Abstract
Rabies is nearly 100% lethal in the absence of treatment, killing an estimated 59,000 people annually. Vaccines and biologics are highly efficacious when administered properly. Sixteen rabies-related viruses (lyssaviruses) are similarly lethal, but some are divergent enough to evade protection from current vaccines and biologics, which are based only on the classical rabies virus (RABV). Here we present the development and characterization of LyssaVax, a vaccine featuring a structurally designed, functional chimeric glycoprotein (G) containing immunologically important domains from both RABV G and the highly divergent Mokola virus (MOKV) G. LyssaVax elicits high titers of antibodies specific to both RABV and MOKV Gs in mice. Immune sera also neutralize a range of wild-type lyssaviruses across the major phylogroups. LyssaVax-immunized mice are protected against challenge with recombinant RABV and MOKV. Altogether, LyssaVax demonstrates the utility of structural modeling in vaccine design and constitutes a broadened lyssavirus vaccine candidate.
Keywords: lyssaviruses, rabies, vaccine, prophylaxis, protein engineering, structural modeling, rhabdoviruses, GLA-SE
Graphical Abstract
Rabies-related viruses cause a similar disease as rabies virus, but protection is not always induced by current vaccines. Fisher et al. design a vaccine featuring a functional, chimeric envelope protein that combines parts of two highly divergent viruses. The vaccine generates robust immune responses and protects against disease in mice.
Introduction
Emerging infectious diseases are on the rise (Smith et al., 2014). The recent outbreaks of Ebola virus, Zika virus, and SARS-CoV-2 highlight our lack of preparedness: despite their relation to known diseases, few therapeutics were available for rapid distribution. Zoonotic diseases are particularly threatening, because they often move to new hosts opportunistically and can rarely be eradicated on a global scale (Cutler et al., 2010; Welburn et al., 2015).
Rabies is a neglected infectious disease that is responsible for an estimated 59,000 global human deaths annually (Hampson et al., 2015; World Health Organization, 2017a), roughly the same number of deaths caused annually by influenza in the United States (Kochanek et al., 2011). Whereas millions of people survive influenza each year, fewer than 30 cases of human rabies survival have been documented (Fooks et al., 2017; Mani et al., 2019). The number of human rabies deaths is likely underestimated, as studies in developing countries with poor health infrastructure suggest (Fooks et al., 2014; Mallewa et al., 2007). Rabies virus (RABV)-induced encephalitis is the most lethal viral infection known to humankind when no intervention is applied prior to symptoms. Less known is that RABV-related lyssaviruses cause the same zoonotic disease, have similar mortality rates as RABV, but are far less studied (Banyard et al., 2014; Evans et al., 2012). The lyssavirus genus is comprised of 17 single-stranded, negative-sense RNA viruses divided into at least three phylogroups (RABV being categorized in phylogroup I) (Markotter and Coertse, 2018). Classical RABV circulates on all continents but Antarctica; non-RABV lyssaviruses are endemic in Europe, Africa, Asia, and Australia (Fisher et al., 2018).
Rabies is highly survivable with prompt administration of vaccines and antiserum. RABV vaccines are touted as one of the lowest cost but highest impact tradeoffs among vaccine-preventable infectious diseases (comparing procurement cost to Gavi, the Vaccine Alliance, and governments per death averted) (Gavi, 2018). Critically, disease from other lyssaviruses is not always prevented by RABV-based vaccines and biologics: protection against phylogroup II and III viruses is minimal (Weyer et al., 2008), and lapses in coverage by post-exposure prophylaxis (PEP) have even been shown within phylogroup I, despite phylogenic proximity to RABV (Liu et al., 2013). Despite this, the available vaccines were developed solely against RABV (Liu et al., 2013; Nel, 2001). Investment in studying lyssaviruses and development of a pan-lyssavirus vaccine is currently lacking but would have a profound impact if or when a divergent lyssavirus emerges.
The fraction of disease burden shared by non-RABV lyssaviruses is unknown: the viral encephalitis and resulting symptoms from lyssaviruses are indistinguishable from RABV infections, and current diagnostic reagents based on the highly conserved nucleoprotein (N) cannot differentiate between lyssaviruses (von Teichman et al., 1998; World Health Organization, 2018). Discriminatory diagnostics are rarely available for either human cases or surveillance in animal populations (Evans et al., 2012; Mallewa et al., 2007). Definitive evidence of a non-RABV lyssavirus infection can only be made in post-mortem analysis, and the methods (sequencing the viral genome or probing with species-specific antibodies) are not yet standardized. Seroprevalence studies in wildlife suggest lyssaviruses circulate in low but steady proportions compared with RABV (Mélade et al., 2016; Orłowska et al., 2020; Suu-Ire et al., 2017). A small number of human deaths caused by six non-RABV lyssaviruses has been confirmed, but the actual number is likely higher (Evans et al., 2012).
Sporadic outbreaks of RABV (Feng et al., 2016; Kuzmin et al., 2012) and the consistent discovery of new lyssaviruses (Aréchiga Ceballos et al., 2013; Hu et al., 2018; Nokireki et al., 2018) challenge our understanding of lyssavirus evolution and host switching (Fisher et al., 2018). RABV likely originated as a bat-derived virus, then spread to terrestrial mammal reservoirs, notably dogs, numerous times (Badrane and Tordo, 2001). Canine RABV is now responsible for 95% of human RABV fatalities (Hampson et al., 2015; World Health Organization, 2017a). Lyssaviruses circulate in bats with two notable exceptions where the reservoirs have not been identified: Ikoma virus (IKOV) (Marston et al., 2012) and Mokola virus (MOKV) (Coertse et al., 2017; Sabeta et al., 2007). The possibility of further terrestrial adaptation and consequent increased risk to humans is of concern. MOKV, a divergent member of phylogroup II, was one of the first non-RABV lyssaviruses to be discovered (Shope et al., 1970), and lack of protection from RABV-based vaccines in animals has been well documented: for example, MOKV has been isolated from rabies-vaccinated domestic cats multiple times (Nel et al., 2000; Sabeta et al., 2010, 2007; von Teichman et al., 1998). Although rare cross-reactivity between RABV and MOKV has been observed, the current RABV vaccine is unlikely to provide protection against MOKV (De Benedictis et al., 2016; Hanlon et al., 2005).
The unique attributes of rabies and its epidemiology call for economic factors to be considered during vaccine development. Despite its favorable cost-to-impact tradeoff, the current rabies vaccine is expensive, especially for rural populations in developing countries where transmission most often occurs. In addition, the current vaccine’s long history of safety and reliability sets a high bar for new iterations. Therefore, in crafting a vaccine with broadened protection, we aimed for a vaccine similar to current vaccines in composition and formulation: inactivated virions of a single strain. The glycoprotein (G) is the sole protein on the surface of the lyssavirus virions, and serum virus neutralizing antibodies (VNAs) against G are considered the primary correlate of protection against rabies disease. Our proposed vaccine therefore focuses on engineering the G.
Creating chimeric protein antigens is a well-established technique for modulating immune responses. The move toward “epitope-based” vaccines is an attractive approach in many efforts to make vaccines with increased safety, potency, and breadth (Bogdanoff et al., 2017; Wei et al., 2010; Xu et al., 2011; Zhou et al., 2009). Previous attempts by other groups to create a chimeric G of RABV G and MOKV G, whether swapping antigenic sites or entire domains, were inconclusive or unsuccessful (Bahloul et al., 1998; Evans et al., 2018a; Jallet et al., 1999; Mebatsion et al., 1995). Site switching is necessarily based on the known antigenic sites of RABV G. Five antigenic regions where VNAs bind were empirically mapped on RABV G, enabling deep understanding of neutralization mechanisms and the humoral response against RABV (Benmansour et al., 1991; Cai et al., 2010; Marissen et al., 2005). However, detailed study of other lyssavirus Gs has not been carried out, so swapping these short regions may miss other important sites. This is evident from recent work in which antigenic sites were swapped between the Gs of RABV and Lagos bat virus (LBV): site II, considered a major site on RABV G, appears to also be immunodominant on LBV G, but the results of other sites are largely inconclusive (Evans et al., 2018a). Swapping entire domains did not reliably produce infectious particles (Bahloul et al., 1998; Jallet et al., 1999; Mebatsion et al., 1995), likely because of the imperfect protein engineering caused by the lack of structural information at the time. Although transported to the cell surface and incorporated into budding virions, the chimeras failed to support viral replication, consistent with the lack of important structural determinants.
The vaccine described here is based on a RABV vaccine strain and features a structurally designed chimeric lyssavirus glycoprotein containing domains from both RABV G and the highly divergent MOKV G. In mice, the inactivated vaccine elicits high titers of antibodies, which neutralize a panel of lyssaviruses, and protects against challenge with RABV and a recombinant MOKV.
Results
Structure-Based Design of Chimeric Lyssavirus Glycoproteins
In designing a more broadly protective lyssavirus vaccine, we initially inserted MOKV G into a RABV vector already containing a native RABV G (BNSP333; Figure S1A). This strategy has been successfully employed with various foreign viral Gs in the BNSP333 vector (Abreu-Mota et al., 2018; Johnson et al., 2016; Kurup et al., 2015). However, the virus containing both Gs lost expression of MOKV G rapidly, as indicated by immunofluorescence (Figure S1B). Furthermore, MOKV G alone or in addition to the native RABV G caused the vector to grow significantly slower (Figure S1C). Therefore, we pursued a more technical strategy to create a single chimeric G, which would serve as the only glycoprotein supporting viral entry.
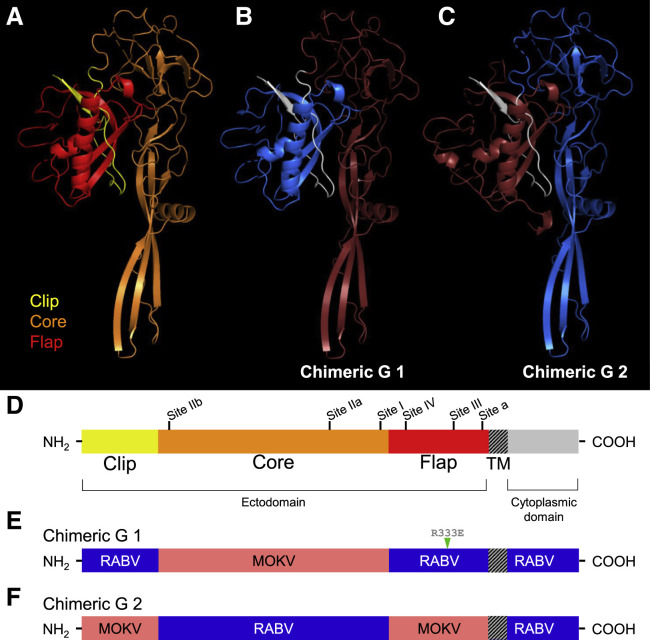
To design a chimeric G, we generated structural models of lyssavirus Gs by threading their amino acid sequences onto the most closely related structure available at the time, that of the vesicular stomatitis virus (VSV) pre-fusion G. We used three structural modeling programs: I-TASSER (Zhang, 2008), SWISS-MODEL (Waterhouse et al., 2018), and Phyre2 (Kelley et al., 2015) (Figure 1 A). Despite sharing only ~18% sequence identity, VSV and RABV Gs appear to share conserved structural features, such as disulfide bonds (Yang et al., 2020), and VSV has previously been used to model RABV G (Fernando et al., 2016). In all of the models, we identified three subdomains in the ectodomain: a “clip” that consists of a small hairpin-shaped region near the amino (N) terminus (yellow); a “core” that forms a large region containing a globular portion, beta sheets, and the putative fusion domain (orange); and a “flap,” the region near the transmembrane (TM) domain that associates closely with the clip and that contains the receptor binding domain (red) (Figures 1A and 1D). The structure of RABV G was recently solved at both low and high pH levels (Yang et al., 2020). Comparison between the high pH (pre-fusion) structure and our model shows similar positioning of the clip, core, and flap (Figure S2), validating the use of structural modeling for designing chimeric proteins.
Figure 1.
Structure-Based Design of Chimeric Lyssavirus Glycoproteins
(A–C) Structural models of lyssavirus glycoprotein (G) ectodomain monomers. (A) Representative structural model of a lyssavirus G with proposed structural domains highlighted (clip in yellow, core in orange, flap in red). Structural model of Chimeric G 1 (B) and Chimeric G 2 (C) clip domains highlighted in white, and core and flap domains highlighted in blue or red, corresponding to patterns in (E) and (F).
(D–F) Linear schematics of a representative lyssavirus G monomer (D) and the RABV G/MOKV G chimeric Gs named Chimeric G 1 (E) and Chimeric G 2 (F). In (D), proposed structural domains of the ectodomain are noted (clip in yellow, core in orange, flap in red), as are the antigenic regions as they are known to exist on the RABV G: site I (residues 224–229), site II (34–42 and 198–200), site III (330–338), site IV (263–264), and minor site “a” (342–343).
R333E, attenuating mutation at RABV G residue 333; TM, transmembrane domain. See also Figures S2 and S3.
The clip, core, and flap subdomains formed the basis for building Chimeric G 1 and Chimeric G 2, which are comprised of alternating subdomains from RABV G (blue) and MOKV G (red) (Figures 1B, 1C, 1E, and 1F). We hypothesized that the design of a functional G protein requires the amino acid sequences of the clip and the flap to be derived from the same virus to reproduce optimal bonding interactions between these two moieties.
Other important features known about the well-studied RABV G were incorporated in the chimeric G designs. Extensive studies have mapped the majority of potently neutralizing monoclonal antibodies (mAbs) to five antigenic sites on RABV G (Benmansour et al., 1991; Dietzschold et al., 1988; Prehaud et al., 1988) (Figure 1D). The chimeric Gs share sites I and II from the same donor G, and sites III, IV, and minor site “a” from the other donor G, resulting in relatively balanced immunogenicity. The short, intracellular carboxy (C) terminal of RABV G interacts with the matrix protein (M) during viral budding and does not contribute to antigenicity (Wirblich et al., 2008), so it was maintained as RABV in both Chimeric G 1 and 2.
