Abstract
The dynamics and organization of the actin cytoskeleton are crucial to many cellular events such as motility, polarization, cell shaping, and cell division. The intracellular and extracellular signaling associated with this cytoskeletal network is communicated through cell membranes. Hence the organization of membrane macromolecules and actin filament assembly are highly interdependent. Although the actin-membrane linkage is known to happen through many routes, the major class of interactions is through the direct interaction of actin-binding proteins with the lipid class containing poly-phosphatidylinositols (PPIs). Among the PPIs, phosphatidylinositol bisphosphate (PI(4,5)P2) acts as a significant factor controlling actin polymerization in the proximity of the membrane by binding to actin-associated proteins. The molecular interactions between these actin-binding proteins and the membrane lipids remain elusive. Here, using molecular modeling, analytical theory, and experimental methods, we investigate the binding of three different actin-binding proteins, mDia2, NWASP, and gelsolin, to membranes containing PI(4,5)P2 lipids. We perform molecular dynamics simulations on the protein-bilayer system and analyze the membrane binding in the form of hydrogen bonds and salt bridges at various PI(4,5)P2 and cholesterol concentrations. Our experimental study with PI(4,5)P2-containing large unilamellar vesicles mimics the computational experiments. Using the multivalencies of the proteins obtained in molecular simulations and the cooperative binding mechanisms of the proteins, we also propose a multivalent binding model that predicts the actin filament distributions at various PI(4,5)P2 and protein concentrations.
Introduction
The lipid composition of a membrane can alter the cell’s ability to carry out different biological functions.1 Phosphatidylinositol 4,5 bisphosphate (PI(4,5)P2) is a poly-phosphoinositide (PPI) that constitutes 1% of the phospholipid content in the plasma membrane2 and is involved in fundamental signaling pathways that regulate cytoskeletal assembly3 and other cellular functions.4,5 Although PI(4,5)P2 is only a small portion of the total membrane, this phospholipid plays fundamental roles in cell biology. Numerous diseases are associated with defects in the enzymes regulating the metabolism of PI(4,5)P2 and other PPIs. For example, cell transformation and the acquisition of invasive or metastatic phenotypes are often associated with defects in the kinases or phosphatases regulating transitions between PI(4,5)P2 and PI(3,4,5)P3.6–10 In contrast, mutations in phosphatases that degrade a different isomer, PI(3,4)P2, are associated with Charcot-Marie-Tooth neuropathies11 and amyotrophic lateral sclerosis (ALS).12,13 PPIs, mainly PI(4,5)P2,14,15 regulate cytoskeletal assembly,3,16 which controls cell motility and other cellular functions that are abnormal in pathologic phenotypes.
Previous works have contributed to defining the effects of PPIs on protein function,17–23 and intracellular signaling related to cytoskeletal assembly3,16,21,24–27 and linked them to mechanisms of formation of functional domains of PPIs,1,28–32 with evidence of localized spatial enrichment of PPIs.31–34 The extensive use of mouse genetics to study PPI-dependent processes over the years has identified several murine genes that play essential roles in cytoskeletal dynamics and phosphoinositide metabolism.35–40 Despite this progress, the limited effectiveness of therapies targeting PPI-directed enzymes can be attributed to insufficient knowledge of the physical chemistry of PPIs and the factors that determine their spatial distribution within membranes. This spatial effect has a direct bearing on PI(4,5)P2’s ability to regulate the activity of numerous actin-binding proteins that regulate actin filament assembly and consequently affect cell integrity, shape, and membrane organization.41,42
PI(4,5)P2 interacts with numerous actin regulating proteins to either inhibit or drive actin filament assembly. Even though the average resting level of PI(4,5)P2 in mammalian cell membranes is 1%,34,43,44 PI(4,5)P2 is known to be distributed heterogeneously with concentrations in sequestered domains of PI(4,5)P2 reaching upwards of 80%.32,42,45 Therefore, the sensitivity of protein binding to PI(4,5)P2 in the range well above the average concentration PI(4,5)P2 could be physiologically relevant, particularly to several processes involving cytoskeletal proteins.33,46
PI(4,5)P2/protein binding interfaces range from being highly specific to relatively non-specific, and the structures of these proteins range in conservation and diversity.2 Some commonly known PI(4,5)P2 domains include PH domains found in a few actin-binding proteins but that are more prominent in proteins that have other functions. The ability of PI(4,5)P2 to regulate actin assembly at the membrane depends not only on PI(4,5)P2 concentrations, but also its distribution and organization within the bilayer. Cholesterol is thought to have effects on PI(4,5)P2 clustering, and bilayer orientation could thereby indirectly mediate PI(4,5)P2/protein interactions. Experimental data have shown that cholesterol depletion leads to various effects such as decreased lateral mobility of membrane proteins,47 decreased PI(4,5)P2/protein binding,48 delocalization of PI(4,5)P2,49 and disruption of PI(4,5)P2 hydrolysis.50 These findings are likely relevant to understanding the mechanism of diseases such as Alzheimer’s disease, Huntington’s disease, or Parkinson’s disease in which cholesterol is essential for neuronal physiology. Atomic-level detail of PI(4,5)P2 distribution and cholesterol-mediated PI(4,5)P2 intracellular regulation will provide insight linking molecular-level specificity to cytoskeletal assembly and dynamics.
Crystallographic studies have focused on structural characterizations of modular domains in signaling and trafficking proteins that recognize specific membrane phospholipids,44,51–56 which have been significant contributors to current understanding of several membrane association domains such as PH domains,44,57 and to how these domains can play effector roles by altering phospholipid distribution and membrane morphology.43,58
In previous experiments,42 we have shown both computationally and experimentally that lipid composition, specifically PI(4,5)P2 concentration and cholesterol presence regulates the ability of PI(4,5)P2 to interact with two actin nucleation promoting proteins: the formin family member mammalian diaphanous-related 2 (mDia2) and neural Wiskott-Aldrich syndrome protein (NWASP).59 In this current study, we show how cholesterol and PI(4,5)P2 distribution alter PI(4,5)P2/protein interactions with structurally unrelated proteins that regulate actin assembly. We chose to investigate three systems, namely, peptides derived from mDia2, NWASP, and gelsolin. PI(4,5)P2 activates mDia2 and NWASP to nucleate straight and branched actin filaments, respectively, but inhibits gelsolin’s ability to cap the fast-growing barbed end of F-actin or to sever the actin filament. mDia2, gelsolin, and NWASP each vary in PI(4,5)P2 binding motifs but contain basic residues that, therefore, can associate with the negatively charged head groups of PI(4,5)P2. Structurally, the PI(4,5)P2-binding peptide of gelsolin contains the most hydrophobic residues, and NWASP contains the most positively charged residues.
Given the shortage of experimentally determined structures for peripherally bound membrane-associated proteins interacting with PI(4,5)P2, we have constructed molecular models of the proteins mentioned above and performed all-atom simulations studies to model how proteins bind to PI(4,5)P2 in membranes and how PI(4,5)P2 distribution alters its binding to these proteins. Since cholesterol is a likely critical factor in PI(4,5)P2 regulation, we undertook computational studies to investigate how changing the concentration of cholesterol in bilayers affects PI(4,5)P2 interactions with proteins. We performed modeling and experiments to study the effect of PI(4,5)P2 concentration on peptide binding. These simulations and experimental results provide us a better understanding of the effects of PI(4,5)P2 and cholesterol as regulators of actin dynamics.
Computational methods
A. The accession of gelsolin structure
A crystal structure that contains a PI(4,5)P2 and F-actin-binding site of gelsolin (residues 150–169 with the sequence KHVVPNEVVVQRLFQVKGRR) was obtained from the protein data bank, PDB ID: 1SOL.
B. Construction and validation of mDia2 and NWASP models
A basic amino acid region in mDia1, amino acids (AA 12–22), was revealed by previous co-sedimentation assays to bind PI(4,5)P2.60 This basic amino acid cluster in mDia1 shares 90% homology with the basic amino acid cluster in mDia2 (AA 25–40 with sequence RGCRESKMPRRKGPQH). The mDia2 (AA 25–40) sequence was employed in the Blast server in order to find a template crystal structure to construct a homology model. Human Cdc37 N-terminal domain, sharing 66% identity with the sequence of human mDia2 (AA 25–40), was selected and aligned using the Clustal Omega Program. The sequences of human Cdc37 N-terminal and human mDia2 were obtained from Uniprot (accessions codes Q16543 and Q9NSV4, respectively).
Similarly, in NWASP, co-sedimentation assays studies showed that NWASP residues 186–200 (sequence KEKKKGKAKKKRLTK) were sufficient to bind PI(4,5)P248 and there is currently no experimental structure of the PI(4,5)P2 binding region of NWASP. The Blast program was used to identify template crystal structure candidates to model NWASP. In order to obtain optimal sequence coverage of NWASP (AA 186–200), three proteins, PBD ID: 1JJ2, 1S72, 3DTP were selected as templates. These template proteins were aligned to NWASP (AA 186–200) and shared 75%, 75%, 82% with NWASP (AA 186–200), respectively.
Molecular models of NWASP (AA 186–200) and mDia2 (AA 25–40) were constructed using both homology modeling and ab initio methods. In order to create homology models, the alignments mentioned above, generated by Clustal Omega of mDia2 and NWASP were input into Modeller,61 and ten homology models of each peptide were constructed. To build ab initio models, NWASP and mDia2 peptide sequences were input to the web server Robetta.62 The discrete optimized protein energy score was used to select the top homology and ab initio models. The models are summarized in Fig. 1.
Fig. 1.
