Abstract
Sup35p is an essential protein in yeast that functions in complex with Sup45p for efficient translation termination. While some may argue that this function is the only important attribute of Sup35p, there are two additional known facets of Sup35p’s biology that may provide equally important functions for yeast, both of which involve various strategies for coping with stress. Recently, the N-terminal and middle regions (NM) of Sup35p, which are not required for translation termination function, have been found to provide stress sensing abilities and facilitate the phase separation of Sup35p into biomolecular condensates in response to transient stress. Interestingly, the same NM domain is also required for Sup35p to misfold and enter into aggregates associated with the [PSI+] prion. Here, we review these three different states or “faces” of Sup35p. For each, we compare the functionality and necessity of different Sup35p domains, including the role these domains play in facilitating interactions with important protein partners, and discuss the potential ramifications that each state affords yeast cells under varying environmental conditions.
Many proteins in yeast have been shown to change folding states in response to environmental stress. One protein, Sup35p, not only plays an essential role as a translation release factor but has been shown to undergo phase separation into biomolecular condensates as well as misfold to form the prion [PSI+]. This budding topic describes the three states of Sup35p (translation termination, phase separation, and prion formation) and sheds light on how short and long-term stress impacts these three states.
Introduction:
Sup35p is an essential protein that corresponds to the eukaryotic translation termination factor 3 (eRF3) in yeast(reviewed in Inge-Vechtomov et al., 2003).In addition toits critical role in translation, this simple protein also has some noteworthy attributes. Recently, it was found that Sup35p can respond to transient stress by undergoing phase transition. In the presence of short heat or pH stress, Sup35ptemporarily assembles into reversible condensates, which are localizedareas that contain high concentrations of protein(Franzmann et al., 2018).Another unique feature of Sup35p is that it can misfold and assemble into self-perpetuating, infectious amyloids that can be propagated for many generations(reviewed in Liebman and Chernoff, 2012). Here, we review the three different states or “faces” of Sup35p, including important interacting partners, the role of Sup35 domains in these protein interactions, and how Sup35p contributes to general cellular functions including the ability to respond to transient and long-term stress.
Sup35p and translation
Two mutants, carrying mutations within SUP35or SUP45, were identified through screens for suppressors of nonsense mutations(Inge-Vechtomov, 1964; Smirnov et al., 1976). Both suppressors were later determined to encode eukaryotic release factors, eRF3 and eRF1, respectively, and are essential proteins required for translation termination(reviewed in Inge-Vechtomov et al., 2003; Nizhnikov et al., 2014). Sup45p plays a role in the recognition of stop codons. Similar to eRF1, Sup45p has a structure resembling a tRNA and initiates peptidyl tRNA hydrolysis (Bertram et al., 2000; Song et al., 2000; Stansfield et al., 1997). Sup35p supplies the GTPase activity, which together with Sup45p, leads to the termination of translation and release of the translational complex.
Sup35p is a modular protein that contains a C-terminal region that is both necessary and sufficient for translation termination, and is required for cell viability. This region, which contains a GTPase fold commonly found in G-proteins, has been shown to have GTPase activity (Salas-Marco and Bedwell, 2004; Stansfield et al., 1993) and interacts with Sup45p(Stansfield et al., 1995). For termination function, both Sup35p and Sup45p interact with the translating ribosome for stop codon recognition (Wada and Ito, 2014). GTP hydrolysis by Sup35p provides the energy for dissociation of the translation complex and release of the newly synthesized polypeptide (reviewed in Dever et al., 2016).
The C-terminus of Sup35p and other eRF3 proteins are important for translational termination function and are highly conserved (Chernoff et al., 1992; Kushnirov et al., 1988). However, regions outside of this C-terminal domain aredispensable for translational termination and are quite diverged (Chernoff et al., 1992; Ter-Avanesyan et al., 1993). In mammals, the non-essential N-terminal region of mammalian eRF3 has been shown to interact with PABP, the poly(A)-binding protein that is required for translation initiation and mRNA stabilization (Hoshino et al., 1999). In vitro translation experiments show that PABP directly affects translational termination activity by enhancing eRF3 binding to the ribosome(Ivanov et al., 2016). Authors have proposed that interaction between PABP and eRF3 plays an important role in positioning of the eRF3/eRF1 complex near stop codons for efficient translational termination (Ivanov et al., 2016).
