HSV infections can cause pathologies ranging from recurrent lesions to significant ocular disease. Initiation of lytic infection and reactivation from latency in sensory neurons are dependent on the induced expression of the viral immediate early genes. Transcription of these genes is controlled at multiple levels, including modulation of the chromatin state of the viral genome and appropriate recruitment of transcription factors and coactivators. Following initiation of transcription, IE genes are subject to a key regulatory stage in which transcriptional elongation rates are controlled by the activity of the super elongation complex. Inhibition of the SEC blocks both lytic infection and reactivation from latency in sensory neurons. In addition to providing insights into the mechanisms controlling viral infection and reactivation, inhibitors of critical components such as the SEC may represent novel antivirals.
KEYWORDS: transcription elongation, HSV reactivation, HCF-1, AFF4, herpes simplex virus, latency, super elongation complex, transcription
ABSTRACT
Induction of herpes simplex virus (HSV) immediate early (IE) gene transcription promotes the initiation of lytic infection and reactivation from latency in sensory neurons. IE genes are transcribed by the cellular RNA polymerase II (RNAPII) and regulated by multiple transcription factors and coactivators. The HCF-1 cellular coactivator plays a central role in driving IE expression at multiple stages through interactions with transcription factors, chromatin modulation complexes, and transcription elongation components, including the active super elongation complex/P-TEFb (SEC-P-TEFb). Here, we demonstrate that the SEC occupies the promoters of HSV IE genes during the initiation of lytic infection and during reactivation from latency. Specific inhibitors of the SEC suppress viral IE expression and block the spread of HSV infection. Significantly, these inhibitors also block the initiation of viral reactivation from latency in sensory ganglia. The potent suppression of IE gene expression by SEC inhibitors indicates that transcriptional elongation represents a determining rate-limiting stage in HSV IE gene transcription and that the SEC plays a critical role in driving productive elongation during both phases of the viral life cycle. Most importantly, this supports the model that signal-mediated induction of SEC-P-TEFb levels can promote reactivation of a population of poised latent genomes.
INTRODUCTION
Following a primary infection, herpes simplex virus (HSV) establishes a quiescent/latent infection in sensory neurons. Upon stimulation, the virus can reenter the lytic replication cycle to produce recurrent disease that ranges from oral and genital lesions to severe ocular disease. In addition, neurological and developmental issues are linked to congenital infections (1, 2).
Regulation of viral infection occurs primarily at the level of transcription of viral genes that are expressed in defined classes in a sequential manner. For HSV, immediate early (IE) genes represent the first wave of viral genes to be expressed and the products of these genes are critical to promote lytic infection and reactivation. Transcription of IE genes is mediated by cellular RNA polymerase II (RNAPII) and is regulated by viral (VP16) and cellular (e.g., GAPB, SP1, Oct-1) transcription factors, transcriptional coactivators, and chromatin modulation complexes that are recruited to IE enhancer/promoters (3). A central component that is essential for IE gene expression is the cellular transcriptional coactivator HCF-1 (4). HCF-1 is a component of multiple chromatin modulation complexes and functions primarily to bridge IE transcription factors and the cellular chromatin modulation machinery. One key HCF-1 complex that controls IE gene expression contains histone demethylases (LSD1/KDM1A and JMJD2s/KDM4s) and histone methyltransferases (SETD1A/KMT2F and MLL1/KMT2A) that function to remove repressive heterochromatin or to prevent its accumulation on viral IE genes (5–9).
HCF-1 also plays a significant role in the control of viral reactivation from latency in sensory neurons. Upon stimulation that promotes viral reactivation, HCF-1 is rapidly transported from the cytoplasm to the nucleus and is assembled on viral IE gene enhancers/promoters (10–12). In addition, inhibitors of the HCF-1-associated histone demethylases (LSD1/KDM1A, JMJD2s/KDM4s) suppress the initiation of viral reactivation (6, 7, 13, 14). Thus, initiation of lytic infection and initiation of reactivation are both critically dependent on this coactivator and its associated components.
In addition to roles in mediating transcriptional initiation via interactions with TFs and chromatin modulation complexes, analyses of HCF-1 complexes in uninfected and infected cells revealed that the protein was associated with a network of transcriptional elongation components (15). For many genes, modulation of transcription elongation rate is a critical regulatory step that may poise genes for rapid activation in response to environmental signals, allow for appropriate transitions in chromatin states, or prevent epigenetic repression of genes (16–20).
Following initiation, RNAPII associates with negative elongation factors (NELF, DSIF), resulting in enhanced pausing and reduced elongation rates. A critical step in the control of pause release is mediated by recruitment of the P-TEFb complex containing cyclin T1 (CCNT1) and CDK9 kinase. CDK9 phosphorylates RNAPII (S2p), NELF, and DSIF, resulting in dissociation of NELF, association of PAF1, and stimulation of polymerase release and elongation (21–24). P-TEFb/CDK9 is itself regulated in a complex manner. More than 50% of P-TEFb is associated with the 7SK-snRNP complex and thus is sequestered in a catalytically inactive configuration. Upon stimulation by stress or environmental signals, P-TEFb is released and the active complex is recruited to specific genes in association with BRD4 or the super elongation complex (SEC) (25–35).
In addition to control of cellular signal-mediated transcriptional induction, regulated transcriptional elongation plays an important role in the life cycle of multiple viruses that utilize the host RNAPII machinery (e.g., adenovirus, human cytomegalovirus [hCMV], Kaposi’s sarcoma-associated herpesvirus [KSHV], hepatitis B virus [HBV], human immunodeficiency virus [HIV], and human T cell leukemia virus [HTLV]) (36–46). In HSV infection, RNAPII pausing and induction of elongation critically regulate viral IE gene expression. Paused polymerase and associated pausing factors (NELF) occupy the promoter-proximal regions of IE genes, and the SEC is specifically required for induction of IE gene transcription (15). Most significantly, compounds that inhibit the competitive adaptor BRD4 or enhance the levels of active SEC-P-TEFb induce HSV reactivation in a mouse model of HSV latency (15). These observations led to the model that stress signal-mediated activation of SEC-P-TEFb can promote IE gene expression and reactivation of a population of latent viral genomes.
Here, the use of recently identified novel inhibitors of the SEC-P-TEFb (47) has allowed for direct probing of the role of this complex in control of HSV lytic infection and reactivation. Inhibition of the SEC suppresses viral IE gene expression, initiation of lytic infection, spread of infection, and viral reactivation from latency in sensory ganglia. These compounds function via reduction in levels of the SEC scaffold protein AFF4, resulting in reduction in SEC occupancy of viral IE promoters, further supporting the model that activation of SEC-P-TEFb directly promotes the initiation of lytic infection and reactivation. Interestingly, levels of the SEC scaffold protein AFF4 are enhanced in HSV-infected cells in an ICP0-dependent manner, suggesting a novel virus-mediated mechanism for modulation of SEC activity.
Translationally, compounds that target cellular components like the SEC that are critical for HSV infection have potential as antivirals, especially for those mutants that are resistant to canonical antivirals that inhibit viral replication machinery (48, 49).
RESULTS
Suppression of HSV IE gene expression and lytic infection by inhibition of the SEC.
Previously, it was demonstrated that transcriptional elongation of the HSV IE genes was a critical rate-limiting step in IE gene expression. Stimulation of elongation specifically required the SEC which was found in association with the transcriptional coactivator HCF-1, an essential component of the HSV IE gene regulatory complex (15).
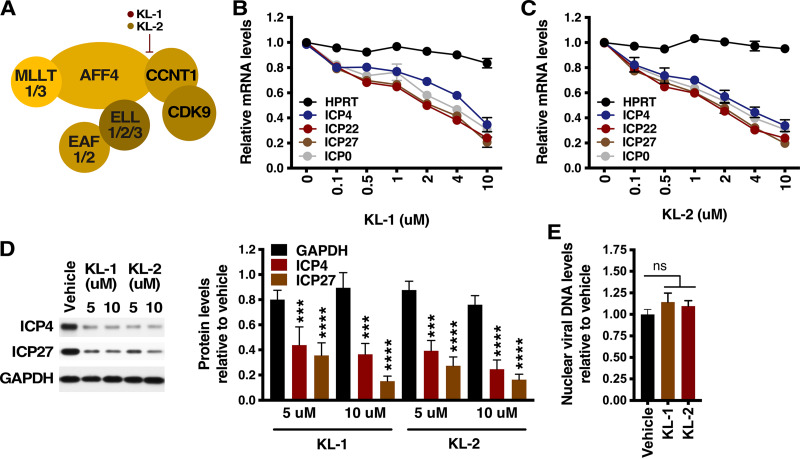
Recently, specific inhibitors of the SEC-P-TEFb (KL-1 and KL-2) that interfere with the assembly/association of P-TEFb subunit CCNT1 (cyclin T1) with the SEC scaffold AFF4 were identified (47) (Fig. 1A). Therefore, to probe the significance of the SEC in HSV IE transcription, primary human foreskin fibroblast (HFF) cells were treated with increasing concentrations of vehicle, KL-1, or KL-2 for 2 h followed by infection with HSV. As shown, treatment with KL-1 or KL-2 resulted in a dose-dependent decrease in levels of viral IE gene mRNAs (Fig. 1B and C) and protein (Fig. 1D) without significantly impacting the expression of the control cellular hypoxanthine phosphoribosyltransferase (HPRT) gene. The 50% inhibitory concentration (IC50) values corresponding to inhibition of IE gene expression by KL-1 (2.25 to 6.98 μM) and KL-2 (1.61 to 2.29 μM) are shown in Table S1.
