Abstract
Alternative polyadenylation (APA) determines stability, localization and translation potential of the majority of mRNA in eukaryotic cells. The heterodimeric mammalian cleavage factor II (CF IIm) is required for pre-mRNA 3′ end cleavage and is composed of the RNA kinase hClp1 and the termination factor hPcf11; the latter protein binds to RNA and the RNA polymerase II carboxy-terminal domain. Here, we used siRNA mediated knockdown and poly(A) targeted RNA sequencing to analyze the role of CF IIm in gene expression and APA in estrogen receptor positive MCF7 breast cancer cells. Identified gene ontology terms link CF IIm function to regulation of growth factor activity, protein heterodimerization and the cell cycle. An overlapping requirement for hClp1 and hPcf11 suggested that CF IIm protein complex was involved in the selection of proximal poly(A) sites. In addition to APA shifts within 3′ untranslated regions (3′-UTRs), we observed shifts from promoter proximal regions to the 3′-UTR facilitating synthesis of full-length mRNAs. Moreover, we show that several truncated mRNAs that resulted from APA within introns in MCF7 cells cosedimented with ribosomal components in an EDTA sensitive manner suggesting that those are translated into protein. We propose that CF IIm contributes to the regulation of mRNA function in breast cancer.
Keywords: gene expression, pre-mRNA 3′ end formation, alternative polyadenylation, breast cancer
INTRODUCTION
Pre-mRNA 3′ end formation is an essential step in the expression of eukaryotic genes. Primary transcripts undergo endonucleolytic cleavage at the poly(A) site followed by addition of a polyadenylate tail to the upstream cleavage product. The core protein machinery responsible for the implementation of these coupled reactions is highly conserved throughout evolution and in human cells includes activities associated with four major multiprotein complexes: cleavage factor I (CF Im), cleavage factor II (CF IIm), cleavage and specificity factor (CPSF) and cleavage stimulation factor (CstF) (Shi et al. 2009). Poly(A) sites are recognized through RNA binding proteins that bind signal sequences located upstream and downstream from a poly(A) site (Zhao et al. 1999; Venkataraman et al. 2005; Tian and Graber 2012; Shi and Manley 2015). 3′ end factors physically interact with the RNA polymerase II (RNAP II) carboxy-terminal domain (CTD) (McCracken et al. 1997; Meinhart and Cramer 2004) and are thought to bind their target sequences cotranscriptionally when the nascent transcript emerges from the polymerase enzyme (Bentley 2014).
Most pre-mRNAs contain multiple poly(A) sites and their differential usage has a profound impact on the function of the derived mature mRNA (Tian and Manley 2017; Turner et al. 2018). Alternative polyadenylation is most frequently observed in the 3′-UTR where usage of proximal and distal poly(A) signals generates short and long 3′-UTRs, respectively. Major determinants of poly(A) site usage are the sequence context of a given poly(A) signal (Gruber et al. 2012; Martin et al. 2012; Li et al. 2015) and the concentration of 3′end factors that bind to the RNA sequence elements (Takagaki et al. 1996). Moreover, the passage of RNAP II along the chromatin template impacts on APA in that pausing at stable nucleosomes promotes the usage of promoter proximal poly(A) sites in a process, which is antagonized by the U1 snRNP (Chiu et al. 2018).
The length of the 3′-UTR is thought to be an important determinant of mRNA function, since it harbors regulatory sequences that are targeted by miRNAs and RNA binding proteins (Tian and Manley 2017; Turner et al. 2018). On the one hand, it has been suggested that enhanced regulation of gene expression during embryonic development is associated with longer 3′-UTRs (Ji et al. 2009). On the other hand, the evasion of regulatory circuits associated with short 3′-UTRs has been correlated with cell proliferation (Sandberg et al. 2008) and transformed cancer cell phenotypes (Mayr and Bartel 2009; Masamha et al. 2014; Xia et al. 2014). Nevertheless, the physiological significance of 3′-UTR length remains unclear (Neve and Furger 2014). For example, 3′-UTR length has only limited influence on mRNA levels and protein synthesis in fibroblasts (Spies et al. 2013) and it does not correlate with protein levels in proliferating T cells (Gruber et al. 2014). 3′-UTR plasticity has been observed in colorectal cancer where both shortening and lengthening of 3′-UTRs has been associated with cancer progression (Morris et al. 2012). In breast cancer, 3′-UTR shortening was observed for estrogen receptor positive MCF7 cells, while estrogen negative and highly invasive MDA-MB-231 cells displayed 3′-UTR lengthening (Fu et al. 2011).
The CF IIm complex was initially isolated from HeLa cell nuclear extract and highly enriched fractions were found to include hClp1 and hPcf11 proteins (de Vries et al. 2000). Reconstitution of the hClp1/hPcf11 heterodimer demonstrated that it indeed equates to CF IIm, since it was sufficient to complement 3′ end cleavage reactions in vitro (Schafer et al. 2018). While the exact role of CF IIm in 3′ end formation remains unclear, it may contribute to poly(A) signal recognition via a nonspecific RNA binding activity (Schafer et al. 2018).
Pcf11 homologs carry conserved CTD interaction domains (Barilla et al. 2001; Licatalosi et al. 2002; Sadowski et al. 2003; Meinhart and Cramer 2004; Noble et al. 2005), bind to RNA (Zhang et al. 2005; Hollingworth et al. 2006; Guegueniat et al. 2017; Schafer et al. 2018) and couple 3′ end formation to RNAP II transcription termination (Grzechnik et al. 2015; Proudfoot 2016). Bridging of both nascent RNA and of the RNAP II CTD by Pcf11 was found to result in dismantling of otherwise highly stable elongation complexes (Zhang et al. 2005; Zhang and Gilmour 2006). Recent analyses of vertebrate PCF11 identified roles in termination of closely spaced full-length genes (Kamieniarz-Gdula et al. 2019), in premature promoter proximal termination (Kamieniarz-Gdula et al. 2019) and the expression of long genes through intronic polyadenylation (Wang et al. 2019). Moreover, hPCF11 was identified as a key factor in defining the APA profiles in neuroblastoma and its down-regulation was associated with spontaneous tumor regression (Ogorodnikov et al. 2018).
Clp1 homologs carry evolutionary conserved ATP binding motifs and mammalian and archaeal proteins exhibit 5′ RNA kinase activity (Weitzer and Martinez 2007; Jain and Shuman 2009), which is required for tRNA splicing and siRNA silencing (Weitzer and Martinez 2007), as well as miRNA activity (Salzman et al. 2016). Loss of hClp1 function results in accumulation of tRNA fragments, which are thought to provoke neurodegenerative disorders (Karaca et al. 2014; Schaffer et al. 2014). Yeast Clp1, which does not display RNA kinase activity (Noble et al. 2007; Ramirez et al. 2008), has been suggested to mediate interactions with Pcf11 and other 3′ end factors and is required for termination at some transcription units (Holbein et al. 2011; Haddad et al. 2012). The kinase activity associated with hClp1 is dispensable for 3′ end cleavage in vitro (Schafer et al. 2018) and it remains unclear which function ATP hydrolysis by hClp1 plays during 3′ end formation.
Here, we used RNAi and a poly(A) tail anchored RNA sequencing approach to analyze the impact of hCLP1 and hPCF11 knockdown on 3′ end formation in MCF7 breast cancer cells. We observed an overlapping requirement for both proteins in proximal poly(A) site selection in the 3′-UTR and promoter proximal regions of target genes, consistent with the idea that hClp1 and hPcf11 participate in APA as components of the CF IIm complex linking its activity to misregulation of mRNA function in breast cancer.
RESULTS AND DISCUSSION
siRNA knockdown of hPCF11 and hCLP1 in MCF7 cells
To analyze the role of CF IIm in pre-mRNA 3′ end formation and APA we used siRNA technology to knock down hPCF11 and hCLP1 in the well characterized estrogen hormone sensitive MCF7 cells (Lee et al. 2015). Gene expression was analyzed using poly(A)-test sequencing (PAT-seq), a targeted poly(A) tail sequencing approach (Harrison et al. 2015). PAT-seq uses biotin-labeled and oligo-dT containing anchor oligonucleotides to enrich on streptavidin beads polyadenylated RNA that was previously subjected to limited RNase T1 digestion. Purified RNA was then modified by 5′ linker ligation and reverse transcribed into cDNA. cDNA libraries were size selected on denaturing 6% urea polyacrylamide gels and fragment lengths in a window of ∼100–300 nt were purified. A final PCR step facilitated indexed library amplification and directional Illumina sequencing.