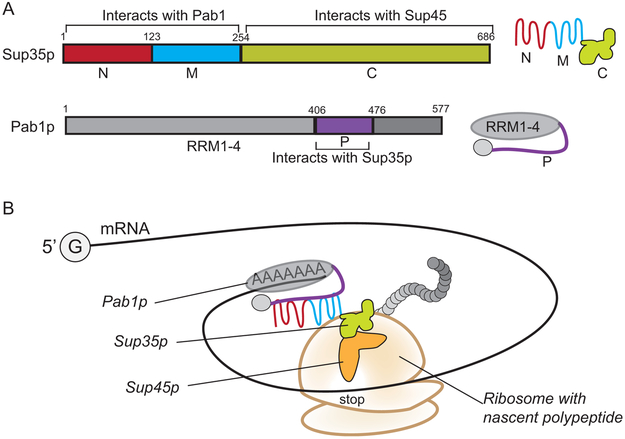
Despite the divergence of non-essential sequences between the yeast Sup35 protein and the mammalian eRF3, the N-terminal region of Sup35p binds to the yeast homolog of the PABP protein, Pab1p(Roque et al., 2015). Yeast Two Hybrid and pull down assays show that a proline rich region in Pab1p (P domain; Fig. 1A)plays an important role in association with the N-terminus of Sup35p through a non-canonical Pab1p binding site (Cosson et al., 2002; Roque et al., 2015). Based on how mammalian PABP influences translational termination (Ivanov et al., 2016), we suspect that a similar mechanism occurs in yeast. It is possible that the interaction between yeast Pab1p and Sup35p positions the Sup35p/Sup45p complex near the stop codon. Such positioning could provide the translation machinery access to release factors in order to stop translation in a timely and efficient manner (Fig. 1B).
Figure 1. Sup35p,Pab1p, and the model of protein interaction during translation.
A. Sup35p is comprised of three regions: The N-terminal (1-123; red) and middle domains (124-254; blue) and the C-terminal region (255-686; light green). A symbol for the folded Sup35 protein and its domains is shown to the right. The C-terminal region of Sup35p interacts with Sup45p and has a GTPase fold necessary for GTPase activity. The N and M regions of Sup35p interact with the poly(A)-binding protein, Pab1p. The Pab1 protein is also modular, containing four RNA recognition motifs (RRM1-4), and a proline rich region (P domain, shown in purple) that interacts with Sup35p.A symbol for Pab1p and its domains are shown to the right. B. During translation, protein interactions between Sup35p NM regions and Pab1pcould position theSup35p/Sup45pcomplex near stop codons. As the translating ribosomes encounter stop codons, the positioned Sup45p is able to recognize the stop codon and initiate tRNA hydrolysis, while Sup35p provides the GTPase activity for the release of the translational complex.
Sup35p and phase separation
Translation is essential for cellular function. However, under times of transient stress, there may be a need to temporarily shut down processes like translation and shift the cellular energy load to other functions. Work in the last few years has shown that Sup35p, along with several other proteins, can change phase states and thus be conditionally pulled from the soluble protein pool into protein-rich biomolecular condensates in a process called phase separation. Phase separation provides the ability to locally organize proteins within the intracellular milieu without the need for distinct membrane organelles. These “membraneles sorganelles” allow proteins to be temporarily sequestered or stored into highly concentrated environments during stress. Upon removal of the stress, proteins compartmentalized through phase separation transition back into the aqueous cellular environment. Proteins containing low-complexity regions that are intrinsically disordered have been found to undergo phase transition (Franzmann and Alberti, 2018; Franzmann et al., 2018; Kato et al., 2012; Kroschwald et al., 2018; Molliex et al., 2015; Riback et al., 2017). It is thought that these low complexity or intrinsically disordered regions are sensitive to changes brought about by transient stress, and mediate the transition between a functional protein in a soluble environment and a protein that is sequestered into the protein dense environment of a membraneless organelle (reviewed in Alberti, 2017).
Stress granules are an example of membraneless organelles, and form in yeast in response to transient heat stress. Proteins move into stress granules through a process called demixing, where a protein population goes from a soluble state to localized regions of high protein concentration. Treatment of yeast cells at 46°C for 8 minutes leads to rapid demixing, yet return to normal temperatures leads to the disappearance of stress granules and release of proteins back to the soluble state (Wallace et al., 2015). 117 yeast proteins have been shown to undergo reversible phase separation in response to heat, many of which are associated with translation, RNA binding, and chaperone function (Wallace et al., 2015). Among the translation associated factors identified on this list were Sup35p, Sup45p, and Pab1p. The identification of these translation associated factors within stress granules is not surprising since there is a correlation between the formation of stress granules and decrease in translational efficiency (Buchan and Parker, 2009). As discussed above, interaction between Sup35p and Pab1p is mediated through the Sup35p N and M regions and the Pab1p hydrophobic P domain(Fig. 1; Cosson et al., 2002; Roque et al., 2015). The hydrophobic P domain of Pab1p has low complexity, and possibly works as a stress sensor (Fig. 2). Reducing the hydrophobicity of this proline-rich region reduces the ability of Pab1pto undergo phase separation. Therefore, this P domain may play an important role in keeping Pab1p soluble in non-stress conditions, but can also sense fluctuations in the environment to induce phase separation(Riback et al., 2017).
