Abstract
Purpose
Patients with SCLC rarely undergo biopsies at relapse. When pursued, tissue obtained can be inadequate for molecular testing, posing a challenge in identifying potentially targetable alterations in a clinically meaningful time frame. We examined the feasibility of ctDNA testing in identifying potentially targetable alterations in SCLC.
Experimental Design
ctDNA test results were prospectively collected from SCLC patients between 2014 and 2017 and analyzed. ctDNA profiles of SCLC at diagnosis and relapse were also compared.
Results
A total of 609 samples collected from 564 patients between 2014 and 2017 were analyzed. The median turnaround time for test results was 14 days. Among patients with data on treatment status, there were 61 samples from 59 patients and 219 samples from 206 patients collected at diagnosis and relapse, respectively. The number of mutations or amplifications detected per sample did not differ by treatment status. Potentially targetable alterations in DNA repair, MAPK and PI3K pathways and genes such as MYC and ARID1A were identifiable through ctDNA testing. Furthermore, our results support that it may be possible to reconstruct the clonal relationship between detected variants through ctDNA testing.
Conclusions
Patients with relapsed SCLC rarely undergo biopsies for molecular testing and often require prompt treatment initiation. ctDNA testing is less invasive and capable of identifying alterations in relapsed disease in a clinically meaningful timeframe. ctDNA testing on an expanded gene panel has the potential to advance our knowledge of the mechanisms underlying treatment resistance in SCLC and aid in the development of novel treatment strategies.
Keywords: Small cell lung cancer, circulating tumor DNA, translational oncology, liquid biopsy
Introduction
Small cell lung cancer (SCLC) accounts for approximately 13% of all lung cancer diagnoses in the United States and is characterized by a tendency for early metastasis and an aggressive clinical course1. Despite initial response to platinum-based chemotherapy, virtually all patients with extensive disease and the majority of those with limited stage disease develop tumor relapse, which is associated with treatment resistance and short survival. Although recent studies have identified potentially targetable alterations in SCLC, patients with relapsed disease generally deteriorate quickly and rarely undergo tissue biopsies at progression to facilitate molecular testing for biomarker directed therapy2–6. Challenges in obtaining tissue biopsies has also limited the ability to comprehensively characterize mechanisms of treatment resistance, which are of critical importance in aiding the discovery of novel therapies.
Circulating tumor DNA (ctDNA) may detect DNA from cancer cells shed into the bloodstream. Several studies have demonstrated the utility of ctDNA in directing therapy, examining mechanisms of treatment resistance, and prognosis in patients with non-small cell lung cancer (NSCLC)7–9. As ctDNA analysis can potentially capture spatial and temporal clonal heterogeneity of cancer without necessitating multiple invasive biopsies, it is an attractive modality for studying the genomic landscape of SCLC, especially at relapse10–13. Here, we report the ctDNA landscape of SCLC using Guardant360®, a targeted hybrid capture-based next-generation sequencing (NGS) platform focused on querying mutations in a set of cancer-related genes, and explore its utility and limitations in identifying alterations that potentially inform therapy.
Methods
Patients
We employed de-identified ctDNA test data from SCLC patients who underwent ctDNA testing at a clinical laboratory (Guardant Health, Redwood City, CA) between September 2014 and 2017. The diagnosis of SCLC was confirmed by clinical information provided by physicians ordering the ctDNA test. Treatment information, date of diagnosis, age, gender, and sample collection details were collected from test requisition forms when available. The study was conducted in accordance with recognized ethical guidelines (e.g., CIOMS, Belmont report, and declaration of Helsinki). Clinical information for patients diagnosed at the Washington University in St. Louis and University of California in San Diego was verified by co-investigators from those institutions after approval by the Institutional Review Board, and the requirement for consent was waived. Samples were classified as having been collected at “diagnosis” or “relapse” based on available clinical information. For samples lacking specific clinical information, treatment status was inferred based on the dates of diagnosis and sample collection. Since ctDNA analysis is typically ordered by clinicians at the time of diagnosis or progression for clinical decision making, samples that were obtained within 3 weeks (duration corresponding to first chemotherapy cycle) of diagnosis were considered to have been collected “at diagnosis” and samples that were collected at least 84 days past date of diagnosis (corresponding to duration of front-line therapy) were considered to be collected “at relapse”.
Sequencing and data analysis
Targeted NGS based ctDNA detection was performed by Guardant Health, Inc. (Guardant360, www.guardanthealth.com/guardant360/), a Clinical Laboratory Improvement Amendment (CLIA)- certified and College of American Pathologists (CAP)-accredited, New York State Department of Health-approved clinical laboratory. The assay’s gene panel was expanded from 54 to 73 genes over the study period. Overall 10, 86, 260, and 253 samples were sequenced using the 54, 68, 70, and 73 gene panels, respectively. Test characteristics differed between gene panels (Supplementary Table 1). The proportion of samples collected at diagnosis and relapse, which were sequenced on each panel was comparable (Supplementary Table 2). Single nucleotide variants, insertion-deletion alterations (indels), fusions, and amplifications within various cancer-related genes were identified through analysis of cell-free (cf) DNA extracted from plasma collected from two 10 ml Streck blood collection tubes. Synonymous alterations were excluded from analysis. The ctDNA assay used in this study had high clinical sensitivity, detecting 85% or more of the variants present in tissue in advanced cancer patients, and analytic specificity greater than 99.9999%7,14. Only somatic alterations were reported and analyzed in this study. SNVs and indels shed into circulation were quantitated using mutant allelic frequency (AF) by taking the number of mutated variants at that position, and dividing by the total number of reads at that position (including wild type). Gene amplifications were reported as absolute gene copy number in plasma14.
