Abstract
Friedreich ataxia (FRDA) is a progressive neurodegenerative disease with developmental features caused by a genetic deficiency of frataxin, a small, nuclear-encoded mitochondrial protein. Frataxin deficiency leads to impairment of iron–sulphur cluster synthesis, and consequently, ATP production abnormalities. Based on the involvement of such processes in FRDA, initial pathophysiological hypotheses focused on reactive oxygen species (ROS) production as a key component of the mechanism. With further study, a variety of other events appear to be involved, including abnormalities of mitochondrially related metabolism and dysfunction in mitochondrial biogenesis. Consequently, present therapies focus not only on free radical damage, but also on control of metabolic abnormalities and correction of mitochondrial biogenesis. Understanding the multitude of abnormalities in FRDA thus offers possibilities for treatment of this disorder.
Keywords: ataxia, biogenesis, reacrive oxygen specie
Friedreich ataxia (FRDA) is an autosomal recessive, neurodegenerative disorder that affects roughly 1 in every 50–100000 people in the United States. FRDA was first described in 1863 as a disease that is primarily early onset, associated with progressive limb and gait ataxia, absent tendon reflexes from the legs, axonal sensory neuropathy, dysarthria, muscle weakness, spasticity in the lower limbs, and loss of position and vibration sense [1–4] (Table 1). Neurodegeneration occurs early in the large proprioceptive sensory neurones of the dorsal root ganglia (DRG) and their axons in the posterior columns, with later atrophy of the corticospinal and spinocerebellar tracts of the spinal cord and the dentate nucleus in the cerebellum [5–9]. There is also loss of pancreatic islet cells and hypertrophic cardiomyopathy, which is the most common cause of death amongst FRDA patients. Patients can also develop scoliosis (curvature of the spine), pes cavus (fixed plantar foot flexion; severely high-arched feet), hearing loss (from auditory neuropathy), and vision loss (from optic neuropathy) [9–12]. In addition, fatigue is a dominating symptom amongst people with FRDA.
Table 1. Clinical features of FRDA.
| System | Pathology | Clinical result |
|---|---|---|
| Neurological | Degeneration of large sensory neurones – proprioception | Loss of balance and coordination |
| Loss of deep tendon reflexes | ||
| Degeneration of spinocerebellar tracts (dorsal) | Loss of balance and coordination | |
| Degeneration of dentate nucleus of the cerebellum | Loss of balance and coordination | |
| Dysarthria (slurred speech) | ||
| Eye movement abnormalities (modest) | ||
| Degeneration of corticospinal tracts | Spasticity, pyramidal weakness | |
| Visual | Degeneration of retinal ganglion cells | Optic neuropathy |
| Auditory | Degeneration of auditory nerve | Auditory neuropathy |
| Cardiac | Hypertrophic cardiomyopathy, with early hypertrophy, later fibrosis | ECG abnormalities |
| Arrhythmias | ||
| Progressive heart failure | ||
| Endocrine | Loss of pancreatic islet cells | Diabetes mellitus |
| Increased insulin resistance | Diabetes mellitus | |
| Orthopedic | Scoliosis | |
| Pes cavus (fixed plantar foot flexion; high arched feet) | ||
Abbreviation: ECG, electrocardiogram.
FRDA results from decreased levels of functional frataxin protein, coded by the FXN gene on chromosome 9 [13,14]. Such decreases in frataxin levels are caused by guanine-adenine-adenine (GAA) trinucleotide repeats within intron 1 of the FXN gene in the vast majority of abnormal alleles. In patients carrying two expanded alleles (96%) in FRDA patients, the length of the allele with the shorter GAA expansion inversely correlates with frataxin levels, age of onset, and rate of disease progression; longer alleles result in earlier onset and faster progression [15–18]. A subset of FRDA patients have GAA expansion in one chromosome and a point mutation in the FXN exon in the other chromosome [19–22]. Most point mutations lead to absence of frataxin production by alterations in the start codon, RNA splice sites, or in residues needed for protein folding. Other mutations do not lower protein levels but instead appear to disrupt the function of frataxin.
Expanded GAA repeats may form unusual triplex structures, disrupting RNA polymerase and preventing transcription elongation [23]. In addition, epigenetic mechanisms decrease frataxin expression as regions flanking GAA repeat expansion exhibit marks of condensed heterochromatin. There is also increased methylation of specific CpG sites, reduction in histone H3 and H4 acetylation levels, and increased histone H3 lysine 9 (H3K9) trimethylation in FRDA lymphoblasts, peripheral blood, brain, and heart [24–28]. Overall, this leads to a decrease in frataxin mRNA synthesis and a decrease (but not absence) in frataxin protein in people with FRDA [29–32]. As the phenotype of FRDA in subjects with point mutations altering frataxin production or stability is almost identical with those with GAA repeats, the clinical syndrome largely if not entirely reflects the loss of frataxin protein rather than the effects on frataxin mRNA levels.
Frataxin protein structure, function, and role in metabolism
FRDA patients’ peripheral tissues typically have less than 10% of the frataxin levels exhibited by unaffected people, and the level of frataxin inversely correlates with disease severity [29–32]. The FXN gene contains seven exons (exons 1–4, 5a, 5b and 6), with exons 4 and 5a being the most conserved across species [33]. Frataxin mRNA is translated by cytoplasmic ribosomes and translocated to the mitochondria based on an N-terminal mitochondrial localization sequence. Upon entry into the mitochondria, frataxin undergoes a two-step proteolytic cleavage by mitochondria processing peptidase (MPP) to generate the mature protein [34–36]. The mature protein forms a twisted, six-stranded β-antiparallel sheet, flanked by N- and C-terminal α helices (α1 and α2) [37]. The negatively charged residues on the helical plane may bind iron, while the uncharged residues on the surface β sheet can lead to protein–protein interactions [38].
Frataxin functions in iron metabolism, iron storage, and iron–sulphur cluster biosynthesis, with resultant effects on many downstream events [39–43] (Table 2). A conserved primary Fe2+-binding site, with a dissociation constant within the micromolar range (3–55 μM), is contained in residues of the acidic ridge in the first α helix. In addition to iron binding, frataxin interacts with mitochondrial aconitase, ferrochelatase, and proteins of the mitochondrial Fe–S cluster synthesis pathway [44]. Iron and Fe–S clusters are essential for metabolic processes including electron transport, DNA synthesis, both redox and non-redox reactions, as well as other cellular functions [45,46]. Iron–sulphur containing proteins play a crucial role in cellular respiration and ATP production; therefore, decreased activity should significantly impair mitochondrial function. Frataxin’s role in iron–sulphur cluster biogenesis makes it essential for enzymatic activity of Fe–S containing aconitase and respiratory chain complexes. Consequently, decreased frataxin levels result in decreased aconitase activity in cell culture models, in vivo, and in heart tissues and biopsies of FRDA patients [47,48]. These effects on key enzymes of energy production lead to a failure of ATP production in FRDA, as observed in humans in muscle spectroscopy [49–51]. This may represent one of the more important pathophysiological events in FRDA, as it is clearly observable in human muscle in FRDA, and is readily linked to one of the most important symptoms of FRDA, fatigue.
