Abstract
Many cellular noncoding RNAs contain chemically modified nucleotides that are essential for their function. The Epstein–Barr virus expresses two highly abundant noncoding RNAs called EBV-encoded RNA 1 (EBER1) and EBER2. To examine whether these viral RNAs contain modified nucleotides, we purified native EBERs from EBV-infected cells and performed mass spectrometry analysis. While EBER2 contains no modified nucleotides at stoichiometric amounts, EBER1 was found to carry 5-methylcytosine (m5C) modification. Bisulfite sequencing indicated that a single cytosine of EBER1 is methylated in ∼95% of molecules, and the RNA methyltransferase NSUN2 was identified as the EBER1-specific writer. Intriguingly, ablation of NSUN2 and thus loss of m5C modification resulted in an increase in EBER1 levels. We further found that EBER1 is a substrate for the RNase Angiogenin and cleavage in vivo is dependent on the presence of m5C, providing an explanation as to why loss of m5C increases EBER1 levels. Taken together, our observations indicate that m5C, a modification previously shown for tRNAs to oppose Angiogenin-mediated degradation, can also adversely affect RNA stability.
Keywords: 5-methylcytosine, EBV noncoding RNA, NSUN2
INTRODUCTION
Epstein–Barr virus (EBV), a human lymphotropic gamma-1 herpesvirus, expresses two noncoding RNAs called EBV-encoded RNA 1 (EBER1) and EBER2, which localize to the nucleus and are ∼170 nt long (Lerner et al. 1981; Howe and Steitz 1986). Upon infection, EBV mainly persists in a latent (dormant) stage by expressing a minimal set of viral genes and only sporadically enters the lytic cycle to produce progeny virions. We have recently shown that EBER2 acts as a guide RNA to recruit the host transcription factor PAX5 to its target sites on the EBV genome, and this recruitment is essential for efficient viral lytic replication (Lee et al. 2015). On the other hand, the molecular function of EBER1 remains poorly understood. The study of EBER1, unlike EBER2, is particularly hindered by the fact that EBER1-RNPs are refractory to antisense oligonucleotide (ASO)-entailing techniques, as no regions are available for hybridization under native conditions, ruling out conventional knockdown or RNA–protein interaction studies to elucidate its function and necessitating the use of aptamers (Lee et al. 2012). Nonetheless, both EBERs must play a fundamental role in the viral life cycle given the fact that both noncoding RNA genes are preserved in all clinical isolates of EBV and exhibit high sequence conservation with only minor nucleotide polymorphisms (Moss and Steitz 2013; Xu et al. 2019).
A remarkable feature of EBERs is their high expression level with 105–106 copies of each viral RNA being present in an infected host cell (Moss and Steitz 2013). It is unclear why this high copy number is necessary for EBV maintenance or how this abundance is achieved. Cellular noncoding RNAs of comparable copy number, such as ribosomal RNAs (rRNAs), small nuclear RNAs, or transfer RNAs (tRNAs), harbor a diverse set of RNA modifications that increases their stability (Roundtree et al. 2017; Bohnsack and Sloan 2018). In recent years, the study of RNA modifications has garnered renewed interest owing to the development of sensitive tools entailing next-generation sequencing, which allowed the search for RNA modifications in a transcriptome-wide manner. Several modifications, such as N6-methyladenosine, N1-methyladenosine, 5-methylcytosine (m5C), or pseudouridine, have thus been shown to not be restricted to abundant noncoding RNAs, but also to be present in messenger RNAs (mRNAs) and to regulate diverse aspects of mRNA processing (Dominissini et al. 2012; Meyer et al. 2012; Squires et al. 2012; Carlile et al. 2014; Schwartz et al. 2014; Li et al. 2017; Safra et al. 2017). In addition, many of the writer, reader, and eraser proteins of RNA modifications have been found to be deregulated in disease tissue, arguing in favor of an essential contribution of these marks to tissue development (Delaunay and Frye 2019).
In addition to tRNAs, rRNAs, and mRNAs, several other functional noncoding RNAs have been shown to contain m5C modifications, such as the lncRNAs HOTAIR and XIST or the RNA polymerase III transcripts H1 and vault RNAs (Squires et al. 2012; Hussain et al. 2013a). The RNA methyltransferases responsible for the deposition of this methyl mark belong to the methyltransferase families of NOP2/Sun (NSUN) and DNA methyltransferase 2 (DNMT2), of which the NSUN2 enzyme has the broadest substrate specificity (Trixl and Lusser 2019). Like other RNA modifications, m5C appears to mediate different functions based on RNA context. For example, m5C in tRNA promotes RNA stability by counteracting endonucleolytic cleavage by the RNase Angiogenin (ANG) (Blanco et al. 2014), while m5C in mRNAs has been shown to promote nuclear export through binding to the RNA-binding protein ALYREF (ALY) (Yang et al. 2017), an adapter protein for nuclear export of mRNAs. The only other reported m5C reader to date is YBX1, a highly abundant general mRNA binding protein (Singh et al. 2015), which in certain biological contexts can enhance mRNA stability (Yang et al. 2019).
Based on the pervasiveness of RNA modifications and the high abundance of EBERs, we asked whether these viral transcripts also contain any of these post-transcriptional marks. We purified native EBERs and subjected them to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis to search for RNA modifications in an unbiased manner. Interestingly, EBER1 was found to carry only m5C modification at stoichiometric levels (i.e., approximately one modification per RNA) at a single cytosine, and abrogation of this mark resulted in an increase in EBER1 levels. We further show that EBER1 is a substrate for the RNase ANG and that ANG-mediated cleavage in vivo is dependent on the presence of m5C. In summary, our results highlight a novel outcome of this modification in RNA regulation by promoting degradation and thus negatively affecting RNA stability.
RESULTS
LC-MS/MS analysis detects m5C modification in EBER1
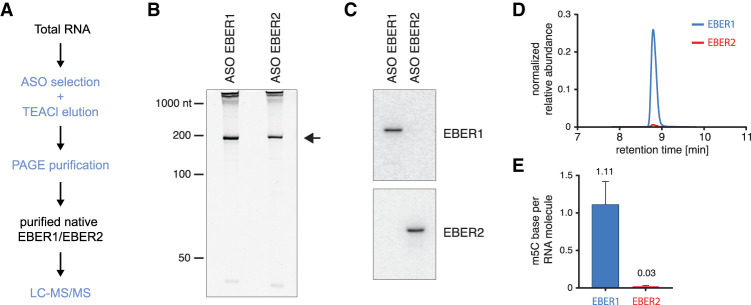
We examined whether EBER1 and EBER2 contain modified nucleotides by LC-MS/MS. This unbiased detection method requires ∼100 ng of highly pure input material devoid of common contaminating cellular RNAs, such as rRNAs or tRNAs, which contain a plethora of RNA modifications that would skew the analysis. We utilized a previously developed protocol to isolate EBERs from total RNA of EBV-positive lymphocytes (BJAB-B1 cells) by combining ASO selection under denaturing conditions and elution of bound RNAs with tetraethylammonium chloride (TEACl)-containing buffer (Nanni and Lee 2018), followed by PAGE purification (Fig. 1A,B). We verified by northern blot analysis that the isolated RNAs were indeed EBER1 and EBER2 (Fig. 1C) and subjected the RNAs to LC-MS/MS analysis as previously described (Thuring et al. 2017). While no RNA modification for EBER2 was detected in stoichiometric amounts, we observed approximately one m5C modification per EBER1 molecule (Fig. 1D,E). Only a few other modifications, such as m7G and m5U, probably stemming from contaminating RNA species, were detected at trace levels for EBER1 (less than 0.03 modifications per RNA molecule; please see Supplemental Table 1 for a complete list of analyzed modifications).