For the purpose of data analysis, genomic coordinates for each variant listed in the clinical test report were obtained using Transvar15. This information was used to analyze data using the suite of functions, maftools16. Amplification information was inputted as custom tables and overlaid onto co-mutation plots using the waterfall plot function where appropriate. Custom code was written in Python utilizing the PANDAS module to calculate alteration frequencies of tested genes (number of samples demonstrating mutation in a gene divided by the total number of samples). Alteration frequencies of tested genes between samples collected at diagnosis and relapse were compared using the mafcompare feature of maftools. The percentage at which various gene alterations were identified in individual samples or AF), was averaged to determine the mean allelic frequency (MAF). Lollipop and clonality plots were also generated using maftools. Clonality of an alteration in a sample was inferred based on its ratio to the maximum AF in that sample (hereby referred to as “ctDNA clonality”), as previously described7. A list of mutations known to be activating in RTK/RAS pathway genes such as KRAS, HRAS, NRAS, BRAF, EGFR, ERBB2 (HER2), MAP2K1, and MET, was curated based on mutation hotspot information available for these genes from NSCLC in cBioportal17,18. Data from previously reported tissue sequencing studies in SCLC were obtained from publications or queried using the cBioportal interface3,4. Graphpad Prism was used for performing Student’s ttests and Fisher’s exact testing.
Results
Sample and test characteristics
A total of 609 samples collected from 564 patients were analyzed (Table 1). The median turnaround time for test results was 14 days (range 6–32). Overall, 61 samples from 59 patients and 219 samples from 206 patients were classified as being collected at diagnosis and at relapse, respectively (Supplementary Table 2). Thirty patients underwent serial testing. At least one non-synonymous mutation or amplification was detected in 552 of 609 samples (90.6%). One or more non-synonymous mutations were detected in 531 samples (average of 3.54, 95% CI 3.03 to 4.05). Non-synonymous mutations were identifiable in 87% of diagnosis (53 out of 61) and relapse (191 out of 219) samples, and amplifications in 43% (26) and 39% (85) of samples, respectively. While the maximum AFs (mean of 25.8% vs. 16.5%, p=0.01) and MAFs (mean of 15.8% vs 7.0%, p=0.002) were significantly higher for samples collected at diagnosis compared to relapse, the number of non-synonymous mutations or amplifications detected per sample did not differ significantly by treatment status (p=0.2 and 0.1 respectively) (Figure 1). Among the 30 patients who underwent serial testing, non-synonymous mutations and amplifications were detectable in more than one sample for 24 and 8 patients, respectively, without significant differences in the number of alterations between first and subsequent collections.
Table 1.
Clinical and demographic characteristics of study cohort
| Characteristic | N (%) |
|---|---|
| Number of patients | 564 |
| • Male | 271 |
| • Female | 293 |
| Average age (range) | 60.5 years (22–99) |
| Number of samples | 609 |
| Median Turn Around Time (range, 95% CI) | 14 days (6–32, 14–15 days) |
| Test details | |
| • 54 gene panel | 10 |
| • 68 gene panel | 86 |
| • 70 gene panel | 260 |
| • 73 gene panel | 253 |
| Treatment status: samples | N (patients) |
| • At diagnosis | 61 (59) |
| • At relapse | 219 (206) |
| • Unable to determine | 329 (296) |
| Number of samples with non-synonymous alterations (patients) | |
| • At diagnosis | 53 (61) |
| • At relapse | 191 (219) |
| Number of samples with amplifications (patients) | |
| • At diagnosis | 26 (61) |
| • At relapse | 85 (219) |
Figure 1. Comparison of the ctDNA mutational profiles of SCLC at diagnosis and relapse.
Panels A and B demonstrate maximum AFs and MAFs, respectively when samples are categorized by treatment status (diagnosis vs. relapse). Panels C and D demonstrate the number of mutations and amplifications respectively in diagnosis and relapse samples. Error bars represent Mean ± SEM. *, p < 0.05. ***, p<0.001. n.s = not significant.