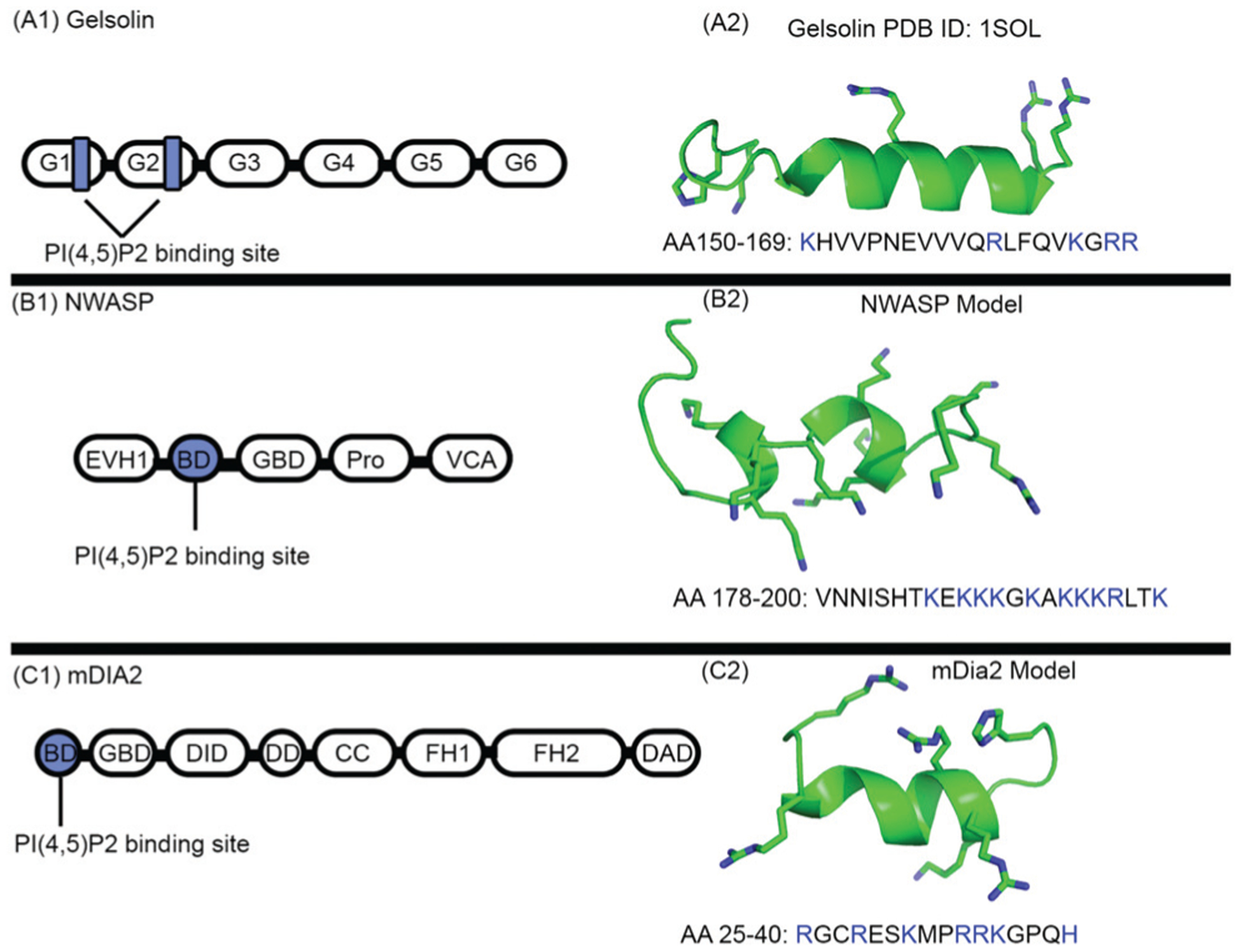
The PI(4,5)P2 binding sites of gelsolin, mDia2, NWASP, require critical basic residues for PI(4,5)P2 association elucidated in Pymol v2.1 software. The basic residues are highlighted in the ball and stick representation. (A1) Schematic representation of gelsolin domain architecture. Black boxes represent the six gelsolin domains, abbreviated as G1-G6. PI(4,5)P2 binding sites are in blue; (A2), the crystal structure of the PI(4,5)P2 binding region of the gelsolin peptide: the basic residues are shown in the stick representation. (B) Schematic representation structure of NWASP domain architecture. Black boxes represent the 5 NWASP domains. The following abbreviations are introduced. EVH1: enabled-VASP homology domain 1, basic region: BD (blue), Pro: proline-rich sequence, V: verprolin a polybasic motif homology domain, connecting region, and A: acidic region. (B2), molecular model of the PI(4,5)P2 binding region of the NWASP peptide; the basic residues are shown in the stick representation. (C1) Schematic representation of the mDia2 domain architecture. Black boxes represent the eight mDia2 domains. The following abbreviations are introduced. GBD: GTPase-binding domain, DID: diaphanous-inhibitory domain, DD: dimerization domain, CC: coiled-coil region, FH: formin homology, DAD: diaphanous-auto-regulatory domain, and BD: N-terminal basic region (blue). (C2) Molecular model of the PI(4,5)P2 binding region of the mDia2 peptide; the basic residues are shown in the stick representation.
C. Preparation of peptides for bilayer binding
The top models of mDia2 and NWASP and the crystal structure of gelsolin were relaxed using all-atom molecular dynamics simulations using the GROMACS package and the CHARMM force field. PROCHECK and SWISS-MODEL SERVER were then employed to assess the quality of the resulting models. The top-scoring models were used for simulations of the peptide binding to the bilayers, and the models were visualized using PYMOL and Visual Molecular Dynamics (VMD).63
D. Molecular dynamics of gelsolin, mDia2, and NWASP with asymmetric bilayers containing varying amounts of PI(4,5)P2 and cholesterol
Molecular dynamics (MD) simulations have been a powerful tool in elucidating atomistic principles governing peripheral protein-membrane binding. We hypothesize that the structural and functional characteristics of each peptide and lipid composition moderate PI(4,5)P2 membrane interactions. We also hypothesize that cholesterol’s presence in bilayers can alter lipid organization that results in altered PI(4,5)P2 clustering and perturbed PI(4,5)P2 peptide interactions. To test these hypotheses, we have performed two replicate simulations for each protein at four separate conditions (24 total simulations), including bilayers with 10%, 20%, or 30% PI(4,5)P2 interspersed with 20% cholesterol and the fourth condition with 20% PI(4,5)P2 and no cholesterol. The bilayer composition of each simulation is listed in Table 1. The outer leaflet in each bilayer contained 1-palmitoyl 2-oleoyl phosphatidylcholine (POPC) and cholesterol (CHL). The inner leaflet of the bilayers contained PI(4,5)P2, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS), and CHL. In order to account for the area change in varying PI(4,5)P2 and CHL concentrations, DOPE concentrations were adjusted to maintain the area, yielding a simulation box of ~62 nm2 for all the bilayers. By maintaining the total surface area in this way, we control for the expected effect of charge on the peptide binding affinity and thereby isolate the effect of cholesterol. Regarding PI(4,5)P2, we note that we use a charge of −4 with protonation at the 5-phosphate position, in line with the different pKa of the two phosphomonoesters.64 The precise characterization of the protonation state depends on both electrostatics and surface pressure, and these have a complex relationship.28,65–67 The model for PI(4,5)P2 consists of 18:0/18:4 acyl chains.
Table 1.
Summary of systems and simulation runs
| Bilayer construction | ||||
|---|---|---|---|---|
| Bilayer | PIP2% | CHL% | DOPE% | DOPS% |
| 1 | 20 | 0 | 55 | 25 |
| 2 | 30 | 20 | 30 | 20 |
| 3 | 20 | 20 | 40 | 20 |
| 4 | 10 | 20 | 50 | 20 |
| MD simulation run GROMACS-CHARMM36 force field | ||||
| Run length | Ns per day | ff of atoms | # water molecules | #CPUS |
| 150 ns | 10 ns | 65 699 | 3845 | 16–32 |
All of the peptide/bilayer molecular simulations were performed using GROMACS version 5.1.2 and the charmm36 force field for all standard protein and lipid parameters.68–70 Long-ranged interactions were considered through the particle-mesh-Ewald method. Each peptide was placed on a pre-equilibrated 252 lipid bilayer within a 3 angstrom distance to a PI(4,5)P2 molecule. The bilayers were solvated with ~4946 water molecules and neutralized using 239 Na+ and 71 Cl− ions. All bond lengths were constrained using the linear constraint solver (LINCS). A time step of 2 fs was used in all simulations. Each system was simulated in the semi-isotropic NPT ensemble, with constant particle number N, normal pressure of 1 atm, and constant temperature of 300 K. The resultant system was energy-minimized, and then the simulations were run for 150 ns. Each run of a simulation typically required 432 hours of computing time on a 16-core CPU. Analysis of all simulations was performed using VMD and in-house analysis codes based on software shared online at http://biophyscode.github.io. Snapshots of the central simulation cell are depicted in Fig. 2. We note that when PI(4,5)P2 is included, the peptides adhere and do not fall off the membrane for the period simulated. The time-frame of the simulation was not sufficient to compute residence times for this reason, but was sufficient for convergence of root-mean-squared deviation and hydrogen bond occupancies.
Fig. 2.
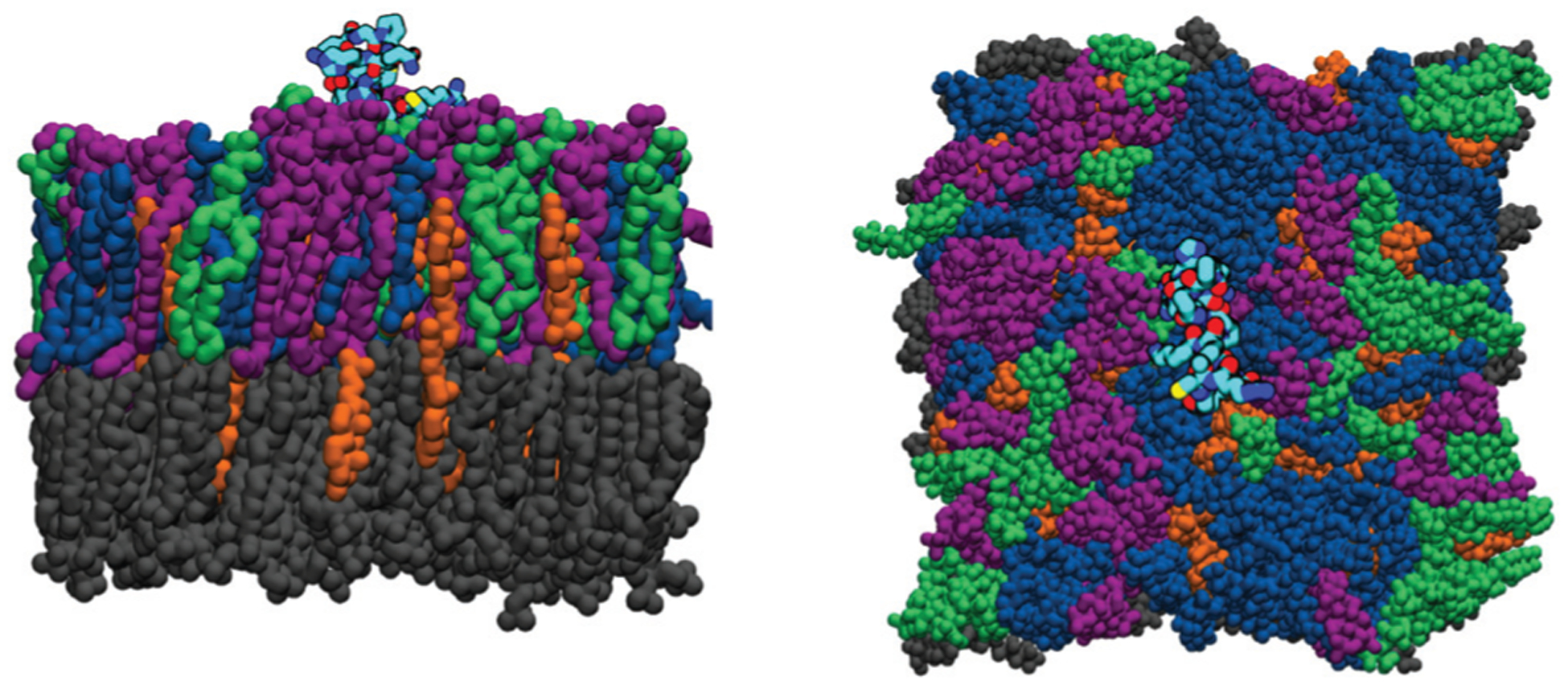
Snapshots of a bilayer containing 20% PI(4,5)P2. Side and front views of the simulation cell. Lipids are rendered in a solid color, where POPC is gray, DOPS is green, DOPE is blue, PI(4,5)P2 is purple, and cholesterol is orange. The peptide structure is given in black along with an atomistic structure colored by atom type, in which nitrogen is red, oxygen is blue, carbon is cyan, and sulfur is yellow. We omit displaying the solvent (water) molecules.