FIG 1.
Inhibitors of the SEC suppress HSV IE gene expression. (A) KL-1 and KL-2 interfere with the interaction of CCNT1/P-TEFb with SEC scaffold subunit AFF4. (B to D) HFF cells were treated with the indicated concentrations of KL-1 (B) or KL-2 (C) and infected with HSV (MOI = 1) for 2.5 h. (B and C) mRNA levels of viral IE genes (ICP4, ICP22, ICP27, and ICP0) and a control cellular gene (HPRT) relative to those in vehicle-treated cells. Sample mRNA levels were normalized based on the levels of cellular GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNAs. Data are means ± SEM of results from at least two experiments. (D) (Left) Western blot of viral IE (ICP4 and ICP27) and cellular control (GAPDH) protein levels. (Right) Quantitation of protein levels in cells treated with KL-1 or KL-2 relative to those in cells treated with vehicle. Data are means ± SEM of results from four replicates (analysis of variance [ANOVA] with Dunnett’s post hoc test). (E) HFF cells were treated with vehicle or 8 μM KL-1 or KL-2 and infected with HSV (MOI = 1) for 2 h. Nuclear HSV DNA levels in cells treated with KL-1 or KL-2 relative to those in cells treated with vehicle are indicated. ns, not significant (ANOVA with Dunnett’s post hoc test).
IC50 values of SEC inhibitors. The IC50 values represent results of KL-1 and KL-2 inhibition of HSV IE gene expression (refer to Fig. 1B and C). Download Table S1, PDF file, 0.03 MB (33KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Neither compound affected the transport of the viral genome to the infected cell nucleus, as the levels of nuclear viral DNA in cells treated with KL-1 or KL-2 were not significantly different from those seen in cells treated with vehicle (Fig. 1E). Importantly, treatment with KL-1 or KL-2 also did not result in apparent cell toxicity (Fig. S1). Thus, disruption of the active SEC results in potent suppression of viral IE gene transcription.
MTT cytotoxicity assays. (A and B) HFF cells were treated with the indicated concentrations of saponin (cytotoxic control), KL-1, or KL-2 for 6 h. Cell viability was measured using MTT assays (Bioassay Systems). Data are means ± SEM of results from 4 replicates at each concentration. Download FIG S1, PDF file, 0.05 MB (50.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
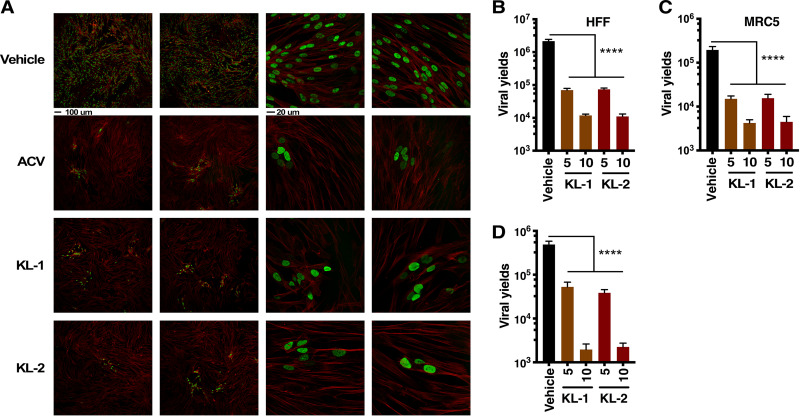
To determine if these compounds would effectively block the spread of HSV infection, HFF and MRC5 cells were infected at a low multiplicity of infection (MOI) for 8 h in the absence of drug to allow one round of replication. Vehicle, KL-1, KL-2, or the DNA replication inhibitor ACV was then added for an additional 16 h. The cultures were stained for the lytic replication protein UL29 or harvested for viral yields. As shown in Fig. 2A, KL-1 and KL-2 both suppressed the spread of infection in the culture in a manner analogous to that seen with the control replication inhibitor ACV. Consistent with this, both inhibitors reduced viral yields 1 to 2 logs in the spread assay (Fig. 2B and C) and in a one-step growth assay (Fig. 2D).
FIG 2.
Inhibitors of the SEC suppress the spread of HSV lytic infection. (A to C) HFF and MRC5 cells were infected with HSV (MOI = 0.025) for 8 h and then treated with vehicle, acyclovir (ACV), or 5 or 10 μM KL-1 or KL-2 for 16 h. (A) HFF cells were stained with anti-UL29 (green) and phalloidin-647 (F-actin, red). Scale bars, 100 μm (left 2 columns) and 20 μm (right two columns). (B and C) Viral yields from HFF-infected cells (B) and MRC5-infected cells (C). Data are means ± SEM of results from 4 replicates (ANOVA with Dunnett’s post hoc test). (D) Viral yields from HFF cells treated with vehicle or 5 or 10 μM KL-1 or KL-2 and infected with HSV (MOI = 3) for 12 h. Data are means ± SEM of results from 8 replicates (ANOVA with Dunnett’s post hoc test).
KL-1 and KL-2 SEC inhibitors reduce the number of transcriptionally active viral genomes.
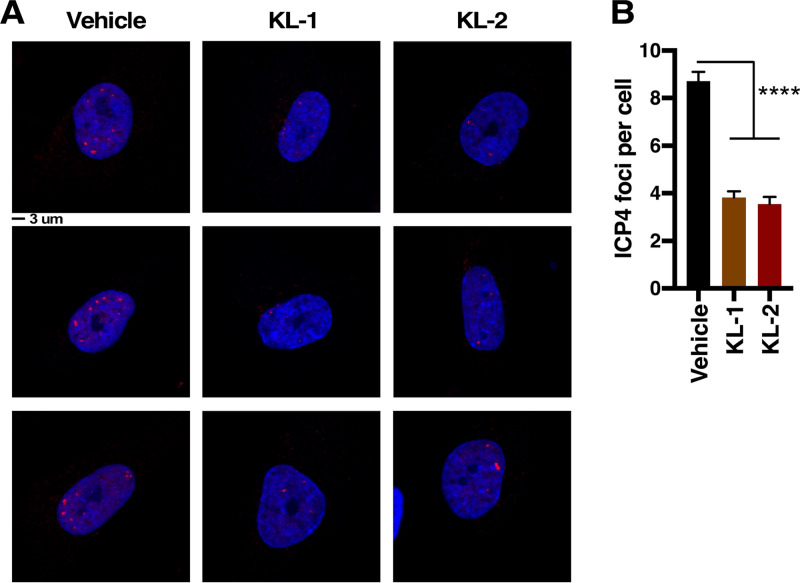
Transcriptionally active viral genomes can be detected by the association with the major viral transcriptional activator (ICP4) early in infection. To assess the impact of the SEC inhibitors on the number of transcriptionally active viral foci, HFF cells were pretreated with vehicle, KL-1, or KL-2 and infected with HSV for 2 h. Cells were stained and the number of ICP4 foci counted. As shown in Fig. 3, both KL-1 and KL-2 significantly reduced the number of transcriptionally active viral foci, a result consistent with early suppression of viral IE gene expression.
FIG 3.
Inhibitors of the SEC reduce the number of transcriptionally active viral genomes. (A and B) HFF cells were treated with vehicle or with KL-1 (10 μM) or KL-2 (10 μM) and infected with HSV (MOI = 5) for 2 h. (A) Cells were stained with anti-ICP4 (viral transcriptional foci; red) and 4′,6-diamidino-2-phenylindole (DAPI; blue). Scale bar = 3 μm. Three representative images are shown for each condition. (B) The number of transcriptionally active foci per cell. Data are means ± SEM (n > 136 cells per group) (ANOVA with Dunnett’s post hoc test).
Inhibition of the SEC reduces the levels of SEC occupancy of viral IE gene promoters.
