Abstract
The underlying mechanism of normal lung organogenesis is not well understood. An increasing number of studies are demonstrating that extracellular vesicles (EVs) play critical roles in organ development by delivering microRNAs (miRNA) to neighboring and distant cells. miRNAs are important for fetal lung growth; however, the role of miRNA-EVs (miRNAs packaged inside the EVs) during fetal lung development is unexplored. The aim of this study was to examine the expression of miRNA-EVs in MLE-12, a murine lung epithelial cell line subjected to mechanical stretch in vitro with the long-term goal to investigate their potential role in fetal lung development. Both cyclic and continuous mechanical stretch regulate miRNA differentially in EVs released from MLE-12 and intracellularly, demonstrating that mechanical signals regulate the expression of miRNA-EVs in lung epithelial cells. These results provide a proof-of-concept for the potential role that miRNA-EVs could play in the development of fetal lung.
Keywords: Mechanical stretch, extracellular vesicles, miRNA, MLE-12
Graphical abstract:
In the current study, we report that mechanical forces differentially regulate the expression of specific miRNAs in lung epithelial cells which provides novel insights into the potential regulatory mechanisms in lung organogenesis mediated by mechanical signals.
Introduction
Pulmonary hypoplasia, or incomplete development of the lung, is an important cause of neonatal morbidity and mortality. Moreover, in the United States, 1 of every 10 infants born prematurely (https://www.cdc.gov/reproductivehealth/maternalinfanthealth/pretermbirth.htm). In addition to the risk of death, these conditions can cause severe respiratory distress at birth and serious long-term morbidities (Wilson-Costello, Friedman, Minich, Fanaroff, & Hack, 2005). Currently, the management of these disorders is primarily supportive with no specific treatment to accelerate the development of the lungs.
Mechanical forces generated in utero by constant distention pressure inside the lumen of the lung and by intermittent breathing-like movements are essential for normal fetal lung development (Joe et al., 1997 & Sanchez-Esteban et al., 1998). Lung fluid composition is also critical for the development of the fetal lung (Luks et al., 2001).
Extracellular vesicles (EVs) are membrane-bound particles released from many cell types, and have been identified in many body fluids including blood, urine, saliva, bronchoalveolar lavage fluid, and amniotic fluid (Gallo, Tandon, Alevizos, & Illei, 2012; Street et al., 2012; Torregrosa Paredes et al., 2012; Wahlgren et al., 2012), and harbor complex cargoes including nucleic acids (all types of RNA and DNA), proteins and lipids (Maas, Breakefield, & Weaver, 2017). According to the International Society for the Extracellular Vesicles, there are three nomenclatures for EVs: i) Apoptotic bodies : vesicles formed by the cell membrane bubbling during the process of apoptosis with a size range of 500–2000 nm in diameter. ii) Microvesicles : vesicles formed directly protruding from cell membranes under physiological condition with a size range of 50–500 nm in diameter. iii) Exosomes : are the smallest vesicles with size a range of 30–100 nm in diameter, formed as multivesicular bodies by endosomal pathway also under physiological condition and released from the cells via fusion with cell membrane (Théry et al., 2018). The two latter types of exctracellular vesicles are the focus of our study, and will be referred to from now on as EVs.
Among the many types of cargoes carried in EVs, miRNAs are getting paid the most attention for their roles in gene expression regulation (Shivdasani, 2006). Rising bodies of research are demonstrating that EVs are increasingly recognized as a mode of cell-to-cell communication by delivering miRNA to neighboring and distant cells (Abels & Breakefield, 2016; Paolicelli, Bergamini, & Rajendran, 2019). These interactions can potentially modify the target’s gene expression, signaling and overall function (Maas et al., 2017; Mittelbrunn & Sanchez-Madrid, 2012).
miRNAs are a subtype of small noncoding RNAs involved in many biological activities including cell proliferation, cell differentiation and apoptosis in normal physiological and pathological conditions (Bartel, 2004; Kota et al., 2009; Tay, Zhang, Thomson, Lim, & Rigoutsos, 2008). miRNAs are important for lung organogenesis (Cushing, Jiang, Kuang, & Lu, 2015; Nardiello & Morty, 2016). However, the role of miRNAs during fetal lung development is unexplored. Given that physiological mechanical signals release EVs, and miRNAs are a key component of EV-cargoes, we hypothesized that mechanical force induced miRNA-EVs have a potential role in fetal lung development. Lung epithelial cells derived from miRNA-EVs determine lung fate in physio-pathological state by interacting with other resident cells (Aliotta et al., 2015; Lee, Zhang, Wu, Otterbein, & Jin, 2017). It has also demonstrated that miR-489 packaged in EVs released from MLE-12 cells participates in alveolar septation (Olave et al., 2016). We therefore setup our present study with the aim of examining the expression of miRNA-EVs in MLE-12 in view of our long-term goal that mechanical force differentially regulates miRNAs in EVs released from lung epithelial cell. We used an in vitro system where MLE-12 cells, a murine alveolar type II epithelial cell line, were exposed to continuous or cyclic mechanical stretch to mimic physiological mechanical forces in fetal lung development. We report differential expression of specific miRNAs in EVs released from MLE-12 in response to both cyclic and continuous stretch. Furthermore, we found differential expression of specific miRNAs intracellularly. Altogether, our data demonstrate that mechanical signals regulate the expression of miRNA-EVs in lung epithelial cells. These results provide a proof-of-concept for the potential role of miRNA-EVs in the signaling and regulation of lung development.
Material and Methods
Bronchoalveolar lavage (BAL)
The experiment was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Lifespan Institutional Animal Care and Use Committee, Providence, RI, USA (Protocol Number: 5066–18). Surgery for collecting the BAL was performed under inhaled isoflurane anesthesia, and all efforts were made to minimize suffering. A Swiss-Webster pregnant mouse was sacrificed at E18.5 of gestation by CO2 inhalation. A total of 7 fetuses were collected and under a stereoscopic microscope, the neck was carefully dissected and the trachea was cannulated with a 26-G neonatal catheter. After cannulation and removal of the needle, aliquots of 100 μl PBS were injected and subsequently aspirated from the lung and collected. This process was repeated three times for each fetus. To confirm that fluid was injected in the lung and to avoid overdistension or rupture of the lung, the chest cavity was opened during the procedure to visualize the inflation and deflation of the lung. BAL was also performed at postnatal day 1.
Cell culture and mechanical stretch experiments
MLE12 cell line was obtained from American Type Culture Collection (ATCC Cat# CRL-2110, RRID: CVCL_3751 Manassas, Virginia). This is a murine cell line that expresses features of type II lung epithelial cells (Wikenheiser et al., 1993). The cells were cultured in complete medium (DMEM/F12 supplemented with insulin (0.005 mg/ml), transferrin (0.01 mg/ml), sodium selenite (30 nM), hydrocortisone (10 nM), β-estradiol (10 nM), HEPES (10 mM), L-glutamine (2 mM) and extracellular-vesicle-depleted 2% fetal bovine serum). Fetal bovine serum was subjected to ultracentrifugation (110,000 rpm, 4 °C, 18 h; rotor Ti 70.1, Beckman Coulter) to deplete EVs and sterilized by filtering using 0.2 μm membrane pore. Cells from passage 2 to 8 were used in the study.
To apply mechanical stretch to the cells, 0.35–0.45 × 106 cells in 2 ml of medium were seeded onto each well of 6-well Bioflex Plates (Flexcell International, Hillsborough, NC, USA) precoated with collagen-1 and kept at 37° C and 5% CO2 inside the incubator. Monolayers were maintained for an additional 24 hour until reached approximately 50–65% confluency, and after fresh medium was replaced, plates were mounted in a Flexcell FX-4000 Stretch Unit (Flexcell International). Regimens of 5% continuous stretch or 10% cyclical stretch, at intervals of 40 cycles/min for 24 hours, were used. Cells grown on non-stretched membranes were treated in an identical manner and served as controls.
Cell Morphology Analysis
The cells were imaged at 20x using an inverted Nikon Eclipse TE2000-E microscope. The images were captured using the MetaView software (Metamorph). Image analysis was performed using ImageJ (v 1.5r) (NIH, Bethesda, MD). Digital images from at least two different wells subjected to 5% continuous stretch, 10% cyclic stretch, or controls were analyzed. To assess cell morphology in the experiments with cyclic stretch, the images were smoothed, and the cells isolated using thresholding (percentile method). A watershed transformation was then applied to segment individual cells (Najrana, Ramos, Abu Eid, & Sanchez-Esteban, 2017). For the experiments with continuous stretch, images were analyzed by smoothing the cells using a 3D Gaussian filter, and an automated watershed (8-connected) was used to segment the field into cells based on the grey level intensity. Particle analysis was applied to all images using the Particle8 Plus plugin (ImageJ) (Landini, 2008). Cell size was compared between static cells and those subjected to stretch, using the area and perimeter of the cells.
