Abstract
Background
The spatial and temporal dynamics of SARS-CoV-2 have been described in case series and retrospective studies. In this study, we provide a coherent overview of the duration of viral detection and viral RNA load in COVID-19 patients, stratified by specimen type, clinical severity, and age.
Method
We systematically searched PubMed/MEDLINE and Cochrane review database for studies published between 1.11.2019 and 23.04.2020. We pooled the data of selected studies (22/7226 (650 patients) for meta-analysis) to estimate duration of viral detection and visualized viral load over time.
Findings
Our analysis showed consistent viral detection from specimen from the upper respiratory tract (URT), the lower respiratory tract (LRT), and faeces, irrespective of the clinical severity of COVID-19. Our analysis suggests that SARS-CoV-2 persists for a longer duration in the LRT compared to the URT in adult patients (5•7 days in mild; 5•9 days in moderate-severe patients). The differences in the duration of viral detection between mild and moderate-severe patients is limited in the LRT, but an indication of longer duration of viral detection for moderate-severe patients was observed in feces (15 days in mild vs. 21 days in moderate-severe patients) and the URT (12 days in mild vs. 16 days in moderate-severe patients). Further, viral load was demonstrated to peak in earlier stages of infection in the URT compared to LRT.
Interpretation
This review may aid mathematical modelling and help in defining appropriate endpoints for clinical trails with antivirals in COVID-19.
Funding
The project has received funding support from Innovation Fund Denmark.
Keywords: SARS-CoV-2, COVID-19, Systematic review, Viral dynamics
1. Introduction
Coronavirus disease 2019 (COVID-19) has caused many deaths and severe suffering worldwide prompting a surge in research and drug development activities. These efforts are revealing clinical and molecular characteristics of SARS-CoV-2 and the infection it causes, but its viral dynamics and spatial shedding patterns in humans remain poorly understood [1]. A variety of case series and retrospectives studies, reporting viral dynamics of SARS-CoV-2 on a patient or patient population level, have been published and illuminate the spatial patterns of SARS-CoV-2’s shedding in different stages of the infection [1]. However, these studies are heterogeneous with regards to patient populations and the specific viral RNA load testing. To provide an overview of our current knowledge of the shedding patterns of SARS-CoV-2, we conducted a review of the temporal and spatial viral dynamics across clinical severities of COVID-19. Firstly, this knowledge can help improve mathematical models of SARS-CoV-2’s replication, as the numerical model can be trained with more real-world data. Secondly, this knowledge can inform the definition of relevant endpoints in clinical trials assessing pharmacological treatments aimed at reducing viral RNA load in patients positive for SARS-CoV-2.
The objective of this paper is to provide a systematic review of the spatial and temporal viral dynamics of SARS-CoV-2 in COVID-19 patients, stratified after clinical severity, location of viral sampling and age, based on individual patient data. The data of the systematic review is then aggregated to report a weighted mean of duration of viral detection, per sampling location for adults and children with mild and moderate-severe symptoms.
2. Materials and methods
This systematic review and subsequent meta-analysis have been conducted according to the PRISMA guidelines [2]. A PRISMA checklist is displayed in Table S1.
2.1. Systematic review
2.1.1. Data sources
We searched the databases MEDLINE/PUBMED and Cochrane Review with the following search terms: “SARS-CoV-2 [MESH]” OR “COVID 19 [MESH]” alone, or in combination with “virology” OR “viral” OR “Epidemio*” AND “clinical”. Studies from November 2019 until 23rd of April 2020 are included in this review.
2.1.2. Eligibility criteria
The inclusion criteria with the corresponding rational is summarized in Table 1. Studies which demonstrated viral data as patient fractions (e.g. as Kaplan Meyer Plots), missed crucial information needed for detailed stratification (e.g. age, clinical severity, individual viral RNA load/dynamic data), contained only asymptomatic patients or could be classified as interventional study were excluded. Given the rather simple study setup (one primary outcome investigated with predefined outcome measure criteria) and precise and narrow eligibility criteria, the study evaluation was performed by only one reviewer. In the rare case of uncertainties, these were discussed and resolved by consensus with all authors of this manuscript or with the support of internal medical specialists.
Table 1.
Inclusion criteria for eligibility of studies
| Inclusion criteria | Rationale |
|---|---|
| English speaking publication or English publicly available manuscript (due to accelerated publishing timelines during COVID19, epub manuscripts, available ahead of publication were included as well) | |
| Individual data (or mean/SD) of viral RNA load or/and time to clearance of infection is reported | Needed to provide coherent summary measures |
| Clinical severity per subject (if diverse patient population) or whole patient population is reported | Needed for stratification |
| Median age for patient population or per subject is reported | Needed for stratification |
| SARS-CoV-2 detection was performed by RT-PCR with defined thresholds to determine negativity of sample | Ensures to have identical molecular methodology across studies; needed to determine time to clearance from graph if not reported in text |
| Location of sampling is defined and can be categorized into lower tract respiratory (LRT) specimen, upper respiratory tract (URT) and Faeces | Needed for stratification |
| Two consecutive measures were obtained to confirm negativity conversion | Ensures quality of measures and reduces risk of false negatives |
| Time to clearance was either reported from day of symptom onset or day from admission | Needed to have coherent and clinically meaningful Day 0 set across studies |
| Study is a case series, systematic review, prospective observational or retrospective study | Reduce risk of reporting bias |
2.1.3. Study selection
Following screening of titles and removing of duplicates, relevant abstracts were selected for screening. Out of the abstracts, studies were selected to be scanned as full text to assess eligibility. Finally, studies were selected to be included in the systematic review.
2.1.4. Data extraction
Data were extracted from the full text/supplemental material or if only available in graphs digitalized using GetData Graph Digitizer © version 2.26.0.20. We used a pre-defined and internally developed extraction protocol to capture identical information across studies (incl. author, title, country, design, sample size, thresholds and genes of RT-PCR, frequency of sampling, patient age (median), COVID-19 severity and classification reference, major clinical characteristics, treatment, location of viral sampling, duration of viral detection, viral RNA load).
2.1.5. Data processing and summary measures
To determine the duration of viral detection from symptom onset, we counted the days from initiation of symptoms (Day 0) to the last positive test (final day was set as Day -1 of first negative test of two consecutive tests for daily sampling). However, in some studies the sampling frequency was not daily, meaning that the last positive test does not equal Day -1 of first negative test. Therefore, we also reported the duration of viral detection until the last positive swab, to not introduce any predictions into the summary measures. In the case that the day of the positive test was not reported for a patient, we excluded this patient in the overall summary measure, as the duration of viral detection becomes predictive. Also, in case a positive test was not reported, we could not be sure that this patient was tested at all positive for the virus at the specific sampling location. Furthermore, for patients that did not clear the virus within the time frame of sampling, we used the day of the last positive test as final day to not introduce bias into the data by excluding these and leaving out patients that needed longer to clear the virus than the average. Finally, we also recorded the duration of viral detection in case recurrence of positivity was observed, meaning a positive test was detected after two negative ones have been obtained. To measure the duration in regard to the “recurrence” endpoint, the final day was also set as Day -1 of the first negative test of two consecutive negative tests.
In case clinical severity was not categorized into mild, moderate or severe by the authors of the publication, we conducted this categorization with the symptoms reported, based on definitions described by Wu et al. [3]. Asymptomatic patients were excluded.
Duration of viral detection is demonstrated as mean (days after symptom or admission onset) with its corresponding standard deviation and viral RNA load was visualized as viral copies/mL [log10] over time (days after symptom onset) stratified after clinical severity, mild or moderate/severe, and location of swab (upper respiratory tract (URT) specimen, lower respiratory tract (LRT) specimen or faeces). Oro-/nasopharyngeal, throat and nasal swabs were combined to “URT”, sputum and tracheal aspirate to “LRT”, anal swabs and stool to “faeces”, and plasma and serum to “Blood”.
