Abstract
Purpose
The purpose of this study was to identify the clinical utility of circulating tumor DNA (ctDNA) from ascites and serial plasma samples from epithelial ovarian cancer (EOC) patients.
Materials and Methods
Using targeted next-generation sequencing, we analyzed a total of 55 EOC samples including ctDNA from ascites and serial plasma and gDNA from tumor tissues. Tumor tissues and ascites were collected during debulking surgeries and plasma samples were collected before and after the surgeries. Because one EOC patient underwent secondary debulking surgery, a total of 11 tumor tissues, 33 plasma samples, and 11 ascites samples were obtained from the 10 patients.
Results
Of the 10 patients, nine (90%) contained somatic mutations in both tumor tissues and ascites ctDNA. This mutational concordance was confirmed through correlation analysis. The mutational concordance between ascites and tumor tissues was valid in recurrent/progressive ovarian cancer. TP53 was the most frequently detected gene with mutations. ctDNA from serial plasma samples identified EOC progression/recurrence at a similar time or even more rapidly than cancer antigen 125, an established serum protein tumor marker for EOC.
Conclusion
Our data suggest that ascites ctDNA can be used to identify the mutational landscape of ovarian cancer for therapeutic strategy planning.
Keywords: Ascites, Circulating tumor DNA, Ovarian neoplasms, Plasma, Next-generation sequencing
Introduction
Epithelial ovarian cancer (EOC) is the most lethal gynecologic malignancy, largely due to late-stage diagnoses [1,2]. The most common histological EOC subtype is high-grade serous adenocarcinoma (52%), followed by endometrioid (10%), mucinous (6%), and clear cell adenocarcinomas (6%) [1]. They are further grouped as type I or type II based on clinicopathologic factors. Molecular characterization has revealed that type I EOC comprises endometrioid, mucinous, and clear cell carcinomas [3]. Endometrioid and clear cell types harbor ARID1A and PI3KCA mutations, whereas the mucinous type harbors KRAS mutations. Type II EOC includes high-grade serous carcinoma and frequently harbors TP53 and BRCA1/2 mutations.
Most EOC patients are diagnosed at stage III (51%) or IV (29%) and have accompanying cancer seed cells in the pelvic/abdominal cavity and malignant ascites [1,4]. In newly diagnosed EOC, ascites is treated by conventional debulking surgery and chemotherapy. However, malignant ascites develops in most advanced or recurrent EOCs. Ascites not only deteriorates quality of life, but also plays a role in the development of pelvic/abdominal metastasis [5]. In a previous study, we discovered that somatic mutations in ovarian cancer were highly conserved in ascitic cell genomes despite the intratumoral heterogeneity. In addition, the data suggested that ascitic cells might originate from multiple regions of solid ovarian cancer, suggesting that ascites may become useful for ovarian cancer diagnosis [6].
Cell-free nucleic acids in human blood were first described 70 years ago and the origins and characteristics of cell-free DNA (cfDNA) were studied in subsequent decades [7]. Thirty years ago, some cfDNA in the plasma of cancer patients was revealed to originate from cancer cells, with later work elucidating that mutations in cfDNA are a highly specific marker for cancer [7], giving rise to the term “circulating tumor DNA” (ctDNA). Compared with tissue biopsy, liquid biopsy using cfDNA is a less invasive method of analyzing genomic profiles of mutations across the entire cancer genome, and serial sampling for monitoring the disease burden and clonal evolution is feasible [7,8]. Besides blood, both cfDNA and ctDNA are detected in various body fluids, including urine, cerebrospinal fluid, pleural fluid, saliva, and ascites. Of them, it was suggested that ctDNA from ascites may provide additional information not detected by tumor and plasma ctDNA molecular characterization in various cancers [9,10]. However, the ctDNA from ovarian cancer ascites has not yet been fully elucidated.
In this study, we performed targeted next-generation sequencing of gDNA from tumor biopsies and ctDNA from serial plasma samples and ascites from 10 EOC patients. The primary aim of the study was to determine the feasibility of analyzing ctDNA from ascites and to elucidate the mutational concordance among the three samples (tumor biopsies and ctDNA from plasma and ascites). The secondary aim was to evaluate whether ctDNA from serial plasma samples can be correlated with the disease burden.
