Abstract
Background:
Previous investigations have illustrated that regulated upon activation, normal T-cell expressed and secreted (RANTES) polymorphisms are linked to susceptibility to childhood asthma; nevertheless, the findings continue to be controversial. Accordingly, we conducted the present meta-analysis to clarify the impact of RANTES genetic polymorphisms (-403G/A and -28C/G) on childhood asthma vulnerability.
Methods:
A search for published literature was performed using the PubMed, EMBASE, Chinese National Infrastructure, Cochrane Library, Scopus, Web of Science, and WanFang databases and selected in the form of PICOS (participants, interventions, comparisons, outcomes, and study design) to identify all eligible research works. The link between RANTES genetic polymorphisms and childhood asthma susceptibility was evaluated by a pooled odds ratio with a 95% confidence interval.
Results:
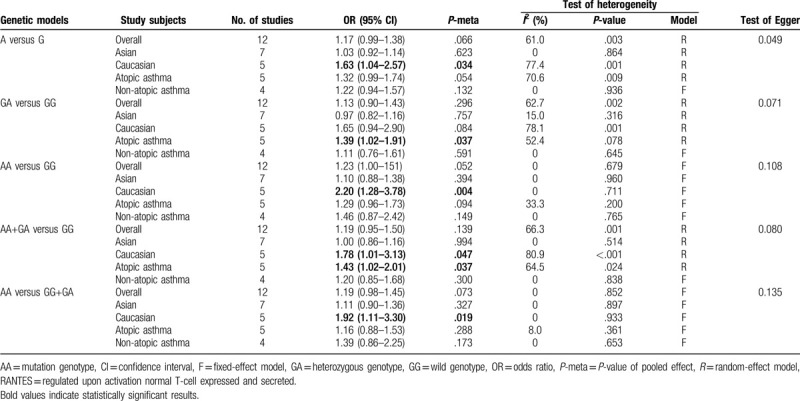
In total, 14 case–control studies were included in the analysis. No significant association existed between risk of childhood asthma and the -403G/A polymorphism subjected to any genetic framework in the overall population. In the stratified analysis, according to ethnicity, the -403G/A polymorphism was linked to augmented vulnerability to childhood asthma in Caucasians (allelic model: odds ratio [OR] = 1.63, 95% confidence interval [CI] = 1.04–2.57, P = .034; codominant model: OR = 2.20, 95% CI = 1.28–3.78, P = .004; dominant model: OR = 1.78, 95% CI = 1.01–3.13, P = .047; and recessive model: OR = 1.92, 95% CI = 1.11–3.30, P = .019). For the stratified analysis by atopic status, the -403G/A polymorphism was linked to augmented childhood asthma in the codominant (OR = 1.39, 95% CI = 1.02–1.91, P = .037) and dominant models (OR = 1.43, 95% CI = 1.02–2.01, P = .037) in atopic asthma. For the -28C/G polymorphism, there was a significant association between childhood asthma and the -28C/G variant (allelic model: OR = 1.33, 95% CI = 1.08–1.65, P = .009; codominant framework: OR = 2.14, 95% CI = 1.47–3.10, P < .001; dominant model: OR = 1.44, 95% CI = 1.07–1.93, P = .017; and recessive model: OR = 2.08, 95% CI = 1.44–3.02, P < .001). Stratified analysis based on ethnicity and the -28C/G polymorphism was linked to augmented vulnerability to childhood asthma in Asian and Caucasian populations. For the subgroup analysis by atopic status, no association was found in atopic and non-atopic asthma.
Conclusion:
The present meta-analysis indicated that the RANTES -403G/A and -28C/G polymorphisms contributed to the development of childhood asthma.
Keywords: asthma; childhood; meta-analysis; polymorphism; regulated upon activation, normal T-cell expressed and secreted
1. Introduction
Asthma is among the most frequent chronic respiratory ailments in children, which is characterized by airway hyper-responsiveness and intermittent airflow obstruction due to chronic inflammation of the airways.[1] The prevalence of pediatric asthma has been increasing in various countries worldwide over the last 2 decades, accompanied by burdensome fiscal issues to not just the family but also to society, resulting in extensive clinical expenses.[2–4] Increasing evidence has revealed that asthma is a result of an intricate interaction between genetic and environmental factors.[5] Recently, a number of studies have placed emphasis on the connection between genetic polymorphisms and susceptibility to childhood asthma, including interleukin-7 (IL-7), a disintegrin and metalloprotease 33 (ADAM33), and regulated upon activation, normal T-cell expressed and secreted (RANTES).[6–8]
RANTES, also known as CC motif chemokine ligand 5 (CCL5), is a potent chemoattractant for eosinophils, lymphocytes, monocytes, and basophils.[9,10] It has been suggested that RANTES stimulates the recruitment of eosinophils to sites of inflammation; accordingly, it is involved in different kinds of allergic and immune disorders.[9,11–14] As indicated, many common single nucleotide polymorphisms in the RANTES gene have been observed to affect promoter activity and increase the expression of RANTES, including -403G/A and -28C/G in the promoter region.[15,16] Therefore, these 2 RANTES genetic polymorphisms are likely to exert an impact on the course of childhood asthma susceptibility by modulating not only the transcription, but also the expression of the RANTES gene.
Although previous case–control investigations have studied the potential contribution of the RANTES gene to childhood asthma susceptibility, the findings of these investigations continue to be controversial and inconclusive. We hypothesize that these inconsistent results are likely to be due to limited sample sizes, clinical heterogeneity, or a combination of the 2. Accordingly, we performed the current meta-analysis to examine the association between RANTES variants and susceptibility to asthma in children.