EV isolation
MLE-12 cells:
Flexercell Strain Unit has the capacity to culture approximately 35 ×106 cells in 4 (6-well Bioflex Plates) plates for each experiment. A total of 48 ml (2 ml/well;4 plates of 6-well Bioflex plate) culture media were collected after exposing the cells to mechanical stretch for 24 h. Collected media were centrifuged at 300g for 10 min at 4°C to sediment floating cells. Supernatant was further centrifuged at 2000g for 20 min at 4°C to pellet cell debris. A subsequent centrifugation at 14,000 rpm for 30 min at 4°C was performed to eliminate cell debris, folded protein and other particles of similar size. Supernatant was then passed through a 1-μm-pore filter to remove remaining apoptotic bodies and the resulting supernatant was stored at −80 °C. These experiments were repeated five times and the culture media collected each time and stored at −80 °C, as described above. The day of EVs isolation, culture media from 5 experiments (equivalent to 240 ml culture medium or 175 ×106 cells) were taken out of the −80 °C, combined together and subjected to ultracentrifugation (110,000 rpm, 4 °C, 18 h; rotor Ti 70.1, Beckman Coulter) for 1h at 4°C to pellet the EVs. EVs were washed with 1 ml of sterile ice cold 1xPBS and resuspended in750 μl of sterile ice cold 1X PBS. The rational to combine 5 experiments is the low yield of EVs from single experiment. EVs isolated from non-stretched monolayers (240 ml culture medium or 175 ×106 cells) were used as control.
Mouse lung:
BALF from E18.5-fetuses and P1-pups mouse lungs were used to isolate EV by sequential centrifugations, as described above.
EV Characterization
Nanoparticle Tracking Analysis:
Concentration and size distribution of the EVs were measured by Nanoparticle Tracking Analysis software (NTA) using NanoSight (NS300, Malvern Panalytical) device following the manufacturer’s recommendation. Briefly, EVs were diluted 1: 1000–3000 in 1xPBS. One ml of the diluted EVs was loaded to measure the concentration of EVs per volume. Negative control was set up by running 1xPBS. The detection concentration sensitivity of NS300 ranges from 106 to 109 particles per ml. Concentration of EV/ml was normalized to the number of cells present in the monolayers. Identification of the EVs was done by Western blot or flow cytometry using the well-accepted markers CD63 and CD9.
Western dot Blot for lung-EV:
Lysate for Western dot blot was prepared in RIPA buffer containing Halt Protease Inhibitor Cocktail (cat. no. 78429, Thermo Scientific). Two μl of the lysate was spotted on PVDF membranes and let them dry. The membranes were blocked with 5% nonfat milk in PBS (Invitrogen) containing 0.1% Tween-20 (PBST) for 1 h at room temperature, and then incubated with anti-CD63 antibody (abcam) at 4oC overnight. The membranes were washed three times in PBST (3 × 5 min) and incubated with HRP conjugated secondary antibody in PBBT for 1 h at room temperature. Membranes were then washed with PBST three times (3X 15 min) followed by chemiluminescent reagent treatment; signals were detected on X-ray film.
Flow cytometry:
Flow cytometry analysis was performed to verify the identification of EVs using Tetraspanin Exo-Flow Combo Capture kit (System Bioscience, cat. no. ExoFlow150A-1) following the manufacturer’s instructions, and standard flow cytometry method (FACS ARIA II, BD). Briefly, tetraspanin (CD9, CD63 and CD81)-biotin antibodies were coupled to the magnetic streptavidin beads of 9.1 μm size. Tetraspanin-coupled magnetic beads were then incubated with isolated EVs (10 μg) to capture EVs, unbound EVs were washed away. Captured EVs were then stained with detection antibodies for CD9 or CD63 conjugated with FITC (System Bioscience, cat. no. ExoFlow 150A-1).
micro BCA protein measurement:
Concentration of EVs was also analyzed using the BCA protein measurement method (Micro BCA protein Assay Kit, ThermoScientific). MicroRNA BCA protein kit is sensitive to measure protein concentration at femto level scale. This method was used to quantify the amount of EVs and based on the quantification 20 μg of EVs was used to isolate total RNA from EVs as described below.
RNA isolation from EVs and cells
Total RNA was isolated from 20 μg of EVs using Total Exosome RNA and Protein Isolation Kit (Invitrogen, cat. no. 447845). Total RNA from 3.5 × 106 cells was isolated using Quick-RNA Mini Prep (Zymo Research) according to the provided protocol. RNA was eluted in Nuclease-Free Water (Ambion, cat no. AM9937). RNA concentration and quality (both wave length ratios for 260/280 and 260/230 are 2.0) were measured by NanoDrop Spectrophotometer (Thermo Scientific Model: ND-2000).
MiRNA microarray assay and data analysis
Genechip™ miRNA_4.0 (Applied Biosystems) was used for miRNA expression analysis. GenechipTM miRNA_4.0 arrays have miRNA probes from 200 species. The annotation date of the GenechipTM miRNA_4.0 array is September 2016; therefore, miRNAs from mouse that are100% identical to other species and reported after September 2016 are absent in the GenechipTMmiRNA_4.0 array. We found 14 miRNAs in our array data that were reported after the annotation date but none of them is significantly differentially regulated. Therefore, heat map was generated using mus musculus miRNAs (mmu-miR) present on the array. The microarray assay was conducted at the Genomic Core Facility at Brown University (Providence, RI, USA). For cyclic stretch experiments, 250 ng of total RNA from the EVs and 650 ng from the cells were used, whereas for continuous stretch experiments, 500 ng of total RNA from both, EVs and cells, were used after optimization according to yield of the EVs. Transcriptome Analysis Console software (ThermoFisher Scientific) was utilized to analyze the microarray data. Log data were used for hierarchical clustering and statistical analysis; log2 to −2 was used as cutoff for the fold change and False Derivative Rate (FDR) adjusted p-value was setup at ≤ 0.05. MiRNA data available to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus database (GEO) (http://www.ncbi.nlm.nih.gov/geo) with GEO accession number GSE131645. GEO repository does not issue DOIs.
Pathway analysis
Gene lists of miRNAs generated by microarray assay from EVs released by MLE-12 cells under mechanical stretch was used to predict downstream effects on biological processes by Ingenuity® Pathway Analysis software (build version: 484108M; content version: 45868156-release date 20–10-09).
Statistics analysis
Student t-test (Graphpad prism software) was used for statistical analysis. P ≤ 0.05 was considered statistically significant.
Results
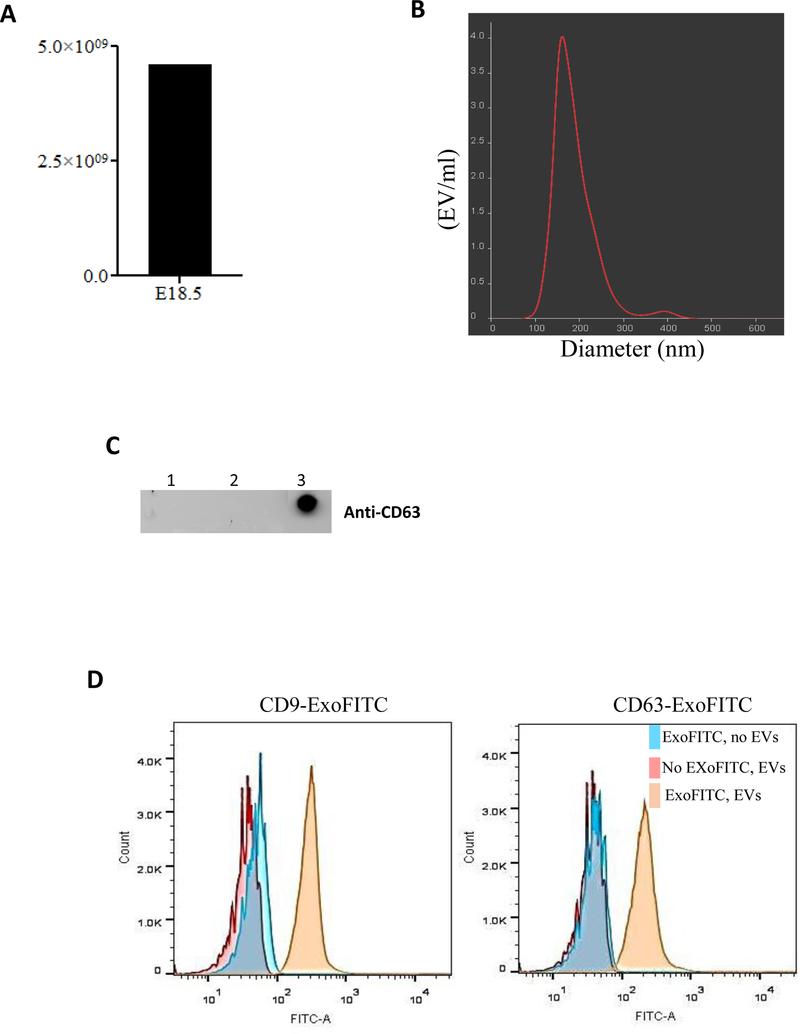
EVs are present in the mouse fetal lung fluid.