Table 2. Selected cellular functions of frataxin.
| Protein | Function |
|---|---|
| Isu1/Nfs1 | Scaffold proteins for Fe–S biogenesis. Frataxin controls iron entry and sulphur production through activation of cysteine desulphurization |
| Aconitase | FXN facilitates and stabilizes transfer of Fe group to Aconitase to convert it into its active form |
| Ferrochelatase | FXN meditates iron delivery to Ferrochelatase in heme synthesis |
| Succinate dehydrogenase | FXN regulates entry of electrons into Complex II of electron transport chain |
| ATP synthase | FXN regulates entry of electrons into Complex II of electron transport chain. Reduced FXN expression is correlated to a reduction in ATP |
| Pyruvate dehydrogenase | Pyruvate dehydrogenase subunit E3 may exhibit proteolytic activity capable of cleaving FXN under certain conditions |
| p38 | FXN deficiency may alter p38 mitogen-activated protein kinase signaling |
| Nrf2 | FXN deficiency impairs Nrf2 translocation to the nucleus |
| Nitric oxide | NO increases as a result of FXN deficiency. This increase is related to the increase in ROS due to iron accumulation. NO increases as a protective effect from Fe-mediated oxidative stress |
| PGC1α | PGCα is the master regulator of mitochondrial biogenesis. FXN deficiency results in dysregulation of PGC1α. This is tissue dependent but is down-regulated in most cell types |
| PDK1 | Frataxin deficiency triggers the activation of PDK1 through increasing phosphorylation levels of S241 and may deactivate pyruvate dehydrogenase and decrease cell metabolism |
| Iron uptake, import, and export protein | Frataxin deficiency causes increased expression of transferrin receptor 1 and mitochondrial iron importer mitoferrin-2, and decreased expression of ferroportin1, contributing to increased iron accumulation in mitochondria |
Abbreviations: Nrf2, nuclear factor E2-related factor 2, PGC1α, peroxisome proliferator-activated receptor γ coactivator 1-α.
Additionally, frataxin deficiency may secondarily affect enzymes of intermediary metabolism. In addition to direct effects on iron–sulphur cluster-containing enzymes, specific cellular and mitochondrial enzymes are regulated through frataxin level or the resultant effects on ATP levels. For example, while FRDA patients have normal pyruvate dehydrogenase (PDH) activity in most tissues [52], under certain conditions, including mitochondrial acidification, the dehydrogenase subunit (E3) of PDH exhibits proteolytic activity that is capable of cleaving frataxin [53]. Although PDH is likely not the only enzyme controlled by frataxin levels, it provides an example of how enzyme-specific regulation at the intersection of multiple mitochondrial metabolic pathways could control cellular phenotype through alteration of metabolism. FRDA patient platelets exhibit significantly decreased acetyl Co-A (Ac-CoA) synthesized through glycolysis than healthy control platelets [54,55] while creating substantially more Ac-CoA and HMG-CoA from palmitate. This emphasizes how the collection of changes in Fe–S containing enzymes alter flux through specific pathways. Recent evidence additionally suggests that frataxin deficiency may alter p38 kinase signaling, providing further evidence of a role for frataxin in signaling and metabolism [56]. Thus, the alterations in Fe–S containing and other enzymes provide a manner for specific frataxin-related changes in metabolism, which may have deleterious effects on cells.
Frataxin deficiency and mitochondrial dysfunction
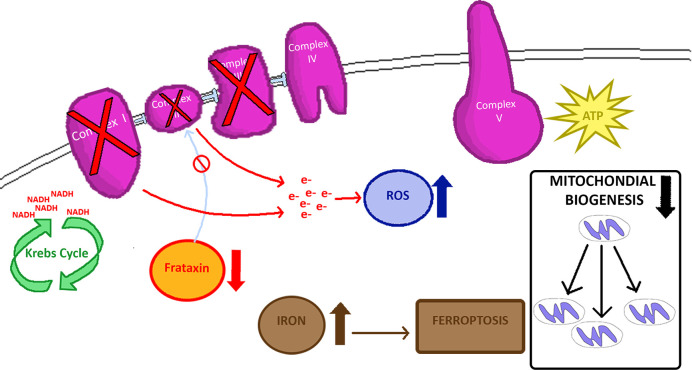
Frataxin overexpression demonstrates this protein’s crucial role in mitochondrial energy conversion and oxidative phosphorylation (OXPHOS), as well as regulation of the Krebs cycle [57] (Figure 1). Frataxin directly interacts with Complex II subunits, suggesting it directly supports the electron transport chain by providing Fe–S complexes [58–60]. Endomyocardial biopsies of FRDA patients exhibit decreased Complexes I, II, and III activity [61], and FRDA mouse models demonstrate mitochondrial biogenesis impairment and OXPHOS dysfunction in respiratory chain complexes I, II, and IV in cerebellum [62].
Figure 1. Mitochondrial features of FRDA.
Frataxin deficiency leads to loss of Fe–S groups in Complexes I, II, III with downstream ROS production and other downstream events.
Frataxin deficiency is also linked to mitochondrial dysfunction through iron accumulation and production of reactive oxygen species (ROS). Although produced throughout the cell, 90% of ROS result from mitochondrial respiration. During the transfer of electrons from the mitochondrial respiratory chain to molecular oxygen (O2) in OXPHOS, a small percentage of electrons will leak, resulting in the production of ROS, such as hydroxyl (HO−) and hydrogen peroxide (H2O2) [63–65]. This leak mainly occurs at Complexes I and II [66]; however, when ROS levels rise too high, oxidative damage, also termed as oxidative stress, can occur in the cell, especially in mitochondria. Oxidative stress damages proteins and DNA, especially mtDNA, as mtDNA lacks the protection from histones and the complex nuclear repair system [66]. Oxidative stress induces apoptosis by opening the mitochondrial permeability transition pore, and has been implicated in a number of neurodegenerative diseases, such as Parkinson’s disease (PD), Alzheimer’s disease (AD), amyotrophic lateral sclerosis, and multiple sclerosis [67,68].
ROS production occurs in multiple models of FRDA [69–73]. In certain Drosophila models with induced frataxin deficiency, H2O2-scavenging enzymes ameliorate features of oxidative stress and restore both ROS-sensitive mitochondrial enzymes and aconitase activities to normal levels. These findings implicate H2O2 as a pathogenic mediator of ROS production in FRDA and suggest that H2O2-scavenging molecules could play a therapeutic role in treating the disease [64]. In fibroblasts from patients with FRDA, treatment with iron-containing compounds or hydrogen peroxide leads to oxidative stress, activation of caspase 3, and apoptosis [74–76]. Analogous results have been identified across many cell types, and treatment with many proposed antioxidant-based therapies restores the healthy phenotypes [77,78]. Consequently, oxidant-induced cell death remains an area of interest for possible FRDA therapies.
One proposed component of increased ROS sensitivity in FRDA patient cells is the accumulation of mitochondrial iron [79–84]. Based on Fenton chemistry, mitochondrial iron accumulation has the potential to dramatically increase susceptibility to ROS [84]. However, FRDA involves iron maldistribution more than iron overload; cells behave as if they are depleted of iron cytosolically while simultaneously having a mitochondrial iron overload [85–87]. Systemic iron indices such as ferritin levels are normal to low in most FRDA patient tissues, except for the heart, where ferritin excess is noted at autopsy [88]. This raises the possibility that the direct effect of iron in FRDA may be tissue-specific.
The components of ROS production and iron overload are combined in a paradigm of cell death referred to as ferroptosis. Ferroptosis is a form of iron-dependent, oxidation-mediated, programmed cell death implicated in a variety of pathological processes, including neurotoxicity, neuroinflammation, and neurodegenerative diseases such as PD, AD, and ischemic stroke [89–92]. Ferroptosis may share some of the same downstream signaling pathways as apoptosis, but this form of cell death differs from classical apoptosis, and the mechanisms that underlie ferroptosis match many of the abnormal findings of FRDA [89–92]. Upon induction of ferroptosis, there is a lack of morphological or biochemical features of apoptosis, such as chromatin condensation and nuclear shrinkage [89,93]. Additionally, there is no cleavage-mediated activation of caspase 3, and caspase inhibitors do not protect against ferroptosis [89]. Oxidative stress releases iron from ferritin in a redox active form, induces lipid peroxidation, particularly of polyunsaturated fatty acids, and leads to accumulation of lipid-based ROS [89,93,94]. Accumulation of lipid peroxidation products and ROS derived from iron metabolism triggers ferroptosis as a response to these harmful metabolic events [92]. Ferroptosis may also be triggered following depletion of intracellular reduced-glutathione (GSH) levels, further leading to increased cellular availability of iron as a ferroptosis catalyst [91].
