Summary
In the current COVID-19 pandemic context, proposing and validating effective treatments represents a major challenge. However, the scarcity of biologically relevant pre-clinical models of SARS-CoV-2 infection imposes a significant barrier for scientific and medical progress, including the rapid transition of potentially effective treatments to the clinical setting. We use reconstituted human airway epithelia to isolate and then characterize the viral infection kinetics, tissue-level remodeling of the cellular ultrastructure, and transcriptional early immune signatures induced by SARS-CoV-2 in a physiologically relevant model. Our results emphasize distinctive transcriptional immune signatures between nasal and bronchial HAE, both in terms of kinetics and intensity, hence suggesting putative intrinsic differences in the early response to SARS-CoV-2 infection. Most important, we provide evidence in human-derived tissues on the antiviral efficacy of remdesivir monotherapy and explore the potential of the remdesivir-diltiazem combination as an option worthy of further investigation to respond to the still-unmet COVID-19 medical need.
Keywords: COVID-19, cell ultrastructure remodeling, innate immune response, remdesivir, diltiazem, repurposing, drug combination, SARS-CoV-2, coronavirus
Graphical Abstract
Highlights
We use reconstituted human airway epithelia to characterize SARS-CoV-2 infection kinetics
SARS-CoV-2 induces characteristic remodeling of the respiratory epithelium cellular ultrastructure
SARS-CoV-2 induces differential early immune responses in nasal and bronchial HAE
We evaluate the antiviral activity of remdesivir and remdesivir-diltiazem in both Vero E6 and HAE models
Pizzorno et al. report the characterization of SARS-CoV-2 infection, tissue-level remodeling of cellular ultrastructure, and transcriptional immune signatures using a model of reconstituted human airway epithelia. This model was advantageously used to evaluate the antiviral activity of remdesivir or a combination of remdesivir-diltiazem against SARS-CoV-2.
Introduction
On December 31, 2019, a cluster of cases of pneumonia of unknown etiology was reported in Wuhan, China. On January 10, 2020, a novel coronavirus, called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and classified into the Betacoronavirus genus, was identified as the causative agent.1 As of April 20, 2020, 5 weeks after the World Health Organization (WHO) declared the COVID-19 a pandemic,2 novel coronavirus disease 2019 (COVID-19) had caused >166,000 deaths among ∼2.5 million confirmed cases reported in at least 185 countries or territories worldwide.3 Compared to the 2 other coronaviruses responsible for epidemic outbreaks in the past, SARS-CoV and Middle East respiratory syndrome-related coronavirus (MERS-CoV), the novel SARS-CoV-2 strain shares ≈79% and ≈50% genome sequence identity, respectively.4 It is not surprising that important differences in terms of the epidemiology and physiopathology between these 3 viruses have also been observed.5, 6, 7
As with most emerging viral diseases, no specific antiviral treatment nor vaccine against any of these three coronaviruses are available, with standard patient management relying mainly on symptom treatment and respiratory support when needed. In that regard, and considering that key features of the biology of SARS-CoV-2 and its induced COVID-19 still require further characterization, the scarce readiness of biologically relevant pre-clinical experimental models of SARS-CoV-2 infection as a complement of the African green monkey Vero E6 cell line represents a major barrier for scientific and medical progress in this area. We and others have previously reported the advantage of using more physiological models such as in-house or commercially available reconstituted human airway epithelia (HAE) to isolate, culture, and study a wide range of respiratory viruses.8,9 Developed from biopsies of nasal or bronchial cells differentiated in the air-liquid interphase, these models reproduce with high fidelity most of the main structural, functional, and innate immune features of the human respiratory epithelium that play a central role in the early stages of infection and constitute robust surrogates to study airway disease mechanisms and for drug discovery.10 In this study, we initially isolated and amplified in Vero E6 cells a SARS-CoV-2 virus directly from a nasal swab from one of the first hospitalized patients with confirmed COVID-19 in France.11 We then advantageously used human reconstituted airway epithelial models of nasal or bronchial origin to characterize viral infection kinetics, tissue-level remodeling of the cellular ultrastructure, and transcriptional immune signatures induced by SARS-CoV-2, and finally evaluated the therapeutic potential of a combined therapy against COVID-19.
Results
SARS-CoV-2 Isolation and Characterization in Vero E6 Cells and HAE
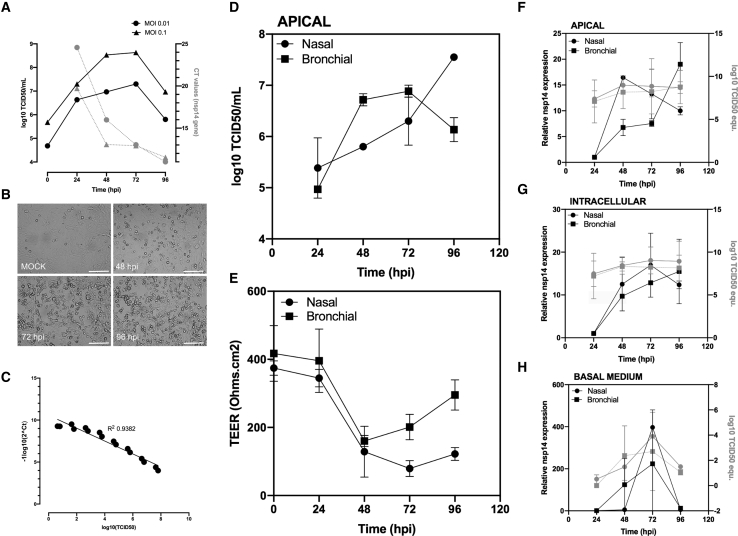
The complete genome sequence of the isolated SARS-CoV-2 virus was deposited in the GISAID EpiCoVTM database under the reference BetaCoV/France/IDF0571/2020 (accession ID EPI_ISL_411218). Phylogenetic analysis confirmed that the isolated virus is representative of the currently circulating strains.12 We characterized the replicative capacities of this viral strain in Vero E6 cells at different multiplicities of infection (MOIs) (Figure 1A) using both classic infectious titer determination in cell culture (TCID50) and molecular semiquantitative methods, the latter based on ORF1b-nsp14-specific primers and probes designed by the School of Public Health at the University of Hong Kong. This double approach was facilitated by the appearance of a clearly observable characteristic cytopathic effect from 48 hpi (Figure 1B) and enabled the validation of a large interval (range 1–8 log10(TCID50)) with high correlation (R2 0.94) between molecular and infectious viral titers (Figure 1C).