Genomic features of samples collected at diagnosis and at relapse
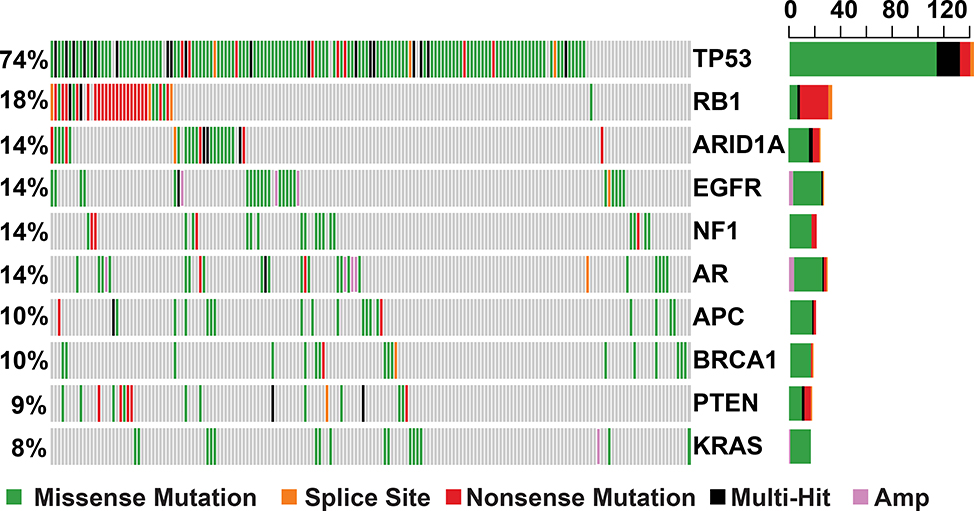
Non-synonymous mutations in TP53 were the most frequent (72%), followed by alterations in RB1 (18%) (Supplementary Figures 1 and 2) across all samples, and this did not considerably differ by treatment status (Supplementary Tables 3 and 4). Following mutations in TP53 and RB1, mutations in ARID1A were the most frequent (14.5%) in samples collected at relapse, with approximately 33% of alterations consisting of nonsense mutations and indels (Figure 2).
Figure 2. Type and Frequency of Mutations in Commonly Altered Genes in Relapsed SCLC.
An oncoplot demonstrating distribution of the ten most frequently altered genes across relapse samples. The right barplot represents the frequency of mutations in each gene. Amp = amplification. Multi-Hit = presence of multiple types of mutation in same gene in a given sample. Each column represents an individual patient sample.
An analysis of differentially altered genes between diagnosis and relapse samples, demonstrated a higher frequency of alterations in the androgen receptor gene, AR, (14% vs. 2%, p<0.05, Fisher’s exact test) among relapsed samples (Figure 2, Supplementary Table 4). AR mutations were observed in 26 samples from 25 patients at relapse, of which 21 were collected from females and 5 from males and all AR amplifications were only seen in samples collected from females. Of the non-synonymous mutations identified in AR across all samples, 44% were in the N-terminal domain Activation Function 1 (NTD AF1) region, 15% in the DNA-Binding (DBD) and hinge Domain, and 21% in the Ligand-Binding Domain (LBD). While none of the identified mutations have been previously reported as activating, most activating AR mutations that have been reported, are typically found in these domains (Figure 3)19. Mutations in APC, a negative regulator of beta-catenin (CTNNB1) and WNT signaling, were also more prevalent among relapse samples (18 patients) compared to those obtained at diagnosis (1 patient) (11% vs. 2% of samples, p<0.05, Fisher’s exact test) (Figures 2,3 and Supplementary Figure 3). In addition to alterations in APC, we also detected known hotspot mutations in CTNNB1 in five samples. While frequencies of alterations in multiple genes appeared to differ by treatment status, none reached statistical significance after correcting for multiple comparisons, possibly owing to inadequate number of samples in each treatment category.
Figure 3. Alterations in AR and APC.
Panel A demonstrates the distribution of AR and APC alterations across different relapse samples in which they are altered. The right barplot represents the frequency of mutations in each gene. Each column represents an individual patient sample. Panel B shows the distribution of AR mutations across different domains of the gene. NTD = N-terminal regulatory domain, AF1 = Activation Function 1, DBD = DNA binding domain, H = Hinge domain, LBD = Ligand binding domain, AR = Androgen Receptor, APC = Adenomatous Polyposis Coli.
Identification of potentially targetable genomic alterations
SCLC is characterized by amplifications in SOX2 and MYC family genes including MYC, MYCL, and MYCN3,4. SOX2, which is not included in the Guardant360 panel, is located on the long arm of chromosome 3 (3q26) adjacent to PIK3CA. Apart from PIK3CA and MYC, amplifications in several other genes were observed (Supplementary Table 3). MYC amplifications, which may be potentially targetable by aurora kinase inhibitors, were observed in approximately 3.5% of the relapse samples20. Despite data supporting a role for MYC in treatment resistant SCLC, the frequency of MYC amplifications did not differ according to treatment status (5% at diagnosis vs 3.5% at relapse)( Supplementary Figure 4)21. Notably, copy number assessment in ctDNA may have lower sensitivity than direct tumor analysis, which could explain the differences in frequency of amplification in genes like MYC between this study and other tissue sequencing studies.