E. Multisite binding model and numerical simulations
To estimate the PI(4,5)P2 bound protein concentration we follow the proposed two-step reaction mechanism of formin in our previous work where the protein first binds to any membrane site (M) and then the bound protein can get activated depending on the PI(4,5)P2 concentration in a Hill reaction.42 We use the notations F for mDia2, P for PI(4,5)P2, N for NWASP, and G for gelsolin. We can write the reactions for mDia2 as F + M ⇌ FM and where and are the association constants for the first and second reaction respectively. Similarly for NWASP: N + M ⇌ NM and where and are the association constants for the first and second reaction respectively and gelsolin: G + M ⇌ GM and where and are the association constants for the first and second reaction respectively. For any association constant Kab with α = f, n or g, and β = m or p, represents corresponding dissociation constants in units of concentration. At equilibrium, the protein concentrations obey the following equations:
where the subscript ‘tot’ represents the total concentration of proteins in the solution. Solving these equations numerically, we obtain the equilibrium concentrations of PI(4,5)P2 bound proteins. The Hill coefficients n1 = 4, n2 = 5, and n3 = 4 are the multivalencies obtained from molecular simulations. The parameters for the binding constants for mDia2 were obtained from our previous work,42 while those for NWASP were determined from the work of ref. 48. The data for gelsolin were not directly available. We note that the association with the membrane occurs through multiple sub-domains and in 3D, while the association with PI(4,5)P2 was considered only through the binding domain included in our molecular model and occurs on the 2D membrane surface. In general, the mapping of the 3D to the 2D binding will require a factor of volume to area ratio. However, in this work, all parameters are reported as the 3D equivalent; i.e., concentrations are always reported based on the total volume. These considerations help to rationalize why the membrane association constants have a larger apparent affinity compared to the association with PI(4,5)P2. The details of the numerical simulations are given in the ESI.†
Experimental materials and methods
NWASP residues AA 186–200 (sequence KEKKKGKAKKKRLTK), mDia2 residues AA 25–40 (sequence RGCRESKMPRRKGPQH) and gelsolin residues AA 150–169 (sequence KHVVPNEVVVQRLFQVKGRR) were purchased from the Lipopharm.pl, Gdańsk, Poland. We prepared PI(4,5)P2-containing large unilamellar vesicles (LUVs) that mimic computational experiments in order to test the PI(4,5)P2 concentration effect on the peptides’ interaction with LUV surfaces. LUV bilayers containing PI(4,5)P2 were prepared based on a known phase diagram for a ternary lipid mixture containing DOPC/DPPC/dCHOL,71 as described in.59 The appropriate lipids were mixed in organic solution, the solvent was evaporated under a stream of N2, and the sample was kept under vacuum to remove solvent traces. LUVs were prepared from the solution containing lipids by subjecting them to 5 freeze/thaw cycles and then extruded using 100 nm pore size filters. The composition of the vesicles used in the experiments are (in mol%):
The hydrodynamic diameters of the LUVs, before and after adding different concentrations of peptides, were determined by dynamic light scattering using a DynaPro99 instrument (Wyatt, formerly Protein Solutions).72
Computational and experimental results
A. Molecular dynamics results
PI(4,5)P2 concentration affects protein interaction with the bilayer.
In order to decipher the atomistic details by which PI(4,5)P2 associates with actin-binding proteins of varying functional and structural properties we studied the truncated PI(4,5)P2 domains of gelsolin, mDia2, and NWASP on bilayer systems through all-atom molecular dynamics simulations. Bilayer systems contained 20% CHOL in both leaflets, POPC on the outer leaflet, and 10, 20, or 30% PI(4,5)P2 on the inner leaflet along with a PE and PS mixture adjusted to control the surface charge density across conditions. In order to measure the effect of cholesterol (CHOL), systems containing 0% CHOL with 20% PI(4,5)P2 were constructed as well. Fig. 2 illustrates a typical peptide adhered to a bilayer system containing 20% PI(4,5)P2. We carried out 150 ns MD simulations for each system in replicates of two and measured the number of bonds between the protein and lipid with respect to time, see Fig. 3. We note that Fig. 3 establishes that our distributions are at steady-state and explicitly show data in both replicates. This result motivates us to combine the data and look at the consolidated distributions. To further analyze the interactions, we constructed histograms of the salt bridges and hydrogen bonds formed between each type of lipid and the protein by combining the replicates in Fig. 4. The charts shown in Fig. 4 display the number of bonds observed between the protein and the lipids on the bilayer with respect to the percentage of the occurrence, labeled by each bilayer system condition; the results reveal that PI(4,5)P2 concentration plays a significant role in the number of bonds formed between the protein and lipids in the bilayer for all three proteins.
Fig. 3.
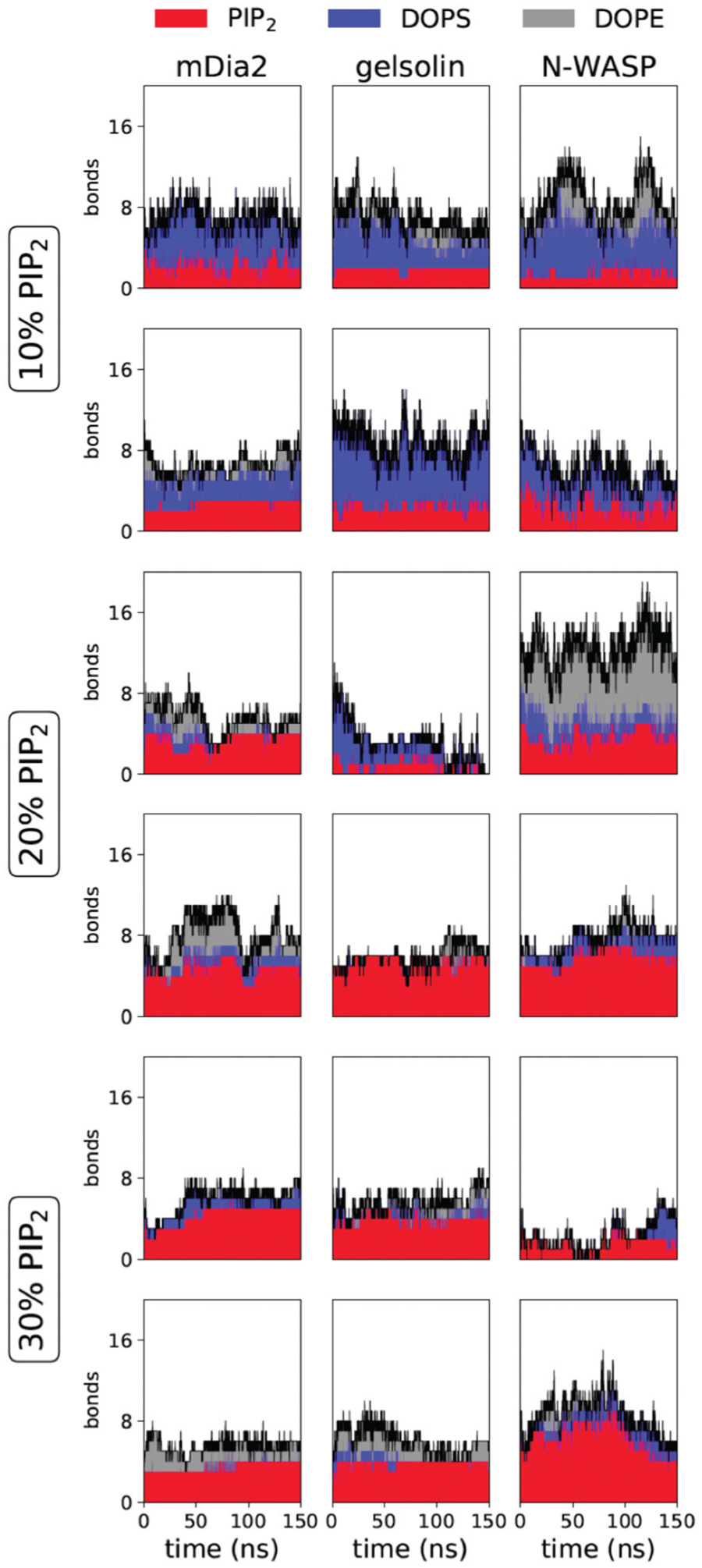
Time series for the number of bound lipids from simulations containing cholesterol. The number of bonds includes both hydrogen bonds and salt bridges. Lipid types are denoted by color, PI(4,5)P2 in red, DOPS in blue, and DOPE in gray, with the total number marked with a black line. The rows include two replicates per condition, while the columns are organized by the peptide.
