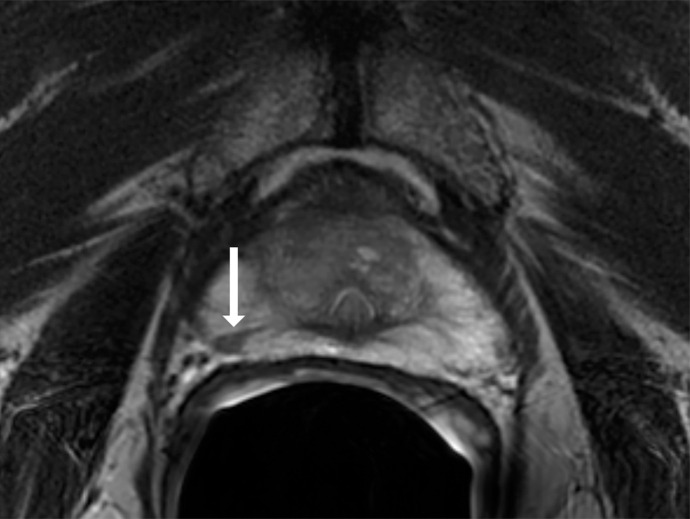
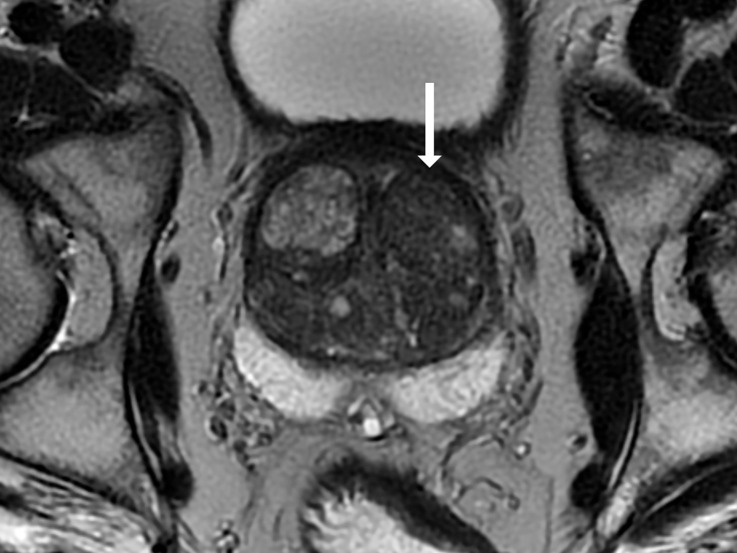
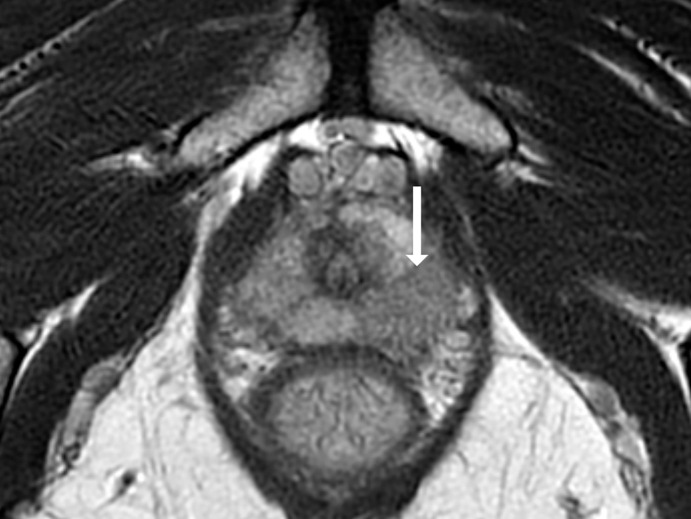
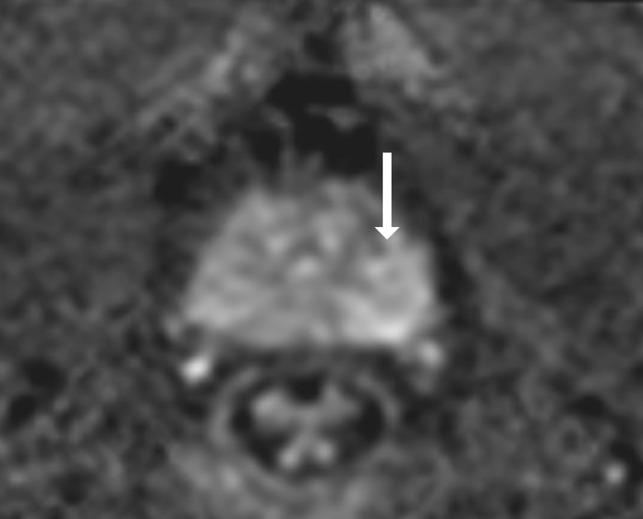
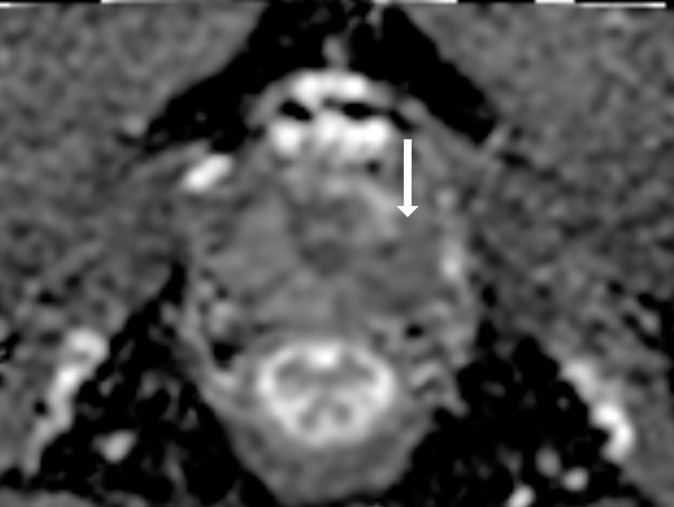
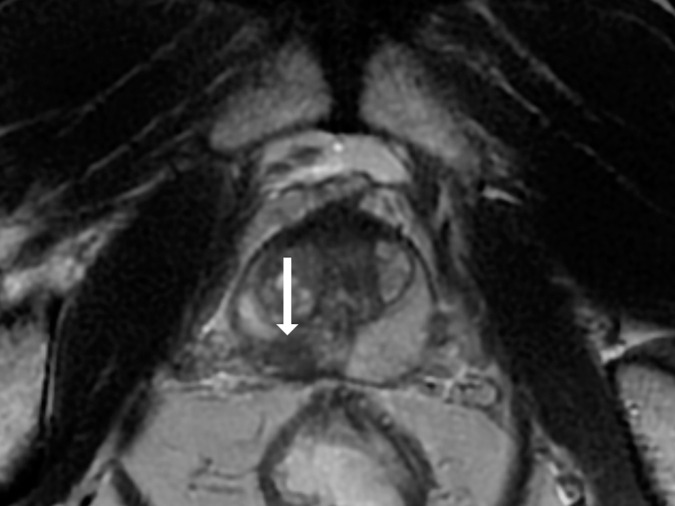
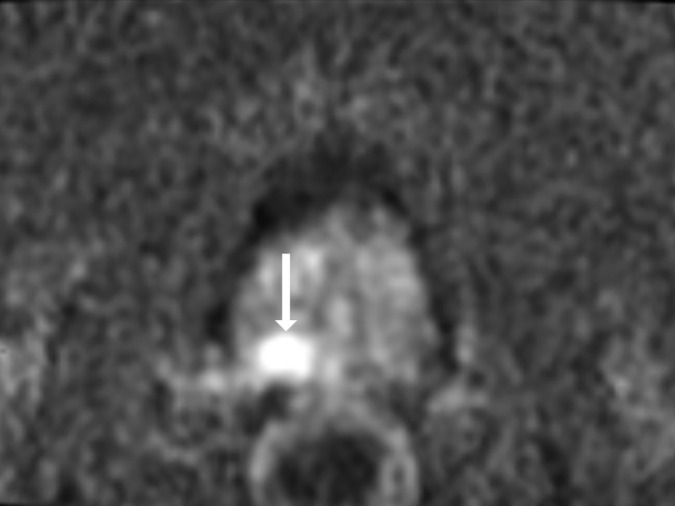
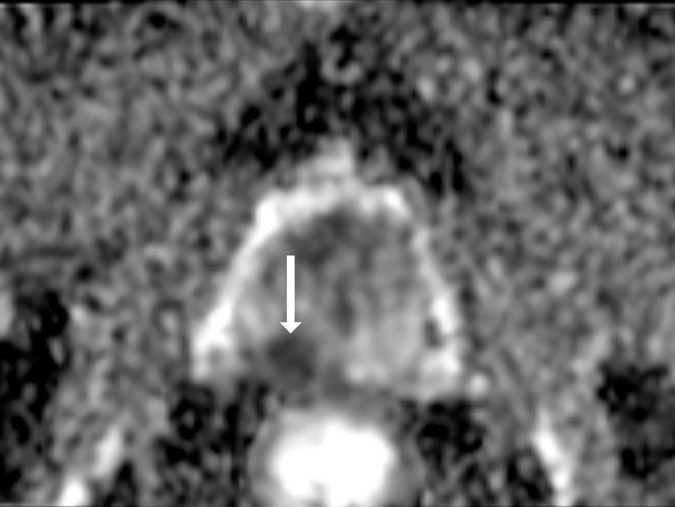
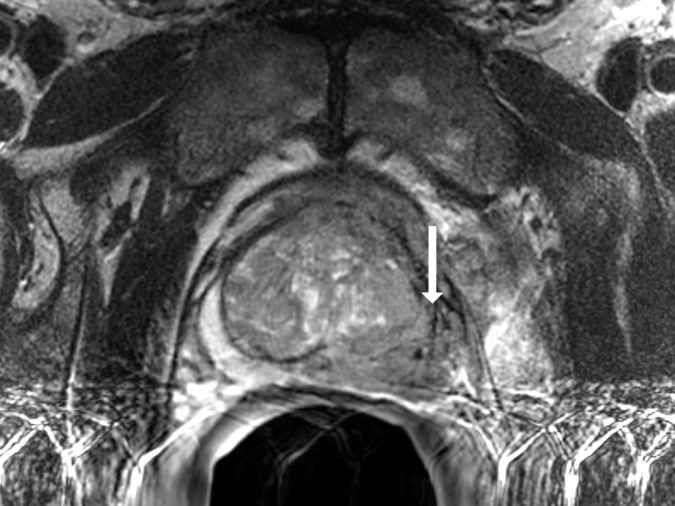
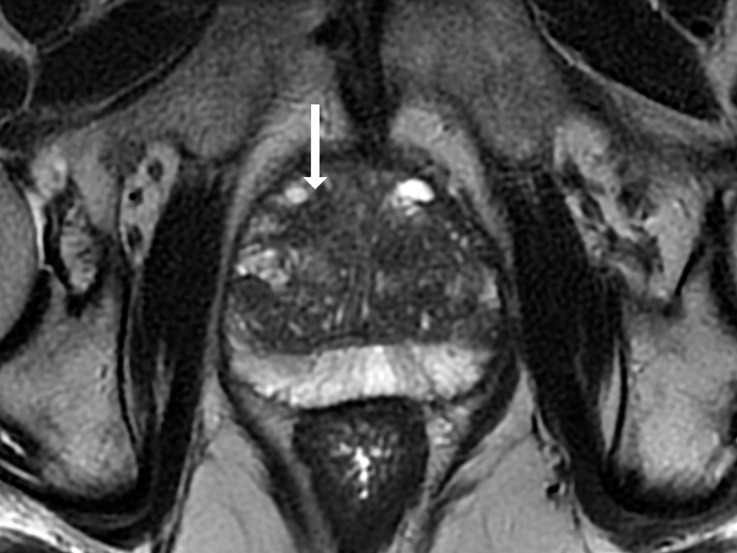
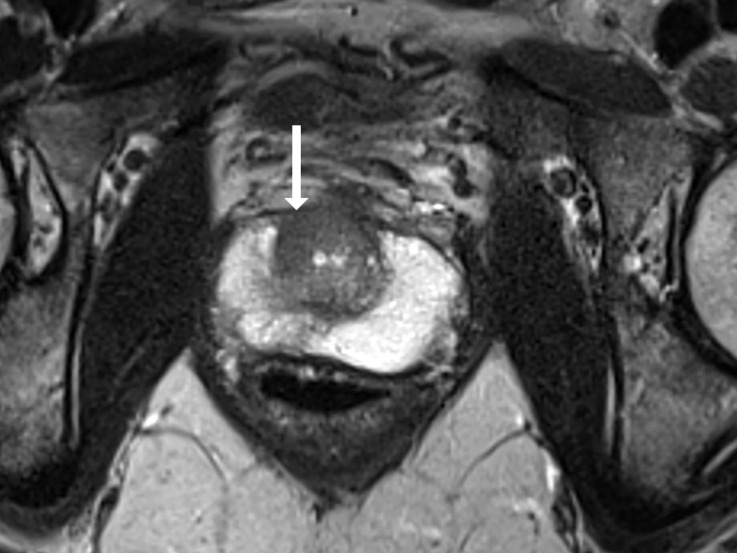
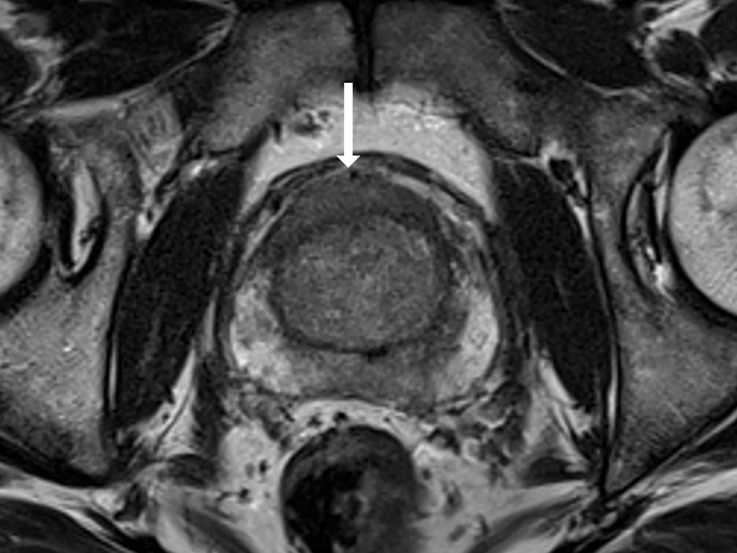
Antonio C Westphalen
Antonio C Westphalen, MD, PhD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,✉,
Charles E McCulloch
Charles E McCulloch, PhD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Jordan M Anaokar
Jordan M Anaokar, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Sandeep Arora
Sandeep Arora, MBBS
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Nimrod S Barashi
Nimrod S Barashi, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Jelle O Barentsz
Jelle O Barentsz, MD, PhD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Tharakeswara K Bathala
Tharakeswara K Bathala, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Leonardo K Bittencourt
Leonardo K Bittencourt, MD, PhD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Michael T Booker
Michael T Booker, MD, MBA
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Vaughn G Braxton
Vaughn G Braxton, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Peter R Carroll
Peter R Carroll, MD, MPH
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
David D Casalino
David D Casalino, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Silvia D Chang