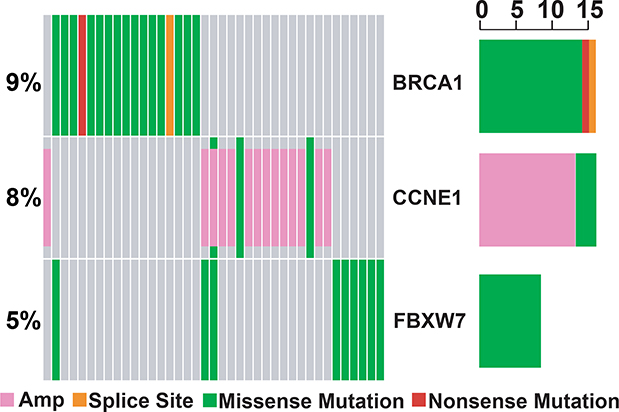
Considering the emerging role of investigational agents targeting DNA repair pathways, genomic instability, and immunotherapies in relapsed SCLC, we examined alterations in DNA repair and damage response pathway genes22,23. Alterations in BRCA1 (9%), BRCA2 (5.5%), ATM (3%), and MLH1 (1%) were detectable in a subset of relapse samples, despite limited coverage of ATM and MLH1 exons. BRCA1, CCNE1, and FBXW7 alterations, which drive genome-wide instability (tandem-duplicator phenotype), were observed in 9%, 8% and 5% of relapse samples, respectively (Figure 4), and mutations in ARID1A, associated with responsiveness to PARP and EZH2 inhibitors, were observed in nearly 15% of patients at relapse24.
Figure 4. DNA repair pathway alterations in relapsed samples.
Oncoplot demonstrating alterations in CCNE1, BRCA1, and FBXW7 in relapse samples. The right barplot represents the frequency of mutations in each gene. Amp = amplification. Each column represents an individual patient sample.
PI3K/MTOR and RAS/RTK pathway alterations
Alterations in PI3K/MTOR signaling genes were observed in approximately a third of all SCLC samples4. Although SCLC is not characterized by RAS/RTK pathway alterations, ctDNA profiling showed an unexpectedly high frequency of these alterations. Activating mutations in RTK/RAS/RAF pathway genes EGFR, KRAS, NRAS, HRAS, BRAF, MAP2K1, and ERBB2 were observed in 96 samples (18%) collected from 77 patients. Mutations in KRAS were the most frequent (n=36), followed by EGFR (n=34) and BRAF (n=9). Amplifications in ERBB2 and MET were detectable in nearly 30% of patients at relapse. All samples demonstrating known activating EGFR mutations were collected at relapse, suggesting that these samples were possibly collected from patients with transformed SCLC following EGFR-directed therapy25. We also observed oncogenic ALK and RET rearrangements in one sample each, but the treatment status of these samples was unknown.
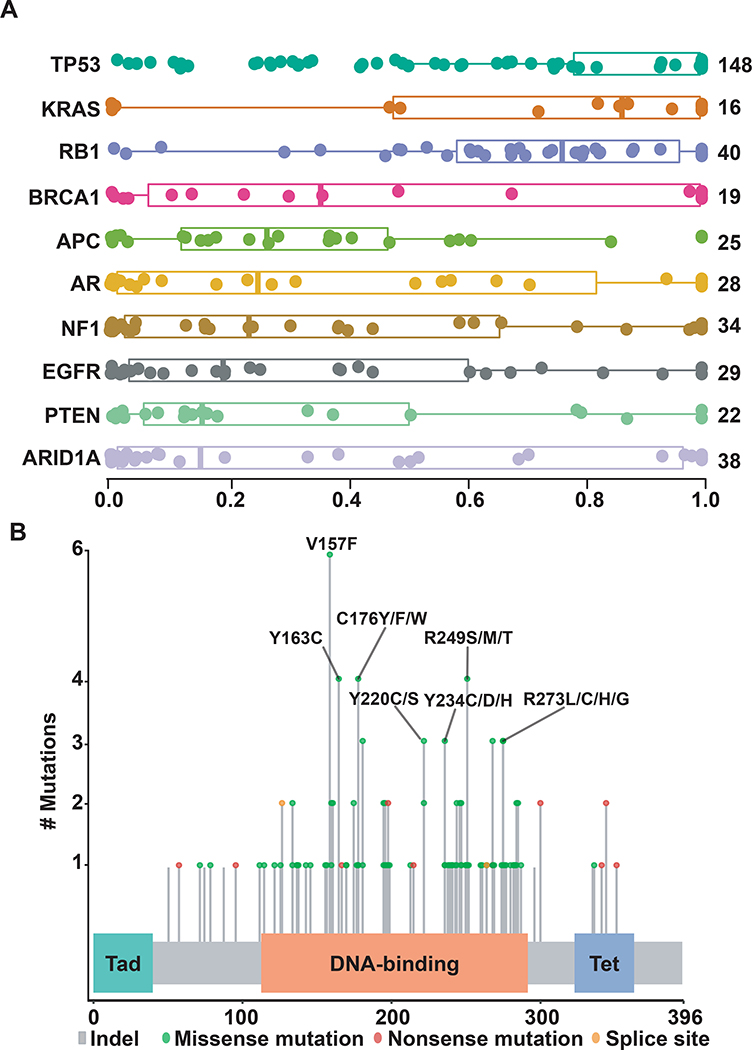
Characteristics of TP53 mutations in SCLC
TP53 alterations are essentially universal and typically clonal in SCLC.4 We analyzed the characteristics of TP53 mutations to obtain a better insight into the ability of ctDNA testing to facilitate disease monitoring. TP53 mutations were detectable in 72.5% of samples with detectable alterations, of which 31% had multiple TP53 mutations. Of the samples collected at relapse, 40% showed multiple TP53 mutations (vs. 31% at diagnosis, p=0.35, Fisher’s exact test). TP53 mutations predominantly clustered in the DNA binding domain irrespective of their AF (Figure 5, Supplementary Figure 5). When only variants with the highest AFs in a sample were considered for each gene, mutations in TP53 and RB1 appeared to be clonal, as expected for SCLC (Figure 5). Among samples with multiple TP53 mutations, the average ctDNA clonality value of TP53 mutations was approximately 4.5-fold higher for mutations with the highest AFs compared to mutations with lower AFs (0.89 vs. 0.22), suggesting a sub-clonal or hematopoetic source for low AF variants.