Data which did not use copies/mL as unit for quantifying viral RNA load were excluded. R Studio ©, Version 1.2.1335 was used for visualization. Raw data can be found in Supplementary Table S2.
2.1.6. Statistical analysis of viral RNA load comparison
Statistical analysis comparing the average load per sampling location across each study was performed using unpaired t test, not assuming consistent SD and using a two stage setup false discovery rate approach of Benjamini, Krieger and Yekutieli, setting the desired FDR to 1% [4]. Statistical analysis was conducted in GraphPad Prism 8.1.2.
2.2. Meta-analysis
2.2.1. Study selection
Out of the studies that were included in the systematic review, we chose studies to be included in the meta-analysis which measured duration of positive detection of SARS-CoV-2 in days after symptom onset, but not after hospitalization ensuring that an identical Day 0 is set across. Patients equal to 18 years or older were classified as adults (if only median was recorded, the median age was considered for categorization). Single case series were excluded. In total, 22 studies were included in the aggregation of data 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26.
2.2.2. Risk of bias assessment
The risk was assessed based on pre-defined criteria and included both the assessment on a study and outcome measure level. The results of the assessment of risk of bias for each study of the meta-analysis is summarized in Table S3. The possibility of publication bias was assessed using funnel-plot asymmetry as suggested by Egger et al. which was analysed in RStudio® with the metafor package [27,28].
2.2.3. Data processing
Mild disease severity was kept as mild and moderate and severe disease severity were merged to moderate-severe in the meta-analysis. Oro-/nasopharyngeal, throat and nasal swabs were combined to “URT”, sputum and tracheal aspirate to “LRT”, anal swabs and stool to “faeces”, and plasma and serum to “Blood”.
2.2.4. Data synthesis and statistical analysis
Aggregation of study data of duration of viral detection was conducted in ReviewManager 5.3. Standard error was computed according to STDEV/(SQRT(N)). We used generic inverse variance as a data type (mean and SE), and random effect as analysis model. Effect measures (pooled estimates) were shown as mean or differences in mean with 95% confidence interval (CI). Mean differences in each study was computed based on a per patient calculation. Heterogeneity was measured by I2. Forest plots are provided for selected data groupings.
3. Results
3.1. Study selection
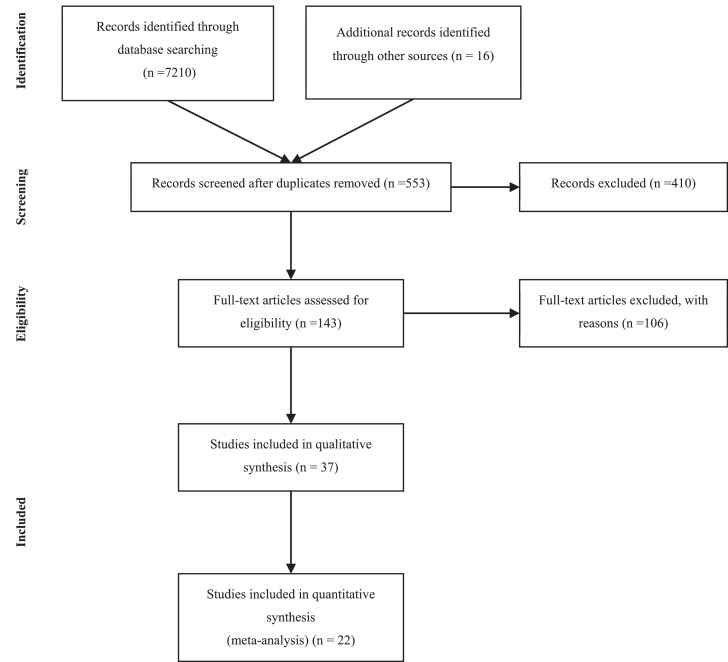
The overall study selection process is displayed in the flow-diagram in Fig. 1. Briefly, we screened a total of 7226 titles and selected 553 for abstract screening. Out of the 553 abstracts, we selected 143 studies to be scanned as full text to investigate whether viral data is generally reported. Out of these studies, 80 studies were chosen for in-depth reading to assess eligibility. Finally, 37 studies were included in the systematic review. Reasons for exclusion were: individual patient data was missing and/or only median of duration of viral detection was demonstrated, clinical severity or age was not reported or only baseline viral data were shown.
Fig. 1.
Study Selection
3.2. Overview of studies describing duration of viral detection in COVID-19 patients
The data summary of the selected studies, describing duration of viral detection as days after symptom onset (as mean and SD) is displayed in Table 2 and days after hospitalization in Table 3. More details of each study, including clinical characteristics can be found in Table S4 in the supplementary material.
Table 2.
Summary of studies describing duration of viral detection as days after symptom onset
| No. | Author | Design | Threshold Gene | Age* | Clinical severity | Reported Treatment | Size | Sampling Location | 2 consecutive negatives Day -1 |
2 consecutive negatives Last positive |
Recurrence Day -1 |
Recurrence Last positive |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | |||||||||
| 1 | Cai J. [5] | Case series | Ct > 35 ORF1ab,N |
6•5 | Mild | NS | 10 | Nasopharyngeal | 12•0 | 4•8 | ||||||
| 6 | Stool | 20•0 | 6•5 | |||||||||||||
| 2 | Caly [33] | Case series | NS | 58•0 | Moderate | None | 1 | Nasopharyngeal | 11•0 | NA | 8•0 | NA | ||||
| 1 | Sputum | 12•0 | NA | 12•0 | NA | |||||||||||
| 3 | Chang [6] | Case series | NS | 35•5 | Mild | aIFN, L/R or other AVs | 16 | Throat | 8•6 | 3•8 | ||||||
| 4 | Chen [17] | RS | Ct ≥ 37 ORF1ab, N |
39•0 | Moderate | NS | 16 | Pharyngeal | 10•9 | 4•9 | 8•3 | 3•9 | 12.6 | 6.7 | 11.0 | 6.5 |
| 9 | Faeces | 16•2 | 7•2 | 12•2 | 4•9 | 19.2 | 9.4 | 15.1 | 5.8 | |||||||
| 14 | Sputum | 26•2 | 13•9 | 24•9 | 13•4 | 30•6 | 14•0 | 30•1 | 13•6 | |||||||
| Severe | 2 | Pharyngeal | 23•0 | 9•9 | 18•5 | 4•9 | 23•0 | 9•9 | 19•5 | 4•9 | ||||||
| 2 | Sputum | 31•5 | 24•7 | 29•0 | 25•5 | 28•0 | 15•6 | 37•0 | 13•5 | |||||||
| 6•5 | Mild | 2 | Faeces | 12•5 | 2•1 | 12•5 | 2•1 | 12•5 | 2•1 | 12•5 | 2•1 | |||||
| 3 | Pharyngeal | 7•7 | 1•2 | 4•0 | 3•0 | 7•7 | 1•2 | 4•0 | 3•0 | |||||||
| 1 | Sputum | 13•0 | NA | 12•0 | NA | |||||||||||
