GSEA analysis confirmed enrichment of interferon response genes in this cluster (Fig. 2E and table S4). Cells in clusters 5, 6, and 7 were enriched for genes linked with cell activation; cluster 5 (THACT1) was enriched in genes linked to ribosomal proteins and RNA translation (RPLx, RPSx, see table S3), cluster 6 (THACT2) was enriched with genes linked to endocytosis and membrane trafficking (ARL6IP5, ARPC5, BIN1, see table S3), and cluster 7 (THACT3) was enriched in genes linked to chromosome maintenance (NPM1, NHP2) and apoptosis (GADD45B, NFKB1, ATF4, PMAIP1, see table S3). Overall, our single-cell transcriptome analysis uncovered substantial heterogeneity among HDM-reactive TH cells.
Proportions of HDM-reactive TH subsets differ between HDM allergic and non-allergic subjects
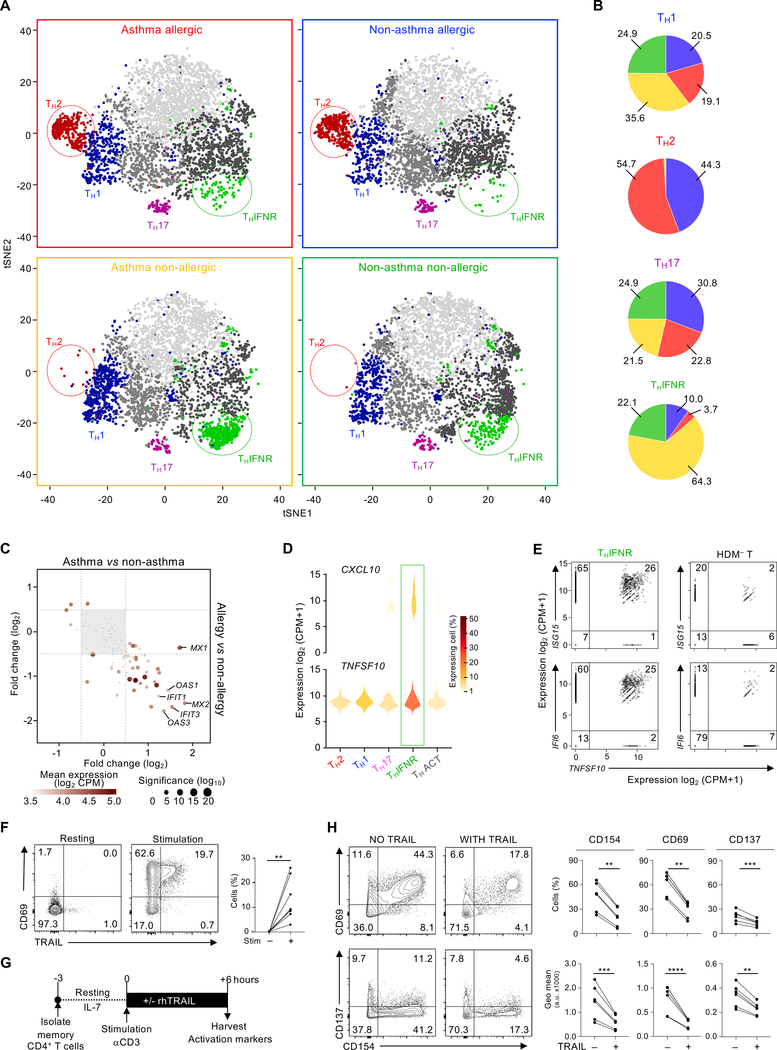
We next asked if the proportions of any of the HDM-reactive subsets varied between subjects with or without HDM allergy or asthma. As expected, the TH2 cluster (cluster 1) was present only in the HDM-allergic groups (Fig. 3, A and B, and fig. S7), consistent with the central role of TH2 cells and type 2 cytokines in IgE class switching and allergy and asthma pathogenesis (55–57). On the other hand, the TH1 cluster, though observed in all subject groups, was present at greater proportions in subjects without HDM allergy, consistent with the reciprocal role of TH1 cells in dampening TH2 differentiation. Several other clusters, including the TH17 cluster, showed no significant differences in their proportions among disease and control groups (Fig. 3, A and B, and fig. S7).
Fig. 3.
Proportions of HDM-reactive TH subsets differ between allergic and non-allergic subjects. (A) t-SNE visualization of Seurat clustering analysis shown in Figure 2b, using equal cell numbers for each disease group (N = 3,720), obtained from all 6 subjects in each group. Cells are colored according to cluster as in Figure 2B. (B) Pie chart illustrating the relative proportion of cells from each disease group in the 4 biologically relevant clusters. (C) Scatter plot shows the log2-fold change of expression of THIFNR signature genes between asthmatic (N = 12) versus non-asthmatic (N = 12) (x-axis) and allergic (N = 12) versus non-allergic (N = 12) (y-axis) subjects among cells in the THIFNR cluster. Equal numbers of cells were sampled per group. Dotted lines indicate the threshold value of fold change for gene filtering. (D) Violin plots show log2(CPM+1) normalized expression of CXCL10 and TNFSF10 in each TH cluster. Cells with no expression were excluded. (E) Scatter plots show co-expression of TNFSF10 with the products of the THINFR signature genes IFI6 and ISG15 by THINFR cells (left) or HDM– T cells (right). Each dot represents one cell. (F) Contour plots show the expression of CD69 versus TRAIL in memory CD4+ T cells before (left) and after 6 h (center) of TCR stimulation with anti-CD3 and anti-CD28. Both plots are representative of 5 independent experiments. Numbers indicate the percentage of cells in each quadrant. Right, quantification of each of the 6 experiments; bars represent the mean and standard error. (G) Diagram of the TRAIL functional assay. (H) Left, contour plots show the expression of the cell-activation markers CD154, CD69, and CD137 in memory CD4+ cells after 6 h of stimulation in the presence or absence of TRAIL. Data shown are from a representative experiment. Right, quantification of each of the 6 experiments; bars represent the mean and standard error. *, P < 0.1; **, P < 0.01, *** P < 0.001.
Subjects without HDM allergy – both the asthmatic and healthy control groups – despite displaying a substantially broad TH response to HDM allergen, failed to generate TH2 cells that respond to HDM ex vivo. Strikingly, the large majority of HDM-reactive TH cells expressing the interferon response signature (THIFNR, cluster 4) were observed in subjects without HDM allergy (Fig. 3, A, B and C). This negative association raised the possibility that the THIFNR subset plays a role in dampening TH2 responses to allergens. Intriguingly, cells in the THIFNR subset expressed the highest levels of CXCL10 and TNFSF10 (Fig. 3D). CXCL10 encodes CXCL10, a chemokine that recruits TH cells expressing the chemokine receptor CXCR3, which mainly comprises TH1 cells (58, 59). Thus, CXCL10 expression by THIFNR cells is likely to promote selective recruitment of TH1 cells (60). TNFSF10 encodes tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), which can drive apoptosis in cells expressing its receptors (TRAIL-R) (61, 62). More recently, both surface-bound and soluble TRAIL have been shown to dampen TCR signaling by inhibiting the phosphorylation of downstream kinases (63–66). Given that activated TH cells express TRAIL-R (65, 66), TRAIL produced by the HDM-reactive THIFNR cells may play an important role in blocking TH cell responses to HDM in vivo. Interestingly, a small fraction of cells expressing THIFNR signature genes (IFI6, ISG15) and TNFSF10 were observed even in resting cells not reactive to HDM, suggesting persistence of this population within PBMC (Fig. 3E). We confirmed that following TCR stimulation TRAIL was expressed by population of TH cells, which is likely to be enriched for the THIFNR subset (Fig. 3F). In published single-cell datasets (67) we found CD4+ T cells in human lungs expressed interferon-response signature genes and TRAIL, which indicated that the THIFNR subset is also present in the human lung tissue (fig. S8). We next experimentally tested TRAIL’s function and found that recombinant TRAIL inhibited TCR-dependent activation of TH cells ex vivo, as measured by the surface expression of the activation markers CD154, CD69, and CD137 (4–1BB) (68) (Fig. 3, G and H). These findings support a potential regulatory role of HDM-reactive THIFNR cells in dampening allergic responses.
A subset of HDM-reactive Treg cells express the interferon response signature
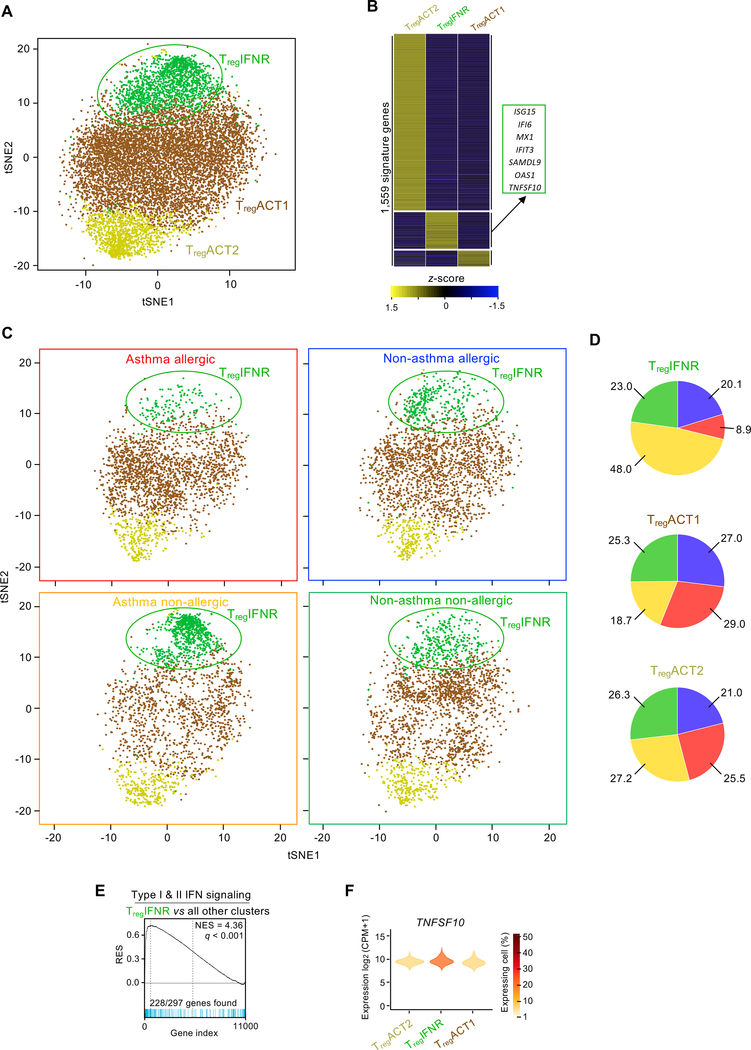
We also investigated whether HDM-reactive Treg cells differed between HDM allergic and non-allergic subjects. As shown previously, we confirmed that the proportion of HDM-reactive Treg cells was not related to HDM allergic status (Fig. 1B) (43). Furthermore, transcriptomic analysis of bulk populations of HDM-reactive Treg cells revealed no major disease-related differences (fig. S9). To determine whether specific subsets of HDM-reactive Treg cells varied with disease state, we performed single-cell transcriptomic analysis of ~10,000 HDM-reactive Treg cells across the 4 subject groups, which separated this population into 3 distinct clusters (Fig. 4, A and B, and table S3). The proportion of cells in cluster 2 was greater in asthmatic subjects without HDM allergy compared with HDM-allergic asthmatics, suggesting preferential expansion of this subset in asthmatic subjects without HDM allergy (Fig. 4, C and D, and fig. S10). GSEA analysis of transcripts enriched in this cluster (N = 248) revealed significant enrichment of interferon response genes (Fig. 4E). The features of this Treg cluster were similar to those of the THIFNR cluster, which was also present at higher proportions in subjects without HDM allergy; for example, interferon-responsive Treg cells (TregIFNR) also expressed higher levels of transcripts encoding for TRAIL (Fig. 4F). Overall, our findings indicate preferential expansion of HDM-reactive Treg and TH cells expressing the interferon response signature in asthmatic subjects without HDM allergy, and that expression of TRAIL by these subsets is likely to play an important role in dampening TH2 responses to HDM allergens, although further studies in animal models would be required to confirm this hypothesis.
