Abstract
Background
Docosahexaenoic acid (DHA) is an important omega-3 long-chain polyunsaturated fatty acid that has a variety of physiological functions for infant development and human health. Although metabolic engineering was previously demonstrated to be a highly efficient way to rapidly increase lipid production, metabolic engineering has seldom been previously used to increase DHA accumulation in Schizochytrium spp.
Results
Here, a sensitive β-galactosidase reporter system was established to screen for strong promoters in Schizochytrium sp. Four constitutive promoters (EF-1αp, TEF-1p, ccg1p, and ubiquitinp) and one methanol-induced AOX1 promoter were characterized by the reporter system with the promoter activity ccg1p> TEF-1p > AOX1p (induced) > EF-1αp > ubiquitinp. With the strong constitutive promoter ccg1p, Schizochytrium ATP-citrate lyase (ACL) and acetyl-CoA carboxylase (ACC) were overexpressed in Schizochytrium sp. ATCC 20888. The cells were cultivated at 28 °C and 250 rpm for 120 h with glucose as the carbon source. Shake-flask fermentation results showed that the overexpression strains exhibited growth curves and biomass similar to those of the wild-type strain. The lipid contents of the wild-type strain and of the OACL, OACC, and OACL-ACC strains were 53.8, 68.8, 69.8, and 73.0%, respectively, and the lipid yields of the overexpression strains were increased by 21.9, 30.5, and 38.3%, respectively. DHA yields of the wild-type strain and of the corresponding overexpression strains were 4.3, 5.3, 6.1, and 6.4 g/L, i.e., DHA yields of the overexpression strains were increased by 23.3, 41.9, and 48.8%, respectively.
Conclusions
Acetyl-CoA and malonyl-CoA are precursors for fatty acid synthesis. ACL catalyzes the conversion of citrate in the cytoplasm into acetyl-CoA, and ACC catalyzes the synthesis of malonyl-CoA from acetyl-CoA. The results demonstrate that overexpression of ACL and ACC enhances lipid accumulation and DHA production in Schizochytrium sp.
Keywords: Schizochytrium sp., Docosahexaenoic acid, ATP-citrate lyase, Acetyl-CoA carboxylase, β-Galactosidase reporter system, Constitutive promoter
Background
Docosahexaenoic acid (DHA, C22:6-∆4,7,10,13,16,19) is an omega-3 long-chain polyunsaturated fatty acid (LC-PUFA). As the principal omega-3 fatty acid in brain gray matter, DHA has neurotrophic and neuroprotective properties that are required for normal perinatal cortical maturation [1]. In addition, DHA supplementation improves human health by increasing cardioprotective, anti-inflammatory, and anticancer activities [2, 3]. DHA is therefore widely used as a nutritional supplement, often as a nutraceutical.
The conventional source of DHA is fish oil obtained from cold-water marine fish. Seasonal variation, overharvest, and population decline, however, prevent the steady supply of DHA that is required to meet the increasing market demands. Other commercial sources of DHA production are thraustochytrids, which are marine microorganisms [4]. Schizochytrium spp., as well as other thraustochytrids (such as species of Thraustochytrium and Ulkenia), are excellent DHA producers [5, 6]. Schizochytrium spp. can produce total fatty acids (TFAs) that represent up to 70% of the cell weight, with DHA representing 25–45% of TFAs [7, 8]. Owing to the increasing demand for DHA, many researchers have attempted to increase DHA production by Schizochytrium spp. [5, 6, 9]. To date, most studies of DHA production by Schizochytrium spp. have focused on the adaptive evolution of the strains [9]; on the optimization of medium composition including sources of carbon and nitrogen and the addition of inorganic salts and antioxidants [5, 6, 10, 11]; and on cultivation conditions and cultivation styles [12, 13]. Only a few studies have employed metabolic engineering to increase DHA accumulation in Schizochytrium. Yan et al. [14], for example, introduced the Escherichia coli acetyl-CoA synthase gene into Schizochytrium sp. TIO1101, which increased the biomass and TFA production of the resulting transformant by 29.9% and 11.3%, respectively. Introduction of an exogenous ω-3 desaturase gene into Schizochytrium sp. converted 3% docosapentaenoic acid (DPA) into DHA [15]. By increasing the number of active ACP domains of PUFA synthase, DHA productivity was increased by 1.8-fold in a recombinant E. coli expressing Schizochytrium PUFA biosynthetic genes [16]. These studies demonstrate that metabolic engineering can increase DHA production by Schizochytrium spp.
Metabolic engineering has also been used with the oleaginous yeast Yarrowia lipolytica, i.e., metabolic engineering efficiently increased the yeast’s production of total lipids and ω-3 PUFAs [17–20]. By rewiring the metabolic pathways of Y. lipolytica, researchers increased lipid accumulation > 60-fold, and caused lipid content to approach 90% of cell mass [18]. Compared to the 10–15% lipid content in wild-type (WT) Y. lipolytica [21], Schizochytrium spp. produces much higher lipid levels, and ω-3 PUFA DHA represents up to 45% of TFAs. Because the genome sequences of several strains of thraustochytrids (Schizochytrium, Thraustochytrium, and Aurantiochytrium) are now available [22–24], metabolic engineering should be an efficient way to rapidly increase their production of DHA and lipids.
Acetyl-CoA is precursor for fatty acid synthesis. ATP-citrate lyase (ACL) catalyzes the conversion of citrate and CoA into acetyl-CoA and oxaloacetate, along with the hydrolysis of ATP [25]. ACL is present in all eukaryotes except non-oleaginous yeasts. In animals and oleaginous basidiomycete yeasts, ACL is encoded by a single gene [26, 27]; in plants and some filamentous fungi, ACL usually consists of two subunits (ACL1 and ACL2) with homology to the N- and C-terminals of the animal ATP-citrate lyase polypeptide [28]. Fatty acid content was increased in Y. lipolytica by overexpression of ACL1 and ACL2 on a non-lipogenic medium in an obese strain [29] or overexpression of ACL from Mus musculus [30]. Acetyl-CoA carboxylase (ACC) catalyzes the synthesis of malonyl-CoA from acetyl-CoA, which is the rate-limiting step in fatty acid synthesis [21]. There are two types of ACCs in nature: in most bacteria and plant chloroplasts, ACC usually consists of multiple subunits, including the biotin carboxylase (BC), the biotin carboxyl carrier protein (BCCP), the α-carboxyltransferase (α-CT) and the β-carboxyltransferase (β-CT); but in mammals, fungi, and the cytoplasm of most plants, ACC is a single multifunctional polypeptide [31]. Overexpression of acetyl-CoA carboxylase in the presence of thioesterase in E. coli led to a sixfold increase in the rate of fatty acid synthesis [32]. ACC overexpression increased lipid content in Y. lipolytica and free fatty acid production in S. cerevisiae [21, 33].
To date, very few studies have attempted to enhance DHA production in Schizochytrium spp. through metabolic engineering. In this study, a sensitive β-galactosidase reporter system was established in Schizochytrium sp. to screen for strong promoters. Because a sufficient supply of acetyl-CoA and malonyl-CoA is a prerequisite for efficient lipid accumulation, Schizochytrium ACL and ACC were overexpressed under the strong constitutive promoter ccg1p in Schizochytrium sp. ATCC 20888 to enhance lipid accumulation and DHA production.