2. Methods and materials
2.1. Search strategy
Literature searches were carried out using the PubMed, EMBASE, Chinese National Knowledge Infrastructure, Cochrane Library, Scopus, Web of Science, and WanFang databases. We have used the following strategy: #1 asthma OR asthmatic OR (bronchial asthma) OR (bronchial hyperreactivity) OR allergy OR atopy; #2 RANTES OR (regulated upon the activation normal T-cell expressed and secreted) OR CCL5 OR (-403 G/A) OR (-28 C/G) OR rs2107538 OR rs2280788; #3 polymorphism∗ OR mutation∗ OR variant∗. #1 AND #2 AND #3. In PubMed, we have used “asthma” as Mesh term. In EMBASE, we have used “asthma” as EMTREE term. All analyses were based on published studies, therefore no ethical approval and patient consent are required.
2.2. Inclusion and exclusion criteria
The qualified research studies adhered to the criteria as follows: they concerned the correlation between RANTES polymorphisms and childhood asthma susceptibility; all patients were diagnosed with asthma; case–control studies were in human beings; and studies provided sufficient information to estimate the odds ratios (ORs) with 95% confidence intervals (CIs). The following were excluded: studies with no control groups; insufficient information for the evaluation of the ORs with 95% CI; abstract, reviews, and animal studies.
In the form of PICOS (P, participants; I, interventions; C, comparisons; O, outcomes; S, study design),[17] the study was described as follows: P, patients with childhood asthma; I, mutant RANTES polymorphisms; C, health control groups; O, RANTES polymorphisms including -403 G/A or -28 C/G; S: case–control study.
2.3. Data extraction and quality assessment
Data extraction from the included studies according to the above-mentioned criteria comprised: first author's name, year of publication, country, ethnicity, atopic status, genotyping method, and genotype numbers in both the cases and controls. The independent extraction of the information from all included studies was carried out by 2 reviewers. Any potential disagreement was settled by discussion. The quality of the included studies was estimated using the Newcastle-Ottawa Scale (NOS).[18]
2.4. Statistical analysis
The evaluation of the P-value of Hardy-Weinberg equilibrium (HWE) among the control groups was carried out by the chi-squared test; a P-value >.001 demonstrated that the population was in genetic equilibrium. ORs with 95% CIs were adopted for the purpose of calculating the robustness of the link between childhood asthma susceptibility and the RANTES polymorphisms. The Z-test was conducted to evaluate the statistical significance of the accumulated ORs. Furthermore, the calculation of the between-study heterogeneity was conducted with the help of the Q test and I2 statistics. The random-effects model was adopted if there was statistical heterogeneity (I2 > 50%)[19]; otherwise, the fixed-effects model was applied.[20] Sensitivity analysis was conducted through the emission of a single study every time to evaluate the robustness of the results. In addition, the assessment of potential publication bias was carried out using Begg funnel plot and Egger linear regression test.[21,22] All statistical analyses were carried out using the STATA version 15.0 software (Stata Corporation, College Station, TX). Furthermore, a P-value <.05 was considered statistically significant.
3. Results
3.1. Study characteristics
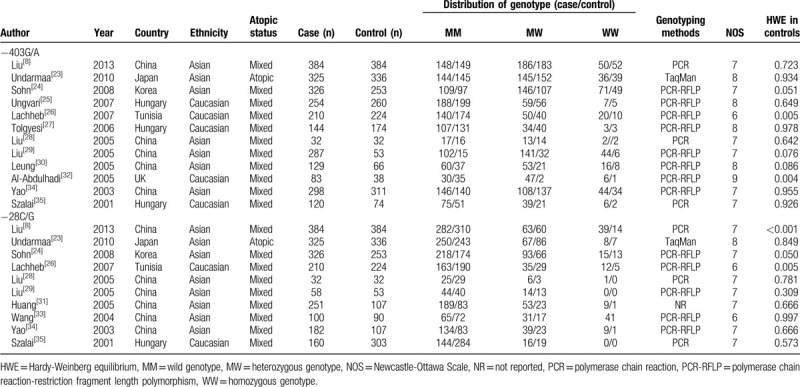
The literature retrieval mechanism is presented in Fig. 1. In total, 297 potentially related studies were identified with the help of a preliminary search of databases. Subsequent to the systematic literature search and in accordance with the inclusion criteria, 14 case–control research studies, which included 2943 asthma patients and 2402 controls subjects, were identified to assess the association of RANTES genetic variants with susceptibility to asthma in children.[8,23–35] Nine investigations were carried out in Asian populations and 5 were conducted in Caucasian populations. Five studies included atopic asthma in children, whereas 4 studies counted non-atopic asthma in children. The quality of the included studies was estimated via the NOS, and the scores for the included studies ranged from 6 to 9, revealing that the enrolled studies were of relatively high quality. The key characteristics of the individual studies are summarized in Table 1.
Figure 1.