Figure 1.
Characterization of a SARS-CoV-2 Infection Model in Vero E6 Cells and in Nasal and Bronchial Reconstituted Human Airway Epithelia (HAE)
(A) SARS-CoV-2 replication kinetics in Vero E6 cells.
(B) Virus-induced cytopathic effects in Vero E6 cells (scale bar, 150 μM).
(C) Correlation between viral quantification methods.
(D) Apical viral production was assessed in washes of the apical pole at 24, 48, 72, and 96 hpi. Vero E6 cells were incubated with serial dilutions of the collected sample for the determination of viral titers (log10 TCID50/mL) at the indicated time points.
(E) Trans-epithelial resistance (TEER in Ω/cm2) between the apical and basal poles was measured at each time point.
(F–H) Relative viral genome quantification at the apical, intracellular, and basal compartments of the HAE was performed after viral or total RNA extraction and qRT-PCR. Results are expressed in fold change of nsp14 expression compared to 24 hpi and also as log10 TCID50 equivalent values.
Presented data (means and SDs) from 3 independent experiments are shown.
In parallel, we successfully inoculated nasal MucilAir HAE on the apical surface directly with nasal swab samples, as confirmed by transmission electron microscopy observations (Figure S1). Characteristic features of coronavirus-induced cell ultrastructure remodeling were easily distinguishable in both the apical and basal sides of the HAE at 48 hpi, notably the high accumulation of progeny virions in mucus-producer goblet cells. Then, we advantageously exploited the MucilAir HAE model and in-house adapted protocols previously optimized for different respiratory viruses13 to perform experimental infections with SARS-CoV-2. Viral replication was monitored through repeated sampling and TCID50 titration at the apical surface of HAE (Figure 1D). Trans-epithelial electrical resistance (TEER), considered a surrogate of epithelium integrity, was also measured during the time course of infection (Figure 1E). In parallel, comparative molecular viral genome quantification was performed at the three levels of the air-liquid HAE interphase: in apical washes (Figure 1F, Apical), total cellular RNA (Figure 1G, Intracellular), and basal medium (Figure 1H, Basal). SARS-CoV-2 viral production at the epithelial apical surface increased sharply at 48 hpi, with the highest virus titers (>7 log10 TCID50/mL) being observed by 72–96 hpi (Figures 1D and 1F). The sharp increase in viral replication correlated with a reduction in epithelium integrity at 48 hpi, reflected by >2.8- and 4-fold decreases in bronchial and nasal HAE TEER values, respectively, followed by a partial recovery in the case of bronchial HAE (Figure 1E). Moreover, viral production at the apical pole was well correlated with intracellular viral genome detection during infection, except for the nasal HAE at 48 hpi, in which a strong relative increase in nsp14 RNA was observed (Figure 1F). Viral genome was detected in the basal medium at 48 hpi and onward (Figure 1H), which indirectly confirms the breach of epithelial integrity caused by the infection at 48 hpi and previously highlighted through TEER measurements.
SARS-CoV-2 Induces Characteristic Remodeling of the Respiratory Epithelium Cellular Ultrastructure
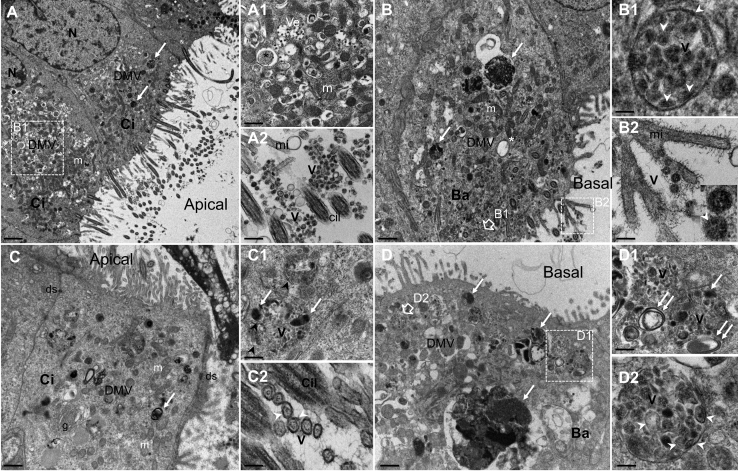
To further characterize the biology of SARS-CoV-2, we inoculated both nasal (Figures 2A and 2B) and bronchial (Figures 2C and 2D) HAE and analyzed the infection-induced remodeling of the cellular ultrastructure using transmission electron microscopy. At 48 hpi, both HAE exhibited a well-established infection, with ciliated, goblet, and to a lesser extent basal cells showing the active production of viral progeny. This observation is in accordance with the viral replication results described in Figure 1 and with a recent study reporting high expression levels of the SARS-CoV-2 cell receptor angiotensin-converting enzyme-2 (ACE2) in both ciliated and goblet respiratory cells.14 As previously observed in structural studies of other coronaviruses, notably SARS-CoV and MERS-CoV,15, 16, 17, 18 we distinguished characteristic clusters in the perinuclear region of infected HAE cells. These clusters are mainly composed of numerous viral single- and double-membrane vesicles (DMVs) and mitochondria (Figures 2A, 2A1, 2B, 2C, and 2D). Large electron-dense structures corresponding to the accumulation of viral material in active virus replication zones as well as typical double-membrane spherules containing pieces of membranes interspaced among virions being formed were also observed at 48 hpi (Figures 2B and 2D). Moreover, double-membraned spherules containing numerous virions are noticeable near the plasmatic membranes (Figures 2B1 and 2D2). These spherules as well as several clusters of virions were observed mostly at the surface of ciliated cells (Figure 2A2). These features are characteristic of the late stages of the viral cycle, hence confirming the capacity of the HAE to reproduce the asynchronous nature of infection.
