Abstract
Purpose of review
This review highlights recent progress in applying genome editing to the study and treatment of cardiovascular disease (CVD).
Recent findings
Recent work has shown that genome editing can be used to determine the pathogenicity of variants of unknown significance in patient-derived induced pluripotent stem cells. These cells can also be used to test therapeutic genome editing approaches in a personalized manner. Somatic genome editing holds great promise for the treatment of CVD, and important proof of concept experiments have already been performed in animal models. Here we briefly review recent progress in patient-derived cells, as well as the development of somatic genome editing therapies for CVD, with a particular focus on liver and heart.
Summary
Translating this technology into the clinic will require precise editing enzymes, efficient delivery systems, and mitigation of off-target events and immune responses.
Further development of these technologies will improve diagnostics and enable permanent correction of some of the most severe forms of CVD.
Keywords: CRISPR/Cas9, Adeno-Associated Virus, genome editing, cardiovascular disease
Introduction
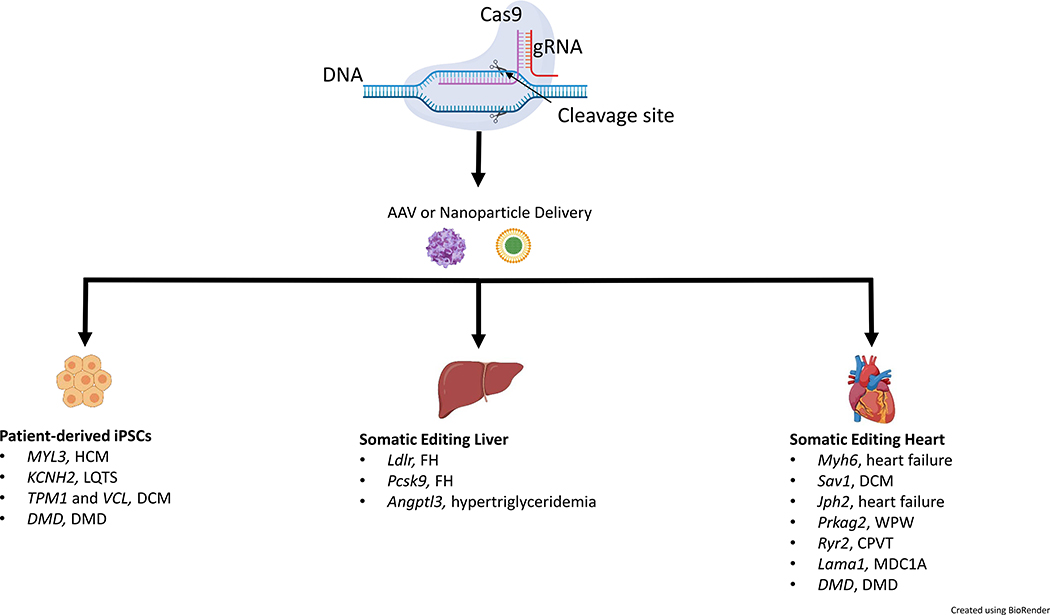
Cardiovascular disease (CVD) research is undergoing a major transformation with the application of CRISPR/Cas9 and other genome engineering systems. The clustered regularly interspaced short palindromic repeats / CRISPR-associated protein 9 (CRISPR/Cas9) system is currently the most widely used genome editing platform. It consists of an RNA-guided nuclease (Cas9) that produces double-stranded breaks in DNA at specific locations based on the sequence of a short guide RNA engineered by the user1. Generating double-stranded breaks with CRISPR/Cas9 can introduce short insertion and deletion mutations (indels), delete large fragments of DNA, or improve the efficiency of homology directed repair (HDR). CRISPR/Cas systems have been cloned from a wide range of bacteria, providing a diversity of targeting options2–5. In addition, Cas9 has been fused with other protein moeities, allowing the user to direct enzymatic activities to specific sites in the genome. These include fluorescent proteins, transcriptional activators or repressors, chromatin modifiers, deaminases, and reverse transcriptases. The development and basic principles of CRISPR/Cas9 genome editing have been covered well in recent literature6–8. This minireview will focus on the application of these emerging technologies to CVD. Specifically, we will highlight genome engineering of patient-derived induced pluripotent stem cells (iPSC), and somatic genome editing approaches targeting the liver and the heart (Figure 1).
