Abstract
Background
COVID-19 is the disease caused by SARS-CoV2, it was identified in Wuhan, China, in 2019. It then extended across the globe and was termed as a pandemic in 2020. Though research work on its vaccine and drugs are carried out across the globe, it is even necessary to look over it through alternative sciences.
Objective
The objective of this study is to look over the disease through Ayurvedic perspective, analyse possible pathologies, select appropriate drugs and to study in-silico screening on these selected drugs.
Materials & Methods
Available symptoms of COVID-19 were thoroughly studied and reviewed through Ayurveda classics, internet, preprints, etc. to understand the nature of the disease with the Ayurvedic perspective. The molecular Docking and Grid were generated through Pyrx Software with Autodock. The Lipinski Rule of Five data generated from Swiss ADME software and Target prediction of selected phytoconstituents were done by Swiss target prediction.
Results
In Ayurveda, COVID-19 can be considered as Janapadaudhwans, Vata-Kaphaja Sannipatika Jwara, Aupasargika Vyadhi, and Dhatupaka Awastha. In the molecular docking study, the binding energy and inhibition of 6 Gingesulphonic acid from Zingiber officinalis (Sunthi) is greater than hydroxychloroquine and quinine. Most of the selected phytoconstituents follow the Lipinski rule of five. Target prediction of selected phytoconstituents was done on target of SARS-CoV-2, humoral immunity, and antiviral activity. Every selected phytoconstituents works on minimum one of the targets.
Conclusion
Thus, from the above results obtained from reviewing Ayurveda classics and after the virtual screening of selected drugs we can conclude that Nagaradi Kashaya (Sunthi, Puskarmoola, Kantakari, Guduchi) may have appreciable results in combating SARS-CoV-2. Thus, Nagaradi Kashayam, a classical formulation can be a trial candidate for conducting further clinical trial.
Keywords: Ayurveda, COVID-19, Molecular docking, Pandemic
Graphical abstract
1. Introduction
The outbreak of COVID-19, caused by the Noval Corona Virus (nCoV) that is now officially designated as Severe Acute Respiratory Syndrome-Related Coronavirus SARS-CoV-2, represents a pandemic threat to global public health. Although the epicentre of the COVID-19 outbreak in December of 2019 was located in Wuhan China, this disease has spread to more than 213 countries with over 1,00,00, 000confirmed cases and still continuing.
Ayurveda is a life science having a holistic approach in dealing with different types of diseases. It has a completely different way of looking at any new diseases. Understanding of any disease in Ayurveda is done by five elements, i.e. Hetu, Purvarupa, Rupa, Upashaya-Anupashaya, Samprapti [1]. By these elements, a physician gets to know about the nature of the diseases (Vikara Prakruti), site of the diseases (Adhisthanam), the course of the disease (Samprapti) and about the aggravating factors of the disease. By understanding these factors in detail, a physician can design the preventive measures and treatment protocol for the disease. Though Ayurveda focuses on the concept of individualism and accepts that each individual is unique but in this situation of the pandemic when many people are affected by the same set of symptoms, one can briefly design a treatment policy by considering the available information about the disease.
Thus, it is the need of an hour to think out of the box so here is an initiation to think about the present pandemic COVID-19 through Ayurveda perspective. The main objective of this study is to look over the disease through Ayurvedic perspective, analyse possible pathologies, select appropriate drugs and to study in-silico screening on these selected drugs. Though, there are many protease of the disease present but this study is limited to virtual screening against main protease (Mpro) of the virus. The present piece of work studies the virtual screening of the phytoconstituents which can help for a further invitro, invivo research for COVID-19.
According to WHO, people who have co-morbidities and those over 60 years of age have a higher risk of developing severe symptoms of COVID-19 [2]. Common symptoms of COVID-19 include fever (Jwar a ), tiredness (Shrama, Klama) and dry cough(Shushka Kasa). Other symptoms include shortness of breath (Shwasa), aches and pains (Anga Marda na), sore throat (Kantha Shoola), very few people will report diarrhoea (Drava Mala), nausea (Hrullas a ) or a running nose (Peenasa).
