To the Editor:
The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the resulting illness, coronavirus disease 2019 (COVID-19), has affected >9 million people globally, with >470,000 deaths. The highest number of cases and fatalities have occurred in the United States, with >120,000 deaths as of the end of June.1 In contrast to other common viral infections, COVID-19 presents unique challenges with high rates of hypoxemic respiratory failure, hyperinflammatory cytokine storm, coagulation abnormalities, and cardiac and renal dysfunction.2 , 3 Identification of COVID-19 subphenotypes could lead to better understanding of the diverse host responses that result in these heterogeneous presentations.
Fever is a common presenting symptom in COVID-19, and the thermoregulatory response to infection operates at the intersection of the immunologic, neurologic, cardiovascular, and other body systems.4 , 5 Thus, longitudinal temperature trajectories could provide unique insights into the multiorgan dysfunction that is seen with COVID-19. We previously published a novel method of identifying subphenotypes in hospitalized patients with all-cause infection with the use of longitudinal body temperature measurements.6 These temperature trajectory subphenotypes had distinct demographics, physiologic and immune markers, and outcomes. We hypothesize that using a similar approach that is specific to patients with COVID-19 would identify subphenotypes with unique clinical characteristics and inflammatory and coagulation abnormalities. Importantly, we hypothesize that these temperature trajectory subphenotypes will have distinct outcomes, with the primary outcome of interest being 30-day inpatient mortality rate.
Methods
We included all adult patients who were admitted to University of Chicago Medicine between March 1 and June 24, 2020, who tested positive for SARS-CoV-2. We excluded patients who had been tested for SARS-CoV-2 more than three days after admission to exclude potential hospital-acquired cases. We also excluded patients who were discharged or died within 24 hours of hospitalization, because these patients may not have adequate temperature data to be classified by the algorithm. We included temperature measurements that had been taken in the first 72 hours of hospitalization in the algorithm. The temperature data from hour 0 to hour 72 were split into one-hour blocks of time. For patients with multiple temperature measurements in a one-hour block, the earliest measurement was used. We applied group-based trajectory modeling (GBTM) to identify the temperature trajectory subphenotypes. GBTM is a finite mixture model used to identify clusters of patients following similar trajectories of a variable of interest (ie, temperature).7 We selected the four-group quadratic model based on our prior work.6 The GBTM algorithm computes a unique quadratic equation of temperature as a function of time for each of the four subphenotypes. Patients are classified into the temperature trajectory subphenotype whose quadratic function most closely matches their temperature measurements. Once patients were classified into subphenotypes, the differences in clinical characteristics between the subphenotypes were compared with the use of analysis of rank (analysis of variance) or chi-squared tests, as appropriate. The primary outcome was 30-day inpatient mortality rate, which was modeled on subphenotypes with the use of Cox regression analysis, controlling for demographics, comorbidities, and severity of illness. Patients who were discharged from the hospital before 30 days were assumed to be alive at 30 days for the regression analysis. GBTM was performed with the traj package command in Stata software (Stata Corp LLC, College Station, TX).
Results
Our final cohort consisted of 696 hospitalized patients who had tested positive for SARS-CoV-2. The median age was 61 years (interquartile range, 47-73 y), with 51% men, and a predominantly black patient population (85%). The ICU admission rate was 35%; the 30-day inpatient mortality rate was 8.8%.