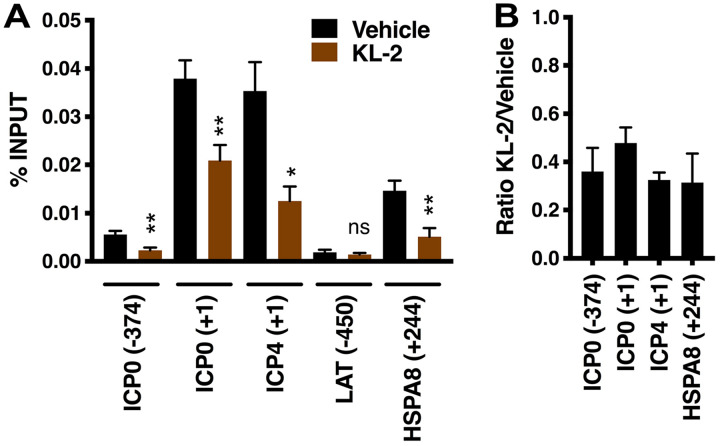
Recruitment of the SEC to viral IE genes early in infection can be detected by association of the SEC scaffold protein AFF4 with viral IE promoters. Given the impact of SEC inhibitors on IE gene expression, the levels of AFF4 occupancy of viral IE promoters were assessed in cells treated with vehicle or KL-2. As shown in Fig. 4, chromatin immunoprecipitation (ChIP) assays illustrate that, in cells treated with vehicle, the SEC was associated primarily with the promoter-proximal regions of the viral IE genes (ICP0 and ICP4) and the positive-control HSPA8 cellular gene whereas little SEC was detected at the ICP0 enhancer region (ICP0 -374) or the viral negative-control promoter (LAT). In cells treated with KL-2, promoter occupancy was significantly reduced at both viral IE and the SEC-responsive HPSA8 promoter-proximal regions. As treatment of cells with KL-1 or KL-2 had previously been shown to result in disruption in the active SEC complex and ultimately in reduced cellular levels of AFF4 (47), this likely accounts for the decreased SEC occupancy of IE promoters. Similar impacts of KL-2 were seen in ChIP assays monitoring the occupancy of a second component of the SEC (AF9/MLLT3; Fig. S2). Together, the data clearly implicate the SEC in mediating transcription of HSV IE genes during the initiation of lytic infection.
FIG 4.
KL-2 reduces the recruitment of the SEC to viral IE promoters. (A and B) HFF cells were treated with vehicle or 8 μM KL-2 and infected with HSV (MOI = 2) for 2 h. (A) ChIP assay data showing the levels of the SEC (AFF4) associated with viral IE (ICP0 and ICP4), control viral (LAT), or control cellular (HSPA8) genes in cells treated with vehicle or KL-2. Data are means ± SEM of results from 4 experiments (paired two-tailed t tests). (B) Ratio of AFF4 occupancy levels in KL-2-treated versus vehicle-treated cells.
KL-2 reduces the levels of SEC occupancy at viral IE genes. (A and B) HFF cells were treated with vehicle or 8 μM KL-2 and infected with HSV (MOI = 2) for 2 h. (A) ChIP assays showing the levels of the SEC subunit AF9 associated with viral IE (ICP0 and ICP4), SEC-responsive cellular positive-control (HSPA8), and cellular negative-control (ZNF554) genes in cells treated with vehicle or KL-2. Data are means ± SEM of results from 2 experiments. (B) Ratios of AF9 occupancy levels in KL-2-treated versus vehicle-treated cells. Download FIG S2, PDF file, 0.5 MB (1MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Suppression of HSV reactivation from latency by inhibition of the SEC.
In addition to control of lytic infection, regulation of transcriptional elongation by the SEC is proposed to also impact the initiation of viral reactivation from latency in sensory neurons. This is based on the induction of viral reactivation by bromodomain and extraterminal (BET) inhibitors that inhibit the competitive adaptor BRD4 and enhance the levels of active SEC-P-TEFb (15). These data led to a model in which a population of poised genomes respond rapidly to signal-mediated induction of active SEC-P-TEFb to promote IE expression and the initiation of reactivation.
In the mouse ganglia explant system, reactivation is stimulated by explant of latently infected mouse trigeminal ganglia into culture. This allows for the inclusion of compounds that target specific components to probe for mechanisms involved in reactivation. Therefore, to directly probe the significance of the SEC in viral reactivation, latently infected trigeminal ganglia were bisected followed by explant of one half of the ganglia in the presence of control vehicle and the other half in the presence of the replication inhibitor ACV or KL-2 for 48 h. JQ1 was included as a control compound that stimulates reactivation via inhibiting the SEC competitor BRD4 and increasing the levels of active P-TEFb (15, 26, 50). Post-explant, viral yields were determined as a measure of reactivation. As shown in Fig. 5A, JQ1 induced reactivation whereas ACV suppressed viral yields as anticipated. Strikingly, KL-2 significantly suppressed reactivation, indicating that the SEC is required for efficient reactivation.
FIG 5.
KL-2 suppresses the initiation of viral reactivation from latency. (A) Viral yields from latently infected trigeminal ganglia explanted in the presence of vehicle, JQ1 (1 μM), KL-2 (10 μM), or ACV (100 μM) for 48 h. Data are from individual ganglia (n ≥ 9; Wilcoxon matched-pairs 2-tailed signed rank test). (B) mRNA levels of viral (ICP27, UL30, and gC) genes and cellular control mHprt genes in latently infected trigeminal ganglia explanted in the presence of vehicle, JQ1 (1 μM), JQ1/iCDK9 (1 μM/150 nM), or JQ1/KL-2 (1 μM/5 μM) for 6 h. (C) mRNA levels of viral (ICP27) and cellular control (mHPRT) genes in latently infected trigeminal ganglia explanted in the presence of vehicle, iCDK9 (150 nM), or KL-2 (5 μM) for 12 h. (B and C) Sample mRNA levels were normalized based on the levels of cellular murine GAPDH (mGapdh) mRNA. Data are means ± SEM of results from 2 experiments (n ≥ 2 pools of five ganglia per group.
To determine if inhibition of the SEC suppressed the initiation of reactivation via inhibition of expression of viral IE genes, latently infected ganglia were explanted in the presence of vehicle, JQ1, or JQ1 in combination with a CDK9/P-TEFb inhibitor (iCDK9 [51]) or the SEC inhibitor KL-2. The levels of viral lytic mRNAs were determined at 6 h post-explant. At this early time point, JQ1 was included to enhance the induction/frequency of reactivation (15). As shown in Fig. 5B, JQ1 effectively induced viral reactivation as evidenced by significant levels of a representative viral IE (ICP27) mRNA and low levels of viral E (UL30) and L (gC) mRNAs. Importantly, both iCDK9 and KL-2 reduced the ability of JQ1 to stimulate viral gene expression.
To determine the impact of KL-2 on viral reactivation in the absence of JQ1 stimulation, latently infected ganglia were explanted in the presence of vehicle, iCDK9, or KL-2 for 12 h prior to determining the levels of viral lytic mRNAs. At this point, in the absence of additional stimulus, only the viral IE gene mRNA was reliably detected and both iCDK9 and KL-2 significantly reduced the mRNA levels (Fig. 5C). No significant change in mRNA levels of control murine HPRT (mHprt) gene was seen at either 6 or 12 h post-explant under any conditions (Fig. 5B and C). Thus, inhibition of the SEC suppresses the initiation of viral reactivation specifically by suppression of viral IE gene expression.
The SEC is recruited to viral IE promoters upon induction of viral reactivation.
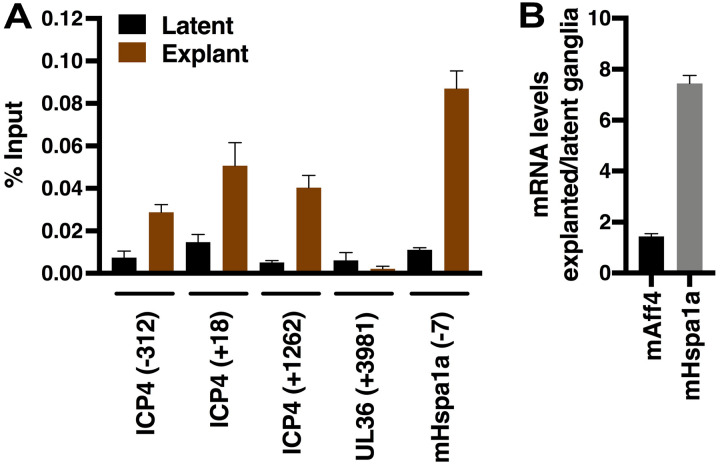
During lytic infection, the SEC is recruited to viral IE gene promoters to stimulate IE transcription (Fig. 4). KL-2-mediated inhibition of IE gene expression during viral reactivation suggested that the SEC may also directly promote IE expression in this context. Therefore, latently infected trigeminal ganglia were harvested immediately (time zero; latent) or were explanted for 6 h and chromatin immunoprecipitations were done to assess SEC occupancy at viral IE and control regions. As shown in Fig. 6A, no significant levels of the SEC were associated with IE genes of the latent viral genome. However, upon explant induction, the SEC was recruited to a representative IE promoter (ICP4). Recruitment was specific, as the complex was not detected at a control region of the viral genome (late gene UL36 coding). Explant also resulted in SEC occupancy of the positive-control mHspa1a gene promoter and correlated with induced levels of mHspa1a mRNA.
FIG 6.
SEC recruitment to viral IE genes during initiation of viral reactivation from latency. (A) Data from ChIP assays showing the levels of the SEC (mAff4) associated with viral (ICP4 and UL36) and cellular (mHspa1a) genes in latently infected trigeminal ganglia at 0 h post-explant (latent) and 6 h post-explant (reactivation). Data are means ± SEM of results from 2 experiments; each immunoprecipitation was performed with a pool of 10 ganglia. (B) mRNA levels of mAff4 and mHspa1a in explanted ganglia relative to latently infected ganglia. mRNA levels were normalized based on the levels of cellular mGapdh mRNA. Data represent means of results from 2 pools of 4 ganglia each.
