Abstract
Introduction
Understanding the feeding behavior and host choice of sand flies provides valuable information on vector-host relationships and elucidates the epidemiological patterns of leishmaniasis transmission. Blood meal analysis studies are essential for estimating the efficiency of pathogen transmission, assessing the relative human disease risk, and assist in identifying the other potential hosts of leishmaniasis. In Sudan and most of East Africa, there are large remaining gaps in knowledge regarding the feeding habits of phlebotomine vectors. The study aimed to identify the blood meal sources and, therefore, the host preferences of the principal vectors Phlebotomus orientalis and Ph. papatasi in leishmaniasis endemic areas of eastern and central Sudan.
Materials and methods
Sand flies were collected from two endemic villages in eastern and central Sudan using CDC light traps and sticky traps. The phlebotomine sand flies were morphologically and then molecularly identified. The source of blood meal of the engorged females was determined using a multiplex PCR methodology and specific primers of cytochrome b gene of mitochondrial DNA for human, goat, cow, and dog. The detection of the Leishmania parasite was done using PCR.
Results
The total number of collected female phlebotomine sand flies was 180. Morphological identification revealed the abundance of Ph. orientalis 103 (57.2%), Ph. papatasi 42 (23.3%), Ph. bergeroti 31 (17.2%), Ph. rodhaini 2 (1.1%) and Ph. duboscqi 2 (1.1%) in the study sites. Out of the 180 collected, 31 (17%) were blood-fed flies. Three species were blood-fed and molecularly identified: Ph. papatasi (N = 7, 22.6%), Ph. bergeroti (N = 9, 26%), and Ph. orientalis (N = 15, 48.4%). Blood meal analysis revealed human DNA in two Ph. orientalis (6.4%), hence, the anthropophilic index was 13.3%.
Conclusions
Multiplex PCR protocol described here allowed the identification of blood meal sources of many vertebrate species simultaneously. The results indicate that wild-caught Ph. orientalis are anthropophilic in the study areas. Further studies on larger blood-fed sample size are required to validate the potential applications of this technique in designing, monitoring and evaluating control programs, particularly in investigating the potential non-human hosts of leishmaniasis.
Introduction
Phlebotomine sand flies (Diptera: Psychodidae: Phlebotominae) are the biological vectors of a group of diseases that includes leishmaniasis, human bartonellosis, and sand fly fever [1, 2]. Leishmaniases are group of diseases caused by protozoan parasites of the genus Leishmania (order: Kinetoplastida; family: Trypanosomatidae) [3]. The diseases are range from self-healing cutaneous leishmaniasis (CL) to disfiguring diffuse cutaneous/post-kala-azar dermal leishmaniasis (DCL/PKDL) and the fatal visceral leishmaniasis (VL, kala-azar) [4]. The diseases are epidemiologically complex, involving multiple vector species and reservoir hosts, and diverse transmission cycles [5]. In Sudan, CL has been endemic since 1910 [6], caused by Le. major and transmitted by Phlebotomus papatasi [7]. CL was endemic in western parts of Sudan before 1970, but after a major epidemic along the River Nile, the disease became endemic in many regions of the country [7, 8]. While in Sudan, VL is caused by Le. donovani and transmitted by Ph. orientalis. The major VL endemic areas in Sudan are in the eastern, central, and southern regions; several other areas of sporadic occurrence are scattered in Kordofan states and in the central and western parts of Darfur states [9]. Transmission of the disease takes place both in Acacia seyal (Taleh) and Balanites aegyptiaca (Higleeg) woodland and inside villages [10]. It is probable that both anthroponotic and zoonotic transmission of Le. donovani takes place in eastern Sudan [3].
Sand fly vectors transmit several etiological agents through feeding on a wide variety of hosts, such as humans, livestock, dogs, and chickens [11]. The blood meal is essential for egg development and various physiological processes, and sand flies can acquire or transmit pathogens by this means [12]. Detailed knowledge of the preferred vertebrate hosts and feeding behavior of sand fly vectors is considered to be a prerequisite for a successful prevention and control program implementation and evaluation of changes in human-vector contact during intervention programs [13].
Blood meal analysis of hematophagous arthropods is considered a practical approach to identifying their preferred hosts under natural conditions [14]. The anthropophilic index (percentage feeding on humans) is a vital component of the vectorial capacity of disease vectors [15–17], and knowledge of animal hosts is also crucial in identifying reservoirs of vector-borne zoonotic or enzootic pathogens [16].
Relatively, limited studies are available regarding blood meal analysis and host preference of different sand fly vectors, despite the variety of available techniques that are available. Methods used for blood meal analysis on sand flies are mostly derived from those used for mosquitoes [13]. However, many factors limit the use of this approach; sand flies are minute insects compared to mosquitoes and ingest less blood volume (0.3–0.6 μl per blood meal) and (2–6 μl per blood meal) respectively [13, 18, 19], and this reduces the active time period which to determine the blood meal source (24–48 hours) post blood meal ingestion [11, 19–22]. These difficulties impose critical challenges from disease ecology perspective and epidemiological assessment of disease transmission [23].
The successful typing of blood meals of wild-caught sand flies requires at least the rapid collection of engorged sand flies after obtaining blood meals. The blood-fed sand flies or their blood meals must be preserved appropriately to avoid degeneration of blood meal, and determination of the optimum concentration of blood meal extracted DNA used in PCR analysis [14, 24].
Most studies of blood meal sources of arthropod and sand fly vectors rely mostly on serological techniques; precipitin test, counter immune-electrophoresis, latex agglutination test and enzyme-linked immunosorbent assay (ELISA) [13, 25–27]. Although serological techniques are informative, they lack sensitivity, and they are also time-consuming [11, 17].
PCR-based methods can be considered as a convenient alternative for the identification of blood meals rather than serological techniques [28]. They can also be used for species confirmation, detection of infection by different pathogens, and population genetic studies, all on an individual specimen [14]. Several PCR-based methods have been used to identify blood meal sources of sand flies including one that targets the prepronociceptin (PNOC) gene [29–31], FTA-based technology [11], PCR-RFLP [32, 33]. Besides, real time PCR assays [34], multiplex PCR [35], barcoding PCR [23, 36, 37], PCR reverse-line blotting (RLB) and ELISA assays [17, 27, 28, 38–40].
The cytochrome b gene of mitochondrial DNA is conserved in all animals, and has distinct characteristics for each species, making it an ideal choice for identifying blood meal sources [41]. Using of cytochrome b gene is a powerful method for identification, due to the high copy number of mitochondrial gene and sufficient genetic variation at the primary sequence level among vertebrate taxa for reliable identification [14, 37]. Multiplex PCR is convenient as it immediately identifies species DNA, offering speed and cost-effectiveness to blood meal identification [42].
This study aimed to identify blood meal sources and host preferences among sand fly vectors (Ph. orientalis and Ph. papatasi) in endemic areas of Sudan using the multiplex PCR method.