FIGURE 1.
Detecting m5C modification on EBER1 by LC-MS/MS analysis. (A) Schematic outline of isolating native EBERs from total RNA followed by mass spec analysis. EBERs are isolated by ASO selection under denaturing conditions and eluted from the beads with TEACl buffer followed by denaturing PAGE. Please note that EBER1-RNPs are refractory to ASO-selection, as no regions are available for hybridization, while naked EBER1 RNA can be purified by ASOs. Size-selected EBERs are subjected to LC-MS/MS analysis. (B) SYBR Gold-stained polyacrylamide gel of purified native EBER1 and EBER2. Arrow indicates the EBERs. (C) Northern blot analysis of the gel-excised EBER1 and EBER2 to confirm correct purification. (D,E) LC-MS/MS trace for m5C modification. EBER1 contains approximately one m5C modification per RNA molecule, while EBER2 carries no chemical modification.
NSUN2 methylates C145 of EBER1
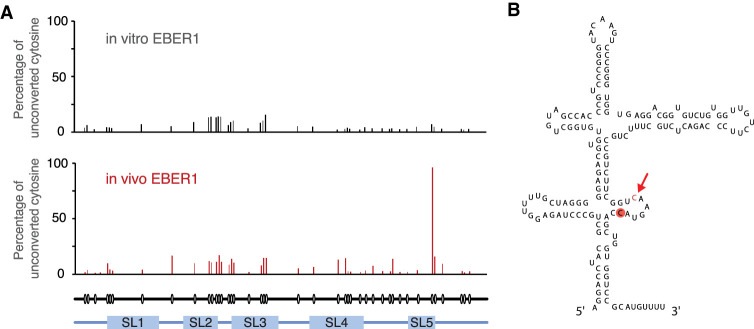
We next performed bisulfite sequencing of native EBER1 to localize the m5C modification within this viral transcript and determine whether a specific site is consistently methylated or whether the methyl mark is deposited randomly throughout EBER1. Cytosine residues become deaminated to uracils in the presence of bisulfite, but m5C-modified bases are refractory to this conversion. Thus, the non-conversion of cytosines as determined by sequencing of cDNA following reverse transcription of bisulfite-treated RNA indicates m5C modification sites. To examine for methylation along the entire EBER1 molecule, we ligated 5′ and 3′ RNA adapters to both ends of EBER1 by splint ligation upon bisulfite treatment and, after reverse transcription, used primers annealing to the adapters to PCR amplify full-length EBER1 cDNA. The EBER1 amplicons were then converted into an Illumina-compatible sequencing library, and deep sequencing of at least 105 reads revealed that ∼95% of all EBER1 molecules are m5C modified at C145 located within stem–loop 5 (Fig. 2A,B). We also included bisulfite sequencing of in vitro transcribed EBER1 to rule out that secondary structure formation is the cause of non-conversion of C145, as C-to-U conversion requires single-strandedness of RNA. As shown in Figure 2A (top panel), no notable peak was observed for synthetic EBER1, indicating that nucleotide 145 is indeed methylated in vivo. Moreover, as almost all EBER1 molecules exhibit m5C modification at this particular site, this modification may play an essential part in EBER1 function.
FIGURE 2.
Verification and localization of m5C modification on EBER1 by bisulfite sequencing. (A) C145 of EBER1 is m5C modified in ∼95% of EBER1 molecules. 5′ and 3′ adapters were ligated to in vitro transcribed or native EBER1 by splint ligation. Primers annealing to the adapters were used to RT-PCR-amplify EBER1, and amplicons were converted into an Illumina-compatible library. Over 105 reads were sequenced per sample and the percentage of unconverted cytosines is shown in the graph (representative of three independent biological replicates). Circles underneath the tracks indicate the position of cytosines within EBER1; the five stem–loops (SL) are indicated. (B) Secondary structure of EBER1. The methylated cytosine is highlighted in red. Arrow indicates the major in vitro ANG cut site.
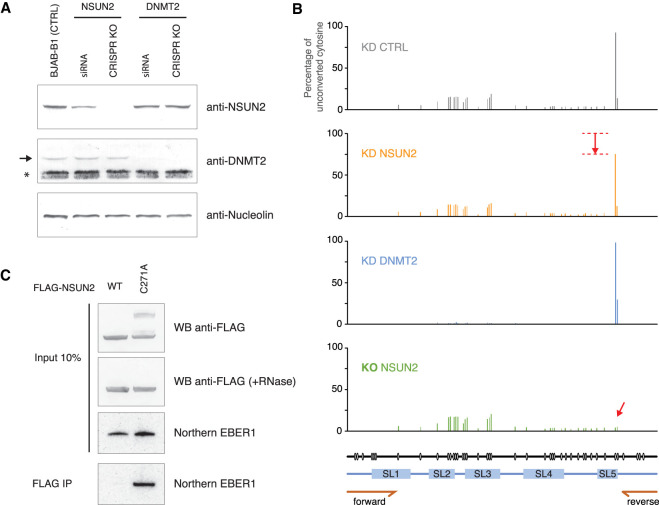
We next sought to identify the RNA methyltransferase that deposits this mark on EBER1. Several m5C RNA methyltransferases have been identified, of which DNMT2 and NSUN2 have the broadest substrate specificity (Motorin et al. 2010; Trixl and Lusser 2019) and were our prime candidates for methylating EBER1. We first used short-interfering (si)RNAs against NSUN2 and DNMT2 to deplete their protein levels and verified their knockdown efficiencies by western blot analysis. RNAi against NSUN2 reduced its abundance to ∼40% of wild-type levels, while DNMT2 knockdown resulted in a highly efficient depletion and a barely detectable signal by western blot (Fig. 3A). Bisulfite sequencing of EBER1 was performed after isolating total RNA from knockdown cells and amplifying EBER1 by RT-PCR using internal primers (location of primer-annealing regions shown at the bottom of Fig. 3B) instead of ligating adapters to both ends to scan the entire RNA, as we were mainly interested in the methylation level of the key nucleotide C145. As determined by deep sequencing of bisulfite-treated EBER1 amplicons, knockdown with control and DNMT2 siRNAs exhibited a comparable m5C modification level at C145 of ∼95% of all EBER1 molecules (Fig. 3B, top and third panel), suggesting that DNMT2 is not the EBER1-specific methyltransferase. On the other hand, knockdown of NSUN2 resulted in a reproducible reduction of C145 methylation to ∼70% (Fig. 3B, second panel), indicating that NSUN2 is responsible for the m5C modification of EBER1. We hypothesized that the modest decrease in m5C methylation is due to the incomplete knockdown of NSUN2 by RNAi and sought to obtain a more pronounced depletion by generating a CRISPR knockout of NSUN2. Previous studies have shown that NSUN2 knockout animals are viable and display phenotypes in epidermis and testis development (Blanco et al. 2011; Hussain et al. 2013b), implying that a NSUN2 knockout could be achieved in EBV-positive lymphocyte cell lines. We succeeded in generating NSUN2 knockout BJAB-B1 clones, as verified by western blot analysis (Fig. 3A; Supplemental Fig. 1A–C), and performed bisulfite deep sequencing of EBER1 with knockout cells. NSUN2 ablation resulted in a complete loss of m5C methylation at nucleotide C145 (Fig. 3B, arrow in bottom panel), demonstrating that NSUN2 is the key EBER1-specific methyltransferase.