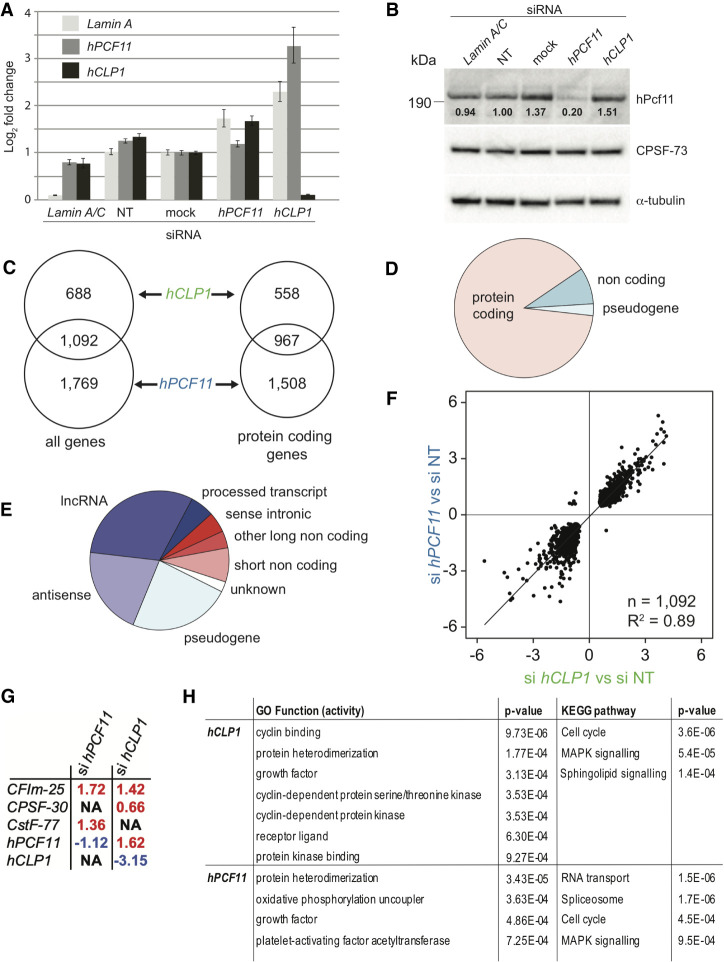
Sequencing data were processed using the tail-tools pipeline (http://rnasystems.erc.monash.edu/software/; Harrison et al. 2015). To establish confidence in reproducibility between replicate samples, we used a multidimensional scaling (MDS) approach to compare the similarity within the samples. While some variability was observed, MDS and heat-map analysis confirmed similarity of knockdown samples (Supplemental Fig. S1A,B). Figure 1A shows representative RT-PCR analyses, which confirmed efficient knockdown of hCLP1 mRNA following treatment with siRNA targeting hCLP1, whereas hPCF11 mRNA and Lamin A control were increased. Use of siRNA targeting hPCF11 revealed stable hPCF11 mRNA levels and slightly increased hCLP1 mRNA and Lamin A control. Efficiency of hPCF11 knockdown was, however, variable and resulted in mRNA reduction in some and mRNA increases in other experiments (data not shown). The apparently variable hPCF11 knockdown efficiency at the level of mRNA may reflect auto-regulation of hPCF11 expression (Kamieniarz-Gdula et al. 2019; Wang et al. 2019). Consistent with this idea, western analysis using commercially available antibodies confirmed fivefold reduction of hPcf11 protein levels following siRNA treatment (Fig. 1B). Attempts to analyze hClp1 protein following knockdown remained inconclusive due to poor performance of commercially available antibodies (data not shown).
FIGURE 1.
Gene expression analysis of MCF7 cells upon knockdown of hPCF11 and hCLP1. (A) Representative RT-PCR analysis of mRNA levels of Lamin A, hPCF11 and hCLP1 following the indicated siRNA treatments of MCF7 cells. mRNA levels were determined relative to the housekeeping gene ß-tubulin. Lamin A/C siRNA was used as a technical control, nontargeting siRNA (NT) and no-siRNA (mock) served as negative controls. Data are shown as mean ± SD. (B) Western blot analysis of proteins following the indicated siRNA treatments of MCF7 cells. Migration of full-length hPcf11, CPSF-73, and α-tubulin is indicated. Signals for hPcf11 and for α-tubulin were derived from the same PVDF membrane, CPSF-73 was detected on a second membrane. Values below the hPcf11 bands indicate quantification of the intensity of the hPcf11 band relative to α-tubulin. (C) Genomic annotations with significant expression changes following siRNA treatment of MCF7 cells. A total of 1780 significantly differentially expressed annotated features (FDR < 0.05) were identified in hCLP1 knockdown samples (n = 2), and 2861 in hPCF11 knockdown samples (n = 2) compared to nontargeting siRNA control samples (n = 3), 1092 were common to both conditions. A total of 1525 features were annotated to protein coding genes in hCLP1 knockdown samples and 2475 in hPCF11 knockdown samples, with 967 common to both conditions. (D) Breakdown of genomic annotations with expression changes observed upon knockdown of hPCF11 and hCLP1. For genes impacted by both knockdown conditions (1092 genes, FDR < 0.05), expression changes were seen primarily in protein coding genes, 8.5% were noncoding RNA, and 2.7% were pseudogenes. (E) Breakdown of non-protein coding annotations with expression changes observed upon knockdown of hPCF11 and hCLP1. For genes impacted by both knockdown conditions, expression changes were seen in the indicated noncoding RNA species (125 genes, FDR < 0.05). (F) Comparison of expression changes observed upon knockdown of hPCF11 (y-axis) and hCLP1 (x-axis). Scatter plot shows 1092 significantly differentially expressed annotated features (FDR < 0.05) that are found relative to nontargeting siRNA control samples (n = 3) in both hCLP1 knockdown samples (n = 2) and in hPCF11 knockdown samples (n = 2). (G) Expression changes observed with subunits of the core 3′ end machinery upon knockdown of hPCF11 and hCLP1. Significant differential expression levels were determined (FDR < 0.05) and fold changes are indicated in red for increased and blue for decreased expression. NA means that no significant change was observed. (H) Gene ontology of molecular function and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways and associated P-values identified through expression changes observed upon knockdown of hPCF11 and hCLP1. Indicated are all top GO function activity terms and KEGG pathway terms (FDR ≤ 0.05) associated with the knockdown conditions.
PAT-seq analyses following siRNA knockdown of hPCF11 and hCLP1
Approximately 29,294 genomics features (genes) were sequenced by PAT-seq in our samples. Knockdown of CF IIm subunits resulted in significant expression changes (FDR < 0.05) for both hCLP1 (1780 features) and hPCF11 samples (2861 features) (Fig. 1C). Among these significant changes, 1092 were common to hPCF11 and hCLP1 samples, in agreement with the proposal that the changes were a consequence of CF IIm complex deficiency, which requires both hPcf11 and hClp1 for proper function. Furthermore, 88.6% of the identified features were associated with protein encoding genes with the remaining 11.4% of features representing noncoding RNAs and pseudogenes (Fig. 1D,E). The bias toward the identification of protein encoding genes is reflecting in part the initial selection of polyadenylated RNA species by the PAT-seq method (Harrison et al. 2015); note that RNA species that lack a poly(A) tail will not be identified by this approach. Figure 1F shows a high degree of correlation (R2 0.89) of effects on genes, which require both hPCF11 and hCLP1 for expression. Overall the observed changes in hPCF11 and hCLP1 samples aligned well, consistent with the idea that they reflected primarily loss of CF IIm function. However, not all genes displayed the same trends for both hPCF11 and hCLP1 samples and differences with respect to the magnitude of the expression changes were also observed. Some of these differences may be attributable to the differences in knockdown efficiencies. Alternatively, some genes may display a specific requirement for hPcf11 and hClp1, respectively.