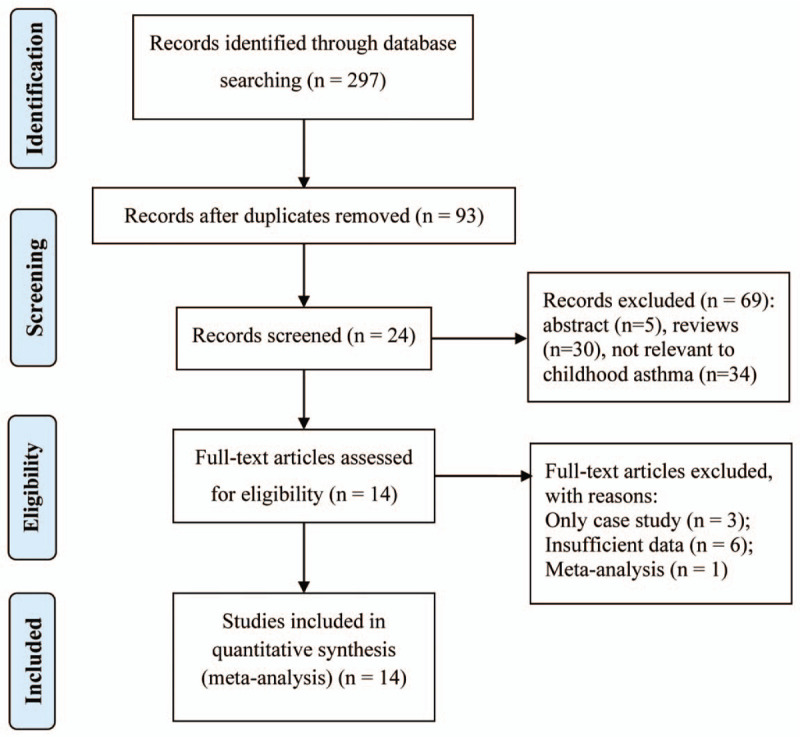
Flow diagram of included and excluded studies.
Table 1.
Main characteristics of included studies in this meta-analysis.
3.2. RANTES -403G/A polymorphism and susceptibility to asthma in children
The correlation between the RANTES −403G/A polymorphism and susceptibility to patients with childhood asthma was evaluated in 12 studies. The results demonstrate that there is no statistically significant connection between the −403G/A polymorphism and susceptibility to asthma in children in any genetic model, as shown in Fig. 2. With regard to the stratified analysis in accordance with ethnicity, as shown in Fig. 3, a substantial link is observed in Caucasian individuals in the allelic genetic model (OR = 1.63, 95% CI = 1.04–2.57, P = .034), dominant genetic model (OR = 1.78, 95% CI = 1.01–3.13, P = .047), codominant genetic model (OR = 2.20, 95% CI = 1.28–3.78, P = .004), and recessive genetic model (OR = 1.92, 95% CI = 1.11–3.30, P = .019). Moreover, the stratification analysis by atopic status reveals that the -403G/A polymorphism has an association with significantly augmented susceptibility to atopic asthma among children in the dominant genetic framework (OR = 1.43, 95% CI = 1.02–2.01, P = .037) as well as the codominant framework (OR = 1.39, 95% CI = 1.02–1.91, P = .037), as shown in Fig. 4. The main results are listed in Table 2.
Figure 2.
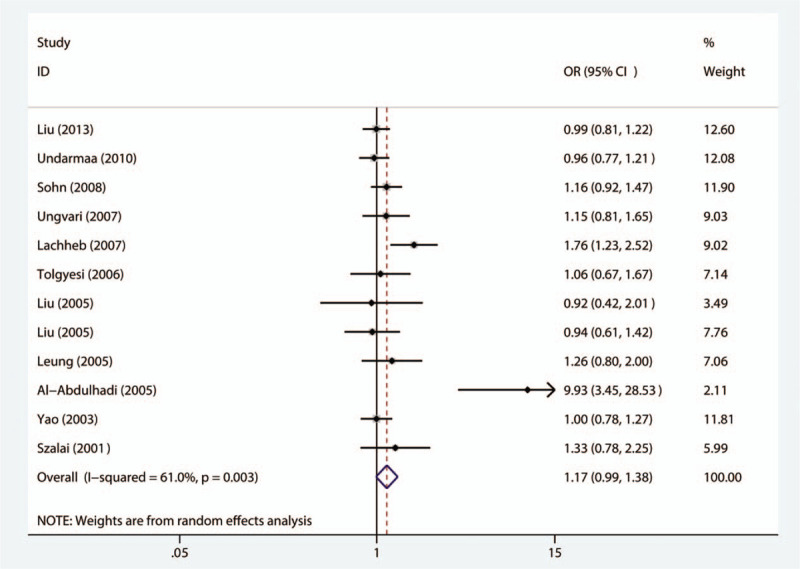
Meta-analysis for the association between asthma susceptibility and the RANTES -403G/A polymorphism among the allelic model (A vs G). RANTES = regulated upon activation normal T-cell expressed and secreted.
Figure 3.
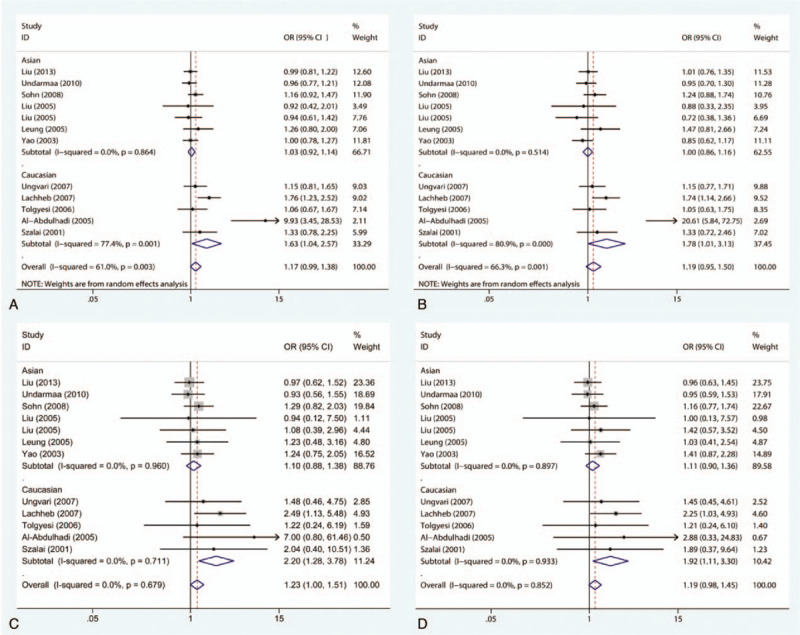
Meta-analysis for the association between asthma susceptibility and the RANTES -403G/A polymorphism: subgroup analysis by ethnicity. (A) For allelic model: A versus G; (B) for dominant model: GA+AA versus GG; (C) for codominant model: AA versus GG; (D) for recessive model: AA versus GG+GA. RANTES = regulated upon activation normal T-cell expressed and secreted.