EVs have been found in many bio-fluid compartments (Gallo et al., 2012; Street et al., 2012; Torregrosa Paredes et al., 2012; Wahlgren et al., 2012) except the fetal lung. Therefore, we first studied whether EVs are present in the fetal lung. Lung fluid was obtained by bronchoalveolar lavage of the fetal lung at E18.5 of gestation. Samples were processed to isolate EVs by ultracentrifugation. Concentration and size distribution were analyzed by NanoSight. Our data in Figure 1A show the presence of >4×109 EVs/ml in the lumen of the lung from the combination of seven fetuses analyzed at E18.5 of gestation (saccular stage of lung development). EVs were also detected in the lumen of the lung from newborn pups (data not shown). We next studied the size distribution of the vesicles by Nanoparticle Tracking Analysis (NTA) (Figure 1B). Our results indicate that > 90% of the EVs preparation consisted of particles around 140 nm in diameter. After that, we investigated the detection of EVs by Western dot blot using anti-CD63 antibody as a specific marker for EVs. Data in Figure 1C show the purity of our preparation by demonstrating the presence of CD63 protein in the pellet after ultracentrifugation. In contrast, we did not see the presence of CD63 protein from pellet samples in lines 1 and 2 after ultracentrifugation containing cell debris and apoptotic bodies, respectively. The predicted molecular size of 26 KD corresponding to the CD63 protein was also confirmed by Western blot (data not shown). Flow cytometry (Figure 1D) was performed to detect CD9 or CD63 proteins on the surface of the EVs as markers for purity and integrity. The data show that the anti-CD9 or anti-CD63 antibodies against CD9 or CD63 antigens did indeed bind to the respective antigens on EVs surface (yellow histogram) and provided a clearly distinguishable fluorescence signal from the negative control samples, tetraspanine-coupled beads only (blue histogram) and the sample without the fluorescence antibody (pink histogram). All together, these studies demonstrate the presence of extracellular vesicles in developing mouse lung.
Figure 1: Isolation and characterization of EVs isolated from the fetal lung fluids.
(A) Lung fluid from seven fetuses at E18.5 of gestation were collected and combined. EVs were isolated by ultracentrifugation. EVs concentration was measured using NanoSight. (B) Graphical representation of size distribution of EVs measured by Nanoparticle Tracking Analysis. (C) Detection of EVs was evaluated by Western dot blot using anti-CD63 antibody. Lanes 1 and 2 represent the pellet containing cell debris and apoptotic bodies after 1,000 and 14,000 rpm whereas lane 3 is the pellet after ultracentrifugation containing EVs. (D) Flow cytometry analysis to assess purity of EVs isolation using anti-CD9 and anti-CD63 antibodies. Blue histograms represent tetraspanin-coupled magnetic beads only (described in methods) and in absence of EVs the detection antibody is unable to bind with magnetic beads which indicate the specificity of detection antibodies for purity and integrity of EVs. Pink histogram represents the negative control in absence of detection fluorescence antibody. Yellow histogram represents EVs that bind to tetraspanin-coupled beads and positive for the detection of the fluorescence antibody. Representative figures are shown for at least two independent experiments.
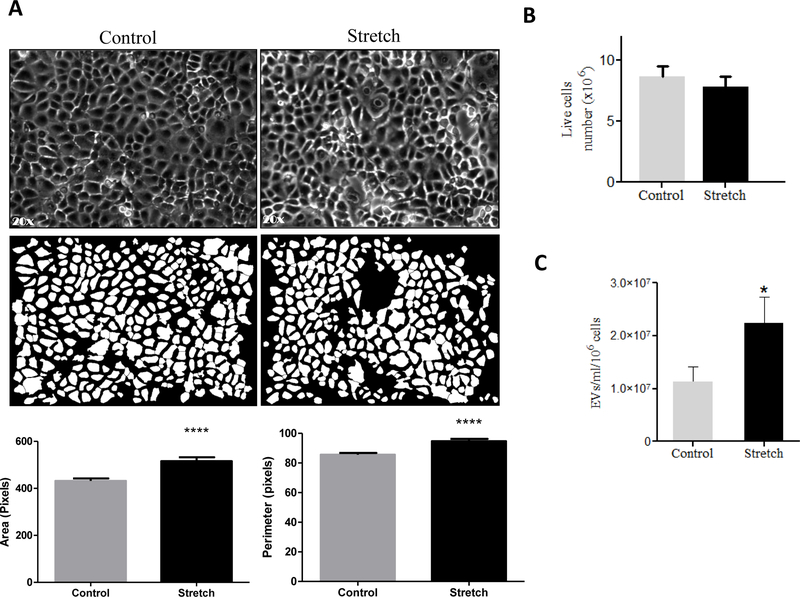
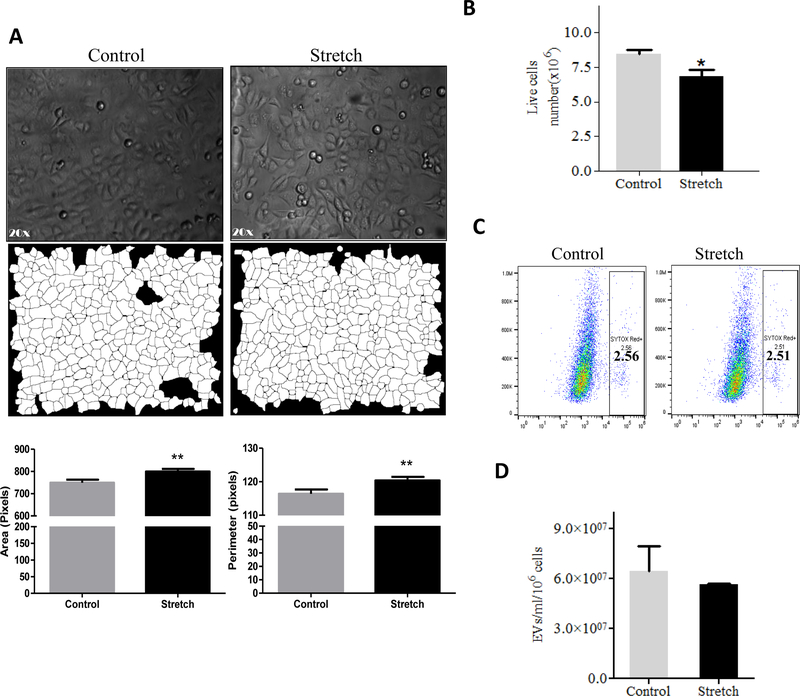
Effect of mechanical stretch on cell size, viability and release of EVs from EC.
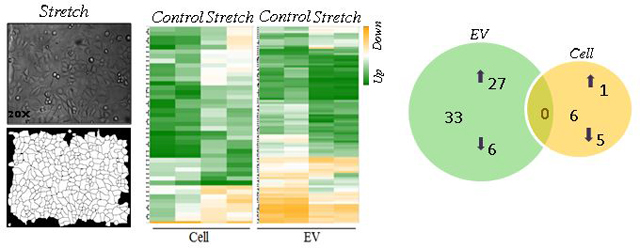
Mechanical signals are critical for normal fetal lung development (Joe et al., 1997; M. Liu & Post, 2000; Sanchez-Esteban et al., 2001; Sanchez-Esteban et al., 1998; Sanchez-Esteban et al., 2002; Torday et al., 1998; Wirtz & Dobbs, 2000). Previous studies have demonstrated that the composition of the fetal lung fluid may be important for cell-to-cell communication during lung development (Luks et al., 2001; Papadakis et al., 1997). Based on this information and our data demonstrates the presence of EVs in the fetal lung fluid, we studied next whether mechanical signals are important to release EVs in the lung using an in vitro cell culture system. We used the murine MLE-12 epithelial cell line, which has been shown to have similar characteristics to primary alveolar epithelial cells (Wikenheiser et al., 1993). MLE-12 cells were exposed to 10% cyclic or 5% continuous mechanical stretch to mimic mechanical signals in fetal lung development. Figure 2A shows bright field microscopic images of the monolayer of MLE-12 cells after 10% cyclic stretch for 24 hours and parallel control samples. We analyzed cellular responses to stretch by measuring changes in cell size using morphometric analysis of digital images. Our data demonstrated a significant increase in the area (P<0.0001) and perimeter (P<0.0001) of the cells exposed to mechanical stretch when compared to controls (Figure 2A). Given that even a small percentage of cell death could release cell membrane particles and confound the harvest of EVs, we wanted to check whether mechanical stretch affects the number of live cells in our experimental conditions. Our data in Figure 2B show that cyclic stretch does not affect the cell numbers when compared to controls. Interestingly, we found that 10% cyclic stretch increased the release of EVs by around 2-fold when compared to controls (112 ×105 ± 2800 ×103 vs 224 ×105 ± 4933 ×103) (>Figure 2C). Likewise, 5% continuous stretch applied to MLE-12 cells shows changes in the size of the cells as demonstrated by an increase in the area (P<0.01) and perimeter (P<0.01) when compared to controls (Figure 3A). Contrary to cyclic stretch, continuous stretch decreased the number of cells by 20% when compared to controls (8.44 ± 0.3 vs 6.87 ± 0.49; P < 0.02) (Figure 3B). These results could be explained by an inhibition of cell proliferation induced by continuous stretch, since cell viability is not affected by the stretch (Figure 3C). We did not observe that continuous stretch affects the number of EVs released by the cells (Figure 3D). In summary, our data demonstrate that application of mechanical forces to epithelial cells induce changes in the size of the cells, cell number, and release of EVs, without compromising the viability of the cells. These effects vary depending on the type of mechanical stimulation applied to the cells.