Fig. 4.
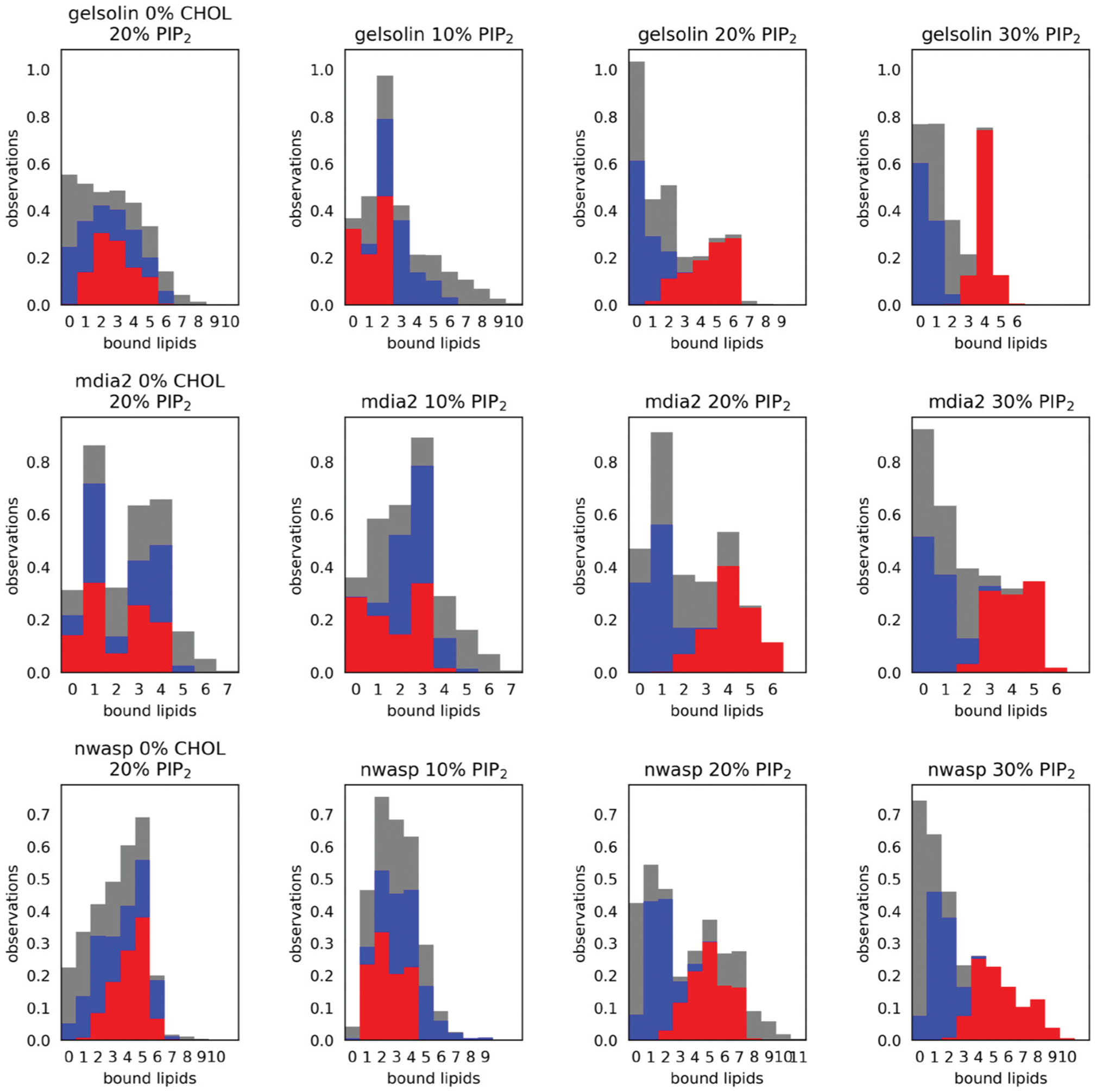
Stacked histograms of the number of hydrogen bonds and salt bridges between the peptides and each lipid type. The vertical axis shows the proportion of simulation frames that contain the number of bonds indicated on the horizontal axis. We count the union of both bond types so that each bond can be either a hydrogen bond or a salt bridge or both. The red bars indicate bonds formed with PI(4,5)P2, while grey represents those formed with DOPE, and blue represents those formed with DOPS. The histograms are stacked for clarity. We have included the “zero” category to express the proportion of simulation frames in which no lipids of a particular type are bound to the peptides. Therefore, each bar color sums to one. The first and third columns identify the effect of cholesterol at 20% PI(4,5)P2, while columns 1–4 describe the effect of concentration of PIP2.
Analysis of the gelsolin peptide (which contains 5 basic residues in its PI(4,5)P2 binding site) simulated on a bilayer containing 10% PI(4,5)P2 showed that 0–3 bonds were observed between gelsolin and PI(4,5)P2, in comparison to 1–6 and 0–10 bonds between and DOPS and DOPE, respectively. When the concentration of PI(4,5)P2 is increased to 20%, the number of observed bonds between gelsolin and PI(4,5)P2 increases to 1–6 bonds, preferentially 6 bonds. When PI(4,5)P2 concentration is further increased to 30%, gelsolin always makes 2–6 bonds with PI(4,5)P2, preferentially 4 bonds with less variation, and does not interact with DOPE for most of the duration of the simulation. Analysis of mDia2 on bilayers containing 10%, 20%, and 30% PI(4,5)P2 displayed an overall increase in the amount of bonds formed between mDia2 and PI(4,5)P2, 0–4, 2–6 and 2–6 bonds respectively. The number of bonds between mDia2 and PI(4,5)P2 did not show much of an increase from a 20% to a 30% PI(4,5)P2 contain bilayer, indicating that 20% PI(4,5)P2 achieved saturation in binding.
Fig. 4 highlights two trends which are common to all three proteins, namely that the number of bound PI(4,5)P2 increases with concentration while the number of bound DOPE and DOPS decreases. For example, we find that the gelsolin peptide co-incubated with 10% PI(4,5)P2 has between zero and two bonds with that lipid, compared to a higher range of 1–6 at the 20% condition. At the highest concentration of 30%, the peptide has a higher average number of bonds and a much tighter range of 3–5 lipids, suggesting more specific and persistent bonding. We see a corresponding dramatic decline in DOPS and DOPE binding at the highest condition, where there is a ceiling of 2 bound DOPS and 3 bound DOPE compared to a maximum of 9 bound DOPE observed at the 10% condition.
While each protein exhibits these trends, four factors distinguish them, including both the floor and ceiling in the number of bound PI(4,5)P2, the variability in the number of bonds, and the overlap between lipid types. For example, we see that even at the 10% PI(4,5)P2 condition, the NWASP peptide always forms at least one bond with that lipid, while mDia2 and gelsolin often have zero. NWASP is also capable of a higher degree of overlap. We see simultaneous bonding with many PI(4,5)P2 and DOPE at the 20% condition, however the DOPE bonds notably almost at the higher 30% condition in favor of PI(4,5)P2.
To summarize the relevant features of the distributions presented in Fig. 4, we have plotted the mean and standard deviation of the number of bonds for each condition and lipid type in Fig. 5. The top row pertains to the hypothesis that peptide binding varies with PI(4,5)P2 concentration. We find an apparent concentration-dependent increase in PI(4,5)P2 binding with a commensurate decline in the preference for DOPE and DOPS. These plots show both the floor, ceiling, and saturation point for each protein. We see that NWASP has a high floor and appears to monotonically increase its PI(4,5)P2 binding, while mDia2 and gelsolin saturate at 20% and a slightly lower average number of bonds.
Fig. 5.
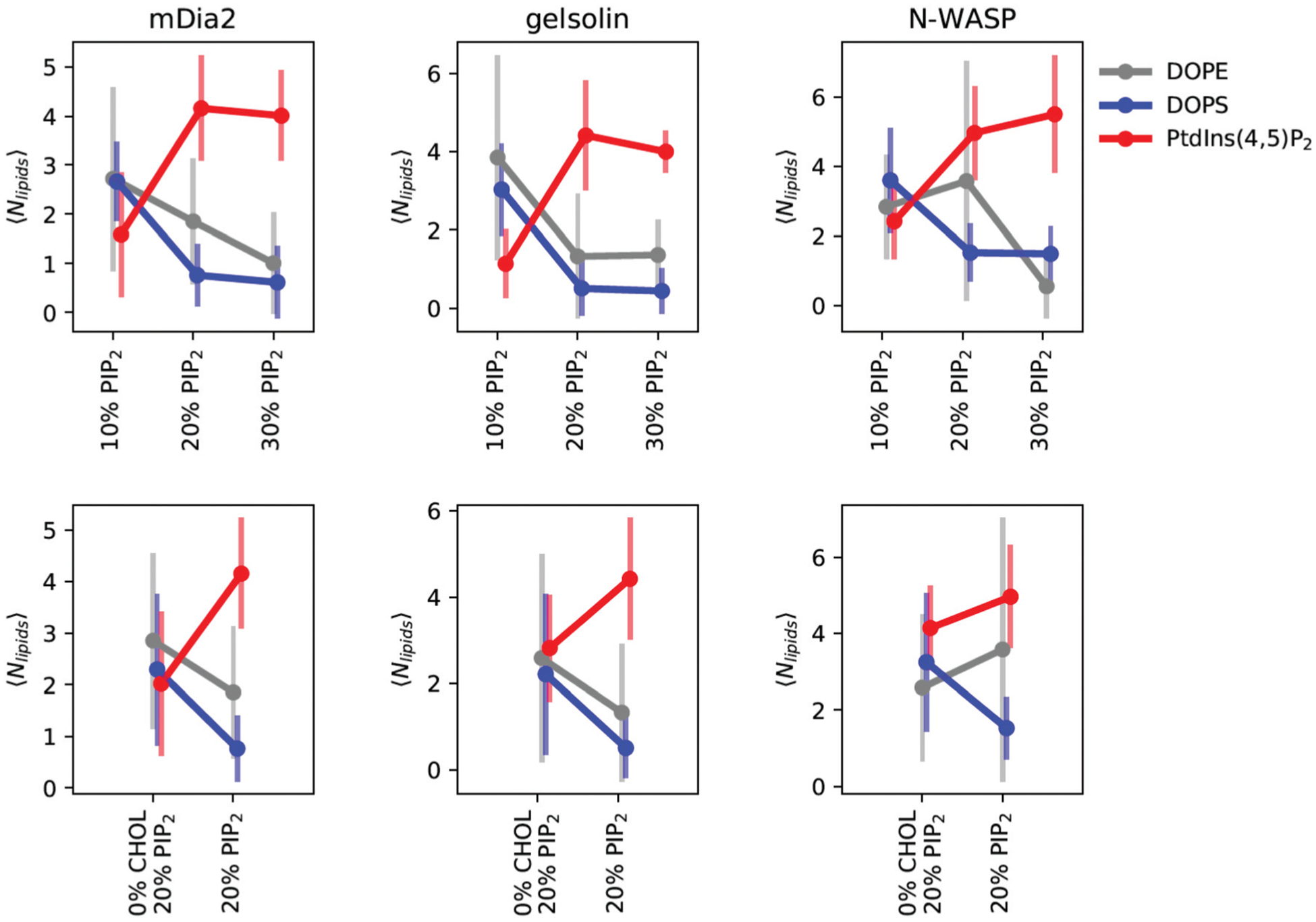
Multivalency summary statistics show the mean and standard deviation of bonds (the union of salt bridges and hydrogen bonds) depicted in Fig. 4. The top row shows the effect of concentration, while the bottom row depicts the effect of cholesterol at the 20% PI(4,5)P2 condition. Lipids are separated by color, with PI(4,5)P2 in red, DOPS in blue, and DOPE in grey. We find that the total number of bonds and the selectivity for PI(4,5)P2 is sensitive to the concentration and presence of cholesterol. The error bars represent one standard deviation from the statistics collected over 200 ns of MD simulations across two replicates.
