Abstract
The genetic hallmark of classic Ewing sarcoma is a recurrent fusion between EWSR1 or FUS gene with a member of the ETS transcription factor family. In contrast, tumors with non-ETS gene partners have been designated until recently ‘Ewing-like sarcoma’, as a provisional molecular entity, as their clinical and pathologic features were still evolving. However, this group was reclassified as ‘round cell sarcoma with EWSR1-non-ETS fusions’ in the latest 2020 WHO classification. Moreover, round cell sarcomas with either CIC or BCOR gene abnormalities, initially classified under Ewing family of tumors, are now regarded as stand-alone pathologic entities based on their distinct features. In this study we investigated the clinical characteristics of 226 confirmed Ewing sarcoma patients [EWSR1-FLI1 (n=176), EWSR1/FUS-ERG (n=35), EWSR1/FUS-FEV (n=12), EWSR1-ETV1/4 (n=3)] and 14 round cell sarcoma patients with EWSR1-non-ETS fusion [EWSR1/FUS-NFATC2 (n=10), EWSR1-PATZ1 (n=3), EWSR1-VEZF1 (n=1)]. The impact on overall survival (OS) was assessed in 90 patients with available follow-up, treated between 2011–2018. Patients with fusions involving FEV and NFATC2 genes showed an older median age at diagnosis, compared to those with EWSR1-FLI1 (p=0.005), while extraskeletal location was more common in tumors with non-canonical EWSR1-FLI1 fusions (p=0.001). Axial and pelvic primary sites were more common in patients with EWSR1-FLI1 (72%), while tumors with NFATC2 fusions were more frequent in the limb (78%, p=0.006). The 3-year OS in patients with EWSR1-FLI1 was 91%, compared to only 60% in patients with alternative fusions (p=0.037); the latter group showing a higher rate of metastases at presentation. However, this OS difference was not significant in patients with localized tumor (p=0.585). Our study demonstrates significant correlations between fusion subtype and age at presentation, primary tumor sites, and OS, in both conventional Ewing sarcoma and round cell sarcoma with EWSR1-non ETS fusions patients. Larger studies are needed to determine survival differences in localized tumors.
Keywords: Ewing sarcoma, Ewing-like sarcoma, EWSR1, FUS, gene fusions
1. INTRODUCTION
Ewing sarcoma is the prototypical round cell sarcoma which occurs with predilection in the bone of children and young adults. The majority of conventional Ewing sarcomas are characterized genetically by recurrent fusions involving EWSR1 and FLI1, the latter encoding a member of the ETS transcription factor family, resulting in an oncogenic transcriptional program1. In less than 10% of cases, the gene fusions of Ewing sarcoma involve other ETS gene members, such as ERG 2, FEV 3, ETV1 4, or ETV4 5. EWSR1 and FUS are members of the FET family of RNA binding proteins, have similar functions and are interchangeable in translocation driven sarcomas6.
In addition to the EWSR1-ETS-positive classic Ewing sarcoma, a rare subset characterized by EWSR1-non-ETS fusions, which was until recently provisionally termed as ‘Ewing-like sarcoma’ tumors has emerged, being characterized by fusions between EWSR1 or FUS with non-ETS partners, such as PATZ17, SP38, NFATC29,10, and SMARCA511. Although preliminary gene expression and epigenetic profiles suggest that at least some of these molecular subsets encountered in Ewing-like sarcomas are distinct from the canonical EWSR1/FUS-ETS fusions12, large clinicopathologic studies are lacking. In this study, taking advantage of our large dataset of Ewing sarcoma and round cell sarcoma with EWSR1-non-ETS fusion cases with well-characterized gene fusions, we sought to correlate the impact of various fusion subtypes on clinical and pathologic findings, as well as survival outcomes.
2. MATERIAL AND METHODS
2.1. Patient selection
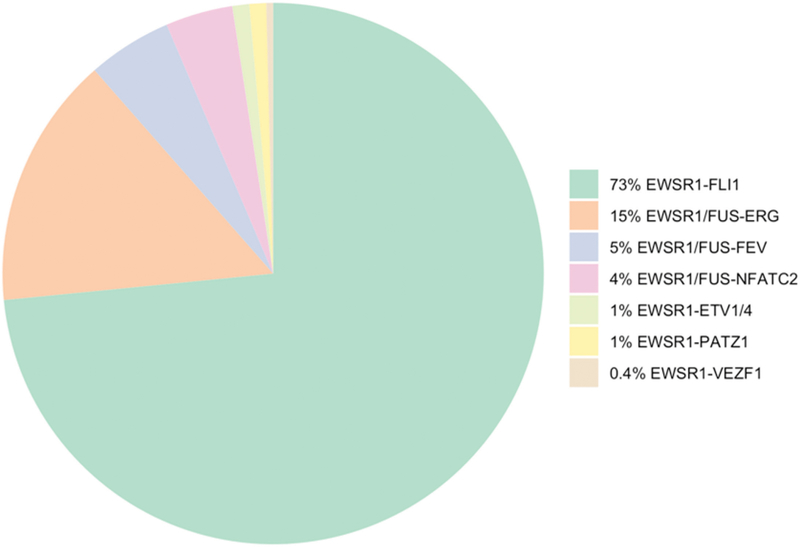
The MSKCC files and personal consultation files of the senior author (CRA) were searched for the diagnosis of Ewing sarcoma and round cell sarcoma with EWSR1-non-ETS fusions with confirmed gene fusion information or with available tissue to determine the molecular subtype. A total of 240 patients diagnosed between 1986 and 2019 were included, showing the following break-down fusions (Fig. 1): EWSR1-FLI1 (n=176), EWSR1/FUS-ERG (n=35), EWSR1/FUS-FEV (n=12), EWSR1/FUS-NFATC2 (n=10), EWSR1-ETV1/4 (n=3), EWSR1-PATZ1 (n=3), or EWSR1-VEZF1 (n=1). Most of the fusion types (n=179) were determined by FISH assay using custom BAC probes for all known gene partners and fusion variants. In a small subset, the translocation partners were obtained from a variety of other molecular methods, including RT-PCR (n=20), karyotype (n=2), targeted RNA sequence (n=12), or MSK-IMPACT assay (n=27). Patients with only evidence of EWSR1/FUS rearrangement but no available tissue for further investigation of the gene partner were excluded from the study (n=37).
Figure 1.
Pie chart showing the distribution of gene fusions in Ewing sarcoma and round cell sarcoma with EWSR1-non-ETS fusion.
For survival analysis, patients with available follow-up data diagnosed between 2011 and 2018 were selected, with a minimum follow-up of one year (n=90). Single patients with rare fusion variants, such as one patient with EWSR1-ETV4, were excluded from further survival analyses. The dose and period of chemotherapy were adjusted based on their age, co-morbidities and toxicities. All patients were followed up according to a standard protocol13.