Results
Developing a β-galactosidase reporter system in Schizochytrium
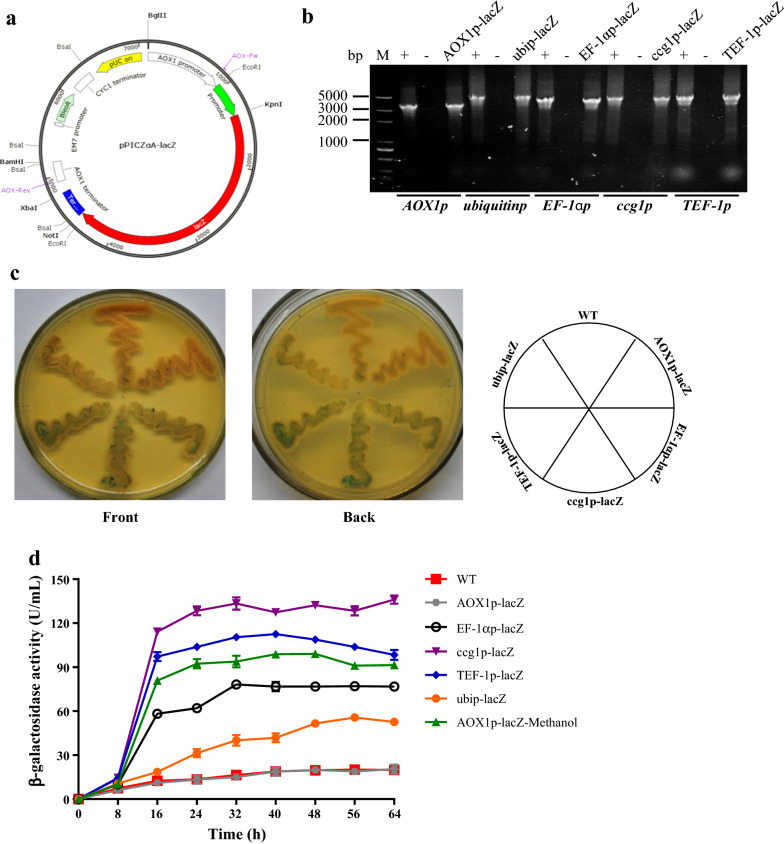
Metabolic engineering involves the rewiring various metabolic pathways to redirect metabolic flux towards the synthesis of target compounds. As a consequence, the expression of relevant pathways must be strictly coordinated to achieve a balanced expression and to avoid metabolic bottlenecks [34]. It is therefore crucial that a reliable reporter system to monitor gene expression levels is established in Schizochytrium. The E. coli β-galactosidase structural gene lacZ has been widely used as a candidate reporter gene, providing convenient methods for qualitative colorimetric detection on agars with 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-gal) and quantitative β-galactosidase activity assays with O-nitrophenyl-β-d-galactopyranoside (ONPG) [35]. To determine whether the β-galactosidase reporter works in Schizochytrium, we constructed the reporter plasmid pPICZαA-ubiquitinp-lacZ, in which the E. coli lacZ gene was driven by a ubiquitin promoter–terminator system (Fig. 1a). pPICZαA containing lacZ without a ubiquitin promoter (termed pPICZαA-AOX1p-lacZ) was also constructed as a control plasmid. The corresponding transformants of Schizochytrium sp. ATCC 20888 were selected on glucose–peptone–yeast extract (GPY) plates with zeocin. DNA fragments of 5.0 and 3.3 kb were amplified from the genomic DNAs of the ubip-lacZ and AOX1p-lacZ transformants, respectively; the sizes of the fragments corresponded with the sizes of the lacZ expression cassettes (Fig. 1b), indicating that both plasmids were integrated into chromosomes.
Fig. 1.
Development of a β-galactosidase reporter system and selection of strong promoters in Schizochytrium sp. a Construction of reporter plasmid pPICZαA-lacZ. b PCR verification of transformants. +: positive control, reporter plasmid was used as template; −: negative control, WT genomic DNA was used as template; AOX1p-lacZ, ubip-lacZ, EF-1αp-lacZ, ccg1p-lacZ, and TEF-1p-lacZ: the transformants of pPICZαA-AOX1p-lacZ, pPICZαA-ubiquitinp-lacZ, pPICZαA-EF-1αp-lacZ, pPICZαA-ccg1p-lacZ, and pPICZαA-TEF-1p-lacZ. c Phenotypes of the corresponding transformants. Cells were grown for 96 h on GPY plates with 40 μg/mL X-gal. d β-galactosidase enzymatic activities of the transformants. Cells were grown in seed medium for 64 h
The transformants of ubip-lacZ and AOX1p-lacZ grew normally on GPY agar. When the substrate X-gal was added to GPY plates, Schizochytrium sp. WT produced orange colonies with a very slight blue color, indicating that the WT possessed endogenous β-galactosidase with very low activity. AOX1p-lacZ produced colonies that were similar to those of the WT, while ubip-lacZ produced blue colonies (Fig. 1c). β-galactosidase enzymatic activity was much higher in the lysate of the ubip-lacZ transformant than in the AOX1p-lacZ transformant (Fig. 1d). These results show that, although Schizochytrium WT possesses endogenous β-galactosidase activity, β-galactosidase is able to serve as a sensitive reporter system in Schizochytrium.
Selection of strong promoters in Schizochytrium
The use of four commonly used eukaryotic promoters (EF-1αp, ccg1p, TEF-1p, and AOX1p) in addition to ubiquitinp was characterized in the β-galactosidase reporter system. In pPICZαA-AOX1p-lacZ, a methanol-induced AOX1p [36] is present upstream of the lacZ gene. On X-gal plates, AOX1p-lacZ produced orange colonies like those of the WT, but the transformants with other promoters produced blue colonies, and the transformants with ccg1p or TEF-1p produced the bluest colonies. β-galactosidase activity in AOX1p-lacZ without methanol induction treatment was similar to that in the WT (Fig. 1c), indicating that AOX1p is inactive without induction by methanol. The enzymatic activities driven by ccg1p, TEF-1p, EF-1αp, or ubiquitinp were much higher than that of WT, with promoter activity of ccg1p > TEF-1p > EF-1αp > ubiquitinp (Fig. 1d). These findings indicate that the four promoters are strong, constitutively expressed promoters. When methanol was added to a 12-h culture of AOX1p-lacZ to a final concentration of 1% (vol/vol), the β-galactosidase activity increased substantially and remained at a high level, with the induction strength between TEF-1p and EF-1αp, indicating that AOX1p can be recognized by Schizochytrium RNA polymerase and induced by methanol. Therefore, AOX1 promoter can serve as a methanol-induced promoter in Schizochytrium, although the induction time and strength require optimization.
Construction of ACL and ACC-overexpression strains in Schizochytrium sp.
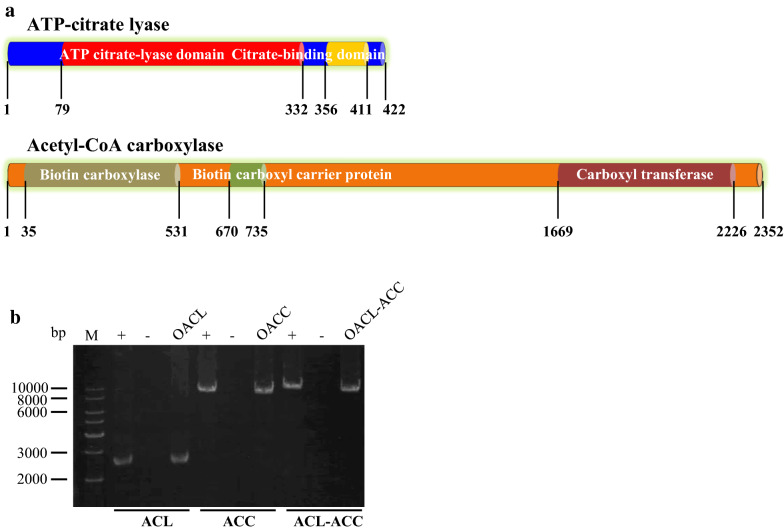
In cytoplasm, ATP-citrate lyase converts intracellular citrate to acetyl-CoA in an ATP-dependent manner, and acetyl-CoA carboxylase catalyzes the synthesis of malonyl-CoA from acetyl-CoA [25]. Acetyl-CoA and malonyl-CoA are the precursors for fatty acids synthesis [21, 29]. A BLAST search of the Schizochytrium sp. CCTCC M209059 genome [22] revealed one putative ACL-encoding gene and one putative ACC-encoding gene (Additional file 2: Table S1). The putative ACL (422 aa) contains an ATP-citrate (pro-S)-lyase domain at the N-terminus, which is homologous to ATP-citrate lyase subunit 1, and a citrate-binding domain at the C-terminus, which is homologous to ATP-citrate lyase subunit 2 (Fig. 2a). Therefore, Schizochytrium ATP-citrate lyase functions as a single-subunit ACL. Like ACC in most fungi, ACC (2352-aa) in Schizochytrium is a single multifunctional polypeptide, containing a biotin carboxylase (BC) domain, a biotin carboxyl carrier protein (BCCP) domain, and a carboxyl transferase (CT) domain (Fig. 2a).