Materials and methods
Study area
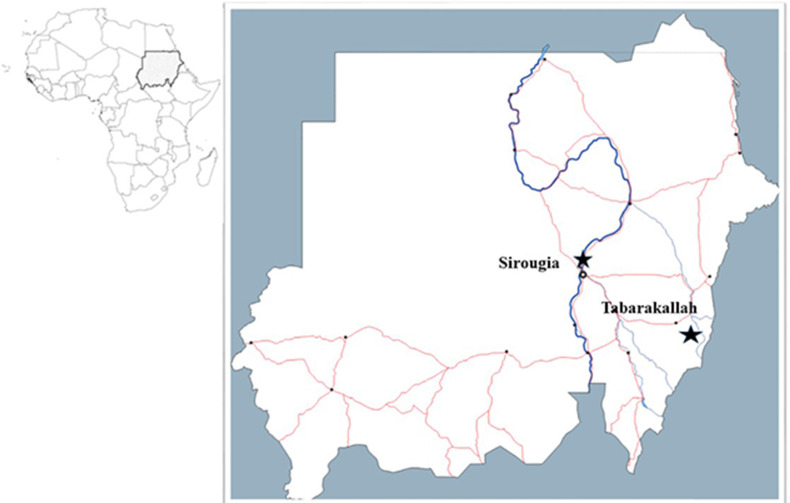
Study samples were collected from two endemic sites of leishmaniasis with different ecologies; Sirougia village (15°45`N, 32°15`E) and Tabarakallah village (13° 37`N, 36° 05`E) (Fig 1). The Sirougia village is a green rich irrigated site with many agricultural schemes on the east bank of the River Nile, approximately 30 km north Khartoum state [7, 8]. It is located in a flat land covered by alluvium of silty clay and sand deposited by the River Nile. The climate is semi-desert with two distinct seasons, rainy season (July-October) and dry season (November-June). Vegetation in the area is composed of desert scrub trees such as A. seyal and A. nilotica (Sunt). The inhabitants of the village live in homes made of clay layers or bricks, roofed with grass and mud or corrugated iron sheets. Several previous CL outbreaks have occurred in the Sirougia village, whilst some of the uncommon CL cases have been reported in the surrounding communities [6, 7].
Fig 1. Regional and local map of the study sites in Sudan.
The areas highlighted with black stars represent the study site villages: Sirougia in central Sudan and Tabarakallah in eastern Sudan.
Tabarakallah village is a known endemic site of VL [43] that lies near the Atbara River. The soil is mainly chromic vertisol soil (black cotton soil), the climate is temperate savannah with a rainy season (May-September) and dry season (October-April). The dominant trees there are A. seyal, B. aegyptiaca and A. senegal (Hashab). The people of the village lived in a typical African hut constructed of wood, bamboo, and grass [44, 45].
Sand fly collection
Sand flies were collected at night from homes and peri-domestic sites in the two villages using the CDC miniature light trap (Model 2836BQ, USA) and sticky traps [46]. Both traps were set up from 6:00 pm—6:00 am for five consecutive nights in the two villages, during March—April 2018 in accordance with the increased numbers of phlebotomine sand flies during dry season [3, 47]. The collected sand flies were sorted out, preserved in absolute ethanol and kept at -20°C for further analysis.
Ethical approval for this study was taken from Noguchi Memorial Institute for Medical Research-IRB, University of Ghana, Ghana (study no. 065/15-16) and informed consent was obtained from owners of homes in villages.
Morphological identification of sand fly vectors
Phlebotomus (blood-fed and unfed) sand flies collected from the wild were sorted out from other Sergentomyia species and then mounted individually in Puri’s media on glass slides, as described by [48]. Using sterilized fine forceps and dissecting micro-needles, the head containing the pharynx and cibarium, and the last four terminal segments of the abdomen were separated under the dissecting microscope and covered with coverslips. The rest of the blood-fed specimen was placed individually in 1.5 ml Eppendorf tubes containing absolute ethanol and labelled with a specific code that denoted the number assigned to the dissected parts mounted on the glass slide, and then stored at -20 ºC for subsequent molecular investigation.
Morphological identification of the wild-caught phlebotomine (blood-fed and unfed) sand fly was done under a binocular microscope at 40X following the keys of [49–51]. The primary morphological features used for the female sand fly identifications were the spermatheca, the pharynx, and the cibarial armature.
DNA extraction and molecular identification of sand fly vectors
The thorax and remaining part of the abdomen of each blood-fed female Phlebotomus specimen was used for genomic DNA extraction as described by [52], with some modifications. Briefly, tissues were first washed three times with distilled water to remove the traces of the ethanol. The washed samples were placed individually in 1.5μl Eppendorf tube and homogenized in 50μl grinding buffer (0.1mM NaCl, 0.1M Tris HCl pH 8.0) using a sterile glass pestle. The homogenates were incubated at 95°C for 30 min, kept at -20°C for 30 min and then centrifuged for 5 min at 14,000 rpm. The supernatants were transferred to fresh Eppendorf tubes, and 30μl of DNA-free water was added and stored at -20°C for further molecular investigations.
Species-specific primers, published by [53], were used for the identification of Ph. (Phlebotomus) ITS2 (rDNA) (Macrogen Inc. Korea); UN-F-JTS4: GCAGCTAACTGTGTGAAAT; UN-R-C1a (Ph. papatasi): CCTGGTTAGTTTCTTTTCCTCCGCT; PdubSSP (Ph. duboscqi): CCATAGCTCCAATGTTTTAC; PbergSSP (Ph. bergeroti): CGTGGTCCATGCAGTTTAAATG, yielding a PCR product of 500 bp (Ph. papatasi); 200 bp (Ph. duboscqi); 100 bp (Ph. bergeroti).
Amplification of the samples was done in a total of 25μl volume by adding 8μl DNA template, PCR water and 10pmol of each primer (UN-F-JTS4, UN-R-C1a, PdubSSP, and PbergSSP) to Maxime PCR premix kit (iNtRON Biotechnology Inc., South Korea) containing (5U/μl) i-Taq DNA polymerase, 2.5mM each of dNTPs, 10x reaction buffer, and 1x gel loading buffer. All PCR reactions were run on a thermocycler (SensoQuest, Germany). The PCR cycling program was as follows: an initial denaturation at 95°C for 2 min, 25 cycles of denaturation at 94°C for 30 second, annealing at 54°C for 30 second, extension at 72°C for 45 second and final extension at 72°C for 7 min. Each reaction included one negative PCR control (no DNA template) and positive controls of Ph. papatasi, Ph. duboscqi, and Ph. bergeroti DNA.
For Ph. orientalis samples, species-specific primers published by [54, 55] based on mtDNA Cytb were used; MM2F: TTTACTCTCTGCTATTCCTTATCTAGG and MM2R: TCTCAGATTTTTGAAATTAGAGGATTT (Macrogen Inc. Korea), yielding amplicon of 675 bp. Amplification of the samples was performed in a total of 25μl volume by adding 8μl DNA template, PCR water and 10pmol of each primer (MM2F and MM2R) to the Maxime PCR premix kit (iNtRON Biotechnology Inc., South Korea). The PCR cycling was modified as follows; an initial denaturation at 95°C for 2 min, 14 cycles of denaturation at 95°C for 30 second, annealing at 61.4°C decreased 0.5°C per cycle for 30 second, extension at 72°C for 70 second, 19 cycles of denaturation at 95°C for 30 second, annealing at 54.4°C for 30 second, extension at 72°C for 70 second and final extension at 72°C for 5 min. Each reaction included one negative PCR control and positive control of Ph. orientalis DNA.
Five μl of the PCR product was electrophoresed on a 1.5% agarose gel stained with Ethidium bromide [56]. A 100 bp DNA ladder was loaded for the determination of band size. The gel was run in 1x TBE buffer (Tris base, boric acid, EDTA pH 8.3) for one hour at 90V. The PCR products were observed under UV illumination and documented using the BDA gel image documentation system (Biometra Analytika Jena Company, Germany).