Hematoxylin and eosin-stained slides and immunohistochemical stains were re-reviewed. The tumors were assessed for growth pattern, cytomorphology (round, oval, spindle, epithelioid, plasmacytoid/rhabdoid phenotype), nuclear features including nuclear contour, chromatin pattern and presence of nucleoli, mitotic activity, necrosis, type of stroma and myxoid change. Review of the CD99 immunohistochemical staining patterns was also assessed. The patients’ charts were retrospectively reviewed. The following clinical data were retrieved: age, gender, primary tumor site, stage at diagnosis (primary versus distant metastasis at diagnosis), tumor size, modality of initial therapy, recurrence, vital status and survival time. In a few patients, information on gender (n=2), skeletal or extraskeletal primary site (n=9), and limb or axial location (n=6) were not available due to the consultation cases. The study was approved by the Institutional Review Board.
2.2. Fluorescence in situ hybridization (FISH)
FISH was conducted for EWSR1/FUS, FLI1, ERG, FEV, NFATC2, ETV1, ETV4, or PATZ1. FISH for break-apart assay was applied on formalin-fixed and paraffin-embedded 4-micron sections as previously described14. Custom probes using bacterial artificial chromosomes (BACs) covering and flanking each gene were utilized15. The BAC clones were selected according to the UCSC genome browser (http://genome.ucsc.edu) and obtained from the BACPAC sources of Children’s Hospital of Oakland Research Institute (CHORI) (Oakland, CA) (http://bacpac.chori.org). DNA from individual BACs was isolated in line with manufacturer’s instructions, labeled with different fluorochromes in a nick translation reaction, denatured, and hybridized to pretreated slides. Slides were then incubated, washed, and mounted with DAPI. Two hundred tumor nuclei were evaluated using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems, Newton, MA). A cut-off of >20% nuclei showing a break-apart signal was considered to be positive for rearrangement. Nuclei with incomplete set of signals were omitted from the score.
2.3. Other molecular methods to determine the fusion type
In a smaller subset of cases the gene fusion was determined either by RT-PCR (n=20), karyotype (n=2), MSK-IMPACT assay (n=27), or targeted RNA sequencing (n=12), as previously described16–18. Targeted RNA sequencing was performed either by using an Archer™ FusionPlex™ platform19,20 or a TruSight RNA Fusion Panel (Illumina, San Diego, CA) on an Illumina MiSeq platform21, using standard protocols. Reads were independently aligned with STAR (version 2.3) against the human reference genome (hg19) and analyzed by STAR-Fusion.
2.4. Statistical analysis
Kaplan-Meier curves were used to estimate overall survival (OS). OS was defined as the time from the diagnosis to death and was censored at the date of the latest follow-up. Categorical variables were compared between groups using chi-square tests. Numerical variables were compared using Kruskal-Wallis Test. Statistical analyses was performed using SPSS version 21 (IBM), with significance set at two-tailed p <0.05.
3. RESULTS
3.1. Correlation between fusion transcript type and clinical parameters
A total of 240 patients diagnosed with Ewing sarcoma or round cell sarcoma EWSR1-non-ETS fusions with complete gene fusion information were included in the study. The most common fusion type was EWSR1-FLI1 (n=176, 73%), followed by tumors with ERG gene rearrangements (n=35, 15%), either EWSR1-ERG (n=26, 11%) or FUS-ERG (n=9, 4%). Other less common fusions were identified, including: EWSR1/FUS-FEV (n=12), EWSR1/FUS-NFATC2 (n=10), EWSR1-ETV1/4 (n=3), EWSR1-PATZ1 (n=3), or EWSR1-VEZF1 (n=1)(Fig. 1).
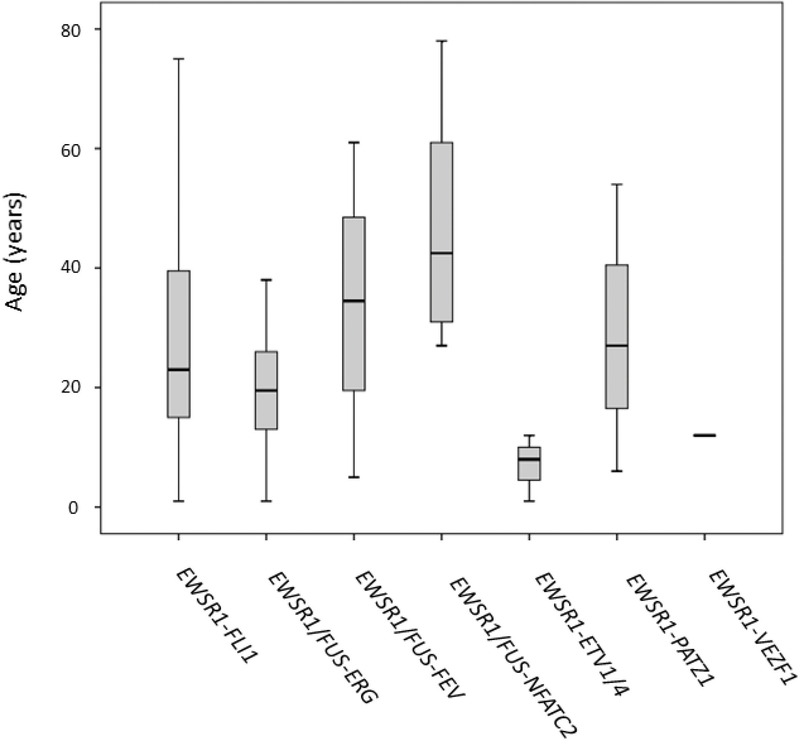
The median age at diagnosis for the entire group was 23 years, with a wide age range of 1–78 years (Table 1). Ewing sarcoma patients with canonical fusions had a median age at diagnosis of 23 years (range, 1–75) in the EWSR1-FLI1-positive cohort, and a median age of 20 years (range, 1–64) in the EWSR1/FUS-ERG group. The Ewing sarcoma patients harboring EWSR1-ETV1/4 fusions had the youngest median age at diagnosis, of 8 years old (range, 1–12) and the single case with EWSR1-VEZF1 fusion occurred in a 12 year-old patient (Fig. 2). In fact, one-third of cases positive for EWSR1-ETV1/4 fusion occurred in very young children (aged <2 years), while only one case each occurred in the more common EWSR1-FLI1 and EWSR1/FUS-ERG molecular groups. In contrast, patients with EWSR1/FUS-FEV and EWSR1/FUS-NFATC2 fusions had an older median age at presentation of 35 (range, 5–61) and 43 (range, 27–78) years, respectively. The distribution of pediatric patients (aged <18 years) was higher in the molecular subsets of EWSR1-ETV1/4 (3/3) and EWSR1-VEZF1 (1/1), compared to EWSR1/FUS-ERG (43%, 15/35) and EWSR1-FLI1 (34%, 63/176) groups. In contrast, the proportion of older adult patients (aged >40 years) was highest in EWSR1/FUS-NFATC2-positive group (50%) compared to that of the canonical EWSR1-FLI1 group (24%).