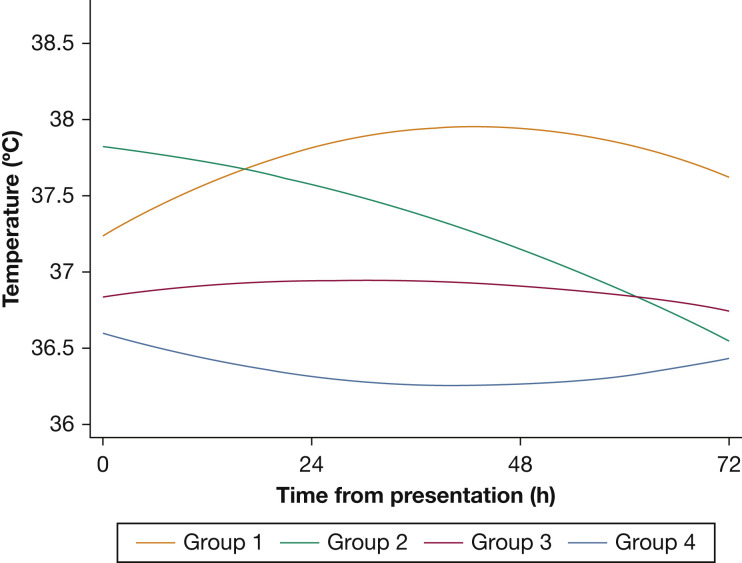
Four subphenotypes were identified: group 1 (n = 139; 20%) had normal presenting temperatures that increased over the subsequent 72 hours; group 2 (n = 97; 14%) had higher presenting temperatures but decreased over time; group 3 (n = 277; 40%) had normal body temperatures throughout without significant changes; group 4 (n = 183; 26%) had low body temperatures (Fig 1 ). Age was significantly different between the subphenotypes: group 1 was the youngest; group 4 was the oldest (57 vs 64 y; P = .04). Group 4 had the highest prevalence of congestive heart failure (32%; P = .002) and chronic pulmonary disease (31%; P = .01). BMI was highest in group 1 and lowest in group 4 (34 vs 29 kg/m2; P < .001). There were significant differences in laboratory results, with group 4 having the highest d-dimer, troponin, lactic acid, creatinine, and total bilirubin (Table 1 ). Although acetaminophen use was significantly different between the subphenotypes, the pattern of acetaminophen administration over time did not suggest that antipyretic medications played a role in the divergent temperature patterns of group 1 and group 2. Specifically, group 1 consistently got more acetaminophen over time compared with group 2, which suggests that the increase in temperature over time was not due solely to inadequate antipyretic therapy.
Figure 1.
Temperature trajectory subphenotypes of patients with coronavirus disease 2019. Group-based trajectory modeling is a finite mixture model that clusters patients following similar trajectories of a variable of interest (ie, temperature). Group 1 had almost five times higher hazard ratio of death compared with group 2, when we controlled for demographics, comorbidities, and severity of illness.
Table 1.
Clinical Characteristics and Outcomes of COVID-19 Temperature Trajectory Subphenotypes
| Characteristics | Total | Group 1 | Group 2 | Group 3 | Group 4 | P Value |
|---|---|---|---|---|---|---|
| No. (%) | 696 | 139 (20) | 97 (14) | 277 (40) | 183 (26) | ... |
| Age, median (IQR), y | 61 (47-73) | 57 (42-71) | 58 (49-73) | 60 (44-72) | 64 (50-78) | .04 |
| Sex, male, No. (%) | 355 (51) | 77 (55.4) | 54 (55.7) | 133 (48) | 91 (49.7) | .4 |
| Race, No. (%) | .08 | |||||
| Black | 588 (84.5) | 121 (87.1) | 81 (83.5) | 235 (84.8) | 151 (82.5) | ... |
| White | 44 (6.3) | 6 (4.3) | 6 (6.2) | 19 (6.9) | 13 (7.1) | ... |
| Other | 64 (9.2) | 12 (8.6) | 10 (10.3) | 23 (8.3) | 19 (10.4) | ... |
| Comorbidity, No. (%) | ... | |||||
| Congestive heart failure | 154 (22.1) | 28 (20.1) | 14 (14.4) | 54 (19.5) | 58 (31.7) | .002 |
| Pulmonary disease | 166 (23.9) | 24 (17.3) | 17 (17.5) | 68 (24.5) | 57 (31.1) | .01 |
| Diabetes mellitus | 92 (13.2) | 20 (14.4) | 11 (11.3) | 38 (13.7) | 23 (12.6) | .9 |
| Hypertension | 233 (33.5) | 48 (34.5) | 35 (36.1) | 94 (33.9) | 56 (30.6) | .8 |
| Renal disease | 41 (5.9) | 7 (5) | 4 (4.1) | 14 (5.1) | 16 (8.7) | .3 |
| Liver disease | 14 (2) | 2 (1.4) | 0 (0) | 8 (2.9) | 4 (2.2) | .3 |
| BMI, kg/m2 | 31 (10) | 34 (11) | 32 (8) | 31 (10) | 29 (8) | < .001 |
| Sequential organ failure assessment score, median (IQR) | 2 (1-5) | 3 (1-5) | 2 (1-5) | 2 (1-5) | 3 (1-6) | .5 |
| Vitals, mean (SD) | ||||||