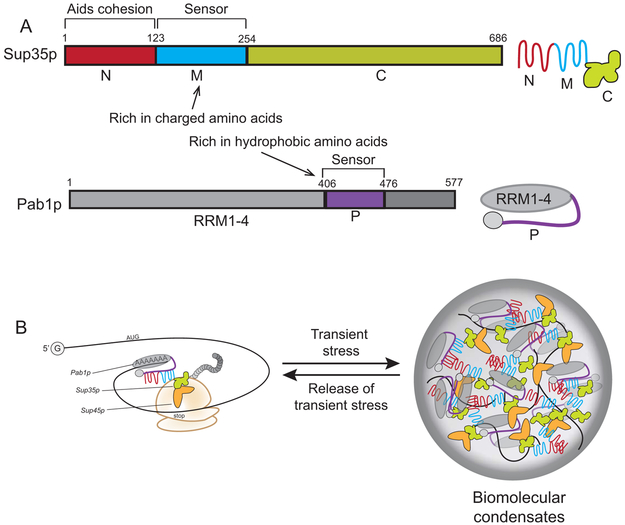
Figure 2. Sup35p, Pab1p, and the model of phase separation.
A. Sup35p and Pab1p are described in figure 1, but the same domains provide different functionalities in phase separation. The charged amino acids within the M region of Sup35pare responsive to changes in pH, allowing Sup35p to enter into biomolecular condensates at low pH. The N-terminal region of Sup35p appears to enhance condensation. The P domain (purple) of Pab1p is hydrophobic, is temperature responsive and can also phase separate. B. During transient stress, the sensor regions of both Sup35p and Pab1p signal the proteins to undergo phase separation into biomolecular condensates. It should be noted that the sensor regions of both proteins also participate in protein interactions between Sup35p and Pab1p as shown in figure 1.
Data supporting the specific phase separation properties of Sup35p has been recently reported by Franzmann et al (2018). Using Sup35-GFP fusion proteins, Sup35p is able to transition into biomolecular condensates upon energy depletion, and return to the cytoplasm upon nutrient enrichment. In parallel, translation stops upon energy depletion, and is restored upon nutrient enrichment. Since pH levels decrease as a result of starvation, authors tested how mitochondrial uncouplers, which transiently lower cytosolic pH, influence Sup35p phase separation. Similar to starvation, pH stress also causes Sup35p to phase separate into protein dense particles along with Pab1p(Fig. 2; Franzmann et al., 2018). The authors showed that acidic amino acids within the M region of Sup35p are important for phase separation in response to pH, since mutation of these acidic amino acids to polar residues reduces the ability to respond to stress. Similar to the proline-rich region of Pab1p, the ionizable nature of the Sup35p M domain may allow the protein to sense intracellular pH changes. While the N-terminal region does not have sensor ability, it appears to enhance the ability of Sup35p to enter into condensates(Franzmann et al., 2018).
The above studies suggest that both Pab1p and Sup35p individually have regions that sense cellular stress and mediate phase transition. These same two sensor regions also mediate the interaction between Sup35p and Pab1p during translation (Fig. 1A and Fig. 2A). The existence of sensor regions that also mediate the interaction of these proteins may not be coincidental. It is possible that while these regions are important for translation efficiency, conformational changes due to transient stress allow these proteins to be temporarily sequestered possibly resulting in translational issues. Yet upon the release of stress, these proteins quickly solubilize and are available for translation (Fig. 2B). It is foreseeable that phase separation can affect other important processes involving proteins with low complexity regions. For example, several proteins associated with transcription and chromatin remodeling contain low complexity regions. One protein, Snf5p, is glutamine and asparagine rich, and also appears to phase separate in response to pH stress (Gutierrez et al., 2018). It is intriguing to speculate that inherent sensing abilities via low complexity regions could provide a wide array of proteins, particularly those involved in global functions such as transcription and translation, with a rapid mechanism for responding to transient stress.
Sup35p as a prion
At the time when Sup35p mutants were being pulled out of the original nonsense suppressor screens in the 1960s, a cytoplasmic element named ψ was also shown to suppress nonsense mutations (Cox, 1965). Years later, this cytoplasmic element was shown to be the misfolded, self-perpetuating, infectious form of the Sup35 protein, called the [PSI+] prion (reviewed in Liebman and Chernoff, 2012). In contrast to transient phase separation of Sup35p into reversible biomolecular condensates, the [PSI+] prionenables the Sup35p protein to assemble into stable amyloid aggregates that can be propagated for many generations. Cell viability is not severely impacted in [PSI+] strains, suggesting that there must be a sufficient amount of functional Sup35p to ensure some basal level of translation termination (Pezza et al., 2014; Zhou et al., 1999). However, toxicity is observed when Sup35p is overexpressed in the presence of [PSI+] (Derkatch et al., 1996). This toxicity appears to be caused by overexpression driving the excess Sup35p into aggregates. Likewise, the excess Sup35p pulled into aggregates sequesters Sup45p away from essential translational functions (Vishveshwara et al., 2009), suggesting that there is a delicate balance between maintaining the prion and ensuring that Sup35p and Sup45p are available for essential translation termination functions.