Figure 4.
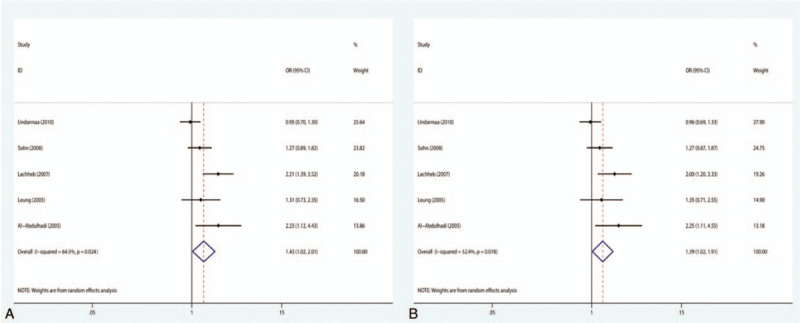
Meta-analysis for the association between asthma susceptibility and the RANTES -403G/A polymorphism: subgroup analysis by atopic status. (A) For dominant model: GA+AA versus GG; (B) for codominant model: GA versus GG. RANTES = regulated upon activation normal T-cell expressed and secreted.
Table 2.
Summary ORs and 95% CI of the association between RANTES -403G/A polymorphism and pediatric asthma susceptibility.
3.3. RANTES -28C/G polymorphism and susceptibility to asthma in children
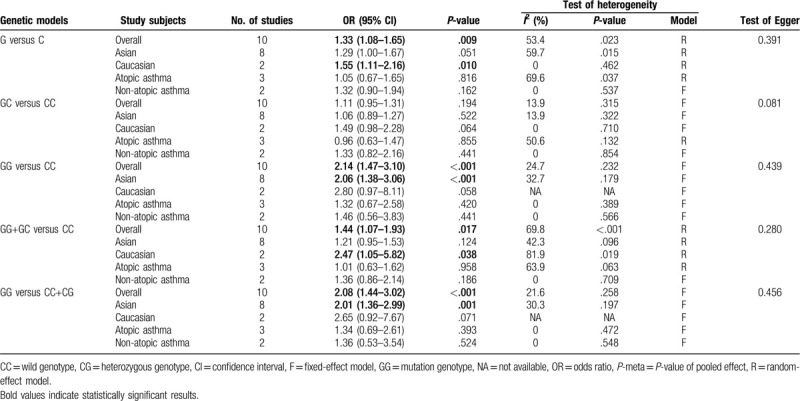
There were 10 case–control studies that investigated the relationship between the −28C/G polymorphism and susceptibility to asthma in children. Among children, there is a substantial link between the −28C/G polymorphism and vulnerability to asthma in the allelic genetic framework (OR = 1.33, 95% CI = 1.08–1.65, P = .009), dominant genetic framework (OR = 1.44, 95% CI = 1.07–1.93, P = .017), codominant genetic model (OR = 2.14, 95% CI = 1.47–3.10, P < .001), and recessive genetic model (OR = 2.08, 95% CI = 1.44–3.02, P < .001), as shown in Fig. 5. With regard to the stratified analysis in accordance with the ethnicity of the research population, the results reveal that the −28C/G polymorphism has an association with augmented susceptibility to childhood asthma in Asian individuals in the codominant genetic model (OR = 2.06, 95% CI = 1.38–3.06, P < .001) and recessive genetic model (OR = 2.01, 95% CI = 1.36–2.99, P = .001), as shown in Fig. 6A and B. However, significantly increased susceptibility to asthma is also observed in Caucasian populations in the allelic genetic framework (OR = 1.55, 95% CI = 1.11–2.16, P = .010) as well as the dominant genetic framework (OR = 2.47, 95% CI = 1.05–5.82, P = .038), as shown in Fig. 6C and D. Additionally, with regard to the subgroup analysis conducted in accordance with atopic status, no substantial link was found between the −28C/G polymorphism and vulnerability to asthma in children with atopic asthma or non-atopic asthma in any of the genetic models.
Figure 5.
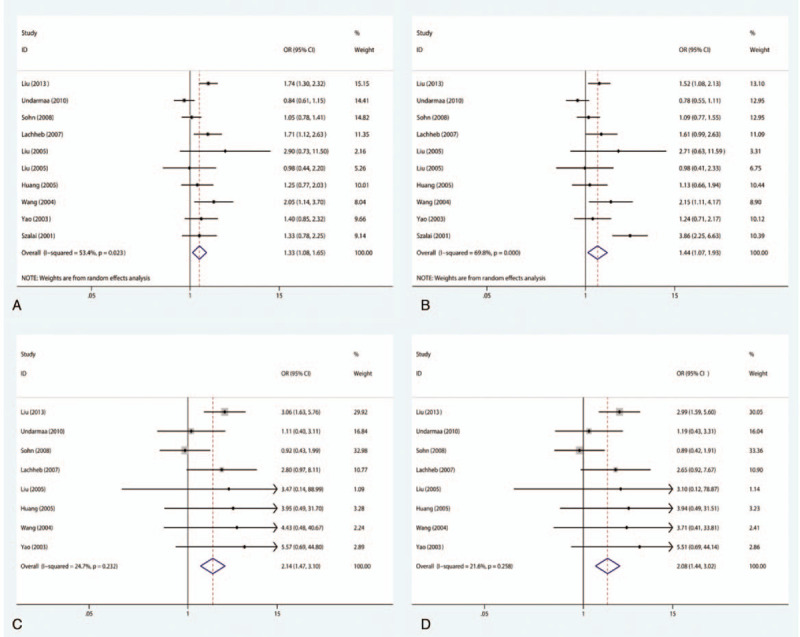
Meta-analysis for the association between asthma susceptibility and the RANTES -28C/G polymorphism. (A) For allelic model: G versus C; (B) for dominant model: CG+CC versus GG; (C) for codominant model: GG versus CC; (D) for recessive model: GG versus CC+CG. RANTES = regulated upon activation normal T-cell expressed and secreted.