FIGURE 1:
Gene editing for cardiovascular disease. Adeno-associated viruses (AAV) and nanoparticles have been used to deliver CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9) components both in vitro and in vivo to edit genes involved in cardiovascular disease (CVD). Patient-derived induced pluripotent stem cells (iPSCs) have been edited with CRISPR/Cas9 to study variants associated with hypertrophic cardiomyopathy (HCM), long QT syndrome (LQTS), dilated cardiomyopathy (DCM), and Duchenne muscular dystrophy (DMD). Somatic genome editing in the liver and heart has been used to target genes that lead to familial hypercholesterolemia (FH), hypertriglyceridemia, heart failure, DCM, Wolff--Parkinson--White syndrome (WPW), catecholaminergic polymorphic ventricular tachycardia (CPVT), muscular dystrophy type 1A (MDC1A), and DMD. Although these studies have shown the CRISPR/Cas9 system’s versatility for treating CVD, significant challenges remain – including delivery efficiency, off-target editing, and host-immune responses.
Disease modeling in patient-derived cells
Rare variants in multiple genes are known to cause cardiomyopathies. However, even for a known gene, it can be difficult to predict whether a given variant is pathogenic. These “variants of uncertain significance” (VUS) present a difficult problem for clinicians and patients. Determination of pathogenicity by traditional means can be difficult, and should ideally be assessed in the genomic context of that particular patient to account for gene-gene and gene- environment interactions. The use of patient-derived pluripotent stem cells (iPSC) makes this possible. These cells can be edited and differentiated into a variety of cell types including cardiomyocytes, macrophages, and hepatocyte-like cells. In a recent example, Ma et al. isolated iPSCs from a healthy individual carrying a VUS in the MYL3 gene (NM_000258.2:c.170C>A, NP_000249.1:p.Ala57Asp) which encodes myosin light chain 3, an important sarcomeric protein9. Other mutations in MYL3 are associated with hypertrophic cardiomyopathy (HCM), and this variant was predicted by in silico algorithms to be damaging10. CRISPR/Cas9 editing was used to generate iPSC lines from this individual, including clones homozygous for either the 170C>A allele or corrected to wild type, which were then differentiated into cardiomyocytes (iPSC-CM)9. The heterozygous and homozygous iPSC-CMs for the 170C>A variant had no discernible phenotype in terms of gene expression, cell size, sarcomere organization, contractility, or calcium handling9. In contrast, conversion of these patient-derived iPSCs to a known pathogenic variant at the same nucleotide (170C>G) did display changes in the β-/α-myosin ratio, faster contraction and relaxation velocities, and proarrhythmic alterations in calcium handling11. This work supports a benign classification for the 170C>A allele9, and demonstrates that genome-edited iPSCs have potential utility for assessing the impact of VUS in a patient-specific manner. This approach has also been used to assess pathogenicity of VUS in KCNH2 as a cause of long QT syndrome, where the T983I variant was deemed pathogenic12.
CRISPR/Cas9 editing of iPSC-CMs can also be used to study gene-gene interactions. Deacon et al. found that the interaction between the sarcomeric gene for tropomyosin1 (TPM1) and the cosarcomeric gene for vinculin (VCL) influenced severity in a family with a dilated cardiomyopathy (DCM) history13. To determine the pathogenicity of the mutations found in TPM1 and VCL, they derived cardiomyocytes from human iPSCs and CRISPR/Cas9 genome-edited H9 human embryonic stem cells (hESCs) with the same variants found in the patients13. TPM1 and VCL variants acted synergistically to impact cardiomyocyte contractility and sarcomeric organization13. Genome editing in iPSC-CMs will also prove useful in testing new therapeutic approaches in a variant- or patient-specific manner. Long et al. recently generated iPSC-CMs from patients with Duchenne muscular dystrophy (DMD), a lethal muscle disease with severe cardiac manifestations14. CRISPR/Cas9 targeting of conserved splice acceptor or donor sites produced DMD alleles with in-frame deletions, restoring dystrophin protein expression and contractile function14. While these are exciting advances, there are important practical considerations to the use of genome-edited iPSC-CMs. Incomplete differentiation to cardiomyocytes in situ is a limitation for assessing functional outcomes in a bulk cell population. Thus far, most applications have focused on sarcomeric organization or expression of key proteins that can be assayed in individual cells through microscopy. Likewise, genome editing of iPSC clones has its own challenges, including the enrichment and selection of edited cells, as well as avoiding unintended indel mutations when performing HDR. Despite these challenges, it is likely that iPSCs will become an important preclinical testing ground for genome editing therapies in the coming years.
