Summary
To explore the biology of lung adenocarcinoma (LUAD) and identify new therapeutic opportunities, we performed comprehensive proteogenomic characterization of 110 tumors and 101 matched normal adjacent tissues (NATs) incorporating genomics, epigenomics, deep-scale proteomics, phosphoproteomics and acetylproteomics. Multi-omics clustering revealed four subgroups defined by key driver mutations, country and gender. Proteomic and phosphoproteomic data illuminated biology downstream of copy number aberrations, somatic mutations and fusions, and identified therapeutic vulnerabilities associated with driver events involving KRAS, EGFR and ALK. Immune subtyping revealed a complex landscape, reinforced the association of STK11 with immune-cold behavior and underscored a potential immunosuppressive role of neutrophil degranulation. Smoking-associated LUADs showed correlation with other environmental exposure signatures and a field effect in NATs. Matched NATs allowed identification of differentially expressed proteins with potential diagnostic and therapeutic utility. This proteogenomics dataset represents a unique public resource for researchers and clinicians seeking to better understand and treat lung adenocarcinomas.
Keywords: Lung Cancer, Adenocarcinoma, Proteogenomics, Proteomics, Genomics, Mass Spectrometry, Protein, Phosphorylation, Acetylation, CPTAC
In Brief
Comprehensive proteogenomic characterization of lung adenocarcinomas and paired normal adjacent tissues from patients of diverse smoking status and country of origin yields insights into cancer taxonomy, oncogenesis and immune response, offers novel candidate biomarkers and therapeutic targets, and provides a community resource for further discovery.
Graphical Abstract
Introduction
Lung cancers are the leading cause of cancer deaths in the United States (Siegel et al., 2019) and worldwide (Bray et al., 2018). Despite therapeutic advances including tyrosine kinase inhibitors and immunotherapy, sustained responses are rare and prognosis remains poor (Herbst et al., 2018), with a 19% overall 5-year survival rate in the United States (Bray et al., 2018) and a worldwide ratio of lung cancer mortality-to-incidence of 0.87. Adenocarcinoma (LUAD), the most common lung malignancy, is strongly related to tobacco smoking, but also the subtype most frequently found in individuals who have reported no history of smoking (“never-smokers”) (Subramanian and Govindan, 2007; Sun et al., 2007). The genetics and natural history of LUAD are strongly influenced by smoking status, gender, and ethnicity, among other variables (Chapman et al., 2016; Okazaki et al., 2016; Subramanian and Govindan, 2007; Sun et al., 2007). However, contemporary large-scale sequencing efforts have typically been based on cohorts of smokers with limited ethnic diversity. Among the major sequencing studies that have helped elucidate the genomic landscape of LUAD (Clinical Lung Cancer Genome Project (CLCGP) and Network Genomic Medicine (NGM), 2013; Ding et al., 2008; Imielinski et al., 2012), only The Cancer Genome Atlas (TCGA) measured a small subset of proteins and phosphopeptides, restricted to a 160-protein reversed phase array (Cancer Genome Atlas Research Network, 2014). As the most frequent genomic aberrations in LUAD involve RAS/RAF/RTK pathway genes that lead to cellular transformation mainly by inducing proteomic and phosphoproteomic alterations (Cully and Downward, 2008), global proteogenomic profiling is needed to provide deeper mechanistic insights. Furthermore, while prior molecular characterization has identified a number of oncologic dependencies and facilitated the development of effective inhibitors for LUAD driven by EGFR mutation (Lynch et al., 2004; Paez et al., 2004) and ALK (Kwak et al., 2010), ROS1 (Shaw et al., 2014) and RET fusions (Gautschi et al., 2017; Kohno et al., 2012; Takeuchi et al., 2012), a substantial proportion of LUADs still lack known or currently targetable mutations.
To further our understanding of LUAD pathobiology and potential therapeutic vulnerabilities, the National Cancer Institute (NCI)’s Clinical Proteomic Tumor Analysis Consortium (CPTAC) undertook comprehensive genomic, deep-scale proteomic and post-translational modifications (PTM) analyses of paired (patient-matched) LUAD tumors and normal adjacent tissues (NATs). Our integrative proteogenomic analyses focused particularly on novel and clinically actionable insights revealed in the proteome and PTMs. The underlying data represent an exceptional resource for further biological, diagnostic and drug discovery efforts.
Results
Proteogenomic landscape and molecular subtypes of LUAD
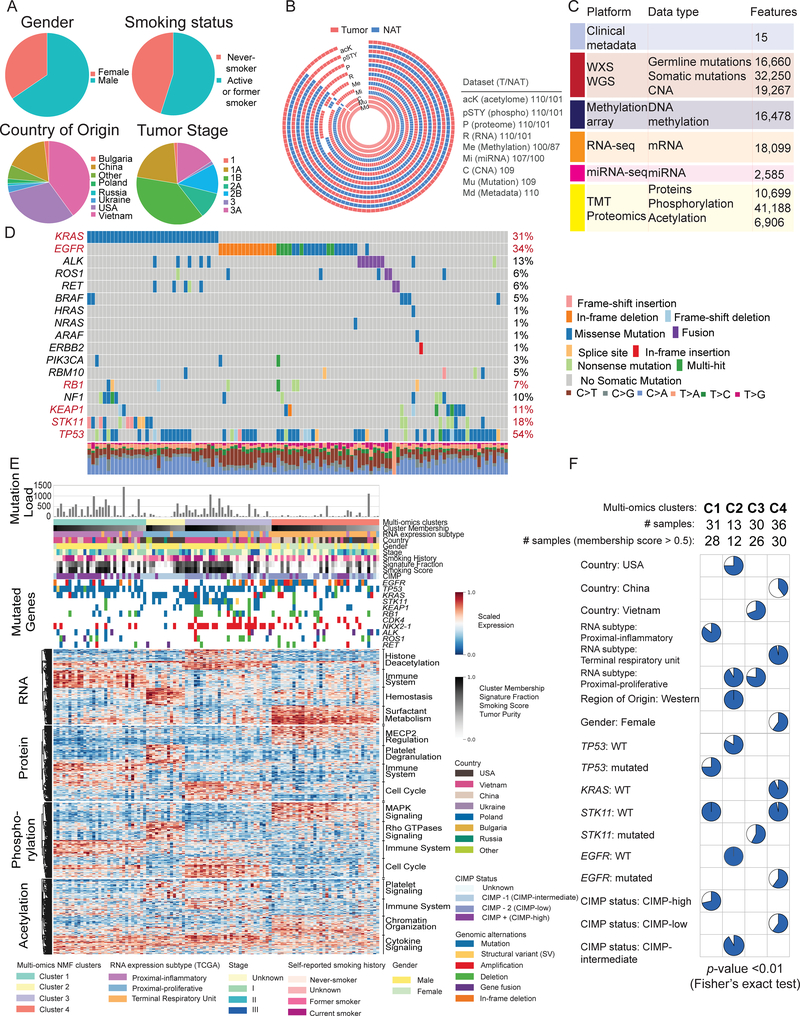
We investigated the proteogenomic landscape of 110 treatment-naïve LUAD tumors and 101 paired normal adjacent tissues (NATs), prospectively collected under strict protocols limiting ischemic time. The samples represented diverse demographic and clinical characteristics including country of origin and smoking status (Figure 1A, Table S1). After confirmation of LUAD histopathology by multiple expert pathologists, aliquots of cryopulverized tissue were profiled by whole exome (WES, nominal 150x coverage), whole genome (WGS, nominal 15x coverage), RNA (RNA-seq) and miRNA sequencing (miRNA-seq), array-based DNA methylation analysis, and in-depth proteomic, phosphoproteomic and acetylproteomic characterization (Figures 1B, S1A, Tables S2, S3), with complete data for 101 tumors and 96 NATs. Tandem mass tags (TMT)-based isobaric labeling was used for precise relative quantification of proteins, phosphosites and acetylsites. Excellent reproducibility and data quality were maintained across the entire dataset (Figure S1C–F). Appropriate filtering resulted in a comprehensive, deepscale proteogenomic dataset allowing extensive integrative analysis (Figure 1C, Tables S2, S3). The general landscape of somatic alterations, focal amplifications and deletions in this study was consistent with prior large-scale profiling efforts including TCGA (Campbell et al., 2016; Cancer Genome Atlas Research Network, 2014; Weir et al., 2007), although with a different distribution likely due to the greater demographic diversity and larger proportion of self-reported never-smokers in the current study (Figure 1D).
Figure 1: Genomic and proteomic landscape of lung adenocarcinoma (LUAD).
(A) Pie charts of key demographic and histologic features, along with self-reported smoking status of LUAD patient samples characterized in this study.
(B) Patient-centric circos plot representing the multi-platform data generated in this study. White gaps in the schematic represent missing data. Numbers to the right indicate samples in each of the categories.
(C) Summary of data and metadata generated in this study.
(D) Oncoplot generated with maftools depicting mutually exclusive driver oncogene somatic mutations in KRAS, EGFR, other RAS/RAF pathway genes and receptor tyrosine kinase gene fusions in the CPTAC LUAD cohort along with their frequencies. Rows represent genes and columns represent samples. Somatic mutations in tumor suppressor genes (NF1, KEAP1, STK11 and TP53) are also depicted. The significantly mutated genes with Benjamini Hochberg (BH) FDR <0.01 are indicated in red. Percentages of transitions/transversions noted in each sample are depicted in the bar plots.
(E) Integrative classification of tumor samples into four NMF-derived clusters (multi-omics cluster-1 (C1) to cluster-4 (C4)). Within each cluster, tumors are sorted by cluster membership scores, decreasing from left to right. “RNA expression subtype” shows classification by previously published RNA-seq-based expression subtypes (TCGA LUAD analysis). The heatmap shows the top 50 differential mRNA transcripts, proteins, phosphoproteins, and acetylated proteins for each multi-omics cluster, annotated for representative pathways.
(F) Pie charts show sample distribution of metadata terms that are significantly overrepresented (Fisher’s exact test) within the most representative “core” cluster members (membership score > 0.5) that define each cluster.
To investigate the intrinsic structure of the proteogenomics data, non-negative matrix factorization (NMF)-based unsupervised clustering was performed on RNA, protein, phosphosites and acetylsites, collectively as “multi-omics clustering” and individually (except RNA) (Figures 1E, S1G–I). The 4 stable clusters (C1–4) (Figure 1E) overlapped with previously characterized mRNA-based proximal-inflammatory, proximal-proliferative and terminal respiratory unit clusters (Cancer Genome Atlas Research Network, 2014; Wilkerson et al., 2012), but subdivided the second of these into two distinct clusters. The core samples of the clusters were significantly associated with distinctive clinical and molecular features (p-value <0.01; Figure 1F, Table S1). Cluster 1 (C1), aligned with proximal-inflammatory, was enriched for TP53 mutants, STK11 wild-type (WT), and CpG island methylator phenotype (CIMP)-high status; C2, a proximal-proliferative subcluster, was distinguished by Western patients (especially from USA), TP53 and EGFR WT status, and intermediate CIMP status; C3, the dominant proximal-proliferative cluster, was enriched for Vietnamese patients and STK11 mutation (including two structural events identified from WGS; Table S1); and C4, aligned with terminal respiratory unit, was enriched for EGFR mutations, female sex and Chinese nationality and was essentially devoid of KRAS or STK11 mutations. Most of the samples harboring EML4-ALK fusions were assigned to C4 and lacked mutations in other key driver genes, consistent with a primary role for EML4-ALK in LUAD tumorigenesis (Gao et al., 2018). Of note, NMF clustering based on sample purity-adjusted protein data matrices led to similar clusters compared to the unadjusted data. While NMF clusters had distinctive biology, linear models did not identify biologically coherent sets of differential markers between sexes, tumor stages or histological subtypes once major covariates were accounted for (Table S3).
To further explore the biology associated with the multi-omics taxonomy, we performed over-representation pathway analysis (Zhang et al., 2016) using differentially regulated genes, proteins, and post-translational modifications (PTMs) in each of the clusters (Figure 1E, Table S3). C1/proximal-inflammatory samples were primarily associated with immune signaling across multiple data types. The C2 subset of the proximal-proliferative subtype demonstrated signaling by Rho GTPases, as well as signatures of hemostasis and platelet activation, signaling and degranulation, suggestive of systematic disturbances in coagulation homeostasis. The dominant proximal-proliferative subtype in C3 had a distinctive histone deacetylase signature but also upregulation of cell cycle pathways. Finally, the terminal respiratory unit subtype in C4 was distinguished by surfactant metabolism, MAPK1/MAPK3 signaling, MECP2 regulation, and chromatin organization in the acetylproteome. Notably, C1, characterized by increased expression of immune system-related genes, included samples with high non-synonymous mutation burden and CIMP-high status. Altogether, the pathway enrichment analysis highlights intrinsic differences in both oncogenic signaling and host response across LUAD subtypes.
To explore the pattern of miRNA expression in LUAD, we performed unsupervised Louvain clustering of 107 tumor samples with available miRNA data based on expression of mature miRNAs. Five subgroups of LUAD patients were identified by their distinctive miRNA expression profiles (Figure S1J, Table S3). Two of the miRNA clusters were markedly enriched for tumors from C1/proximal-inflammatory and C3/proximal-proliferative multi-omics clusters, while the remaining three miRNA clusters had mixed composition. One miRNA cluster included all 5 EML4-ALK as well as the HMBOX1-ALK fusion tumors, and featured high expression of miR-494, miR-495, and miR-496, the first two previously implicated in NSCLC (Romano et al. 2012; Chen et al. 2017). The vast majority of patients with STK11 mutations were categorized into another subgroup in which well-documented cancer-associated miRNAs such as miR-106b-5p, miR-20a-5p, and miR-17–5p were highly expressed (Lu et al., 2017; Shi et al., 2018).
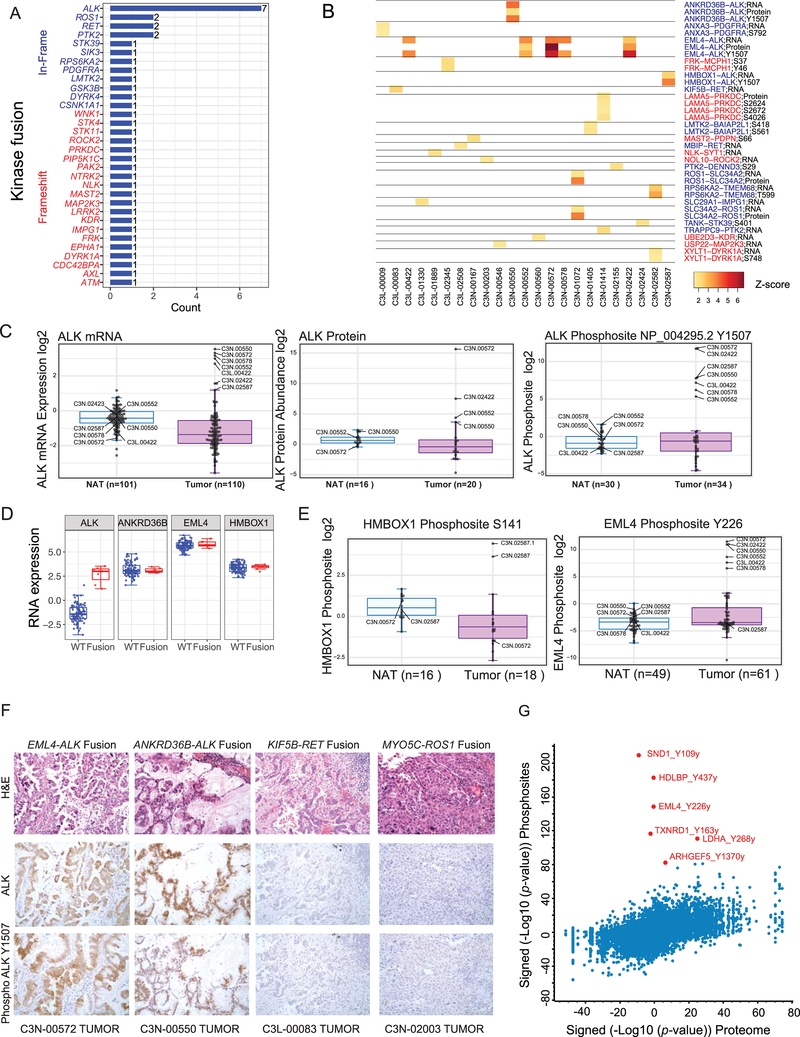
The relationships between epigenetic and genomic events and downstream expression of RNA, proteins, and PTMs were explored in detail. Cross-referencing gene fusions in the cohort with a curated kinase fusion database (Gao et al., 2018) allowed identification of all rearrangements involving kinases (Figure 2A). While fusions involving ALK, ROS1, RET, and PTK2 genes were most recurrent, several novel, potentially oncogenic kinase fusions were also discovered. Generally, such oncogenic kinases contained in-frame fusions, while kinases with a tumor suppressive role (such as STK11, STK4, ATM, FRK, and EPHA1) exhibited disruptive out-of-frame events (Figure 2A). Several kinase fusions showed commensurate differential RNA, protein, and phosphosite expression of the index cases (Figure 2B). Besides ALK, instances of ROS1, RET, PRKDC, and PDGFRA overexpression were found in tumors but not in paired NAT samples. Investigation of the fusion architecture of the highly recurrent in-frame ALK gene fusions (n=7) identified multiple 5’ partners including the well-established EML4 as well as novel HMBOX1 and ANKRD36B genes (Figure S2A). WGS data provided precise genomic breakpoints in the intron proximal to exon-20 (e20) underlying ALK rearrangements in 5 cases (Figure S2B). All ALK gene fusion cases showed outlier expression of ALK mRNA and all in which the protein was detected (4/7) showed outlier ALK total protein abundance. However, the most dramatic difference was seen in the specific increase in ALK phosphosite Y1507 (Figure 2C). While RNA expression levels of the 5’ partner genes were uniformly high and did not differ between fusion-positive and -negative samples (Figure 2D), both EML4-Y226 and HMBOX-S141 showed increased phosphorylation only in the corresponding gene fusion-positive tumor samples (Figure 2E). We employed IHC to validate observation of the fusion-specific ALK phosphosite Y1507 using commercially available ALK and phospho (Y1507) ALK antibodies. We noted tumor-specific positive staining in all available ALK fusion-positive cases, whereas no detectable staining was observed in either samples with ROS1/RET fusions or paired NATs (Figures 2F, S2C). To assess phosphorylation of canonical and possible novel targets by mislocalized ALK fusion proteins (Ducray et al., 2019), we identified all protein phosphorylation events associated with ALK fusion. This analysis identified tyrosine phosphorylation of multiple proteins such as SND1, HDLBP, and ARHGEF5 (Figure 2G), providing new potential insights into oncogenic ALK fusion protein signaling, pending further validation to establish direct functional connections. SND1, for instance, has previously been described as an oncogene (Jariwala et al., 2017), impacts biological processes such as angiogenesis and invasion, and regulates expression of oncogenic miRNAs (Chidambaranathan-Reghupaty et al., 2018), suggesting a novel role in ALK fusion-mediated tumorigenesis.
Figure 2: Novel phosphoproteomic aberrations associated with ALK gene fusions.
(A) Summary of all kinase gene fusions identified from RNA-seq analysis.
(B) RNA expression, protein abundance and specific phosphosite modifications noted to be outliers in the index fusion event sample relative to all other samples.
(C) Boxplot showing outlier expression of ALK RNA, protein and the ALK Y1507 phosphosite in tumors with ALK fusion. Blue: Normal adjacent tissues (NAT); Pink: Tumor samples. Sample IDs of outlier cases are indicated.
(D) Boxplot showing overexpression of ALK mRNA observed in fusion-positive (Red) versus - negative (Blue) tumors. The three 5’ partners show comparably high expression in both fusion-positive and -negative tumors, as expected.
(E) Boxplot showing the phosphorylation of two ALK fusion partners, HMBOX1 and EML4, in the indicated index cases.
(F) Immunohistochemistry reveals upregulation of both total ALK and the ALK Y1507 phosphosite specifically in the tumor epithelia of ALK fusion-positive samples. No staining was seen in RET or ROS1 fusion samples or in matched NATs (Figure S2C).
(G) Scatterplot of significantly regulated phosphosites and their corresponding protein expression in tumors with and without ALK fusion. Phosphosites showing distinct upregulation in ALK fusion samples are highlighted in red.
See also Figure S2
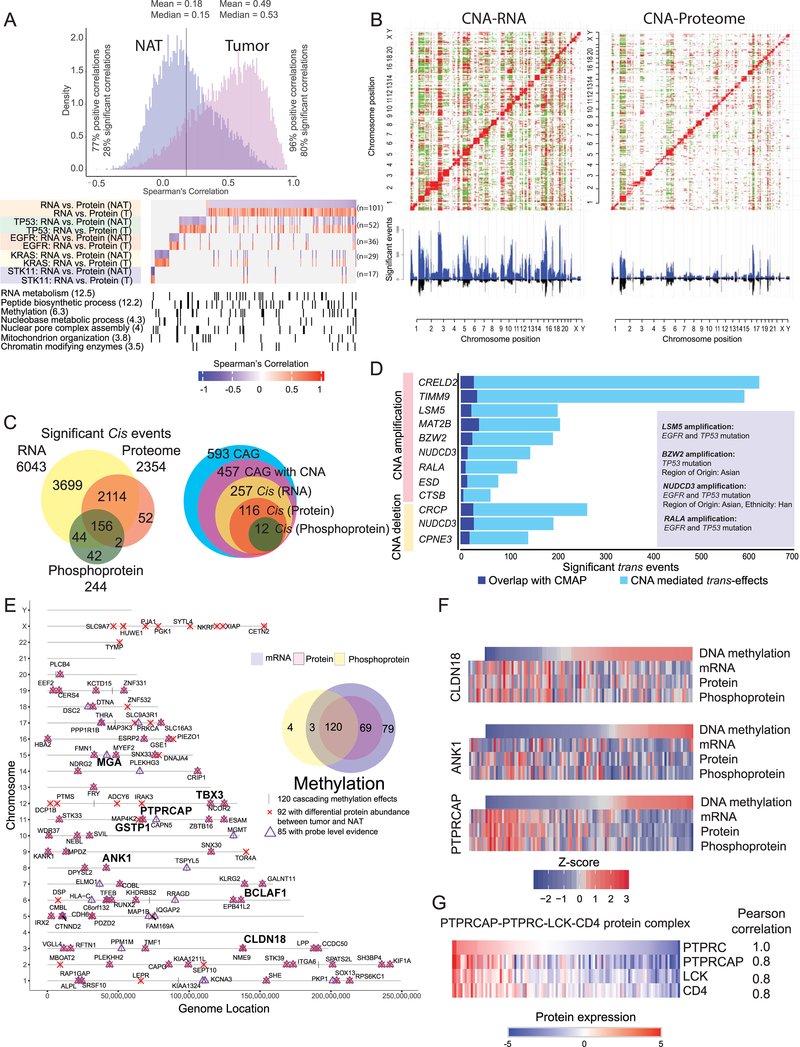
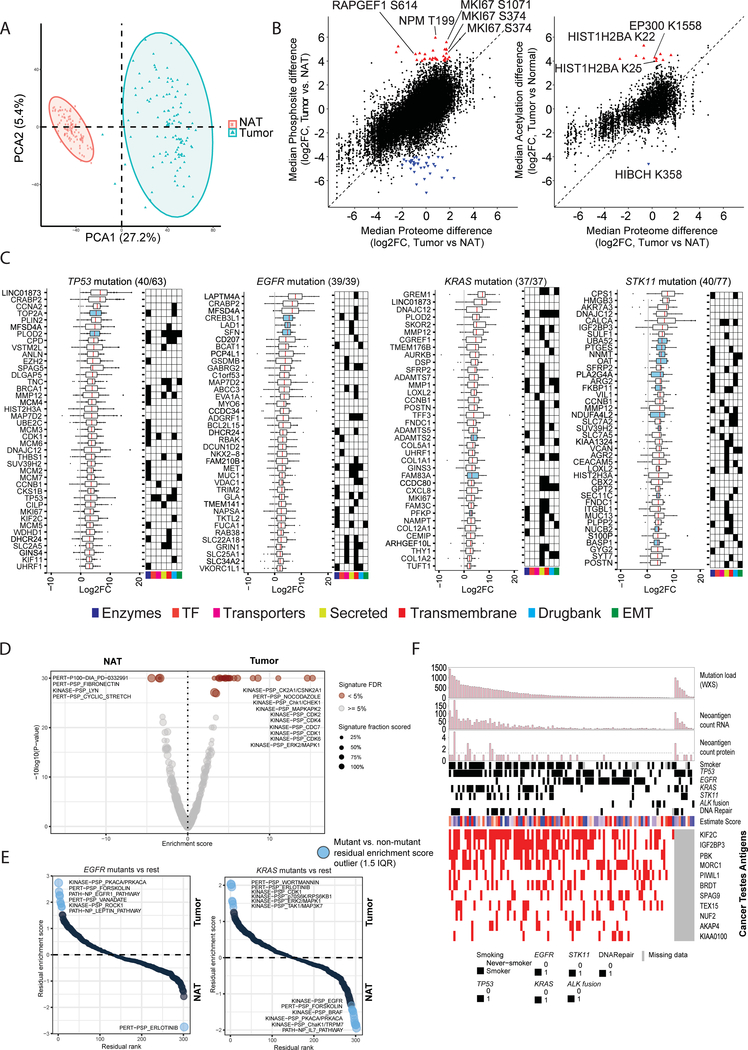
While sample-wise mRNA-protein correlations were fairly consistent between tumors and NATs (Figure S3A, Table S4), gene-wise correlations displayed striking differences (Figure 3A), results unchanged after adjusting for immune and stromal infiltration. We identified a total of 227 transcript / protein pairs differentially correlated (FDR < 0.01) between tumors and NAT pairs, globally or within 4 major mutational subtypes (Figure 3A, Table S4). The identified gene products were markedly enriched for RNA metabolism, peptide biosynthesis, methylation, mRNA splicing, nuclear processing, mitochondrial organization, and chromatin modifiers (p-value <10−3), suggesting tighter or more active translational control of proteins involved in proliferation, cell cycle events and survival in tumors (Figure S3B).
Figure 3: Impact of copy number alteration (CNA) and DNA methylation on protein and phosphoprotein expression.
(A) Correlation between steady-state mRNA and protein abundances in tumors and NATs (n=101 pairs) for genes with discrepant tumor/normal mRNA-protein correlations. Bottom panel represents enriched biological terms, with -Log10 (p-value) in brackets.
(B) Correlation plots between CNA and RNA expression and between CNA and protein abundance. Significant (FDR <0.05) positive and negative correlations are indicated in red and green, respectively. CNA-driven cis-effects appear as the red diagonal line; trans-effects appear as vertical red and green lines. The accompanying histograms show the number of significant (FDR <0.05) cis- and trans-events corresponding to the indicated genomic loci (upward plot) as well as the overlap between CNA-RNA and CNA-protein events (downward plot).
(C) Venn diagrams depicting the cascading effects of CNAs. The Venn diagram on the left shows the overlap between significant cis-events across the transcriptome, proteome and phosphoproteome. The Venn diagram on the right shows the same analysis restricted to cancer-associated genes (CAG) with significant cis-effects across multiple data types.
(D) Genes with CNA events that show significant similarity (BH FDR <0.1) between their significant trans-effects (FDR <0.05) and the Connectivity Map (CMAP) genomic perturbation profiles. Inset shows significant enrichment (Fisher’s exact test, FDR <0.1) for specific mutational or demographic features for 4 genes.
(E) Genes whose DNA methylation was associated with cascading cis-regulation of their cognate mRNA expression, global protein level and phosphopeptide abundance. Bold type highlights a few known cancer genes.
(F) Methylation-driven cis-regulation of selected genes (n = 109 samples). Gene-level methylation scores, RNA expression levels and protein/phosphopeptide abundances were converted into Z-scores and the tumor samples were ordered by methylation levels.
(G) Coordinated expression of proteins associated with PTPRC (CD45) complex in tumors.
The impact of CNAs on RNA and protein abundance in both cis and trans was characterized (Figure 3B, Table S4). CNA correlations were broadly comparable but considerably dampened at the levels of protein and PTMs (Figures 3C, S3C). A total of 6,043, 2,354, and 244 significant positive correlations (cis-effects) were observed for RNA, proteins, and phosphoproteins, respectively, with only 156 significant cis-effects overlapping between all 3 (Figure 3C; Table S4). A similar trend was observed within 593 cancer-associated genes (CAG) (Figure 3C, Table S4); the 12 CAGs showing significant overlapping regulation were CREBBP, KMT2B, PSIP1, AKT2, EGFR, GMPS, IL6ST, IRF6, NFKB2, PHF6, YES1, and ZBTB7B. In addition, numerous genes associated with recurrent LUAD-specific CNA events (Campbell et al., 2016) showed downstream expression effects, including significant cis-regulation at RNA and protein levels for CDK4, RB1, SMAD4, ARID2, MET, ZMYND11 and ZNF217.
To help nominate functionally important genes within CNA regions, we compared protein-level trans-effects to approximately half a million genomic perturbation signatures contained in the Connectivity Map database (https://clue.io/cmap). Trans-effects significantly paralleled the associated gene perturbation profiles for 12 CNA events (FDR <0.1) (Figure 3D, Table S4). Ras-related protein Ral-A (RALA) is a GTPase that has been shown to mediate oncogenic signaling and regulate EGFR and KRAS mutation-mediated tumorigenesis (Gildea et al., 2002; Kashatus, 2013; Peschard et al., 2012). Our data suggests that amplification of RALA may affect the biology of EGFR mutant tumors. The role of basic leucine zipper and W2 domain 2 (BZW2) in LUAD has not been elaborated, but BZW2 stimulates AKT/mTOR/PI3K signaling and cell growth in bladder and hepatocellular carcinoma (Gao et al., 2019; Jin et al., 2019), and has also been shown to interact with EGFR (Foerster et al., 2013). The lysosomal cysteine proteinase cathepsin B (CTSB) has long been described as a marker of poor prognosis in LUAD (Fujise et al., 2000; Inoue et al., 1994) with mechanistic association with metastasis (Erdel et al., 1990; Higashiyama et al., 1993). Protein-level trans-effects thus provide testable mechanistic hypotheses for the tumorigenic impact of CNAs.
DNA methylation analyses showed LUAD tumors to be much more highly methylated than their counterpart NATs (p-value <0.0001) (Figure S3D, Table S2). Unsupervised clustering of the tumor methylome revealed CIMP-high, -intermediate, and -low clusters, with CIMP-low clusters nevertheless having focal areas of increased methylation (Figure S3E). Figure 3E shows the landscape of 120 methylation-driven cis-effects that were associated with coordinated differential expression at the RNA, protein and phosphoprotein levels, increasing their likelihood of functional significance (Song et al., 2019); Table S4). The majority (85/120) were directly supported by probe-level data in the promoter region of the gene. While many of these were novel, others, including CLDN18, ANK1 and PTPRCAP (Figure 3F) have strong associations with LUAD biology. CLDN18 is highly expressed in lung alveolar epithelium; its knockdown leads to increased lung parenchyma, expansion of lung epithelial progenitor populations, and increased propensity for lung adenocarcinoma development (Zhou et al., 2018). ANK1 promoter CpG islands are hypomethylated in normal lung but methylated in more than half of lung adenocarcinomas, especially with positive smoking history. ANK1 knockdown affects cancer-relevant pathways; furthermore, miR-486–3p and miR-486–5p, both strongly associated with lung adenocarcinoma oncogenesis, are located within ANK1 introns and are co-expressed with their host gene. PTPRCAP (CD45 associated protein), together with the three other members of its supramolecular complex, PTPRC (phosphatase CD45), co-receptor CD4, and kinase LCK, is implicated in regulation of lymphocyte function (Kruglova et al., 2017; Matsuda et al., 1998). While methylation probe positions did not allow us to determine whether the complex partners of PTPRCAP are regulated by methylation, they showed coordinated expression at the protein level (Figure 3G). Notably, PTPRCAP was included in a 5-gene methylation-based immune signature associated with survival in multiple malignancies including lung cancer (Jeschke et al., 2017). Other cancer-related genes with “cascading” methylation effects include BCLAF1, GSTP1, MGA, and TBX3, all of which have established roles in tumorigenesis or cancer prognosis (Cancer Genome Atlas Research Network, 2014)(Chen et al., 2013; Gurioli et al., 2018).
Connecting driver mutations to proteome, phosphoproteome and pathways
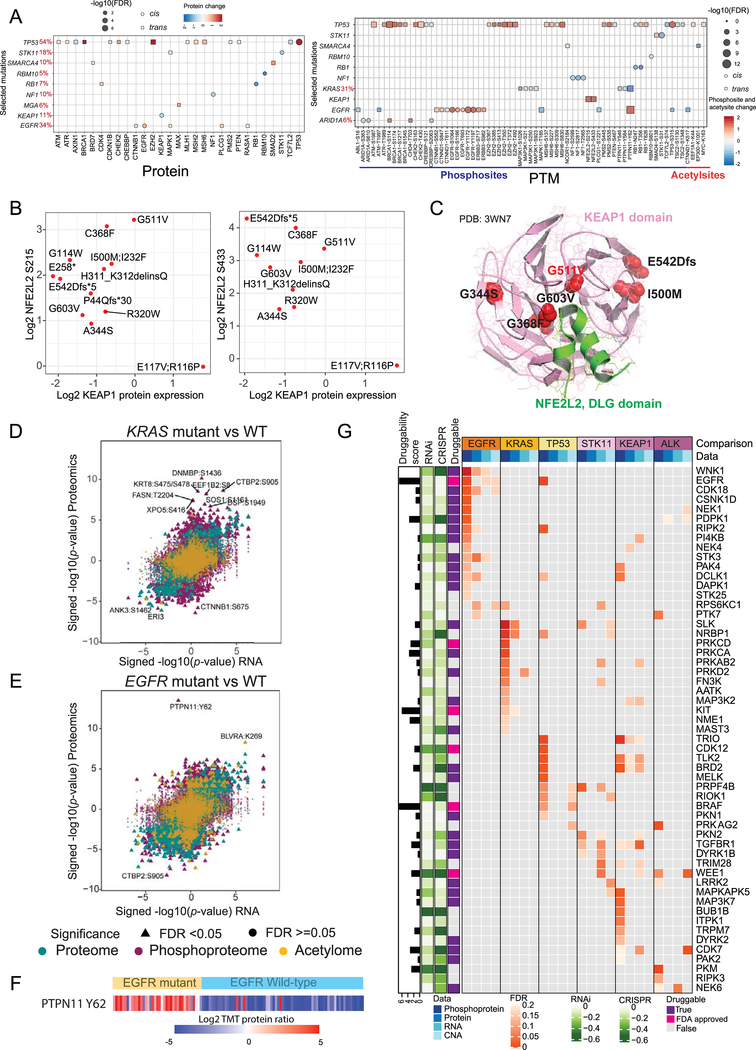
We examined how selected mutated genes that were significant in prior large-scale LUAD genomics studies (Cancer Genome Atlas Research Network, 2014; Ding et al., 2008); (Table S5) influenced expression of either the cognate gene product (cis-effects), or other gene products (trans-effects), specifically of a defined set of cancer-related genes (Bailey et al., 2018). We identified 11 genes with significant (FDR < 0.05) cis- or trans-effects in RNA, protein or phosphoprotein data (Figures 4A, S4A). TP53 and EGFR mutations resulted in elevated cognate protein and phosphosite abundance, whereas STK11, RBM10, RB1, NF1 and KEAP1 mutations reduced both cognate protein and phosphosite abundance. TP53 showed evidence of post-translational regulation, while TP53 mutant tumors showed upregulation of proteins in the mismatch repair (MMR) pathway, such as MLH1, MSH2, MSH6, and PSM2, and proteins involved in the DNA damage response (DDR) pathway, including ATM, ATR, and BRCA1. TP53 mutant tumors also showed significantly elevated EZH2 protein relative to RNA expression, as observed in TP53 mutant cell lines (Kuser-Abali et al., 2018), and downregulation of proteins involved in Wnt signaling (e.g. AXIN1 and TCF7L2) (Rother et al., 2004; Sanchez-Vega et al., 2018). Mutations in RB1, another key cell cycle-related gene, were associated with increased CDK4 protein abundance, which may contribute to resistance to CDK4/6 inhibitors in RB1-mutated LUAD samples. SMARCA4 mutation led to increased SMAD2 protein expression, while STK11 mutation was associated with increased phosphorylation of SMAD4 (S138). SMADs 2 and 4 are key elements in the transcriptional regulation of epithelial-mesenchymal transition (EMT) induced by TGF-β signaling (Xu et al., 2009). EGFR mutant samples showed decreased CTNNB1 expression at the level of RNA but elevated expression both at the level of proteome and phosphoproteome. CTNNB1 has been shown to play a critical role in EGFR-driven LUAD (Nakayama et al., 2014), and the trans-regulated phosphosite S552 on CTNNB1 induces its transcriptional activity (Fang et al., 2007). Altered phosphorylation and decreased acetylation were also observed for CTNND1, which has been implicated in NF-KB and RAC1-mediated signaling but not previously described in EGFR-mediated LUAD (Mizoguchi et al., 2017; Perez-Moreno et al., 2006).
Figure 4: Impact of somatic mutation on the proteogenomic landscape.
(A) Significant (Wilcoxon rank-sum test) cis- and trans-effects of selected mutations (x-axis) on the expression of cancer-associated proteins (left) and PTMs (right).
(B) Scatterplots showing the relationship between log2 KEAP1 protein and log2 NFE2L2 phosphosite (S215 and S433) expression in KEAP1 mutant samples. Only significant sites (Wilcoxon rank-sum test) are shown.
(C) Ribbon/Richardson diagram (Protein Data Bank crystal structure 3WN7) representing 3D protein structure of KEAP1 (Pink) and NFE2L2 DLG motif (green) interaction. Positions of various KEAP1 amino acid residues affected by somatic mutations observed in this cohort are indicated.
(D, E) Scatterplots showing significance of RNA, protein (green), phosphorylation site (purple), and acetylation site (yellow) abundance changes between KRAS mutant (D) or EGFR mutant(E) and WT tumors as determined using the Wilcoxon rank sum test. All identified sites are represented, with significant PTMs (FDR < 0.05) specified by triangles. Identities of the most extreme outliers are designated.
(F) Heatmap showing phosphorylation of PTPN11 Y62 in EGFR mutant and WT samples.
(G) Heatmap showing the outlier kinases enriched (FDR < 0.2) at the phosphoprotein, protein, RNA and CNA levels and their association with mutations in select genes. Cancer Dependency Map-supported (https://depmap.org) panels on the left show log2-transformed relative survival averaged across all available lung cell lines after depletion of the indicated gene (rows) by RNAi or CRISPR. Druggability based on the Drug Gene Interaction Database (http://www.dgidb.org/) is indicated alongside the availability of FDA-approved drugs. The log-transformed druggability score indicates the sum of PubMed journal articles that support the drug-gene relationship.
