Abstract
Infectious diseases, biodiversity loss and livestock expansion are increasing globally, and examining patterns that link them is important for both public health and conservation. This study is a first attempt to analysis globally these patterns using General additive modelling and Structural equation modelling. A positive association between the number of infectious and parasitic diseases recorded in humans and the total number of animal species between nations was observed. A similar positive association between the number of outbreaks of human infectious diseases, corrected for the number of surveys, and the number of threatened animal species, corrected for the number of animal species, suggests that outbreaks of human infectious diseases are linked with threatened biodiversity. Results of the analyses over the longest period of the dataset (2000–2019) showed a positive correlation between the increasing number of cattle and the number of threatened species, a positive correlation between the increasing number of cattle and the number of outbreaks of human diseases, and a lack of correlation between the number of outbreaks and the number of threatened animal species. As a result, the growing importance of livestock on the planet, while threatening biodiversity, increasingly puts human and animal health at risk. This study calls for further analyses on the consequences of livestock expansion, which depends on several factors that vary by country, namely the growth of human population, changes in diet linked to the westernization of habits, agricultural industrialization and the integration into the world trade, but also the cultural values of livestock.
Keywords: Biodiversity, Infectious diseases, Outbreaks, Livestock, Public health
1. Introduction
Bat species are suspected reservoir species of the novel coronavirus (SARS-CoV-2) agent of the outbreak of coronavirus disease 2019 (COVID-19). Some studies have identified pangolin species as natural reservoirs of SARS-CoV-2-like CoVs (Zhang et al., 2020), without forgetting the potential role of the cat civet associated with the emergence of the SARS-CoV in 2002–2003, or the camel for the emergence of the MERS-CoV in 2012. Whatever the association of the bat coronavirus with wildlife reservoirs and intermediate hosts (bats, pangolins, civets), the COVID-19 pandemic calls into question the role of biodiversity in emerging zoonotic diseases. This is particularly relevant in the context of the current biodiversity crisis, underlined by the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services (IPBES). Many scientific articles and books have been devoted to studying or examining the link between biodiversity and infectious diseases (Young et al., 2017) or more broadly between biodiversity and health (Chivian, 2003; Morand and Lajaunie, 2017).
Several scientific controversies, namely the “dilution effect” hypothesis (Ostfeld and Keesing, 2000) also named the “negative diversity–disease” relationship (Magnusson et al., 2020), may have somewhat obscured rather than clarified the contribution of ecology to both the public health and animal health sectors. The “dilution effect” was initially introduced by.
Hamilton (1971) for predator–prey relationships when an increase in the number of individuals in a prey group leads to a decrease in an individual's probability to be attacked by a predator Ostfeld and Keesing (2000) redefined the “dilution effect” for host–parasite relationships when high host diversity diluted the impact of the main reservoir of Lyme disease. The “dilution effect”, or the “negative diversity–disease”, postulates that biodiversity losses may promote disease transmission (Keesing et al., 2006). Most of the hard discussion and sometimes dispute among scholars were theoretical, technical and methodological arguments. Few case studies, although important, have been used in these debates, with the exception of Lyme's disease or West Nile disease (see Johnson et al., 2015). Meta-analyses performed to test the dilution effect hypothesis have yielded conflicting results. A first meta-analysis of 13 studies showed that biodiversity had a weak influence on disease prevalence suggesting that disease transmission depends on local idiosyncratic factors (Salkeld et al., 2013). The review of Johnson et al. (2015) considered 90 studies and most of them supported a dilution effect for various diseases affecting humans, wildlife, livestock or plants. A third study of 61 parasite species found consistent support for dilution effects in various host communities (Civitello et al., 2015). Finally, a recent meta-analysis took into account the scale effect on the negative diversity–disease relationship (i.e. the dilution effect) and showed a significant negative diversity–disease relationships across spatial scales from global to intermediate landscapes to small sites (Magnusson et al., 2020). Therefore, as pointed out by Johnson et al. (2015), two paradoxical perspectives have emerged. The first one, the ‘diversity begets diversity’ hypothesis postulates that any increase in host diversity is positively correlated with overall parasite diversity and then the risk of disease transmission (Hechinger and Lafferty, 2005; Dunn et al., 2010). The second one, the ‘negative diversity–disease’ hypothesis, postulates that biodiversity losses promote the transmission of pathogens (Keesing et al., 2006).