Genome editing of the liver
The liver plays a central role in lipid metabolism and is a major target organ for genome editing to treat CVD. Initial proof of concept studies have involved disruption of the Ldlr, Pcsk9, and Angptl3 genes. The low density lipoprotein receptor (LDLR) binds to ApoB-containing lipoproteins and mediates their clearance from the circulation15. Loss-of-function mutations in LDLR cause Familial Hypercholesterolemia, a severe genetic disorder characterized by xanthomas, excessively high plasma cholesterol, and premature atherosclerotic CVD16. Jarrett et al. showed that an all-in-one Adeno-Associated Virus (AAV) vector packaged with Staphylococcus aureus Cas9 (SaCas9) and an Ldlr-targeting gRNA efficiently knocks out Ldlr in mouse liver and causes atherosclerosis in just 20 weeks17. This study underscores the importance of hepatic LDLR expression in clearance of LDL cholesterol, and paves the way for AAV-CRISPR correction of mutations in LDLR. One drawback to gene editing of LDLR is the sheer number of mutations that have been linked to elevated circulating cholesterol - over 120018. Ultimately, a successful gene editing therapy would need to be generalizable, and would most likely involve integration of a full-length LDLR coding sequence. There is currently a gene therapy trial in progress to correct Familial Hypercholesterolemia through AAV delivery of an LDLR transgene to the liver19. The success of this therapy remains to be determined, and it is possible that genome editing approaches may also be worth pursuing.
PCSK9 is a liver-secreted protein that promotes lysosomal degradation of LDLR20. Loss of function mutations in PCSK9 are associated with low levels of LDL-C and confer protection against CVD21–25. PCSK9 is a major target for LDL lowering in CVD for patients who are resistant to statin therapy, and there are several highly successful monoclonal antibodies currently in use26. In 2014, Ding et al. demonstrated in vivo editing of the Pcsk9 gene with the CRISPR/Cas9 system using adenovirus to target mouse liver27. Pcsk9 was also disrupted by Ran et al. who identified a Cas9 ortholog from Staphylococcus aureus that is small enough for delivery with AAV vectors3. Since then, Pcsk9 has been a frequent target for proof of concept studies using different CRISPR/Cas editors and delivery systems. For example, base editors that can convert cytosine bases to thymine have been used in multiple reports to edit Pcsk9 in the liver28–30. Chadwick et al. used a base editing strategy to inactivate mouse Pcsk928. The base editor gRNAs were designed to alter codons for glutamine, arginine, or tryptophan, leading to a premature stop codon and PCSK9 truncation28. Wang et al. tested the feasibility of targeting human hepatocytes using CRISPR/Cas9 in a chimeric liver-humanized mouse model30. They demonstrated successful gene editing of human PCSK9, reduced blood levels of human PCSK9 protein, and a lack of detectable off-target mutagenesis at predicted sites30. More recently, Carreras et al. presented the use of base editors to correct hypercholesterolemia in a human PCSK9 overexpression mouse model29.
Angiopoietin-like proteins are a family of secreted proteins linked with regulation of various physiological and pathophysiological processes, including triglyceride-rich lipoprotein metabolism and CVD31. Chadwick et al. targeted hepatic Angptl3 in 5 week old mice with a CRISPR/Cas base editor 3 (BE3) delivered via adenovirus31. They found that BE3-Angptl3-treated mice had lower circulating ANGPTL3, triglycerides, and cholesterol compared to control mice31. In Ldlr KO mice, targeting Angptl3 substantially reduced plasma triglyceride and cholesterol levels (56% and 51%, respectively) at 14 days post treatment, whereas Pcsk9 knockout did not31. Hypertriglyceridemia-induced CVD has proven a difficult target for drug development, but these studies have implicated Angptl3 as a promising candidate- either for knockdown, inhibition, or genome editing.