In addition to ROS generation, ferroptosis is associated with the loss of mitochondrial integrity [89–92]. EM shows cells treated with ferroptosis inducers exhibit obvious changes in mitochondrial morphology [89]. Investigators have found that a protein originally characterized during pro-apoptotic signaling, BID, translocates to the mitochondria during ferroptotic signaling. BID can act as a sensor of oxidative stress in an iron-dependent manner and its translocation to mitochondria mediates the loss of mitochondrial integrity and function [90]. Induced ferroptosis in neurones leads to loss of mitochondrial membrane potential, increased mitochondrial fragmentation, reduced ATP levels, and permeabilization of the outer mitochondrial membrane [90]. Distinct morphological alterations are also apparent, including decreased mitochondrial size, condensed mitochondrial membranes, reduction in mitochondrial cristae, and outer mitochondrial membrane rupture [90–92].
Lipid peroxidation, elevated ROS generation, GSH depletion, and increased iron availability are all pathogenic alterations found in many neurologic diseases, and interestingly, they are also common features of ferroptosis [91]. The dysregulated iron metabolism of FRDA suggests that ferroptosis may also play a role in cell death in FRDA.
ROS production is difficult to demonstrate in humans with FRDA. Although some studies find elevated urinary oxidative stress biomarker levels, specifically the isoprostanes dihydroguanosine and malondialdehyde, isoprostanes are not elevated in FRDA and only a single study has found abnormalities in DNA oxidation [95–99]. Moreover, confounding factors, including the overwhelming use of antioxidant supplements by FRDA patients and the relative inactivity of such patients leading to a lack of ongoing OXPHOS and an absence of ROS, result in further challenges to demonstrate ROS accumulation in FRDA patients [98]. It is also possible, however, that significantly increased ROS production is not continually occurring in FRDA. Not all cell death in animal models of FRDA is associated with detectable ROS production or iron accumulation. In mouse models of FRDA, cell death occurs without detectable accumulation of ROS or iron [100]. Such data provide evidence that in these models, other events such as loss of specific enzymatic activities, failure of ATP production, or other processes may be sufficient to induce cell death in FRDA without inducing ferroptotic pathways.
Failure of nuclear factor E2-related factor 2 and mitochondrial biogenesis pathways
Nuclear factor E2-related factor 2 (Nrf2) is a transcription factor that regulates cellular antioxidant response under oxidative stress conditions. Under normal conditions, the interaction between Nrf2 and Keap1 leads to the degradation of Nrf2 through the ubiquitin-proteasome pathway [101]. Typical oxidative stress conditions inhibit the interaction between Nrf2 and Keap1, leading to increased levels of active Nrf2 [102,103]; however, Nrf2 is degraded in FRDA patients and laboratory models, which is unexpected in an environment of oxidative stress [102,104].
In the presence of ROS, Nrf2 induces the expression of ROS-response antioxidant genes such as heme oxygenase-1 (HO-1), NAD(P)H quinone oxidoreductase 1 (NQO1), Cu/Zn and Mn-superoxide dismutases (SOD 1,2), glutathione synthetic enzymes, and others by binding to the antioxidant response element (ARE) on nuclear DNA, including an ARE site within FXN [104,105]. In a healthy state, oxidative stress causes Nrf2 translocation to the nucleus, resulting in the expression of antioxidant genes to protect cells from damage. In FRDA models, Nrf2 translocation to the nucleus is compromised in response to oxidative insults, thus leading to reduced expression of antioxidant genes such as NQO1 and SOD-1,2 [101,106]. This may increase vulnerability to oxidative stress and lead to a cascade of oxidant-induced damage in neurons and other cell types. Interestingly, studies to find compounds that induce Nrf2 lead to identifying compounds that up-regulate frataxin gene expression [101]. Thus, Nrf2 expression correlates with frataxin expression. Nrf2 also regulates synthesis of GSH, a tripeptide antioxidant that moderates ROS production and ferroptosis [107]. In FRDA, the altered homeostasis between reduced and oxidized glutathione, increases cells’ susceptibility to oxidative stress [62,104,107].
In addition to increased ROS production and paradoxical loss of Nrf-2, frataxin deficiency is also associated with other components of mitochondrial dysfunction in both FRDA patients and animal models. Mitochondrial biogenesis deficits appear in multiple models of FRDA, including human lymphocytes and mouse models such as the frataxin knockin/knockout (KIKO) mouse [108–110]. Interestingly, the levels of PGC-1a, the master regulator of mitochondrial biogenesis, are significantly decreased in cerebellar homogenates of KIKO mice, even when mice are behaviorally asymptomatic [62]. This suggests early impairment of mitochondrial biogenesis pathways as a potential mediator of mitochondrial loss and dysfunction in FRDA. Parallel dysfunction in downstream genes of the entire PGC-1a/NRF1/Tfam pathway in KIKO mouse cerebellum confirms mitochondrial biogenesis impairment as an early event in this model.
Other markers of mitochondrial number fusion are also altered in FRDA. The mitochondrial chaperone, glucose-related protein 75 (GRP75), which physically interacts with frataxin, and the mitochondrial fusion protein mitofusin-1 (MFN1), are lower in cerebellar homogenates of FRDA KIKO mice [62]. Human FRDA fibroblast and PBMCs also show decreased GRP75 levels [111,112]. Furthermore, in KIKO mice, this decrease is associated with a long-term deficit in mitochondrial number, suggesting that in some brain regions, FRDA may give rise not only to abnormal mitochondria, but also lead to decrease in numbers of mitochondria [62]. Although the correlation between GRP75 levels and the severity of FRDA remains to be determined, GRP75 reduction should result in further decreases in frataxin levels and iron–sulphur cluster biogenesis and may thus impact mitochondrial function. Alternatively, GRP75 reduction could potentially lead to mtDNA damage, thereby contributing to the progression of FRDA.
Clinical trials and therapeutic strategies
At present, there is no cure or effective treatment for FRDA [113]. Current strategies aim to increase frataxin expression or target downstream pathways affected secondary to frataxin deficiency [114–120]. High-throughput screening with different cellular models is also being used to search for new drugs. Even when restorative therapies for frataxin achieve success, mitochondria-based therapies are still likely to be useful covering the deficiencies in restoration of frataxin levels.
Antioxidants and OXPHOS
Frataxin deficiency potentiates cellular damage from oxidative stress, suggesting that antioxidants might present a therapeutic approach for FRDA. For example, idebenone is a short-chain Coenzyme Q10 (CoQ10) analog that acts as an antioxidant by protecting membrane lipids from peroxidation and stimulating OXPHOS and ATP production by carrying electrons from Complexes I and II to Complex III in the electron transport chain [121]. Initial enthusiasm for idebenone was highly based on its ability to protect respiratory Complex II from iron inactivation and decreased lipoperoxidation; however, neither idebenone nor other similar agents have proven effective in double-blind trials as compared with placebo [122–125]. Other antioxidants like CoQ10 with vitamin E, and VP20629 have also shown no benefit in clinical trials [126].
Iron chelating strategy
As the pathogenesis of FRDA involves an imbalance in the intracellular accumulation of iron, with mitochondrial accumulation and relative cytosolic depletion, targetted iron chelation could be beneficial in restoring a healthy iron balance. Deferiprone, an iron chelator that localizes to the mitochondria, rapidly distributes in the CNS, crossing membranes, and can penetrate mitochondria to remove excess iron [127]. Deferiprone has a lower affinity for iron than other iron chelators (pFe3+ log stability constant of 19.9 compared with deferoxamine (26.6) and less tendency to cause overall iron depletion, leading to an improved safety profile over other iron chelators in patients with low iron overload [128]. It restores mitochondrial redox potential, reduces ROS, and increases aconitase activity, without affecting frataxin levels [129–133]. The drug is typically well tolerated and can be administered orally. However, exacerbation of tremor occurred at high doses and the risk of agranulocytosis remains a threat of deferiprone treatment [133].