| Mean arterial pressure, min, mm Hg | 71 (13) | 71 (11) | 72 (11) | 72 (12) | 69 (16) | .3 |
| Respiratory rate, max, breaths/min | 31 (9) | 34 (8) | 33 (7) | 31 (11) | 29 (7) | < .001 |
| Heart rate, max, beats/min | 113 (22) | 116 (22) | 112 (17) | 113 (21) | 109 (25) | .008 |
| Glasgow Coma Scale, min | 13 (3) | 13 (4) | 14 (3) | 13 (3) | 13 (4) | .6 |
| Saturation of oxygen, min | 87 (10) | 86 (8) | 86 (10) | 87 (11) | 88 (10) | .1 |
| Admission temperature >38°C, mean (SD) | 36.9 (0.8) | 37.2 (0.8) | 37.4 (0.9) | 36.8 (0.6) | 36.6 (0.6) | < .001 |
| Fever, No. (%) | 345 (49.6) | 138 (99.3) | 94 (96.9) | 96 (34.7) | 17 (9.3) | < .001 |
| Laboratory results | ... | |||||
| Leukocytosis, No. (%) | 136 (19.5) | 19 (13.7) | 12 (12.4) | 64 (23.1) | 41 (22.4) | .02 |
| Leukopenia, No. (%) | 160 (23) | 43 (30.9) | 21 (21.6) | 64 (23.1) | 32 (17.5) | .04 |
| Platelet count, mean (SD), count/μL (in thousands) | 205 (92) | 179 (66) | 189 (71) | 215 (98) | 218 (104) | < .001 |
| Creatinine, mean (SD), mg/dL | 2.2 (2.8) | 1.9 (2.3) | 1.9 (2.1) | 2.1 (2.6) | 2.6 (3.5) | .03 |
| Total bilirubin, mean (SD), mg/dL | 0.8 (1.5) | 0.6 (0.4) | 0.6 (0.3) | 0.7 (0.7) | 1.2 (2.9) | .001 |
| Lactic acid, mean (SD), mmol/L | 2.7 (3) | 1.9 (1.1) | 1.9 (1) | 2.7 (2.6) | 3.7 (4.5) | < .001 |
| D-dimer, mean (SD), μg/mL | 4 (5) | 4 (5) | 3 (4) | 3 (5) | 5 (6) | .01 |
| Ferritin, mean (SD), ng/mL | 1,478 (2,931) | 1,372 (2,514) | 1,597 (1,993) | 1,244 (1,867) | 1,871 (4,619) | .3 |
| Lactate dehydrogenase, mean (SD), units/L | 460 (467) | 461 (221) | 487 (230) | 425 (226) | 502 (845) | .8 |
| Creatine kinase, mean (SD), units/L | 1,031 (5,615) | 1,228 (2,238) | 664 (1,010) | 571 (1,597) | 1,811 (10,928) | .5 |
| Troponin, mean (SD), ng/L | 64 (147) | 50 (111) | 32 (46) | 62 (157) | 96 (184) | .003 |
| Erythrocyte sedimentation rate, mm/h | 84 (33) | 82 (31) | 88 (32) | 84 (34) | 85 (35) | .6 |
| C-reactive protein, mean (SD), mg/L | 120 (96) | 150 (102) | 150 (90) | 110 (90) | 92 (91) | < .001 |
| IL-6, mean (SD), pg/mL | 43 (73) | 51 (71) | 34 (36) | 35 (57) | 60 (112) | .7 |
| Outcomes | ... | |||||
| Acetaminophen doses, mean (SD), No. | 3.4 (3.5) | 6.5 (3.2) | 4.3 (2.7) | 2.7 (3.2) | 1.5 (2.5) | < .001 |
| Length of hospital stay, d | 7 (4-11) | 8 (6-14) | 7 (5-11) | 6 (4-12) | 6 (4-9) | .002 |
| ICU admission, No. (%) | 242 (34.8) | 55 (39.6) | 33 (34) | 90 (32.5) | 64 (35) | .6 |
| Mechanical ventilation, No. (%) | 101 (14.5) | 28 (20.1) | 11 (11.3) | 34 (12.3) | 28 (15.3) | .1 |
| Deaths, No. (%) | 61 (8.8) | 17 (12.2) | 3 (3.1) | 23 (8.3) | 18 (9.8) | .1 |
Group 1 had the highest 30-day inpatient mortality rate at 12.2%; group 2 had the lowest mortality rate at 3.1%. On Cox regression that controlled for demographics, comorbidities, and sequential organ failure assessment score, patients’ subphenotype was associated significantly with death (P < .05). Group 1 had almost five times higher hazard ratio of death than patients in group 2 (hazard ratio, 4.98; 95% CI, 1.41-17.6; P = .01). Groups 3 and 4 had higher hazard ratio of deaths compared with group 2 but did not reach statistical significance (group 3: hazard ratio, 3.12; P = .07; group 4: hazard ratio, 2.38; P = .2).