Fig. 4.
A subset of HDM-reactive Treg cells express the interferon response signature. (A) t-SNE visualization of Seurat clustering analysis of the transcriptomes of 10,526 single HDM-activated Treg cells obtained from all 24 subjects. (B) Heatmap showing hierarchical clustering of row-wise z-score-normalized mean expression of cluster-specific differentially expressed genes (N = 1,559) Columns represent each Treg cluster. Right - lists of biologically relevant examples genes for the TregIFNR cluster. Equal numbers of cells were sampled per disease group. (C) t-SNE visualization of Seurat clustering analysis shown in Figure 4A, using equal cell numbers for each subject group (n = 2,180) obtained from all 6 subjects. (D) Pie charts illustrate the relative proportion of cells from each subject group within each of the 3 Treg clusters. (E) GSEA between each TregIFNR and the 2 other Treg clusters. q, false discovery rate; NES, normalized enrichment score; RES, relative enrichment score; list of genes provided in table S4. (F) Violin plots show log2(CPM+1) normalized expression of TNFSF10 in each Treg cluster. Cells with no expression are excluded (see Materials and Methods).
HDM-reactive TH2 cells are enriched for transcripts linked to enhanced functionality
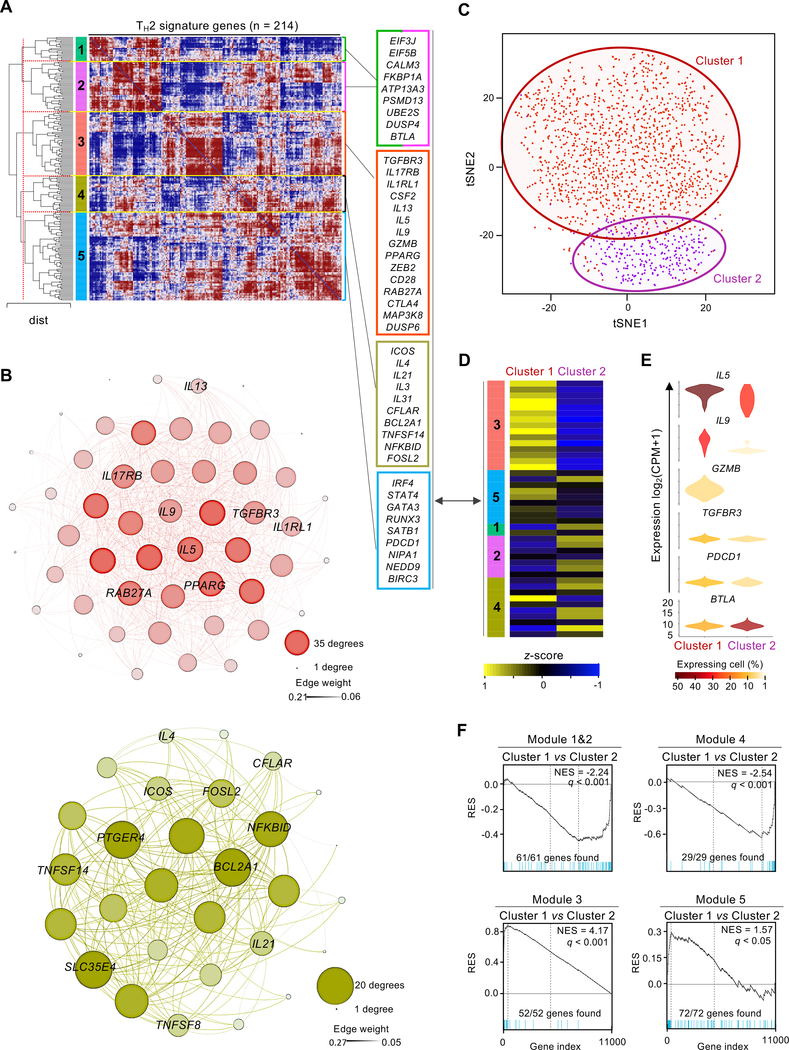
Given the important role of TH2 cells in the pathogenesis of allergy and asthma, we analyzed the genes enriched in the TH2 cluster to gain insights into their functional properties. Gene co-expression analysis is a powerful method to discover new genes that are likely to play important role in the differentiation or function of a given cell type (69, 70). Hierarchical gene clustering (Fig. 3A) and weighted gene co-expression network analysis (WGCNA) (71) (Materials and Methods, and Fig. 3B) of the 214 ‘TH2 enriched’ transcripts, defined from single-cell transcriptome analysis (Fig. 2, A and B), revealed 5 modules of highly co-expressed genes (Fig. 5A). Among these 5 modules, 2 modules (green and purple) contained genes linked to cellular metabolism, protein trafficking, active transcription and oxidative phosphorylation (EIF3J, EIF5B, CALM3, FKBP1A, PTPN11, ATP13A3, PSMD13, UBE2S, DUSP4), indicating increased metabolic and transcriptional activity in TH2 cells; a third (blue) module contained genes encoding for important transcription factors linked to TH2 cell differentiation such as GATA3, IRF4, and SATB1 (72–74).
Fig. 5.
HDM-reactive TH2 cells are enriched for transcripts linked to enhanced functionality. (A) Hierarchical clustering of Spearman correlation coefficient matrix for saver-imputed expression values of the 214 genes uniquely up-regulated in the TH2 cluster. Values are clustered with complete linkage. Dotted red line indicates Euclidian distance threshold value used to define the 5 modules of co-expressed genes. Right – list of example genes for each module (modules 1 and 2 merged). (B) Gephi visualization of weighted correlation network analysis (WGCNA) for genes co-expressed in modules 3 (top) and module 4 (bottom) from Figure 5A. Nodes correspond to a given gene and are sized based on the number of edges (connections); edges thickness correlates to strength degree of correlation. (C) t-SNE visualization of Seurat clustering analysis of single TH2 cluster cell transcriptomes (N = 1,751) obtained from 12 allergic subjects regardless of asthma status (red, TH2-cluster 1 (N = 1,440); purple, TH2-cluster 2 (N = 311)). Red and purple circling lines represent limits of each TH2 clusters 1 and 2, respectively. (D) Heatmap showing row-wise z-score-normalized mean expression of genes shown in Figure 5A between both TH2 sub-clusters (columns). (E) Violin plots show log2(CPM+1) normalized expression for genes biologically relevant between both TH2 sub-clusters. Color code represents the fraction of cells expressing the given gene in each TH2 sub-cluster; cells with no expression are not included. (F) GSEA between TH2 sub-clusters. Plots illustrate significative enrichment of module genes shown in Figure 5A between both TH2 sub-clusters. q, false discovery rate; NES, normalized enrichment score; RES, relative enrichment score; list of genes provided in table S4.
The module including the canonical type 2 cytokine genes (IL5, IL13) (red in Fig. 5, A and B), likely includes other genes that play an important role in driving the effector functions of TH2 cells. One of the most highly co-expressed transcripts encodes for the effector cytokine IL-9, which has recently been shown to be produced by a subset of TH2 cells that expressed PPAR-γ following TGF-β signaling (75). We found that transcripts encoding for PPAR-γ and the TGF-β receptor 3 (TGFBR3) were also enriched in TH2 cells and highly co-expressed with IL9 (Fig. 5, A and B), suggesting that the IL-9 differentiation pathway is active in TH2 cells. Finally, transcripts linked to cytotoxicity function (GZMB, RAB27A (76–78)) and differentiation of cytotoxic TH cells (ZEB2, RUNX3 (46, 77, 79)) were also highly co-expressed with IL5 and IL13, indicating that HDM-reactive cells may include cytotoxic TH2 cells (Fig. 5, A and B). Cytotoxic TH cells are known to contribute to antiviral immunity (77, 80) and autoimmunity (81), and our findings bring up the possibility that they may also play a role in asthma pathogenesis.