Figure 6.
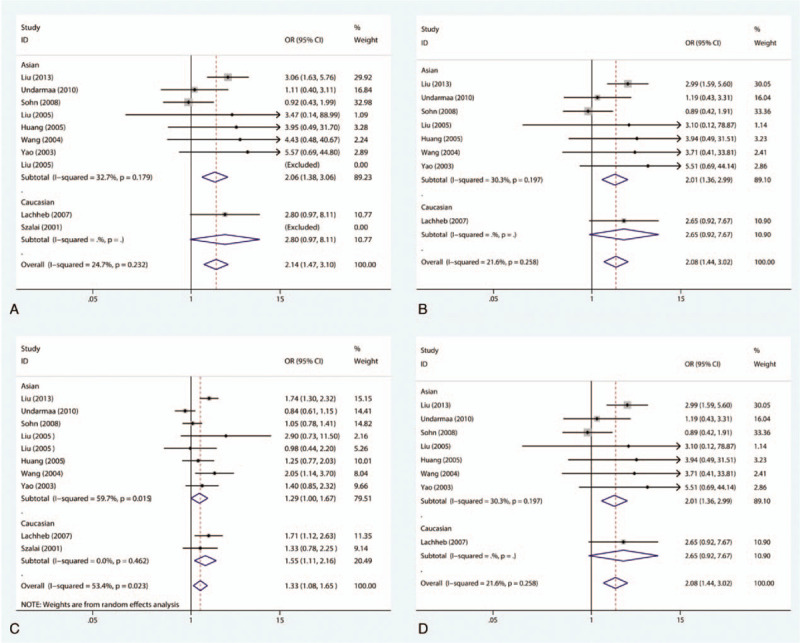
Meta-analysis for the association between asthma susceptibility and the RANTES -28C/G polymorphism: subgroup analysis by ethnicity. (A) For codominant model in Asian: GG versus CC; (B) for recessive model in Asian: GG versus CC+CG; (C) for allelic model in Caucasian: G versus C; (D) for recessive model in Caucasian: GG versus CC+CG. RANTES = regulated upon activation normal T-cell expressed and secreted.
3.4. Sensitivity analysis and publication bias
Sensitivity analysis was carried out to assess the stability of the findings through the sequential removal of individuals. As the findings suggest, the accumulated ORs do not have a significant impact, as shown in Fig. 7. Moreover, both the Begg funnel plot and Egger test were applied to evaluate potential publication bias. The shape of the funnel plot shows symmetry, which suggests that no significant publication bias is present, as shown in Fig. 8. The Egger test was also used to evaluate potential publication bias (Tables 2 and 3); there was no publication bias in the present meta-analysis.
Figure 7.
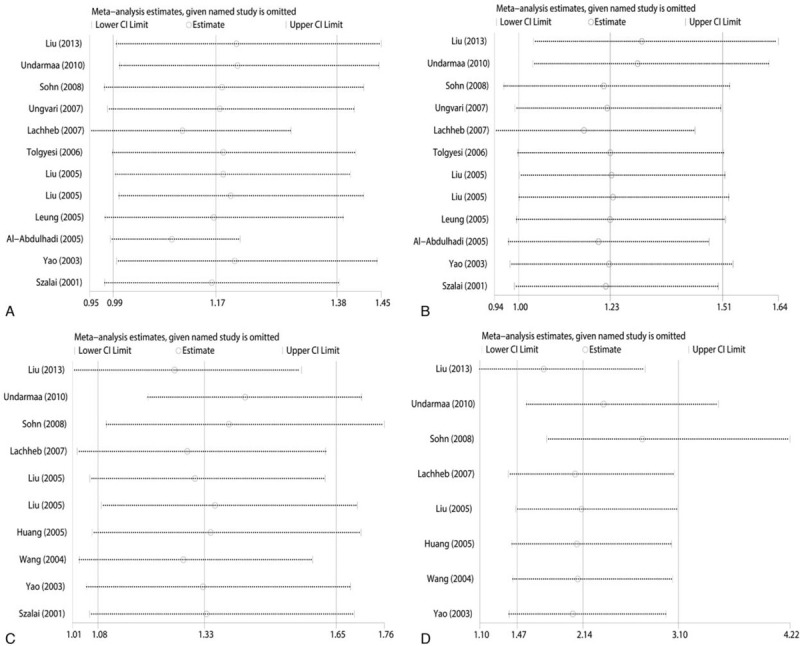
Sensitivity analysis on the association between RANTES polymorphisms and susceptibility asthma. For allelic (A) and codominant (B) model of -403G/A polymorphism; and for allelic (C) and codominant (D) model of -28C/G polymorphism. RANTES = regulated upon activation normal T-cell expressed and secreted.
Figure 8.