Viral delivery systems have been very effective for genome editing in animal models. However, these vectors have limitations for clinical translation. Adenoviral vectors elicit potent innate as well as adaptive immune responses32. Although these vectors have certain niche applications (i.e. oncolytic Adenovirus), serious safety concerns preclude the use of high doses of Adenoviral vectors for tissue-directed gene therapy in humans33. In the case of AAV, limitations include a high prevalence of neutralizing antibodies, and persistent expression of Cas9 which could increase the risk of off-target mutagenesis or immune responses. Therefore, there is a compelling need to develop complementary and alternative delivery systems. Recently, other groups have produced lipid nanoparticles to deliver CRISPR/Cas. Zhang et al. used gold nanoclusters modified with cationic HIV-1-transactivating transcriptor peptide complexed with Cas9 and a gRNA to disrupt Pcsk934. The entire structure was further encapsulated by a galactose-modified lipid layer, which targets the gold nanoclusters to the liver34. They showed Pcsk9 editing efficiency of 60%, and a 30% reduction in plasma LDL-C in mice34. Liu et al. developed a bioreducible nanoparticle that packages the Cas9 mRNA and gRNA together, and delivered this into mice intravenously35. They showed a 20% reduction in plasma PCSK9 as compared to controls35. While these studies show that lipid nanoparticles can deliver CRISPR/Cas9 and effectively target Pcsk9 in the liver, further studies are required to analyze off-target mutagenesis and nanoparticle safety.
Genome editing of the heart
The heart is also an attractive target for genome editing. Carroll et al. were the first to report cardiac-specific Cas9 transgenic mice as a means of ablating gene expression in the heart36. The authors used AAV9 delivery of a gRNA to disrupt the Mhy6 gene, which encodes cardiac alpha myosin heavy chain36. CRISPR/Cas9-deletion of Myh6 at postnatal day 10 resulted in atria and ventricle dilation, ventricle wall thinning, cardiac dilatation and heart failure, which was later reproduced by Johansen et al37. Johansen et al. also targeted Sav1 and Tbx20 using AAV delivery of gRNA37. Even with efficient editing there was no significant effect on protein levels with one gRNA, but using two gRNAs targeting Sav1 resulted in a modest increase in heart size and hypertrophic markers37. Guo et al. used AAV9 to deliver gRNAs targeting junctaphilin 2 (Jph2), which tethers the T-tubule membrane to the sarcoplasmic reticulum (SR)38. High dose delivery of AAV-gRNA to Jph2 in neonatal mice (P1) resulted in ventricular dilatation, cardiomyocyte hypertrophy, heart failure, and death38. However, lower doses produced mosaic removal of Jph2, allowing the authors to study its role in T-tubule formation in isolated cardiomyocytes38. The authors then used this “CASAAV” approach to screen 8 candidate genes, and uncovered a cell autonomous role for the SR calcium release channel (Ryr2) in this process38. These studies all observed effective but mosaic editing, and demonstrate the utility of this approach to study cardiac physiology.
Significant progress has been made towards therapeutic genome editing for the heart. In particular, autosomal dominant diseases present a unique opportunity for allele-specific gene disruption. Patients with familial Wolff-Parkinson-White (WPW) syndrome suffer from a conduction system abnormality that often manifests as paroxysmal supraventricular tachycardia. If not corrected with surgical ablation, certain cases of WPW can result in progressive heart failure or sudden cardiac death from ventricular tachyarrhythmia39. A knock-in mouse model for familial WPW was generated harboring the (c.1589A>G;H530R) mutation in the PRKAG2 gene39. These mice were successfully rescued by delivery of AAV9-Cas9 and gRNA targeting the 1589A>G sequence as neonates, revealing a role for H530R PRKAG2 allele in WPW syndrome39. Pan et al. used a similar approach to correct catecholaminergic polymorphic ventricular tachycardia in heterozygous animals harboring the a R176Q mutation in the Ryr2 gene40. Targeted mice displayed reduced total Ryr2 mRNA and protein levels, normalized calcium handling, and were protected from pacing-induced arrhythmias40.