Moreover, knockdown of CF IIm components significantly increased expression levels of other core 3′ end factors (Fig. 1G). hCLP1 knockdown resulted in increased levels of hPCF11, CF Im-25 and CPSF-30, and knockdown of hPCF11 resulted in increased levels of CF Im-25 and CstF-77. The reasons for these changes remain unclear but may reflect cross-regulation of 3′ end factors in response to the reduced cellular levels of hClp1 and hPcf11, respectively.
Gene ontology (GO) and KEGG (Kyoto Encyclopedia of Genes and Genomes) (Kanehisa and Goto 2000) pathway analyses for significant differentially expressed genes (FDR < 0.05) uncovered an involvement of CF IIm in “protein hetero-dimerization activity,” “cell growth” and the “cell cycle” (Fig. 1H). Annotations specific for hCLP1 also included “receptor ligand,” “protein kinase binding” and signaling pathways, pointing toward an involvement of the protein in signal transduction mechanisms, but the nature of such a connection remains currently unknown. For hPCF11, annotation of “RNA transport,” “spliceosome” and “RNA degradation” aligned well with its known role in mRNA synthesis, its partial copurification with splicing factors (de Vries et al. 2000) and its link to nuclear mRNA export (Volanakis et al. 2017).
Knockdown of CF IIm favors distal poly(A) sites
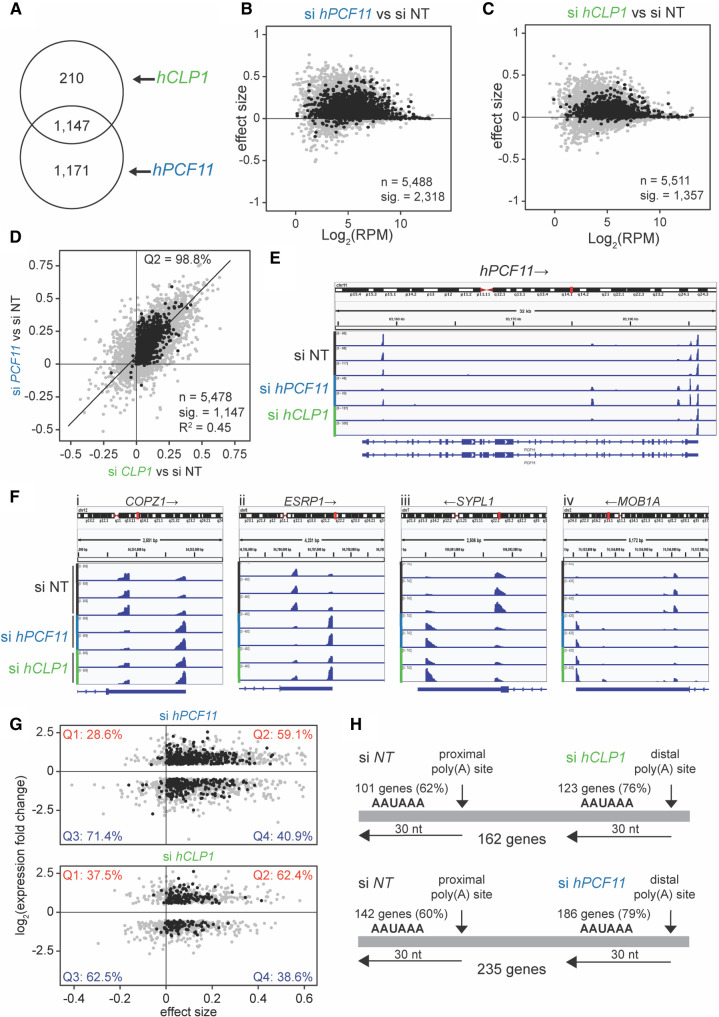
Next, we analyzed the use of alternative poly(A) sites following CF IIm impairment via hCLP1 and hPCF11 knockdown. For this purpose, 12,063 genomic annotations were identified as having two or more poly(A) sites. The sites were then given a confident inner bound on effect size after adjusting for multiple testing, a “confect,” which refers to the observed shift between two alternative poly(A) sites in our samples (see Materials and Methods). We found significant 3′-UTR shifts within 2318 annotations for hPCF11 knockdown and within 1357 annotations for hCLP1 knockdown, with 1147 annotations across both conditions (Fig. 2A). The majority of observed shifts generated longer 3′-UTRs for both hCLP1 and hPCF11 knockdowns (Fig. 2B,C) and displayed a high degree of correlation between the two conditions (Fig. 2D). The plot of Figure 2D compares the shifts at two levels: (i) it shows that 98.8% of shifts for hPCF11 and hCLP1 knockdown occur in the same direction because they locate in the Q2 region of the graph; and (ii) it reveals the “strength” of a shift. A shift will have the value “1” if the usage of a poly(A) site has fully shifted from one site to an alternative site and the strength of the shifts observed in the two samples will be identical if located on the diagonal of the quadrant. The observed correlation coefficient for the shifts for hPCF11 and hCLP1 knockdown was 0.45, revealing that both conditions resulted in a well comparable strength of the observed shift consistent with both proteins participating in alternative polyadenylation as a complex.
FIGURE 2.
Analysis of alternative polyadenylation upon knockdown of hPCF11 and hCLP1. (A) Genomic annotations with significant 3′ end changes following siRNA treatment of MCF7 cells. A total of 1357 significant 3′ end changes (FDR < 0.05) were identified in hCLP1 knockdown samples (n = 2), and 2318 in hPCF11 knockdown samples (n = 2) compared to nontargeting siRNA control samples (n = 3), 1147 were common to both conditions. (B) 3′ end shift effects in annotated features observed upon knockdown of hPCF11. A positive effect size indicates shifts toward longer 3′-UTRs, and a negative effect size indicates shifts toward shorter 3′UTRs. Gray dots indicate the estimated effect size, and where this is significant corresponding black dots show the “confect,” a confident inner bound of this effect size found using the Limma and TopConfects R packages. A total of 2318 of these effects in hPCF11 samples (n = 2) were significant (FDR < 0.05). Log2 (RPM) indicates the average number of reads per million. si NT (n = 3) indicates the nontargeting siRNA control that served as reference. (C) 3′ end shift effects in annotated features as described above for B, however observed upon knockdown of hCLP1. A total of 1357 of these effects in hCLP1 samples (n = 2) were significant (FDR < 0.05). (D) Comparison of 3′ end shift effects in annotated features as described above for B observed upon knockdown of hPCF11 (y-axis) and hCLP1 (x-axis). Gray dots indicate shift effects for all analyzed features (n = 5478); black dots indicate shift effects that are significant (n = 1147; FDR < 0.05). si NT indicates the nontargeting siRNA control that served as reference. (E) Integrative genomics viewer (IGV) representation of PAT-seq results for the hPCF11 gene. Shown is the entire gene including 5′ and 3′ regulatory regions. si hPCF11 (n = 2) and si hCLP1 (n = 2) samples had significant (FDR < 0.05) shifts in poly(A) site usage compared to nontargeting siRNA control samples (si NT) (n = 3). Arrows indicate direction of gene transcription. The representation uses different y-axes for each horizontal panel to highlight the relative ratio of poly(A) site usage within a gene in a replicate sample. An alternative representation of the data with normalization to the highest peak in all shown panels is shown in Supplemental Figure S2B. (F) IGV representation of PAT-seq results for (i) COPZ1 (coatomer protein complex subunit zeta 1), (ii) ESRP1 (epithelial splicing regulator protein 1), (iii) SYPL1 (synaptophysin like 1) and (iv) MOB1A (MOB kinase activator 1A). Shown are only relevant 3′-UTR regions. si hPCF11 (n = 2) and si hCLP1 (n = 2) samples had significant (FDR < 0.05) shifts in poly(A) site usage compared to nontargeting siRNA control samples (si NT) (n = 3). Arrows indicate direction of gene transcription. (G) Comparison between changes in expression (fold change; y-axis) and 3′-UTR length (effect size of shift) upon hPCF11 (upper panel) and hCLP1 (lower panel) knockdown. This analysis included for hPCF11 samples (n = 2) a total of 1291 features (gray dots); for 603 of those features the shift effects were significant (FDR < 0.05; black dots) compared to nontargeting siRNA control samples (n = 3). For hCLP1 samples (n = 2), a total of 791 features (gray dots) were included; for 237 of those features the shift effects were significant (FDR < 0.05; black dots) compared to nontargeting siRNA control samples (n = 3). A positive effect size indicates UTR lengthening, a negative effect size indicates UTR shortening. Percentages shown in each quadrant indicate how many genes have an increased or decreased expression relative to lengthening of the 3′UTR (on the right) or shortening of the 3′UTR (on the left). (H) Comparison of canonical poly(A) signal usage in APA following knockdown of hPCF11 and hCLP1. A representative set of 162 genes identified usage of the AAUAAA signal within 30 bp of the proximal poly(A) site in 62% of cases in the nontargeting siRNA control (n = 3) and switched to usage of distal AAUAAA in 79% of cases following knockdown of hCLP1 (n = 2). Accordingly, a representative set of 142 genes identified usage of the AAUAAA signal within 30 bp of the proximal poly(A) site in 60% of cases in the nontargeting siRNA control and switched to usage of distal AAUAAA in 79% of cases following knockdown of hPCF11 (n = 2).