Abbreviations
- Ac-CoA
acetyl Co-A
- AD
Alzheimer’s disease
- ARE
antioxidant response element
- CoQ10
coenzyme Q10
- FRDA
Friedreich ataxia
- GAA
guanine-adenine-adenine
- GRP
glucose related protein
- GSH
glutathione
- KIKO
knockin/knockout
- Nrf2
nuclear factor E2-related factor 2
- NQO1
NAD(P)H quinone oxidoreductase 1
- OXPHOS
oxidative phosphorylation
- PD
Parkinson’s disease
- PDH
pyruvate dehydrogenase
- ROS
reactive oxygen species
- SOD
superoxide dismutase
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
David R. Lynch receives funding from the National Institutes of Health, the Food and Drug Administration, the Friedreich’s Ataxia Research Alliance, and the Muscular Dystrophy Association. Hong Lin receives funding from the Friedreich’s Ataxia Research Alliance. Elizabeth Mercado Ayon receives funding from The California State University Sally Casanova Pre-Doctoral Program.
Author contribution
E.C., J.J., Y.N.D., E.M.-A., N.W., M.Z., E.M., A.S., H.L., and D.R.L. all wrote portions of the first draft and provided critical review.
References
- 1.Friedreich N.U. (1863) ber degenerative Atrophie der spinalen Hinterstrange. Virchows Arch. Pathol. Anat. 26, 433–459 10.1007/BF01878006 [DOI] [Google Scholar]
- 2.Delatycki M., Williamson R. and Forrest S. (2000) Friedreich ataxia: an overview. J. Med. Genet. 37, 1–8 10.1136/jmg.37.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harding A.E. (1981) Friedreich’s ataxia: a clinical and genetic study of 90 families with an analysis of early diagnostic criteria and intrafamilial clustering of clinical features. Brain 104, 589–620 10.1093/brain/104.3.589 [DOI] [PubMed] [Google Scholar]
- 4.Lynch D.R., Farmer J.M., Balcer L.J. and Wilson R.B. (2002) Friedreich ataxia: effects of genetic understanding on clinical evaluation and therapy. Arch. Neurol. 59, 743–747 10.1001/archneur.59.5.743 [DOI] [PubMed] [Google Scholar]
- 5.Koeppen A.H., Becker A.B., Qian J., Gelman B.B. and Mazurkiewicz J.E. (2017) Friedreich ataxia: developmental failure of the dorsal root entry zone. J. Neuropathol. Exp. Neurol. 76, 969–977 10.1093/jnen/nlx087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koeppen A.H., Becker A.B., Qian J. and Feustel P.J. (2017) Friedreich ataxia: hypoplasia of spinal cord and dorsal root ganglia. J. Neuropathol. Exp. Neurol. 76, 101–108 [DOI] [PubMed] [Google Scholar]
- 7.Koeppen A.H. and Mazurkiewicz J.E. (2013) Friedreich ataxia: neuropathology revised. J. Neuropathol. Exp. Neurol. 72, 78–90 10.1097/NEN.0b013e31827e5762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koeppen A.H., Davis A.N. and Morral J.A. (2011) The cerebellar component of Friedreich’s ataxia. Acta Neuropathol. 122, 323–330 10.1007/s00401-011-0844-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rance G., Corben L., Barker E., Carew P., Chisari D. and Rogers M. (2010) Auditory perception in individuals with Friedreich’s ataxia. Audiol. Neurotol. 15, 229–240 10.1159/000255341 [DOI] [PubMed] [Google Scholar]
- 10.Montermini L., Richter A., Morgan K., Justice C.M., Julien D., Castellotti B. et al. (1997) Phenotypic variability in Friedreich ataxia: role of the associated GAA triplet repeat expansion. Ann. Neurol. 41, 675–682 10.1002/ana.410410518 [DOI] [PubMed] [Google Scholar]
- 11.Parkinson M.H., Boesch S., Nachbauer W., Mariotti C. and Giunti P. (2013) Clinical features of Friedreich’s ataxia: classical and atypical phenotypes. J. Neurochem. 1, 103–117 10.1111/jnc.12317 [DOI] [PubMed] [Google Scholar]
- 12.Seyer L.A., Galetta K., Wilson J., Sakai R., Perlman S., Mathews K. et al. (2013) Analysis of the visual system in Friedreich ataxia. J. Neurol. 260, 2362–2369 10.1007/s00415-013-6978-z [DOI] [PubMed] [Google Scholar]
- 13.Dürr A., Cossee M., Agid Y., Campuzano V., Mignard C., Penet C. et al. (1996) Clinical and genetic abnormalities in patients with Friedreich’s ataxia. N. Engl. J. Med. 335, 1169–1175 10.1056/NEJM199610173351601 [DOI] [PubMed] [Google Scholar]
- 14.Campuzano V., Montermini L., Moltò M.D., Pianese L., Cossée M., Cavalcanti F. et al. (1996) Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271, 1423–1427 10.1126/science.271.5254.1423 [DOI] [PubMed] [Google Scholar]
- 15.Sharma R., De Biase I., Gómez M., Delatycki M.B., Ashizawa T. and Bidichandani S.I. (2004) Friedreich ataxia in carriers of unstable borderline GAA triplet-repeat alleles. Ann. Neurol. 56, 898–901 10.1002/ana.20333 [DOI] [PubMed] [Google Scholar]
- 16.Patel M., Isaacs C., Seyer L., Brigatti K., Gelbard S., Strawser C. et al. (2016) Progression of Friedrich ataxia: quantitative characterization over five years. Ann. Clin. Transl. Neurol. 3, 684–694 10.1002/acn3.332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filla A., De Michele G., Cavalcanti F., Pianese L., Monticelli A., Campanella G. et al. (1996) The relationship between trinucleotide (GAA) repeat length and clinical features in Friedreich ataxia. Am. J. Hum. Genet. 59, 554–560 [PMC free article] [PubMed] [Google Scholar]
- 18.Bidichandani S., Delatycki M. (2014, Friedreich ataxia, GeneReviews, https://www.ncbi.nlm.nih.gov/books/NBK1281/ [Google Scholar]
- 19.Becker A.B., Qian J., Gelman B.B., Yang M., Bauer P. and Koeppen A.H. (2017) Heart and nervous system pathology in compound heterozygous Friedreich ataxia. J. Neuropathol. Exp. Neurol. 76, 665–675 10.1093/jnen/nlx047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galea C.A., Huq A., Lockhart P.J., Tai G., Corben L.A., Yiu E.M. et al. (2016) Compound heterozygous FXN mutations and clinical outcome in friedreich ataxia. Ann. Neurol. 79, 485–495 10.1002/ana.24595 [DOI] [PubMed] [Google Scholar]
- 21.McCormack M.L., Guttmann R.P., Schumann M., Farmer J.M., Stolle C.A., Campuzano V. et al. (2000) Frataxin point mutations in two patients with Friedreich’s ataxia and unusual clinical features. J. Neurol. Neurosurg. Psychiatry 68, 661–664 10.1136/jnnp.68.5.661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cossée M., Dürr A., Schmitt M., Dahl N., Trouillas P., Allinson P. et al. (1999) Friedreich’s ataxia: point mutations and clinical presentation of compound heterozygotes. Ann. Neurol. 45, 200–206 [DOI] [PubMed] [Google Scholar]
- 23.Li Y., Lu Y., Polak U., Lin K., Shen J., Farmer J. et al. (2015) Expanded GAA repeats impede transcription elongation through the FXN gene and induce transcriptional silencing that is restricted to the FXN locus. Hum. Mol. Genet. 24, 6932–6943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al-Mahdawi S., Pinto R., Ismail O., Varshney D., Lymperi S., Sandi C. et al. (2008) The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum. Mol. Genet. 17, 735–746 10.1093/hmg/ddm346 [DOI] [PubMed] [Google Scholar]
- 25.Castaldo I., Pinelli M., Monticelli A., Acquaviva F., Giacchetti M., Filla A. et al. (2008) DNA methylation in intron 1 of the frataxin gene is related to GAA repeat length and age of onset in Friedreich ataxia patients. J. Med. Genet. 45, 808–812 10.1136/jmg.2008.058594 [DOI] [PubMed] [Google Scholar]