The cis- and trans-effects identified above (Figure 4A) helped reveal the detailed regulatory network of the KEAP1/NFE2L2 (NRF2) complex. KEAP1 interacts with NFE2L2 through two distinct binding domains, DLG and ETGE (Canning et al., 2015; Fukutomi et al., 2014), and undergoes conformational change under oxidative stress allowing NFE2L2 to execute the antioxidant response vital to lung cancer progression and metastasis (Lignitto et al., 2019; Wiel et al., 2019). Twelve LUAD tumors harbored KEAP1 mutations (Figure S4B) that did not impact expression of KEAP1 or NFE2L2 RNA (Figure S4C), but generally resulted in downregulation of KEAP1 protein expression and increased phosphorylation of NFE2L2 on S215 and S433 (FDR <0.05) (Figures S4C, 4B). One BTB domain missense mutation (G511V) did not downregulate KEAP1 protein expression but had amongst the highest levels of NFE2L2 phosphorylation (Figure 4B), suggesting a novel mechanism of action. Superposition of the site on the KEAP1 crystal structure showed that the G511V mutation fell close to the KEAP1/NFE2L2 binding domain (Figure 4C). We hypothesize that this mutation functions to disrupt KEAP1-NFE2L2 interaction rather than to impact protein stability. Most proteins and phosphosites upregulated in samples with KEAP1 mutations (Figures S4D, E) are members of the NFE2L2 oncogenic signatures and associated with antioxidant responses cytoprotective to cancer cells (Figure S4F) (Taguchi and Yamamoto, 2017).
Identification of therapeutic strategies from proteogenomics analyses
Comparison of global differential regulation of RNA, proteins, phosphosites and acetylsites revealed extreme phosphosite outliers in both KRAS and EGFR mutant tumors (Figures 4D, E, Table S4). KRAS mutant tumors showed significant upregulation of numerous cancer-associated phosphosites, including SOS1 phosphorylation on S1161. SOS1 is a guanine exchange factor (GEF) that activates KRAS (Vigil et al., 2010), and inhibition of SOS1 and KRAS is an emerging therapeutic strategy for KRAS mutant cancers (Hillig et al., 2019; O’Bryan, 2019). The observed C-terminal phosphorylation of SOS1 (Kamioka et al., 2010) likely relieves its constitutive interaction with GRB2 (Giubellino et al., 2008) allowing its recruitment to the membrane for KRAS activation in a GRB2-independent manner (Aronheim et al., 1994; Rojas et al., 2011). Interestingly, we also observed C-terminal phosphorylation of another GEF containing protein, DNMBP (TUBA), the role of which is not yet established in LUAD or KRAS mutant cancers.
EGFR mutant tumors showed highly significant and remarkably consistent tyrosine phosphorylation of PTPN11/Shp2 at Y62, but no effect was observed at the RNA or protein levels (Figures 4E, F). While prior studies have associated PTPN11/Shp2 phosphorylation with important biological consequences in non-small cell lung cancer (NSCLC) cell lines and xenograft models, this is, to our knowledge, the first report of such phosphorylation in a large set of primary treatment-naïve LUADs. In its basal state, PTPN11/Shp2 is inactive in a closed conformation due to the interaction between the N-terminal Src homology 2 (N-SH2) domain and the active site of the phosphatase (PTP) domain. Upon active conformational change induced by growth factor receptor and cytokine signaling, the phosphatase regulates cell survival and proliferation chiefly through RAS and ERK activation (Matozaki et al., 2009). Elevated PTPN11/Shp2 mRNA and protein expression have been associated with metastasis and decreased overall and progression-free survival in EGFR-positive NSCLC patients (Tang et al., 2013, Karachaliou et al., 2019). Importantly, residue Y62 falls in the interface between the N-SH2 and PTP domains, where its phosphorylation is thought to stabilize the active protein conformation (Ren et al., 2010). Notably, ALK fusion-driven tumors also showed outlier phosphorylation of PTPN11/Shp2, albeit at the C-terminal tyrosine phosphorylation sites Y546 and Y584 (Figure S4G).
Irrespective of the mode of activation, multiple lines of evidence suggest that PTPN11/Shp2 inactivation can suppress tumorigenesis (Aceto et al., 2012; Prahallad et al., 2015; Ren et al., 2010; Schneeberger et al., 2015), making it among the highest priority PTP targets for anticancer drug development (Ostman et al., 2006). PTPN11/Shp2 inhibitors have shown great promise in preclinical trials (Chen et al., 2016b) and targeted agents from multiple companies are now in clinical trials. Our data suggest that EGFR mutant- and ALK fusion-driven LUADs would be particularly promising target populations for such therapy.
Protein-level pathway comparison of tumors driven by EGFR and KRAS mutations showed remarkable disparity in complement and clotting cascades, with upregulation of coagulation in KRAS and downregulation in EGFR mutant samples (Figure S4I and hemostasis signature, Figure 1E). The increased risk of venous thromboembolism (VTE) in patients with primary lung cancer is well-established (Chew et al., 2008), as are the risks of prophylactic anticoagulation (Key et al., 2019). Our data suggest that VTE management might be stratified by mutation type, a concept supported by a recent NSCLC study in which the likelihood of VTE was significantly lower in patients without EGFR mutations (Dou et al., 2018).
To systematically nominate druggable targets specific to groups of LUADs characterized by key driver events, we assessed hyperphosphorylation of kinases as a proxy for abnormal kinase activity (Blumenberg et al.; Dou et al., 2020; Mertins et al., 2016) (Figure 4G) and annotated outliers for the degree to which shRNA- or CRISPR-mediated depletion reduced survival and proliferation in lung cancer cell lines (Barretina et al., 2012; Tsherniak et al., 2017). Multiple significantly hyperphosphorylated kinases (FDR <0.20) were identified in samples with EGFR, KRAS, TP53, STK11, KEAP1 or EML4-ALK alterations, the majority of which lacked any associated aberration in CNA, RNA or protein expression. Importantly, several driver-specific outlier kinases have interactions with FDA-approved drugs. In addition to EGFR in EGFR mutants, we saw outliers in PRKCD in KRAS mutants, BRAF in TP53 mutants, and WEE1 in EML4-ALK fusions. Furthermore, we identified 27 putatively druggable kinases with known but as yet non-FDA approved inhibitors (Cotto et al., 2018). Similar phosphorylation outlier analyses were performed for phosphatases, ubiquitinases, and deubiquitinases (Figure S4J), though the role of phosphorylation in these protein classes is not fully established.
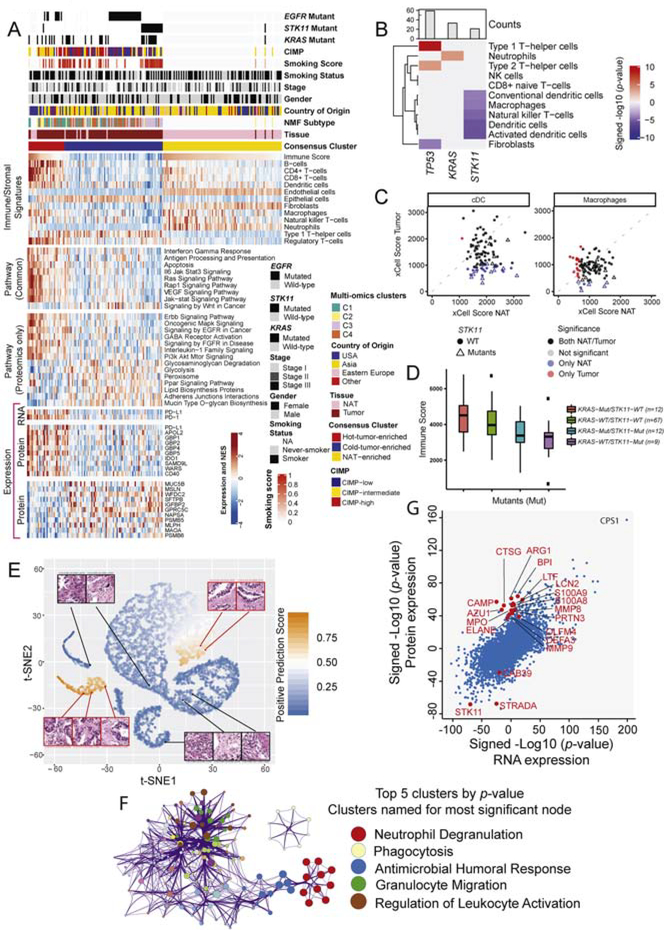
Immune landscape of lung adenocarcinoma
The composition of the tumor microenvironment in our cohort was studied using xCell (Aran et al., 2017) on the RNAseq data of both tumors and NATs. 64 different cell types were identified, spanning immune, stromal and other groups (Table S5). Consensus clustering identified three major immune clusters, designated “Hot”- (HTE), and “Cold”-tumor-enriched (CTE) and NAT-enriched (Figure 5A, upper panel, Table S5). Associations were observed between immune and multi-omics clusters, with enrichment of multi-omics cluster C1 in HTE and of clusters C3 and C4 in CTE immune clusters (p-value < 0.0003). CIMP-low status also associated with HTE (Figure 5A). HTE were distinguished from CTE tumors by their stronger signatures for B-cells, CD4+ and CD8+ T-cells, dendritic cells and macrophages. The HTE proteome was characterized by upregulation of multiple immune-related, oncogenic, and signaling pathways (Figure 5A, middle panels, Table S5), many of which were significantly enriched (FDR <0.01) exclusively in the proteomics dataset. PD1 RNA and PD-L1 RNA and protein were also upregulated in the immune HTE cluster (FDR < 0.01, Figure 5A, lower panel, Table S5). Notably, however, the HTE subtype also revealed the presence of immune inhibitory cells such as regulatory T-cells, and showed RNA upregulation of key markers of T-reg function such as CTLA4 (FDR < 1E-10) and FOXP3 (FDR < 0.0001) (Table S5). Transcripts for cytokines including TGF-beta and IL-10, known to enhance T-reg suppressive mechanisms, were upregulated in HTE tumors. As tumors with high T-reg infiltration are typically associated with poor prognosis (Shimizu et al., 2010), anti-CTLA4 therapy may benefit this population (Wing et al., 2008).
Figure 5: Immune landscape in LUAD.
(A) Heatmaps show three consensus clusters based on immune/stromal signatures identified from xCell, together with derived relative abundance of immune and stromal cell types. The pathway heatmap panels show some key upregulated pathways in HTE and CTE clusters based on multi-omics (“Common”) or global protein abundance only (FDR <0.01, Fisher’s exact test). The expression heatmap panel depicts the RNA and protein levels of various markers involved in immune evasion mechanisms.
(B) Association between mutation profiles and immune/stromal signatures from xCell. Only associations significant at FDR < 0.05 are shown.
(C) xCell scores for conventional dendritic cells (cDC) and macrophages for NAT samples (x-axis) and tumor samples (y-axis). Scatterplots indicate if a given sample shows significant infiltration by either dendritic cells (left) or macrophages (right) (xCell p-value < 0.05) in both NAT and tumor (black), only in NAT (blue), only in tumor (red), or in neither NAT nor tumor (light-gray). Samples with STK11 mutations are displayed with a triangle. STK11 mutation was found enriched in the subset of samples with infiltration of macrophages and dendritic cells only in NATs (Fisher’s exact test, FDR <0.1).
(D) Boxplots show association between STK11 mutation and immune score (ESTIMATE).
(E) t-SNE (t-Distributed Stochastic Neighbor Embedding) plot provides a two-dimensional representation of the activation scores of individual STK11 mutated (orange) and WT (blue) tumor histopathology tiles submitted to a deep learning algorithm. Examples of true positive (red outline) and negative (black outline) tiles exhibit different histologic features. STK11 WT tiles correctly recognized by the model harbor abundant inflammatory cells, whereas STK11 mutant tiles showed typical adenocarcinoma characteristics without inflammation.
(F) Cluster diagram representing pathways significantly associated with STK11 mutation-enriched cluster IC-068 (Figure S5F) in protein-based unsupervised ICA clustering. The Metascape output represents enriched biological concepts as nodes, aggregates those nodes into clusters based on the similarity of their protein membership, and names the clusters based on their most significant node. Node size represents the number of differentially expressed gene products. Amongst the top 20 clusters, the one representing neutrophil degranulation showed highest significance (Q value < 10−14). The top 5 clusters by p-value are highlighted.
G) Scatterplot shows differentially regulated protein and RNA expression (signed -log 10 p-value) in tumors with and without STK11 mutation. Proteins associated with neutrophil degranulation are highlighted in red.
Various metabolic pathways were upregulated in CTE cluster tumors (Figure 5A, Table S5). Glycolysis, which has been implicated in immune evasive mechanisms in many solid tumors but only marginally in LUAD (Ganapathy-Kanniappan, 2017) (Giatromanolaki et al., 2019), was significantly upregulated only in proteomics data, as were “Peroxisome” and “PPAR Signaling Pathway” activities (both FDR < .001) (Figure 5A, middle panel, Table S5). Several studies have shown that IFN gamma (IFNG) promoter activity can be inhibited by PPAR-gamma activation (Marx et al., 2000), and that suppression of the inflammatory immune response by PPAR-gamma activation may be achieved through induction of immune cell apoptosis. PPAR-gamma activation was shown to impair T-cell proliferation through an IL-2 dependent mechanism, while PPAR-beta activation was shown to favor oxidation of fatty acids and glucose in developing T-cells (Le Menn and Neels, 2018). In addition, CTE tumors showed upregulation of cell-cell junction and other proteins that provide barrier functions for epithelium, suggesting a mechanical barrier against immune cell infiltration (Figure 5A, Table S5; cf Figure 1E) (Salerno et al., 2016) (Streeck et al., 2011).
As an orthogonal assessment of the immune landscape of LUAD, we ranked tumors by activity of the IFNG axis, which is responsible for activation of the adaptive immune system (Abril-Rodriguez and Ribas, 2017), and assessed regulation of established protein markers of immune evasion (Achyut and Arbab, 2016; Allard et al., 2016a; Liu et al., 2018). The protein abundance of some important immune evasion markers (Jerby-Arnon et al., 2018), including IDO1, was upregulated in both the HTE and INFG-high clusters (Figures 5A, S5A). IDO1 has well-documented roles in angiogenesis, EMT (Zhang et al., 2019a), and cancer immunosuppression (Liu et al., 2018); hence IDO1 inhibition may represent an additional therapeutic opportunity in immune hot LUAD tumors (Kozuma et al., 2018a; Takada et al., 2019). Other important immune evasive or immune-related markers were also observed. The pulmonary epithelium is a physical barrier that produces antimicrobial mucus and surfactant proteins, facilitates host-microbiota interactions to control mucosal immunity, and is critical for tumor development (Whitsett and Alenghat, 2015). Upregulation of immunosuppressive components of the pulmonary epithelium barrier, including MUC5B and WFDC2 (HE4), was observed in the CTE cluster of lung tumors (Figure 5A, lower panel) (Parikh et al., 2019; Roy et al., 2014), and surfactants SFTPB, DMBT1, SFTPA1, and SFTPD were increased in tumors with low IFNG axis scores (Figure S5B) (Nayak et al., 2012; Seifart et al., 2005; Wang et al., 2009).
Notably, the NAT-enriched cluster had immune infiltration signatures that were intermediate between the HTE and CTE subtypes (Figure 5A), suggesting bi-directional regulation, with pro-inflammatory mechanisms in HTE and immune-evasive mechanisms in CTE tumors. The most dramatic down-regulation of immune activation was in STK11 mutant tumors, with marked reductions in xCell-derived Dendritic cell, Natural Killer T-cell and Macrophage signatures (Figure 5B, Table S5, FDR < 0.1). In striking contrast, STK11 mutant- associated NATs were enriched for dendritic cell and macrophage infiltration (Figure 5C, FDR < 0.1). ESTIMATE immune scores (Yoshihara et al., 2013), reduced for all STK11 mutants, were particularly low for those wild-type for KRAS (Figure 5D, Table S5). This immune downregulation was not due to low mutation burden, as NMF cluster C3, strongly enriched for STK11 mutants (Figure 1E), was second only to cluster C1 in somatic mutation burden (Figures S5C, D). The immune-cold landscape of STK11 mutant tumors proved to be the dominant feature in a deep-learning-based predictive algorithm for determining LUAD mutational status from histopathology that achieved 94% accuracy at the slide level (Figure 5E). The defining histopathologic features of STK11 mutant samples related to tumor epithelium, whereas STK11 WT samples were predominantly characterized by immune cells (Figure 5C).
To understand the mechanisms underlying the immune-cold phenotype of STK11 mutants, we examined differential RNA, protein and phosphoprotein expression between STK11 WT and mutant samples. Pathway enrichment identified neutrophil degranulation to be the signature most strongly associated with STK11 mutation. Notably, neutrophils did not appear to be either specifically enriched or depleted in STK11 mutant tumors (Figures 5A, 5B). Nevertheless, the robustness of this association was apparent even in unsupervised approaches. Independent component analysis (Liu et al., 2019) identified a cluster strongly enriched for STK11 mutant tumors, the defining proteomic pathway feature of which was neutrophil degranulation (Figures 5F, S5F, Table S5). All 16 of the measured proteins strongly associated with neutrophil degranulation were coherently overexpressed in STK11 mutant tumors (Figure S5G). This signal was not detectable at the RNA level as the proteins, following translation, are stored in the granules until later release (Figures 5G, S5G). Most of these proteins, including CAMP, LTF, BPI, MMP8, MMP9, MPO, LCN2, ELANE and ARG1, have established immune modulatory functions, collectively suggesting a compelling hypothetical mechanism that may account for some of the immunologic effects of STK11 mutation.
Characterization of smoking-related phenotype in tumors and NATs
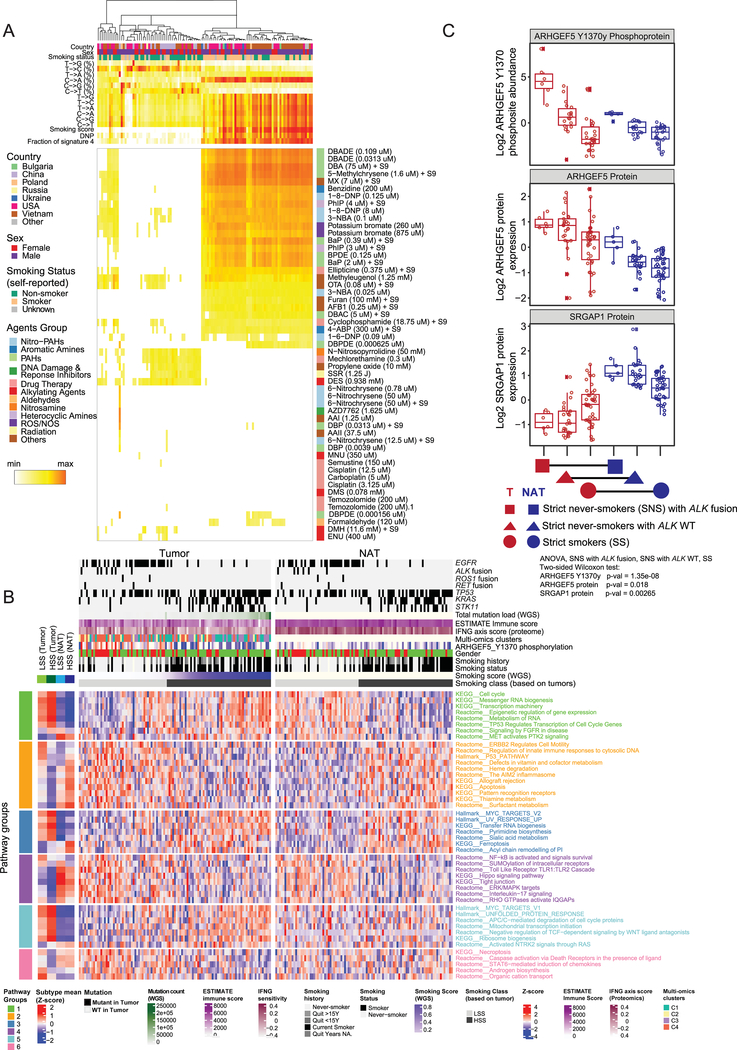
In order to better characterize the influence of smoking as a major contributor to LUAD, we used SignatureAnalyzer (Kim et al., 2016) (Figure S6A, Table S6) to identify the dominant di-nucleotide polymorphisms (DNP) GG->TT or CC->AA (~50%) associated with smoking status. We then integrated tumor purity estimates, counts of total mutations, and percentages that are smoking-signature mutations and smoking-signature DNPs into a continuous smoking signature score, and defined High and Low Smoking Scores (HSS, LSS) (Figure S6B, Table S6). No fully independent smoking effect emerged from linear models adjusted for known confounders including mutation status, sex and place of origin. However, conventional differential protein and pathway analysis to identify potential carcinogenic or tumor-supportive mechanisms specific to never-smokers (NS) identified a set of proteins with prior evidence of relevance to LUAD biology (Table S6). Regression of the 96 possible trinucleotide mutation combinations between the samples in our cohort and the environmental signatures reported by Kucab and colleagues (Kucab et al., 2019) found strong correlations in many samples of signatures of polycyclic aromatic hydrocarbons (PAHs) known to be present in cigarette smoke, including DBADE, DBA, and 5-Methylchrysene (Figure 6A, Table S6). Moreover, these cases correlated highly with our smoking score and with self-reported smoker status (Figure 6A). Other environmental contributors, evidently unrelated to cigarette smoking, were nevertheless also strongly correlated (Figure S6C), suggesting caution in interpreting these mutational associations and emphasizing the need for comprehensive clinical annotation including details on environmental and occupational exposures and dietary habits.
Figure 6: Environmental and smoking-related molecular signatures.
(A) Heatmap showing correlation coefficients between the mutational signatures of LUAD tumor samples and 53 signatures of environmental exposure (Kucab et al., 2019). Self-reported smoking status, derived smoking score, di-nucleotide polymorphism (DNP) status, and the fraction of Cosmic signature 4 are shown.
(B) Impact of tumor-derived high or low smoking score (HSS; >0.1; LSS; <0.1) on pathways associated with protein expression in tumors and paired NATs. The heatmaps show protein expression-derived, differentially regulated (FDR <0.05) pathways associated with LSS and HSS, separately in tumors (left) and NATs (right). Pathway Groups (PG1–6) are defined according to the patterns of differential HSS/LSS expression in tumors and NATs. A complete list of differentially activated pathways is provided in Table S6.
C. Boxplots showing log2 relative abundance of ARHGEF5 phosphosite Y1370, ARHGEF5 and SRGAP1 protein expression in tumors and NATs from strict never-smokers (SNS) with and without ALK fusion and from strict smokers (SS). None of the SS tumors had ALK fusion. ANOVA test was performed on tumor samples only.
As reported for other cancers (Malta et al., 2018), tumors showed significantly higher RNA-based stemness index compared to NATs (Figure S6D). Within both tumors and NATs, samples with HSS showed higher stemness than samples with LSS (Figure S6E), consistent with the known field cancerization effect of tobacco exposure (Walser et al., 2008).
We identified 6 patterns of differential pathway regulation between tumor and paired NAT samples with HSS and LSS (Figure 6B, Table S6). Pathways including cell cycle and transcription machinery were reduced in NATs with HSS compared to LSS, but this pattern was reversed in tumors (Pathway Group (PG)1). Contrariwise, the AIM2 inflammasome, P53 pathway activity, and apoptosis were higher in NATs with HSS than LSS, but lower in HSS tumors, consistent with smoking-related tumors more effectively inactivating tumor suppressors and overcoming immune surveillance and apoptosis (PG2). HSS had parallel effects on tumors and NATs in higher MYC target activity and ferroptosis, and lower Hippo pathway signaling and NF-kB and IL-17 activity (PG3 and 4). Finally, pathways including the unfolded protein response and RAS signaling through NTRK2 were higher in tumors but not NATs with HSS, while necroptosis and caspase signaling through death receptors were lower (PG5 and 6). Notably, the smoking signature-associated pathway-level differences that defined pathway groups 1–4 were more prominent on the protein than RNA level (Figure S6F).
Among the proteins differentially regulated in smokers and never-smokers were Rho GTPase signaling pathway members ARHGEF5 and its phosphosite ARHGEF5_Y1370y, elevated in SNS, and SRGAP1, suppressed in SNS (Figures 6C and PG4 in 6B). ARHGEF5_Y1370y levels were highest in patients with ALK fusion, consistent with its extreme outlier status (Figure 2F). Activating phosphorylation of ARHGEF5 by tyrosine kinases (e.g. EML4-ALK), accompanied by downregulation of the negative Rho GTPase regulator SRGAP1, may lead to hyperactivation of Rho GTPase signaling and tumorigenesis in a subset of non-smoking patients. Auto-inhibitory peptides blocking the activity of ARHGEF5 have been described (He et al., 2015; Huang et al., 2015) and represent a potential therapeutic intervention in this population. Differential pathway analysis also provided evidence that, in non-smokers, the cytoprotective and anti-inflammatory stress response Heme oxygenase system might contribute to tumor survival (see also PG2, Figure 6B). This process can potentially be inhibited by metalloporphyrins or imidazole-based drugs (Podkalicka et al., 2018).
Tumor-NAT comparisons reveal tumorigenic changes and biomarker candidates
Proteogenomic profiles were derived for both tumors and paired NATs, presenting a unique opportunity to explore proteogenomic remodeling upon tumorigenesis (Table S7). Protein-level PCA showed tumor and much more homogenous NAT populations to be completely distinct (Figures 7A, S7A). Enrichment analysis of differential protein abundance between paired tumor and NAT samples (Figure S7B, Table S7) revealed that tumorigenic processes including cell cycle progression, MYC targets and glycolysis were upregulated in tumor samples (FDR < 0.001) (Figure S7C, Table S7). We observed 70 phosphosites [31 up, 39 down] and 11 acetyl-sites [10 up, 1 down]) for which abundance in tumors was markedly differential relative to associated protein expression, indicating a change in site stoichiometry (Table S7). NPM1 T199 showed the highest level of phosphorylation in tumors (log2 FC >5, FDR < 0.01); phosphorylation of the T199 residue is known to be critical for NPM1-mediated DNA damage repair (Koike et al., 2010) (Table S7). Of note, proliferation marker MKI67 phosphorylation was dramatically upregulated in tumors (log2 FC>5) relative to its protein abundance (log2 FC<2) (Figure 7B). Acetylsite regulation included hyper-acetylation of the EP300 substrate, Histone 2B (HIST1H2BA K22/K25, log2 FC >4–5) (Weinert et al., 2018). Interestingly, we also observed significant acetylation of EP300 K1558 (log 2 FC >4), a key acetylation site in the protein activation loop that may be indicative of its activity (Thompson et al., 2004). HIBCH, associated with valine metabolism, was the only protein distinctly hypoacetylated in tumors (K358; -log2 FC >4).
Figure 7: Summary of global proteogenomic alterations in tumors and paired NATs.
(A) Principal component analysis of protein expression shows distinct separation of tumor samples (n=110) and NATs (n=101). The larger rectangle and triangle represent the centroids of the distributions.
(B) Scatterplots show the median log2 fold-change between tumors and paired NATs in the proteome vs phosphosites (left) and acetylsites (right). The dashed line shows equivalence with intercept 0. Red triangles indicate sites with at least log2 4-fold site-level increased abundance compared to associated protein changes between log2, +2 and −2-fold. Blue triangles represent downregulated sites using symmetric parameters (Full list in Table S7).
(C) Proteomics-based biomarker candidates (log2 fold change (log2FC) > 2 and FDR <0.01 in ≥ 80% of tumor-NAT pairs) for tumors with any of 4 frequently mutated genes. Numbers in parentheses show candidates displayed / identified. Each dot represents a tumor sample. Blue-colored boxplots highlight proteins with overexpression in more than 99% of tumor samples with the associated mutation. Protein functional groups and relevant clinical trial drug targets of the biomarker candidates are shown in the accompanying schematic.
(D) Volcano plot showing the enrichment score (x-axis) and associated log p-value (y-axis) of differentially regulated phosphosite-driven signatures between tumors and matched NATs as assessed by PTM Signature Enrichment Analysis (Krug et al., 2018). Significant (FDR <0.05) signatures are highlighted in shades of brown. The size of the circles shows the overlap between phosphosites detected in our dataset and the phosphosite-specific signatures in PTMsigDB (Krug et al., 2018).
(E) Rank plots depicting differential phosphosite-driven signatures (1.5 x interquartile range, IQR) between tumor and paired NATs in tumors with mutations in EGFR (N=38) or KRAS (N=33). Residual enrichment scores (y-axis) were calculated between mutated tumors (EGFR or KRAS) and all other tumors in order to highlight tumor / NAT differences in tumors harboring each specific mutation.
(F) Heatmap representing tumor antigens including neoantigens (top panel) and cancer testes (CT) antigens (downloaded from CT database (Almeida et al., 2009)). “DNA repair” indicates mutation in DNA repair genes (POLE, MLH1, MLH3, MSH3, MSH4, MSH6, BRCA1, BRCA2). Displayed CT antigen proteins were overexpressed at least 2-fold in tumors compared to paired NATs in more than 10% of samples.
Deep proteogenomics characterization of LUAD tumors and paired NATs also provided a powerful dataset to nominate candidate biomarkers. Using stringent cutoffs for quantitative difference, significance and consistency (log2 FC >2, FDR <0.01, and differential in ≥90% of all Tumor-NAT pairs), we identified 289 proteins upregulated at the protein level (Table S7). The potential clinical utility of these protein markers is annotated in Figure S7D, with orthogonal support provided by the proportions of tumors in the Human Protein Atlas (HPA) showing high, medium or low IHC staining. Sixty of these proteins (Figure S7D: Pan-LUAD) were also significantly differential at the RNA level, of which 5 (GFPT1, BZW2, PDIA4, P4HB, PMM2) were upregulated in all tumor samples compared to their paired NATs, extending data implicating these metabolic enzymes in cancer (Chen et al., 2002; Tufo et al., 2014; Yang et al., 2016). Gremlin 1 (GREM1) protein, highly overexpressed in tumors (log2 FC >5, FDR <0.01) in our study, is a known marker of poor prognosis in lung cancer (Mulvihill et al., 2012), and implicated in EMT and metastasis processes (Figure S7D, Table S7) (Cleynen et al., 2007; Friedman et al., 2004; Tang et al., 2019). Ovarian cancer immunoreactive antigen domain containing 2 (OCIAD2), highly overexpressed in tumors (log2 FC > 4, FDR <0.01), is a known poor prognosis marker (Sakashita et al., 2018), as are stress-related marker candidates DHFR, HYOU1, LDHA, and CBX8 (Fahrmann et al., 2016; Llado et al., 2009; Takei et al., 2017). Significantly hyperphosphorylated and hyperacetylated sites are described in Table S7. While only a few amongst these marker candidates are currently targeted by therapeutics in clinical trials, their strong and consistent differential expression and associations with lung cancer biology and decreased survival support potential utility in early detection and prognostic stratification (Kim et al., 2018a; Mulvihill et al., 2012; Sakashita et al., 2018; Wang et al., 2015).
We also explored mutation-specific tumor - NAT differential expression in TP53, EGFR, KRAS and STK11 mutant phenotypes (Figures 7C, S7D, Table S7). Patients with TP53 mutant tumors show high expression of TP53, CCNA2, TOP2A, PLOD2, ANLN, and MMP12 (Figure 7C), all shown to have roles in tumorigenesis (Chen et al., 2015; Hosgood et al., 2008; Konofaos et al., 2013; Qu et al., 2009; Song et al., 2013). The observed elevated CDK1 and CCNB1 protein expression and CDK1 phosphorylation in TP53 mutants have been associated with resistance in preclinical models modulated by p53 status (Schwermer et al., 2015). Significant overexpression of the proto-oncogene MET was noted in EGFR mutants. Extracellular glycoproteins, collagens and enzymes were enriched in KRAS mutant tumors, as were the well-described KRAS-associated chemokine CXCL8 and immune target THY1 (Sunaga et al., 2012). STK11 mutant tumors were enriched for amino acid metabolism proteins, which are associated with nitric oxide metabolic processes, suggesting perturbation of the urea cycle in the context of STK11 mutation (Kim et al., 2017; Lam et al., 2019).
Phosphosite-specific pathway analyses (Krug et al., 2018) of the entire population of tumor/NAT pairs showed upregulated phosphosite-driven signatures chiefly of checkpoint control and cell cycle progression in tumors (Figure 7D, Table S7) compared to extracellular matrix-focused signatures in paired NATs. Phosphosite-driven signatures that were differential between NATs and paired tumors with EGFR (N=38) or KRAS (N=33) mutations yielded near-mirror image plots (Figure 7D, Table S7). KRAS mutant tumors showed site-driven activation of pathways downstream of RAS, including MAPK1, as well as of TAK1, the hub at which IL1, TGF-β and Wnt signaling pathways converge (Santoro et al., 2017). Pathways upregulated in EGFR mutant tumors included ROCK1, a Rho-associated protein kinase that has been shown to enhance EGFR activation in some cancer types (Nakashima et al., 2011).
Cancer testis (CT) antigens and tumor neoantigens can serve both diagnostic and therapeutic roles, including as targets for potential cancer vaccines. Of 44 CT antigens recurrently over-expressed in tumors (fold-change ≥2), 9 were observed in ≥10% of samples (Figure 7F). KIF2C was the most ubiquitous, being highly expressed in 63% of samples. Seven of these 9 common CT antigens have been previously associated with lung cancer (Bai et al., 2019; Lei et al., 2015; Loriot et al., 2003; Scanlan et al., 2000; Xie et al., 2018; Zhao et al., 2017), although their specific roles in tumorigenesis and progression are unclear. IGF2BP3 is associated with tumor progression and poor prognosis in colorectal, lung and hepatocellular carcinomas (Jiang et al., 2008; Lochhead et al., 2012; Xu et al., 2012), while AKAP4 has been proposed to be a potential biomarker in NSCLC (Loriot et al., 2003). To our knowledge, MORC1 and NUF2 are novel CT antigens in LUAD tumors, covering 38% and 16% of patients, respectively. To identify additional predicted tumor neoantigens, we also searched for both RNA transcripts and peptides containing evidence of somatic mutations. We identified a total of 2481 mRNA-validated and 49 peptide-validated somatic mutations, corresponding to 104 patients (Figure 7F, Table S7). Overall, 97 samples had evidence of either CT antigens or neoantigens, holding promise for the future of immunotherapy-based approaches to LUAD management.
Discussion
In this study, we report comprehensive proteogenomic characterization of 110 LUAD tumors and 101 matched NATs. Unlike TCGA, which included primarily smoking-related LUAD, our cohort included roughly equal numbers of current or former smokers and never-smokers, as well as a geographically diverse population. Multi-omics unsupervised clustering showed that previously-described terminal respiratory unit and proximal-inflammatory clusters translate to the protein level, while proximal-proliferative samples showed substructure based on TP53 status and place of origin. miRNA taxonomy included clusters enriched for STK11 mutant and ALK fusion-driven tumors. We observed consistent differential phosphorylation of ALK Y1507 in samples with ALK fusion, in addition to multiple other proteins exclusively regulated at the level of phosphoproteome, underscoring their likely relevance to ALK-associated biology.
The inclusion of deep-scale proteomic and PTM data allowed us to track the downstream signaling consequences of epigenetic and genomic alterations and identify putative methylation cis-effects and a novel KEAP1/NFE2L2 regulatory mechanism. Extreme phosphorylation events implied therapeutic possibilities including SOS1 inhibition in KRAS mutant and PTPN11/Shp2 inhibition in both ALK fusion-and EGFR mutant tumors, the latter amenable to inhibitors already in clinical trials. We also systematically identified and annotated outlier kinases, some unique to major mutational subtypes, many of which have known inhibitors or appear to be druggable. Outliers were predominantly phosphorylation events, reinforcing the value of post-translational modification analysis. Paired tumor-NAT analysis illuminated elements of oncogenesis and nominated biomarker candidates and potential drug development targets.
Integrated proteogenomics further allowed extensive characterization of the immune landscape of LUADs and identification of a number of potential therapeutic vulnerabilities, including anti-CTLA4 therapy and IDO1 inhibition in immune-hot tumors. We highlighted the particular association of STK11 mutation with immune-cold behavior, and implicated neutrophil degranulation as a potential immunosuppressive mechanism in STK11 mutant LUAD evident only in the proteomics space. The combination of proteogenomic data, balanced representation of smokers and never-smokers, and paired tumor / NAT analyses enabled us to capture the impact of cancerization in both tumors and adjacent tissues, and highlighted a potential oncogenic mechanism centered on ARHGEF5 in never-smokers.
There are inherent limitations to a study of this type. The interdependence of variables including mutational status, ethnicity or geography, gender and smoking status require that comparisons based on any one of these be interpreted with caution. Furthermore, given the large number of confounders, efforts to adjust for this by linear modeling may not be effective in a dataset of this size, frustrating association analyses such as for gender and smoking effects. This effort shares with all bulk tumor analyses the lack of spatial and cellular resolution that might add orthogonal insights into tumor biology, such as by disambiguating the contributions of tumor epithelium and microenvironment. Approaches geared to more spatially resolved proteogenomics, such as we and others have recently described (Hunt et al., 2019; Satpathy et al., 2020), or integration of single cell genomics and proteomics, might add nuance to our understanding of crosstalk between tumor and the microenvironment or of tumor evolution. Most importantly, associations of the sort described throughout this manuscript are hypothesis-generating, and generally cannot be understood as providing firm biological conclusions. The integration of deep-scale proteomic and PTM data nevertheless represents a substantial advance over prior genomics studies of LUAD, and paired with microscaling methods (Satpathy et al., 2020) points the way to improved characterization of clinical cohorts. We hope that both the specific observations and hypotheses delineated in this manuscript, and the data that underlie them, will be a rich resource for those investigating LUAD and for the larger research community, including for the development of targeted chemo- or immuno-therapies.
STAR⋆METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests should be directed to and will be fulfilled by the lead author, M.A.G. (gillette@broadinstitute.org).
Material availability
This study did not generate new unique reagents.
Data and Code Availability
Proteomics raw datasets are publicly available though the CPTAC data portal https://cptac-data-portal.georgetown.edU/cptac/s/S056
Genomic and transcriptomic data files can be accessed at the Genomic Data Commons (GDC); https://portal.gdc.cancer.gov/, via dbGaP Study Accession: phs001287.v5.p4 https://www.ncbi.nlm.nih.gov/proiects/gap/cgi-bin/study.cgi7studvid=phs001287.v5.p4
Sample annotation, processed and normalized data files are provided as Tables S1–S3.