FIGURE 3.
NSUN2 is the RNA methyltransferase for EBER1. (A) Western blot analysis following depletion of NSUN2 and DNMT2 by siRNA-treatment or CRISPR knockout. RNAi against NSUN2 results in a reduction of NSUN2 protein levels to ∼40% of wild-type levels. Arrow indicates the DNMT2 band; asterisk indicates a major cross-reactive band recognized by the anti-DNMT2 antibody used in this study in lymphocyte lysate. Anti-Nucleolin antibody was used as a loading control. (B) Bisulfite sequencing of EBER1 following RNAi against NSUN2 and DNMT2, or knockout of NSUN2. Depletion of DNMT2 does not affect m5C modification of EBER1 C145, while RNAi against NSUN2 decreases the abundance of C145 methylation (second panel). Knockout of NSUN2 completely eliminates C145 methylation (bottom panel, arrow). EBER1 was amplified using internal primers whose annealing locations are shown at the bottom (half arrows marked forward and reverse). (C) Coimmunoprecipitation of EBER1 by C271A NSUN2 mutant. Western blot using anti-FLAG antibody showing expression of FLAG-tagged wild-type or C271A mutant of NSUN2 (top panel). The C271A mutant shows several higher molecular weight bands, which disappear upon RNase A treatment (second panel). Northern blot analysis of EBER1 for the input and immunoprecipitated material is shown (bottom two panels).
To show that the loss of m5C modification on EBER1 following NSUN2 depletion is not due to indirect effects, we leveraged the NSUN2 C271A mutant to demonstrate that EBER1 is indeed a substrate for NSUN2. The methyltransferase activity of NSUN2 requires two cysteine residues within the active site, of which C271 mediates the release of the methylated substrate from the enzyme upon completion of the methylation reaction; the C271A mutation causes covalent tethering of RNA substrates to NSUN2 mutant protein (Hussain et al. 2013a). We expressed either FLAG-tagged wild-type or C271A mutant NSUN2 in CRISPR knockout BJAB-B1 cells and examined whether EBER1 would coprecipitate with the C271A mutant. Western blot analysis confirmed that the C271A mutant covalently bound RNA substrates, as higher molecular weight bands were observed, which disappeared after RNase A treatment of cell lysate and were not detected for the wild-type enzyme (Fig. 3C, top). Importantly, EBER1 was coimmunoprecipitated only by the C271A mutant and not by wild-type NSUN2 (Fig. 3C, bottom), demonstrating that EBER1 is indeed a substrate of NSUN2. Taken together, our results demonstrate that NSUN2 is responsible for the m5C modification of nucleotide C145 of EBER1.
m5C-modified EBER1 is targeted by ANG for degradation
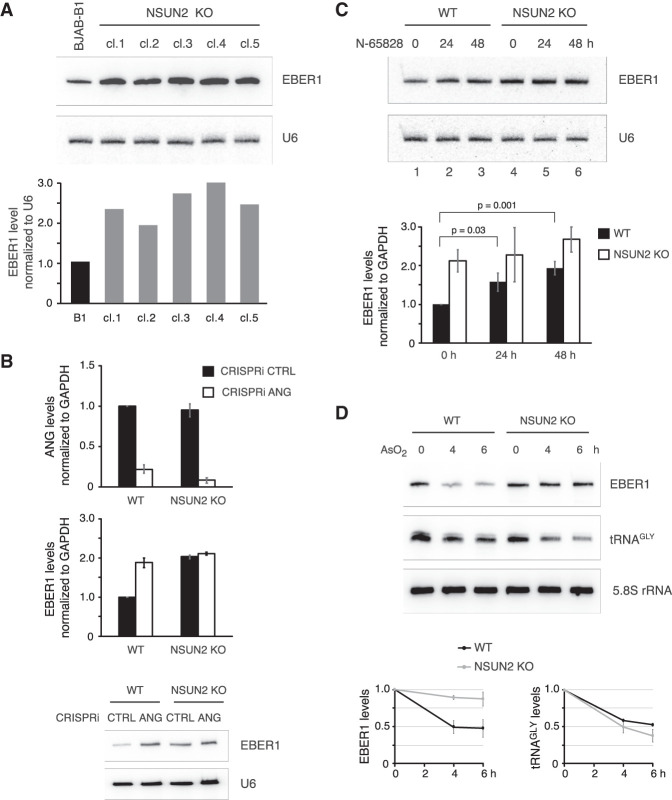
While characterizing NSUN2 knockout clones of the BJAB-B1 cell line, we observed that all studied clones exhibited an elevated EBER1 level that exceeded wild-type abundance by approximately two- to threefold (Fig. 4A). This increase could not be explained by a higher EBV genome copy number inside host cells and consequently higher transcription of EBER1, as all clones had a comparable EBV genome content, except for knockout clone 4, which had a 1.8-fold higher copy number (Supplemental Fig. 1D). As m5C modification of tRNAs has previously been shown to affect RNA stability through degradation by ANG (Schaefer et al. 2010; Blanco et al. 2014), we examined whether this RNase also modulates the stability of EBER1. To this end, we knocked down ANG by CRISPR interference (CRISPRi), which uses a catalytically inactive Cas9 endonuclease (dCas9) that retains the ability to be targeted by single guide (sg)RNAs and acts as a road block for RNA polymerase II (Gilbert et al. 2013), as our attempts of using RNAi with siRNAs were unsuccessful in BJAB-B1 lymphocytes. We were able to identify a sgRNA that targets a promoter-proximal region of ANG and efficiently knocks down its expression (Fig. 4B, top). Notably, EBER1 levels were increased following ANG depletion to a similar extent as observed upon NSUN2 knockout (Fig. 4B, bottom), while ANG depletion had no apparent further effect on EBER1 levels in NSUN2 knockout cells. To corroborate the ANG-mediated regulation of EBER1, we next pharmacologically inhibited ANG by treating both wild-type and NSUN2 knockout cells with the ANG inhibitor N-65828 (Kao et al. 2002) and subsequently measured EBER1 levels by qRT-PCR. Inhibition of ANG did not significantly alter EBER1 levels in NSUN2 knockout cells, whereas in wild-type cells EBER1 levels were gradually increased to approximately twofold by 48 h of N-65828 treatment (Fig. 4C). To further substantiate the notion that ANG cleaves EBER1 in an m5C-dependent manner, wild-type and NSUN2 knockout cells were treated with arsenite, as previous studies have shown that such oxidative stress activates ANG-mediated degradation of target RNAs, that is, tRNA, and their decrease is enhanced in the absence of m5C modification (Blanco et al. 2014). While tRNA levels were expectedly reduced upon arsenite treatment and even more so in NSUN2 knockout cells given the stabilizing effect of m5C modification on tRNAs, (Fig. 4D, middle panel), EBER1 levels were greatly unaffected when unmodified and exhibited a lower stability only when m5C-modified (Fig. 4D, top). Taken together, our results indicate that m5C modification on EBER1 decreases its stability through ANG-mediated degradation.