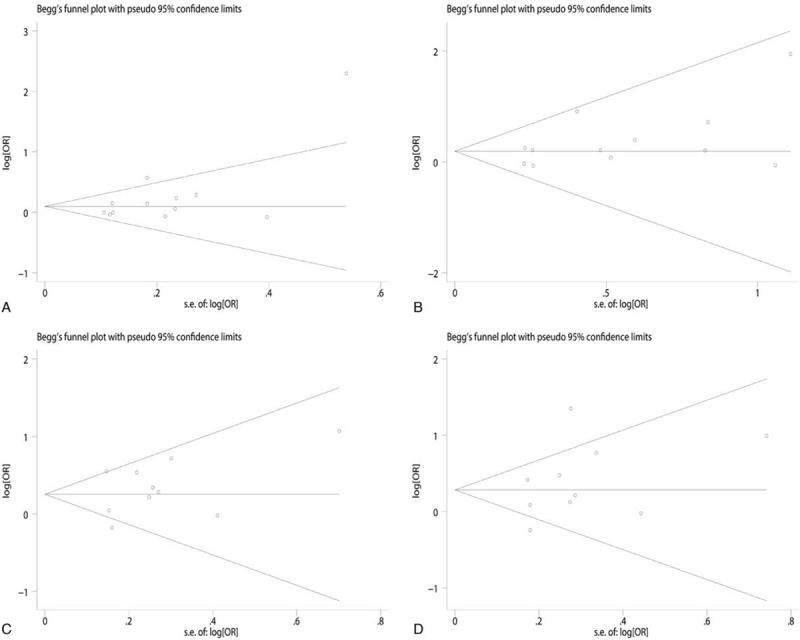
Funnel plot analysis to examine publication bias. For allelic (A) and codominant (B) model of -403G/A polymorphism; and for allelic (C) and codominant (D) model of -28C/G polymorphism.
Table 3.
Summary ORs and 95% CI of the association between RANTES -28C/G polymorphism and pediatric asthma susceptibility.
4. Discussion
Earlier research has shown that asthma vulnerability is determined not only by the infectious agent and environmental determinants, but also by host genetics.[3,5] Several candidate genes have been studied for accessing the likely connection between the modulations of asthma risk across various populations. Despite comprehensively stating the recruitment criteria, sample size, characteristics of participants, and genotyping methodologies, most of the studies were short of the proper conclusion. Hence, to improve the statistical power, together with determining the effect size of the RANTES -403G/A and -28C/G polymorphisms, a meta-analysis was carried out with the updated data of 14 studies to determine a more concrete correlation between the RANTES -403G/A and -28C/G polymorphisms and the occurrence of asthma susceptibility.
As far we know, this is the most detailed and systematic meta-analysis performed to date, meant to assess the potential link between the RANTES -403G/A and -28C/G polymorphisms and asthma susceptibility in childhood. Meta-analysis data revealed that the RANTES -28C/G polymorphism was significantly associated with augmented asthma susceptibility in childhood. Compared with the wild type C allele and homozygous CC genotype, the subjects with a G allele and 2 variant GG alleles had 1.33- and 2.14-fold increased risk of the development of asthma, respectively. With regard to the stratified analysis based on ethnicity, the C28G polymorphism was linked to augmented susceptibility to childhood asthma in Asian and Caucasian populations. We also carried out a subgroup analysis in accordance with atopic status; no significant association was found in atopic and non-atopic asthma. In addition, the data showed that the RANTES -403G/A polymorphism did not have a statistical association with asthma susceptibility in childhood. Nevertheless, for the stratified analysis according to ethnicity, the subjects with an A allele, together with 2 variant AA alleles, had 1.63- and 2.20-fold augmented susceptibility to the development of asthma, compared with the wild type G allele and homozygous GG genotype in Caucasian children, respectively. In the same manner, with regard to the stratified analysis by atopic status, dominant and codominant models indicated the increased risk of atopic asthma. Based on the above data and the significance of RANTES polymorphisms in the pathogenesis of asthma in children, it is biologically plausible that both the -403G/A and -28C/G polymorphisms are likely to modulate the risk of asthma, in addition to being a genetic determinant for the inter-individual discrepancies in vulnerability to asthma in children.
From previous reports, we know that RANTES constitutes a key chemotactic determinant created through asthmatic feedback, and contributes to the deterioration of allergic airway inflammation.[36–38] There are substantially high concentrations of RANTES in asthmatic patients, together with asthma severity, demonstrating the pivotal function of RANTES in the pathogenesis of this disorder.[37,39,40] Serum RANTES is likely to be a helpful noninvasive diagnostic marker for monitoring asthma severity. Identifying and hampering RANTES and/or its receptor are likely to constitute a potential therapeutic methodology for asthmatic patients.[41] In the study by Leung et al,[30] the -403G/A polymorphism is linked to allergen sensitization and forced expiratory volume in 1-s (FEV1), and there is an association with the -28C/G polymorphism. In contrast, Liu et al[8] found that the -28C/G polymorphism has a substantial association with increased asthma susceptibility in childhood. Contrary to these results, -403G/A and -28C/G polymorphisms have no detectable impact on asthma susceptibility.[35] Several previous meta-analyses have shown controversial findings regarding the association between these 2 polymorphisms and asthma susceptibility.[42–44] In comparison with the earlier research works, the current meta-analysis offered a comprehensive and systematic meta-analysis to assess the potential association between the RANTES -403G/A and -28C/G polymorphisms and asthma susceptibility in childhood.
The genetic susceptibility to asthma in children is polygenic; hence, a single genetic variant is typically not suitable for the prediction of the susceptibility of this disease. The most pivotal attribute of these gene polymorphisms suggests that their occurrence has the potential to vary significantly between various racial or ethnic populations. Before reaching a conclusion, limitations of this study should be addressed. First, just research works, authored in English or Chinese, were included in the meta-analysis. This suggests that qualified research works published in other languages were likely to have been neglected, which likely introduced selection bias. Second, the sample size of some research works had limitations; in addition, the findings required careful interpretation. Third, the current research work had statistical heterogeneity, despite the fact that this is extensively frequent in the meta-analysis of genetic association research. Accordingly, we carried out a subgroup analysis for the purpose of identifying all factors that contributed to the heterogeneity. Ultimately, other determinants, such as sex, environment, and lifestyle, which were likely to exert an impact on the contact of RANTES polymorphisms with asthma susceptibility in children, could not be analyzed due to the lack of data.
