Abstract
Coronavirus disease 2019 (COVID-19) emerged in December 2019 in Wuhan, China; it has since caused a pandemic, with more than 10,000 confirmed cases (> 800,000 tests) in Korea as of May 2020. Real-time reverse transcription polymerase chain reaction (RT-PCR) is currently the most commonly used method for the diagnosis of COVID-19 worldwide. The Korean Society for Laboratory Medicine and Korea Centers for Disease Prevention and Control regularly update the guidelines for COVID-19 diagnosis. Emergency use authorization for some laboratory diagnostic kits has been granted, enabling the timely diagnosis and treatment of COVID-19, and the isolation of infected patients. Due to the collective efforts of the government, medical professionals, local authorities, and the public, Korea’s response to the COVID-19 outbreak has been accepted widely as a model. Here, we summarize the currently available laboratory tests for COVID-19 diagnosis. Although RT-PCR tests are used widely to confirm COVID-19, antibody tests could provide information about immune responses to the virus.
Keywords: COVID-19, Diagnosis, Real-time reverse transcription polymerase chain reaction, Coronavirus, Serology
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), is a large, enveloped, single-strand ribonucleic acid (RNA) genome virus consisting of four structural proteins: nucleocapsid, spike, envelope, and membrane proteins. The receptor-binding domain of the spike protein binds to human and bat angiotensin-converting enzyme 2 receptors, mediating virus entry into host cells [1]. The World Health Organization (WHO) named the novel respiratory disease caused by SARS-CoV-2 to COVID-19 on February 11, 2020; it declared that this disease had caused a public health emergency of international concern on January 30, 2020, and finally, it was a pandemic on March 11, 2020.
Based on previous experience with Middle East Respiratory Syndrome, Korea responded promptly to the COVID-19 outbreak. The Korea Centers for Disease Control and Prevention (KCDC) and the Ministry of Food and Drug Safety (MFDS) of Korea developed diagnostic kits for COVID-19, and emergency use authorization (EUA) for these kits was granted. The timely distribution of these diagnostic tests to medical institutions enabled early detection and self-isolation of confirmed COVID-19 cases. Based on cooperation among the government, local authorities, and the public, effective contact tracing and quarantine policies were established in the early stage of the disease outbreak.
Clinical laboratories and in vitro diagnostic tests played crucial roles in this health crisis, providing for largescale patient screening, diagnosis and monitoring, as well as epidemiological surveillance (Table 1) [2]. In this article, we provide an overview of laboratory tests used for COVID-19 diagnosis and summarize current practices for laboratory data interpretation.
Table 1.
The laboratory tests used most commonly to monitor patients with coronavirus disease 2019
| Role | Tests |
|---|---|
| Diagnosis | RT-PCR (gold standard), serologic tests |
| Staging, prognostication, therapeutic monitoring | Various in vitro diagnostic tests (↑WBC, ↑neutrophil counts, ↓lymphocytes count, ↓platelet count, ↓albumin, ↑LDH, ↑AST & ALT, ↑total bilirubin, ↑urea & creatinine, ↑cardiac troponin, ↑D-dimer & prothrombin time, ↑procalcitonin & CRP, ↑ferritin, ↑cytokines |
| Epidemiological surveillance | SARS-CoV-2 antibodies |
↑, increased; ↓, decreased.
RT-PCR, real-time reverse transcription polymerase chain reaction; WBC, white blood cell; LDH, lactate dehydrogenase; AST, aspartate transaminase; ALT, alanine transaminase; CRP, C-reactive protein; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
CASE CLASSIFICATION FOR LABORATORY TESTING
In Korea, COVID-19 case definition is currently based on the 8th (May 15, 2020) edition of the KCDC’s Guidelines on Response to Coronavirus Disease 2019. According to these guidelines, cases are classified as confirmed, suspected, and patient under investigation (PUI). The definitions of suspected and PUI cases are revised regularly based on the incidence of confirmed cases, results of epidemiological studies, and extent of the epidemic [3,4]. In the United States, according to the Centers for Disease Control and Prevention guidelines, cases are classified as high priority and priority (Table 2) [5].