In addition to allele-specific disruption, others have explored targeted activation of disease-modifier genes that are too large for conventional gene therapy. Kemaladewi et al. used AAV9, to deliver a catalytically inactive version of Cas9 (dCas) fused to a VP64 transactivation domain, and a gRNA targeting the promoter for Lama1 in a mouse model of muscular dystrophy type 1A (MDC1A)41. Treated mice showed Lama1 upregulation in skeletal muscle and peripheral nerves, preventing muscle fibrosis and paralysis41. Disease progression was halted and reversed, showing for the first time a mutation-independent approach to treat MDC1A41. These studies show that it is possible to correct severe genetic diseases through somatic genome editing in the heart. In the case of autosomal dominant diseases, disruption of the disease-causing allele is now feasible in cardiomyocytes, but care must be taken to ensure that haploinsufficiency does not create new pathology. Likewise, off-target editing and unintended editing events at the on-target site (i.e. large insertions and deletions) will need to be avoided. The report of Kemaladewi et al. demonstrates the power of CRISPR/Cas9 transcriptional activation to upregulate expression of modifier genes, and possibly other targets that are too large for gene therapy vectors. However, this approach involves persistent expression of the Cas9 fusion proteins in the heart, where the long-term efficacy and safety could be limited by pre-existing or acquired immunity to the Cas9 protein.
Landmark papers in Science in 2016 showed it is possible to correct DMD in the mdx mouse model through in-frame deletion of exon 23 with AAV9 delivery of CRISPR/Cas942–44. In addition to severe muscle weakness and degeneration, DMD patients also suffer from dilated cardiomyopathy, and heart failure is the major cause of death. Refaey et al. used a similar editing strategy in the mdx/Utr+/− mice45. Mice that received high doses of AAV-CRISPR showed 23±5.1% restored dystrophin protein relative to WT by densitometry analysis, along with improved heart muscle fiber architecture and contractile strength45. Recently, the long-term safety and side effects of AAV-CRISPR editing for DMD has been examined by several groups46–48. Hakim et al. followed mdx mice for 18 months, and reported selective depletion of the AAV-gRNA vector47. In the same study, substantially higher doses succeeded in partially restoring dystrophin protein, reduced fibrosis, and improved hemodynamics47. Xu et al. used an AAVrh.74 vector to deliver CRISPR/Cas9 to the mdx mouse at age P3, and followed the animals for 19 months48. Genome editing was efficient and cardiac function improved without serious adverse events48. Nelson et al. reported humoral and cellular immune responses when AAV-CRISPR vectors were administered to adult mice, which was not seen with delivery to neonates46. Unintended on-target editing events were also found, including a high frequency of AAV vector insertions46, as previously described in liver21,49, and heart40. Efficacy has also been assessed in a canine model of DMD, where AAV9 was used to deliver SpCas9 and a gRNA targeting a region close to the exon 51 splice acceptor site50. Off-target analysis showed no unwanted gene editing, although only three predicted off-target sites were analyzed by targeted deep sequencing. Remarkably, dystrophin protein expression was restored to 92% of normal levels along with improved muscle histology50. These studies highlight the potential of CRISPR/Cas9 to achieve long-term editing for otherwise untreatable genetic diseases, but also raise concerns about host immune responses and unintended genetic alterations.
Conclusion
Genome editing is beginning to change the landscape of cardiovascular research and therapy. It is now possible to introduce specific variants in patient-derived cells, and determine their effects in an isogenic setting. Cellular phenotyping of genome-edited iPSC derived cardiomyocytes and other cell types will be an invaluable platform for assessing the safety and efficacy of gene-editing therapeutics. In parallel with these advances, delivery with viral vectors and nanoparticles can edit genes with high efficiency in the liver and heart. Somatic genome editing has been used in animal models to correct several disorders including hypercholesterolemia, hypertriglyceridemia, Wolff-Parkinson-White syndrome, Catecholaminergic Polymorphic Ventricular Tachycardia, and Duchenne muscular dystrophy. While these early successes are very promising, they have also identified important challenges. Off-target editing with CRISPR/Cas systems will vary depending on the specific editing enzyme, design, target cell type, and delivery method. Efficient delivery to somatic tissues is achievable in mice, but not all delivery systems will scale effectively to humans. Unintended editing events at the on-target site, such as large deletions and insertions, require further study and risk assessment. Likewise, pre-existing immunity and subsequent development of immune response to the bacterially-derived Cas9 protein will need special attention. Despite these concerns, CRISPR/Cas therapeutic potential is expansive, and is expected to dramatically change the future of research and patient care.