Software and code used in this study are referenced in their corresponding STAR Method sections and also the Key Resource Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-CD8 (C8/144B) | Cellmarque | Catalog #108M |
| Rabbit monoclonal anti-CD4 (SP35) | Roche | Catalog #790-4423 |
| Liquid Concentrated Monoclonal Antibody anti-CD163 | Leica Biosystems | Catalog #NCL-L-163 |
| PTMScan Acetyl-lysine Kit | Cell Signaling Technology | Catalog: 13416 |
| Biological Samples | ||
| Primary tumor samples | See Experimental Model and Subject Details | N/A |
| Chemicals and Reagents | ||
| HPLC-grade water | J.T. Baker | Catalog: 4218-03 |
| Urea | Sigma | Catalog: U0631 |
| Sodium chloride | Sigma | Catalog: 71376 |
| 1M Tris, pH 8.0 | Invitrogen | Catalog: AM9855G |
| Ethylenediaminetetraacetic acid | Sigma | Catalog: E7889 |
| Aprotinin | Sigma | Catalog: A6103 |
| Leupeptin | Roche | Catalog: 11017101001 |
| Phenylmethylsulfonyl fluoride | Sigma | Catalog: 78830 |
| Sodium fluoride | Sigma | Catalog: S7920 |
| Phosphatase inhibitor cocktail 2 | Sigma | Catalog: P5726 |
| Phosphatase inhibitor cocktail 3 | Sigma | Catalog: P0044 |
| Dithiothretiol, No-Weigh Format | Fisher Scientific | Catalog: 20291 |
| Iodoacetamide | Sigma | Catalog: A3221 |
| Lysyl endopeptidase | Wako Chemicals | Catalog: 129-02541 |
| Sequencing-grade modified trypsin | Promega | Catalog: V511X |
| Formic acid | Sigma | Catalog: F0507 |
| Acetonitrile | Honeywell | Catalog: 34967 |
| Trifluoroacetic acid | Sigma | Catalog: 302031 |
| Tandem Mass Tag reagent kit – 11plex | ThermoFisher | Catalog: A34808 |
| 0.5M HEPES, pH 8.5 | Alfa Aesar | Catalog: J63218 |
| Hydroxylamine solution, 50% (vol/vol) in H2O | Aldrich | Catalog: 467804 |
| Methanol | Honeywell | Catalog: 34966 |
| Ammonium hydroxide solution, 28% (wt/vol) in H2O | Sigma | Catalog: 338818 |
| Ni-NTA agarose beads | Qiagen | Catalog: 30410 |
| Iron (III) chloride | Sigma | Catalog: 451649 |
| Acetic acid, glacial | Sigma | Catalog: AX0073 |
| Potassium phosphate, monobasic | Sigma | Catalog: P0662 |
| Potassium phosphate, dibasic | Sigma | Catalog: P3786 |
| MOPS | Sigma | Catalog: M5162 |
| Sodium hydroxide | VWR | Catalog: BDH7225 |
| Sodium phosphate, dibasic | Sigma | Catalog: S9763 |
| Phosphate-buffered saline | Fisher Scientific | Catalog: 10010023 |
| iVIEW DAB Detection Kit | Roche | Catalog: 760-091 |
| Equipment | ||
| Reversed-phase tC18 SepPak, 3cc 200mg | Waters | Catalog: WAT054925 |
| Solid-phase C18 disk, for Stage-tips | Empore | Catalog: 66883-U |
| Stage-tip needle | Cadence | Catalog: 7928 |
| Stage-tip puncher, PEEK tubing | Idex Health & Science | Catalog: 1581 |
| PicoFrit LC-MS column | New Objective | Catalog: PF360-75-10-N-5 |
| ReproSil-Pur, 120 Å, C18-AQ, 1.9-μm resin | Dr. Maisch | Catalog: r119.aq |
| Nanospray column heater | Phoenix S&T | Catalog: PST-CH-20U |
| Column heater controller | Phoenix S&T | Catalog: PST-CHC |
| 300 μL LC-MS autosampler vial and cap | Waters | Catalog: 186002639 |
| Offline HPLC column, 3.5-μm particle size, 4.6 um × 250 mm | Agilent | Catalog: Custom order |
| Offline 96-well fractionation plate | Whatman | Catalog: 77015200 |
| 700 μL bRP fractionation autosampler vial | ThermoFisher | Catalog: C4010-14 |
| 700 μL bRP fractionation autosampler cap | ThermoFisher | Catalog: C4010-55A |
| 96-well microplate for BCA | Greiner | Catalog: 655101 |
| Microplate foil cover | Corning | Catalog: PCR-AS-200 |
| Vacuum centrifuge | ThermoFisher | Catalog: SPD121P-115 |
| Centrifuge | Eppendorf | Catalog: 5427 R |
| Benchtop mini centrifuge | Corning | Catalog: 6765 |
| Benchtop vortex | Scientific Industries | Catalog: SI-0236 |
| Incubating shaker | VWR | Catalog: 12620-942 |
| 15 mL centrifuge tube | Corning | Catalog: 352097 |
| 50 mL centrifuge tube | Corning | Catalog: 352070 |
| 1.5 mL microtube w/o cap | Sarstedt | Catalog: 72.607 |
| 2.0 mL microtube w/o cap | Sarstedt | Catalog: 72.608 |
| Microtube caps | Sarstedt | Catalog: 72.692 |
| 1.5 mL snapcap tube | ThermoFisher | Catalog: AM12450 |
| 2.0 mL snapcap tube | ThermoFisher | Catalog: AM12475 |
| Instrumentation | ||
| Microplate Reader | Molecular Devices | Catalog: M2 |
| Offline HPLC System for bRP fractionation | Agilent 1260 | Catalog: G1380-90000 |
| Online LC for LC-MS | ThermoFisher | Catalog: LC140 |
| Q Exactive Plus Mass Spectrometer | ThermoFisher | Catalog: IQLAAEGA APFALGMBDK |
| Q Exactive HF-X Mass Spectrometer | ThermoFisher | Catalog: 0726042 |
| Orbitrap Fusion Lumos Tribrid Mass Spectrometer | ThermoFisher | Catalog: IQLAAEGA APFADBMBHQ |
| Critical Commercial Assays | ||
| TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Gold | Illumina | Catalog: RS-122-2301 |
| Infinium MethylationEPIC Kit | Illumina | Catalog: WG-317-1003 |
| Nextera DNA Exosome Kit | Illumina | Catalog: 20020617 |
| KAPA Hyper Prep Kit, PCR-free | Roche | Catalog: 07962371001 |
| BCA Protein Assay Kit | ThermoFisher | Catalog: 23225 |
| Deposited Data | ||
| PhosphoSitePlus | (Hornbeck et al., 2012) | https://www.phosphosite.org |
| Connectivity Map (CMAP) | (Lamb et al., 2006; Subramanian et al., 2017) | https://www.broadinstitute.org/connectivity-map-cmap |
| Human Protein Atlas (HPA) | (Uhlén et al., 2005) | https://www.proteinatlas.org |
| CT Antigen database | (Almeida et al., 2009) | http://www.cta.lncc.br |
| Software and Algorithms | ||
| methylationArrayAnalysis (version 3.9) | (Maksimovic et al., 2016) | https://master.bioconductor.org/packages/release/workflows/html/methylationArrayAnalysis.html |
| Illumina EPIC methylation array (3.9) | Hansen KD, 2019 | https://bioconductor.org/packages/release/data/annotation/html/IlluminaHumanMethylationEPICanno.ilm10b2.hg19.html |
| Methylation array analysis pipeline for CPTAC | Li Ding Lab | https://github.com/ding-lab/cptac_methylation |
| miRNA-Seq analysis pipeline for CPTAC | Li Ding Lab | https://github.com/ding-lab/CPTAC_miRNA |
| Somatic variant calling pipeline for CPTAC | Li Ding Lab | https://github.com/ding-lab/somaticwrapper |
| VarDict | (Lai et al., 2016) | https://github.com/AstraZeneca-NGS/VarDict |
| Strelka2 | (Kim et al., 2018b) | https://github.com/Illumina/strelka |
| MUTECT1.1.7 | (Cibulskis et al., 2013) | https://software.broadinstitute.org/gatk/download/archive |
| VarScan2.3.8 | (Koboldt et al., 2012) | http://varscan.sourceforge.net |
| Pindel0.2.5 | (Ye et al., 2009) | http://gmt.genome.wustl.edu/packages/pindel/ |
| SignatureAnalyzer | (Kim et al., 2016) | https://software.broadinstitute.org/cancer/cga/msp |
| Fusion calling pipeline for CPTAC | Li Ding Lab | https://github.com/cuidaniel/Fusion_hg38 |
| CNVEX | Marcin Cieslik Lab | https://github.com/mctp/cnvex |
| CRISP | Marcin Cieslik Lab | https://github.com/mcieslik-mctp/crisp-build |
| Spectrum Mill | Karl R. Clauser, Steven Carr Lab | https://proteomics.broadinstitute.org/ |
| ComBat (v3.20.0) | (Johnson et al., 2007) | https://bioconductor.org/packages/release/bioc/html/sva.html |
| DreamAI | Pei Wang Lab | https://github.com/WangLab-MSSM/DreamAI |
| GISTIC2.0 | (Mermel et al., 2011) | ftp://ftp.broadinstitute.org/pub/GISTIC2.0/GISTIC_2_0_23.tar.gz |
| iProFun | (Song et al., 2019) | https://github.com/WangLab-MSSM/iProFun |
| ESTIMATE | (Yoshihara et al., 2013) | https://bioinformatics.mdanderson.org/public-software/estimate/ |
| WebGestaltR | (Wang et al., 2017) | http://www.webgestalt.org/ |
| Joint Random Forest | (Petralia et al., 2016) | https://github.com/WangLab-MSSM/ptmJRF |
| GSVA | (Hänzelmann et al., 2013) | https://bioconductor.org/packages/release/bioc/html/GSVA.html |
| TCGAbiolinks | (Colaprico et al., 2016) | http://bioconductor.org/packages/release/bioc/html/TCGAbiolinks.html |
| TSNet | (Petralia et al., 2018) | https://github.com/WangLab-MSSM/TSNet |
| xCell | (Aran et al., 2017) | http://xcell.ucsf.edu/ |
| CPTAC LUAD Data Viewer | Steven Carr lab | http://prot-shiny-vm.broadinstitute.org:3838/CPTAC-LUAD2020/ |
| MODMatcher | (Yoo et al., 2014) | https://github.com/integrativenetworkbiology/Modmatcher |
| ConsensusClusterPlus | (Wilkerson and Hayes, 2010) | http://bioconductor.org/packages/release/bioc/html/CancerSubtypes.html |
| pyQUILTS (v1.0) | (Ruggles et al., 2016) | http://openslice.fenyolab.org/cgi-bin/pyquilts_cgi.pl |
| MS-GF+ | (Kim and Pevzner, 2014) | https://github.com/MSGFPlus/msgfplus |
| NeoFlow | Bing Zhang lab | https://github.com/bzhanglab/neoflow |
| netMHCpan | (Jurtz et al., 2017) | http://www.cbs.dtu.dk/services/NetMHCpan/ |
| Optitype | (Szolek et al., 2014) | https://github.com/FRED-2/OptiType |
| Customprodbj | (Wang and Zhang, 2013) | https://github.com/bzhanglab/customprodbj |
| PDV | (Li et al., 2019) | https://github.com/wenbostar/PDV |
| PepQuery | (Wen et al., 2019) | http://pepquery.org |
| PTM-SEA | (Krug et al., 2018) | https://github.com/broadinstitute/ssGSEA2.0 |
| Terra | Broad Institute data science platform. | https://terra.bio/ |
| CMap | (Lamb et al., 2006; Subramanian et al., 2017) | https://due.io/cmap |
| PTM-SEA | (Krug et al., 2018) | https://github.com/broadinstitute/ssGSEA2.0 |
| LIMMA v3.36 (R Package) | (Ritchie et al., 2015) | https://bioconductor.org/packages/release/bioc/html/limma.html |
| FactoMineR v1.41NMF (R - package) | (Gaujoux and Seoighe, 2010; Lê et al., 2008) | https://cran.r-project.org/web/packages/FactoMineR/index.html |
| MClust v5.4 (R package) | (Scrucca, Fop, Murphy and Raftery, 2017) | https://cran.r-project.org/web/packages/mclust/index.html |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Subjects
A total of 111 participants (73 males, 38 females, 35–81 years old) were included in this study, collected by 13 different tissue source sites from 8 different countries (Table S1). Only histopathologically-defined adult lung adenocarcinoma tumors were considered for analysis, with an age range of 35–81. Institutional review boards at tissue source sites, reviewed protocols and consent documentation adhering to the Clinical Proteomic Tumor Analysis Consortium (CPTAC) guidelines.
Clinical Data Annotation
Clinical data were obtained from tissue source sites and aggregated by an internal database called the CDR (Comprehensive Data Resource) that synchronizes with the CPTAC DCC. Clinical data can be accessed and downloaded from the DCC (Data Coordinating Center) at https://cptac-data-portal.georgetown.edu/cptac/s/S046. Demographics, histopathologic information, and treatment details were collected. LUAD histopathology was confirmed for all cases by at least 2 expert pathologists based on high resolution images of H&E sections. All histologic https://www.cancerimagingarchive.net/datascope/cptac/home/ and radiologic https://public.cancerimagingarchive.net/nbia-search/ details can be accessed from the listed webportals. The genotypic, clinical, geographical and other associated metadata is summarized in Table S1.
METHOD DETAILS
Specimen Acquisition
The tumor, normal adjacent tissue (NAT), and whole blood samples used in this manuscript were prospectively collected for the CPTAC project. Biospecimens were collected from newly diagnosed patients with LUAD who underwent surgical resection and had received no prior treatment for their disease, including chemotherapy or radiotherapy. All cases had to be of acceptable LUAD histology but were collected regardless of surgical stage or histologic grade. Cases were staged using the AJCC cancer staging system 7th edition (Edge et al., 2010). The tumor specimen weights ranged from 125 to 715 milligrams. The average tissue mass was 238 mg. For most cases, three to four tumor specimens were collected. Paired histologically-normal adjacent lung tissues (NATs) were collected from the same patient at tumor resection. Each tissue specimen endured cold ischemia for less than 40 minutes prior to freezing in liquid nitrogen; the average ischemic time was 13 minutes from resection/collection to freezing. Specimens were either flash frozen in liquid nitrogen or embedded in optimal cutting temperature (OCT) medium. Histologic sections obtained from top and bottom portions from each case were reviewed by a board-certified pathologist to confirm the assigned pathology. For samples to be deemed acceptable, the top and bottom sections had to contain an average of 50% tumor cell nuclei with less than 20% necrosis. Specimens were shipped overnight from the tissue source sites to the biospecimen core resource (BCR) located at Van Andel Research Institute, Grand Rapids, MI using a cryoport that maintained an average temperature of less than −140°C. At the biospecimen core resource, specimens were confirmed for pathology qualification and prepared for genomic, transcriptomic, and proteomic analyses. Selected specimens were cryopulverized using a Covaris CryoPREP instrument and material aliquoted for subsequent molecular characterization. Genomic DNA and total RNA were extracted and sent to the genome sequencing centers. The whole exome and whole genome DNA sequencing and methylation EPIC array analyses were performed at the Broad Institute, Cambridge, MA and RNA and miRNA sequencing was performed at the University of North Carolina, Chapel Hill, NC. Material for proteomic analyses were sent to the Proteomic Characterization Center (PCC) at the Broad Institute, Cambridge, MA.
Sequencing sample preparation
Our study sampled a single site of the primary tumor from surgical resections, with an internal requirement to process a minimum of 125mg of tumor issue and 50mg of NAT. DNA and RNA were extracted from tumor and NAT specimens in a co-isolation protocol using Qiagen’s QIAsymphony DNA Mini Kit and QIAsymphony RNA Kit. Genomic DNA was also isolated from peripheral blood (3–5mL) to serve as matched normal reference material. The Qubit™ dsDNA BR Assay Kit was used with the Qubit® 2.0 Fluorimeter to determine the concentration of dsDNA in an aqueous solution. Any sample that passed quality control and produced enough DNA yield to go through the multiple planned genomic assays was sent for genomic characterization. RNA quality was quantified using the NanoDrop 8000 and quality assessed using an Agilent Bioanalyzer. A sample of sufficient quantity that passed RNA quality control and had a minimum RIN (RNA integrity number) score of 7 was subjected to RNA sequencing. Identity matches for germline, normal adjacent tissue, and tumor tissue were confirmed at the BCR using the Illumina Infinium QC array. This beadchip contains 15,949 markers designed to prioritize sample tracking, quality control, and stratification.
Whole Exome Sequencing (WES)
Library construction and Hybrid Selection
Library construction was performed as described in (Fisher et al., 2011), with the following modifications: initial genomic DNA input into shearing was reduced from 3μg to 20–250ng in 50μL of solution. For adapter ligation, Illumina paired-end adapters were replaced with palindromic forked adapters, purchased from Integrated DNA Technologies (IDT), with unique dual-indexed molecular barcode sequences to facilitate downstream pooling. Kapa HyperPrep reagents in 96-reaction kit format were used for end repair/A-tailing, adapter ligation, and library enrichment PCR. In addition, during the post-enrichment SPRI cleanup, elution volume was reduced to 30μL to maximize library concentration, and a vortexing step was added to maximize the amount of template eluted. After library construction, libraries were pooled into groups of up to 96 samples. Hybridization and capture were performed using the relevant components of Illumina’s Nextera Exome Kit and following the manufacturer’s suggested protocol, with the following exceptions: First, all libraries within a library construction plate were pooled prior to hybridization. Second, the Midi plate from Illumina’s Nextera Exome Kit was replaced with a skirted PCR plate to facilitate automation. All hybridization and capture steps were automated on the Agilent Bravo liquid handling system.
Cluster Amplification and Sequencing
After post-capture enrichment, library pools were quantified using qPCR (KAPA Biosystems) using an automated assay on the Agilent Bravo with probes specific to the ends of the adapters. Based on qPCR quantification, libraries were normalized to 2nM. Cluster amplification of DNA libraries was performed following manufacturer’s protocol (Illumina) using exclusion amplification chemistry and flowcells. Flowcells were sequenced utilizing sequencing-by-synthesis chemistry. The flow cells were then analyzed using RTA v.2.7.3 or later. Each pool of whole exome libraries was sequenced on paired 76-cycle runs with two 8-cycle index reads across the number of lanes needed to meet coverage for all libraries in the pool. Pooled libraries were run on HiSeq4000 paired-end runs to achieve a minimum of 150x on-target coverage per library. The raw Illumina sequence data were demultiplexed and converted to FASTQ files; adapter and low-quality sequences were trimmed. The raw reads were mapped to the GRCh38/hg38 human reference genome and the validated BAMs were used for downstream analysis and variant calling.
Whole Genome Sequencing (WGS)
Cluster Amplification and Sequencing
An aliquot of genomic DNA (350ng in 50μL) was used as the input into DNA fragmentation (aka shearing). Shearing was performed acoustically using a Covaris focused-ultrasonicator, targeting 385bp fragments. Following fragmentation, additional size selection was performed using SPRI cleanup. Library preparation was performed using a commercially available KAPA Hyper Prep without amplification module kit (KAPA Biosystems) and with palindromic forked adapters with unique 8-base index sequences embedded within the adapter (IDT). Following sample preparation, libraries were quantified using quantitative PCR (KAPA Biosystems), with probes specific to the ends of the adapters using the automated Agilent’s Bravo liquid handling platform. Based on qPCR quantification, libraries were normalized to 1.7nM and pooled into 24-plexes.
Sample pools were combined with HiSeqX Cluster Amp Reagents EPX1, EPX2, and EPX3 into single wells on a strip tube using the Hamilton Starlet Liquid Handling system. Cluster amplification of the templates was performed according to the manufacturer’s protocol (Illumina) with the Illumina cBot. Flowcells were sequenced to a minimum of 15x on HiSeqX utilizing sequencing-by-synthesis kits to produce 151 bp paired-end reads. Output from Illumina software was processed by the Picard data processing pipeline to yield BAM files containing demultiplexed, aggregated, aligned reads. All sample information tracking was performed by automated LIMS messaging.
Array Based Methylation Analysis
The Methylation EPIC array uses an 8-sample version of the Illumina Beadchip capturing >850,000 methylation sites per sample. Two hundred and fifty nanograms of DNA was used for the bisulfite conversion using Infinium MethylationEPIC BeadChip Kit (Illumina). The EPIC array includes sample plating, bisulfite conversion, and methylation array processing. After scanning, the data was processed through an automated genotype-calling pipeline. Data output consisted of raw idats and a sample sheet.
RNA and miRNA sequencing
Quality Assurance and Control of RNA Analytes
All RNA analytes were assayed for RNA integrity, concentration, and fragment size. Samples for total RNA-seq were quantified on a TapeStation system (Agilent, Inc. Santa Clara, CA). Samples with RINs >7.0 were considered high quality and were considered for sequencing.
Total RNA-seq libraries were generated using 300 nanograms of total RNA using the TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Gold and bar-coded with individual tags following the manufacturer’s instructions (Illumina). Total RNA Libraries were prepared on an Agilent Bravo automated liquid handling system. Quality control was performed at every step, and the libraries were quantified using a TapeStation system.
Total RNA Sequencing
Indexed libraries were prepared and run on HiSeq4000 paired-end 75 base pairs to generate a minimum of 120 million reads per sample library with a target of greater than 90% mapped reads. The raw Illumina sequence data were demultiplexed and converted to FASTQ files, and adapter and low-quality sequences were trimmed. Samples were then assessed for quality by mapping reads to GRCh38/hg38, estimating the total number of mapped reads, amount of RNA mapping to coding regions, amount of rRNA in the sample, number of genes expressed, and relative expression of housekeeping genes. Samples passing this QA/QC were then clustered with other expression data from similar and distinct tumor types to confirm expected expression patterns. Atypical samples were then SNP typed from the RNA data to confirm source analyte. FASTQ files of all reads were then uploaded to the GDC repository.
miRNA-seq Library Construction
miRNA-seq library construction was performed from the RNA samples using the NEXTflex Small RNA-Seq Kit (v3, PerkinElmer, Waltham, MA) and barcoded with individual tags following the manufacturer’s instructions. Libraries were prepared on a Sciclone Liquid Handling Workstation. Quality control was performed at every step, and the libraries were quantified using a TapeStation system and an Agilent Bioanalyzer using the Small RNA analysis kit. Pooled libraries were then size selected according to NEXTflex kit specifications using a Pippin Prep system (Sage Science, Beverly, MA).
miRNA Sequencing
Indexed libraries were loaded on the HiSeq4000 to generate a minimum of 10 million reads per library with a minimum of 90% reads mapped. The raw Illumina sequence data were demultiplexed and converted to FASTQ files for downstream analysis. Resultant data were analyzed using a variant of the small RNA quantification pipeline developed for TCGA (Chu et al., 2016). Data from samples were assessed for the number of miRNAs called, species diversity, and total abundance before uploading to the GDC repository.
Mass Spectrometry methods
The protocols below for protein extraction, tryptic digestion, TMT-10 labeling of peptides, peptide fractionation by basic reversed-phase liquid chromatography, phosphopeptide enrichment using immobilized metal affinity chromatography, and LC-MS/MS were performed as previously described in depth (Mertins et al., 2018). Acetyl-enrichment was performed as described before (Svinkina et al., 2015; Udeshi et al., 2020) with modifications as indicated below.
Protein Extraction and Tryptic Digestion
Fifty milligrams (wet weight) of cryopulverized human LUAD and NAT samples were homogenized in lysis buffer at a ratio of about 200 uL lysis buffer for every 50 mg wet weight tissue. The lysis buffer consisted of 8 M urea, 75 mM NaCl, 1 mM EDTA, 50 mM Tris HCl (pH 8), 10 mM NaF, phosphatase inhibitor cocktail 2 (1:100; Sigma, P5726) and cocktail 3 (1:100; Sigma, P0044), 2 μg/mL aprotinin (Sigma, A6103), 10 μg/mL leupeptin (Roche, 11017101001), and 1 mM PMSF (Sigma, 78830). Lysates were centrifuged at 20,000 g for 10 minutes and protein concentrations of the clarified lysates were measured by BCA assay (Pierce). Protein lysates were subsequently reduced with 5 mM dithiothreitol (Thermo Scientific, 20291) for an hour at 37C and alkylated with 10 mM iodoacetamide (Sigma, A3221) for 45 minutes in the dark at room temperature. Prior to digestion, samples were diluted 4-fold to achieve 2 M urea with 50mM Tris HCl (pH 8). Digestion was performed with LysC (Wako, 100369–826) for 2 hours and with trypsin (Promega, V511X) overnight, both at a 1:50 enzyme-to-protein ratio and at room temperature. Digested samples were acidified with formic acid (FA; Fluka, 56302) to achieve a final volumetric concentration of 1 % (final pH of ~3), and centrifuged at 1,500 g for 15 minutes to clear precipitated urea from peptide lysates. Samples were desalted on C18 SepPak columns (Waters, 100mg, WAT036820) and dried down using a SpeedVac apparatus.
Construction of the Common Reference Pool
The proteomic and phosphoproteomic analyses of lung cancer samples were structured as TMT-10 plex experiments. To facilitate quantitative comparison between all samples across experiments, a common reference (CR) sample was included in each 10-plex. A common physical, rather than in silico reference was used for this purpose for optimal quantitative precision between TMT10-plex experiments. Considerations prior to creating the reference sample were that this sample needed to be of adequate quantity to cover all planned experiments for both the current “discovery” and future “confirmatory” sets with overhead for additional possible experiments. The CR includes nearly all the samples analyzed in the TMT experiments, yielding an internal reference that is representative of all the samples in the study. Making the CR as representative of the study as a whole was particularly important since, by definition, only analytes represented in the reference sample would be included in the final ratio-based data analyses.
111 unique tumor samples with 102 paired NAT samples were distributed amongst 25 10-plex experiments, with 9 individual samples occupying the first 9 channels of each experiment and the 10th channel being reserved for the CR sample. The first 8 channels of each experiment contained 4 tumor/NAT pairs, with each pair of patient samples adjacent to each other. All the tumors were in the C channels and all the NAT samples were in the N channels. Of the 25 130C channels, 9 contained unpaired tumors, 4 contained tumor-only CRs, 4 had NAT-only CRs, 2 were LUAD-derived CRs from a seperate study (unpublished, Taiwan LUAD study), 2 were replicate tumor samples, and 4 samples were 2 tumor/NAT paired patients, split for the purpose of confirming high-fidelity replication in the project.
To ensure capacity for additional experiments given a target input of 300 μg protein per channel per experiment, 30 mg total was targeted for reference material. To meet these collective requirements, after reserving 300 μg peptide / sample for individual sample analysis, an additional 150 μg for each sample with adequate remaining quantity was used for pooled CR generation. In total, 203 samples were selected for the combined tumor/NAT CR. To make the CR, tumor-only and NAT-only CRs were first created separately. 103 tumor samples and 100 NAT samples contributed to their respective pooled reference samples. After creating individual CRs, a pool of combined CR was made, consisting of 4.8 mg tumor-only reference and 4.8 mg NAT-only reference. The 9.6 mg pooled reference material was divided into 300 μg aliquots and frozen at −80°C until use. 3.9 mg of tumor-only and 3.9 mg of NAT-only reference pools were set aside for future combined tumor/NAT CR generation. The remaining tumor-only and NAT-only references were aliquoted into 300 pg amounts, dried down, and stored at −80C for future use.
Construction and utilization of the CR Sample
As a quality control measure, two “comparative reference” (“CompRef”) samples were generated as previously described (Li et al., 2013; Mertins et al., 2018) and used to monitor the longitudinal performance of the proteome, phosphoproteome, and acetylproteome workflows throughout the course of the project. Briefly, patient-derived xenograft tumors from established basal (WHIM2) and luminal-B (WHIM16) breast cancer intrinsic subtypes (Li et al., 2013) were raised subcutaneously in 8 week old NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice (Jackson Laboratories, Bar Harbor, ME) using procedures reviewed and approved by the institutional animal care and use committee at Washington University in St. Louis. All PDX models are available through the application to the Human and Mouse-Linked Evaluation of Tumors core at http://digitalcommons.wustl.edu/hamlet/. Xenografts were grown in multiple mice, pooled, and cryopulverized to provide a sufficient amount of material for the duration of the project. Using the same analysis protocol as for the patient samples, four proteome, phosphoproteome, and acetylproteome process replicates of each of the two xenografts were prepared as described below and run as TMT 10-plex experiments (5 aliquots of each PDX model/plex) at the beginning and end of the 25 patient plexes and interposed after patient plexes 8 and 16. Interstitial samples were evaluated for depth of coverage and for consistency in quantitative comparison between the basal and luminal models.
TMT-10 Labeling of Peptides
Desalted peptides, 300 μg per sample (based on peptide-level BCA after digestion), were labeled with 10-plex TMT reagents according to the manufacturer’s instructions (Thermo Scientific; Pierce Biotechnology, Germany). For each 300 μg peptide aliquot of an individual tumor sample, 2.4 mg of labeling reagent was used. Peptides were dissolved in 300 μL of 50 mM HEPES (pH 8.5) solution and labeling reagent was added in 123 μL of acetonitrile. After 1 h incubation with shaking and after confirming good label incorporation, 24 μL of 5% hydroxylamine was added to quench the unreacted TMT reagents. Good label incorporation was defined as having a minimum of 95% fully labeled MS/MS spectra in each sample, as measured by LC-MS/MS after taking out a 2.8 μg aliquot from each sample and analyzing 1.25 μg. If a sample did not have sufficient label incorporation, additional TMT was added to the sample and another 1 h incubation was performed with shaking. At the time that the labeling efficiency quality control samples were taken, an additional 4 μg of material from each sample was removed and combined as a mixing control. After analyzing the mixing control sample by LC-MS/MS, intensity values of the individual TMT reporter ions were summed across all peptide-spectrum matches and compared to ensure that the total reporter ion intensity of each sample met a threshold of +/− 15% of the common reference. If necessary, adjustments were made by either labeling additional material or reducing an individual sample’s contribution to the mixture, and analyzing a subsequent mixing control, until all samples met the threshold and were thus approximately 1:1:1. Differentially labeled peptides were then mixed (10 × 300 μg), dried down via vacuum centrifuge, and quenched, prior to desalting on a 200 mg C18 SepPak column.
Peptide Fractionation
To reduce sample complexity, peptide samples were separated by high-pH reversed-phase (RP) separation as described previously (Mertins et al., 2018). A desalted 3 mg, 10-plex TMT-labeled experiment (based on protein-level BCA prior to digestion) was reconstituted in 900 μL 5mM ammonium formate (pH 10) and 2% acetonitrile, loaded on a 4.6 mm x 250 mm RP Zorbax 300 A Extend-C18 column (Agilent, 3.5 μm bead size), and separated on an Agilent 1260 Series HPLC instrument using basic reversed-phase chromatography. Solvent A (2% acetonitrile, 4.5 mM ammonium formate, pH 10) and a nonlinear increasing concentration of solvent B (90% acetonitrile, 4.5 mM ammonium formate, pH 10) were used to separate peptides. The 4.5 mM ammonium formate solvents were made by 40-fold dilution of a stock solution of 180 mM ammonium formate, pH 10. To make 1L of stock solution, 25 mL of 28% (wt/vol) ammonium hydroxide (28%, density 0.9 g/ml, Sigma-Aldrich) was added to ~850ml of HPLC grade water, then ~35 mL of 10 % (vol/vol) formic acid (>95% Sigma-Aldrich) was added to titrate the pH to 10.0 before bringing the final volume to 1 liter with HPLC-grade water. The 96-minute separation LC gradient followed this profile: (min: %B) 0:0; 7:0; 13:16; 73:40; 77:44; 82:60; 96:60. The flow rate was 1 mL/min. Per 3 mg separation, 82 fractions were collected into a 96 deep-well x 2mL plate (Whatman, #7701– 5200), with fractions combined in a stepwise non-contiguous concatenation strategy and acidified to a final concentration of 0.1% FA as reported previously. An additional 14 fractions were collected from the 96 deep-well plate for fraction A, consisting of early-eluting fractions that tend to contain multi-phosphorylated peptides. 5% of the volume of each of the 24+A proteome fractions was allocated for proteome analysis, dried down, and re-suspended in 3% MeCN/0.1% FA (MeCN; acetonitrile) to a peptide concentration of 0.25 μg/μL for LC-MS/MS analysis. The remaining 95% of 24 concatenated fractions were further combined into 12 fractions, with fraction A as a separate fraction. These 13 fractions were then enriched for phosphopeptides as described below.
Phosphopeptide Enrichment
Ni-NTA agarose beads were used to prepare Fe3+-NTA agarose beads. In each phosphoproteome fraction, ~237.5 μg peptides (based on peptide-level BCA after digestion with uniformly distributed fractionation presumed) were reconstituted in 475 μL 80% MeCN/0.1% TFA (trifluoroacetic acid) solvent and incubated with 10 μL of the IMAC beads for 30 minutes on a shaker at RT. After incubation, samples were briefly spun down on a tabletop centrifuge; clarified peptide flow-throughs were separated from the beads; and the beads were reconstituted in 200 μL IMAC binding/wash buffer (80 MeCN/0.1% TFA) and loaded onto equilibrated Empore C18 silica-packed stage tips (3M, 2315). Samples were then washed twice with 50 μL of IMAC binding/wash buffer and once with 50 uL 1% FA, and were eluted from the IMAC beads to the stage tips with 3 × 70 uL washes of 500 mM dibasic sodium phosphate (pH 7.0, Sigma S9763). Stage tips were then washed once with 100 μL 1% FA and phosphopeptides were eluted from the stage tips with 60 μL 50% MeCN/0.1% FA. Phosphopeptides were dried down and re-suspended in 9 μL 50% MeCN/0.1%FA for LC-MS/MS analysis, where 4 μL was injected per run.
Acetylpeptide Enrichment
Acetylated lysine peptides were enriched using an antibody against the acetyl-lysine motif (CST PTM-SCAN Catalogue No. 13416). IMAC eluents were concatenated into 4 fractions (~750 μg peptides per fraction) and dried down using a SpeedVac apparatus. Peptides were reconstituted with 1.4ml of IAP buffer (5 mM MOPS pH 7.2, 1 mM Sodium Phosphate (dibasic), 5 mM NaCl) per fraction and incubated for 2 hours at 4°C with pre-washed (4 times with IAP buffer) agarose beads bound to acetyl-lysine motif antibody. Peptide-bound beads were washed 4 times with ice-cold PBS followed by elution with 100ul of 0.15% TFA. Eluents were desalted using C18 stage tips, eluted with 50% ACN and dried down. Acetylpeptides were suspended in 7ul of 0.1% FA and 3% ACN and 4ul were injected per run.
LC-MS/MS for Proteomics Analyses
Online separation was done with a nanoflow Proxeon EASY-nLC 1200 UHPLC system (Thermo Fisher Scientific). In this set up, the LC system, column, and platinum wire used to deliver electrospray source voltage were connected via a stainless steel cross (360μm, IDEX Health & Science, UH-906x). The column was heated to 50°C using a column heater sleeve (Phoenix-ST) to prevent over-pressuring of columns during UHPLC separation. From each peptide fraction, ~1ug (based on protein-level BCA prior to digestion with uniformly-distributed fractionation presumed), the equivalent of 12% of each global proteome sample in a 2 ul injection volume or 50% of each phosphoproteome sample in a 4 ul injection volume, was injected onto an in-house packed 22cm x 75um internal diameter C18 silica picofrit capillary column (1.9 μm ReproSil-Pur C18-AQ beads, Dr. Maisch GmbH, r119.aq; Picofrit 10um tip opening, New Objective, PF360–75-10-N-5). Mobile phase flow rate was 200 nL/min, comprised of 3% acetonitrile/0.1% formic acid (Solvent A) and 90% acetonitrile /0.1% formic acid (Solvent B). The 110-minute LC-MS/MS method consisted of a 10-min column-equilibration procedure; a 20-min sample-loading procedure; and the following gradient profile: (min:%B) 0:2; 1:6; 85:30; 94:60; 95;90; 100:90; 101:50; 110:50 (the last two steps at 500 nL/min flow rate). For acetylproteome analysis, the same LC and column setup was used, but the gradient was extended to 260 minutes with the following gradient profile: (min:%B) 0:2; 1:6; 235:30; 244:60; 245;90; 250:90; 251:50; 260:50 (the last two steps at 500 nL/min flow rate).
For proteome analysis, samples were analyzed with a benchtop Q Exactive HF-X mass spectrometer (Thermo Fisher Scientific) equipped with a nanoflow ionization source (James A. Hill Instrument Services, Arlington, MA). Data-dependent acquisition was performed using Q Exactive HF-X Orbitrap v 2.9 software in positive ion mode at a spray voltage of 1.5 kV. MS1 Spectra were measured with a resolution of 60,000, an AGC target of 3e6 and a mass range from 350 to 1800 m/z. The data-dependent mode cycle was set to trigger MS/MS on up to the top 20 most abundant precursors per cycle at an MS2 resolution of 45,000, an AGC target of 5e4, an isolation window of 0.7 m/z, a maximum injection time of 105 msec, and an HCD collision energy of 31%. Peptides that triggered MS/MS scans were dynamically excluded from further MS/MS scans for 45 sec. Peptide match was set to preferred for monoisotopic peak determination, and charge state screening was enabled to only include precursor charge states 2–6, with an intensity threshold of 9.5e4. Advanced precursor determination feature (APD) (Myers et al., 2018) was turned off using a software patch provided to us by Thermo Fisher Scientific allowing us to turn APD off in the tune file, Tune version 2.9.0.2926 (later versions of Exactive Tune 2.9 sp2 for the HFX have this option as standard).
For phosphoproteome and acetylproteome analysis, samples were analyzed with a benchtop Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) equipped with a NanoSpray Flex NG ion source. Data-dependent acquisition was performed using Xcalibur Orbitrap Fusion Lumos v3.0 software in positive ion mode at a spray voltage of 1.8 kV. MS1 Spectra were measured with a resolution of 60,000, an AGC target of 4e5 and a mass range from 350 to 1800 m/z. The data-dependent mode cycle time was set at 2 seconds with a MS2 resolution of 50,000, an AGC target of 6e4, an isolation window of 0.7 m/z, a maximum injection time of 105 msec, and an HCD collision energy of 36%. Peptide mode was selected for monoisotopic peak determination, and charge state screening was enabled to only include precursor charge states 2–6, with an intensity threshold of 1e4. Peptides that triggered MS/MS scans were dynamically excluded from further MS/MS scans for 45 sec, with a +/− 10 ppm mass tolerance. “Perform dependent scan on single charge state per precursor only” was enabled for phosphoproteome analysis and disabled for acetylproteome analysis.
Immunohistochemistry
Total ALK and phospho-ALK (Y1507) immunostainings were performed on representative tumor and matched NATs from the available cases that contained ALK, ROS1 or RET gene fusions. The antibodies used included anti-ALK primary rabbit monoclonal antibody (ALK(D5F3) XP, Cell Signaling Technology, cat #3633 at 1 in 250 dilution) and anti-phospho ALK rabbit monoclonal antibody (D6F1V, Cell Signaling Technology, cat#14678 at 1:500 dilution). Briefly, 5-micron formalin fixed, paraffin sections were rehydrated and a heat-induced epitope retrieval was performed with citrate buffer (pH 6). Incubations with the respective antibodies were carried out overnight at 4 degrees C followed by buffer washes. For total-ALK, post-incubation with secondary antibody was done for 30 minutes and for phospho-ALK (Y1507), post-incubation was done initially with amplifier antibody (goat anti-rabbit IgG) for 15 minutes followed by secondary for 30 minutes. After buffer washes for total-ALK the signal was developed using DAB Peroxidase Substrate Kit (SK-4100; Vector laboratories) and for phospho-ALK using equal volumes of ImmPACT DAB EqV Reagent 1 (chromogen) and ImmPACT DAB EqV Reagent 2 (Diluent) for 5 minutes. Slides were counterstained with 50% Hematoxylin for 2 minutes, dehydrated, and cover-slipped. IHC was assessed for nuclear and cytoplasmic expression on tumor cells and the background was assessed in NATs (R.M. and R.M.).