Fig. 2.
Overexpression of ATP-citrate lyase and acetyl-CoA carboxylase in Schizochytrium sp. a Schematic diagram of Schizochytrium sp. ACL and ACC. The proposed enzymatic domains are marked in different colors. b PCR verification of the transformants. +: positive control, the plasmid was used as template; −: negative control, WT genomic DNA was used as template; OACL, OACC, and OACL-ACC: genomic DNA from the transformants of pPICZαA-ACL, pPICZαA-ACC, and pPICZαA-ACL-ACC was used as template
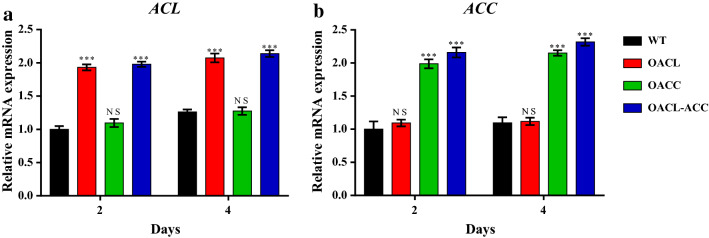
To promote lipid biosynthesis and DHA accumulation in Schizochytrium sp. ATCC 20888, we developed transformants that overexpressed Schizochytrium ATP-citrate lyase and acetyl-CoA carboxylase. ACL and ACC genes were amplified from the cDNA of Schizochytrium sp. ATCC 20888 and were cloned separately or together into pPICZαA, in which the cloned gene was driven by the ccg1 promoter and terminator (Additional file 1: Figure S1). After transformation, 2.8-, 8.7-, and 12.0-kb PCR fragments containing the corresponding expression cassettes (ccg1p-ACL-ccg1t, ccg1p-ACC-ccg1t, and ccg1p-ACL-ccg1t-ccg1p-ACC-ccg1t) were amplified from the genomic DNAs of pPICZαA-ACL, pPICZαA-ACC, and pPICZαA-ACL-ACC transformants (termed OACL, OACC, and OACL-ACC) (Fig. 2b), indicating that the plasmids were integrated into the chromosomes. The transcription levels of ACL and ACC were examined by qRT-PCR in WT, OACL, OACC, and OACL-ACC cultivated in fermentation broth for 2 and 4 d. Compared to WT, the expression of ACL were increased in OACL and OACL-ACC, and the expression of ACC were increased in OACC and OACL-ACC at both time points, indicating that transcription levels of ACL and ACC were increased in the corresponding overexpression strains (Fig. 3).
Fig. 3.
qRT-PCR analysis of the transcription levels of ACL and ACC in WT, OACL, OACC, and OACL-ACC. RNAs were isolated from WT, OACL, OACC, and OACL-ACC grown in fermentation media for 2 and 4 days. P values were determined by Student’s t test. ***P < 0.001; NS, not significant
ACL and ACC overexpression enhanced lipid accumulation
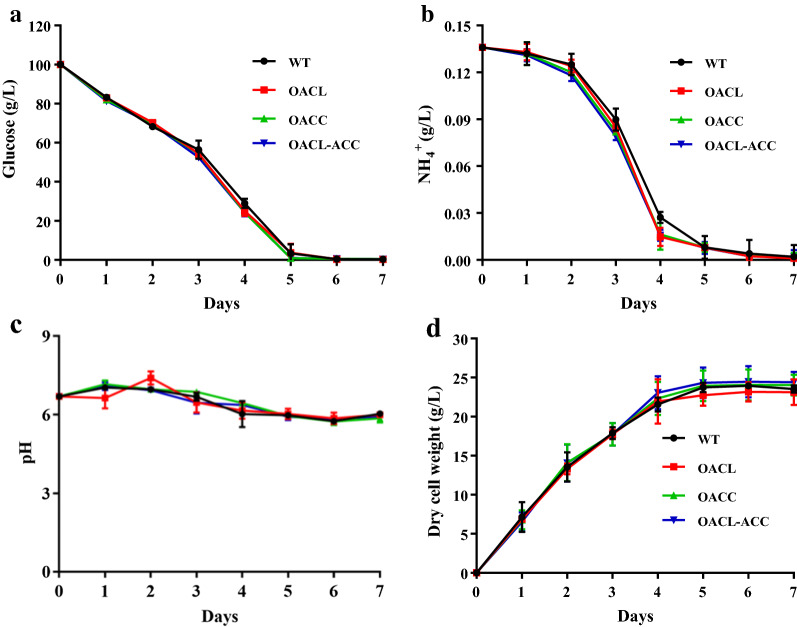
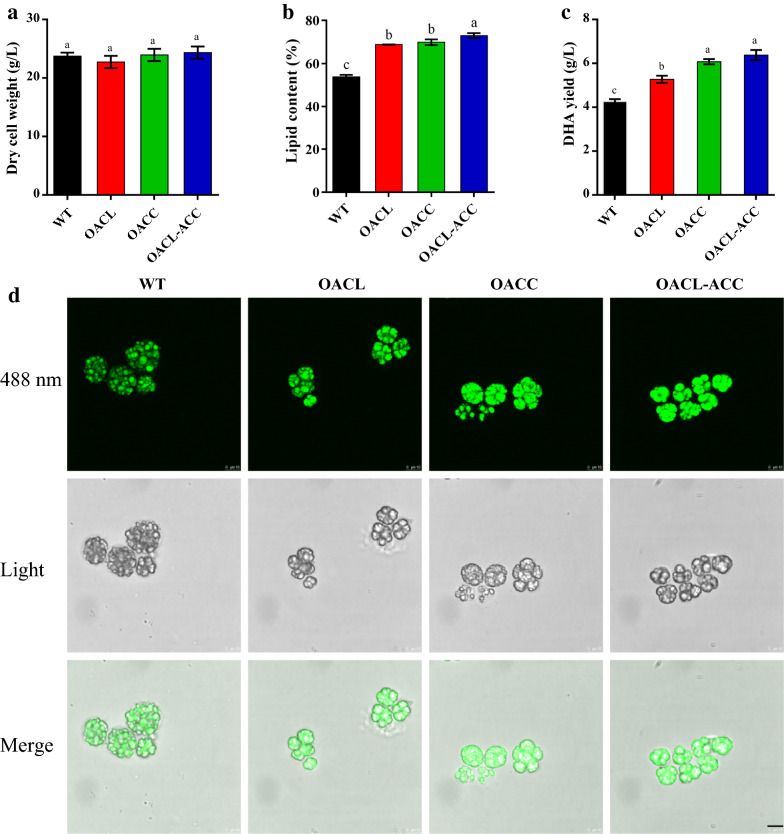
Shake-flask fermentation results showed that the WT strain and the overexpression transformants did not significantly differ in their consumption of carbon and nitrogen sources, pH values, or dry cell weights (DCW) (Fig. 4). DCW reached their maximum values on day 5, which were 23.7, 22.7, 23.9, and 24.3 g/L for WT, OACL, OACC, and OACL-ACC, and then decreased slightly with further cultivation (Figs. 4d, 5a). Thus, overexpression of ACL and/or ACC did not affect cell growth of Schizochytrium.