An immunofluorescence assay showed the chimeric Gs to successfully traffic to the cell surface and exhibit the anticipated antibody staining patterns (Figure S3). Cells transiently expressing Chimeric G 1 or 2 were positively stained by polyclonal sera generated against RABV G or MOKV G, whereas cells expressing a wild-type (WT) G were stained only by their cognate sera. The human anti-RABV G mAb 4C12 binds in the flap region of RABV G and thus stains only Chimeric G 1.
Chimeric Glycoprotein Is Functional and Supports Viral Replication
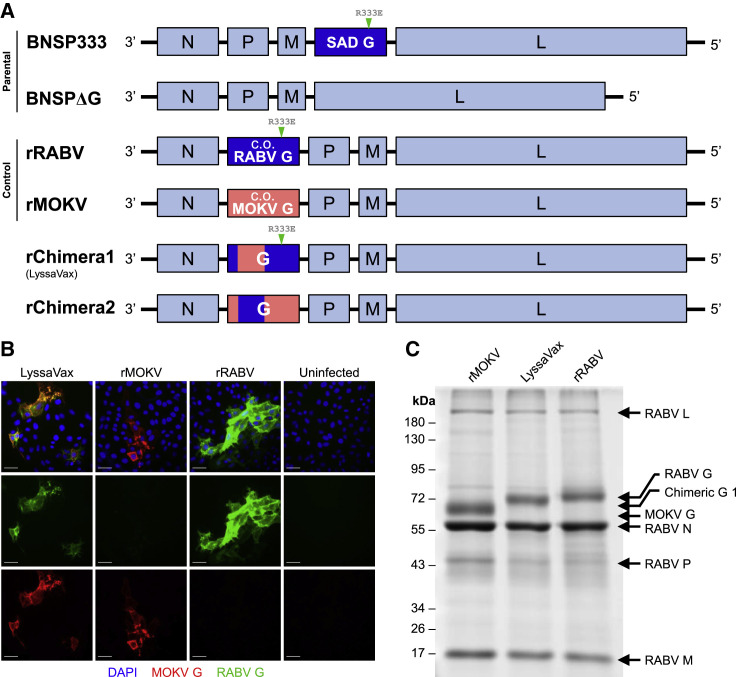
Four vaccines were then constructed in the BNSP RABV vector lacking its native G in the fourth position (BNSPΔG; Figure 2 A). The G gene of interest was inserted into the second position: the vaccines rRABV and rMOKV contain the codon-optimized genes of RABV G or MOKV G, respectively, and the vaccines rChimera1 and rChimera2 contain the respective chimeric Gs 1 and 2 (Figure 2A). Placing of G in the second position of the genome instead of its native fourth position increases expression levels because of the transcription gradient exhibited by rhabdoviruses (McGettigan et al., 2003). This increase in expression also contributes attenuation, which, despite proposed administration in an inactivated form, renders the vaccine safer to work with.
Figure 2.
Construction and Recovery of a Chimeric Lyssavirus G Vaccine
(A) Viral genome schematics. BNSP333 is the parent vaccine vector based on RABV strain SAD B19. Its G is located in the native fourth position and contains the attenuating R333E mutation. BNSPΔG is based on BNSP333 but lacks the native G. All of the following experimental constructs are based on BNSPΔG: rRABV contains a human codon-optimized (c.o.) RABV G with the attenuating mutation R333E at the second position; rMOKV contains human c.o. MOKV G at the second position; rChimera1 (LyssaVax) contains Chimeric G 1, with the attenuating R333E mutation, at the second position; and rChimera2 contains Chimeric G 2 at the second position.
(B) Infection immunofluorescence. VERO cells infected with either LyssaVax (left column), rMOKV (second column), rRABV (third column), or uninfected (right column) were fixed and stained with a DyLight 488-conjugated human anti-RABV G mAb 4C12 (green) and mouse anti-MOKV G sera (red). Nuclei labeled in blue by DAPI. Scale bars represent 50 μm.
(C) Analysis of purified virions. 3-μg viral particles denatured and resolved by SDS-PAGE, then total protein stained with SYPRO Ruby. Viral proteins are indicated.
Multiple attempts to recover rChimera2 did not yield appreciable titers of infectious virus, suggesting that the chimeric G did not efficiently support viral spread. By contrast, rChimera1 was successfully recovered, demonstrating the functionality of this G. An immunofluorescence assay confirmed that only cells infected with rChimera1, but not with either rRABV or rMOKV, exhibited dual staining with both anti-RABV G and anti-MOKV G reagents (Figure 2B). Furthermore, the presence of foci indicates the virus’s ability to spread from cell to cell mediated solely by the Chimeric G. Purified virions were analyzed by SDS-PAGE, which shows comparable incorporation of Chimeric G 1 into virions as compared with the control vaccines (Figure 2C). rChimera1 is henceforth referred to as LyssaVax.
LyssaVax Is Apathogenic by Intramuscular and Intranasal Routes
Even though rabies vaccines are administered in an inactivated (killed) form to humans, safety is a necessary priority during production when the virus is live and concentrated. We therefore analyzed live LyssaVax for any pathogenicity, comparing it with similar vectors containing the wild-type (WT) G protein from RABV or MOKV. LyssaVax was administered live by two inoculation routes, intranasal (i.n.) and intramuscular (i.m.), to assess potential pathogenicity in Swiss Webster mice (Figure S4). Both male and female mice were used to ensure sex did not affect pathogenicity. LyssaVax was apathogenic both i.n., compared with the SPBN strain of RABV (McGettigan et al., 2001) (Figure S4A), and i.m., compared with the CVS-N2c strain (Morimoto et al., 1999) (Figure S4B).
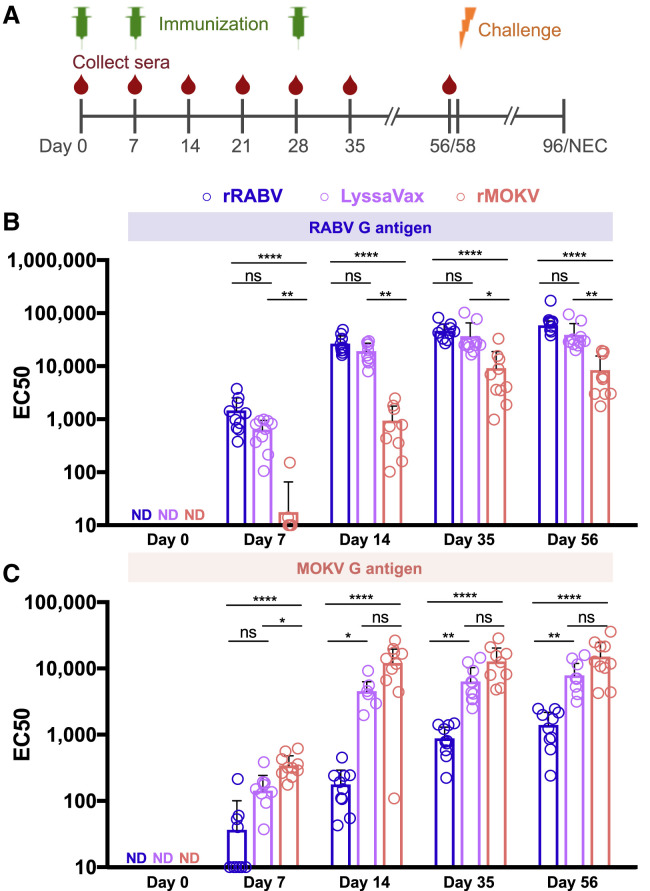
LyssaVax Elicits High Titers of Antibodies against Both RABV and MOKV
To assess immunogenicity, we administered inactivated LyssaVax to groups of 10 Swiss Webster mice. Inactivated rRABV and rMOKV were administered individually as control vaccines. Figure 3 A displays the immunization and blood draw schedule (including challenge, discussed in the next section). Sera were analyzed by enzyme-linked immunosorbent assay (ELISA) against RABV G and MOKV G antigens. To avoid cross-reactivity with other RABV proteins, soluble Gs were produced, stripped, and purified from a recombinant VSV, which either expressed RABV G instead of VSV G or which lacked a G gene and was trans-complemented with MOKV G. mAbs against each protein were used to validate the antigen: the mouse anti-RABV G mAb 1C5 and the mouse mAb 1409-7, which cross-reacts with MOKV G (Dietzschold et al., 1988) (Figure S5).
Figure 3.
Humoral Response to LyssaVax
(A) Schematic timeline of immunization (green syringe), sera collection (red drop), and challenge (orange bolt) through necropsy (NEC).
(B and C) Development of antibodies over time in groups of mice (n = 10 mice per group, analyzed in triplicate, mean ± SD) immunized three times with either LyssaVax (purple), rRABV (blue), or rMOKV (red). Graphs compare half-maximal responses (EC50s) between sera from immune mice probed against RABV G (B) or MOKV G (C) antigens in ELISA format. Day 0 samples did not seroconvert, so EC50 values were not calculated. Analysis within time points by Kruskal-Wallis test and Dunn’s multiple comparisons test (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 for adjusted p values; n.s. is not significant).
See also Figure S5.
Sera were tested to assess immunogenicity before immunization (day 0), 7 days following each immunization (days 7, 14, and 35) and just prior to challenge (day 56). Individual mouse half-maximal responses (EC50s) are compared against RABV G (Figure 3B) and MOKV G (Figure 3C). Dilution curves of group averages are displayed in Figure S5. Sera from mice immunized with LyssaVax reacted strongly against both RABV and MOKV G antigens, nearly matching sera from cognate immunizations. EC50s of RABV G-specific antibodies were not significantly different between LyssaVax and rRABV immune sera (Figure 3B). EC50s of MOKV G-specific antibodies from LyssaVax-immunized mice were significantly lower than rMOKV only at day 7 (p = 0.0412; Figure 3C). Sera from mock-immunized mice did not seroconvert to either antigen (Figure S5). Together, these data suggest that the chimeric G in LyssaVax is highly immunogenic and broadens the antigenicity of a single lyssavirus glycoprotein.
Interestingly, sera from rMOKV immune mice reacted to RABV G (red dots/bars in Figure 3B) and sera from rRABV-immune mice reacted to MOKV G (blue dots/bars in Figure 3C), although both at significantly lower levels than LyssaVax sera. In both cases, titers increased over time (compared with mock-immunized sera), suggesting that these antibody responses were specific and being boosted by each subsequent immunization.
LyssaVax Elicits RABV Neutralizing Antibodies
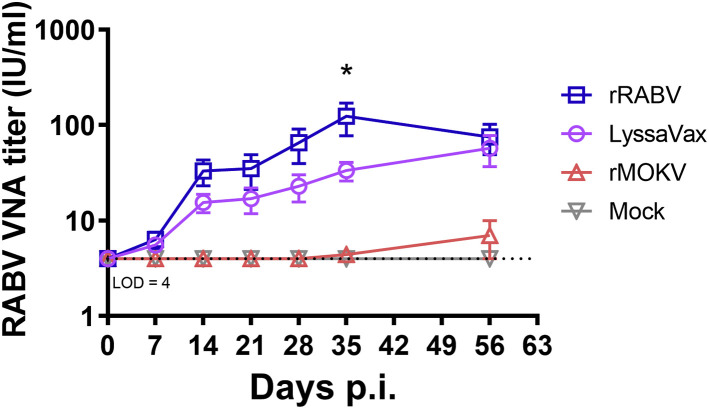
Sera from mice immunized with LyssaVax neutralized RABV strain CVS-11 at comparable levels with rRABV control immune sera, as determined by the rapid fluorescent focus inhibition test (RFFIT) (Figure 4 ; Table S1). When normalized to a rabies immunoglobulin standard, serum containing greater than 0.5 international units per milliliter (IU/mL) VNAs is considered adequate for protection (Bogel, 1978; Keates, 2010). Neutralizing titers in all 10 LyssaVax-immunized mice reached >4 IU/mL by day 14 post-immunization (p.i.; after two vaccine inoculations on days 0 and 7). Titers continued to climb after the final immunization on day 28, peaking at an average of 57.2 IU/mL on day 56 p.i. Sera from mice immunized with the control vaccine, rRABV, yielded higher titers at each time point, but differed significantly (p = 0.0342) only on day 35 p.i., when titers peaked, averaging 103.5 IU/mL. None of the mock-immunized groups exhibited virus neutralization capability, as determined by assaying pooled sera. These data demonstrate the potency of the chimeric G vaccine, because VNA titers are considered the most important correlate of protection (Both et al., 2012; World Health Organization, 2017b).
Figure 4.
RABV Neutralizing Titers over Time
Development of RABV neutralizing antibodies over time, averaged from mice in groups of mice (n = 10 mice per group, analyzed in duplicate, mean ± SD) immunized on days 0, 7, and 28 with either rRABV (blue), rMOKV (red), or LyssaVax (purple), or mock immunized (gray). See Figure 3A for full immunization scheme. Titers were calculated in international units (IU) per milliliter by comparison with the US standard rabies immune globulin. Level of detection (LOD) was 4 IU/mL. Two-way ANOVA and Tukey’s multiple comparisons tests were performed. ∗p = 0.034, comparing rRABV and LyssaVax. See also Table S1 and Figure S6.
rMOKV immune sera did not neutralize CVS-11 above the 4 IU/mL level of detection initially used in the RFFIT, despite the high titers of cross-reactive antibodies observed in the RABV G ELISA. To address the possibility of VNA titers below 4 IU/mL, we performed a follow-up assay with larger dilutions of sera from days 28, 56, and 96 p.i., enabling a 0.2 IU/mL level of detection (Figure S6). At day 28, only two mice exhibited titers above this new level of detection at 0.6 and 2 IU/mL, respectively. At day 56, 9/10 sera exhibited low levels of neutralization, with four mice achieving titers above the 0.5 IU/mL threshold for protection.