In order to inspect the precise binding interfaces quantified by the histograms in Fig. 4, we have collected a series of representative snapshots of the simulations depicted in Fig. 6. These snapshots were taken from simulation frames containing the most common number of hydrogen bonds and salt bridges (the mode) from each replicate. They, therefore, represent the most typical lipid neighborhood that facilitates the bonds. We can glean some of the same common trends from these snapshots, namely that the presence of PI(4,5)P2 increases with concentration. The features which distinguish the peptides are also present, including the diverse set of lipids bound to NWASP at the 20% condition explained above. These images also highlight the dynamics in the structure of the peptide, which may generate the variability in the number and composition of lipid bonds.
Fig. 6.
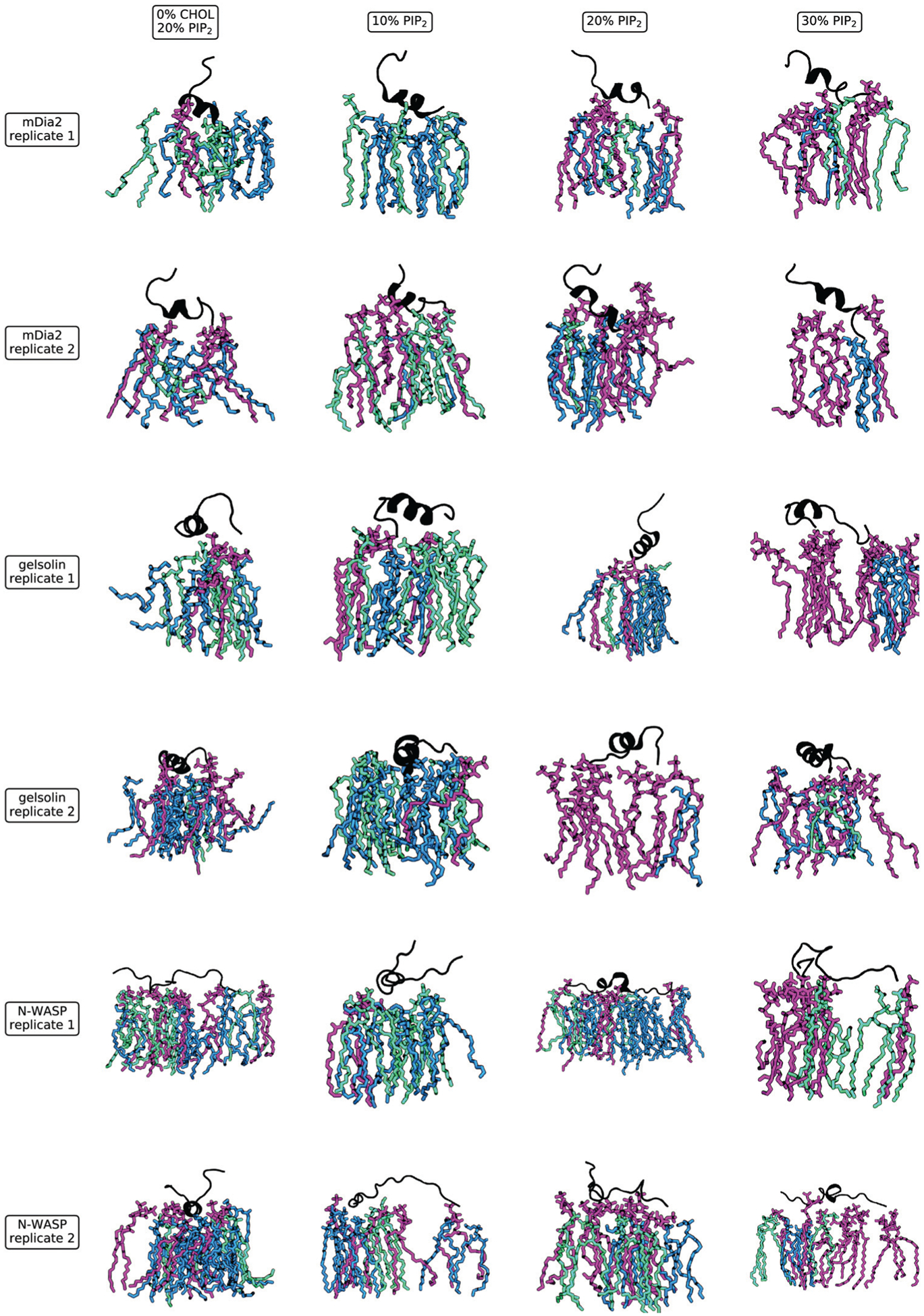
Snapshots of the most representative lipid configurations. These snapshots show all lipids within 5 Å of the protein. We select one representative frame from each replicate such that the frame includes the most common (mode) of the bonds tabulated in Fig. 4. Lipids are colored by type, with PI(4,5)P2 shown in purple, DOPS in green, and DOPE in blue (with one orange cholesterol molecule shown for NWASP). We see a larger number of bound PI(4,5)P2 (purple) with increasing concentration and fewer bonds with PI(4,5)P2 when cholesterol is absent.
The membrane cholesterol affects interactions of PI(4,5)P2-binding peptides with the bilayer.
The bond distributions and snapshots described in the previous section also include a comparison to the cholesterol-free condition at 20% PI(4,5)P2. Since we have controlled for the surface charge, this comparison isolates the effect of cholesterol on the binding. A comparison of the first and third columns in Fig. 4 indicates that simulations containing cholesterol include more peptide bonds with PI(4,5)P2 as well as less competing bonding to the other lipids. The most-representative snapshots in Fig. 6 likewise show a more-dense cluster of bound lipids when cholesterol is present. The summary statistics presented in the bottom row of Fig. 6 confirm this observation by showing that cholesterol-free simulations have no lipid preference, while those with cholesterol show a clear preference for PI(4,5)P2 over DOPE and DOPS. These results also indicate that when compared to the other two proteins, NWASP is less sensitive to cholesterol. When cholesterol is present, NWASP is less selective for PI(4,5)P2 because it readily binds the other lipids as well.
We note that in this study, we have not explicitly quantified the clustering of PI(4,5)P2 induced by the peptide binding due to small system sizes relevant to cluster formation. However, related studies reported recently have given insight into the mechanisms of nanocluster formation in PI(4,5)P2 containing bilayers using both theoretical and experimental approaches.73,74 The mechanism by which cholesterol influences and enhances the multivalency of peptide binding to PI(4,5)P2 is an open question. A direct effect of cholesterol is to induce phase separation into lipid-ordered (LO)/lipid-disordered (LD) phases. Under the current experimental conditions, we expect such a phase separation to occur.75 The phase separation has the effect of doubling the PI(4,5)P2 concentration as it distributes primarily in the LD phase. However, another effect of cholesterol also manifests by altering the PI(4,5)P2 distribution within the LD phase. In an earlier study focused on mDia2, we had ruled out specific interactions such as hydrogen bonding and had alluded to packing interactions (as quantified through the calculations of radial distribution functions) as the main driver;42 however, as future work, it would be insightful to pursue a more detailed study of the cholesterol mediated mechanisms of PI(4,5)P2 clustering.
B. Experimental results
As shown in Fig. 7 interaction of the peptides from gelsolin (residue 150–169 with the sequence KHVVPNEVVVQRLFQVKGRR; panel A/B), mDia2 (AA 25–40 with sequence RGCRESKMPRRKGPQH; panel C/D) and NWASP residues 186–200 (sequence KEKKKGKAKKKRLTK E/F) with phospholipids organized in bilayer membranes caused changes in vesicle size. In this system peptide-LUV interaction involves association of variably amphipathic positively charged peptides with negatively charged membrane phospholipids that might result in a conformational change of the peptides, changes in membrane fluidity and membrane penetration followed by membrane aggregation or fusion. LUV aggregation is a fast process and usually 10–15 minutes is enough to reach its maximum. The three tested peptides caused LUV aggregation to different extents. Of the three peptides, the one from gelsolin has the strongest dependence on cholesterol, with vesicle aggregation strongly suppressed when the vesicles have 20% PI(4,5)P2 but no CHL (Fig. 7A). Vesicle aggregation also depends on the mole fraction of PI(4,5)P2 in the vesicle, but not monotonically, with 30% PI(4,5)P2 vesicles relatively resistant to aggregation, perhaps because the packing density is too high to permit the much larger gelsolin to fully bind all the lipids and neutralize the negative charge density. Alternatively, the high affinity of the peptides for PI(4,5)P2, and the tendency of PI(4,5)P2 to make micelles rather than bilayers at high mole fractions could lead to extraction of PI(4,5)P2 out of the LUVs by the peptides, rather than the peptides aggregating the LUVs as they do at lower PI(4,5)P2 mole fractions. mDia has somewhat higher apparent affinity to PI(4,5)P2 containing vesicles, and a similar dependence on cholesterol (Fig. 7B). NWASP has the highest apparent affinity for the vesicles, but little dependence on either cholesterol or PI(4,5)P2 mole fraction. The vesicle size assay cannot provide accurate binding constants, since multiple mechanisms determine vesicle aggregate size, but the qualitative differences of these three PI(4,5)P2 binding sites on cholesterol clearly show that NWASP is less dependent on cholesterol than either gelsolin or mDia2.