- 26.Greene E., Mahishi L., Entezam A., Kumari D. and Usdin K. (2007) Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucleic Acids Res. 35, 3383–3390 10.1093/nar/gkm271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herman D., Jenssen K., Burnett R., Soragni E., Perlman S.L. and Gottesfeld J.M. (2006) Histone deacetylase inhibitors reverse gene silencing in Friedreich’s ataxia. Nat. Chem. Biol. 2, 551–558 10.1038/nchembio815 [DOI] [PubMed] [Google Scholar]
- 28.Evans-Galea M.V., Carrodus N., Rowley S.M., Corben L.A., Tai G., Saffery R. et al. (2012) FXN methylation predicts expression and clinical outcome in Friedreich ataxia. Ann. Neurol. 71, 487–497 10.1002/ana.22671 [DOI] [PubMed] [Google Scholar]
- 29.Campuzano V., Montermini L., Lutz Y., Cova L., Hindelang C., Jiralerspong S. et al. (1997) Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum. Mol. Genet. 6, 1771–1780 10.1093/hmg/6.11.1771 [DOI] [PubMed] [Google Scholar]
- 30.Lazaropulos M., Dong Y., Clark E., Greeley N.R., Seyer L.A., Brigatti K.W. et al. (2015) Measurement of frataxin levels in peripheral tissue in Friedreich ataxia: analysis using repeated measures. Ann. Clin. Transl. Neurol. 2, 831–842 10.1002/acn3.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deutsch E.C., Santani A.B., Perlman S.L., Farmer J.M., Stolle C.A., Marusich M.F. et al. (2010) A rapid, noninvasive immunoassay for frataxin: utility in assessment of Friedreich ataxia. Mol. Genet. Metab. 101, 238–245 10.1016/j.ymgme.2010.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deutsch E.C., Oglesbee D., Greeley N.R. and Lynch D.R. (2014) Usefulness of frataxin immunoassays for the diagnosis of Friedreich ataxia. J. Neurol. Neurosurg. Psychiatry 85, 994–1002 10.1136/jnnp-2013-306788 [DOI] [PubMed] [Google Scholar]
- 33.Abruzzo P.M., Marini M., Bolotta A., Malisardi G., Manfredini S., Ghezzo A. et al. (2013) Frataxin mRNA isoforms in FRDA patients and normal subjects: effect of tocotrienol supplementation. Biomed. Res. Int. 2013, 276808 10.1155/2013/276808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Branda S.S., Cavadini P., Adamec J., Kalousek F., Taroni F. and Isaya G. (1999) Yeast and human frataxin are processed to mature form in two sequential steps by the mitochondrial processing peptidase. J. Biol. Chem. 274, 22763–22769 10.1074/jbc.274.32.22763 [DOI] [PubMed] [Google Scholar]
- 35.Cavadini P., Adamec J., Taroni F., Gakh O. and Isaya G. (2000) Two-step processing of human frataxin by mitochondrial processing peptidase. Precursor and intermediate forms are cleaved at different rates. J. Biol. Chem. 275, 41469–41475 10.1074/jbc.M006539200 [DOI] [PubMed] [Google Scholar]
- 36.Koutnikova H., Campuzano V. and Koenig M. (1998) Maturation of wild-type and mutated frataxin by the mitochondrial processing peptidase. Hum. Mol. Genet. 7, 1485–1489 10.1093/hmg/7.9.1485 [DOI] [PubMed] [Google Scholar]
- 37.Dhe-Paganon S., Shigeta R., Chi Y., Ristow M. and Shoelson S.E. (2000) Crystal structure of human frataxin. J. Biol. Chem. 275, 30753–30756 10.1074/jbc.C000407200 [DOI] [PubMed] [Google Scholar]
- 38.Bencze K.Z., Yoon T., Millan-Pacheco C., Bradley P.B., Pastor N., Cowan J.A. et al. (2007) Human frataxin: iron and ferrochelatase binding surface. Chem. Commun. 14, 1798–1800 10.1039/B703195E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bulteau A.L., O’neill H.A., Kennedy M.C., Ikeda-Saito M., Isaya G. and Szweda L.I. (2004) Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science 305, 242–245 10.1126/science.1098991 [DOI] [PubMed] [Google Scholar]
- 40.Adinolfi S., Trifuoggi M., Politou A.S., Martin S. and Pastore A. (2002) A structural approach to understanding the iron-binding properties of phylogenetically different frataxins. Hum. Mol. Genet. 11, 1865–1877 10.1093/hmg/11.16.1865 [DOI] [PubMed] [Google Scholar]
- 41.Cavadini P., Gellera C., Patel P. and Isaya G. (2009) Human frataxin maintains mitochondrial iron homeostasis in Saccharomyces cerevisiae. Hum. Mol. Genet. 9, 2523–2530 10.1093/hmg/9.17.2523 [DOI] [PubMed] [Google Scholar]
- 42.Gerber J., Muhlenhoff U. and Lill R. (2003) An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep. 4, 906–911 10.1038/sj.embor.embor918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon T. and Cowan J.A. (2004) Frataxin-mediated iron delivery to ferrochelatase in the final step of heme biosynthesis. J. Biol. Chem. 279, 25943–25946 10.1074/jbc.C400107200 [DOI] [PubMed] [Google Scholar]
- 44.Martelli A. and Puccio H. (2014) Dysregulation of cellular iron metabolism in Friedreich ataxia: from primary iron-sulfur cluster deficit to mitochondria iron accumulation. Front. Pharmacol. 5, 130 10.3389/fphar.2014.00130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gonzalez-Cabo P. and Palau F. (2013) Mitochondrial pathophysiology in Friedreich’s ataxia. J. Neurochem. 126, 53–64 10.1111/jnc.12303 [DOI] [PubMed] [Google Scholar]
- 46.Pastore A. and Puccio H. (2013) Frataxin: a protein in search for a function. J. Neurochem. 126, 43–52 10.1111/jnc.12220 . [DOI] [PubMed] [Google Scholar]
- 47.Rotig A., de Lonlay P., Chretien D., Foury F., Koenig M., Sidi D. et al. (1997) Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat. Genet. 17, 215–217 10.1038/ng1097-215 [DOI] [PubMed] [Google Scholar]
- 48.Walden W.E. (2002) From bacteria to mitochondria: aconitase yields surprises. Proc. Natl. Acad. Sci. U.S.A. 99, 4138–4140 10.1073/pnas.082108799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lynch D.R., Lech G., Farmer J.M., Balcer L.J., Bank W., Chance B. et al. (2002) Near infrared muscle spectroscopy in patients with Friedreich’s ataxia. Muscle Nerve 25, 664–673 10.1002/mus.10077 [DOI] [PubMed] [Google Scholar]
- 50.DeBrosse C., Nanga R.P., Wilson N., D’Aquilla K., Elliott M., Hariharan H. et al. (2016) Muscle oxidative phosphorylation quantitation using creatine chemical exchange saturation transfer (CrCEST) MRI in mitochondrial disorders. JCI Insight 1, e88207 10.1172/jci.insight.88207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lodi R., Cooper J.M., Bradley J.L., Manners D., Styles P., Taylor D.J. et al. (1999) Deficit of in vivo mitochondrial ATP production in patients with Friedreich ataxia. Proc. Natl. Acad. Sci. U.S.A. 96, 11492–11495 10.1073/pnas.96.20.11492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Evans O.B. (1983) Human muscle pyruvate dehydrogenase activity. Neurology 33, 51–56 10.1212/WNL.33.1.51 [DOI] [PubMed] [Google Scholar]