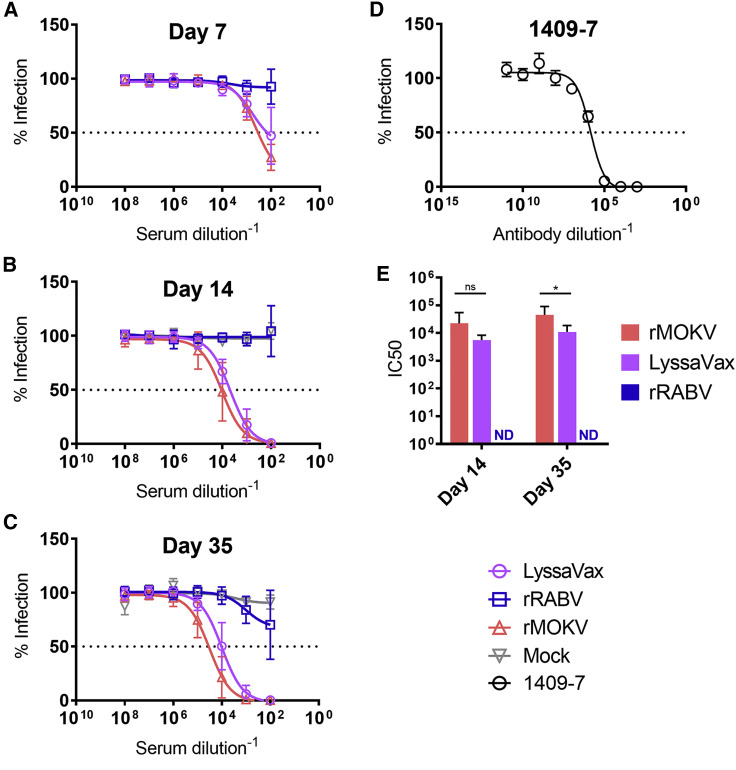
Antibodies Elicited by LyssaVax Neutralize MOKV G Pseudotypes
Unlike RABV, neither reference assays nor standards have been established for non-RABV lyssaviruses. Therefore, to assess the functionality of anti-MOKV G VNAs within the immunized mouse sera, we tested sera in a pseudotype neutralization assay. VSV lacking G and expressing NanoLuciferase and EGFP (VSVΔG-NanoLuc-EGFP) was trans-complemented with MOKV G to create MOKV G pseudotype viruses (MOKV G PTVs; see Figure S7 for schematic). Cells express NanoLuc and EGFP when infected with these single round infectious particles; thus, neutralization was measured as a reduction in luminescence. To account for background, we normalized luminescence to day 0 sera. Three time points were tested, each 1 week following a boost: days 7, 14, and 35 (Figure 5 ). Similar to the RFFIT, LyssaVax-immune sera neutralized MOKV G PTVs at low concentrations, nearly as low as the control rMOKV. Of the two time points for which half-maximal inhibitory concentrations (IC50) could be calculated, LyssaVax differed from rMOKV significantly only at day 35 (p = 0.0133) (Figure 5D). Sera from mice immunized with rRABV did not neutralize the MOKV G PTVs. Similar to the rMOKV sera in the RFFIT assay, high titers of cross-reactive sera seen in the ELISA did not correlate with functional VNA by day 35. Pooled sera from mock-immunized mice showed no neutralization.
Figure 5.
MOKV G Pseudotype Neutralizing Titers
VNA titers against MOKV G pseudotype viruses (PTVs). PTVs made by trans-complementing VSVΔG-NanoLuc-EGFP with MOKV G (Figure S7). VNA titers measured in sera from mice immunized with either LyssaVax (purple open circle), rRABV (blue open square), or rMOKV (red open triangle), or mock immunized with PBS (gray open inverted triangle).
(A–C) Average titers shown over time: day 7 (A), day 14 (B), and day 35 (C) (n = 10 mice per group, analyzed in triplicate, mean ± SD).
(D) Pseudotype neutralization by the mAb 1409-7. Luminescence data background subtracted using paired sera from day 0 and normalized to 100% infection in no-sera controls.
(E) Serum IC50 data analyzed by the Mann-Whitney test (∗p = 0.0133).
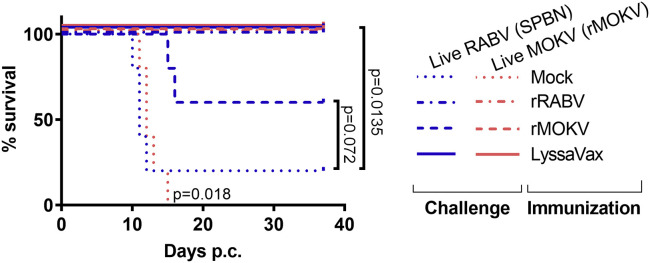
LyssaVax Protects against Lethal Challenge of Both RABV and rMOKV
The vaccinated mice were challenged at 58 days p.i. (see schedule in Figure 3A). The 10 mice per immunization group (LyssaVax, rRABV, rMOKV, and PBS mock) were split into two subgroups and challenged with 105 focus-forming units (FFUs) of either live RABV (SPBN strain) or live rMOKV i.n. (Figures 6 and S8). Mock-immunized mice lost weight and were euthanized by day 12 post-challenge (p.c.) for SPBN and day 15 p.c. for rMOKV. One animal (mouse 1-4) challenged with SPBN survived and developed RABV neutralizing titers (Table S1). By contrast, all mice immunized with LyssaVax maintained weight and were protected against the live virus challenges. Mice immunized with the control vaccines were also protected against challenge with their cognate live virus: rRABV immune mice survived challenge by SPBN, and rMOKV immune mice survived live rMOKV challenge. Strikingly, some mice survived non-cognate challenge as well: three rMOKV immune mice survived SPBN challenge, and all five rRABV immune mice survived rMOKV challenge, although two mice (3-6 and 3-9) lost weight and recovered. Survival of these mice with low or negligible titers of cross-neutralizing antibodies may suggest alternate mechanisms of protection.
Figure 6.
Survival after Challenge with Pathogenic RABV and rMOKV
Overall survival data post-challenge (p.c.) with either live RABV (SPBN) or live rMOKV (n = 5 mice per immunization group per challenge virus). Survival was analyzed using the log rank Mantel-Cox test. See Figure S8 for weight loss curves.
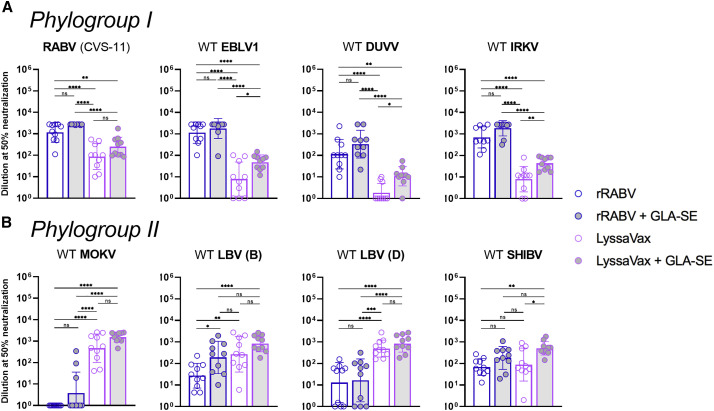
Antibodies Elicited by LyssaVax Neutralize Diverse WT Lyssaviruses
Because LyssaVax is composed of two component lyssavirus Gs, we sought to test whether sera elicited by LyssaVax cross-neutralized non-component viruses. We also included the TLR-4 agonist glucopyranosyl lipid adjuvant-stable emulsion (GLA-SE) as an adjuvant in some groups. GLA-SE has been shown to increase the magnitude and breadth of humoral immune responses and is currently in clinical trials (Coler et al., 2010; Patton et al., 2015; Sirima et al., 2017). We immunized four groups of mice with either rRABV or LyssaVax, with or without GLA-SE, following the same schedule in Figure 3A. Sera from day 47 p.i. were tested in a micro-neutralization assay against a panel of WT lyssaviruses spanning two phylogroups: RABV, European bat lyssavirus 1 (EBLV1), Irkut virus (IRKV), and Duvenhage virus (DUVV) from phylogroup I (Figure 7 A); and MOKV, Shimoni bat virus (SHIBV), Lagos bat virus B (LBV-B), and LBV-D from phylogroup II (Figure 7B).
Figure 7.
Microneutralization Assay with Panel of WT Lyssaviruses
(A and B) Neutralizing antibody 50% endpoint titers in sera from mice 47 days after first immunization with rRABV (blue/white), rRABV with the adjuvant GLA-SE (blue/gray), LyssaVax (purple/white), or LyssaVax with GLA-SE (purple/gray) (n = 10 mice per group, analyzed in duplicate, mean ± SD). See Figure 3A for immunization scheme. Lyssaviruses tested span two phylogroups: (A) RABV, EBLV-1, DUVV, and IRKV; and (B) MOKV, LBV-D, LBV-D, and SHIBV. All titers normalized to day 0 sera were pooled within groups. Upper limit endpoint: 2,795. Microneutralizations were analyzed using an ordinary one-way ANOVA test with Tukey’s multiple comparisons (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 for adjusted p values; n.s. is not significant).
Among phylogroup I viruses (Figure 7A), all sera from the four groups neutralized RABV, as expected. Immune sera from rRABV with or without GLA-SE also cross-neutralized EBLV1, DUVV, and IRKV, consistent with cross-reactivity within phylogroups previously reported. Neutralizing titers from LyssaVax with or without GLA-SE were significantly lower than those of rRABV with or without adjuvant. However, including GLA-SE with LyssaVax raised the average neutralizing titer for all four viruses tested (significantly so for DUVV and IRKV), as compared with unadjuvanted LyssaVax. For DUVV, only 3/10 of the unadjuvanted LyssaVax-immune samples were neutralizing, whereas 9/10 of LyssaVax + GLA-SE samples neutralized.
Among phylogroup II viruses (Figure 7B), sera from LyssaVax, both with and without adjuvant, neutralized WT viruses at significantly higher titers than sera from rRABV (with or without adjuvant). Strikingly, of the phylogroup II viruses, only WT MOKV was not neutralized by rRABV-immune sera, and only three samples exhibited neutralizing titers above baseline in the rRABV + GLA-SE group. However, rRABV with and without GLA-SE induced neutralizing titers against WT LBV (B), WT LBV (D), and WT SHIBV.
Overall, LyssaVax stimulates superior titers of VNAs against the phylogroup II viruses tested but has lost some capability in stimulating VNAs against non-RABV phylogroup I viruses. GLA-SE raised the average VNA titer against all viruses tested when administered with both LyssaVax and rRABV.
Discussion
Chimeric G Vaccine Rationale
There are a variety of ways to broaden a vaccine’s protective breadth, some of which have been attempted for lyssaviruses. A straightforward approach is to create multiple vaccine constructs, each expressing a separate lyssavirus G (Evans et al., 2018b), but this strategy would multiply the cost of a vaccine already deemed too expensive for the regions that need it most (van de Burgwal et al., 2017). Furthermore, one lyssavirus G might be immunodominant over others. Some have proposed lyssavirus vaccines in live vectors (Kgaladi et al., 2017; Weyer et al., 2008), which, although less expensive and successful in wildlife RABV vaccination campaigns (Mähl et al., 2014), are unlikely ever to be approved for use in humans for safety reasons (Vuta et al., 2016). Another approach is to add multiple Gs to a single vaccine construct (Kgaladi et al., 2017). Foreign viral Gs have been successfully added to the RABV genome, and their Gs expressed to comparable amounts as the native RABV G (Abreu-Mota et al., 2018; Keshwara et al., 2019; Willet et al., 2015). However, the stability of multiple lyssavirus Gs has not been rigorously tested. In our experience, lyssavirus Gs individually exhibit different growth speeds and expression levels. Our preliminary data suggest that when combined in a single vector, the less efficient G confers a disadvantage to the virus and puts selective pressure on the virus to lose expression of the G conferring slower growth. We therefore advocate for a single-G lyssavirus vaccine construct.
Prior to the recent publication of the RABV G crystal structure (Yang et al., 2020), our protein design effort benefitted the pre-fusion G structure of the related rhabdovirus, VSV (Roche et al., 2007). The VSV G structure enabled us to revisit the chimera strategy (Mebatsion et al., 1995), design an updated chimeric lyssavirus G, and generate functional virus. Additionally, despite a lack of detailed knowledge about antigenic regions on non-RABV lyssavirus Gs, care was taken to design a chimeric G in which potential antigenic sites were “balanced” between the two major domains (Figure 1D). Sites II and III are likely of highest importance, because they share binding sites with two of the putative RABV cellular receptors, nicotinic acetylcholine receptor (nAChR) and the low-affinity neurotrophin receptor (p75NTR), respectively, (Lafon, 2005). Although site II has often been considered the most immunogenic based on the high proportions of G-specific mAbs that bind it (Benmansour et al., 1991; Dietzschold et al., 1983), many of the mAbs being developed to replace the immune sera in PEP bind to site I (Both et al., 2013). The immunogenicity of site IV has been demonstrated in mice, humans, and dogs (Cai et al., 2010; Johnson et al., 2002; Ni et al., 1995). Antibody responses to RABV from different species are not thought to vary significantly (Horton et al., 2014). Altogether, we believe the domain-based approach to generating chimeric Gs is a superior option.
Glycosylation sites differ between lyssavirus Gs: RABV G has three predicted N-linked glycosylation sites at residues 37, 247, and 319 (Shakin-Eshleman et al., 1992); MOKV G shares the N319 site but has only one other predicted site at N202. Chimeric G 1 therefore has two predicted sites: N202 and N319. The N319 site is conserved across lyssaviruses and is suggested to be the minimal site needed for maturation and trafficking through the endoplasmic reticulum and Golgi apparatus (Badrane and Tordo, 2001). N37 has been shown not to be efficiently glycosylated and is likely dispensable for proper G folding and function (Wojczyk et al., 1998). Finally, it has been suggested that the N202 site in MOKV G is not glycosylated in vivo (Badrane and Tordo, 2001). Although further study should define the glycosylation sites of the chimeric G, our data are consistent with the cited works because we did not observe evidence of glycosylation affecting the antigenicity of LyssaVax.
Recovery of Viruses with Chimeric Gs
It is unclear why the Chimeric G 2 did not enable viral recovery. As the single surface protein, the G carries out numerous tasks, including trimerization, engaging with cellular receptors, and mediating fusion between membranes, any of which may have been disturbed by the newly engineered protein. The immunofluorescence of transfected cells (Figure S3) demonstrates that Chimeric G 2 is successfully produced, trafficked to the cell surface, and exhibits the anticipated antigenicity, suggesting that functional rather than structural issues hampered recovery.