The ability of Sup35p to become a prion is dependent upon the region that is dispensable for translation termination but contributes to phase separation, the N-terminal region (Fig. 3A; Ter-Avanesyan et al., 1994). The N-terminus of Sup35p contains a region rich in glutamine and asparagine, followed by a repeat region that contains several degenerate tandem repeats required for stabilizing the intermolecular interactions between Sup35p molecules within the aggregate during prion formation (Liu and Lindquist, 1999; Parham et al., 2001). While the M region and the C-terminus are not required for prion formation, the M region has been shown to foster the formation of specific Sup35p conformations called prion strains or variants(Bradley and Liebman, 2004), and shown to be important for [PSI+] stabilization (Liu and Lindquist, 1999). [PSI+] variants have different size and biochemical characteristics, as well as stabilities and translational termination efficiencies. For example,without M sequences, strong [PSI+] variants are able to stably propagate for many generations,whereas weaker [PSI+] variants are quickly lost from the population (Bradley and Liebman, 2004).
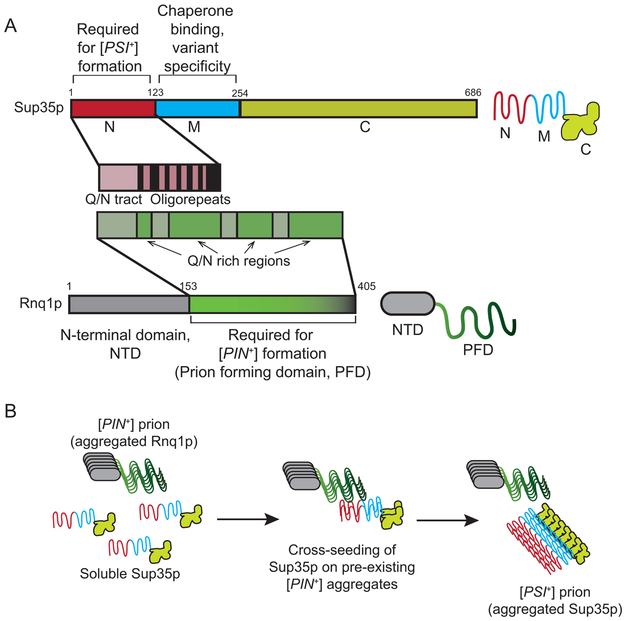
Figure 3. Sup35p, Rnq1p, and the model of prion formation through cross seeding.
A. The N-terminal region of Sup35p contains both aglutamine and asparagine (Q/N) rich region and oligopeptide repeats that are required for prion formation. The M region interacts with chaperone machinery and fosters the formation of certain [PSI+] variants. The Rnq1 protein has a dispensable N-terminal region and a C-terminal domain that contains 4 Q/N rich regions involved [PIN+] prion maintenance. A symbol for the Rnq1 protein and its domains is shown to the right. B. [PSI+] formation is proposed to be enhanced through a cross seeding mechanism in which the[RNQ+] prion is able to nucleate Sup35p assembly into the [PSI+] prion. It is thought that interactions between Q/N rich regions mediate cross seeding.
The propagation of [PSI+] over many generations requires protein quality control factors such as chaperones. A complex of molecular chaperones, such as Hsp104p, Sis1p, and Ssa1p, work together to recognize the prion particles and ultimately shear the prion into smaller particles. The transmission of these smaller prion particles to daughter cells ensures that the prion is propagated to the next generation(reviewed in Liebman and Chernoff, 2012).It has been shown that the M region of Sup35p plays a role in the interaction with chaperone machinery to enhance the propagation of the prion (Helsen and Glover, 2012). The ability to propagate [PSI+] over many generations may have some adaptive value to cells, potentially due to the phenotypic variation facilitated by the readthrough of nonsense mutations or subtle variation in translation termination efficiency (True and Lindquist, 2000). However, [PSI+] does not cause dramatic global proteomic changes (Chan et al., 2017). Instead, ribosomal profiling studies show that approximately 100 genes are susceptible to stop codon readthrough in the presence of [PSI+], and [PSI+] appears to impact reading frame selection for a subset of genes (Baudin-Baillieu et al., 2014).