Materials and Methods
1. Samples
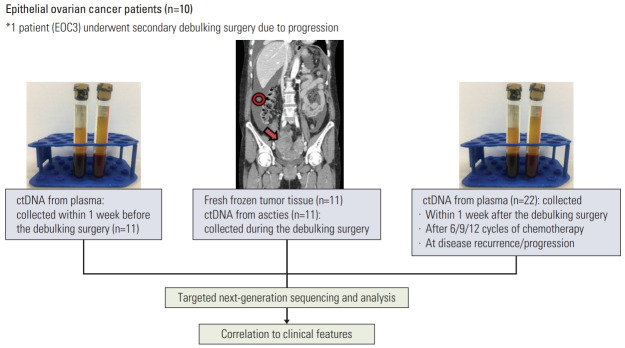
All specimens from the EOC patients (stages III and IV) in this study were obtained with appropriate consent and approval of the institutional review board. Fresh frozen tumor tissues and malignant ascites were collected during primary debulking surgery. The tumor tissue from each EOC patient was ≥ 1×1×1 cm3 and malignant ascites was collected in volumes of ≥ 10 mL. From each patient, 10 mL of blood was collected within 1 week before the debulking surgery and within 1 week after the surgery. From six of the 10 EOC patients, an additional 10 mL of blood was collected after 6/9/12 cycles of chemotherapy and at disease recurrence/progression (S1 Table). Thus, a total of 11 tumor tissues, 33 plasma samples, and 11 ascites samples were obtained from the 10 EOC patients (Fig. 1).
Fig. 1.
Study design. Of 10 epithelial ovarian cancer patients, one (EOC3) underwent secondary debulking surgery. Two tissue samples and two vials of ascites were collected from each debulking surgery. ctDNA, circulating tumor DNA.
2. Preparation of ctDNA
ctDNA was extracted from plasma and ascites using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Initial QC checks of eluted ctDNA were performed using the Qubit dsDNA HS Assay Kit and the Qubit 2.0 fluorometer (Life Technologies, Carlsbad, CA) and the 2100 Bioanalyzer with High-Sensitivity DNA chips (Agilent Technologies, Santa Clara, CA) according to the manufacturers’ instructions.
3. Sequencing library construction and target enrichment
gDNA libraries were prepared with the SureSelect XT protocol (Agilent Technologies), whereas ctDNA libraries were prepared with the SureSelect XT low-input protocol (Agilent Technologies). Prepared libraries were captured by Axen Cancer Panel 1, which includes 88 genes (Macrogen, Seoul, Korea) (S2 Table). For each sample, libraries were individually indexed and molecular-barcoded. Their quality was checked using the 2100 Bioanalyzer (Agilent Technologies) and a product size of 200-400 bp was required. The libraries were then quantified using the Qubit dsDNA HS Assay Kit and the Qubit 2.0 fluorometer (Life Technologies). The paired-end (2×150 bp) sequencing was performed on a NextSeq500 instrument (Illumina, San Diego, CA) with high output using Sequencing by Synthesis chemistry to a depth of approximately 5,000× coverage.
4. Targeted next-generation sequencing and data analysis
The adapter sequences of gDNA libraries and ctDNA libraries were removed by fastp [11] and Agilent SurecallTrimmer v4.0.1 (Agilent Technologies), respectively. Trimmed reads were aligned to the reference genome (GRCh37/hg19) using BWA-MEM [12]. Poorly mapped reads with a mapping quality below 20 were removed using Samtools ver. 1.3.1 [13]. Duplicated reads of gDNA libraries and ctDNA libraries were discarded using Picard MarkDuplicates (ver. 2.2.4) and Agilent LocatIt v4.0.1 (Agilent Technologies), respectively.
The base quality of deduplicated reads was recalibrated using GATK4 BaseRecalibrator. Somatic mutations were detected using MuTect2 [14], and mutations were examined using GATK4 FilterMutectCalls, FilterAlignmentArtifacts, CollectSequencingArtifactMetrics, and FilterByOrientationBias to filter out false-positive calls. A frequency of 0.1% was applied to assess mutations in ctDNA since the majority of ctDNA originates from dying non-cancerous cells. ANNOVAR (Annotate Variation) was used for functional annotation of each variant in coding regions [15]. Germline variants were excluded when the minor allele frequency was ≥ 1% in the Genome Aggregation Database (East Asian) and KOVA (Korean Variant Archive) [16]. PROVEAN and SIFT algorithms were used to predict the possible impact of an amino acid substitution on protein function and structure [17,18].