Table 2.
Coronavirus disease 2019 case classification in Korea and the United States
| Korea (KCDC) | United States (CDC) | |||
|---|---|---|---|---|
| Diagnostic test | Nucleic acid (RT-PCR) | Nucleic acid or antigen | ||
| Case definition | Suspected case | High priority | ||
| Cases with clinical symptom of COVID-19 (major symptoms: fever, cough, shortness of breath, chills, muscle pain, headache, sore throat, loss of smell or taste or pneumonia, etc.) within 14 days of being in close contact with a confirmed case. | Hospitalized patients with symptoms | |||
| Healthcare facility workers, workers in congregate living settings, and first responders with symptoms | ||||
| Residents in long-term care facilities or other congregate living settings, including prisons and shelters, with symptoms | ||||
| PUI (patient under investigation) | Priority | |||
| Cases suspected of having a symptom of COVID-19 based on a physician’s opinion (e.g., pneumonia of unknown origin). | Persons with symptoms of potential COVID-19 infection, including: fever, cough, shortness of breath, chills, muscle pain, new loss of taste or smell, vomiting or diarrhea, and/or sore throat. | |||
| Cases with history of visiting overseas and having a symptom of COVID-19 within 14 days after return to homeland | Persons without symptoms who are prioritized by health departments or clinicians, for any reason, including but not limited to: public health monitoring, sentinel surveillance, or screening of other asymptomatic individuals according to state and local plans. | |||
| Cases showing epidemiological correlation with the domestic mass outbreak of COVID-19 and exhibiting a symptom of COVID-19 within 14 days. | ||||
KCDC, Korea Centers for Disease Control and Prevention; CDC, Centers for Disease Control and Prevention; RT-PCR, real-time reverse transcription polymerase chain reaction; COVID-19, coronavirus disease 2019.
SPECIMENS USED FOR TESTING
Specimen types
For real-time reverse transcription polymerase chain reaction (RT-PCR) tests, upper respiratory specimens (including nasopharyngeal and oropharyngeal swabs) and lower respiratory specimens (including sputum, bronchoalveolar lavage [BAL] specimens, and tracheal aspirates) are used. The use of induced sputum specimens for RT-PCR testing is not recommended. When necessary, additional specimens, such as blood, urine, and feces, may be collected; however, the diagnostic value of these specimens remains unclear. For serological tests, whole blood and serum samples are used.
Specimen selection according to patient status
For asymptomatic individuals and those with mild symptoms, both nasopharyngeal and oropharyngeal swabs are recommended. When collecting only one specimen, a nasopharyngeal swab is recommended first. Although sputum can also be collected for examination, sputum induction is not recommended for these patients. For patients with severe symptoms, productive cough, and intubation, lower respiratory specimens, such as sputum, BAL specimens, and tracheal aspirates, are collected. When possible, nasopharyngeal and oropharyngeal swabs should also be tested. For patients referred for additional testing, including those with pneumonia whose nasopharyngeal and/or oropharyngeal swab tests yielded negative results, lower respiratory specimens, such as sputum, BAL specimens, and tracheal aspirates, should be collected.
CONFIRMATORY TESTING BY RT-PCR
RT-PCR procedures
For SARS-CoV-2 detection using RT-PCR, the following steps are required: sample collection, sample preparation, nucleic acid extraction, reverse transcription, and PCR using real-time fluorescence signal detection. Currently, RT-PCR is the gold-standard laboratory test for COVID-19 confirmation in suspected cases. RT-PCR detects the nucleic acid of the virus with high specificity and sensitivity.
Sometimes it can be used to quantify the target gene amount. It is a similar method to PCR, but the amplified nucleic acid is measured by real-time monitoring fluorescence signal RT-PCR yields cycle threshold (Ct) values, which are inversely proportional to the amounts of the target nucleic acid in samples (Fig. 1). RT-PCR tests should comply with the standard operating procedures of each laboratory and the manufacturers’ instructions. Currently, clinical laboratories in Korea are using various nucleic acid extraction methods and reagents for RT-PCR with EUA for COVID-19 diagnosis (Table 3).