| Moderate | 1 | Faeces | 9•0 | NA | 9•0 | NA | ||||||||||
| 1 | Pharyngeal | 8•0 | NA | 6•0 | NA | |||||||||||
| 5 | Chen [31] | Case series | NS | 46•0 | Moderate | NS | 1 | Oropharyngeal | 11•0 | NA | 8•0 | NA | 19•0 | NA | 17•0 | NA |
| 6 | Cheng [34] | Case series | TCDC | 55•0 | Moderate | NS | 1 | Sputum | 15•0 | NA | 14•0 | NA | ||||
| 1 | Oropharyngeal | 19•0 | NA | 10•0 | NA | |||||||||||
| 7 | Colavita [29] | Case series | Ct > 45 ND |
65•0 | Mild | NS | 1 | Nasal | 20•0 | NA | 16•0 | NA | ||||
| 1 | Ocular | 27 | NA | |||||||||||||
| 8 | Grace Lui [20] | Prospective Cohort Study | < 694 copies/mL; Ct > 39•9 N |
58•0 | Mild | L/R | 1 | Nasopharyngeal | 10•0 | NA | 8•0 | NA | ||||
| 1 | Stool | 7•0 | NA | 7•0 | NA | |||||||||||
| Moderate | 5 | Nasopharyngeal | 14•6 | 5•6 | 14•0 | 5•8 | ||||||||||
| 1 | Plasma | 5•0 | NA | 3•0 | NA | |||||||||||
| 5 | Stool | 12•2 | 4•1 | 10•6 | 2•6 | |||||||||||
| 2 | Tracheal aspirate/Sputum | 20•5 | 0•7 | 20•5 | 0•7 | |||||||||||
| Severe | 5 | Nasopharyngeal | 15•4 | 5•7 | 13•0 | 4•5 | 16•8 | 8•3 | 14•6 | 9•2 | ||||||
| 1 | Plasma | 16•0 | NA | 14•0 | 5•9 | 15•7 | 6•8 | 14•3 | 6•8 | |||||||
| 5 | Stool | 18•6 | 2•5 | 18•6 | 7•8 | |||||||||||
| 5 | Tracheal aspirate/Sputum | 22•6 | 5•0 | 21•6 | 4•9 | |||||||||||
| 9 | Han [21] | Case series | [35] | 44•0 | Mild | NS | 22 | Sputum | 38•4 | 8•8 | ||||||
| 10 | Holshue [36] | Case series | CDC | 35•0 | Moderate | NS | 1 | Oropharyngeal | 10•0 | NA | ||||||
| 1 | Nasopharyngeal | 11•0 | NA | |||||||||||||
| 11 | Hu [15] | Case series | Ct > 38 ORF1ab, N | 28•0 | Mild | NS | 3 | Nasopharyngeal | 11•7 | 2•3 | 11•0 | 2•6 | ||||
| 3 | Anal | 18•3 | 8•4 | 18•0 | 8•4 | |||||||||||
| 12 | Hu [23] | RS | NS, ORF1ab, N |
53•0 | Mild | aIFN | 4 | Pharyngeal | 10•5 | 6•0 | 10•0 | 6•4 | ||||
| 10•0 | 1 | Pharyngeal | 12•0 | NA | 11•0 | NA | ||||||||||
| 13 | Huang [22] | RS | Ct > 40 ORF1ab, N | 59•5 | Severe | Mechanic ventilation | 13 | Anal swab/ Faeces | 24•6 | 11•4 | 24•2 | 11•7 | 29•7 | 12•3 | 28•7 | 12•2 |
| 12 | Nasal swab | 21•0 | 8•9 | 20•9 | 8•7 | 21•8 | 11•0 | 22•8 | 11•0 | |||||||
| 2 | Blood | 10•5 | 0•7 | |||||||||||||
| 9 | Throat | 20•6 | 9•4 | 19•7 | 9•4 | 20•6 | 9•4 | 19•8 | 9•4 | |||||||
| 16 | Sputum | 25•4 | 8•8 | 25•1 | 8•8 | 33•53 | 12•3 | 33•2 | 12•4 | |||||||
| 14 | Kujawski [24],+ | Case series | NS | 53•0 | Mild | None | 10 | Oro/Naso-pharyngeal# | 18•1 | 4•8 | 17•3 | 5•2 | ||||
| 2 | Stool | 20•0 | 0 | 17•0 | 0 | |||||||||||
| Severe | None | 3 | Stool | 20•0 | 4•0 | 17•3 | 5•8 | |||||||||
| 8 | Oro/Naso-pharyngeal# | 16•9 | 7•1 | 13•5 | 4•3 | |||||||||||
| Remdesivir | 2 | Stool | 13•0 | 1•4 | 9•0 | 4•2 | ||||||||||
| 6 | Oro/Naso-pharyngeal# | 11•8 | 9•7 | 9•8 | 7•3 | |||||||||||
| 15 | Lee [37] | Case series | NA, RdRp, E, N |
46•0 | Moderate | AV | 1 | Oropharyngeal | 19•0 | NA | 6•0 | NA | ||||
| 1 | Nasopharyngeal | 18•0 | NA | 18•0 | NA | |||||||||||
| 16 | Lescure [25] | Case series | < 101 copies/mL, RdRP-IP1, RdRp, E |
38•5 | Mild | None | 4 | Nasopharyngeal | 8•6 | 0•6 | ||||||
| 2 | Stool | 16•5 | 3•5 | |||||||||||||
| 80•0 | Severe | Remdesivir | 1 | Blood | 15 | NA | ||||||||||
| 1 | Nasopharyngeal | 24•0 | NA | |||||||||||||
| 17 | Lim [30] | Case series | NS | 54•0 | Mild | L/R + other | 1 | Sputum | 10•0 | NA | 10•0 | NA | 16•0 | NA | 16•0 | NA |
| 18 | Liu [19] | Case series | Ct > 43 E, RdRP, N |
42•5 | Mild | L/R + aIFN | 5 | Nasal/Throat | 12•4 | 3•8 | ||||||
| Severe | 5 | Nasal Throat | 12•2 | 6•5 | ||||||||||||
| 19 | Marchand-Senéca [38] | Case series | Ct ≥ 40 RdRp, E, N, ORF3a |
56•0 | Moderate | NS | 1 | Mid-turbinate | 8•0 | NA | ||||||
| 1 | Throat | 6•0 | NA | |||||||||||||
| 1 | Nasopharyngeal | 22•0 | NA | |||||||||||||
| 20 | Pongpirul [14] | Case series | WHO | 61•0 | Mild | Antimicrobials | 11 | Oro/naso-pharyngeal | 16•5 | 9•3 | ||||||
| 21 | Scott [39] | Case series | CDC | 26•0 | Mild | None | 1 | Nasopharyngeal | 18•0 | NA | ||||||
| 1 | Sputum | 8 | NA | |||||||||||||
| 1 | Oropharyngeal | 16•0 | NA | |||||||||||||
| 22 | Tan [26] | RS | NS | 7•0 | Mild | NS | 3 | Throat | 15•0 | 1•7 | 14•3 | 1•2 | ||||
| Moderate | 2 | Stool | 17•5 | 7•8 | 16•5 | 5•0 | ||||||||||
| 4 | Throat | 14•5 | 6•0 | 13•5 | 6•0 | |||||||||||
| 23 | Tan [40] | Case series | Ct = 45 ORF1 |
73•0 | Severe | Yes, NS | 1 | Throat | 15•0 | NA | ||||||
| 1 | Rectal | 22•0 | NA | |||||||||||||
| 24 | Thevarajan[41] | Case series | Ct ≥ 45 E |
47•0 | Moderate | NS | 1 | Nasopharyngeal | 6•0 | NA | ||||||
| 1 | Faeces | 6•0 | NA | |||||||||||||
| 1 | Faeces | 6•0 | NA | |||||||||||||
| 25 | Woelfel [7] | Case series | < 102 copies/mL E, RdRp |
Adult NS |
Mild | None | 8 | Stool | 15•3 | 7•0 | ||||||
| 9 | Sputum | 16•9 | 4•9 | |||||||||||||
| 9 | Oro/naso-pharyngeal | 11•2 | 3•4 | |||||||||||||
| 26 | Wu [8] | Case series | NS N, E |
41 (mean) |
Moderate | AV (88%), aIFN (12%) | 33 | Throat | 15•2 | 5•9 | ||||||
| 33 | Faeces | 26•7 | 12•3 | |||||||||||||
| 27 | Xu [16] | Prospective Obser-vational Study | Ct > 40 ORFab1 N |
|
Mild | aIFN | 7 | Nasopharyngeal | 5•7 | 4•7 | 4•3 | 4•8 | ||||
| 6 | Rectal | 16•7 | 6•0 | 16•5 | 6•3 | |||||||||||
| 28 | Young [9] | Case series | Ct > 38 N, S, ORF1ab |