The potential links between infectious diseases and biodiversity have sometimes been poorly presented to be understandable by the public health and animal health sectors, but see the recent effort to clarify these links by Rohr et al. (2020). A first pitfall lies in what is measured. Is it a static measure of the presence of an agent (or the diversity of agents), or the disease it causes (or they cause), or the prevalence in a given reservoir or vector? Or is it a dynamic measure like an epidemic, which reveals a chain of transmission of a given noticed disease? A second pitfall is the ecological scale as Magnusson et al. (2020) recently pointed out. Is it a local study that analysed the disease transmission in a specific socio-ecosystem? Or is it a study that is more concerned on the global dynamics of infectious diseases between nations? The comparison of local studies has led to a third pitfall because studies on the ecology of disease transmission are too few in number but diverse in host-pathogen systems investigated, which may affect the results of meta-analyses (Bordes et al., 2015). All of these pitfalls raised above call into question the translation of scientific results in the sectoral fields of conservation, public health and animal health at a time when the integration of approaches is advocated, i.e. One Health (FAO-OIE-WHO, 2010; Destoumieux-Garzón et al., 2018) or Planetary Health (Whitmee et al., 2015).
Rather than giving another review, the approach taken by this study was to comprehensively analyse infectious diseases affecting humans and livestock using available open data. In order to contribute to the renewal of studies on biodiversity loss and infectious diseases, the main objective was to describe the global patterns of biodiversity, biodiversity loss and infectious diseases across countries and over the years that can help draw verifiable hypotheses and develop new collaborations between the biodiversity conservation sector and the public and animal health sectors.
A first piece of evidence is that the number of emerging infectious diseases has increased over the last decades as already warned several years ago by Wilcox and Gubler (2005), Wolfe et al. (2007), Jones et al. (2008) among many others. Concurrently, the number of reported infectious disease outbreaks has also dramatically increased during the last few decades (Morand et al., 2013; Smith et al., 2014; Poisot et al., 2015). The likely causes were often attributed to the growing human-induced environmental changes (Morand and Waret-Szkuta, 2012) and biodiversity loss (Keesing et al., 2010a, Keesing et al., 2010b).
A second piece of evidence was provided by Dunn et al. (2010) who analysed the burden of human diseases by nation. Their results showed that the diversity of human pathogens is positively associated with the species richness of birds and mammals between nations, confirming that the density of human pathogens increases towards the equator (Guernier et al., 2004) in the same as mammal species richness (Schipper et al., 2008).
A third piece of evidence relates to the importance of livestock and pets for the sharing infectious diseases with humans and wildlife (Lloyd-Smith et al., 2009). Archaeological studies suggest the occurrence of a large-scale domestication of animals starting around 12,000 years ago. Environmental historian William McNeill hypothesized long ago about the role played by animal domestication in increasing the diversity of human pathogens, who was the first to suggest a positive relationship between sharing pathogens between domestic animals and humans throughout their history of domestication (McNeill, 1976). The hypothesis has been quantitatively examined on a small number of domestic mammals by the study of Morand et al. (2014a), which confirmed McNeill's original hypothesis by showing a significant positive relationship between the time elapsed since domestication and the number of infectious and parasitic diseases shared between domesticated mammals and humans. Recently, Wells et al. (2020) assessed the sharing patterns of viruses both DNA and RNA, among a wide variety of mammals, including humans, wildlife and domestic species. Using network analysis, they have shown that domestic mammals occupy the most central positions in networks of known mammalian–virus associations. Cattle, pigs, horses and sheep occupy the highest centrality position in terms of sharing DNA viruses. Moreover, domestic animals act as epidemiological bridges between wildlife and humans favouring amplification of pathogens originated from wildlife. These results suggest that these abundant and economically important mammalian livestock species (Thornton, 2010) strongly contribute to the sharing of viruses between wildlife and humans (see also Johnson et al., 2020).
The question on how the loss of biodiversity may favour the emergence and the spread (outbreaks) of infectious diseases, when a good correlation between biodiversity and human pathogen diversity is observed, should be re-examined in the light of evidence listed above. For this, it is first necessary to consider separately the presence of a disease in a nation, that is to say its endemicity, and the patterns of its outbreaks. Second, it must take into account the growing importance of livestock on the planet and their likely role as reservoirs and intermediate hosts of many infectious diseases. Those steps must account for spatial autocorrelation given the results gained by the study of Dunn et al. (2010). Third, the potential links between threatened biodiversity, outbreaks of infectious diseases and livestock expansion should be investigated over time.