Fig. 4.
Time course of fermentation profiles of Schizochytrium sp. WT, OACL, OACC, and OACL-ACC. a Glucose (g/L); b NH4+ (g/L); c pH; d dry cell weight (DCW, g/L)
Fig. 5.
Effect of ACL and ACC overexpression on lipid accumulation and DHA production by Schizochytrium sp. a Dry cell weight (DCW, g/L). b Total lipids (% DCW). c DHA yield (g/L). Cells were cultured in fermentation medium for 120 h. d Imaging analyses of WT, OACL, OACC, and OACL-ACC. The 48-h cultured cells were stained with Nile red dye for neutral lipid staining. Scale bar, 10 µm. Error bars: SD from three independent experiments. Data were analyzed by one-way ANOVAs and Duncan’s multiple range tests in SPSS version 23.0. In a–c, columns with different lowercase letters are significantly different at P < 0.05
After 5 days of cultivation, lipid yields were lowest for the WT (12.8 g/L), highest for OACL-ACC (17.7 g/L), and intermediate for OACL (15.6 g/L), OACC (16.7 g/L) (Table 1). A similar pattern was evident for lipid content, i.e., lipid content was substantially higher in the overexpression strains than in the WT (Fig. 5b). Compared to the WT strain, the lipid yields were increased by 21.9% in OACL, by 30.5% in OACC, and by 38.3% in OACL-ACC. Microscopic observation revealed an increased intensity of lipid droplet staining in the overexpression strains (Fig. 5d). The findings indicated that overexpression of ACL and ACC increased lipid production in Schizochytrium sp., probably by increasing the supply of acetyl-CoA and malonyl-CoA.
Table 1.
Fermentation characteristics of strains of Schizochytrium sp.
| Strains | DCW (g/L) | Lipid yield (g/L) | Lipid content (%) | DHA yield (g/L) | DHA content (%) |
|---|---|---|---|---|---|
| WT | 23.7 ± 0.6a | 12.8 ± 0.5d | 53.8c | 4.3 ± 0.1c | 36.4b |
| OACL | 22.7 ± 1.0a | 15.6 ± 0.4c | 68.8b | 5.3 ± 0.2b | 36.2b |
| OACC | 23.9 ± 1.1a | 16.7 ± 0.7b | 69.8b | 6.1 ± 0.1a | 37.6a |
| OACL-ACC | 24.3 ± 1.1a | 17.7 ± 0.3a | 73.0a | 6.4 ± 0.2a | 37.9a |
Cells were cultured in fermentation medium for 120 h. Data were analyzed by one-way ANOVAs and Duncan’s multiple range tests in SPSS version 23.0. Values with different lowercase letters are significantly different at P < 0.05
ACL and ACC overexpression promoted DHA production
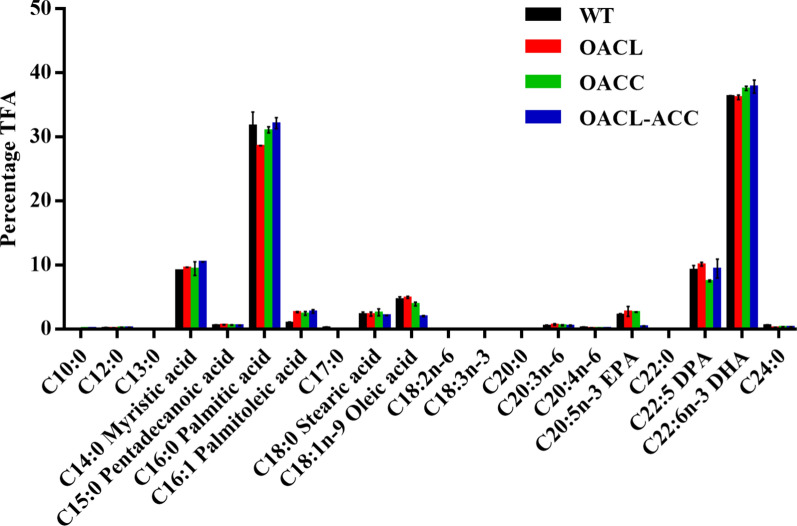
Gas chromatography analysis showed that the main fatty acid components of Schizochytrium sp. ATCC 20888 were DHA, palmitic acid (C16:0), myristic acid (C14:0), and docosapentaenoic acid (DPA, C22:5) (Fig. 6). Overexpression of ACL in Schizochytrium sp. WT did not significantly affect the percentage of TFAs represented by DHAs (36.4% for the WT, and 36.2% for OACL) (Fig. 6), but slightly decreased the percentage represented by palmitic acid and slightly increased the percentage represented by C16:1. In ACC-overexpression strains, the percentage of TFAs represented by DHAs increased slightly for OACC (37.6%) and OACL-ACC (37.9%), and the percentage represented by oleic acid and DPA decreased slightly (Fig. 6). Compared to the DHA yield of the WT (4.3 g/L), the DHA yields of OACL, OACC, and OACL-ACC strains were 5.3, 6.1, and 6.4 g/L, increased by 23.3, 41.9, and 48.8%, respectively (Fig. 5c). These results indicated that overexpression of ACL and ACC greatly increased DHA production in Schizochytrium sp. ATCC 20888.
Fig. 6.
Effect of ACL and ACC overexpression on fatty acid composition (TFA, %) of Schizochytrium sp. Cells were cultured in fermentation medium for 120 h
Discussion
In this study, a sensitive β-galactosidase reporter system in Schizochytrium was developed and used to compare the strengths of some commonly used eukaryotic promoters. Although endogenous β-galactosidase is present in Schizochytrium, the reporter system was able to detect different levels of β-galactosidase activity with the LacZ-reporter driven by different promoters. ccg1p, TEF-1p, EF-1αp, and ubiquitinp promoters are constitutive promoters and the AOX1p promoter is inducible by methanol in Schizochytrium. More work can be carried out for subsequent research, such as screening for more endogenous constitutive promoters with different expression intensities and optimization of the induction conditions for the methanol-induced AOX1p promoter. The β-galactosidase reporter system will facilitate characterization of novel genetic elements and will help identify promoters for fine-tuning gene expression in Schizochytrium.
Acetyl-CoA and malonyl-CoA are precursors for fatty acid synthesis [37]. Previous studies have shown that increasing the substrates supply significantly enhanced the synthesis of fatty acids and lipids in bacterium, yeast and fungi [30, 32, 38]. In the study, lipid and DHA production were greatly increased by overexpression of ACL and ACC. ACL converts intracellular citrate to acetyl-CoA [29], and ACC catalyzes the synthesis of malonyl-CoA from acetyl-CoA [21]. It follows that overexpression of ACL and ACC evidently promoted production of intracellular acetyl-CoA and malonyl-CoA, resulting in enhanced biosynthesis of fatty acids, which in turn promoted lipid accumulation. The percentage of TFAs represented by DHAs in ACL-overexpression strain was similar to that in the WT, indicating that overexpression of the ACL gene led to increased intracellular acetyl-CoA pools, which promoted the production of both saturated fatty acids and polyunsaturated fatty acids. In ACC-overexpression strains, the percentage of TFAs represented by DHAs improved slightly, while the percentage of TFAs represented by oleic acid and DPA decreased slightly. Thus, the increased malonyl-CoA pool in ACC-overexpression strains increased DHA production more than saturated fatty acid production. The findings suggested that the supply of malonyl-CoA is the limiting factor of DHA overproduction in Schizochytrium. In oleaginous microorganisms, efficient fatty acid synthesis requires not only an abundant supply of acetyl-CoA and malonyl-CoA, but also an ample NADPH supply [38, 39]. Therefore, combining this strategy with other strategies rewiring the metabolic flux of Schizochytrium towards precursors and NADPH accumulation might significantly improve its lipogenesis capability and DHA productivity.