1.1. Possible ayurvedic concepts
With the help of the available data regarding COVID-19 and after going through the Ayurveda classics thoroughly, the following possible pathological models can be drawn.
1.1.1. Janapada-Udhwamsa Vikara
COVID-19 has evolved itself into a pandemic, affecting a large population irrespective of their physical features, dietary patterns, psychological attributes, etc., Ayurveda considers it as a Janapada-Udhwamsa Vikara [3].
1.1.2. Vata Kaphaja Jwara
By looking at its symptomatology, it resembles with the classical features of Va ta Ka phaja Jwaram mentioned in the classics [4].
1.1.3. Bhutabhishangaja Jwara
It can also be grouped under the class of Aagantuja Vika ra with special reference to the class of Bhu tabhishangaja Jwara. The management of Aagantuja Vik ara should follow the lines of Nija Vika ra [5].
1.1.4. Aupasargika Jwara
COVID-19 is transmitting from one human to others by direct contact (Gatra Sansparshat), air droplet (Ni-shwa sat). This type of the disease is termed as Aup a sargika in Ayurveda [6].
1.1.5. Dhatu Paka Awastha
Few severe conditions show symptoms of Sannipata Jwar a leading to Dhatupaka Awsatha , where all three Doshas get vitiated and suppress the Dhatu Bala (immunity) of an individual leading to Oja-Kshaya and ultimately death [7].
1.2. Etiology of disease in ayurveda terms
Dosha – Va ta, Kapha Pradhana
Dushya- Rasa, Rakta, Mansa, Oja
Adhisthana- Koshtha (Aamashaya and Ura Pradesha)
Strotas a - Pranavaha Strotas a , Annavaha Strotas a
Awastha- Stage 1 : Va t a Kaphaj a Jwara
Stage 2 : Va t a Ka ph aj a Sannipatik a Jwara
Stage 3 : Sannipatik a Jwara, Dhatupaka Awastha.
Upashaya- Pravar a Bala- Uttam a Upashaya
Anupashaya: Vrudha Vaya – Anupashaya.
2. Material and methods
2.1. Proteins/macromolecules
COVID-19 3clpro/Mpro (PDB ID: 6LU7) structures was obtained from PDB in.pdb format. PDB is an archive for the crystal structures of biological macromolecules, worldwide. The 6LU7 protein contains two chains, A and B, which form a homodimer. Chain A was used for macromolecule preparation [8]. The coronavirus main protease (Mpro), which plays a pivotal role in viral gene expression and replication through the proteolytic processing of replicase polyproteins, is an attractive target for anti CoV drug design [9].
2.2. Ligand and drug scan
The 160 3-dimensional (3D) structures were obtained from PubChem in.sdf format. PubChem is a chemical substance and biological activities repository consisting of three databases, including substance, compound, and bioassay databases. The compounds used in the present study is shown in Table 2 , along with its binding energy with protein [10].
Table 2.
Properties of COVID-19 Mpro potential inhibitor candidates.