This study examines the global patterns between (1) biodiversity, using IUCN global data (total species, species at threat by countries and over years), (2) outbreaks of human infectious diseases, using the GIDEON global database (which had already been used in previous studies, see Dunn et al., 2010; Smith et al., 2014; Morand and Walther, 2018), (3) outbreaks of animal infectious diseases, using the WAHIS global database of OIE (Organization of Animal Health), and (4) livestock abundance by country and by year, using the FAOSTAT global database from FAO (United Nations Food and Agriculture Organization).
2. Material and methods
Data on human infectious diseases were obtained from GIDEON (www.gideononline.com), which contains information on the presence of endemic diseases and the occurrence of epidemics of human infectious diseases in each nation. The 1960–2019 extracted dataset contains 16,994 infectious disease outbreak of 252 human infectious diseases as well as the number of surveys that has been conducted in a country as measure of investigation effort (see Morand et al., 2013).
Data on animal infectious diseases were obtained from the WAHIS database (https://www.oie.int/en/animal-health-in-the-world/wahis-portal-animal-health-data/) curated by the World Organization for Animal Health Organization (OIE). The WAHIS database lists 180 diseases of livestock, poultry, and all farming animals such as crustaceans, amphibian, fishes and bees (https://www.oie.int/en/animal-health-in-the-world/oie-listed-diseases-2020/). The entire 2006–2019 were extracted using the package package ‘httr’ (Wickham 2019a and ‘rvest’ (Wickham, 2019b).
Data on livestock from 1961 to 2016 were obtained from the FAOSTAT database (http://www.fao.org/faostat/en/) of the Food and Agricultural Organization (FAO) of the United Nations, focusing on one cattle.
Data on species richness and species at threat were obtained from the International Union for Conservation of Nature (IUCN) and the IUCN Red List version 2020 (www.iucnredlist.org). Extracted data were (1) changes in numbers of species in the threatened Red List categories from 1996 to 2020 for the major taxonomic groups on the Red List, and summary totals for animal species in each nation: (2) total least concern and data deficient species; (3) total critically endangered species, endangered and vulnerable species; (4) total endangered mammal and bird species; (5) total assessed species.
2.1. Statistical analyses
Because of the time limitation of the datasets, analyses were limited to the period from 1960 to 2019 for human disease outbreaks (GIDEON), to the period from 2006 to 2019 for animal disease outbreaks (WAHIS-FAO), to the period from 1960 to 2016 for livestock (FAOSTAT), to the period from 2000 to 2016 for animal species at threat (IUCN).
All data and their representations were performed in R (R Development Core Team, 2019). Smooth regression was use to visualize the patterns of changes over time (Harrell, 2015). Correlation was analysed using the packages ‘Hmisc’ (Harrell, 2019) and ‘correlation’ (Makowski and Lüdecke, 2020) in R (R Development Core Team, 2019).
Maps were drawn using the packages ‘raster’ (Hijmans, 2020) and ‘rworldmap’ (South, 2011).
Moran's I test was performed to test the significance of spatial autocorrelation using ‘spatialEco’ (Evans, 2015). The level of spatial autocorrelation was investigated for all investigated variables using the centroid of each country.
2.2. Spatial analyses
General additive modelling (GAM), an extension of the generalized linear models, was used to investigate the relationships between human infectious diseases and their outbreaks with wildlife diversity and endangered wildlife taking into account the spatial autocorrelation. The model assumes that the response variable is dependent on the univariate smooth-terms of independent variables (Hastie and Tibshirani, 1990). All models were fitted using the ‘MGCV’ package (Wood, 2017). We used the function gam.check to choose the basis dimension for each predictor according to estimated degrees of freedom value in the main effect. Outputs of GAM models were obtained using the packages ‘gratia’ (Simpson, 2019) and ‘mgcViz’ (Fasiolo et al., 2018).