Fig. 7.
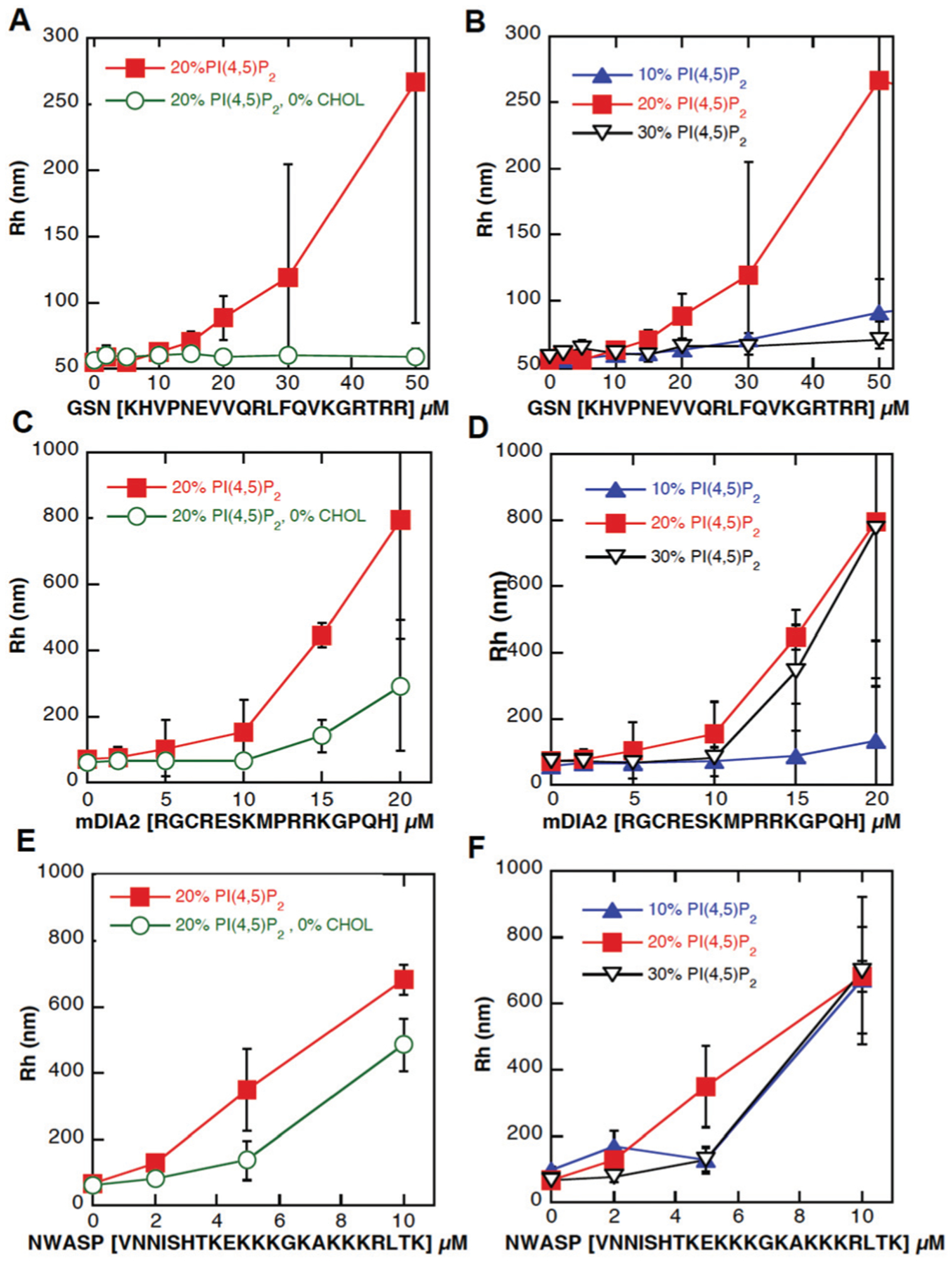
Interaction of peptides from gelsolin (GSN), (panels A and B); mDia2 (panels C and D), and NWASP (panels E and F) with artificial bilayers of LUV membranes containing 20% PI(4,5)P2 and 20% PI(4,5)P2 with presence of cholesterol (left panels) or increasing concentration of PI(4,5)P2 (10–30%) with cholesterol (right panels). Error bars represent standard deviations (n = 3).
Discussion and conclusions
Unravelling the cytoskeleton-membrane interaction is critical to understanding various cellular pathways, biochemical mechanisms, and disease progressions. In this study, we combine the methods of molecular modeling, analytical theory, and experimental techniques to decipher the biochemical mechanisms that alter the binding of cytoskeletal associated proteins with the membrane. Specifically, we focus on the interaction between three proteins-mDia2, NWASP, and gelsolin with cholesterol-containing membranes at various PI(4,5)P2 concentrations. We first constructed the molecular models for the proteins: for NWASP and mDia2, the models are constructed from both homology and ab initio methods, while for gelsolin, we obtained the crystal structure from the protein data bank. To simulate the binding of protein with the membrane, we relax the structures using all-atom molecular dynamics simulations. The resulting protein configurations are used to study the adhesion interaction of mDia2, NWASP, and gelsolin with asymmetric bilayers containing various amounts of PI(4,5)P2 and cholesterol using molecular dynamics. Our analysis of binding interactions in the form of hydrogen bonds and salt bridges between the proteins and the membrane lipids shows that PI(4,5)P2 concentration plays a significant role in the number of bonds established between these proteins and the bilayer membrane. The simulation study by varying cholesterol concentration shows a significant increase in bond formation between the proteins and the PI(4,5)P2 in the presence of cholesterol, with NWASP being the least sensitive to changes in PI(4,5)P2 concentration.
The experimental study of binding interaction using vesicle size assay provides quantitative differences in binding of the three proteins with PI(4,5)P2 and in general agreement with simulation results. This study also suggests a lower dependence of NWASP-PI(4,5)P2 binding on cholesterol concentration compared to the other two proteins, in agreement with the molecular dynamics simulations. A direct comparison of the modeling and experiments is possible with measurements of actin polymerization rate versus PI(4,5)P2 concentration as was reported for mDia2 in our previous study.42 Since these data are not available for the other two systems (NWASP and gelsolin), we utilize our theoretical model for multisite binding to describe how our results may translate across the three systems, as we describe below.
The multivalency of the actin-related proteins to the bilayer membrane in the presence of PI(4,5)P2 obtained from the molecular dynamics simulations and experiments allows us to propose a multivalent binding model that predicts actin distribution at given PI(4,5)P2 and protein concentration. The multivalent binding model utilizes reaction mechanisms based on the cooperative binding of actin-associated proteins proposed in our previous study and the binding information (multivalency of proteins) obtained from the current molecular dynamics study.42 Given the role of mDia2, NWASP, and gelsolin in actin filament assembly, this minimum model predicts the length distribution and branching of filaments at the membrane interface at various PI(4,5)P2 and protein concentration.
Knowing the concentration of PI(4,5)P2 bound proteins allows us to investigate the required concentration of each protein for various actin filament distributions through the formulation of a multisite binding model.76 Upon binding to PI(4,5)P2, mDia2 promotes linear actin polymerization, and NWASP promotes the branching of filaments. The free gelsolin in the solution (not bound to PI(4,5)P2) binds to the actin filament side or barbed end and promotes capping or severing of actin filaments. Hence an increase in PI(4,5)P2 bound gelsolin indicates an increase in linear filament formation. Here we examine four different filament configurations that are dependent on these PI(4,5)P2-protein interactions.
Long linear actin filaments
Fig. 8A shows the concentration of PI(4,5)P2 bound gelsolin and mDia2 in the first and second columns, respectively. These observations are made in the absence of NWASP. As the formation of long linear filaments requires a high concentration of PI(4,5)P2 bound mDia2 and gelsolin, and less free gelsolin, we use the product [mDia2]bound × [Gelsolin]bound, shown in the third column, as a measure of the presence of linear filaments. A higher value of this product indicates a higher probability of forming long filaments.
Fig. 8.