Identification of blood meal source using multiplex PCR
A set of primers that amplify segments of cytochrome b gene of vertebrate mtDNA were used following [32, 35]; UNREV1025A “universal”: GGTTGTCCTCCAATTCATGTTA; UNFOR403 “mammals”: TGAGGACAAATATCATTCTGAGG; Human 741F: GGCTTACTTCTCTTCATTCTCTCCT; Goat 894F: CCTAATCTTAGTACTTGTACCCTTCCTC; Cow121F: CATCGGCACAAATTTAGTCG; Dog 368F: GGAATTGTACTATTATTCGCAACCAT (Macrogen Inc. Korea), yielding PCR products of 623bp, 334bp, 132bp, 561bp, and 680bp respectively. PCR amplification was performed in a total of 25μl volume that comprised of 8μl DNA template, PCR water, and 10pmol of each primer (UNFOR403 and UNREV1025A; UNREV1025A, Human 741F, Goat 894F, Cow121F, and Dog 368F) added to the Maxime PCR premix kit (iNtRON Biotechnology Inc., South Korea). PCR reaction was run on a thermocycler (SensoQuest, Germany). The multiplex PCR cycling condition was modified from previous publications [32, 35] as follows: an initial denaturation at 95°C for 5 min, 35 cycles of denaturation at 95°C for 60 seconds, annealing at 58°C for 60 seconds, extension at 72°C for 60 seconds, and final extension at 72°C for 7 min. To each amplification test, one negative PCR control and positive controls of human, goat, cow, and dog DNA were included. The PCR products electrophoresed on 1.5% agarose gel and analyzed as described above.
Detection of Leishmania parasite in the blood-fed phlebotomine vectors
Leishmania DNA was detected among blood-fed sand fly vectors using the method of [57]. The primers 18S-LEISH forward: GCTGTGCAGGTTTGTTCCTG′3 and 18S-LEISH reverse: GGACGCACTAAACCCCTCAA (Macrogen Inc. Korea), were used to amplify DNA amplicon of 357bp within the 18S rRNA gene of Le. donovani. PCR was performed in 25μl reaction volume using Maxime PCR premix (iNtRON Biotechnology Inc., South Korea) according to the manufacturer’s instructions. Cycling conditions were done with minor modification from previous report [57]; an initial denaturation at 95°C for 2 min, 30 cycles of denaturation at 95°C for 30 second, annealing at 59.3°C for 30 seconds, extension at 72°C for 40 second, and a final extension at 72°C for 5 min. One negative PCR control and positive control of Le. donovani DNA were included in every PCR run. PCR products were electrophoresed on 1.5% agarose gel and analyzed as described above.
Results
The total number of collected phlebotomine (blood-fed and unfed) sand flies from Sirougia was 50 females, while in Tabarakallah it was 130 females. The morphological identification revealed 103 (57.2%) Ph. orientalis, 42 (23.3%) Ph. papatasi and 31 (17.2%) Ph. bergeroti were found in both Sirougia and Tabarakallah sites, while Ph. rodhaini 2 (1.1%) and Ph. duboscqi 2 (1.1%) were found only in Tabarakallah site (Table 1). Details of abundance of each species per site are reported in Table 1.
Table 1. Numbers and percentages of female phlebotomine sand fly vectors collected from two study sites in Sudan during 2018.
| Sand fly species | Sirougia village | Tabarakallah village | Total | ||
|---|---|---|---|---|---|
| Blood-fed | Unfed | Blood-fed | Unfed | ||
| Phlebotomus orientalis | 0 (0%) | 1 (2%) | 15 (11.5%) | 87 (66.9%) | 103 (57.2%) |
| Ph. rodhaini | 0 (0%) | 0 (0%) | 0 (0%) | 2 (1.5%) | 2 (1.1%) |
| Ph. papatasi | 4 (8%) | 21 (42%) | 3 (2.3%) | 14 (10.8%) | 42 (23.3%) |
| Ph. bergeroti | 5 (10%) | 19 (38%) | 4 (3.1%) | 3 (2.3%) | 31 (17.2%) |
| Ph. Duboscqi | 0 (0%) | 0 (0%) | 0 (0%) | 2 (1.5%) | 2 (1.1%) |
| Total | 9 (18%) | 41(82%) | 22 (16.9%) | 108 (83.1%) | 180 |
The total number of blood-fed phlebotomine females in both sites was 31 (17%) specimens. Out of the total blood-fed females, 9 (18%) were found on Sirougia site, while 22 (16.9%) were found on Tabarakallah site (Table 1).
Results of molecular identification of blood-fed specimens confirmed the presence of Ph. bergeroti (N = 5, 10%) and Ph. papatasi (N = 4, 8%) in Sirougia site, while in Tabarakallah, Ph. orientalis (N = 15, 11.5%) was confirmed in addition to Ph. papatasi (N = 3, 2.3%) and Ph. bergeroti (N = 4, 3.1%) (Table 1).
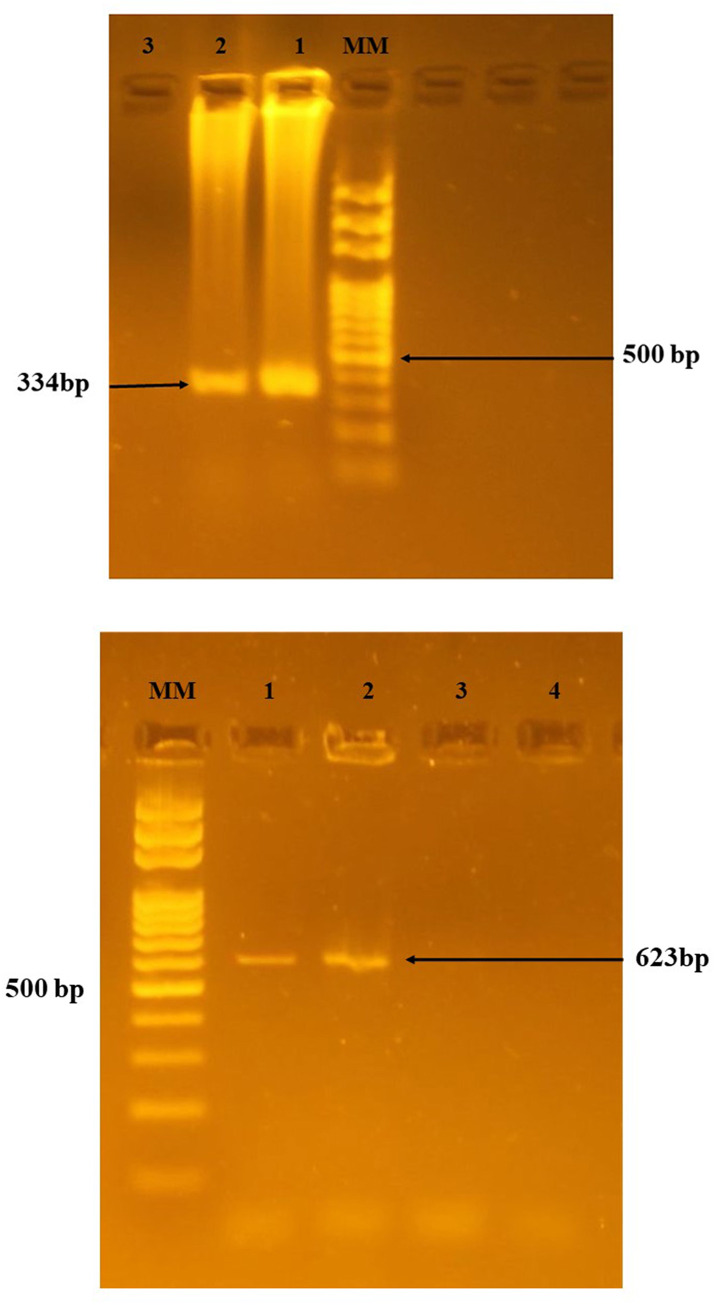
The results of blood meal sources showed that 2 (7%) Ph. orientalis specimens from Tabarakallah site had fed on humans (Fig 2A), and the anthropophilic index was 13.3%, while the other vertebrate hosts have not been detected in the rest of the samples. The results of human DNA were confirmed by using the universal primers of the mammalian host (Fig 2B). No Leishmania parasite DNA was detected in all blood-fed specimens.
Fig 2.