Levels of the SEC scaffold AFF4 protein are regulated in HSV-infected cells.
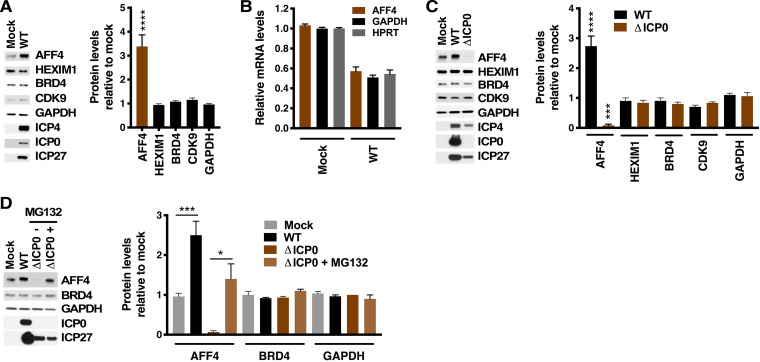
Overall, the data presented here highlight the significance of AFF4/SEC as a regulatory complex that is important for the expression of HSV IE genes during lytic infection and reactivation from latency. Strikingly, in HFF cells infected with HSV, the levels of AFF4 protein were increased (Fig. 7A) in contrast to the levels of other related proteins (HEXIM1, BRD4, and CDK9). This increase in AFF4 protein levels was enhanced with increasing MOI (Fig. S3A) and was seen in several cell types (HFF, MRC5, and Vero; Fig. S3C). The increased levels of AFF4 protein were not due to enhanced transcription of the AFF4 gene as the relative mRNA levels did not change upon infection (Fig. 7B; see also Fig. S3B).
FIG 7.
HSV infection increases the levels of AFF4 protein in an ICP0-dependent manner. (A) (Left) Western blot of AFF4 and control cellular proteins (HEXIM1, BRD4, CDK9, and GAPDH) and viral IE proteins (ICP4, ICP0, and ICP27) in extracts from mock-infected HFF cells or cells infected with wild-type HSV (MOI = 5) for 4 h. (Right) Quantitation of protein levels in HSV-infected relative to mock-infected cells. Data are means ± SEM of results from 4 experiments (unpaired two-tailed t tests). (B) mRNA levels of AFF4 and cellular control genes (GAPDH and HPRT) in cells infected with HSV (MOI = 5) relative to those in mock-infected cells. Data are means ± SEM of results from at least 2 experiments and 4 to 10 replicates (ANOVA with Dunnett’s post hoc test). (C) (Left) Western blot of AFF4 and control cellular proteins (HEXIM1, BRD4, CDK9, and GAPDH) and viral IE proteins (ICP4, ICP0, and ICP27) in extracts from mock-infected cells or cells infected with wild-type (WT) HSV or ΔICP0 HSV (MOI = 5) for 4 h. (Right) Quantitation of protein levels in HSV-infected relative to mock-infected cells. Data are means ± SEM of results from 2 experiments and 3 to 6 replicates (ANOVA with Dunnett’s post hoc test). (D) (Left) Western blot of AFF4 and control cellular proteins (BRD4 and GAPDH) and viral IE proteins (ICP0 and ICP27) in extracts from mock-infected cells or cells infected with wild-type (WT) HSV or ΔICP0 HSV (MOI = 5) for 4 h in the absence or presence of 10 μM MG132. (Right) Quantitation of protein levels in HSV-infected relative to mock-infected cells. Data are means ± SEM of results from 3 replicates (unpaired two-tailed t tests).
Increased AFF4 protein levels in HSV-infected cells. (A) Western blot of AFF4, viral IE ICP4, and control cellular GAPDH (glyceraldehyde-3-phosphate dehydrogenase) in HFF cells that were mock infected or infected with HSV at the indicated MOI. The graph represents the quantitation of protein levels in infected cells relative to mock-infected cells. Data are means ± SEM of results from 2 experiments (analysis of variance [ANOVA] with Dunnett’s post hoc test). (B) mRNA levels of AFF4 and control cellular genes (GAPDH and HPRT) in cells infected with HSV at the indicated MOI relative to levels in mock-infected cells. Data are means ± SEM of results from 3 replicates. (C) Western blot of AFF4 and control GAPDH in HFF, MRC5, and Vero cells that were mock infected or infected with HSV (MOI = 5). The graph represents the quantitation of protein levels in infected cells relative to mock-infected cells. Data are means ± SEM of results from 3 replicates (paired two-tailed t tests). Download FIG S3, PDF file, 0.1 MB (121.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
In cells, the levels of active SEC are modulated by targeted degradation of the AFF4 scaffold and ELL1/2 subunits by the E3 ubiquitin ligase SIAH1 (52–55). Consistent with this, depletion of SIAH1 in MRC5 cells resulted in strongly (∼5-fold) enhanced levels of AFF4 protein and a concomitant increase in transcription of HSV IE (ICP4 and ICP27) genes (Fig. S4A-B). Similar increases in AFF4 and HSV IE (ICP4) protein levels were also seen in HFF cells depleted for SIAH1 (Fig. S4C).
Depletion of SIAH1 enhances the levels of AFF4 and HSV IE proteins. (A and B) MRC5 cells were transfected with control siRNA or SIAH1 siRNAs. Cells were infected with HSV (MOI = 3) for 4 h. (A) Western blot of AFF4 and control cellular proteins (BRD4 and GAPDH) and viral IE proteins (ICP4). (B) Quantitation of protein levels and mRNA levels relative to those in cells transfected with control siRNA. Data are means ± SEM of results from 2 experiments. (C) HFF cells were transfected with control siRNA or SIAH1 siRNAs and infected with HSV (MOI = 3) for 4 h. Western blotting of AFF4 and control cellular proteins (BRD4 and GAPDH) and viral IE proteins (ICP4). Data representing quantitation of protein levels are relative to those in cells transfected with control siRNA. Data are means ± SEM of results from 2 experiments. Download FIG S4, PDF file, 0.08 MB (82.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Interestingly, previous studies identified SIAH1 as an interaction partner for the HSV IE protein ICP0 and demonstrated that a recombinant virus with a mutant ICP0 that did not interact with SIAH1 was defective in growth in vitro and in vivo (56, 57). Therefore, to determine if ICP0 was involved in the regulation of AFF4 protein levels upon infection, cells were infected with wild-type (WT) HSV or a recombinant lacking ICP0 (ΔICP0 [58, 59]). In contrast to the stabilization of AFF4 seen upon infection with WT virus, infection with the ΔICP0 virus resulted in reduced levels of AFF4 relative to the levels in mock-infected cells (Fig. 7C). The reduction in AFF4 protein levels in ΔICP0-infected cells could be reversed, at least partially, by treatment with the proteasome inhibitor MG132, suggesting that turnover of AFF4 was enhanced in the absence of ICP0 in HSV-infected cells (Fig. 7D). Thus, HSV infection resulted in increased levels of the SEC scaffold AFF4 protein in an ICP0-dependent manner.
DISCUSSION
Expression of HSV IE genes is a critical stage that determines the progression of lytic infection and the initiation of reactivation from latency in sensory neurons. Regulation of IE expression has been extensively studied, but the mechanisms involved in induction of IE transcription early in infection and, more importantly, during the initiation of reactivation from latency remain to be fully described.
IE transcription is mediated by cellular RNAPII and is controlled primarily via a cellular transcriptional coactivator, HCF-1, that mediates activation of transcription factors assembled on the viral IE enhancer/promoter regions. One of key functions of this protein is to prevent the accumulation of repressive histone modifications (H3K9me3) and to promote the installation of activating marks (H3K4me3) on nucleosomes associated with viral IE genes. This is accomplished via recruitment of an HCF-1 complex that contains histone H3K9 demethylases (LSD1 and JMJD2s) and H3K4 methyltransferases (SETD1A/MLL1). Ultimately, the transition from repressed to active chromatin promotes the initiation of IE transcription. Importantly, HCF-1 and its associated chromatin modulation components also appear to be critical components of the viral latency-reactivation cycle. HCF-1 is rapidly transported to the nucleus upon induction of reactivation, and inhibition of the HCF-1-associated histone demethylases blocks the initiation of viral reactivation (Fig. 8).
FIG 8.