Table 1.
Demographic and primary site correlations with gene fusion type
| Total | % | EWSR1-FLI1 | % | EWSR1/FUS-ERG | % | EWSR1/FUS-FEV | % | EWSR1/FUS-NFATC2 | % | EWSR1-ETV1/4 | % | EWSR1-PATZ1 | % | EWSR1-VEZF1 | % | p value | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total (n = 240) | 240 | 176 | 73 | 35 | 15 | 12 | 5 | 10 | 4 | 3 | 1 | 3 | 1 | 1 | 0.4 | |||
| Age (median, range, years) | 23 (1–78) | 23 (1–75) | 20 (1–64) | 35 (5–61) | 43 (27–78) | 8 (1–12) | 27 (6–54) | 12 (NA) | 0.005 | |||||||||
| Sex (n =238) | Male | 144 | 60 | 108 | 61 | 17 | 50 | 8 | 67 | 6 | 67 | 2 | 67 | 3 | 100 | 0 | 0 | 0.482 |
| Female | 94 | 40 | 68 | 39 | 17 | 50 | 4 | 33 | 3 | 33 | 1 | 33 | 0 | 0 | 1 | 100 | ||
| Primary site (n = 231) | Skeletal | 127 | 55 | 104 | 61 | 19 | 54 | 0 | 0 | 4 | 44 | 0 | 0 | 0 | 0 | 0 | 0 | 0.001 |
| Extraskeletal | 104 | 45 | 67 | 39 | 15 | 46 | 12 | 100 | 5 | 56 | 3 | 100 | 1 | 100 | 1 | 100 | ||
| Primary site (n = 234) | Limb | 79 | 34 | 48 | 28 | 16 | 46 | 4 | 33 | 7 | 78 | 2 | 67 | 1 | 100 | 1 | 100 | 0.006 |
| Axial and pelvis | 155 | 66 | 125 | 72 | 19 | 54 | 8 | 67 | 2 | 22 | 1 | 33 | 0 | 0 | 0 | 0 |
NA, not available
Figure 2.
Box plot showing age distribution of patients with round cell sarcomas with EWSR1/FUS gene rearrangements in relationship to gene fusions types.
Only 39% of patients with EWSR1-FLI1 canonical fusion presented at extraskeletal sites, in contrast to all patients harboring EWSR1/FUS-FEV, EWSR1-ETV1/4, EWSR1-PATZ1, and EWSR1-VEZF1 fusions (Fig 3). The limb location was a frequent primary site in the round cell sarcoma group with EWSR1-non-ETS fusion patients, encompassing all cases with EWSR1-PATZ1 and EWSR1-VEZF1 fusions, and 7/9 EWSR1/FUS-NFATC2-positive cases (Fig 3). In contrast, axial and pelvis presentation was more common in patients with canonical fusions such as EWSR1-FLI1 (72%) and EWSR1/FUS-ERG (54%). Overall, statistically significant differences with fusion type were detected in age at presentation (p=0.005), skeletal versus extraskeletal location (p=0.001), and limb versus axial primary sites (p=0.006).
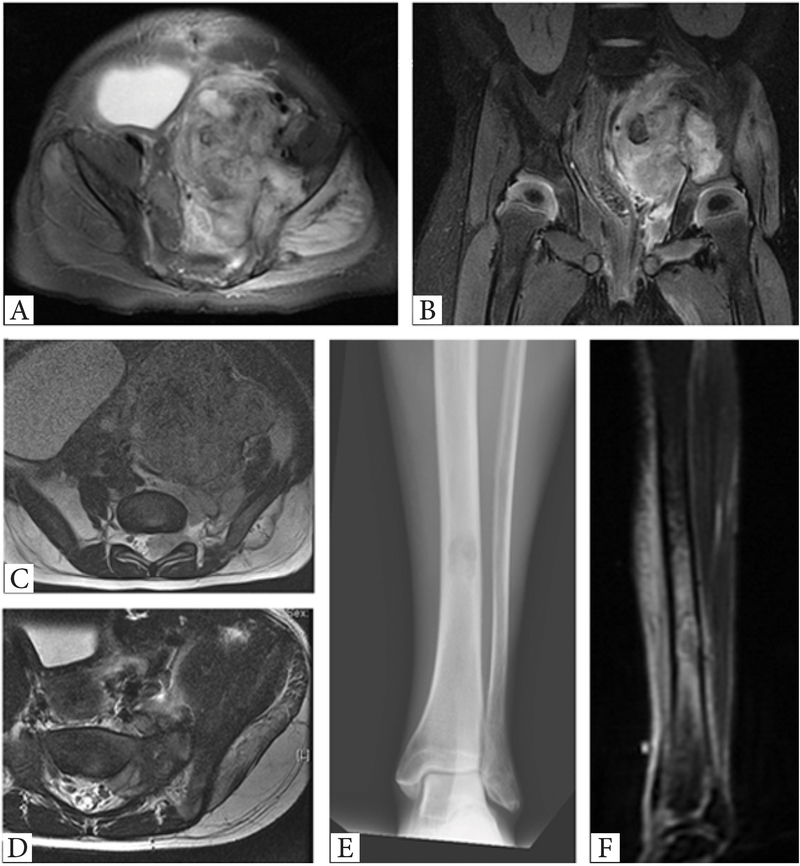
Figure 3. Radiographic findings in rare Ewing sarcoma and round cell sarcoma with EWSR1-non-ETS fusion subsets.
A-D. Large paraspinal/pelvic soft tissue mass in a one-year old female with a rare EWSR1-ETV4 fusion, showing a favorable chemotherapy response to Ewing sarcoma regimen. A,B. MRI T2 weighted image showing a left pelvic side wall soft tissue mass extending to paraspinal area. C. MRI T1 weighted image showing tumor extension to the left L5/S1 neural foramen. D. MRI T2 weighted image showing a substantial decrease in tumor size after preoperative chemotherapy. E,F. Permeative tibial lesion in a 78 year-old male with a round cell sarcoma with an EWSR1-NFATC2 fusion.