Schizochytrium spp. are excellent producers of ω-3 PUFA: they can synthesize DHA de novo and DHA represents up to 45% of TFAs [7, 8]. To date, most studies of Schizochytrium spp. have focused on the optimization of fermentation media and cultivation conditions and the adaptive evolution of the strains [5, 6, 9–12]. Very few studies have employed metabolic engineering to improve DHA production of Schizochytrium spp. [14–16], mainly because the genetic background of Schizochytrium remains poorly understood. Metabolic engineering has been proven to be a highly efficient way to increase total lipids and ω-3 PUFA accumulation in the yeast Y. lipolytica [17–20]. In the current investigation, we demonstrated that metabolic engineering is an efficient way for increasing lipid accumulation and DHA production in Schizochytrium. The study provided a sensitive reporter system to monitor gene expression levels in Schizochytrium and a genetically engineered Schizochytrium sp. for industrial production of DHA.
Conclusions
A strain of Schizochytrium sp. ATCC 20888 that overexpressed ACL and ACC under the strong constitutive promoter ccg1p was constructed and thereby produced high quantities of DHA. Under shake-flask culture conditions, OACL, OACC, and OACL-ACC strains attained a dry cell weight of 22.7, 23.9, and 24.3 g/L, respectively. Compared to the WT, total lipid content of OACL, OACC, and OACL-ACC strains reached 68.8, 69.8, and 73.0%, respectively, and the lipid yields of the overexpression strains were increased by 21.9, 30.5, and 38.3%, respectively. A final DHA yield of 6.4 g/L in OACL-ACC was achieved, which was 48.8% higher than that of the WT. Next, fermentation control of OACL-ACC in fermentors will be optimized to make it more suitable for industrial application.
Methods
Microorganisms and culture conditions
Strains and plasmids used in the study are listed in Table 2. Media and growth conditions of Schizochytrium sp. were according to Ling et al. [5] with modifications. Schizochytrium sp. was cultured at 28 °C on solid GPY medium containing per liter 20 g of glucose, 10 g of peptone, 5 g of yeast extract, 20 g of sea crystal, and 20 g of agar. Transformants were selected and cultured on GPY supplemented with 40 μg/mL zeocin. For lipid and DHA production, 250-mL flasks containing 50 mL of seed medium (containing per liter 30 g of glucose, 10 g of peptone, 5 g of yeast extract, and 20 g of sea crystal) were inoculated with Schizochytrium sp. cells and incubated for 24 h at 28 °C on a rotary shaker (230 rpm). The seed culture was inoculated at 5% (vol/vol) into 50 mL of fermentation medium (containing per liter 100 g of glucose, 5 g of yeast extract, 3.94 g of NaCl, 0.264 g of KCl, 0.5 g of (NH4)2SO4, 1 g of KH2PO4, 1.43 g of MgSO4, 0.04 g of CaCl2, 10 g of sodium glutamate, 0.001 g of vitamin B1, and 0.001 g of vitamin B12) and was then incubated at 28 °C on a rotary shaker (250 rpm) for 168 h. For qualitative colorimetric and quantitative detection of β-galactosidase, cells were cultured on GPY plates with 40 μg/mL X-gal and in seed medium, respectively. E. coli was grown in LB medium at 37 °C [35].
Table 2.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Schizochytrium sp. | ||
| ATCC 20888 | Wild-type strain (WT) | American Type Culture Collection |
| AOX1p-lacZ | WT strain carrying pPICZαA-AOX1p-lacZ | This study |
| ubip-lacZ | WT strain carrying pPICZαA-ubiquitinp-lacZ | This study |
| TEF-1p-lacZ | WT strain carrying pPICZαA-TEF-1p –lacZ | This study |
| EF-1αp-lacZ | WT strain carrying pPICZαA-EF-1αp-lacZ | This study |
| ccg1p-lacZ | WT strain carrying pPICZαA-ccg1p-lacZ | This study |
| OACL | ACL overexpression strain | This study |
| OACC | ACC overexpression strain | This study |
| OACL-ACC | ACL and ACC co-overexpression strain | This study |
| E. coli | ||
| JM109 | General cloning host for plasmid manipulation | Laboratory stock |
| Plasmids | ||
| pPICZαA | Yeast expression vector | [42] |
| pPICZαA-AOX1p-lacZ | lacZ reporter vector using AOX1 promoter and terminator | This study |
| pPICZαA-ubiquitinp-lacZ | lacZ reporter vector using ubiquitin promoter and terminator | This study |
| pPICZαA-TEF-1p-lacZ | lacZ reporter vector using TEF-1 promoter and terminator | This study |
| pPICZαA-EF-1αp-lacZ | lacZ reporter vector using EF-1α promoter and terminator | This study |
| pPICZαA-ccg1p-lacZ | lacZ reporter vector using ccg1 promoter and terminator | This study |
| pPICZαA-ACL | ACL overexpression vector based on pPICZαA | This study |
| pPICZαA-ACC | ACC overexpression vector based on pPICZαA | This study |
| pPICZαA-ACL-ACC | ACL and ACC co-overexpression vector based on pPICZαA | This study |
Construction of β-galactosidase reporter plasmids and of ACL-, and ACC-overexpression plasmids
To construct β-galactosidase reporter plasmids, a 3075-bp fragment containing the coding sequence of the lacZ gene was amplified from pMC1403 [35] by PCR using primer pair lac-Fw and lac-Rev (Additional file 2: Table S2). The promoters and terminators of EF-1α [40] and ubiquitin [15] were amplified from Schizochytrium sp. ATCC 20888; ccg1 promoter and terminator were amplified from the Neurospora expression vector pCCG.N-3xMyc [41]; and the TEF-1 promoter and CYC-1 terminator were amplified from the yeast expression vector pPICZαA [42] using the primer pairs listed in Additional file 2: Table S2. After purification, the lacZ gene was digested with KpnI/NotI, the promoters were digested with EcoRI/KpnI, and the terminators were digested with NotI/XbaI; the digested gene, promoters, and terminators were then simultaneously ligated into EcoRI/XbaI-digested pPICZαA to generate reporter plasmids. Ligation reactions were performed overnight at 16 °C using T4 DNA Ligase (TaKaRa, Japan).
To construct the ACL overexpression plasmid, a 1269-bp fragment of the ACL gene was amplified from Schizochytrium sp. ATCC 20888 cDNA. After purification, the KpnI/NotI-digested ACL gene, EcoRI/KpnI-digested ccg1p, and NotI/XbaI-digested ccg1t were simultaneously ligated into EcoRI/XbaI-digested pPICZαA to generate the overexpression plasmid pPICZαA-ACL. For construction of the ACC overexpression plasmid, a 7059-bp ACC fragment was amplified from the same cDNA and purified. The ACC gene and the ccg1 promoter and terminator were inserted into EcoRI-digested pPICZαA to generate pPICZαA-ACC using the Seamless assembly cloning kit (Clone Smarter, USA) following the manufacture’s protocol. To construct the ACL and ACC co-overexpression plasmid, the 2611-bp ccg1p-ACL-ccg1t expression cassette was amplified from plasmid pPICZαA-ACL and was inserted into EcoRI-digested pPICZαA-ACC to generate pPICZαA-ACL-ACC by Seamless assembly cloning.