The gene for another canonical type 2 cytokine, IL-4, also linked to the function of T follicular helper cells (TFH) and IgE class switching, was present in a fourth module (yellow). Important molecules encoded by genes in this module include IL-31, a member of the IL-6 family of cytokines that is produced by activated TH2 cells and leads to itching in skin inflammation (Fig. 5, A and B) (82, 83), and IL-3, which is linked to hematopoietic progenitor proliferation and recruitment (84–86). We recently showed that IL-3 plays an important role in the activation and survival of eosinophils (87). Other genes in this module encode for products such as ICOS and IL-21, which is linked to B cell help and immunoglobulin isotype class switching (Fig. 5, A and B), suggesting that this module was enriched for genes linked to TFH cell function. The presence of gene modules with distinct co-expression patterns indicated potential heterogeneity in the TH2 population. To address this issue, we re-clustered cells only from the TH2 population; this analysis revealed 2 distinct sub-clusters (Fig. 5C), each highly enriched for genes in modules 3 and 4 (yellow and red) (Fig. 5, D, E and F).
Overall, these results show that cells in the TH2 cluster were enriched for the expression of transcripts encoding for several co-stimulatory and inhibitory receptors as well as transcription factors and molecules known to promote T cell survival. The expression of the first class of molecules, including CD28, ICOS, BTLA, CTLA-4, PD-1, HVEM receptor (LIGHT, TNFSF14) (88), suggests that these molecules could be targeted to dampen the pro-inflammatory function of TH2 cells in asthma. Pro-survival factors included several in the NF-κB signaling pathway, including NFKBID, NIPAI, MAP3K8, FOSL2, NEDD9 (89), ZEB2, BCL2A1 (90), BIRC3 (91), DUSP4/MKP-2 (92, 93), CFLAR (cFLIP/CASPER) (94). Together, these expression patterns suggest that these cells are endowed with properties that allow them to exert sustained and strong type 2 inflammatory responses in asthma.
IL-9-expressing HDM-reactive TH2 cells are increased in asthma
We next sought to identify potential asthma-specific changes in HDM-reactive TH2 cells from subjects with HDM allergy. Single-cell differential gene expression analysis of HDM-reactive TH2 cells from HDM-allergic asthmatics versus non-asthmatics revealed that among the TH2-enriched transcripts, IL9 was the most upregulated gene in asthmatic subjects (Fig. 6A). Furthermore, as shown in Figure 5, D and E, sub-clustering of the TH2 subset showed that IL9-expressing cells were highly enriched in the larger TH2 sub-cluster (Fig. 6B). The relative proportion of cells in the IL9-enriched TH2 subset (TH2-cluster 1) was greater in asthmatics compared with (75) non-asthmatic subjects (Fig. 6C). Thus, enrichment of IL9 expression in TH2 cells appears to be associated in the development of asthma.
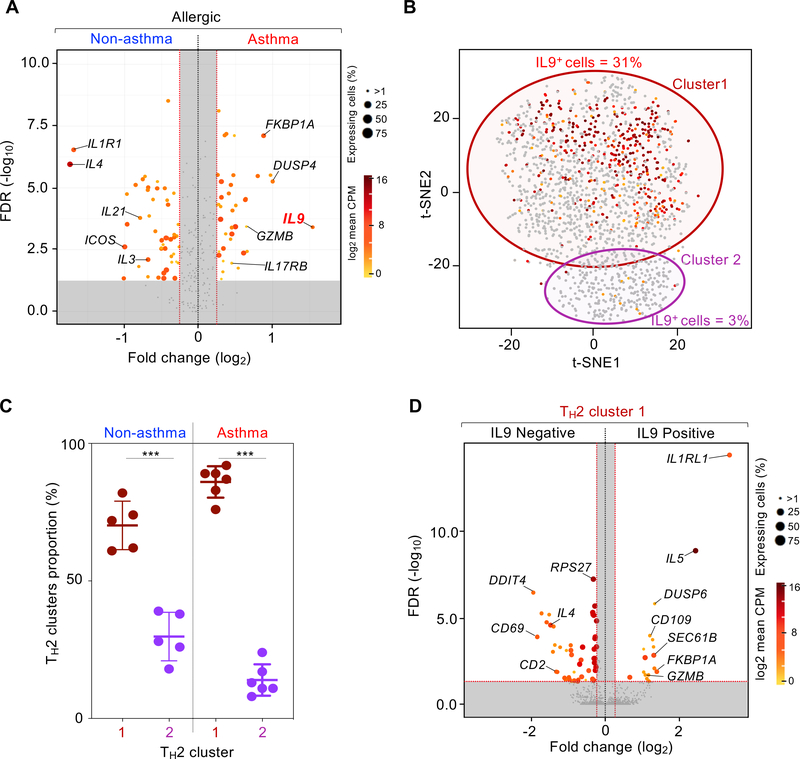
Fig. 6.
Asthma-specific TH2 single cells analysis. (A) Volcano plot shows statistical significance (-log10 adjusted P-value) in function of the log2-fold difference in expression for filtered genes (see Materials and Methods), when comparing expression between TH2 cells from asthma allergic (N = 6) versus non-asthma allergic (N = 6) subjects. Dots are colored accordingly to the average of expression (log2) and sized based on the fraction of cells expressing the given gene, both derived from the group in which the gene is upregulated. Equal numbers of cells where sampled in each group (N = 661). Grey dotted lines represent the threshold value for fold change (vertical, log2(|FC|) > 0.5-fold) and significance (horizontal, -log10(adjusted P-value) > 2). (B) t-SNE visualization of TH2 cluster cell transcriptomes shown in Figure 5C in which each dot is a cell is cell colored according to expression for IL9 (grey, none). Outlines represent TH2 sub-cluster limits. (C) Box and whisker plot shows percentage of TH2 cells in each sub-cluster in asthma allergic (N = 6) and non-asthma allergic (N = 5) subjects. Center line, median value; box, quartiles; whisker lines, 10th and 90th percentiles. **; P < 0.01. (D) Volcano plot similar to Figure 6A comparing expression between IL9-positive cells (N = 444) and IL9-negative cells (N = 444) in the asthma allergic TH2 cluster 1.
To determine the properties of IL9-expressing HDM-reactive TH2 cells in asthmatic subjects, we compared the single-cell gene expression profiles of IL9-expressing and non-expressing cells contained within the IL9-enriched TH2-cluster 1. We were surprised to find that expression of several transcripts encoding products linked to pathogenicity and survival of TH2 cells was increased in IL9-expressing cells (Fig. 6D and table S3). These included transcripts encoding canonical TH2 cytokine IL-5, cytotoxicity molecules (granzyme B, ZEB2, EFHD2), TH2 polarizing and survival-related signaling receptor (IL-33R) (95, 96), and CD109, a membrane-anchored molecule described as negative regulator of TGF-β signaling (97, 98) but also as a co-activator of the JAK/STAT3 signaling pathway (98–100) that is important for TH2 cell development (98, 101) (Fig. 6D, fig. S11, and table S3). Overall, these findings suggest that IL9-expressing HDM-reactive TH2 cells displayed greater pathogenic properties that could play an important role in driving asthma pathogenesis.
Discussion
In this study, we present large-scale single-cell transcriptome analysis of allergen-reactive TH and Treg cells (N ~ 50,000) from subjects with asthma and/or allergy and healthy controls. Our characterization of HDM-reactive TH cells identified substantial heterogeneity as well as quantitative and qualitative changes related to allergy and asthma and revealed a unique subset of TH cells with a strong interferon response signature.
Our single-cell study addresses some of the important unanswered questions in the field of allergy and asthma research. Most fundamentally, exposure to common allergens, such as HDM, is nearly ubiquitous and TH responses to these allergens are seen in both allergic and non-allergic subjects (33–35), yet why do only some people develop TH2 responses to allergens? By comparing HDM-reactive T cells from asthmatics with and without HDM allergy (TH2 responses versus no TH2 responses), we found that in subjects without HDM allergy (but sensitized to other allergens, see table S1), a subset of TH and Treg cells expressing the interferon response signature was expanded. These cells expressed higher levels of TRAIL, a molecule that can inhibit TCR signaling, activation of TH cells and inflammation in model systems (102). Therefore, we hypothesize that these TRAIL-expressing HDM-reactive T cells could play an important role in dampening TH2 inflammation in allergy and asthma. A recent single-cell transcriptomic study identified TH cells expressing the interferon response signature in the lung tissue of mice sensitized and challenged with HDM (20). This finding implies that THIFNR cells can be generated and sustained in vivo, and that HDM sensitization of mice may be an appropriate system to test the role of these cells in allergic inflammation. Future studies are also required to determine the molecular mechanisms and signals that drive the differentiation, maintenance, and persistence of THIFNR cells, ideally through the analysis of their molecular and chromatin landscape.
Another important question is why only some patients with allergy develop asthma? Do TH cells from allergic patients with asthma display distinct molecular features from those of allergic patients without asthma? By comparing HDM-reactive TH cells from allergic subjects with and without asthma, we defined a subset of IL9-expressing TH2 cells that are enriched in asthmatic subjects. We show that IL9-expressing HDM-reactive cells display several features likely to enhance their pathogenicity and persistence, which may contribute to asthma pathogenesis. Notably, in the context of peanut allergy, IL-9 was shown to best differentiate children with peanut allergy from children with peanut sensitization, who tolerate peanut, suggesting a potentially important role in food allergy (103). Studies blocking IL-9 activity in animal models of asthma have indicated that this may be a promising treatment approach (104, 105), but the relative importance of IL-9-producing T cells has not been fully explored. A recent report in cancer showed that IL-9-producing murine TH cells are more cytolytic, hyperproliferative, and less exhausted (106, 107); these properties conferred potent antitumor activity for these cells when tested in adoptive transfer experiments. Studies in mouse models of allergic airway inflammation are required to demonstrate the relative pathogenicity and persistence of IL-9 producing TH2 cells in vivo. Moreover, this cell population should also be characterized in human subjects with severe asthma, including those that reside in the airways. In summary, our single-cell transcriptomic study of HDM allergen-specific T cells has identified TH subsets that may contribute to the pathogenesis of allergy and asthma.
Materials and Methods
Study design
The goal of this study was to use bulk and single-cell RNA-seq assay to capture the transcriptome of HDM allergen-reactive CD4+ T cell memory subsets from peripheral blood mononuclear cells (PBMC) of 6 asthmatic subjects allergic to HDM, 6 asthmatic subjects non-allergic to HDM, 6 non-asthmatic subjects allergic to HDM, and 6 non-asthmatic non-allergic to HDM. To isolate HDM-reactive CD4+ cells, PBMC were stimulated with HDM and CD4+ memory cells were sorted based on CD154 and CD137 expression: CD154+ (HDM-reactive TH), CD154– CD137+ (HDM-reactive Treg), and CD154– CD137– (HDM-non-reactive T cells). For bulk RNA-seq, we collected 200 cells in triplicates, and for single-cell RNA-seq assay, we collected between 1,500 to 2,000 cells per cell type per patient (see table S1).