Representative 3′-UTR shifts associated with impairment of CF IIm are shown in Figure 2F. The COPZ1 (coatomer protein complex subunit zeta 1) gene, the ESRP1 (epithelial splicing regulator protein 1) gene, the SYPL1 (synaptophysin like 1) gene and the MOB1A (MOB kinase activator 1A) gene require both hCLP1 and hPCF11 for efficient use of a proximal poly(A) site in the 3′-UTR. Interestingly, we observed APA also within promoter proximal regions. For example, poly(A) site usage associated within the hPCF11 and METTL4 genes was dominated in control samples by the usage of promoter proximal poly(A) sites, indicating that a significant amount of transcription was terminated before synthesis of the main gene body took place (Fig. 2E; Supplemental Fig. S1A,B). Knock-down of hCLP1 and hPCF11 suppressed the usage of promoter proximal poly(A) sites, facilitating productive transcription elongation and usage of poly(A) signals in the 3′-UTR. These observations are consistent with the idea that CF IIm acted to attenuate transcription via early termination of transcription, which participates in the cross-regulation of 3′ end factor expression (Fig. 1G). In agreement with this we observed up-regulation of hPCF11 expression following hCLP1 knockdown (Fig. 1A). Similarly, hCLP1 or hPCF11 knockdown resulted in the increased usage of distal sites relative to a proximal site in Lamin A (Supplemental Fig. S2C) consistent with increased expression of this mRNA (Fig. 1A).
A role for hPCF11 in transcription termination has been established (West and Proudfoot 2008), but no such role has been suggested for hCLP1. While this manuscript was in preparation, an involvement of vertebrate Pcf11 in the expression of closely spaced genes and of long full-length genes has been reported (Kamieniarz-Gdula et al. 2019; Wang et al. 2019). Substoichiometric association of hPcf11 with the pre-mRNA 3′ end formation machinery in addition to auto-regulation of Pcf11 expression was proposed to highlight a specific requirement for Pcf11 in these analyses (Kamieniarz-Gdula et al. 2019; Wang et al. 2019). Our observation that CF IIm was required for APA in MCF7 cells indicates, however, that the hClp1/hPcf11 heterodimer may represent the relevant functional entity. This idea is supported by the observation that hPcf11 remained insoluble in the absence of hClp1 in vitro (Schafer et al. 2018). siRNA knockdown of hClp1 may destabilize hPcf11 in vivo and cause the overlapping requirement for CF IIm that was observed in this work. This idea is, however, not supported by western analysis of hCLP1 knockdown samples, which revealed stable hPcf11 levels (Fig. 1B). Alternatively, ATP hydrolysis or RNA kinase activity associated with hClp1, while dispensable for 3′ end cleavage in vitro (Schafer et al. 2018), may contribute to the role of CF IIm in APA. Such a potential function may be associated with changes in the structure of CF IIm or its association with components of the 3′ end machinery, RNAP II or other factors involved in termination of transcription.
Next, we compared the observed shifts in poly(A) site usage with changes in mRNA levels, applying a FDR cut-off value of 0.05 (Fig. 2G). For hPCF11, 59.1% of genes that showed up-regulation of gene expression were associated with an increase in 3′-UTR length, while 40.9% were associated with a decreased 3′-UTR length. For hCLP1, 62.4% of genes that showed up-regulation of gene expression were associated with an increase in 3′-UTR length, while 38.6% were associated with a decreased 3′-UTR length. These observations indicated that for both hPCF11 and hCLP1 the usage of longer 3′-UTRs was biased toward higher mRNA expression levels.
Analysis of cleavage site signals present in genes with an associated 3′-UTR shift revealed an increase in usage of the canonical AAUAAA hexamer sequence following knockdown of hCLP1 (14% increase) and hPCF11 (19% increase), respectively (Fig. 2H). These observations indicated that usage of proximal poly(A) sites was biased toward a CF IIm requirement and that stronger distal poly(A) signals facilitated usage of poly(A) sites when CF IIm was absent or deficient.
APA shifts in breast cancer cells
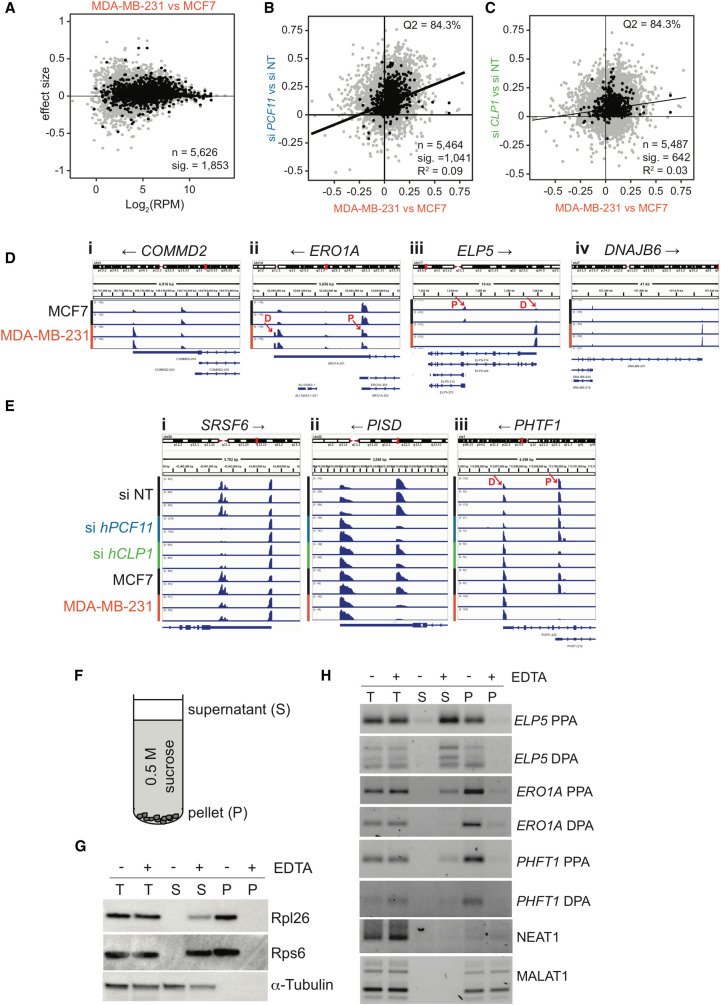
Previous analyses of APA in breast cancer cells revealed a shortening of 3′UTRs in estrogen receptor-positive MCF7 cells relative to a nontransformed mammary epithelial cell line, whereas a lengthening of 3′-UTRs was observed for estrogen receptor-negative and highly invasive MDA-MB-231 cells (Fu et al. 2011). The disparity of 3′-UTR shifts in the two cancer cell lines was striking and was suggested to reflect variability in gene expression due to evolutionary adaptations in response to driver-mutations (Fu et al. 2011). We therefore replicated the main finding of Fu and coworkers and compared poly(A) site usage in the two cancer cell lines by PAT-seq to ensure bioinformatic compatibility. Figure 3A shows that genes with short 3′UTRs in MCF7 cells indeed shifted toward long 3′UTRs in MDA-MB-231 cells. To evaluate the role played by CF IIm in APA in breast cancer we correlated APA shifts that occurred following siRNA knockdown of hPCF11 and hCLP1, respectively, to the poly(A) shifts that occurred in MDA-MB-231 cells relative to MCF7 cells. Figure 3B,C show that there was a high degree of overlap of the direction of APA shifts (proximal vs distal) as evidenced by the location of shifts in the Q2 region of the graph with 84.3% of transcripts containing longer 3′-UTRs in both cases. However, the overall R2 values of the correlation were low (0.09 and 0.03, respectively). These data may indicate that shifts that occurred in MDA-MB-231 cells relative to MCF7 cells are not indicative of impaired CFIIm function. An alternative interpretation could be that low correlation values reflect mechanistic differences resulting on the one hand from dysregulation of APA following siRNA knockdown and on the other hand from cancer cell endogenous misregulation of APA. The mechanisms for the latter remain unclear and may not be due to loss of function as is assumed for the siRNA conditions.