3.2. Morphologic Features and Immunohistochemistry
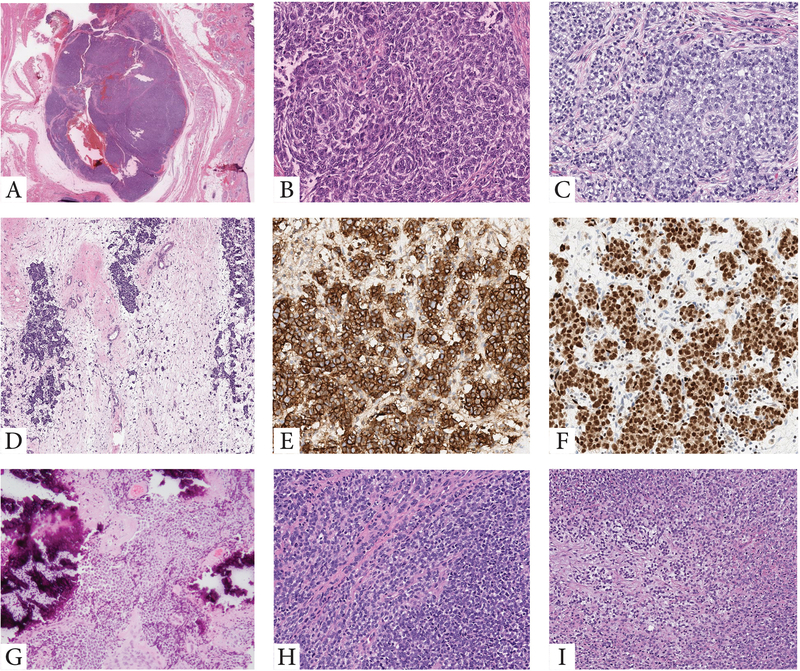
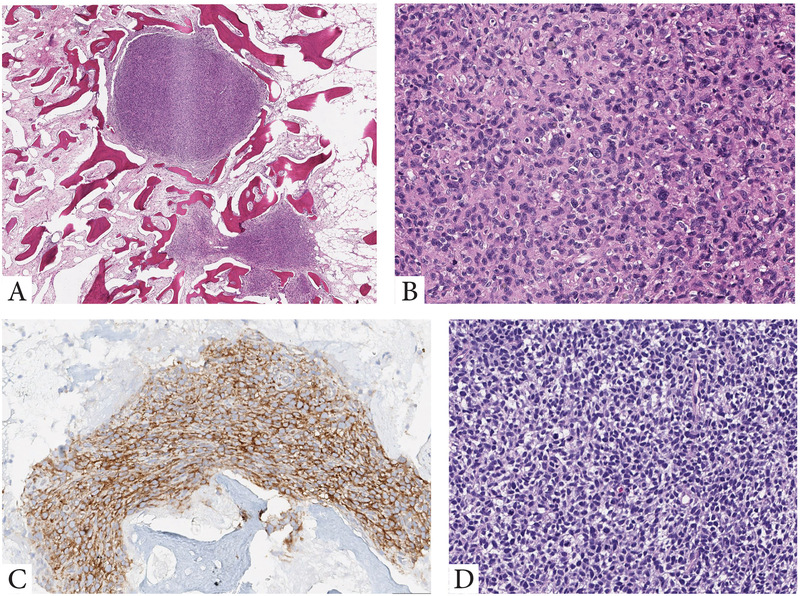
Although all tumors included in this comprehensive series were classified as round cell malignancies, only Ewing sarcomas with the canonical EWSR1/FUS-FLI1 and variant EWSR1/FUS-FEV showed the classic features typically recognized in Ewing sarcoma. The latter features including solid sheets of uniform round cells with ill-defined cell borders, scant, often clear, cytoplasm, and monomorphic round nuclei, with smooth contours and fine chromatin. Tumors in these 2 molecular groups were uniformly positive for CD99, while Ewing sarcoma cases with canonical fusions also showed consistent nuclear positivity for FLI1. However, FLI1 reactivity was not limited to this genetic subset being positive in other ETS-related fusions. In contrast, tumors with EWSR1/FUS-ERG fusions showed in general a more variable spectrum of histologies, which spanned from round, ovoid, short spindle and even epithelioid cells (Fig 4). Immunohistochemically, this group was consistently positive for CD99 and ERG markers (Fig 4). Rare EWSR1-ETS fusion variants included cases with unusual histologic features. One of the EWSR1-ETV4 tumor occurring in an infant, showed extensive tumoral calcifications, while the single case harboring an EWSR1-VEZF1 fusions showed a primitive round cell phenotype with an increased myxoid and fibrous stromal components (Fig. 4). Moreover, round cell sarcomas with EWSR1-non-ETS fusions with either NFATC2 or PATZ1 gene abnormalities showed increased variability (Fig 5), compared to the uniform cytomorphology or immunoprofile of tumors with the EWSR1-FLI1 canonical fusion.
Figure 4. Morphologic features of tumors with variant fusions in the Ewing sarcoma family.
A-F. Morphologic spectrum of tumors harboring EWSR1-ERG fusions. A,B. Superficial nodule within the subcutis showing at high power primitive round to ovoid cells with a peculiar whorling-like growth (9/M, orbit). C. Omental Ewing sarcoma in a 21 year-old female showing solid sheets of round to epithelioid cells with moderate amount of clear to amphophilic cytoplasm. D-F. Post-therapy lesion showing islands of residual viable tumor cells within the fibrotic marrow spaces of the rib. Viable cells remained strongly and diffusely positive for CD99 (E) and ERG (F). G-I. Less common EWSR1-ETS variants in Ewing sarcomas. G. A paraspinal lesion with extensive calcifications arising in an infant with EWSR1-ETV4 (same patient as in Fig 3). H. Primitive round cells in solid sheets in a tumor with EWSR1-FEV (10/M). I. Round cell malignancy with increased myxoid or fibrous stromal component in a tumor positive for EWSR1-VEZF1 (12/F, calf).
Figure 5. Histologic spectrum of round cell sarcoma with EWSR1-non-ETS fusion.
A-C. Intra-osseous nodules of primitive cells harboring an EWSR1-NFATC2 fusion, which at high power were composed of a mixture of round, ovoid and scattered slightly pleomorphic cells with enlarged nuclei, and ill-defined cell borders. The tumor cells were diffusely positive for CD99 (membranous pattern) (78/M, tibia, same patient as in Fig 3). D. Primitive round cell tumor with an EWSR1-PATZ1 fusion (37/M, shoulder).
The CD99 immunohistochemical findings showed strong and diffuse membranous immunoreactivity for all cases with EWSR1-FLI1, EWSR1/FUS-ERG, EWSR1/FUS-FEV, and EWSR1-ETV1/4 fusions. Moreover, all EWSR1-NFATC2 tumors with available material were positive for CD99, however, in 2 of the 6 cases the reactivity was only focal, while remaining showed diffuse strong membranous staining. The single case with EWSR1-VEZF1 was negative for CD99. Desmin and MyoD1 were positive in one tumor with EWSR1-PATZ1.