The spontaneous formation of [PSI+] in yeast cells occurs at a very low rate of approximately 10−7 per generation (Allen et al., 2007; Lancaster et al., 2010; Lund and Cox, 1981). Overexpression of Sup35p or its N- and M- regions (Sup35NM) can marginally increase prion formation, while prion formation can be dramatically enhanced by overexpression in the presence of a second prion. For example, the presence of the prion form of the Rnq1p protein called [PIN+], also known as [RNQ+], can enhance the conversion of Sup35p to the prion state(Fig. 3B; Derkatch et al., 2001; Osherovich and Weissman, 2001; Sondheimer and Lindquist, 2000). Two models have been suggested to mediate the process by which [PIN+] induces [PSI+] formation. The first is the inhibitor titration model, which suggests that the [PIN+] prion sequesters or titrates a factor such as a chaperone that normally inhibits the aggregation of Sup35p(Derkatch et al., 2001; Osherovich and Weissman, 2001). However, there is little evidence to support or negate this model. The second is the cross seeding model in which a pre-existing aggregated protein is used as a template to enhance the misfolding and aggregation of a second heterologous protein (Derkatch et al., 2001; Osherovich and Weissman, 2001). It has been shown from in vitro studies that Rnq1p fibers can cross seed the formation of Sup35p fibers (Keefer et al., 2017; Sharma and Liebman, 2013; Vitrenko et al., 2007), and in vivo studies have provided both co-localization and genetic support for the cross seeding model (Derkatch et al., 2004; Keefer et al., 2017). Within the sequence of Rnq1p, the long glutamine/asparagine (Q/N) rich region near the C-terminus is not only essential for the propagation of [PIN+], but even a Q-R substitution in this region can dramatically reduce [PSI+] induction (Fig. 3B; Derkatch et al., 2001; Keefer et al., 2017).It has been suggested that the heterotypic interactions between [PIN+] and the high concentration of Sup35NM through overexpression fosters amyloid nucleation and bypasses the formation of condensates. When [PIN+] is absent, it is well established that prion formation is diminished and the overexpression of Sup35NM leads to condensate formation rather than the prion state (Khan et al., 2018).
The process of [PIN+] dependent prion formation can be monitored both biochemically and visually using fluorescently tagged Sup35NM fusion proteins. During formation, Sup35p is initially converted from a monomericform to small SDS-resistant oligomeric complexes(Salnikova et al., 2005; Sharma et al., 2017). At the time of this initial SDS-resistant oligomer detection, Sup35NM-GFP visibly exhibits diffuse cytoplasmic fluorescence indicating that Sup35p is associated with extremely small aggregates. As the oligomers assemble into larger SDS-stable aggregates over time, small highly mobile fluorescent foci are visually detected. These foci quickly coalesce into a single fluorescent dot before being sequestered near the cell periphery (Lyke and Manogaran, 2017; Sharma et al., 2017). Once residing at the periphery, the majority of the diffuse Sup35NM quickly converges into the aggregate within 20 minutes. This quick sequestration coincides with the growth of the aggregate into dots, or ring and line-like structures(Sharma et al., 2017; Zhou et al., 2001). Cells containing these structures are considered to be hallmarks of [PSI+] since cells that contain aggregates give rise to [PSI+] cells but diffuse cells do not (Ganusova et al., 2006; Sharma et al., 2017).
The de novo formation of [PSI+] has been shown to be influenced by both the actin cytoskeleton and protein quality control systems. Loss of genes that influence the formation of endocytic cortical actin patches leads to decreased dot, ring, and line formation (Ganusova et al., 2006; Manogaran et al., 2011). It has been proposed that the glutamine-rich proteins of the endocytic actin patch may provide a location near the periphery of the cell that allows the cross seeding of Sup35p into dot, ring, and line aggregates (Ganusova et al., 2006). Protein quality control also influences prion formation. Mutations that disrupt autophagy, oxidative stress response, or the ubiquitin proteasome system results in increased prion formation (Allen et al., 2007; Chernova et al., 2011; Doronina et al., 2015; Speldewinde et al., 2015), suggesting that under normal conditions, protein quality control mechanisms actively limit prion formation. Conversely, long-termstress can also enhance prion formation. Exposure to high salt or hydrogen peroxide stress for 12–24 hours leads to high levels of cell death. However, of the small surviving population, prion formation was shown to be enhanced by approximately 60 fold (Tyedmers et al., 2008). The enhancement of prion formation in response to long-term stress may provide a means to generate phenotypic variation through the readthrough of nonsense mutations or alteration of translation termination efficiency. Therefore, this variation may lead to a small subset of the population that is well suited for the extreme environment.
The three faces of Sup35
Here, we have discussed three separate states for Sup35p: a functional translation termination state, a phase separation state, and a prion state.The role of Sup35p in translation termination is widely accepted, however the purpose of Sup35p’s ability to be sequestered into biomolecular condensates and the ability to form a prion is highly debated. It has been proposed that the ancestral function of the N- and M- regions of Sup35p is to sense acute stress and foster reversible phase separation into condensates rather than encourage prion formation (Franzmann and Alberti, 2018; Franzmann et al., 2018).This argument is to some extent supported by the fact that [PSI+] can be detrimental, as only those variants with translation termination function are able to survive (Wickner, 2011). However, the ancestral function of the N and M regions of Sup35p is likely much more complicated than simply preferring phase separation over prion formation. [PSI+] formation is enhanced by cross seeding and through chronic stress. While [PSI+] is not abundant in nature, other prions such as [PIN+] are found in both wild and commercial strains(Halfmann et al., 2012; Nakayashiki et al., 2005). It is possible the existence of [PIN+] in the wild provides a means to efficiently induce [PSI+] in response to chronic stress. This formation of [PSI+] could be advantageous, since it has been shown that [PSI+] provides growth benefits under distinct stress conditions and genetic backgrounds (True and Lindquist, 2000). It is possible that cells need both phase transition and prion formation to serve specific purposes. Under acute stress, phase transition allows for the short-term sequestration of Sup35p in order to transiently shut down translation termination. Under chronic conditions, prion formation could allow for phenotypic variation in populations. Those cells that endure the stress while retaining the prion would be able to divide and propagate the prion form for many generations (Fig. 4).