Genomic Data Analysis
Copy Number Calling
Copy-number analysis was performed jointly leveraging both whole-genome sequencing (WGS) and whole-exome sequencing (WES) data of the tumor and germline DNA, using CNVEX (https://github.com/mctp/cnvex). CNVEX uses whole-genome aligned reads to estimate coverage within fixed genomic intervals, and whole-genome and whole-exome variant calls to compute B-allele frequencies at variable positions (we used TNScope germline calls). Coverages were computed in 10kb bins, and the resulting log coverage ratios between tumor and normal samples were adjusted for GC bias using weighted LOESS smoothing across mappable and non-blacklisted genomic intervals within the GC range 0.3–0.7, with a span of 0.5 (the target, blacklist, and configuration files are provided with CNVEX). The adjusted log coverage ratios (LR) and B-allele frequencies (BAF) were jointly segmented by custom algorithm based on Circular Binary Segmentation (CBS). Alternative probabilistic algorithms were implemented in CNVEX, including algorithms based on recursive binary segmentation (RBS) (Gey and Lebarbier, 2008), and dynamic programming (Bellman, 1961), as implemented in the R-package jointseg (Pierre-Jean et al., 2014). For the CBS-based algorithm, first LR and mirrored BAF were independently segmented using CBS (parameters alpha=0.01, trim=0.025) and all candidate breakpoints collected. The resulting segmentation track was iteratively “pruned” by merging segments that had similar LR, BAFs and short lengths. For the RBS- and DP-based algorithms, joint-breakpoints were “pruned” using a statistical model selection method (Lebarbier, 2005). For the final set of CNV segments, we chose the CBS-based results as they did not require specifying a prior on the number of expected segments (K) per chromosome arm, were robust to unequal variances between the LR and BAF tracks, and provided empirically the best fit to the underlying data.
Somatic Variant Calling
We called somatic variants for GDC-aligned WES BAMs by using the SomaticWrapper pipeline (https://github.com/ding-lab/somaticwrapper), which includes four different callers, i.e., Strelka v.2 (Saunders et al., 2012), MUTECT v1.7 (Cibulskis et al., 2013), VarScan v.2.3.8 (Koboldt et al., 2012), and Pindel v.0.2.5 (Ye et al., 2009). We kept SNVs called by any 2 callers among MUTECT v1.7, VarScan v.2.3.8, and Strelka v.2 and indels called by any 2 callers among VarScan v.2.3.8, Strelka v.2, and Pindel v.0.2.5. For the merged SNVs and indels, we applied a 14X and 8X coverage cutoff for tumor and normal, separately. We also filtered SNVs and indels by a minimal variant allele frequency (VAF) of 0.05 in tumors and a maximal VAF of 0.02 in normal samples. Finally, we filtered any SNV that was within 10bp of an indel found in the same tumor sample.
In step 13 of the SomaticWrapper pipeline, it combined adjacent SNVs into DNP (di-nucleotide polymorphisms) by using COCOON: As input, COCOON takes a MAF file from standard variant calling pipeline. First, it extracts variants within a 2bp window as DNP candidate sets. Next, if the corresponding BAM files used for variant calling are available, it extracts the reads (denoted as n_t) spanning all candidate DNP locations in each variant set, and then counts the number of reads with all the co-occurring variants (denoted as n_c) to calculate co-occurrence rate (r_c=n_c/n_t); If r_c ≥ 0.8, the nearby SNVs will be combined into DNP and annotation updated for the DNPs from the same codon based on the transcript and coordinates information in the MAF file. Among a total 32,250 somatic variants identified from the SomaticWrapper pipeline, there were 437 DNPs, in which 228 fell in the dominant smoking-related DNP type (CC->AA or GG->TT).
GISTIC and MutSig analysis
The Genomic Identification of Significant Targets in Cancer (GISTIC2.0) algorithm (Mermel et al., 2011) was used to identify significantly amplified or deleted focal-level and arm-level events, with Q value <0.25 considered significant. The following parameters were used:
Amplification Threshold = 0.1
Deletion Threshold = 0.1
Cap Values = 1.5
Broad Length Cutoff = 0.98
Remove X-Chromosome = 0
Confidence Level = 0.99
Join Segment Size = 4
Arm Level Peel Off = 1
Maximum Sample Segments = 2000
Gene GISTIC = 1
Each gene of every sample is assigned a thresholded copy number level that reflects the magnitude of its deletion or amplification. These are integer values ranging from −2 to 2, where 0 means no amplification or deletion of magnitude greater than the threshold parameters described above. Amplifications are represented by positive numbers: 1 means amplification above the amplification threshold; 2 means amplification larger than the arm level amplifications observed in the sample. Deletions are represented by negative numbers: −1 means deletion beyond the threshold; −2 means deletions greater than the minimum arm-level copy number observed in the sample.
The somatic variants were filtered through a panel of normals to remove potential sequencing artifacts and undetected germline variants. MutSig2CV (Lawrence et al. 2014) was run on these filtered results to evaluate the significance of mutated genes and estimate mutation densities of samples. These results were constrained to genes in the Cancer Gene Census (Sondka et al. 2018), with false discovery rates (q values) recalculated. Genes of q value < 0.1 were declared significant.
RNAseq and miRNAseq Quantification
RNAseq Quantification
Transcriptome data have been analyzed as described previously (Robinson et al., 2017), using the Clinical RNA-seq Pipeline (CRISP) developed at the University of Michigan (https://github.com/mcieslik-mctp/crisp-build). Briefly, raw sequencing data was trimmed, merged using BBMap, and aligned to GRCh38/hg38 using STAR. The resulting BAM files were analyzed for expression using feature counts against a transcriptomic reference based on Gencode 26. The resulting gene-level counts for protein-coding genes were upper-quartile normalized, transformed into RPKMs using edgeR, and log2 transformed. Genes quantified in fewer than 30% of all samples were removed from the data matrix. Data rows of redundant gene symbols were aggregated by calculating the average log2(RPKM).
For integrative multi-omics subtyping we normalized each gene by the median log2(RPKM) across all tumors (gene-centering) and applied the same per-sample normalization strategy used to normalize proteomics data tables (see below: Two-component normalization of TMT ratio distributions).
miRNA-Seq Data Analysis
miRNA-seq FASTQ files were downloaded from the CPTAC GDC API (https://docs.gdc.cancer.gov). TPM (Transcripts per million) values of mature miRNA and precursor miRNA were reported after adapter trimming, quality check, alignment, annotation, and reads counting (https://github.com/ding-lab/CPTAC_miRNA/blob/master/cptac_mirna_analysis.md). The mature miRNA expression was calculated irrespective of its gene of origin by summing the expression from its precursor miRNAs.
Unsupervised miRNA expression subtype identification was performed on mature miRNAs expression (log2 TPM) from 106 LUAD patients using Louvain clustering. (https://doi.org/10.5281/zenodo.595481). The expression of top 50 differentially expressed miRNAs from each miRNA-based subtype was shown in the heatmap (Figure S3J). For consistency, miRNA expression, RNA expression and protein expression were scaled to 0–1.
Proteomics Data Analysis
Spectrum quality filtering and searching
All MS data were interpreted using the Spectrum Mill software package v7.0 pre-release (Agilent Technologies, Santa Clara, CA) co-developed by Karl Clauser of the Carr laboratory (https://www.broadinstitute.org/proteomics). Similar MS/MS spectra acquired on the same precursor m/z within +/− 45 sec were merged. MS/MS spectra were excluded from searching if they failed the quality filter by not having a sequence tag length > 0 (i.e., minimum of two masses separated by the in-chain mass of an amino acid) or did not have a precursor MH+ in the range of 800–6000. MS/MS spectra were searched against a RefSeq-based sequence database containing 41,457 proteins mapped to the human reference genome (GRCh38/hg38) obtained via the UCSC Table Browser (https://genome.ucsc.edu/cgi-bin/hgTables) on June 29, 2018, with the addition of 13 proteins encoded in the human mitochondrial genome, 264 common laboratory contaminant proteins, and 553 non-canonical small open reading frames. Scoring parameters were ESI-QEXACTIVE-HCD-v2, for whole proteome datasets, and ESI-QEXACTIVE-HCD-v3, for phosphoproteome and acetylproteome datasets. All spectra were allowed +/− 20 ppm mass tolerance for precursor and product ions, 30% minimum matched peak intensity, and “trypsin allow P” enzyme specificity with up to 4 missed cleavages. Allowed fixed modifications included carbamidomethylation of cysteine and selenocysteine. TMT labeling was required at lysine, but peptide N-termini were allowed to be either labeled or unlabeled. Allowed variable modifications for whole proteome datasets were acetylation of protein N-termini, oxidized methionine, deamidation of asparagine, hydroxylation of proline in PG motifs, pyro-glutamic acid at peptide N-terminal glutamine, and pyro-carbamidomethylation at peptide N-terminal cysteine with a precursor MH+ shift range of −18 to 97 Da. For the phosphoproteome dataset the allowed variable modifications were revised to allow phosphorylation of serine, threonine, and tyrosine, allow deamidation only in NG motifs, and disallow hydroxylation of proline with a precursor MH+ shift range of −18 to 272 Da. For the acetylproteome dataset the allowed variable modifications were revised to allow acetylation of lysine, allow deamidation only in NG motifs, and disallow hydroxylation of proline with a precursor MH+ shift range of −400 to 70 Da.
Protein grouping, and localization of PTMs
Identities interpreted for individual spectra were automatically designated as confidently assigned using the Spectrum Mill autovalidation module to use target-decoy based false discovery rate (FDR) estimates to apply score threshold criteria. For the whole proteome dataset thresholding was done in 3 steps: at the peptide spectrum match (PSM) level, the protein level for each TMT-plex, and the protein level for all 25 TMT-plexes. For the phosphoproteome and acetylproteome datasets thresholding was done in two steps: at the PSM and variable modification (VM) site levels.
In step 1 for all datasets, PSM-level autovalidation was done first and separately for each TMT-plex experiment consisting of either 25 LC-MS/MS runs (whole proteome), 13 LC-MS/MS runs (phosphoproteome), or 4 LC-MS/MS runs (acetylproteome) using an auto-thresholds strategy with a minimum sequence length of 7; automatic variable range precursor mass filtering; and score and delta Rankl - Rank2 score thresholds optimized to yield a PSM-level FDR estimate for precursor charges 2 through 4 of <0.8% for each precursor charge state in each LC-MS/MS run. To achieve reasonable statistics for precursor charges 5–6, thresholds were optimized to yield a PSM-level FDR estimate of <0.4% across all runs per TMT-plex experiment (instead of per each run), since many fewer spectra are generated for the higher charge states.
In step 2 for the whole proteome dataset, protein-polishing autovalidation was applied separately to each TMTplex experiment to further filter the PSMs using a target protein-level FDR threshold of zero. The primary goal of this step was to eliminate peptides identified with low scoring PSMs that represent proteins identified by a single peptide, so-called “one-hit wonders”. After assembling protein groups from the autovalidated PSMs, protein polishing determined the maximum protein level score of a protein group that consisted entirely of distinct peptides estimated to be false-positive identifications (PSMs with negative delta forward-reverse scores). PSMs were removed from the set obtained in the initial peptide-level autovalidation step if they contributed to protein groups that had protein scores below the maximum false-positive protein score. Step 3 was then applied, consisting of protein-polishing autovalidation across all TMT plexes together using the protein grouping method “expand subgroups, top uses shared” to retain protein subgroups with either a minimum protein score of 25 or observation in at least 4 TMT plexes. The primary goal of this step was to eliminate low scoring proteins that were infrequently detected in the sample cohort. As a consequence of these two protein-polishing steps, each identified protein reported in the study was comprised of multiple peptides, unless a single excellent scoring peptide was the sole match and that peptide was observed in at least 4 TMT-plexes. In calculating scores at the protein level and reporting the identified proteins, peptide redundancy was addressed in Spectrum Mill as follows: The protein score was the sum of the scores of distinct peptides. A distinct peptide was the single highest scoring instance of a peptide detected through an MS/MS spectrum. MS/MS spectra for a particular peptide may have been recorded multiple times (e.g. as different precursor charge states, in adjacent bRP fractions, modified by deamidation at Asn or oxidation of Met, or with different phosphosite localization), but were still counted as a single distinct peptide. When a peptide sequence of >8 residues was contained in multiple protein entries in the sequence database, the proteins were grouped together and the highest scoring one and its accession number were reported. In some cases when the protein sequences were grouped in this manner, there were distinct peptides that uniquely represent a lower scoring member of the group (isoforms, family members, and different species). Each of these instances spawned a subgroup. Multiple subgroups were reported, counted towards the total number of proteins, and given related protein subgroup numbers (e.g. 3.1 and 3.2 for group 3, subgroups 1 and 2). For the whole proteome datasets the above criteria yielded false discovery rates (FDR) for each TMT-plex experiment of <0.6% at the peptide-spectrum match level and <0.8% at the distinct peptide level. After assembling proteins with all the PSMs from all the TMT-plex experiments together, the aggregate FDR estimates were 0.57% at the peptide-spectrum match level, 2.6% at the distinct peptide level, and <0.01% (1/11,015) at the protein group level. Since the protein level FDR estimate neither explicitly required a minimum number of distinct peptides per protein nor adjusted for the number of possible tryptic peptides per protein, it may underestimate false positive protein identifications for large proteins observed only on the basis of multiple low scoring PSMs.
In step 2 for the phosphoproteome and acetylproteome datasets, variable modification (VM) site polishing autovalidation was applied across all 25 TMT plexes to retain all VM-site identifications with either a minimum id score of 8.0 or observation in at least 4 TMT plexes. The intention of the VM site polishing step is to control FDR by eliminating unreliable VM site-level identifications, particularly low scoring VM sites that are only detected as low scoring peptides that are also infrequently detected across all of the TMT plexes in the study. In calculating scores at the VM-site level and reporting the identified VM sites, redundancy was addressed in Spectrum Mill as follows: A VM-site table was assembled with columns for individual TMT-plex experiments and rows for individual VM-sites. PSMs were combined into a single row for all non-conflicting observations of a particular VM-site (e.g. different missed cleavage forms, different precursor charges, confident and ambiguous localizations, and different sample-handling modifications). For related peptides, neither observations with a different number of VM-sites nor different confident localizations were allowed to be combined. Selecting the representative peptide from the combined observations was done such that once confident VM-site localization was established, higher identification scores and longer peptide lengths were preferred. While a Spectrum Mill identification score was based on the number of matching peaks, their ion type assignment, and the relative height of unmatched peaks, the VM site localization score was the difference in identification score between the top two localizations. The score threshold for confident localization, >1.1, essentially corresponded to at least 1 b or y ion located between two candidate sites that has a peak height >10% of the tallest fragment ion (neutral losses of phosphate from the precursor and related ions as well as immonium and TMT reporter ions were excluded from the relative height calculation). The ion type scores for b-H3PO4, y-H3PO4, b-H2O, and y-H2O ion types were all set to 0.5. This prevented inappropriate confident localization assignment when a spectrum lacked primary b or y ions between two possible sites but contained ions that could be assigned as either phosphate-loss ions for one localization or water loss ions for another localization. VM-site polishing yielded 65,103 phosphosites with an aggregate FDR of 0.74% at the phosphosite level. In aggregate, 71% of the reported phosphosites in this study were fully localized to a particular serine, threonine, or tyrosine residue. VM-site polishing yielded 13,480 acetylsites with an aggregate FDR of 0.89% at the acetylsite level. In aggregate, 99% of the reported acetylsites in this study were fully localized to a particular lysine residue.
Quantitation using TMT ratios
Using the Spectrum Mill Protein/Peptide Summary module, a protein comparison report was generated for the proteome dataset using the protein grouping method “expand subgroups, top uses shared” (SGT). For the phosphoproteome and acetylproteome datasets a Variable Modification site comparison report limited to either phospho or acetyl sites, respectively, was generated using the protein grouping method “unexpand subgroups”. Relative abundances of proteins and VM-sites were determined in Spectrum Mill using TMT reporter ion intensity ratios from each PSM. TMT reporter ion intensities were corrected for isotopic impurities in the Spectrum Mill Protein/Peptide summary module using the afRICA correction method, which implements determinant calculations according to Cramer’s Rule (Shadforth et al., 2005) and correction factors obtained from the reagent manufacturer’s certificate of analysis (https://www.thermofisher.com/order/catalog/product/90406) for TMT10_lot number SE240163. A protein-level, phosphosite-level, or acetylsite-level TMT ratio is calculated as the median of all PSM-level ratios contributing to a protein subgroup, phosphosite, or acetylsite. PSMs were excluded from the calculation if they lacked a TMT label, had a precursor ion purity < 50% (MS/MS has significant precursor isolation contamination from co-eluting peptides), or had a negative delta forward-reverse identification score (half of all false-positive identifications). Lack of TMT label led to exclusion of PSMs per TMT plex with a range of 1.4 to 3.3% for the proteome, 1.2 to 3.9% for the phosphoproteome, and 1.3 to 6.6% for the acetylproteome datasets. Low precursor ion purity led to exclusion of PSMs per TMT plex with a range of 1.2 to 1.6% for the proteome, 2.0 to 2.9% for the phosphoproteome, and 4.6 to 7.5% for the acetylproteome datasets.
Two-component normalization of TMT ratios
It was assumed that for every sample there would be a set of unregulated proteins or phosphosites that have abundance comparable to the common reference (CR) sample. In the normalized sample, these proteins, phosphosites, or acetylsites should have a log TMT ratio centered at zero. In addition, there were proteins, phosphosites, and acetylsites that were either up- or down-regulated compared to the CR. A normalization scheme was employed that attempted to identify the unregulated proteins phosphosites or acetylsites, and centered the distribution of these log-ratios around zero in order to nullify the effect of differential protein loading and/or systematic MS variation. A 2-component Gaussian mixture model-based normalization algorithm was used to achieve this effect. The two Gaussians N(μi1,δi1) and N(μi2,δi2) for a sample i were fitted and used in the normalization process as follows: the mode mi of the log-ratio distribution was determined for each sample using kernel density estimation with a Gaussian kernel and Shafer-Jones bandwidth. A two-component Gaussian mixture model was then fit with the mean of both Gaussians constrained to be mi, i.e., μi1=μi2=mi. The Gaussian with the smaller estimated standard deviation was assumed to represent the unregulated component of proteins/phosphosites/acetylsites, and was used to normalize the sample. The sample was standardized using (mi), by subtracting the mean mi from each protein/phosphosite/acetylsite and dividing by the standard deviation δi.
Comparative reference sample
To better dissect the tumor/stroma (human/mouse) origin of orthologous proteins in the CompRef xenograft samples, a few divergences were made in the data analysis described above. The sequence database used for searching MS/MS spectra was expanded to include 30,608 mouse proteins, mapped to the mouse reference genome (mm10) obtained via the UCSC Table Browser (https://genome.ucsc.edu/cgi-bin/hgTables) on the same date as the corresponding human reference genome June 29, 2018, along with the addition of 13 proteins encoded in the mouse mitochondrial genome. For the proteome dataset, autovalidation step 3 consisted of protein-polishing autovalidation across all 4 TMT plexes together using the protein grouping method “unexpand subgroups”, to retain protein groups with either a minimum protein score of 25 or observation in at least 2 TMT plexes. The subsequent protein comparison report generated for the proteome dataset employed the subgroup-specific (SGS) protein grouping option, which omitted peptides that are shared between subgroups, and included only subgroup specific peptide sequences toward each subgroup’s count of distinct peptides and protein level TMT quantitation. If evidence for both human and mouse peptides from an orthologous protein were observed, then peptides that cannot distinguish the two (shared) were ignored. However, the peptides shared between species were retained if there was specific evidence for only one of the species, thus yielding a single subgroup attributed to only the single species consistent with the specific peptides. Furthermore, if all peptides observed for a protein group were shared between species, thus yielding a single subgroup composed of indistinguishable species, then all peptides were retained. For the proteome dataset, only PSMs from subgroup-specific peptide sequences contributed to the protein level quantification. A protein detected with all contributing PSMs shared between human and mouse was considered to be human. For the phosphoproteome and acetylproteome datasets, a phosphosite or acetylsite was considered to be mouse if the contributing PSMs were distinctly mouse and human if they were either distinctly human or shared between human and mouse.
Systems Biology analysis
Sample exclusion
To ensure that poor quality or questionable samples were not included in the final dataset, we performed principal component analysis (PCA) on the RNA-seq, global proteome and phosphosite expression data. In the input to PCA (Figure 7A), we excluded any genes, proteins and phosphosites (in the respective datasets) missing in 50% or more of the samples. For each dataset, we plotted the 95% confidence ellipse in the PC1 vs PC2 plot for the tumor and normal groups. Any samples falling outside these ellipses were deemed to be outliers. Samples that were outliers in all three datasets (RNA-seq, proteome and phosphosite) and had inconsistent pathology reviews were excluded. Only sample C3N.00545 satisfied all exclusion criteria and was removed from the final dataset.
Dataset filtering
Genes (RNA-seq), proteins (global proteome), phosphosites and acetylsites present in fewer than 30% of samples (i.e., missing in >70% of samples) were removed from the respective datasets. Furthermore:
Proteins were required to have at least two observed TMT ratios in >25% of samples in order to be included in the proteome dataset. Phosphosites and acetylsites were required to have at least one observed TMT ratio in >25% of samples.
Proteins, phosphosites and acetylsites were required to have TMT ratios with an overall standard deviation >0.5 across all the samples where they were observed. This ensured that a small number of proteins, phosphosites and acetylsites that did not vary much over the set of samples were excluded to minimize noise.
Replicate samples in the dataset were merged by taking the mean of the respective expression values or ratios.
Some of the filtering steps were modified for specific analyses in the study. For many of the marker selection and gene set enrichment analyses, at least 50% of samples were required to have non-missing values for proteins/phosphosites/acetyl sites, since missing values were imputed, and excessive missing values can result in poor imputation. Alternate filtering has been noted in descriptions of the relevant methods.
Unsupervised multi-omics clustering using NMF
We used non-negative matrix factorization (NMF) implemented in the NMF R-package (Gaujoux and Seoighe, 2010) to perform unsupervised clustering of tumor samples and to identify proteogenomic features (proteins, phosphosites, acetylsites and RNA transcripts) that show characteristic expression patterns for each cluster. Briefly, given a factorization rank k (where k is the number of clusters), NMF decomposes a p x n data matrix V into two matrices W and H such that multiplication of W and H approximates V. Matrix H is a k x n matrix whose entries represent weights for each sample (1 to N) to contribute to each cluster (1 to k), whereas matrix W is a p x k matrix representing weights for each feature (1 to p) to contribute to each cluster (1 to k). Matrix H was used to assign samples to clusters by choosing the k with maximum score in each column of H. For each sample we calculated a cluster membership score as the maximal fractional score of the corresponding column in matrix H. We defined a “cluster core” as the set of samples with cluster membership score > 0.5. Matrix W containing the weights of each feature to a certain cluster was used to derive a list of representative features separating the clusters using the method proposed in (Kim and Park, 2007).
To enable integrative multi-omics clustering we enforced all data types (and converted if necessary) to represent ratios to either a common reference measured in each TMT plex (proteome, phosphoproteome, acetylproteome) or an in silico common reference calculated as the median abundance across all samples (mRNA, see “RNA Quantification”). All data tables were then concatenated and filtered to contain a maximum of 30% missing values across all tumors. The remaining missing values were imputed via k-nearest neighbor (kNN) imputation implemented in the impute R-package (DOI: 10.18129/B9.bioc.impute) using the 5 nearest neighbors. To remove uninformative features from the dataset prior to NMF clustering we removed features with the lowest standard deviation (bottom 5th percentile) across all samples. Each row in the data matrix was further scaled and standardized such that all features from different data types were represented as z-scores.
Since NMF requires a non-negative input matrix we converted the z-scores in the data matrix into a non-negative matrix as follows:
Create one data matrix with all negative numbers zeroed.
Create another data matrix with all positive numbers zeroed and the signs of all negative numbers removed.
Concatenate both matrices resulting in a data matrix twice as large as the original, but containing only positive values and zeros and hence appropriate for NMF.
The resulting matrix was then subjected to NMF analysis leveraging the NMF R-package (Gaujoux and Seoighe, 2010) and using the factorization method described in (Brunet et al., 2004). To determine the optimal factorization rank k (number of clusters) for the multi-omic data matrix we tested a range of clusters between k=2 and 8. For each k we factorized matrix V using 50 iterations with random initializations of W and H. To determine the optimal factorization rank we calculated cophenetic correlation coefficients measuring how well the intrinsic structure of the data was recapitulated after clustering and chose the k with maximal cophenetic correlation for cluster numbers between k=3 and 8. (Figure S1G).
Having determined the optimal factorization rank k, in order to achieve robust factorization of the multi-omics data matrix V, we repeated the NMF analysis using 200 iterations with random initializations of W and H and performed the partitioning of samples into clusters as described above. Due to the non-negative transformation applied to the z-scored data matrix as described above, matrix W of feature weights contained two separate weights for positive and negative z-scores of each feature, respectively. In order to revert the non-negative transformation and to derive a single signed weight for each feature, we first normalized each row in matrix W by dividing by the sum of feature weights in each row, aggregated both weights per feature and cluster by keeping the maximal normalized weight and multiplication with the sign of the z-score in the initial data matrix. Thus, the resulting transformed version of matrix Wsigned contained signed cluster weights for each feature in the input matrix.
For Functional characterization of clustering results by single sample Gene Set Enrichment Analysis (ssGSEA), we calculated normalized enrichment scores (NES) of cancer-relevant gene sets by projecting the matrix of signed multi-omic feature weights (Wsigned) onto Hallmark pathway gene sets (Liberzon et al., 2015) using ssGSEA (Barbie et al., 2009). To derive a single weight for each gene measured across multiple omics data types (protein, RNA, phosphorylation site, acetylation site) we retained the weight with maximal absolute amplitude. We used the ssGSEA implementation available on https://github.com/broadinstitute/ssGSEA2.0 using the following parameters:
gene.set.database-’h.all.v6.2.symbols.gmt”
sample.norm.type-’rank”
weight=1
statistic=“area.under.RES”
output.score.type=“NES”
nperm=1000
global.fdr=TRUE
min.overlap=5
correl.type-’z.score”
To test the association of the resulting clusters to clinical variables we used Fisher’s exact test (R function fisher.tesf) to test for overrepresentation in the set of samples defining the cluster core as described above. The following variables were included in the analysis: RNA.Expression.Subtype.TCGA, Region.of.Origin, Stage, Gender, Smoking.Status (self reported), TP53.mutation.status, KRAS.mutation.status, STK11.mutation.status, EGFR.mutation.status, KEAP1.mutation.status, ALK.fusion, CIMP.status.
In order to adjust for tumor purity, for each omic data type (i.e., gene expression, global protein, phosphoproteome and acetylproteome abundance), each marker was modeled as a function of tumor purity from TSNet (Petralia et al., 2018) via a linear regression. Then, residuals from linear regression were considered to perform multi-omic clustering.
The entire workflow described above has been implemented as a module for Broad’s Cloud platform Terra (https://app.terra.bio/). The docker containers encapsulating the source code and required R-packages for NMF clustering and ssGSEA have been submitted to Dockerhub (broadcptac/pgdac_mo_nmf:9, broadcptac/pgdac_ssgsea:5). The source code for ssGSEA is available on GitHub: https://github.com/broadinstitute/ssGSEA2.0.
RNA subtvping
Starting with RNA expression data for the CPTAC LUAD cohort, the top 5,000 most variable genes were subjected to clustering using ConsensusClusterPlus (Wilkerson and Hayes, 2010). The resulting three clusters were mapped to TCGA RNA expression subtypes (Cancer Genome Atlas Research Network, 2014; Wilkerson et al., 2012) by associating enriched clinical features and gene mutations. The association of subtype and features were compared using Fisher’s exact test.
Pathway over-representation analysis
To designate the representative pathways of multi-omics subtypes, we used the Wilcoxon rank sum test to select the top 250 differentially expressed features (mRNA, proteins and phosphosites), or features with p-value less than 0.05 (acetylsites) for each subtype. We then performed hierarchical clustering on these 1000 features and 573 acetylsites. Each set of clustered features underwent pathway enrichment analysis using Reactome (Fabregat et al., 2017). Pathways with p-value smaller than 0.05 were manually reviewed and highlighted in Figure 1E. For visualization purposes, only the top 50 differentially expressed features for each subtype were displayed. In total, 200 features were shown for each data type in the heatmap.
Fusion detection and analysis
Structural variants in WGS samples were called with Manta 1.3.2, retaining variants where sample site depth was less than 3x the median chromosome depth near one or both variant breakends, somatic score was greater than 30, and for small variants (<1000 bases) in the normal sample, the fraction of reads with MAPQ0 around either breakend did not exceed 0.4.
Fusions in RNA-Seq samples were called using three callers: STAR-Fusion, EricScript, and Integrate, with fusions reported by at least 2 callers or reported by STAR-Fusion being retained. Fusions present in the following databases were then excluded: 1) uncharacterized genes, immunoglobulin genes, mitochondrial genes, etc., 2) fusions from the same gene or paralog genes, and 3) fusions reported in TCGA normal samples, GTEx tissues, and non-cancer cell studies. Finally, normal fusions were filtered out from the tumor fusions.
mRNA and Protein correlation
To compare mRNA expression and protein abundance across samples we focused on the RNAseq data with 18,099 genes, and global proteome with 10,316 quantified proteins. Only genes or proteins with <50% NAs (missing values) were considered for the analysis, and protein IDs were mapped to gene names. In total, 9,616 genes common to both RNAseq and proteome data spanning 110 tumor samples were used in the analysis. The analyses were carried out on normalized data - RNAseq data were log2 transformed, upper quartile normalized RPKM values, which were median-centered by row (i.e. gene); proteome data was two-component normalized as described earlier. Correlation was calculated by Spearman’s correlation method using cor.test (Bioconductor, version 3.5.2) function in R. Both correlation coefficient and p-value were computed. Further, adjusted p-value was calculated using the Benjamini-Hochberg procedure. Similarly, mRNA-protein correlation among NAT samples was carried out with overlapping genes over the 101 NAT samples.
To identify genes that reverse their direction in tumors relative to NATs, we selected significant (Benjamini-Hochberg multiple test, FDR <0.1) mRNA-protein pairs in NATs and Tumors, respectively, that changed from negative correlation to positive correlation or vice-versa. Significant genes identified in the global tumor-NAT comparison and individual mutant categories were merged together and are shown in Figure 3A with corresponding correlation coefficients. For paired tumor-NAT analysis, we considered 101 out of 110 samples for which we have paired NATs, out of which 52, 36, 29, and 17 samples had TP53, EGFR, KRAS and STK11 mutations, respectively.
CNA-driven cis and trans effects
Correlations between copy number alterations (CNA) and RNA, proteome, phosphoproteome and acetylproteome (with proteome and PTM data mapped to genes, by choosing the most variable protein isoform/PTM site as the gene-level representative) were determined using Pearson correlation of common genes present in CNA-RNA-proteome (9,341 genes), CNA-RNA-phosphoproteome (5,244 genes) and CNA-RNA-acetylproteome (1,313 genes). In addition, p-values (corrected for multiple testing using Benjamini-Hochberg FDR) for assessing the statistical significance of the correlation values were also calculated. CNA trans-effects for a given gene were determined by identifying genes with statistically significant (FDR < 0.05) positive or negative correlations.
CMAP analysis
Candidate genes driving response to copy number alterations were identified using large-scale Connectivity Map (CMAP) queries. The CMAP (Lamb et al., 2006; Subramanian et al., 2017) is a collection of about 1.3 million gene expression profiles from cell lines treated with bioactive small molecules (~20,000 drug perturbagens), shRNA gene knockdowns (~4,300) and ectopic expression of genes. The CMAP dataset is available on GEO (Series GSE92742). For this analysis, we use the Level 5 (signatures from aggregating replicates) TouchStone dataset with 473,647 total profiles, containing 36,720 gene knock-down profiles, with measurements for 12,328 genes. See https://clue.io/GEO-guide for more information.
To identify candidate driver genes, proteome profiles of copy number-altered samples were correlated with gene knockdown mRNA profiles in the above CMAP dataset, and enrichment of up/down-regulated genes was evaluated. Normalized log2 copy number values less than −0.3 defined deletion (loss), and values greater than +0.3 defined copy number amplifications (gains). In the copy number-altered samples (separately for CNA amplification and CNA deletion), the trans-genes (identified by significant correlation in “CNA driven cis and trans effects” above) were grouped into UP and DOWN categories by comparing the protein ratios of these genes to their ratios in the copy number neutral samples (normalized log2 copy number between −0.3 and +0.3). The lists of UP and DOWN trans-genes were then used as queries to interrogate CMAP signatures and calculate weighted connectivity scores (WTCS) using the single-sample GSEA algorithm (Krug et al., 2018). The weighted connectivity scores were then normalized for each perturbation type and cell line to obtain normalized connectivity scores (NCS). See (Subramanian et al., 2017) for details on WTCS and NCS. For each query we then identified outlier NCS scores, where a score was considered an outlier if it lay beyond 1.5 times the interquartile range of score distribution for the query. The query gene was designated a candidate driver if (i) the score outliers were statistically cis-enriched (Fisher test with BH-FDR multiple testing correction) and (ii) the gene had statistically significant and positive cis-correlation.
For a gene to be considered for inclusion in a CMAP query it needed to i) have a copy number change (amplification or deletion) in at least 15 samples; ii) have at least 20 significant trans genes; and iii) be on the list of shRNA knockdowns in the CMAP. 501 genes satisfied these conditions and resulted in 737 queries (CNA amplification and deletion combined) that were tested for enrichment. Twelve (12) candidate driver genes were identified with Fisher’s test FDR < 0.1, using this process.
In order to ensure that the identified candidate driver genes were not a random occurrence, we performed a permutation test to determine how many candidate driver genes would be identified with random input (Mertins et al., 2016). For the 737 queries used, we substituted the bona-fide trans-genes with randomly chosen genes, and repeated the CMAP enrichment process. To determine FDR, each permutation run was treated as a Poisson sample with rate λ, counting the number of identified candidate driver genes. Given the small n (=10) and λ, a Score confidence interval was calculated (Barker, 2002) and the midpoint of the confidence interval used to estimate the expected number of false positives. Using 10 random permutations, we determined the overall false discovery rate to be FDR=0.13, with a 95% CI of (0.06, 0.19).
To identify how many trans-correlated genes for all candidate regulatory genes could be directly explained by gene expression changes measured in the CMAP shRNA perturbation experiments, knockdown gene expression consensus signature z-scores (knockdown/control) were used to identify regulated genes with α = 0.05, followed by counting the number of trans-genes in this list of regulated genes.
To obtain biological insight into the list of candidate driver genes, we performed (i) enrichment analysis on samples with extreme CNA values (amplification or deletion) to identify statistically enriched sample annotation subgroups; and (ii) GSEA on cis/trans-correlation values to find enriched pathways.
Defining cancer-associated genes
Cancer-associated genes (CAG) were compiled from genes defined by Bailey et al. (Bailey et al., 2018) and cancer-associated genes listed in Mertins et. al (Mertins et al., 2016) and adapted from Vogelstein et al.(Vogelstein et al., 2013). The list of genes is provided in Table S4.
DNA methylation data preprocessing
Raw methylation image files were downloaded from the CPTAC DCC (See data availability). We calculated and analyzed methylated (M) and unmethylated (U) intensities for LUAD samples as described previously (Fortin et al., 2014). We flagged locus as NA where probes did not meet a detection p-value of 0.01. Probes with MAF more than 0.1 were removed, and samples with more than 85% NA values were removed. Resulting beta values of methylation were utilized for subsequent analysis.
Gene-level methylation scores were generated by taking the mean beta values of probes in the CpG islands of promoters and 5’ UTR regions of the gene. Methylation profiles (i.e., density plots) of some samples had unexpectedly skewed distributions of methylation beta values, in addition to significantly more missing values. To systematically determine the subset of methylation samples with these evident data QC issues, we subjected all the samples to model-based clustering using the Mclust package (Scrucca et al., 2016) in R, using the median beta value over all the genes as the representative metric. The clustering automatically determined the optimal number of clusters, and identified 3 clusters. Two of these clusters (with centroids at 0.036 and 0.045) captured the bulk of the samples (187). The third cluster (centroid at 0.211, significantly higher than the other two clusters) consisted of 19 samples, each of which had a skewed distribution of beta values with a mean of 5,704 missing values per sample (in contrast to 2.7 missing values per sample for clusters 1 and 2 combined). Based on this analysis, we concluded that the 19 samples in cluster 3 represent samples with poor data quality. These have been excluded from all methylation analysis.
CpG Island Methylator Phenotype
To classify the 100 tumor samples with high-quality DNA methylation data into the CpG island methylator phenotypes (CIMP), we performed consensus clustering of the methylation data. Specifically, we first generated the gene-level methylation score, by taking the average beta values of all probes harboring in the CpG islands of promoter or 5’ UTR regions of the gene. Then, we considered all genes that were hypermethylated in tumor, i.e. had gene-level methylation scores >0.2, transformed the score into M-values (Du et al. 2010), normalized the transformed score, and then imputed the missing values as zero (mean of normalized data). We then performed consensus clustering 1000 times, each time taking 80% of the samples and all genes, and calculated the consensus matrix (probabilities of two samples clustering together) for each predetermined number of clusters K. For each value of K, we visualized the consensus matrix using hierarchical clustering with Pearson correlation as the distance metric. Finally, we determined the optimal number of clusters by considering the relative change in area under the consensus cumulative density function (CDF) curve. In the end, three distinct clusters were identified: One was hypermethylated with mean M value 0.3, and two were hypomethylated with mean M value −0.17 and −0.18, respectively. We labeled these three clusters as CIMP-high, CIMP-intermediate, and CIMP-low groups.
iProFun Based Cis Association Analysis
We used iProFun, an integrative analysis tool to identify multi-omic molecular quantitative traits (QT) perturbed by DNA-level variations (Song et al., 2019). In comparison with analyzing each molecular trait separately, the joint modeling of multi-omics data via iProFun provided enhanced power for detecting significant cis-associations shared across different omics data types, and achieved better accuracy in inferring cis-associations unique to certain type(s) of molecular trait(s). Specifically, we considered three functional molecular quantitative traits (mRNA expression levels, global protein abundances, and phosphopeptide abundances) for their associations with DNA methylation. We also adjusted for cis somatic mutations, cis CNAs measured by log ratio and b-allele frequency, age, gender, smoking status, country of origin and tumor purity when assessing the associations.
We analyzed the tumor sample data from 100 cases with high quality of methylation data in the current cohort collected by CPTAC. The mRNA expression levels measured with RNA-seq were available for 19,267 genes, the global protein abundance measurements were available for 10,699 isoforms of 10,316 genes, and the phosphopeptide abundance was available for 41,188 peptides from 7650 genes. The log ratio and b-allele frequency of CNAs using a segmentation method combining whole genome sequencing and whole exome sequencing was obtained for 19,267 genes. The DNA methylation levels (beta values) averaging the CpG islands located in the promoter and 5’ UTR regions were available for 16,479 genes. Somatic mutations were called using whole exome sequencing (See Somatic variant calling section above).
Proteomics and phosphoproteomics data were preprocessed with TMT outlier filtering and missing data imputation to increase number of features in the Cis Association Analysis. Due to the quantification of extremely small values on the spectrum level, some extreme values with either positive or negative sign were generated after log2 transform of the TMT ratios. We were concerned those extreme values would lead to instability in imputation of the data set since missing values are dependent on the observed values of the same samples or same protein/phosphosite. To identify TMT ratio outliers with extreme values, we performed an inter-TMT-plex t-test for each individual protein/phosphosite. For each protein/phosphosite, the TMT ratios of samples within a single TMT-plex were compared against the TMT ratios of samples in all the other 24 TMT-plexes using a Spearman two-sample t-test assuming equal variance. In the proteomics data, 344 TMT ratios were identified as outliers with inter-TMT t-test p values lower than 10e-6; 3053 data points (0.122% of all observations) were removed from the data sets. And in phosphoproteomics 729 TMT ratios were identified as outliers with inter TMT t-test p value lower than 10e-7; 6458 data points (0.088% of all observations) were removed from the data sets. Imputation was performed after outlier filtering. We selected proteins/phosphosites with missing rates less than 50%, and imputed with an algorithm tailored for proteomics data using the DreamAI tool (https://github.com/WangLab-MSSM/DreamAI).