Figure 1.
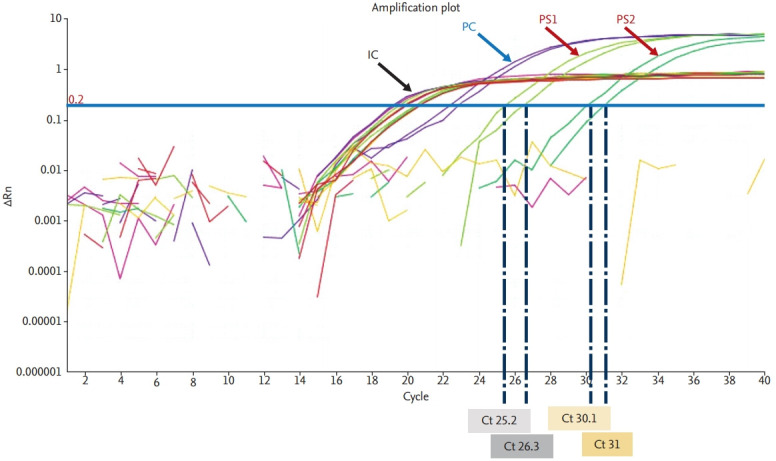
Coronavirus disease 2019 diagnosis using real-time reverse transcription polymerase chain reaction. IC, internal control; PC, positive control; PS, positive specimen; Rn, normalized reporter (normalized fluorescence signal of reporter dye); Ct, cycle threshold.
Table 3.
Target genes of seven RT-PCR kits currently used for SARS-CoV-2 detection in Korea (as of May 29, 2020)
| Approval date | Kit name (manufacturer) | Target genes |
|---|---|---|
| Feb 4, 2020 | PowerCheck 2019-nCoV real-time PCR kit (Kogene biotech) | RdRp, E |
| Feb 12, 2020 | Allplex 2019-nCoV Assay (Seegene) | RdRp, E, N |
| Feb 27, 2020 | DiaPlexQ Novel Coronavirus (2019-nCoV) Detection Kit (Solgent) | ORF1a, N |
| STANDARD M n-CoV Real-Time Detection Kit (SD biosensors) | RdRp, E | |
| Mar 13, 2020 | Real-Q 2019-nCoV Detection Kit (BioSewoom) | RdRp, E |
| May 11, 2020 | BioCore 2019-nCoV Real Time PCR Kit (BioCore) | RdRp, N |
| May 29, 2020 | careGENE N-CoV RT-PCR Kit (Wells Bio) | RdRp, E |
RT-PCR, real-time reverse transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Interpretation of RT-PCR results
For RT-PCR–based viral nucleic acid detection methods, Ct values of the target genes below the cut-off Ct value indicate positivity and Ct values above the cut-off Ct value indicate negativity. Positive and negative controls are included in each reaction to assess method performance and the presence of contaminants (Fig. 1). In specimens with low viral loads, Ct values can be close to the cut-off and may yield false-negative or false-positive results. Thus, RT-PCR results should be interpreted carefully, with consideration of patients’ histories and clinical symptoms. In some cases, retesting should be performed using residual or newly collected specimens. All EUA kits currently available in Korea can detect two or three viral genes (Table 3). It is recommended that samples are considered to be positive only when all target genes are detected. When only one gene is detected, retesting or consultation with the reference laboratory is recommended.
The optimal timing of viral genome detection in COVID-19 cases remains unknown. COVID-19 cannot be ruled out safely based on one negative result from the testing of upper respiratory specimens alone. Thus, highly suspicious COVID-19 cases with negative results from single upper respiratory specimens, should be retested using additional upper respiratory specimens or lower respiratory specimens. False-negative results could be obtained for samples with low viral loads and those collected too early or too late during the clinical course of the disease. Mutations in the viral genome, the presence of PCR inhibitors, the receipt of antiviral therapy prior to testing, and poor specimen quality could also lead to false-negative results. Thus, for patients with negative results from upper respiratory specimens, lower respiratory specimens should be tested. Positive and negative control samples should be tested simultaneously with patient specimens, and internal controls should be included in all reactions to monitor the performance of PCR. For highly suspicious cases that repeatedly yield negative test results, specimens should be submitted to a reference laboratory for further confirmatory testing.