5. Ethical statement
All specimens from the EOC patients (stages III and IV) in this study were obtained with appropriate consent and approval of the institutional review board of Seoul St. Mary’s Hospital, The Catholic University of Korea, College of Medicine (Development of liquid biopsy-based genomic method for gynecologic malignancy diagnosis, KC17TNSI0215).
Results
1. Patients, samples, and clinical data
We analyzed 11 tumor tissues, 33 plasma samples, and 11 ascites samples from 10 EOC patients (Fig. 1). The patients had FIGO (International Federation of Gynecology and Obstetrics) stage IIIC-IVB with histopathological diagnosis of high-grade serous adenocarcinoma (n=9) and clear cell adenocarcinoma (n=1). The clinicopathological features are described in Table 1. All of the patients underwent debulking surgery and chemotherapy. Of the 10 EOC patients, nine were treatment-naïve ovarian cancer patients and 1 (EOC10) had been treated with neoadjuvant chemotherapy before the debulking surgery. In addition, three of the 10 EOC patients carried BRCA1/2 germline mutations (Table 1, S3 Fig.) and one patient had a family history of ovarian cancer; five patients developed recurrence or progression, with one (EOC3) undergoing secondary debulking surgery at progression. Variants with 30%-70% mutant allele frequency (MAF) were considered germline, and BRCA1/2 germline mutations were confirmed by Sanger sequencing. Serum protein tumor marker cancer antigen (CA125) was routinely monitored during the treatment course (upper limit of normal ≤ 35 UI/mL) (S1 Table).
Table 1.
Clinicopathological features of the 10 epithelial ovarian cancer patients
| Patient ID | Age (yr) | FIGO staging | Pathologic diagnosis | BRCA1/2 germline mutation | Prognosis | Remarks |
|---|---|---|---|---|---|---|
| EOC1 | 51 | IVB | High-grade serous adenocarcinoma | BRCA1 nonsynonymous SNV (c.T4588C) | No recurrence for 33 mo | - |
| EOC2 | 45 | IIIC | High-grade serous adenocarcinoma | - | Recurrence after 10 mo | Concurrent thyroid cancer |
| EOC3 | 56 | IVB | High-grade serous adenocarcinoma | - | Progression after 6 mo | Secondary debulking surgery was performed |
| EOC4 | 61 | IIIC | High-grade serous adenocarcinoma | BRCA2 nonsynonymous SNV (c.A4376G) | Recurrence after 15 mo | - |
| EOC5 | 59 | IIIC | High-grade serous adenocarcinoma | - | No recurrence for 27 mo | - |
| EOC6 | 44 | IIIC | High-grade serous adenocarcinoma | - | Recurrence after 12 mo | Sister: ovarian cancer |
| EOC7 | 65 | IIIC | Clear cell adenocarcinoma | - | Recurrence after 10 mo | - |
| EOC8 | 46 | IIIC | High-grade serous adenocarcinoma | - | No recurrence after 7 mo | - |
| EOC9 | 61 | IVA | High-grade serous adenocarcinoma | BRCA2 nonsynonymous SNV (c.G6325A) | Expired after 3 mo due to septic shock | - |
| EOC10 | 50 | IIIC | High-grade serous adenocarcinoma | - | No recurrence after 7 mo | Three cycles of neoadjuvant chemotherapy |
FIGO, International Federation of Gynecology and Obstetrics; EOC, epithelial ovarian cancer; SNV, single nucleotide variant.
2. Genomic profiling of EOC from ascites ctDNA, preoperative plasma ctDNA, and tumor DNA
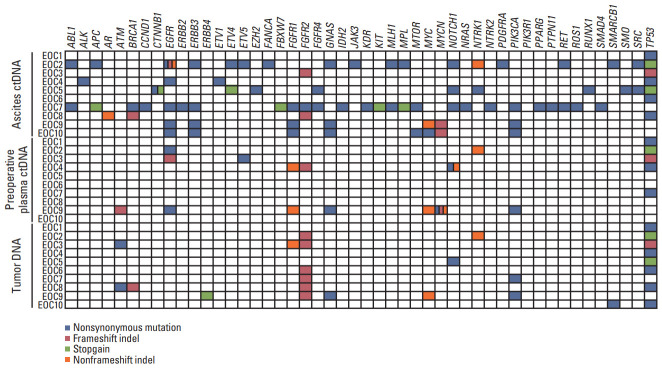
All EOC samples underwent targeted next-generation sequencing (Axen Cancer Panel 1). The median coverage depths for tumor tissues, ascites samples, and plasma samples were 1,232-3,153×, 513-3,820×, and 181-7,937×, respectively (S1 Table). The ascites ctDNA, preoperative ctDNA, and tumor DNA of the 10 EOC patients harbored 81 (range, 1 to 26), 19 (range, 0 to 7), and 26 (range, 1 to 5) genes with mutations, respectively (Fig. 2, S4 Table). Of them, TP53 was the most frequently mutated gene in all of the ascites ctDNA, preoperative ctDNA, and tumor DNA samples. In addition, 15 mutations (somatic and germline mutations) were selected and Sanger sequencing was performed for validation (S3 Fig.).