Key Points.
Genome editing of patient-derived induced pluripotent stem cells can be used to determine the pathogenicity of variants as well as to study gene-gene and gene-environment interactions.
Liver-directed gene editing shows promise for correcting familial hypercholesterolemia and hypertriglyceridemia.
Somatic gene editing in the heart is a useful approach for studying cardiac physiology in the whole heart as well as isolated cardiomyocytes ex vivo.
Therapeutic editing strategies are in development to treat Wolff-Parkinson White syndrome, Catecholaminergic Polymorphic Ventricular Tachycardia, and Duchenne muscular dystrophy.
Acknowledgments
Funding for this work was received from the National Institutes of Health (NIH)
Financial support and sponsorship: This research was supported by NIH grant HL132840 to W.R.L., as well as an American Heart Association predoctoral fellowship 19PRE34380467 to A.M.D.
Footnotes
Conflicts of interest: none.
References
- 1.Wiedenheft B, Sternberg SH & Doudna JA RNA-guided genetic silencing systems in bacteria and archaea. Nature 482, 331–338 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Kleinstiver BP et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ran FA et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zetsche B et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 163, 759–771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slaymaker IM et al. Rationally engineered Cas9 nucleases with improved specificity. Science (80-. ). 351, 84–88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doudna JA & Charpentier E The new frontier of genome engineering with CRISPR-Cas9. Science (80-. ). 346, (2014). [DOI] [PubMed] [Google Scholar]
- 7.Adli M The CRISPR tool kit for genome editing and beyond. Nat. Commun. 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pickar-Oliver A & Gersbach CA The next generation of CRISPR–Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 20, 490–507 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma N et al. Determining the pathogenicity of a genomic variant of uncertain significance using CRISPR/Cas9 and human-induced pluripotent stem cells. Circulation 138, 2666–2681 (2018).** This study shows that genome-edited iPSCs can be used to investigate variants of uncertain significance in a patient specific manner.
- 10.Andersen PS et al. A novel myosin essential light chain mutation causes hypertrophic cardiomyopathy with late onset and low expressivity. Biochem. Res. Int. 2012, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee WH et al. Different expressivity of a ventricular essential myosin light chain gene Ala57Gly mutation in familial hypertrophic cardiomyopathy. Am. Heart J. 141, 184–189 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Garg P et al. Genome Editing and Induced Pluripotent Stem Cells in Cardiac Channelopathy. J. Am. Coll. Cardiol. 72, 62–75 (2018).* This group describes the use of patient-dreived iPSCs to describe the pathogenicity of a variant of unknown significance in long QT syndrome.
- 13.Deacon DC et al. Combinatorial interactions of genetic variants in human cardiomyopathy. Nat. Biomed. Eng. 3, 147–157 (2019).** This study is the first to use iPSC derived cardiomyocytes to delineate the role of two genes in a family with a history for dilated cariomyopathy. Tropomyosin1 and vinculin acted synergistically to impact cardiomyocyte contractility and sarcomeric organization.
- 14.Long C et al. Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci. Adv. 4, 1–11 (2018).*This group generated iPSC cardiomyocytes from patients with Duchenne muscular dystrophy, then used CRISPR/Cas9 to target a conserved splice acceptor or donor site to restore dystrophin protein expression.
- 15.Yamamoto T et al. The human LDL receptor: A cysteine-rich protein with Multiple Alu Sequences in its mRNA. Proc. Am. Math. Soc. 39, 27–38 (1984). [DOI] [PubMed] [Google Scholar]
- 16.Brown MS & Goldstein JL A Receptor-Mediated Pathway for Cholesterol Homeostasis Author ( s ): Brown Michael S. and Joseph L. Goldstein Published by: American Association for the Advancement of Science Stable URL: http://www.jstor.org/stable/1697034 Science (80-. ). 232, 34–47 (1986). [DOI] [PubMed] [Google Scholar]
- 17.Jarrett KE et al. Somatic genome editing with CRISPR/Cas9 generates and corrects a metabolic disease. Sci. Rep. 7, 1–12 (2017).* This is the first study to somatically target the low density lipoprotein receptor in mouse liver with AAV and CRISPR/Cas9 resulting in hypercholesterolemia and atherosclerosis. It also highlights the importance of hepatic LDLR expression in clearance of LDL cholesterol.