- 53.Vaubel R.A., Rustin P. and Isaya G. (2011) Mutations in the dimer interface of dihydrolipoamide dehydrogenase promote site-specific oxidative damages in yeast and human cells. J. Biol. Chem. 286, 40232–40245 10.1074/jbc.M111.274415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Basu S.S., Deutsch E.C., Schmaier A.A., Lynch D.R. and Blair I.A. (2013) Human platelets as a platform to monitor metabolic biomarkers using stable isotopes and LC-MS.. Bioanalysis 5, 3009–3021 10.4155/bio.13.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Worth A.J., Basu S.S., Deutsch E.C., Hwang W.T., Snyder N.W., Lynch D.R. et al. (2015) Stable isotopes and LC-MS for monitoring metabolic disturbances in Friedreich’s ataxia platelets. Bioanalysis 7, 1843–1855 10.4155/bio.15.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cotticelli M.G., Xia S., Kaur A., Lin D., Wang Y., Ruff E. et al. (2018) Identification of p38 MAPK as a novel therapeutic target for Friedreich’s ataxia. Sci. Rep. 8, 5007 10.1038/s41598-018-23168-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ristow M., Pfister M., Yee A., Schubert M., Michael L., Zhang C.Y. et al. (2000) Frataxin activates mitochondrial energy conversion and oxidative phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 97, 12239–12243 10.1073/pnas.220403797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vazquez-Manrique R.P., Gonzalez-Cabo P., Ros S., Aziz H., Baylis H.A. and Palau F. (2006) Reduction of Caenorhabditis elegans frataxin increases sensitivity to oxidative stress, reduces lifespan, and causes lethality in a mitochondrial complex II mutant. FASEB J. 20, 172–174 10.1096/fj.05-4212fje [DOI] [PubMed] [Google Scholar]
- 59.Yoon T. and Cowan J.A. (2003) Iron-sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. J. Am. Chem. Soc. 125, 6078–6084 10.1021/ja027967i [DOI] [PubMed] [Google Scholar]
- 60.Schmucker S., Martelli A., Colin F., Page A., Wattenhofer-Donzé M., Reutenauer L. et al. (2011) Mammalian frataxin: an essential function for cellular viability through an interaction with a preformed ISCU/NFS1/ISD11 iron-sulfur assembly complex. PLoS ONE 6, e16199 10.1371/journal.pone.0016199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gonzalez-Cabo P., Vazquez-Manrique R.P., Garcia-Gimeno M.A., Sanz P. and Palau F. (2005) Frataxin interacts functionally with mitochondrial electron transport chain proteins. Hum. Mol. Genet. 14, 2091–2098 10.1093/hmg/ddi214 [DOI] [PubMed] [Google Scholar]
- 62.Lin H., Magrane J., Rattelle A., Stepanova A., Galkin A., Clark E.M. et al. (2017) Early cerebellar deficits in mitochondrial biogenesis and respiratory chain complexes in the KIKO mouse model of Friedreich ataxia. Dis. Model Mech. 10, 1343–1352 10.1242/dmm.030502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anderson P.R., Kirby K., Orr W.C., Hilliker A.J. and Phillips J.P. (2008) Hydrogen peroxide scavenging rescues frataxin deficiency in a Drosophila model of Friedreich’s ataxia. Proc. Natl. Acad. Sci. U.S.A. 105, 611–616 10.1073/pnas.0709691105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chiang S., Kalinowski D.S., Jansson P.J., Richardson D.R. and Huang M.L. (2018) Mitochondrial dysfunction in the neuro-degenerative and cardio-degenerative disease, Friedreich’s ataxia. Neurochem. Int. 117, 35–48 10.1016/j.neuint.2017.08.002 [DOI] [PubMed] [Google Scholar]
- 65.Cadenas E. and Davies K.J. (2000) Mitochondrial free radical generation, oxidative stress, and aging. Free Radic. Biol. Med. 29, 222–230 10.1016/S0891-5849(00)00317-8 [DOI] [PubMed] [Google Scholar]
- 66.Grimm A. and Eckert A. (2017) Brain aging and neurodegeneration: from a mitochondrial point of view. J. Neurochem. 143, 418–431 10.1111/jnc.14037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Skulachev V.P. (1996) Why are mitochondria involved in apoptosis? Permeability transition pores and apoptosis as selective mechanisms to eliminate superoxide-producing mitochondria and cell. FEBS Lett. 397, 7–10 10.1016/0014-5793(96)00989-1 [DOI] [PubMed] [Google Scholar]
- 68.Calabrese V., Lodi R., Tonon C., D’Agata V., Sapienza M., Scapagnini G. et al. (2005) Oxidative stress, mitochondrial dysfunction and cellular stress response in Friedreich’s ataxia. J. Neurol. Sci. 233, 145–162 10.1016/j.jns.2005.03.012 [DOI] [PubMed] [Google Scholar]
- 69.Al-Mahdawi S., Pinto R.M., Varshney D., Lawrence L., Lowrie M.B., Hughes S. et al. (2006) GAA repeat expansion mutation mouse models of Friedreich ataxia exhibit oxidative stress leading to progressive neuronal and cardiac pathology. Genomics 88, 580–590 10.1016/j.ygeno.2006.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Calmels N., Schmucker S., Wattenhofer-Donze M., Martelli A., Vaucamps N., Reutenauer L. et al. (2009) The first cellular models based on frataxin missense mutations that reproduce spontaneously the defects associated with Friedreich ataxia. PLoS ONE 4, e6379 10.1371/journal.pone.0006379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Herrero E., Ros J., Bellı G. and Cabiscol E. (2008) Redox control and oxidative stress in yeast cells. Biochim. Biophys. Acta 1780, 1217–1235 10.1016/j.bbagen.2007.12.004 [DOI] [PubMed] [Google Scholar]
- 72.Jauslin M.L., Meier T., Smith R.A. and Murphy M.P. (2003) Mitochondria-targeted antioxidants protect Friedreich ataxia fibroblasts from endogenous oxidative stress more effectively than untargeted antioxidants. FASEB J. 17, 1972–1974 10.1096/fj.03-0240fje [DOI] [PubMed] [Google Scholar]
- 73.Puccio H., Simon D., Cossee M., Criqui-Filipe P., Tiziano F., Melki J. et al. (2001) Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat. Genet. 27, 181–186 10.1038/84818 [DOI] [PubMed] [Google Scholar]
- 74.Tan G., Chen L.S., Lonnerdal B., Gellera C., Taroni F.A. and Cortopassi G.A. (2001) Frataxin expression rescues mitochondrial dysfunctions in FRDA cells. Hum. Mol. Genet. 10, 2099–2107 10.1093/hmg/10.19.2099 [DOI] [PubMed] [Google Scholar]
- 75.Cotticelli M.G., Rasmussen L., Kushner N.L., McKellip S., Sosa M.I., Manouvakhova A. et al. (2012) Primary and secondary drug screening assays for Friedreich ataxia. J. Biomol. Screen 17, 303–313 10.1177/1087057111427949 [DOI] [PubMed] [Google Scholar]
- 76.Calmels N., Schmucker S., Wattenhofer-Donzé M., Martelli A., Vaucamps N., Reutenauer L. et al. (2009) The first cellular models based on frataxin missense mutations that reproduce spontaneously the defects associated with Friedreich ataxia. PLoS ONE 4, e6379 10.1371/journal.pone.0006379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Madathil M.M., Khdour O.M., Jaruvangsanti J. and Hecht S.M. (2012) Synthesis and biological activities of N-(3-carboxylpropyl)-5-amino-2-hydroxy-3-tridecyl-1,4-benzoquinone and analogues. J. Nat. Prod. 75, 2209–2215 10.1021/np3007099 [DOI] [PubMed] [Google Scholar]
- 78.Armstrong J.S., Khdour O. and Hecht S.M. (2010) Does oxidative stress contribute to the pathology of Friedreich’s ataxia? A radical question. FASEB J. 24, 2152–2163 10.1096/fj.09-143222 [DOI] [PubMed] [Google Scholar]