Model of epigenetic modulation and SEC-mediated induction of viral IE gene expression during initiation of HSV reactivation. Stress signaling promotes viral reactivation at multiple rate-limiting stages. Signaling induces the transport of HCF-1 to the nucleus of latently infected neurons where HCF-1 complexes containing histone H3K9 demethylases (LSD1 and JMJD2s) and histone H3K4 methyltransferases (SETD1A and MLLs) are involved in promoting the transition of heterochromatic viral genomes to accessible euchromatic genomes. This transition can be blocked by inhibitors (LSDi and ML324) of these HCF-1-associated H3K9 demethylases, resulting in suppression of viral reactivation. Signaling also results in the release of P-TEFb from the 7SK-snRNP complexes, which increases the levels of active SEC-P-TEFb. Along with HCF-1, recruitment of SEC-P-TEFb to viral IE gene promoters stimulates productive expression of IE genes and reactivation of a population of accessible genomes. Compounds such as BET inhibitors (JQ1) enhance the levels of active P-TEFb and induce reactivation, while SEC inhibitors (KL-1/2) disrupt the active SEC and suppress reactivation. Following induction, the accumulation of the viral IE ICP0 protein may further stabilize the SEC, enhance active SEC-P-TEFb levels, and contribute to reactivation. Note that while part of this model reflects the described roles of HCF-1 and its associated chromatin modulation factors in viral reactivation, the epigenetic regulation of latent viral genomes is complex. Other repressive histone marks (e.g., H3K27me3 [72, 77–81]) are also associated with latent viral genomes, and it is likely that additional pathways and components contribute to the epigenetic regulation of latency and reactivation.
For many genes, following RNAPII initiation, transcriptional elongation becomes a critical rate-limiting stage. Regulated RNAPII pausing may poise genes for rapid expression in response to signaling, allow for coordinated chromatin modulation and assembly of RNA processing complexes, and/or prevent genes from being inactivated during periods of low-level transcription. One important step in mediating pause release and efficient elongation is the recruitment of an active P-TEFb kinase complex which phosphorylates RNAPII and the pausing factors NELF and DSIF. This complex is itself regulated at several levels. It is sequestered in an inactive state by association with the 7SK-snRNP. Upon stress signaling, P-TEFb is released and the active enzyme associates with the specificity adaptors BRD4 or the SEC.
A number of viruses that utilize host cell RNAPII transcriptional machinery also exhibit regulated pausing and elongation. For HIV, this is a well-characterized mechanism that appears to play an essential role in the control of viral latency/reactivation. During latency, low-level transcription is mediated by BRD4-P-TEFb whereas reactivation is induced upon TAT-dependent recruitment and stabilization of the SEC that promotes enhanced transcription of the viral genome (60, 61).
For HSV, IE genes also exhibit characteristics of genes that are highly dependent upon regulated transcriptional elongation. RNAPII and associated pausing factors (NELF) are detected at the promoter-proximal regions of IE genes during the initiation of lytic infection, and, as in the case of HIV, the SEC is required for efficient pause-release and transcriptional elongation. Consistent with these results, PRO-Seq analyses defined distinct RNAPII pause sites in IE genes during infection (62). Interestingly, in addition to functions in the modulation of chromatin and initiation of IE gene transcription, the coactivator HCF-1 also associates with multiple transcriptional elongation components, including PAF1, SETD2, FACT, and the SEC-P-TEFb, suggesting that HCF-1 may coordinate multiple stages of IE transcription.
In addition to the control of lytic infection, regulation of elongation of HSV IE transcription may also be a critical rate-limiting step for viral reactivation from latency as compounds (e.g., BET inhibitors) that enhance the levels of active P-TEFb also potently induce viral reactivation in sensory ganglia (15, 26, 27, 63). These data are consistent with a model where induction of active P-TEFb promotes IE expression and initiation of reactivation in a population of latent viral genomes (Fig. 8).
Recently, inhibitors of the SEC (KL-1, KL-2) were identified in a screen for compounds that disrupt the interaction of the SEC scaffold AFF4 with the cyclin T1 component of the p-TEFb kinase subcomplex. Disruption results in enhanced turnover of AFF4 and in reduced levels of active SEC, ultimately impacting the rate of transcriptional elongation of the population of genes that normally exhibit SEC occupancy (47). These compounds also suppressed Tat-mediated induction of HIV transcription in a reporter-based assay (47).
Given the indication that the SEC-P-TEFb plays a critical role in driving expression of HSV IE genes, the impact of inhibition of the SEC was investigated during lytic infection and reactivation from latency. These SEC inhibitors potently suppressed HSV IE gene expression and spread of a primary infection and reduced SEC occupancy of viral IE gene promoters during the initiation of viral lytic infection. However, as the SEC is required for IE expression and these IE gene products are required for expression of subsequent E/L genes, it remains unknown if the SEC is also involved in regulation of later classes of HSV genes or if alternative components or mechanisms are involved. The viral IE protein ICP22 has been shown to interact with and modulate P-TEFb/CDK9 activity as well as promote the recruitment of elongation factors FACT, SPT5, and SPT6 to the viral genome (64–67). Moreover, ICP22 is required for efficient transcriptional elongation of some HSV genes later in infection (65). With respect to the alternative P-TEFb adaptor BRD4, while this protein does not play a significant role in mediating IE expression, depletion of this adaptor does reduce the accumulation of viral proteins and viral yields late in infection (68). Nonetheless, the potential involvement of the SEC in regulation of multiple gene classes could account for the potent suppression of viral spread by the SEC inhibitors.
Importantly, inhibitors of specific components that may regulate viral IE gene expression allow probing of mechanisms that control the induction of IE expression during the initiation of viral reactivation. Here, inhibition of the SEC suppressed reactivation of HSV in explants of latently infected sensory ganglia as measured by viral yields and the reduction in viral IE gene expression at an early point in reactivation. Most significantly, the SEC was detected at viral IE genes during the early stage of reactivation and correlated with expression of viral IE mRNAs. Together, the data strongly support a model that, as in the case of HIV, the initiation of HSV lytic infection and reactivation from latency are critically regulated by SEC-dependent transcriptional elongation.
The levels of active SEC-P-TEFb are regulated by multiple mechanisms. In addition to signal-mediated modulation of P-TEFb availability, the levels of the SEC scaffold AFF4 and the ELL2 subunit proteins are regulated by the ubiquitin ligase SIAH1, which induces turnover of these critical SEC components. More recently, SIAH1 itself was shown to be targeted for degradation by poly(ADP-ribose) polymerase 1 (PARP1)-mediated PARylation-dependent ubiquitination, resulting in enhanced levels of ELL2 and induction of HIV transcription (55).
Strikingly, levels of AFF4 protein were induced in cells infected with HSV, suggesting that a virus-mediated mechanism might enhance SEC levels for HSV gene expression. Interestingly, reports from the Hauber laboratory have shown that the ICP0 viral IE protein interacts with SIAH1 through SIAH1-binding consensus motifs and that viruses with mutations in these motifs are defective for growth in culture and in vivo (56, 57). The authors propose several models, including one in which ICP0 functions to sequester SIAH1, thus blocking SIAH1’s interactions with its targets. Consistent with this, infection with an ICP0 null virus resulted in the loss of AFF4 induction that was partially recovered by proteasomal inhibition (Fig. 7C and D). These results suggest that ICP0 may modulate SIAH1 activity and thus ultimately regulate the stability of AFF4. This may be a particularly important function of ICP0 that would serve to amplify SEC-P-TEFb activity for robust induction of HSV IE gene transcription. In addition, as ICP0 can also promote reactivation from latency (69–71), it is possible that ICP0-dependent modulation of SEC levels is also important in this context (Fig. 8).
Finally, it is important that targeting of cellular components that critically modulate viral infection and/or reactivation can lead to novel approaches to control viral infection, especially in cases where resistance has evolved to the current antiherpesvirus pharmaceuticals that target the viral replication machinery. For example, studies have demonstrated that (i) inhibition of cellular histone demethylases (LSD1/KDM1A, JMJD2/KDM4, UTX/JMJD6A) prevented the removal of repressive (H3K9me3, H3K27me3) chromatin associated with the viral genome, resulting in a block to viral infection and reactivation from latency (6, 7, 13, 14, 72); (ii) inhibition of Jun N-terminal protein kinase (JNK) blocked stress-mediated phosphorylation of histones associated with the viral genome and suppressed viral reactivation from latency (73); (iii) inhibition of AKT phosphorylation resulted in suppression of viral protein synthesis and reduced lytic ocular infection in vivo (74); and (iv) inhibition of the proteasome suppressed infection via a block in the transport of the viral capsid to the nucleus (75). In addition to targeting components required for viral infection, compounds that inhibit the repressive histone methyltransferase EZH1/2 enhanced the host antiviral responses, resulting in suppression of infection in vitro and in vivo (76). Given that inhibition of the SEC robustly suppressed HSV lytic infection and reactivation from latency, SEC inhibitors clearly exhibit antiviral potential.
MATERIALS AND METHODS
Cell culture, viral infections, and drug treatments.
Telomerase-immortalized HFF (tert-human foreskin fibroblast), MRC5, and Vero cell stocks were grown and maintained according to standard procedures. Viral infections were done by incubating cells with HSV-1 (strains F, 17, and 17-ΔICP0) at the indicated multiplicity of infection (MOI) in Dulbecco’s modified Eagle medium (DMEM) containing 1% fetal bovine serum (FBS) for 1 h at 37°C. After virus adsorption, the inoculum was removed and fresh medium containing 1% FBS was added for 3 to 20 h. For drug treatments, the indicated concentrations of control vehicle (dimethyl sulfoxide [DMSO]) or SEC inhibitors (KL-1 and KL-2) were added 2.5 h before viral infection. These inhibitors were removed during virus adsorption and were added back postadsorption for the indicated time. For MG132, the inhibitor was added after virus adsorption. The cell lines, viruses, and inhibitors/compounds used are listed in Table S2. Cytotoxicity of KL-1 and KL-2 was measured using 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assays (Bioassay Systems) according to the manufacturer’s recommendations.