Figure 4. Sup35p response to different stresses.
The ability of Sup35p to enter phase transition could be dependent upon the type of stress. Transient (acute) stress could foster Sup35p to condense into biomolecular condensates, whereas extreme, long-term (chronic) stress could foster Sup35p into the prion form.
The three separate states of Sup35p are reminiscent of the 1957 film, The Three Faces of Eve. A story of a woman with three separate personalities: Eve White, a modest unassuming woman who takes care of the home, Eve Black, a tempestuous woman who quickly appears and disappears during conflict, and Jane, who emerges after many years as a stable and consistent individual. Similarly Sup35p could be viewed as having multiple personalities: the first involved in translational termination, the second who seamlessly phase separates into and out of biomolecular condensates in response to acute stress, and third, the stable prion that can be propagated for many generations.Given that intriguing facets of Sup35p have continued to be uncovered over the 50+ years of study, experiments over the next decade will further define these Sup35p states, and may reveal new and unexpected Sup35p states in the future.
Acknowledgements:
We thank Adam Knier for critical comments on the manuscript. This work was supported by a grant from the National Institutes of Health (GM109336) to A.L.M. and D.R.L. was supported by the M.U. Fellowship.
Literature Cited
- Alberti S (2017). Phase separation in biology. Curr Biol 27, R1097–R1102. [DOI] [PubMed] [Google Scholar]
- Allen KD, Chernova TA, Tennant EP, Wilkinson KD, and Chernoff YO (2007). Effects of ubiquitin system alterations on the formation and loss of a yeast prion. JBiolChem 282, 3004–3013. [DOI] [PubMed] [Google Scholar]
- Baudin-Baillieu A, Legendre R, Kuchly C, Hatin I, Demais S, Mestdagh C, Gautheret D, and Namy O (2014). Genome-wide translational changes induced by the prion [PSI+]. Cell Rep 8, 439–448. [DOI] [PubMed] [Google Scholar]
- Bertram G, Bell HA, Ritchie DW, Fullerton G, and Stansfield I (2000). Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA 6, 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley ME, and Liebman SW (2004). The Sup35 domains required for maintenance of weak, strong or undifferentiated yeast [PSI+] prions. Mol Microbiol 51, 1649–1659. [DOI] [PubMed] [Google Scholar]
- Buchan JR, and Parker R (2009). Eukaryotic stress granules: the ins and outs of translation. Mol Cell 36, 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PHW, Lee L, Kim E, Hui T, Stoynov N, Nassar R, Moksa M, Cameron DM, Hirst M, Gsponer J, et al. (2017). The [PSI (+)] yeast prion does not wildly affect proteome composition whereas selective pressure exerted on [PSI (+)] cells can promote aneuploidy. Sci Rep 7, 8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Ptyushkina MV, Samsonova MG, Sizonencko GI, Pavlov YI, Ter-Avanesyan MD, and Inge-Vechtomov SG (1992). Conservative system for dosage-dependent modulation of translational fidelity in eukaryotes. Biochimie 74, 455–461. [DOI] [PubMed] [Google Scholar]
- Chernova TA, Romanyuk AV, Karpova TS, Shanks JR, Ali M, Moffatt N, Howie RL, O’Dell A, McNally JG, Liebman SW, et al. (2011). Prion induction by the short-lived, stress-induced protein Lsb2 is regulated by ubiquitination and association with the actin cytoskeleton. Mol Cell 43, 242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson B, Couturier A, Chabelskaya S, Kiktev D, Inge-Vechtomov S, Philippe M, and Zhouravleva G (2002). Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol Cell Biol 22, 3301–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox BS (1965). [PSI], a cytoplasmic suppressor of super-suppressors in yeast. Heridity 20. [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY, and Liebman SW (2001). Prions affect the appearance of other prions: the story of [PIN(+)]. Cell 106, 171–182. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, and Liebman SW (1996). Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 144, 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, and Liebman SW (2004). Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci U S A 101, 12934–12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Kinzy TG, and Pavitt GD (2016). Mechanism and Regulation of Protein Synthesis in Saccharomyces cerevisiae. Genetics 203, 65–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doronina VA, Staniforth GL, Speldewinde SH, Tuite MF, and Grant CM (2015). Oxidative stress conditions increase the frequency of de novo formation of the yeast [PSI+] prion. Mol Microbiol 96, 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann T, and Alberti S (2018). Prion-like low-complexity sequences: Key regulators of protein solubility and phase behavior. J Biol Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann TM, Jahnel M, Pozniakovsky A, Mahamid J, Holehouse AS, Nuske E, Richter D, Baumeister W, Grill SW, Pappu RV, et al. (2018). Phase separation of a yeast prion protein promotes cellular fitness. Science 359. [DOI] [PubMed] [Google Scholar]