Concerns regarding RT-PCR testing
To ensure infection control during a COVID-19 outbreak, a strict sample transportation protocol should be implemented. All specimens must be considered to be potentially infectious, and appropriate personal protective equipment must be used during specimen collection [6]. The molecular diagnosis of COVID-19 typically begins with the collection of a nasopharyngeal or an oropharyngeal swab and its transport to a laboratory in a universal/viral transport medium. According to a recent report on the SARS-CoV-2 detection rate in 1,070 clinical specimens collected from 205 patients with COVID-19, the positivity rate is highest for BAL fluid (14 of 15; 93%), followed by sputum (72 of 104; 72%), nasal swabs (5 of 8; 63%), fibrobronchoscope brush biopsy (6 of 13; 46%), pharyngeal swabs (126 of 398; 32%), feces (44 of 153; 29%), and blood (3 of 307; 1%). None of the 72 urine specimens was found to be positive [7]. The exact sensitivity and specificity of RT-PCR tests for COVID-19 are unknown, but a positive test result appears to be strongly suggestive of true COVID-19 whereas a negative result does not rule out COVID-19 [8]. Hence, multiple specimens should be tested in highly suspicious cases.
Potential RT-PCR test pitfalls include pre-analytical errors, such as inappropriate identification, collection, handling, transport, and storage of samples, as well as the collection of inappropriate or inadequate material. Additionally, the presence of interfering substances, manual errors, sample contamination, and patients’ receipt of antiretroviral therapies can further interfere with the accuracy of RT-PCT tests [9]. Furthermore, most RT-PCR tests have relatively long turnaround times, ranging from 4 to 6 hours from specimen preparation to RT-PCR; thus, further improvement is required such as shorten TAT for testing in the current COVID-19 outbreak [10].
In Korea, as of May 15, 2020, 447 (approximately 4.5%) cases reported reactivated or positive result after recovered COVID-19. It has been implementing measures equivalent to confirmed patients since April 14, but an epidemiological investigation into the case of reactivated case has not confirmed the evidence that the cases of positive result after recovery are infectious. Current guidelines regarding hospital discharge and the discontinuation of quarantine may need to be reevaluated [11]. Although the presence of residual viral nucleic acid after recovery is likely, more research is needed because it shows various progress in COVID-19 patients.
ALTERNATIVE TESTS FOR COVID-19
Antibody detection using serological tests
The urgent needs for the standard of procedures of sampling from different anatomic sites, sample transportation, optimization of RT‐PCR, serology diagnosis/screening for SARS‐CoV‐2 infection, and distinct diagnosis from other respiratory diseases such as influenza infections as well [12]. A wide range of immunoassays for the detection of SARS-CoV-2 proteins and antibodies in serum and plasma has been developed. Using chemiluminescent immunoassay (CLIA), manual enzyme-linked immunosorbent assay (ELISA), and fast lateral flow immunoassay (LFIA) complementing molecular diagnosis using RT-PCR [13].
Antibodies against SARS-CoV-2 can be detected in the middle and late stages of the disease [14]. Immunoglobulin M (IgM) and IgA antibodies were detected 3 to 6 days (median 5 days) after the onset of symptoms, whereas IgG antibodies were detected 10 to 18 days (median 14 days) after symptom development, with positivity rates of 85.4%, (IgM and IgA 92.7%, and IgG 77.9%, respectively) [15]. IgG and IgM seroconversion occurred simultaneously or sequentially, with the plateauing of titers within 6 days thereafter [16,17]. Neutralizing antibodies (NAbs) play essential roles in protection against viral diseases and viral clearance. Virus-specific NAbs, induced through natural infection or vaccination, can block viral infection. The efficacy of passive antibody therapy is associated with the NAb concentration in the plasma of recovered donors. As the COVID-19 pandemic proceeds, the transfusion of convalescent plasma or serum from recovered patients has been considered as a promising approach to prevent infection and treat the disease [18].