| Chemical constituent | Binding energy | Inhibition | Lipinski rule | Target Prediction | |
|---|---|---|---|---|---|
| Hydroxychloroquine | −3.85 kcal/mol | 1.50 mM | Molecular weight | 335.87 | Tyrosine-protein kinase SYK |
| #H-bond acceptors | 3 | Tyrosine-protein kinase LCK | |||
| #H-bond donors | 2 | Macrophage colony stimulating factor receptor | |||
| iLOGP | 3.58 | ||||
| Lipinski #violations | 0 | ||||
| Quinine | −5.62 kcal/mol | 0.0761 mM | Molecular weight | 324.42 | Tyrosine-protein kinase ABL |
| #H-bond acceptors | 4 | Tyrosine-protein kinase LCK | |||
| #H-bond donors | 1 | Tyrosine-protein kinase SYK | |||
| iLOGP | 3.36 | Macrophage stimulating protein receptor | |||
| Lipinski #violations | 0 | ||||
| Sunthi (Zingiber officinale Roscoe) | |||||
| 6gingesulphonic acid | −6.20 kcal/mol | 5.36 mM | Molecular weight | 358.45 | Interferon-induced, double-stranded RNA-activated protein kinase |
| #H-bond acceptors | 6 | Angiotensin converting enzyme | |||
| #H-bond donors | 2 | Tyrosine-protein kinase LCK- | |||
| iLOGP | 2.48 | Tyrosine-protein kinase HCK | |||
| Lipinski #violations | 0 | ||||
| Pushkarmool (Inula racemose Hook.F.) | |||||
| Alantolactum | −6.70 kcal/mol | 0.01540 mM | Molecular weight | 232.32 | Inhibitor of nuclear factor kappa B kinase beta subunit |
| #H-bond acceptors | 2 | T-cell protein-tyrosine phosphatase p | |||
| #H-bond donors | 0 | Protein-tyrosine phosphatase 1B | |||
| iLOGP | 2.75 | ||||
| Lipinski #violations | 0 | ||||
| Guduchi (Tinospora cordifolia Miers) | |||||
| Berberine | −7.00 kcal/mol | 0.01757 mM | Molecular weight | 336.36 | Inhibitor of nuclear factor kappa B kinase beta subunit |
| #H-bond acceptors | 0 | ||||
| #H-bond donors | 0 | ||||
| iLOGP | 0 | ||||
| Lipinski #violations | 0 | ||||
| Kantakari (Solanum xanthocarpum L.) | |||||
| Stigmasterol | −6.90 kcal/mol | 0.01083 mM | Molecular weight | 412.69 | Angiotensin converting enzyme |
| #H-bond acceptors | 1 | Indoleamine 2,3dioxygenase | |||
| #H-bond donors | 1 | Protein-tyrosine phosphatase 1B | |||
| iLOGP | 4.76 | ||||
| Lipinski #violations | 1 | ||||
| Pippali (Piper longum L.) | |||||
| Bisdemethoxycurcumin | −6.90 kcal/mol | 0.10335 mM | Molecular weight | 308.33 | Glycogen synthase kinase-3 beta |
| #H-bond acceptors | 4 | Inhibitor of NFkappa-B kinase (IKK) | |||
| #H-bond donors | 2 | Macrophage colony stimulating factor receptor | |||
| iLOGP | 1.75 | ||||
| Lipinski #violations | 0 | ||||
| Haridra (Curcuma longa L.) | |||||
| Demethoxycurcumin | −6.20 kcal/mol | 0.188.74 mM | Molecular weight | 338.35 | Inhibitor of NFkappa-B kinase (IKK) |
| #H-bond acceptors | 5 | Toll-like receptor (TLR7/TLR9) | |||
| #H-bond donors | 2 | Tyrosine-protein kinase Lyn | |||
| iLOGP | 2.78 | Tyrosine-protein kinase BTK | |||