Figure 2.
Ultrastructure of SARS-CoV-2-Infected Nasal and Bronchial Reconstituted HAE
MucilAir HAE were infected on the apical surface with SARS-CoV-2 (MOI 0.1). Forty-eight hours post-inoculation, HAE were fixed and processed for transmission electron microscopy analysis, as described in Method Details.
(A and B) Section of apical ciliated (Ci) and basal (Ba) cells from nasal HAE showing numerous viral vesicles (DMVs) clustered in the perinuclear region in areas with mitochondria (m) and electron-dense accumulation of viral material (white arrow). Scale bars, 2 μm (A) and 1 μm (B).
(A1) Enlargement of cytoplasmic area with smooth-walled secretory vesicles containing virions (Ve) and virus-induced DMVs (asterisk). Scale bar, 1 μm.
(A2) Enlargement of ciliated cell surface showing virion clusters (V). Microvilli (mi) and transverse sections of cilia (Cil) are also observed. Scale bar, 0.05 μm.
(B1) Enlargement of double-membraned spherules containing electron-dense material and pieces of double membranes interspaced among virions (V). Scale bar, 0.1 μm.
(B2) Enlargement of and virons (V). The white arrows point to viral double membranes seen at high magnification (inset). Scale bar, 0.2 μm.
(C and D) Section of Ci and Ba cells from bronchial HAE showing numerous viral vesicles (DMVs) clustered in the perinuclear region in areas with m and electron-dense accumulation of viral materials (white arrow). Scale bar, 1 μm.
(C1) Enlargement of cytoplasmic area with spherules containing virions being formed (V), electron-dense accumulation of viral material (white arrow), and pieces of membranes (black arrowhead). Scale bar, 0.5 μm.
(C2) High magnification of transverse section of virions (V) at the cell surface with Ci and mi. Their double membrane (white arrowhead) and spikes at their outer edge are visible. Scale bar, 0.1 μm.
(D1) Section of Ba showing virus-induced DMVs, large electron-dense accumulation of viral materials (white arrow), and double-membrane vesicles containing virions near the plasmatic membrane. Scale bar, 1 μm.
(E) Enlargements of the cytoplasmic area containing viral replication sites (double white arrows) and virions being formed (V). Scale bar, 0.1 μm.
(D2) Enlargement of a double-membraned spherule containing virions (V), double-membrane vesicles, and electron-dense viral materials. Scale bar, 0.1 μm. N, nucleus; DMV, cytoplasmic double-membrane vesicles; m, mitochondria; and ds, desmosome.
Representative micrographs are shown from 2 independent experiments.
See also Figure S1.
SARS-CoV-2 Induces Differential Early Immune Responses in Nasal and Bronchial HAE
Recent reports associate COVID-19 with high plasma levels of certain immunostimulant and proinflammatory cytokines (e.g., IL6), notably in patients with severe disease,6 suggesting a potential link with a poor prognosis.19 However, such an inflammatory state has been much less characterized in the respiratory microenvironment thus far. To investigate the effect of SARS-CoV-2 infection on gene expression, we used Nanostring hybridization-based technology on both nasal and bronchial HAE for multiplex mRNA detection and the relative quantification of two complementary panels of genes20 involved in the immune response (Data S1). Heatmap and hierarchical analyses identified two distinct levels of clustering. Regardless of the nasal or bronchial nature of the HAE model, the gene expression profile at 24 hpi highlights a marked upregulation of a subset of ∼14% of the studied genes, which are subsequently downregulated at 48, 72, and 96 hpi (Figure 3A). This observation was substantiated by unsupervised analysis of the data using the full gene panel. The first component of the principal-component analysis (PCA) that accounts for 63% of the variance is mainly driven by the time of infection, with a clear discrimination between 24 hpi and the other time points (Figure 3B, red triangles/dots). The immune transcriptomic signatures seem at least partially driven by the nature of the HAE beyond 24 hpi (Figure 3A). This is in agreement with the second component analysis, gathering 12.6% of total variance, which allowed a clear differentiation between the nasal and bronchial compartments (Figure 3B, green/purple triangles versus dots). In line with a recent report by Blanco-Melo and collaborators,21 specific interferon (IFN) and interferon-stimulated gene (ISG) responses were almost undetectable in the first 24 hpi. Nevertheless, the innate immune expression signature after 24 hpi (peak at 72–96 hpi) is driven by a strong upregulation of type I and type III IFNs (IFNB1, IFNL1, and IFNL2, -3, and -4), as well as other immunity-related genes, notably in the nasal HAE. Whereas only a subset of these genes (CXCL10/IP10, CXCL2/MIP2A, IL1A, IL1B, Mx1, and ZBP1) follow the same pattern in the bronchial HAE, although at overall lower expression levels, the initial modulation of IFNB1, IFNL1, CCL2/MCP1, and IL-6 in this tissue seems to fade at 96 hpi. Moreover, the relative expression of a subset of genes associated with the nuclear factor κB (NF-κB) and tumor necrosis factor α (TNF-α) pathways (e.g., IL-18, IL-18R1, NFKB2, NFKBIA, TNFA, and TNFAIP3) is mostly unchanged all throughout the infection in bronchial HAE, but it is highly upregulated in nasal HAE at 48 hpi and onward (Figure 3C; Data S1). Our results highlight distinctive transcriptional immune signatures between nasal and bronchial HAE, both in terms of kinetics and intensity, hence suggesting potential intrinsic differences in the early response to SARS-CoV-2 infection between the upper and lower respiratory tracts. These results are in accordance with the first clinical reports describing in some patients a rapid worsening of the respiratory condition and overall clinical state by days 7–10 after symptom onset,22 most probably related to a cytokine storm syndrome.23
Figure 3.
Nasal and Bronchial Innate Immune Transcriptional Signature during the Time Course of SARS-CoV-2 Infection
Differential expression of both immune response (96 genes) and type III IFNs (12 genes) panels was evaluated in infected nasal and bronchial HAE using the Nanostring technology at the indicated time points. Data processing and normalization were performed with nSolver analysis software (version 4.0, NanoString Technologies), and the results are expressed in fold change induction compared to the mock condition.