Figure 2: Effects of 10% cyclic mechanical stretch on cell size, viability and release of EVs from MLE-12 cells.
MLE-12 cells grown in culture media containing EV-depleted FBS were exposed to 10% cyclic stretch for 24 hours. (A) Bright field microscopic images in the upper captured at x 20 magnification and corresponding binary images of the digitally isolated cells are shown in the lower panels. Area and perimeter were used as determinants for the cell-size and are shown in the bar diagrams. (B) Cells from control and stretched experiments were recovered from the plates by trypsinization and counted for live cells number using cellometer. (C) EVs were isolated from the culture media after experimental condition; the concentration of EVs was evaluated by NanoSight. Data were normalized to the number of live cells recovered from the plate after experiments. Results represent the average of three independent experiments ± SEM, each done at least in duplicate. *p<0.05 and ****p <0.0001
Figure 3: Effects of 5% continuous stretch on cell size, viability and release of EVs from MLE-12 cell.
A. MLE-12 cells grown in culture media containing EV-depleted FBS were exposed to 5% continuous stretch for 24 hours. (A) Bright field microscopic images in the upper captured at x 20 magnification and corresponding binary images of the digitally isolated cells are shown in the lower panels. Area and perimeter were used as determinants for the cell-size and are shown in the bar diagrams. (B) Cells from control and stretched experiments were recovered from the plates by trypsinization and counted for live cells number using a cellometer. (C) Flow cytometry was done to analyze cell viability using SYTOX Red dye (D) EVs were isolated from the culture media after experimental condition; the concentration of EVs was evaluated by NanoSight. Data were normalized to the number of live cells recovered from the plate after experiments. Results represent the average of three independent experiments ± SEM, each done at least in duplicate. ** p<0.01.
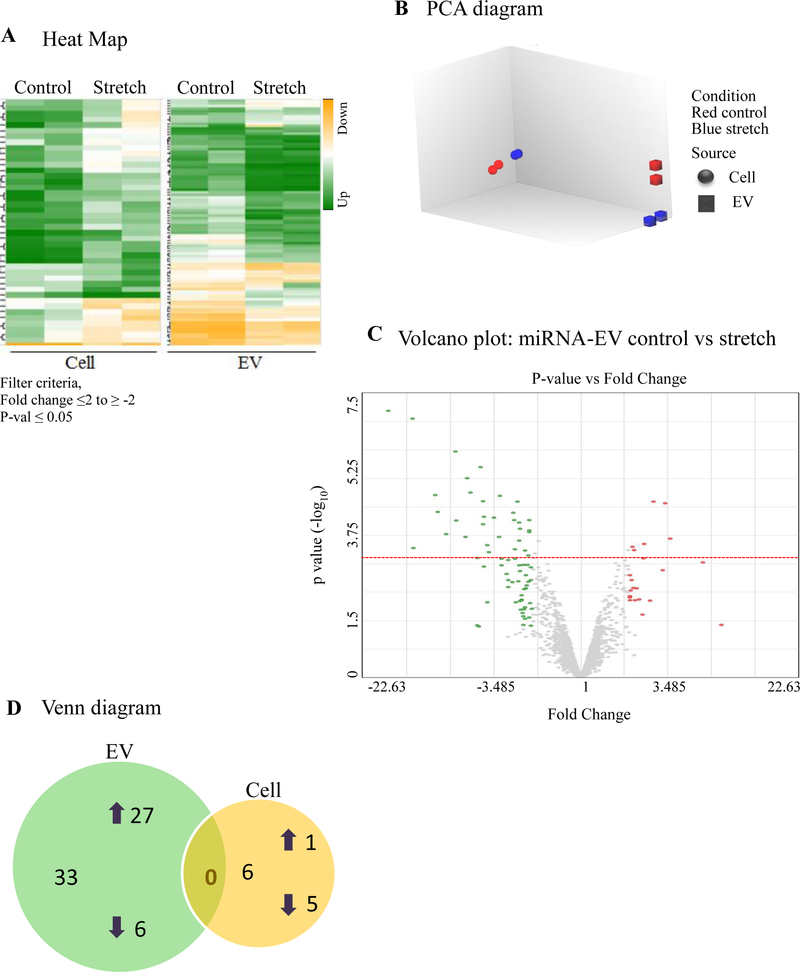
Effect of continuous stretch on miRNA-EVs expression released from MLE-12 cells.
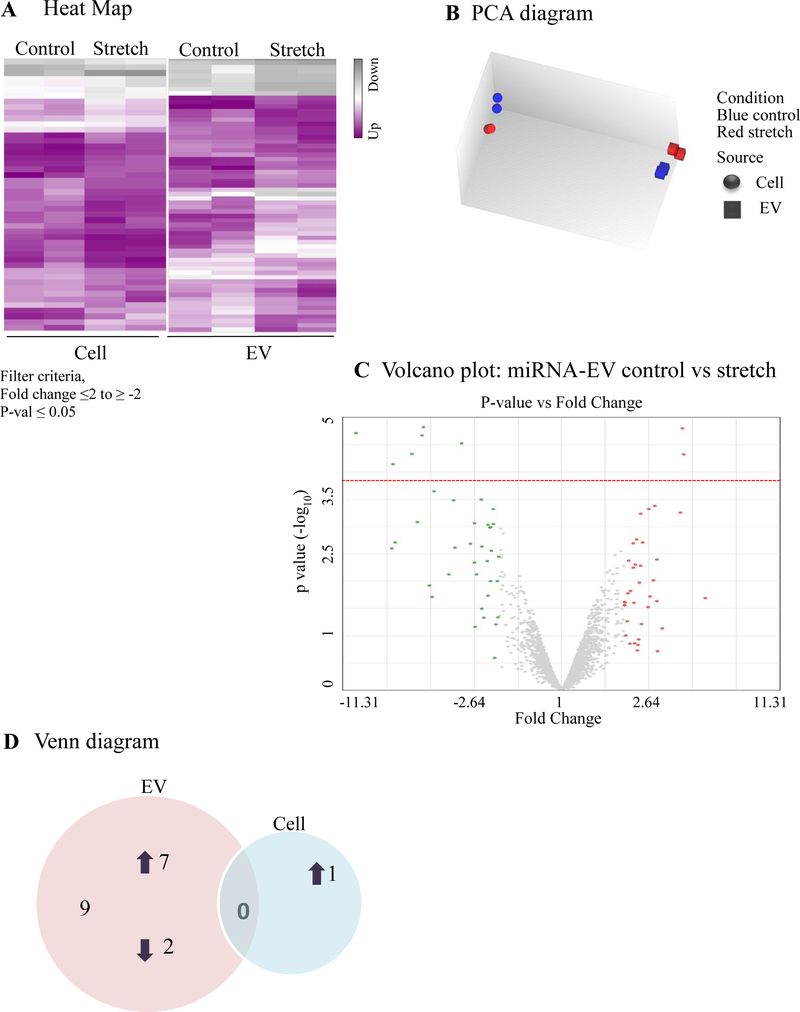
miRNAs play a critical role in fetal lung morphogenesis (Cushing et al., 2015; Dong et al., 2010). To test the hypothesis that mechanical stretch differentially regulates miRNA-EVs in mouse lung epithelial cells, we exposed MLE-12 monolayers to 5% continuous stretch for 24 h. EVs were isolated from the culture media as described above and total RNA was extracted from both EVs and cells. The microarray assay was done using total RNA to generate a miRNAs expression profile. Expression data were analyzed using Transcriptome Analysis Console software (ThermoFisher scientific). Expression profiles of miRNA in EVs released from MLE-12 cells and inside the cells are shown in Figure 4A. Figure 4B represents the Principle Component Analysis (PCA) graphs under control and stretch conditions to show the intrinsic cluster within the data sets where blue and red colors represent control and stretch condition respectively; circle and square represent cell and EV. Differentially expressed miRNAs with fold changes (x- axis) and statistical significance (-log10 of p value, Y axis) are shown by volcano graph in Figure 4C. Table-1 shows 33 miRNAs that were differentially regulated in EVs from MLE-12 cells after exposure to 5% continuous stretch when compared to control; from those miRNAs, 27 were upregulated and 6 were downregulated. Inside the cells (Table-2), 6 miRNAs were differentially expressed with 1 miRNA upregulated and 5 downregulated. Venn diagram was depicted in Figure 4D to show the distribution of stretch-responsive miRNAs in both EVs and cells. Notice that no single stretch-response miRNA expression is common to both compartments.