FIGURE 3.
APA shifts in breast cancer cells. (A) 3′ end shift effects in annotated features (n = 5626) observed in MDA-MB-231 cells relative to MCF7 cells. The interpretation is as described in the legend to Figure 2B. 1853 of these effects were significant (FDR < 0.05). Log2 (RPM) indicates the number of reads per million. n = 2. (B) Comparison of 3′ end shift effects observed upon knockdown of hPCF11 (n = 2) relative to nontargeting siRNA samples (n = 3) (y-axis) and MDA-MB-231 cells (n = 2) relative to MCF7 cells (n = 2) (x-axis). Gray dots indicate shift effects for all analyzed features (n = 5464); black dots indicate shift effects that are significant (n = 1041; FDR < 0.05). The majority of genes experienced a shift into the same direction (long versus short) as indicated by 84.3% of all features located to the top right quadrant. (C) Comparison of 3′ end shift effects observed upon knockdown of hCLP1 (n = 2) relative to nontargeting siRNA samples (n = 3) (y-axis) and MDA-MB-231 cells (n = 2) relative to MCF7 cells (n = 2) (x-axis). Gray dots indicate shift effects for all analyzed features (n = 5487); black dots indicate shift effects that are significant (n = 642; FDR < 0.05). The majority of genes experienced a shift into the same direction (long versus short) as indicated by 84.3% of all features located in the top right quadrant. (D) IGV representation of PAT-seq data for MCF7 and MDA-MB-231 cells for (i) COMMD2 (COMM domain-containing protein 2), (ii) ERO1A (endoplasmic reticulum oxidoreductase 1 alpha), (iii) ELP5 (elongator acetyltransferase complex subunit 5), and (iv) DNAJB6 (DnaJ heat shock protein family [Hsp40] member B6). Shown are only relevant 3′-UTR regions and poly(A) site usage in MCF7 and MDA-MB-231 cells, respectively. Overlapping noncoding transcripts are shown at the bottom of the IGV window. Black arrows indicate direction of gene transcription. n = 2. Red arrows indicate the approximate position of primers designed to amplify ∼100 bases of unique sequence within the peak region of PAT-seq reads as used for Figure 3H. D and P indicate the location of a distal and proximal poly(A) sites, respectively. (E) IGV representation of PAT-seq data for (i) SRSF6 (serine and arginine rich splicing factor 6), (ii) PISD (phosphatidylserine decarboxylase), and (iii) PHTF1 (putative homeodomain transcription factor 1). Shown are relevant 3′-UTR regions and poly(A) site usage in nontargeting siRNA control (n = 3) and hPCF11 (n = 2) and hCLP1 (n = 2) knockdown samples as well as MCF7 (n = 2) and MDA-MB-231 (n = 2) cells, respectively. Overlapping noncoding transcripts are shown at the bottom of the IGV window. Black arrows indicate direction of gene transcription. Red arrows indicate the approximate position of primers designed to amplify ∼100 bases of unique sequence within the peak region of PAT-seq reads as used for Figure 3H. D and P indicate the location of a distal and proximal poly(A) sites, respectively. The representation uses different y-axes for each horizontal panel to highlight the relative ratio of poly(A) site usage within a gene in a replicate sample. An alternative representation of the data with normalization to the highest peak in all shown panels is shown in Supplemental Figure S2C. (F) Schematic representation of sucrose cushion analysis. Centrifugation yielded a supernatant fraction (S) and a pellet fraction (P), which was analyzed for protein and RNA content. (G) Western analysis of protein obtained from sucrose cushion centrifugation of MCF7 lysates in the absence and presence of EDTA. Supernatant (S) and pellet fractions (P) were analyzed for Rpl26 and Rps6, which are components of the large and small ribosomal subunits, respectively. α-tubulin was used as cytosolic control. (H) RT-qPCR analysis of RNA obtained from sucrose cushion centrifugation of MCF7 lysates in the absence and presence of EDTA. Supernatant (S) and pellet fractions (P) were analyzed for the presence of the indicated RNA species. PPA indicates RNA resulting from usage of a major proximal, UTR or intronic poly(A) site, and DPA indicates RNA resulting from usage of a distal poly(A) site located in the 3′-UTR, respectively. Approximate location of primers used to amplify ∼100 bases of unique sequence is indicated in D and E. NEAT1 and MALAT1 are nontranslated lncRNA controls.
Representative 3′-UTR shifts are shown in Figure 3D. In MCF7 cells COMMD2 (COMM domain-containing protein 2), ERO1A (endoplasmic reticulum oxidoreductase 1 Alpha), ELP5 (elongator acetyltransferase complex subunit 5) and DNAJB6 (DnaJ heat shock protein family [Hsp40] member B6) displayed more sequencing products that aligned with proximal poly(A) sites compared to distal poly(A) sites. In contrast, the poly(A) site profile was normalized to a more distal site choice in MDA-MB-231 cells. These analyses agreed well with conclusions drawn by Fu and coworkers who interpreted the observed poly(A) site profile such that MCF7 cells preferentially used short 3′-UTRs and MDA-MB-231 cells preferentially used long 3′-UTRs (Fu et al. 2011). Figure 3E and Supplemental Figure S2D highlight examples where APA shifts associated with hPCF11 or hCLP1 knockdown are mirrored in shifts observed in MDA-MB-231 cells relative to MCF7. Usage of a proximal poly(A) site in the SRSF6 (serine and arginine rich splicing factor 6) 3′UTR was sensitive to knockdown of both CFIIm subunits, while proximal poly(A) sites in PISD (phosphatidylserine decarboxylase) and PHTF1 (putative homeodomain transcription factor 1) appeared more dependent on either hPCF11 or hCLP1, respectively.
Truncated transcripts in MCF7 cells co-sediment with polysomes in an EDTA sensitive manner
Recent updates of the reference genome included numerous additional truncated transcripts that overlap with full-length protein coding genes and have unclear functional significance. These apparent noncoding RNAs are abundant throughout the genome and are generally thought to be polyadenylated and targeted for degradation by the exosome (Ogami et al. 2018). Notably, such noncoding RNA can display high stability, like CstF-77 intronic polyadenylation products that have been found to exhibit half-lives similar to protein coding transcripts (Luo et al. 2013). Due to the prevalence of annotated noncoding RNA species in MCF7, we reasoned that exosome function may have been compromised. Supplemental Figure S3A shows that MCF7 cells indeed exhibited a concerted down-regulation of mRNA encoding exosome components. In order to correlate reduced mRNA expression levels with exosome activity, we analyzed levels of lncRNAs, which are known to be degraded by the exosome (Ogami et al. 2018). Supplemental Figure S3B shows that lncRNAs were 2.58 more likely to be stabilized in MCF7 compared to MDA-MB-231. This included the well-characterized lncRNA members MALAT1 and NEAT1, which were significantly more stable in MCF7, consistent with the idea that exosome activity was reduced in MCF7 cells. It seems possible, therefore, that stabilization and accumulation of noncoding transcripts relative to distal poly(A) sites associated with protein coding mRNAs contributed to instances where apparent shifts toward proximal poly(A) sites occurred in MCF7 cells.