A first GAM was developed to investigate the number of infectious and parasitic diseases in humans (in log) as a function of the total number of wildlife (in log) and survey effort taking into account their spatial distribution (using the country centroids) and using a standard Gaussian link function. This first model was:
A second GAM was developed to investigate the association between (a) the ratio of the total number of outbreaks of infectious diseases (in log) to the number of infectious and parasitic diseases in humans (in log) and (b) the ratio of the number of threatened wildlife specie (in log) to the number of wildlife species, and the number of cattle taking into account their spatial distribution (using the country centroids) and using a standard Gaussian link function. The ratios were used a way to control the number of diseases outbreaks or the number of threatened species for the number of reported diseases or wildlife species in a country. The second model was:
2.3. Temporal analysis
Structural Equation Modelling (SEM) was used to investigate the temporal relationships between outbreaks of human infectious diseases, wildlife species at threat, wildlife species assessed, and cattle number. SEM combines measurement models (e.g., reliability) with structural models (e.g., regression). SEM was performed using the package ‘piecewiseSEM’ (Lefcheck, 2016). We used the number of species assessed to control the potential bias of the increase of number threatened species. The following structural equation model was tested for the longest period of the dataset (2000–2016):
3. Results
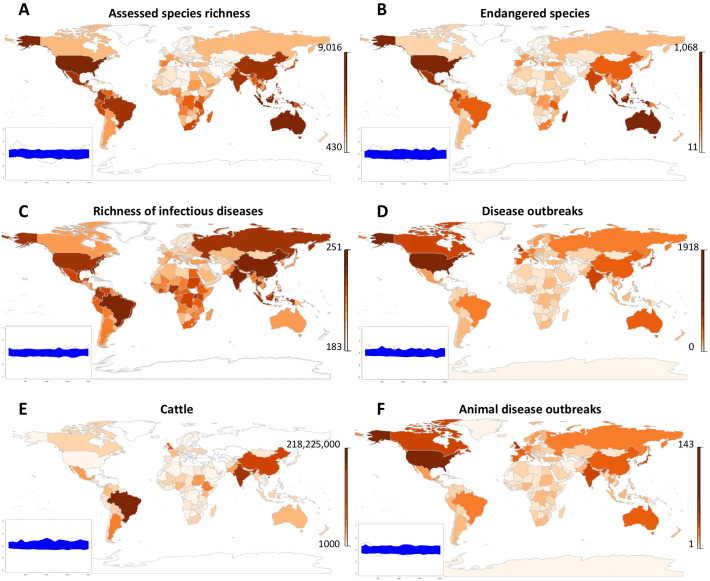
Significant spatial autocorrelation was detected for total number of wildlife species and wildlife species at threat (from 2000 to 5000 kms from each country centroid), total number outbreaks of animal diseases (from 1000 to 3000 kms) (Fig. 1 ). Relatively limited or no significant spatial autocorrelation were detected for human diseases richness and for cattle number (Fig. 1).
Fig. 1.
Maps and spatial autocorrelation (small insert for each map) of (A) the number of assessed species, (B) the number of endangered species; (C) the richness of infectious diseases reported; (D) the number of outbreaks of human infectious diseases; (E) the number of cattle; (F) the number of outbreaks of animal infectious diseases (data from GIDEON, ICUN and FAOSTAT).
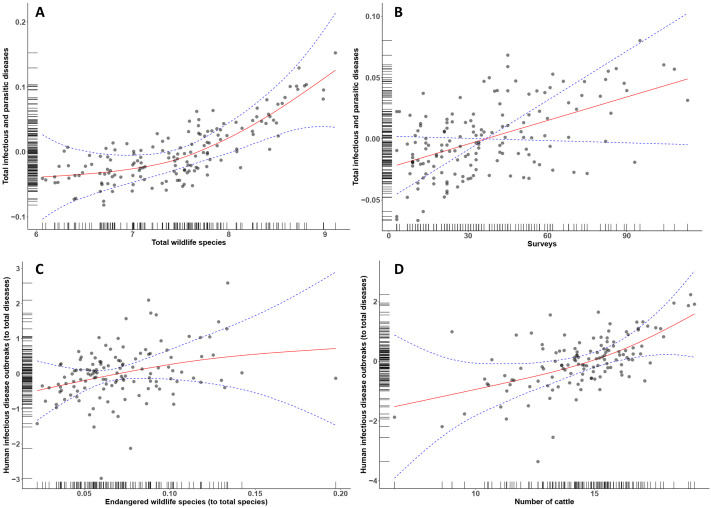
The first GAM investigating the number of infectious and parasitic diseases in humans explained 84.6% of the deviance (R2 (adj) = 0.82) (Table 1 ) with a significant positive influence of the number of wildlife species (P < 0.00001, Table 1, Fig. 2A), taking into account the effect of surveys (P < 0.00001, Table 1, Fig. 2B) and the spatial autocorrelation (P < 0.00001, Table 1).
Table 1.