Figure 4: Effect of 5% continuous stretch on miRNA expression in EVs from lung epithelial cells.
MiRNA expression profile was generated by microarray assay, using total RNA extracted from EVs released from MLE-12 cells after exposed to 5% continuous stretch for 24 hours and controls (unstretched). (A) Heat map was generated using a clustering method in which miRNAs was grouped based on their similarity of expression patterns. Data are shown in a grid where each row represents a gene and each column represents a sample. Fold change for gene expression was set up at ≤2 to ≥ −2 and ≤ 0.05 for p-values. Green and yellow colors indicate up regulation and down regulation of miRNA expression, respectively. (B) Intrinsic cluster within the data set was shown by Principle Component analysis (PCA) graph. Samples are colored and shaped by experimental conditions and sources in which red-control, blue-stretch, circle-cell and square for EVs respectively. Each shape represents a single experiment. (C) Volcano graph demonstrates differentially expressed miRNAs with fold changes (x- axis) and statistical significance (-log10 of p value, Y axis). The dashed red line shows where p=0.05 with points above the line having p <0.05. Gray color points represent the miRNAs with fold changes < 2. (D) Venn diagram showing the number and overlap between differential expressed miRNAs identified by 5% continuous stretch in the EVs and cells.
Table 1.
Shown are the fold changes, p values and FDR p-values for each miRNA differentially regulated when MLE-12 exposed with 5% continuous stretch
| miRNA | Fold Change | P-value | FDR P-value |
|---|---|---|---|
| mmu-miR-297c-5p | 11.1 | 5.37E-08 | 0.0004 |
| mmu-miR-6769b-5p | 6 | 4.70E-07 | 0.0007 |
| mmu-miR-467h | 4.19 | 1.32E-06 | 0.0016 |
| mmu-miR-7058–3p | 5.07 | 2.56E-06 | 0.0026 |
| mmu-miR-7049–3p | 4.85 | 6.52E-06 | 0.0054 |
| mmu-miR-6958–3p | 8.08 | 7.54E-06 | 0.0056 |
| mmu-miR-669d-5p | 3.18 | 8.40E-06 | 0.0059 |
| mmu-miR-6918–3p | 4.08 | 1.14E-05 | 0.007 |
| mmu-miR-2861 | −3.36 | 1.33E-05 | 0.0072 |
| mmu-miR-5110 | −2.84 | 1.25E-05 | 0.0072 |
| mmu-miR-708–3p | 2.48 | 1.30E-05 | 0.0072 |
| mmu-miR-711 | 7.78 | 2.25E-05 | 0.01 |
| mmu-miR-669a-5p | 2.63 | 2.53E-05 | 0.01 |
| mmu-miR-669p-5p | 2.63 | 2.53E-05 | 0.01 |
| mmu-miR-7021–3p | 4 | 3.10E-05 | 0.01 |
| mmu-miR-6966–3p | 3.48 | 3.39E-05 | 0.01 |
| mmu-miR-669l-5p | 2.61 | 4.05E-05 | 0.02 |
| mmu-miR-297b-5p | 5.98 | 3.92E-05 | 0.02 |
| mmu-miR-7084–3p | 2.42 | 4.85E-05 | 0.02 |
| mmu-miR-6933–3p | 4.06 | 5.04E-05 | 0.02 |
| mmu-miR-383–3p | 2.1 | 8.25E-05 | 0.02 |
| mmu-miR-7087–5p | 2.1 | 9.20E-05 | 0.02 |
| mmu-miR-93–5p | 3.21 | 0.0001 | 0.03 |
| mmu-miR-7079–5p | 5.24 | 0.0001 | 0.03 |
| mmu-miR-211–3p | −3.6 | 0.0001 | 0.03 |
| mmu-miR-185–3p | 2.42 | 0.0002 | 0.03 |
| mmu-miR-5130 | −2.47 | 0.0002 | 0.03 |
| mmu-miR-7036–5p | 3.8 | 0.0002 | 0.03 |
| mmu-miR-383–5p | −2.1 | 0.0002 | 0.04 |
| mmu-miR-7052–5p | 10.99 | 0.0002 | 0.04 |
| mmu-miR-103–1-5p | 2.56 | 0.0003 | 0.04 |
| mmu-miR-365–1-5p | −2.16 | 0.0003 | 0.04 |
| mmu-miR-7010–3p | 3.74 | 0.0003 | 0.05 |
Table 2:
Shown are the fold changes, p values and FDR p-values for each miRNA differentially regulated when MLE-12 exposed with 5% continuous stretch
| miRNA | Fold change | p value | FDR p value |
|---|---|---|---|
| mmu-miR-192–5p | −5.74 | 1.29E-05 | 0.0162 |
| mmu-miR-7234–5p | 2.72 | 2.29E-05 | 0.0259 |
| mmu-miR-3473a | −2.87 | 4.42E-05 | 0.0382 |
| mmu-miR-30b-5p | −4.02 | 8.57E-05 | 0.0443 |
| mmu-miR-674–3p | −2.12 | 6.65E-05 | 0.0443 |
| mmu-miR-378b | −6.22 | 9.30E-05 | 0.0468 |
Effect of cyclic stretch on EVs-miRNA expression released from MLE-12 cells.
Similar to continuous stretch, data in Figure 5A show the heat map of the miRNA profile in the EVs and cells under cyclic stretch and respective control conditions. PCA graph in Figure 5B was used to show the intrinsic cluster within the data set. Differentially expressed miRNAs with fold changes (x- axis) and statistical significance (-log10 of p value, Y axis) are shown by volcano graph in Figure 5C. Our data in Table-3 shows 9 miRNAs that were differentially regulated in EVs from MLE-12 cells after exposure to 10% cyclic stretch; from those, 7 were upregulated and 2 were downregulated. Inside the cells (Table-4), only mmu-miR-15a-3p was differentially regulated after cyclic stretch. Venn diagram (Figure 5D) shows the graphical representation of the distribution of the stretch-responsive miRNAs in both EVs and cells.
Figure 5: Effect of 10% cyclic stretch on miRNA expression in EVs from lung epithelial cells.
MiRNA expression profile was generated by microarray assay, using total RNA extracted from EVs released from MLE-12 cells after exposed to 10% cyclic stretch for 24 hours and unstretched controls. (A) Heat map was generated using a clustering method in which miRNAs was grouped based on their similarity of expression patterns. Data are shown in a grid where each row represents a gene and each column represents a sample. Fold change for gene expression was set up at ≤2 to ≥ −2 and ≤ 0.05 for p-values. Purple and grey colors indicate up regulation and down regulation of miRNA expression respectively. (B) Intrinsic cluster within the data set was shown by Principle Component analysis (PCA) graph. Samples are colored and shaped by experimental conditions and sources in which blue-control, red-stretch, circle-cell and square for EVs respectively. Each shape represents a single experiment. (C). Volcano graph demonstrates differentially expressed miRNAs with fold changes (x- axis) and statistical significance (-log10 of p value, Y axis). The dashed red line shows where p=0.05 with points above the line having p <0.05. Gray color points represent the miRNAs with fold changes < 2. (D) Venn diagram showing the number and overlap between differential expressed miRNAs identified by 10% cyclic stretch in the EVs and cells.