Discussion
We report the use of longitudinal temperature measurements to identify novel subphenotypes in COVID-19 illness. Group 1 were the youngest subphenotype with the highest BMI; group 4 were the oldest with a high prevalence of pulmonary disease and congestive heart failure. Group 1 had the highest mortality rate; group 4 had the most significant laboratory abnormalities with elevated creatinine, total bilirubin, and lactic acid levels. The high mortality rate that was seen in group 1 and the organ dysfunction that was seen in group 4 suggest that both subphenotypes have a dysregulated response to COVID-19. Conversely, group 2 had the lowest mortality rate, which suggests a potentially favorable host response to infection.
Given the heterogeneity of COVID-19 and the diversity of potential therapeutics, identification of clinically relevant subphenotypes is necessary for a precision approach to treatment.8 For instance, it is unclear which patients benefit from therapies that block elements of the cytokine storm response.8 , 9 Fever is a hallmark of cytokine storm; thus, group 1 with rising body temperatures and elevated C-reactive protein levels may benefit from this type of therapy. Similarly, prolonged time to normalization of body temperature correlates with SARS-2-COV shedding.10 Therefore, group 1 may have prolonged viral shedding and could require extended antiviral therapy. Group 4 comprised the oldest patients and had 32% prevalence of congestive heart failure; given the age, high-risk comorbidity, and elevated D-dimer levels, group 4 may be at higher risk for VTE and may benefit from aggressive screening or treatment, which are hypotheses that should be tested in future work. The limitations of the study include that it was retrospective and at a single center. Further research is needed with the use of multicenter data to investigate the prognostic and phenotypic potential of temperature trajectory subphenotypes in COVID-19.
In conclusion, we found four distinct subphenotypes of patients with COVID-19 with markedly different clinical characteristics and mortality rates. Our results suggest that patients in these subphenotypes may have differing risks for experiencing cytokine storm, coagulopathy, and cardiac and renal injury and may require targeted management.
Acknowledgments
Role of sponsors: The sponsor had no role in the design of the study, the collection and analysis of the data, or the preparation of the manuscript.
Footnotes
FINANCIAL/NONFINANCIAL DISCLOSURES: None declared.
FUNDING/SUPPORT: S. V. B. is funded by National Institutes of Health (NIH) grant T32HL007605; E. S. H. is funded by NIH grant K24DK105340; P. A. V. is funded by NIH grant R03HL148295; and M. M. C. is funded by NIH grant R01GM123193.
References
- 1.World Health Organization Novel coronavirus (COVID-19) situation. https://covid19.who.int/ Updated June 26, 2020.
- 2.Ziehr DR, Alladina J, Petri CR, et al. Respiratory pathophysiology of mechanically ventilated patients with COVID-19: a cohort study. Am J Respir Crit Care Med. 2020;201(12):1560-1564. [DOI] [PMC free article] [PubMed]
- 3.Richardson S, Hirsch JS, Narasimhan M, et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA. 2020;323(20):2052-2059. [DOI] [PMC free article] [PubMed]
- 4.Wynants L., Van Calster B., Bonten M.M.J. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ (Clinical Res) 2020;369:m1328. doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamura K. Central circuitries for body temperature regulation and fever. Am J Physiol Regul Integr Comp Physiol. 2011;301(5):R1207–R1228. doi: 10.1152/ajpregu.00109.2011. [DOI] [PubMed] [Google Scholar]
- 6.Bhavani S.V., Carey K.A., Gilbert E.R., Afshar M., Verhoef P.A., Churpek M.M. Identifying novel sepsis subphenotypes using temperature trajectories. Am J Respir Crit Care Med. 2019;200(3):327–335. doi: 10.1164/rccm.201806-1197OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagin D.S., Odgers C.L. Group-based trajectory modeling in clinical research. Annu Rev Clin Psychol. 2010;6:109–138. doi: 10.1146/annurev.clinpsy.121208.131413. [DOI] [PubMed] [Google Scholar]
- 8.Sanders JM, Monogue ML, Jodlowski TZ, Cutrell JB. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;323(18):1824-1836. [DOI] [PubMed]
- 9.Cao X. COVID-19: immunopathology and its implications for therapy. Nat Rev Immunol. 2020;20(5):269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu K, Chen Y, Yuan J, et al. Factors associated with prolonged viral RNA shedding in patients with Coronavirus Disease 2019 (COVID-19). Clin Infect Dis. 2020;71(15):799-806. [DOI] [PMC free article] [PubMed]