(A) Heatmap and hierarchical clustering of differentially expressed genes compared to the mock-infected condition.
(B) Principal-component analysis (PCA).
(C) mRNA expression ratio of selected genes compared to the mock infected condition.
Data from 2 independent experiments are shown.
Remdesivir-Diltiazem Combined Treatment Potentiates the Efficacy of Remdesivir Monotherapy
The current absence of specific treatments against COVID-19 results in the empirical repurposing of approved or experimental drugs designed for other diseases. These treatments are usually based on limited clinical or preclinical data. Remdesivir (GS-5734) is a prodrug of an adenosine nucleotide analog with demonstrated broad antiviral activity against several RNA viruses in different preclinical models.24 Remdesivir has recently shown very promising results in animal models for the treatment of different coronaviruses, including MERS-CoV,25 as well as one in vitro study against SARS-CoV-2.26 Various clinical trials on the use of remdesivir for the treatment of patients with COVID-19 have already started in China and the United States. Diltiazem is a voltage-gated Ca2+ channel antagonist commonly used as an antihypertensive for the control of angina pectoris and cardiac arrhythmia.27 In a recent study, we repurposed diltiazem as an effective host-directed influenza inhibitor due to its thus far undescribed capacity of inducing the IFN antiviral response, particularly IFN-1β and type III IFNs (Figure S2),8 henceforth prompting an ongoing Phase IIb clinical trial to assess the efficacy of diltiazem-oseltamivir bitherapy in patients with severe influenza (FLUNEXT TRIAL PHRC #15-0442, NCT03212716). The rationale behind testing this kind of virus-directed plus host-directed drug combination is consistent with the evaluation of the potential benefits of a remdesivir-diltiazem combined treatment against COVID-19. In that regard, a recent study describing hypertension as a potential risk factor observed among a cohort of inpatients with COVID-19,19 as well as two reports not anticipating potential adverse effects of diltiazem28 or negative pharmacological interactions between remdesivir and diltiazem for the treatment of COVID-19,29 further support our strategy.
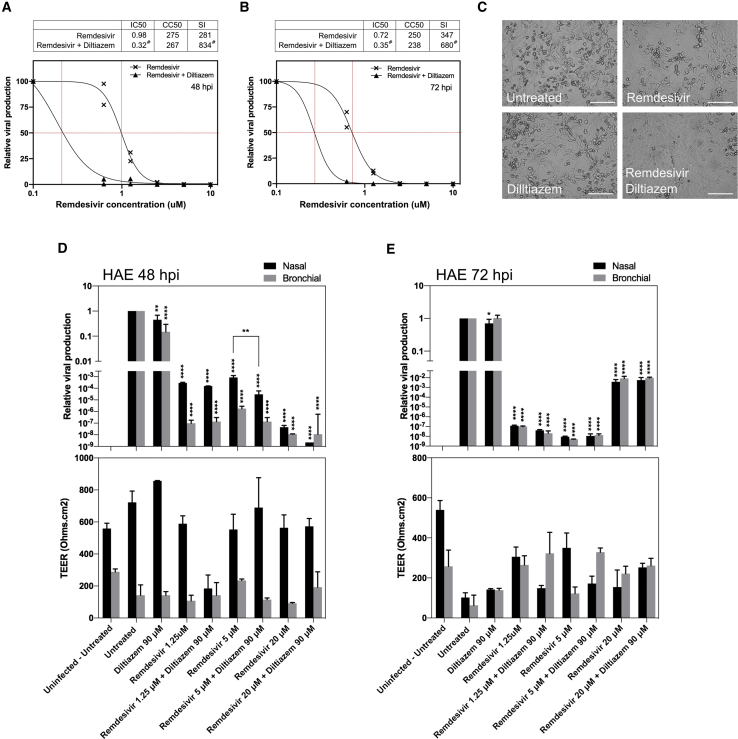
We therefore explored the antiviral potential against SARS-CoV-2 of remdesivir monotherapy, but also in combination with diltiazem in both Vero E6 and HAE. Figures 4A–4C shows a very strong antiviral effect of post-infection treatment with remdesivir in Vero E6 cells, with 50% inhibitory concentration (IC50) values of 0.98 ± 0.07 μM at 48 hpi and 0.72 ± 0.03 μM at 72 hpi and a selectivity index (SI) of 281 and 347, respectively. Although the IFN-λ1 response in Vero E6 cells has been proved functional, this cell line cannot produce type I IFNs.30,31 This incomplete IFN response most likely accounts for the lack of significant antiviral effect observed with diltiazem monotherapy in our experimental conditions. Nonetheless, the addition of 11.5 μM diltiazem significantly potentiated the antiviral effect of remdesivir, as evidenced by the shift on dose-response curves toward IC50 values at least 3- and 2-fold lower than those observed for remdesivir monotherapy at 48 and 72 hpi, respectively (Figures 4A–4C).
Figure 4.
Evaluation of Antiviral Activity of Remdesivir-Diltiazem in Vero E6 Cells and in HAE
(A and B) Dose-response curves of remdesivir and remdesivir-diltiazem combination (11.5 μM fixed diltiazem concentration) at 48 hpi (A) and 72 hpi (B) in Vero E6 cells. #Estimated IC50 and SI values.
(C) Effect of antiviral treatment on virally induced cytopathic effects in Vero E6 cells (scale bar, 150 μM).
(D and E) Relative intracellular viral genome quantification and trans-epithelial resistance (TEER in Ω/cm2) between the apical and basal poles in nasal and bronchial HAE at (D) 48 and (E) 72 hpi. Results are expressed in relative viral production compared to the infected untreated control and relative TEER compared to t = 0 (before infection).
Data (means ± SDs) from 3 independent experiments are shown. Statistical significance was calculated by 1-way ANOVA; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗∗p < 0.0001 versus untreated group.
See also Figure S2.