Table 3:
Shown are the fold changes, p values and FDR p-values for each miRNA differentially regulated when MLE-12 exposed with 10% cyclic stretch
| miRNA | Fold Change | p value | FDR p value |
|---|---|---|---|
| mmu-miR-6985–5p | −3.83 | 1.02E-05 | 0.0071 |
| mmu-let-7a-5p | 4.66 | 1.04E-05 | 0.0071 |
| mmu-miR-23b-3p | 4.72 | 1.51E-05 | 0.0071 |
| mmu-miR-23a-3p | 9.81 | 1.47E-05 | 0.0071 |
| mmu-miR-1946a | −3.9 | 3.33E-05 | 0.02 |
| mmu-miR-690 | 5.28 | 3.48E-05 | 0.02 |
| mmu-let-7c-5p | 6.5 | 5.58E-05 | 0.02 |
| mmu-let-7e-5p | 4.12 | 0.0002 | 0.04 |
| mmu-miR-182–5p | 3.31 | 0.0003 | 0.05 |
Table 4:
Shown are the fold changes, p values and FDR p-values for each miRNA differentially regulated when MLE-12 exposed with 10% cyclic stretch
| miRNA | Fold Change | p value | FDR p value |
|---|---|---|---|
| mmu-miR-15a-3p | 5.97 | 1.93E-06 | 0.0116 |
Discussion
Lung underdevelopment due to extreme prematurity or pulmonary hypoplasia can cause significant morbidity and mortality to newborns. An obligatory first step to design therapies to accelerate lung development is to understand how the fetal lung develops and specifically, how mechanical signals regulate fetal lung development. Using a murine lung EC line, our data show that mechanical stretch differentially regulates the expression of specific miRNAs including miR-let-7c and miR-690 that have been identified in the developing mouse fetal lung (Dong et al., 2010). Given the importance of miRNA in fetal lung development, our study provides a proof-of-concept for the potential role of miRNA in fetal lung development mediated by mechanical signals.
EVs are important for cell-to-cell communication. However, the role of EVs in fetal lung development is unknown. We provide the first evidence that EVs are present in the murine fetal lung. These findings suggest a potential implication for EV’s in fetal lung development. It has been, for example, that exosomes purified from mesenchymal stroma cells significantly improved lung morphology and pulmonary development in a neonatal injury model (Willis et al., 2018). Identification of critical cargo components of the EVs at different stages of lung development could be used to develop therapies to rescue underdeveloped lungs.
The mechanisms by which mechanical forces accelerate lung development are poorly understood. Mechanical signals trigger the release of EVs in lung endothelial and bronchial epithelial cells exposed to injurious stretch (Letsiou et al., 2015; Park et al., 2012). Our data, applying mechanical forces to MLE-12 cells, also demonstrate that mechanical signals influence not only the number of vesicles released by stretch, but also the expression of specific miRNAs inside the vesicles. These effects were even more significant after continuous stretch, where we observed a variety of miRNAs differentially regulated, even in the absence of an increase in the number of vesicles release after mechanical stimulation. Cell viability was not affected by continuous stretched even though we found number of live cells was decreased by 20% which may result from the inhibition of proliferation. These results support the concept that both, continuous and cyclic stretch could play essential roles during normal lung development.
miRNAs are critical for normal lung development (Cushing et al., 2015; Sessa & Hata, 2013). Recent study has found miRNA in the exosomes released by the MLE-12 (Olave et al., 2016), suggesting that exosomes released by EC may be important for lung development. We provide novel information that mechanical signals not only stimulate the release of EVs from epithelial cells, but also are able to differentially regulate the expression of specific miRNA in lung epithelial cells. Searching the lungMAP database (https://www.lungmap.net), our results found for example that lung type 1 cell membrane-associated glycoprotein (T1 alpha) is a predicted target of mmu-miR-669a-5p and mmu-miR-182–5p, both miRNAs were upregulated in the EVs after continuous and cyclic stretch, respectively (Table 5 & 6). T1alpha, a lung type I cell differentiation gene, is developmentally regulated and critical for normal lung development. T1α knockout mice die at birth of respiratory failure and histologic analyses show underdeveloped lungs (Ramirez et al., 2003). During fetal lung development, alveolar type I and type II epithelial cells are derived from a bipotent progenitor cell (Desai, Brownfield, & Krasnow, 2014). Previous studies found that the phenotypes of type-I and type-II alveolar EC are strongly influenced by the basal degree of lung expansion and observed a rise in the number of type I cells after increase of pressure induced by tracheal occlusion, and the opposite was found when fluid was drained from the lung (Flecknoe, Wallace, Harding, & Hooper, 2002). Based on our results, we speculate that one of the potential mechanisms by which mechanical signals promote type 1 phenotype expression is by interacting with specific miRNAs.
Table 5:
Shown are the predicted targets and functions of differentially regulated miRNAs when MLE-12 was exposed to continuous stretch from web-based search.
| miRNA | Predicted target | Function |
|---|---|---|
| mmu-miR-669–5p | T1 alpha | Lung development |
| mmu-miR-467h | Endomucin | Angiogenesis |
| mmu-miR-30b-5p | Expressed in type II cells at E16.5 gestation mouse fetal lung | Unknown |
Table 6:
Shown are the predicted targets and functions of differentially regulated miRNAs when MLE-12 was exposed to cyclic stretch from web-based search.
| miRNA | Predicted target | Function |
|---|---|---|
| mmu-miR-182–5p | T1alpha | Lung development |
| mmu-miR-let-7c-5p mmu-miR-690 | Expressed in endothelial and type II cells at E16.5 gestation | Unknown |
LungMAP also identified endomucin as a predictive target of miRNA-467h (upregulated in the EVs after continuous stretch in our study). Endomucin is present in the endothelium of the developing lung (Table 5) and it has been suggested to have a novel role as a regulator of angiogenesis (Park-Windhol et al., 2017). Cross-talk between epithelium and endothelium is critical for distal lung development. Based on these data, we speculate that miRNA present in the EVs after mechanical stimulation could also play an important role in this process during lung development.
We also identified that other miRNAs modified by stretch, are present in the fetal lung. For instance, miR-30b-5p, found to be downregulated inside the cells after continuous stretch, is expressed in isolated murine type II epithelial cells starting at E16.5 of gestation (Table 5). Likewise, miR-690 and miR-let-7c-5p, both upregulated in the EVs after cyclic stretch, have been detected in epithelial and endothelial cells from E16.5 of gestation (Table 6). These results, when combined with the identification of T1alpha and endomucin as predicted targets of some miRNAs, suggest that mechanical signals via release of miRNA could participate in cell-to-cell communication via epithelial-endothelial interactions.
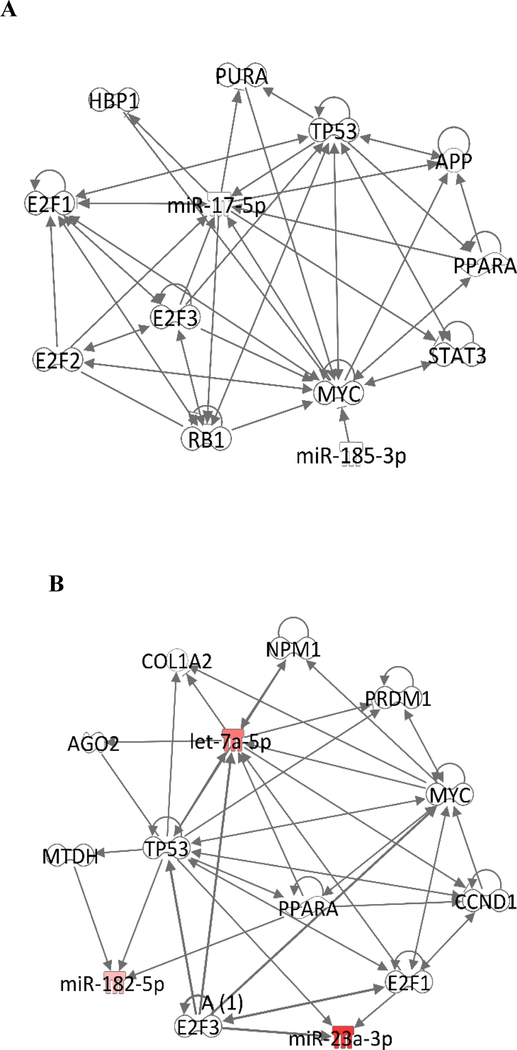
Using the ingenuity pathway analysis (IPA), we identified potential interactions between some of stretch-induced miRNAs and transcriptional regulators (Figure 6). For example, miR-185–3p, upregulated by continuous stretch in the EVs, has direct interactions with MYC, a family of transcription factors related to cell proliferation. Moreover, 5% continuous stretch differentially regulates miRNAs that interact with other transcription factors such as the E2F family of transcription factors that control cell cycle, the STAT3 that is involved in cell growth and apoptosis, the retinoblastoma protein that inhibits cell cycle, and PPARA, a nuclear receptor that participates in fatty acid metabolism. In summary, IPA has identified possible interactions of miRNAs differentially modulated by stretch with transcription factors that play critical roles in cellular function such as proliferation, differentiation and apoptosis. These results emphasize the potential role of miRNAs released by mechanical signals in cell-to-cell communication in the lung.
Figure 6:
Network of molecules identified by ingenuity pathway analysis (IPA) to be associated with miRNAs modified stretch. (A) Network was generated for the miRNA-EVs differentially regulated in response to 5% continuous stretch. (B) Network was generated for the miRNA-EVs differentially regulated in response to 10% cyclic stretch.