| Lipinski #violations | 0 | ||||
| Shatavari (Asparagus racemosus Willd) | |||||
| Kaempferol | −6.70 kcal/mol | 0.06181 mM | Molecular weight | 286.24 | Tyrosine-protein kinase SRC |
| #H-bond acceptors | 6 | Tyrosine-protein kinase receptor UFO | |||
| #H-bond donors | 4 | Interleukin-8 receptor A | |||
| iLOGP | 1.7 | ||||
| Lipinski #violations | 0 | ||||
| Gokshura (Tribulus terrestris L.) | |||||
| Gitogenin | −8.00 kcal/mol | 0.00366 mM | Molecular weight | 432.64 | Tyrosine-protein kinase JAK3 |
| #H-bond acceptors | 4 | Nuclear receptor ROR-gamma | |||
| #H-bond donors | 2 | ||||
| iLOGP | 4.19 | ||||
| Lipinski #violations | 1 | ||||
| Yashtimadhu (Glycyrriza glabra L.) | |||||
| Glycerol | −8.47 kcal/mol | 0.00062273 mM | Molecular weight | 92.09 | Glycogen synthase kinase-3 beta |
| #H-bond acceptors | 3 | Tyrosine-protein kinase ABL | |||
| #H-bond donors | 3 | Tyrosine-protein kinase JAK1 | |||
| iLOGP | 0.45 | Tyrosine-protein kinase JAK3 | |||
| Lipinski #violations | 0 | Tyrosine-protein kinase LCK | |||
| Musta (Cyprus rotundus L.) | |||||
| Patcholane | −6.12 kcal/mol | 0.03258 mM | Molecular weight | 206.37 | Interleukin-6 receptor subunit beta |
| #H-bond acceptors | 0 | Tyrosine-protein kinase JAK1 | |||
| #H-bond donors | 0 | Tyrosine-protein kinase JAK2 | |||
| iLOGP | 3.22 | Tyrosine-protein kinase JAK3 | |||
| Lipinski #violations | 1 | Tyrosine-protein kinase TYK2 | |||
2.3. Molecular docking
The Ligand was docked against the protein and the interactions were analyzed by using Pyrx 0.8. For the docking of ligands into protein active site and to estimate the binding affinities of docked compounds and for generation of grid, studies were carried out using pyrx AutoDock wizard with MGL tools 1.5.6 installed in a Pentium ®Dual –Core CPU T4200 machine running on a 2.0 GHz Intel Core processor with 2 GB RAM by using Lamarkian Algorithm. The scoring function gives score based on best docked ligand complex is picked out [11].
2.4. Swiss ADME
Drug-like properties were calculated using Lipinski’s rule of five, which proposes that molecules with poor permeation and oral absorption have molecular weights >500, C logP >5, more than 5 hydrogen-bond donors, and more than 10 acceptor groups. Adherence with Lipinski’s rule of five were calculated using SWISS ADME prediction [[12], [13], [14]].
2.5. Swiss target prediction
The active site of a protein was determined using the Swiss Target Prediction. The unique engine behind swiss target prediction, extensively detailed elsewhere, calculate the similarity between the users query compound and those compiled in curated, cleansed collection of known actives in well defined experimental binding assays. The quantification of similarity is 2 folds. It gives the active site bind to the molecule [15,16].