FIGURE 4.
Presence of m5C modification on EBER1 elicits a reduction in RNA levels in vivo. (A) Five BJAB-B1 clones generated by CRISPR knockout of NSUN2 (cl.1–cl.5) were probed by northern blot for EBER1. Please see Supplemental Figure 1 for further characterization of knockout cell lines. U6 was probed as a loading control. Quantification of a representative northern blot is shown in the graph below. In the absence of m5C modification, EBER1 levels are elevated approximately two- to threefold. (B) CRISPRi against ANG in wild-type or NSUN2 knockout BJAB-B1 cells. ANG mRNA and EBER1 levels were measured by qRT-PCR following ANG knockdown. A representative northern blot probing for EBER1 is also shown. ANG depletion in wild-type cells results in an increase in EBER1 levels, but not in NSUN2 knockout cells. Values are the average of three independent biological replicates; error bars indicate standard deviation. (C) Wild-type or NSUN2 knockout BJAB-B1 cells were treated with the ANG inhibitor N-65828 at 100 µM for the indicated time periods. A representative northern blot analysis probing for EBER1 and U6 as a loading control is shown. The graph underneath depicts the EBER1 levels measured by qRT-PCR normalized to GAPDH expression. Values are the average of three independent biological replicates; error bars indicate standard deviation. In wild-type cells, inhibition of ANG results in a statistically significant increase in EBER1 levels (P-values from Student's t-test are indicated), while in NSUN2 knockout cells EBER1 abundance does not change significantly. (D) Stress-induced ANG-mediated cleavage of EBER1 is enhanced by m5C modification. EBV-positive wild-type and NSUN2 knockout cells were treated with 0.5 mM arsenite for the indicated time periods followed by northern blot analysis to quantify EBER1 and tRNA levels. While tRNAGLY is less stable upon stress in NSUN2 KO cells compared to wild-type cells, EBER1 levels show the opposite trend by being largely unaffected when unmodified and prone to degradation in the presence of m5C. Quantification of EBER1 and tRNAGLY northern blots normalized to 5.8S rRNA levels, which remained unchanged upon arsenite treatment, from three independent biological replicates is shown; error bars indicate standard deviation.
EBER1 is an in vitro substrate for ANG
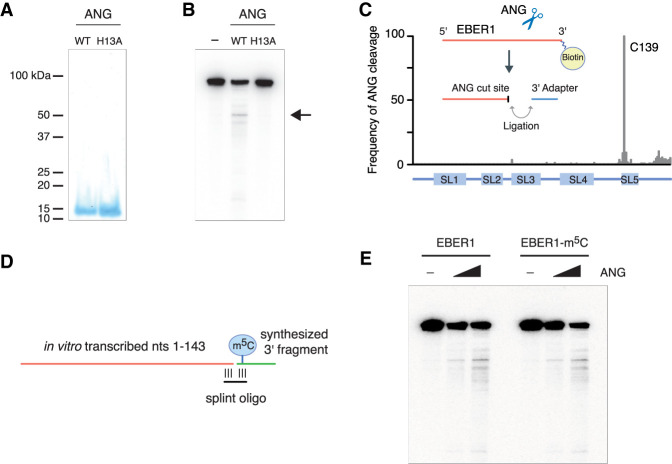
To show that EBER1 is a direct substrate of ANG, we performed in vitro RNase digestion assays with recombinant ANG and synthetic EBER1 as a substrate. We recombinantly expressed and purified from E. coli both wild-type and a catalytically inactive ANG enzyme to near homogeneity (Fig. 5A). Mutating a single histidine in the catalytic core of ANG to alanine (H13A) has been shown to greatly abolish its RNase activity (Shapiro and Vallee 1989). Incubation of EBER1 with ANG revealed a predominant cleavage site (Fig. 5B, arrow), indicating specific processing and EBER1 to be a bona fide substrate for ANG in vitro. This specific degradation product was not generated when EBER1 was incubated with the H13A ANG mutant, thus ruling out the possibility that the cleavage was mediated by a contaminating nuclease. We next mapped the ANG-specific cleavage site on EBER1 by attaching a 3′ linker to all ANG-mediated internal cut sites and subsequently identifying computationally the junction sites with EBER1 after deep sequencing the degradation products. Consistent with the homology to RNase A and the fact that RNase A cuts after cytosines and uracils, the majority of cleavage occurred after C139 of EBER1 (45.4%, 1.66 × 105 of 3.66 × 105 reads) (Fig. 5C), which is located in stem–loop 5 and adjacent to the m5C modification site at C145 (Fig. 2B), suggesting that ANG cleavage may be potentially linked to the recognition of m5C modification in vivo. To examine whether m5C modification confers substrate preference also in vitro, unmodified or m5C-modified EBER1 molecules were generated by splint ligation of an in vitro transcribed 5′ fragment consisting of nucleotides 1–143 and a chemically synthesized 3′ fragment either containing or lacking m5C at nucleotide 145 (Fig. 5D). Both EBER1 substrates were subjected to ANG cleavage. However, the presence of m5C modification had no effect on substrate specificity or preference (Fig. 5E), suggesting that additional factors may govern EBER1 turnover in vivo.