Interestingly, proximal poly(A) site usage in MCF7 cells generated numerous truncated mRNAs, e.g., for ELP5, ERO1A (both Fig. 3D) and PHFT1 (Fig. 3E). To evaluate the physiological relevance of such transcripts with locus internal 3′UTRs, we performed sucrose cushion centrifugation. In these experiments, lysates obtained from cycloheximide treated MCF7 cells were layered on 0.5 M sucrose and centrifuged to separate polysomes from cytosolic fractions (Fig. 3F). This procedure facilitated the efficient removal of ribosomal components (Rpl26 and Rps6) from the supernatant that contains cytosolic proteins (α-Tubulin), and enrichment of ribosomes in the pellet fraction (Fig. 3G). In the presence of EDTA, which disrupts polysomes, the ribosomal proteins were undetectable in the pellet fraction and were partially retained in the supernatant. RT-PCR analysis of RNA contained within supernatant and pellet fractions revealed that ELP5, ERO1A and PHFT1 transcripts that originated from usage of either distal poly(A) sites (DPA) located within introns or proximal poly(A) sites located within the 3′UTR (PPA) were (i) strongly reduced in the supernatant, (ii) associated with the pellet fraction, and (iii) pellet association was sensitive to the presence of EDTA. In contrast, bona fide noncoding RNAs NEAT1 and MALAT1 were found associated with the pellet fraction, indicating that they are associated with large structures, which were, however, not sensitive to the presence of EDTA. These observations suggested that truncated mRNA transcripts derived from ELP5, ERO1A and PHFT1 genes are indeed being translated. APA in MCF7 may thus give rise to the synthesis of truncated polypeptides with aberrant protein function and potential dominant negative impact. It seems possible that such truncated polypeptides may contribute to the transformed phenotypes associated with these cells.
Taken together, our results implicate CF IIm in APA in breast cancer cells. It will be important to delineate in future work the activities associated with CF IIm, which contribute to APA in healthy and diseased cells and to understand the regulatory mechanisms, which define the transcription units targeted by CF IIm.
MATERIALS AND METHODS
Cell culture
MCF7 adenocarcinoma and MDA-MB-231 cells were grown in T75 flasks with Dulbecco's modified Eagle's medium (DMEM; Gibco) containing 10% fetal bovine serum (FBS) in the presence of 5% carbon dioxide (CO2) at 37°C.
siRNA knockdown
RNAi reaction mixtures included siRNA (Dharmacon) in 250 µL opti-MEM (Gibco) and 245 µL of Lipofectamine RNAiMAX (Invitrogen) and were added to six well plates and incubated in 5% CO2 at 37°C for 48 h. Final concentration of siRNAs was 60 nM for Lamin A/C and nontargeting controls and 75 nM for hPCF11 and hCLP1 treatments. siRNAs included Lamin A/C and nontargeting siRNA controls (siGENOME Controls basic, K-002800-C4-01), PCF11#1 GAUACAAAUCAGCGACUUA and PCF11#2 GUGUGCAAAUUUAACGAAA (On-Target plus, DHALQ-015381-01), CLP1#1 GGGCAAGUCUACAGUGUGU, CLP1#2, GCAUGUCUCAAGAGGAUAA, CLP1#3 GAUUACAUCUCGUUUAGCA, CLP1#4 GGAGGCAUCUCAGUCAGUU (On-Target plus, DHALQ-019895-00). Medium was removed and wells were washed with 1 mL 1% PBS. RNA was extracted using 750 µL TRIzol and purified using Qiagen RNeasy spin columns.
RT-PCR
cDNA synthesis was done using SuperScript IV VILO (Invitrogen). An amount of 10 ng of cDNA was included in PCR reactions using Applied Biosystems Power SYBR Green according to manufacturers instructions. Oligonucleotides used were: LAMINA-Forward TGAGTCCATTCTCCCAGGTA, LAMINA-Reverse CTCACCTTTCTTC TCCCTTCTT, β-tubulin-Forward TGGACTCTGTTCGCTCAGGT, β-tubulin-Reverse TGCCTCCTTCCGTACCACAT, PCF11-Forward AGCAAGCTAAGGCACAGTTGGC, PCF11-Reverse TTGGATGGTGCAGATCCAGGAC, CLP1-Forward ACTGTGCTGCTCCCTAAATC, CLP1-Reverse AACAGCCTCGGAATCCATAAA
Protein extraction and western blotting
For analysis of knockdown cultures cells were incubated in 400 µL lysis buffer (50 mM HEPES pH 7.5, 0.5% NP-40, 5% Triton X-100, 50 mM NaCl, 5% Glycerol, 1 mM EDTA, and protease inhibitor cocktail without EDTA [Roche]) on ice for 10 min. Lysates were sonicated for 30 sec and cleared by centrifugation at 13,000 rpm. An amount of 30 µg total protein was separated on 4%–12% NuPAGE gels and transferred on poly-vinylidene difluoride (PVDF) membrane with 1× transfer buffer (25 mM Tris Base and 192 mM Glycine) supplemented with 5% methanol. Antibody dilution for primary rabbit anti-hPcf11 (Abcam) was 1:2000, rat anti-α-tubulin (Abcam ab 6160) was 1:2000, rabbit anti-CPSF73 (Abcam) was 1:2000; secondary HRP coupled anti-rabbit antibody (Abcam) was used 1:5000 and HRP coupled anti-rat antibody (Abcam) was used 1:5000.
Sucrose cushion analysis
MCF7 cells were grown in T75 flasks in DMEM supplemented with 10% FBS at 37°C, 5% CO2. Once 80%–90% confluency was reached cells were incubated for 5 min with 10 μg/mL cycloheximide at 37°C. Cells were washed with PBS + 10 μg/mL cycloheximide and scraped directly from the plate using a total of 3 ml lysis buffer (10 mM NaCl, 10 mM MgCl2, 10 mM Tris-HCl pH 7.5, 1% Triton X-100, 1% sodium deoxycholate, 10 μg/mL cycloheximide, and 1 mM dithiothreitol) and transferred to two 1.5 mL eppendorf tubes. After 15 min of incubation on ice with occasional vortexing cellular debris were removed by centrifugation for 5 min at 14,000 rpm in an Eppendorf 5417R centrifuge at 4°C. Cleared lysate was incubated on ice for 20 min in the absence and presence of 0.1 M EDTA, respectively. An amount of 250 μL of the treated lysate was layered onto 2150 μL of 0.5 M sucrose in lysis buffer in polyallomer tubes. Centrifugation was done at 34,000 rpm at 4°C for 1 h using the TLS-55 swing-out rotor in a Beckman Coulter TL-100 benchtop ultracentrifuge. Following centrifugation, 200 μL of the supernatant was collected from the very top of the tube and the pellet was resuspended in 200 μL lysis buffer. The sucrose cushion fraction was not analyzed. An amount of 30 µL of supernatant and pellet fractions (∼30 µg total protein) was separated on 4%–12% NuPAGE gels and transferred on PVDF membrane as described above.