Results of General additive models (GAM) to explain: (1) the number of infectious and parasitic diseases in humans with the number of wildlife species, the number of surveys of infectious and parasitic diseases in humans, and the matrix of longitude/latitude of country centroids as independent variables; and (2) the ratio of the number of outbreaks of human infectious diseases (to the number of infectious parasitic diseases in humans) with the ratio of the number of wildlife species at threat (to the number of wildlife species), the number of cattle, and the matrix of longitude/latitude of country centroids as independent variables (edf = estimated degrees of freedom for the model terms, df = estimated residual degrees of freedom) (see Methods for the model, data from GIDEON, IUCN and FAOSTAT).
| Dependant variable | Explanatory variables | edf | F (df) | P-value | Deviance (R2 adjusted) |
|---|---|---|---|---|---|
| 1. Infectious and parasitic diseases | (Longitude, latitude) | 20.78 | 7.69 (29) | <0.00001 | |
| Wildlife species | 3.123 | 8.69(9) | <0.00001 | ||
| Surveys | 0.95 | 9 2.32 (9) | <0.00001 | 84.6% (0.82) | |
| 2. Outbreak of human infectious diseases | (Lon, lat) | 11.88 | 2.14 (29) | <0.00001 | |
| Wildlife species at threat | 1.31 | 1.19 (9) | 0.0005 | ||
| Cattle number | 2.31 | 6.56 (9) | <0.00001 | 61.9% (0.57) |
Fig. 2.
Results of General additive modelling (GAM). The first model explained the number of infectious and parasitic infectious diseases in humans with the number of wildlife species (A) and the number of surveys investigating human infectious diseases (B) by country (see Fig. 1) taking into account the spatial dependancy (using the country centroids) (see Table 1, model 1). The second model explained the ratio of the number of outbreaks of infectious in humans, to the number of infectious and parasitic infectious diseases, with the ratio of the number of wildlife species (C), to the number of wildlife species, and the number of cattle (D) by country (see Fig. 1) taking into account the spatial dependancy (using the country centroids) (see Table 1, model 2) (data from GIDEON, ICUN and FAOSTAT).
The second GAM model investigating the number of outbreaks of infectious diseases in humans explained 61.9% of the deviance (R2 (adj) = 0.57) (Table 1) with a significant positive influence of the number of threatened wildlife species (P = 0.0005, Table 1, Fig. 2C) and the number of cattle (P < 0.00001, Table 1, Fig. 2D), taking into account the spatial autocorrelation (P < 0.00001, Table 1).
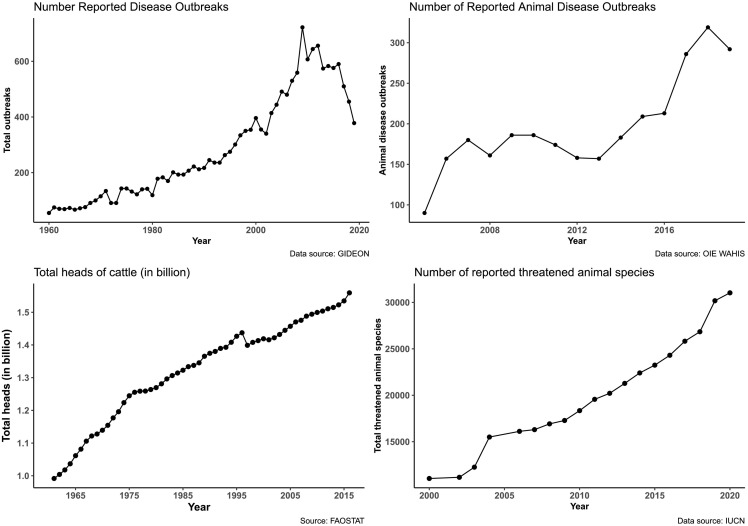
From 1960 to 2019, the number of outbreaks of human infectious diseases dramatically increased, although a slight decrease was observed for the last years (2015 to 2019) (Fig. 3A). A similar increase in the number of outbreaks of animal diseases was observed from 2006 to 2019 (Fig. 3B), although a plateau was observed from 2008 to 2012. The number of heads of cattle was below 500 millions in 1960 to reach 1200 millions in 2016 (Fig. 3C). During the years 2000–2019, the number of threatened wildlife species (all categories of IUCN) increased from 11,041 to 30,178 (Fig. 3D).
Fig. 3.
A. Number of outbreaks of human diseases from 1960 to 2019 (data obtained from GIDEON).
B. Number of outbreaks of animal diseases from 2006 to 2019 (data obtained from WAHIS – OIE).
C. Number of heads of cattle from 1960 to 2016 (data obtained from FAOSTAT).
D. Number of the number of threatened wildlife species (all categories of IUCN) from 2000 to 2019 (data obtained from IUCN).