DIANA TOOLS, a web service at DIANA-LAB identify miRNAs predicted to target selected genes or gene targets of selected miRNAs (Paraskevopoulou et al., 2013; Reczko, Maragkakis, Alexiou, Grosse, & Hatzigeorgiou, 2012), identified signaling pathways such as TGF-beta, Wnt, Notch, Erb, and MAPK associated with several miRNAs modified by stretch including miR-23b-3p, miR-297c-5p, miR-30b-5p, miR-93–5p, miR-211–3p, miR-103–1-5p, miR-185–3p, miR-711, miR-5130, miR-let-7c-5p, miR-690, miR-182–5p, miR-467h, etc. These pathways have important roles in lung development. Specifically, TGF-beta is a key player in lung branching morphogenesis and alveolarization (Bartram & Speer, 2004) and was found to respond to mechanical signals (Vuckovic et al., 2016; Wang, Thampatty, Lin, & Im, 2007). TGF-beta signaling has been previously found to be associated with miRNA 29 family and type II cell differentiation (Guo, Benlhabib, & Mendelson, 2016). A vast and accumulating amount of data indicates that canonical Wnt signaling is a main regulator of lung development. Wnt/β‐catenin signaling regulates respiratory specification and epithelial and mesenchymal differentiation during lung development (Volckaert & De Langhe, 2015). Loss of Wnt2 (Wnt2‐/‐ null mice) results in severe lung hypoplasia and reduced cell proliferation in both epithelial and mesenchymal compartments (Goss et al., 2009). Notch signaling (associated with some of our identified miRNAs) is crucial for differentiation of the airway epithelium and for epithelial-mesenchymal interactions that lead to alveolar formation in the developing lung. (Tsao et al., 2016). The importance of ErbB receptors in lung development is well documented. EGFR is critical for branching morphogenesis, alveolarization, and differentiation of type II epithelial cells (Miettinen et al., 1995; Miettinen et al., 1997; Sibilia et al., 2007; Sibilia & Wagner, 1995). ErbB2 and ErbB3 promote fetal lung epithelial cell proliferation (Patel et al., 2000) and ErbB4 regulates type II cell differentiation (W. Liu et al., 2010). Previous studies from our laboratory have shown that mechanical strain, simulating mechanical forces in utero, activates these receptors and ERK pathway in fetal type II epithelial cells (Huang, Wang, Nayak, Dammann, & Sanchez-Esteban, 2012). In summary, these studies show the importance of these pathways in fetal lung development. Our data demonstrate that cyclic and continuous stretch differentially regulates miRNAs predicted to be associated with these pathways, suggesting a potential role of miRNA in mechanical signaling during lung development. However, these findings will need to be validated in other experimental systems.
Our study raises the question whether the percentage of stretch applied to MLE-12 cells reflects the actual distension pressure that fetal lungs are exposed in utero and whether MLE-12 cells have the same mechanical properties as the fetal epithelial cells. It has been clearly established from experimental models that mechanical forces generated in utero by constant distention pressure and by intermittent breathing-like movements are essential for normal fetal lung development (Joe et al., 1997; Sanchez-Esteban et al., 1998; Torday et al., 1998). However, the actual pressure that the fetal cells “sense” in utero is not clearly defined. In our experiments, we tried to mimic these two forces. The rational to use 5% constant distention pressure and 10% cyclic pressures were based on preliminary experiments demonstrating changes in the morphology of the cells and release of EVs under these experimental conditions. In either case, the main focus of our studies is to demonstrate that each mechanical signal induces a different and specific miRNA-EVs profile, further supporting that both types of mechanical signals are critical for normal lung development.
In conclusion, our data show that cyclic and continuous stretch differentially regulates the expression of specific miRNAs-EVs in lung epithelial cells. These studies serve as a proof-of-concept that one of the potential mechanisms by which miRNAs inside the EVs regulate lung development is via mechanical signals. Information derived from these studies will provide the bases for future investigations testing the role of specific miRNAs and/or miRNAs-EVs in fetal lung development.
Acknowledgments
This work was supported from the National Institute of Health (NIGMS grant Number P30GM114750 & P30GM103410, NCRR grant Numbers P30RR031153, P20RR018728 & S10RR02763); National Science Foundation (EPSCoR grant No 0554548); Oh–Zopfi for Perinatal Research Award, Women & Infants Hospital of Rhode Island. We thank Brenda Vecchio for her support in manuscript formatting and Quanfu Mao for his support to use the instruments.
Funding Statement
This work was supported from the National Institute of Health (NIGMS grant Number P30GM114750 & P30GM103410, NCRR grant Numbers P30RR031153, P20RR018728 & S10RR02763); National Science Foundation (EPSCoR grant No 0554548); Oh–Zopfi for Perinatal Research Award, Women & Infants Hospital of Rhode Island.
Footnotes
Data Availability Statement
MiRNA data available to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus database (GEO) (http://www.ncbi.nlm.nih.gov/geo) with GEO accession number GSE131645. GEO repository does not issue DOIs.
Conflict of Interest Statement: The authors declare that there are no conflicts of interest in the authorship or publication of this manuscript.
References
- Abels ER, & Breakefield XO (2016). Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell Mol Neurobiol, 36(3), 301–312. doi: 10.1007/s10571-016-0366-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliotta JM, Pereira M, Sears EH, Dooner MS, Wen S, Goldberg LR, & Quesenberry PJ (2015). Lung-derived exosome uptake into and epigenetic modulation of marrow progenitor/stem and differentiated cells. J Extracell Vesicles, 4, 26166. doi: 10.3402/jev.v4.26166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116(2), 281–297. doi: 10.1016/s0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bartram U, & Speer CP (2004). The role of transforming growth factor beta in lung development and disease. Chest, 125(2), 754–765. [DOI] [PubMed] [Google Scholar]
- Cushing L, Jiang Z, Kuang P, & Lu J (2015). The roles of microRNAs and protein components of the microRNA pathway in lung development and diseases. Am J Respir Cell Mol Biol, 52(4), 397–408. doi: 10.1165/rcmb.2014-0232RT [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai TJ, Brownfield DG, & Krasnow MA (2014). Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature, 507(7491), 190–194. doi: 10.1038/nature12930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Jiang G, Asmann YW, Tomaszek S, Jen J, Kislinger T, & Wigle DA (2010). MicroRNA networks in mouse lung organogenesis. PLoS One, 5(5), e10854. doi: 10.1371/journal.pone.0010854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flecknoe SJ, Wallace MJ, Harding R, & Hooper SB (2002). Determination of alveolar epithelial cell phenotypes in fetal sheep: evidence for the involvement of basal lung expansion. J Physiol, 542(Pt 1), 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo A, Tandon M, Alevizos I, & Illei GG (2012). The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One, 7(3), e30679. doi: 10.1371/journal.pone.0030679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss AM, Tian Y, Tsukiyama T, Cohen ED, Zhou D, Lu MM, Morrisey EE (2009). Wnt2/2b and beta-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Dev Cell, 17(2), 290–298. doi: 10.1016/j.devcel.2009.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Benlhabib H, & Mendelson CR (2016). The MicroRNA 29 Family Promotes Type II Cell Differentiation in Developing Lung. Mol Cell Biol, 36(16), 2141. doi: 10.1128/MCB.00096-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Wang Y, Nayak PS, Dammann CE, & Sanchez-Esteban J (2012). Stretch-induced fetal type II cell differentiation is mediated via ErbB1-ErbB4 interactions. J Biol Chem, 287(22), 18091–18102. doi: 10.1074/jbc.M111.313163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joe P, Wallen LD, Chapin CJ, Lee CH, Allen L, Han VK, Kitterman JA (1997). Effects of mechanical factors on growth and maturation of the lung in fetal sheep. Am J Physiol, 272(1 Pt 1), L95–105. [DOI] [PubMed] [Google Scholar]
- Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, … Mendell JT (2009). Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell, 137(6), 1005–1017. doi: 10.1016/j.cell.2009.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landini G (2008). Advanced shape analysis with imageJ. In The Second ImageJ User and Developer Conference (pp. 116–131). Luxembourg. [Google Scholar]