A. Electrophoresis of DNA multiplex polymerase chain reaction profile (1.5% agarose gel) after amplification of cytochrome b gene of vertebrate mtDNA fragments among blood-fed Phlebotomus orientalis using UNREV1025A, Human 741F, Goat 894F, Cow121F, and Dog 368F primers. MM: 100 bp DNA molecular marker; lane 1: human blood; lane 2: human blood; lane 3: negative control (PCR water). B. Electrophoresis of DNA multiplex polymerase chain reaction profile (1.5% agarose gel) after amplification of cytochrome b gene of mammalian mtDNA fragments among blood-fed Phlebotomus orientalis using UNFOR403 and UNREV1025A primers. MM: 100 bp DNA molecular marker; lane 1: mammalian blood; lane 2: mammalian blood; lanes 3, 4: negative control (PCR water).
Discussion
An improved understanding of the feeding behaviors of sand flies could contribute to new, more effective strategies for the control of sand fly populations. The present study of host preference of sand fly vectors using multiplex PCR is the first to be conducted in leishmaniasis endemic sites in Sudan. In this study, sand fly blood meals were examined by multiplex PCR using specific primers for cytochrome b gene, distinguishing between human, goat, cow, and dog. The multiplex PCR protocol described here, allowed identification of blood meal sources of many vertebrate species in one assay.
Most collected specimens in the current study were Ph. orientalis (57.2%; Table 1), and the majority were from Tabarakallah site, the well-known endemic site of VL in eastern Sudan [43]. Ph. orientalis is a confirmed biological vector of VL in Sudan, parts of Ethiopia and Kenya [3, 10, 27, 44, 47, 58–68]. Recent studies in nearby endemic villages of VL in Ethiopia found that Ph. orientalis was the predominant feeder [17, 27, 67–69].
The presence of Ph. papatasi, Ph. duboscqi and Ph. bergeroti in the study sites are of epidemiological interest. The three species exist sympatrically in many places in central and eastern Sudan [7, 53, 70–72]. Ph. papatasi is a known vector transmitting CL in different parts of the country [6], while Ph. duboscqi and Ph. bergeroti have been involved in the transmission of Le. major in places, such as Ethiopia [73], Kenya [74], and Sahara [75].
Few specimens were identified as Ph. rodhaini (1.1%; Table 1). Generally, Ph. rodhaini is considered a rare species, some studies from Sudan reported small numbers of Ph. rodhaini captured from endemic sites [66, 68, 72, 76, 77] indicating its involvement in maintaining the transmission of VL between wild animals and dogs [66].
Hundreds of Sergentomyia sand flies were collected during the study period, and has been excluded from the study, because the genus Sergentomyia is known vector of the subgenus Sauroleishmania that infects reptiles and edentates [78–80] but not a proven biological vector of human leishmaniasis [79, 81]. However, some studies have reported the detection of different Leishmania species DNA in species of Sergentomyia [82–88]. It may be worth examining the biological role of this species in Sudan.
The results showed that 7% of wild-caught Ph. orientalis had fed on a human host in the VL community (Table 1), corroborating the role of this species as a proven vector of leishmaniasis in Sudan and eastern Africa [1, 2, 17]. The host preferences of Ph. orientalis collected from an area nearby Tabarakallah village with a similar environment revealed that 8.3% of meals from a human host [17], other studies showed that Ph. orientalis prefer feeding on humans and bovines [17, 27, 64, 67, 68, 69], these findings supported the fact that Ph. orientalis is a vector of VL in the area.
Ph. orientalis feed on wide range of hosts, this is remarkably affecting disease transmission, not only by offering alternative reservoir for the Leishmania parasites, but also by supporting survival of sand fly population through providing a continuous access to blood meal [64]. Reports from Sudan and New World showed that high sero-prevalence of Le. donovani and Le. braziliensis were found in donkeys, respectively [89, 90]. Also, dogs have been shown to have high sero-positivity for Le. donovani in Sudan and Ethiopia [91–95], and owning dogs is considered a risk factor for VL [96–98].
The identification of blood meal source of sand fly vectors is a challenge, as observed in this study, however, only two blood meal sources were successfully analyzed, while the rest failed, since the positive controls used during the tests showed the respective bands for each host. The justification for this failure could be attributed to the degraded and insufficient amounts of quality DNA from the tiny sand flies [14, 19], time of collection in response to blood digestion (more than 48 hrs) [22], and due to the limited set of primers which were used, implying blood sources other the studied animals.
No Leishmania DNA was detected among the screened low number samples, this result is not surprising, and is consistent with recent study done in northwestern Ethiopia [17], where the expected infection rate among phlebotomine vectors was 1–5 out of 1,000 flies [17, 44, 67]. PCR-based methods have been used previously for the detection of Leishmania in phlebotomine sand flies [17, 38, 99–102]. DNA prepared from the whole body of Phlebotomus sand flies not only can be used for blood meal identification, but can also be used for parasite detection by PCR simultaneously [11]. This simple method could be advantageous in epidemiological studies for parasite species identification in sand fly vectors. Also, it could be helpful in epidemic scenarios when and where robust, reliable results are urgently needed.
The correct identification of vertebrate hosts of sand flies is vital because it might reveal the nature of host preference and additional pathogen reservoirs to consider in the disease control. This knowledge can help in developing an effective disease control strategy that is considering patterns for the potential spread of vector-borne disease within and among communities. The details of sand fly blood-feeding patterns can also be used as a surveillance tool that assesses the efficacy and effectiveness of various prevention and control interventions, and to estimate the degree of coverage required for various leishmaniasis vaccines when developed [14].
Further studies investigating the host preference and feeding patterns of phlebotomine sand fly vectors should directly measure species composition of larger blood-fed sample size collected among dry and wet seasons from different leishmaniasis communities in Sudan to understand their vectorial capacity and to clarify natural transmission cycles.
Conclusions
The blood meal identification in wild-caught sand flies can confirm a strong association between sand flies and reservoir hosts; in this case, humans in rural areas and help to improve our understanding of the role of reservoir host in the disease transmission. The rapid and higher sensitivity of the multiplex PCR would warrant its use in vector incrimination and reservoir determination of vector-borne diseases.
Standardization of blood meal study protocols, including the incorporation of vertebrate surveys and collection of sand fly vectors during dry and wet seasons, would improve prediction of the impact of vector-host interactions on disease ecology. The present study used analysis could be used to explore sand fly-vertebrate host association, in which host availability data are unavailable.
Supporting information
(PDF)
Acknowledgments
Thanks are extended to Mr. Talal Ahmed from University of Khartoum for assistance in field collection of the sand fly, Dr. Anwar Banaga from Ministry of Health, Gedarif state, Sudan for logistic assistance, to staff of MSF Switzerland at Tabarakallah site for providing accommodation to the study team during field visits, to Dr. Beirag Nazar from Brunel University London for revising the English language of the manuscript and to people of Sirougia and Tabarakallah for their cooperation during field collection. This work is part of the PhD fellowship for Arwa Elaagip from OWSD (Organization for Women in Science for the Developing World) and SIDA (Swedish International Development Cooperation Agency).
List of abbreviations
- Ph
Phlebotomus.
- Le
Leishmania.
- PCR
Polymerase chain reaction
- DNA
Deoxyribonucleic acid
- ELISA
Enzyme-linked immunosorbent assay
- PNOC
Prepronociceptin
- RFLP
Restriction fragment length polymorphism
- RLB
Reverse-line blotting
- VL
Visceral leishmaniasis
- CL
Cutaneous leishmaniasis
- ITS2
Internal transcribed spacer 2
- rDNA
Ribosomal DNA
- bp
Base pair.