Figure 5. Distribution plots for ctDNA clonality and TP53 mutations across SCLC samples.
Panel A shows distribution of inferred AFs for non-synonymous mutations in different genes across relapse samples, with inferred AF values shown on X-axis. Genes near the top of the Y-axis with higher inferred AF values are likely to represent clonal alterations, while genes below them are more likely to be sub-clonal. Boxes represent interquartile range around the median. Panel B demonstrates pattern of mutations in TP53 that were shed at highest allelic frequency in each individual sample. Tad = Transactivating domain. Tet = Tetrameriization domain.
Discussion
We examined the ctDNA profiles of nearly 600 SCLC patients, making this the largest study to examine the genomic features of SCLC to date, whether in tissue or in plasma. This large sample size enabled us to make several important observations and investigate the utility of ctDNA in identifying potentially targetable alterations in SCLC. TP53 and RB1 alterations play an essential role in SCLC tumorigenesis, and are also retained at relapse, a feature that is consistently reproducible through ctDNA analysis4,26. The frequency of RB1 mutations was notably lower compared to their nearly universal presence in tumor biopsies. Of note, the test used herein cannot easily identify biallelic loss or loss of heterzygosity. Notably, the frequency of RB1 mutations in our data (12%) was comparable to previously published studies when only missense and nonsense variants were considered (18% to 35% in tissue sequencing studies), suggesting that these differences could be explained by differences in RB1 coverage (for type of mutation) between tissue sequencing in previous studies and earlier versions of the ctDNA test3,4,17,18.
The number of mutations detectable per sample, a surrogate for tumor mutation burden, did not differ by treatment status. The MAF of detected variants was significantly higher across samples collected at diagnosis compared with those collected at relapse. Previous studies suggest a correlation between MAF of variants detected on ctDNA and radiographic disease burden27. Therefore, our observation may be explained by the fact that SCLC patients often present with a higher burden of disease at diagnosis.
We also observed a higher percentage of alterations in APC and AR in relapse samples. Although identification of APC alterations in our study is consistent with a recent study highlighting the role of WNT activation in chemoresistant SCLC, a more systematic evaluation of the impact of these alterations in SCLC is needed5. The identification of AR alterations is provocative since it may be potentially targetable. In SCLC, both testosterone and dihydrotestosterone stimulated growth of AR-positive SCLC cell lines which was reversed by the addition of anti-androgens, including fludamide28. Additionally, AR manipulation has an established role in patients with AR positive tumors, and there are anecdotal cases describing responses with this approach in other solid tumors29,30. Response to anti-AR therapy may also depend on whether AR copy number gain is focal or a result of aneuploidy. Although not addressable in this study, it may be possible to adopt approaches such as comparing the number of copies of ARAF and AR genes, as both reside on the X chromosome, to filter out aneuploidy. Further studies are warranted to explore the role of AR signaling in SCLC, especially considering the role this pathway plays in driving chemoresistance in prostate cancer31.
The role of RTK/RAS signaling in SCLC is controversial32. We observed known RTK/RAS pathway activating mutations in nearly 18% of our patients, including therapeutically targetable alterations in EGFR, BRAF, ERBB2, and MET. While it is possible that these findings are a consequence of misclassification of NSCLC or large cell neuroendocrine cancers as SCLC, presence of mixed histology tumors, or identification of hematopoietic variants by ctDNA testing, alterations in RAS pathway genes and/or pathway activation in SCLC have been previously reported33,34. Cancer initiation and progression are complex processes, and preclinical studies in murine models suggest that the final histology of lung cancer is likely to be influenced by the cell of origin and the combination and sequence in which these cells acquire specific mutations35,36. It is therefore possible that these findings reflect heterogeneity in the cell of origin, tumorigenesis, and inter- or intra-tumoral heterogeneity37,38. The higher inferred VAFs of some of the KRAS mutations supports this hypothesis. The ability to target RAS signaling has been reported in SCLC with combination therapies39. We also observed amplifications in ERBB2 and MET in 12% of the samples. Although there is no data in SCLC, studies have reported benefit from targeting these alterations in other solid tumors 40–45.