The mRNA expression levels, global protein and phosphoprotein abundances were also normalized on each gene/phosphosite, to align the mean to 0 and standard deviation to 1. Tumor purity was determined using ESTIMATE (Yoshihara et al., 2013) from RNA-seq data.
The iProFun procedure was applied to a total of 4992 genes, including 12 genes measured across all seven data types (mRNA, global protein, phosphoprotein, CNA – lr, CNA – baf, mutation, DNA methylation) and the rest 4980 genes measured across all six data types (without mutation data due to mutation rate <5%) for their cis regulatory patterns in tumors. Specifically, for each gene, we considered the following regressions:
When multiple isoform data was available for a protein or multiple peptide level data was available for a phosphoprotein, we selected one with the most significant test statistics across all DNA-level alterations (mutation, CNA and methylation) to denote the gene. The association summary statistics of methylation was applied to iProFun to call posterior probabilities of belonging to each of the eight possible configurations (“None”, “mRNA only” “global only”, “phospho only” “mRNA & global”, “mRNA & phospho”, “global & phospho” and “all three”) and to determine the significance of associations (Table S4). The significant genes needed to pass three criteria: (1) the satisfaction of biological filtering procedures, (2) posterior probabilities > 75%, and (3) empirical false discovery rates (eFDR)<10%. Specifically, the biological filtering criterion for DNA methylations was that only DNA methylations with negative associations with all the types of molecular QTs were considered for a significance call. Secondly, significance was called only for posterior probabilities > 75% of a predictor being associated with a molecular QT, by summing over all configurations consistent with the association of interest. For example, the posterior probability of a DNA methylation being associated with mRNA expression levels was obtained by summing up the posterior probabilities in the following four association patterns – “mRNA only”, “mRNA & global”, “mRNA & phospho” and “all three”, all of which were consistent with DNA methylation being associated with mRNA expression. Lastly, we calculated eFDR by considering 100 permutations per molecular QT. In each permutation, we shuffled the label of the molecular QTs and re-calculated the posterior probabilities of associations via iProFun. For any pre-selected posterior probability cutoff alpha, eFDR could be calculated by: eFDR= (Averaged no. of genes with posterior probabilities > alpha in permuted data) / (Averaged no. of genes with posterior probabilities > alpha in original data). We considered a grid of potential alpha values in the range of 75%−100%, and selected the minimal alpha that satisfied eFDR<10%. Associations with posterior probabilities > alpha were thus significant at eFDR 10%.
Among all the genes whose methylation levels were significantly associated with all three molecular traits, Figure 3E annotated those whose protein abundances significantly differed between tumor and NAT, protein clusters, and immune clusters.
Differential marker analysis
RNA, protein, and PTM abundance were compared between mutated and WT tumor samples using the Wilcoxon rank-sum test. P-values were adjusted within a data type using the Benjamini-Hochberg method. Signed −log10 (p-value) was used to indicate quantitative differences between mutated and WT tumors where signs “+” and “−” indicated upregulated and downregulated mRNA, proteins, phosphosites, and acetylsites, respectively.
We developed linear models to identify differential markers between several key variables, such as gender, tumor stage and histological subtype, accounting for major covariates such as smoking status, region of origin, and mutational status (EGFR, KRAS, STK11, TP53 and ALK fusions). The 22 differentially expressed gender-specific proteins (FDR <0.05, Table S3) showed no coherent functional annotations, while tumor stage, whether examined at the individual level or aggregated into stages 1, 2 and 3, revealed no significant markers (FDR <0.05). Most tumors had typical glandular/acinar morphology; of the remaining six dominant histologic subtypes, solid and true papillary had numbers permitting statistical comparison. Twelve RNA species, some with established relevance to cancer, were differential between these subtypes, including elevation of Krebs cycle enzyme IDH3A in the solid and tyrosine kinase PTK7 in the papillary subtype, but no proteins were differential after adjustment for confounding variables.
Deriving mutation based signatures
Non-negative matrix factorization (NMF) was used in deciphering mutation signatures in cancer somatic mutations stratified by 96 base substitutions in tri-nucleotide sequence contexts. To obtain a reliable signature profile, we used SomaticWrapper to call mutations from WGS data. SignatureAnalyzer exploited the Bayesian variant of the NMF algorithm and enabled an inference for the optimal number of signatures from the data itself at a balance between the data fidelity (likelihood) and the model complexity (regularization) (Kasar et al., 2015; Kim et al., 2016; Tan and Fevotte, 2013). After decomposing into three signatures, signatures were compared against known signatures derived from COSMIC (Tate et al., 2019) and cosine similarity was calculated to identify the best match.
Continuous Smoking Score
We also sought to integrate count of total mutations, t, percentage that are signature mutations, c, and count of DNPs, n, into a continuous score, 0 < S < 1, to quantify the degree of confidence that a sample was associated with smoking signature. We referred to these quantities as the data, namely D = C ∩ T ∩ N, and used A and A’ to indicate smoking signature or lack thereof, respectively. In a Bayesian framework, it is readily shown that a suitable form is S = 1 / (1 + R), where R is the ratio of the joint probability of A’ and D to the joint probability of A and D. For example, the latter can be written P(A) ・ P(C|A) ・ P(T|A) ・ P(N|A) and the former similarly, where each term of the former is the complement of its respective term in this expression. Common risk statistics are invoked as priors, i.e. P(A) = 0.9 (Walser et al., 2008).
We consider S to be a score because rigorous conditioned probabilities are difficult to establish. (For example, the data types themselves are not independent of one another and models using common distributions like the Poisson do not recapitulate realistic variances.) Instead, we adopted a data-driven approach of estimating contributions of each data type based on 2-point fitting of the extremes using shape functions based on the Gaussian error function, erf. The general model for the data type G is P(G|A) = [x・erf (g/y) + 1] /(x + 2), with the resulting fitted values being the following: for total mutations G = T and (x,y) = (4028, 1000) when g = t; for percentage that are signature mutations G = C and (x,y) = (200, 50) when g = c; and for number of DNPs G = N and (x,y) = (30, 4) when g = n. Each of these parametric combinations adds significant weight above a linear contribution as the count for its respective data type increases above the average. For example, for g/y ≈ 0.6, weights for each data type are around 50% higher than their corresponding linear values would be.
The shape function for T includes an expected-value correction for purity, u. (Correction for C is implicit, as it is a percentage of T.) Namely, assuming mutation-calling does not capture all mutations because of impurities, t is taken as the observed number of mutations divided by a purity shape function, f, where f ≤ 1. Although one might model f according to common characteristics of mutation callers, e.g. close to 100% sensitivity for pure samples and very low calling rate for low variant allele fractions (VAFs), the purity estimates for these data are based on RNA-seq and are not highly correlated with total mutation counts. Consequently, we use a weaker, linear shape function, f = 0.3 ・ u + 0.7, which does not strongly impact the adjustment of low-purity samples.
Determination of Sternness score
Stemness scores were calculated as previously described (Malta et al., 2018). To calculate the stemness scores based on mRNA expression, we built a predictive model using one-class logistic regression (OCLR) (Sokolov et al., 2016) on the pluripotent stem cell samples (ESC and iPSC) from the Progenitor Cell Biology Consortium (PCBC) dataset (Daily et al., 2017; Salomonis et al., 2016). For mRNA expression-based signatures, to ensure compatibility with the CPTAC LUAD cohort, we first mapped the gene names from Ensembl IDs to Human Genome Organization (HUGO), dropping any genes that had no such mapping. The resulting training matrix contained 12,945 mRNA expression values measured across all available PCBC samples. To calculate the mRNA-based stemness index (mRNASi) we used RPKM mRNA expression values for all CPTAC LUAD and NAT samples (uq-rpkm-log2-NArm-row-norm.gct). We used the function TCGAanalyze_Stemness from the package TCGAbiolinks (Colaprico et al., 2016) and followed our previously-described workflow (Ho et al., 1987), with “stemSig” argument set to PCBC_stemSig.
Immune Subtyping and downstream analysis
The abundances of 64 different cell types for lung tumors and NAT samples were computed via xCell (Aran et al., 2017; (http://xCell.ucsf.edu/) using log2 (UQ) RPKM expression values. Table S5 contains the final score computed by xCell of different cell types for all tumor and NAT samples. Consensus clustering on xCell signatures performed in order to identify groups of samples with the same immune/stromal characteristics. Only cells that were detected in at least 5 patients (FDR < 1%) were utilized. Consensus clustering was performed using the R package ConsensusClusterPlus (Monti et al., 2003; Wilkerson and Hayes, 2010). Specifically, 80% of the original samples were randomly subsampled without replacement and partitioned into 3 major clusters using the K-Means algorithm.
For estimating Tumor Purity, Stromal and Immune Scores, in addition toXcell, we utilized ESTIMATE (Yoshihara et al., 2013) on RNA-seq to infer immune and stromal scores and TSNet for tumor purity (Petralia et al., 2018).
ssGSEA (Barbie et al., 2009) was utilized to obtain pathway scores based on RNA-seq and global proteomics data using the R package GSVA (Hänzelmann et al., 2013). A Wilcoxon test was performed subsequently to find pathways differentially expressed between cold-tumor-enriched and hot-tumor-enriched subgroups. P-values were adjusted via the Benjamini-Hochberg procedure. Table S5 shows genes/proteins and pathways differentially expressed based on RNA-seq and global proteomics abundance.
To determine mutations that are associated with xCell signatures, raw xCells signatures were modeled as a linear function of mutation status. For this analysis, only mutations that occur in more than 15 samples across all tumor samples were considered (i.e., 66 genes). P-values were adjusted for multiple comparisons using Benjamini-Hochberg correction and the association test results are listed in Table S5.
In addition to exploring the effect of STK11 mutation itself, we assessed whether any other mutation was associated with immune infiltration given STK11 status. A linear model was developed in which the immune score from ESTIMATE was modeled as a function of STK11 mutation and the mutation status of the 66 genes carrying more than 15 mutations each. P-values were corrected using the Benjamini-Hochberg adjustment. The only mutation significantly associated (positively) with immune score given STK11 mutation status was KRAS mutation at FDR 10%.
Determining Immune evasive mechanisms
Immune evasion is a process wherein tumor cells employ multiple mechanisms to evade anti-tumor immune response, facilitating tumor cell survival and evolution. Immune checkpoint blockade therapy has emerged as a treatment strategy for cancer patients, based on harnessing the anti-tumor immune response genes (Abril-Rodriguez and Ribas, 2017). However, a significant number of patients have failed to respond to immunomodulation strategies such as checkpoint inhibitors, likely due to tumor-specific immunosuppressive mechanisms and incomplete restoration of adaptive immunity (Achyut and Arbab, 2016; Allard et al., 2016b; Jerby-Arnon et al., 2018; Kozuma et al., 2018b). We postulate that two main factors contribute to the failure of immune therapy: (i) the insufficient activation of the immune response, and (ii) the evolutionarily selected mechanisms of immune evasion. We also hypothesized that activation of the adaptive immune system and sensitivity to checkpoint therapy principally depends on upregulation or downregulation of IFNG axis – a pathways of 15 genes, which is composed of proteins expressed primarily in cancer cells: IFNG receptors (IFNGR1, IFNGR2); JAK/STAT-signaling component (JAK1, JAK2, STAT1, STAT3, IRF1); antigen presenting (HLA-A, HLA-B, HLA-C, HLA-E, HLA-F, HLA-G); and checkpoint proteins (PD-L1/PD1). Thus, non-responder tumors are either those that are invisible to immune cells because of a suppressed IFNG axis, or those with the IFNG axis activated along with activated immune evasion that prevents leukocyte-driven cancer cell death. Following this idea, we arrived at a general protocol to reveal proteins involved in immune evasion and determine potential targets for combination therapy. First, we inferred relative activation of the IFNG axis pathway across tumors. We ranked tumor proteins in descending order of abundance, then determined for each IFNG pathway protein the probability that it would by random chance occupy its observed or a higher position in that list. An individual protein would therefore have a smaller probability (be enriched towards the top of the list) the higher it was on the list. To assess whether the set of IFNG pathway proteins were significantly overrepresented in a sample, the enrichment probabilities for individual constituent proteins were geometrically averaged using Fisher’s exact test. The process was then repeated, this time combining individual probabilities that a protein was enriched towards the bottom of the abundance list to assess for significant underrepresentation of the IFNG pathway in a sample. The inferred pathway activation score was defined as the negative log of the ratio of these two probabilities. This score is positive when pathway proteins occur in the top half of the abundance list, and negative when confined to the bottom. Secondly, we determined proteins that are significantly upregulated with inferred activation of the IFNG axis and have known immune evasion role (markers of MDSC (Achyut and Arbab, 2016), adenosine signaling signature (Allard et al., 2016b), IDO1 pathway (Kozuma et al., 2018b; Liu et al., 2018; Takada et al., 2019; Zhang et al., 2019b) or have potential therapeutic value as targets of drugs from Drug Bank (Frolkis et al., 2010; Jewison et al., 2014).
Identifying histological features
LUAD tissue histopathology slides were first downloaded from The Cancer Imaging Archive (TCIA) database. The slides and their corresponding per-slide level labels were then separated into training (80%), validation (10%), and test sets (10%) at the per-patient level. Each slide was then tiled into 299-by-299-pixel pieces with overlapping areas of 49 pixels from each edge, omitting those with over 30% background. Tiles of each set were packaged into a TFrecord file. Then, the InceptionV3-architectured convolutional neural network (CNN) was trained from scratch and the best performing model was picked based on validation set performance. The performance of the model was evaluated by statistical metrics (area under ROC, area under PRC, and accuracy) on per-slide and per-tile levels. Lastly, the trained model was applied to the test set, and the per-tile prediction scores were aggregated by slides and shown as heatmaps. 10,000 tiles were randomly selected for visualization from the test set of 137,990 tiles cropped from 36 slides of 11 individual patients. The test data were propagated through the trained model to obtain positive prediction scores, the probability of being a STK11 mutation positive case estimated by the deep learning model. Additionally, for each test example, activation scores of the fully-connected layer immediately before the output layer, a vector of 2,048 elements, were extracted as representation of the input sample in perspective of the predictive model. The activation scores of 10,000 sample tiles were further reduced to two-dimensional representations by tSNE. Overlay of positive prediction scores on sample points showed distinct clusters for predicted positive (orange) and predictive negative (blue) cases. Examples of true positive (red outline) and true negative (black outline) tiles exhibited different histologic features (Figure 5E), such that the STK11 WT tiles correctly recognized by the model harbored abundant inflammatory cells, and STK11 mutant tiles showed typical adenocarcinoma characteristics.
Independent component analysis
As previously described (Liu et al., 2019), Independent component analysis (ICA) was run 100 times with random initial values on 110 tumor samples. In each run, 110 independent components (equal to the number of samples) were extracted to obtain as much information as possible. All components were then pooled and grouped into 110 clusters using K-medoids method and Spearman correlation as dissimilarity measures. Each independent component (and a sample point submitted to the clustering algorithm) was a vector comprising weights of all genes in the original data. Genes that contributed heavily to a component were assigned large coefficients that could serve as a pathway-level molecular signature. Consistent clusters of independent components would exhibit large intra-group homogeneity (average silhouette width>0.8) and are composed of members generated in different runs (>90), indicating that similar signals were extracted recurrently when the algorithm was initiated from different values. The centroids of the clusters were considered as representative of a stable signature, and mean mixing scores (activity of each signature over all samples) of each cluster were used to represent the activity levels of the corresponding signature in each sample. To investigate the correlation between blindly extracted features and known clinical characteristics, the corresponding mixing scores for all members of a component cluster were regressed against 46 clinical variables, and the count of significant correlations (P<10−5, linear regression, P value controlled for multiple testing at the 0.01 level) indicated association between the particular molecular signature and clinical variable pair. Signatures that showed a high percentage of significant correlations for all members and large average -log10(p-value) values within the cluster were considered to be associated with the clinical feature. Genes heavily weighted in the cluster centroid coefficients vector may thus shed light on molecular mechanisms underlying the clinical feature. One highly consistent signature (average cluster silhouette width 0.97, 100 members produced by 100 different runs) was found to be significantly associated with STK11 mutation status, with an average -logP value of 5.7.
Mutation-based cis- and trans-effects
We examined the cis- and trans-effects of 18 mutations that were significant in a previous large-scale TCGA LUAD study (Cancer Genome Atlas Research Network, 2014) on the RNA, proteome, and phosphoproteome of cancer-related genes (Bailey et al., 2018). After excluding silent mutations, samples were separated into mutated and WT groups. We used the Wilcoxon rank-sum test to report differentially expressed features (RNA, proteins, phosphosites and acetylsites) between the two groups. Differentially enriched features passing an FDR <0.05 cut-off were separated into two categories based on cis- and trans-effects.
Multi-omic Outlier Analysis
We calculated the median and interquartile range (IQR) values for phosphopeptide, protein, gene expression and copy number alterations of known kinases (N=701), phosphatases (N=135), E3 ubiquitin ligases (N=377) and de-ubiquitin ligases (N=87) using TMT-based global phosphoproteomic and proteomic data, RNA-Seq expression data or CNA data. Outliers were defined as any value higher than the median plus 1.5x IQR. Phosphopeptide data was aggregated into genes by summing outlier and non-outlier values per sample. Outlier counts were used to determine enriched genes in a group of samples at each data level. First, genes without an outlier value in at least 10% of samples in the group of interest were filtered out. Additionally, only genes where the frequency of outliers in the group of interest was higher than the frequency in the outgroup were considered in the analysis. The group of interest was compared to the rest of the samples using Fisher’s exact test on the count of outlier and non-outlier values per group. Resulting p-values were corrected for multiple comparisons using the Benjamini-Hochberg correction. Druggability was determined for each gene using the drug-gene interaction database (DGIdb)(Cotto et al., 2018).The mean impact of shRNA- or CRISPR-mediated depletion of each gene on survival and proliferation in lung cancer cell lines was also visualized based on previous studies (Barretina et al., 2012; Tsherniak et al., 2017).
Pathway analysis reported in Figure 6
In the set of tumor samples, the high smoking score (HSS) subset consists of 58 samples, while the low smoking score (LSS) subset contains 49 samples. There are 52 NAT samples with paired HSS tumor samples, and 46 NAT samples with paired LSS tumor samples.
We used gene sets of molecular pathways from KEGG (Kanehisa and Goto, 2000), Hallmark (Liberzon et al., 2015) and Reactome (Croft et al., 2014) databases to compute single sample gene set enrichment scores (Barbie et al., 2009) for each sample. To compute pathway HSS vs LSS differential scores for both tumor and NAT, we ran two one-sided Wilcoxon rank-sum tests (greater than, and lesser than) on HSS vs LSS sets of samples and performed Benjamini-Hochberg correction on computed p-values (FDR). The differential score (Q) is obtained as signed -log10(FDR) from the lower of the two p-values derived from two one-sided Wilcoxon rank-sum tests. The signs “+” and “−” indicated upregulated and downregulated pathways respectively, in HSS. Differential scores were computed for both proteome (for the set of 7,136 proteins with no missing values) and transcriptome (18,099 genes).
To select the six groups of pathways with characteristic HSS vs LSS proteome behavior in tumor and NAT, we used the FDR < 0.05 for differential behavior and FDR > 0.3 for the absence of differential behavior. For specific pathway groups, this amounted to the following conditions: group 1: Q(Tumor) > 1.301 & Q(NAT) < −1.301; group 2: Q(Tumor) < −1.301 & Q(NAT) > 1.301; group 3: Q(Tumor) > 1.301 & Q(NAT) > 1.301; group 4: Q(Tumor) < −1.301 & Q(NAT) < −1.301; group 5: Q(Tumor) > 1.301 & |Q(NAT)| < 0.523; group 6: Q(Tumor) < −1.301 & |Q(NAT)| < 0.523.
Tumor-NAT related analysis
PCA was performed on RNA (18,099), protein (10,165), phosphosites (40,845), and acetylsites (6,984) datasets using the factoextra (Bioconductor, version 1.0.5) package in R (3.1.2). Features with no variance were removed.
To identify Tumor vs NAT differential markers, a Wilcoxon rank sum test was applied to TMT-based global proteomic data to determine differential abundance of proteins between tumor and NAT samples. Proteins with log2-fold-change (FC)> 1 in tumors and Benjamini-Hochberg FDR < 0.01 were considered to be tumor-associated proteins. Biomarker candidate selection was more stringent, requiring both protein log2 FC > 2 and overexpression at the RNA level (log2FC > 1, FDR < 0.05). Immunohistochemistry-based antibody-specific staining scores in lung tumors were obtained from the Human Protein Atlas (HPA, https://www.proteinatlas.org), in which tumor-specific staining is reported in four levels, i.e. high, medium, low, and not detected. The protein-specific annotations such as protein class, found in plasma, or ontology were obtained from HPA, Uniprot and GO. Proteins of specific type or function such as transcription factors, enzymes, transporters, and transmembranes were identified. “Plasma proteins” represent proteins found in plasma, whereas “secreted” have been annotated as secreted/exported outside the cell. FDA-approved drugs targeting the protein or drugs under clinical trial were so designated. Given the role of epithelial-to-mesenchymal transition (EMT) in metastasis, proteins overlapping with hallmarks of EMT gene sets were shown separately. Proteins differentially expressed between tumors and NATs (Benjamini-Hochberg FDR < 0.01, Wilcoxon signed rank test) and having <50% missing values were used for pathway enrichment analysis with GSEA (Subramanian et al., 2005) as implemented in WebGestalt (Wang et al., 2017). Similar analyses were performed on the phosphoproteome and acetylproteome to detect tumor-specific phosphosites and acetylsites, respectively.
To identify, mutant phenotype-specific protein biomarkers, four driver mutant phenotypes were considered; TP53 (n=52), EGFR (n=36), KRAS (n=29), and STK11 (n=17). A Wilcoxon rank sum test was performed between tumor and paired NAT samples using only samples with mutations. Similar analyses were performed on samples with wild type (WT) phenotype only (TP53WT=49, EGFRWT=65, KRASWT=72, STK11WT=84). Differentially expressed proteins in a given mutant phenotype were selected based on >4-fold difference and Benjamini-Hochberg adjusted p-value (FDR) < 0.01. Further, mutant-specific proteins were filtered using log2 (median difference between mutant and WT) >1.5 to remove noise from corresponding WT samples. The filtered proteins were nominated as mutant-specific biomarkers if their expression was upregulated in 80% of tumor samples compared to matched normal samples. The fold changes between tumor and matched normal are shown in heatmaps for identified protein biomarkers in each mutant phenotype.
Phosphorylation-driven signature analysis
Based on the results of the Tumor-NAT related analysis described above, we performed phosphosite-specific signature enrichment analysis (PTM-SEA) (Krug et al., 2018) to identify dysregulated phosphorylation-driven pathways in tumors compared to paired normal adjacent tissues (NATs). To adequately account for both magnitude and variance of measured phosphosite abundance, we used p-values derived from application of the Wilcoxon rank-sum test to phosphorylation data as ranking for PTM-SEA. To that end, p-values were log-transformed and signed according to the fold change (signed -log10 (p-value)) such that large positive values indicated tumor-specific phosphosite abundance and large negative values NAT-specific phosphosite abundance.
PTM-SEA relies on site-specific annotation provided by PTMsigDB and thus a single site-centric data matrix data is required such that each row corresponds to a single phosphosite. We note that in this analysis the data matrix comprised a single data column (log transformed and signed p-values of the tumor vs. NAT comparison) and each row represented a confidently localized phosphosite assigned by Spectrum Mill software.
We employed the heuristic method introduced by Krug et al. (Krug et al., 2018) to deconvolute multiple phosphorylated peptides to separate data points (log-transformed and signed p-values). Briefly, phosphosites measured on different phospho-proteoform peptides were resolved by using the p-value derived from the least modified version of the peptide. For instance, if a site T4 measured on a doubly phosphorylated (T4, S8) peptide (PEPtIDEsR) was also measured on a mono-phosphorylated version (PEPtIDESR), we assigned the p-value derived from the mono-phosphorylated peptide proteoform to T4, and the p-value derived from PEPtIDEsR to S8. If only the doubly phosphorylated proteoform was present in the dataset, we assigned the same p-value to both sites T4 and S8.
We queried the PTM signatures database (PTMsigDB) v1.9.0 downloaded from http://prot-shiny-vm.broadinstitute.org:3838/ptmsigdb-app/ using the flanking amino acid sequence (+/− 7 aa) as primary identifier. We used the implementation of PTM-SEA available on GitHub (https://github.com/broadinstitute/ssGSEA2.0) using the command interface R-script (ssgsea-cli.R). The following parameters were used to run PTM-SEA:
weight: 1 statistic: “area.under.RES” output.score.type: “NES” nperm:1000 min.overlap: 5 correl.type: “rank”
The sign of the normalized enrichment score (NES) calculated for each signature corresponds to the sign of the tumor-NAT log fold change. P-values for each signature were derived from 1,000 random permutations and further adjusted for multiple hypothesis testing using the method proposed by Benjamini & and Hochberg (Benjamini and Hochberg, 1995). Signatures with FDR-corrected p-values < 0.05 were considered to be differential between tumor and NAT.
For mutational subtype analysis (EGFR, KRAS, TP53, STK11) we derived a residual enrichment score between mutated and WT samples by separately applying PTM-SEA to mutated and WT samples to derive signature enrichment scores from which we calculated the residuals via linear regression (mut ~ non-mut). From the resulting distribution of residual enrichment scores we identified outliers using the +/− 1.5*IQR definition used in box and whisker plots.
Variant Peptide Identification
We used NeoFlow (https://github.com/bzhanglab/neoflow) for neoantigen prediction (Wen et al., 2020). Specifically, Optitype (Szolek et al., 2014) was used to find human leukocyte antigens (HLA) in the WES data. Then we used netMHCpan (Jurtz et al., 2017) to predict HLA peptide binding affinity for somatic mutation-derived variant peptides with a length between 8–11 amino acids. The cutoff of IC50 binding affinity was set to 150 nM. HLA peptides with binding affinity higher than 150 nM were removed. Variant identification was also performed at both mRNA and protein levels using RNA-Seq data and MS/MS data, respectively. To identify variant peptides, we used a customized protein sequence database approach (Wang et al., 2012). We derived customized protein sequence databases from matched WES data and then performed database searching using the customized databases for individual TMT experiments. We built a customized database for each TMT experiment based on somatic variants from WES data. We used Customprodbj (Wen et al., 2020) (https://github.com/bzhanglab/customprodbj) for customized database construction. MS-GF+ was used for variant peptide identification for all global proteome and phosphorylation data. Results from MS-GF+ were filtered with 1% FDR at PSM level. Remaining variant peptides were further filtered using PepQuery (http://www.pepquery.org) (Wen et al., 2019) with the p-value cutoff <= 0.01. The spectra of variant peptides were annotated using PDV (http://www.zhang-lab.org/) (Li et al., 2019).
Cancer/testis Antigen Prediction
Cancer/testis (CT) antigens were downloaded from the CTdatabase (Almeida et al., 2009). CT antigens with a 2-fold increase in tumor from NAT in at least 10% of the samples were highlighted.
QUANTIFICATION AND STATISTICAL ANALYSIS
RNA and Protein quantification
Transciptome and proteome quantification has been described under “RNAseq Gene Expression and miRNAseq Quantification and Analysis” and “Proteomics Data Analysis: Protein-peptide identification, phosphosite / acetylsite localization, and quantification”. The details of statistical analysis are presented within the text and the corresponding STAR Method sections.
ADDITIONAL RESOURCES
The CPTAC program website, detailing program initiatives, investigators, and datasets, is found at https://proteomics.cancer.gov/programs/cptac.
A website for interactive visualization of the multi-omics dataset is available at: http://prot-shiny-vm.broadinstitute.org:3838/CPTAC-LUAD2020/. The heatmap depicts somatic copy number aberrations, mRNA, protein, phosphosite and acetylsite abundances across 100 tumor-NAT pairs for which all data types were available. Copy number alterations are relative to matched normal blood samples and are on log2(CNA)-1 scale. For other data types the heatmap depicts abundances relative to paired normal adjacent tissue (NAT).
All processed data matrices will also be available at LinkedOmics (Vasaikar et al., 2018) (http://www.linkedomics.org), where computational tools are available for further exploration of this dataset.
Supplementary Material
Figure S1, Related to Figure 1: Experimental workflow and data quality metrics.
A. Schematic representation showing sample processing steps. Fresh frozen tumors and their matched normal-adjacent tissues (NATs) were cryopulverized and aliquoted for genomics and proteomics analyses before undergoing comprehensive proteogenomic characterization, facilitating uniformity in input samples.
B. Schematic representation of the workflows used for proteome, phosphoproteome and acetylproteome analyses. Tandem mass tags (TMT) were used to multiplex 9 samples (4 tumors and their matched NATs, in addition to a 9th sample, an unpaired tumor) and 1 common reference (pool of all tumors and NATs) that was used to link multiple TMT10 plexes. Matched tumor / NAT pairs were included in the same TMT plex.
C. Pearson similarity matrices showing intra- and inter-plex reproducibility across 4 interspersed comparative reference (CompRef) process replicates for proteome, phosphoproteome and acetylproteome. The CompRef process replicates demonstrated excellent reproducibility (Pearson Correlation, Proteome: R=0.91, Phosphoproteome: R=0.88, Acetylproteome: R=0.73) and consistent identifications across several months of data acquisition time.
D. Bar plot showing consistent numbers of identified and quantified proteins, phosphosites and acetylsites across the 25 plexes used for analyzing 212 tumors and NATs.
E. Principal component analysis (PCA) plot representation of proteome, phosphoproteome and acetylproteome separately for tumors and NATs, colored by TMT plex (n=25). PCA was based on features that were fully quantified across all 25 TMT plexes.
F. Sample-wise Pearson correlation between copy number alteration (CNA) and RNA, and between CNA and Proteome. The dark red-colored diagonal demonstrates the absence of sample swaps.
G. Cophenetic correlation coefficient (y-axis) calculated for a range of factorization ranks (x-axis). The maximal cophenetic correlation coefficient was observed for rank K=4 as shown in red.
H. Silhouette plot for K=4. This plot indicates the quality of cluster separation.
I. Non-negative matrix factorization (NMF) clustering applied individually to proteome, phosphoproteome and acetylproteome. Each heatmap shows the maximum-normalized membership score for each sample (x-axis) in each cluster (y-axis) - essentially, the strength of a sample’s “belongingness” to each of the clusters. The proteome cluster overlaps substantially with the multi-omics clusters depicted in Figure 1E, but divergence is seen in both the phosphoproteome and acetylproteome, with additional substructure in the phosphoproteome. Color schematics for the different annotations and data rows are detailed in the bottom panel.
J. Louvain clustering of miRNA showed parallels with NMF results but identified five clusters. miRNA cluster 2 was markedly enriched for tumors from multi-omics cluster C1, in turn aligned with proximal-inflammatory RNA signatures, while miRNA cluster 3 was enriched for the STK11 mutant subset of the NMF C3, proximal-proliferative cluster. While the remaining three miRNA clusters had mixed composition, miRNA cluster 5 was markedly enriched for ALK fusion-driven tumors, including all 5 EML4-ALK as well as the HMBOX1-ALK fusions.
Figure S2, Related to Figure 2: Genomic sequence evidence for ALK rearrangements and ALK immunohistochemistry
A. ALK gene fusion transcript architecture constructed from RNAseq data and fusion evidence for ALK fusion transcripts. Red arrows on the ALK and various 5’ partner genes’ schematic diagrams indicate fusion breakpoints observed in the respective index samples. Blue arrows indicate gene orientation and numbers indicate genomic coordinates from GRCh38/hg38 assembly.
B. Identification of the precise genomic breakpoints from whole genome sequencing (WGS) data for ALK gene fusions. WGS evidence supporting the underlying genomic rearrangements in the ALK locus is indicated in red and blue; numbers indicate genomic coordinates from GRCh38/hg38 assembly.
C. Immunohistochemistry reveals upregulation of both total ALK and the ALK Y1507 phosphosite specifically in the tumor epithelia of ALK fusion-positive samples. No staining was seen in RET or ROS1 fusion samples or in matched NATs.
Figure S3, Related to Figure 3: Multi-omics integration.
A. Density plots showing distribution of sample-wise RNA-protein Spearman correlations separately for tumors (red) and NATs (blue).
B. Differential RNA and protein correlation between tumors and paired NATs is seen in gene products involved in Cell proliferation and transcriptional regulation, RNA splicing, Cell division, Beta catenin signaling and Chromosomal condensation. We hypothesize that, in NATs, homeostatic biological activities such as cell maintenance and homeostasis, circadian rhythm and survival predominate and are mediated by proteins the abundances of which reflect mRNA transcript levels, post-transcriptional processes, and post-translational stability. While the same components are at play in tumors, their more dynamic context and highly proliferative state leads to more consistent kinetics and coherent expression of RNA and proteins (Carpy et al., 2014; Jovanovic et al., 2015; Komili and Silver, 2008; Lu et al., 2007; Marguerat et al., 2012).
C. Correlation plots of CNA vs Phosphoprotein and CNA vs Acetylprotein expression. Significant (FDR <0.05) positive and negative correlations are indicated in red and green, respectively. CNA-driven cis-effects (consequence of CNA on the same locus) appear as the red diagonal line; trans-effects (consequence of CNA on genes encoded elsewhere) appear as vertical red and green lines. The accompanying histograms show the number of significant (FDR <0.05) cis- and trans-events corresponding to the indicated genomic loci (upward plot) as well as the overlap between CNA-RNA and CNA-protein events (downward plot).
D. Heatmap showing 2 dominant clusters of DNA methylation, defined primarily by tumors and NATs. LUAD tumors tend to be significantly more highly methylated than their counterpart NATs (two-sided Wilcoxon rank-sum test).
E. Consensus clustering of CpG island methylator phenotype (CIMP) defines three stable clusters representing high (red), intermediate (yellow) and low (blue) CIMP phenotypes (respectively referred to as CIMP+, −1, and −2 in Table S4). Both the overall tripartite structure and the highlighted genes also showed a pattern consistent with a previous report (Cancer Genome Atlas Research Network, 2014)
Figure S4, Related to Figure 4: Impact of somatic mutations on proteogenomic landscape of tumors.
A. The cis- (circles) and trans- (squares) effects of select mutated genes on the RNA expression of cancer-associated genes. The red and blue scale represents the median difference in RNA expression between samples with and without mutations. Size represents significance.
B. Lollipop plot showing KEAP1 mutations identified in this LUAD cohort. Twelve LUAD tumors harbored KEAP1 mutations, including missense mutations and truncations distributed across the entire length of the protein. The color of the lollipops indicate the type of mutation and numbers represent amino acid positions. Protein domains are indicated by different colors.
C. Boxplots showing KEAP1 and NFE2L2 RNA expression and KEAP1 protein expression in KEAP1 wild-type (WT) and mutant samples. Also shown is the downregulation of KEAP1 in KEAP1 mutant samples, seen at the protein but not at the RNA level.
D. Volcano plot showing differentially regulated proteins in KEAP1 WT vs mutant samples. These differential proteins underlie the pathway analysis shown in Figure S4F.
E. Volcano plot showing differentially regulated phosphosites in KEAP1 WT vs mutant samples.
F. Pathways enriched among proteins differentially expressed between KEAP1 mutant and WT tumors. Significant enrichment of the oxidative stress response supports activation of NRF2 signaling in these samples.
G. Boxplots showing unchanged PTPN11 protein and significantly (FDR <0.05) elevated phosphopeptide expression (Y546 and Y584) in ALK fusion relative to wild-type samples. Various activating functions have been proposed for these phosphorylation sites, including directly driving the active conformation (Bennett et al., 1994; Lu et al., 2001) and serving as GRB2 docking sites (Bennett et al., 1994; Cunnick et al., 2002; Okazaki et al., 2013; Vogel and Ullrich, 1996). The PTPN11/Shp2 adaptor protein Grb2-associated binder-1 (GAB1) (Montagner et al., 2005) was also significantly upregulated in ALK fusion- driven tumors (Figure S4H).
H. GAB1 protein expression in samples with and without ALK fusion.
I. Protein-level gene-set enrichment (GSEA) pathway comparison of EGFR- and KRAS-driven LUAD tumors showing disparity in complement and clotting cascades, with upregulation of both in KRAS and downregulation in EGFR mutant samples. This analysis compares gene-set enrichment in KRAS mutant versus wildtype to EGFR mutant versus wildtype, where “wildtype” in each case excludes both KRAS and EGFR mutants.
J. Heatmaps show the phosphatase, ubiquitinase and deubiquitinase outliers enriched (FDR <0.2) at the phosphoprotein, protein, RNA and CNA levels (represented by columns under each gene name) and their association with mutations in select genes (EGFR, KRAS, TP53, STK11, KEAP1, EML4-ALK). Cancer Dependency Map-supported (https://depmap.org) panels on the left show log2 transformed relative survival averaged across all available lung cell lines after depletion of the indicated gene (rows) by RNAi or CRISPR. Druggability based on the Drug Gene Interaction Database (http://www.dgidb.org/) is indicated alongside the availability of FDA-approved drugs. The log-transformed druggability score indicates the sum of PubMed journal articles that support the drug-gene relationship. This implementation of outlier detection complements other analytic approaches by identifying potentially druggable alterations that occur in personalized fashion; hence, for example, PTPN11 Y62 did not appear as an “outlier” phosphatase in EGFR mutant tumors (N=38) because of its uniform high expression in that group.
Figure S5, Related to Figure 5: Immune landscape in LUAD
A. Heatmap of expression levels of proteins most correlated with inferred activation of the Interferon gamma (IFNG) axis. Proteins involved in immune evasion signatures and proteins annotated as drug targets (as defined by :https://www.drugbank.ca) are highlighted by vertical bars on the left side of the figure. Important immune-related markers observed include WARS, LCK, CD4, TYMP, B2M (upregulated in the HTE cluster) and PTGR2, PDE4D, MAOA (upregulated in the CTE cluster). LCK and CD4 are members of the supramolecular lymphocyte regulatory complex that includes PTPRCAP, which showed differential DNA méthylation in our analysis (Figure 3F, G). Whether these derive from cancer cells or immune infiltrates is unclear.
B. The heatmap shows abundance levels (converted to Z-scores) of proteins of the IFNG axis pathway (Abril-Rodriguez and Ribas, 2017) and the Surfactant Metabolism pathway from Reactome database (Fabregat et al., 2018). The IFNG axis pathway determines activation of the adaptive immune system; 11 proteins of the IFNG axis were detected in global proteomics as presented in the heatmap (red vertical bar). The Surfactant Metabolism pathway was identified among the top three top pathways anti-correlated with inferred activation of the IFNG axis, with 14 of 30 pathway proteins, including 5 of 6 primary surfactant proteins (SFTP-A1, A2, B, C, and D), detected by global proteomics. Lung surfactant, responsible for preventing alveolar collapse at end-expiration, can also regulate pulmonary innate immunity (Whitsett, 2014), increasing immunosuppression (Pastva et al., 2007, Nayak et al, 2012). Prior studies have shown an association between genetic polymorphisms of surfactant proteins and lung carcinoma (Seifart et al., 2005) and bronchopulmonary dysplasia (Pavlovic et al., 2006), with genetic defects in SFTPA2 especially associated with lung cancer development (Wang et al., 2009). The observed upregulation of surfactant proteins in CTE lung tumors supports their immune-suppressive effects in lung cancer.