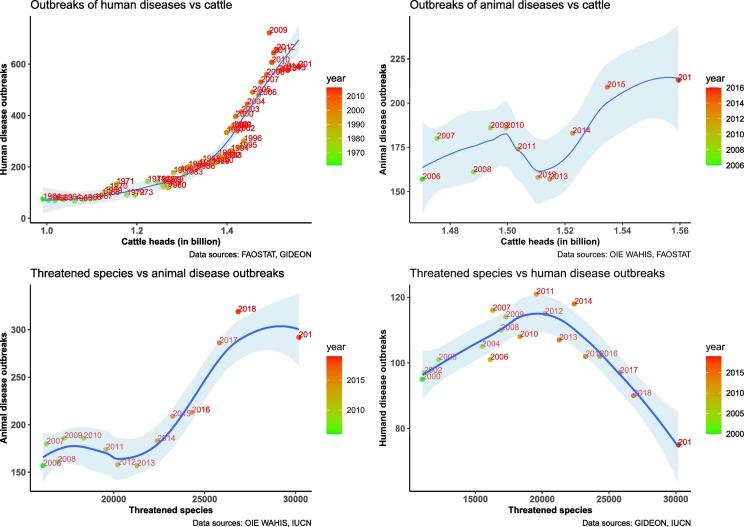
There were positive relationships between the increase of cattle heads and the number of outbreaks of human diseases from the period 1960–2019 (Fig. 4A), and the number of animal diseases 2006–2019 (Fig. 4B).
Fig. 4.
A. Association between the number of head of cattle and the number of outbreaks of human diseases from 1960 to 2019.
B. Association between the number of head of cattle and the number of outbreaks of animal diseases from 2006 to 2019.
C. Association between the number of threatened wildlife species and the number of outbreaks of human diseases from 2000 to 2019.
C. Association between the number of threatened wildlife species and the number of outbreaks of animal diseases from 2006 to 2019.
The relationships between the number of threatened wildlife species (all IUCN categories) showed different patterns according to the number outbreaks of human diseases or the number of outbreaks of animal diseases. An inverted U relationship was observed between the number of outbreaks of human diseases and the number of threatened wildlife species (Fig. 4C), with a peak of outbreaks recorded around 2011 for a value of 20,000 threatened wildlife species. A positive association was observed between the number outbreaks of animal diseases and the number of threatened wildlife species (Fig. 3D), although the period of analysis was limited to 2006–2019.
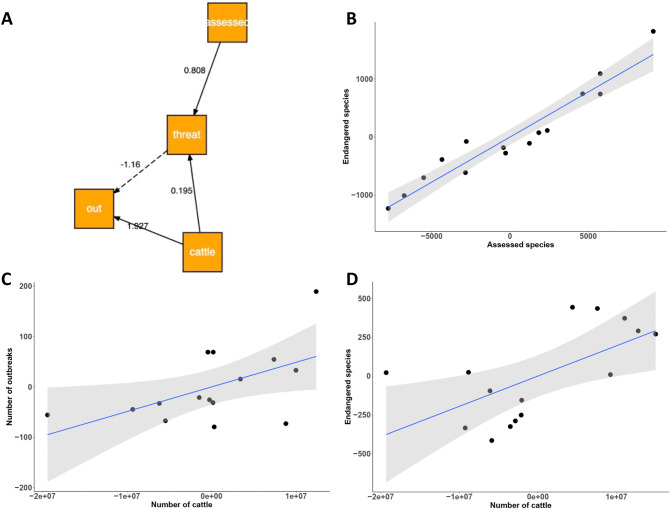
By performing temporal cross-correlation analyses over the longest period of the dataset (2000–2019), the results showed (1) a positive correlation between the increasing number of cattle and the number of endangered wildlife species (P < 0.0001), (2) a positive correlation between the increasing number of cattle and the number of outbreaks of human diseases (P < 0.0001), and (3) a lack of correlation between the number of outbreaks of human diseases and the number of endangered wildlife species (P = 0.90).
The SEM confirmed the above results (Table 2 , Fig. 5A), with a positive correlation between the number of outbreaks of human diseases and the increasing number of cattle and (P = 0.043, R2 = 0.67, Fig. 5C), and a lack of association between the number of outbreaks of human diseases with the increasing number of endangered wildlife species from 2000 to 2016. The number of endangered wildlife species assessed by IUCN was positively associated with an increase in the number of cattle (P < 0.0001, Fig. 5D) from 2000 to 2019 (R2 = 0.99).
Table 2.