3.3. Comparison of fusion type with overall survival
Ninety patients with available follow-up, treated during 2011 and 2018, were selected for survival analysis. Patients characteristics are shown in Table 2. The fusion distribution among this smaller cohort of patients was the following: EWSR1-FLI1 (n=67, 74%), EWSR1/FUS-ERG (n=16, 18%), EWSR1/FUS-FEV (n=4, 4%), and EWSR1/FUS-NFATC2 (n=3, 3%), which mirrored the overall distributions in age and primary location observed in the entire cohort (Table 2). The median age at diagnosis for this smaller group of patients was 23 years of age, which was identical to the group of patients with EWSR1-FLI1 canonical fusion (range, 2–78); while for the EWSR1/FUS-ERG group was 19 years (range, 2–64). In contrast, patients with EWSR1/FUS-NFATC2 fusion had an older age at diagnosis (median 61 years, range, 27–78). Extraskeletal primary sites were more common in the EWSR1/FUS-FEV (100%) and EWSR1/FUS-NFATC2 (67%) group of tumors, compared to that of the EWSR1-FLI1 group (45%). Similar to the entire cohort, all cases in this selected subset with EWSR1/FUS-NFATC2 fusion presented in the limb, while axial and pelvis location was more common in patients with EWSR1-FLI1 fusions (66%).
Table 2.
Demographic, clinical characteristics and survival correlations with fusion type
| Total | % | EWSR1-FLI1 | % | EWSR1/FUS-ERG | % | EWSR1/FUS-FEV | % | EWSR1/FUS-NFATC2 | % | p value | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | 90 | 67 | 74 | 16 | 18 | 4 | 4 | 3 | 3 | |||
| Age (median, range, years) | 23 (2–78) | 23 (3–64) | 19 (2–64) | 28 (16–40) | 61 (27–78) | 0.099 | ||||||
| Sex | Male | 49 | 56 | 40 | 60 | 6 | 38 | 3 | 75 | 2 | 67 | 0.346 |
| Female | 39 | 44 | 27 | 40 | 10 | 62 | 1 | 25 | 1 | 33 | ||
| Primary site | Skeletal | 45 | 51 | 37 | 55 | 7 | 44 | 0 | 0 | 1 | 33 | 0.150 |
| Extraskeletal | 43 | 49 | 30 | 45 | 9 | 56 | 4 | 100 | 2 | 67 | ||
| Primary site | Limb | 31 | 35 | 23 | 34 | 6 | 38 | 0 | 0 | 3 | 100 | 0.052 |
| Axial and pelvis | 57 | 65 | 44 | 66 | 10 | 62 | 4 | 100 | 0 | 0 | ||
| Tumor size (median, range) | 8 (1–26) | 8 (1–18) | 7 (3–22) | 8 (5–18) | 3 (3–26) | 0.889 | ||||||
| AJCC stage at diagnosis* | IIa | 28 | 33 | 22 | 35 | 4 | 29 | 0 | 0 | 2 | 67 | 0.226 |
| IIb | 24 | 29 | 20 | 32 | 3 | 21 | 1 | 25 | 0 | 0 | ||
| IVa | 18 | 21 | 14 | 22 | 2 | 14 | 1 | 25 | 1 | 33 | ||
| IVb | 14 | 17 | 7 | 11 | 5 | 36 | 2 | 50 | 0 | 0 | ||
| Metastasis at presentation | 31 | 34 | 20 | 30 | 7 | 44 | 3 | 75 | 1 | 33 | 0.245 | |
| Definitive surgery | 61 | 69 | 47 | 70 | 12 | 75 | 1 | 25 | 3 | 100 | 0.149 | |
| (Neo)adjuvant chemotherapy | 58 | 66 | 46 | 69 | 9 | 56 | 2 | 50 | 2 | 67 | 0.722 | |
| Survival status | DOD | 15 | 17 | 8 | 12 | 4 | 25 | 2 | 50 | 1 | 33 | |
| DOOD | 2 | 2 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
DOD, dead of disease; DOOD, dead of other diseases
AJCC stage is not shown in six patients due to lack of size data
Overall, treatment for the primary disease included surgical resection in 61 of 90 (68%) patients. Of 59 patients presenting with localized disease, 48 patients underwent definitive surgical resection for the primary tumor, while 10 patients presenting with localized disease underwent radiation therapy as a definitive treatment for the primary tumor and one patient died during preoperative chemotherapy. Among the 59 patients presenting with localized disease, 54 (92%) patients underwent neoadjuvant and/or adjuvant chemotherapy. The majority of patients (n=48, 89%) received a chemotherapy regimen including vincristine, doxorubicin, cyclophosphamide with ifosfamide and etoposide (VDC/IE) ± irinotecan and temozolomide. Patients with localized disease harboring rare fusion variants, such as EWSR1-FEV (n=1) or EWSR1-NFATC2 (n=2), were also treated with neoadjuvant/adjuvant chemotherapy involving VDC/IE. Of 28 patients who received neoadjuvant/adjuvant chemotherapy with available data, 12 (43%) patients had more than 90% chemotherapy-induced necrosis in their resected specimens. The proportions of patients with favorable chemotherapy response, defined as > 90% necrosis, were not significantly different among the different fusion types (EWSR1-FLI1, 42%, 10 of 24; EWSR1/FUS-ERG, 67%, 2 of 3, p=0.482, however, this analysis did not have enough power due to small sample size. One such example of favorable pathologic response was encountered in a 1-year-old female who presented with a left pelvic sidewall soft tissue tumor harboring an EWSR1-ETV4 fusion (Fig. 3). The patient was treated with neoadjuvant/adjuvant chemotherapy with VDC/IE and irinotecan/temozolomide per our Ewing sarcoma regimen, as well as with radiation therapy after surgical resection. The patient remains alive with no evidence of disease 60 months follow-up.
Thirty-one patients (34%) presented with metastatic disease at diagnosis. The proportion of patients with metastasis at presentation was higher in patients with EWSR1/FUS-ERG (44%), EWSR1/FUS-FEV (75%) and EWSR1/FUS-NFATC2 (33%), compared to EWSR1-FLI1 (30%).