C. Heatmap depicting normalized enrichment scores (NES) of Hallmark gene sets (Liberzon et al., 2015) in each multi-omic cluster. To calculate cluster-specific NES we projected the matrix of multi-omic feature weights (W) derived by non-negative matrix factorization (NMF) onto gene sets using single-sample Gene Set Enrichment Analysis (ssGSEA) (Barbie et al., 2009). To derive a single weight for each gene measured across multiple omics data types (protein, RNA, phosphosite, acetylsite) we retained the weight with maximal absolute amplitude. Only gene sets significant in at least one cluster are shown (FDR <0.01).
D. Boxplot showing the distribution of the non-synonymous somatic mutations in each multi-omic cluster.
E. Flow diagram showing the workflow for developing and testing a deep learning algorithm to identify STK11 mutant samples based on hematoxylin and eosin stained histopathology slides. (I) LUAD tumor histopathology slides corresponding to analyzed tissue fragments were downloaded from The Cancer Imaging Archive (TCIA) database; (II) slides and corresponding per-slide level labels were separated into training (80%), validation (10%), and test sets (10%) at the per-patient level; (III) slides were tiled into 299-by-299-pixel pieces with overlapping areas of 49 pixels from each edge, omitting those with over 30% background. Tiles of each set were packaged into a TFrecord file; (IV) the InceptionV3-architectured convolutional neural network (CNN) was trained from scratch and the best performing model was picked based on validation set performance; (V) the model was applied to the test set, and the per-tile prediction scores were aggregated by slides and shown as heatmaps. The last-layer activations of 10000 randomly sampled tiles were exported for feature visualization on t-Distributed Stochastic Neighbor Embedding (t-SNE) (Figure 5E); (VI) counts and statistical metrics for area under receiver operating characteristic (AUROC), area under Precision-Recall Curve (AUPRC), and accuracy on per-slide and per-tile level were calculated with bootstrapped 95% confidence interval in parentheses. Table: the model achieved per-slide level AUROC of 0.961 and per-tile level AUROC of 0.892 in predicting STK11 mutation. Slide-level predictive accuracy was 94%.
F. To extract pathway-level proteomic features in an unsupervised manner, protein abundance measurements of 110 tumor samples were submitted to independent component analysis following the method proposed in (Liu et al., 2019). One signature (IC_068) showed significant associations with STK11 mutation status (average log10 nominal P-values within component cluster: −5.7). Average mixing scores for IC_068 represented ‘activity’ of the meta-gene level signature in each of the samples. Raw protein abundance values of genes contributing heavily to the signature (coefficient larger than 3) were shown in a heatmap.
G. Heatmaps showing protein (upper) and RNA expression (lower) of 16 gene products associated with neutrophil degranulation. STK11 and its partner STRADA are also shown.
Figure S6, Related to Figure 6: Impact of smoking on somatic mutations.
A. Bar plots showing distinct mutational signatures identified in 110 LUAD tumors. Somatic single nucleotide variants (SNVs) were determined from WGS data using SomaticWrapper, and 10 distinct mutation signatures were subsequently identified using SignatureAnalyzer (Kim et al., 2016); STAR Methods). We further combined two adjacent SNVs into a unitary di-nucleotide polymorphism (DNP) mutation if they were in the same haplotype (Table S6, STAR Methods). GG->TT or CC->AA were the dominant DNP types (~50%) and were associated with smoking status.
B. Schematic showing the approach used for determining the smoking score used in this study. Tumor purity estimates, counts of total mutations, and percentages that are smoking signature mutations and smoking-signature DNPs were used to derive a continuous smoking signature score.
C. Shown are the pairwise cosine similarities of each pair of the substitution signature probabilities for the 53 environmental mutagen exposures reported in Kucab et al. (Kucab et al., 2019). MX is a chlorine disinfection byproduct and a known DNA mutagen suspected of increasing cancer risk when present at sufficient levels in drinking water (McDonald and Komulainen, 2005). The MX signature was highly co-correlated to smoking signatures three PAHs DBADE, DBA, and 5-Methylchrysene. Benzidine, a chemical once heavily used in the dyeing industry and suspected to play a role in lung cancer (Tomioka et al., 2016), and PhIP, present in cooked meat and linked to various cancers (Tang et al., 2007), were also highly co-correlated to these PAHs (>0.5 and >0.7, respectively).
D. Boxplot showing significant difference (p = 2.2e-16) in RNA-based stemness index (mRNASI) between tumors and NATs. See also table S1.
E. Boxplot showing decrease of RNA-based stemness index (mRNASI) in both tumors and NATs with high smoking score (HSS) compared to corresponding samples with low smoking score (LSS). Differences are significant in NATs and approach significance in tumors. Within both tumors and NATs, samples with HSS showed higher mRNASI than samples with LSS (tumors: t test p = 0.069; NATs: t test, p = 0.038), consistent with the known field cancerization effect of tobacco exposure (Walser et al., 2008).
F. Upper: Scatterplot showing direction and significance of pathway-level protein differences between samples with high and low smoking scores (HSS and LSS) in tumors and NATs. Pathways are color-coded according to their pathway group in Figure 6B. Group 1 pathways are upregulated in HSS in tumors and downregulated in HSS in NATs. Group 2 pathways are downregulated in HSS in tumors and upregulated in HSS in NATs. Groups 3 and 4 are upregulated and downregulated, respectively, in both tumors and NATs. Signed -log 10 FDR represents Benjamini-Hochberg corrected p-values from the one sided-Mann-Whitney-Wilcoxon (MWW) test and the direction (+ or −) indicates activation or suppression (i.e. MWW test side with lower p-value). Lower: HSS vs LSS differential pathway scores in tumors and NATs at the transcriptome level. The group separations clearly defined by protein-based pathways (Figure 6B) are less evident at the RNA level. Smoking and mutation status are inextricably interwoven, so it is likely that these smoking score-related differentials represent a complex interplay between direct effects of combustion-related carcinogen exposure and effects mediated by mutational differences related to that exposure.
Figure S7, Related to Figure 7: Proteogenomic differences between tumors and matched adjacent normals.
A. PCA plots showing RNA, protein, phosphosite and acetylsite abundance in 110 tumor samples (triangles: cadet blue) and 101 NATs (circles: coral red).
B. Schematic showing differentially regulated RNA, proteins, phosphosites and acetylsites between tumors and paired NATs (upregulated sites, FDR <0.01, log2 FC >1; downregulated sites, FDR <0.01, log2 FC <−1). Most quantified proteins (76%) had differential expression between tumor and NAT (FDR < 0.01, Wilcoxon signed rank test); amongst those with at least 2-fold differential expression, a slight majority (64%) were higher in NAT (Table S7).
C. Gene-set enrichment analysis (GSEA) revealing pathways differentially expressed between tumor and paired NATs. Cell cycle progression, MYC Targets Upregulation, Unfolded Protein Response, Glycolysis and TCA cycle (adjusted p <0.001) were upregulated in tumor samples whereas KRAS Signaling (FDR <0.001), STAT3 Signaling (FDR <0.001), and Muscle Differentiation (FDR <0.001) were downregulated in tumor samples compared to NATs.
D. Using stringent cutoffs for quantitative difference, significance and consistency (log2 FC >2, FDR <0.01, and differential in >90% of all Tumor-NAT pairs (Pan-LUAD)), we identified 289 proteins upregulated at the protein level in tumors, 60 of which were supported by RNA and are shown in the figure (see also Table S7). “HPA staining proportions” indicate the proportion of lung adenocarcinoma sections staining positive for the specific marker in the Human Proteome Atlas database (https://www.proteinatlas.org/). This global tumor / NAT comparison revealed 18 enzymes, 3 transcription factors (TF), SOX4, TCF3, and HMGA1,2 transporters, 23 secreted, and 21 transmembrane proteins as candidate biomarkers. GREM1, SOX4, SPINT1, ST14, SPINT2, CTHRC1, KDF1, MDK, SFN, HMGA1, ESRP1, NME1, SERPINH1 and CBX8 are implicated in EMT and metastasis. Highly upregulated metabolic proteins included GFPT1, P4HB, PLOD2, PYCR1, SHMT2, PSAT1, ERO1A, IL411, DHFR, and LDHA. Stress-related marker candidates with prognostic significance included ERO1A, DHFR, MANF, HYOU1, LDHA, and CBX8. Remainder of figure: Proteomics-based tumor biomarker candidates (fold change>4 and adjusted p-value <0.01 in ≥80% of tumor / NAT pairs) for 4 frequently mutated genes: TP53 (40/63 shown), EGFR (39), KRAS (37) and STK11 (40/77 shown) (Table S7). Each dot in the box plot represents a tumor sample. Blue-colored boxplots highlight proteins with overexpression in more than 99% of tumor samples with the associated mutation. HPA proportions indicate the proportion of LUAD sections staining positive for the specific marker in the Human Proteome Atlas. Relevant characteristics of the biomarker candidates and relevant targeted drugs in clinical trials are shown in the accompanying plot. Condensed representations of these plots are shown in Figure 7C.
E. Rank plots depicting differential phosphosite-driven signatures between tumor and paired NATs in tumors with mutations in STK11 or TP53. Residual enrichment scores (y-axis) were calculated between mutated tumors (STK11 or TP53) and all other tumors in order to highlight tumor / NAT differences in tumors harboring the indicated mutation.
Table S1, Related to Figures 1–7, Figures S1–7: Metadata associated with LUAD patients and tumors related to STAR methods, Figures 1–7.
A. Sample annotations and associated genotype, clinical, geographical and pathological metadata.
Table S2, Related to Figures 1–7, Figures S1–7: Datasets pertaining to copy number alterations, somatic mutations, DNA methylation, protein coding RNA expression and microRNA expression obtained from tumors and NATs, related to Figures 1–7.
A. Data matrix of somatic copy number alterations (CNA) in tumors. CNAs are represented as log2(copy_number)-1 ratio. Refseq GRCh38/hg38 database was used as a reference.
B. Data matrix of site-level somatic mutations in tumors. Refseq Hg38 database was used as a reference.
C. Gene-level methylation data obtained by taking the median of beta values of all probes within the genomic location encompassing the promoter and gene body of each gene.
D. Log2-transformed RNA expression matrix obtained by upper-quartile normalization of “Reads Per Kilobase of transcript per Million mapped reads” (RPKM).
E. Row-median normalization applied to RNA expression matrix from Table S2D.
F. Log2-transformed “Transcript per Kilobase Million” (TPM)-values of mature miRNAs.
G. Luad-v3.1-mirna-mature-tpm-log2-row-norm.gct: Row-median normalization applied to miRNA expression from Table S2F.
H. Focal amplification in the CPTAC LUAD cohort obtained from GISTIC analysis.
I. Focal deletion in the CPTAC LUAD cohort obtained from GISTIC analysis.
Table S3, Related to Figures 1–7, Figures S1–7: Filtered and unfiltered proteomics datasets, pathway analysis and differentially regulated miRNA for tumors and NATs, related to Figures 1–7.
A. Two-component-normalized log2-transformed protein expression. The dataset was filtered as described in the STAR methods.
B. Two-component-normalized log2-transformed phosphosite expression. The dataset was filtered as described in the STAR methods.
C. Two-component-normalized log2-transformed acetylsite expression. The dataset was filtered as described in the STAR methods.
D. Unfiltered version of Table S3A.
E. Unfiltered version of Table S3B.
F. Unfiltered version of Table S3C.
G. Multi-omics pathway analysis and list of differentially regulated genes used for pathway analysis.
H. List summarizing 22 differentially expressed proteins obtained after adjustment for known confounding factors including smoking status, region of origin and mutation status of commonly mutated genes (EGFR, KRAS, STK11, TP53) and ALK fusions.
I. Differentially regulated miRNA in tumors. Histological subtype analysis was constrained by availability, as a substantial majority of cases in this cohort were typical adenocarcinomas with glandular/acinar formation. Of the remaining 6 dominant histomorphological subtypes represented in the study, solid (10 cases) and true papillary (9 cases) had enough samples for statistical comparison. Solid tumors segregated exclusively into NMF clusters C1 and C4 (Proximal inflammatory and Terminal respiratory unit), while all but one papillary fell into C1 or C3 (Proximal proliferative.) Twelve RNA species, some with established relevance to cancer, were differential between subtypes. For instance, IDH3A, a rate-limiting enzyme in the Krebs cycle, was elevated in the solid subtype, while PTK7, a tyrosine kinase thought to affect cell proliferation and survival in LUAD, was elevated in the papillary subtype. However, no proteins were identified as differential after adjustment for confounding variables.
Table S4, Related to Figures 3,4, Figures S3,S4: Multi-omics correlation, and additional analyses including pathways, Connectivity Map (CMAP), CIMP classification, iProFun, and differential regulation between KRAS and EGFR mutants versus WT samples, related to Figures 3 and 4.
A. Gene-wise mRNA-Protein Spearman correlation in tumors
B. Gene-wise mRNA-Protein Spearman correlation in NATs
C. Sample-wise mRNA-Protein Spearman correlation in tumors
D. Sample-wise mRNA-Protein Spearman correlation in NATs
E. mRNA-protein pairs that change their direction between tumors and NATs
F. CNA-RNA, CNA-Protein, CNA-Phosphoprotein and CNA-Acetylprotein correlations
G. List of cancer-associated genes and selected mutated genes used (see STAR methods)
H. CMAP-nominated genes and their significant associations with metadata (Table S1)
I. CMAP-nominated genes and the significant trans-events that overlapped with CMAP signatures
J. Results for classification of samples with CIMP (CpG Island Methylator Phenotype)
K. iProFun results with mRNA, protein and phosphoprotein as outcomes, and methylation as the primary predictor, adjusted for cis mutation, cis CNA, age, gender, smoking status, country of origin and tumor purity. Results include identified methylation effects at the gene level at FDR<0.1 and their proteomic associations with tumor and NAT status.
L. Differential RNA, global protein, phosphorylation, and acetylation between tumors with KRAS mutations vs WT tumors. Wilcoxon rank sum test was performed requiring at least 4 non-missing values in each group.
M. Differential RNA, global protein, phosphorylation, and acetylation between LUAD tumors with EGFR mutations vs WT tumors. Wilcoxon rank sum test was performed requiring at least 4 non-missing values in each group.
Table S5, Related to Figure 5, Figure S5: Immune landscape of tumors and NATs, related to Figure 5.
A. Immune composition of LUAD tumors and NATs as judged by deconvolution of RNA expression using xCell.
B. Immune clusters based on xCell signatures.
C. Pathways upregulated in immune groups based on RNAseq data.
D. Pathways upregulated in immune groups based on proteomics data. (D1) Pathways based on proteomics dataset (D2) Pathways exclusive to proteomics datasets (D3) Pathways exclusive to RNAseq dataset
E. Genes differentially expressed in immune groups based on RNAseq and proteomics data.
F. Association of xCell signatures with mutations with frequency >15 samples
G. ESTIMATE scores based on RNAseq data and tumor purity computed via TSNet (Petralia et al, 2018)
H. Supplementary data containing an STK11 mutation related proteomic signature extracted by ICA (independent component analysis)
Table S6, Related to Figure 6, Figure S6: Factors used for calculating smoking scores and environmental exposure, related to Figure 6.
A. Smoking score and the information integrated therein, which includes tumor purity estimates, counts of total mutations, percentages of smoking signature mutations, counts of smoking-signature DNPs, and adjustment to the strong alternative/non-smoking signatures.
B. Links to environmental factors based on regression of the 96 possible trinucleotide mutation combinations between the samples in the CPTAC LUAD cohort and the environmental signatures reported by Kucab et al. (Kucab et al., 2019). Correlations were calculated by least-squares fit, with significance assessed by T-testing and subsequent multiple test correction via FDR. A total of 5,830 hypothesis tests were conducted (53 rows/environmental factors * 110 columns/LUAD samples).
C. RNA- and proteomics-driven differentially regulated pathways between HSS (high smoking score) versus LSS (low smoking score) in both tumors and paired NATs
D. Differential protein markers in strict never-smokers (SNS) vs strict smokers (SS) and SNS tumors vs their paired NATs
Table S7, Related to Figure 7, Figure S7: Tumor-normal analysis and neoantigen analysis, related to Figure 7.
A. Differential RNA expression between tumors and paired NATs. Differential expression is indicated as log2-transformed fold-change
B. Differential protein expression between tumors and paired NATs (log2 FC>2 and FDR <0.01 in >80% of samples). Differential expression is indicated as log2-transformed fold-change.
C. Differential phosphosite expression between tumors and paired NATs. Differential expression is indicated as log2 transformed fold-change
D. Differential acetylsite expression between tumors and paired-NATs. Differential expression is indicated as log2-transformed fold-change
E. Proteome-driven differential pathway analysis between tumors and paired NATs
F. Differential phosphosites analysis between tumors and paired NATs and their corresponding protein expression
G. Differential acetylsite analysis between tumors and paired NATs and their corresponding protein expression
H. Protein biomarkers in pan-LUAD (all tumor-NAT pairs) and major mutation-driven tumor subsets (EGFR, TP53, KRAS and STK11)
I. PTM-SEA applied to phosphosite-specific differential tumor vs NAT analysis
J. Mutational subtype-specific tumor vs NAT analysis using PTM-SEA
K. Predicted tumor neoantigens identified in the CPTAC LUAD cohort at the level of RNA transcripts and peptides containing evidence of somatic mutations
Highlights.
Comprehensive LUAD proteogenomics exposes multi-omic clusters and immune subtypes
Phosphoproteomics identifies candidate ALK-fusion diagnostic markers and targets
Candidate drug targets: PTPN11 (EGFR), SOS1 (KRAS), neutrophil degranulation (STK11)
Phospho and acetyl modifications denote tumor-specific markers and druggable proteins
Integrated proteomics and genomics analysis of lung adenocarcinoma reveals multi-layered insights into potential drug targets and tumor markers
Acknowledgements
This work was supported by grants U24CA210986, U01CA214125, U24CA210979, U24CA210954, U24CA210972, U24CA210967, U24CA210993 and P30ES017885 from the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC).
Declaration of interest
BZ has received research funding from Bristol-Myers Squibb. All other authors have no conflict of interests to declare.
Footnotes
SECONDARY AUTHORS
Alex Webster, Alicia Francis, Alyssa Charamut, Amanda G. Paulovich, Amy M. Perou, Andrew K. Godwin, Andrii Karnuta, Annette Marrero-Oliveras, Barbara Hindenach, Barbara Pruetz, Bartosz Kubisa, Brian J. Druker, Chet Birger, Corbin D. Jones, Dana R. Valley, Daniel C. Rohrer, Daniel C. Zhou, Daniel W. Chan, David Chesla, David J. Clark, Dmitry Rykunov, Donghui Tan, Elena V. Ponomareva, Elizabeth Duffy, Eric Burks, Eric E. Schadt, Erik J. Bergstrom, Eugene S. Fedorov, Ewa Malc, George D. Wilson, Hai-Quan Chen, Halina M. Krzystek, Hongwei Liu, Houston Culpepper, Hua Sun, Hui Zhang, Jacob Day, James Suh, Jeffrey R. Whiteaker, Jennifer Eschbacher, John McGee, Karen A. Ketchum, Karin D. Rodland, Karna Robinson, Katherine A. Hoadley, Kei Suzuki, Ki Sung Um, Kim Elburn, Liang-Bo Wang, Lijun Chen, Linda Hannick, Liqun Qi, Lori J. Sokoll, Matgorzata Wojtyś, Marcin J. Domagalski, Marina A. Gritsenko, Mary B. Beasley, Matthew E. Monroe, Matthew J. Ellis, Maureen Dyer, Meghan C. Burke, Melissa Borucki, Meng-Hong Sun, Michael H. Roehrl, Michael J. Birrer, Michael Noble, Michael Schnaubelt, Michael Vernon, Michelle Chaikin, Mikhail Krotevich, Munziba Khan, Myvizhi E. Selvan, Nancy Roche, Nathan J. Edwards, Negin Vatanian, Olga Potapova, Pamela Grady, Peter B. McGarvey, Piotr Mieczkowski, Pushpa Hariharan, Rashna Madan, Ratna R. Thangudu, Richard D. Smith, Robert J. Welsh, Robert Zelt, Rohit Mehra, Ronald Matteotti, Sailaja Mareedu, Samuel H. Payne, Sandra Cottingham, Sanford P. Markey, Seema Chugh, Shaleigh Smith, Shirley Tsang, Shuang Cai, Simina M. Boca, Sonya Carter, Stacey Gabriel, Stephanie De Young, Stephen E. Stein, Sunita Shankar, Tanya Krubit, Tao Liu, Tara Skelly, Thomas Bauer, Uma Velvulou, Umut Ozbek, Vladislav A. Petyuk, Volodymyr Sovenko, William E. Bocik, William W. Maggio, Xi Chen, Yan Shi, Yige Wu, Yingwei Hu, Yuxing Liao, Zhen Zhang, Zhiao Shi
Consortia
Alex Webster, Alexey Nesvizhskii, Alicia Francis, Alyssa Charamut, Amanda Paulovich, Amy Perou, Ana Robles, Andrew Godwin, Andrii Karnuta, Annette Marrero-Oliveras, Antonio Colaprico, Arul Chinnaiyan, Azra Krek, Barbara Hindenach, Barbara Pruetz, Bartosz Kubisa, Bing Zhang, Bo Wen, Boris Reva, Brian Druker, Chandan Kumar-Sinha, Chelsea Newton, Chet Birger, Christopher Kinsinger, Corbin Jones, D. R. Mani, Dana Valley, Daniel Rohrer, Daniel Zhou, Daniel Chan, David Chesla, David Fenyö, David Heiman, David Clark, Dmitry Rykunov, Donghui Tan, Elena Ponomareva, Elizabeth Duffy, Emily Boja, Emily Kawaler, Eric Burks, Eric Schadt, Erik Bergstrom, Eugene Fedorov, Ewa Male, Felipe da Veiga Leprevost, Francesca Petralia, Gad Getz, Galen Hostetter, George Wilson, Gilbert Omenn, Hai-Quan Chen, Halina Krzystek, Harry Kane, Henry Rodriguez, Hongwei Liu, Houston Culpepper, Hua Sun, Hui Zhang, Jacob Day, James Suh, Jeffrey Whiteaker, Jennifer Eschbacher, Jiayi Ji, John McGee, Kai Li, Karen Ketchum, Karin Rodland, Karl Clauser, Karna Robinson, Karsten Krug, Katherine Hoadley, Kei Suzuki, Kelly Ruggles, Ki Sung Um, Kim Elburn, Lauren Tang, Li Ding, Liang-Bo Wang, Lijun Chen, Lili Blumenberg, Linda Hannick, Liqun Qi, Lori Sokoll, Maciej Wiznerowicz, Macintosh Cornwell, Matgorzata Wojtys, Marcin Cieslik, Marcin Domagalski, Marina Gritsenko, Mary Beasley, Mathangi Thiagarajan, Matthew. Wyczalkowski, Matthew Monroe, Matthew Ellis, Maureen Dyer, Meghan Burke, Mehdi Mesri, Melanie MacMullan, Melissa Borucki, Meng-Hong Sun, Michael Gillette, Michael Roehrl, Michael Birrer, Michael Noble, Michael Schnaubelt, Michael Vernon, Michelle Chaikin, Mikhail Krotevich, Munziba Khan, Myvizhi Selvan, Nancy Roche, Nathan Edwards, Negin Vatanian, Olga Potapova, Pamela Grady, Pankaj Vats, Pei Wang, Peter McGarvey, Piotr Mieczkowski, Pushpa Hariharan, Qing Kay Li, Rahul Mannan, Rajwanth R. Veluswamy, Ramani Bhupendra Kothadia, Ramaswamy Govindan, Rashna Madan, Ratna R. Thangudu, Richard Smith, Robert Welsh, Robert Zelt, Rohit Mehra, Ronald Matteotti, Runyu Hong, Sailaja Mareedu, Samuel H. Payne, Sandra Cottingham, Sanford P. Markey, Sara Savage, Saravana M. Dhanasekaran, Scott Jewell, Seema Chugh, Seungyeul Yoo, Shaleigh Smith, Shankha Satpathy, Shayan Avanessian, Shirley Tsang, Shuang Cai, Simina M. Boca, Song Cao, Sonya Carter, Stacey Gabriel, Stephanie De Young, Stephen Stein, Steven Carr, Suhas Vasaikar, Sunita Shankar, Tanya Krubit, Tao Liu, Tara Hiltke, Tara Skelly, Thomas Bauer, Uma Velvulou, Umut Ozbek, Vladislav Petyuk, Volodymyr Sovenko, Wenke Liu, Wen-Wei Liang, William E. Bocik, William W. Maggio, Xi Chen, Xiaoyu Song, Yan Shi, Yifat Geffen, Yige Wu, Yingwei Hu, Yize Li, Yosef Maruvka, Yuxing Liao, Zeynep Gümüş, Zhen Zhang, Zhiao Shi
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abril-Rodriguez G, and Ribas A (2017). Snapshot: Immune Checkpoint Inhibitors. Cancer Cell 31, 848–848.e1. [DOI] [PubMed] [Google Scholar]
- Aceto N, Sausgruber N, Brinkhaus H, Gaidatzis D, Martiny-Baron G, Mazzarol G, Confalonieri S, Quarto M, Hu G, Balwierz PJ, et al. (2012). Tyrosine phosphatase SHP2 promotes breast cancer progression and maintains tumor-initiating cells via activation of key transcription factors and a positive feedback signaling loop. Nat. Med 18, 529–537. [DOI] [PubMed] [Google Scholar]
- Achyut BR, and Arbab AS (2016). Myeloid cell signatures in tumor microenvironment predicts therapeutic response in cancer. Onco. Targets. Ther 9, 1047–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard D, Allard B, Gaudreau P-O, Chrobak P, and Stagg J (2016a). CD73-adenosine: a next-generation target in immuno-oncology. Immunotherapy 8, 145–163. [DOI] [PubMed] [Google Scholar]
- Allard D, Allard B, Gaudreau P-O, Chrobak P, and Stagg J (2016b). CD73-adenosine: a next-generation target in immuno-oncology. Immunotherapy 8, 145–163. [DOI] [PubMed] [Google Scholar]
- Almeida LG, Sakabe NJ, deOliveira AR, Silva MCC, Mundstein AS, Cohen T, Chen Y-T, Chua R, Gurung S, Gnjatic S, et al. (2009). CTdatabase: a knowledge-base of high- throughput and curated data on cancer-testis antigens. Nucleic Acids Res. 37, D816–D819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao X, Jiang M, Zhou J, Liang H, Xia H, and Chen G (2019). lincRNA-p21 inhibits the progression of non-small cell lung cancer via targeting miR-17–5p. Oncol. Rep 41, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D, Hu Z, and Butte AJ (2017). xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 18, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronheim A, Engelberg D, Li N, al-Alawi N, Schlessinger J, and Karin M (1994). Membrane targeting of the nucleotide exchange factor Sos is sufficient for activating the Ras signaling pathway. Cell 78, 949–961. [DOI] [PubMed] [Google Scholar]
- Bai Y, Xiong L, Zhu M, Yang Z, Zhao J, and Tang H (2019). Co-expression network analysis identified KIF2C in association with progression and prognosis in lung adenocarcinoma. Cancer Biomark. 24, 371–382. [DOI] [PubMed] [Google Scholar]
- Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, Colaprico A, Wendl MC, Kim J, Reardon B, et al. (2018). Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 173, 371–385.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. (2009). Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker L (2002). A comparison of nine confidence intervals for a Poisson parameter when the expected number of events is≤ 5. Am. Stat 56, 85–89. [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. (2012). The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, and Hochberg Y (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Series B Stat. Methodol 57, 289–300. [Google Scholar]
- Bennett AM, Tang TL, Sugimoto S, Walsh CT, and Neel BG (1994). Protein-tyrosine-phosphatase SHPTP2 couples platelet-derived growth factor receptor beta to Ras. Proc. Natl. Acad. Sci. U. S. A 91, 7335–7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenberg L, Kawaler E, Cornwell M, Smith S, Ruggles K, and Fenyo D BlackSheep: A Bioconductor and Bioconda package for differential extreme value analysis. [DOI] [PMC free article] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, and Jemal A (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin 68, 394–424. [DOI] [PubMed] [Google Scholar]
- Brunet J-P, Tamayo P, Golub TR, and Mesirov JP (2004). Metagenes and molecular pattern discovery using matrix factorization. Proc. Natl. Acad. Sci. U. S. A 101, 4164–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, et al. (2016). Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet 48, 607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2014). Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canning P, Sorrell FJ, and Bullock AN (2015). Structural basis of Keap1 interactions with Nrf2. Free Radic. Biol. Med 88, 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman AM, Sun KY, Ruestow P, Cowan DM, and Madl AK (2016). Lung cancer mutation profile of EGFR, ALK, and KRAS: Meta-analysis and comparison of never and ever smokers. Lung Cancer 102, 122–134. [DOI] [PubMed] [Google Scholar]
- Chen C, Lu Z, Yang J, Hao W, Qin Y, Wang H, Xie C, and Xie R (2016a). MiR-17–5p promotes cancer cell proliferation and tumorigenesis in nasopharyngeal carcinoma by targeting p21. Cancer Med. 5, 3489–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Gharib TG, Huang C-C, Thomas DG, Shedden KA, Taylor JMG, Kardia SLR, Misek DE, Giordano TJ, lannettoni MD, et al. (2002). Proteomic analysis of lung adenocarcinoma: identification of a highly expressed set of proteins in tumors. Clin. Cancer Res 8, 2298–2305. [PubMed] [Google Scholar]
- Chen H, Wang X, Bai J, and He A (2017). Expression, regulation and function of miR-495 in healthy and tumor tissues. Oncol. Lett. 13, 2021–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Ren S, Meng T, Aguilar J, and Sun Y (2013). Impact of glutathione-S-transferases (GST) polymorphisms and hypermethylation of relevant genes on risk of prostate cancer biochemical recurrence: a meta-analysis. PLoS One 8, e74775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Sun Y, Ji P, Kopetz S, and Zhang W (2015). Topoisomerase lla in chromosome instability and personalized cancer therapy. Oncogene 34, 4019–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-NP, LaMarche MJ, Chan HM, Fekkes P, Garcia-Fortanet J, Acker MG, Antonakos B, Chen CH-T, Chen Z, Cooke VG, et al. (2016b). Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature 535, 148–152. [DOI] [PubMed] [Google Scholar]
- Chew HK, Davies AM, Wun T, Harvey D, Zhou H, and White RH (2008). The incidence of venous thromboembolism among patients with primary lung cancer. J. Thromb. Haemost 6, 601–608. [DOI] [PubMed] [Google Scholar]
- Chidambaranathan-Reghupaty S, Mendoza R, Fisher PB, and Sarkar D (2018). The multifaceted oncogene SND1 in cancer: focus on hepatocellular carcinoma. Hepatoma Res 4, 10–20517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu A, Robertson G, Brooks D, Mungall AJ, Birol I, Coope R, Ma Y, Jones S, and Marra MA (2016). Large-scale profiling of microRNAs for The Cancer Genome Atlas. Nucleic Acids Res. 44, e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, and Getz G (2013). Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol 31, 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleynen I, Huysmans C, Sasazuki T, Shirasawa S, Van de Ven W, and Peeters K (2007). Transcriptional control of the human high mobility group A1 gene: basal and oncogenic Ras-regulated expression. Cancer Res. 67, 4620–4629. [DOI] [PubMed] [Google Scholar]
- Clinical Lung Cancer Genome Project (CLCGP), and Network Genomic Medicine (NGM) (2013). A genomics-based classification of human lung tumors. Sci. Transl. Med 5, 209ra153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N, Brown MK, Steptoe AL, Wani S, Chan WL, Forrest ARR, Kolle G, Gabrielli B, and Grimmond SM (2008). The miR-17–5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol. 9, R127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM, Castiglioni I, et al. (2016). TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 44, e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotto KC, Wagner AH, Feng Y-Y, Kiwala S, Coffman AC, Spies G, Wollam A, Spies NC, Griffith OL, and Griffith M (2018). DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 46, D1068–D1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, Caudy M, Garapati P, Gillespie M, Kamdar MR, et al. (2014). The Reactome pathway knowledgebase. Nucleic Acids Res. 42, D472–D477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cully M, and Downward J (2008). Snapshot: Ras Signaling. Cell 133, 1292–1292.e1. [DOI] [PubMed] [Google Scholar]
- Cunnick JM, Meng S, Ren Y, Desponts C, Wang H-G, Djeu JY, and Wu J (2002). Regulation of the mitogen-activated protein kinase signaling pathway by SHP2. J. Biol. Chem 277, 9498–9504. [DOI] [PubMed] [Google Scholar]
- Daily K, Ho Sui SJ, Schriml LM, Dexheimer PJ, Salomonis N, Schroll R, Bush S, Keddache M, Mayhew C, Lotia S, et al. (2017). Molecular, phenotypic, and sample-associated data to describe pluripotent stem cell lines and derivatives. Sci Data 4, 170030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. (2008). Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455, 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou F, Li H, Zhu M, Liang L, Zhang Y, Yi J, and Zhang Y (2018). Association between oncogenic status and risk of venous thromboembolism in patients with non-small cell lung cancer. Respir. Res 19, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Kawaler EA, Cui Zhou D, Gritsenko MA, Huang C, Blumenberg L, Karpova A, Petyuk VA, Savage SR, Satpathy S, et al. (2020). Proteogenomic Characterization of Endometrial Carcinoma. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducray SP, Natarajan K, Garland GD, Turner SD, and Egger G (2019). The Transcriptional Roles of ALK Fusion Proteins in Tumorigenesis. Cancers 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdel M, Trefz G, Spiess E, Habermaas S, Spring H, Lah T, and Ebert W (1990). Localization of cathepsin B in two human lung cancer cell lines. J. Histochem. Cytochem 38, 1313–1321. [DOI] [PubMed] [Google Scholar]
- Fabregat A, Sidiropoulos K, Viteri G, Forner O, Marin-Garcia P, Arnau V, D’Eustachio P, Stein L, and Hermjakob H (2017). Reactome pathway analysis: a high-performance inmemory approach. BMC Bioinformatics 18, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, et al. (2018). The Reactome Pathway Knowledgebase. Nucleic Acids Res. 46, D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrmann JF, Grapov D, Phinney BS, Stroble C, DeFelice BC, Rom W, Gandara DR, Zhang Y, Fiehn O, Pass H, et al. (2016). Proteomic profiling of lung adenocarcinoma indicates heightened DNA repair, antioxidant mechanisms and identifies LASP1 as a potential negative predictor of survival. Clin. Proteomics 13, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D, Hawke D, Zheng Y, Xia Y, Meisenhelder J, Nika H, Mills GB, Kobayashi R, Hunter T, and Lu Z (2007). Phosphorylation of beta-catenin by AKT promotes beta-catenin transcriptional activity. J. Biol. Chem 282, 11221–11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, Young G, Fennell TJ, Allen A, Ambrogio L, et al. (2011). A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 12, R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerster S, Kacprowski T, Dhople VM, Hammer E, Herzog S, Saafan H, Bien-Möller S, Albrecht M, Völker U, and Ritter CA (2013). Characterization of the EGFR interactome reveals associated protein complex networks and intracellular receptor dynamics. Proteomics 13, 3131–3144. [DOI] [PubMed] [Google Scholar]
- Fortin J-P, Labbe A, Lemire M, Zanke BW, Hudson TJ, Fertig EJ, Greenwood CM, and Hansen KD (2014). Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 15, 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RS, Bangur CS, Zasloff EJ, Fan L, Wang T, Watanabe Y, and Kalos M (2004). Molecular and immunological evaluation of the transcription factor SOX-4 as a lung tumor vaccine antigen. J. Immunol 172, 3319–3327. [DOI] [PubMed] [Google Scholar]
- Frolkis A, Knox C, Lim E, Jewison T, Law V, Hau DD, Liu P, Gautam B, Ly S, Guo AC, et al. (2010). SMPDB: The Small Molecule Pathway Database. Nucleic Acids Res. 38, D480–D487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujise N, Nanashim A, Taniguchi Y, Matsuo S, Hatano K, Matsumoto Y, Tagawa Y, and Ayabe H (2000). Prognostic impact of cathepsin B and matrix metalloproteinase-9 in pulmonary adenocarcinomas by immunohistochemical study. Lung Cancer 27, 19–26. [DOI] [PubMed] [Google Scholar]
- Fukutomi T, Takagi K, Mizushima T, Ohuchi N, and Yamamoto M (2014). Kinetic, thermodynamic, and structural characterizations of the association between Nrf2-DLGex degron and Keap1. Mol. Cell. Biol 34, 832–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathy-Kanniappan S (2017). Linking tumor glycolysis and immune evasion in cancer: Emerging concepts and therapeutic opportunities. Biochim. Biophys. Acta Rev. Cancer 1868, 212–220. [DOI] [PubMed] [Google Scholar]
- Gao H, Yu G, Zhang X, Yu S, Sun Y, and Li Y (2019). BZW2 gene knockdown induces cell growth inhibition, G1 arrest and apoptosis in muscle-invasive bladder cancers: A microarray pathway analysis. J. Cell. Mol. Med 23, 3905–3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Liang W-W, Foltz SM, Mutharasu G, Jayasinghe RG, Cao S, Liao W-W, Reynolds SM, Wyczalkowski MA, Yao L, et al. (2018). Driver Fusions and Their Implications in the Development and Treatment of Human Cancers. Cell Rep. 23, 227–238.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaujoux R, and Seoighe C (2010). A flexible R package for nonnegative matrix factorization. BMC Bioinformatics 11, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi O, Milia J, Filleron T, Wolf J, Carbone DP, Owen D, Camidge R, Narayanan V, Doebele RC, Besse B, et al. (2017). Targeting RET in Patients With RET-Rearranged Lung Cancers: Results From the Global, Multicenter RET Registry. J. Clin. Oncol 35, 1403–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giatromanolaki A, Koukourakis IM, Balaska K, Mitrakas AG, Harris AL, and Koukourakis MI (2019). Programmed death-1 receptor (PD-1) and PD-ligand-1 (PD-L1) expression in non-small cell lung cancer and the immune-suppressive effect of anaerobic glycolysis. Med. Oncol 36, 76. [DOI] [PubMed] [Google Scholar]
- Gildea JJ, Harding MA, Seraj MJ, Gulding KM, and Theodorescu D (2002). The role of Ral A in epidermal growth factor receptor-regulated cell motility. Cancer Res. 62, 982–985. [PubMed] [Google Scholar]
- Giubellino A, Burke TR Jr, and Bottaro DP (2008). Grb2 signaling in cell motility and cancer. Expert Opin. Ther. Targets 12, 1021–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurioli G, Martignano F, Salvi S, Costantini M, Gunelli R, and Casadio V (2018). GSTP1 methylation in cancer: a liquid biopsy biomarker? Clin. Chem. Lab. Med 56, 702–717. [DOI] [PubMed] [Google Scholar]
- Hänzelmann S, Castelo R, and Guinney J (2013). GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Tan D-L, Liu H-X, Lv F-L, and Wu W (2015). The auto-inhibitory state of Rho guanine nucleotide exchange factor ARHGEF5/TIM can be relieved by targeting its SH3 domain with rationally designed peptide aptamers. Biochimie 111, 10–18. [DOI] [PubMed] [Google Scholar]
- Herbst RS, Morgensztern D, and Boshoff C (2018). The biology and management of non-small cell lung cancer. Nature 553, 446–454. [DOI] [PubMed] [Google Scholar]
- Higashiyama M, Doi O, Kodama K, Yokouchi H, and Tateishi R (1993). Cathepsin B expression in tumour cells and laminin distribution in pulmonary adenocarcinoma. J. Clin. Pathol 46, 18–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillig RC, Sautier B, Schroeder J, Moosmayer D, Hilpmann A, Stegmann CM, Werbeck ND, Briem H, Boemer U, Weiske J, et al. (2019). Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction. Proc. Natl. Acad. Sci. U. S. A 116, 2551–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho EE, Atwood JR, and Meyskens FL Jr (1987). Methodological development of dietary fiber intervention to lower colon cancer risk. Prog. Clin. Biol. Res 248, 263–281. [PubMed] [Google Scholar]
- Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, and Sullivan M (2012). PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosgood HD 3rd, Menashe I, Shen M, Yeager M, Yuenger J, Rajaraman P, He X, Chatterjee N, Caporaso NE, Zhu Y, et al. (2008). Pathway-based evaluation of 380 candidate genes and lung cancer susceptibility suggests the importance of the cell cycle pathway. Carcinogenesis 29, 1938–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang O, Wu D, Xie F, Lin L, Wang X, Jiang M, Li Y, Chen W, Shen K, and Hu X (2015). Targeting rho guanine nucleotide exchange factor ARHGEF5/TIM with auto-inhibitory peptides in human breast cancer. Amino Acids 47, 1239–1246. [DOI] [PubMed] [Google Scholar]
- Hunt AL, Bateman NW, Hood BL, Conrads KA, Zhou M, Litzi TJ, Oliver J, Mitchell D, Gist G, Blanton B, et al. (2019). Extensive Intratumor Proteogenomic Heterogeneity Revealed by Multiregion Sampling in a High-Grade Serous Ovarian Tumor Specimen.
- Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. (2012). Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150, 1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Ishida T, Sugio K, and Sugimachi K (1994). Cathepsin B expression and laminin degradation as factors influencing prognosis of surgically treated patients with lung adenocarcinoma. Cancer Res. 54, 6133–6136. [PubMed] [Google Scholar]
- Jariwala N, Rajasekaran D, Mendoza RG, Shen X-N, Siddiq A, Akiel MA, Robertson CL, Subler MA, Windle JJ, Fisher PB, et al. (2017). Oncogenic Role of SND1 in Development and Progression of Hepatocellular Carcinoma. Cancer Res. 77, 3306–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerby-Arnon L, Shah P, Cuoco MS, Rodman C, Su M-J, Melms JC, Leeson R, Kanodia A, Mei S, Lin J-R, et al. (2018). A Cancer Cell Program Promotes T Cell Exclusion and Resistance to Checkpoint Blockade. Cell 175, 984–997.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeschke J, Bizet M, Desmedt C, Calonne E, Dedeurwaerder S, Garaud S, Koch A, Larsimont D, Salgado R, Van den Eynden G, et al. (2017). DNA methylation-based immune response signature improves patient diagnosis in multiple cancers. J. Clin. Invest 127, 3090–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewison T, Su Y, Disfany FM, Liang Y, Knox C, Maciejewski A, Poelzer J, Huynh J, Zhou Y, Arndt D, et al. (2014). SMPDB 2.0: big improvements to the Small Molecule Pathway Database. Nucleic Acids Res. 42, D478–D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Lohse CM, Chu PG, Wu C-L, Woda BA, Rock KL, and Kwon ED (2008). Oncofetal protein IMP3: a novel molecular marker that predicts metastasis of papillary and chromophobe renal cell carcinomas. Cancer 112, 2676–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Liao M, Zhang L, Yang M, and Zhao J (2019). Role of the novel gene BZW2 in the development of hepatocellular carcinoma. J. Cell. Physiol [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, and Rabinovic A (2007). Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127. [DOI] [PubMed] [Google Scholar]
- Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, and Nielsen M (2017). NetMHCpan-4.0: Improved Peptide-MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J. Immunol 199, 3360–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamioka Y, Yasuda S, Fujita Y, Aoki K, and Matsuda M (2010). Multiple decisive phosphorylation sites for the negative feedback regulation of SOS1 via ERK. J. Biol. Chem 285, 33540–33548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, and Goto S (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasar S, Kim J, Improgo R, Tiao G, Polak P, Haradhvala N, Lawrence MS, Kiezun A, Fernandes SM, Bahl S, et al. (2015). Whole-genome sequencing reveals activation-induced cytidine deaminase signatures during indolent chronic lymphocytic leukaemia evolution. Nat. Commun 6, 8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashatus DF (2013). Ral GTPases in tumorigenesis: emerging from the shadows. Exp. Cell Res 319, 2337–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key NS, Khorana AA, Kuderer NM, Bohlke K, Lee AYY, Arcelus JI, Wong SL, Balaban EP, Flowers CR, Francis CW, et al. (2019). Venous Thromboembolism Prophylaxis and Treatment in Patients With Cancer: ASCO Clinical Practice Guideline Update. J. Clin. Oncol JCO1901461. [DOI] [PubMed] [Google Scholar]
- Kim H, and Park H (2007). Sparse non-negative matrix factorizations via alternating non-negativity-constrained least squares for microarray data analysis. Bioinformatics 23, 1495–1502. [DOI] [PubMed] [Google Scholar]
- Kim S, and Pevzner PA (2014). MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun 5, 5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Mouw KW, Polak P, Braunstein LZ, Kamburov A, Kwiatkowski DJ, Rosenberg JE, Van Allen EM, D’Andrea A, and Getz G (2016). Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat. Genet 48, 600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Hu Z, Cai L, Li K, Choi E, Faubert B, Bezwada D, Rodriguez-Canales J, Villalobos P, Lin Y-F, et al. (2017). CPS1 maintains pyrimidine pools and DNA synthesis in KRAS/LKB1-mutant lung cancer cells. Nature 546, 168–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KM, An AR, Park HS, Jang KY, Moon WS, Kang MJ, Lee YC, Ku JH, and Chung MJ (2018a). Combined expression of protein disulfide isomerase and endoplasmic reticulum oxidoreductin 1-a is a poor prognostic marker for non-small cell lung cancer. Oncol. Lett 16, 5753–5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Scheffler K, Halpern AL, Bekritsky MA, Noh E, Kallberg M, Chen X, Kim Y, Beyter D, Krusche P, et al. (2018b). Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods 15, 591–594. [DOI] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, and Wilson RK (2012). VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno T, Ichikawa H, Totoki Y, Yasuda K, Hiramoto M, Nammo T, Sakamoto H, Tsuta K, Furuta K, Shimada Y, et al. (2012). KIF5B-RET fusions in lung adenocarcinoma. Nat. Med 18, 375–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike A, Nishikawa H, Wu W, Okada Y, Venkitaraman AR, and Ohta T (2010). Recruitment of phosphorylated NPM1 to sites of DNA damage through RNF8-dependent ubiquitin conjugates. Cancer Res. 70, 6746–6756. [DOI] [PubMed] [Google Scholar]
- Kong W, Cheng Y, Liang H, Chen Q, Xiao C, Li K, Huang Z, and Zhang J (2018). Prognostic value of miR-17–5p in cancers: a meta-analysis. Onco. Targets. Ther 11, 3541–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konofaos P, Kontzoglou K, Parakeva P, Kittas C, Margari N, Giaxnaki E, Pouliakis M, Kouraklis G, and Karakitsos P (2013). The role of ThinPrep cytology in the investigation of ki-67 index, p53 and HER-2 detection in fine-needle aspirates of breast tumors. J. BUON 18, 352–358. [PubMed] [Google Scholar]
- Kozuma Y, Takada K, Toyokawa G, Kohashi K, Shimokawa M, Hirai F, Tagawa T, Okamoto T, Oda Y, and Maehara Y (2018a). Indoleamine 2,3-dioxygenase 1 and programmed cell death-ligand 1 co-expression correlates with aggressive features in lung adenocarcinoma. European Journal of Cancer 101, 20–29. [DOI] [PubMed] [Google Scholar]
- Kozuma Y, Takada K, Toyokawa G, Kohashi K, Shimokawa M, Hirai F, Tagawa T, Okamoto T, Oda Y, and Maehara Y (2018b). Indoleamine 2,3-dioxygenase 1 and programmed cell death-ligand 1 co-expression correlates with aggressive features in lung adenocarcinoma. Eur. J. Cancer 101, 20–29. [DOI] [PubMed] [Google Scholar]
- Krug K, Mertins P, Zhang B, Hornbeck P, Raju R, Ahmad R, Szucs M, Mundt F, Forestier D, Jane-Valbuena J, et al. (2018). A curated resource for phosphosite-specific signature analysis. Mol. Cell. Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglova NA, Meshkova TD, Kopylov AT, Mazurov DV, and Filatov AV (2017). Constitutive and activation-dependent phosphorylation of lymphocyte phosphatase-associated phosphoprotein (LPAP). PLoS One 12, e0182468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucab JE, Zou X, Morganella S, Joel M, Nanda AS, Nagy E, Gomez C, Degasperi A, Harris R, Jackson SP, et al. (2019). A Compendium of Mutational Signatures of Environmental Agents. Cell 177, 821–836.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuser-Abali G, Gong L, Yan J, Liu Q, Zeng W, Williamson A, Lim CB, Molloy ME, Little JB, Huang L, et al. (2018). An EZH2-mediated epigenetic mechanism behind p53-dependent tissue sensitivity to DNA damage. Proc. Natl. Acad. Sci. U. S. A 115, 3452–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak EL, Bang Y-J, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou S-HI, Dezube BJ, Jänne PA, Costa DB, et al. (2010). Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med 363, 1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Markovets A, Ahdesmaki M, Chapman B, Hofmann O, McEwen R, Johnson J, Dougherty B, Barrett JC, and Dry JR (2016). VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 44, e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S-K, Yan S, Xu S, U K-P, Cheng PN-M, and Ho JC-M (2019). Endogenous arginase 2 as a potential biomarker for PEGylated arginase 1 treatment in xenograft models of squamous cell lung carcinoma. Oncogenesis 8, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet J-P, Subramanian A, Ross KN, et al. (2006). The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935. [DOI] [PubMed] [Google Scholar]
- Le S, Josse J, and Husson F (2008). FactoMineR: AnRPackage for Multivariate Analysis. Journal of Statistical Software 25. [Google Scholar]
- Lei B, Qi W, Zhao Y, Li Y, Liu S, Xu X, Zhi C, Wan L, and Shen H (2015). PBK/TOPK expression correlates with mutant p53 and affects patients’ prognosis and cell proliferation and viability in lung adenocarcinoma. Hum. Pathol 46, 217–224. [DOI] [PubMed] [Google Scholar]
- Le Menn G, and Neels JG (2018). Regulation of Immune Cell Function by PPARs and the Connection with Metabolic and Neurodegenerative Diseases. Int. J. Mol. Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Vaudel M, Zhang B, Ren Y, and Wen B (2019). PDV: an integrative proteomics data viewer. Bioinformatics 35, 1249–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Shen D, Shao J, Crowder R, Liu W, Prat A, He X, Liu S, Hoog J, Lu C, et al. (2013). Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 4, 1116–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, and Tamayo P (2015). The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lignitto L, LeBoeuf SE, Homer H, Jiang S, Askenazi M, Karakousi TR, Pass HI, Bhutkar AJ, Tsirigos A, Ueberheide B, et al. (2019). Nrf2 Activation Promotes Lung Cancer Metastasis by Inhibiting the Degradation of Bach1. Cell 178, 316–329.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Wang X, Wang L, Ma X, Gong Z, Zhang S, and Li Y (2018). Targeting the IDO1 pathway in cancer: from bench to bedside. J. Hematol. Oncol 11, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Payne SH, Ma S, and Fenyö D (2019). Extracting Pathway-level Signatures from Proteogenomic Data in Breast Cancer Using Independent Component Analysis. Mol. Cell. Proteomics 18, S169–S182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lladó V, Terés S, Higuera M, Alvarez R, Noguera-Salva MA, Halver JE, Escribá PV, and Busquets X (2009). Pivotal role of dihydrofolate reductase knockdown in the anticancer activity of 2-hydroxyoleic acid. Proc. Natl. Acad. Sci. U. S. A 106, 13754–13758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochhead P, Imamura Y, Morikawa T, Kuchiba A, Yamauchi M, Liao X, Qian ZR, Nishihara R, Wu K, Meyerhardt JA, et al. (2012). Insulin-like growth factor 2 messenger RNA binding protein 3 (IGF2BP3) is a marker of unfavourable prognosis in colorectal cancer. Eur. J. Cancer 48, 3405–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loriot A, Boon T, and De Smet C (2003). Five new human cancer-germline genes identified among 12 genes expressed in spermatogonia. Int. J. Cancer 105, 371–376. [DOI] [PubMed] [Google Scholar]
- Lu J, Wei J-H, Feng Z-H, Chen Z-H, Wang Y-Q, Huang Y, Fang Y, Liang Y-P, Cen J-J, Pan Y-H, et al. (2017). miR-106b-5p promotes renal cell carcinoma aggressiveness and stem-cell-like phenotype by activating Wnt/β-catenin signalling. Oncotarget 8, 21461–21471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Gong D, Bar-Sagi D, and Cole PA (2001). Site-specific incorporation of a phosphotyrosine mimetic reveals a role for tyrosine phosphorylation of SHP-2 in cell signaling. Mol. Cell 8, 759–769. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. (2004). Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med 350, 2129–2139. [DOI] [PubMed] [Google Scholar]
- Maksimovic J, Phipson B, and Oshlack A (2016). A cross-package Bioconductor workflow for analysing methylation array data. F1000Res. 5, 1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN, Kaminska B, Huelsken J, Omberg L, Gevaert O, et al. (2018). Machine Learning Identifies Sternness Features Associated with Oncogenic Dedifferentiation. Cell 173, 338–354.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx N, Mach F, Sauty A, Leung JH, Sarafi MN, Ransohoff RM, Libby P, Plutzky J, and Luster AD (2000). Peroxisome proliferator-activated receptor-gamma activators inhibit IFN-gamma-induced expression of the T cell-active CXC chemokines IP-10, Mig, and I-TAC in human endothelial cells. J. Immunol 164, 6503–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda A, Motoya S, Kimura S, McInnis R, Maizel AL, and Takeda A (1998). Disruption of lymphocyte function and signaling in CD45-associated protein-null mice. J. Exp. Med 187, 1863–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, and Getz G (2011). GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P, Wang X, Qiao JW, Cao S, Petralia F, et al. (2016). Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534, 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P, Tang LC, Krug K, Clark DJ, Gritsenko MA, Chen L, Clauser KR, Clauss TR, Shah P, Gillette MA, et al. (2018). Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat. Protoc 13, 1632–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Ikeda S, Watanabe S, Sugawara M, and Itoh M (2017). Mib1 contributes to persistent directional cell migration by regulating the Ctnnd1-Rac1 pathway. Proc. Natl. Acad. Sci. U. S. A 114, E9280–E9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagner A, Yart A, Dance M, Perret B, Salles J-P, and Raynal P (2005). A novel role for Gab1 and SHP2 in epidermal growth factor-induced Ras activation. J. Biol. Chem 280, 5350–5360. [DOI] [PubMed] [Google Scholar]
- Mulvihill MS, Kwon Y-W, Lee S, Fang LT, Choi H, Ray R, Kang HC, Mao J-H, Jablons D, and Kim I-J (2012). Gremlin is overexpressed in lung adenocarcinoma and increases cell growth and proliferation in normal lung cells. PLoS One 7, e42264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SA, Klaeger S, Satpathy S, Viner R, Choi J, Rogers J, Clauser K, Udeshi ND, and Carr SA (2018). Evaluation of Advanced Precursor Determination for Tandem Mass Tag (TMT)-Based Quantitative Proteomics across Instrument Platforms. J. Proteome Res [DOI] [PubMed] [Google Scholar]
- Nakashima M, Adachi S, Yasuda I, Yamauchi T, Kawaguchi J, Hanamatsu T, Yoshioka T, Okano Y, Hirose Y, Kozawa O, et al. (2011). Inhibition of Rho-associated coiled-coil containing protein kinase enhances the activation of epidermal growth factor receptor in pancreatic cancer cells. Mol. Cancer 10, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama S, Sng N, Carretero J, Welner R, Hayashi Y, Yamamoto M, Tan AJ, Yamaguchi N, Yasuda H, Li D, et al. (2014). β-catenin contributes to lung tumor development induced by EGFR mutations. Cancer Res. 74, 5891–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak A, Dodagatta-Marri E, Tsolaki AG, and Kishore U (2012). An Insight into the Diverse Roles of Surfactant Proteins, SP-A and SP-D in Innate and Adaptive Immunity. Frontiers in Immunology 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Bryan JP (2019). Pharmacological targeting of RAS: Recent success with direct inhibitors. Pharmacol. Res 139, 503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki I, Ishikawa S, Ando W, and Sohara Y (2016). Lung Adenocarcinoma in Never Smokers: Problems of Primary Prevention from Aspects of Susceptible Genes and Carcinogens. Anticancer Res. 36, 6207–6224. [DOI] [PubMed] [Google Scholar]
- Okazaki T, Chikuma S, Iwai Y, Fagarasan S, and Honjo T (2013). A rheostat for immune responses: the unique properties of PD-1 and their advantages for clinical application. Nat. Immunol 14, 1212–1218. [DOI] [PubMed] [Google Scholar]
- Ostman A, Hellberg C, and Bohmer FD (2006). Protein-tyrosine phosphatases and cancer. Nat. Rev. Cancer 6, 307–320. [DOI] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. (2004). EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500. [DOI] [PubMed] [Google Scholar]
- Parikh K, Antanaviciute A, Fawkner-Corbett D, Jagielowicz M, Aulicino A, Lagerholm C, Davis S, Kinchen J, Chen HH, Alham NK, et al. (2019). Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature 567, 49–55. [DOI] [PubMed] [Google Scholar]
- Pavlovic J, Papagaroufalis C, Xanthou M, Liu W, Fan R, Thomas NJ, Apostolidou I, Papathoma E, Megaloyianni E, DiAngelo S, et al. (2006). Genetic variants of surfactant proteins A, B, C, and D in bronchopulmonary dysplasia. Dis. Markers 22, 277–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Moreno M, Davis MA, Wong E, Pasolli HA, Reynolds AB, and Fuchs E (2006). p120-catenin mediates inflammatory responses in the skin. Cell 124, 631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschard P, McCarthy A, Leblanc-Dominguez V, Yeo M, Guichard S, Stamp G, and Marshall CJ (2012). Genetic deletion of RALA and RALB small GTPases reveals redundant functions in development and tumorigenesis. Curr. Biol 22, 2063–2068. [DOI] [PubMed] [Google Scholar]
- Petralia F, Song W-M, Tu Z, and Wang P (2016). New Method for Joint Network Analysis Reveals Common and Different Coexpression Patterns among Genes and Proteins in Breast Cancer. J. Proteome Res 15, 743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia F, Wang L, Peng J, Yan A, Zhu J, and Wang P (2018). A new method for constructing tumor specific gene co-expression networks based on samples with tumor purity heterogeneity. Bioinformatics 34, i528–i536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podkalicka P, Mucha O, Józkowicz A, Dulak J, and Łoboda A (2018). Heme oxygenase inhibition in cancers: possible tools and targets. Contemp. Oncol 22, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahallad A, Heynen GJJE, Germano G, Willems SM, Evers B, Vecchione L, Gambino V, Lieftink C, Beijersbergen RL, Di Nicolantonio F, et al. (2015). PTPN11 Is a Central Node in Intrinsic and Acquired Resistance to Targeted Cancer Drugs. Cell Rep. 12, 1978–1985. [DOI] [PubMed] [Google Scholar]
- Qu P, Du H, Wang X, and Yan C (2009). Matrix metalloproteinase 12 overexpression in lung epithelial cells plays a key role in emphysema to lung bronchioalveolar adenocarcinoma transition. Cancer Res. 69, 7252–7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Chen Z, Chen L, Fang B, Win-Piazza H, Haura E, Koomen JM, and Wu J (2010). Critical role of Shp2 in tumor growth involving regulation of c-Myc. Genes Cancer 1, 994–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas JM, Oliva JL, and Santos E (2011). Mammalian son of sevenless Guanine nucleotide exchange factors: old concepts and new perspectives. Genes Cancer 2, 298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano G, Acunzo M, Garofalo M, Di Leva G, Cascione L, Zanca C, Bolon B, Condorelli G, and Croce CM (2012). MiR-494 is regulated by ERK1/2 and modulates TRAIL-induced apoptosis in non-small-cell lung cancer through BIM down-regulation. Proc. Natl. Acad. Sci. U. S. A 109, 16570–16575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rother K, Johne C, Spiesbach K, Haugwitz U, Tschop K, Wasner M, Klein-Hitpass L, Möröy T, Mössner J, and Engeland K (2004). Identification of Tcf-4 as a transcriptional target of p53 signalling. Oncogene 23, 3376–3384. [DOI] [PubMed] [Google Scholar]
- Roy MG, Livraghi-Butrico A, Fletcher AA, McElwee MM, Evans SE, Boerner RM, Alexander SN, Bellinghausen LK, Song AS, Petrova YM, et al. (2014). Muc5b is required for airway defence. Nature 505, 412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggles KV, Tang Z, Wang X, Grover H, Askenazi M, Teubl J, Cao S, McLellan MD, Clauser KR, Tabb DL, et al. (2016). An Analysis of the Sensitivity of Proteogenomic Mapping of Somatic Mutations and Novel Splicing Events in Cancer. Mol. Cell. Proteomics 15, 1060–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakashita M, Sakashita S, Murata Y, Shiba-Ishii A, Kim Y, Matsuoka R, Nakano N, Sato Y, and Noguchi M (2018). High expression of ovarian cancer immunoreactive antigen domain containing 2 (OCIAD2) is associated with poor prognosis in lung adenocarcinoma. Pathol. Int 68, 596–604. [DOI] [PubMed] [Google Scholar]
- Salerno EP, Bedognetti D, Mauldin IS, Deacon DH, Shea SM, Pinczewski J, Obeid JM, Coukos G, Wang E, Gajewski TF, et al. (2016). Human melanomas and ovarian cancers overexpressing mechanical barrier molecule genes lack immune signatures and have increased patient mortality risk. Oncoimmunology 5, e1240857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonis N, Dexheimer PJ, Omberg L, Schroll R, Bush S, Huo J, Schriml L, Ho Sui S, Keddache M, Mayhew C, et al. (2016). Integrated Genomic Analysis of Diverse Induced Pluripotent Stem Cells from the Progenitor Cell Biology Consortium. Stem Cell Reports 7, 110–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia S, et al. (2018). Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 173, 321–337.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Carbone C, Piro G, Chiao PJ, and Melisi D (2017). TAK-ing aim at chemoresistance: The emerging role of MAP3K7 as a target for cancer therapy. Drug Resist. Updat 33–35, 36–42. [DOI] [PubMed] [Google Scholar]
- Satpathy S, Jaehnig EJ, Krug K, Kim B-J, Saltzman AB, Chan DW, Holloway KR, Anurag M, Huang C, Singh P, et al. (2020). Microscaled proteogenomic methods for precision oncology. Nat. Commun 11, 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CT, Wong WSW, Swamy S, Becq J, Murray LJ, and Cheetham RK (2012). Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28, 1811–1817. [DOI] [PubMed] [Google Scholar]
- Scanlan MJ, Altorki NK, Gure AO, Williamson B, Jungbluth A, Chen YT, and Old LJ (2000). Expression of cancer-testis antigens in lung cancer: definition of bromodomain testis-specific gene (BRDT) as a new CT gene, CT9. Cancer Lett. 150, 155–164. [DOI] [PubMed] [Google Scholar]
- Schneeberger VE, Ren Y, Luetteke N, Huang Q, Chen L, Lawrence HR, Lawrence NJ, Haura EB, Koomen JM, Coppola D, et al. (2015). Inhibition of Shp2 suppresses mutant EGFR-induced lung tumors in transgenic mouse model of lung adenocarcinoma. Oncotarget 6, 6191–6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwermer M, Lee S, Koster J, van Maerken T, Stephan H, Eggert A, Morik K, Schulte JH, and Schramm A (2015). Sensitivity to cdk1-inhibition is modulated by p53 status in preclinical models of embryonal tumors. Oncotarget 6, 15425–15435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrucca L, Fop M, Murphy TB, and Raftery AE (2016). mclust 5: Clustering, Classification and Density Estimation Using Gaussian Finite Mixture Models. R J. 8, 289–317. [PMC free article] [PubMed] [Google Scholar]
- Seifart C, Lin H-M, Seifart U, Plagens A, DiAngelo S, Von Wichert P, and Floros J (2005). Rare SP-A alleles and the SP-A1–6A4 allele associate with risk for lung carcinoma. Clinical Genetics 68, 128–136. [DOI] [PubMed] [Google Scholar]
- Shadforth IP, Dunkley TPJ, Lilley KS, and Bessant C (2005). i-Tracker: for quantitative proteomics using iTRAQ. BMC Genomics 6, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AT, Ou S-HI, Bang Y-J, Camidge DR, Solomon BJ, Salgia R, Riely GJ, Varella-Garcia M, Shapiro GI, Costa DB, et al. (2014). Crizotinib in ROS1-rearranged non-small-cell lung cancer. N. Engl. J. Med 371, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D-M, Bian X-Y, Qin C-D, and Wu W-Z (2018). miR-106b-5p promotes stem cell-like properties of hepatocellular carcinoma cells by targeting PTEN via PI3K/Akt pathway. Onco. Targets. Ther 11, 571–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Nakata M, Hirami Y, Yukawa T, Maeda A, and Tanemoto K (2010). Tumor-Infiltrating Foxp3 Regulatory T Cells are Correlated with Cyclooxygenase-2 Expression and are Associated with Recurrence in Resected Non-small Cell Lung Cancer. Journal of Thoracic Oncology 5, 585–590. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, and Jemal A (2019). Cancer statistics, 2019. CA Cancer J. Clin 69, 7–34. [DOI] [PubMed] [Google Scholar]
- Sokolov A, Paull EO, and Stuart JM (2016). ONE-CLASS DETECTION OF CELL STATES IN TUMOR SUBTYPES. Pac. Symp. Biocomput 21, 405–416. [PMC free article] [PubMed] [Google Scholar]
- Song N, Liu B, Wu J-L, Zhang R-F, Duan L, He W-S, and Zhang C-M (2013). Prognostic value of HMGB3 expression in patients with non-small cell lung cancer. Tumour Biol. 34, 2599–2603. [DOI] [PubMed] [Google Scholar]
- Song X, Ji J, Gleason KJ, Yang F, Martignetti JA, Chen LS, and Wang P (2019). Insights into Impact of DNA Copy Number Alteration and Methylation on the Proteogenomic Landscape of Human Ovarian Cancer via a Multi-omics Integrative Analysis. Mol. Cell. Proteomics 18, S52–S65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeck H, Kwon DS, Pyo A, Flanders M, Chevalier MF, Law K, Julg B, Trocha K, Jolin JS, Anahtar MN, et al. (2011). Epithelial adhesion molecules can inhibit HIV-1-specific CD8+ T-cell functions. Blood 117, 5112–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian J, and Govindan R (2007). Lung cancer in never smokers: a review. J. Clin. Oncol 25, 561–570. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, et al. (2017). A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 171, 1437–1452.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Schiller JH, and Gazdar AF (2007). Lung cancer in never smokers--a different disease. Nat. Rev. Cancer 7, 778–790. [DOI] [PubMed] [Google Scholar]
- Sunaga N, Imai H, Shimizu K, Shames DS, Kakegawa S, Girard L, Sato M, Kaira K, Ishizuka T, Gazdar AF, et al. (2012). Oncogenic KRAS-induced interleukin-8 overexpression promotes cell growth and migration and contributes to aggressive phenotypes of non-small cell lung cancer. Int. J. Cancer 130, 1733–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svinkina T, Gu H, Silva JC, Mertins P, Qiao J, Fereshetian S, Jaffe JD, Kuhn E, Udeshi ND, and Carr SA (2015). Deep, Quantitative Coverage of the Lysine Acetylome Using Novel Anti-acetyl-lysine Antibodies and an Optimized Proteomic Workflow. Mol. Cell. Proteomics 14, 2429–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szolek A, Schubert B, Mohr C, Sturm M, Feldhahn M, and Kohlbacher O (2014). OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics 30, 3310–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi K, and Yamamoto M (2017). The KEAP1-NRF2 System in Cancer. Front. Oncol 7, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada K, Kohashi K, Shimokawa M, Haro A, Osoegawa A, Tagawa T, Seto T, Oda Y, and Maehara Y (2019). Co-expression of IDO1 and PD-L1 in lung squamous cell carcinoma: Potential targets of novel combination therapy. Lung Cancer 128, 26–32. [DOI] [PubMed] [Google Scholar]
- Takei N, Yoneda A, Sakai-Sawada K, Kosaka M, Minomi K, and Tamura Y (2017). Hypoxia-inducible ERO1 promotes cancer progression through modulation of integrin-p1 modification and signalling in HCT116 colorectal cancer cells. Sci. Rep 7, 9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, Hatano S, Asaka R, Hamanaka W, Ninomiya H, Uehara H, et al. (2012). RET, ROS1 and ALK fusions in lung cancer. Nat. Med 18, 378–381. [DOI] [PubMed] [Google Scholar]
- Tan VYF, and Fevotte C (2013). Automatic relevance determination in nonnegative matrix factorization with the p-divergence. IEEE Trans. Pattern Anal. Mach. Intell 35, 1592–1605. [DOI] [PubMed] [Google Scholar]
- Tang B, Tian Y, Liao Y, Li Z, Yu S, Su H, Zhong F, Yuan G, Wang Y, Yu H, et al. (2019). CBX8 exhibits oncogenic properties and serves as a prognostic factor in hepatocellular carcinoma. Cell Death Dis. 10, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, et al. (2019). COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 47, D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J, et al. (2004). Regulation of the p300 HAT domain via a novel activation loop. Nat. Struct. Mol. Biol 11, 308–315. [DOI] [PubMed] [Google Scholar]
- Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM, et al. (2017). Defining a Cancer Dependency Map. Cell 170, 564–576.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufo G, Jones AWE, Wang Z, Hamelin J, Tajeddine N, Esposti DD, Martel C, Boursier C, Gallerne C, Migdal C, et al. (2014). The protein disulfide isomerases PDIA4 and PDIA6 mediate resistance to cisplatin-induced cell death in lung adenocarcinoma. Cell Death Differ. 21, 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udeshi ND, Mani DC, Satpathy S, Fereshetian S, Gasser JA, Svinkina T, Olive ME, Ebert BL, Mertins P, and Carr SA (2020). Rapid and deep-scale ubiquitylation profiling for biology and translational research. Nature Communications 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Björling E, Agaton C, Szigyarto CA-K, Amini B, Andersen E, Andersson A-C, Angelidou P, Asplund A, Asplund C, et al. (2005). A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteomics 4, 1920–1932. [DOI] [PubMed] [Google Scholar]
- Vasaikar SV, Straub P, Wang J, and Zhang B (2018). LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 46, D956–D963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigil D, Cherfils J, Rossman KL, and Der CJ (2010). Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat. Rev. Cancer 10, 842–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel W, and Ullrich A (1996). Multiple in vivo phosphorylated tyrosine phosphatase SHP-2 engages binding to Grb2 via tyrosine 584. Cell Growth Differ. 7, 1589–1597. [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, and Kinzler KW (2013). Cancer genome landscapes. Science 339, 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser T, Cui X, Yanagawa J, Lee JM, Heinrich E, Lee G, Sharma S, and Dubinett SM (2008). Smoking and lung cancer: the role of inflammation. Proc. Am. Thorac. Soc 5, 811–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, and Zhang B (2013). customProDB: an R package to generate customized protein databases from RNA-Seq data for proteomics search. Bioinformatics 29, 3235–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Hao T, Pan Y, Qian X, and Zhou D (2015). Increased expression of SOX4 is a biomarker for malignant status and poor prognosis in patients with non-small cell lung cancer. Mol. Cell. Biochem 402, 75–82. [DOI] [PubMed] [Google Scholar]
- Wang J, Vasaikar S, Shi Z, Greer M, and Zhang B (2017). WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 45, W130–W137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Slebos RJC, Wang D, Halvey PJ, Tabb DL, Liebler DC, and Zhang B (2012). Protein identification using customized protein sequence databases derived from RNA-Seq data. J. Proteome Res 11, 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kuan PJ, Xing C, Cronkhite JT, Torres F, Rosenblatt RL, DiMaio JM, Kinch LN, Grishin NV, and Garcia CK (2009). Genetic defects in surfactant protein A2 are associated with pulmonary fibrosis and lung cancer. Am. J. Hum. Genet 84, 52–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert BT, Narita T, Satpathy S, Srinivasan B, Hansen BK, Scholz C, Hamilton WB, Zucconi BE, Wang WW, Liu WR, et al. (2018). Time-Resolved Analysis Reveals Rapid Dynamics and Broad Scope of the CBP/p300 Acetylome. Cell 174, 231–244.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. (2007). Characterizing the cancer genome in lung adenocarcinoma. Nature 450, 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B, Wang X, and Zhang B (2019). PepQuery enables fast, accurate, and convenient proteomic validation of novel genomic alterations. Genome Res. 29, 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B, Li K, Zhang Y, and Zhang B (2020). Cancer neoantigen prioritization through sensitive and reliable proteogenomics analysis. Nat. Commun 11, 1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitsett JA (2014). The molecular era of surfactant biology. Neonatology 105, 337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitsett JA, and Alenghat T (2015). Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat. Immunol 16, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiel C, Le Gal K, Ibrahim MX, Jahangir CA, Kashif M, Yao H, Ziegler DV, Xu X, Ghosh T, Mondal T, et al. (2019). BACH1 Stabilization by Antioxidants Stimulates Lung Cancer Metastasis. Cell 178, 330–345.e22. [DOI] [PubMed] [Google Scholar]
- Wilkerson MD, and Hayes DN (2010). ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26, 1572–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson MD, Yin X, Walter V, Zhao N, Cabanski CR, Hayward MC, Miller CR, Socinski MA, Parsons AM, Thorne LB, et al. (2012). Differential pathogenesis of lung adenocarcinoma subtypes involving sequence mutations, copy number, chromosomal instability, and methylation. PLoS One 7, e36530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, Nomura T, and Sakaguchi S (2008). CTLA-4 control over Foxp3+ regulatory T cell function. Science 322, 271–275. [DOI] [PubMed] [Google Scholar]
- Xie K, Zhang K, Kong J, Wang C, Gu Y, Liang C, Jiang T, Qin N, Liu J, Guo X, et al. (2018). Cancer-testis gene PIWIL1 promotes cell proliferation, migration, and invasion in lung adenocarcinoma. Cancer Med. 7, 157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Lamouille S, and Derynck R (2009). TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 19, 156–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q-W, Zhao W, Wang Y, Sartor MA, Han D-M, Deng J, Ponnala R, Yang J-Y, Zhang Q-Y, Liao G-Q, et al. (2012). An integrated genome-wide approach to discover tumor-specific antigens as potential immunologic and clinical targets in cancer. Cancer Res. 72, 6351–6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Peng P, Li L, Shao M, Zhao J, Wang L, Duan F, Song S, Wu H, Zhang J, et al. (2016). High expression of GFAT1 predicts poor prognosis in patients with pancreatic cancer. Sci. Rep 6, 39044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, and Ning Z (2009). Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25, 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S, Huang T, Campbell JD, Lee E, Tu Z, Geraci MW, Powell CA, Schadt EE, Spira A, and Zhu J (2014). MODMatcher: multi-omics data matcher for integrative genomic analysis. PLoS Comput. Biol 10, e1003790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Trevino V, Shen H, Laird PW, Levine DA, et al. (2013). Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun 4, 2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu T, Zhang Z, Payne SH, Zhang B, McDermott JE, Zhou J-Y, Petyuk VA, Chen L, Ray D, et al. (2016). Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell 166, 755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhang J, Zhang Z, Guo Y, Wu Y, Wang R, Wang L, Mao S, and Yao X (2019a). Overexpression of Indoleamine 2,3-Dioxygenase 1 Promotes Epithelial-Mesenchymal Transition by Activation of the IL-6/STAT3/PD-L1 Pathway in Bladder Cancer. Translational Oncology 12, 485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhang J, Zhang Z, Guo Y, Wu Y, Wang R, Wang L, Mao S, and Yao X (2019b). Overexpression of Indoleamine 2,3-Dioxygenase 1 Promotes Epithelial-Mesenchymal Transition by Activation of the IL-6/STAT3/PD-L1 Pathway in Bladder Cancer. Transl. Oncol 12, 485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Lu D, Liu L, Cai J, Zhou Y, Yang Y, Zhang Y, and Zhang J (2017). Insulin-like growth factor 2 mRNA binding protein 3 (IGF2BP3) promotes lung tumorigenesis via attenuating p53 stability. Oncotarget 8, 93672–93687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Flodby P, Luo J, Castillo DR, Liu Y, Yu F-X, McConnell A, Varghese B, Li G, Chimge N-O, et al. (2018). Claudin-18-mediated YAP activity regulates lung stem and progenitor cell homeostasis and tumorigenesis. J. Clin. Invest 128, 970–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, Related to Figure 1: Experimental workflow and data quality metrics.
A. Schematic representation showing sample processing steps. Fresh frozen tumors and their matched normal-adjacent tissues (NATs) were cryopulverized and aliquoted for genomics and proteomics analyses before undergoing comprehensive proteogenomic characterization, facilitating uniformity in input samples.
B. Schematic representation of the workflows used for proteome, phosphoproteome and acetylproteome analyses. Tandem mass tags (TMT) were used to multiplex 9 samples (4 tumors and their matched NATs, in addition to a 9th sample, an unpaired tumor) and 1 common reference (pool of all tumors and NATs) that was used to link multiple TMT10 plexes. Matched tumor / NAT pairs were included in the same TMT plex.
C. Pearson similarity matrices showing intra- and inter-plex reproducibility across 4 interspersed comparative reference (CompRef) process replicates for proteome, phosphoproteome and acetylproteome. The CompRef process replicates demonstrated excellent reproducibility (Pearson Correlation, Proteome: R=0.91, Phosphoproteome: R=0.88, Acetylproteome: R=0.73) and consistent identifications across several months of data acquisition time.
D. Bar plot showing consistent numbers of identified and quantified proteins, phosphosites and acetylsites across the 25 plexes used for analyzing 212 tumors and NATs.
E. Principal component analysis (PCA) plot representation of proteome, phosphoproteome and acetylproteome separately for tumors and NATs, colored by TMT plex (n=25). PCA was based on features that were fully quantified across all 25 TMT plexes.
F. Sample-wise Pearson correlation between copy number alteration (CNA) and RNA, and between CNA and Proteome. The dark red-colored diagonal demonstrates the absence of sample swaps.