Subject recruitment, ethical approval and characteristics
Recruitment of subjects included in this study followed Institutional Review Board (La Jolla Institute for Immunology, La Jolla, CA) approval, and study participants gave written informed consent. Twelve non-smoking subjects with mild asthma treated only with inhaled bronchodilators (mild asthma), six subjects with allergic rhinitis but no asthma, meeting established diagnostic criteria, and 6 healthy nonatopic subjects were studied (Fig. 1A and table S1). Subjects with asthma underwent pulmonary function tests and/or methacholine challenge to establish diagnosis (bronchodilator response of ≥12 %, or ≥200 ml, and/or methacholine challenge with a provocative concentration causing a drop of the forced expiratory volume in 1 s [FEV1] of 20 %, 8 mg/ml). All subjects were classified as allergic to HDM based on skin test reactivity to HDM allergens (Der p and Der f, table S1).
Sample processing
We used peripheral blood mononuclear cells (PBMC) obtained from blood samples by density gradient centrifugation according to the manufacturer’s instructions and cryopreserved in liquid nitrogen.
Antigen-reactive T cell enrichment (ARTE) assay
The HDM peptide pool was generated as described and contained 75 peptides at a total concentration of 20 mg/ml (188.7 ng/ml for each peptide) (108, 109). The assay to isolate HDM-reactive T cells based on HDM peptide pool stimulation, MACS-based enrichment and FACS sorting of CD154+ memory CD4+ T cells from PBMC was adapted from Bacher et al. 2016 (43), and is outlined in fig. S1. For each donor, PBMC cryovials were thawed, washed and, plated overnight in 6-well culture plates at a concentration of 10 × 106 cells/ml in 2 ml of serum-free TexMACS medium (Miltenyi Biotec) (5% CO2, 37˚C). In presence of a blocking CD40 antibody (1 μg/ml; Miltenyi Biotec), cells were then stimulated by addition of HDM peptide pool (1 μg/ml) for 6 h. Subsequently, cells were stained with fluorescence-labeled antibodies and a biotinylated CD154 antibody (clone 5C8; Miltenyi Biotec). Anti-biotin microbeads (Miltenyi Biotec) were added to allow MACS-based enrichment of CD154+ cells using MS columns (Miltenyi Biotec). 5% of cells were kept as control sample (‘input’) and used for FACS sorting of HDM– T cells and analysis of cell frequencies before enrichment. Positively selected cells (CD154+) were eluted from the column and used for FACS sorting of CD154+ memory CD4+ T cells. The flow-through from the MACS column was collected, stained with a biotinylated CD137 antibody (clone REA765; Miltenyi Biotec) and anti-biotin MicroBeads and applied to a second MS column. Positively selected cells (CD137+) were used for FACS sorting of CD137+ cells. All cell populations were FACS-sorted using a FACSAria II (Becton Dickinson); the gating strategy is shown in fig S1. All flow cytometry data were analyzed using FlowJo software (version 10).
Cell isolation for bulk and single-cell RNA-seq assay
For bulk assays, cells of interest were directly collected by sorting 200 cells into 0.2 ml PCR tubes (low-retention, Axygen) containing 8 μl of ice-cold lysis buffer (Triton X-100 [0.1 %, Sigma-Aldrich] containing RNase inhibitor (1:100, Takara). Once collected, tubes were vortexed for 10 seconds, spun for 1 minute at 3000 g and stored at −80 ˚C. For single cell RNA-seq assays (10x Genomics), 1000 to 2000 HDM-reactive T cells per subject were collected by sorting in low retention and sterile ice-cold 1.5 mL collection tubes containing 500 μL of PBS:FBS (1:1 vol:vol) with RNase inhibitor (1:100). HDM-reactive T cells from 6 subjects in each of the 4 groups (asthma with HDM-allergy, asthma without HDM allergy, HDM-allergy without asthma and healthy without HDM-allergy) were collected in the same tube. Collection tubes with ~9,000 to 12,000 sorted cells/study group were inverted a few times, ice-cold PBS was added to reach a volume of 1400 μl, and centrifugated for 5 minutes at a speed of 600 g at 4 ˚C. Supernatant was cautiously removed leaving 5 to 10 μl of volume. Pellets were then resuspended with 25 μl of 10x Genomics resuspension buffer (0.22 μm filtered ice-cold PBS supplemented with ultra-pure bovine serum albumin (0.04 %, Sigma-Aldrich). 33 μl of cell suspension were transferred to an 8 PCR-tube strip for downstream steps as per manufacturer’s instructions (10x Genomics, San Francisco).
Bulk RNA library preparation for sequencing
For full-length bulk transcriptome analyses, we used the Smart-Seq2 protocol (110), adapted for samples with small cell numbers (111, 112). We followed the protocol as described previously (111, 112) with following modifications: (i) the pre-amplification PCR cycle for T cells was set at 22 cycles; (ii) to eliminate any traces of primer-dimers, the PCR pre-amplified cDNA product was purified using 0.8x Ampure-XP beads (Beckman Coulter) before using the DNA for sequencing library preparation. One ng of pre-amplified cDNA was used to generate barcoded Illumina sequencing libraries (Nextera XT library preparation kit, Illumina) in 8 μl reaction volume. Samples failing any quality control step (DNA quality assessed by capillary electrophoresis (Fragment analyzer, Advance analytical) and quantity (Picogreen quantification assay, Thermo Fisher) were eliminated from further downstream steps. Libraries were then pooled at equal molar concentration and sequenced using the HiSeq 2500 Illumina platform to obtain 50-bp single-end reads (HiSeq SBS Kit v4; Illumina). In total, 1.7 billion uniquely mapped reads we generated with a median ± standard deviation of 17.8 ± 3.8 million uniquely mapped reads per sample.
10x Genomics single-cell RNA library preparation for sequencing
Samples were processed using 10x Genomics 3’TAG v2 chemistry as per manufacturer’s recommendations; 11 cycles were used for cDNA amplification and library preparation respectively (77). Barcoded RNA was collected and processed following manufacturer’s recommendations. After quantification, equal molar concentration of each libraries was pooled and sequenced using the HiSeq2500 Illumina sequencing platform to obtain 26- and 100-bp paired-end reads using the following read length: read 1, 26 cycles; read 2, 100 cycles; and i7 index, 8 cycles.
Bulk RNA-seq analysis
Bulk RNA-seq data were mapped against the hg19 reference using TopHat (113) (v1.2.1 (--library-type fr-unstranded --no-coverage-search) with FastQC (v0.11.2), Bowtie (114) (v1.1.2), Samtools 0.1.18.0) (115) and we employed htseq-count -m union -s no -t exon -i gene_name (part of the HTSeq framework, version 0.7.1) (71, 116). Cutadapt (v1.3) was used to remove adapters (117). Values throughout are displayed as log2 TPM (transcripts per million) counts; a value of 1 was added prior to log transformation (pseudo-count). We performed principal component analysis and clustering analysis using t-distributed stochastic neighbor embedding dimensional reduction algorithm (tSNE) (118) (based on 3 PC using the top 200 most variable genes). To identify genes expressed differentially between groups, we performed negative binomial tests for paired comparisons by employing DESeq2 (119) (1.16.1) with default parameters. We considered genes to be expressed differentially by any comparison when the DESeq2 analysis resulted in a Benjamini-Hochberg–adjusted P-value of at most 0.01 and a log2 fold change of at least 2. Gene set enrichment analysis (GSEA) were performed as previously described (112, 120) using the Qlucore visualization software (version 3.5) (121). Gene lists used for GSEA analysis are shown in table S4.
Single-cell RNA-seq analysis
Analysis of 3’ single-cell transcriptomes using the 10x Genomics platform
Raw data was processed as previously described (70, 77), merging multiple sequencing runs using count function from Cell Ranger (table S3), then aggregating multiple cell types with cell ranger aggr (v3.0.2). The merged data was transferred to the R statistical environment for analysis using the package Seurat (v3.0.2) (122).
Doublet cell filtering
Barcoded single-cell RNA-seq was demultiplexed patient-wise using Demuxlet (123) with the following parameters: alpha = 0, 0.5 and --geno-error = 0.05. Cells called as doublet by Demuxlet were removed from downstream analyses (fig. S3A and table S3). Identities were inferred by analyzing VCF files from the genotyping analysis containing the corresponding individual for each particular library. Each cell was assigned a donor ID or marked as a doublet, and then incorporated to the annotation table. We did not observe major changes in singlets/doublets proportion between the different 10x Genomics libraries (reflecting cell type and subject groups), suggesting optimal processing of cells during 10x (Gel Bead-In Emulsions) GEM generation and downstream steps (fig. S3A). All downstream analyses were performed using singlet cells.
Activation score and cell filtering
To filter out cells with low level of activation or no activation by the HDM-peptide pool i.e., HDM non-reactive cells, we performed pair-wise single-cell differential gene expression analysis using MAST algorithm (124) between HDM-reactive TH cells (CD154+ cells, N = 3075, random sampled) and HDM– T cells (CD154neg cells, N = 3075). We defined a gene set (N = 110 genes, called HDM+ TH activation genes) that captured the transcripts upregulated in the CD154+ versus CD154neg cells (table S3) using the following parameters (fig. S3B): Benjamini-Hochberg–adjusted P-value ≤ 0.05, log2 fold change ≥ 2, log2 mean of expression ≥ 0.75 CPM and, percentage of expressing cells (> 0 CPM) in HDM+ TH cells > 37.5 % (fig. S3B). We scored each cell using AddModuleScore from Seurat (122). Briefly, the module score is calculated by binning the genes by the average expression level, then the average expression of each gene is subtracted by the aggregated expression of the control gene sets (100) randomly selected per bin. Finally, based on the distribution of cells based on their activation score (Fig. 2A and fig. S3C), we applied a threshold for defining activated cells. The proportions of cells expressing important canonical genes such as IL4, IL5, IFNG, IL17A pre- and post-activation filtering indicated cell eliminated due to low-activation score did not upregulate transcripts for these genes (data not shown). Similarly, an independent HDM+ Treg activation score was also calculated using similar approach to analyze HDM+ Treg single-cell datasets (table S3 and fig. S3, right panels). In total, 3505 cells with low-activation score were eliminated from downstream analysis.