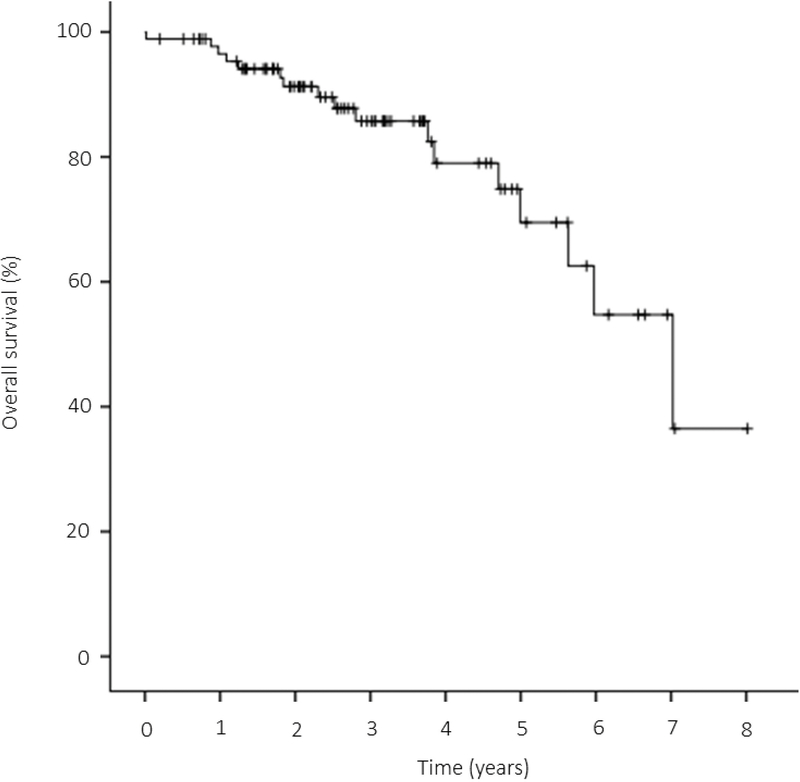
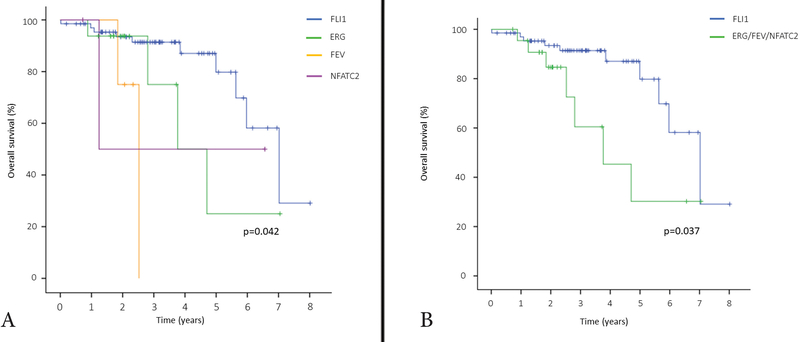
At the time of last follow-up, 73 patients (81%) were alive, including 41 (47%) without evidence of disease and 32 (34%) alive with disease. Fifteen patients (17%) died of disease and two patients (2%) experienced deaths of other causes. The 3-year and 5-year OS for the entire cohort were 85.7% (95% CI: 74.7% to 92.2%) and 69.5% (95% CI: 50.0% to 82.6%), respectively (Fig 6). Overall, the OS was significantly different among fusion types (p=0.042, Fig. 7A). The classic Ewing sarcoma group (EWSR1-FLI1, EWSR1/FUS-ERG, and EWSR1/FUS-FEV) showed a better outcome compared to round cell sarcoma with EWSR1-non-ETS fusions (EWS/FUS-NFATC2), with a 3-year OS of 86.6% vs 50.0%, respectively, however, the difference was not statistically significant (p=0.657). OS was also not significantly different between Ewing sarcoma patients with EWSR1-FLI1 versus EWSR1/FUS-ERG canonical fusions (p=0.167). However, when patients with EWSR1-FLI1 positive tumors were compared to all other fusion types in this cohort (EWSR1/FUS-ERG, EWSR1/FUS-FEV, and EWSR1/FUS-NFATC2) this subset was associated with a significantly better OS (3-year OS, 91.4% vs 60.5%, p=0.037, Fig. 7B).
Figure 6.
Kaplan-Meier curve depicting the overall survival of our cohort.
Figure 7. Kaplan-Meier curve illustrating the overall survival stratified by gene fusion type.
A. EWSR1-FLI1 (Blue); EWSR1/FUS-ERG (Green); EWSR1/FUS-FEV (Orange); EWSR1/FUS-NFATC2 (Purple). B. EWSR1-FLI1 (Blue); other fusion types including EWSR1/FUS-ERG, EWSR1/FUS-FEV, and EWSR1/FUS-NFATC2 (Green).
The overall 3-year and 5-year OS of the patients presenting with localized disease at diagnosis (n=59) were 93.2% (95% CI: 79.7%−97.8%) and 80.7% (95% CI: 55.0%−92.6%), respectively. In this clinical subset, no significant differences in OS were noted when comparing Ewing sarcoma (EWSR1-FLI1, EWSR1/FUS-ERG, and EWSR1/FUS-FEV) and round cell sarcomas with EWS/FUS-NFATC2 fusion (p=0.450), nor comparing EWSR1-FLI1 and EWSR1/FUS-ERG fusion variants (p=0.872). Although the 3-year OS appeared to be longer in the EWSR1-FLI1 genetic group compared to other fusion variants overall (EWSR1/FUS-ERG, EWSR1/FUS-FEV, and EWS/FUS-NFATC2), the difference was not statistically significant (95.3% vs 66.7%, respectively, p=0.585). In 36 patients with available EWSR1-FLI1 exonic composition, there was no significant difference in OS among type 1 (EWSR1 exon7 fused to FLI1 exon6) versus other variants (3-year OS, 100% vs 90.8%, respectively, p=0.377).
4. DISCUSSION
The advances and wide application of molecular studies in routine clinical practice have allowed a detailed genetic subclassification of Ewing sarcoma and round cell sarcomas with EWSR1-non-ETS fusions based on their recently described gene fusions9–12. As these novel molecular subsets are quite rare, large studies assessing their clinical presentation, pathologic features and survival are unavailable. Although methylation classifiers or/and gene expression clustering have suggested that lesions formerly known as Ewing-like sarcoma subsets, such as tumors with EWSR1-NFATC2 and EWSR1-PATZ1, are epigenetically and genomically distinct from conventional Ewing sarcoma with canonical fusions12,22,23, clinical and survival comparisons of these two genetic subgroups are lacking. Moreover, it is likely that a number of prior (pre-NGS) clinical studies of Ewing sarcoma, selected based on EWSR1 and FUS gene rearrangements by FISH, may have combined Ewing sarcoma and round cell sarcoma with EWSR1-non-ETS fusion patients under one group, and thus the clinical features and survival information might be biased.
Our results provide information regarding the incidence of round cell sarcoma with EWSR1-non-ETS fusions among a large group of Ewing family of tumors, which accounts for 6% (14/240) of the patients. Among the classic Ewing sarcoma group, the two most common fusion variants were EWSR1-FLI1 (73%; 176/240) and EWSR1/FUS-ERG fusions (15%; 35/240), followed by EWSR1/FUS-FEV (5%; 12/240), and EWSR1-ETV1/4 (1%; 3/240).
By comparing the clinical and pathologic features with the gene fusion type in this large cohort of Ewing sarcoma and round cell sarcoma with EWSR1-non-ETS fusion patients some interesting correlations emerged particularly related to age at diagnosis and primary anatomic site. Patients with round cell sarcomas harboring EWSR1/FUS-NFATC2 fusion had an older median age at diagnosis (43 years), while sarcoma patients harboring EWSR1-ETV1/4 fusions had the youngest median age at diagnosis, of 8 years old (range, 1–12). In fact, one-third of cases positive for EWSR1-ETV1/4 fusion occurred in very young children (aged <2 years). The single case with EWSR1-VEZF1 fusion occurred in a 12 year-old patient. Other smaller series have suggested similar correlations, with one infant patient harboring an EWSR1-ETV4 fusion5, and four patients with EWSR1/FUS-NFATC2 showed relatively increased median age (38 years), all presenting in the extremity24.