G. Cophenetic correlation coefficient (y-axis) calculated for a range of factorization ranks (x-axis). The maximal cophenetic correlation coefficient was observed for rank K=4 as shown in red.
H. Silhouette plot for K=4. This plot indicates the quality of cluster separation.
I. Non-negative matrix factorization (NMF) clustering applied individually to proteome, phosphoproteome and acetylproteome. Each heatmap shows the maximum-normalized membership score for each sample (x-axis) in each cluster (y-axis) - essentially, the strength of a sample’s “belongingness” to each of the clusters. The proteome cluster overlaps substantially with the multi-omics clusters depicted in Figure 1E, but divergence is seen in both the phosphoproteome and acetylproteome, with additional substructure in the phosphoproteome. Color schematics for the different annotations and data rows are detailed in the bottom panel.
J. Louvain clustering of miRNA showed parallels with NMF results but identified five clusters. miRNA cluster 2 was markedly enriched for tumors from multi-omics cluster C1, in turn aligned with proximal-inflammatory RNA signatures, while miRNA cluster 3 was enriched for the STK11 mutant subset of the NMF C3, proximal-proliferative cluster. While the remaining three miRNA clusters had mixed composition, miRNA cluster 5 was markedly enriched for ALK fusion-driven tumors, including all 5 EML4-ALK as well as the HMBOX1-ALK fusions.
Figure S2, Related to Figure 2: Genomic sequence evidence for ALK rearrangements and ALK immunohistochemistry
A. ALK gene fusion transcript architecture constructed from RNAseq data and fusion evidence for ALK fusion transcripts. Red arrows on the ALK and various 5’ partner genes’ schematic diagrams indicate fusion breakpoints observed in the respective index samples. Blue arrows indicate gene orientation and numbers indicate genomic coordinates from GRCh38/hg38 assembly.
B. Identification of the precise genomic breakpoints from whole genome sequencing (WGS) data for ALK gene fusions. WGS evidence supporting the underlying genomic rearrangements in the ALK locus is indicated in red and blue; numbers indicate genomic coordinates from GRCh38/hg38 assembly.
C. Immunohistochemistry reveals upregulation of both total ALK and the ALK Y1507 phosphosite specifically in the tumor epithelia of ALK fusion-positive samples. No staining was seen in RET or ROS1 fusion samples or in matched NATs.
Figure S3, Related to Figure 3: Multi-omics integration.
A. Density plots showing distribution of sample-wise RNA-protein Spearman correlations separately for tumors (red) and NATs (blue).
B. Differential RNA and protein correlation between tumors and paired NATs is seen in gene products involved in Cell proliferation and transcriptional regulation, RNA splicing, Cell division, Beta catenin signaling and Chromosomal condensation. We hypothesize that, in NATs, homeostatic biological activities such as cell maintenance and homeostasis, circadian rhythm and survival predominate and are mediated by proteins the abundances of which reflect mRNA transcript levels, post-transcriptional processes, and post-translational stability. While the same components are at play in tumors, their more dynamic context and highly proliferative state leads to more consistent kinetics and coherent expression of RNA and proteins (Carpy et al., 2014; Jovanovic et al., 2015; Komili and Silver, 2008; Lu et al., 2007; Marguerat et al., 2012).
C. Correlation plots of CNA vs Phosphoprotein and CNA vs Acetylprotein expression. Significant (FDR <0.05) positive and negative correlations are indicated in red and green, respectively. CNA-driven cis-effects (consequence of CNA on the same locus) appear as the red diagonal line; trans-effects (consequence of CNA on genes encoded elsewhere) appear as vertical red and green lines. The accompanying histograms show the number of significant (FDR <0.05) cis- and trans-events corresponding to the indicated genomic loci (upward plot) as well as the overlap between CNA-RNA and CNA-protein events (downward plot).
D. Heatmap showing 2 dominant clusters of DNA methylation, defined primarily by tumors and NATs. LUAD tumors tend to be significantly more highly methylated than their counterpart NATs (two-sided Wilcoxon rank-sum test).
E. Consensus clustering of CpG island methylator phenotype (CIMP) defines three stable clusters representing high (red), intermediate (yellow) and low (blue) CIMP phenotypes (respectively referred to as CIMP+, −1, and −2 in Table S4). Both the overall tripartite structure and the highlighted genes also showed a pattern consistent with a previous report (Cancer Genome Atlas Research Network, 2014)
Figure S4, Related to Figure 4: Impact of somatic mutations on proteogenomic landscape of tumors.
A. The cis- (circles) and trans- (squares) effects of select mutated genes on the RNA expression of cancer-associated genes. The red and blue scale represents the median difference in RNA expression between samples with and without mutations. Size represents significance.
B. Lollipop plot showing KEAP1 mutations identified in this LUAD cohort. Twelve LUAD tumors harbored KEAP1 mutations, including missense mutations and truncations distributed across the entire length of the protein. The color of the lollipops indicate the type of mutation and numbers represent amino acid positions. Protein domains are indicated by different colors.
C. Boxplots showing KEAP1 and NFE2L2 RNA expression and KEAP1 protein expression in KEAP1 wild-type (WT) and mutant samples. Also shown is the downregulation of KEAP1 in KEAP1 mutant samples, seen at the protein but not at the RNA level.
D. Volcano plot showing differentially regulated proteins in KEAP1 WT vs mutant samples. These differential proteins underlie the pathway analysis shown in Figure S4F.
E. Volcano plot showing differentially regulated phosphosites in KEAP1 WT vs mutant samples.
F. Pathways enriched among proteins differentially expressed between KEAP1 mutant and WT tumors. Significant enrichment of the oxidative stress response supports activation of NRF2 signaling in these samples.
G. Boxplots showing unchanged PTPN11 protein and significantly (FDR <0.05) elevated phosphopeptide expression (Y546 and Y584) in ALK fusion relative to wild-type samples. Various activating functions have been proposed for these phosphorylation sites, including directly driving the active conformation (Bennett et al., 1994; Lu et al., 2001) and serving as GRB2 docking sites (Bennett et al., 1994; Cunnick et al., 2002; Okazaki et al., 2013; Vogel and Ullrich, 1996). The PTPN11/Shp2 adaptor protein Grb2-associated binder-1 (GAB1) (Montagner et al., 2005) was also significantly upregulated in ALK fusion- driven tumors (Figure S4H).
H. GAB1 protein expression in samples with and without ALK fusion.
I. Protein-level gene-set enrichment (GSEA) pathway comparison of EGFR- and KRAS-driven LUAD tumors showing disparity in complement and clotting cascades, with upregulation of both in KRAS and downregulation in EGFR mutant samples. This analysis compares gene-set enrichment in KRAS mutant versus wildtype to EGFR mutant versus wildtype, where “wildtype” in each case excludes both KRAS and EGFR mutants.
J. Heatmaps show the phosphatase, ubiquitinase and deubiquitinase outliers enriched (FDR <0.2) at the phosphoprotein, protein, RNA and CNA levels (represented by columns under each gene name) and their association with mutations in select genes (EGFR, KRAS, TP53, STK11, KEAP1, EML4-ALK). Cancer Dependency Map-supported (https://depmap.org) panels on the left show log2 transformed relative survival averaged across all available lung cell lines after depletion of the indicated gene (rows) by RNAi or CRISPR. Druggability based on the Drug Gene Interaction Database (http://www.dgidb.org/) is indicated alongside the availability of FDA-approved drugs. The log-transformed druggability score indicates the sum of PubMed journal articles that support the drug-gene relationship. This implementation of outlier detection complements other analytic approaches by identifying potentially druggable alterations that occur in personalized fashion; hence, for example, PTPN11 Y62 did not appear as an “outlier” phosphatase in EGFR mutant tumors (N=38) because of its uniform high expression in that group.
Figure S5, Related to Figure 5: Immune landscape in LUAD
A. Heatmap of expression levels of proteins most correlated with inferred activation of the Interferon gamma (IFNG) axis. Proteins involved in immune evasion signatures and proteins annotated as drug targets (as defined by :https://www.drugbank.ca) are highlighted by vertical bars on the left side of the figure. Important immune-related markers observed include WARS, LCK, CD4, TYMP, B2M (upregulated in the HTE cluster) and PTGR2, PDE4D, MAOA (upregulated in the CTE cluster). LCK and CD4 are members of the supramolecular lymphocyte regulatory complex that includes PTPRCAP, which showed differential DNA méthylation in our analysis (Figure 3F, G). Whether these derive from cancer cells or immune infiltrates is unclear.
B. The heatmap shows abundance levels (converted to Z-scores) of proteins of the IFNG axis pathway (Abril-Rodriguez and Ribas, 2017) and the Surfactant Metabolism pathway from Reactome database (Fabregat et al., 2018). The IFNG axis pathway determines activation of the adaptive immune system; 11 proteins of the IFNG axis were detected in global proteomics as presented in the heatmap (red vertical bar). The Surfactant Metabolism pathway was identified among the top three top pathways anti-correlated with inferred activation of the IFNG axis, with 14 of 30 pathway proteins, including 5 of 6 primary surfactant proteins (SFTP-A1, A2, B, C, and D), detected by global proteomics. Lung surfactant, responsible for preventing alveolar collapse at end-expiration, can also regulate pulmonary innate immunity (Whitsett, 2014), increasing immunosuppression (Pastva et al., 2007, Nayak et al, 2012). Prior studies have shown an association between genetic polymorphisms of surfactant proteins and lung carcinoma (Seifart et al., 2005) and bronchopulmonary dysplasia (Pavlovic et al., 2006), with genetic defects in SFTPA2 especially associated with lung cancer development (Wang et al., 2009). The observed upregulation of surfactant proteins in CTE lung tumors supports their immune-suppressive effects in lung cancer.
C. Heatmap depicting normalized enrichment scores (NES) of Hallmark gene sets (Liberzon et al., 2015) in each multi-omic cluster. To calculate cluster-specific NES we projected the matrix of multi-omic feature weights (W) derived by non-negative matrix factorization (NMF) onto gene sets using single-sample Gene Set Enrichment Analysis (ssGSEA) (Barbie et al., 2009). To derive a single weight for each gene measured across multiple omics data types (protein, RNA, phosphosite, acetylsite) we retained the weight with maximal absolute amplitude. Only gene sets significant in at least one cluster are shown (FDR <0.01).
D. Boxplot showing the distribution of the non-synonymous somatic mutations in each multi-omic cluster.
E. Flow diagram showing the workflow for developing and testing a deep learning algorithm to identify STK11 mutant samples based on hematoxylin and eosin stained histopathology slides. (I) LUAD tumor histopathology slides corresponding to analyzed tissue fragments were downloaded from The Cancer Imaging Archive (TCIA) database; (II) slides and corresponding per-slide level labels were separated into training (80%), validation (10%), and test sets (10%) at the per-patient level; (III) slides were tiled into 299-by-299-pixel pieces with overlapping areas of 49 pixels from each edge, omitting those with over 30% background. Tiles of each set were packaged into a TFrecord file; (IV) the InceptionV3-architectured convolutional neural network (CNN) was trained from scratch and the best performing model was picked based on validation set performance; (V) the model was applied to the test set, and the per-tile prediction scores were aggregated by slides and shown as heatmaps. The last-layer activations of 10000 randomly sampled tiles were exported for feature visualization on t-Distributed Stochastic Neighbor Embedding (t-SNE) (Figure 5E); (VI) counts and statistical metrics for area under receiver operating characteristic (AUROC), area under Precision-Recall Curve (AUPRC), and accuracy on per-slide and per-tile level were calculated with bootstrapped 95% confidence interval in parentheses. Table: the model achieved per-slide level AUROC of 0.961 and per-tile level AUROC of 0.892 in predicting STK11 mutation. Slide-level predictive accuracy was 94%.
F. To extract pathway-level proteomic features in an unsupervised manner, protein abundance measurements of 110 tumor samples were submitted to independent component analysis following the method proposed in (Liu et al., 2019). One signature (IC_068) showed significant associations with STK11 mutation status (average log10 nominal P-values within component cluster: −5.7). Average mixing scores for IC_068 represented ‘activity’ of the meta-gene level signature in each of the samples. Raw protein abundance values of genes contributing heavily to the signature (coefficient larger than 3) were shown in a heatmap.
G. Heatmaps showing protein (upper) and RNA expression (lower) of 16 gene products associated with neutrophil degranulation. STK11 and its partner STRADA are also shown.
Figure S6, Related to Figure 6: Impact of smoking on somatic mutations.
A. Bar plots showing distinct mutational signatures identified in 110 LUAD tumors. Somatic single nucleotide variants (SNVs) were determined from WGS data using SomaticWrapper, and 10 distinct mutation signatures were subsequently identified using SignatureAnalyzer (Kim et al., 2016); STAR Methods). We further combined two adjacent SNVs into a unitary di-nucleotide polymorphism (DNP) mutation if they were in the same haplotype (Table S6, STAR Methods). GG->TT or CC->AA were the dominant DNP types (~50%) and were associated with smoking status.
B. Schematic showing the approach used for determining the smoking score used in this study. Tumor purity estimates, counts of total mutations, and percentages that are smoking signature mutations and smoking-signature DNPs were used to derive a continuous smoking signature score.
C. Shown are the pairwise cosine similarities of each pair of the substitution signature probabilities for the 53 environmental mutagen exposures reported in Kucab et al. (Kucab et al., 2019). MX is a chlorine disinfection byproduct and a known DNA mutagen suspected of increasing cancer risk when present at sufficient levels in drinking water (McDonald and Komulainen, 2005). The MX signature was highly co-correlated to smoking signatures three PAHs DBADE, DBA, and 5-Methylchrysene. Benzidine, a chemical once heavily used in the dyeing industry and suspected to play a role in lung cancer (Tomioka et al., 2016), and PhIP, present in cooked meat and linked to various cancers (Tang et al., 2007), were also highly co-correlated to these PAHs (>0.5 and >0.7, respectively).
D. Boxplot showing significant difference (p = 2.2e-16) in RNA-based stemness index (mRNASI) between tumors and NATs. See also table S1.
E. Boxplot showing decrease of RNA-based stemness index (mRNASI) in both tumors and NATs with high smoking score (HSS) compared to corresponding samples with low smoking score (LSS). Differences are significant in NATs and approach significance in tumors. Within both tumors and NATs, samples with HSS showed higher mRNASI than samples with LSS (tumors: t test p = 0.069; NATs: t test, p = 0.038), consistent with the known field cancerization effect of tobacco exposure (Walser et al., 2008).
F. Upper: Scatterplot showing direction and significance of pathway-level protein differences between samples with high and low smoking scores (HSS and LSS) in tumors and NATs. Pathways are color-coded according to their pathway group in Figure 6B. Group 1 pathways are upregulated in HSS in tumors and downregulated in HSS in NATs. Group 2 pathways are downregulated in HSS in tumors and upregulated in HSS in NATs. Groups 3 and 4 are upregulated and downregulated, respectively, in both tumors and NATs. Signed -log 10 FDR represents Benjamini-Hochberg corrected p-values from the one sided-Mann-Whitney-Wilcoxon (MWW) test and the direction (+ or −) indicates activation or suppression (i.e. MWW test side with lower p-value). Lower: HSS vs LSS differential pathway scores in tumors and NATs at the transcriptome level. The group separations clearly defined by protein-based pathways (Figure 6B) are less evident at the RNA level. Smoking and mutation status are inextricably interwoven, so it is likely that these smoking score-related differentials represent a complex interplay between direct effects of combustion-related carcinogen exposure and effects mediated by mutational differences related to that exposure.
Figure S7, Related to Figure 7: Proteogenomic differences between tumors and matched adjacent normals.
A. PCA plots showing RNA, protein, phosphosite and acetylsite abundance in 110 tumor samples (triangles: cadet blue) and 101 NATs (circles: coral red).
B. Schematic showing differentially regulated RNA, proteins, phosphosites and acetylsites between tumors and paired NATs (upregulated sites, FDR <0.01, log2 FC >1; downregulated sites, FDR <0.01, log2 FC <−1). Most quantified proteins (76%) had differential expression between tumor and NAT (FDR < 0.01, Wilcoxon signed rank test); amongst those with at least 2-fold differential expression, a slight majority (64%) were higher in NAT (Table S7).
C. Gene-set enrichment analysis (GSEA) revealing pathways differentially expressed between tumor and paired NATs. Cell cycle progression, MYC Targets Upregulation, Unfolded Protein Response, Glycolysis and TCA cycle (adjusted p <0.001) were upregulated in tumor samples whereas KRAS Signaling (FDR <0.001), STAT3 Signaling (FDR <0.001), and Muscle Differentiation (FDR <0.001) were downregulated in tumor samples compared to NATs.
D. Using stringent cutoffs for quantitative difference, significance and consistency (log2 FC >2, FDR <0.01, and differential in >90% of all Tumor-NAT pairs (Pan-LUAD)), we identified 289 proteins upregulated at the protein level in tumors, 60 of which were supported by RNA and are shown in the figure (see also Table S7). “HPA staining proportions” indicate the proportion of lung adenocarcinoma sections staining positive for the specific marker in the Human Proteome Atlas database (https://www.proteinatlas.org/). This global tumor / NAT comparison revealed 18 enzymes, 3 transcription factors (TF), SOX4, TCF3, and HMGA1,2 transporters, 23 secreted, and 21 transmembrane proteins as candidate biomarkers. GREM1, SOX4, SPINT1, ST14, SPINT2, CTHRC1, KDF1, MDK, SFN, HMGA1, ESRP1, NME1, SERPINH1 and CBX8 are implicated in EMT and metastasis. Highly upregulated metabolic proteins included GFPT1, P4HB, PLOD2, PYCR1, SHMT2, PSAT1, ERO1A, IL411, DHFR, and LDHA. Stress-related marker candidates with prognostic significance included ERO1A, DHFR, MANF, HYOU1, LDHA, and CBX8. Remainder of figure: Proteomics-based tumor biomarker candidates (fold change>4 and adjusted p-value <0.01 in ≥80% of tumor / NAT pairs) for 4 frequently mutated genes: TP53 (40/63 shown), EGFR (39), KRAS (37) and STK11 (40/77 shown) (Table S7). Each dot in the box plot represents a tumor sample. Blue-colored boxplots highlight proteins with overexpression in more than 99% of tumor samples with the associated mutation. HPA proportions indicate the proportion of LUAD sections staining positive for the specific marker in the Human Proteome Atlas. Relevant characteristics of the biomarker candidates and relevant targeted drugs in clinical trials are shown in the accompanying plot. Condensed representations of these plots are shown in Figure 7C.
E. Rank plots depicting differential phosphosite-driven signatures between tumor and paired NATs in tumors with mutations in STK11 or TP53. Residual enrichment scores (y-axis) were calculated between mutated tumors (STK11 or TP53) and all other tumors in order to highlight tumor / NAT differences in tumors harboring the indicated mutation.
Table S1, Related to Figures 1–7, Figures S1–7: Metadata associated with LUAD patients and tumors related to STAR methods, Figures 1–7.
A. Sample annotations and associated genotype, clinical, geographical and pathological metadata.
Table S2, Related to Figures 1–7, Figures S1–7: Datasets pertaining to copy number alterations, somatic mutations, DNA methylation, protein coding RNA expression and microRNA expression obtained from tumors and NATs, related to Figures 1–7.
A. Data matrix of somatic copy number alterations (CNA) in tumors. CNAs are represented as log2(copy_number)-1 ratio. Refseq GRCh38/hg38 database was used as a reference.
B. Data matrix of site-level somatic mutations in tumors. Refseq Hg38 database was used as a reference.
C. Gene-level methylation data obtained by taking the median of beta values of all probes within the genomic location encompassing the promoter and gene body of each gene.
D. Log2-transformed RNA expression matrix obtained by upper-quartile normalization of “Reads Per Kilobase of transcript per Million mapped reads” (RPKM).
E. Row-median normalization applied to RNA expression matrix from Table S2D.
F. Log2-transformed “Transcript per Kilobase Million” (TPM)-values of mature miRNAs.
G. Luad-v3.1-mirna-mature-tpm-log2-row-norm.gct: Row-median normalization applied to miRNA expression from Table S2F.
H. Focal amplification in the CPTAC LUAD cohort obtained from GISTIC analysis.
I. Focal deletion in the CPTAC LUAD cohort obtained from GISTIC analysis.
Table S3, Related to Figures 1–7, Figures S1–7: Filtered and unfiltered proteomics datasets, pathway analysis and differentially regulated miRNA for tumors and NATs, related to Figures 1–7.
A. Two-component-normalized log2-transformed protein expression. The dataset was filtered as described in the STAR methods.
B. Two-component-normalized log2-transformed phosphosite expression. The dataset was filtered as described in the STAR methods.
C. Two-component-normalized log2-transformed acetylsite expression. The dataset was filtered as described in the STAR methods.
D. Unfiltered version of Table S3A.
E. Unfiltered version of Table S3B.
F. Unfiltered version of Table S3C.
G. Multi-omics pathway analysis and list of differentially regulated genes used for pathway analysis.
H. List summarizing 22 differentially expressed proteins obtained after adjustment for known confounding factors including smoking status, region of origin and mutation status of commonly mutated genes (EGFR, KRAS, STK11, TP53) and ALK fusions.
I. Differentially regulated miRNA in tumors. Histological subtype analysis was constrained by availability, as a substantial majority of cases in this cohort were typical adenocarcinomas with glandular/acinar formation. Of the remaining 6 dominant histomorphological subtypes represented in the study, solid (10 cases) and true papillary (9 cases) had enough samples for statistical comparison. Solid tumors segregated exclusively into NMF clusters C1 and C4 (Proximal inflammatory and Terminal respiratory unit), while all but one papillary fell into C1 or C3 (Proximal proliferative.) Twelve RNA species, some with established relevance to cancer, were differential between subtypes. For instance, IDH3A, a rate-limiting enzyme in the Krebs cycle, was elevated in the solid subtype, while PTK7, a tyrosine kinase thought to affect cell proliferation and survival in LUAD, was elevated in the papillary subtype. However, no proteins were identified as differential after adjustment for confounding variables.
Table S4, Related to Figures 3,4, Figures S3,S4: Multi-omics correlation, and additional analyses including pathways, Connectivity Map (CMAP), CIMP classification, iProFun, and differential regulation between KRAS and EGFR mutants versus WT samples, related to Figures 3 and 4.
A. Gene-wise mRNA-Protein Spearman correlation in tumors
B. Gene-wise mRNA-Protein Spearman correlation in NATs
C. Sample-wise mRNA-Protein Spearman correlation in tumors
D. Sample-wise mRNA-Protein Spearman correlation in NATs
E. mRNA-protein pairs that change their direction between tumors and NATs
F. CNA-RNA, CNA-Protein, CNA-Phosphoprotein and CNA-Acetylprotein correlations
G. List of cancer-associated genes and selected mutated genes used (see STAR methods)
H. CMAP-nominated genes and their significant associations with metadata (Table S1)
I. CMAP-nominated genes and the significant trans-events that overlapped with CMAP signatures
J. Results for classification of samples with CIMP (CpG Island Methylator Phenotype)
K. iProFun results with mRNA, protein and phosphoprotein as outcomes, and methylation as the primary predictor, adjusted for cis mutation, cis CNA, age, gender, smoking status, country of origin and tumor purity. Results include identified methylation effects at the gene level at FDR<0.1 and their proteomic associations with tumor and NAT status.
L. Differential RNA, global protein, phosphorylation, and acetylation between tumors with KRAS mutations vs WT tumors. Wilcoxon rank sum test was performed requiring at least 4 non-missing values in each group.
M. Differential RNA, global protein, phosphorylation, and acetylation between LUAD tumors with EGFR mutations vs WT tumors. Wilcoxon rank sum test was performed requiring at least 4 non-missing values in each group.
Table S5, Related to Figure 5, Figure S5: Immune landscape of tumors and NATs, related to Figure 5.
A. Immune composition of LUAD tumors and NATs as judged by deconvolution of RNA expression using xCell.
B. Immune clusters based on xCell signatures.
C. Pathways upregulated in immune groups based on RNAseq data.
D. Pathways upregulated in immune groups based on proteomics data. (D1) Pathways based on proteomics dataset (D2) Pathways exclusive to proteomics datasets (D3) Pathways exclusive to RNAseq dataset
E. Genes differentially expressed in immune groups based on RNAseq and proteomics data.
F. Association of xCell signatures with mutations with frequency >15 samples
G. ESTIMATE scores based on RNAseq data and tumor purity computed via TSNet (Petralia et al, 2018)
H. Supplementary data containing an STK11 mutation related proteomic signature extracted by ICA (independent component analysis)
Table S6, Related to Figure 6, Figure S6: Factors used for calculating smoking scores and environmental exposure, related to Figure 6.
A. Smoking score and the information integrated therein, which includes tumor purity estimates, counts of total mutations, percentages of smoking signature mutations, counts of smoking-signature DNPs, and adjustment to the strong alternative/non-smoking signatures.
B. Links to environmental factors based on regression of the 96 possible trinucleotide mutation combinations between the samples in the CPTAC LUAD cohort and the environmental signatures reported by Kucab et al. (Kucab et al., 2019). Correlations were calculated by least-squares fit, with significance assessed by T-testing and subsequent multiple test correction via FDR. A total of 5,830 hypothesis tests were conducted (53 rows/environmental factors * 110 columns/LUAD samples).
C. RNA- and proteomics-driven differentially regulated pathways between HSS (high smoking score) versus LSS (low smoking score) in both tumors and paired NATs
D. Differential protein markers in strict never-smokers (SNS) vs strict smokers (SS) and SNS tumors vs their paired NATs
Table S7, Related to Figure 7, Figure S7: Tumor-normal analysis and neoantigen analysis, related to Figure 7.
A. Differential RNA expression between tumors and paired NATs. Differential expression is indicated as log2-transformed fold-change
B. Differential protein expression between tumors and paired NATs (log2 FC>2 and FDR <0.01 in >80% of samples). Differential expression is indicated as log2-transformed fold-change.
C. Differential phosphosite expression between tumors and paired NATs. Differential expression is indicated as log2 transformed fold-change
D. Differential acetylsite expression between tumors and paired-NATs. Differential expression is indicated as log2-transformed fold-change
E. Proteome-driven differential pathway analysis between tumors and paired NATs
F. Differential phosphosites analysis between tumors and paired NATs and their corresponding protein expression
G. Differential acetylsite analysis between tumors and paired NATs and their corresponding protein expression
H. Protein biomarkers in pan-LUAD (all tumor-NAT pairs) and major mutation-driven tumor subsets (EGFR, TP53, KRAS and STK11)
I. PTM-SEA applied to phosphosite-specific differential tumor vs NAT analysis
J. Mutational subtype-specific tumor vs NAT analysis using PTM-SEA
K. Predicted tumor neoantigens identified in the CPTAC LUAD cohort at the level of RNA transcripts and peptides containing evidence of somatic mutations
Data Availability Statement
Proteomics raw datasets are publicly available though the CPTAC data portal https://cptac-data-portal.georgetown.edU/cptac/s/S056
Genomic and transcriptomic data files can be accessed at the Genomic Data Commons (GDC); https://portal.gdc.cancer.gov/, via dbGaP Study Accession: phs001287.v5.p4 https://www.ncbi.nlm.nih.gov/proiects/gap/cgi-bin/study.cgi7studvid=phs001287.v5.p4
Sample annotation, processed and normalized data files are provided as Tables S1–S3.
Software and code used in this study are referenced in their corresponding STAR Method sections and also the Key Resource Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-CD8 (C8/144B) | Cellmarque | Catalog #108M |
| Rabbit monoclonal anti-CD4 (SP35) | Roche | Catalog #790-4423 |
| Liquid Concentrated Monoclonal Antibody anti-CD163 | Leica Biosystems | Catalog #NCL-L-163 |
| PTMScan Acetyl-lysine Kit | Cell Signaling Technology | Catalog: 13416 |
| Biological Samples | ||
| Primary tumor samples | See Experimental Model and Subject Details | N/A |
| Chemicals and Reagents | ||
| HPLC-grade water | J.T. Baker | Catalog: 4218-03 |
| Urea | Sigma | Catalog: U0631 |
| Sodium chloride | Sigma | Catalog: 71376 |
| 1M Tris, pH 8.0 | Invitrogen | Catalog: AM9855G |
| Ethylenediaminetetraacetic acid | Sigma | Catalog: E7889 |
| Aprotinin | Sigma | Catalog: A6103 |
| Leupeptin | Roche | Catalog: 11017101001 |
| Phenylmethylsulfonyl fluoride | Sigma | Catalog: 78830 |
| Sodium fluoride | Sigma | Catalog: S7920 |
| Phosphatase inhibitor cocktail 2 | Sigma | Catalog: P5726 |
| Phosphatase inhibitor cocktail 3 | Sigma | Catalog: P0044 |
| Dithiothretiol, No-Weigh Format | Fisher Scientific | Catalog: 20291 |
| Iodoacetamide | Sigma | Catalog: A3221 |
| Lysyl endopeptidase | Wako Chemicals | Catalog: 129-02541 |
| Sequencing-grade modified trypsin | Promega | Catalog: V511X |
| Formic acid | Sigma | Catalog: F0507 |
| Acetonitrile | Honeywell | Catalog: 34967 |
| Trifluoroacetic acid | Sigma | Catalog: 302031 |
| Tandem Mass Tag reagent kit – 11plex | ThermoFisher | Catalog: A34808 |
| 0.5M HEPES, pH 8.5 | Alfa Aesar | Catalog: J63218 |
| Hydroxylamine solution, 50% (vol/vol) in H2O | Aldrich | Catalog: 467804 |
| Methanol | Honeywell | Catalog: 34966 |
| Ammonium hydroxide solution, 28% (wt/vol) in H2O | Sigma | Catalog: 338818 |
| Ni-NTA agarose beads | Qiagen | Catalog: 30410 |
| Iron (III) chloride | Sigma | Catalog: 451649 |
| Acetic acid, glacial | Sigma | Catalog: AX0073 |
| Potassium phosphate, monobasic | Sigma | Catalog: P0662 |
| Potassium phosphate, dibasic | Sigma | Catalog: P3786 |
| MOPS | Sigma | Catalog: M5162 |
| Sodium hydroxide | VWR | Catalog: BDH7225 |
| Sodium phosphate, dibasic | Sigma | Catalog: S9763 |
| Phosphate-buffered saline | Fisher Scientific | Catalog: 10010023 |
| iVIEW DAB Detection Kit | Roche | Catalog: 760-091 |
| Equipment | ||
| Reversed-phase tC18 SepPak, 3cc 200mg | Waters | Catalog: WAT054925 |
| Solid-phase C18 disk, for Stage-tips | Empore | Catalog: 66883-U |
| Stage-tip needle | Cadence | Catalog: 7928 |
| Stage-tip puncher, PEEK tubing | Idex Health & Science | Catalog: 1581 |
| PicoFrit LC-MS column | New Objective | Catalog: PF360-75-10-N-5 |
| ReproSil-Pur, 120 Å, C18-AQ, 1.9-μm resin | Dr. Maisch | Catalog: r119.aq |
| Nanospray column heater | Phoenix S&T | Catalog: PST-CH-20U |
| Column heater controller | Phoenix S&T | Catalog: PST-CHC |
| 300 μL LC-MS autosampler vial and cap | Waters | Catalog: 186002639 |
| Offline HPLC column, 3.5-μm particle size, 4.6 um × 250 mm | Agilent | Catalog: Custom order |
| Offline 96-well fractionation plate | Whatman | Catalog: 77015200 |
| 700 μL bRP fractionation autosampler vial | ThermoFisher | Catalog: C4010-14 |
| 700 μL bRP fractionation autosampler cap | ThermoFisher | Catalog: C4010-55A |
| 96-well microplate for BCA | Greiner | Catalog: 655101 |
| Microplate foil cover | Corning | Catalog: PCR-AS-200 |
| Vacuum centrifuge | ThermoFisher | Catalog: SPD121P-115 |
| Centrifuge | Eppendorf | Catalog: 5427 R |
| Benchtop mini centrifuge | Corning | Catalog: 6765 |
| Benchtop vortex | Scientific Industries | Catalog: SI-0236 |
| Incubating shaker | VWR | Catalog: 12620-942 |
| 15 mL centrifuge tube | Corning | Catalog: 352097 |
| 50 mL centrifuge tube | Corning | Catalog: 352070 |
| 1.5 mL microtube w/o cap | Sarstedt | Catalog: 72.607 |
| 2.0 mL microtube w/o cap | Sarstedt | Catalog: 72.608 |
| Microtube caps | Sarstedt | Catalog: 72.692 |
| 1.5 mL snapcap tube | ThermoFisher | Catalog: AM12450 |
| 2.0 mL snapcap tube | ThermoFisher | Catalog: AM12475 |
| Instrumentation | ||
| Microplate Reader | Molecular Devices | Catalog: M2 |
| Offline HPLC System for bRP fractionation | Agilent 1260 | Catalog: G1380-90000 |
| Online LC for LC-MS | ThermoFisher | Catalog: LC140 |
| Q Exactive Plus Mass Spectrometer | ThermoFisher | Catalog: IQLAAEGA APFALGMBDK |
| Q Exactive HF-X Mass Spectrometer | ThermoFisher | Catalog: 0726042 |
| Orbitrap Fusion Lumos Tribrid Mass Spectrometer | ThermoFisher | Catalog: IQLAAEGA APFADBMBHQ |
| Critical Commercial Assays | ||
| TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Gold | Illumina | Catalog: RS-122-2301 |
| Infinium MethylationEPIC Kit | Illumina | Catalog: WG-317-1003 |
| Nextera DNA Exosome Kit | Illumina | Catalog: 20020617 |
| KAPA Hyper Prep Kit, PCR-free | Roche | Catalog: 07962371001 |
| BCA Protein Assay Kit | ThermoFisher | Catalog: 23225 |
| Deposited Data | ||
| PhosphoSitePlus | (Hornbeck et al., 2012) | https://www.phosphosite.org |
| Connectivity Map (CMAP) | (Lamb et al., 2006; Subramanian et al., 2017) | https://www.broadinstitute.org/connectivity-map-cmap |
| Human Protein Atlas (HPA) | (Uhlén et al., 2005) | https://www.proteinatlas.org |
| CT Antigen database | (Almeida et al., 2009) | http://www.cta.lncc.br |
| Software and Algorithms | ||
| methylationArrayAnalysis (version 3.9) | (Maksimovic et al., 2016) | https://master.bioconductor.org/packages/release/workflows/html/methylationArrayAnalysis.html |
| Illumina EPIC methylation array (3.9) | Hansen KD, 2019 | https://bioconductor.org/packages/release/data/annotation/html/IlluminaHumanMethylationEPICanno.ilm10b2.hg19.html |
| Methylation array analysis pipeline for CPTAC | Li Ding Lab | https://github.com/ding-lab/cptac_methylation |
| miRNA-Seq analysis pipeline for CPTAC | Li Ding Lab | https://github.com/ding-lab/CPTAC_miRNA |
| Somatic variant calling pipeline for CPTAC | Li Ding Lab | https://github.com/ding-lab/somaticwrapper |
| VarDict | (Lai et al., 2016) | https://github.com/AstraZeneca-NGS/VarDict |
| Strelka2 | (Kim et al., 2018b) | https://github.com/Illumina/strelka |
| MUTECT1.1.7 | (Cibulskis et al., 2013) | https://software.broadinstitute.org/gatk/download/archive |
| VarScan2.3.8 | (Koboldt et al., 2012) | http://varscan.sourceforge.net |
| Pindel0.2.5 | (Ye et al., 2009) | http://gmt.genome.wustl.edu/packages/pindel/ |
| SignatureAnalyzer | (Kim et al., 2016) | https://software.broadinstitute.org/cancer/cga/msp |
| Fusion calling pipeline for CPTAC | Li Ding Lab | https://github.com/cuidaniel/Fusion_hg38 |
| CNVEX | Marcin Cieslik Lab | https://github.com/mctp/cnvex |
| CRISP | Marcin Cieslik Lab | https://github.com/mcieslik-mctp/crisp-build |
| Spectrum Mill | Karl R. Clauser, Steven Carr Lab | https://proteomics.broadinstitute.org/ |
| ComBat (v3.20.0) | (Johnson et al., 2007) | https://bioconductor.org/packages/release/bioc/html/sva.html |
| DreamAI | Pei Wang Lab | https://github.com/WangLab-MSSM/DreamAI |
| GISTIC2.0 | (Mermel et al., 2011) | ftp://ftp.broadinstitute.org/pub/GISTIC2.0/GISTIC_2_0_23.tar.gz |
| iProFun | (Song et al., 2019) | https://github.com/WangLab-MSSM/iProFun |
| ESTIMATE | (Yoshihara et al., 2013) | https://bioinformatics.mdanderson.org/public-software/estimate/ |
| WebGestaltR | (Wang et al., 2017) | http://www.webgestalt.org/ |
| Joint Random Forest | (Petralia et al., 2016) | https://github.com/WangLab-MSSM/ptmJRF |
| GSVA | (Hänzelmann et al., 2013) | https://bioconductor.org/packages/release/bioc/html/GSVA.html |
| TCGAbiolinks | (Colaprico et al., 2016) | http://bioconductor.org/packages/release/bioc/html/TCGAbiolinks.html |
| TSNet | (Petralia et al., 2018) | https://github.com/WangLab-MSSM/TSNet |
| xCell | (Aran et al., 2017) | http://xcell.ucsf.edu/ |
| CPTAC LUAD Data Viewer | Steven Carr lab | http://prot-shiny-vm.broadinstitute.org:3838/CPTAC-LUAD2020/ |
| MODMatcher | (Yoo et al., 2014) | https://github.com/integrativenetworkbiology/Modmatcher |
| ConsensusClusterPlus | (Wilkerson and Hayes, 2010) | http://bioconductor.org/packages/release/bioc/html/CancerSubtypes.html |
| pyQUILTS (v1.0) | (Ruggles et al., 2016) | http://openslice.fenyolab.org/cgi-bin/pyquilts_cgi.pl |
| MS-GF+ | (Kim and Pevzner, 2014) | https://github.com/MSGFPlus/msgfplus |
| NeoFlow | Bing Zhang lab | https://github.com/bzhanglab/neoflow |
| netMHCpan | (Jurtz et al., 2017) | http://www.cbs.dtu.dk/services/NetMHCpan/ |
| Optitype | (Szolek et al., 2014) | https://github.com/FRED-2/OptiType |
| Customprodbj | (Wang and Zhang, 2013) | https://github.com/bzhanglab/customprodbj |
| PDV | (Li et al., 2019) | https://github.com/wenbostar/PDV |
| PepQuery | (Wen et al., 2019) | http://pepquery.org |
| PTM-SEA | (Krug et al., 2018) | https://github.com/broadinstitute/ssGSEA2.0 |
| Terra | Broad Institute data science platform. | https://terra.bio/ |
| CMap | (Lamb et al., 2006; Subramanian et al., 2017) | https://due.io/cmap |
| PTM-SEA | (Krug et al., 2018) | https://github.com/broadinstitute/ssGSEA2.0 |
| LIMMA v3.36 (R Package) | (Ritchie et al., 2015) | https://bioconductor.org/packages/release/bioc/html/limma.html |
| FactoMineR v1.41NMF (R - package) | (Gaujoux and Seoighe, 2010; Lê et al., 2008) | https://cran.r-project.org/web/packages/FactoMineR/index.html |
| MClust v5.4 (R package) | (Scrucca, Fop, Murphy and Raftery, 2017) | https://cran.r-project.org/web/packages/mclust/index.html |