- 79.Campanella A., Rovelli E., Santambrogio P., Cozzi A., Taroni F. and Levi S. (2009) Mitochondrial ferritin limits oxidative damage regulating mitochondrial iron availability: hypothesis for a protective role in Friedreich ataxia. Hum. Mol. Genet. 18, 1–11 10.1093/hmg/ddn308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Colin F., Martelli A., Clemancey M., Latour J.M., Gambarelli S., Zeppieri L. et al. (2013) Mammalian frataxin controls sulfur production and iron entry during de novo Fe4S4 cluster assembly. J. Am. Chem. Soc. 135, 733–740 10.1021/ja308736e [DOI] [PubMed] [Google Scholar]
- 81.Huang M.L., Becker E.M., Whitnall M., Suryo Rahmanto Y., Ponka P. and Richardson D.R. (2009) Elucidation of the mechanism of mitochondrial iron loading in Friedreich’s ataxia by analysis of a mouse mutant. Proc. Natl. Acad. Sci. U.S.A. 106, 16381–16386 10.1073/pnas.0906784106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramirez R.L., Qian J., Santambrogio P., Levi S. and Koeppen A.H. (2012) Relation of cytosolic iron excess to cardiomyopathy of Friedreich’s ataxia. Am. J. Cardiol. 110, 1820–1827 10.1016/j.amjcard.2012.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Richardson D.R., Lane D.J., Becker E.M., Huang M.L., Whitnall M., Suryo Rahmanto Y. et al. (2010) Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol. Proc. Natl. Acad. Sci. U.S.A. 107, 10775–10782 10.1073/pnas.0912925107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vaubel R.A. and Isaya G. (2013) Iron-sulfur cluster synthesis, iron homeostasis and oxidative stress in Friedreich ataxia. Mol. Cell. Neurosci. 55, 50–61 10.1016/j.mcn.2012.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lobmayr L., Brooks D.G. and Wilson R.B. (2005) Increased IRP1 activity in Friedreich ataxia. Gene. 354, 157–161 [DOI] [PubMed] [Google Scholar]
- 86.Wilson R.B., Lynch D.R., Farmer J.M., Brooks D.G. and Fischbeck K.H. (2000) Increased serum transferrin receptor concentrations in Friedreich ataxia. Ann. Neurol. 47, 659–661 [DOI] [PubMed] [Google Scholar]
- 87.Wilson R.B., Lynch D.R. and Fischbeck K.H. (1998) Normal serum iron and ferritin concentrations in patients with Friedreich’s ataxia. Ann. Neurol. 44, 132–134 10.1002/ana.410440121 [DOI] [PubMed] [Google Scholar]
- 88.Ramirez R.L., Qian J., Santambrogio P., Levi S. and Koeppen A.H. (2012) Relation of cytosolic iron excess to cardiomyopathy of Friedreich’s ataxia. Am. J. Cardiol. 110, 1820–1827 10.1016/j.amjcard.2012.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lewerenz J., Ates G., Methner A., Conrad M. and Maher P. (2018) Oxytosis/ferroptosis—(Re-) emerging roles for oxidative stress-dependent non-apoptotic cell death in diseases of the central nervous system. Front. Neurosci. 12, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Neitemeier S., Jelinek A., Laino V., Hoffmann L., Eisenbach I. and Eying R. (2017) BID links ferroptosis to mitochondrial cell death pathways. Redox Biol. 12, 558–570 10.1016/j.redox.2017.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guiney S.J., Adlard P.A., Bush A.I., Finkelstein D.I. and Ayton S. (2017) Ferroptosis and cell death mechanisms in Parkinson’s disease. Neurochem. Int. 104, 34–48 10.1016/j.neuint.2017.01.004 [DOI] [PubMed] [Google Scholar]
- 92.Xie Y., Hou W., Song X., Yu Y., Huang J., Sun X. et al. (2016) Ferroptosis: process and function. Cell Death Differ. 23, 369–379 10.1038/cdd.2015.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fricker M., Tolkovsky A.M., Borutaite V., Coleman M. and Brown G.C. (2018) Neuronal cell death. Physiol. Rev. 98, 813–880 10.1152/physrev.00011.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang W.S. and Stockwell B.R. (2016) Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 26, 165–176 10.1016/j.tcb.2015.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Emond M., Lepage G., Vanasse M. and Pandolfo M. (2000) Increased levels of plasma malondialdehyde in Friedreich ataxia. Neurology 55, 1752–1753 10.1212/WNL.55.11.1752 [DOI] [PubMed] [Google Scholar]
- 96.Schulz J., Dehmer T., Schöls L., Mende H., Hardt C. and Vorgerd M. (2000) Oxidative stress in patients with Friedreich ataxia. Neurology 55, 1719–1721 10.1212/WNL.55.11.1719 [DOI] [PubMed] [Google Scholar]
- 97.Myers L.M., Lynch D.R., Farmer J.M., Friedman L.S., Lawson J.A. and Wilson R.B. (2008) Urinary isoprostanes in Friedreich ataxia: lack of correlation with disease features. Mov. Disord. 23, 1920–1922 10.1002/mds.22038 [DOI] [PubMed] [Google Scholar]
- 98.Myers L., Farmer J.M., Wilson R.B., Friedman L., Tsou A. and Perlman S.L. (2008) Antioxidant use in Friedreich ataxia. J. Neurol. Sci. 267, 174–176 10.1016/j.jns.2007.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Haugen A.C., Di Prospero N.A., Parker J.S., Fannin R.D., Chou J. and Meyer J.N. (2010) Altered gene expression and DNA damage in peripheral blood cells from Friedreich’s ataxia patients: cellular model of pathology. PLoS Genet. 6, e1000812 10.1371/journal.pgen.1000812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Puccio H., Simon D., Cossée M., Criqui-Filipe P., Tiziano F., Melki J. et al. (2001) Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat. Genet. 27, 181–186 10.1038/84818 [DOI] [PubMed] [Google Scholar]
- 101.Kobayashi A., Kang M.I., Okawa H., Ohtsuji M., Zenke Y., Chiba T. et al. (2004) Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 24, 7130–7139 10.1128/MCB.24.16.7130-7139.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shan Y., Schoenfeld R.A., Hayashi G., Napoli E., Akiyama T., Iodi Carstens M. et al. (2013) Frataxin deficiency leads to defects in expression of antioxidants and Nrf2 expression in dorsal root ganglia of the Friedreich’s ataxia YG8R mouse model. Antioxid. Redox Signal. 19, 1481–1493 10.1089/ars.2012.4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sahdeo S., Scott B.D., McMackin M.Z., Jasoliya M., Brown B., Wulff H. et al. (2014) Dyclonine rescues frataxin deficiency in animal models and buccal cells of patients with Friedreich’s ataxia. Hum. Mol. Genet. 23, 6848–6862 10.1093/hmg/ddu408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Anzovino A., Chiang S., Brown B.E., Hawkins C.L., Richardson D.R. and Huang M.L. (2017) Molecular alterations in a mouse cardiac model of friedreich ataxia: an impaired Nrf2 response mediated via upregulation of Keap1 and activation of the Gsk3beta axis. Am. J. Pathol. 187, 2858–2875 10.1016/j.ajpath.2017.08.021 [DOI] [PubMed] [Google Scholar]