Patients with rare variant fusions such as EWSR1/FUS-FEV, EWSR1-ETV1/4, EWSR1-PATZ1, and EWSR1-VEZF1 showed the highest incidence of extraskeletal sites. In contrast Ewing sarcoma with canonical fusions showed a predilection for skeletal sites (58%) and pelvic and trunk (68%). Other smaller series have shown tumors with EWSR1-ETV4, EWSR1-PATZ1, or EWSR1-VEZF1 mainly arise from extraskeletal sites5,23,24. Previous studies of Ewing sarcoma with canonical fusions demonstrated a prevalence for skeletal primary sites in 73% of cases and of axial location in 62% of cases25,26, which was consistent with our results.
Our study highlights certain differences in the overall survival among various gene fusion groups, likely attributable to the higher metastatic rate at presentation in the patients with non-EWSR1-FLI1 fusions. Specifically, patients with EWSR1/FUS-ERG, EWSR1/FUS-FEV, and EWSR1/FUS-NFATC2 fusions were associated with higher rates of metastasis at presentation and worse overall survival. Prior data from the literature reveal discrepant results between outcome and fusion type/fusion transcript25–28. Although retrospective studies initially suggested that the EWSR1–FLI1 type 1 variant (EWSR1 exon 7 fused to FLI1 exon 6) was associated with better survival26,27, two subsequent prospective studies did not validate this observation25,26. In keeping with latter prospective data, patients with tumors harboring EWSR1-FLI1 type 1 variant was not associated with better survival outcome in our small dataset. In the de Alava study29, the median survival regardless of stage in the group of patients with fusion transcripts other than type 1 was 27 ± 3.8 months compared to 113 ± 2.2 months for the group with type 1 transcripts. In contrast, the survival data of our cohort demonstrates a significant improvement and the difference in outcome based on fusion type has diminished, presumably due to the improved chemotherapeutic regimens applied. Moreover, previous reports showed that Ewing sarcomas with either EWSR1–FLI1 or EWSR1–ERG have similar survival outcomes29. Restricting our survival analysis on patients presenting with localized disease at diagnosis, our results did not show a significant difference in overall survival between fusion subsets, although the number of evaluated patients in each group was relatively limited. Further studies using a large/international cohort are needed to determine a survival difference based on fusion type, particularly in patients with localized disease.
Previous studies have demonstrated that older age at diagnosis was associated with a worse outcome in Ewing sarcoma30,31. Whether this disparity in survival outcomes is attributable to differences in treatment modalities or sensitivity, tumor biology, or host microenvironment remains an area of active investigation. Moreover, our results point to a different layer of complexity, that of a variable predilection of certain fusion types for certain age groups, which may translate into different tumor biology and survival differences among age groups.
Some of the potential biological differences among various molecular Ewing sarcoma subsets have been investigated in a few recent studies12,23,32. Using a comprehensive whole-genome and transcriptome sequencing analysis, one study demonstrated that Ewing sarcomas with ERG gene rearrangements occur through an unbalanced, chromoplexy pattern of fusion, likely due to the opposite directions of transcriptions of the EWSR1 and ERG genes32. In that study, Ewing sarcoma patients harboring gene fusions through the chromoplexy process were more likely to relapse and had a high incidence of p53 mutations, compared to patients showing balanced EWSR1-FLI1 fusions32. These results correlate with our findings of a higher incidence of metastatic rate at presentation and worse OS in EWSR1-ERG fusions. Moreover, recent molecular studies described that EWSR1-NFATC2 or EWSR1-PATZ1 positive round cell sarcomas have distinct genomic and/or methylation signatures12,23 Unsupervised hierarchical clustering of DNA methylation data revealed a homogeneous methylation cluster for undifferentiated small round cell sarcoma with EWSR1-NFATC2 fusion, which clearly segregated from canonical Ewing sarcoma12. In addition, copy number profiles of EWSR1-NFATC2 cases showed characteristic recurrent losses on chromosome 9q and segmental gains on 20q13 and 22q12 involving the EWSR1 and NFATC2 loci, respectively12. In EWSR1-PATZ1-positive sarcomas, secondary driver mutations in cell-cycle genes, in particular CDKN2A (71%), were common23. Our current results, showing distinct clinicopathologic features of these two molecular subsets from classic Ewing sarcoma, provide further support in favor of them being stand-alone pathologic entities.
Our study had several limitations, most importantly the relatively small number of patients and events in some of the molecular subsets, which precluded conducting a multivariate survival analysis and hampered our ability to make definitive conclusions. Moreover, the patient cohort was limited to a single quaternary institution, which is likely to infer a referral bias towards more rare molecular subsets or patients with more aggressive behavior. Despite these limitations, the study’s strengths rely in providing detailed clinical and therapeutic information of a large, molecularly well-characterized, cohort of Ewing sarcoma and round cell sarcoma with EWSR1-non-ETS fusion group.
In summary, we outline a number of important correlations between the EWSR1/FUS gene fusion types and clinicopathologic features. Our data reveal a less favorable overall survival in patients with non-canonical fusions, such as EWSR1/FUS-ERG, EWSR1/FUS-FEV and EWSR1/FUS-NFATC2. This survival difference is likely due to the higher incidence of metastatic spread at presentation in these less common subsets. Larger, multi-institutional studies are needed to definitively evaluate if these differences in survival outcomes may persist in patients with localized disease.