FIGURE 5.
EBER1 is an in vitro substrate of Angiogenin. (A) Recombinant wild-type and catalytically inactive (H13A mutant) recombinant ANG was purified from E. coli. Purified fractions were resolved on a 15% SDS gel and stained with Coomassie. (B) In vitro transcribed EBER1 was incubated with either wild-type or H13A mutant ANG followed by northern blot analysis to detect degradation products of EBER1. While a smear of bands is visible to a minor extent, indicative of random degradation, one preferential cut site within EBER1 is detected (arrow). This degradation product is not observed when incubated with the H13A ANG mutant, demonstrating an ANG-specific RNase activity. (C) EBER1 degradation products were attached to a 3′ linker, converted into an Illumina-compatible library, and deep-sequenced to identify ANG-mediated cut sites. ANG primarily digests EBER1 after C139, which is located in stem–loop 5 adjacent to the m5C site. (D,E) In vitro ANG digestion assay with either unmodified or m5C-modified EBER1 as substrate. Schematic for generating either unmodified or m5C-modified EBER1 by splint ligation. An RNA fragment consisting of EBER1 nucleotides 1–143 was in vitro transcribed using T7 polymerase. A 3′ fragment either lacking or containing m5C145 was chemically synthesized and ligated to the 5′ fragment using a splint oligonucleotide (D). m5C modification of EBER1 does not confer substrate preference in vitro (E).
We next examined whether the reported m5C readers ALY and YBX1 also recognize this modification on EBER1. To this end, we performed HITS-CLIP (high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation) for ALY and YBX1 in wild-type BJAB-B1 cells to map their binding sites within EBER1 with nucleotide resolution. After immunoprecipitating ALY or YBX1 from EBV-positive cell lysate, higher molecular weight bands corresponding to RNA–protein adducts were observed in the UV-crosslinked sample (Supplemental Fig. 2A, red boxes). These bands were excised, converted into an Illumina-compatible library, and following deep sequencing were mapped to the EBV reference genome. While YBX1 did not bind over the m5C modification site of EBER1 and was ruled out as an EBER1-specific m5C reader, three distinct peaks were observed for ALY, one of which overlapped with nucleotide C145 (Supplemental Fig. 2B). To examine whether ALY is a bona fide m5C reader for EBER1, we repeated the HITS-CLIP assay with NSUN2 knockout cells, which do not contain any m5C modification at C145. However, the same binding profile of ALY was observed over EBER1 (Supplemental Fig. 2B, middle track), demonstrating that ALY binding to EBER1 is independent of m5C modification. Consistent with the fact that neither ALY nor YBX1 are EBER1-specific m5C readers, knockdown of neither protein had an effect on EBER1 levels (Supplemental Fig. 2C–F).
m5C modification of EBER1 is not required for viral lytic replication
The related EBER2 viral noncoding RNA has recently been shown to be required for efficient viral lytic replication (Lee et al. 2015). We thus asked whether methylation of EBER1 could play a role in this process as well. For this, we generated NSUN2 knockout clones of the replication-permissive cell line HH514–16 (Rabson et al. 1983) and treated both wild-type and NSUN2 knockout cells with sodium butyrate to induce lytic reactivation. Replication of the viral genome inside host cells as well as the viral titer in the culture supernatant were not affected by the absence of NSUN2 and thus loss of m5C modification in EBER1 under these reactivation conditions in this cell line (Supplemental Fig. 3). This observation suggests that the relevance of EBER1 methylation is restricted to the latent stage of the EBV life cycle.
DISCUSSION
As many cellular noncoding RNAs expressed at high copy number have been shown to contain various RNA modifications, we probed EBER1 and EBER2 by LC-MS/MS for the presence of any type of modified nucleotide. We found that EBER1 contains m5C at a single site in almost all RNA molecules (Figs. 1, 2), suggesting that this methyl mark may play an essential role in the as-yet unknown molecular function of EBER1. The small percentage (∼5%) of non-methylated EBER1 could represent nascent transcripts that have yet to undergo RNA maturation. In terms of secondary structure, the methylation site in EBER1 is somewhat reminiscent of the nucleotides in the T loop of tRNAs abutting the variable loop, where nucleotides 47/48 are m5C-modified by the RNA methyltransferase NSUN2 (Motorin et al. 2010). Consistent with the similarity in secondary structure of both RNAs, we identified the writer enzyme for EBER1 to be NSUN2 as well (Fig. 3). However, a major difference between tRNAs and EBER1 is the consequence of m5C modification. Whereas the deposition of m5C in tRNAs by NSUN2 decreases ANG-mediated endonucleolytic cleavage (Tuorto et al. 2012; Blanco et al. 2014), incorporation of m5C in EBER1 has a stimulatory effect on ANG-mediated degradation in vivo, as depletion of NSUN2 results in an increase in EBER1 levels (Fig. 4A). Moreover, this increase is observed upon inhibition of ANG as well, but only in wild-type cells and not in NSUN2-depleted cells, which completely lack methylation at C145 and are non-responsive to ANG depletion or inhibition in terms of EBER1 stabilization (Fig. 4B,C). Our in vitro RNase assays with recombinant ANG confirm EBER1 to be a direct substrate based on the generation of a specific cleavage product, rather than random degradation along the entire length of the RNA (Fig. 5A–C). We further observed that ANG-mediated cleavage in vitro is not stimulated by the incorporation of m5C within EBER1, as the extent of cleavage was comparable between the unmodified and m5C-modified substrates (Fig. 5E). Additional studies are required to reconcile this apparent inconsistency between the in vivo and in vitro observations. One possibility could be that an as-yet unknown reader protein of methylated C145 may bind to act in a stimulatory manner for ANG-mediated cleavage. Overall, this turnover mechanism may have been implemented to fine-tune EBER1 levels and prevent the accumulation of EBER1 beyond a certain intracellular threshold (approximately 106 copies per cell).
We examined whether the reported m5C reader proteins ALY or YBX1 bind this methyl mark on EBER1. While both factors interact with distinct regions on EBER1, consistent with previous reports that these proteins are promiscuous general RNA-binding proteins, the YBX1 footprint in our CLIP assay did not overlap with the methylation site. On the other hand, we observed ALY to bind EBER1 over three regions, one of which overlaps the methylation site (Supplemental Fig. 2B). However, the same binding profile was observed in NSUN2 knockout cells, which completely lack m5C methylation within EBER1, suggesting that ALY can also be ruled out as the EBER1-specific m5C reader, as we would have expected a complete abrogation of interaction. In line with the fact that neither ALY nor YBX1 are EBER1-specific m5C readers, depletion of neither factor resulted in a change in EBER1 levels. Thus, our results suggest that another as-yet unknown m5C reader protein may exist and bind in the context of EBER1.