5. Conclusions
To summarize, the present meta-analysis revealed that the RANTES -403G/A and -28C/G polymorphisms appear to be associated with the susceptibility to childhood asthma. However, prospective, well-designed, large research studies will likely be beneficial for the validation of this association in various populations, including consideration of environmental factors responsible for the vulnerability to asthma in children.
Author contributions
Conceptualization: Yan-Qin Zhang, Xiuxiang Gao.
Data curation: Yan-Qin Zhang.
Formal analysis: Yan-Qin Zhang.
Funding acquisition: Yan-Qin Zhang.
Investigation: Yan-Qin Zhang.
Methodology: Yan-Qin Zhang.
Project administration: Yan-Qin Zhang, Xiuxiang Gao.
Resources: Xiuxiang Gao.
Software: Xiuxiang Gao.
Supervision: Xiuxiang Gao.
Validation: Xiuxiang Gao.
Visualization: Xiuxiang Gao.
Writing – original draft: Xiuxiang Gao.
Writing – review & editing: Xiuxiang Gao.
Footnotes
Abbreviations: ADAM33 = a disintegrin and metalloprotease 33, CCL5 = CC motif chemokine ligand 5, CI = confidence interval, CNKI = Chinese National Knowledge Infrastructure, FEV1 = forced expiratory volume in 1-s, HWE = Hardy-Weinberg equilibrium, IL-7 = Interleukin-7, OR = odds ratio, RANTES = regulated upon activation normal T-cell expressed and secreted.
How to cite this article: Zhang YQ, Gao XX. Association of RANTES Gene Polymorphisms with Susceptibility to Childhood Asthma: A meta-analysis. Medicine. 2020;99:29(e20953).
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are publicly available.
References
- [1].Asher MI, Montefort S, Bjorksten B, et al. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood: ISAAC Phases One and Three repeat multicountry cross-sectional surveys. Lancet 2006;368:733–43. [DOI] [PubMed] [Google Scholar]
- [2]. Global Initiative for Asthma (GINA). Global strategy for asthma management and prevention. National Institute of Health (NIH) Publication, Updated 2017. Available at: http://www.ginasthma.org. Accessed January 28, 2019. [Google Scholar]
- [3].Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev 2011;242:10–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ferrante G, La Grutta S. The burden of pediatric asthma. Front Pediatr 2018;6:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bouzigon E, Nadif R, Le Moual N, et al. Genetic and environmental factors of asthma and allergy: results of the EGEA study. Rev Mal Respir 2015;32:822–40. [DOI] [PubMed] [Google Scholar]
- [6].Cao J, Tian L, Li Z, et al. Interleukin-7 gene polymorphism rs766736182 associates with the risk of asthma in children. J Clin Lab Anal 2018;33:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Saba N, Yusuf O, Rehman S, et al. Single nucleotide polymorphisms in asthma candidate genes TBXA2R, ADAM33 FCER1B and ORMDL3 in Pakistani asthmatics a case control study. Asthma Res Pract 2018;4:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liu Q, Hua L, Fang D, et al. Interleukin-13 and RANTES polymorphisms in relation to asthma in children of Chinese Han nationality. Asian Pac J Allergy Immunol 2013;31:247–52. [DOI] [PubMed] [Google Scholar]
- [9].Castan L, Magnan A, Bouchaud G. Chemokine receptors in allergic diseases. Allergy 2017;72:682–90. [DOI] [PubMed] [Google Scholar]
- [10].Schall TJ, Bacon K, Toy KJ, et al. Selective attraction of monocytes and T lymphocytes of the memory phenotype by cytokine RANTES. Nature 1990;347:669–71. [DOI] [PubMed] [Google Scholar]
- [11].Venge P. The eosinophil and airway remodelling in asthma. Clin Respir J 2010;4: suppl: 15–9. [DOI] [PubMed] [Google Scholar]
- [12].Sato E, Simpson KL, Grisham MB, et al. Effects of reactive oxygen and nitrogen metabolites on RANTES- and IL-5-induced eosinophil chemotactic activity in vitro. Am J Pathol 1999;155:591–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fichna M, Zurawek M, Budny B, et al. Elevated serum RANTES chemokine levels in autoimmune Addison disease. Pol Arch Intern Med 2018;128:216–21. [DOI] [PubMed] [Google Scholar]
- [14].Johnstone S, Barsova J, Campos I, et al. Equine herpesvirus type 1 modulates inflammatory host immune response genes in equine endothelial cells. Vet Microbiol 2016;192:52–9. [DOI] [PubMed] [Google Scholar]
- [15].Muro M, Marin L, Torio A, et al. CCL5/RANTES chemokine gene promoter polymorphisms are not associated with atopic and nonatopic asthma in a Spanish population. Int J Immunogenet 2008;35:19–23. [DOI] [PubMed] [Google Scholar]
- [16].Nickel RG, Casolaro V, Wahn U, et al. Atopic dermatitis is associated with a functional mutation in the promoter of the C-C chemokine RANTES. J Immunol 2000;164:1612–6. [DOI] [PubMed] [Google Scholar]
- [17].Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg 2010;8:336–41. [DOI] [PubMed] [Google Scholar]
- [18]. Wells GA, Shea B, O’Connell D, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality if nonrandomized studies in meta-analyses; 2019. Available at: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. Accessed January 28, 2019. [Google Scholar]