Reagents. Download Table S2, PDF file, 0.05 MB (48.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Viral yields.
For viral spread assays, HFF or MRC5 cells were infected at a low MOI (0.025) and the indicated concentrations of inhibitors were added at 8 h postinfection (hpi). Supernatants were collected at 16 h after addition of drugs. For one-step growth assays, HFF cells were infected at an MOI of 3 for 1 h, washed extensively with phosphate-buffered saline (PBS), and overlaid with fresh media containing inhibitors. Supernatants were collected at 12 hpi. Viral titers were determined by plaque assays.
Western blotting.
HFF cells were mock infected or infected with HSV-1 at the indicated MOI for 3 to 4 h. Cells were lysed, and protein extracts were produced using modified radioimmunoprecipitation assay (RIPA) lysis buffer (150 mM NaCl, 0.5 mM EDTA, 50 mM Tris-HCl [pH 8.0], 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 100 U/ml Benzonase, cOmplete protease inhibitor, and phosphatase inhibitors). Western blotting was performed using the indicated antibodies (Table S2), and bands were quantitated using GeneTools (Syngene G:Box) or Image J.
Immunofluorescence microscopy.
HFF cells were grown on coverslips and subjected to mock infection or HSV-1 infection at a low MOI (MOI = 0.025, spread assays) for 24 h or at a high MOI (MOI = 5, viral transcriptional focus assays) for 2 h. After infection, cells were fixed in 4% paraformaldehyde–PBS, permeabilized in 0.1% Triton X-100, and stained with the indicated primary and fluorescent secondary antibodies using standard procedures. Coverslips were mounted in Fluoromount-G containing 4′,6-diamidino-2-phenylindole (DAPI) and visualized using a Leica SP8 confocal microscope with LASAF software. Images were assembled from sequential Z-sections using Imaris software (version 8.2.0; Bitplane).
HSV-1 reactivation in explanted trigeminal ganglia.
BALB/c mice were infected with 1 × 106 PFU HSV-1 (strain F) via the ocular route. Trigeminal ganglia from latently infected mice (45 to 60 days postinfection) were bisected, and the paired halves were explanted into media containing control vehicle (DMSO) or inhibitor. Viral yields were quantified by determining the titers of ganglion homogenates at 48 h post-explant. mRNA levels of viral and control cellular genes were determined following explantation of latently infected ganglia in the presence of vehicle or an inhibitor(s) for 6 or 12 h. Primer sequences are in Table S3. All animal care and handling were done in accordance with the U.S. National Institutes of Health Animal Care and Use Guidelines and as approved by the NIAID Animal Care and Use Committee.
RNA isolation, cDNA synthesis, and quantification.
Quantitation of RNA levels in tissue culture cells and trigeminal ganglia was done as detailed in Text S1.
Supplemental materials and methods. Download Text S1, DOCX file, 0.02 MB (16.4KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Chromatin immunoprecipitations.
ChIP assays of cultured cells and of trigeminal ganglia were done as detailed in Text S1. Primer sequences are in Table S3.
siRNA depletions.
MRC5 or HFF cells (1 × 106) were transfected with 30 nM siSIAH1 small interfering RNA (siRNA) or siControl pools using TransIT-X2 reagent (Mirus) per the manufacturer’s recommendations. The siRNAs used are listed in Table S2.
Statistical analyses.
Results are presented as means ± standard errors of the means (SEM). Parameters are stated in the figure legends and detailed in Table S4. Analyses were done using GraphPad Prism 8. Where indicated, asterisks denote statistical significance as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Primers. Download Table S3, PDF file, 0.04 MB (43.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Statistics. Download Table S4, PDF file, 0.05 MB (52.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
ACKNOWLEDGMENTS
We thank the NIAID Bld33 Vivarium staff for excellent technical support. We thank T. Shenk for TERT-HFF cells, B. Roizman for HSV (strain F), and C. Boutell for HSV (strains 17 and 17-ΔICP0).
This work was supported with funds from the NIAID Division of Intramural Research (T.M.K.).
Footnotes
This article is a direct contribution from Thomas M. Kristie, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Richard Longnecker, Northwestern University Feinberg School of Medicine, and David C. Bloom, University of Florida College of Medicine.
Citation Alfonso-Dunn R, Arbuckle JH, Vogel JL, Kristie TM. 2020. Inhibition of the super elongation complex suppresses herpes simplex virus immediate early gene expression, lytic infection, and reactivation from latency. mBio 11:e01216-20. https://doi.org/10.1128/mBio.01216-20.
REFERENCES
- 1.Roizman B, Knipe DM, Whitley RJ. 2013. Herpes simplex viruses, p 2501–2601. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Whitley R, Kimberlin DW, Prober CG. 2007. Pathogenesis, clinical disease, host response, and epidemiology: alphaherpes viruses: pathogenesis and disease, p 589–601. In Arvin A, Whitley R (ed), Human herpesviruses biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 3.Vogel JL, Kristie TM. 2013. The dynamics of HCF-1 modulation of herpes simplex virus chromatin during initiation of infection. Viruses 5:1272–1291. doi: 10.3390/v5051272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narayanan A, Nogueira ML, Ruyechan WT, Kristie TM. 2005. Combinatorial transcription of herpes simplex virus and varicella zoster virus immediate early genes is strictly determined by the cellular coactivator HCF-1. J Biol Chem 280:1369–1375. doi: 10.1074/jbc.M410178200. [DOI] [PubMed] [Google Scholar]
- 5.Kristie TM. 2015. Dynamic modulation of HSV chromatin drives initiation of infection and provides targets for epigenetic therapies. Virology 479–480:555–561. doi: 10.1016/j.virol.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang Y, Vogel JL, Arbuckle JH, Rai G, Jadhav A, Simeonov A, Maloney DJ, Kristie TM. 2013. Targeting the JMJD2 histone demethylases to epigenetically control herpesvirus infection and reactivation from latency. Sci Transl Med 5:167ra5. doi: 10.1126/scitranslmed.3005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. 2009. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat Med 15:1312–1317. doi: 10.1038/nm.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narayanan A, Ruyechan WT, Kristie TM. 2007. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc Natl Acad Sci U S A 104:10835–10840. doi: 10.1073/pnas.0704351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. 2003. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev 17:896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JY, Mandarino A, Chao MV, Mohr I, Wilson AC. 2012. Transient reversal of episome silencing precedes VP16-dependent transcription during reactivation of latent HSV-1 in neurons. PLoS Pathog 8:e1002540. doi: 10.1371/journal.ppat.1002540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kristie TM, Vogel JL, Sears AE. 1999. Nuclear localization of the C1 factor (host cell factor) in sensory neurons correlates with reactivation of herpes simplex virus from latency. Proc Natl Acad Sci U S A 96:1229–1233. doi: 10.1073/pnas.96.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitlow Z, Kristie TM. 2009. Recruitment of the transcriptional coactivator HCF-1 to viral immediate-early promoters during initiation of reactivation from latency of herpes simplex virus type 1. J Virol 83:9591–9595. doi: 10.1128/JVI.01115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill JM, Quenelle DC, Cardin RD, Vogel JL, Clement C, Bravo FJ, Foster TP, Bosch-Marce M, Raja P, Lee JS, Bernstein DI, Krause PR, Knipe DM, Kristie TM. 2014. Inhibition of LSD1 reduces herpesvirus infection, shedding, and recurrence by promoting epigenetic suppression of viral genomes. Sci Transl Med 6:265ra169. doi: 10.1126/scitranslmed.3010643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang Y, Quenelle D, Vogel JL, Mascaro C, Ortega A, Kristie TM. 2013. A novel selective LSD1/KDM1A inhibitor epigenetically blocks herpes simplex virus lytic replication and reactivation from latency. mBio 4:e00558-12. doi: 10.1128/mBio.00558-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alfonso-Dunn R, Turner AW, Jean Beltran PM, Arbuckle JH, Budayeva HG, Cristea IM, Kristie TM. 2017. Transcriptional elongation of HSV immediate early genes by the super elongation complex drives lytic infection and reactivation from latency. Cell Host Microbe 21:507–517.e5. doi: 10.1016/j.chom.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Core L, Adelman K. 2019. Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev 33:960–982. doi: 10.1101/gad.325142.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. 2008. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev 22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. 2007. RNA polymerase is poised for activation across the genome. Nat Genet 39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheidegger A, Nechaev S. 2016. RNA polymerase II pausing as a context-dependent reader of the genome. Biochem Cell Biol 94:82–92. doi: 10.1139/bcb-2015-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams LH, Fromm G, Gokey NG, Henriques T, Muse GW, Burkholder A, Fargo DC, Hu G, Adelman K. 2015. Pausing of RNA polymerase II regulates mammalian developmental potential through control of signaling networks. Mol Cell 58:311–322. doi: 10.1016/j.molcel.