3. Results and Discussion
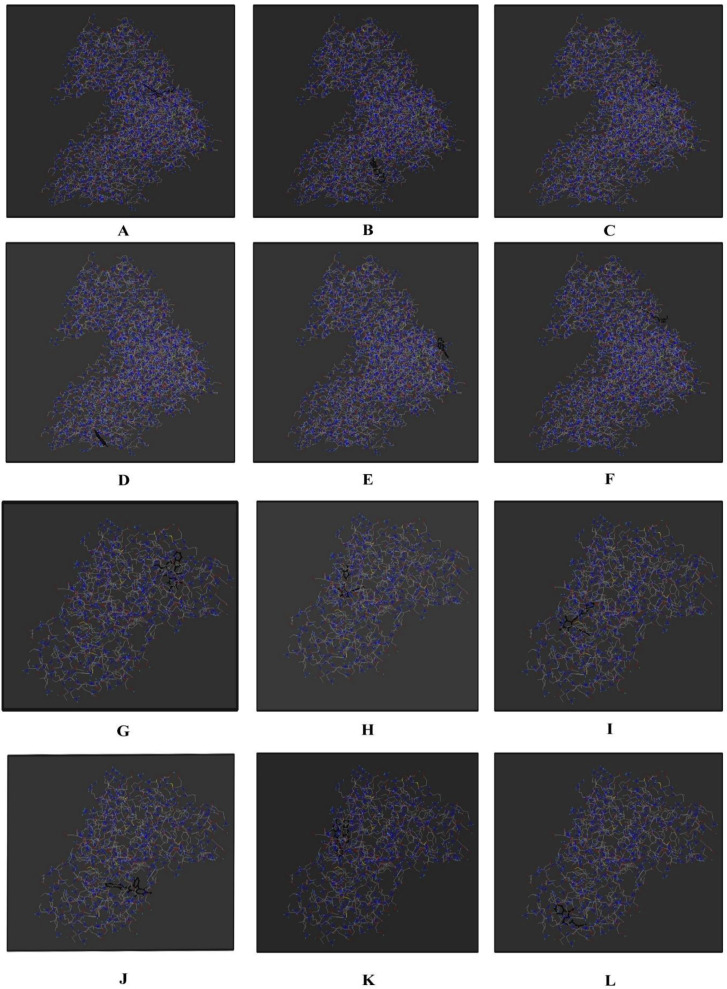
Within weeks from when the genetic sequence of the SARS-CoV-2 was made available, structural biologists have used these techniques to see proteins that make up the SARS-CoV-2 virus. In this study, as shown in Table 1, we have selected 10 drugs on the basis of their ayurvedic properties which can probably be used for SARS-CoV-2. For every plant we carried out molecular docking on each phytoconstituent present in the plant. From this, we selected one phytoconstituent from every plant which is having higher binding energy and inhibition to protein molecule of SARS-CoV-2. The protein molecule used for the study is 6lu7. At present, the hydroxychloroquine and quinine are being used for the treatment of COVID-19 [17]. Thus, we are comparing the efficacy of the following drugs with the hydroxychloroquine and quinine. The molecular docking, Lipinski Rule of Five and Target prediction were calculated. Target prediction for SARS-CoV-2, Humoral immunity and antiviral activity were taken and searched on the selected phytoconstituents. The data is shown in Table 2. Grid of the phytoconcstituents is shown in Fig. 1 .
Table 1.
Ayurvedic properties of certain potential drugs [18].
| Drug name | Latin name | Family | Rasa | Guna | Virya | Doshaghnata | Karma |
|---|---|---|---|---|---|---|---|
| Sunthi | Zingiber officinale Roscoe | Zingiberaceae | Katu | Snigdha, Teekshna | Ushna | Vatakapha hara | Deepana, Rochana, Shoolaprashamana |
| Pushkarmoola | Inula racemose Hook.F. | Asteraceae | Katu Tikta | Laghu | Ushna | Vatakaphahara | Shwasahara, Parshwashool Nut, Jwarghna, Shwaas |
| Guduchi | Tinospra cordifolia Miers | Menispermaceae | Tikta, Katu | Laghu, Snighdha, | Ushna | Tridoshahara, | Jwaraghna, Rasayana, Kasa, Pandu. |
| Kantakari | Solanum zanthocarpum L. | Solanaceae | Katu Tikta | Deepana, Laghu, Sara, Ruksha, | Ushna | Kapha-vataghna | Shwasa –Kasa Jit, Pachani, Parshwaroga |
| Pippali | Piper longum L. | Piperaceae | Katu | Deepana, Snighdha, Laghu | Anushna | Vata-Shleshmahara | Vrushya, Rasayana, Shwasa-Kasa Hara |
| Haridra | Curcuma longa L. | Zingibraceae | Katu, Tikta | Ruksha | Ushna | Kapha-Pittahara | Prameha-Hara, Kushthaghna |
| Shatavari | Asparagys racemosus Willd. | Liliaceae | Tikta, Madhura | Snighdha, Guru | Sheeta | Vata-Pittaghna | Rasayani, Balya, |
| Gokshura | Tribulus terrestris L. | Zygophyllaceae | Madhura | Snighdha, Guru | Sheeta | Vatahara | Balya, Basti-Shodhana, Vrushya, Shwaas-Kaas-Ghna |
| Yashtimadhu | Glycyrrhiza glabra L. | Fabaceae | Madhu | Snighdha, Guru | Sheeta | Pitta-Vata-Ghna. | Balya, Shukral |
| Musta | Cyperus rotundus L. | Cyperaceae | Katu, Tikta, Kashaya | Laghu, Ruksha. | Sheeta | Kapha-Pitta Hara | Deepana- Pachana, Grahi, Jwaraghna |
Fig. 1.