Fig. 2.
Somatic mutations detected in ascites circulating tumor DNA (ctDNA), preoperative plasma ctDNA, and tumor DNA. Genes with somatic mutations are listed on the x-axis and samples are shown on the y-axis. Nonsynonymous mutations, frameshift indels, stop-gain mutations, and nonframeshift indels are shown in blue, red, green and orange, respectively.
Next, we identified the concordance of somatic mutations among ascites ctDNA, preoperative plasma ctDNA, and tumor DNA. We found that five of the 10 EOC patients harbored at least one gene with a mutation among the three DNA sources (Fig. 3A, S4 Table). Of them, TP53 (n=4) was the most frequently mutated gene, followed by NTRK1 (n=1), PIK3CA (n=1), and MYC (n=1). No somatic mutation was identified in the preoperative ctDNA of three of the 10 EOC patients (EOC5, EOC6, and EOC8) but all of their ascites ctDNA and tumor DNA harbored the TP53 mutation (Table 1).
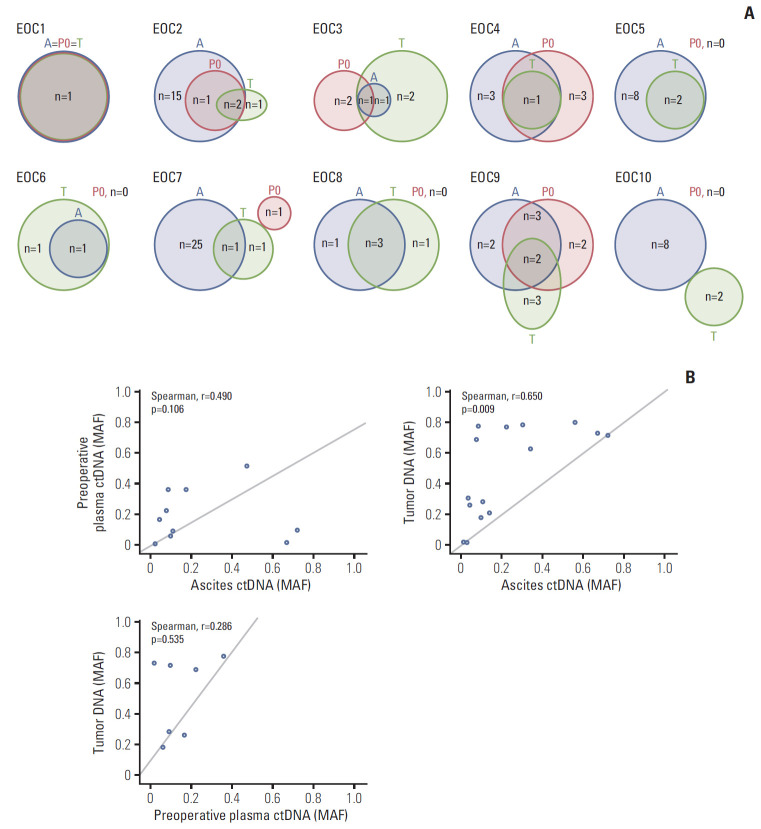
Fig. 3.
Mutational concordance between ascites circulating tumor DNA (ctDNA), preoperative plasma ctDNA, and tumor DNA. (A) Numbers of somatic mutations co-detected in ascites ctDNA, preoperative plasma ctDNA, and tumor DNA are shown in red, blue, and black, respectively. (B) Comparison of mutant allele frequencies of the shared mutations between preoperative plasma ctDNA and ascites ctDNA, tumor DNA and ascites ctDNA, and tumor DNA and preoperative ctDNA. The mutant allele frequencies (MAF) are shown on the x- and y-axes.