Results of the Structural equation modelling (SEM) to explore the temporal association between outbreaks of human diseases, cattle number, endangered wildlife species and their assessment from 2000 to 2016 (see Methods for the model, data from GIDEON, IUCN and FAOSTAT).
| Response variable | Predictor variable | Estimate (Std Err), df | P | R2 |
|---|---|---|---|---|
| Outbreaks of human infectious diseases | Endangered wildlife species | −0.029 (0.022), 12 | 0.197 | |
| Cattle | 0.00 (0.00), 12 | 0.043 | 0.67 | |
| Endangered wildlife species | Cattle | 0.00 (0.00), 12 | <0.0001 | |
| Assessed wildlife species | 0.15 (0.01), 12 | 0.016 | 0.99 |
Fig. 5.
Results of Structural equation modelling (A, Table 2) on temporal trends from 2000 to 2016, with significant positive partial correlations between: (B) the number of endangered wildlife species and assessed species; (C) the number of outbreaks of human diseases and the number of cattle; (D) the number of endangered wildlife species and number of cattle (data from GIDEON, ICUN and FAOSTAT).
4. Discussion
The results of present analysis are in agreement with previous studies that showed positive correlation between the number of wildlife species and the number of recorded infectious and parasitic diseases in humans globally (Dunn et al., 2010; Morand et al., 2013) or regionally (Morand and Waret-Szkuta, 2012; Morand et al., 2014b). All of these studies have shown that the diversity of human pathogens among nations is positively associated with the diversity of wildlife species.
The results of the present analysis using GAM also show that the number of outbreaks of infectious diseases (corrected for the burden of infectious diseases) is positively associated with the relative number of endangered wildlife species (corrected from the wildlife species richness) among nations taking into account their spatial dependency. Loss of biodiversity appears to favour epidemics of human infectious diseases. The plateaus observed for the burden of infectious diseases or the number of outbreaks with high diversity of wildlife species or high relative number of threatened wildlife species confirm the meta-analysis by Magnusson et al. (2020), in which a strong negative diversity-disease relationship was found in the temperate region, characterized by moderately rich biodiversity, while no effect was found in the subtropical and tropical regions, characterized by highly rich biodiversity.
Over the periods analysed, all studied variables increased dramatically in number: the outbreaks of human diseases (1960–2018), the outbreaks of animal diseases (2006 to 2019), the heads of cattle (1960 to 2016) and the threatened wildlife species (2000–2018).
The number of human disease outbreaks was positively associated with the increase in the number of endangered wildlife species up to a peak reached in 2011, suggesting that a further increase in the number of threatened species was no longer associated with the number of outbreaks. Worse, the observed trend suggests an association between the increase in endangered species and the decrease the number of outbreaks of human infectious diseases recorded after a threshold reached around 2011. This observation is confirmed using SEM, which showed a positive association between the number of outbreaks of human infectious diseases and the number of wildlife species at threat, taking into account the number of assessed species, over 17 years of available data. This correlative observation needs to further explored in detail with each country and for a longer period of time. However, the data are not yet available. The last few years have so far been characterized by increasing biodiversity at risk, which contributes less and less to human infectious diseases. The recent emergence of the bat coronavirus could represent the latest viral explosion of declining biodiversity (Morand, 2016).
The pattern observed between endangered species and the expansion of cattle provides a more clear picture. The continuous increase of in head of cattle seems to contribute positively to the number of infectious outbreaks registered and to the increase in number of threatened wildlife species at least for the period 2000–2016. This observation confirms studies that have investigated the disproportionate effects of livestock expansion on biodiversity (Steinfeld et al., 2006; Reid et al., 2010) and on natural and traditionally managed habitats (Alkemade et al., 2013).
Several potential biases may limit the analyzes presented in this study. First, the registration of infectious diseases depends on several factors such as expenditure on public health services or the quality of the animal health surveillance system, which obviously vary from country to country but also from year to year in relation to financial priorities, development aid, private sector participation, among others. Many epidemics are transboundary epidemics, such as pandemics, which require effective recording of the time, place and geographic spread of each disease. For example, the plateau observed in the number of infectious diseases from 2008 to 2012 can surely be explained by the global financial crisis of 2008, which may have negatively affected agricultural production and trade or by a drop in public investment in animal health surveillance. Either the decrease in agricultural production and trade or the notification by the animal health surveillance would have reduced the recorded number of outbreaks of animal diseases. Another limitation concerns the causes of the expansion of animal husbandry, which depends on several factors which also vary from country to country, namely the growth of human population, changes in diet linked to economic growth and westernization of habits, agricultural industrialization and the integration into world trade, but also the cultural values of livestock. Last but not least, a historical and comparative analysis of the links between biodiversity losses and the rise of emerging diseases and global epidemics will always suffer from the impossibility of carrying out experimental manipulations.