Transformation of Schizochytrium sp.
Transformation of Schizochytrium was performed as described previously with modification [15]. Schizochytrium sp. cells were cultured in seed medium for 24 h to the logarithmic growth phase, and were harvested by centrifugation (5900g, 4 °C, 10 min) (HITACHI CF16RXII, Japan), washed with ice-cold sterile water, washed with 1 M sorbitol, and then suspended in 1 M sorbitol. The plasmids were linearized with restriction enzyme BamHI before transformation. The competent cells and 5 μg of linearized plasmid DNA were placed in a 0.1-cm-gap cuvette. The parameters of electroporation were 0.75 kV, 200 Ω, 50 μF, twice. After electroporation, 1 mL of seed medium was added to the mixture, which was incubated at 28 °C for 4 h. The transformants were spread on GPY plates with 40 μg/mL zeocin and grown at 28 °C.
Genomic PCR analysis of transformants
Genomic DNAs of putative transformants were extracted according to Lippmeier et al. [43]. To confirm Schizochytrium sp. transformants, the incorporation of the expression cassette into the genome was verified by PCR using primers AOX-Fw and AOX-Rev (Additional file 2: Table S2). PCR reactions were set up using Taq DNA polymerase (TaKaRa, Japan) following the manufacture’s protocol. PCR amplification parameters were as follows: 5 min at 95 °C; followed by 30 cycles of 50 s at 95 °C, 50 s at 55 °C, and 5 min at 72 °C; and a final extension for 10 min at 72 °C.
β-Galactosidase activity assay
Schizochytrium sp. cells cultured in seed medium were collected by centrifugation at the indicated time, washed with phosphate buffer saline solution (PBS, 38.7 mM Na2HPO4∙12H2O, 11.3 mM NaH2PO4∙2H2O, and 150 mM NaCl), resuspended in 1 mL PBS, and disrupted with Mini Bead Beater (Biospec Mini-Bead-Beater-16 Model 607EUR, USA) for 50 s each for 5 times. After centrifugation, 500 μL of supernatant was transferred to a new tube. A 500-μL volume of buffer Z (60.0 mM Na2HPO4∙12H2O, 39.7 mM NaH2PO4∙2H2O, 10.0 mM KCl, 1.0 mM MgSO4∙7H2O, and 2.7 mL/L β-mercaptoethanol) and 200 μL of 13.3 mM ONPG were added to the supernatant, and the mixture was incubated at 37 °C for 15 min; the reaction was stopped by adding 500 μL of 1 M Na2CO3. The OD420 was recorded with an ultraviolet spectrophotometer for the determination of β-galactosidase activity. The amount of enzyme that releases 1 µmol of ONP per minute is defined as one unit of enzyme activity.
Determination of dry cell weight, pH and glucose and nitrogen concentrations
Determination of dry cell weight was performed as described previously with modification [10]. A 40-mL volume of fermentation broth was centrifuged at 5900g for 10 min, and dry cell weight was determined after freeze-drying for 24 to 48 h to a constant weight. For measurement of pH, glucose and nitrogen concentrations, 1 mL of broth was centrifuged (Heraeus BIOFUGE pico, Germany) at 13,523g for 10 min, and the supernatant was used for determination. The pH was measured by a laboratory pH meter (METTLER TOLEDO FiveEasy, Switzerland). The concentration of glucose was determined by the 3,5-dinitrosalicylic acid (DNS) method [44, 45]. NH4+ concentration was measured by the indophenol blue spectrophotometric method [46].
Microscopic analysis
Nile red staining of cells was conducted as described previously with modification [18]. A 1-mL volume of a culture grown in fermentation medium for 48 h was collected by centrifugation, washed twice with PBS solution, and resuspended in 1 mL of PBS. Cells were stained with Nile red dye (0.5 mg/L) and were incubated for 5 min in the dark. Fluorescence images were captured with a LEICA TCS SP8 microscope equipped with an oil immersion objective (×1000 magnification).
Lipid extraction and fatty acid composition analysis
Lipids were extracted as described previously with some modification [47–49]. About 0.3 g of a freeze-dried Schizochytrium sp. pellet was mixed with 6 mL of 4 M HCl for 30 min and then incubated in boiling water for 8 min before 16 mL of methanol/chloroform (1:1, vol/vol) was added. The preparation was mixed vigorously, and then centrifuged at 129g for 10 min. The lower phase was transferred to a pre-weighed glass tube and evaporated under a stream of nitrogen.
Fatty acid methyl esters (FAMEs) were prepared according to Ren et al. [50] with some modifications. About 30 mg of lipid sample was transferred to a glass tube before 1 mL of internal standard (methyl nonadecanoate, C19:0, 1 mg/mL) and 1 mL of 0.5 M KOH in methanol were added; the mixture was incubated at 65 °C in a water bath for 15 min. After the mixture had cooled to room temperature, 2.1 mL of methanol and 0.9 mL of 45% BF3–ether were added to the tube, which was incubated at 65 °C for 5 min. Then 1 mL of hexane and 2 mL of saturated sodium chloride solution were added; the preparation was mixed vigorously and allowed to stand for 10 min. The upper layer of the solution was transferred to a new tube and used for analysis of fatty acid composition. FAMEs were separated by gas chromatography (WUFENG GC522) with an Agilent J & W DB23 capillary column (30 m × 0.25 mm i.d.). Nitrogen was used as the carrier gas at a flow rate of 2 mL/min. The injector was at 250 °C. The column temperature was increased from 150 to 200 °C at the rate of 5 °C per min, was kept at 200 °C for 1 min, was then raised to 230 °C at the rate of 4 °C per min, and was maintained at 230 °C an additional 9 min.
RNA preparation and quantitative real-time PCR analysis (qRT-PCR)
Schizochytrium sp. cells cultured in fermentation medium were collected at 2 and 4 days, frozen in liquid nitrogen, and ground to fine powder. Total RNA was extracted with TRIzol reagent (Tiangen, China) according to the manufacturer’s protocol. cDNA was synthesized by M-MLV (RNase H−; TaKaRa) with oligo-dT18 from 4 µg of total RNA. qRT-PCR analysis was performed using FastStart Universal SYBR Green Master (ROX) with primers listed in Additional file 2: Table S2. PCR included a 10-min preincubation at 95 °C, followed by 40 cycles of denaturation at 95 °C for 10 s, and annealing and extension at 60 °C for 30 s. The relative expression levels were determined according to the comparative Ct method, using actin as the internal control.
Statistical analysis
All experiments were performed with three biological replicates. Statistical analyses were performed using one-way ANOVAs and Duncan’s multiple range tests or two-tailed Student’s t-tests. And it was considered indicative of statistical significance at p < 0.05.
Supplementary information
Additional file 1: Figure S1. Physical map of overexpression plasmid pPICZαA-ACL.
Additional file 2: Table S1. The open reading frames of ACL and ACC genes in Schizochytrium sp. Table S2. Primers used in this study.
Acknowledgements
The authors are grateful to Prof. B. Jaffee for English editing of the manuscript.