37•0 | Mild | None | 12 | Nasopharyngeal | 15•2 | 5•5 | 14•9 | 5•7 | 15•4 | 5•4 | 15•3 | 5•5 |
| 3 | Stool | 6•7 | 0•6 | |||||||||||||
| 56•0 | Moderate | L/R (5/6) | 1 | Stool | 8•0 | NA | ||||||||||
| 4 | Nasopharyngeal | 7•0 | 5•3 | 6•0 | 5•6 | 9•8 | 5•3 | 9•0 | 5•9 | |||||||
| Severe | 1 | Blood | 13 | NA | ||||||||||||
| 2 | Nasopharyngeal | 18•0 | 11•3 | 18•0 | 11•3 | 20•0 | 8•5 | 20•0 | 8•5 | |||||||
| 29 | Yu [10] | RS | < 25 copies/mL | 37•0 | Mild | NS | 17 | Nasopharyngeal | 15•1 | 5•0 | 15•0 | 5•0 | ||||
| 30 | Zhang [11] | Case series | CDC China | 8•0 | Mild | aIFN | 3 | Throat | 17•0 | 6•1 | ||||||
| 31 | Zheng [12] | RS | < 1000 copies/mL ORFab1 |
47•5 | Mild | aIFN, L/R, arbidol, favipiravir, darunavir-cobicistat | 21 | Sputum/saliva | 17•0 | 10•0 | ||||||
| 7 | Blood | 17•1 | 10•4 | |||||||||||||
| 13 | Stool | 21•8 | 8•3 | |||||||||||||
| 57•0 | Severe | 73 | Sputum/saliva | 22•8 | 10•8 | |||||||||||
| 34 | Blood | 17•5 | 7•3 | |||||||||||||
| 40 | Stool | 25•3 | 12•0 | |||||||||||||
| 32 | Zhou [18] | Case series | CDC China | 58•0 | Severe | NS | 41 | Throat | 30•8 | 8•7 | ||||||
| 33 | Zou [13] | Case series | Ct ≥ 40 N, Orf1b |
59•0 | Mild | NS | 12 | Nasal | 8•6 | 3•7 | 8•2 | 3•4 | 9•2 | 5•0 | 9•0 | 5•1 |
| 10 | Throat | 7•0 | 3•9 | 6•6 | 3•4 | 8•1 | 4•3 | 7•6 | 3•9 | |||||||
| Severe | 3 | Nasal | 10•3 | 3•8 | 10•3 | 3•8 | 12•0 | 1•0 | 12•0 | 1•0 | ||||||
| 3 | Throat | 9•3 | 4•5 | 9•3 | 4•5 | 13•0 | 1•7 | 13•0 | 1•7 | |||||||
Blank cells apply if sampling was daily (value of Day -1 equals value of last positive) or no recurrence occurred. Samples that were not tested positive are excluded in this overview.
aIFN: Alpha Interferon, AV: Antiviral, CDC: Center for Disease Control and Prevention, CT: Threshold Cycle, E: Envelope, L/R: Lopinavir/Ritonavir, N: Truncated nucleocapsid, NA: Not applicable, ND: Not determined, NS: Not specified, ORF = Open reading frame, RdRp: RNA dependent RNA polymerase, RS: Retrospective, S: Spike, SD: Standard deviation, WHO: World health organization;
Age in years and displayed as median (if not otherwise specified),
Oro- and nasopharyngeal combined,
sputum samples from this study were excluded as the majority captured only one baseline sample.
Table 3.
Summary of studies describing duration of viral detection as days after hospitalization.
| No | Author | Design | Threshold Gene | Age * | Clinical severity | Reported Treatment | Size | Sampling Location | 2 Consecutive negatives Day -1 | 2 Consecutive negatives Last positive | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||||||||
| 34 | Chen [42] | Retrospective | WHO | 51•0 | Mild | NS | 2 | Pharyngeal | 4•5 | 2•1 | 3•5 | 2•1 |
| 2 | Stool | 10•5 | 3•5 | 9•5 | 3•5 | |||||||
| Moderate | 30 | Pharyngeal | 6•6 | 4•3 | 5•6 | 4•3 | ||||||
| 17 | Stool | 11•7 | 7•2 | 9•9 | 7•2 | |||||||
| Severe | 10 | Pharyngeal | 13•4 | 3•2 | 10• | 3 | ||||||
| 9 | Stool | 8 | 4•9 | 14•2 | 6•1 | |||||||
| 35 | Lo [43] | Case series | Ct > 38 ORF1ab, N |
54•0 | Mild | L/R | 2 | Faeces | 16•5 | 0•7 | 15•0 | 1•4 |
| 2 | Nasopharyngeal | 13•0 | 5•7 | 12•0 | 7•1 | |||||||
| Moderate-Severe | 7 | Faeces | 13•7 | 3•5 | 12•1 | 3•6 | ||||||
| 8 | Nasopharyngeal | 12•5 | 6•6 | 12•0 | 6•9 | |||||||
| 36 | Xing [44] | Case series | [45] | 5•0 | Mild | aIFN +Ribavirin | 3 | Faeces | 12•7 | 2•5 | ||
| 3 | Throat | 18•7 | 12•1 | |||||||||
| 37 | Xu [46] | Case series | Ct > 40 ORFab1, N |
51•0 | Mild | NS | 47 | Throat | 11•4 | 5•6 | ||
No recurrence was reported for these studies. Blank columns apply if sampling was daily (value of Day -1 equals last positive) or no recurrence occurred.
aIFN: Alpha interferon, Ct = Threshold cycle, L/R: Lopinavir/Ritonavir, N: Truncated nucleocapsid, ORF: Open reading frame, SD: Standard deviation, WHO: World health organization;
Age displayed as median in years
The SARS-CoV-2 virus is detected in both in the lower and upper respiratory tract, and faeces, irrespective of the severity of the disease.
The average duration of positive SARS-CoV-2 viral detection (Day -1) from URT specimens ranged from 7•9 days to 20 days after symptom onset in mild adult patients, and from 6 to 30•8 days in moderate-severe patients. The average duration of positive SARS-CoV-2 viral detection from LRT specimens ranged from 8 days to 38•4 days after symptom onset in mild adult patients, and from 6 days to 26•9 days in adults with moderate-severe symptoms. In children with mild COVID-19 symptoms, average duration of positive SARS-CoV-2 viral detection ranged from 5•7 days to 17 days after symptom onset in the URT and was reported as 13 days in the LRT in one study [17]. Duration of positive SARS-CoV-2 viral detection in children with moderate symptoms was observed in two studies reporting viral detection for 8 days and 14•5 days after symptom onset in the URT [17,26]. Furthermore, several studies have reported positive SARS-CoV-2 viral detection in the faeces and blood in adults, and in faeces only in children. One adult patient was reported to have viral positivity in ocular fluids, whereas other studies did not find evidence of positive SARS-CoV-2 viral detection in the eye [29]. Only one adult patient was reported to have a positive viral sample in the urine [22].