The related viral noncoding RNA EBER2 plays a role in EBV lytic replication (Lee et al. 2015), and given the many parallels between both viral RNAs, we examined whether the m5C modification on EBER1 is also required for viral lytic replication. However, our results rule out a function for EBER1 methylation during the lytic stage of the EBV life cycle, as lytic reactivation assays indicated that viral DNA replication in NSUN2 knockout cells is indistinguishable from wild-type cells (Supplemental Fig. 3). While the molecular function of EBER1 remains unclear, the relevance of EBER1 m5C modification is thus likely restricted to the latency stage of EBV infection.
MATERIALS AND METHODS
Isolation of EBERs for LC-MS/MS analysis
Total RNA from EBV-infected BJAB-B1 cells was isolated with TRIzol. An amount of 250 µg of total RNA was resuspended in 100 µL TE buffer, incubated for 3 min at 95°C to disrupt secondary structures, and placed on ice. Biotinylated DNA ASOs against EBER1 (5′- TCACCACCCGGGACTTGTACCCGGGAC/3BioTEG-3′, complementary to nucleotides 61–87) and EBER2 (5′-CCTGACTTGCAAATGCTCTAGGCG/3BioTEG-3′, complementary to nucleotides 101–124) were complexed with MyOne Streptavidin C1 Dynabeads (ThermoFisher). An amount of 10 µL of ASO-beads were added together with 100 µL H2O, 100 µL Denaturant buffer (100 mM HEPES pH 7.5, 8 M urea, 200 mM NaCl, 2% SDS), and 300 µL 2× Hybridization buffer (1.12 M urea, 1.5 M NaCl, 10× Denhardt's solution, 10 mM EDTA), and incubated for 4 h at RT on a rotator. Beads were washed three times with Wash buffer (10 mM HEPES pH 7.5, 250 mM NaCl, 2 mM EDTA, 1 mM EGTA, 0.1% N-lauroylsarcosine, 0.2% SDS), and RNA was eluted from the beads by adding 200 µL TEACl buffer (10 mM Tris pH 7.4, 2.4 M TEACl, 0.05% Tween-20) and incubating the beads for 5 min at 40°C. RNA in the supernatant was extracted with phenol–chloroform and resolved on a 10% denaturing polyacrylamide gel. RNA running at the expected size were gel-extracted and verified by northern blot analysis to be EBER1 or EBER2. The following DNA oligonucleotides were used as northern blot probes after radioactive labeling with gamma-32P-ATP and PNK treatment: 5′-CCAGCTGGTACTTGACCGAAGAC-3′ (EBER1), 5′-CAGAGGGATTAGAGAATCCTGACTTG-3′ (EBER2). Approximately 100 ng of native purified EBER1 or EBER2 were obtained after PAGE purification and subjected to LC-MS/MS analysis as previously described (Thuring et al. 2017).
Bisulfite treatment of RNA and preparation of deep sequencing libraries
For localizing m5C modification within EBER1, total RNA or purified native and in vitro transcribed EBER1 were bisulfite-treated using the Methylamp RNA Bisulfite Conversion Kit (Epigentek) according to manufacturer's instructions. After C-to-U conversion, RNA was phosphatase-treated, followed by PNK-treatment to convert the triphosphate 5′ end of EBER1 into a monophosphate group for subsequent ligation. 5′ and 3′ adapters (5′-OH-AGGGAGGACGAUGCGG-OH-3′ and 5′-P-GUGUCAGUCACUUCCAGCGG-Puromycin-3′, respectively) were ligated specifically to EBER1 using the following splint oligonucleotides and T4 RNA Ligase 2: 5′-tcctCCGCATCGTCC-3′, 5′-TGACTGACACaaaacat-3′ (nucleotides complementary to EBER1 are in lower case). RNA was reverse-transcribed using SuperScript IV RT (ThermoFisher), and EBER1 was PCR-amplified using primers annealing to the adapter sequences (5′-AGGGAGGACGATGCGG-3′ and 5′-CCGCTGGAAGTGACTGACAC-3′). For measuring localized methylation level only, internal primers annealing to the very 5′ and 3′ ends of EBER1 were used in PCR: 5′-AGGATTTATGTTGTTTTAGAGGTTTTG-3′; 5′-AAAACATACAAACCACCAACT-3′).
EBER1 amplicons were converted into an Illumina-compatible deep sequencing library using the NEBNext Ultra II DNA Library Prep Kit (NEB). Following deep sequencing, raw reads from paired-end runs were paired using the program PEAR with default parameters (Zhang et al. 2014). Assembled reads were trimmed of 5′ and 3′ adapters using the program cutadapt with parameters ‘–m 15 –no-indels –g Adp1…Adp2’. Reads containing both adapters were mapped to the EBV genome (GenBank AJ507799.2), and alignment was performed using the program Bismark with parameters ‘–non-directional –bowtie2 -N 1 -L 10’ (Krueger and Andrews 2011). Bismark’s methylation extractor (v0.19.1_dev) was run with parameters ‘–s –comprehensive –bedgraph –CX_context’ to identify methylated cytosines. The output was used to generate histograms of methylation at each nucleotide position with Matplotlib.
RNAi, CRISPRi, and CRISPR knockout in EBV-infected BJAB-B1 cells
RNAi against NSUN2, DNMT2, and ANG were carried out by nucleofecting BJAB-B1 cells with siGENOME siRNAs from Dharmacon (NSUN2 cat no. M-018217-01-0005; DNMT2 cat no. M-006671-01-0005; ANG cat. no. M-011206-01-0005) at a concentration of 100 nM, unless stated otherwise, using the 4D-Nuclefector System (Lonza) in combination with SF solution and program EN-150. As control siRNA, the siGENOME Non-Targeting siRNA Pool #2 (Dharmacon cat. no. D-001206-14-05) was used. Two days after the initial nucleofection, BJAB-B1 cells were nucleofected again with siRNAs, and cells were harvested after 4 d of siRNA treatment. To monitor knockdown efficiencies, the following antibodies were used for western blot analysis: rabbit anti-NSUN2 (Proteintech cat. no. 20854-1-AP) at 1:2000 dilution; mouse anti-DNMT2 (Santa Cruz cat. no. sc365001) at 1:100 dilution, which performs suboptimally for lymphocyte lysates due to the recognition of a strong cross-reactive band. Three distinct anti-ANG antibodies were used in western blot analysis (Abcam cat. no. ab202829; Proteintech cat. no. 20854-1-AP; ThermoFisher cat. no. PA5-34422), but in our hands none detected a specific band in whole lymphocyte lysate. Instead, qRT-PCR was used to verify knockdown of ANG mRNA using primers 5′-GTTGGAAGAGATGGTGATGG-3′ and 5′-CATAGTGCTGGGTCAGGAAG-3′ as described (Sheng et al. 2014).