Silvia D Chang, MD, FRCPC
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Fergus V Coakley
Fergus V Coakley, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Ravjot Dhatt
Ravjot Dhatt, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Steven C Eberhardt
Steven C Eberhardt, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Bryan R Foster
Bryan R Foster, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Adam T Froemming
Adam T Froemming, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Jurgen J Fütterer
Jurgen J Fütterer, MD, PhD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Dhakshina M Ganeshan
Dhakshina M Ganeshan, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Mark R Gertner
Mark R Gertner, PhD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Lori Mankowski Gettle
Lori Mankowski Gettle, MD, MBA
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Sangeet Ghai
Sangeet Ghai, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Rajan T Gupta
Rajan T Gupta, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Michael E Hahn
Michael E Hahn, MD, PhD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Roozbeh Houshyar
Roozbeh Houshyar, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Candice Kim
Candice Kim, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Chan Kyo Kim
Chan Kyo Kim, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Chandana Lall
Chandana Lall, MD, MBA
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Daniel J A Margolis
Daniel J A Margolis, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Stephen E McRae
Stephen E McRae, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Aytekin Oto
Aytekin Oto, MD, MBA
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Rosaleen B Parsons
Rosaleen B Parsons, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Nayana U Patel
Nayana U Patel, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Peter A Pinto
Peter A Pinto, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Thomas J Polascik
Thomas J Polascik, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Benjamin Spilseth
Benjamin Spilseth, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Juliana B Starcevich
Juliana B Starcevich, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Varaha S Tammisetti
Varaha S Tammisetti, MBBS, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Samir S Taneja
Samir S Taneja, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Baris Turkbey
Baris Turkbey, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Sadhna Verma
Sadhna Verma, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
John F Ward
John F Ward, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Christopher A Warlick
Christopher A Warlick, MD, PhD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Andrew R Weinberger
Andrew R Weinberger, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Jinxing Yu
Jinxing Yu, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Ronald J Zagoria
Ronald J Zagoria, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1,
Andrew B Rosenkrantz
Andrew B Rosenkrantz, MD