Abbreviations
- ACC
Acetyl-CoA carboxylase
- ACL
ATP-citrate lyase
- DCW
Dry cell weight
- DHA
Docosahexaenoic acid
- DNS
3,5-Dinitrosalicylic acid
- DPA
Docosapentaenoic acid
- FAMEs
Fatty acid methyl esters
- LC-PUFA
Long chain polyunsaturated fatty acid
- ONPG
O-Nitrophenyl-β-d-galactopyranoside
- PBS
Phosphate buffer saline
- TFAs
Total fatty acids
- WT
Wild type
- X-gal
5-Bromo-4-chloro-3-indolyl β-d-galactopyranoside
Authors’ contributions
ZC and XH designed the study. XH and ZZ performed the experiments. YW helped with analysis and discussion of results. XH and ZC wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (No. 31470190).
Availability of supporting data
All data supporting the conclusions of this article are included in the manuscript and in the additional information.
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13068-020-01767-z.
References
- 1.McNamara RK. DHA deficiency and prefrontal cortex neuropathology in recurrent affective disorders. J Nutr. 2010;140:864–868. doi: 10.3945/jn.109.113233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hashimoto M, Hossain S, Al Mamun A, Matsuzaki K, Arai H. Docosahexaenoic acid: one molecule diverse functions. Crit Rev Biotechnol. 2017;37:579–597. doi: 10.1080/07388551.2016.1207153. [DOI] [PubMed] [Google Scholar]
- 3.Zarate R, El Jaber-Vazdekis N, Tejera N, Perez JA, Rodriguez C. Significance of long chain polyunsaturated fatty acids in human health. Clin Transl Med. 2017;6:25. doi: 10.1186/s40169-017-0153-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan KW, Chen F, Jones EB, Vrijmoed LL. Eicosapentaenoic and docosahexaenoic acids production by and okara-utilizing potential of thraustochytrids. J Ind Microbiol Biotechnol. 2001;27:199–202. doi: 10.1038/sj.jim.7000169. [DOI] [PubMed] [Google Scholar]
- 5.Ling X, Guo J, Liu X, Zhang X, Wang N, Lu Y, et al. Impact of carbon and nitrogen feeding strategy on high production of biomass and docosahexaenoic acid (DHA) by Schizochytrium sp. LU310. Bioresour Technol. 2015;184:139–147. doi: 10.1016/j.biortech.2014.09.130. [DOI] [PubMed] [Google Scholar]
- 6.Ren LJ, Sun XM, Ji XJ, Chen SL, Guo DS, Huang H. Enhancement of docosahexaenoic acid synthesis by manipulation of antioxidant capacity and prevention of oxidative damage in Schizochytrium sp. Bioresour Technol. 2017;223:141–148. doi: 10.1016/j.biortech.2016.10.040. [DOI] [PubMed] [Google Scholar]
- 7.Ashford A, Barclay WR, Weaver CA, Giddings TH, Zeller S. Electron microscopy may reveal structure of docosahexaenoic acid-rich oil within Schizochytrium sp. Lipids. 2000;35:1377–1386. doi: 10.1007/s11745-000-0655-2. [DOI] [PubMed] [Google Scholar]
- 8.Raghukumar S. Thraustochytrid marine protists: production of PUFAs and other emerging technologies. Mar Biotechnol. 2008;10:631–640. doi: 10.1007/s10126-008-9135-4. [DOI] [PubMed] [Google Scholar]
- 9.Sun XM, Ren LJ, Ji XJ, Chen SL, Guo DS, Huang H. Adaptive evolution of Schizochytrium sp. by continuous high oxygen stimulations to enhance docosahexaenoic acid synthesis. Bioresour Technol. 2016;211:374–381. doi: 10.1016/j.biortech.2016.03.093. [DOI] [PubMed] [Google Scholar]
- 10.Ganuza E, Anderson AJ, Ratledge C. High-cell-density cultivation of Schizochytrium sp. in an ammonium/pH-auxostat fed-batch system. Biotechnol Lett. 2008;30:1559–1564. doi: 10.1007/s10529-008-9723-4. [DOI] [PubMed] [Google Scholar]
- 11.Hu XC, Ren LJ, Chen SL, Zhang L, Ji XJ, Huang H. The roles of different salts and a novel osmotic pressure control strategy for improvement of DHA production by Schizochytrium sp. Bioprocess Biosyst Eng. 2015;38:2129–2136. doi: 10.1007/s00449-015-1452-1. [DOI] [PubMed] [Google Scholar]
- 12.Zeng Y, Ji XJ, Lian M, Ren LJ, Jin LJ, Ouyang PK, et al. Development of a temperature shift strategy for efficient docosahexaenoic acid production by a marine fungoid protist, Schizochytrium sp. HX-308. Appl Biochem Biotechnol. 2011;164:249–255. doi: 10.1007/s12010-010-9131-9. [DOI] [PubMed] [Google Scholar]
- 13.Chi ZY, Liu Y, Frear C, Chen SL. Study of a two-stage growth of DHA-producing marine algae Schizochytrium limacinum SR21 with shifting dissolved oxygen level. Appl Microbiol Biotechnol. 2009;81:1141–1148. doi: 10.1007/s00253-008-1740-7. [DOI] [PubMed] [Google Scholar]
- 14.Yan J, Cheng R, Lin X, You S, Li K, Rong H, et al. Overexpression of acetyl-CoA synthetase increased the biomass and fatty acid proportion in microalga Schizochytrium. Appl Microbiol Biotechnol. 2013;97:1933–1939. doi: 10.1007/s00253-012-4481-6. [DOI] [PubMed] [Google Scholar]
- 15.Ren LJ, Zhuang XY, Chen SL, Ji XJ, Huang H. Introduction of omega-3 desaturase obviously changed the fatty acid profile and sterol content of Schizochytrium sp. J Agric Food Chem. 2015;63:9770–9776. doi: 10.1021/acs.jafc.5b04238. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi S, Satoh Y, Ujihara T, Takata Y, Dairi T. Enhanced production of polyunsaturated fatty acids by enzyme engineering of tandem acyl carrier proteins. Sci Rep. 2016;6:35441. doi: 10.1038/srep35441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue Z, Sharpe PL, Hong SP, Yadav NS, Xie D, Short DR, et al. Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nat Biotechnol. 2013;31:734–740. doi: 10.1038/nbt.2622. [DOI] [PubMed] [Google Scholar]
- 18.Blazeck J, Hill A, Liu L, Knight R, Miller J, Pan A, et al. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat Commun. 2014;5:3131. doi: 10.1038/ncomms4131. [DOI] [PubMed] [Google Scholar]
- 19.Yuzbasheva EY, Mostova EB, Andreeva NI, Yuzbashev TV, Laptev IA, Sobolevskaya TI, et al. Co-expression of glucose-6-phosphate dehydrogenase and acyl-CoA binding protein enhances lipid accumulation in the yeast Yarrowia lipolytica. N Biotechnol. 2017;39:18–21. doi: 10.1016/j.nbt.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Xu P, Qiao K, Stephanopoulos G. Engineering oxidative stress defense pathways to build a robust lipid production platform in Yarrowia lipolytica. Biotechnol Bioeng. 2017;114:1521–1530. doi: 10.1002/bit.26285. [DOI] [PubMed] [Google Scholar]
- 21.Tai M, Stephanopoulos G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab Eng. 2013;15:1–9. doi: 10.1016/j.ymben.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 22.Ji XJ, Mo KQ, Ren LJ, Li GL, Huang JZ, Huang H. Genome sequence of Schizochytrium sp. CCTCC M209059, an effective producer of docosahexaenoic acid-rich lipids. Genome Announc. 2015;3:e00819-15. doi: 10.1128/genomeA.00819-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao X, Dauenpen M, Qu C, Qiu X. Genomic analysis of genes involved in the biosynthesis of very long chain polyunsaturated fatty acids in Thraustochytrium sp. 26185. Lipids. 2016;51:1065–1075. doi: 10.1007/s11745-016-4181-6. [DOI] [PubMed] [Google Scholar]