Daily basolateral treatment of HAE with 1.25, 5, and 20 μM remdesivir resulted in 3.5, 3.1, and 7.3 log10 reductions of intracellular SARS-CoV-2 nasal HAE viral titers at 48 hpi, respectively. Comparably, 7.0, 5.8, and 7.9 log10 reductions of bronchial HAE viral titers were observed at the same time point (Figure 4D, upper panel). Nasal and bronchial HAE viral titers were also reduced at 72 hpi following treatment with 1.25 μM (6.9 and 7.0 log10), 5 μM (8.0 and 8.3 log10), and 20 μM (2.4 and 2.0 log10) remdesivir, respectively (Figure 4E, upper panel). It is not surprising for a model with a completely functional IFN response that daily treatment with 90 μM diltiazem resulted in moderate reductions of intracellular viral titers in nasal (40% and 69%) and bronchial (80% and 24%) HAE at 48 and 72 hpi, respectively (Figures 4D and 4E, upper panel). We observed additional 1.45 and 1.3 log10 reductions in nasal HAE viral titers at 48 hpi for the remdesivir-diltiazem combination when compared with 5 and 20 μM remdesivir monotherapies, respectively, although only the former was statistically significant (p = 0.0066). Moreover, TEER analysis revealed that the antiviral effects induced by remdesivir, diltiazem, or the remdesivir-diltiazem combination mostly translated into a protection of the nasal HAE barrier integrity at 48 and 72 hpi (Figures 4D and 4E, lower panels). Regardless of their effect on viral reduction, the 3 remdesivir-diltiazem combinations tested were particularly effective in protecting the integrity of bronchial HAE at 72 hpi, as evidenced by TEER values comparable to those of non-infected controls (Figure 4E, lower panel).
Discussion
Here, we report the relevance of reconstituted HAE as an efficient model for the study of respiratory viral infections and virus-host interactions in highly biologically relevant experimental conditions. The results we obtained in this complex model provide meaningful contributions to the characterization of the kinetics of viral infection and on the tissue-level remodeling of the cellular ultrastructure and local innate immune responses induced by SARS-CoV-2. Nanostring results highlight a differential effect of SARS-CoV-2 infection on the early innate immune response of nasal and bronchial HAE. These responses are in line with viral replication kinetics, and in some cases,6 but not in others,32 follow trends similar to what has been described in blood samples from patient cohorts. We expect that our results will provide a benchmark for future studies aimed at further characterizing the local pathophysiology and immune response to SARS-CoV-2 infection, particularly in the lower respiratory tract, with the ultimate objective of providing insight in terms of putative prognostic biomarkers and/or patient management.
Finally, the HAE model of SARS-CoV-2 infection described in this study also constitutes an advantageous physiologic model to evaluate candidate therapeutic approaches, provided that in many cases the inhibitory effects observed in classic reductionist models of immortalized cell lines do not necessarily translate into a real clinical setting.33 Most important, we provide valuable evidence on the antiviral efficacy of remdesivir in both the upper and lower human respiratory tracts and on the potential of the remdesivir-diltiazem combination as an option worthy of further investigation to respond to the current medical unmet need imposed by COVID-19. The combination of a virus-directed plus a host-directed drug could result in enhanced antiviral and/or immunomodulatory effects, including during the immunopathologic phase frequently observed during the second week of infection. This association could also reduce the therapeutic doses of chemotherapeutic agents targeting nucleic acid synthesis and therefore minimize putative adverse side effects. This two-pronged approach would be of special interest for the treatment of at-risk patients within the first 10 days from symptom onset, when the effective clearance of viral load still plays a major role in the clinical outcome.34
Limitations of Study
Besides the above-mentioned physiological advantages of the HAE model, the absence of white blood cells (e.g., neutrophils, monocytes, dendritic cells) may partially bias the analysis of the innate immune response induced by infection. Moreover, although this study suggests the potential of the remdesivir-diltiazem combination in the treatment of certain SARS-CoV-2 infections, its validation and implementation in the clinic need to be explored further.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| SARS-CoV-2 BetaCoV/France/IDF0571/2020 | This paper | EPI_ISL_411218 |
| Biological Samples | ||
| SARS-CoV-2-positive human nasal swab sample | Bichat Claude Bernard Hospital, Paris | Lescure et al. 11 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Remdesivir | Med Chem Tronica | Ref: HY-104077, CAS: 1809249-37-3 |
| (+)-cis-Diltiazem hydrochloride | Sigma-Aldrich | Ref: D2521, CAS: 33286-22-5 |
| Critical Commercial Assays | ||
| NanoString nCounter® Custom Gene Expression panels | NanoString Technologies | MAN-10083-01 |
| CellTiter 96® AQueous One Solution Cell Proliferation Assay | Promega | Ref: G3580 |
| Deposited Data | ||
| NanoString source data | This paper | Data S1 |
| Experimental Models: Cell Lines | ||
| VeroE6 cells | ATCC | CRL-1586 |
| MucilAir Nasal HAE | Epithelix SARL | Ref: EP02MP |
| MucilAir Bronchial HAE | Epithelix SARL | Ref: EP01MD |
| Oligonucleotides | ||
| Forward primer HKU-ORF1b-nsp14F: 5′-TGGGGYTTTACRGGTAACCT-3′ | Eurogentec | N/A |
| Reverse primer HKU-ORF1b-nsp14R: 5′-AACRCGCTTAACAAAGCACTC-3′ | Eurogentec | N/A |
| Probe HKU-ORF1b-nsp141P: 5′-FAM-TAGTTGTGATGCWATCATGACTAG-TAMRA-3′ | Eurogentec | N/A |
| Software and Algorithms | ||
| nSolver | NanoString Technologies | https://www.nanostring.com/products/analysis-software/nsolver |
| Genomics Suite 7 | Partek | https://documentation.partek.com/ |
| DigitalMicrograph | Gatan | https://www.gatan.com/products/tem-analysis/gatan-microscopy-suite-software |
| Quest Graph IC50 calculator | AAT Bioquest | https://www.aatbio.com/tools/ic50-calculator |
| Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Olivier Terrier (olivier.terrier@univ-lyon1.fr).