Regardless, Chimeric G 1 is a preferable choice for a chimeric G vaccine because it includes the attenuating mutation R333E within the flap domain contributed by RABV G (Figure 1E). The R333 residue in RABV G is critical for association with a putative RABV cellular co-receptor, the low-affinity neurotrophin receptor, p75NTR (Langevin and Tuffereau, 2002). The R333E mutation alone abrogates pathogenicity by peripheral infection routes in adult mice (Mebatsion, 2001) and likely contributed to LyssaVax’s apathogenicity by both routes tested (Figure S4).
Vaccine-Generated Antibody Responses
We were interested in the antibody responses generated against LyssaVax, because antibodies are indicative of a strong vaccine response. LyssaVax elicited high titers of IgG antibodies against both MOKV G and RABV G, as seen by ELISA (Figures 3 and S5). Sera from rRABV and rMOKV immunizations also contained appreciable titers of antibodies, which bound to the heterologous antigen (e.g., sera from mouse immunized with rMOKV binding to RABV G) (Figure 3) by day 14 p.i. However, ELISAs detect a wide array of antibodies, regardless of function (e.g., neutralizing and non-neutralizing). Furthermore, the antigens used in the ELISA are detergent solubilized, which may expose epitopes otherwise inaccessible on live, intact virions.
A smaller subset of antibodies function in neutralizing virus, and high titers of these VNAs are the best-established correlate of protection against RABV infection (World Health Organization, 2017b). As such, administration of rabies immune globulin (RIG) is a critical component of current PEP providing short-term passive immunity in addition to a vaccine course. LyssaVax-immune mouse sera neutralized both CVS-11 and MOKV G pseudotypes at nearly the same levels as control immunizations for either rRABV or rMOKV, respectively (Figures 4 and 5). Although RABV VNAs from LyssaVax were lower than controls at days 28 and 35 (Figure 4), they were matched by day 56. Furthermore, LyssaVax titers at day 35 averaged over 60-fold higher than the 0.5 IU/mL threshold for protection, demonstrating the robust functionality of the VNAs induced by LyssaVax. Sera from rRABV and rMOKV controls were only marginally cross-neutralizing in the RFFIT and PTV neutralization assay (Figures 4 and 5), and only by late time points.
After establishing robust functional antibody responses against the component viruses, we designed an immunogenicity study to assess cross-neutralization with additional lyssaviruses. Anticipating the possibility of lower VNA titers against non-component viruses, we adjuvanted LyssaVax and the control vaccine, rRABV, with the Toll-like receptor 4 (TLR4) agonist GLA-SE (Figure 7). LyssaVax-immune sera neutralized all viruses tested; of phylogroup I viruses, LyssaVax-induced sera neutralized significantly less strongly than that of the rRABV control vaccine and, in the case of DUVV, required GLA-SE to achieve neutralization in the majority of samples. Of phylogroup II viruses, VNA titers induced by LyssaVax + GLA-SE were highest, and in the case of MOKV and LBV D, unadjuvanted LyssaVax was significantly higher than even rRABV + GLA-SE. Two results of the micro-neutralization panel were surprising: the relatively low VNA titers that LyssaVax generated against non-RABV phylogroup I viruses and that rRABV, with and without GLA-SE, induced cross-neutralizing VNAs against LBV-B, LBV-D, and SHIBV.
Regarding low phylogroup I VNA titers, it is possible that antigenic sites located in the core domain, which is contributed by MOKV G in LyssaVax, are more important for neutralizing non-RABV phylogroup I viruses. In a study testing anti-RABV mAbs against a panel of strains and lyssaviruses, none of the five mAbs bound to EBLV-1 or DUVV (Hanlon et al., 2001), and VNA titers against EBLV-1 and DUVV were also lowest in a previously reported RABV G/MOKV G chimeric vaccine (Bahloul et al., 1998). These data suggest that, to provide comprehensive coverage across phylogroups I and II, LyssaVax may need components from divergent phylogroup I viruses. It has also been shown that higher concentrations of anti-RABV sera are necessary for neutralizing non-RABV phylogroup I viruses (Hanlon et al., 2005), so the RABV G-specific titers from LyssaVax may not have been high enough. The ability of GLA-SE to boost phylogroup I VNA titers when added to LyssaVax supports this.
Although division of the lyssavirus genus into phylogroups is based on genetic and antigenic clustering, there are many examples of less discrete patterns of antigenicity. For example, inter-phylogroup neutralization has been observed in RABV-vaccinated laboratory workers with exceptionally high titers (De Benedictis et al., 2016; Hanlon et al., 2005), and there are at least two examples of anti-RABV G mAbs reported to cross-neutralize MOKV: 1409-7 (Dietzschold et al., 1988) and JA-3.3A5 (Hanlon et al., 2001). Furthermore, antigenic cartography studies have determined that only 67% of antigenic differences among lyssavirus Gs are predictable from the amino acid sequence (Horton et al., 2010).
In the cases where LyssaVax is cross-neutralizing, it is unknown whether the sera contain individual cross-reactive antibodies or whether discrete populations of antibodies bind to each antigen; both possibilities likely contribute. This question can be answered only by isolating and characterizing mAbs, a goal of future studies. Knowledge of where physiologically relevant antibodies bind on non-RABV lyssavirus Gs will be important for detailed study of how LyssaVax elicits protective antibodies against multiple lyssaviruses.
Protection from Rabies Disease
Based on the high titers of VNAs against the two component viruses, which contributed to the chimeric G, we anticipated full protection. LyssaVax indeed protected all mice challenged with either RABV or rMOKV, with no weight loss or clinical symptoms observed. Although lyssaviruses are typically administered intracranially, the i.n. route was chosen in this study for several reasons. First, we observed uniform pathogenicity in female mice during pathogenicity studies (Figure S4). Second, rMOKV is not pathogenic by the i.m. route (Figure S4), consistent with WT MOKV studies (Badrane et al., 2001; Percy et al., 1973; Perrin et al., 1996). Third, the i.n. route has been shown to be an acceptable alternative to intracranial injection for RABV challenge (Lafay et al., 1991; Lewis et al., 2013; Rosseels et al., 2011). Finally, i.n. inoculation poses a lesser risk to laboratory personnel.
Among the control groups, mice immunized and challenged with homologous vaccines/viruses survived, as expected (Figures S8C and S8H), whereas some mice survived challenge with heterologous virus (Figures S8D and S8G). This was surprising because, despite appreciable titers of antibodies against heterologous Gs detected in the ELISA (Figure 3), mice immunized with rMOKV had marginal RABV-neutralizing titers (Figure 4) and rRABV did not neutralize MOKV G pseudotypes (Figure 6). However, in light of the cross-neutralization of other phylogroup I viruses that rMOKV sera exhibited in Figure 7, the survival is less exceptional.
It is notable that two mice immunized with rMOKV lost weight and were euthanized, and two mice immunized with rRABV lost weight after rMOKV challenge and recovered (Figure S8). The atypical challenge model (attenuated strains administered i.n.) may be responsible; this would be addressed by the WT challenge experiment. Given that 9/10 mock-immunized mice were euthanized (Figure S8A) and the 10th mouse indeed survived after infection, as evidenced by RABV VNAs detected at necropsy (Table S1, mouse ID 1-4), we are confident that the mice in Figures S8D and S8G were successfully infected.
The mechanism for developing broadly neutralizing lyssavirus VNAs has not been studied and raises important questions about the antigenic relationships between lyssaviruses and how protection is conferred. There may be additional, uncharacterized immune mechanisms that contribute to protection in the absence of neutralizing antibodies. This possibility warrants further investigation.
Conclusions
We present a lyssavirus vaccine featuring a single chimeric glycoprotein that was designed based on observations of predicted lyssavirus G structures. The chimeric G retains antigenic qualities of component Gs (RABV and MOKV) and cell-infecting functionality. When administered as an inactivated vaccine formulation, it stimulates high titers of neutralizing antibodies against component viral Gs and some additional lyssaviruses. Finally, LyssaVax was shown to protect against challenge with RABV and a recombinant MOKV. Development is needed to improve VNA titer responses against phylogroup I viruses. We propose that, with further development, this vaccine could be employed during a lyssavirus outbreak or supplant current rabies vaccines in areas where non-RABV lyssaviruses are endemic.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Fluorescein isothiocyanate (FITC)-conjugated anti-rabies virus nucleoprotein (RABV N) monoclonal globulin | Fujirebio | Cat#800-092; RRID: AB_2802166 |
| U.S. standard rabies immune globulin | Center for Biologics Evaluation & Research, US Food and Drug Administration | Lot R-3 |
| Mouse anti-RABV G mAb 1C5 | Abcam | Cat#Ab82460; RRID: AB_1658373 |
| Mouse anti-Lyssavirus G mAb 1409-7 | Provided by Dr. Todd Smith, US Centers for Disease Control and Prevention | (Dietzschold et al., 1988) |
| Human anti-RABV G mAb 4C12 | Provided by Dr. Scott Dessain, Lankenau Institute for Medical Research | N/A |
| Pooled mouse anti-MOKV G sera | Provided by Dr. Gene Tan, J. Craig Venter Institute | N/A |
| HRP-conjugated goat anti-mouse IgG (H+L) | Jackson Immunoresearch | Cat#115-035-146; RRID: AB_2307392 |
| Cy3-conjugated goat anti-mouse IgG | Jackson Immunoresearch | Cat#115-165-146; RRID: AB_2338690 |
| Bacterial and Virus Strains | ||
| Stellar competent cells | Clontech/Takara | Cat#636763 |
| VSVΔG-GFP-RABVG | Schnell Laboratory | (Abreu-Mota et al., 2018) |
| VSVΔG-NanoLuc-EGFP | Schnell Laboratory | (Abreu-Mota et al., 2018) |
| MOKV G pseudotype viruses (MOKV G PTVs) | This paper | N/A |
| Irkut virus (IRKV, RFFIT) | Provided by Dr. Todd Smith, US Centers for Disease Control and Prevention | N/A |
| European bat lyssavirus 1 (EBLV1, RFFIT) | Provided by Dr. Todd Smith, US Centers for Disease Control and Prevention | N/A |
| Duvenhage virus (DUVV, 86132SA) | Provided by Dr. Todd Smith, US Centers for Disease Control and Prevention | GenBank: EU293119.1 |
| Lagos bat virus (LBV, lineage B, isolate 8619NGA) | Provided by Dr. Todd Smith, US Centers for Disease Control and Prevention | GenBank: EU293110.1 |
| Lagos bat virus (LBV, lineage D, isolate KE576) | Provided by Dr. Todd Smith, US Centers for Disease Control and Prevention | GenBank: GU170202.1 |
| Shimoni bat virus (SHIBV, Kenya, 2009) | Provided by Dr. Todd Smith, US Centers for Disease Control and Prevention | GenBank: GU170201.1 |
| Mokola virus (MOKV, isolate 252/97) | Provided by Dr. Todd Smith, US Centers for Disease Control and Prevention | GenBank: GQ500112.1 |
| rRABV | This paper | N/A |
| RABV (CVS-11 strain) | Schnell Laboratory | N/A |
| RABV (N2c) | Schnell Laboratory | N/A |
| RABV (SPBN) | Schnell Laboratory | (McGettigan et al., 2001) |
| Biological Samples | ||
| Soluble RABV G | Schnell Laboratory | (Blaney et al., 2013) |
| Soluble MOKV G | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| β-propiolactone (BPL) | Sigma | Cat#P5648 |
| SYPRO Ruby Protein Gel Stain | Invitrogen | Cat#S12000 |
| Glucopyranosyl lipid adjuvant-stable emulsion (GLA-SE) | Infectious Disease Research Institute | Cat#EM-082 |
| octyglucopyranoside (OGP) | Fisher | Cat#BP585-5 |
| Vectashield Hard Set containing 4′,6-diamidino-2-phenylindole (DAPI) | Vector Laboratories | Cat#H-1500 |
| SIGMAFAST o-phenylenediamine dihydrochloride (OPD) tablets | Sigma | Cat#P9187-50SET |
| Passive cell culture lysis buffer | Promega | Cat#E1941 |
| Critical Commercial Assays | ||
| SuperScript II Reverse Transcriptase | Invitrogen | Cat#10928-034 |
| Bicinchoninic acid (BCA) assay kit | Pierce | Cat#23250 |
| Nano-Glo Luciferase Assay System | Promega | Cat#N113 |
| Experimental Models: Cell Lines | ||
| Mouse neuroblastoma (NA) cells | Schnell Laboratory | N/A |
| BSR cells (derivative of BHK-21) | Schnell Laboratory | N/A |
| Vero cells | ATCC | CCL81 |
| BEAS-2b cells | ATCC | CRL-9609 |
| Experimental Models: Organisms/Strains | ||
| Swiss Webster mice | Charles River | 24107540 |
| Oligonucleotides | ||
| MOKV G forward oligo (CO-041) 5′- TAACACCCCTCCCGTACGACCA TGAACCTACCCTGTCTGAC-3′ |
IDT | N/A |
| MOKV G reverse oligo (CO-042) 5′ GTGTTAGTTTTTTTCATGGCTAGC TCATGTACCAGGGAGC-3′ |
IDT | N/A |
| Fragment 1 forward oligo for Chimeric G 1 (CO-062) 5′-TTGGCAAAGAATTCG AGCTCCGTACGGCCGCCACCATG-3′ |
IDT | N/A |
| Fragment 1 reverse oligo for Chimeric G 1 (CO-063) 5′-GTTGCAGCCCTCCTC CTCGACCACCAGATTGTTA-3′ |
IDT | N/A |
| Fragment 2 forward oligo for Chimeric G 1 (CO-064) 5′- GAGGAGGGCTGCA ACGCC-3′ |
IDT | N/A |
| Fragment 2 reverse oligo for Chimeric G 1 (CO-065) 5′- CCTGAAGTCGTGCAGA TTGACCAGCTGGTTGGG-3′ |
IDT | N/A |
| Fragment 3 forward oligo for Chimeric G 1 (CO-066) 5′- CTGCACGACTTCA GGAGC-3′ |
IDT | N/A |
| Fragment 3 reverse oligo for Chimeric G 1 and Fragment 4 reverse oligo for Chimeric G 2 (CO-067) 5′- AAAAAGATCTGCTAGCTC ACAGCCTGGTCTCGCC-3′ |
IDT | N/A |
| Fragment 1 forward oligo for Chimeric G 2 (CO-068) 5′- TTGGCAAAGAATTCGAG CTCCGTACGAAAAAGCGGCCG-3′ |
IDT | N/A |
| Fragment 1 reverse oligo for Chimeric G 2 (CO-069) 5′- GGTACACCCTTCGTCTTG GGAGAGCAGATTATTCG-3′ |
IDT | N/A |
| Fragment 2 forward oligo for Chimeric G 2 (CO-070) 5′- GACGAAGGGTGTA CCAAC-3′ |
IDT | N/A |
| Fragment 2 reverse oligo for Chimeric G 2 (CO-071) 5′- GCGGTCATTGTGTAT ATTCACCAGTTTGTCAGGG-3′ |
IDT | N/A |
| Fragment 3 forward oligo for Chimeric G 2 (CO-072) 5′- ATACACAATGACC GCCTTG-3′ |
IDT | N/A |
| Fragment 3 reverse oligo for Chimeric G 2 (CO-073) 5′- CACTGTGGAGGGATCA ATCAGAGGGTGTCGCAAT-3′ |
IDT | N/A |
| Fragment 4 forward oligo for Chimeric G 2 (CO-074) 5′- GATCCCTCCACAGTGTTC-3′ | IDT | N/A |
| Forward RT-PCR primer for amplifying G in RABV vectors 5′- GGAGGTCGACTAA AGAGATCTCACATAC-3′ |
IDT | N/A |
| Reverse RT-PCR primer for amplifying G in RABV vectors 5′- TTCTTCAGCCATCTCA AGATCGGCCAGAC |
IDT | N/A |
| Recombinant DNA | ||
| BNSPΔG cDNA | Schnell Laboratory | (Blaney et al., 2011) |
| BNSP333 cDNA | Schnell Laboratory | (Blaney et al., 2011) |
| rRABV cDNA | This paper | N/A |
| rMOKV cDNA | This paper | N/A |
| rChimera1 cDNA | This paper | N/A |
| rChimera2 cDNA | This paper | N/A |
| pCAGGS-coMOKVG | Provided by Dr. Gene Tan, J. Craig Venter Institute | MOKV.NIG68-RV4 strain (Wright et al., 2010), GenBank accession number HM623780 |
| pCAGGS-coRABVG | This paper | N/A |
| pCAGGS-ChimericG1 | This paper | N/A |
| pCAGGS-ChimericG2 | This paper | N/A |
| pTIT-RABVN | Schnell Laboratory | (Gomme et al., 2010) |
| pTIT-RABVP | Schnell Laboratory | (Gomme et al., 2010) |
| pTIT-RABVL | Schnell Laboratory | (Gomme et al., 2010) |
| pCAGGS-T7 | Schnell Laboratory | (Gomme et al., 2010) |
| Software and Algorithms | ||
| Fiji | https://imagej.net/Fiji | (Schindelin et al., 2012) |
| I-TASSER | https://zhanglab.ccmb.med.umich.edu/I-TASSER/ | (Zhang, 2008) |
| SWISS-MODEL | https://swissmodel.expasy.org/ | (Waterhouse et al., 2018) |
| Phyre2 | http://www.sbg.bio.ic.ac.uk/∼phyre2/html/page.cgi?id=index | (Kelley et al., 2015) |
| GraphPad Prism for macOS (Version 8) | https://www.graphpad.com/ | N/A |
| The PyMOL Molecular Graphics System, Version 2 by Schrödinger, LLC. | https://pymol.org/2/ | N/A |
| Other | ||
| InFusion Cloning Kit | Clontech/Takara | Cat#639649 |
| X-tremeGENE 9 transfection reagent | Millipore Sigma | Cat#06365809001 |
| PureLink RNA Mini Kit | Ambion | Cat#12183018A |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Matthias Schnell (Matthias.Schnell@jefferson.edu).