- 18.Varret M & Rabes J-P Missense Mutation in the LDLR gene: a Wide Spectrum in the Severity of Familial Hypercholesterolemia. IntechOpen 55–74 (2012). doi: 10.5772/57353 [DOI] [Google Scholar]
- 19.Ajufo E & Cuchel M Recent Developments in Gene Therapy for Homozygous Familial Hypercholesterolemia. Curr. Atheroscler. Rep. 18, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagor WR & Millar JS Overview of the LDL receptor: relevance to cholesterol metabolism and future approaches for the treatment of coronary heart disease Overview of the LDL receptor. J. Receptor. Ligand Channel Res. 3, 1–14 (2010). [Google Scholar]
- 21.Jarrett KE et al. Somatic editing of Ldlr with adeno-associated viral-CRISPR is an efficient tool for atherosclerosis research. Arterioscler. Thromb. Vasc. Biol. 38, 1997–2006 (2018).* This group developed an all-in-one AAV-CRISPR vector as a tool to disrupt Ldlr as a new method for performing atherosclerosis studies in mice
- 22.Lu H et al. Hypercholesterolemia induced by a PCSK9 gain-of-function mutation augments angiotensin II-induced abdominal aortic aneurysms in C57BL/6 mice-brief report. Arterioscler. Thromb. Vasc. Biol. 36, 1753–1757 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar S, Kang DW, Rezvan A & Jo H Accelerated atherosclerosis development in C57Bl6 mice by overexpressing AAV-mediated PCSK9 and partial carotid ligation. Lab. Investig. 97, 935–945 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao R et al. A single injection of gain-of-function mutant PCSK9 adeno- associated virus vector induces cardiovascular calcification in mice with no genetic modification. Atherosclerosis 48, 1119–1130 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abifadel M et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 34, 154–156 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Nicholls SJ The New Face of Hyperlipidemia and the Role of PCSK9 Inhibitors. Curr. Cardiol. Rep. 21, 19–23 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Qiurong D et al. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ. Res. 115, 488–492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chadwick AC, Wang X & Musunuru K In Vivo Base Editing of PCSK9 as a Therapeutic Alternative to Genome Editing. 37, 1741–1747 (2017).** This is the first study to use base editing to knock out Pcsk9 in the liver.
- 29.Carreras A et al. In vivo genome and base editing of a human PCSK9 knock-in hypercholesterolemic mouse model. BMC Biol. 17, 1–14 (2019).** This is the first study to use base editing to disrupt human PCSK9 in a chimeric mouse model, thus correcting hypercholesterolemia.
- 30.Wang X et al. CRISPR/Cas9 Targeting of Pcsk9 in Human Hepatocytes In Vivo- Brief Report. Arterioscler. Thromb. Vasc. Biol. 136, 783–786 (2017).* This group showed successful targeting of human PCSK9 in a chimeric liver-humanized mouse model.
- 31.Chadwick AC, Evitt NH, Lv W & Musunuru K Reduced blood lipid levels with in vivo CRISPR-Cas9 base editing of ANGPTL3. Circulation 137, 975–977 (2018).** This is the first study used CRISPR/Cas9 base editing to target hepatic Angptl3. This study opens a door for potentially treating hypertriglyceridemia-induced cardiovascular disease.
- 32.Atasheva S, Yao J & Shayakhmetov DM Innate immunity to adenovirus: lessons from mice. 1–23 (2019). doi: 10.1002/1873-3468.13696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raper SE et al. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol. Genet. Metab. 80, 148–158 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Zhang L et al. Triple Targeting Delivery of CRISRP/Cas9 to Reduce the Risk of Cardiovascular Diseases. Angew. Chemie Int. Ed. 1–6 (2019). doi: 10.1002/anie.201903618* This group used lipid nanoclusters to deliver CRISPR/Cas9 and a gRNA to successfully knock down Pcsk9
- 35.Liu J et al. Fast and Efficient CRISPR/Cas9 Genome Editing In Vivo Enabled by Bioreducible Lipid and Messenger RNA Nanoparticles. Adv. Mater. 1902575, 1–7 (2019).* This group developed bioreducible lipid nanoparticles to package Cas9 mRNA and gRNA together targeting PCSK9.