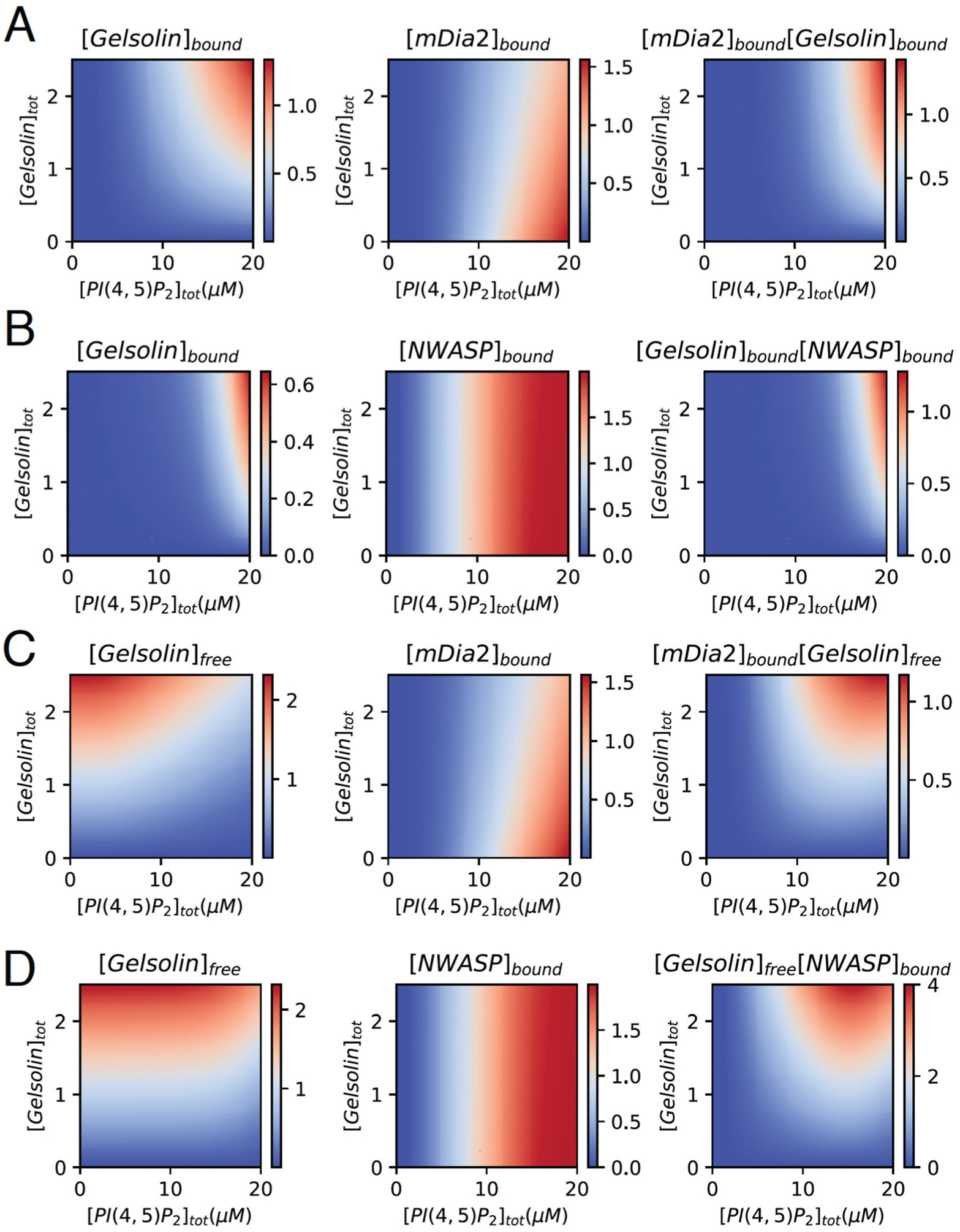
(A) Concentration of bound gelsolin and mDia2 as a function of PI(4,5)P2 and gelsolin in the absence of NWASP when [mDia2] = 2 μM, , , , and Mtot = 10. (B) Concentration of bound gelsolin and NWASP as a function of PI(4,5)P2 and gelsolin in the absence of mDia2 when [NWASP] = 2 μM, , , , and Mtot = 10. (C) Concentration of free gelsolin and bound mDia2 as a function of PI(4,5)P2 and gelsolin in the absence of NWASP when [mDia2] = 2 μM, , , , and Mtot = 10. (D) Concentration of free gelsolin and bound NWASP as a function of PI(4,5)P2 and gelsolin in the absence of mDia2 when [NWASP] = 2 μM, , , , and Mtot = 10. Detailed figure is given in ESI,† Fig. S3.
Long branched filaments
Fig. 8B shows the concentration of PI(4,5)P2 bound gelsolin and NWASP in the first and second columns, respectively. These observations are made in the absence of mDia2. As long branched filaments require a high concentration of bound NWASP and gelsolin, and less free gelsolin, we compute the product [NWASP]bound × [Gelsolin]bound in the third column. A higher value of this product indicates a higher probability of forming long, branched filaments.
Short linear actin filaments
Fig. 8C shows the concentration of free gelsolin in the solution and PI(4,5)P2-bound mDia2 in the first and second columns, respectively. These observations are made in the absence of NWASP. As short linear filaments require a high concentration of bound mDia2 and high free gelsolin in the solution, we compute the product [mDia2]bound × [Gelsolin]free in the third column. A higher value of this product indicates a higher probability of forming short linear actin filaments.
Short branched actin filaments
Fig. 8D shows the concentration of free gelsolin and PI(4,5)P2-bound mDia2 in the first and second columns, respectively. These observations are made in the absence of mDia2. As short branched filaments require a high concentration of bound mDia2 and free gelsolin, we compute the product [mDia2]bound × [Gelsolin]free in the third column. A higher value of this product indicates a higher probability of short branched filaments.
These results suggest a new understanding of the underlying mechanisms of peptide binding to the membrane and subsequent mechanisms of actin nucleation in a physiologically composed bilayer environment mediated by PI(4,5)P2 and cholesterol. A recent study reported that despite using different domains for lipid binding, cytoskeletal proteins associate with membranes through similar multivalent electrostatic interactions, but display enormous differences in the dynamics of membrane interactions and in the ranges of PI(4,5)P2 densities that they sense.77 Some proteins display transient, low-affinity interactions with PI(4,5)P2-rich membranes, whereas others reside on PI(4,5)P2-rich membranes for longer periods to perform their functions. Our multiscale model provides a quantitative framework to rationalize how the molecular interactions lead to PI(4,5)P2 sensitivity and control of cytoskeletal assembly. In particular, the validation of the mapping of different modes of actin assembly described by our model is a good candidate for future experimental investigations.
Supplementary Material
Acknowledgements
We acknowledge valuable discussions from Tatyana Svitkina and the members of PAJ and RR laboratories. We acknowledge support from the University of Pennsylvania Vice Provost Post-doctoral Fellowships for Academic Diversity. We also acknowledge supercomputing resources from extreme science and engineering discovery environment, Penn Materials Research Center, and Penn Institute for Computational Sciences.
Funding
This work was supported in part by NIH National Cancer Institute through grant CA227550 (RR), NIH National Institute for General Medical Sciences through grant GM111942 and GM136259 (PAJ), National Science Center, Poland under Grant: UMO-2015/17/B/NZ6/03473 (to RB).
Footnotes
Conflicts of interest
There are no conflicts to declare.
Electronic supplementary information (ESI) available: ESI methods. See DOI: 10.1039/d0sm00267d
References
- 1.Levental I, et al. , Calcium-dependent lateral organization in phosphatidylinositol 4,5-bisphosphate (PIP2)- and cholesterol-containing monolayers, Biochemistry, 2009, 48(34), 8241–8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McLaughlin S, et al. , PIP(2) and proteins: interactions, organization, and information flow, Annu. Rev. Biophys. Biomol. Struct, 2002, 31, 151–175. [DOI] [PubMed] [Google Scholar]
- 3.Yin HL and Janmey PA, Phosphoinositide regulation of the actin cytoskeleton, Annu. Rev. Physiol, 2003, 65, 761–789. [DOI] [PubMed] [Google Scholar]
- 4.Czech MP, PIP2 and PIP3: complex roles at the cell surface, Cell, 2000, 100(6), 603–606. [DOI] [PubMed] [Google Scholar]
- 5.Gambhir A, et al. , Electrostatic sequestration of PIP2 on phospholipid membranes by basic/aromatic regions of proteins, Biophys. J, 2004, 86(4), 2188–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katso R, et al. , Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer, Annu. Rev. Cell Dev. Biol, 2001, 17, 615–675. [DOI] [PubMed] [Google Scholar]
- 7.Engelman JA, The role of phosphoinositide 3-kinase pathway inhibitors in the treatment of lung cancer, Clin. Cancer Res, 2007, 13(15 Pt 2), s4637–s4640. [DOI] [PubMed] [Google Scholar]
- 8.Liu P, et al. , Targeting the phosphoinositide 3-kinase pathway in cancer, Nat. Rev. Drug Discovery, 2009, 8(8), 627–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang X, et al. , Phosphoinositide 3-kinase pathway activation in phosphate and tensin homolog (PTEN)-deficient prostate cancer cells is independent of receptor tyrosine kinases and mediated by the p110beta and p110delta catalytic subunits, J. Biol. Chem, 2010, 285(20), 14980–14989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wojtalla A and Arcaro A, Targeting phosphoinositide 3-kinase signalling in lung cancer, Crit. Rev. Oncol. Hematol, 2011, 80(2), 278–290. [DOI] [PubMed] [Google Scholar]
- 11.Chow CY, et al. , Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J, Nature, 2007, 448(7149), 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chow CY, et al. , Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS, Am. J. Hum. Genet, 2009, 84(1), 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volpicelli-Daley L and De Camilli P, Phosphoinositides’ link to neurodegeneration, Nat. Med, 2007, 13(7), 784–786. [DOI] [PubMed] [Google Scholar]
- 14.Sun M, et al. , Cancer-derived mutations in the regulatory subunit p85alpha of phosphoinositide 3-kinase function through the catalytic subunit p110alpha, Proc. Natl. Acad. Sci. U. S. A, 2010, 107(35), 15547–15552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaguchi H, et al. , Phosphatidylinositol 4,5-bisphosphate and PIP5-kinase Ialpha are required for invadopodia formation in human breast cancer cells, Cancer Sci, 2010, 101(7), 1632–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janmey PA and Lindberg U, Cytoskeletal regulation: rich in lipids, Nat. Rev. Mol. Cell Biol, 2004, 5(8), 658–666. [DOI] [PubMed] [Google Scholar]
- 17.Janmey PA and Stossel TP, Modulation of gelsolin function by phosphatidylinositol 4,5-bisphosphate, Nature, 1987, 325(6102), 362–364. [DOI] [PubMed] [Google Scholar]
- 18.Janmey PA, et al. , Polyphosphoinositide micelles and polyphosphoinositide-containing vesicles dissociate endogenous gelsolin-actin complexes and promote actin assembly from the fast-growing end of actin filaments blocked by gelsolin, J. Biol. Chem, 1987, 262(25), 12228–12236. [PubMed] [Google Scholar]
- 19.Janmey PA and Matsudaira PT, Functional comparison of villin and gelsolin. Effects of Ca2 +, KCl, and polyphosphoinositides, J. Biol. Chem, 1988, 263(32), 16738–16743. [PubMed] [Google Scholar]
- 20.Lamb JA, et al. , Modulation of gelsolin function. Activation at low pH overrides Ca2+ requirement, J. Biol. Chem, 1993, 268(12), 8999–9004. [PubMed] [Google Scholar]
- 21.Janmey PA, Protein regulation by phosphatidylinositol lipids, Chem. Biol, 1995, 2(2), 61–65. [DOI] [PubMed] [Google Scholar]
- 22.Stock A, et al. , Domain analysis of cortexillin I: actin-bundling, PIP(2)-binding and the rescue of cytokinesis, EMBO J, 1999, 18(19), 5274–5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rando OJ, et al. , Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex, Proc. Natl. Acad. Sci. U. S. A, 2002, 99(5), 2824–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janmey PA, Phosphoinositides and calcium as regulators of cellular actin assembly and disassembly, Annu. Rev. Physiol, 1994, 56, 169–191. [DOI] [PubMed] [Google Scholar]
- 25.Hartwig JH, et al. , Thrombin receptor ligation and activated Rac uncap actin filament barbed ends through phosphoinositide synthesis in permeabilized human platelets, Cell, 1995, 82(4), 643–653. [DOI] [PubMed] [Google Scholar]