Materials Availability
Upon request, further information, resources, and reagents are available from the authors pending an executed MTA as well as biosafety approval of the requesting institution(s).
Data and Code Availability
All data are available upon request to the lead contact author. No proprietary software was used in the data analysis.
Experimental Model and Subject Details
Swiss Webster mice (Charles River), age 6-10 weeks, were used in this study. All mice used were female except where noted. Mice used in this study were handled in adherence to the recommendations described in the Guide for the Care and Use of Laboratory Animals, and work was approved by the Institutional Animal Care and Use Committee (IACUC) of Thomas Jefferson University (TJU) under protocols 01886 and 01940. Mice were housed with up to five individuals per cage, under controlled conditions of humidity, temperature, and light (12 h light, 12 h dark cycles). Food and water were available ad libitum. Animal procedures were conducted under 3% isoflurane/O2 gas anesthesia.
The following cell lines and their culture conditions were used in this work: mouse neuroblastoma (NA) cells were grown in RPMI (Corning) with 5% fetal bovine serum (FBS, Atlanta Biologicals) and 1X Penicillin/Streptomycin (Corning). The other cell lines were grown in DMEM (Corning) with 5% FBS and 1X Penicillin/Streptomycin: BSR cells (a derivative of the baby hamster kidney cell line BHK-21), the African green monkey cell line VERO, and the human lung cell line BEAS-2b. Cells were kept at 37°C and 5% CO2 during non-infectious growth and 34°C with 5% CO2 during infectious growth. Infected cell cultures were cultured in OptiPRO SFM (Life Technologies) unless otherwise noted.
Method Details
Structural modeling and chimeric glycoprotein design
To generate structural models of RABV and MOKV G, their amino acid sequences were threaded onto the pre-fusion structure of vesicular stomatitis virus (VSV) G (Roche et al., 2007). Three different modeling programs were used to increase the likelihood of producing an accurate model: I-TASSER (Zhang, 2008), SWISS-MODEL (Waterhouse et al., 2018), and Phyre2 (Kelley et al., 2015). After analysis and delineation of the clip, core, and flap regions of the glycoproteins, the proposed chimeric G were also threaded onto VSV G to confirm the placement of the ectodomain regions.
cDNA construction of vaccine vectors
To make the rRABV vector, we inserted a human codon-optimized RABV G (SAD B19 strain with R333E mutation, synthesized by Genscript USA) into the BNSPΔG vector (Blaney et al., 2011) using the BsiWI and NheI restriction sites. Human codon optimization was selected in anticipation of downstream vaccine production in primate cells. To make the BNSP333-coMOKVG vector, we inserted MOKV G (MOKV.NIG68-RV4 strain (Wright et al., 2010), GenBank accession number HM623780, provided by Gene Tan) into the BNSP333 vector using the InFusion cloning kit (Clontech) and oligos CO-041 and CO-042. To make the rMOKV vector, we inserted the human codon-optimized MOKV G sequence into the BNSPΔG vector using the NotI and NheI restriction sites, the NotI site having been cloned into the vector previously. To make the chimeric glycoproteins 1 and 2, fragments of codon optimized RABV G and MOKV G were first amplified by PCR using the primers listed in the table below and cloned into a pCAGGS expression vector via InFusion cloning (Clontech). Three fragments were combined to make Chimeric G 1 (amplified using oligos CO-062 through CO-067) and four fragments were combined to make Chimeric G 2 (amplified using oligos CO-067 through CO-074). The chimeric Gs were then cloned into the BNSPΔG vector using the NotI and NheI restriction sites to create rChimera1 (later termed LyssaVax) and rChimera2. The correct sequences of all four plasmids were confirmed by Sanger sequencing.
Recovery of recombinant vectors
Recombinant RABV were recovered as described previously (Abreu-Mota et al., 2018; Kurup et al., 2015; Papaneri et al., 2013). Briefly, X-tremeGENE 9 transfection reagent (Millipore Sigma) in Opti-MEM reduced serum medium (Life Technologies) was used to co-transfect the respective full-length viral cDNA clones along with the plasmids encoding RABV N, P, and L and the T7 RNA polymerase into BSR cells in T25 flasks. We harvested the supernatants of transfected cells after 7 days and analyzed the supernatants for the presence of infectious virus by infecting fresh BSR cell cultures and immunostaining with FITC-conjugated anti-RABV N mAb (Fujirebio).
To confirm the glycoprotein sequence in the recovered viruses, the viruses were sequenced by the following method: BSR cells were infected at an MOI of 1 then incubated for 2 days. Media was removed and the PureLink RNA Mini Kit (Ambion) was used to lyse the cells and extract RNA. Using the SuperScript II Reverse Transcriptase (Invitrogen), sections of the viral genomes containing G were amplified out of the total RNA (primers RP951 and RP952). RT-PCR products were run on an 1% agarose gel and bands were excised and analyzed by Sanger sequencing using the same primers.
Immunofluorescence
To analyze broadened reactivity of the chimeric glycoproteins, VERO cells grown on 15 mm coverslips (Fisherbrand) were transfected with pCAGGS vectors containing either RABV G, MOKV G, Chimeric G 1 or Chimeric G 2 using XtremeGene 9. Two days post-transfection, cells were fixed with 4% paraformaldehyde (PFA), blocked with PBS containing 5% FBS, and stained with either the human anti-RABV G mAb 4C12 conjugated to DyLight 488, mouse anti-MOKV G sera (from G. Tan), or mouse anti-RABV G sera (generated against BNSP333), each at 1:400 dilution and incubated for 2 h at RT. Coverslips were washed with PBS and samples stained with mouse sera were then stained with Cy3-conjugated goat anti-mouse IgG secondary at 1:200. After a 2 h incubation at RT, coverslips were washed and mounted onto glass slides with Vectashield Hard Set containing DAPI (Vector Laboratories). Images of slides were analyzed in ProgRes (Jenoptic) and Fiji software (Schindelin et al., 2012).
Immunofluorescence assays on infected cells were carried out in a similar manner, with the following difference: VERO cells were infected at MOI 0.01 with live virus (rRABV, rMOKV or LyssaVax in Figure 2; rRABV, rMOKV, BNSP333-MOKVG or BNSP333-RABVG in Figure S1) then fixed with 4% PFA 2 days post-transfection.
Viral growth curve
Related to Figure S1. BSR cells were seeded in 6-well cell culture plates and incubated until 70% confluent. Cells were then infected at a MOI of 0.01 for 3 hours, washed 2x with PBS, and replenished with OptiPRO media (GIBCO). Samples of each well were collected every 24 h, stored at 4°C, then titered in triplicate.
Purification and inactivation of the virus particles
Large volumes of rRABV- and LyssaVax-containing supernatants were concentrated in a stirred 300 mL ultrafiltration cell (Millipore) and then purified over a 20% sucrose cushion in an SW32 Ti rotor (Beckman, Inc.) at 25,000 rpm for 1.5 h ay 4°C. rMOKV was purified similarly but without prior concentration in ultrafiltration cells. Virion pellets were resuspended in phosphate-buffered saline (PBS), and protein concentrations were determined using a bicinchoninic acid (BCA) assay kit (Pierce). The virus particles were inactivated with 50 μL per mg of particles of a 1:100 dilution of β-propiolactone (BPL) in cold water. The absence of infectivity was verified by inoculating BSR cells with 10 μg of BPL-inactivated viruses. After 4 days of incubation at 34°C, the cells were subcultured and 500 μL of supernatant was passaged on fresh BSR cells. Cultures were split 3 times, every 3 days. After the final growth period, cells were fixed and stained with a FITC-conjugated anti-RABV N mAb to confirm the absence of live virus.
Protein gel
To examine their purity, inactivated virus particles were diluted 1:1 in urea buffer (200 mM Tris-HCl [pH 6.8], 8 M urea, 5% sodium dodecyl sulfate (SDS), 0.1 mM ethylenediaminetetraacetic acid [pH 8], 0.03% bromophenol blue, and 0.5 M dithiothreitol) and denatured at 95°C for 5 m. Three micrograms of protein were resolved on a 10% SDS–polyacrylamide gel and stained with SYPRO Ruby Protein Gel Stain (Invitrogen) according to the manufacturer’s specifications. The gel was then exposed under UV light for 430 ms.
Pathogenicity experiments
Related to Figure S4. Four groups of Swiss Webster mice (Charles River, 5 male and 5 female per group, age 6 to 10 weeks) were intranasally (i.n.) infected with 105 focus-forming units (FFU) of live virus diluted in 20 μL phosphate-buffered saline (PBS). The mice were weighed and monitored daily until day 21 post-infection and further monitored until day 30. Mice exhibiting signs of disease or that lost greater than 25% weight were euthanized. To assess a peripheral route of infection, 4 groups of Swiss Webster mice were intramuscularly (i.m.) infected with 105 FFU of live virus diluted in 100 μL PBS, distributed equally to muscle of both hind limbs. The mice were weighed and monitored daily until day 21 post-infection and further monitored until day 28. Mice exhibiting signs of disease or that lost greater than 25% weight were euthanized. Survival was analyzed using the log-rank Mantel-Cox test in GraphPad Prism.
Immunization and challenge
Swiss Webster mice (Charles River) were used in this study: groups of female mice, age 6 to 10 weeks, were immunized i.m. with 10 μg BPL-inactivated virus diluted in 100 μL phosphate-buffered saline (PBS) and distributed equally to muscle of both hind limbs. In groups which received glucopyranosyl lipid adjuvant-stable emulsion (GLA-SE, IDRI), 20 μL of 0.25 mg/ml adjuvant were included in the 100 μL total per mouse. Mice were immunized on days 0, 7, and 28. Blood was drawn (100 μL via the retro-orbital route) weekly and centrifuged at 10,000 rpm for 10 m for serum collection. Serum was analyzed from individual mice (unless noted). One set of mice (Figures 3, 4, 5, and 6) was challenged on day 58 post-immunization (p.i.) with either SPNB or rMOKV. 105 FFU of live virus were diluted in 20 μL PBS was administered i.n. The mice were weighed and monitored daily until day 21 post-infection and further monitored until day 37. Mice exhibiting signs of disease or that lost greater than 25% weight were euthanized. Survival was analyzed using the log-rank Mantel-Cox test in GraphPad Prism. The other set of mice (Figure 7) was terminally bled via heart puncture on day 47 p.i. and euthanized.