The Food and Drug Administration has granted EUA for a serological test that is intended for use by clinical laboratories. This test might aid determination of whether individuals have immunity to the virus and identify those who could donate plasma for the treatment of seriously ill patients with COVID-19 [19]. Test results can be obtained in 15 to 20 minutes [20,21]. Numerous LFIA-based rapid point-of-care (POC) tests have been developed by several companies; they enable the detection of IgM and IgG antibodies produced in response to SARS-COV-2 infection. Updates on the Foundation for Innovative New Diagnostics (FIND) independent evaluation of SARS-CoV-2 immunoassays are provided regularly; five currently available antigen-detection rapid diagnostic tests (RDTs), 26 serological-antibody-detection RDTs, and eight serological ELISA/automated immunoassays are currently available [22].
Although the usefulness of serological tests remains under debate, these can clearly aid the identification of asymptomatic patients with COVID-19 who may transmit the virus. They can also be used to determine the seroprevalence in different populations, assess previous exposure, and contact trace. Moreover, serology can be used to diagnose COVID-19 in the late stages of the disease, even in viral RNA-negative patients. Serological RDTs could be used to complement currently used IVD assays, improving COVID-19 diagnosis and providing additional information about immune status in suspected cases [23]. However, a recent study showed that IgM and IgG could be detected only about 2 weeks after the onset of infection [24], and the overall testing sensitivity and specificity have been found to be 88.66% and 90.63%, respectively [23]. Although the rapid acquisition of results could reduce the lengths of emergency department stays, test performance should not be compromised to achieve this goal [25]. Current WHO guidelines recommend the use of new POC immunodiagnostic tests only in research settings until evidence supporting their clinical use becomes available [26]. Although RT-PCR testing of respiratory samples is currently the gold standard for COVID-19 diagnosis, molecular testing is time consuming and requires experienced professionals, increasing the need for the development of novel rapid diagnostics. Rapid LFIA and automated CLIA tests for IgM and IgG are believed to be promising emerging methods; however, their clinical performance needs to be evaluated thoroughly [27].
As the COVID-19 outbreak begins to subside, serological testing will enable the identification of individuals with immunity against SARS-CoV-2, who thus might be able to return to work [28].
Antigen detection
RDTs detect the presence of viral proteins (antigens) in respiratory specimens. The accurate detection of SARS-CoV-2 is essential to curb the global spread of COVID-19. Nevertheless, currently available RT-PCR–based diagnostic assays are not robust as they are still missing several infected cases [6,8,9]. Additionally, they are not widely available in developing countries and remote locations, which lack well-equipped laboratories and experienced professionals. The MFDS of Korea has approved the export of rapid IVD tests; however, their use is strictly regulated.
Nasopharyngeal and oropharyngeal swabs are commonly used for RDTs. However, the collection of these specimens requires close contact with patients, which entails the risk of virus transmission to healthcare workers. In contrast, saliva specimens can be obtained easily and noninvasively as sputum samples, minimizing the risk of SARS-CoV-2 exposure for healthcare workers. Hence, saliva is considered to be a promising, noninvasively collected specimen type for COVID-19 diagnosis, monitoring, and infection control [29].
Other tests for the monitoring of patients with COVID-19
SARS-CoV-2 infection can cause mild to severe respiratory, enteric, cardiovascular, and neurological symptoms. The laboratory tests used most commonly to monitor patients with COVID-19 are listed in Table 1. The serum levels of certain markers can be used to predict the course of the disease and patient prognosis [30-32]. Careful monitoring of the myocardial enzyme profiles is of great importance in reducing the complications and mortality in patients with COVID‐19 [33].