For RNA analysis 1 mL TRIzol was added to 150 μL supernatant and pellet samples and total RNA was prepared using the Direct-zol RNA Miniprep Kit (ZYMO RESEARCH) and concentration was determined with nanodrop ND-1000 spectrophotometer. An amount of 150 ng of RNA was reverse transcribed using SuperScript III (Invitrogen) following the TVN-PAT assay method (Janicke et al. 2012). Genes were amplified and reactions were resolved on 2% high-resolution agarose gels, prestained with SYBR safe. Amersham Imager 680 was used to image gels against a 100-bp ladder (New England Biolabs). Primers used were: H.s ELP5_PPA_PAT CCCTCTGCCAAGGAGTGTG, H.s ELP5_DPA_PAT GCGTCTCATCAGGACAGAAGG, H.s PHTF1_PPA_PAT CCATTGAGTTGCTTTTGTTCTC, H.s PHTF1_DPA_PAT CTCTATCAAAGAGAGATCAAGCG, H.s MALAT_PAT CGCAGACGAAAATGGAAAG, H.s NEAT1_PAT GTACTGGTATGTTGCTCTGTATGG, H.s ERO1A_PPA_PAT GCATAATAGCAATGACAGTCTTAAGC, H.s ERO1A_DPA_PAT CAAGGGTCAGTAGAGTCCCAAG, TVN-PAT primer GCGAGCTCCGCGGCCGCGTTTTTTTTTTTTVN.
PAT-seq library preparation
For RNA-sequencing analysis, 1 µg of total RNA was processed for 3′end focused RNA-seq by the poly(A) tail sequencing (PAT-seq) approach as described in Harrison et al. (2015). All data has been uploaded to GEO, accession number GSE133473.
Sequencing, data processing, and statistical methods
Next-generation sequencing for the PAT-seq was performed at the Monash Health Translation Precinct medial genomic facility in Clayton, Victoria, on the Illumina HiSeq 1500 instrument. Reads were processed using the Tail Tools pipeline (Harrison et al. 2015). Read counts were produced at the level of whole genes and at the levels of peaks detected by the pipeline. The reference annotation used was Ensembl Homo Sapiens version 93. These counts were then used to test for differential gene expression and APA.
Differential gene expression was tested for between experimental groups, using the Fitnoise library (https://github.com/pfh/fitnoise) as part of the Tail Tools pipeline, after log transforming and weighting peak counts with voom (Law et al. 2014) from the limma R package after TMM library size normalization (Robinson and Oshlack 2010). Fitnoise is an implementation of Empirical Bayes moderated t-tests on weighted linear models as described in Smyth (2004). Genes for which no relevant sample had at least 10 reads were removed before testing.
To detect shifts in APA usage, peaks were first assigned to genes. Peaks up to 2 kb down-strand of the gene (but not proceeding into another gene on the same strand) were counted as belonging to that gene. Next, peak counts were log2 transformed and linear models were fitted for each peak, using TMM normalization, voom, and limma, producing as coefficients estimates of the log2 abundance of reads at each peak in each of two experimental conditions. For each gene, a “shift score” was then estimated using these fitted coefficients, as follows.
Considering a single gene, for two conditions i = A, B, and n peaks j = 1.n, there is a fitted coefficient βi,j, and the proportion of reads for each peak within a condition is
A shift score s is then calculated, ranging from −1 to 1 where −1 indicates all reads in condition B are up-strand of condition A and 1 indicates all reads in condition B are down-strand of condition A.
The fitted coefficients βi,j have an associated error covariance matrix, which can be propagated through these steps by the delta method to obtain an approximate standard error associated with s. Shift scores and associated standard errors are calculated for each gene with two or more peaks, then the topconfects R package (Harrison et al. 2019) is used to provide “confect” inner confidence bounds on the shift scores with correction for multiple testing. Confect values are only given for significantly nonzero shifts, with a False Discovery Rate of 0.05. Ordering results by absolute confect values and reading down this list only as far as is desired, a False Coverage-statement Rate of 0.05 is guaranteed for these confidence bounds.
Gene ontology analysis
Pathways and gene ontology were analyzed using DAVID (https://david.ncifcrf.gov) and GO-Rilla (http://cbl-gorilla.cs.technion.ac.il) website tools.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Beilharz laboratory for constructive comments on the manuscript, Michael Ming See from the Monash Bioinformatics Platform for technical assistance and Yasmin Saab for help with the sucrose cushion analysis. The Victorian Centre for Functional Genomics (K.J.S.) is funded by the Australian Cancer Research Foundation (ACRF), the Australian Phenomics Network (APN) through funding from the Australian Government's National Collaborative Research Infrastructure Strategy (NCRIS) program and the Peter MacCallum Cancer Centre Foundation. T.H.B. was supported by a Monash Bio Discovery Fellowship and research in the Beilharz Laboratory was funded by grants from the NHMRC (APP1128250) and the ARC (DP170100569 and FT180100049).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.075226.120.
REFERENCES
- Barilla D, Lee BA, Proudfoot NJ. 2001. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc Natl Acad Sci 98: 445–450. 10.1073/pnas.98.2.445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. 2014. Coupling mRNA processing with transcription in time and space. Nat Rev Genet 15: 163–175. 10.1038/nrg3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu AC, Suzuki HI, Wu X, Mahat DB, Kriz AJ, Sharp PA. 2018. Transcriptional pause sites delineate stable nucleosome-associated premature polyadenylation suppressed by U1 snRNP. Mol Cell 69: 648–663 e647. 10.1016/j.molcel.2018.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries H, Ruegsegger U, Hubner W, Friedlein A, Langen H, Keller W. 2000. Human pre-mRNA cleavage factor IIm contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J 19: 5895–5904. 10.1093/emboj/19.21.5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sun Y, Li Y, Li J, Rao X, Chen C, Xu A. 2011. Differential genome-wide profiling of tandem 3′-UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res 21: 741–747. 10.1101/gr.115295.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Martin G, Keller W, Zavolan M. 2012. Cleavage factor Im is a key regulator of 3′-UTR length. RNA Biol 9: 1405–1412. 10.4161/rna.22570 [DOI] [PubMed] [Google Scholar]
- Gruber AR, Martin G, Muller P, Schmidt A, Gruber AJ, Gumienny R, Mittal N, Jayachandran R, Pieters J, Keller W, et al. 2014. Global 3′-UTR shortening has a limited effect on protein abundance in proliferating T cells. Nat Commun 5: 5465 10.1038/ncomms6465 [DOI] [PubMed] [Google Scholar]
- Grzechnik P, Gdula MR, Proudfoot NJ. 2015. Pcf11 orchestrates transcription termination pathways in yeast. Genes Dev 29: 849–861. 10.1101/gad.251470.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guegueniat J, Dupin AF, Stojko J, Beaurepaire L, Cianferani S, Mackereth CD, Minvielle-Sebastia L, Fribourg S. 2017. Distinct roles of Pcf11 zinc-binding domains in pre-mRNA 3'-end processing. Nucleic Acids Res 45: 10115–10131. 10.1093/nar/gkx674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad R, Maurice F, Viphakone N, Voisinet-Hakil F, Fribourg S, Minvielle-Sebastia L. 2012. An essential role for Clp1 in assembly of polyadenylation complex CF IA and Pol II transcription termination. Nucleic Acids Res 40: 1226–1239. 10.1093/nar/gkr800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PF, Powell DR, Clancy JL, Preiss T, Boag PR, Traven A, Seemann T, Beilharz TH. 2015. PAT-seq: a method to study the integration of 3′-UTR dynamics with gene expression in the eukaryotic transcriptome. RNA 21: 1502–1510. 10.1261/rna.048355.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PF, Pattison AD, Powell DR, Beilharz TH. 2019. Topconfects: a package for confident effect sizes in differential expression analysis provides a more biologically useful ranked gene list. Genome Biol 20: 67 10.1186/s13059-019-1674-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbein S, Scola S, Loll B, Dichtl BS, Hubner W, Meinhart A, Dichtl B. 2011. The P-loop domain of yeast Clp1 mediates interactions between CF IA and CPF factors in pre-mRNA 3′ end formation. PLoS ONE 6: e29139 10.1371/journal.pone.0029139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth D, Noble CG, Taylor IA, Ramos A. 2006. RNA polymerase II CTD phosphopeptides compete with RNA for the interaction with Pcf11. RNA 12: 555–560. 10.1261/rna.2304506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Shuman S. 2009. Characterization of a thermostable archaeal polynucleotide kinase homologous to human Clp1. RNA 15: 923–931. 