Production of soluble RABV and MOKV G
To produce soluble RABV G, BEAS-2b cells were inoculated with VSVΔG-GFP-RABVG at MOI 0.01. Three days post-infection, supernatant was collected, filtered through a 0.45 μm filter and concentrated by tangential flow filtration. Concentrated virus was purified over 20% sucrose cushion in a SW 32 Ti rotor (Beckman) at 25,000 rpm for 2 h. Pellets were resuspended in TEN buffer with 5% sucrose. To solubilize the glycoprotein, octyglucopyranoside (OGP, Fisher) was added to 2% final concentration and solution was incubated at room temperature with constant mixing for 30 m. The suspension was spun at max speed in a benchtop centrifuge for 3 m to pellet debris. Pellets were treated with OGP in the same manner 2 more times. Pooled supernatants from all 3 extractions were spun in a SW 55 Ti rotor (Beckman) at 45,000 rpm for 1.5 h. Supernatant containing soluble G was analyzed for protein concentration by BCA (Pierce), and for purity by SDS-PAGE and western blot.
To produce soluble MOKV G, BEAS-2b cells were transfected with pCAGGS-coMOKVG using the XtremeGene 9 transfection reagent (Millipore-Sigma). Two days post-transfection, cells were infected with MOKV G PTVS (described below), and 2 days post-infection, supernatant containing MOKVG pseudotyped VSV was collected. MOKV G was solubilized from the pseudotype virions in the same manner as RABV G.
ELISA
To assess vaccine immunogenicity, mouse sera were analyzed by enzyme-linked immunosorbent assay (ELISA), probing for reactivity against either soluble RABV G or MOKV G (production described above). Mouse sera from days 0, 7, 14, 35, and 56 were analyzed individually in triplicate, except mock infected sera which was pooled. Immulon 96-well plates (Nunc) were coated with soluble G diluted in carbonate buffer (15 mM Na2CO3, 35 mM NaHCO3 [pH 9.5]). For RABV G, 50 ng in 100 μL buffer was used per well and for MOKV G, 25 ng in 50 μL per well. Plates were incubated overnight at 4°C. Plates were then washed 3 times with 300 μL per well of PBS containing 0.05% Tween 20 (PBST), then blocked with 5% milk in PBST (250 μL per well) for 2 h at RT, shaking. Plates were washed again, then coated with primary buffer (PBS with 0.5% bovine serum albumen), either 100 μL per well (RABV G plates) or 50 μL per well (MOKV G plates). Serum was diluted 3-fold down the plate in triplicate, starting at either 1:100 or 1:300, then plates were incubated overnight at 4°C. Plates were then washed 3 times and coated with PBST containing HRP-conjugated Goat anti-mouse IgG (H+L) at 1:10,000. After incubation for 2 h at RT, plates were washed and 200 μL of o-phenylenediamine dihydrochloride (OPD) substrate (Sigma) was added to each well. After a 15 m incubation, the reaction was stopped by the addition of 50 μL 3M H2SO4 per well. Optical density was determined at 490 nm (OD490). Individual mouse data were analyzed in GraphPad Prism using a sigmoidal dose-response fit (variable slope) to determine 50% effective concentration [EC50]. Data from groups were also averaged (n = 10) and plotted.
RFFIT neutralization assays
Serum was first heat inactivated at 56°C for 30 m. Mouse sera from days 0, 7, 14, 21, 28, 35, 56 and at necropsy (surviving mice only) were analyzed individually in duplicate, except mock infected sera which was pooled. Rabies virus neutralizing activity was determined using the rapid fluorescent focus inhibition test assay (RFFIT) (Wunner and Jackson, 2010). Mouse neuroblastoma (NA) cells were seeded in 96-well plates 2 days prior to the assay (30,000 cells per well). Serum samples were 2-fold serially diluted in duplicate in 96-well plates, starting from at a dilution of 1:50 (unless otherwise noted) in 50 μL Opti-MEM (Life Technologies). The U.S. standard rabies immune globulin was used at 2 IU/ml. Working dilutions of RABV strain CVS-11 were prepared in Opti-MEM, and 5 μL the working dilution was added to each well containing diluted serum. Plates were incubated for 1 h at 34°C. Medium was removed from NA cells and diluted serum/virus mixtures were transferred to the cell plates. After 2 h incubation at 34°C, media was aspirated, replaced with fresh Opti-MEM, and plates were incubated for 22 h at 34°C with 5% CO2. Plates were then fixed with 80% acetone and stained with FITC-conjugated anti-RABV N antibody. 50% endpoint titers were calculated using the Reed-Muench method and converted to international units (IU) per milliliter by comparing to the standard.
Generation of MOKV G PTVs
MOKV G pseudotype viruses (MOKV G PTVs) are single-round infectious particles comprised of MOKV Gs on the surface of the virion and a VSV genome lacking G and containing Nanoluciferase and EGFP (VSVΔG-NanoLuc-EGFP) packaged within the virion (Figure S7). To generate MOKV G PTVs, the human lung cell line BEAS-2B was first transfected with an expression vector containing human codon-optimized MOKV G (pCAGGS-coMOKVG) using X-tremeGENE 9 transfection reagent (Millipore Sigma). Two days post-transfection, cells were infected at an MOI of 1 with VSVΔG-NanoLuc-EGFP pseudotyped with an irrelevant glycoprotein (Lassa fever virus glycoprotein complex) for initial infection (Abreu-Mota et al., 2018). Two h post-infection, the inoculum was removed and cells were washed 3 times with PBS before media was replaced. Two days post-infection, cells were inspected for GFP expression under a fluorescent microscope and supernatant containing MOKV G PTVs was collected. MOKV G PTVs were passaged 3 times prior to use in the neutralization assay to ensure a pure population of virions.
Pseudotype virus neutralization assay
Serum was first heat inactivated at 56°C for 30 m. Individual mouse sera were analyzed in triplicate. Serum was diluted 10-fold starting at 1:100 dilution in Opti-MEM (Life Technologies) and 104 MOKV G PTV particles were added to each dilution. The mix of sera/antibody plus virus was incubated for 1 h at 34°C with 5% CO2 and transferred to a previously seeded monolayer of VERO cells in a 96-well plate and further incubated for 2 h at 34°C with 5% CO2. Next, the virus/serum mix was replaced with DMEM. At 18–22 h post-infection, media was removed and cells were lysed with 40 μL 1X cell culture lysis buffer (Promega) and transferred to a white, opaque 96-well plate. The Nano-Glo Luciferase Assay System (Promega) was then used according to the manufacturer’s recommendations. Relative luminescence units (RLU) were normalized to 100% infectivity signal as measured by no sera control (maximum signal) and signal from naive samples were background subtracted from experimental samples. Values that were above 100% infectivity were converted to 100%. Half maximal inhibition (IC50) values were calculated by GraphPad Prism 7 using a nonlinear fit model (Log (inhibitor) versus normalized response — variable slope). IC50 data analyzed for statistical significance by the Mann-Whitney test.
Microneutralization assay
Sera from vaccinated mice were tested for VNAs against wild-type lyssaviruses using a micro-neutralization test (Kuzmin et al., 2008; Smith and Gilbert, 2017). Briefly, serum was heat inactivated at 56°C for 30 m, diluted 5-fold starting at 1:10 dilution in MEM supplemented with 10% FBS (CDC Division of Scientific Resources or Atlanta Biologics), and incubated at 37°C for 90 m with 50 FFD50 of each of the following non-RABV lyssaviruses: RABV (CVS-11 strain), Irkut virus (IRKV), European bat lyssavirus 1 (EBLV1) Duvenhage virus (DUVV), Lagos bat virus (LBV, lineage B and lineage D), Shimoni bat virus (SHIBV), Mokola virus (MOKV). Individual mouse sera were analyzed in duplicate. Assays were performed either on 6mm Teflon-coated slides or in 96-well cell culture plates. After incubation ~50,000 cells/ml BSR (a clone of BHK-21) cells were added and mixtures were incubated at 37°C, 0.5% CO2 for 20 to 44 h (depending on the virus used). Cells were then fixed with cold acetone and stained with FITC-conjugated anti-RABV N antibody (Fujirebio). Ten microscopic fields were observed for each dilution under fluorescent microscopy and 50% endpoint titers were calculated using the Reed-Muench method. Titers from pooled, naive (day 0) sera from each group were background subtracted from immune serum titers. Titers > 1:10 were considered positive for VNAs. Micro-neutralization tests for LBV, MOKV, and SHIBV were performed under biosafety level 3 (BL3) conditions. The other tests were performed under BL2 conditions.
Quantification and Statistical Analysis
All statistical analysis was performed using GraphPad Prism software (version 8). ELISA EC50 values were compared by Kruskal-Wallis tests and Dunn’s multiple comparisons tests. RABV VNA titers were compared by two-way ANOVA and Tukey’s multiple comparisons tests. MOKV pseudotype IC50 values were compared using the Mann-Whitney test. Survival was analyzed using the log-rank Mantel-Cox test. Microneutralization data were analyzed using an ordinary one-way ANOVA test with Tukey’s multiple comparisons. Where applicable, data was analyzed in a D’Agostino-Pearson omnibus normality test to check for normal distribution. Additional details of data processing are detailed in respective methods descriptions and additional details of statistical tests can be found in the figure legends.
Acknowledgments
This work was supported in part by NIH grant R21 AI128175 (to M.J.S.) and by the Jefferson Vaccine Center. We thank Gene S. Tan (J. Craig Venter Institute, La Jolla, CA, USA) for MOKV G cDNA and pooled mouse anti-MOKV G and Rachael Lambert (Thomas Jefferson University) for technical assistance in vaccine production. Publication made possible in part by support from the Thomas Jefferson University + Philadelphia University Open Access Fund. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Author Contributions
Conceptualization, C.R.F. and M.J.S.; Methodology, C.R.F., C.W., and G.C.; Investigation, C.R.F., D.E.L., T.G.S., and Y.Y.; Resources, C.W., D.E.L., T.G.S., and C.L.H.; Writing – Original Draft, C.R.F.; Writing – Review & Editing, C.R.F., D.E.L., T.G.S., C.L.H., G.C., and M.J.S.; Supervision, M.J.S.; Funding Acquisition, M.J.S.
Declaration of Interests
C.R.F., C.W., and M.J.S. are inventors on the U.S. Patent Application No. WO2018231974A1 (“Composition and Administration of Chimeric Glycoprotein Lyssavirus Vaccines for Coverage against Rabies”).
Published: July 21, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107920.