Transcriptome-based clustering analysis
The merged data was transferred to the R (v3.5) statistical environment for analysis using the package Seurat (v3.0.2) (122). Only cells expressing more than 200 genes and with a total mitochondrial gene expression less than 5 %, and genes expressed in at least 3 cells were included in the analysis. The data was then log-normalized per cell and a list of the most variable genes with a mean expression > 0.1 (UMI) and explaining 30 % of the cumulative standardized variance given by the FindVariableFeatures function were used for clustering analysis (fig. S4). We performed clustering analysis using distinct lists of most variable genes for HDM+ TH, HDM+ TREG and HDM+ TH2 clusters (table S3). In regard to TH2 sub-clustering, because of the limited number of cells, we considered most variable genes that were expressed by more than 10 % of the cells and with a standardized variance greater than 2. We also limited the selection of the most variable genes to 10 % of the cumulative standardized variance (fig. S4, right panels). Normalized single-cell transcriptomic data was then further scaled by the number of UMI-detected and percentage of mitochondrial reads. We then performed principal component (PC) analysis with RunPCA algorithm (122) using the determined most variable gene lists. We followed Seurat procedure to determine the number of PCs to select for downstream analyses, using the standard deviation of PCs. We applied FindNeighbors and FindClusters functions from Seurat with default settings (table S3 and fig. S4) to identify clusters. All clusters had more than 50 cells and none were excluded from the downstream analysis. Cluster specific markers were obtained by the FindAllMarkers function with default parameters, test.use = MAST (124). Further visualizations of exported normalized data such has “violin” plots were generated using the Seurat package and custom R scripts. Notably, our violin plots show Seurat normalized expression for a particular gene (log2 (CPM + 1 pseudo-count)) only for the cells expressing the gene of interest. Violin shape represents the distribution of cell expressing transcript of interest (based on a Gaussian Kernel density estimation model) and are colored according to the percentage of cell expressing the transcript of interest.
Single-cell differential gene expression analysis
Pairwise single-cell differential gene expression analysis was conducted after conversion of data to count per million base-pairs (CPM + 1) using MAST algorithm (q < 0.05, v1.2.1) (R package) (124). We used equal number of cells from each subject group, and random sampling performed when necessary. A gene was considered differentially expressed when Benjamini-Hochberg–adjusted P-value was < 0.05 and a log2 fold change was more than 0.25. For cluster-specific signatures, a gene was considered significantly different (unique to a group), only if the gene was enriched in every pair-wise comparison for a particular cluster with other clusters.
Single-cell co-expression analysis and weighted gene correlation analysis (WGCNA)
In order to perform co-expression analysis, given the high levels of genes drop-out associated with single cell analysis, we performed a data imputation using SAVER imputation algorithm (125). Briefly, SAVER analysis was implemented on the Cell Ranger UMI matrix output for HDM+ TH using the function saver (v1.1.1) with pred.genes.only = TRUE. Then, we calculated Spearman correlations coefficients using the cor function and determined the cluster-modules through hclust on Euclidean distances and cutree functions k = 5 according to the within groups sums of squares elbow (similar to TH single-cell clustering analysis, fig. S4). We also performed weighted correlation analysis using WGCNA algorithm (v1.66) (71) using the function TOMsimilarityfromExpr, power = 3, and exportNetworkToCytoscape, weighted = TRUE, threshold = 50th quantile of the topological overlap matrix. Network plots were generated by Gephi (0.9.2) using Fruchterman-Reingold and Noverlap layouts (126). The size and color of the nodes were defined according to the degree, while the edge width and color were scaled according to the weight value.
Genotyping
For each patient, genomic DNA was isolated from PBMC using the DNeasy Blood and Tissue Kit (Qiagen) and utilized for genotyping using the Infinium Multi-Ethnic Global-8 Kit (Illumina) following the manufacturer’s instructions. Raw data from the genotyping analysis, data quality assessment and SNPs identification were performed as previously described (127).
Stimulation of memory CD4+ T cells with human recombinant TRAIL
Memory CD4+ T cells were isolated from PBMC using the Memory CD4+ T Cell Isolation Kit (Miltenyi Biotec) and cultured in Iscove’s Modified Dulbecco’s Medium (IMDM; Invitrogen) supplemented with 5 % (vol/vol) heat-inactivated fetal bovine serum (FBS) and 2 % (vol/vol) human AB serum (CellGro). The memory CD4+ T cells were stimulated with pre-coated human recombinant TRAIL (5 μg/ml), anti-CD3 antibodies (2.5 μg/ml) and soluble anti-CD28 antibodies (1 μg/ml) in presence of IL-7 (5 ng/ml; Miltenyi Biotec). The expression of surface markers (CD69, CD154, CD137) was analyzed by flow cytometry after 6 h.
Expression of TRAIL on memory CD4+ T cells
Total PBMC were thawed, washed and plated overnight in serum-free TexMACS medium (Miltenyi Biotec) / complete IMDM (as described above). In presence of a blocking CD40 antibody (1 μg/ml in culture; clone HB14; Miltenyi Biotec), cells were then left untreated or stimulated by addition of the control reagent CytoStim (1:500 dilution of stock; Miltenyi Biotec). The expression of surface markers (CD69, CD154 and TRAIL) was analyzed by FACS after 6 h.
Statistical analysis
We used non-parametric Kruskal-Wallis one-way analysis of variance test (ANOVA) to compare unpaired data for more than 2 conditions and Kolmogoroz-Smirnov test when comparing 2 groups of data. We used paired Student’s t-test for time-course flow cytometry analysis. We used GraphPad Prism 7.0. All source datasets and statistical used are detailed in table S5.
Supplementary Material
Table S1. Study subject details (Excel spreadsheet)
Table S2. Differential gene expression analysis in bulk populations (Excel spreadsheet).
Table S4. Gene signatures lists (Excel spreadsheet).
Table S5. Raw data file with statistics (Excel spreadsheet).
Table S3. Single cell analysis (Excel spreadsheet).
Fig. S1. Experimental design and gating strategy to isolate HDM-reactive memory TH and Treg cells.
Fig. S2. Expression of differentially expressed genes in HDM-reactive cells.
Fig. S3. Single cell filtering of doublets and low HDM-reactive T cells.
Fig. S4. Single-cell clustering analysis using Seurat.
Fig. S5. Distribution of cell frequency for the 7 HDM+ TH clusters for the 24 subjects.
Fig. S6. Co-expression of TH1- or TH17-specific signature genes.
Fig. S7. Donor and disease-groups cell distribution in each of HDM+ TH clusters.
Fig. S8. Co-expression of THIFNR-specific signature genes in publicly available dataset.
Fig. S9. TREG disease-related differences.
Fig. S10. Proportions of HDM-reactive Treg subsets amongst disease groups.
Fig. S11. TH2 single-cell analysis.
Acknowledgments
We thank Drs. C. Kim, D. Hinz and members of the flow cytometry core facility; Dr. J. Greenbaum and members of the Bioinformatics Core at La Jolla Institute for Immunology (LJI); Drs. A. Sette and B. Peters and their teams for graciously providing us with the pool of House Dust Mite peptides (108, 109); Mrs. J. Moore for assistance with manuscript and figure preparation; and the members of the Vijayanand, Sette and Peters labs for constructive intellectual and technical support. Finally, we thank all donors for their charitable contribution to academic research.
Funding: Supported by (i) NIH research grants: U19AI100275, U19AI135731, R01HL114093; (ii) NIH equipment grants: S10RR027366 (BD FACSAria II), S10 RR027366 (Illumina HiSeq 2500); (iii) the William K. Bowes Jr. Foundation (P.V.).
Footnotes
Competing interests: P.V received research funding unrelated to this work from Pfizer. G.S. received a career development fellowship award from Kyowa Kirin Pharmaceutical Research Inc. to independently pursue research on IL-9 in severe asthma. P.V. and G.S. are listed as inventors on a provisional patent application covering findings reported in this manuscript. The other authors declare that they have no competing interests.
Data availability: Scripts are available in our repository on GitHub (https://github.com/vijaybioinfo/hdm_2019). Sequencing data for this study has been deposited into the Gene Expression Omnibus under GSE146172. including GSE146046 for bulk-RNA-seq and GSE146170 for single-cell datasets.