- 105.Petrillo S., Piermarini E., Pastore A., Vasco G., Schirinzi T., Carrozzo R. et al. (2017) Nrf2-inducers counteract neurodegeneration in frataxin-silenced motor neurons: disclosing new therapeutic targets for Friedreich’s ataxia. Int. J. Mol. Sci. 18, 10.3390/ijms18102173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.D’Oria V., Petrini S., Travaglini L., Priori C., Piermarini E., Petrillo S. et al. (2013) Frataxin deficiency leads to reduced expression and impaired translocation of NF-E2-related factor (Nrf2) in cultured motor neurons. Int. J. Mol. Sci. 14, 7853–7865 10.3390/ijms14047853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Carletti B., Piermarini E., Tozzi G., Travaglini L., Torraco A., Pastore A. et al. (2014) Frataxin silencing inactivates mitochondrial Complex I in NSC34 motoneuronal cells and alters glutathione homeostasis. Int. J. Mol. Sci. 15, 5789–5806 10.3390/ijms15045789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Coppola G., Marmolino D., Lu D., Wang Q., Cnop M., Rai M. et al. (2009) Functional genomic analysis of frataxin deficiency reveals tissue-specific alterations and identifies the PPAR gamma pathway as a therapeutic target in Friedreich’s ataxia. Hum. Mol. Genet. 18, 2452–2461 10.1093/hmg/ddp183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Marmolino D., Manto M., Acquaviva F., Vergara P., Ravella A., Monticelli A. et al. (2010) PGC-1alpha down-regulation affects the antioxidant response in Friedreich’s ataxia. PLoS ONE 5, e10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jasoliya M.J., McMackin M.Z., Henderson C.K., Perlman S.L. and Cortopassi G.A. (2017) Frataxin deficiency impairs mitochondrial biogenesis in cells, mice and humans. Hum. Mol. Genet. 26, 2627–2633 10.1093/hmg/ddx141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shan Y., Napoli E. and Cortopassi G. (2007) Mitochondrial frataxin interacts with ISD11 of the NFS1/ISCU complex and multiple mitochondrial chaperones. Hum. Mol. Genet. 16, 929–941 10.1093/hmg/ddm038 [DOI] [PubMed] [Google Scholar]
- 112.Selak M.A., Lyver E., Micklow E., Deutsch E.C., Onder O., Selamoglu N. et al. (2011) Blood cells from Friedreich ataxia patients harbor frataxin deficiency without a loss of mitochondrial function. Mitochondrion 11, 342–350 10.1016/j.mito.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Strawser C.J., Schadt K.A. and Lynch D.R. (2014) Therapeutic approaches for the treatment of Friedreich’s ataxia. Expert Rev. Neurother. 14, 949–957 10.1586/14737175.2014.939173 [DOI] [PubMed] [Google Scholar]
- 114.Santoro A., Anjomani Virmouni S., Paradies E., Villalobos Coa V.L., Al-Mahdawi S. and Khoo M. (2018) Effect of diazoxide on Friedreich ataxia models. Hum. Mol. Genet. 27, 992–1001 10.1093/hmg/ddy016 [DOI] [PubMed] [Google Scholar]
- 115.Tomassini B., Arcuri G., Fortuni S., Villalobos Coa V.L., Al-Mahdawi S. and Khoo M. (2012) Interferon gamma upregulates frataxin and corrects the functional deficits in a Friedreich ataxia model. Hum. Mol. Genet. 21, 2855–2861 10.1093/hmg/dds110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mariotti C., Nachbauer W., Panzeri M., Poewe W., Taroni F. and Boesch S. (2013) Erythropoietin in Friedreich ataxia. J. Neurochem. 126, 80–87 10.1111/jnc.12301 [DOI] [PubMed] [Google Scholar]
- 117.Soragni E., Miao W., Iudicello M., Jacoby D., De Mercanti S., Clerico M. et al. (2014) Epigenetic therapy for Friedreich ataxia. Ann. Neurol. 76, 489–508 10.1002/ana.24260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gottesfeld J.M., Rusche J.R. and Pandolfo M. (2013) Increasing frataxin gene expression with histone deacetylase inhibitors as a therapeutic approach for Friedreich’s ataxia. J. Neurochem. 126, 147–154 10.1111/jnc.12302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Perdomini M., Belbellaa B., Monassier L., Reutenauer L., Messaddeq N., Cartier N. et al. (2014) Prevention and reversal of severe mitochondrial cardiomyopathy by gene therapy in a mouse model of Friedreich’s ataxia. Nat. Med. 20, 542–547 [DOI] [PubMed] [Google Scholar]
- 120.Piguet F., de Montigny C., Vaucamps N., Reutenauer L., Eisenmann A. and Puccio H. (2018) Rapid and complete reversal of sensory ataxia by gene therapy in a novel model of Friedreich ataxia. Mol. Ther. 26, 1940–1952 10.1016/j.ymthe.2018.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Di Prospero N.A., Baker A., Jeffries N. and Fischbeck K.H. (2007) Neurological effects of high-dose idebenone in patients with Friedreich’s ataxia: a randomised, placebo-controlled trial. Lancet Neurol. 6, 878–886 10.1016/S1474-4422(07)70220-X [DOI] [PubMed] [Google Scholar]
- 122.Meier T. and Buyse G. (2009) Idebenone: an emerging therapy for Friedreich ataxia. J. Neurol. 256, 25–30 10.1007/s00415-009-1005-0 [DOI] [PubMed] [Google Scholar]
- 123.Rustin P., von Kleist-Retzow J.C., Chantrel-Groussard K., Sidi D., Munnich A. and Rötig A. (1999) Effect of idebenone on cardiomyopathy in Friedreich’s ataxia: a preliminary study. Lancet 354, 477–479 10.1016/S0140-6736(99)01341-0 [DOI] [PubMed] [Google Scholar]
- 124.Meier T., Perlman S.L., Rummey C., Coppard N.J. and Lynch D.R. (2012) Assessment of neurological efficacy of idebenone in pediatric patients with Friedreich’s ataxia: data from a 6-month controlled study followed by a 12-month open-label extension study. J. Neurol. 259, 284–291 10.1007/s00415-011-6174-y [DOI] [PubMed] [Google Scholar]
- 125.Lynch D.R., Perlman S.L. and Meier T. (2010) A phase 3, double-blind, placebo-controlled trial of idebenone in Friedreich ataxia. Arch. Neurol. 67, 941–947 10.1001/archneurol.2010.168 [DOI] [PubMed] [Google Scholar]
- 126.Hart P.E., Lodi R., Rajagopalan B., Taylor D.J., Crilley J.G., Bradley J.L. et al. (2005) Antioxidant treatment of patients with Friedreich ataxia: four-year follow-up. Arch. Neurol. 62, 621–626 10.1001/archneur.62.4.621 [DOI] [PubMed] [Google Scholar]
- 127.Boddaert N., Le Quan Sang K.H., Rotig A., Leroy-Willig A., Gallet S., Brunelle F. et al. (2007) Selective iron chelation in Friedreich ataxia: biologic and clinical implications. Blood 110, 401–408 10.1182/blood-2006-12-065433 [DOI] [PubMed] [Google Scholar]
- 128.Daar S., Al-Khabori M.K., Al-Huneini M. I, Al-Hashim A. II and Al-Kemyani N. III (2016) Long-term iron chelation therapy with deferiprone in patients with thalassemia major and low iron load. Blood 128, 3626 [Google Scholar]
- 129.Goncalves S., Paupe V., Dassa E.P. and Rustin P. (2008) Deferiprone targets aconitase: implication for Friedreich’s ataxia treatment. BMC Neurol. 8, 20 10.1186/1471-2377-8-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kakhlon O., Manning H., Breuer W., Melamed-Book N., Lu C., Cortopassi G. et al. (2008) Cell functions impaired by frataxin deficiency are restored by drug mediated iron relocation. Blood 112, 5219–5227 10.1182/blood-2008-06-161919 [DOI] [PubMed] [Google Scholar]
- 131.Richardson D.R. (2003) Friedreich’s ataxia: iron chelators that target the mitochondrion as a therapeutic strategy? Expert Opin. Invest. Drugs 12, 235–245 10.1517/13543784.12.2.235 [DOI] [PubMed] [Google Scholar]
- 132.Sohn Y., Breuer W., Munnich A. and Cabantchik Z.I. (2008) Redistribution of accumulated cell iron: a modality of chelation with therapeutic implications. Blood 111, 1690–1699 10.1182/blood-2007-07-102335 [DOI] [PubMed] [Google Scholar]
- 133.Pandolfo M., Arpa J., Delatycki M.B., Le Quan Sang K.H., Mariotti C., Munnich A. et al. (2014) Deferiprone in Friedreich ataxia: a 6-month randomized controlled trial. Ann. Neurol. 76, 509–521 10.1002/ana.24248 [DOI] [PubMed] [Google Scholar]