Furthermore, the MAFs of the shared mutations between ascites ctDNA and preoperative plasma ctDNA, between ascites ctDNA and tumor DNA, and between preoperative plasma ctDNA and tumor DNA were identified (Fig. 3B). Spearman’s correlation analysis revealed a strong and significant positive correlation between ascites ctDNA and tumor DNA (Spearman r=0.650, p=0.009). Although the associations were not significant, moderate and weak positive correlations were found between ascites ctDNA and preoperative plasma ctDNA and between preoperative plasma ctDNA and tumor DNA.
3. Longitudinal monitoring of somatic mutations in plasma ctDNA and CA125
We performed serial monitoring of the detected somatic mutations in 33 plasma samples from the 10 EOC patients. All serial plasma samples of patients without preoperative ctDNA mutation identified before debulking surgery were also sequenced to track the potential emergence of mutations (S4 Table). For the six patients with identified preoperative ctDNA mutations before debulking surgery, the subsequent monitoring of MAF in consecutive plasma samples was compared with CA125 levels (upper limit of normal ≤ 35 UI/mL) (S1 Table, S5 Fig.). Preoperative and postoperative plasma ctDNA was obtained from all patients. The data showed that the MAF from preoperative plasma ctDNA decreased after debulking surgery, similar to the CA125 level. From six patients, we identified 23 somatic mutations (in 19 genes) in preoperative plasma ctDNA; most of their MAF values (17 of 23) decreased to zero postoperatively, whereas the CA125 level decreased to 20% of the preoperative value, on average. In addition, we found that the MAF of TP53, the most commonly detected gene with mutations, from plasma ctDNA had a strong positive correlation with CA125 (Spearman r=0.658, p=0.001) (Fig. 4).
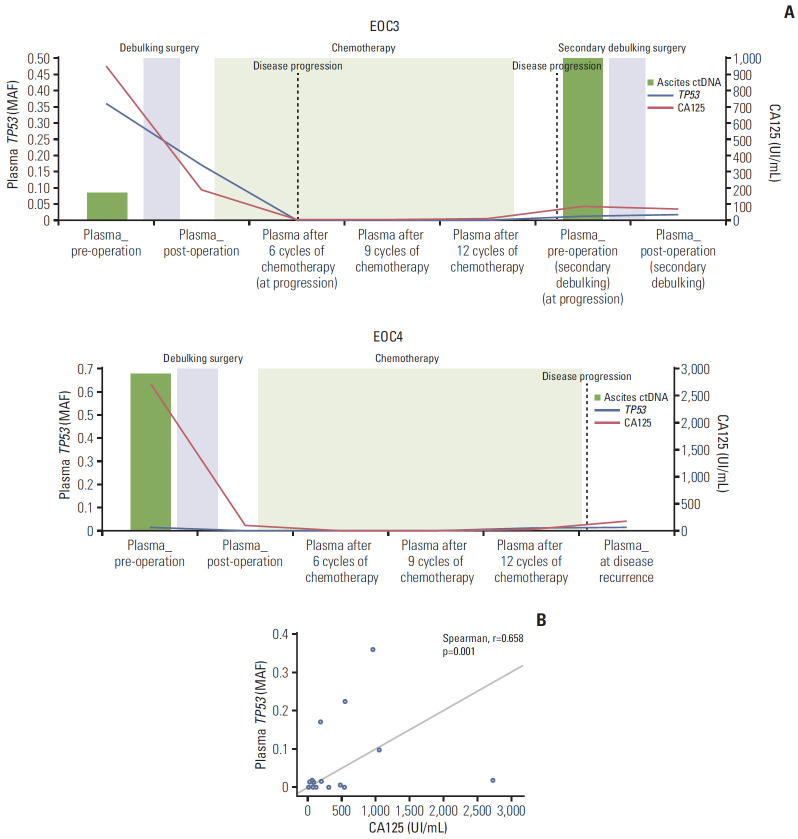
Fig. 4.
Circulating tumor DNA (TP53 mutant allele frequencies [MAF]) from ascites and serial plasma samples and cancer antigen 125 (CA125) kinetics during primary/secondary debulking surgeries and chemotherapy. (A) Epithelial ovarian cancer (EOC) 3 and EOC4. (B) Correlation between TP53 MAF and CA125 in the 10 EOC patients. CA125 levels (UI/mL) are shown on the x-axis, and TP53 MAF are shown on the y-axis.
Plasma ctDNA was obtained after progression/recurrence from two of the 10 EOC patients (EOC3 and EOC4); one underwent secondary debulking surgery, with ascites ctDNA and tumor DNA obtained during the surgery (EOC3).