- mtDNA
Mitochondrial DNA
- Cytb
Cytochrome b
- TBE
Tris base EDTA
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Maroli M, Feliciangeli M, Bichaud L, Charrel R, Gradon L. Phlebotomine sandflies and the spreading of leishmaniases and other diseases of public health concern. Medical and Veterinary Entomology. 2013;27(2):123–147. 10.1111/j.1365-2915.2012.01034.x [DOI] [PubMed] [Google Scholar]
- 2.Bates P, Depaquit J, Galati E, Kamhawi S, Maroli M, McDowell M, et al. Recent advances in phlebotomine sand fly research related to leishmaniasis control. Parasites & Vectors. 2015;8:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elnaiem D. Ecology and control of the sand fly vectors of Leishmania donovani in East Africa, with special emphasis on Phlebotomus orientalis. Journal of Vector Ecology. 2011;36(Suppl 1):S23–31. [DOI] [PubMed] [Google Scholar]
- 4.Khalil E. Vaccines for visceral leishmaniasis: Hopes and hurdles. Leishmaniases as Re-emerging Diseases. 2018. Pp: 95–104. 10.5772/intechopen.75184. [DOI] [Google Scholar]
- 5.World Health Organization. Report of the scientific working group meeting on Insect Vectors and Human Health. 2003;TDR/SWG/VEC/03.1. World Health Organization; Geneva. Switzerland. [Google Scholar]
- 6.EL Hassan A, Zijlstra E. Leishmaniasis in Sudan. Cutaneous leishmaniasis. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2001;95(suppl 1):1–17. [DOI] [PubMed] [Google Scholar]
- 7.Khalid N, Aboud M, Alrabba F, Elnaiem D, Tripet F. Evidence for genetic differentiation at the microgeographic scale in Phlebotomus papatasi populations from Sudan. Parasites & Vectors. 2012;5:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hassan M, Widaa S, Osman O, Numiary M, Ibrahim M, Abushama H. Insecticide resistance in the sand fly, Phlebotomus papatasi from Khartoum State, Sudan. Parasites & Vectors. 2012;5:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahamoud A, Osman H, Abass E, el Agib A, Madid R, Semiao-Santose S, et al. Identification of an area predominantly endemic for childhood and adolescent visceral leishmaniasis in central Sudan. Acta Tropica. 2018;178:142–147. 10.1016/j.actatropica.2017.11.010 [DOI] [PubMed] [Google Scholar]
- 10.Elnaiem D, Osman O. Evidence for active transmission of visceral leishmaniasis within a village in eastern Sudan. Acta Tropica. 1998;71:305–309. 10.1016/s0001-706x(98)00067-9 [DOI] [PubMed] [Google Scholar]
- 11.Sant’Anna M, Jones N, Hindley J, Mendes-Sousa A, Dillon R, Cavalcante R, et al. Blood meal identification and parasite detection in laboratory-fed and field-captured Lutzomyia longipalpis by PCR using FTA databasing paper. Acta Tropica. 2008;107:230–237. 10.1016/j.actatropica.2008.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramalho-Ortigão M, Jochim R, Anderson J, Lawyer P, Pham V, Kamhawi S, et al. Exploring the midgut transcriptome of Phlebotomus papatasi: comparative analysis of expression profiles of sugar-fed, blood-fed and Leishmania major-infected sandflies. BMC Genomics. 2007;8:300 10.1186/1471-2164-8-300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ian B, Lalita R, Albert P, Murari D, Suman R, Matthew R, et al. Development of an enzyme-linked immunosorbent 1 assay (ELISA) to identify host feeding preferences of Phlebotomus species (Diptera: Psychodidae) in endemic foci of visceral leishmaniasis in Nepal. Journal of Medical Entomology. 2010;47(5):902–906. 10.1603/me09184 [DOI] [PubMed] [Google Scholar]
- 14.Mukabana W, Takken W, Knols B. Analysis of arthropod bloodmeals using molecular genetic markers. TRENDS in Parasitology. 2002;18(11):505–509. 10.1016/s1471-4922(02)02364-4 [DOI] [PubMed] [Google Scholar]
- 15.MacDonald G. The epidemiology and control of malaria. Oxford University Press, London: 1957. [Google Scholar]
- 16.Boakye D, Tang J, Truc P, Merriweather A, Unnasch T. Identification of bloodmeals in haematophagous Diptera by cytochrome B heteroduplex analysis. Medical and Veterinary Entomology. 1999;13:282–287. 10.1046/j.1365-2915.1999.00193.x [DOI] [PubMed] [Google Scholar]
- 17.Yared S, Gebresilassie A, Abbasi I, Aklilu E, Kirstein O, Balkew M, et al. A molecular analysis of sand fly blood meals in a visceral leishmaniasis endemic region of northwestern Ethiopia reveals a complex host-vector system. Heliyon. 2019;5(7):e02132 10.1016/j.heliyon.2019.e02132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clements A. The biology of mosquitoes: development, nutrition, and reproduction. Chapman & Hall; 1992. – Nature. [Google Scholar]
- 19.Pruzinova K, Sadlova J, Seblova V, Homola M, Votypka J, Volf P. Comparison of bloodmeal digestion and the peritrophic matrix in four sand fly species differing in susceptibility to Leishmania donovani. PLoS ONE. 2015;10(6):e0128203 10.1371/journal.pone.0128203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dillon R, Lane R. Bloodmeal digestion in the midgut of Phlebotomus papatasi and Phlebotomus langeroni. Medical and Veterinary Entomology. 1993;7:225–232. 10.1111/j.1365-2915.1993.tb00681.x [DOI] [PubMed] [Google Scholar]
- 21.Gomes L, Duarte R, Lima D, Diniz B, Serrão M, Labarthe N. Comparison between precipitin and ELISA tests in the bloodmeal detection of Aedes aegypti (Linnaeus) and Aedes fluviatilis (Lutz) mosquitoes experimentally fed on feline, canine and human hosts. Mem. Inst. Oswaldo Cruz. 2001;96:693–695. 10.1590/s0074-02762001000500020 [DOI] [PubMed] [Google Scholar]
- 22.Pruzinova K, Sadlova J, Myskova J, Lestinova T, Janda J, Volf P. Leishmania mortality in sand fly blood meal is not species-specific and does not result from direct effect of proteinases. Parasites & Vectors. 2018;11:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzâlez C, Leoân C, Paz A, Loâpez M, Molina G, Toro D, et al. Diversity patterns, Leishmania DNA detection, and bloodmeal identification of phlebotominae sand flies in villages in northern Colombia. PLoS ONE. 2018;13(1):e0190686 10.1371/journal.pone.0190686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kent R, Norris D. Identification of mammalian blood meals in mosquitoes by a multiplexed Polymerase Chain Reaction targeting cytochrome b. American Journal of Tropical Medicine and Hygiene. 2005;73(2):336–342. [PMC free article] [PubMed] [Google Scholar]
- 25.Gomez B, Sanchez E, Feliciangeli M. Man-vector contact of phlebotomine sand flies (Diptera: Psychodidae) in north-central Venezuela as assessed by blood meal identification using dot-ELISA. Journal of American Mosquito Control Association. 1998;14:28–32. [PubMed] [Google Scholar]
- 26.Tanure A, Peixoto J, Afonso M, Duarte R, Pinheiro A, Coelho S, et al. Identification of sandflies (Diptera: Psychodidae: Phlebotominae) blood meals in an endemic leishmaniasis area in Brazil. Revista do Instituto de Medicina Tropical de Sao Paulo. 2015;57(4):321–324. 10.1590/S0036-46652015000400008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gebresilassie A, Abbasi I, Aklilu E, Yared S, Kirstein O, Moncaz A, et al. Host-feeding preference of Phlebotomus orientalis (Diptera: Psychodidae) in an endemic focus of visceral leishmaniasis in northern Ethiopia. Parasites & Vectors. 2015. a;8:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abbasi I, Cunio R, Warburg A. Identification of blood meals imbibed by phlebotomine sand flies using cytochrome b PCR and reverse line blotting. Vector-Borne and Zoonotic Diseases. 2009; 9(1). [DOI] [PubMed] [Google Scholar]