1From the Departments of Radiology and Biomedical Imaging (A.C.W., R.J.Z.), Urology (A.C.W., P.R.C.), and Epidemiology and Biostatistics (C.E.M.) and the Clinical and Translational Science Institute (C.E.M.), University of California, San Francisco, 505 Parnassus Ave, M-392, Box 0628, San Francisco, CA 94143; Department of Diagnostic Imaging, Fox Chase Cancer Center, Philadelphia, Pa (J.M.A., R.B.P.); Departments of Radiology and Radiological Sciences (S.A., V.G.B) and Urologic Surgery (S.A.), Vanderbilt University Medical Center, Nashville, Tenn; Departments of Radiology (A.O.) and Urology (N.S.B), University of Chicago, Chicago, Ill; Departments of Radiology (J.O.B) and Nuclear Medicine (J.J.F.), Radboud University Nijmegen Medical Center, Nijmegen, the Netherlands; Departments of Diagnostic Radiology (T.K.B., D.M.G), Interventional Radiology (S.E.M.), and Urology (J.F.W.), University of Texas MD Anderson Cancer Center, Houston, Tex; Diagnósticos da América S/A, Rio de Janeiro, Brazil (L.K.B); and Department of Radiology, Fluminense Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.K.B.); Department of Radiology, University of California, San Diego, San Diego, Calif (M.T.B., M.E.H.); UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, Calif (P.R.C.); Department of Radiology, Northwestern University, Feinberg School of Medicine, Chicago, Ill (D.D.C., A.R.W.); Department of Radiology, University of British Columbia, Vancouver, Canada (S.D.C., R.D.); Department of Diagnostic Radiology, Oregon Health Science University, Portland, Ore (F.V.C., B.R.F.); Department of Radiology, University of New Mexico Health Sciences Center, Albuquerque, NM (S.C.E., B.S., J.B.S.); and Department of