1) Patient EOC3
A 56-year-old female patient with high-grade serous adenocarcinoma had TP53, EGFR, and ETV5 mutations in preoperative plasma ctDNA with TP53 concurrently detected in tumor DNA and ascites ctDNA from debulking surgery (Fig. 4, S5 Fig.). Chemotherapy was performed after the primary surgery, but computed tomography (CT) imaging revealed that the disease progressed after 5 months. Thus, the patient underwent secondary debulking surgery 17 months after the primary debulking surgery. Preoperative/postoperative plasma ctDNA, ascites ctDNA, and tumor DNA were obtained during the secondary debulking surgery, as with the primary debulking surgery. Particular attention was paid to TP53 mutations, which were detected in all of the plasma ctDNA/ascites ctDNA and tumor DNA. The data showed that the TP53 MAF (p.G113fs) decreased after primary debulking surgery but was re-identified in preoperative/postoperative plasma ctDNA during secondary debulking surgery. The TP53 MAF from plasma showed a similar pattern of an increase compared with CA125 during disease progression. During the secondary debulking surgery, the TP53 mutation was also detected in ascites ctDNA with a higher MAF than during the primary debulking surgery (0.495 and 0.084 during the secondary and primary debulking surgeries, respectively); this mutation was also found in tumor tissue (Fig. 4, S4 Table).
2) Patient EOC4
A 61-year-old female patient with high-grade serous adenocarcinoma had FGFR1, FGFR2, NOTCH1 (c.C6767T and c.4732_4734del), and TP53 mutations in preoperative plasma ctDNA, with TP53 concurrently detected in tumor DNA and ascites ctDNA from debulking surgery (Fig. 4, S5 Fig.). After the surgery, the patient underwent chemotherapy. CT imaging showed that the disease recurred after 17 months (at P5). The TP53 MAF (0.014) was increased at P4 (12 months after the primary debulking surgery) before the diagnosis of recurrence by CT imaging (at P5), but the CA125 (upper limit of normal ≤ 35 UI/mL) was not increased.
Discussion
In this study, we attempted to determine the clinical utility of ctDNA from ascites and serial plasma samples of ovarian cancer using targeted next-generation sequencing. First, we observed that ctDNA mutations from ascites are correlated with DNA mutations from tumor biopsies. Second, our data showed that ctDNA from serial plasma samples is a promising approach to monitor disease progression.
Our data support previous studies indicating that cfDNA from ascites may be clinically useful and provide additional information that is not available from plasma cfDNA [9,10]. Of the 10 EOC patients, nine (90%) contained somatic mutations in both tumor tissues and ascites ctDNA and this mutational concordance was confirmed with correlation analysis (Fig. 3). The remaining patient (EOC10) who did not harbor somatic mutations in either ascites ctDNA or tumor tissues underwent neoadjuvant chemotherapy before the ascites ctDNA was obtained. This finding suggests that ascites sampling through ascites tapping before neoadjuvant chemotherapy may increase the detection rate of somatic mutations in ascites ctDNA.
In the present study, there is strong mutational concordance between ascites and tumor tissues in recurrent/progressive ovarian cancer. One patient (EOC3) underwent secondary debulking surgery due to disease progression, with ascites ctDNA obtained at this time. We found that the ascites ctDNA re-identified the TP53 MAF (p.G113fs) detected in the ascites ctDNA and tumor tissue from the primary debulking surgery and serial plasma samples. Notably, the TP53 MAF was re-identified in the tissue from the secondary debulking surgery. Taken together, the data may suggest that ascites ctDNA can be a valuable tool for identifying overall heterogeneous ovarian cancer genomes as genomes of ascitic cells [6].
TP53 mutations were the most frequently co-occurring genomic alterations among ascites ctDNA, preoperative plasma ctDNA, and tumor DNA from EOC patients. Many studies have shown that TP53 mutations are significantly associated with EOCs because TP53 mutations are identified in 97% of the most aggressive high-grade serous ovarian carcinomas, which account for more than two-thirds of all ovarian cancers [19]. We also found that NTRK1, PIK3CA, and MYC mutations co-occurred among ascites ctDNA, preoperative plasma ctDNA, and tumor DNA from our EOC patients. In a previous study, somatic mutations in the PIK3CA gene were found in 12% of ovarian cancer patients [20] and over 90% of ovarian cancers show mutations in the MYC pathway network, indicating MYC as a potential molecular marker in ovarian cancer [21]. Thus, it is necessary to evaluate mutations in genes other than TP53.