- 29.Haouas N, Pesson B, Boudabous R, Dedet J, Babba H, Ravel C. Development of a molecular tool for the identification of Leishmania reservoir hosts by blood meal analysis in the insect vectors. American Journal of Tropical Medicine and Hygiene. 2007;77:1054–1059. [PubMed] [Google Scholar]
- 30.Jaouadi K, Haouas N, Chaara D, Boudabous R, Gorcii M, Kidar A, et al. Phlebotomine (Diptera, Psychodidae) bloodmeal sources in Tunisian cutaneous leishmaniasis foci: could Sergentomyia minuta, which is not an exclusive herpetophilic species, be implicated in the transmission of pathogens? Ann. Entomol. Soc. Am. 2013;106(1):79–85. [Google Scholar]
- 31.Bauma M, de Castroa E, Pintob M, Goulartc T, Bauraa W, Klisiowicza D, et al. Molecular detection of the blood meal source of sand flies (Diptera: Psychodidae) in a transmission area of American cutaneous leishmaniasis, Paraná State, Brazil. Acta Tropica. 2015;143:8–12. 10.1016/j.actatropica.2014.11.006 [DOI] [PubMed] [Google Scholar]
- 32.Maleki-Ravasan N, Oshaghi M, Javadian E, Rassi Y, Sadraei J, Mohtarami F. Blood meal identification in field-captured sand flies: Comparison of PCR-RFLP and ELISA assays. Iranian Journal of Arthropod-Borne Diseases. 2009;3(1):8–18. [PMC free article] [PubMed] [Google Scholar]
- 33.Rocha Soares V, da Silva J, da Silva K, Cruz M, Santos M, Ribolla P, et al. Identification of blood meal sources of Lutzomyia longipalpis using polymerase chain reaction-restriction fragment length polymorphism analysis of the cytochrome B gene. Mem Inst Oswaldo Cruz, Rio de Janeiro. 2014;109(3):379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.da Silva Sales K, Costa P, de Morais R, Otranto D, Brandão-Filho S, de Paiva Cavalcanti M, et al. Identification of phlebotomine sand fly blood meals by real-time PCR. Parasites & Vectors. 2015;8:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pengsakul T, Sudsom N, Foakes G, Bhatt K, Eisenberg M, Siriyasatien P. Molecular DNA identification of blood sources fed on, for culicine mosquitoes (Diptera: Culicidae) collected in the Songkhla province, southern Thailand. Songklanakarin Journal of Science Technology. 2017;39 (6):731–737. [Google Scholar]
- 36.Chaskopoulou A, Giantsis I, Demir S, Bon M. Species composition, activity patterns and blood meal analysis of sand fly populations (Diptera: Psychodidae) in the metropolitan region of Thessaloniki, an endemic focus of canine leishmaniasis. Acta Tropica. 2016;158:170–176. 10.1016/j.actatropica.2016.03.006 [DOI] [PubMed] [Google Scholar]
- 37.Inbar E, Lawyer P, Sacks D, Podini D. The potential use of forensic DNA methods applied to sand fly blood meal analysis to identify the infection reservoirs of anthroponotic visceral leishmaniasis. PLoS Neglected Tropical Diseases. 2016;10(5):e0004706 10.1371/journal.pntd.0004706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oshaghi M, Maleki Ravasan N, Javadian E, Mohebali M, Hajjaran H, Zare Z, et al. Vector incrimination of sand flies in the most important visceral leishmaniasis focus in Iran. American Journal of Tropical Medicine and Hygiene. 2009;81(4):572–577. 10.4269/ajtmh.2009.08-0469 [DOI] [PubMed] [Google Scholar]
- 39.Garlapati R, Abbasi I, Warburg A, Poche D, Poche R. Identification of bloodmeals in wild caught blood fed Phlebotomus argentipes (Diptera: Psychodidae) using cytochrome b PCR and reverse line blotting in Bihar, India. Journal of Medical Entomology. 2012;49(3):515–521. 10.1603/me11115 [DOI] [PubMed] [Google Scholar]
- 40.Maia C, Parreira R, Cristóvão J, Freitas F, Afonso M, Campino L. Molecular detection of Leishmania DNA and identification of blood meals in wild caught phlebotomine sand flies (Diptera: Psychodidae) from southern Portugal. Parasites & Vectors. 2015;8:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jain S, Brahmbhatt M, Rank D, Joshi C, Solanki J. Use of cytochrome b gene variability in detecting meat species by multiplex PCR assay. The Indian Journal of Animal Sciences. 2007;77(9):880. [Google Scholar]
- 42.Lee J, Hassan H, Hill G, Cupp E, Higazi T, Mitchell C, et al. Identification of mosquito avian-derived blood meals by polymerase chain reaction-heteroduplex analysis. American Journal of Tropical Medicine and Hygiene. 2002;66:599–604. 10.4269/ajtmh.2002.66.599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zijlstra E, El-Hassan A. Leishmaniasis in Sudan. Visceral leishmaniasis. Transactions of Royal Society of Tropical Medicine and Hygiene. 2001;95 Suppl. 1:S27–S58. [DOI] [PubMed] [Google Scholar]
- 44.Elnaiem D, Conners S, Thomson M, Hassan M, Hassan K, Aboud M, et al. Environmental determinants of the distribution of Phlebotomus orientalis in Sudan. Annals of Tropical Medicine & Parasitology. 1998;92:2877–2887. [DOI] [PubMed] [Google Scholar]
- 45.Elnaiem D, Schorscher J, Obsomer V, Osman M, Mekkawi A, Connor S, et al. Risk mapping of visceral leishmaniasis: the role of local variation in rainfall and altitude on the presence and incidence of kala-azar in eastern Sudan. American Journal of Tropical Medicine & Hygiene. 2003;68(1):10–17. [PubMed] [Google Scholar]
- 46.Alten B, Ozbel Y, Ergunay K, Kasap O, Cull B, Antoniou M, et al. Sampling strategies for phlebotomine sand flies (Diptera: Psychodidae) in Europe. Bulletin of Entomological Research. 2015:1–15. [DOI] [PubMed] [Google Scholar]
- 47.Quate L. Phlebotomus sandflies of the Paloich area in the Sudan (Diptera: Psychodidae). Journal of Medical Entomology. 1964;1:213–268. 10.1093/jmedent/1.3.213 [DOI] [PubMed] [Google Scholar]
- 48.Killick-Kendrick R, Killick-Kendrick M. Honey-drew of aphids as a source of sugar for Phlebotomus ariasi. Medical & Veterinary Entomology. 1987;1:297–302. [DOI] [PubMed] [Google Scholar]
- 49.Kirk R, Lewis D. The phlebotomine of the Ethiopian region. Transactions of Royal Entomological Society of London. 1951;102:383–510. [Google Scholar]
- 50.Abonnenc E, Minter D. Bilingual key for the identification of sandflies of the Ethiopian region. (French and English). Cahiers ORSTOM, Sreie Entomologie Medicale. 1965;5:1–63. [Google Scholar]
- 51.El Hossary S. Identification key to genera, subgenera and species of phlebotomine sandflies in Sudan. Vector Biology Research Program. NAMRU3. 2007. [Google Scholar]
- 52.Balbino V, Coutinho-Abreu I, Sonoda I, Melo M, Andrade P, Castro J. Genetic structure of natural populations of the sandfly Lutzomyia longipalpis (Diptera: Psychodidae) from the Brazilian northeastern region. Acta Tropica. 2006;98:15–24. 10.1016/j.actatropica.2006.01.007 [DOI] [PubMed] [Google Scholar]
- 53.Khalid N, El Naiem D, Aboud M, Al Rabba F, Tripet F. Morphometric and molecular differentiation of Phlebotomus (Phlebotomus) sandflies. Medical and Veterinary Entomology. 2010;24:352–360. 10.1111/j.1365-2915.2010.00893.x [DOI] [PubMed] [Google Scholar]
- 54.Numairy M. Genetic diversity and population structure of Phlebotomus orientalis (Diptera: Psychodidae) from selected regions in Sudan. A thesis submitted to University of Khartoum for master degree in Science. 2009.