References and Notes
- 1.Caminati M, Pham DL, Bagnasco D, Canonica GW, Type 2 immunity in asthma. World Allergy Organ J 11, 13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller JD, The Role of Dust Mites in Allergy. Clin Rev Allergy Immunol 57, 312–329 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Larche M, Akdis CA, Valenta R, Immunological mechanisms of allergen-specific immunotherapy. Nat Rev Immunol 6, 761–771 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Kay AB, Allergy and allergic diseases. Second of two parts. N Engl J Med 344, 109–113 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Kay AB, Allergy and allergic diseases. First of two parts. N Engl J Med 344, 30–37 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Gregory LG, Lloyd CM, Orchestrating house dust mite-associated allergy in the lung. Trends Immunol 32, 402–411 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sykes A, Johnston SL, Etiology of asthma exacerbations. J Allergy Clin Immunol 122, 685–688 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Oddera S, Silvestri M, Penna R, Galeazzi G, Crimi E, Rossi GA, Airway eosinophilic inflammation and bronchial hyperresponsiveness after allergen inhalation challenge in asthma. Lung 176, 237–247 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Arshad SH, Hamilton RG, Adkinson NF Jr., Repeated aerosol exposure to small doses of allergen. A model for chronic allergic asthma. Am J Respir Crit Care Med 157, 1900–1906 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Maestrelli P, Zanolla L, Puccinelli P, Pozzan M, Fabbri LM, Regione Veneto Study G, Low domestic exposure to house dust mite allergens (Der p 1) is associated with a reduced non-specific bronchial hyperresponsiveness in mite-sensitized asthmatic subjects under optimal drug treatment. Clin Exp Allergy 31, 715–721 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Ali FR, Oldfield WL, Higashi N, Larche M, Kay AB, Late asthmatic reactions induced by inhalation of allergen-derived T cell peptides. Am J Respir Crit Care Med 169, 20–26 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Muehling LM, Lawrence MG, Woodfolk JA, Pathogenic CD4(+) T cells in patients with asthma. J Allergy Clin Immunol 140, 1523–1540 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raemdonck K, Baker K, Dale N, Dubuis E, Shala F, Belvisi MG, Birrell MA, CD4(+) and CD8(+) T cells play a central role in a HDM driven model of allergic asthma. Respir Res 17, 45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibeon D, Menzies-Gow AN, Targeting interleukins to treat severe asthma. Expert Rev Respir Med 6, 423–439 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Kim AS, Doherty TA, New and emerging therapies for asthma. Ann Allergy Asthma Immunol 116, 14–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maltby S, Gibson PG, Powell H, McDonald VM, Omalizumab Treatment Response in a Population With Severe Allergic Asthma and Overlapping COPD. Chest 151, 78–89 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Drick N, Seeliger B, Welte T, Fuge J, Suhling H, Anti-IL-5 therapy in patients with severe eosinophilic asthma - clinical efficacy and possible criteria for treatment response. BMC Pulm Med 18, 119 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulten V, Tripple V, Seumois G, Qian Y, Scheuermann RH, Fu Z, Locci M, Rosales S, Vijayanand P, Sette A, Alam R, Crotty S, Peters B, Allergen-specific immunotherapy modulates the balance of circulating Tfh and Tfr cells. J Allergy Clin Immunol 141, 775–777 e776 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noval Rivas M, Chatila TA, Regulatory T cells in allergic diseases. J Allergy Clin Immunol 138, 639–652 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tibbitt CA, Stark JM, Martens L, Ma J, Mold JE, Deswarte K, Oliynyk G, Feng X, Lambrecht BN, De Bleser P, Nylen S, Hammad H, Henriksson M. Arsenian, Saeys Y, Coquet JM, Single-Cell RNA Sequencing of the T Helper Cell Response to House Dust Mites Defines a Distinct Gene Expression Signature in Airway Th2 Cells. Immunity 51, 169–184 e165 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Lewkowich IP, Herman NS, Schleifer KW, Dance MP, Chen BL, Dienger KM, Sproles AA, Shah JS, Kohl J, Belkaid Y, Wills-Karp M, CD4+CD25+ T cells protect against experimentally induced asthma and alter pulmonary dendritic cell phenotype and function. J Exp Med 202, 1549–1561 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leech MD, Benson RA, De Vries A, Fitch PM, Howie SE, Resolution of Der p1-induced allergic airway inflammation is dependent on CD4+CD25+Foxp3+ regulatory cells. J Immunol 179, 7050–7058 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Strickland DH, Stumbles PA, Zosky GR, Subrata LS, Thomas JA, Turner DJ, Sly PD, Holt PG, Reversal of airway hyperresponsiveness by induction of airway mucosal CD4+CD25+ regulatory T cells. J Exp Med 203, 2649–2660 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wambre E, Bajzik V, DeLong JH, O’Brien K, Nguyen QA, Speake C, Gersuk VH, DeBerg HA, Whalen E, Ni C, Farrington M, Jeong D, Robinson D, Linsley PS, Vickery BP, Kwok WW, A phenotypically and functionally distinct human TH2 cell subpopulation is associated with allergic disorders. Sci Transl Med 9, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith KA, Gray NJ, Saleh F, Cheek E, Frew AJ, Kern F, Tarzi MD, Characterisation of CD154+ T cells following ex vivo allergen stimulation illustrates distinct T cell responses to seasonal and perennial allergens in allergic and non-allergic individuals. BMC Immunol 14, 49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Upadhyaya B, Yin Y, Hill BJ, Douek DC, Prussin C, Hierarchical IL-5 expression defines a subpopulation of highly differentiated human Th2 cells. J Immunol 187, 3111–3120 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Januszyk M, Rennert RC, Sorkin M, Maan ZN, Wong LK, Whittam AJ, Whitmore A, Duscher D, Gurtner GC, Evaluating the Effect of Cell Culture on Gene Expression in Primary Tissue Samples Using Microfluidic-Based Single Cell Transcriptional Analysis. Microarrays (Basel) 4, 540–550 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nestor CE, Ottaviano R, Reinhardt D, Cruickshanks HA, Mjoseng HK, McPherson RC, Lentini A, Thomson JP, Dunican DS, Pennings S, Anderton SM, Benson M, Meehan RR, Rapid reprogramming of epigenetic and transcriptional profiles in mammalian culture systems. Genome Biol 16, 11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazzatti DJ, White A, Forsey RJ, Powell JR, Pawelec G, Gene expression changes in long-term culture of T-cell clones: genomic effects of chronic antigenic stress in aging and immunosenescence. Aging Cell 6, 155–163 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seumois G, Zapardiel-Gonzalo J, White B, Singh D, Schulten V, Dillon M, Hinz D, Broide DH, Sette A, Peters B, Vijayanand P, Transcriptional Profiling of Th2 Cells Identifies Pathogenic Features Associated with Asthma. J Immunol 197, 655–664 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geginat J, Paroni M, Maglie S, Alfen JS, Kastirr I, Gruarin P, De Simone M, Pagani M, Abrignani S, Plasticity of human CD4 T cell subsets. Front Immunol 5, 630 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DuPage M, Bluestone JA, Harnessing the plasticity of CD4(+) T cells to treat immune-mediated disease. Nat Rev Immunol 16, 149–163 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Hinz D, Seumois G, Gholami AM, Greenbaum JA, Lane J, White B, Broide DH, Schulten V, Sidney J, Bakhru P, Oseroff C, Wambre E, James EA, Kwok WW, Peters B, Vijayanand P, Sette A, Lack of allergy to timothy grass pollen is not a passive phenomenon but associated with the allergen-specific modulation of immune reactivity. Clin Exp Allergy 46, 705–719 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Birrueta G, Tripple V, Pham J, Manohar M, James EA, Kwok WW, Nadeau KC, Sette A, Peters B, Schulten V, Peanut-specific T cell responses in patients with different clinical reactivity. PLoS One 13, e0204620 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schulten V, Westernberg L, Birrueta G, Sidney J, Paul S, Busse P, Peters B, Sette A, Allergen and Epitope Targets of Mouse-Specific T Cell Responses in Allergy and Asthma. Front Immunol 9, 235 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akdis M, Verhagen J, Taylor A, Karamloo F, Karagiannidis C, Crameri R, Thunberg S, Deniz G, Valenta R, Fiebig H, Kegel C, Disch R, Schmidt-Weber CB, Blaser K, Akdis CA, Immune responses in healthy and allergic individuals are characterized by a fine balance between allergen-specific T regulatory 1 and T helper 2 cells. J Exp Med 199, 1567–1575 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gandhi VD, Davidson C, Asaduzzaman M, Nahirney D, Vliagoftis H, House dust mite interactions with airway epithelium: role in allergic airway inflammation. Curr Allergy Asthma Rep 13, 262–270 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Calderon MA, Kleine-Tebbe J, Linneberg A, De Blay F, Hernandez D Fernandez de Rojas, J. C. Virchow, P. Demoly, House Dust Mite Respiratory Allergy: An Overview of Current Therapeutic Strategies. J Allergy Clin Immunol Pract 3, 843–855 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Dullaers M, Schuijs MJ, Willart M, Fierens K, Van Moorleghem J, Hammad H, Lambrecht BN, House dust mite-driven asthma and allergen-specific T cells depend on B cells when the amount of inhaled allergen is limiting. J Allergy Clin Immunol 140, 76–88 e77 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Salo PM, Arbes SJ Jr., Crockett PW, Thorne PS, Cohn RD, Zeldin DC, Exposure to multiple indoor allergens in US homes and its relationship to asthma. J Allergy Clin Immunol 121, 678–684 e672 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frentsch M, Arbach O, Kirchhoff D, Moewes B, Worm M, Rothe M, Scheffold A, Thiel A, Direct access to CD4+ T cells specific for defined antigens according to CD154 expression. Nat Med 11, 1118–1124 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Chattopadhyay PK, Yu J, Roederer M, A live-cell assay to detect antigen-specific CD4+ T cells with diverse cytokine profiles. Nat Med 11, 1113–1117 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Bacher P, Heinrich F, Stervbo U, Nienen M, Vahldieck M, Iwert C, Vogt K, Kollet J, Babel N, Sawitzki B, Schwarz C, Bereswill S, Heimesaat MM, Heine G, Gadermaier G, Asam C, Assenmacher M, Kniemeyer O, Brakhage AA, Ferreira F, Wallner M, Worm M, Scheffold A, Regulatory T Cell Specificity Directs Tolerance versus Allergy against Aeroantigens in Humans. Cell 167, 1067–1078 e1016 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Arlehamn CL, Seumois G, Gerasimova A, Huang C, Fu Z, Yue X, Sette A, Vijayanand P, Peters B, Transcriptional profile of tuberculosis antigen-specific T cells reveals novel multifunctional features. J Immunol 193, 2931–2940 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serroukh Y, Gu-Trantien C, Hooshiar Kashani B, Defrance M, Vu Manh TP, Azouz A, Detavernier A, Hoyois A, Das J, Bizet M, Pollet E, Tabbuso T, Calonne E, van Gisbergen K, Dalod M, Fuks F, Goriely S, Marchant A, The transcription factors Runx3 and ThPOK cross-regulate acquisition of cytotoxic function by human Th1 lymphocytes. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kroczek AL, Hartung E, Gurka S, Becker M, Reeg N, Mages HW, Voigt S, Freund C, Kroczek RA, Structure-Function Relationship of XCL1 Used for in vivo Targeting of Antigen Into XCR1(+) Dendritic Cells. Front Immunol 9, 2806 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dorner BG, Scheffold A, Rolph MS, Huser MB, Kaufmann SH, Radbruch A, Flesch IE, Kroczek RA, MIP-1alpha, MIP-1beta, RANTES, and ATAC/lymphotactin function together with IFN-gamma as type 1 cytokines. Proc Natl Acad Sci U S A 99, 6181–6186 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshida T, Imai T, Kakizaki M, Nishimura M, Takagi S, Yoshie O, Identification of single C motif-1/lymphotactin receptor XCR1. J Biol Chem 273, 16551–16554 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Haider AS, Cardinale IR, Whynot JA, Krueger JG, Effects of etanercept are distinct from infliximab in modulating proinflammatory genes in activated human leukocytes. J Investig Dermatol Symp Proc 12, 9–15 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, Fouser LA, Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med 203, 2271–2279 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, Kleinewietfeld M, Kunder S, Hafler DA, Sobel RA, Regev A, Kuchroo VK, Induction and molecular signature of pathogenic TH17 cells. Nat Immunol 13, 991–999 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramesh R, Kozhaya L, McKevitt K, Djuretic IM, Carlson TJ, Quintero MA, McCauley JL, Abreu MT, Unutmaz D, Sundrud MS, Pro-inflammatory human Th17 cells selectively express P-glycoprotein and are refractory to glucocorticoids. J Exp Med 211, 89–104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu SY, Sanchez DJ, Aliyari R, Lu S, Cheng G, Systematic identification of type I and type II interferon-induced antiviral factors. Proc Natl Acad Sci U S A 109, 4239–4244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee AJ, Ashkar AA, The Dual Nature of Type I and Type II Interferons. Front Immunol 9, 2061 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Georas SN, Guo J, De Fanis U, Casolaro V, T-helper cell type-2 regulation in allergic disease. Eur Respir J 26, 1119–1137 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Corry DB, Kheradmand F, Induction and regulation of the IgE response. Nature 402, B18–23 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Oettgen HC, Geha RS, IgE in asthma and atopy: cellular and molecular connections. J Clin Invest 104, 829–835 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li J, Ge M, Lu S, Shi J, Li X, Wang M, Huang J, Shao Y, Huang Z, Zhang J, Nie N, Zheng Y, Pro-inflammatory effects of the Th1 chemokine CXCL10 in acquired aplastic anaemia. Cytokine 94, 45–51 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Gauthier M, Chakraborty K, Oriss TB, Raundhal M, Das S, Chen J, Huff R, Sinha A, Fajt M, Ray P, Wenzel SE, Ray A, Severe asthma in humans and mouse model suggests a CXCL10 signature underlies corticosteroid-resistant Th1 bias. JCI Insight 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Groom JR, Richmond J, Murooka TT, Sorensen EW, Sung JH, Bankert K, von Andrian UH, Moon JJ, Mempel TR, Luster AD, CXCR3 chemokine receptor-ligand interactions in the lymph node optimize CD4+ T helper 1 cell differentiation. Immunity 37, 1091–1103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beyer K, Baukloh AK, Stoyanova A, Kamphues C, Sattler A, Kotsch K, Interactions of Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand (TRAIL) with the Immune System: Implications for Inflammation and Cancer. Cancers (Basel) 11, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peteranderl C, Herold S, The Impact of the Interferon/TNF-Related Apoptosis-Inducing Ligand Signaling Axis on Disease Progression in Respiratory Viral Infection and Beyond. Front Immunol 8, 313 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lehnert C, Weiswange M, Jeremias I, Bayer C, Grunert M, Debatin KM, Strauss G, TRAIL-receptor costimulation inhibits proximal TCR signaling and suppresses human T cell activation and proliferation. J Immunol 193, 4021–4031 (2014). [DOI] [PubMed] [Google Scholar]
- 64.Chyuan IT, Tsai HF, Wu CS, Sung CC, Hsu PN, TRAIL-Mediated Suppression of T Cell Receptor Signaling Inhibits T Cell Activation and Inflammation in Experimental Autoimmune Encephalomyelitis. Front Immunol 9, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roberts AI, Devadas S, Zhang X, Zhang L, Keegan A, Greeneltch K, Solomon J, Wei L, Das J, Sun E, Liu C, Yuan Z, Zhou JN, Shi Y, The role of activation-induced cell death in the differentiation of T-helper-cell subsets. Immunol Res 28, 285–293 (2003). [DOI] [PubMed] [Google Scholar]
- 66.Zhang XR, Zhang LY, Devadas S, Li L, Keegan AD, Shi YF, Reciprocal expression of TRAIL and CD95L in Th1 and Th2 cells: role of apoptosis in T helper subset differentiation. Cell Death Differ 10, 203–210 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, Kang B, Liu Z, Jin L, Xing R, Gao R, Zhang L, Dong M, Hu X, Ren X, Kirchhoff D, Roider HG, Yan T, Zhang Z, Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med 24, 978–985 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Bacher P, Schink C, Teutschbein J, Kniemeyer O, Assenmacher M, Brakhage AA, Scheffold A, Antigen-reactive T cell enrichment for direct, high-resolution analysis of the human naive and memory Th cell repertoire. J Immunol 190, 3967–3976 (2013). [DOI] [PubMed] [Google Scholar]
- 69.van Dam S, Vosa U, van der Graaf A, Franke L, de Magalhaes JP, Gene co-expression analysis for functional classification and gene-disease predictions. Brief Bioinform 19, 575–592 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clarke J, Panwar B, Madrigal A, Singh D, Gujar R, Wood O, Chee SJ, Eschweiler S, King EV, Awad AS, Hanley CJ, McCann KJ, Bhattacharyya S, Woo E, Alzetani A, Seumois G, Thomas GJ, Ganesan AP, Friedmann PS, Sanchez-Elsner T, Ay F, Ottensmeier CH, Vijayanand P, Single-cell transcriptomic analysis of tissue-resident memory T cells in human lung cancer. J Exp Med 216, 2128–2149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Langfelder P, Horvath S, WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huber M, Lohoff M, IRF4 at the crossroads of effector T-cell fate decision. Eur J Immunol 44, 1886–1895 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Ahlfors H, Limaye A, Elo LL, Tuomela S, Burute M, Gottimukkala KV, Notani D, Rasool O, Galande S, Lahesmaa R, SATB1 dictates expression of multiple genes including IL-5 involved in human T helper cell differentiation. Blood 116, 1443–1453 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.O’Garra A, Gabrysova L, Transcription Factors Directing Th2 Differentiation: Gata-3 Plays a Dominant Role. J Immunol 196, 4423–4425 (2016). [DOI] [PubMed] [Google Scholar]
- 75.Micosse C, von Meyenn L, Steck O, Kipfer E, Adam C, Simillion C, Seyed Jafari SM, Olah P, Yawlkar N, Simon D, Borradori L, Kuchen S, Yerly D, Homey B, Conrad C, Snijder B, Schmidt M, Schlapbach C, Human “TH9” cells are a subpopulation of PPAR-gamma(+) TH2 cells. Sci Immunol 4, (2019). [DOI] [PubMed] [Google Scholar]
- 76.Prakash MD, Munoz MA, Jain R, Tong PL, Koskinen A, Regner M, Kleifeld O, Ho B, Olson M, Turner SJ, Mrass P, Weninger W, Bird PI, Granzyme B promotes cytotoxic lymphocyte transmigration via basement membrane remodeling. Immunity 41, 960–972 (2014). [DOI] [PubMed] [Google Scholar]
- 77.Patil VS, Madrigal A, Schmiedel BJ, Clarke J, O’Rourke P, de Silva AD, Harris E, Peters B, Seumois G, Weiskopf D, Sette A, Vijayanand P, Precursors of human CD4(+) cytotoxic T lymphocytes identified by single-cell transcriptome analysis. Sci Immunol 3, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stinchcombe JC, Barral DC, Mules EH, Booth S, Hume AN, Machesky LM, Seabra MC, Griffiths GM, Rab27a is required for regulated secretion in cytotoxic T lymphocytes. J Cell Biol 152, 825–834 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Omilusik KD, Best JA, Yu B, Goossens S, Weidemann A, Nguyen JV, Seuntjens E, Stryjewska A, Zweier C, Roychoudhuri R, Gattinoni L, Bird LM, Higashi Y, Kondoh H, Huylebroeck D, Haigh J, Goldrath AW, Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J Exp Med 212, 2027–2039 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marshall NB, Swain SL, Cytotoxic CD4 T cells in antiviral immunity. J Biomed Biotechnol 2011, 954602 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thewissen M, Somers V, Hellings N, Fraussen J, Damoiseaux J, Stinissen P, CD4+CD28null T cells in autoimmune disease: pathogenic features and decreased susceptibility to immunoregulation. J Immunol 179, 6514–6523 (2007). [DOI] [PubMed] [Google Scholar]
- 82.Takamori A, Nambu A, Sato K, Yamaguchi S, Matsuda K, Numata T, Sugawara T, Yoshizaki T, Arae K, Morita H, Matsumoto K, Sudo K, Okumura K, Kitaura J, Matsuda H, Nakae S, IL-31 is crucial for induction of pruritus, but not inflammation, in contact hypersensitivity. Sci Rep 8, 6639 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gibbs BF, Patsinakidis N, Raap U, Role of the Pruritic Cytokine IL-31 in Autoimmune Skin Diseases. Front Immunol 10, 1383 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schroeder JT, Chichester KL, Bieneman AP, Human basophils secrete IL-3: evidence of autocrine priming for phenotypic and functional responses in allergic disease. J Immunol 182, 2432–2438 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Williams GT, Smith CA, Spooncer E, Dexter TM, Taylor DR, Haemopoietic colony stimulating factors promote cell survival by suppressing apoptosis. Nature 343, 76–79 (1990). [DOI] [PubMed] [Google Scholar]