Mutations in genes involved in DNA repair and damage signaling were observed in a subset of relapse samples, and potentially predict for responsiveness to therapeutic strategies targeting genomic instability and immunotherapies22,24. In particular, we observed alterations in BRCA1, FBXW7, CCNE1, and ARID1A in relapse samples. Alterations in these genes may predict response to DNA damaging therapies, PARP inhibitors and/or immunotherapy raising the possibility for more targeted therapies for a subset of SCLC patients46,47,48.
Finally, nearly a third of samples in this analysis demonstrated multiple TP53 mutations. In many instances, these mutations were detectable at very low percentages in plasma, raising the possibility of them being sub-clonal or hematopoietic in origin. Recent data have shown the ability of ctDNA and tissue molecular testing to pick up variants from clonal hematopoiesis (CH)49,50. Incorporation of strict quality control measures, a high level of clinician suspicion, and/or bioinformatic filtering is critical when calling mutations to ensure that they are related to the tumor of interest and not CH. Our results suggest that CH identified on ctDNA testing could potentially lead to the identification of spurious variants, incorrect estimation of tumor mutation burdens, or pose a challenge in interpreting presence of minimal residual disease. Delineating potential hematopoietic variants from ctDNA variants may require sequencing of matched PBMCs as shown by some groups27. Utilizing ctDNA clonality values may aid in the identification of variants that are likely to have originated from the tumor compartment. Nevertheless, this approach will require further validation.
Given the novelty of ctDNA analysis in SCLC, it is important to recognize the limitations of our study. First, we were unable to perform independent verification of the histology or treatment status of a significant number of samples analyzed in this study. Second, samples collected at diagnosis and relapse were also not typically patient matched. Therefore, we cannot rule out the possibility that the observed variant frequencies are a consequence of sample misclassification or lack of pairing. Third, samples in this study were sequenced using different versions of a commercial assay that differed in their abilities to identify mutations and amplifications in different genes and the extent to which some genes were sequenced (limited exons vs. full coding sequence). Fourth, treatment status of several samples was inferred based on date of diagnosis and sample collection, which can be inaccurate. Some or all of these factors could have altered the frequency at which various mutations were identified in samples collected at diagnosis and at relapse. Fifth, information related to disease burden at time of collection, specific treatment received by individual patients and response to treatment were not available for a more informed clinical-correlative analysis. Additionally, hematopoietic variants may have been misclassified as tumor derived in our analysis. However, this limitation is not exclusive to ctDNA analyses, since leukocyte infiltration of solid tumors can also lead to the detection of these variants with tissue-based sequencing50.
Despite the noted limitations, our findings demonstrate that ctDNA profiling holds promise for examining mechanisms of treatment resistance and directing clinical care in SCLC by facilitating the rapid identification of potentially targetable alterations. Furthermore, unlike prior studies that predominantly sequenced surgically resected early stage SCLC samples, xenografts, or cell lines, our study included samples collected from a large cohort of patients who most likely received treatment for advanced stage disease, shedding light on alterations that could play important roles in SCLC progression and serve as potential new candidates for therapy. Further studies of ctDNA in patients with SCLC may help accelerate the discovery and development of new drugs against this relentless disease.
Supplementary Material
Translational relevance.
Patients with relapsed SCLC rarely undergo biopsies for molecular testing and often require prompt treatment initiation. Circulating tumor DNA (ctDNA) testing is less invasive and is capable of identifying alterations in relapsed disease that can be potentially targeted. ctDNA profiles of nearly 600 patients who underwent testing on a commercially available targeted gene panel were analyzed in this study. Our results demonstrate that it is feasible to detect potentially targetable alterations in patients with SCLC in a clinically meaningful time frame. Additionally, ctDNA testing has the ability to provide insights into the biology of SCLC at relapse.
Conflicts of interest statement and acknowledgements
S Devarakonda receives research funding from the International Association for Study of Lung Cancer and serves on the steering committee for Guardant 360 and receives travel support. K Gold receives consulting/advisory fees from Takeda, AstraZeneca, and Regeneron, and research support from Pharmacyclics, AstraZeneca, Pfizer, BerGenBio, and AbbVie. J Ward is supported by the National Cancer Institute of the National Institutes of Health Paul Calabresi Career Development Award in Clinical Oncology (K12CA167540). AC is a scientific advisor for Geneoscopy, and for Roche Sequencing Solutions, receives speaker honoraria and travel support from Varian Medical Systems, Roche Sequencing Solutions, and Foundation Medicine, research support from Roche Sequencing Solutions, and has served as a consultant for Tempus Labs and for Oscar Health. TKO receives research support from Pfizer, Regeneron/Sanofi, BMS, AstraZeneca, G1 Therapeutics, Novartis, Amgen, AbbVie, Corvus, Adaptimmune, serves on the advisory board of AbbVie, Amgen, AstraZeneca, BMS, Lilly/Armo, PharmaMar, Boehringer Ingelheim; Xcovery, Amgen, DSMB of EMD Serono and Roche/Genetech, and is the co-founder of Cambium Oncology. VR and RBL are employees and shareholders of Guardant Health. RTL serves on the advisory board for Guardant Health, Genentech and ThermoFisher Scientific. CMR has consulted regarding oncology drug development with AbbVie, Amgen, Ascentage, Astra Zeneca, BMS, Celgene, Daiichi Sankyo, Genentech/Roche, Ipsen, Johnson & Johnson, Loxo, and Pharmar, and is on the scientific advisory boards of Elucida and Harpoon. R Govindan consults for Abbvie, Adaptimmune, AstraZeneca, Celgene, Ignyta, Inivata, Merck, Nektar, Pfizer, and Roche, and is supported by the National Institutes of Health/National Cancer Institute (1U01CA231844-01). DM consults for Abbvie, PharmaMar, Takeda and Bristol Myers Squibb. SS, BH, SW, JTP report no conflicts of interest.