- Ganusova EE, Ozolins LN, Bhagat S, Newnam GP, Wegrzyn RD, Sherman MY, and Chernoff YO (2006). Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol Cell Biol 26, 617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez JI, Brittingham G, Wang X, Fenyo D, and Holt LJ (2018). The largest SWI/SNF polyglutamine domain is a pH sensor. bioRxiv 10.1101/165043. [DOI] [Google Scholar]
- Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, and Lindquist S (2012). Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature 482, 363–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsen CW, and Glover JR (2012). Insight into molecular basis of curing of [PSI+] prion by overexpression of 104-kDa heat shock protein (Hsp104). J Biol Chem 287, 542–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino S, Imai M, Kobayashi T, Uchida N, and Katada T (1999). The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3’-Poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J Biol Chem 274, 16677–16680. [DOI] [PubMed] [Google Scholar]
- Inge-Vechtomov S, Zhouravleva G, and Philippe M (2003). Eukaryotic release factors (eRFs) history. Biol Cell 95, 195–209. [DOI] [PubMed] [Google Scholar]
- Inge-Vechtomov SG (1964). Reversions to prototrophy in adenineless yeast. Vestnik LGU (Russ), 112–116. [Google Scholar]
- Ivanov A, Mikhailova T, Eliseev B, Yeramala L, Sokolova E, Susorov D, Shuvalov A, Schaffitzel C, and Alkalaeva E (2016). PABP enhances release factor recruitment and stop codon recognition during translation termination. Nucleic Acids Res 44, 7766–7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. (2012). Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefer KM, Stein KC, and True HL (2017). Heterologous prion-forming proteins interact to cross-seed aggregation in Saccharomyces cerevisiae. Sci Rep 7, 5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan T, Kandola TS, Wu J, Venkatesan S, Ketter E, Lange JJ, Rodriguez Gama A, Box A, Unruh JR, Cook M, et al. (2018). Quantifying Nucleation In Vivo Reveals the Physical Basis of Prion-like Phase Behavior. Mol Cell 71, 155–168 e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Munder MC, Maharana S, Franzmann TM, Richter D, Ruer M, Hyman AA, and Alberti S (2018). Different Material States of Pub1 Condensates Define Distinct Modes of Stress Adaptation and Recovery. Cell Rep 23, 3327–3339. [DOI] [PubMed] [Google Scholar]
- Kushnirov VV, Ter-Avanesyan MD, Telckov MV, Surguchov AP, Smirnov VN, and Inge-Vechtomov SG (1988). Nucleotide sequence of the SUP2 (SUP35) gene of Saccharomyces cerevisiae. Gene 66, 45–54. [DOI] [PubMed] [Google Scholar]
- Lancaster AK, Bardill JP, True HL, and Masel J (2010). The spontaneous appearance rate of the yeast prion [PSI+] and its implications for the evolution of the evolvability properties of the [PSI+] system. Genetics 184, 393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman SW, and Chernoff YO (2012). Prions in yeast. Genetics 191, 1041–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, and Lindquist S (1999). Oligopeptide-repeat expansions modulate ‘protein-only’ inheritance in yeast. Nature 400, 573–576. [DOI] [PubMed] [Google Scholar]
- Lund PM, and Cox BS (1981). Reversion analysis of [psi-] mutations in Saccharomyces cerevisiae. Genetical research 37, 173–182. [DOI] [PubMed] [Google Scholar]
- Lyke DR, and Manogaran AL (2017). Spatial sequestration and oligomer remodeling during de novo [PSI+] formation. Prion 11, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manogaran AL, Hong JY, Hufana J, Tyedmers J, Lindquist S, and Liebman SW (2011). Prion formation and polyglutamine aggregation are controlled by two classes of genes. PLoS genetics 7, e1001386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, and Taylor JP (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki T, Kurtzman CP, Edskes HK, and Wickner RB (2005). Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci U S A 102, 10575–10580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizhnikov AA, Antonets KS, Inge-Vechtomov SG, and Derkatch IL (2014). Modulation of efficiency of translation termination in Saccharomyces cerevisiae. Prion 8, 247–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherovich LZ, and Weissman JS (2001). Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell 106, 183–194. [DOI] [PubMed] [Google Scholar]