Materials availability
The SARS-CoV-2 isolate generated in is study will be made available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
The complete genome sequence of the isolated SARS-CoV-2 is available at the GISAID EpiCoVTM database under the reference BetaCoV/France/IDF0571/2020 (accession ID EPI_ISL_411218). Original data have been deposited to Mendeley Data: http://dx.doi.org/10.17632/cxs7xwjpfj.1. Source data for Figures 1 and 4 are available from the corresponding author upon request.
Experimental Model and Subject Details
Cells
Vero E6 cells (ATCC CRL-1586) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Lonza, MD, USA) with 4.5 g/L glucose, 10% Fetal Calf Serum (FCS), 1 mM L-Glutamine and 100 U/mL penicillin-streptomycin at 37°C and 5% CO2.
HAE
MucilAir™ HAE reconstituted from human primary cells obtained from nasal or bronchial biopsies were provided by Epithelix SARL (Geneva, Switzerland) and maintained in air-liquid interphase with specific culture medium in Costar Transwell inserts (Corning, NY, USA) according to the manufacturer’s instructions. The nasal HAE used in this study were derived from a pool of 14 donors (gender not specified) whereas the bronchial HEA were derived from a single donor (56 y-o Caucasian female). In both cases, no existing pathologies were identified. All samples provided by Epithelix SARL have been obtained with informed consent as part of studies or other processes that have had ethical review and approval. These studies were conducted according to the declaration of Helsinki on biomedical research (Hong Kong amendment, 1989), and received approval from local ethics commission.
Virus
The SARS-CoV-2 strain used in this study was isolated from a 47 y-o female patient enrolled in a French clinical cohort assessing patients with COVID-19 (NCT04262921). This study was conducted according to the declaration of Helsinki and received approval from local ethics commission. The viral strain was sequenced with Illumina MiSeq and deposited in the GISAID EpiCoVTM database under the reference BetaCoV/France/IDF0571/2020 (accession ID EPI_ISL_411218).
Method Details
Clinical samples, viral isolation, and sequencing
All experiments involving clinical samples and the manipulation of infectious SARS-CoV-2 were performed in biosafety level 3 (BSL-3) facilities, using appropriate protocols. The SARS-CoV-2 strain used in this study was isolated by directly inoculating Vero E6 cell monolayers with a nasal swab sample collected from a one of the first COVID-19 cases confirmed in France: a 47 y-o female patient hospitalized in January 2020 in the Department of Infectious and Tropical Diseases, Bichat Claude Bernard Hospital, Paris11. Once characteristic CPE was observable in more than 50% of the cell monolayer, supernatants were collected and immediately stored at −80°C for subsequent viral RNA extraction using the QiAmp viral RNA Kit (QIAGEN). The complete viral genome sequence was obtained using Illumina MiSeq sequencing technology, was then deposited after assembly on the GISAID EpiCoV platform (Accession ID EPI_ISL_411218) under the name BetaCoV/France/IDF0571/2020.
Viral quantification
Viral stocks and collected samples were titrated by classic tissue culture infectious dose 50% (TCID50/ml) in Vero E6 cells, using the Reed & Muench statistical method. In parallel, relative quantification of viral genome was performed by one-step real-time quantitative reverse transcriptase and polymerase chain reaction (RT-qPCR) from viral or total RNA extracted using QiAmp viral RNA or RNeasy Mini Kit (QIAGEN) in the case of supernatants/apical washings or cell lysates, respectively. Primer and probe sequences targeting the ORF1b-nsp14 (forward primer HKU-ORF1b-nsp14F: 5′-TGGGGYTTTACRGGTAACCT-3′; reverse primer HKU-ORF1b-nsp14R: 5′-AACRCGCTTAACAAAGCACTC-3′; probe HKU-ORF1b-nsp141P: 5′-FAM-TAGTTGTGATGCWATCATGACTAG-TAMRA-3′) were selected from those designed by the School of Public Health/University of Hong Kong (Leo Poon, Daniel Chu and Malik Peiris) and synthetized by Eurogentec. Real-time one-step RT-qPCR was performed using the EXPRESS One-Step Superscript qRT-PCR Kit (Invitrogen, reference 1178101K), in a 20 μl reaction volume containing 10 μl of Express qPCR supermix at 2X, 1 μl of forward primer at 10 μM, 1 μl of reverse primer at 10 μM, 0.5 μl of probe at 10 μM, 3.1 μl of PCR-water (QIAGEN, reference 17000-10), 0.4 μl of Rox dye at 25 μM, and 2 μl of vRNA template. Thermal cycling was performed in a StepOnePlus Real-Time PCR System (Applied Biosystems) in MicroAmp Fast Optical 96-well reaction plates (Applied Biosystems, reference 4346907). Cycling conditions were as follows: reverse transcription at 50°C during 15 min, followed by initial polymerase activation at 95°C for 2 min, and then 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 1 minute. The investigator in charge of TCID50 or RT-qPCR titration was blinded to group allocation.
Viral replication kinetics and antiviral treatment in Vero E6 cells
Vero E6 cells were seeded 24 h in advance in multi-well 6 plates, washed twice with PBS and then infected with a 1 mL suspension of SARS-CoV-2 in infection medium (DMEM 4.5 g/L glucose) at the indicated MOIs. For replication kinetics studies, supernatant samples were collected at different time-points and separated into 2 tubes: one for TCID50 viral titration and one RT-qPCR. For treatment studies, the inoculum of infected Vero E6 was removed 1 hpi and cells were immediately treated with 2 mL of serial dilutions of a solution in treatment medium (DMEM 4.5 g/L glucose, 2% FCS) of candidate molecules alone or in combination, and then incubated at 37°C and 5% CO2. Treatment medium alone (for diltiazem) or containing a DMSO concentration equivalent to that of the highest remdesivir dose tested was used as mock-treatment control. Supernatants were collected at 48 and 72 hpi and stored at −80°C for RNA extraction and viral titration. Cell toxicity induced by remdesivir and diltiazem was evaluated using the colorimetric CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega) following the manufacturer instructions. Drug 50% cytotoxic concentration (CC50), 50% inhibitory concentration (IC50) and selectivity index (SI) values were calculated using the Quest Graph IC50 calculator (AAT Bioquest).