- Lee H, Zhang D, Wu J, Otterbein LE, & Jin Y (2017). Lung Epithelial Cell-Derived Microvesicles Regulate Macrophage Migration via MicroRNA-17/221-Induced Integrin beta1 Recycling. J Immunol, 199(4), 1453–1464. doi: 10.4049/jimmunol.1700165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letsiou E, Sammani S, Zhang W, Zhou T, Quijada H, Moreno-Vinasco L, Garcia JG (2015). Pathologic mechanical stress and endotoxin exposure increases lung endothelial microparticle shedding. Am J Respir Cell Mol Biol, 52(2), 193–204. doi: 10.1165/rcmb.2013-0347OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Purevdorj E, Zscheppang K, von Mayersbach D, Behrens J, Brinkhaus MJ, … Dammann CE (2010). ErbB4 regulates the timely progression of late fetal lung development. Biochim Biophys Acta, 1803(7), 832–839. doi:S0167-4889(10)00072-8 [pii] 10.1016/j.bbamcr.2010.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luks FI, Roggin KK, Wild YK, Piasecki GJ, Rubin LP, Lesieur-Brooks AM, & De Paepe ME (2001). Effect of lung fluid composition on type II cellular activity after tracheal occlusion in the fetal lamb. J Pediatr Surg, 36(1), 196–201. [DOI] [PubMed] [Google Scholar]
- Maas SL, Breakefield XO, & Weaver AM (2017). Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol, 27(3), 172–188. doi: 10.1016/j.tcb.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen PJ, Berger JE, Meneses J, Phung Y, Pedersen RA, Werb Z, & Derynck R (1995). Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature, 376(6538), 337–341. [DOI] [PubMed] [Google Scholar]
- Miettinen PJ, Warburton D, Bu D, Zhao JS, Berger JE, Minoo P, Derynck R (1997). Impaired lung branching morphogenesis in the absence of functional EGF receptor. Dev Biol, 186(2), 224–236. [DOI] [PubMed] [Google Scholar]
- Mittelbrunn M, & Sanchez-Madrid F (2012). Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol, 13(5), 328–335. doi: 10.1038/nrm3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najrana T, Ramos LM, Abu Eid R, & Sanchez-Esteban J (2017). Oligohydramnios compromises lung cells size and interferes with epithelial-endothelial development. Pediatr Pulmonol, 52(6), 746–756. doi: 10.1002/ppul.23662 [DOI] [PubMed] [Google Scholar]
- Nardiello C, & Morty RE (2016). MicroRNA in late lung development and broncho pulmonary dysplasia: the need to demonstrate causality. Mol Cell Pediatr, 3(1), 19. doi: 10.1186/s40348-016-0047-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olave N, Lal CV, Halloran B, Pandit K, Cuna AC, Faye-Petersen OM, … Ambalavanan N (2016). Regulation of alveolar septation by microRNA-489. Am J Physiol Lung Cell Mol Physiol, 310(5), L476–487. doi: 10.1152/ajplung.00145.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli RC, Bergamini G, & Rajendran L (2019). Cell-to-cell Communication by Extracellular Vesicles: Focus on Microglia. Neuroscience, 405, 148–157. doi: 10.1016/j.neuroscience.2018.04.003 [DOI] [PubMed] [Google Scholar]
- Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Hatzigeorgiou AG (2013). DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res, 41(Web Server issue), W169–173. doi: 10.1093/nar/gkt393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park-Windhol C, Ng YS, Yang J, Primo V, Saint-Geniez M, & D’Amore PA (2017). Endomucin inhibits VEGF-induced endothelial cell migration, growth, and morphogenesis by modulating VEGFR2 signaling. Sci Rep, 7(1), 17138. doi: 10.1038/s41598-017-16852-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JA, Sharif AS, Tschumperlin DJ, Lau L, Limbrey R, Howarth P, & Drazen JM (2012). Tissue factor-bearing exosome secretion from human mechanically stimulated bronchial epithelial cells in vitro and in vivo. J Allergy Clin Immunol, 130(6), 1375–1383. doi: 10.1016/j.jaci.2012.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NV, Acarregui MJ, Snyder JM, Klein JM, Sliwkowski MX, & Kern JA (2000). Neuregulin-1 and human epidermal growth factor receptors 2 and 3 play a role in human lung development in vitro. Am J Respir Cell Mol Biol, 22(4), 432–440. [DOI] [PubMed] [Google Scholar]
- Ramirez MI, Millien G, Hinds A, Cao Y, Seldin DC, & Williams MC (2003). T1alpha, a lung type I cell differentiation gene, is required for normal lung cell proliferation and alveolus formation at birth. Dev Biol, 256(1), 61–72. [DOI] [PubMed] [Google Scholar]
- Reczko M, Maragkakis M, Alexiou P, Grosse I, & Hatzigeorgiou AG (2012). Functional microRNA targets in protein coding sequences. Bioinformatics, 28(6), 771–776. doi: 10.1093/bioinformatics/bts043 [DOI] [PubMed] [Google Scholar]
- Sanchez-Esteban J, Tsai SW, Sang J, Qin J, Torday JS, & Rubin LP (1998). Effects of mechanical forces on lung-specific gene expression. Am J Med Sci, 316(3), 200–204. [PubMed] [Google Scholar]
- Sessa R, & Hata A (2013). Role of microRNAs in lung development and pulmonary diseases. Pulm Circ, 3(2), 315–328. doi: 10.4103/2045-8932.114758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani RA (2006). MicroRNAs: regulators of gene expression and cell differentiation. Blood, 108(12), 3646–3653. doi: 10.1182/blood-2006-01-030015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibilia M, Kroismayr R, Lichtenberger BM, Natarajan A, Hecking M, & Holcmann M (2007). The epidermal growth factor receptor: from development to tumorigenesis. Differentiation, 75(9), 770–787. doi:S0301–4681(09)60170–5 [pii] 10.1111/j.1432-0436.2007.00238.x [DOI] [PubMed] [Google Scholar]
- Sibilia M, & Wagner EF (1995). Strain-dependent epithelial defects in mice lacking the EGF receptor. Science, 269(5221), 234–238. [DOI] [PubMed] [Google Scholar]
- Street JM, Barran PE, Mackay CL, Weidt S, Balmforth C, Walsh TS, … Dear JW (2012). Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J Transl Med, 10, 5. doi: 10.1186/1479-5876-10-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, & Rigoutsos I (2008). MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature, 455(7216), 1124–1128. doi: 10.1038/nature07299 [DOI] [PubMed] [Google Scholar]
- Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, … Zuba-Surma EK (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7(1), 1535750. doi: 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torregrosa Paredes P, Esser J, Admyre C, Nord M, Rahman QK, Lukic A, … Gabrielsson S (2012). Bronchoalveolar lavage fluid exosomes contribute to cytokine and leukotriene production in allergic asthma. Allergy, 67(7), 911–919. doi: 10.1111/j.1398-9995.2012.02835.x [DOI] [PubMed] [Google Scholar]
- Tsao PN, Matsuoka C, Wei SC, Sato A, Sato S, Hasegawa K, … Morimoto M (2016). Epithelial Notch signaling regulates lung alveolar morphogenesis and airway epithelial integrity. Proc Natl Acad Sci U S A, 113(29), 8242–8247. doi: 10.1073/pnas.1511236113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckaert T, & De Langhe SP (2015). Wnt and FGF mediated epithelial-mesenchymal crosstalk during lung development. Dev Dyn, 244(3), 342–366. doi: 10.1002/dvdy.24234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuckovic A, Herber-Jonat S, Flemmer AW, Ruehl IM, Votino C, Segers V, … Jani JC (2016). Increased TGF-beta: a drawback of tracheal occlusion in human and experimental congenital diaphragmatic hernia? Am J Physiol Lung Cell Mol Physiol, 310(4), L311–327. doi: 10.1152/ajplung.00122.2015 [DOI] [PubMed] [Google Scholar]
- Wahlgren J, De LKT, Brisslert M, Vaziri Sani F, Telemo E, Sunnerhagen P, & Valadi H (2012). Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res, 40(17), e130. doi: 10.1093/nar/gks463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JH, Thampatty BP, Lin JS, & Im HJ (2007). Mechanoregulation of gene expression in fibroblasts. Gene, 391(1–2), 1–15. doi: 10.1016/j.gene.2007.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikenheiser KA, Vorbroker DK, Rice WR, Clark JC, Bachurski CJ, Oie HK, & Whitsett JA (1993). Production of immortalized distal respiratory epithelial cell lines from surfactant protein C/simian virus 40 large tumor antigen transgenic mice. Proc Natl Acad Sci U S A, 90(23), 11029–11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis GR, Fernandez-Gonzalez A, Anastas J, Vitali SH, Liu X, Ericsson M, … Kourembanas S (2018). Mesenchymal Stromal Cell Exosomes Ameliorate Experimental Bronchopulmonary Dysplasia and Restore Lung Function through Macrophage Immunomodulation. Am J Respir Crit Care Med, 197(1), 104–116. doi: 10.1164/rccm.201705-0925OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Costello D, Friedman H, Minich N, Fanaroff AA, & Hack M (2005). Improved survival rates with increased neurodevelopmental disability for extremely low birth weight infants in the 1990s. Pediatrics, 115(4), 997–1003. doi:115/4/997 [pii] 10.1542/peds.2004-0221 [DOI] [PubMed] [Google Scholar]