- 55.Bari A. Morphometric and molecular study of the sand fly species Phlebotomus orientalisr Parrot, 1936 (Diptera: Psychodidae) collected from three geographical areas in Sudan. A thesis submitted to University of Khartoum for master degree in Medical Entomology and Vector Control. 2013.
- 56.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: a laboratory Manual, 2nd edition Cold Spring Harbor laboratory press, Cold Spring Harbor, New York: 1989. p. 484–515. [Google Scholar]
- 57.Mohamed N, Osman H, Muneer M, Samy A, Ahmed A, Mohammed A, et al. Identifying asymptomatic Leishmania infections in non-endemic villages in Gedaref state, Sudan. BMC Research Notes. 2019;12:566 10.1186/s13104-019-4608-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoogstraal J, Heyneman D. Leishmaniasis in the Sudan Republic. American Journal of Tropical Medicine and Hygiene. 1969;18:1091–1210. [Google Scholar]
- 59.Schorscher J, Goris M. Incrimination of Phlebotomus (Larroussius) orientalis as a vector of visceral leishmaniasis in western Upper Nile Province, southern Sudan. Transactions of Royal Society of Tropical Medicine and Hygiene. 1992;86:622–623. [DOI] [PubMed] [Google Scholar]
- 60.Gebre-Michael T, Lane R. The roles of Phlebotomus martini and P. celiae (Diptera: Phlebotominae) as vectors of visceral leishmaniasis in the Aba Roba focus, southern Ethiopia. Medical and Veterinary Entomology. 1996;10:53–62. 10.1111/j.1365-2915.1996.tb00082.x [DOI] [PubMed] [Google Scholar]
- 61.Elnaiem D, Hassan M, Maingon R, Nureldin G, Mekawi A, Miles M, et al. The Egyptian Mongoose, Herpestes ichneumon, is a probable reservoir host of visceral leishmaniasis in eastern Sudan. Parasitology. 2001;122:531–536. 10.1017/s0031182001007594 [DOI] [PubMed] [Google Scholar]
- 62.Hassan M, Elraba’a F, Ward R, Maingon R, Elnaiem D. Detection of high in-village transmission of Leishmania donovani in eastern Sudan. Acta Tropica. 2004;92:77–82. 10.1016/j.actatropica.2004.06.001 [DOI] [PubMed] [Google Scholar]
- 63.Hassan M, Elamin E, Mukhtar M. Isolation and identification of Leishmania donovani from Phlebotomus orientalis, in an area of eastern Sudan with endemic visceral leishmaniasis. Annals of Tropical Medicine and Parasitology. 2008;102:1–3. [DOI] [PubMed] [Google Scholar]
- 64.Gebre-Michael T, Balkew M, Berhe N, Hailu A, Mekonnen Y. Further studies on the phlebotomine sandflies of the kala-azar endemic lowlands of Humera-Metema (north-west Ethiopia) with observations on their natural blood meal sources. Parasites & Vectors. 2010;3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ngumbia P, Kaburia J, Anjilia C, Haas F. Phlebotomus (Larroussius) orientalis (Diptera: Psychodidae) as a probable secondary vector of visceral leishmaniasis in Kenya. Journal of Vector Borne Diseases. 2010;47:58–60. [PubMed] [Google Scholar]
- 66.Elnaiem D, Hassan H, Osman O, Maingon R, Killick-Kendrick R, Ward R. A possible role for Phlebotomus (Anaphlebotomus) rodhaini (Parrot, 1930) in transmission of Leishmania donovani. Parasites & Vectors. 2011;4:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gebresilassie A, Abbasi I, Kirstein O, Aklilu E, Yared S, Tekie H, et al. Physiological age structure and Leishmania spp. detection in Phlebotomus (Larroussius) orientalis (Parrot, 1936) (Diptera: Psychodidae) at an endemic focus of visceral leishmaniasis in Northern Ethiopia. Journal of Tropical Medicine. 2015. b;710528 10.1155/2015/710528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gebresilassie A, Yared S, Aklilu E, Kirstein O, Moncaz A, Tekie H, et al. Host choice of Phlebotomus orientalis (Diptera: Psychodidae) in animal baited experiments: a field study in Tahtay Adiyabo district, northern Ethiopia. Parasites & Vectors. 2015. c;8:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yared S, Gebresilassie A, Akililu E, Balkew M, Warburg A, Hailu A, et al. Habitat preference and seasonal dynamics of Phlebotomus orientalis in urban and semi-urban areas of kala-azar endemic district of Kafta Humera, northwest Ethiopia. Acta Tropica. 2017;166:25–34. 10.1016/j.actatropica.2016.10.011 [DOI] [PubMed] [Google Scholar]
- 70.Elnaiem D, Hassan H, Ward R. Phlebotomine sandflies in a focus of visceral leishmaniasis in a border area of eastern Sudan. Annals of Tropical Medicine and Parasitology. 1997;91:307–318. 10.1080/00034989761157 [DOI] [PubMed] [Google Scholar]
- 71.Lambert M, Dereure J, El-Safi S, Bucheton B, Dessein A, Boni M, et al. The sandfly fauna in the visceral-leishmaniasis focus of Gedaref in the Atbara-River area of eastern Sudan. Annals of Tropical Medicine and Parasitology. 2002;96:631–636. 10.1179/000349802125001474 [DOI] [PubMed] [Google Scholar]
- 72.Hassan M, Widaa S, Ibrahim M, Abu Shara R, Osman O, Numairy M, et al. Studies on the ecology of sandflies (Diptera: Psychodidae) in Sudan: the first records of Phlebotomus orientalis and P. rodhaini in northern Sudan. Annals of Tropical Medicine & Parasitology. 2007;101(7):653–655. [DOI] [PubMed] [Google Scholar]
- 73.Balkew M, Gebre-Michael T, Berhe N, Ali A, Hailu A. Leishmaniasis in the middle course of the Ethiopian Rift Valley. II. Entomological observations. Ethiopian Medical Journal. 2002;40:271–282. [PubMed] [Google Scholar]
- 74.Beach R, Kiilu G, Kendricks L, Oster C, Leeuwenberg J. Cutaneous leishmaniasis in Kenya. Transmission of Leishmania major to man by the bite of Phlebotomus duboscqi. Transactions of Royal Society of Tropical Medicine and Hygiene. 1984;78:747–751. [DOI] [PubMed] [Google Scholar]
- 75.Seccombe A, Ready P, Huddleston L. A catalogye of the Old World phlebotominae sandflies (Diptera, Psychodidae, Phlebotominae). Occasional Papers on Systematic Entomology. 1993;8:1–57. [Google Scholar]
- 76.Widaa S, Ahmed K, Bari A, Ali M, Ibrahim M, Bashir M, et al. Sandflies (Diptera: Psychodidae) in a focus of visceral leishmaniasis in White Nile, Sudan. Mem Inst Oswaldo Cruz, Rio de Janeiro. 2012;107(4):470–475. [DOI] [PubMed] [Google Scholar]
- 77.Adam B, Hassan M, Abdelnour O, Awadallah A. Identification and classification of sand flies species and it’s habitats in El-Kadaba village, White Nile state, Sudan. International Journal of Infectious Diseases and Therapy. 2017;2(1):15–21. [Google Scholar]
- 78.Ready P. Biology of phlebotomine sand flies as vectors of disease agents. Annual Review of Entomology. 2013;58:227–250. 10.1146/annurev-ento-120811-153557 [DOI] [PubMed] [Google Scholar]
- 79.Polanska N, Rohousova I, Volf P. The role of Sergentomyia schwetzi in epidemiology of visceral leishmaniasis in Ethiopia. Parasites & Vectors. 2014;7(Suppl 1):O5. [Google Scholar]