- [19].DerSimonian R, Laird N. Meta-analysis in clinical trials revisited. Contemp Clin Trials 2015;45:139–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959;22:719–48. [PubMed] [Google Scholar]
- [21].Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994;50:1088–101. [PubMed] [Google Scholar]
- [22].Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Undarmaa S, Mashimo Y, Hattori S, et al. Replication of genetic association studies in asthma and related phenotypes. J Hum Genet 2010;55:342–9. [DOI] [PubMed] [Google Scholar]
- [24].Sohn MH, Kim SH, Kim KW, et al. RANTES gene promoter polymorphisms are associated with bronchial hyperresponsiveness in Korean children with asthma. Lung 2008;186:37–43. [DOI] [PubMed] [Google Scholar]
- [25].Ungvari I, Tolgyesi G, Semsei AF, et al. CCR5 Delta 32 mutation, Mycoplasma pneumoniae infection, and asthma. J Allergy Clin Immunol 2007;119:1545–7. [DOI] [PubMed] [Google Scholar]
- [26].Lachheb J, Chelbi H, Hamzaoui K, et al. Association between RANTES polymorphisms and asthma severity among Tunisian children. Hum Immunol 2007;68:675–80. [DOI] [PubMed] [Google Scholar]
- [27].Tolgyesi G, Keszei M, Ungvari I, et al. Involvement of TNFalpha -308A promoter polymorphism in the development of asthma in children infected with Chlamydophila pneumoniae. Pediatr Res 2006;60:543–8. [DOI] [PubMed] [Google Scholar]
- [28].Liu M, Hai-Lin LI, Huang YK, et al. The SNPs of chemokine RANTES promoter in children with asthma. Chin J Birth Health Herd 2005;13:20–3. [Google Scholar]
- [29].Liu CH, Chen H, Hu LP, et al. Association between the genetic polymorphism of chemokine genes and asthma in Chinese children. Zhonghua Er Ke Za Zhi 2005;43:462–3. [PubMed] [Google Scholar]
- [30].Leung TF, Tang NLS, Lam CWK, et al. RANTES G-401A polymorphism is associated with allergen sensitization and FEV1 in Chinese children. Respir Med 2005;99:216–9. [DOI] [PubMed] [Google Scholar]
- [31].Huang JL. Asthma severity and genetics in Taiwan. J Microbiol Immunol Infect 2005;38:158–63. [PubMed] [Google Scholar]
- [32].Al-Abdulhadi SA, Helms PJ, Main M, et al. Preferential transmission and association of the -403 G --> A promoter RANTES polymorphism with atopic asthma. Genes Immun 2005;6:24–30. [DOI] [PubMed] [Google Scholar]
- [33].Wang LJ, Li YR, Chen JH. Polymorphism of regulated upon activation, normal T cell expressed and secreted promoter region -28 position in Chinese allergic asthmatic children. Chin J Tuberc Respir Dis 2004;27:394–7. [PubMed] [Google Scholar]
- [34].Yao T-C, Kuo M-L, See L-C, et al. The RANTES promoter polymorphism: a genetic risk factor for near-fatal asthma in Chinese children. J Allergy Clin Immunol 2003;111:1285–92. [DOI] [PubMed] [Google Scholar]
- [35].Szalai C, Kozma GT, Nagy A, et al. Polymorphism in the gene regulatory region of MCP-1 is associated with asthma susceptibility and severity. J Allergy Clin Immunol 2001;108:375–81. [DOI] [PubMed] [Google Scholar]
- [36].Kim CK, Kita H, Callaway Z, et al. The roles of a Th2 cytokine and CC chemokine in children with stable asthma: potential implication in eosinophil degranulation. Pediatr Allergy Immunol 2010;21:e697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Larose MC, Archambault AS, Provost V, et al. Regulation of eosinophil and Group 2 innate lymphoid cell trafficking in asthma. Front Med (Lausanne) 2017;4:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lukacs NW, Standiford TJ, Chensue SW, et al. C-C chemokine-induced eosinophil chemotaxis during allergic airway inflammation. J Leukoc Biol 1996;60:573–8. [DOI] [PubMed] [Google Scholar]
- [39].Giuffrida MJ, Valero N, Mosquera J, et al. Increased cytokine/chemokines in serum from asthmatic and non-asthmatic patients with viral respiratory infection. Influenza Other Respir Viruses 2014;8:116–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Isgro M, Bianchetti L, Marini MA, et al. The C-C motif chemokine ligands CCL5, CCL11, and CCL24 induce the migration of circulating fibrocytes from patients with severe asthma. Mucosal Immunol 2013;6:718–27. [DOI] [PubMed] [Google Scholar]
- [41].Saad-El-Din Bessa S, Abo El-Magd GH, Mabrouk MM. Serum chemokines RANTES and monocyte chemoattractant protein-1 in Egyptian patients with atopic asthma: relationship to disease severity. Arch Med Res 2012;43:36–41. [DOI] [PubMed] [Google Scholar]
- [42].Huang H, Nie W, Zang Y, et al. Association between CC motif chemokine ligand 5 (CCL5) polymorphisms and asthma risk: an updated meta-analysis. J Investig Allergol Clin Immunol 2015;25:26–33. [PubMed] [Google Scholar]
- [43].Xie ZK, Zhao H, Huang J, et al. The regulated upon activation normal T-cell expressed and secreted (RANTES) -28C/G and -403G/A polymorphisms and asthma risk: a meta-analysis. Mol Diagn Ther 2014;18:523–31. [DOI] [PubMed] [Google Scholar]
- [44].Zhang YG, Huang J, Zhang J, et al. RANTES gene polymorphisms and asthma risk: a meta-analysis. Arch Med Res 2010;41:50–8. [DOI] [PubMed] [Google Scholar]