Gene knockout of NSUN2 and DNMT2 was carried out using the CRISPR Cas9 system as described (Ran et al. 2013) and obtaining the required Cas9 plasmid from Addgene. For large deletions of genomic regions, two sgRNAs were designed for each gene, one at the relative 5′ and the other at the 3′ end (see Supplemental Fig. 1 for genome location). The sgRNA sequences for NSUN2 were 5′-GAGCTCAAGATCGTGCCCGA-3′ and 5′-GCCAGACAATGACGTGACTG-3′, and for DNMT2 5′-AAGCTGTATACCTGCACAAG-3′ and 5′-GTATCCCACATACCGAACTC-3′.
CRISPRi against ANG was carried out with a deactivated Cas9 fused to the KRAB protein (dCas9-KRAB) as previously described (Lee et al. 2015) using a sgRNA (5′-AGCAGCAGCGAATAAGTACG-3′) targeting a promoter-proximal sequence of the ANG locus. RNAi with siGENOME siRNAs against ANG did not result in a successful knockdown.
Overexpression of NSUN2 and coimmunoprecipitation of RNA substrates
Constructs consisting of either wild-type or C271A mutant NSUN2 cDNA with an amino-terminal FLAG-tag cloned into the pcDNA3 vector were obtained from Dr. Bryan Cullen (Duke University) (Courtney et al. 2019). These plasmids were nucleofected into NSUN2-depleted BJAB-B1 cells using the protocol outlined above. Cells were subsequently lysed in Lysis buffer (10 mM Tris pH 7.4, 100 mM NaCl, 1 mM EDTA, 1% Triton X-100) in the presence of protease inhibitor, and immunoprecipitation was carried out by adding anti-FLAG M2 magnetic beads (SIGMA) and rotating at 4°C for 3 h. The beads were washed three times in Lysis buffer by rotating for 5 min at 4°C. Prior to western blot analysis, input material was treated with 10 µg RNase A for 15 min at 37°C where indicated. Prior to northern blot analysis, input and immunoprecipitated materials were resuspended in PBS containing 0.5% SDS and 1 mM EDTA and treated with 100 µg Proteinase K at 37°C for 1 h before RNA was extracted using TRIzol.
Induction of oxidative stress using arsenite
EBV-positive wild-type BJAB-B1 or NSUN2 KO cells were treated with 0.5 mM sodium arsenite (SIGMA S7400) at a cell density of 5 × 105 cells/mL for the indicated time periods. RNA was extracted using TRIzol followed by northern blot analysis. The following probe was used to detect tRNAGLY as previously described (Yamasaki et al. 2009): 5′-GGCAGGCGAGAATTCTACCACTGAACCACCAA-3′;
Generating m5C-modified EBER1 substrate for RNase digestion assay with ANG
ANG inhibitor N-65828 (Kao et al. 2002) was obtained from the NCI, resuspended in DMSO, and added to the culture medium at a final concentration of 100 µM for the indicated time periods. For in vitro RNase assays, recombinant wild-type and catalytically inactive H13A mutant ANG was expressed as a GST fusion protein in E. coli BL21 cells after cloning the cDNA encompassing amino acids 25–147 (lacking the putative leader sequence) into the pGEX-2K vector. After induction with IPTG at 0.5 mM overnight at 16°C, GST-ANG was purified from bacterial lysate with Glutathione Sepharose 4B resin (GE Healthcare) by incubating for 3 h at 4°C, followed by cleavage with thrombin for 2 h at RT. The supernatant was then loaded onto a MonoS column to isolate recombinant ANG.
To generate unmodified or m5C-modified EBER1 as a substrate for in vitro digestion assays, nucleotides 1–143 of EBER1 were in vitro transcribed with T7 polymerase and subsequently PAGE-purified. A 3′ fragment encompassing nucleotides 144–167 including or lacking m5C at nucleotide C145 was obtained from Dharmacon. These synthetic 3′ fragments also contain a biotin moiety at their 3′ ends. Both parts were ligated to each other using a splint oligonucleotide (5′-GGACCACCAGCTGGTACTTGACCGAAGACG-3′) and T4 RNA Ligase 2 (NEB). The full-length EBER1 molecules were PAGE-purified prior to RNase digestion. Indicated amounts of recombinant ANG were added to 50 ng of unmodified or m5C-modified EBER1 in PBS and incubated at 37°C for 10 min. Digestion reactions were terminated by the addition of phenol followed by extraction of RNA and northern blot analysis for EBER1.
Mapping ANG-mediated in vitro cleavage site in EBER1
EBER1 digestion products from an ANG cleavage reaction (starting material 50 ng) were purified by phenol–chloroform extraction. RNA was dephosphorylated by T4 polynucleotide kinase-treatment to remove 2′–3′ cyclic phosphate groups at the 3′ ends, followed by phenol–chloroform extraction. A 3′-linker (5′-P-GUGUCAGUCACUUCCAGCGG-Puromycin-3′) was ligated to the EBER1 cleavage products using T4 RNA ligase. The puromycin modification prevents linker self-ligation. Please note that in vitro transcribed EBER1 contains a biotin moiety at its 3′ end, thus preventing linker ligation to full-length EBER1 and only to internal degradation products. Unligated linkers were removed by purifying the reaction using RNAClean XP beads (Beckman Coulter). A 5′-linker was then added by template-switching reverse transcription by adding to the RNA 2 µL of 10 µM RT-primer (5′-CCGCTGGAAGTGACTGACAC-3′), 1 µL of 100 µM template-switch oligo (5′-AAGCAGTGGTATCAACGCAGAGTGAATrGrGrG-3′), 10 µL of 5x RT-buffer, 2.5 µL of 10 mM dNTPs, 1 µL of RNase inhibitor, and 2 µL of Maxima RT (ThermoFisher) in a 50 µL reaction. The RT-reaction was subsequently amplified by PCR using following primer pair: 5′-CCGCTGGAAGTGACTGACAC-3′ and 5′-AAGCAGTGGTATCAACGCAGAGT-3′, and the amplicon was converted into an Illumina-compatible deep sequencing library using NEBNext Ultra DNA Library Prep Kit (NEB).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Dr. Bryan Cullen (Duke University) for sharing NSUN2 plasmids; Dr. Michaela Frye (German Cancer Research Center, DKFZ) for reagents; the National Cancer Institute (NCI)/Division of Cancer Treatment and Diagnosis (DCTD)/Developmental Therapeutics Program (DTP) (http://dtp.cancer.gov) for compounds. This work was supported by the National Institutes of Health (grant number R21 AI151073).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.075275.120.