Recurrence of viral positivity after two consecutive negative tests were obtained, was observed in 7 out of 37 studies. Recurrence of positivity occurred in adult patients and URT specimen with mild symptoms (1 patient in LRT), and in mild-severe adult patients and URT, LRT and faecal specimen.
However, the actual risk of a reinfection with SARS-CoV-2 after confirmed negativity remains to be elucidated. Among the seven studies which reported recurrence of viral positivity, five studies reported only sporadic cases which are most likely a technical false-positive due to the short time between SARS-CoV-2 negativity and reinfection and lack of validation of viral positivity with a second test [9,13,20,30,31]. In contrary, in two studies the duration of recurred positivity of SARS-CoV-2 was reported to last for approximately 2 weeks after having a confirmed negative test, implying true recurrence [17,22]. However, as these studies did neither capture the antibody response nor reported clinical signs throughout the study and in the follow-up period, the clinical significance of the observed recurred viral positivity cannot be unambiguously determined.
Generally, the average duration of viral detection only differed minimally when setting the final day as Day -1 of first negative test (results described in text above) or as day of last positive test in the above patient populations.
3.3. Aggregation of study data
We then aggregated the data of 22 selected studies to report a pooled estimate of the duration of positive SARS-CoV-2 viral detection. We report the weighted mean for both outcome measures, negative conversion defined by obtaining two consecutive negative samples and negative conversion after recurrence (positive sample was detected after two consecutive negatives were obtained). As sampling frequency varied and was not daily for all the studies, we also report the results obtained when setting different endpoint measures: 1) final day of positive detection of virus is day -1 of first negative of two consecutive measurement and 2) final day of positive detection of virus is the day on which last positive test was obtained, in case Day -1 and last positive day were not identical, to avoid introducing predictions into the outcome measures. Notably, there was a portion of patients that did not reach viral negativity within the time of sampling for which we reported the day of the last positive test as final day. Considering studies that were included in the aggregation analysis, the share of mild patients (children and adults) that did not reach negativity were 27/139 for URT specimen, 6/52 for LRT specimen and 26/45 for faecal specimen. Regarding moderate-severe patients (children and adults), these were 14/154 for URT specimen, 6/112 for LRT specimen, 0/38 for blood specimen and 10/110 for faecal specimen. For the overall patient population, the patients that did not clear within the sampling time accounted for 14% of the whole dataset.
We aggregated the data of selected studies to report a weighted mean of duration of positive SARS-CoV-2 viral detection in both URT, LRT, and faeces for mild and moderate-severe patients. We chose to display the results of the aggregated data for which the final day was set as Day -1 of first negative in the main text of this manuscript, as this is the most conservative approach (Figs. 2 and 3). Also, the result of this measure differed only marginally to the result defining the final day as last positive (maximum 1 Day). The pooled estimates for all data groupings, including recurrence, can be found in the supplementary Table S5.
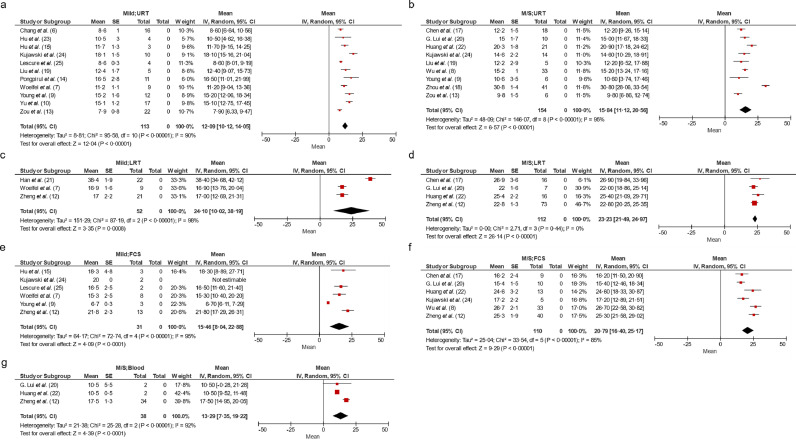
Fig. 2.
Forest plots of selected aggregations reporting weighted means of duration of viral detection as days after symptom onset in adults.
Pooled estimate for (a) all mild patients (non-treated and treated) and URT specimens, (b) all moderate-severe patients and URT specimen, (c) all mild patients and LRT specimen, (d) all moderate-severe and LRT specimen, (e) all mild patients and faecal specimen, (f) moderate-severe and faecal specimen and (g) moderate-severe patients and blood specimen. The forest plots include data that describe duration of viral detection defining the final day as day -1 of first negative day of two consecutive negatives. CI: Confidence Interval; D-1: Day -1 of first negative test of two consecutive; FCS: Faeces, LRT: Lower respiratory tract, SE: Standard error, URT: Upper respiratory tract. Numbers in brackets refer to listing in reference list.
Fig. 3.
Forest plots of selected aggregations reporting weighted means of duration of viral detection as days after symptom onset in children
Pooled estimate for (a) all mild patients (non-treated and treated) and URT specimens, (b) all mild patients and faecal specimen. The forest plots include data that describe duration of viral detection defining the final day as day -1 of first negative day of two consecutive negatives. CI: Confidence Interval; D-1: Day -1 of first negative test of two consecutive; FCS: Faeces, SE: Standard error, URT: Upper respiratory tract. Number in brackets refer to listing in reference list.
In total we included data from 650 patients in this analysis. Notably, a high heterogeneity (I2 ∼ 80–90%) was observed in the majority of the subgroup analysis indicating that these data should be interpreted cautiously and only considered as trends. Significant publication bias was found by Egger test only for the subgroup of adult, mild patients with URT specimen (p < 0•05).
In mild adult patients, the pooled estimate of the mean duration of positive SARS-CoV-2 viral detection after symptom onset in the URT was 12•1 days (CI: 10•12, 14•05) (Fig. 2a), 24•1 days (CI: 10•02, 38•19) in LRT (Fig. 2c), and 15•5 days (CI: 8•04, 22•88) in faeces (Fig. 2e). There was only one study describing duration of viral detection in the blood (17•1 days) in mild adult patients [12].
In moderate-severe adult patients, the pooled estimate of mean duration of positive SARS-CoV-2 viral detection after symptom onset in the URT was 15•8 days (CI: 11•12, 20•56) (Fig. 2b), 23•2 days (CI: 21•49, 24•97) in the LRT (Fig. 2d), 20•8 days (CI: 16•40, 25•17) in faeces (Fig. 2f), and 13•3 days (CI: 7•35, 19•22) in blood (Fig. 2g).
In children with mild symptoms, the pooled estimate of the mean duration of positive SARS-CoV-2 viral detection after symptom was 11•1 days (CI: 7•14, 15•11) in URT and 16•0 days (CI: 11•49, 20•47) in the faeces (Fig. 3a and b). No viral detection was reported in the blood for children.
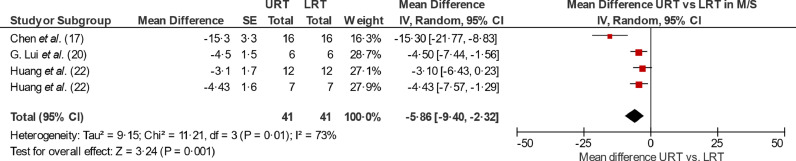
We then directly compared the duration of viral detection of URT and LRT specimen in each patient per study. Regarding mild adult patients, only one study sampled both URT and LRT specimen in the same patient and found that the mean difference of viral detection of SARS-CoV-2 in URT compared to LRT is −5•7 days (SD: 4 days) (7). Regarding moderate-severe adult patients, the pooled estimate of the mean difference of duration of positive SARS-CoV-2 viral detection in URT compared to LRT was −5•9 days (CI: −9•40, −2•32) (see Fig. 4). For children, a direct comparison was not performed due to lack of data.