Acknowledgments
Disclosures: Supported in part by: P50 CA140146 (CRA), P50 CA217694 (CRA), P30 CA008748, Cycle for Survival (CRA), Kristin Ann Carr Foundation (CRA), St. Baldrick’s Foundation (CRA), Alan Slifka Foundation (CRA)
Footnotes
Conflicts of Interests: none
Data Availability Statement: The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1.Delattre O, Zucman J, Plougastel B, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. [DOI] [PubMed] [Google Scholar]
- 2.Delattre O, Zucman J, Melot T, et al. The Ewing family of tumors--a subgroup of small-round-cell tumors defined by specific chimeric transcripts. NEJM. 1994;331:294–299. [DOI] [PubMed] [Google Scholar]
- 3.Peter M, Couturier J, Pacquement H, et al. A new member of the ETS family fused to EWS in Ewing tumors. Oncogene. 1997;14:1159–1164. [DOI] [PubMed] [Google Scholar]
- 4.Jeon IS, Davis JN, Braun BS, et al. A variant Ewing’s sarcoma translocation (7;22) fuses the EWS gene to the ETS gene ETV1. Oncogene. 1995; 10: 1229–1234. [PubMed] [Google Scholar]
- 5.Kaneko Y, Yoshida K, Handa M, et al. Fusion of an ETS-family gene, EIAF, to EWS by t(17;22)(q12;q12) chromosome translocation in an undifferentiated sarcoma of infancy. Genes Chromosomes & Cancer. 1996;15:115–121. [DOI] [PubMed] [Google Scholar]
- 6.Antonescu C Round cell sarcomas beyond Ewing: emerging entities. Histopathology. 2014;64:26–37. [DOI] [PubMed] [Google Scholar]
- 7.Mastrangelo T, Modena P, Tornielli S, et al. A novel zinc finger gene is fused to EWS in small round cell tumor. Oncogene. 2000;19:3799–3804. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Bhargava R, Zheng T, et al. Undifferentiated small round cell sarcomas with rare EWS gene fusions: identification of a novel EWS-SP3 fusion and of additional cases with the EWS-ETV1 and EWS-FEV fusions. J Mol Diagnostics. 2007;9:498–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szuhai K, Ijszenga M, de Jong D, Karseladze A, Tanke HJ, Hogendoorn PC. The NFATc2 gene is involved in a novel cloned translocation in a Ewing sarcoma variant that couples its function in immunology to oncology. Clin Cancer Res. 2009;15:2259–2268. [DOI] [PubMed] [Google Scholar]
- 10.Wang GY, Thomas DG, Davis JL, et al. EWSR1-NFATC2 Translocation-associated Sarcoma Clinicopathologic Findings in a Rare Aggressive Primary Bone or Soft Tissue Tumor. Am J Surg Pathol. 2019. [DOI] [PubMed] [Google Scholar]
- 11.Sumegi J, Nishio J, Nelson M, Frayer RW, Perry D, Bridge JA. A novel t(4;22)(q31;q12) produces an EWSR1-SMARCA5 fusion in extraskeletal Ewing sarcoma/primitive neuroectodermal tumor. Mod Pathol. 2011;24:333–342. [DOI] [PubMed] [Google Scholar]
- 12.Koelsche C, Kriegsmann M, Kommoss FKF, et al. DNA methylation profiling distinguishes Ewing-like sarcoma with EWSR1-NFATc2 fusion from Ewing sarcoma. J Cancer Res Clin Oncol. 2019;145:1273–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biermann JS, Chow W, Reed DR, et al. NCCN Guidelines Insights: Bone Cancer, Version 2.2017. J Natl Compr Canc Netw. 2017;15:155–167. [DOI] [PubMed] [Google Scholar]
- 14.Chen S, Deniz K, Sung YS, Zhang L, Dry S, Antonescu CR. Ewing sarcoma with ERG gene rearrangements: A molecular study focusing on the prevalence of FUS-ERG and common pitfalls in detecting EWSR1-ERG fusions by FISH. Genes Chromosomes & Cancer. 2016;55:340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antonescu CR, Zhang L, Chang NE, et al. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes & Cancer. 2010;49:1114–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Molec Diagnostics.. 2015;17:251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nature Medicine. 2017;23:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nature Medicine. 2014;20:1479–1484. [DOI] [PubMed] [Google Scholar]
- 19.Zhu G, Benayed R, Ho C, et al. Diagnosis of known sarcoma fusions and novel fusion partners by targeted RNA sequencing with identification of a recurrent ACTB-FOSB fusion in pseudomyogenic hemangioendothelioma. Mod Pathol. 2019;32:609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dickson BC, Sung YS, Rosenblum MK, et al. NUTM1 Gene Fusions Characterize a Subset of Undifferentiated Soft Tissue and Visceral Tumors. Am J Surg Pathol. 2018;42:636–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosquera JM, Sboner A, Zhang L, et al. Recurrent NCOA2 gene rearrangements in congenital/infantile spindle cell rhabdomyosarcoma. Genes Chromosomes & Cancer. 2013;52:538–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watson S, Perrin V, Guillemot D, et al. Transcriptomic definition of molecular subgroups of small round cell sarcomas. J Pathol. 2018;245:29–40. [DOI] [PubMed] [Google Scholar]
- 23.Bridge JA, Sumegi J, Druta M, et al. Clinical, pathological, and genomic features of EWSR1-PATZ1 fusion sarcoma. Mod Pathol. 2019. [DOI] [PubMed] [Google Scholar]
- 24.Bode-Lesniewska B, Fritz C, Exner GU, Wagner U, Fuchs B. EWSR1-NFATC2 and FUS-NFATC2 Gene Fusion-Associated Mesenchymal Tumors: Clinicopathologic Correlation and Literature Review. Sarcoma. 2019;2019: 9386390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Deley MC, Delattre O, Schaefer KL, et al. Impact of EWS-ETS fusion type on disease progression in Ewing’s sarcoma/peripheral primitive neuroectodermal tumor: prospective results from the cooperative Euro-E.W.I.N.G. 99 trial. J Clinical Oncol. 2010;28:1982–1988. [DOI] [PubMed] [Google Scholar]
- 26.van Doorninck JA, Ji L, Schaub B, et al. Current treatment protocols have eliminated the prognostic advantage of type 1 fusions in Ewing sarcoma: a report from the Children’s Oncology Group. J Clinical Oncol. 2010;28:1989–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ginsberg JP, de Alava E, Ladanyi M, et al. EWS-FLI1 and EWS-ERG gene fusions are associated with similar clinical phenotypes in Ewing’s sarcoma. J Clinical Oncol. 1999;17:1809–1814. [DOI] [PubMed] [Google Scholar]
- 28.Zoubek A, Dockhorn-Dworniczak B, Delattre O, et al. Does expression of different EWS chimeric transcripts define clinically distinct risk groups of Ewing tumor patients? J Clinical Oncol. 1996;14:1245–1251. [DOI] [PubMed] [Google Scholar]
- 29.de Alava E, Kawai A, Healey JH, et al. EWS-FLI1 fusion transcript structure is an independent determinant of prognosis in Ewing’s sarcoma. J Clinical Oncol. 1998;16:1248–1255. [DOI] [PubMed] [Google Scholar]
- 30.Lee J, Hoang BH, Ziogas A, Zell JA. Analysis of prognostic factors in Ewing sarcoma using a population-based cancer registry. Cancer. 2010;116:1964–1973. [DOI] [PubMed] [Google Scholar]
- 31.Cotterill SJ, Ahrens S, Paulussen M, et al. Prognostic factors in Ewing’s tumor of bone: analysis of 975 patients from the European Intergroup Cooperative Ewing’s Sarcoma Study Group. J Clinical Oncol. 2000;18:3108–3114. [DOI] [PubMed] [Google Scholar]
- 32.Anderson ND, de Borja R, Young MD, et al. Rearrangement bursts generate canonical gene fusions in bone and soft tissue tumors. Science. 2018;361. [DOI] [PMC free article] [PubMed] [Google Scholar]