- 80.Giantsis I, Chaskopoulou A, Bon M. Direct multiplex PCR (dmPCR) for the identification of six phlebotomine sand fly species (Diptera: Psychodidae), including major Leishmania vectors of the Mediterranean. Journal of Economic Entomology. 2016;1–5. 10.1093/jee/tov238 [DOI] [PubMed] [Google Scholar]
- 81.Sadlova J, Dvorak V, Seblova V, Warburg A, Votypka J, Volf P. Sergentomyia schwetzi is not a competent vector for Leishmania donovani and other Leishmania species pathogenic to humans. Parasites & Vectors. 2013;6:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mutinga M, Massamba N, Basimike M, Kamau C, Amimo F, Onyido A, et al. Cutaneous leishmaniasis in Kenya: Sergentomyia garnhami (Diptera: Psychodidae), a possible vector of Leishmania major in Kitui District: a new focus of the disease. East African Medical Journal. 1994;71(7):424–428. [PubMed] [Google Scholar]
- 83.Mukherjee S, Hassan M, Ghosh A, Ghosh K, Bhattacharya A, Adhya S. Leishmania DNA in Phlebotomus and Sergentomyia species during a kala-azar epidemic. American Journal of Tropical Medicine and Hygiene. 1997;57:423–425. 10.4269/ajtmh.1997.57.423 [DOI] [PubMed] [Google Scholar]
- 84.Parvizi P, Amirkhani A. Mitochondrial DNA characterization of Sergentomyia sintoni populations and finding mammalian Leishmania infections in this sand fly by using ITS-rDNA. Iranian Journal of Veterinary Research. 2008;22:9–18. [Google Scholar]
- 85.Berdjane-Brouk Z, Kone A, Djimde A, Charrel R, Ravel C, Delaunay P, et al. First detection of Leishmania major DNA in Sergentomyia (Spelaeomyia) darlingi from cutaneous leishmaniasis foci in Mali. PloS one. 2012;7:e28266 10.1371/journal.pone.0028266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Campino L, Cortes S, Dionísio L, Neto L, Afonso M, Maia C. The first detection of Leishmania major in naturally infected Sergentomyia minuta in Portugal. Mem Inst Oswaldo Cruz Rio de Janeiro. 2013;108:516–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kanjanopas K, Siripattanapipong S, Ninsaeng U, Hitakarun A, Jitkaew S, Kaewtaphaya P, et al. Sergentomyia (Neophlebotomus) gemmea, a potential vector of Leishmania siamensis in southern Thailand. BMC Infectious Diseases. 2013;13:333 10.1186/1471-2334-13-333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nzelu C, Kato H, Puplampu N, Desewu K, Odoom S, Wilson M, et al. First detection of Leishmania tropica DNA and Trypanosoma species in Sergentomyia sand flies (Diptera: Psychodidae) from an outbreak area of cutaneous leishmaniasis in Ghana. PLoS Neglected Tropical Diseases. 2014;8(2):e2630 10.1371/journal.pntd.0002630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mukhtar M, Sharief A, el Saffi S, Harith A, Higazi T, Adam A, et al. Detection of antibodies of Leishmania donovani in animals in Kala azar endemic regions in eastern Sudan: preliminary report. Transactions of Royal Society of Tropical Medicine and Hygiene. 2000;94:33–36. [DOI] [PubMed] [Google Scholar]
- 90.Truppel J, Otomura F, Teodoro U, Massafera R, da Costa-Ribeiro M, Catarino C, et al. Can equids be a reservoir of Leishmania braziliensis in endemic areas? PLoS One. 2014;9:e93731 10.1371/journal.pone.0093731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dereure J, El-Safi S, Bucheton B, Boni M, Kheir M, Davoust B, et al. Visceral leishmaniasis in eastern Sudan: parasite identification in humans and dogs; host-parasite relationships. Microbiology Infections. 2003;5:1103–1108. [DOI] [PubMed] [Google Scholar]
- 92.Hassan M, Osman F, Elraba'a F, Schallig D, Elnaiem D. Role of the domestic dog as a reservoir host of Leishmania donovani in Eastern Sudan. Parasites & Vectors. 2009;2:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kalayou S, Tadelle H, Bsrat A, Abebe N, Haileselassie M, Schallig H. Serological evidence of Leishmania donovani infection in apparently healthy dogs using direct agglutination test (DAT) and rk39 dipstick tests in Kafta Humera, northwest Ethiopia. Transboundary Emerging Diseases. 2011;58:255–262. 10.1111/j.1865-1682.2011.01209.x [DOI] [PubMed] [Google Scholar]
- 94.Kenubih A, Dagnachew S, Almaw G, Abebe T, Takele Y, Hailu A, et al. Preliminary survey of domestic animal visceral leishmaniasis and risk factors in north-west Ethiopia. Tropical Medicine and International Health. 2015;20:205–210. 10.1111/tmi.12418 [DOI] [PubMed] [Google Scholar]
- 95.Rohousova I, Talmi-Frank D, Kostalova T, Polanska N, Lestinova T, Kassahun A, et al. Exposure to Leishmania spp. and sand flies in domestic animals in northwestern Ethiopia. Parasites & Vectors. 2015;8:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bashaye S, Nombela N, Argaw D, Mulugeta A, Herrero M, Nieto J, et al. Risk factors for visceral leishmaniasis in a new epidemic site in Amhara Region, Ethiopia. American Journal of Tropical Medicine and Hygiene. 2009;81:34–39. [PubMed] [Google Scholar]
- 97.Argaw D, Mulugeta A, Herrero M, Nombela N, Teklu T, Tefera T, et al. Risk factors for visceral leishmaniasis among residents and migrants in Kafta-Humera, Ethiopia. PLoS Neglected Tropical Diseases. 2013;7:e2543 10.1371/journal.pntd.0002543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yared S, Deribe K, Gebreselassie A, Lemma W, Akililu E, Kirstein O, et al. Risk factors of visceral leishmaniasis: a case control study in north-western Ethiopia. Parasites & Vectors. 2014;7:470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cabrera O, Munsterman L, Cárdenas R, Gutiérrez R, Ferro C. Definition of appropriate temperature and storage conditions in the detection of Leishmania DNA with PCR in phlebotomine flies. Biomedica. 2002;22:296–302. [PubMed] [Google Scholar]
- 100.Paiva B, Secundino N, Nascimento J, Pimenta P, Galati E, Junior H, et al. Detection and identification of Leishmania species in field-captured phlebotomine sandflies based on mini-exon gene PCR. Acta Tropica. 2006;99:252–259. 10.1016/j.actatropica.2006.08.009 [DOI] [PubMed] [Google Scholar]
- 101.Myskova J, Votypka J, Volf P. Leishmania in sand flies: comparison of quantitative polymerase chain reaction with other techniques to determine the intensity of infection. Journal of Medical Entomology. 2008;45:133–138. 10.1603/0022-2585(2008)45[133:lisfco]2.0.co;2 [DOI] [PubMed] [Google Scholar]
- 102.Ranasinghe S, Rogers M, Hamilton J, Bates P, Maingon R. A real time PCR assay to estimate Leishmania chagasi load in its natural sand fly vector Lutzomyia longipalpis. Transactions of Royal Society of Tropical Medicine and Hygiene. 2008;102(9):875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.