Fig. 4.
Forest plots of selected aggregations reporting mean difference of duration of viral detection in URT compared to LRT in moderate severe patients.
Pooled estimate for all moderate-severe patients (non-treated and treated) comparing mean difference of duration of positive viral detection in URT relative to LRT per patient. The forest plots include data that describe duration of viral detection defining the final day as day -1 of first negative day of two consecutive negatives. CI: Confidence Interval; D-1: Day -1 of first negative test of two consecutive, SE: Standard error, URT: Upper respiratory tract. Number in brackets refer to listing in reference list. For Huang et al. two comparisons were separated fed in the analysis: nasal swab vs. sputum (upper) and throat swab vs. sputum (lower).
Generally, the mean duration of positive SARS-CoV-2 viral detection accounting for recurrence of positivity did not differ substantially in mild adults or children (if different, less than 1 Day difference). But, in moderate-severe patients the pooled estimate of the mean duration of positive SARS-CoV-2 viral detection was on average approximately 3•4 days longer for LRT specimen and 1•2 days for faeces, accounting for recurrence of viral positivity (see Table S5).
3.4. Spatial and temporal dynamics of viral RNA load
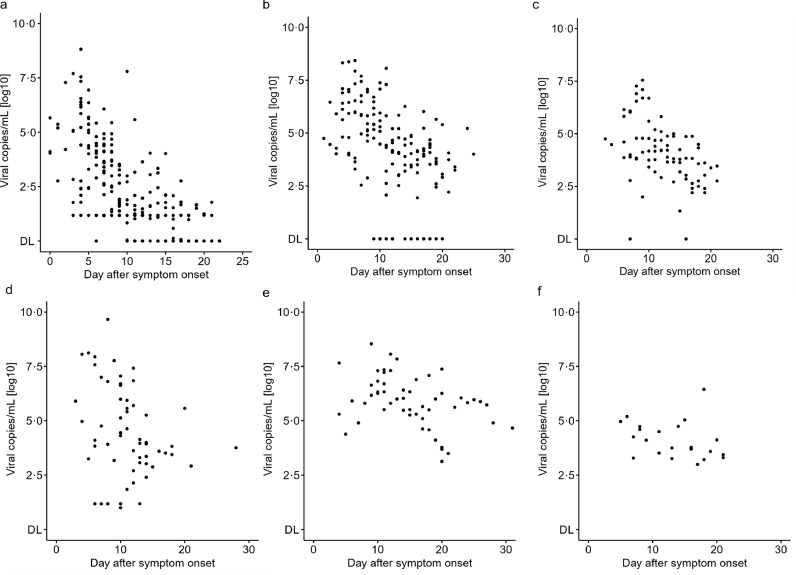
To investigate whether the viral RNA load differs between disease severity and sampling location, we combined data from four published studies and analysed the viral RNA load per patient as a function of time, stratified after clinical severity (mild and moderate-severe) and location of sampling (URT, LRT and faeces) (Fig. 5). No quantitative viral data was available for children and only one study reported limited quantitative data of blood specimen (four datapoints) [20].
Fig. 5.
Spatial and temporal viral RNA load in mild and moderate-severe adult COVID-19 patients.
Viral RNA load (viral copies/mL) as a function of time (days after symptom onset) in (a) upper respiratory tract (URT) specimen in mild patients, (b) lower respiratory tract (LRT) specimen in mild patients, (c) faecal specimen in mild patients, (d) URT specimen in moderate-severe patients, (e) LRT specimen in moderate-severe patients, (f) faecal specimen in moderate-severe patients. Four Studies were included [7,13,20,47]. DL: limit of detection. Detailed viral load data are displayed in Table S2.
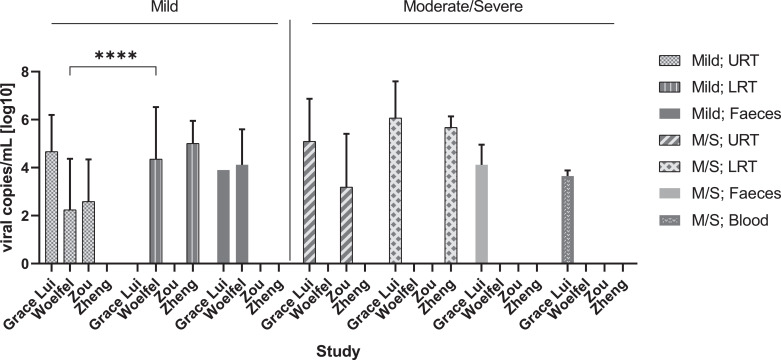
In mild patients, viral RNA load was highest during the first week in the URT having a maximum viral RNA load of ∼ 6•61 × 108 viral copies/mL on Day 4, whereas the maximum viral RNA load reported for LRT was ∼ 2•69 × 108 copies/mL on Day 6 (Table 4 and Fig. 5a vs. b). The maximum viral RNA load in faeces was reported as ∼ 3•55 × 107 copies/mL on Day 9 in mild patients (Fig. 5c). The average viral RNA load was found to be significantly higher in LRT relative to URT in the Woelfel et al. study (t test, q < 0•001) (Fig. 6) [7]. However, as this was the only study which allowed this comparison, additional studies of quantitative viral dynamics are needed to assess the viral RNA load of the URT compared to LRT.
Table 4.
Summary of maximum viral RNA load per location and COVID19 clinical severity
| Author | URT | LRT | Faeces | Blood |
|---|---|---|---|---|
| Maximum viral copies/mL [log 10] on Day after symptom onset | ||||
| Mild | ||||
| Woelfel et al. [7]a | ∼ 6•61 × 108 on Day 4 (6•66 × 108 in publ.) | ∼ 2•69 × 108 on Day 6 (7•11 × 108 copies/swab in publ.) | ∼ 3•55 × 107 on Day 9 | ND |
| Zou [13]# | ∼ 2•19 × 107 on Day 4 | ND | ND | ND |
| G. Lui et al. [20],* | 2•50 × 106 on Day 4 | ND | 7•94 × 103 on Day 7 | ND |
| Zheng et al.[12],a | ND | ∼ 2•00 × 107 on Day 11 | ND | ND |
| Moderate-Severe | ||||
| Woelfel et al [7]a | ND | ND | ND | ND |
| Zou [13]a | ∼ 1•32 × 108 on Day 5 | ND | ND | ND |
| G. Lui et al. [20],* | 4•60 × 109 on Day 8 | 3•45 × 108 on Day 11 | 2•76 × 106 on Day 18 | 1 × 104 on Day 3 |
| Zheng et al. [12]a | ND | ∼1•82 × 106 on Day 4 | ND | ND |
LRT: Lower respiratory tract, ND: Not determined, URT: Upper respiratory tract;
all subjects in this study received Lopinavir/Ritonavir; LRT: Lower respiratory tract, ND: Not determined, publ.: Publication; URT: Upper respiratory tract;
estimated data as digitalized from graph. Number in brackets refer to listing in reference list.
Fig. 6.
Average viral RNA load per location and clinical severity
Bar graph displaying mean with SD. **** q < 0•001, unpaired t-test using false discovery approach of Benjamini, Krieger and Yekutieli; M/S: Moderate-severe, URT: upper respiratory tract, LRT: lower respiratory tract. Four studies were included [7,12,13,20].
In moderate-severe patients, the maximum viral RNA load was reported as 4•60 × 109 copies/mL on Day 8 in URT and 3•45 × 108 copies/mL on Day 11 in LRT (Table 4, Fig. 5d vs. e). Maximum viral RNA load in faeces was shown to be 2•76 × 106 copies/mL on Day 18 and 1 × 104 copies/mL on Day 3 in blood specimen (Table 4). No significant difference was found in the average viral RNA load comparing URT and LRT samples in this disease population.