The CA125 serum protein marker is used not only for ovarian cancer screening, but also as a response marker for the disease. Although it is sensitive for the detection of ovarian cancer, it lacks specificity. In addition, because the CA125 level may not change rapidly enough to reflect the effect of the treatment, ctDNA from plasma has been studied as a promising alternative [22,23]. Previous studies demonstrated that ctDNA in plasma from ovarian cancer is a prognostic biomarker and associated with the time to progression [24-26]. Using targeted next-generation sequencing of serial plasma ctDNA, we found that ctDNA from plasma detects ovarian cancer progression/recurrence at a similar time or even more rapidly than the CA125 level (EOC3 and EOC4). TP53 mutations were the most frequently co-occurring genomic alterations among ascites ctDNA, preoperative plasma ctDNA, and tumor DNA in EOC patients. In addition, we found that the TP53 MAF from plasma ctDNA had a strong positive correlation with the CA125 level (Fig. 4). This finding supports the recent evidence suggesting that a combination assay of ctDNA from plasma and a protein marker may become a valuable tool with higher diagnostic sensitivity for ovarian cancer [27].
The present study has several limitations to consider. First, our relatively small sample size, compared to recent targeted next-generation sequencing studies, may cause our study to be underpowered. Although our sample groups are small compared with other studies, we designed our study to provide an accurate initial diagnosis as well as continual monitoring of EOC patients. Second, we only investigated genetic variants in the coding regions of 88 genes. However, previous studies found that variants located in the intronic and non-coding regions of genomes may play an important role in identifying individuals at genetic risk for developing ovarian cancer [28,29]. Thus, this targeted-sequencing study has limitations in identifying genetic variants associated with EOC in non-coding regions. Further studies are required to elucidate our findings.
This is the first study to not only identify the clinical utility of ascites ctDNA, but also its mutational concordance with tumor tissues. We also showed that plasma ctDNA was associated with CA125, a serum protein marker. In this regard, our data strongly suggest that ascites ctDNA might be more suitable than plasma ctDNA for identifying the mutational landscape of ovarian cancer and assisting therapeutic strategy planning. In addition, a combination assay of plasma ctDNA and CA125 could be used to increase the diagnostic sensitivity of ovarian cancer.
Acknowledgments
This research was supported by Incheon National University Research Grant in 2019 and National Research Foundation of Korea (2016R1D1A1B04932862).
Footnotes
Conflict of interest relevant to this article was not reported.
Electronic Supplementary Material
Supplementary materials are available at Cancer Research and Treatment website (https://www.e-crt.org).
Description of obtained samples from ten epithelial ovarian cancer patients
Genes analyzed by Axen Cancer Panel 1
Validation of the mutations by Sanger sequencing. EOC, epithelial ovarian cancer.
Detected mutations of the 10 epithelial ovarian cancer (EOC) patients
Circulating tumor DNA from serial plasma samples and cancer antigen 125 (CA125) kinetics during primary/secondary debulking surgeries and chemotherapy. EOC, epithelial ovarian cancer.
References
- 1.Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68:284–96. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clarke-Pearson DL. Clinical practice. Screening for ovarian cancer. N Engl J Med. 2009;361:170–7. doi: 10.1056/NEJMcp0901926. [DOI] [PubMed] [Google Scholar]
- 3.Jayson GC, Kohn EC, Kitchener HC, Ledermann JA. Ovarian cancer. Lancet. 2014;384:1376–88. doi: 10.1016/S0140-6736(13)62146-7. [DOI] [PubMed] [Google Scholar]
- 4.Smolle E, Taucher V, Haybaeck J. Malignant ascites in ovarian cancer and the role of targeted therapeutics. Anticancer Res. 2014;34:1553–61. [PubMed] [Google Scholar]
- 5.Ahmed N, Stenvers KL. Getting to know ovarian cancer ascites: opportunities for targeted therapy-based translational research. Front Oncol. 2013;3:256. doi: 10.3389/fonc.2013.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi YJ, Rhee JK, Hur SY, Kim MS, Lee SH, Chung YJ, et al. Intraindividual genomic heterogeneity of high-grade serous carcinoma of the ovary and clinical utility of ascitic cancer cells for mutation profiling. J Pathol. 2017;241:57–66. doi: 10.1002/path.4819. [DOI] [PubMed] [Google Scholar]
- 7.Wan JC, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223–38. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 8.Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–12. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 9.Husain H, Nykin D, Bui N, Quan D, Gomez G, Woodward B, et al. Cell-free DNA from ascites and pleural effusions: molecular insights into genomic aberrations and disease biology. Mol Cancer Ther. 2017;16:948–55. doi: 10.1158/1535-7163.MCT-16-0436. [DOI] [PubMed] [Google Scholar]
- 10.Zhou S, Xu B, Qi L, Zhu D, Liu B, Wei J. Next-generation sequencing reveals mutational accordance between cell-free DNA from plasma, malignant pleural effusion and ascites and directs targeted therapy in a gastric cancer patient. Cancer Biol Ther. 2019;20:15–20. doi: 10.1080/15384047.2018.1504720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–90. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997v2.