Radiology, Mayo Clinic, Rochester, Minn (A.T.F.). Joint Department of Medical Imaging, University Health Network–Mount Sinai Hospital–Women’s College Hospital, Toronto, Canada (M.R.G., S.G.); Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, Wis (L.M.G.); Departments of Radiology (R.T.G.) and Surgery (R.T.G., T.J.P.), Duke University Medical Center and Duke Cancer Institute, Durham, NC; Department of Radiological Sciences and Urology, University of California, Irvine, Orange, Calif (R.H.); Virginia Commonwealth University School of Medicine, Richmond, Va (C.K.); Department of Radiology and Center for Imaging Sciences, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea (C.K.K.); Department of Radiology, University of Florida College of Medicine, Jacksonville, Fla (C.L.); Department of Radiology, Weill Cornell Medicine, New York, NY (D.J.A.M.); Department of Radiology, University of Colorado at Denver, Denver, Colo (N.U.P.); Molecular Imaging Program (B.T.) and Urologic Oncology Branch (P.A.P.), National Cancer Institute, National Institutes of Health, Bethesda, Md; Department of Diagnostic and Interventional Imaging, University of Texas Health Science Center, Houston, Tex (V.S.T.); Departments of Radiology (A.B.R.) and Urologic Oncology (S.S.T.), New York University Langone Health, New York, NY; Department of Radiology, University of Cincinnati Medical Center, Cincinnati, Ohio (S.V.); Department of Urology, University of Minnesota Institute for Prostate and Urologic Cancers, Minneapolis, Minn (C.A.W.); and Department of Radiology, Virginia Commonwealth University, Richmond, Va (J.Y.).
1