4. Discussion
In this study we conduct a systematic review of the spatial and temporal viral dynamics of SARS-CoV-2 stratified after age and clinical severity. We report the per study data of duration of viral detection and viral RNA load over time, but also show pooled estimates of these, providing a better basis for interpretation. While viral trophism likely extends beyond the specimens described in this study, the lack of data on viral RNA load from other locations has limited our analysis to faeces, blood, URT and LRT [32].
Additionally, this study has several limitations which need to be considered. First, different genes and thresholds were used to assess negative conversion of SARS-CoV-2 hampering the direct comparison of studies. Second, the classification of clinical symptoms into the severity categories, mild, moderate, severe was based on different guidelines across the studies and we were not able to unify and verify all (mostly due to missing individual symptom data). Third, studies that only reported fractions or median of duration of viral shedding were excluded, which might introduce a selection bias into this review. Fourth, the viral RNA load was measured via RT-PCR, which cannot differentiate dead virus particle, and hence data presented here might not necessarily reflect active viral replication, which is likely of shorter duration. However, this technique is currently used worldwide to measure the quantities of SARS-CoV-2. Fifth, a portion of the patients included in this review (14 %) did not clear the virus in the time frame of sampling and were not followed until clearance of infection, hence the here presented data might be an underestimation of the duration of virus detection which might introduce a reporting bias in the outcome measure. Sixth, only a selection of studies investigated the duration of SARS-CoV-2’s positivity in more than one location, e.g. LRT sampling was more often performed in moderate-severe patients than mild patients, which might further introduce a bias in the reported outcome measure. Also, it further limits the direct comparison of duration of viral detection in LRT relative to URT samples on a per patient level to only specific disease severities and age groups. Finally, our aggregation analysis exhibited a high heterogeneity (I2 ∼ 80–90%) and included too few studies to make statements about statistical significance, which highlights that these data should be interpreted cautiously and only considered as trends. There are several variables that could explain the high heterogeneity of data: first, there was no uniform cycle threshold (Ct) value of one distinct gene used to define negativity of SARS-CoV-2 across the studies; second, different RT-PCR/RNA extraction kits were used (if reported) for sample analysis; thirdly, samples were combined to URT and LRT specimen which might mean that the analysis misses granularity regarding specific subgroups of sampling locations. However, given the lack of big data sets, these three points are still not uniformly defined across currently ongoing studies highlighting the complexity of having a universal defined viral endpoint.
Regardless of these limitations, some trends can be extracted from this analysis. Firstly, we consistently find that SARS-CoV-2 is detected in LRT, URT and faecal specimens, irrespective of clinical severity of disease. Second, our data indicates that the duration of detection of SARS-CoV-2 is longer in LRT than the URT in adult patients. Third, there seems to be little difference in the duration of detection of SARS-CoV-2 in mild patients as opposed to moderate-severe patients in the LRT, but an indication of longer duration of viral detection in faeces and the URT for moderate-severe patients was shown. Fourth, viral RNA load in URT peaks within the first week of infection, whereas viral RNA load in LRT and faeces within the second week of infection.
In conclusion, we believe that this systematic review and meta-analysis can support refinement of mathematical modelling as well as aid in the definition of appropriate endpoints for clinical trials testing therapeutic intervention aimed at reducing viral RNA load of SARS-CoV-2. Furthermore, it highlights the importance of testing for viral presence in the LRT, which may clear more slowly compared to the URT.
5. Research in context
5.1. Evidence before this study
The temporal and spatial dynamics of SARS-Cov-2 have been mainly summarized in case series and retrospective studies. We systematically searched PubMed/MEDLINE and Cochrane review database for studies published between 1. November 2019 and 23rd of April 2020 using SARS-CoV-2 [MESH]” OR “COVID 19 [MESH]” alone, or in combination with “virology” OR “viral” OR “Epidemio*” AND “clinical as search terms. We included studies that reported individual viral data over time measuring negative conversion by two consecutive negative tests, individual clinical severity, and age. We excluded studies that reported viral data as patient fraction, reported only baseline data, included solely asymptomatic patients or were interventional studies. Extracted data included author, title, design, sample size, thresholds and genes of RT-PCR, patient age, COVID-19 severity, clinical characteristics, treatment, location of viral sampling, duration of viral detection, and viral load. We pooled the data of selected studies to determine effect estimates of duration of viral detection and conducted subgroup analyses per disease severity and age. Viral load was visualized over time.
5.2. Added value of this study
Our analysis demonstrated consistent viral detection from specimen from the URT, the LRT, and faeces in patients with mild and moderate-severe COVID-19. We showed that SARS-CoV-2 persists for a longer duration in the LRT compared to the URT in moderate-severe and mild adult patients. However, we found that the duration of viral detection between mild and moderate-severe adult patients is similar in the LRT. But, an indication of longer duration of viral detection in faeces and the URT for moderate-severe patients was shown. Further, viral load was demonstrated to peak in the URT within first week of infection, whereas maximum viral load has been observed to occur later and within the second week of infection in the LRT.
5.3. Implications of all the available evidence
This systematic review and meta-analysis can support the improvement of mathematical modelling of SARS-CoV-2’s infection as well as aid in the definition of appropriate endpoints for clinical trials which are testing therapeutic intervention aimed at reducing viral load of SARS-CoV-2.
Funding sources
The project has received funding support from Innovation Fund Denmark. The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Author contribution
A.W. and M.O.A.S wrote the manuscript and conceived the study. M. J. and M.O.A.S. provided input to the analysis and interpretation of study. A.W. analysed the data. All authors reviewed the manuscript.
Declaration of Competing Interest
A.W., M. J., and M.S. have nothing to disclose.
Acknowledgment
We would like to thank Dr. Philippe Andres for critical revision of the manuscript and Dr. Rasmus Toft-Kehler and Morten Boesen for their support to secure funding for this study.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102916.