- 86.Weber GF, Chousterman BG, He S, Fenn AM, Nairz M, Anzai A, Brenner T, Uhle F, Iwamoto Y, Robbins CS, Noiret L, Maier SL, Zonnchen T, Rahbari NN, Scholch S, Klotzsche-von Ameln A, Chavakis T, Weitz J, Hofer S, Weigand MA, Nahrendorf M, Weissleder R, Swirski FK, Interleukin-3 amplifies acute inflammation and is a potential therapeutic target in sepsis. Science 347, 1260–1265 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nelson RK, Brickner H, Panwar B, Ramirez-Suastegui C, Herrera-de la Mata S, Liu N, Diaz D, Alexander LEC, Ay F, Vijayanand P, Seumois G, Akuthota P, Human Eosinophils Express a Distinct Gene Expression Program in Response to IL-3 Compared with Common beta-Chain Cytokines IL-5 and GM-CSF. J Immunol 203, 329–337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen L, Flies DB, Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol 13, 227–242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Oh H, Ghosh S, NF-kappaB: roles and regulation in different CD4(+) T-cell subsets. Immunol Rev 252, 41–51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mandal M, Borowski C, Palomero T, Ferrando AA, Oberdoerffer P, Meng F, Ruiz-Vela A, Ciofani M, Zuniga-Pflucker JC, Screpanti I, Look AT, Korsmeyer SJ, Rajewsky K, von Boehmer H, Aifantis I, The BCL2A1 gene as a pre-T cell receptor-induced regulator of thymocyte survival. J Exp Med 201, 603–614 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sarkar SA, Kutlu B, Velmurugan K, Kizaka-Kondoh S, Lee CE, Wong R, Valentine A, Davidson HW, Hutton JC, Pugazhenthi S, Cytokine-mediated induction of anti-apoptotic genes that are linked to nuclear factor kappa-B (NF-kappaB) signalling in human islets and in a mouse beta cell line. Diabetologia 52, 1092–1101 (2009). [DOI] [PubMed] [Google Scholar]
- 92.Lawan A, Al-Harthi S, Cadalbert L, McCluskey AG, Shweash M, Grassia G, Grant A, Boyd M, Currie S, Plevin R, Deletion of the dual specific phosphatase-4 (DUSP-4) gene reveals an essential non-redundant role for MAP kinase phosphatase-2 (MKP-2) in proliferation and cell survival. J Biol Chem 286, 12933–12943 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Al-Mutairi MS, Cadalbert LC, McGachy HA, Shweash M, Schroeder J, Kurnik M, Sloss CM, Bryant CE, Alexander J, Plevin R, MAP kinase phosphatase-2 plays a critical role in response to infection by Leishmania mexicana. PLoS Pathog 6, e1001192 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tseveleki V, Bauer J, Taoufik E, Ruan C, Leondiadis L, Haralambous S, Lassmann H, Probert L, Cellular FLIP (long isoform) overexpression in T cells drives Th2 effector responses and promotes immunoregulation in experimental autoimmune encephalomyelitis. J Immunol 173, 6619–6626 (2004). [DOI] [PubMed] [Google Scholar]
- 95.Lohning M, Stroehmann A, Coyle AJ, Grogan JL, Lin S, Gutierrez-Ramos JC, Levinson D, Radbruch A, Kamradt T, T1/ST2 is preferentially expressed on murine Th2 cells, independent of interleukin 4, interleukin 5, and interleukin 10, and important for Th2 effector function. Proc Natl Acad Sci U S A 95, 6930–6935 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alvarez F, Fritz JH, Piccirillo CA, Pleiotropic Effects of IL-33 on CD4(+) T Cell Differentiation and Effector Functions. Front Immunol 10, 522 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Finnson KW, Tam BY, Liu K, Marcoux A, Lepage P, Roy S, Bizet AA, Philip A, Identification of CD109 as part of the TGF-beta receptor system in human keratinocytes. FASEB J 20, 1525–1527 (2006). [DOI] [PubMed] [Google Scholar]
- 98.Stritesky GL, Muthukrishnan R, Sehra S, Goswami R, Pham D, Travers J, Nguyen ET, Levy DE, Kaplan MH, The transcription factor STAT3 is required for T helper 2 cell development. Immunity 34, 39–49 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chuang CH, Greenside PG, Rogers ZN, Brady JJ, Yang D, Ma RK, Caswell DR, Chiou SH, Winters AF, Gruner BM, Ramaswami G, Spencley AL, Kopecky KE, Sayles LC, Sweet-Cordero EA, Li JB, Kundaje A, Winslow MM, Molecular definition of a metastatic lung cancer state reveals a targetable CD109-Janus kinase-Stat axis. Nat Med 23, 291–300 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Litvinov IV, Bizet AA, Binamer Y, Jones DA, Sasseville D, Philip A, CD109 release from the cell surface in human keratinocytes regulates TGF-beta receptor expression, TGF-beta signalling and STAT3 activation: relevance to psoriasis. Exp Dermatol 20, 627–632 (2011). [DOI] [PubMed] [Google Scholar]
- 101.Gavino AC, Nahmod K, Bharadwaj U, Makedonas G, Tweardy DJ, STAT3 inhibition prevents lung inflammation, remodeling, and accumulation of Th2 and Th17 cells in a murine asthma model. Allergy 71, 1684–1692 (2016). [DOI] [PubMed] [Google Scholar]
- 102.Chyuan IT, Tsai HF, Wu CS, Hsu PN, TRAIL suppresses gut inflammation and inhibits colitogeic T-cell activation in experimental colitis via an apoptosis-independent pathway. Mucosal Immunol 12, 980–989 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brough HA, Cousins DJ, Munteanu A, Wong YF, Sudra A, Makinson K, Stephens AC, Arno M, Ciortuz L, Lack G, Turcanu V, IL-9 is a key component of memory TH cell peanut-specific responses from children with peanut allergy. J Allergy Clin Immunol 134, 1329–1338 e1310 (2014). [DOI] [PubMed] [Google Scholar]
- 104.Gong F, Pan YH, Huang X, Zhu HY, Jiang DL, From bench to bedside: Therapeutic potential of interleukin-9 in the treatment of asthma. Exp Ther Med 13, 389–394 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Oh CK, Leigh R, McLaurin KK, Kim K, Hultquist M, Molfino NA, A randomized, controlled trial to evaluate the effect of an anti-interleukin-9 monoclonal antibody in adults with uncontrolled asthma. Respir Res 14, 93 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lu Y, Hong B, Li H, Zheng Y, Zhang M, Wang S, Qian J, Yi Q, Tumor-specific IL-9-producing CD8+ Tc9 cells are superior effector than type-I cytotoxic Tc1 cells for adoptive immunotherapy of cancers. Proc Natl Acad Sci U S A 111, 2265–2270 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xue G, Jin G, Fang J, Lu Y, IL-4 together with IL-1beta induces antitumor Th9 cell differentiation in the absence of TGF-beta signaling. Nat Commun 10, 1376 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hinz D, Oseroff C, Pham J, Sidney J, Peters B, Sette A, Definition of a pool of epitopes that recapitulates the T cell reactivity against major house dust mite allergens. Clin Exp Allergy 45, 1601–1612 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oseroff C, Christensen LH, Westernberg L, Pham J, Lane J, Paul S, Greenbaum J, Stranzl T, Lund G, Hoof I, Holm J, Wurtzen PA, Meno KH, Frazier A, Schulten V, Andersen PS, Peters B, Sette A, Immunoproteomic analysis of house dust mite antigens reveals distinct classes of dominant T cell antigens according to function and serological reactivity. Clin Exp Allergy 47, 577–592 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R, Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 10, 1096–1098 (2013). [DOI] [PubMed] [Google Scholar]
- 111.Rosales SL, Liang S, Engel I, Schmiedel BJ, Kronenberg M, Vijayanand P, Seumois G, A Sensitive and Integrated Approach to Profile Messenger RNA from Samples with Low Cell Numbers. Methods Mol Biol 1799, 275–301 (2018). [DOI] [PubMed] [Google Scholar]
- 112.Engel I, Seumois G, Chavez L, Samaniego-Castruita D, White B, Chawla A, Mock D, Vijayanand P, Kronenberg M, Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat Immunol 17, 728–739 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Trapnell C, Pachter L, Salzberg SL, TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Langmead B, Trapnell C, Pop M, Salzberg SL, Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li H, Durbin R, Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Anders S, Pyl PT, Huber W, HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Martin M, Cutadapt Removes Adapter Sequences From High-Throughput Sequencing Reads. EMBnet.Journal 17, 10–12 (2011). [Google Scholar]
- 118.van der Maaten L, Accelerating t-SNE using Tree-Based Algorithms. Journal of Machine Learning Research 15, 3221–3245 (2014). [Google Scholar]
- 119.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ganesan AP, Clarke J, Wood O, Garrido-Martin EM, Chee SJ, Mellows T, Samaniego-Castruita D, Singh D, Seumois G, Alzetani A, Woo E, Friedmann PS, King EV, Thomas GJ, Sanchez-Elsner T, Vijayanand P, Ottensmeier CH, Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat Immunol 18, 940–950 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R, Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kang HM, Subramaniam M, Targ S, Nguyen M, Maliskova L, McCarthy E, Wan E, Wong S, Byrnes L, Lanata CM, Gate RE, Mostafavi S, Marson A, Zaitlen N, Criswell LA, Ye CJ, Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol 36, 89–94 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, Linsley PS, Gottardo R, MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 16, 278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Huang M, Wang J, Torre E, Dueck H, Shaffer S, Bonasio R, Murray JI, Raj A, Li M, Zhang NR, SAVER: gene expression recovery for single-cell RNA sequencing. Nat Methods 15, 539–542 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bastian M, Heymann S, Jacomy M, Gephi: An Open Source Software for Exploring and Manipulating Networks. 2009 (2009). [Google Scholar]
- 127.Schmiedel BJ, Singh D, Madrigal A, Valdovino-Gonzalez AG, White BM, Zapardiel-Gonzalo J, Ha B, Altay G, Greenbaum JA, McVicker G, Seumois G, Rao A, Kronenberg M, Peters B, Vijayanand P, Impact of Genetic Polymorphisms on Human Immune Cell Gene Expression. Cell 175, 1701–1715 e1716 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Study subject details (Excel spreadsheet)
Table S2. Differential gene expression analysis in bulk populations (Excel spreadsheet).
Table S4. Gene signatures lists (Excel spreadsheet).
Table S5. Raw data file with statistics (Excel spreadsheet).
Table S3. Single cell analysis (Excel spreadsheet).
Fig. S1. Experimental design and gating strategy to isolate HDM-reactive memory TH and Treg cells.
Fig. S2. Expression of differentially expressed genes in HDM-reactive cells.
Fig. S3. Single cell filtering of doublets and low HDM-reactive T cells.
Fig. S4. Single-cell clustering analysis using Seurat.
Fig. S5. Distribution of cell frequency for the 7 HDM+ TH clusters for the 24 subjects.
Fig. S6. Co-expression of TH1- or TH17-specific signature genes.
Fig. S7. Donor and disease-groups cell distribution in each of HDM+ TH clusters.
Fig. S8. Co-expression of THIFNR-specific signature genes in publicly available dataset.
Fig. S9. TREG disease-related differences.
Fig. S10. Proportions of HDM-reactive Treg subsets amongst disease groups.
Fig. S11. TH2 single-cell analysis.