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo J, Price DH. 2013. RNA polymerase II transcription elongation control. Chem Rev 113:8583–8603. doi: 10.1021/cr400105n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jonkers I, Lis JT. 2015. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schier AC, Taatjes DJ. 2020. Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev 34:465–488. doi: 10.1101/gad.335679.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Q, Li T, Price DH. 2012. RNA polymerase II elongation control. Annu Rev Biochem 81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bacon CW, D'Orso I. 2019. CDK9: a signaling hub for transcriptional control. Transcription 10:57–75. doi: 10.1080/21541264.2018.1523668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartholomeeusen K, Xiang Y, Fujinaga K, Peterlin BM. 2012. Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein. J Biol Chem 287:36609–36616. doi: 10.1074/jbc.M112.410746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Contreras X, Barboric M, Lenasi T, Peterlin BM. 2007. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog 3:1459–1469. doi: 10.1371/journal.ppat.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 29.Lin C, Garrett AS, De Kumar B, Smith ER, Gogol M, Seidel C, Krumlauf R, Shilatifard A. 2011. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes Dev 25:1486–1498. doi: 10.1101/gad.2059211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu X, Zhu X, Li Y, Liu M, Yu B, Wang Y, Rao M, Yang H, Zhou K, Wang Y, Chen Y, Chen M, Zhuang S, Chen LF, Liu R, Chen R. 2016. Multiple P-TEFbs cooperatively regulate the release of promoter-proximally paused RNA polymerase II. Nucleic Acids Res 44:6853–6867. doi: 10.1093/nar/gkw571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo Z, Lin C, Shilatifard A. 2012. The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol 13:543–547. doi: 10.1038/nrm3417. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen VT, Kiss T, Michels AA, Bensaude O. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 33.Quaresma AJ, Bugai A, Barboric M. 2016. Cracking the control of RNA polymerase II elongation by 7SK snRNP and P-TEFb. Nucleic Acids Res 44:7527–7539. doi: 10.1093/nar/gkw585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 35.Yang Z, Zhu Q, Luo K, Zhou Q. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 36.Cary DC, Fujinaga K, Peterlin BM. 2016. Molecular mechanisms of HIV latency. J Clin Invest 126:448–454. doi: 10.1172/JCI80565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho WK, Jang MK, Huang K, Pise-Masison CA, Brady JN. 2010. Human T-lymphotropic virus type 1 Tax protein complexes with P-TEFb and competes for Brd4 and 7SK snRNP/HEXIM1 binding. J Virol 84:12801–12809. doi: 10.1128/JVI.00943-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Francisco JC, Dai Q, Luo Z, Wang Y, Chong RH, Tan YJ, Xie W, Lee GH, Lin C. 12 September 2017, posting date Transcriptional elongation control of hepatitis B virus covalently closed circular DNA transcription by super elongation complex and BRD4. Mol Cell Biol doi: 10.1128/MCB.00040-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu H, Li Z, Xue Y, Zhou Q. 2013. Viral-host interactions that control HIV-1 transcriptional elongation. Chem Rev 113:8567–8582. doi: 10.1021/cr400120z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mbonye U, Karn J. 2014. Transcriptional control of HIV latency: cellular signaling pathways, epigenetics, happenstance and the hope for a cure. Virology 454–455:328–339. doi: 10.1016/j.virol.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ott M, Geyer M, Zhou Q. 2011. The control of HIV transcription: keeping RNA polymerase II on track. Cell Host Microbe 10:426–435. doi: 10.1016/j.chom.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parida M, Nilson KA, Li M, Ball CB, Fuchs HA, Lawson CK, Luse DS, Meier JL, Price DH. 2019. Nucleotide resolution comparison of transcription of human cytomegalovirus and host genomes reveals universal use of RNA polymerase II elongation control driven by dissimilar core promoter elements. mBio 10:e02047-18. doi: 10.1128/mBio.02047-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perng YC, Campbell JA, Lenschow DJ, Yu D. 2014. Human cytomegalovirus pUL79 is an elongation factor of RNA polymerase II for viral gene transcription. PLoS Pathog 10:e1004350. doi: 10.1371/journal.ppat.1004350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toth Z, Brulois KF, Wong LY, Lee HR, Chung B, Jung JU. 2012. Negative elongation factor-mediated suppression of RNA polymerase II elongation of Kaposi’s sarcoma-associated herpesvirus lytic gene expression. J Virol 86:9696–9707. doi: 10.1128/JVI.01012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vijayalingam S, Chinnadurai G. 2013. Adenovirus L-E1A activates transcription through mediator complex-dependent recruitment of the super elongation complex. J Virol 87:3425–3434. doi: 10.1128/JVI.03046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zaborowska J, Isa NF, Murphy S. 2016. P-TEFb goes viral. Bioessays 38(Suppl 1):S75–S85. doi: 10.1002/bies.201670912. [DOI] [PubMed] [Google Scholar]
- 47.Liang K, Smith ER, Aoi Y, Stoltz KL, Katagi H, Woodfin AR, Rendleman EJ, Marshall SA, Murray DC, Wang L, Ozark PA, Mishra RK, Hashizume R, Schiltz GE, Shilatifard A. 2018. Targeting processive transcription elongation via SEC disruption for MYC-induced cancer therapy. Cell 175:766–779.e17. doi: 10.1016/j.cell.2018.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piret J, Boivin G. 2016. Antiviral resistance in herpes simplex virus and varicella-zoster virus infections: diagnosis and management. Curr Opin Infect Dis 29:654–662. doi: 10.1097/QCO.0000000000000288. [DOI] [PubMed] [Google Scholar]
- 49.van Velzen M, van de Vijver DA, van Loenen FB, Osterhaus AD, Remeijer L, Verjans GM. 2013. Acyclovir prophylaxis predisposes to antiviral-resistant recurrent herpetic keratitis. J Infect Dis 208:1359–1365. doi: 10.1093/infdis/jit350. [DOI] [PubMed] [Google Scholar]
- 50.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. 2010. Selective inhibition of BET bromodomains. Nature 468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu H, Xue Y, Yu GK, Arias C, Lin J, Fong S, Faure M, Weisburd B, Ji X, Mercier A, Sutton J, Luo K, Gao Z, Zhou Q. 2015. Compensatory induction of MYC expression by sustained CDK9 inhibition via a BRD4-dependent mechanism. Elife 4:e06535. doi: 10.7554/eLife.06535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Basu S, Barad M, Yadav D, Nandy A, Mukherjee B, Sarkar J, Chakrabarti P, Mukhopadhyay S, Biswas D. 2020. DBC1, p300, HDAC3, and Siah1 coordinately regulate ELL stability and function for expression of its target genes. Proc Natl Acad Sci U S A 117:6509–6520. doi: 10.1073/pnas.1912375117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Izumi K, Nakato R, Zhang Z, Edmondson AC, Noon S, Dulik MC, Rajagopalan R, Venditti CP, Gripp K, Samanich J, Zackai EH, Deardorff MA, Clark D, Allen JL, Dorsett D, Misulovin Z, Komata M, Bando M, Kaur M, Katou Y, Shirahige K, Krantz ID. 2015. Germline gain-of-function mutations in AFF4 cause a developmental syndrome functionally linking the super elongation complex and cohesin. Nat Genet 47:338–344. doi: 10.1038/ng.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu M, Hsu J, Chan C, Li Z, Zhou Q. 2012. The ubiquitin ligase Siah1 controls ELL2 stability and formation of super elongation complexes to modulate gene transcription. Mol Cell 46:325–334. doi: 10.1016/j.molcel.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu D, Liu R, Yang G, Zhou Q. 2018. The PARP1-Siah1 axis controls HIV-1 transcription and expression of Siah1 substrates. Cell Rep 23:3741–3749. doi: 10.1016/j.celrep.2018.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Czechowicz JS, Nagel CH, Voges M, Spohn M, Eibl MM, Hauber J. 2018. Interaction between the cellular E3 ubiquitin ligase SIAH-1 and the viral immediate-early protein ICP0 enables efficient replication of herpes simplex virus type 2 in vivo. PLoS One 13:e0201880. doi: 10.1371/journal.pone.0201880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagel CH, Albrecht N, Milovic-Holm K, Mariyanna L, Keyser B, Abel B, Weseloh B, Hofmann TG, Eibl MM, Hauber J. 2011. Herpes simplex virus immediate-early protein ICP0 is targeted by SIAH-1 for proteasomal degradation. J Virol 85:7644–7657. doi: 10.1128/JVI.02207-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Everett RD, Sourvinos G, Orr A. 2003. Recruitment of herpes simplex virus type 1 transcriptional regulatory protein ICP4 into foci juxtaposed to ND10 in live, infected cells. J Virol 77:3680–3689. doi: 10.1128/jvi.77.6.3680-3689.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Everett RD, Boutell C, Orr A. 2004. Phenotype of a herpes simplex virus type 1 mutant that fails to express immediate-early regulatory protein ICP0. J Virol 78:1763–1774. doi: 10.1128/jvi.78.4.1763-1774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Asamitsu K, Fujinaga K, Okamoto T. 2018. HIV Tat/P-TEFb interaction: a potential target for novel anti-HIV therapies. Molecules 23:E933. doi: 10.3390/molecules23040933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, Zhou Q. 2010. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell 38:428–438. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Birkenheuer CH, Baines JD. 14 February 2019, posting date RNA polymerase II promoter-proximal pausing and release to elongation are key steps regulating herpes simplex virus 1 transcription. J Virol doi: 10.1128/JVI.02035-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nilsson LM, Green LC, Muralidharan SV, Demir D, Welin M, Bhadury J, Logan DT, Walse B, Nilsson JA. 2016. Cancer differentiating agent hexamethylene bisacetamide inhibits BET bromodomain proteins. Cancer Res 76:2376–2383. doi: 10.1158/0008-5472.CAN-15-2721. [DOI] [PubMed] [Google Scholar]