Appendix. Supplementary materials
References
- 1.Bar-On YM, Flamholz A, Phillips R, Milo R. Sars-cov-2 (Covid-19) by the numbers. Elife. 2020;9:e57309. doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moher D, Shamseer L, Clarke M, Ghersi D, Liberati A, Petticrew M. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev. 2015;4.1(1):1. doi: 10.1186/2046-4053-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu J, Liu J, Zhao X, Liu C, Wang W, Wang D. Clinical characteristics of imported cases of COVID-19 in Jiangsu Province: a multicenter descriptive study. Clin Infect Dis. 2020;10 doi: 10.1093/cid/ciaa199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benjamini Y, Krieger AM, Yekutieli D. Adaptive linear step-up procedures that control the false discovery rate. Biometrika. 2006;93(3):491–507. [Google Scholar]
- 5.Jiehao C, Jing X, Daojiong L, Lei X, Zhenghai Q, Yuehua Z. A Case Series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang D, Mo G, Yuan X, Tao Y, Peng X, Wang F. Time kinetics of viral clearance and resolution of symptoms in novel coronavirus infection. Am J Respir Crit Care Med. 2020:1–12. doi: 10.1164/rccm.202003-0524LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woelfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Mueller MA. Virological assessment of hospitalized cases of coronavirus disease 2019. Nature. 2020 doi: 10.1038/s41586-020-2196-x. 2020.03.05.20030502. [DOI] [PubMed] [Google Scholar]
- 8.Wu Y, Guo C, Tang L, Hong Z, Zhou J, Dong X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5(5):434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young BE, Ong SWX, Kalimuddin S, Low JG, Tan SY, Loh J. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA – J Am Med Assoc. 2020;323(15):1488–1494. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu Jun IS, Anderson DE, Zheng Kang AE, Wang L-F, Rao P, Young BE. Assessing viral shedding and infectivity of tears in coronavirus disease 2019 (COVID-19) Patients. Ophthalmology. 2020;2019:2019–2021. doi: 10.1016/j.ophtha.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang T, Cui X, Zhao X, Wang J, Zheng J, Zheng G. Detectable SARS-CoV-2 Viral RNA in feces of three children during recovery period of COVID-19 pneumonia. J Med Virol. 2020 doi: 10.1002/jmv.25795. (March) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng S, Fan J, Yu F, Feng B, Lou B, Zou Q. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369(March):m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1175–1177. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pongpirul WA, Mott JA, Woodring JV., Uyeki TM, MacArthur JR, Vachiraphan A. Clinical characteristics of patients hospitalized with coronavirus disease, Thailand. Emerg Infect Dis. 2020;26(7) doi: 10.3201/eid2607.200598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Y, Shen L, Yao Y, Xu Z, Zhou J, Zhou H. A report of three COVID-19 cases with prolonged viral RNA detection in anal swabs. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y, Li X, Zhu B, Liang H, Fang C, Gong Y. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020;26(April) doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C, Gao G, Xu Y, Pu L, Wang Q, Wang L. SARS-CoV-2–Positive Sputum and Feces After Conversion of Pharyngeal Samples in Patients With COVID-19. Ann Intern Med. 2020;172(1) doi: 10.7326/M20-0991. ITC1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou The duration of viral shedding of discharged patients with severe COVID-19. J Gerontol Ser A Biol Sci Med Sci. 2020:1–11. doi: 10.1093/cid/ciaa451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu F, Xu A, Zhang Y, Xuan W, Yan T, Pan K. Patients of COVID-19 may benefit from sustained lopinavir-combined regimen and the increase of eosinophil may predict the outcome of COVID-19 progression. Int J Infect Dis. 2020 doi: 10.1016/j.ijid.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lui G. Viral dynamics of SARS-CoV-2 across a spectrum of disease severity in COVID-19. J Infect. 2020;1(1):1–11. doi: 10.1016/j.jinf.2020.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han C, Duan C, Zhang S, Spiegel B, Shi H. Digestive symptoms in COVID-19 patients with mild disease severity: clinical presentation, stool viral RNA testing, and outcomes. Am J Gastroenterol. 2020:1–8. doi: 10.14309/ajg.0000000000000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Y, Chen S, Yang Z, Guan W, Liu D, Lin Z. SARS-CoV-2 viral load in clinical samples of critically Ill patients. Am J Respir Crit Care Med. 2020;(2017):1–15. doi: 10.1164/rccm.202003-0572LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Z, Song C, Xu C, Jin G, Chen Y, Xu X. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci China Life Sci. 2020;63(5):706–711. doi: 10.1007/s11427-020-1661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kujawski SA, Wong KK, Collins JP, Epstein L, Killerby ME, Midgley CM. First 12 patients with coronavirus disease 2019 (COVID-19) in the United States. medRxiv. 2020 doi: 10.1038/s41591-020-0877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lescure F-X, Bouadma L, Nguyen D, Parisey M, Wicky P-H, Behillil S. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis. 2020;2(20):1–10. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.pin Tan Y, yu Tan B, Pan J, Wu J, Zeng S zhen, Wei H yan. Epidemiologic and clinical characteristics of 10 children with coronavirus disease 2019 in Changsha, China. J Clin Virol. 2020 doi: 10.1016/j.jcv.2020.104353. 104353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Br Med J. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viechtbauer W. Conducting meta-analyses in R with the metafor. J Stat Softw. 2010 [Google Scholar]
- 29.Colavita F, Lapa D, Carletti F, Lalle E, Bordi L, Marsella P. SARS-CoV-2 isolation from ocular secretions of a patient with COVID-19 in Italy with prolonged viral RNA detection. Ann Intern Med. 2020 doi: 10.7326/M20-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim J, Jeon S, Shin HY, Kim MJ, Seong YM, Lee WJ, Choe KW, Kang YM, Lee B PS. Case of the index patient who caused tertiary transmission of coronavirus disease 2019 in Korea: the application of lopinavir/ritonavir for the treatment of COVID-19 pneumonia monitored by quantitative RT-PCRNo title. J Korean Med Sci. 2020;35(6):e79. doi: 10.3346/jkms.2020.35.e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen D, Xu W, Lei Z, Huang Z, Liu J, Gao Z. Recurrence of positive SARS-CoV-2 RNA in COVID-19: a case report. Int J Infect Dis. 2020 doi: 10.1016/j.ijid.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Victor G, Puelles M.D.PD, Marc Lütgehetmann MD, Maja T, Lindenmeyer PD, Jan P, Sperhake MD, Milagros N, Wong MD, Lena Allweiss PD. Multiorgan and renal tropism of SARS-CoV-2. NEJM. 2020 doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caly L, Druce J, Roberts J, Bond K, Tran T, Kostecki R. Isolation and rapid sharing of the 2019 novel coronavirus (SAR-CoV-2) from the first patient diagnosed with COVID-19 in Australia. Med J Aust. 2020 doi: 10.5694/mja2.50569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng SC, Chang YC, Fan Chiang YL, Chien YC, Cheng M, Yang CH. First case of Coronavirus Disease 2019 (COVID-19) pneumonia in Taiwan. J Formos Med Assoc. 2020 doi: 10.1016/j.jfma.2020.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee NY, Li CW, Tsai HP, Chen PL, Syue LS, Li MC. A case of COVID-19 and pneumonia returning from Macau in Taiwan: Clinical course and anti-SARS-CoV-2 IgG dynamic. J Microbiol Immunol Infect. 2020 doi: 10.1016/j.jmii.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marchand-Senécal X, Kozak R, Mubareka S, Salt N, Gubbay JB, Eshaghi A. Diagnosis and management of first case of COVID-19 in Canada: lessons applied from SARS. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott SE, Zabel K, Collins J, Hobbs KC, Kretschmer MJ, Lach M. First Mildly Ill, non-hospitalized case of coronavirus disease 2019 (COVID-19) without viral transmission in the United States – Maricopa County, Arizona, 2020. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Tan L, NM Ngoc, That BTT, Uyen LTT, Hong NTT, Dung NTP. Duration of viral detection in throat and rectum of a patient with COVID-19. medRxiv. 2020 [Google Scholar]
- 41.Thevarajan I, Nguyen THO, Koutsakos M, Druce J, Caly L, van de Sandt CE. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat Med. 2020;26(4):453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y, Chen L, Deng Q, Zhang G, Wu K, Ni L. The Presence of SARS-CoV-2 RNA in Feces of COVID-19 Patients. J Med Virol. 2020 doi: 10.1002/jmv.25825. (March) [DOI] [PubMed] [Google Scholar]
- 43.Lo IL, Lio CF, Cheong HH, Lei CI, Cheong TH, Zhong X. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int J Biol Sci. 2020;16(10):1698–1707. doi: 10.7150/ijbs.45357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xing Y, Ni W, Wu Q, Li W, Li G, Wang W. Dynamics of faecal SARS-CoV-2 in infected children during the convalescent phase. J Infect. 2020 doi: 10.1016/j.jinf.2020.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xing Q, Li G, Xing Y, Chen T, Li W, Ni W, et al. Precautions are needed for COVID-19 patients with coinfection of common respiratory pathogens. medRxiv. 2020.
- 46.Xu T, Chen C, Zhu Z, Cui M, Chen C, Dai H. Clinical features and dynamics of viral load in imported and non-imported patients with COVID-19. Int J Infect Dis. 2020;94(300):68–71. doi: 10.1016/j.ijid.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng S, Fan J, Yu F, Feng B, Lou B, Zou Q. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020:369. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.