CONCLUSIONS
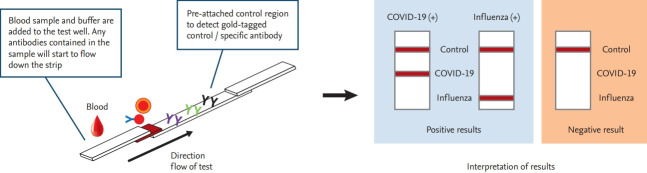
RT-PCR testing has played a vital role in COVID-19 diagnosis and infection control. New, more accurate, and rapid RT-PCR tests are expected to become the gold standard for SARS-CoV-2 detection in the near future. Furthermore, serological tests could complement currently used RT-PCR tests, and can be used to identify individuals who have developed immunity against SARS-CoV-2. As the need for rapid testing is increasing continuously, and with consideration of the possibility that COVID-19 may become seasonal, similar to influenza, diagnostic kits that can differentiate the two diseases are urgently needed (Fig. 2).
Figure 2.
Diagram of a proposed point-of-care testing procedure for coronavirus disease 2019 (COVID-19) and influenza diagnosis.
Footnotes
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Tai W, He L, Zhang X, et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Version 2. Cell Mol Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lippi G, Plebani M. The critical role of laboratory medicine during coronavirus disease 2019 (COVID-19) and other viral outbreaks. Clin Chem Lab Med. doi: 10.1515/cclm-2020-0240. 2020 Mar 19 [Epub]. https://doi.org/10.1515/cclm-2020-0240. [DOI] [PubMed] [Google Scholar]
- 3.Korea Centers for Disease Control and Prevention . Guidelines in Response to Coronavirus Disease 2019. 8th ed for local government. Cheongju (KR): KCDC; 2020. [Google Scholar]
- 4.Hong KH, Lee SW, Kim TS, et al. Guidelines for laboratory diagnosis of coronavirus disease 2019 (COVID-19) in Korea. Ann Lab Med. 2020;40:351–360. doi: 10.3343/alm.2020.40.5.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention . Atlanta (GA): CDC; 2020. Evaluating testing persons for coronavirus disease 2019 (COVID-19): priorities for COVID-19 testing [Internet] [cited 2020 Jun 16]. Available from: http://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-criteria.html. [Google Scholar]
- 6.Tan SS, Yan B, Saw S, et al. Practical laboratory considerations amidst the COVID-19 outbreak: early experience from Singapore. J Clin Pathol. doi: 10.1136/jclinpath-2020-206563. 2020 Mar 20 [Epub]. [DOI] [PubMed] [Google Scholar]
- 7.Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zitek T. The Appropriate use of testing for COVID-19. West J Emerg Med. 2020;21:470–472. doi: 10.5811/westjem.2020.4.47370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lippi G, Simundic AM, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19) Clin Chem Lab Med. doi: 10.1515/cclm2020-0285. 2020 Mar 16 [Epub]. [DOI] [PubMed] [Google Scholar]
- 10.Kim YJ, Sung H, Ki CS, Hur M. COVID-19 testing in South Korea: current status and the need for faster diagnostics. Ann Lab Med. 2020;40:349–350. doi: 10.3343/alm.2020.40.5.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lan L, Xu D, Ye G, et al. Positive RT-PCR test results in patients recovered from COVID-19. JAMA. 2020;323:1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Yao L, Li J, et al. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J Med Virol. 2020;92:903–908. doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Padoan A, Cosma C, Sciacovelli L, Faggian D, Plebani M. Analytical performances of a chemiluminescence immunoassay for SARS-CoV-2 IgM/IgG and antibody kinetics. Clin Chem Lab Med. doi: 10.1515/cclm-2020-0443. 2020 Apr 16 [Epub]. [DOI] [PubMed] [Google Scholar]
- 14.Xiang F, Wang X, He X, et al. Antibody detection and dynamic characteristics in patients with COVID-19. Clin Infect Dis. doi: 10.1093/cid/ciaa461. 2020 Apr 19 [Epub]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo L, Ren L, Yang S, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin Infect Dis. doi: 10.1093/cid/ciaa310. 2020 Mar 21 [Epub]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. doi: 10.1038/s41591-020-0897-1. 2020 Apr 29 [Epub]. [DOI] [PubMed] [Google Scholar]