Supplemental Information
References
- Abreu-Mota T., Hagen K.R., Cooper K., Jahrling P.B., Tan G., Wirblich C., Johnson R.F., Schnell M.J. Non-neutralizing antibodies elicited by recombinant Lassa-Rabies vaccine are critical for protection against Lassa fever. Nat. Commun. 2018;9:4223. doi: 10.1038/s41467-018-06741-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aréchiga Ceballos N., Vázquez Morón S., Berciano J.M., Nicolás O., Aznar López C., Juste J., Rodríguez Nevado C., Aguilar Setién A., Echevarría J.E. Novel lyssavirus in bat, Spain. Emerg. Infect. Dis. 2013;19:793–795. doi: 10.3201/eid1905.121071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badrane H., Tordo N. Host switching in Lyssavirus history from the Chiroptera to the Carnivora orders. J. Virol. 2001;75:8096–8104. doi: 10.1128/JVI.75.17.8096-8104.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badrane H., Bahloul C., Perrin P., Tordo N. Evidence of two Lyssavirus phylogroups with distinct pathogenicity and immunogenicity. J. Virol. 2001;75:3268–3276. doi: 10.1128/JVI.75.7.3268-3276.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahloul C., Jacob Y., Tordo N., Perrin P. DNA-based immunization for exploring the enlargement of immunological cross-reactivity against the lyssaviruses. Vaccine. 1998;16:417–425. doi: 10.1016/s0264-410x(97)00204-1. [DOI] [PubMed] [Google Scholar]
- Banyard A.C., Evans J.S., Luo T.R., Fooks A.R. Lyssaviruses and bats: emergence and zoonotic threat. Viruses. 2014;6:2974–2990. doi: 10.3390/v6082974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmansour A., Leblois H., Coulon P., Tuffereau C., Gaudin Y., Flamand A., Lafay F. Antigenicity of rabies virus glycoprotein. J. Virol. 1991;65:4198–4203. doi: 10.1128/jvi.65.8.4198-4203.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaney J.E., Wirblich C., Papaneri A.B., Johnson R.F., Myers C.J., Juelich T.L., Holbrook M.R., Freiberg A.N., Bernbaum J.G., Jahrling P.B. Inactivated or live-attenuated bivalent vaccines that confer protection against rabies and Ebola viruses. J. Virol. 2011;85:10605–10616. doi: 10.1128/JVI.00558-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaney J.E., Marzi A., Willet M., Papaneri A.B., Wirblich C., Feldmann F., Holbrook M., Jahrling P., Feldmann H., Schnell M.J. Antibody quality and protection from lethal Ebola virus challenge in nonhuman primates immunized with rabies virus based bivalent vaccine. PLoS Pathog. 2013;9:e1003389. doi: 10.1371/journal.ppat.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanoff W.A., Perez E.I., López T., Arias C.F., DuBois R.M. Structural Basis for Escape of Human Astrovirus from Antibody Neutralization: Broad Implications for Rational Vaccine Design. J. Virol. 2017;92:e01546-17. doi: 10.1128/JVI.01546-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogel K. Proposed International Reference Rabies Vaccine (HDC-Origin) and the potency tests used to test these products. In: Hennessen W., Regamey R.H., editors. Joint WHO/IABS Symposium on the Standardization of Rabies Vaccines for Human Use Produced in Tissue Culture [Rabies III] Karger; 1978. pp. 267–271. [Google Scholar]
- Both L., Banyard A.C., van Dolleweerd C., Horton D.L., Ma J.K.C., Fooks A.R. Passive immunity in the prevention of rabies. Lancet Infect. Dis. 2012;12:397–407. doi: 10.1016/S1473-3099(11)70340-1. [DOI] [PubMed] [Google Scholar]
- Both L., van Dolleweerd C., Wright E., Banyard A.C., Bulmer-Thomas B., Selden D., Altmann F., Fooks A.R., Ma J.K.C. Production, characterization, and antigen specificity of recombinant 62-71-3, a candidate monoclonal antibody for rabies prophylaxis in humans. FASEB J. 2013;27:2055–2065. doi: 10.1096/fj.12-219964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai K., Feng J.N., Wang Q., Li T., Shi J., Hou X.J., Gao X., Liu H., Tu W., Xiao L., Wang H. Fine mapping and interaction analysis of a linear rabies virus neutralizing epitope. Microbes Infect. 2010;12:948–955. doi: 10.1016/j.micinf.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Coertse J., Markotter W., le Roux K., Stewart D., Sabeta C.T., Nel L.H. New isolations of the rabies-related Mokola virus from South Africa. BMC Vet. Res. 2017;13:37. doi: 10.1186/s12917-017-0948-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coler R.N., Baldwin S.L., Shaverdian N., Bertholet S., Reed S.J., Raman V.S., Lu X., DeVos J., Hancock K., Katz J.M. A synthetic adjuvant to enhance and expand immune responses to influenza vaccines. PLoS ONE. 2010;5:e13677. doi: 10.1371/journal.pone.0013677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S.J., Fooks A.R., van der Poel W.H.M. Public health threat of new, reemerging, and neglected zoonoses in the industrialized world. Emerg. Infect. Dis. 2010;16:1–7. doi: 10.3201/eid1601.081467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedictis P., Minola A., Rota Nodari E., Aiello R., Zecchin B., Salomoni A., Foglierini M., Agatic G., Vanzetta F., Lavenir R. Development of broad-spectrum human monoclonal antibodies for rabies post-exposure prophylaxis. EMBO Mol. Med. 2016;8:407–421. doi: 10.15252/emmm.201505986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzschold B., Wunner W.H., Wiktor T.J., Lopes A.D., Lafon M., Smith C.L., Koprowski H. Characterization of an antigenic determinant of the glycoprotein that correlates with pathogenicity of rabies virus. Proc. Natl. Acad. Sci. USA. 1983;80:70–74. doi: 10.1073/pnas.80.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzschold B., Rupprecht C.E., Tollis M., Lafon M., Mattei J., Wiktor T.J., Koprowski H. Antigenic diversity of the glycoprotein and nucleocapsid proteins of rabies and rabies-related viruses: implications for epidemiology and control of rabies. Rev. Infect. Dis. 1988;10(Suppl 4):S785–S798. doi: 10.1093/clinids/10.supplement_4.s785. [DOI] [PubMed] [Google Scholar]
- Evans J.S., Horton D.L., Easton A.J., Fooks A.R., Banyard A.C. Rabies virus vaccines: is there a need for a pan-lyssavirus vaccine? Vaccine. 2012;30:7447–7454. doi: 10.1016/j.vaccine.2012.10.015. [DOI] [PubMed] [Google Scholar]
- Evans J.S., Selden D., Wu G., Wright E., Horton D.L., Fooks A.R., Banyard A.C. Antigenic site changes in the rabies virus glycoprotein dictates functionality and neutralizing capability against divergent lyssaviruses. J. Gen. Virol. 2018;99:169–180. doi: 10.1099/jgv.0.000998. [DOI] [PubMed] [Google Scholar]
- Evans J.S., Wu G., Selden D., Buczkowski H., Thorne L., Fooks A.R., Banyard A.C. Utilisation of chimeric lyssaviruses to assess vaccine protection against highly divergent lyssaviruses. Viruses. 2018;10:130. doi: 10.3390/v10030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Shi Y., Yu M., Xu W., Gong W., Tu Z., Ding L., He B., Guo H., Tu C. Livestock rabies outbreaks in Shanxi province, China. Arch. Virol. 2016;161:2851–2854. doi: 10.1007/s00705-016-2982-9. [DOI] [PubMed] [Google Scholar]
- Fernando B.-G.G., Yersin C.-T.T., José C.-B.B., Paola Z.-S.S. Predicted 3D model of the rabies virus glycoprotein trimer. BioMed Res. Int. 2016;2016:1674580. doi: 10.1155/2016/1674580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher C.R., Streicker D.G., Schnell M.J. The spread and evolution of rabies virus: conquering new frontiers. Nat. Rev. Microbiol. 2018;16:241–255. doi: 10.1038/nrmicro.2018.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fooks A.R., Banyard A.C., Horton D.L., Johnson N., McElhinney L.M., Jackson A.C. Current status of rabies and prospects for elimination. Lancet. 2014;384:1389–1399. doi: 10.1016/S0140-6736(13)62707-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fooks A.R., Cliquet F., Finke S., Freuling C., Hemachudha T., Mani R.S., Müller T., Nadin-Davis S., Picard-Meyer E., Wilde H., Banyard A.C. Rabies. Nat. Rev. Dis. Primers. 2017;3:17091. doi: 10.1038/nrdp.2017.91. [DOI] [PubMed] [Google Scholar]
- Gavi . Gavi; 2018. 06a—Annex C: Rabies Investment Case: Vaccine Investment Strategy Programme and Policy Committee Meeting.https://www.gavi.org/sites/default/files/document/ppc-meeting-18-19-october-2018---vis-06a---annex-c--rabies-investment-casepdf.pdf [Google Scholar]
- Gomme E.A., Faul E.J., Flomenberg P., McGettigan J.P., Schnell M.J. Characterization of a single-cycle rabies virus-based vaccine vector. J. Virol. 2010;84:2820–2831. doi: 10.1128/JVI.01870-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson K., Coudeville L., Lembo T., Sambo M., Kieffer A., Attlan M., Barrat J., Blanton J.D., Briggs D.J., Cleaveland S., Global Alliance for Rabies Control Partners for Rabies Prevention Estimating the global burden of endemic canine rabies. PLoS Negl. Trop. Dis. 2015;9:e0003709. doi: 10.1371/journal.pntd.0003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon C.A., DeMattos C.A., DeMattos C.C., Niezgoda M., Hooper D.C., Koprowski H., Notkins A., Rupprecht C.E. Experimental utility of rabies virus-neutralizing human monoclonal antibodies in post-exposure prophylaxis. Vaccine. 2001;19:3834–3842. doi: 10.1016/s0264-410x(01)00135-9. [DOI] [PubMed] [Google Scholar]
- Hanlon C.A., Kuzmin I.V., Blanton J.D., Weldon W.C., Manangan J.S., Rupprecht C.E. Efficacy of rabies biologics against new lyssaviruses from Eurasia. Virus Res. 2005;111:44–54. doi: 10.1016/j.virusres.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Horton D.L., McElhinney L.M., Marston D.A., Wood J.L.N., Russell C.A., Lewis N., Kuzmin I.V., Fouchier R.A.M., Osterhaus A.D.M.E., Fooks A.R., Smith D.J. Quantifying antigenic relationships among the lyssaviruses. J. Virol. 2010;84:11841–11848. doi: 10.1128/JVI.01153-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton D.L., Banyard A.C., Marston D.A., Wise E., Selden D., Nunez A., Hicks D., Lembo T., Cleaveland S., Peel A.J. Antigenic and genetic characterization of a divergent African virus, Ikoma lyssavirus. J. Gen. Virol. 2014;95:1025–1032. doi: 10.1099/vir.0.061952-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S.-C., Hsu C.-L., Lee M.-S., Tu Y.-C., Chang J.-C., Wu C.-H., Lee S.-H., Ting L.-J., Tsai K.-R., Cheng M.-C. Lyssavirus in Japanese Pipistrelle, Taiwan. Emerg. Infect. Dis. 2018;24:782–785. doi: 10.3201/eid2404.171696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallet C., Jacob Y., Bahloul C., Drings A., Desmezieres E., Tordo N., Perrin P. Chimeric lyssavirus glycoproteins with increased immunological potential. J. Virol. 1999;73:225–233. doi: 10.1128/jvi.73.1.225-233.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N., Mansfield K.L., Fooks A.R. Canine vaccine recipients recognize an immunodominant region of the rabies virus glycoprotein. J. Gen. Virol. 2002;83:2663–2669. doi: 10.1099/0022-1317-83-11-2663. [DOI] [PubMed] [Google Scholar]
- Johnson R.F., Kurup D., Hagen K.R., Fisher C., Keshwara R., Papaneri A., Perry D.L., Cooper K., Jahrling P.B., Wang J.T. An Inactivated Rabies Virus-Based Ebola Vaccine, FILORAB1, Adjuvanted With Glucopyranosyl Lipid A in Stable Emulsion Confers Complete Protection in Nonhuman Primate Challenge Models. J. Infect. Dis. 2016;214(Suppl 3):S342–S354. doi: 10.1093/infdis/jiw231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keates L., WHO Publication Rabies vaccines: WHO position paper—recommendations. Vaccine. 2010;28:7140–7142. doi: 10.1016/j.vaccine.2010.08.082. [DOI] [PubMed] [Google Scholar]
- Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshwara R., Hagen K.R., Abreu-Mota T., Papaneri A.B., Liu D., Wirblich C., Johnson R.F., Schnell M.J. A Recombinant Rabies Virus Expressing the Marburg Virus Glycoprotein Is Dependent Upon Antibody-Mediated Cellular Cytotoxicity for Protection Against Marburg Virus Disease in a Murine Model. J. Virol. 2019;93:e01865-18. doi: 10.1128/JVI.01865-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kgaladi J., Faber M., Dietzschold B., Nel L.H., Markotter W. Pathogenicity and Immunogenicity of Recombinant Rabies Viruses Expressing the Lagos Bat Virus Matrix and Glycoprotein: Perspectives for a Pan-Lyssavirus Vaccine. Trop. Med. Infect. Dis. 2017;2:37. doi: 10.3390/tropicalmed2030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochanek K.D., Xu J., Murphy S.L., Miniño A.M., Kung H.-C. Deaths: final data for 2009. Natl. Vital Stat. Rep. 2011;60:1–116. [PubMed] [Google Scholar]
- Kurup D., Wirblich C., Feldmann H., Marzi A., Schnell M.J. Rhabdovirus-based vaccine platforms against henipaviruses. J. Virol. 2015;89:144–154. doi: 10.1128/JVI.02308-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin I.V., Niezgoda M., Franka R., Agwanda B., Markotter W., Beagley J.C., Urazova O.Y., Breiman R.F., Rupprecht C.E. Lagos bat virus in Kenya. J. Clin. Microbiol. 2008;46:1451–1461. doi: 10.1128/JCM.00016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin I.V., Shi M., Orciari L.A., Yager P.A., Velasco-Villa A., Kuzmina N.A., Streicker D.G., Bergman D.L., Rupprecht C.E. Molecular inferences suggest multiple host shifts of rabies viruses from bats to mesocarnivores in Arizona during 2001-2009. PLoS Pathog. 2012;8:e1002786. doi: 10.1371/journal.ppat.1002786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafay F., Coulon P., Astic L., Saucier D., Riche D., Holley A., Flamand A. Spread of the CVS strain of rabies virus and of the avirulent mutant AvO1 along the olfactory pathways of the mouse after intranasal inoculation. Virology. 1991;183:320–330. doi: 10.1016/0042-6822(91)90145-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon M. Rabies virus receptors. J. Neurovirol. 2005;11:82–87. doi: 10.1080/13550280590900427. [DOI] [PubMed] [Google Scholar]
- Langevin C., Tuffereau C. Mutations Conferring Resistance to Neutralization by a Soluble Form of the Neurotrophin Receptor (p75NTR) Map outside of the Known Antigenic Sites of the Rabies Virus Glycoprotein. J. Virol. 2002;76:10756–10765. doi: 10.1128/JVI.76.21.10756-10765.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis C.E., Reising M.M., Fry A.M., Conrad S.K., Siev D., Gatewood D.M., Hermann J.R. Evaluation of a non-invasive, inhalational challenge method for rabies vaccine potency assay. J. Virol. Methods. 2013;190:49–52. doi: 10.1016/j.jviromet.2013.03.020. [DOI] [PubMed] [Google Scholar]