- 13.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S, Seo J, Park J, Nam JY, Choi A, Ignatius JS, et al. Korean Variant Archive (KOVA): a reference database of genetic variations in the Korean population. Sci Rep. 2017;7:4287. doi: 10.1038/s41598-017-04642-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC. SIFT missense predictions for genomes. Nat Protoc. 2016;11:1–9. doi: 10.1038/nprot.2015.123. [DOI] [PubMed] [Google Scholar]
- 18.Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–7. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Cao L, Nguyen D, Lu H. TP53 mutations in epithelial ovarian cancer. Transl Cancer Res. 2016;5:650–63. doi: 10.21037/tcr.2016.08.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levine DA, Bogomolniy F, Yee CJ, Lash A, Barakat RR, Borgen PI, et al. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin Cancer Res. 2005;11:2875–8. doi: 10.1158/1078-0432.CCR-04-2142. [DOI] [PubMed] [Google Scholar]
- 21.Zeng M, Kwiatkowski NP, Zhang T, Nabet B, Xu M, Liang Y, et al. Targeting MYC dependency in ovarian cancer through inhibition of CDK7 and CDK12/13. Elife. 2018;7:e39030. doi: 10.7554/eLife.39030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lheureux S, Gourley C, Vergote I, Oza AM. Epithelial ovarian cancer. Lancet. 2019;393:1240–53. doi: 10.1016/S0140-6736(18)32552-2. [DOI] [PubMed] [Google Scholar]
- 23.Soletormos G, Duffy MJ, Othman Abu Hassan S, Verheijen RH, Tholander B, Bast RC Jr, et al. Clinical use of cancer biomarkers in epithelial ovarian cancer: updated huidelines from the European group on tumor markers. Int J Gynecol Cancer. 2016;26:43–51. doi: 10.1097/IGC.0000000000000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamat AA, Baldwin M, Urbauer D, Dang D, Han LY, Godwin A, et al. Plasma cell-free DNA in ovarian cancer: an independent prognostic biomarker. Cancer. 2010;116:1918–25. doi: 10.1002/cncr.24997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giannopoulou L, Kasimir-Bauer S, Lianidou ES. Liquid biopsy in ovarian cancer: recent advances on circulating tumor cells and circulating tumor DNA. Clin Chem Lab Med. 2018;56:186–97. doi: 10.1515/cclm-2017-0019. [DOI] [PubMed] [Google Scholar]
- 26.Parkinson CA, Gale D, Piskorz AM, Biggs H, Hodgkin C, Addley H, et al. Exploratory analysis of TP53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: a retrospective study. PLoS Med. 2016;13:e1002198. doi: 10.1371/journal.pmed.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–30. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X, Paranjape T, Stahlhut C, McVeigh T, Keane F, Nallur S, et al. Targeted resequencing of the microRNAome and 3'UTRome reveals functional germline DNA variants with altered prevalence in epithelial ovarian cancer. Oncogene. 2015;34:2125–37. doi: 10.1038/onc.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans DGR, van Veen EM, Byers HJ, Wallace AJ, Ellingford JM, Beaman G, et al. A dominantly inherited 5' UTR variant causing methylation-associated silencing of BRCA1 as a cause of breast and ovarian cancer. Am J Hum Genet. 2018;103:213–20. doi: 10.1016/j.ajhg.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of obtained samples from ten epithelial ovarian cancer patients
Genes analyzed by Axen Cancer Panel 1
Validation of the mutations by Sanger sequencing. EOC, epithelial ovarian cancer.
Detected mutations of the 10 epithelial ovarian cancer (EOC) patients
Circulating tumor DNA from serial plasma samples and cancer antigen 125 (CA125) kinetics during primary/secondary debulking surgeries and chemotherapy. EOC, epithelial ovarian cancer.