- 17.Acarturk TO. Treatment of large ischial ulcers communicating with the hip joint with proximal femoral resection and reconstruction with a combined vastus lateralis, vastus intermedius and rectus femoris musculocutaneous flap. J Plast Reconstr Aesthet Surg. 2009;62:1497–1502. doi: 10.1016/j.bjps.2008.04.063. [DOI] [PubMed] [Google Scholar]
- 18.Casadevall A, Pirofski LA. The convalescent sera option for containing COVID-19. J Clin Invest. 2020;130:1545–1548. doi: 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han G, Zhou YH. Possibly critical role of wearing masks in general population in controlling COVID-19. J Med Virol. doi: 10.1002/jmv.25886. 2020 Apr 15 [Epub]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maggi F, Pistello M, Antonelli G. Future management of viral diseases: role of new technologies and new approaches in microbial interactions. Clin Microbiol Infect. 2019;25:136–141. doi: 10.1016/j.cmi.2018.11.015. [DOI] [PubMed] [Google Scholar]
- 21.Sultana S, Alzahrani N, Alzahrani R, et al. Stability issues and approaches to stabilised nanoparticles based drug delivery system. J Drug Target. 2020;28:468–486. doi: 10.1080/1061186X.2020.1722137. [DOI] [PubMed] [Google Scholar]
- 22.Find Because Diagnosis Matters et al. Geneva (CH): FIND Because Diagnosis Matters; COVID-19 diagnostics resource centre [Internet] c2019 [cited 2020 Jun 16]. Available from: https://www.finddx.org/covid-19/sarscov2-eval-immuno. [Google Scholar]
- 23.Li Z, Yi Y, Luo X, et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. doi: 10.1002/jmv.25727. 2020 Feb 27 [Epub]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dohla M, Boesecke C, Schulte B, et al. Rapid point-of-care testing for SARS-CoV-2 in a community screening setting shows low sensitivity. Public Health. 2020;182:170–172. doi: 10.1016/j.puhe.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cassaniti I, Novazzi F, Giardina F, et al. Performance of VivaDiag COVID-19 IgM/IgG rapid test is inadequate for diagnosis of COVID-19 in acute patients referring to emergency room department. J Med Virol. doi: 10.1002/jmv.25800. 2020 Mar 30 [Epub]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.World Health Organization . Geneva (CH): WHO; 2020. Advice on the use of pointof-care immunodiagnostic tests for COVID-19 [Internet] [cited 2020 Jun 16]. Available from: https://apps.who.int/iris/handle/10665/331713. [Google Scholar]
- 27.Vashist SK. In vitro diagnostic assays for COVID-19: recent advances and emerging trends. Diagnostics (Basel) 2020;10:202. doi: 10.3390/diagnostics10040202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu H, Stratton CW, Tang YW. An evolving approach to the laboratory assessment of COVID-19. J Med Virol. doi: 10.1002/jmv.25954. 2020 Apr 29 [Epub]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.To KK, Tsang OT, Chik-Yan Yip C, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. doi: 10.1093/cid/ciaa149. 2020 Feb 12 [Epub]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mardani R, Ahmadi Vasmehjani A, Zali F, et al. Laboratory parameters in detection of COVID-19 patients with positive RT-PCR; a diagnostic accuracy study. Arch Acad Emerg Med. 2020;8:e43. [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Y, Li T, Han M, et al. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. J Med Virol. 2020;92:791–796. doi: 10.1002/jmv.25770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frater JL, Zini G, d’Onofrio G, Rogers HJ. COVID-19 and the clinical hematology laboratory. Int J Lab Hematol. doi: 10.1111/ijlh.13229. 2020 Apr 20 [Epub]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han H, Xie L, Liu R, et al. Analysis of heart injury laboratory parameters in 273 COVID-19 patients in one hospital in Wuhan, China. J Med Virol. 2020;92:819–823. doi: 10.1002/jmv.25809. [DOI] [PMC free article] [PubMed] [Google Scholar]