Viral infection and treatment in reconstituted human airway epithelia (HAE)
For infection experiments, the apical poles of HAE were gently washed twice with warm OptiMEM medium (GIBCO, ThermoFisher Scientific) and then infected directly with patient nasal swab samples or a 150 μL dilution of virus in OptiMEM medium, at a multiplicity of infection (MOI) of 0.1. For mock infection, the same procedure was followed using OptiMEM as inoculum. Samples collected from apical washes or basolateral medium at different time-points after incubation at 37°C and 5% CO2 were separated into 2 tubes: one for TCID50 viral titration and one RT-qPCR. HAE cells were harvested in RLT buffer (QIAGEN) and total ARN was extracted using the RNeasy Mini Kit (QIAGEN) for subsequent RT-qPCR and Nanostring assays. Treatments with specific dilutions of candidate molecules alone or in combination in 700 μL of MucilAir® culture medium were applied through basolateral poles. All treatments were initiated on day 0 (1h after viral infection) and continued by replacing the basal medium once daily at 24 and 48 hpi (2 and 3 treatments in total for samples collected at 48 and 72 hpi, respectively). Variations in transepithelial electrical resistance (Δ TEER) were measured using a dedicated volt-ohm meter (EVOM2, Epithelial Volt/Ohm Meter for TEER) and expressed as Ohm.cm2.
NanoString gene expression analysis
After on-column mRNA extraction, gene expression levels were evaluated using two customized NanoString nCounter Gene Expression Panels: “immune response” (96 genes) and “type III IFNs” (12 genes). Data processing and normalization were performed with nSolver analysis software (version 4.0, NanoString technologies) and results are expressed in fold change induction compared to the mock condition. Heatmap and Principal Component Analysis (PCA) were produced using Genomics Suite 7 (Partek, St Louis, MO, USA).
Transmission electron microscopy
Infected nasal and bronchial HAE were fixed with 2% glutaraldehyde (EMS) in 0.1 M sodium cacodylate (pH 7.4) buffer at room temperature for 30 min. After washing three times in 0.2 M sodium cacodylate buffer, cell cultures were post-fixed with 2% osmium tetroxide (EMS) at room temperature for 1 h and deshydrated in a graded series of ethanol at room temperature and embedded in Epon. After polymerization, ultrathin sections (100 nm) were cut on a UCT (Leica) ultramicrotome and collected on 200 mesh grids. Sections were stained with uranyl acetate and lead citrate before observations on a Jeol 1400JEM (Tokyo, Japan) transmission electron microscope, equipped with an Orius 600 camera and Digital Micrograph Software (Gatan).
Quantification and Statistical Analysis
No statistical methods were used to predetermine sample size. All experimental assays were performed in triplicate at a minimum, except for NanoString analyses that were performed in duplicate. Statistical details of experiments can be found in the figure legends. The testing level (α) was 0.05. Statistical analyses were performed on all available data using GraphPad Prism 8.
Acknowledgments
The authors want to acknowledge Denis Ressnikoff and the CIQLE team (Université Claude Bernard Lyon 1) for their technical assistance with electron microscopy image acquisition and analysis, as well as Dr. Guy Boivin for his critical reading of the manuscript. This study was funded by INSERM REACTing (REsearch & ACtion emergING infectious diseases), CNRS, and Mérieux research grants. The sponsors had no role in study design, collection, analysis and interpretation of data, manuscript writing, or in the decision to submit the article for publication.
Author Contributions
Conceptualization, A.P., O.T., and M.R.-C. Formal Analysis, A.P., S.T.-A., J.F., and O.T. Funding Acquisition, B.L., O.T., and M.R.-C. Investigation, A.P., B.P., T.J., S.T.-A., A.T., E.E.-C., V.D., J.D., and O.T. Methodology, S.C. (MucilAir HAE). Project Administration, O.T. and M.R.-C. Resources, A.G., S.C., B.L., and Y.Y. Supervision, A.P., O.T., and M.R.-C. Validation, B.L., O.T., and M.R.-C. Visualization, A.P., S.T.-A., O.T., and M.R.-C. Writing – Original Draft, A.P., O.T., and M.R.-C. Writing – Review & Editing, S.C., J.P., B.L., Y.Y., and M.R.-C.
Declaration of Interests
A.P., B.P., T.J., A.T., O.T., and M.R.-C. are co-inventors of a patent application filed by INSERM, CNRS, Université Claude Bernard Lyon 1, and Signia Therapeutics for the repurposing of diltiazem for the treatment of SARS-CoV-2 infections (FR 20/02351). A.P., O.T., and M.R.-C. are co-founders of Signia Therapeutics SAS. S.C. is the founder of Epithelix, the developer and provider of MucilAir HAE. F.-X.L. has received funding for lectures from Gilead Sciences and Merck Sharp & Dohme France, and funding for travel to meetings from Merck Sharp & Dohme France, Astellas, and Eumedica. B.L. is the co-chair of the Global Influenza and RSV Initiative and the chair of the scientific committee of the Global Influenza Hospital Surveillance Network. B.L. received no personal remuneration for these activities. B.L. received travel grants to attend meetings by Abbott, Seegene, Sanofi, and bioMérieux. All of the other authors declare no competing interests.
Published: July 21, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xcrm.2020.100059.
Contributor Information
Olivier Terrier, Email: olivier.terrier@univ-lyon1.fr.
Manuel Rosa-Calatrava, Email: manuel.rosa-calatrava@univ-lyon1.fr.
Supplemental Information
Briefly, after on-columns mRNA extraction, expression was evaluated using two different gene panels (composed of 96 and 12 genes respectively) using the Nanostring technology. Data treatment and normalization ware performed with nSolver analysis software (version 4.0, NanoString technologies). Results are expressed in counts and fold change induction compared to the mock condition.