10.1261/rna.1492809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicke A, Vancuylenberg J, Boag PR, Traven A, Beilharz TH. 2012. ePAT: a simple method to tag adenylated RNA to measure poly(A)-tail length and other 3′ RACE applications. RNA 18: 1289–1295. 10.1261/rna.031898.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. 2009. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci 106: 7028–7033. 10.1073/pnas.0900028106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamieniarz-Gdula K, Gdula MR, Panser K, Nojima T, Monks J, Wisniewski JR, Riepsaame J, Brockdorff N, Pauli A, Proudfoot NJ. 2019. Selective roles of vertebrate PCF11 in premature and full-length transcript termination. Mol Cell 74: 158–172.e9. 10.1016/j.molcel.2019.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaca E, Weitzer S, Pehlivan D, Shiraishi H, Gogakos T, Hanada T, Jhangiani SN, Wiszniewski W, Withers M, Campbell IM, et al. 2014. Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell 157: 636–650. 10.1016/j.cell.2014.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. 2014. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15: R29 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AV, Oesterreich S, Davidson NE. 2015. MCF-7 cells–changing the course of breast cancer research and care for 45 years. J Natl Cancer Inst 107: djv073 10.1093/jnci/djv073 [DOI] [PubMed] [Google Scholar]
- Li W, You B, Hoque M, Zheng D, Luo W, Ji Z, Park JY, Gunderson SI, Kalsotra A, Manley JL, et al. 2015. Systematic profiling of poly(A)+ transcripts modulated by core 3′ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genet 11: e1005166 10.1371/journal.pgen.1005166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell 9: 1101–1111. 10.1016/S1097-2765(02)00518-X [DOI] [PubMed] [Google Scholar]
- Luo W, Ji Z, Pan Z, You B, Hoque M, Li W, Gunderson SI, Tian B. 2013. The conserved intronic cleavage and polyadenylation site of CstF-77 gene imparts control of 3′ end processing activity through feedback autoregulation and by U1 snRNP. PLoS Genet 9: e1003613 10.1371/journal.pgen.1003613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Gruber AR, Keller W, Zavolan M. 2012. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′-UTR length. Cell Rep 1: 753–763. 10.1016/j.celrep.2012.05.003 [DOI] [PubMed] [Google Scholar]
- Masamha CP, Xia Z, Yang J, Albrecht TR, Li M, Shyu AB, Li W, Wagner EJ. 2014. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature 510: 412–416. 10.1038/nature13261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684. 10.1016/j.cell.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361. 10.1038/385357a0 [DOI] [PubMed] [Google Scholar]
- Meinhart A, Cramer P. 2004. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature 430: 223–226. 10.1038/nature02679 [DOI] [PubMed] [Google Scholar]
- Morris AR, Bos A, Diosdado B, Rooijers K, Elkon R, Bolijn AS, Carvalho B, Meijer GA, Agami R. 2012. Alternative cleavage and polyadenylation during colorectal cancer development. Clin Cancer Res 18: 5256–5266. 10.1158/1078-0432.CCR-12-0543 [DOI] [PubMed] [Google Scholar]
- Neve J, Furger A. 2014. Alternative polyadenylation: less than meets the eye? Biochem Soc Trans 42: 1190–1195. 10.1042/BST20140054 [DOI] [PubMed] [Google Scholar]
- Noble CG, Hollingworth D, Martin SR, Ennis-Adeniran V, Smerdon SJ, Kelly G, Taylor IA, Ramos A. 2005. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nat Struct Mol Biol 12: 144–151. 10.1038/nsmb887 [DOI] [PubMed] [Google Scholar]
- Noble CG, Beuth B, Taylor IA. 2007. Structure of a nucleotide-bound Clp1-Pcf11 polyadenylation factor. Nucleic Acids Res 35: 87–99. 10.1093/nar/gkl1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogami K, Chen Y, Manley JL. 2018. RNA surveillance by the nuclear RNA exosome: mechanisms and significance. Noncoding RNA 4: 8 10.3390/ncrna4010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogorodnikov A, Levin M, Tattikota S, Tokalov S, Hoque M, Scherzinger D, Marini F, Poetsch A, Binder H, Macher-Goppinger S, et al. 2018. Transcriptome 3'end organization by PCF11 links alternative polyadenylation to formation and neuronal differentiation of neuroblastoma. Nat Commun 9: 5331 10.1038/s41467-018-07580-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ. 2016. Transcriptional termination in mammals: stopping the RNA polymerase II juggernaut. Science 352: aad9926 10.1126/science.aad9926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez A, Shuman S, Schwer B. 2008. Human RNA 5'-kinase (hClp1) can function as a tRNA splicing enzyme in vivo. RNA 14: 1737–1745. 10.1261/rna.1142908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski M, Dichtl B, Hubner W, Keller W. 2003. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J 22: 2167–2177. 10.1093/emboj/cdg200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman DW, Nakamura K, Nallur S, Dookwah MT, Metheetrairut C, Slack FJ, Weidhaas JB. 2016. miR-34 activity is modulated through 5'-end phosphorylation in response to DNA damage. Nat Commun 7: 10954 10.1038/ncomms10954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647. 10.1126/science.1155390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer P, Tuting C, Schonemann L, Kuhn U, Treiber T, Treiber N, Ihling C, Graber A, Keller W, Meister G, et al. 2018. Reconstitution of mammalian cleavage factor II involved in 3′ processing of mRNA precursors. RNA 24: 1721–1737. 10.1261/rna.068056.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer AE, Eggens VR, Caglayan AO, Reuter MS, Scott E, Coufal NG, Silhavy JL, Xue Y, Kayserili H, Yasuno K, et al. 2014. CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell 157: 651–663. 10.1016/j.cell.2014.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Manley JL. 2015. The end of the message: multiple protein-RNA interactions define the mRNA polyadenylation site. Genes Dev 29: 889–897. 10.1101/gad.261974.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR III, Frank J, Manley JL. 2009. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell 33: 365–376. 10.1016/j.molcel.2008.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- Spies N, Burge CB, Bartel DP. 2013. 3′-UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome Res 23: 2078–2090. 10.1101/gr.156919.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL. 1996. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87: 941–952. 10.1016/S0092-8674(00)82000-0 [DOI] [PubMed] [Google Scholar]
- Tian B, Graber JH. 2012. Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip Rev RNA 3: 385–396. 10.1002/wrna.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Manley JL. 2017. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol 18: 18–30. 10.1038/nrm.2016.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner RE, Pattison AD, Beilharz TH. 2018. Alternative polyadenylation in the regulation and dysregulation of gene expression. Semin Cell Dev Biol 75: 61–69. 10.1016/j.semcdb.2017.08.056 [DOI] [PubMed] [Google Scholar]
- Venkataraman K, Brown KM, Gilmartin GM. 2005. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev 19: 1315–1327. 10.1101/gad.1298605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volanakis A, Kamieniarz-Gdula K, Schlackow M, Proudfoot NJ. 2017. WNK1 kinase and the termination factor PCF11 connect nuclear mRNA export with transcription. Genes Dev 31: 2175–2185. 10.1101/gad.303677.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Zheng D, Wei L, Ding Q, Tian B. 2019. Regulation of intronic polyadenylation by PCF11 impacts mRNA expression of long genes. Cell Rep 26: 2766–2778 e2766. 10.1016/j.celrep.2019.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzer S, Martinez J. 2007. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature 447: 222–226. 10.1038/nature05777 [DOI] [PubMed] [Google Scholar]
- West S, Proudfoot NJ. 2008. Human Pcf11 enhances degradation of RNA polymerase II-associated nascent RNA and transcriptional termination. Nucleic Acids Res 36: 905–914. 10.1093/nar/gkm1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ, Li W. 2014. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3'-UTR landscape across seven tumour types. Nat Commun 5: 5274 10.1038/ncomms6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Gilmour DS. 2006. Pcf11 is a termination factor in Drosophila that dismantles the elongation complex by bridging the CTD of RNA polymerase II to the nascent transcript. Mol Cell 21: 65–74. 10.1016/j.molcel.2005.11.002 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Fu J, Gilmour DS. 2005. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev 19: 1572–1580. 10.1101/gad.1296305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445. 10.1128/MMBR.63.2.405-445.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.