Visualisation of selected phytoconstituents through pyrx. A represents visualisation of hydroxychloroquine with protein 6lu7. B represents visualisation of quinine with protein 6lu7. C represents visualisation of Sunthi (Zingiber officinale Roscoe) with protein 6lu7. D represents visualisation of Pushkarmool (Inula racemose Hook.F.) with protein 6lu7. E represents visualisation of Guduchi (Tinospora cordifolia Miers) with protein 6lu7. F represents visualisation of Kantakari (Solanum xanthocarpum L.) with protein 6lu7. G represents visualisation of Pippali (Piper longum L.) with protein 6lu7. H represents visualisation of Haridra (Curcuma longa L.) with protein 6lu7. I represents visualisation of Shatavari (Asparagus racemosus Willd) with protein 6lu7. J represents visualisation of Gokshura (Tribulus terrestris L.) with protein 6lu7. K represents visualisation of Yashtimadhu (Glycyrriza glabra L.) with protein 6lu7. L represents visualisation of Musta (Cyprus rotundus L.) with protein 6lu7.
From studying the patho-physiology of the disease through Ayurveda perspective, we have selected 10 Ayurvedic drugs based upon their properties. The molecular docking study gives encouraging results for the selected phytoconstituents. 6 gingesulphonic acid which is present in Sunthi is having higher binding energy and inhibition constant than hydroxychloroquine and quinine whereas other phytoconstituents having less binding energy and inhibition constant than 6 gingesulphonic acid. Most of the selected compound follows Lipinski Rule of Five so they are pharmacologically active compounds. Also target prediction of selected phytoconstituents were carried out in which we got the target which are effectable on SARS-CoV-2, Humoral immunity and Antiviral activity.
From overviewing the Ayurvedic perspective, etiology of the disease and studying the phytoconstituents of the drugs, we hypothesized to use Nagaradi Kashaya which includes Sunthi ( Zingiber officinale Roscoe), Pushkarmool (Inula racemose Hook.F.), Guduchi (Tinospora cordifolia Miers.), Kantakari (Solanum virginianum L.) for combating COVID-19. The above combination is mentioned in Jwara Chikitsa. As most of the drugs are having Tikta Rasa, Laghu Guna and Pachana Karma . It helps in Aama Pachana . It also has Ka s a-Shwa s a -ghna properties and are indicated for Jwara, Ka s a , Shw as a and Parshawashoola i.e. the symptoms similar to SARS-CoV-2. Primary evidence for the usage of these drugs, virtual screening of the drugs shows positive results.
As though we use phytoconstituents for this study but further research is necessary to investigate the potential uses of the medicinal plants as a whole.
4. Limitations of the study
Although computer-aided drug design and network pharmacology have been widely used and developed, there still have deficiencies and limitations: (1) The model maturity and computational accuracy of computer docking algorithms need to be further improved. (2) Due to the structure-based methodology, several compounds are not suitable for computer-aided design because of their special structure characteristics. (3) A large number of databases can improve different information for the obtained potential targets, the progress of the selection of these databases and their effective information annotation, still requires continuously practical activities to optimize. With the advancement of computer science, and the constant optimization of algorithms, including the maturity of the protein model. Through more practical researches and development examples are available to upgrade the entire process of in silico methodology, we believe that in the future, this methodological process will enable the discovery of new drugs more efficiently, accurately and quickly. This methodology will be more widely useable in future work on revealing and predicting the basis of medicinal materials.
5. Conclusion
Currently, COVID-19 has emerged in the human population and is a potential threat to global health worldwide. Many drug research and developments projects as well as vaccine development research work is in progress. The aim of this study is to examine several medicinal plant-derived compounds that may be used to inhibit the COVID-19 infection pathway.