- 26.Funaki M, DiFransico L and Janmey PA, PI 4,5-P2 stimulates glucose transport activity of GLUT4 in the plasma membrane of 3T3-L1 adipocytes, Biochim. Biophys. Acta, 2006, 1763(8), 889–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El Sayegh TY, et al. , Phosphatidylinositol-4,5 bisphosphate produced by PIP5KIgamma regulates gelsolin, actin assembly, and adhesion strength of N-cadherin junctions, Mol. Biol. Cell, 2007, 18(8), 3026–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levental I, Janmey PA and Cebers A, Electrostatic contribution to the surface pressure of charged monolayers containing polyphosphoinositides, Biophys. J, 2008, 95(3), 1199–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levental I, Cebers A and Janmey PA, Combined electrostatics and hydrogen bonding determine intermolecular interactions between polyphosphoinositides, J. Am. Chem. Soc, 2008, 130(28), 9025–9030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christian DA, et al. , Spotted vesicles, striped micelles and Janus assemblies induced by ligand binding, Nat. Mater, 2009, 8(10), 843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ellenbroek WG, et al. , Divalent Cation-Dependent Formation of Electrostatic PIP2 Clusters in Lipid Monolayers, Biophys. J, 2011, 101(9), 2178–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang YH, et al. , Divalent cation-induced cluster formation by polyphosphoinositides in model membranes, J. Am. Chem. Soc, 2012, 134(7), 3387–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kapus A and Janmey P, Plasma membrane-cortical cytoskeleton interactions: a cell biology approach with biophysical considerations, Compr. Physiol, 2013, 3(3), 1231–1281. [DOI] [PubMed] [Google Scholar]
- 34.van den Bogaart G, et al. , Membrane protein sequestering by ionic protein-lipid interactions, Nature, 2011, 479(7374), 552–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, et al. , Platelets lacking PIP5KIgamma have normal integrin activation but impaired cytoskeletal-membrane integrity and adhesion, Blood, 2013, 121(14), 2743–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki A, et al. , RhoA is essential for maintaining normal megakaryocyte ploidy and platelet generation, PLoS One, 2013, 8(7), e69315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Min SH and Abrams CS, Regulation of platelet plug formation by phosphoinositide metabolism, Blood, 2013, 122(8), 1358–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lian L, et al. , Loss of pleckstrin defines a novel pathway for PKC-mediated exocytosis, Blood, 2009, 113(15), 3577–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, et al. , Loss of PIP5KIbeta demonstrates that PIP5KI isoform-specific PIP2 synthesis is required for IP3 formation, Proc. Natl. Acad. Sci. U. S. A, 2008, 105(37), 14064–14069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, et al. , Loss of PIP5KIgamma, unlike other PIP5KI isoforms, impairs the integrity of the membrane cytoskeleton in murine megakaryocytes, J. Clin. Invest, 2008, 118(2), 812–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janmey PA, Bucki R and Radhakrishnan R, Regulation of actin assembly by PI(4,5)P2 and other inositol phospholipids: An update on possible mechanisms, Biochem. Biophys. Res. Commun, 2018, 506(2), 307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bucki R, et al. , Lateral distribution of phosphatidylinositol 4,5-bisphosphate in membranes regulates formin- and ARP2/3-mediated actin nucleation, J. Biol. Chem, 2019, 294(12), 4704–4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLaughlin S, et al. , PIP2 AND PROTEINS: Interactions, Organization, and Information Flow, Annu. Rev. Biophys. Biomol. Struct, 2002, 31(1), 151–175. [DOI] [PubMed] [Google Scholar]
- 44.Lemmon MA, Membrane recognition by phospholipid-binding domains, Nat. Rev. Mol. Cell Biol, 2008, 9(2), 99–111. [DOI] [PubMed] [Google Scholar]
- 45.Slochower DR, et al. , Counterion-mediated pattern formation in membranes containing anionic lipids, Adv. Colloid Interface Sci, 2014, 208, 177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janmey PA, Bucki R and Radhakrishnan R, Regulation of actin assembly by PI(4,5)P2 and other inositol phospholipids: An update on possible mechanisms, Biochem. Biophys. Res. Commun, 2018, 506(2), 307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwik J, et al. , Membrane cholesterol, lateral mobility, and the phosphatidylinositol 4,5-bisphosphate-dependent organization of cell actin, Proc. Natl. Acad. Sci. U. S. A, 2003, 100(24), 13964–13969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Papayannopoulos V, et al. , A polybasic motif allows NWASP to act as a sensor of PIP(2) density, Mol. Cell, 2005, 17(2), 181–191. [DOI] [PubMed] [Google Scholar]
- 49.Pike LJ and Miller JM, Cholesterol depletion delocalizes phosphatidylinositol bisphosphate and inhibits hormone-stimulated phosphatidylinositol turnover, J. Biol. Chem, 1998, 273(35), 22298–22304. [DOI] [PubMed] [Google Scholar]
- 50.Hong Z, et al. , How cholesterol regulates endothelial biomechanics, Front. Physiol, 2012, 3, 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lemmon MA, Pleckstrin homology (PH) domains and phosphoinositides, Biochem. Soc. Symp, 2007, 74, 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Narayan K and Lemmon MA, Determining selectivity of phosphoinositide-binding domains, Methods, 2006, 39(2), 122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lemmon MA, Pleckstrin homology domains: two halves make a hole?, Cell, 2005, 120(5), 574–576. [DOI] [PubMed] [Google Scholar]
- 54.Lemmon MA, Pleckstrin homology domains: not just for phosphoinositides, Biochem. Soc. Trans, 2004, 32(Pt 5), 707–711. [DOI] [PubMed] [Google Scholar]
- 55.Lemmon MA, Ferguson KM and Abrams CS, Pleckstrin homology domains and the cytoskeleton, FEBS Lett, 2002, 513(1), 71–76. [DOI] [PubMed] [Google Scholar]
- 56.Kutateladze TG, Translation of the phosphoinositide code by PI effectors, Nat. Chem. Biol, 2010, 6(7), 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moravcevic K, Oxley CL and Lemmon MA, Conditional peripheral membrane proteins: facing up to limited specificity, Structure, 2012, 20(1), 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bethoney KA, et al. , A possible effector role for the pleckstrin homology (PH) domain of dynamin, Proc. Natl. Acad. Sci. U. S. A, 2009, 106(32), 13359–13364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bucki R, et al. , Lateral distribution of phosphatidylinositol 4,5-bisphosphate in membranes regulates formin and ARP2/3-mediated actin nucleation, J. Biol. Chem, 2019, 294(12), 4704–4722, DOI: 10.1074/jbc.RA118.005552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramalingam N, et al. , Phospholipids regulate localization and activity of mDia1 formin, Eur. J. Cell Biol, 2010, 89(10), 723–732. [DOI] [PubMed] [Google Scholar]
- 61.Fiser A and Sali A, Modeller: generation and refinement of homology-based protein structure models, Methods Enzymol, 2003, 374, 461–491. [DOI] [PubMed] [Google Scholar]
- 62.Kim DE, Chivian D and Baker D, Protein structure prediction and analysis using the Robetta server, Nucleic Acids Res, 2004, 32(Web Server issue), W526–W531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Humphrey W, Dalke A and Schulten K, VMD: visual molecular dynamics, J. Mol. Graphics, 1996, 14(1), 33–38, 27–28. [DOI] [PubMed] [Google Scholar]
- 64.van Paridon PA, et al. , Polyphosphoinositides undergo charge neutralization in the physiological pH range: a 31PNMR study, Biochim. Biophys. Acta, Lipids Lipid Metab, 1986, 877(1), 216–219. [DOI] [PubMed] [Google Scholar]
- 65.Graber ZT, et al. , Phosphatidylinositol-4,5-bisphosphate ionization and domain formation in the presence of lipids with hydrogen bond donor capabilities, Chem. Phys. Lipids, 2012, 165(6), 696–704. [DOI] [PubMed] [Google Scholar]
- 66.Slochower DR, et al. , Quantum and all-atom molecular dynamics simulations of protonation and divalent ion binding to phosphatidylinositol 4,5-bisphosphate (PIP2), J. Phys. Chem. B, 2013, 117(28), 8322–8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kooijman EE, et al. , Ionization properties of phosphatidylinositol polyphosphates in mixed model membranes, Biochemistry, 2009, 48(40), 9360–9371. [DOI] [PubMed] [Google Scholar]
- 68.Bjelkmar P, et al. , Implementation of the CHARMM Force Field in GROMACS: Analysis of Protein Stability Effects from Correction Maps, Virtual Interaction Sites, and Water Models, J. Chem. Theory Comput, 2010, 6(2), 459–466. [DOI] [PubMed] [Google Scholar]
- 69.Van Der Spoel D, et al. , GROMACS: fast, flexible, and free, J. Comput. Chem, 2005, 26(16), 1701–1718. [DOI] [PubMed] [Google Scholar]
- 70.Pastor RW and Mackerell AD Jr., Development of the CHARMM Force Field for Lipids, J. Phys. Chem. Lett, 2011, 2(13), 1526–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Veatch SL and Keller SL, Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol, Biophys. J, 2003, 85(5), 3074–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bucki R, et al. , Antibacterial activities of rhodamine B-conjugated gelsolin-derived peptides compared to those of the antimicrobial peptides cathelicidin LL37, magainin II, and melittin, Antimicrob. Agents Chemother, 2004, 48(5), 1526–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wen Y, Vogt VM and Feigenson GW, Multivalent Cation-Bridged PI(4,5)P2 Clusters Form at Very Low Concentrations, Biophys. J, 2018, 114(11), 2630–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bradley RP, et al. , Divalent cations bind to phosphoinositides to induce ion and isomer specific propensities for nano-cluster initiation in bilayer membranes, R. Soc. Open Sci, 2020, DOI: 10.1074/jbc.RA118.005552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Veatch SL and Keller SL, Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol, Biophys. J, 2003, 85(5), 3074–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dill KA, Molecular driving forces, Garland Science, New York, 2003. [Google Scholar]
- 77.Senju Y, et al. , Mechanistic principles underlying regulation of the actin cytoskeleton by phosphoinositides, Proc. Natl. Acad. Sci. U. S. A, 2017, 114(43), E8977–E8986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