- 36.Carroll KJ et al. A mouse model for adult cardiac-specific gene deletion with CRISPR/Cas9. Proc. Natl. Acad. Sci. U. S. A. 113, 338–343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johansen AK et al. Postnatal cardiac gene editing using CRISPR/Cas9 with AAV9-mediated delivery of short guide RNAs results in mosaic gene disruption. Circ. Res. 121, 1168–1181 (2017).** This study used CRISPR/Cas9 and AAV9 to deliver gRNAs targeting Myh6, Sav1, and Tbx20 in the heart. They found that using two gRNAs against Sav1 increased somatic editing efficiency and resulting protein knockdown as well as a modest increase in heart size and hypertrophic markers.
- 38.Guo Y et al. Analysis of Cardiac Myocyte Maturation Using CASAAV, A Platform for Rapid Dissection of Cardiac Myocyte Gene Function In Vivo. 120, 1874–1888 (2017).** This group used AAV9 to deliver gRNAs targeting junctophilin 2 and other candiate genes to study their roles in T-tubule formation in mice. This study demonstrates mosaic but effective genome editing in the heart, and how this approach can be applied to study cardiac physiology at the cellular level.
- 39.Xie C et al. Genome editing with CRISPR/Cas9 in postnatal mice corrects PRKAG2 cardiac syndrome. Cell Res. 26, 1099–1111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan X et al. In Vivo Ryr2 editing corrects catecholaminergic polymorphic ventricular tachycardia. Circ. Res. 123, 953–963 (2018).*This group used AAV9 to deliver SaCas9 targeting the Ryr2 mutation R176Q which results in polmorphic ventricular tachycardia. Allele-specific disruption of the disease-causing allele with AAV-CRISPR normalized calcium handling and protected the mice from pacing induced arrhythmias.
- 41.Kemaladewi DU et al. A mutation-independent approach for muscular dystrophy via upregulation of a modifier gene. Nature 572, 125–130 (2019).** This group used a catalytically inactive version of Cas9 fused to a VP64 transactivation domain to activate the expression of the Lama1 gene in a mouse model for MDC1A. Activation of this modifier gene protected the treated mice from muscle fibrosis and paralysis.
- 42.Tabebordbar M et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science (80-. ). 351, 407–411 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Long C et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science (80-. ). 351, 400–403 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson CE et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. 351, 403–407 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Refaey M et al. In vivo genome editing restores dystrophin expression and cardiac function in dystrophic mice. Circ. Res. 121, 923–929 (2017).*This group used AAV9 delivery of CRISPR/Cas9 targeting exon 23 in the mdx/Utr+/− mouse model for Duchenne muscular dystrophy, and showed that the mice had restored dystrophin protein expression along with improved muscle fiber architecture and contractile strength.
- 46.Nelson CE et al. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat. Med. 4, 427–432 (2019).**This study showed that AAV-CRISPR treatment for DMD in adult mice provoked humoral and cellular immune responses, but these were not seen when the vectors were delivered to neonates. Unintended on-target editing events were seen, along with a high frequency of AAV vector insertions, in both adult and neonatally injected mice.
- 47.Hakim CH et al. AAV CRISPR editing rescues cardiac and muscle function for 18 months in dystrophic mice. JCI Insight 3, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu L, Lau YS, Gao Y, Li H & Han R Life-Long AAV-Mediated CRISPR Genome Editing in Dystrophic Heart Improves Cardiomyopathy without Causing Serious Lesions in mdx Mice. Mol. Ther. 27, 1407–1414 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li A et al. A Self-Deleting AAV-CRISPR System for In Vivo Genome Editing. Mol. Ther. - Methods Clin. Dev. 12, 111–122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amoasii L et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science (80-. ). 362, 86–91 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]