REFERENCES
- Blanco S, Kurowski A, Nichols J, Watt FM, Benitah SA, Frye M. 2011. The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet 7: e1002403 10.1371/journal.pgen.1002403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, et al. 2014. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J 33: 2020–2039. 10.15252/embj.201489282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Sloan KE. 2018. Modifications in small nuclear RNAs and their roles in spliceosome assembly and function. Biol Chem 399: 1265–1276. 10.1515/hsz-2018-0205 [DOI] [PubMed] [Google Scholar]
- Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. 2014. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515: 143–146. 10.1038/nature13802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney DG, Tsai K, Bogerd HP, Kennedy EM, Law BA, Emery A, Swanstrom R, Holley CL, Cullen BR. 2019. Epitranscriptomic addition of m(5)C to HIV-1 transcripts regulates viral gene expression. Cell Host Microbe 26: 217–227 e216. 10.1016/j.chom.2019.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay S, Frye M. 2019. RNA modifications regulating cell fate in cancer. Nat Cell Biol 21: 552–559. 10.1038/s41556-019-0319-0 [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485: 201–206. 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154: 442–451. 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe JG, Steitz JA. 1986. Localization of Epstein-Barr virus-encoded small RNAs by in situ hybridization. Proc Natl Acad Sci 83: 9006–9010. 10.1073/pnas.83.23.9006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J, et al. 2013a. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep 4: 255–261. 10.1016/j.celrep.2013.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Tuorto F, Menon S, Blanco S, Cox C, Flores JV, Watt S, Kudo NR, Lyko F, Frye M. 2013b. The mouse cytosine-5 RNA methyltransferase NSun2 is a component of the chromatoid body and required for testis differentiation. Mol Cell Biol 33: 1561–1570. 10.1128/MCB.01523-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao RY, Jenkins JL, Olson KA, Key ME, Fett JW, Shapiro R. 2002. A small-molecule inhibitor of the ribonucleolytic activity of human angiogenin that possesses antitumor activity. Proc Natl Acad Sci 99: 10066–10071. 10.1073/pnas.152342999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572. 10.1093/bioinformatics/btr167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Pimienta G, Steitz JA. 2012. AUF1/hnRNP D is a novel protein partner of the EBER1 noncoding RNA of Epstein-Barr virus. RNA 18: 2073–2082. 10.1261/rna.034900.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Moss WN, Yario TA, Steitz JA. 2015. EBV noncoding RNA binds nascent RNA to drive host PAX5 to viral DNA. Cell 160: 607–618. 10.1016/j.cell.2015.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner MR, Andrews NC, Miller G, Steitz JA. 1981. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc Natl Acad Sci 78: 805–809. 10.1073/pnas.78.2.805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Xiong X, Zhang M, Wang K, Chen Y, Zhou J, Mao Y, Lv J, Yi D, Chen XW, et al. 2017. Base-resolution mapping reveals distinct m1A methylome in nuclear- and mitochondrial-encoded transcripts. Mol Cell 68: 993–1005 e1009. 10.1016/j.molcel.2017.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. 2012. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149: 1635–1646. 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss WN, Steitz JA. 2013. Genome-wide analyses of Epstein-Barr virus reveal conserved RNA structures and a novel stable intronic sequence RNA. BMC Genomics 14: 543 10.1186/1471-2164-14-543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y, Lyko F, Helm M. 2010. 5-methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res 38: 1415–1430. 10.1093/nar/gkp1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanni AV, Lee N. 2018. Identification of host RNAs that interact with EBV noncoding RNA EBER2. RNA Biol 15: 1181–1191. 10.1080/15476286.2018.1518854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabson M, Heston L, Miller G. 1983. Identification of a rare Epstein-Barr virus variant that enhances early antigen expression in Raji cells. Proc Natl Acad Sci 80: 2762–2766. 10.1073/pnas.80.9.2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8: 2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, He C. 2017. Dynamic RNA modifications in gene expression regulation. Cell 169: 1187–1200. 10.1016/j.cell.2017.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safra M, Sas-Chen A, Nir R, Winkler R, Nachshon A, Bar-Yaacov D, Erlacher M, Rossmanith W, Stern-Ginossar N, Schwartz S. 2017. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551: 251–255. 10.1038/nature24456 [DOI] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. 2010. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 24: 1590–1595. 10.1101/gad.586710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. 2014. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159: 148–162. 10.1016/j.cell.2014.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro R, Vallee BL. 1989. Site-directed mutagenesis of histidine-13 and histidine-114 of human angiogenin. Alanine derivatives inhibit angiogenin-induced angiogenesis. Biochemistry 28: 7401–7408. 10.1021/bi00444a038 [DOI] [PubMed] [Google Scholar]
- Sheng J, Luo C, Jiang Y, Hinds PW, Xu Z, Hu GF. 2014. Transcription of angiogenin and ribonuclease 4 is regulated by RNA polymerase III elements and a CCCTC binding factor (CTCF)-dependent intragenic chromatin loop. J Biol Chem 289: 12520–12534. 10.1074/jbc.M114.551762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Pratt G, Yeo GW, Moore MJ. 2015. The clothes make the mRNA: past and present trends in mRNP fashion. Annu Rev Biochem 84: 325–354. 10.1146/annurev-biochem-080111-092106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. 2012. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res 40: 5023–5033. 10.1093/nar/gks144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuring K, Schmid K, Keller P, Helm M. 2017. LC-MS analysis of methylated RNA. Methods Mol Biol 1562: 3–18. 10.1007/978-1-4939-6807-7_1 [DOI] [PubMed] [Google Scholar]
- Trixl L, Lusser A. 2019. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip Rev RNA 10: e1510 10.1002/wrna.1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. 2012. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol 19: 900–905. 10.1038/nsmb.2357 [DOI] [PubMed] [Google Scholar]
- Xu M, Yao Y, Chen H, Zhang S, Cao SM, Zhang Z, Luo B, Liu Z, Li Z, Xiang T, et al. 2019. Genome sequencing analysis identifies Epstein-Barr virus subtypes associated with high risk of nasopharyngeal carcinoma. Nat Genet 51: 1131–1136. 10.1038/s41588-019-0436-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF, Anderson P. 2009. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185: 35–42. 10.1083/jcb.200811106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, Li A, Wang X, Bhattarai DP, Xiao W, et al. 2017. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res 27: 606–625. 10.1038/cr.2017.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang L, Han X, Yang WL, Zhang M, Ma HL, Sun BF, Li A, Xia J, Chen J, et al. 2019. RNA 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol Cell 75: 1188–1202 e1111. 10.1016/j.molcel.2019.06.033 [DOI] [PubMed] [Google Scholar]
- Zhang J, Kobert K, Flouri T, Stamatakis A. 2014. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30: 614–620. 10.1093/bioinformatics/btt593 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.