- 24.Liu B, Ertesvag H, Aasen IM, Vadstein O, Brautaset T, Heggeset TM. Draft genome sequence of the docosahexaenoic acid producing thraustochytrid Aurantiochytrium sp. T66. Genom Data. 2016;8:115–116. doi: 10.1016/j.gdata.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, He X, Geng H, Liu H. Physiological characterization of ATP-citrate lyase in Aspergillus niger. J Ind Microbiol Biotechnol. 2014;41:721–731. doi: 10.1007/s10295-014-1418-3. [DOI] [PubMed] [Google Scholar]
- 26.Zhao S, Torres A, Henry RA, Trefely S, Wallace M, Lee JV, et al. ATP-citrate lyase controls a glucose-to-acetate metabolic switch. Cell Rep. 2016;17:1037–1052. doi: 10.1016/j.celrep.2016.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chavez-Cabrera C, Marsch R, Bartolo-Aguilar Y, Flores-Bustamante ZR, Hidalgo-Lara ME, Martinez-Cardenas A, et al. Molecular cloning and characterization of the ATP citrate lyase from carotenogenic yeast Phaffia rhodozyma. FEMS Yeast Res. 2015;15. [DOI] [PubMed]
- 28.Gawryluk RM, Eme L, Roger AJ. Gene fusion, fission, lateral transfer, and loss: not-so-rare events in the evolution of eukaryotic ATP citrate lyase. Mol Phylogenet Evol. 2015;91:12–16. doi: 10.1016/j.ympev.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Dulermo T, Lazar Z, Dulermo R, Rakicka M, Haddouche R, Nicaud JM. Analysis of ATP-citrate lyase and malic enzyme mutants of Yarrowia lipolytica points out the importance of mannitol metabolism in fatty acid synthesis. Biochim Biophys Acta. 2015;1851:1107–1117. doi: 10.1016/j.bbalip.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H, Zhang L, Chen H, Chen YQ, Chen W, Song Y, et al. Enhanced lipid accumulation in the yeast Yarrowia lipolytica by over-expression of ATP:citrate lyase from Mus musculus. J Biotechnol. 2014;192(Pt A):78–84. doi: 10.1016/j.jbiotec.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Gerhardt EC, Rodrigues TE, Muller-Santos M, Pedrosa FO, Souza EM, Forchhammer K, et al. The bacterial signal transduction protein GlnB regulates the committed step in fatty acid biosynthesis by acting as a dissociable regulatory subunit of acetyl-CoA carboxylase. Mol Microbiol. 2015;95:1025–1035. doi: 10.1111/mmi.12912. [DOI] [PubMed] [Google Scholar]
- 32.Davis MS, Solbiati J, Cronan JE., Jr Overproduction of acetyl-CoA carboxylase activity increases the rate of fatty acid biosynthesis in Escherichia coli. J Biol Chem. 2000;275:28593–28598. doi: 10.1074/jbc.M004756200. [DOI] [PubMed] [Google Scholar]
- 33.Zhou YJ, Buijs NA, Zhu Z, Qin J, Siewers V, Nielsen J. Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat Commun. 2016;7:11709. doi: 10.1038/ncomms11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu T, Zhou YJ, Huang M, Liu Q, Pereira R, David F, et al. Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis. Cell. 2018;174:1549–1558. doi: 10.1016/j.cell.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 35.Casadaban MJ, Martinezarias A, Shapira SK, Chou J. Beta-galactosidase gene fusions for analyzing gene-expression in Escherichia coli and yeast. Method Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- 36.Sasagawa T, Matsui M, Kobayashi Y, Otagiri M, Moriya S, Sakamoto Y, et al. High-throughput recombinant gene expression systems in Pichia pastoris using newly developed plasmid vectors. Plasmid. 2011;65:65–69. doi: 10.1016/j.plasmid.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 37.Krivoruchko A, Zhang YM, Siewers V, Chen Y, Nielsen J. Microbial acetyl-CoA metabolism and metabolic engineering. Metab Eng. 2015;28:28–42. doi: 10.1016/j.ymben.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Adams IP, Ratledge C. Malic enzyme: the controlling activity for lipid production? Overexpression of malic enzyme in Mucor circinelloides leads to a 2.5-fold increase in lipid accumulation. Microbiology. 2007;153:2013–2025. doi: 10.1099/mic.0.2006/002683-0. [DOI] [PubMed] [Google Scholar]
- 39.Wasylenko TM, Ahn WS, Stephanopoulos G. The oxidative pentose phosphate pathway is the primary source of NADPH for lipid overproduction from glucose in Yarrowia lipolytica. Metab Eng. 2015;30:27–39. doi: 10.1016/j.ymben.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 40.Sakaguchi K, Matsuda T, Kobayashi T, Ohara J, Hamaguchi R, Abe E, et al. Versatile transformation system that is applicable to both multiple transgene expression and gene targeting for thraustochytrids. Appl Environ Microbiol. 2012;78:3193–3202. doi: 10.1128/AEM.07129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong Q, Wang Y, Qi S, Gai K, He Q, Wang Y. Histone variant H2A.Z antagonizes the positive effect of the transcriptional activator CPC1 to regulate catalase-3 expression under normal and oxidative stress conditions. Free Radic Biol Med. 2018;121:136–148. doi: 10.1016/j.freeradbiomed.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 42.Huang J, Xia J, Yang Z, Guan F, Cui D, Guan G, et al. Improved production of a recombinant Rhizomucor miehei lipase expressed in Pichia pastoris and its application for conversion of microalgae oil to biodiesel. Biotechnol Biofuels. 2014;7:111. doi: 10.1186/1754-6834-7-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lippmeier JC, Crawford KS, Owen CB, Rivas AA, Metz JG, Apt KE. Characterization of both polyunsaturated fatty acid biosynthetic pathways in Schizochytrium sp. Lipids. 2009;44:621–630. doi: 10.1007/s11745-009-3311-9. [DOI] [PubMed] [Google Scholar]
- 44.Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. [Google Scholar]
- 45.Enshaeieh M, Nahvi I, Madani M. Improving microbial oil production with standard and native oleaginous yeasts by using Taguchi design. Int J Environ Sci Technol. 2014;11:597–604. [Google Scholar]
- 46.Gao XF, Yang Y, Liang ZL, Zheng JJ. A comparison of three indophenol blue methods for measuring ammonium in five types of soils in China. Adv Mater Res-Switz. 2013;634–638:59–63. [Google Scholar]
- 47.Li Z, Yuan H, Yang J, Li B. Optimization of the biomass production of oil algae Chlorella minutissima UTEX2341. Bioresour Technol. 2011;102:9128–9134. doi: 10.1016/j.biortech.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 48.Enshaeieh M, Abdoli A, Madani M, Bayat M. Recycling of lignocellulosic waste materials to produce high-value products: single cell oil and xylitol. Int J Environ Sci Technol. 2015;12:837–846. [Google Scholar]
- 49.Madani M, Enshaeieh M, Abdoli A. Single cell oil and its application for biodiesel production. Process Saf Environ. 2017;111:747–756. [Google Scholar]
- 50.Ren LJ, Huang H, Xiao AH, Lian M, Jin LJ, Ji XJ. Enhanced docosahexaenoic acid production by reinforcing acetyl-CoA and NADPH supply in Schizochytrium sp. HX-308. Bioprocess Biosyst Eng. 2009;32:837–843. doi: 10.1007/s00449-009-0310-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Physical map of overexpression plasmid pPICZαA-ACL.
Additional file 2: Table S1. The open reading frames of ACL and ACC genes in Schizochytrium sp. Table S2. Primers used in this study.
Data Availability Statement
All data supporting the conclusions of this article are included in the manuscript and in the additional information.