- 64.Durand LO, Advani SJ, Poon AP, Roizman B. 2005. The carboxyl-terminal domain of RNA polymerase II is phosphorylated by a complex containing cdk9 and infected-cell protein 22 of herpes simplex virus 1. J Virol 79:6757–6762. doi: 10.1128/JVI.79.11.6757-6762.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fox HL, Dembowski JA, DeLuca NA. 2017. A herpesviral immediate early protein promotes transcription elongation of viral transcripts. mBio 8:e00745-17. doi: 10.1128/mBio.00745-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo L, Wu WJ, Liu LD, Wang LC, Zhang Y, Wu LQ, Guan Y, Li QH. 2012. Herpes simplex virus 1 ICP22 inhibits the transcription of viral gene promoters by binding to and blocking the recruitment of P-TEFb. PLoS One 7:e45749. doi: 10.1371/journal.pone.0045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zaborowska J, Baumli S, Laitem C, O'Reilly D, Thomas PH, O'Hare P, Murphy S. 2014. Herpes simplex virus 1 (HSV-1) ICP22 protein directly interacts with cyclin-dependent kinase (CDK)9 to inhibit RNA polymerase II transcription elongation. PLoS One 9:e107654. doi: 10.1371/journal.pone.0107654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ren K, Zhang W, Chen X, Ma Y, Dai Y, Fan Y, Hou Y, Tan RX, Li E. 2016. An epigenetic compound library screen identifies BET inhibitors that promote HSV-1 and -2 replication by bridging P-TEFb to viral gene promoters through BRD4. PLoS Pathog 12:e1005950. doi: 10.1371/journal.ppat.1005950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Halford WP, Kemp CD, Isler JA, Davido DJ, Schaffer PA. 2001. ICP0, ICP4, or VP16 expressed from adenovirus vectors induces reactivation of latent herpes simplex virus type 1 in primary cultures of latently infected trigeminal ganglion cells. J Virol 75:6143–6153. doi: 10.1128/JVI.75.13.6143-6153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Halford WP, Schaffer PA. 2001. ICP0 is required for efficient reactivation of herpes simplex virus type 1 from neuronal latency. J Virol 75:3240–3249. doi: 10.1128/JVI.75.7.3240-3249.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leib DA, Coen DM, Bogard CL, Hicks KA, Yager DR, Knipe DM, Tyler KL, Schaffer PA. 1989. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J Virol 63:759–768. doi: 10.1128/JVI.63.2.759-768.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Messer HG, Jacobs D, Dhummakupt A, Bloom DC. 2015. Inhibition of H3K27me3-specific histone demethylases JMJD3 and UTX blocks reactivation of herpes simplex virus 1 in trigeminal ganglion neurons. J Virol 89:3417–3420. doi: 10.1128/JVI.03052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cliffe AR, Arbuckle JH, Vogel JL, Geden MJ, Rothbart SB, Cusack CL, Strahl BD, Kristie TM, Deshmukh M. 2015. Neuronal stress pathway mediating a histone methyl/phospho switch is required for herpes simplex virus reactivation. Cell Host Microbe 18:649–658. doi: 10.1016/j.chom.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jaishankar D, Yakoub AM, Yadavalli T, Agelidis A, Thakkar N, Hadigal S, Ames J, Shukla D. 14 February 2018, posting date An off-target effect of BX795 blocks herpes simplex virus type 1 infection of the eye. Sci Transl Med doi: 10.1126/scitranslmed.aan5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schneider SM, Pritchard SM, Wudiri GA, Trammell CE, Nicola AV. 2019. Early steps in herpes simplex virus infection blocked by a proteasome inhibitor. mBio 10:e00732-19. doi: 10.1128/mBio.00732-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arbuckle JH, Gardina PJ, Gordon DN, Hickman HD, Yewdell JW, Pierson TC, Myers TG, Kristie TM. 2017. Inhibitors of the histone methyltransferases EZH2/1 induce a potent antiviral state and suppress infection by diverse viral pathogens. mBio 8:e01141-17. doi: 10.1128/mBio.01141-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bloom DC, Giordani NV, Kwiatkowski DL. 2010. Epigenetic regulation of latent HSV-1 gene expression. Biochim Biophys Acta 1799:246–256. doi: 10.1016/j.bbagrm.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cliffe AR, Coen DM, Knipe DM. 2013. Kinetics of facultative heterochromatin and polycomb group protein association with the herpes simplex viral genome during establishment of latent infection. mBio 4:e00590-12. doi: 10.1128/mBio.00590-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cliffe AR, Garber DA, Knipe DM. 2009. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J Virol 83:8182–8190. doi: 10.1128/JVI.00712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Knipe DM, Cliffe A. 2008. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol 6:211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- 81.Kwiatkowski DL, Thompson HW, Bloom DC. 2009. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J Virol 83:8173–8181. doi: 10.1128/JVI.00686-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
IC50 values of SEC inhibitors. The IC50 values represent results of KL-1 and KL-2 inhibition of HSV IE gene expression (refer to Fig. 1B and C). Download Table S1, PDF file, 0.03 MB (33KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
MTT cytotoxicity assays. (A and B) HFF cells were treated with the indicated concentrations of saponin (cytotoxic control), KL-1, or KL-2 for 6 h. Cell viability was measured using MTT assays (Bioassay Systems). Data are means ± SEM of results from 4 replicates at each concentration. Download FIG S1, PDF file, 0.05 MB (50.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
KL-2 reduces the levels of SEC occupancy at viral IE genes. (A and B) HFF cells were treated with vehicle or 8 μM KL-2 and infected with HSV (MOI = 2) for 2 h. (A) ChIP assays showing the levels of the SEC subunit AF9 associated with viral IE (ICP0 and ICP4), SEC-responsive cellular positive-control (HSPA8), and cellular negative-control (ZNF554) genes in cells treated with vehicle or KL-2. Data are means ± SEM of results from 2 experiments. (B) Ratios of AF9 occupancy levels in KL-2-treated versus vehicle-treated cells. Download FIG S2, PDF file, 0.5 MB (1MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Increased AFF4 protein levels in HSV-infected cells. (A) Western blot of AFF4, viral IE ICP4, and control cellular GAPDH (glyceraldehyde-3-phosphate dehydrogenase) in HFF cells that were mock infected or infected with HSV at the indicated MOI. The graph represents the quantitation of protein levels in infected cells relative to mock-infected cells. Data are means ± SEM of results from 2 experiments (analysis of variance [ANOVA] with Dunnett’s post hoc test). (B) mRNA levels of AFF4 and control cellular genes (GAPDH and HPRT) in cells infected with HSV at the indicated MOI relative to levels in mock-infected cells. Data are means ± SEM of results from 3 replicates. (C) Western blot of AFF4 and control GAPDH in HFF, MRC5, and Vero cells that were mock infected or infected with HSV (MOI = 5). The graph represents the quantitation of protein levels in infected cells relative to mock-infected cells. Data are means ± SEM of results from 3 replicates (paired two-tailed t tests). Download FIG S3, PDF file, 0.1 MB (121.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Depletion of SIAH1 enhances the levels of AFF4 and HSV IE proteins. (A and B) MRC5 cells were transfected with control siRNA or SIAH1 siRNAs. Cells were infected with HSV (MOI = 3) for 4 h. (A) Western blot of AFF4 and control cellular proteins (BRD4 and GAPDH) and viral IE proteins (ICP4). (B) Quantitation of protein levels and mRNA levels relative to those in cells transfected with control siRNA. Data are means ± SEM of results from 2 experiments. (C) HFF cells were transfected with control siRNA or SIAH1 siRNAs and infected with HSV (MOI = 3) for 4 h. Western blotting of AFF4 and control cellular proteins (BRD4 and GAPDH) and viral IE proteins (ICP4). Data representing quantitation of protein levels are relative to those in cells transfected with control siRNA. Data are means ± SEM of results from 2 experiments. Download FIG S4, PDF file, 0.08 MB (82.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Reagents. Download Table S2, PDF file, 0.05 MB (48.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Supplemental materials and methods. Download Text S1, DOCX file, 0.02 MB (16.4KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Primers. Download Table S3, PDF file, 0.04 MB (43.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Statistics. Download Table S4, PDF file, 0.05 MB (52.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.