References
- 1.Govindan R, Page N, Morgensztern D, et al. : Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol 24:4539–44, 2006 [DOI] [PubMed] [Google Scholar]
- 2.Peifer M, Fernández-Cuesta L, Sos ML, et al. : Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 44:1104–10, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudin CM, Durinck S, Stawiski EW, et al. : Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 44:1111–6, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.George J, Lim JS, Jang SJ, et al. : Comprehensive genomic profiles of small cell lung cancer. Nature 524:47–53, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner AH, Devarakonda S, Skidmore ZL, et al. : Recurrent WNT pathway alterations are frequent in relapsed small cell lung cancer. Nat Commun 9:3787, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pietanza MC, Byers LA, Minna JD, et al. : Small cell lung cancer: will recent progress lead to improved outcomes? Clin Cancer Res 21:2244–55, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zill OA, Banks KC, Fairclough SR, et al. : The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer Patients. Clin Cancer Res 24:3528–3538, 2018 [DOI] [PubMed] [Google Scholar]
- 8.Abbosh C, Birkbak NJ, Wilson GA, et al. : Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545:446–451, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blakely CM, Watkins TBK, Wu W, et al. : Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet 49:1693–1704, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Almodovar K, Iams WT, Meador CB, et al. : Longitudinal Cell-Free DNA Analysis in Patients with Small Cell Lung Cancer Reveals Dynamic Insights into Treatment Efficacy and Disease Relapse. J Thorac Oncol 13:112–123, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nong J, Gong Y, Guan Y, et al. : Circulating tumor DNA analysis depicts subclonal architecture and genomic evolution of small cell lung cancer. Nat Commun 9:3114, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCoach CE, Blakely CM, Banks KC, et al. : Clinical Utility of Cell-Free DNA for the Detection of ALK Fusions and Genomic Mechanisms of ALK Inhibitor Resistance in Non-Small Cell Lung Cancer. Clin Cancer Res 24:2758–2770, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blackhall F, Frese KK, Simpson K, et al. : Will liquid biopsies improve outcomes for patients with small-cell lung cancer? Lancet Oncol 19:e470–e481, 2018 [DOI] [PubMed] [Google Scholar]
- 14.Odegaard JI, Vincent JJ, Mortimer S, et al. : Validation of a Plasma-Based Comprehensive Cancer Genotyping Assay Utilizing Orthogonal Tissue- and Plasma-Based Methodologies. Clin Cancer Res 24:3539–3549, 2018 [DOI] [PubMed] [Google Scholar]
- 15.Zhou W, Chen T, Chong Z, et al. : TransVar: a multilevel variant annotator for precision genomics. Nat Methods 12:1002–3, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayakonda A, Koeffler H: Maftools: efficient analysis, visualization and summarization of MAF files from large-scale cohort based cancer studies, bioRxiv 2016 [Google Scholar]
- 17.Gao J, Aksoy BA, Dogrusoz U, et al. : Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:pl1, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerami E, Gao J, Dogrusoz U, et al. : The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401–4, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottlieb B, Beitel LK, Nadarajah A, et al. : The androgen receptor gene mutations database: 2012 update. Hum Mutat 33:887–94, 2012 [DOI] [PubMed] [Google Scholar]
- 20.Mollaoglu G, Guthrie MR, Böhm S, et al. : MYC Drives Progression of Small Cell Lung Cancer to a Variant Neuroendocrine Subtype with Vulnerability to Aurora Kinase Inhibition. Cancer Cell 31:270–285, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drapkin BJ, George J, Christensen CL, et al. : Genomic and Functional Fidelity of Small Cell Lung Cancer Patient-Derived Xenografts. Cancer Discov 8:600–615, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sen T, Gay CM, Byers LA: Targeting DNA damage repair in small cell lung cancer and the biomarker landscape. Transl Lung Cancer Res 7:50–68, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hellmann MD, Callahan MK, Awad MM, et al. : Tumor Mutational Burden and Efficacy of Nivolumab Monotherapy and in Combination with Ipilimumab in Small-Cell Lung Cancer. Cancer Cell 33:853–861.e4, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menghi F, Barthel FP, Yadav V, et al. : The Tandem Duplicator Phenotype Is a Prevalent Genome-Wide Cancer Configuration Driven by Distinct Gene Mutations. Cancer Cell 34:197–210.e5, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oser MG, Niederst MJ, Sequist LV, et al. : Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol 16:e165–72, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sutherland KD, Proost N, Brouns I, et al. : Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell 19:754–64, 2011 [DOI] [PubMed] [Google Scholar]