- Parham SN, Resende CG, and Tuite MF (2001). Oligopeptide repeats in the yeast protein Sup35p stabilize intermolecular prion interactions. EMBO J 20, 2111–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezza JA, Villali J, Sindi SS, and Serio TR (2014). Amyloid-associated activity contributes to the severity and toxicity of a prion phenotype. Nat Commun 5, 4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riback JA, Katanski CD, Kear-Scott JL, Pilipenko EV, Rojek AE, Sosnick TR, and Drummond DA (2017). Stress-Triggered Phase Separation Is an Adaptive, Evolutionarily Tuned Response. Cell 168, 1028–1040 e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque S, Cerciat M, Gaugue I, Mora L, Floch AG, de Zamaroczy M, Heurgue-Hamard V, and Kervestin S (2015). Interaction between the poly(A)-binding protein Pab1 and the eukaryotic release factor eRF3 regulates translation termination but not mRNA decay in Saccharomyces cerevisiae. RNA 21, 124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Marco J, and Bedwell DM (2004). GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol Cell Biol 24, 7769–7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikova AB, Kryndushkin DS, Smirnov VN, Kushnirov VV, and Ter-Avanesyan MD (2005). Nonsense suppression in yeast cells overproducing Sup35 (eRF3) is caused by its non-heritable amyloids. J Biol Chem 280, 8808–8812. [DOI] [PubMed] [Google Scholar]
- Sharma J, and Liebman SW (2013). Exploring the basis of [PIN(+)] variant differences in [PSI(+)] induction. J Mol Biol 425, 3046–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma J, Wisniewski BT, Paulson E, Obaoye JO, Merrill SJ, and Manogaran AL (2017). De novo [PSI +] prion formation involves multiple pathways to form infectious oligomers. Sci Rep 7, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov VN, Surguchov AP, Fominykch ES, Lizlova LV, Saprygina TV, and Inge-Vechtomov SG (1976). Recessive nonsense-suppression in yeast: further characterization of a defect in translation. FEBS Lett 66, 12–15. [DOI] [PubMed] [Google Scholar]
- Sondheimer N, and Lindquist S (2000). Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell 5, 163–172. [DOI] [PubMed] [Google Scholar]
- Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, and Barford D (2000). The crystal structure of human eukaryotic release factor eRF1--mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100, 311–321. [DOI] [PubMed] [Google Scholar]
- Speldewinde SH, Doronina VA, and Grant CM (2015). Autophagy protects against de novo formation of the [PSI+] prion in yeast. Mol Biol Cell 26, 4541–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfield I, Grant CM, Akhmaloka, and Tuite MF (1993). Errors in stop codon recognition in a temperature sensitive mutation of yeast. Biochemical Society transactions 21, 329S. [DOI] [PubMed] [Google Scholar]
- Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, and Tuite MF (1995). The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. Embo J 14, 4365–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfield I, Kushnirov VV, Jones KM, and Tuite MF (1997). A conditional-lethal translation termination defect in a sup45 mutant of the yeast Saccharomyces cerevisiae. Eur J Biochem 245, 557–563. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Dagkesamanskaya AR, Kushnirov VV, and Smirnov VN (1994). The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics 137, 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Kushnirov VV, Dagkesamanskaya AR, Didichenko SA, Chernoff YO, Inge-Vechtomov SG, and Smirnov VN (1993). Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol 7, 683–692. [DOI] [PubMed] [Google Scholar]
- True HL, and Lindquist SL (2000). A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407, 477–483. [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Madariaga ML, and Lindquist S (2008). Prion switching in response to environmental stress. PLoS Biol 6, e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishveshwara N, Bradley ME, and Liebman SW (2009). Sequestration of essential proteins causes prion associated toxicity in yeast. Mol Microbiol 73, 1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitrenko YA, Gracheva EO, Richmond JE, and Liebman SW (2007). Visualization of aggregation of the Rnq1 prion domain and cross-seeding interactions with Sup35NM. J Biol Chem 282, 1779–1787. [DOI] [PubMed] [Google Scholar]
- Wada M, and Ito K (2014). A genetic approach for analyzing the co-operative function of the tRNA mimicry complex, eRF1/eRF3, in translation termination on the ribosome. Nucleic Acids Res 42, 7851–7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace EW, Kear-Scott JL, Pilipenko EV, Schwartz MH, Laskowski PR, Rojek AE, Katanski CD, Riback JA, Dion MF, Franks AM, et al. (2015). Reversible, Specific, Active Aggregates of Endogenous Proteins Assemble upon Heat Stress. Cell 162, 1286–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB (2011). Prion diseases: Infectivity versus toxicity. Nature 470, 470–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Derkatch IL, and Liebman SW (2001). The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI(+)] and [PIN(+)]. Mol Microbiol 39, 37–46. [DOI] [PubMed] [Google Scholar]
- Zhou P, Derkatch IL, Uptain SM, Patino MM, Lindquist S, and Liebman SW (1999). The yeast non-Mendelian factor [ETA+] is a variant of [PSI+], a prion-like form of release factor eRF3. EMBO J 18, 1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]