References
- 1.Hui D.S., I Azhar E., Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., Mchugh T.D., Memish Z.A., Drosten C. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . 2020. WHO Director-General’s opening remarks at the media briefing on COVID-19 - 11 March 2020.https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 [Google Scholar]
- 3.World Health Organization . 2020. Novel Coronavirus (2019-nCoV) Situation Report - 3, 23 January 2020.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200123-sitrep-3-2019-ncov.pdf?sfvrsn=d6d23643_8 [Google Scholar]
- 4.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. 2020. Features, Evaluation and Treatment Coronavirus (COVID-19)https://www.ncbi.nlm.nih.gov/books/NBK554776/ [PubMed] [Google Scholar]
- 6.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y., Wang Y., Chen Y., Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J. Med. Virol. 2020;92:568–576. doi: 10.1002/jmv.25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pizzorno A., Terrier O., Nicolas de Lamballerie C., Julien T., Padey B., Traversier A., Roche M., Hamelin M.-E., Rhéaume C., Croze S. Repurposing of Drugs as Novel Influenza Inhibitors From Clinical Gene Expression Infection Signatures. Front. Immunol. 2019;10:60. doi: 10.3389/fimmu.2019.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farsani S.M., Deijs M., Dijkman R., Molenkamp R., Jeeninga R.E., Ieven M., Goossens H., van der Hoek L. Culturing of respiratory viruses in well-differentiated pseudostratified human airway epithelium as a tool to detect unknown viruses. Influenza Other Respir. Viruses. 2015;9:51–57. doi: 10.1111/irv.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boda B., Benaoudia S., Huang S., Bonfante R., Wiszniewski L., Tseligka E.D., Tapparel C., Constant S. Antiviral drug screening by assessing epithelial functions and innate immune responses in human 3D airway epithelium model. Antiviral Res. 2018;156:72–79. doi: 10.1016/j.antiviral.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lescure F.-X., Bouadma L., Nguyen D., Parisey M., Wicky P.-H., Behillil S., Gaymard A., Bouscambert-Duchamp M., Donati F., Le Hingrat Q. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect. Dis. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.GISAID . 2020. Genomic epidemiology of hCoV-19.https://www.gisaid.org/epiflu-applications/next-hcov-19-app/ [Google Scholar]
- 13.Nicolas de Lamballerie C., Pizzorno A., Dubois J., Julien T., Padey B., Bouveret M., Traversier A., Legras-Lachuer C., Lina B., Boivin G. Characterization of cellular transcriptomic signatures induced by different respiratory viruses in human reconstituted airway epithelia. Sci. Rep. 2019;9:11493. doi: 10.1038/s41598-019-48013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sungnak, W., Huang, N., Bécavin, C., Berg, M., and HCA Lung Biological Network. SARS-CoV-2 Entry Genes Are Most Highly Expressed in Nasal Goblet and Ciliated Cells within Human Airways. arXiv 10.1038/s41591-020-0868-6, arXiv:2003.0612.
- 15.Knoops K., Kikkert M., Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLOS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldsmith C.S., Tatti K.M., Ksiazek T.G., Rollin P.E., Comer J.A., Lee W.W., Rota P.A., Bankamp B., Bellini W.J., Zaki S.R. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 2004;10:320–326. doi: 10.3201/eid1002.030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Afzelius B.A. Ultrastructure of human nasal epithelium during an episode of coronavirus infection. Virchows Arch. 1994;424:295–300. doi: 10.1007/BF00194614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Wilde A.H., Raj V.S., Oudshoorn D., Bestebroer T.M., van Nieuwkoop S., Limpens R.W.A.L., Posthuma C.C., van der Meer Y., Bárcena M., Haagmans B.L. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment. J. Gen. Virol. 2013;94:1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mouton W., Albert-Vega C., Boccard M., Bartolo F., Oriol G., Lopez J., Pachot A., Textoris J., Mallet F., Brengel-Pesce K., Trouillet-Assant S. Towards standardization of immune functional assays. Clin. Immunol. 2020;210:108312. doi: 10.1016/j.clim.2019.108312. [DOI] [PubMed] [Google Scholar]
- 21.Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., HLH Across Speciality Collaboration, UK COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheahan T.P., Sims A.C., Leist S.R., Schäfer A., Won J., Brown A.J., Montgomery S.A., Hogg A., Babusis D., Clarke M.O. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.PubChem . 2019. Compound Summary: Diltiazem.https://pubchem.ncbi.nlm.nih.gov/compound/39186 [Google Scholar]
- 28.Fang L., Karakiulakis G., Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir. Med. 2020;8:e21. doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.University of Liverpool . 2020. COVID-19 Interactions.https://www.covid19-druginteractions.org/ [Google Scholar]
- 30.Emeny J.M., Morgan M.J. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 1979;43:247–252. doi: 10.1099/0022-1317-43-1-247. [DOI] [PubMed] [Google Scholar]
- 31.Prescott J., Hall P., Acuna-Retamar M., Ye C., Wathelet M.G., Ebihara H., Feldmann H., Hjelle B. New World hantaviruses activate IFNlambda production in type I IFN-deficient vero E6 cells. PLOS ONE. 2010;5:e11159. doi: 10.1371/journal.pone.0011159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thevarajan I., Nguyen T.H.O., Koutsakos M., Druce J., Caly L., van de Sandt C.E., Jia X., Nicholson S., Catton M., Cowie B. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 2020;26:453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y., Yan L.-M., Wan L., Xiang T.-X., Le A., Liu J.-M., Peiris M., Poon L.L.M., Zhang W. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect. Dis. 2020;20:656–657. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Briefly, after on-columns mRNA extraction, expression was evaluated using two different gene panels (composed of 96 and 12 genes respectively) using the Nanostring technology. Data treatment and normalization ware performed with nSolver analysis software (version 4.0, NanoString technologies). Results are expressed in counts and fold change induction compared to the mock condition.
Data Availability Statement
The complete genome sequence of the isolated SARS-CoV-2 is available at the GISAID EpiCoVTM database under the reference BetaCoV/France/IDF0571/2020 (accession ID EPI_ISL_411218). Original data have been deposited to Mendeley Data: http://dx.doi.org/10.17632/cxs7xwjpfj.1. Source data for Figures 1 and 4 are available from the corresponding author upon request.