- 27.Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. : Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov 7:1394–1403, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maasberg M, Rotsch M, Jaques G, et al. : Androgen receptors, androgen-dependent proliferation, and 5 alpha-reductase activity of small-cell lung cancer cell lines. Int J Cancer 43:685–91, 1989 [DOI] [PubMed] [Google Scholar]
- 29.Boon E, van Boxtel W, Buter J, et al. : Androgen deprivation therapy for androgen receptor-positive advanced salivary duct carcinoma: A nationwide case series of 35 patients in The Netherlands. Head Neck 40:605–613, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munoz J, Wheler JJ, Kurzrock R: Androgen receptors beyond prostate cancer: an old marker as a new target. Oncotarget 6:592–603, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komura K, Jeong SH, Hinohara K, et al. : Resistance to docetaxel in prostate cancer is associated with androgen receptor activation and loss of KDM5D expression. Proc Natl Acad Sci U S A 113:6259–64, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cristea S, Sage J: Is the Canonical RAF/MEK/ERK Signaling Pathway a Therapeutic Target in SCLC? J Thorac Oncol 11:1233–1241, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wakuda K, Kenmotsu H, Serizawa M, et al. : Molecular profiling of small cell lung cancer in a Japanese cohort. Lung Cancer 84:139–44, 2014 [DOI] [PubMed] [Google Scholar]
- 34.Kodaz H, Taştekin E, Erdoğan B, et al. : KRAS Mutation in Small Cell Lung Carcinoma and Extrapulmonary Small Cell Cancer. Balkan Med J 33:407–10, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park JW, Lee JK, Sheu KM, et al. : Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage. Science 362:91–95, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang D, Denny SK, Greenside PG, et al. : Intertumoral Heterogeneity in SCLC Is Influenced by the Cell Type of Origin. Cancer Discov 8:1316–1331, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calbo J, van Montfort E, Proost N, et al. : A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell 19:244–56, 2011 [DOI] [PubMed] [Google Scholar]
- 38.Lim JS, Ibaseta A, Fischer MM, et al. : Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature 545:360–364, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmid K, Bago-Horvath Z, Berger W, et al. : Dual inhibition of EGFR and mTOR pathways in small cell lung cancer. Br J Cancer 103:622–8, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choudhury NJ, Campanile A, Antic T, et al. : Afatinib Activity in Platinum-Refractory Metastatic Urothelial Carcinoma in Patients With ERBB Alterations. J Clin Oncol 34:2165–71, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ross JS, Fakih M, Ali SM, et al. : Targeting HER2 in colorectal cancer: The landscape of amplification and short variant mutations in ERBB2 and ERBB3. Cancer 124:1358–1373, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Camidge D, Ou S, Shapiro G, et al. : Efficacy and safety of crizotinib in patients with advanced c-MET-amplified non-small cell lung cancer (NSCLC). J ClinOncol 32:5s, 2014. (suppl; abstr 8001), 2014 [Google Scholar]
- 43.Angevin E, Spitaleri G, Rodon J, et al. : A first-in-human phase I study of SAR125844, a selective MET tyrosine kinase inhibitor, in patients with advanced solid tumours with MET amplification. Eur J Cancer 87:131–139, 2017 [DOI] [PubMed] [Google Scholar]
- 44.Schöffski P, Wozniak A, Escudier B, et al. : Crizotinib achieves long-lasting disease control in advanced papillary renal-cell carcinoma type 1 patients with MET mutations or amplification. EORTC 90101 CREATE trial. Eur J Cancer 87:147–163, 2017 [DOI] [PubMed] [Google Scholar]
- 45.Shitara K, Kim TM, Yokota T, et al. : Phase I dose-escalation study of the c-Met tyrosine kinase inhibitor SAR125844 in Asian patients with advanced solid tumors, including patients with. Oncotarget 8:79546–79555, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen J, Peng Y, Wei L, et al. : ARID1A Deficiency Impairs the DNA Damage Checkpoint and Sensitizes Cells to PARP Inhibitors. Cancer Discov 5:752–67, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katagiri A, Nakayama K, Rahman MT, et al. : Loss of ARID1A expression is related to shorter progression-free survival and chemoresistance in ovarian clear cell carcinoma. Mod Pathol 25:282–8, 2012 [DOI] [PubMed] [Google Scholar]
- 48.Bitler BG, Aird KM, Garipov A, et al. : Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med 21:231–8, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swanton C, Venn O, Aravanis A, et al. : Prevalence of clonal hematopoiesis of indeterminate potential (CHIP) measured by an ultra-sensitive sequencing assay: Exploratory analysis of the Circulating Cancer Genome Atlas (CCGA) study., DOI: 10.1200/JCO.2018.36.15_suppl.12003 Journal of Clinical Oncology 36, no. 15_suppl (May 20 2018) 12003–12003. [DOI] [Google Scholar]
- 50.Ptashkin RN, Mandelker DL, Coombs CC, et al. : Prevalence of Clonal Hematopoiesis Mutations in Tumor-Only Clinical Genomic Profiling of Solid Tumors. JAMA Oncol 4:1589–1593, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.