Thus, from the above results obtained from reviewing Ayurveda classics and after the virtual screening of selected drugs we can conclude that Nagara di Kashaya may have appreciable results in combating SARS-CoV-2. Nagar adi Kashaya can be a trial candidate for conducting further clinical trial.
Sources of funding
None declared.
Conflicts of interest
None.
Footnotes
Peer review under responsibility of Transdisciplinary University, Bangalore.
References
- 1.Acharya Y.T. Ashtanga Hrudaya of Vagbhata, nidana sthana; sarvaroganidanam : chapter 1, Verse 2. Chowkhambha Sanskrit Series; Varanasi: 2006. p. 441. [Google Scholar]
- 2.World Health Organization (WHO) WHO Bull; 2020. Novel coronavirus (2019-nCoV) pp. 1–7. no. Jan. [Google Scholar]
- 3.Acharya Y.T. Charak samhita of agnivesha, Vimana sthana; janapadaudhwansaniyam Vimanam: chapter 3. Choukhambha Subharati Prakashan; Varanasi: 2013. p. 240. [Google Scholar]
- 4.Acharya Y.T. Charak samhita of agnivesha, nidana sthana; Jwara nidanam: chapter 1, Verse 29. Choukhambha Subharati Prakashan; Varanasi: 2013. p. 201. [Google Scholar]
- 5.Acharya Y.T. Charak samhita of agnivesha, nidana sthana; Jwara nidanam: chapter 1, Verse 30. Choukhambha Subharati Prakashan; Varanasi: 2013. p. 201. [Google Scholar]
- 6.Acharya Y.T. Sushruta samhita of sushruta, nidana sthana; kushtha nidanam: chapter 5, ver. 34. Choukhambha Orientalia; Prakashan; Varanasi: 2013. p. 289. [Google Scholar]
- 7.Acharya Y.T. Charak samhita of agnivesha, chikitsa sthana; Jwara chikitsa: chapter 3, Verse 89. Choukhambha Subharati Prakashan; Varanasi: 2013. p. 406. [Google Scholar]
- 8.Worldwide Protein Data Bank Foundation. 2020. http://www.rcsb.org/ Available form: [cited 2020 May 6] [Google Scholar]
- 9.Liu X., Wang X.J. Potential inhibitors against 2019-nCoV coronavirus M protease from clinically approved medicines. J Gene Genom. 2020 Feb 20;47(2):119. doi: 10.1016/j.jgg.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dallakyan S., Olson A.J. Humana Press; New York, NY: 2015. Small-molecule library screening by docking with PyRx. Chemical biology; pp. 243–250. [DOI] [PubMed] [Google Scholar]
- 11.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 1997 Jan 15;23(1–3):3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 12.Gimenez B.G., Santos M.S., Ferrarini M., Fernandes J.P. Evaluation of blockbuster drugs under the rule-of-five Die Pharmazie. Int J Pharma Sci. 2010 Feb 1;65(2):148–152. [PubMed] [Google Scholar]
- 13.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017 Mar 3;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gfeller D., Grosdidier A., Wirth M., Daina A., Michielin O., Zoete V. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014 Jul 1;42(W1):W32–W38. doi: 10.1093/nar/gku293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swiss Institute of Bioinformatics. 2020. http://www.swissadme.ch/ Available form: [cited 2020 May 6] [Google Scholar]
- 16.Singh A.K., Singh A., Shaikh A., Singh R., Misra A. Choloroquinine and hydroxychloroquinine in the treatment of COVID -19 with or without diabetes: a systematic search and a narrative review with a special reference to India and other developing countries. Diabe Metobol Synd: Clin Res Rev. 2020 Mar 26;14(3):241–246. doi: 10.1016/j.dsx.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhavmisra S., Pandey G. Bhavprakashnighantu. Choukhambha Bharati Academy; Varanasi: 2010. [Google Scholar]
- 18.Acharya Y.T. Ashtanga Hrudaya of Vagbhata, chikitsa sthana; Jwara chikitsa : chapter 1, Verse 66. Chowkhambha Sanskrit Series; Varanasi: 2006. [Google Scholar]