- Liu Y., Chen Q., Zhang F., Zhang S., Li N., Lian H., Wang Y., Zhang J., Hu R. Evaluation of rabies biologics against Irkut virus isolated in China. J. Clin. Microbiol. 2013;51:3499–3504. doi: 10.1128/JCM.01565-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähl P., Cliquet F., Guiot A.-L., Niin E., Fournials E., Saint-Jean N., Aubert M., Rupprecht C.E., Gueguen S. Twenty year experience of the oral rabies vaccine SAG2 in wildlife: a global review. Vet. Res. (Faisalabad) 2014;45:77. doi: 10.1186/s13567-014-0077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallewa M., Fooks A.R., Banda D., Chikungwa P., Mankhambo L., Molyneux E., Molyneux M.E., Solomon T. Rabies encephalitis in malaria-endemic area, Malawi, Africa. Emerg. Infect. Dis. 2007;13:136–139. doi: 10.3201/eid1301.060810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani R.S., Damodar T., S D., Domala S., Gurung B., Jadhav V., Konanki R., Lingappa L., Loganathan S.K., Salagare R., Tambi P. Case Reports: Survival from rabies: Case series from India. Am. J. Trop. Med. Hyg. 2019;100:165–169. doi: 10.4269/ajtmh.18-0711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marissen W.E., Kramer R.A., Rice A., Weldon W.C., Niezgoda M., Faber M., Slootstra J.W., Meloen R.H., Clijsters-van der Horst M., Visser T.J. Novel rabies virus-neutralizing epitope recognized by human monoclonal antibody: fine mapping and escape mutant analysis. J. Virol. 2005;79:4672–4678. doi: 10.1128/JVI.79.8.4672-4678.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markotter W., Coertse J. Bat lyssaviruses. Rev. Sci. Tech. 2018;37:385–400. doi: 10.20506/rst.37.2.2809. [DOI] [PubMed] [Google Scholar]
- Marston D.A., Horton D.L., Ngeleja C., Hampson K., McElhinney L.M., Banyard A.C., Haydon D., Cleaveland S., Rupprecht C.E., Bigambo M. Ikoma lyssavirus, highly divergent novel lyssavirus in an African civet. Emerg. Infect. Dis. 2012;18:664–667. doi: 10.3201/eid1804.111553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGettigan J.P., Sarma S., Orenstein J.M., Pomerantz R.J., Schnell M.J. Expression and immunogenicity of human immunodeficiency virus type 1 Gag expressed by a replication-competent rhabdovirus-based vaccine vector. J. Virol. 2001;75:8724–8732. doi: 10.1128/JVI.75.18.8724-8732.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGettigan J.P., Pomerantz R.J., Siler C.A., McKenna P.M., Foley H.D., Dietzschold B., Schnell M.J. Second-generation rabies virus-based vaccine vectors expressing human immunodeficiency virus type 1 gag have greatly reduced pathogenicity but are highly immunogenic. J. Virol. 2003;77:237–244. doi: 10.1128/JVI.77.1.237-244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mebatsion T. Extensive attenuation of rabies virus by simultaneously modifying the dynein light chain binding site in the P protein and replacing Arg333 in the G protein. J. Virol. 2001;75:11496–11502. doi: 10.1128/JVI.75.23.11496-11502.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mebatsion T., Schnell M.J., Conzelmann K.K. Mokola virus glycoprotein and chimeric proteins can replace rabies virus glycoprotein in the rescue of infectious defective rabies virus particles. J. Virol. 1995;69:1444–1451. doi: 10.1128/jvi.69.3.1444-1451.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mélade J., McCulloch S., Ramasindrazana B., Lagadec E., Turpin M., Pascalis H., Goodman S.M., Markotter W., Dellagi K. Serological Evidence of Lyssaviruses among Bats on Southwestern Indian Ocean Islands. PLoS ONE. 2016;11:e0160553. doi: 10.1371/journal.pone.0160553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto K., Hooper D.C., Spitsin S., Koprowski H., Dietzschold B. Pathogenicity of different rabies virus variants inversely correlates with apoptosis and rabies virus glycoprotein expression in infected primary neuron cultures. J. Virol. 1999;73:510–518. doi: 10.1128/jvi.73.1.510-518.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nel L.H. Proceedings of the Southern and Eastern African Rabies Group / World Health Organization Meeting. Editions Fondation Marcel Mérieux; Lyon, France: 2001. Mokola virus: A brief review of the status quo; pp. 80–86. [Google Scholar]
- Nel L., Jacobs J., Jaftha J., von Teichman B., Bingham J., Olivier M. New cases of Mokola virus infection in South Africa: a genotypic comparison of Southern African virus isolates. Virus Genes. 2000;20:103–106. doi: 10.1023/a:1008120511752. [DOI] [PubMed] [Google Scholar]
- Ni Y., Tominaga Y., Honda Y., Morimoto K., Sakamoto S., Kawai A. Mapping and characterization of a sequential epitope on the rabies virus glycoprotein which is recognized by a neutralizing monoclonal antibody, RG719. Microbiol. Immunol. 1995;39:693–702. doi: 10.1111/j.1348-0421.1995.tb03258.x. [DOI] [PubMed] [Google Scholar]
- Nokireki T., Tammiranta N., Kokkonen U.M., Kantala T., Gadd T. Tentative novel lyssavirus in a bat in Finland. Transbound. Emerg. Dis. 2018;65:593–596. doi: 10.1111/tbed.12833. [DOI] [PubMed] [Google Scholar]
- Orłowska A., Smreczak M., Freuling C.M., Müller T., Trębas P., Rola J. Serological Survey of Lyssaviruses in Polish Bats in the Frame of Passive Rabies Surveillance Using an Enzyme-Linked Immunosorbent Assay. Viruses. 2020;12:271. doi: 10.3390/v12030271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaneri A.B., Wirblich C., Marissen W.E., Schnell M.J. Alanine scanning of the rabies virus glycoprotein antigenic site III using recombinant rabies virus: implication for post-exposure treatment. Vaccine. 2013;31:5897–5902. doi: 10.1016/j.vaccine.2013.09.038. [DOI] [PubMed] [Google Scholar]
- Patton K., Aslam S., Shambaugh C., Lin R., Heeke D., Frantz C., Zuo F., Esser M.T., Paliard X., Lambert S.L. Enhanced immunogenicity of a respiratory syncytial virus (RSV) F subunit vaccine formulated with the adjuvant GLA-SE in cynomolgus macaques. Vaccine. 2015;33:4472–4478. doi: 10.1016/j.vaccine.2015.07.025. [DOI] [PubMed] [Google Scholar]
- Percy D.H., Bhatt P.N., Tignor G.H., Shope R.E. Experimental infection of dogs and monkeys with two rabies serogroup viruses, Lagos bat and Mokola (IbAn 27377). Gross pathologic and histopathologic changes. Vet. Pathol. 1973;10:534–549. doi: 10.1177/030098587301000607. [DOI] [PubMed] [Google Scholar]
- Perrin P., Tino de Franco M., Jallet C., Fouque F., Morgeaux S., Tordo N., Colle J.H. The antigen-specific cell-mediated immune response in mice is suppressed by infection with pathogenic Lyssoviruses. Res. Virol. 1996;147:289–299. doi: 10.1016/0923-2516(96)82287-4. [DOI] [PubMed] [Google Scholar]
- Prehaud C., Coulon P., LaFay F., Thiers C., Flamand A. Antigenic site II of the rabies virus glycoprotein: structure and role in viral virulence. J. Virol. 1988;62:1–7. doi: 10.1128/jvi.62.1.1-7.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche S., Rey F.A., Gaudin Y., Bressanelli S. Structure of the Prefusion Form of vesicular stomatitis virus glycoprotein G. Science. 2007;315:843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- Rosseels V., Nazé F., De Craeye S., Francart A., Kalai M., Van Gucht S. A non-invasive intranasal inoculation technique using isoflurane anesthesia to infect the brain of mice with rabies virus. J. Virol. Methods. 2011;173:127–136. doi: 10.1016/j.jviromet.2011.01.019. [DOI] [PubMed] [Google Scholar]
- Sabeta C.T., Markotter W., Mohale D.K., Shumba W., Wandeler A.I., Nel L.H. Mokola virus in domestic mammals, South Africa. Emerg. Infect. Dis. 2007;13:1371–1373. doi: 10.3201/eid1309.070466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeta C., Blumberg L., Miyen J., Mohale D., Shumba W., Wandeler A. Mokola virus involved in a human contact (South Africa) FEMS Immunol. Med. Microbiol. 2010;58:85–90. doi: 10.1111/j.1574-695X.2009.00609.x. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakin-Eshleman S.H., Remaley A.T., Eshleman J.R., Wunner W.H., Spitalnik S.L. N-linked glycosylation of rabies virus glycoprotein. Individual sequons differ in their glycosylation efficiencies and influence on cell surface expression. J. Biol. Chem. 1992;267:10690–10698. [PubMed] [Google Scholar]
- Shope R.E., Murphy F.A., Harrison A.K., Causey O.R., Kemp G.E., Simpson D.I.H., Moore D.L. Two African viruses serologically and morphologically related to rabies virus. J. Virol. 1970;6:690–692. doi: 10.1128/jvi.6.5.690-692.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirima S.B., Durier C., Kara L., Houard S., Gansane A., Loulergue P., Bahuaud M., Benhamouda N., Nebié I., Faber B., AMA1-DiCo Study Group Safety and immunogenicity of a recombinant Plasmodium falciparum AMA1-DiCo malaria vaccine adjuvanted with GLA-SE or Alhydrogel® in European and African adults: A phase 1a/1b, randomized, double-blind multi-centre trial. Vaccine. 2017;35:6218–6227. doi: 10.1016/j.vaccine.2017.09.027. [DOI] [PubMed] [Google Scholar]
- Smith T.G., Gilbert A.T. Comparison of a Micro-Neutralization Test with the Rapid Fluorescent Focus Inhibition Test for Measuring Rabies Virus Neutralizing Antibodies. Trop. Med. Infect. Dis. 2017;2:24. doi: 10.3390/tropicalmed2030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.F., Goldberg M., Rosenthal S., Carlson L., Chen J., Chen C., Ramachandran S. Global rise in human infectious disease outbreaks. J. R. Soc. Interface. 2014;11:20140950. doi: 10.1098/rsif.2014.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suu-Ire R.D., Fooks A.R., Banyard A.C., Selden D., Amponsah-Mensah K., Riesle S., Ziekah M.Y., Ntiamoa-Baidu Y., Wood J.L.N., Cunningham A.A. Lagos Bat Virus Infection Dynamics in Free-Ranging Straw-Colored Fruit Bats (Eidolon helvum) Trop. Med. Infect. Dis. 2017;2:25. doi: 10.3390/tropicalmed2030025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Burgwal L.H.M., Neevel A.M.G., Pittens C.A.C.M., Osterhaus A.D.M.E., Rupprecht C.E., Claassen E. Barriers to innovation in human rabies prophylaxis and treatment: A causal analysis of insights from key opinion leaders and literature. Zoonoses Public Health. 2017;64:599–611. doi: 10.1111/zph.12352. [DOI] [PubMed] [Google Scholar]
- von Teichman B.F., de Koker W.C., Bosch S.J.E., Bishop G.C., Meredith C.D., Bingham J. Mokola virus infection: description of recent South African cases and a review of the virus epidemiology. J. S. Afr. Vet. Assoc. 1998;69:169–171. doi: 10.4102/jsava.v69i4.847. [DOI] [PubMed] [Google Scholar]
- Vuta V., Picard-Meyer E., Robardet E., Barboi G., Motiu R., Barbuceanu F., Vlagioiu C., Cliquet F. Vaccine-induced rabies case in a cow (Bos taurus): Molecular characterisation of vaccine strain in brain tissue. Vaccine. 2016;34:5021–5025. doi: 10.1016/j.vaccine.2016.08.013. [DOI] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J.C., Huang Y.Z., Zhong D.K., Kang L., Ishag H., Mao X., Cao R.B., Zhou B., Chen P.Y. Design and evaluation of a multi-epitope peptide against Japanese encephalitis virus infection in BALB/c mice. Biochem. Biophys. Res. Commun. 2010;396:787–792. doi: 10.1016/j.bbrc.2010.04.133. [DOI] [PubMed] [Google Scholar]
- Welburn S.C., Beange I., Ducrotoy M.J., Okello A.L. The neglected zoonoses—the case for integrated control and advocacy. Clin. Microbiol. Infect. 2015;21:433–443. doi: 10.1016/j.cmi.2015.04.011. [DOI] [PubMed] [Google Scholar]
- Weyer J., Kuzmin I.V., Rupprecht C.E., Nel L.H. Cross-protective and cross-reactive immune responses to recombinant vaccinia viruses expressing full-length lyssavirus glycoprotein genes. Epidemiol. Infect. 2008;136:670–678. doi: 10.1017/S0950268807008965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willet M., Kurup D., Papaneri A., Wirblich C., Hooper J.W., Kwilas S.A., Keshwara R., Hudacek A., Beilfuss S., Rudolph G. Preclinical Development of Inactivated Rabies Virus-Based Polyvalent Vaccine Against Rabies and Filoviruses. J. Infect. Dis. 2015;212(Suppl 2):S414–S424. doi: 10.1093/infdis/jiv251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirblich C., Tan G.S., Papaneri A., Godlewski P.J., Orenstein J.M., Harty R.N., Schnell M.J. PPEY motif within the rabies virus (RV) matrix protein is essential for efficient virion release and RV pathogenicity. J. Virol. 2008;82:9730–9738. doi: 10.1128/JVI.00889-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojczyk B.S., Stwora-Wojczyk M., Shakin-Eshleman S., Wunner W.H., Spitalnik S.L. The role of site-specific N-glycosylation in secretion of soluble forms of rabies virus glycoprotein. Glycobiology. 1998;8:121–130. doi: 10.1093/glycob/8.2.121. [DOI] [PubMed] [Google Scholar]
- World Health Organization Human rabies: 2016 updates and call for data. Wkly. Epidemiol. Rec. 2017;92:77–86. [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; 2017. The Immunological Basis for Immunization Series: Module 17: Rabies Vaccines. [Google Scholar]
- World Health Organization . Fifth Edition. World Health Organization; 2018. Laboratory Techniques in Rabies. [Google Scholar]
- Wright E., Hayman D.T.S., Vaughan A., Temperton N.J., Wood J.L.N., Cunningham A.A., Suu-Ire R., Weiss R.A., Fooks A.R. Virus neutralising activity of African fruit bat (Eidolon helvum) sera against emerging lyssaviruses. Virology. 2010;408:183–189. doi: 10.1016/j.virol.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunner W.H., Jackson A.C. Academic Press; 2010. Rabies: Scientific Basis of the Disease and Its Management. [Google Scholar]
- Xu H., Hu C., Gong R., Chen Y., Ren N., Xiao G., Xie Q., Zhang M., Liu Q., Guo A., Chen H. Evaluation of a novel chimeric B cell epitope-based vaccine against mastitis induced by either Streptococcus agalactiae or Staphylococcus aureus in mice. Clin. Vaccine Immunol. 2011;18:893–900. doi: 10.1128/CVI.00066-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Lin S., Ye F., Yang J., Qi J., Chen Z., Lin X., Wang J., Yue D., Cheng Y. Structural Analysis of Rabies Virus Glycoprotein Reveals pH-Dependent Conformational Changes and Interactions with a Neutralizing Antibody. Cell Host Microbe. 2020;27:441–453.e7. doi: 10.1016/j.chom.2019.12.012. [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W.-Y., Shi Y., Wu C., Zhang W.-J., Mao X.-H., Guo G., Li H.-X., Zou Q.-M. Therapeutic efficacy of a multi-epitope vaccine against Helicobacter pylori infection in BALB/c mice model. Vaccine. 2009;27:5013–5019. doi: 10.1016/j.vaccine.2009.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available upon request to the lead contact author. No proprietary software was used in the data analysis.