5. A call for further contributive studies
This study highlights the importance of expanding livestock farming both as a threat to biodiversity and as increasingly putting human and animal health at risk (Rohr et al., 2019). The complex global and spatio-temporal patterns linking livestock, biodiversity, animal and human health call for more detailed studies performed between and within countries over a long time series. Livestock and the diversity of wild species are not uniformly distributed on the planet and are not managed equally by societies, according to the values associated with wild and domestic animals, which may depend on various socio-economic trajectories. New studies should provide a better understanding of these complex and adaptative relationships in order to make an effective contribution to the conservation (wild species and domestic breeds), animal health and public health sectors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study is part of the French ANR project FutureHealthSEA (ANR-17-CE35-0003-01). S.M. is supported by the Thailand International Cooperation Agency (TICA) with the project “Innovative Animal Health”.
References
- Alkemade R., Reid R.S., van den Berg M., de Leeuw J., Jeuken M. Assessing the impacts of livestock production on biodiversity in rangeland ecosystems. Proceedings National Academy of Sciences USA. 2013;110:20900–20905. doi: 10.1073/pnas.1011013108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordes F., Blasdell K., Morand S. Transmission ecology of rodent-borne diseases: new frontiers. Integrative Zoology. 2015;10:424–435. doi: 10.1111/1749-4877.12149. [DOI] [PubMed] [Google Scholar]
- Chivian E. Harvard Med School; Boston: 2003. Biodiversity: Its Importance to Human Health. [Google Scholar]
- Civitello D.J., Cohen J., Fatima H., Halstead N.T., Liriano J., McMahon T.A., Ortega C.N., Sauer E.L., Sehgal T., Young S., Rohr J.R. Biodiversity inhibits parasites: broad evidence for the dilution effect. Proceedings of the National Academy of Sciences USA. 2015;112:8667–8671. doi: 10.1073/pnas.1506279112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destoumieux-Garzón D., Mavingui P., Boetch G., Boissier J., Darriet F., Duboz P., Fritsch C., Giraudoux P., Le Roux F., Morand S. The one health concept: 10 years old and a long road ahead. Frontiers in Veterinary Science. 2018;5 doi: 10.3389/fvets.2018.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn R.R., Davies T.J., Harris N.C., Gavin M.C. Global drivers of human pathogen richness and prevalence. Proceedings of the Royal Society of London B. 2010;277:2587–2595. doi: 10.1098/rspb.2010.0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J. R package version 2.0-0. 2015. spatialEco. [Google Scholar]
- FAO-OIE-WHO . A Tripartite Concept Note; Hanoi: 2010. The FAO-OIE-WHO Collaboration: Sharing Responsibilities and Coordinating Global Activities to Address Health Risks at the Animal-Human-Ecosystems Interfaces.https://www.who.int/foodsafety/areas_work/zoonose/concept-note/en/ [Google Scholar]
- Fasiolo M., Nedellec R., Goude Y., Wood S.N. 2018. Scalable Visualisation Methods for moDern Generalized Additive Models. arXiv:1809.10632. [Google Scholar]
- Guernier V., Hochberg M.E., Guégan J.F. Ecology drives the worldwide distribution of human diseases. PLoS Biol. 2004;2:740–746. doi: 10.1371/journal.pbio.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton W.D. Geometry for the selfish herd. Journal Theoretical Biology. 1971;3:295–311. doi: 10.1016/0022-5193(71)90189-5. [DOI] [PubMed] [Google Scholar]
- Harrell F. Springer; 2015. Regression Modeling Strategies: With Applications to Linear Models, Logistic and Ordinal Regression, and Survival Analysis. [Google Scholar]
- Harrell F. R package version 4.2-0. 2019. Hmisc: Harrell miscellaneous.https://CRAN.R-project.org/package=Hmisc [Google Scholar]
- Hastie T.J., Tibshirani R.J. Chapman and Hall – CRC; New-York: 1990. Generalized Additive Models. [DOI] [PubMed] [Google Scholar]
- Hechinger R.F., Lafferty K.D. Host diversity begets parasite diversity: bird final hosts and trematodes in snail intermediate hosts. Proceedings of the Royal Society of London B. 2005;272:1059–1066. doi: 10.1098/rspb.2005.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans R.J. R package version 3.0-12. 2020. Raster: geographic data analysis and modeling.https://CRAN.R-project.org/package=raster [Google Scholar]
- Johnson P.T.J., Ostfeld R.S., Keesing F. Frontiers in research on biodiversity and disease. Trends in Ecology and Evolution. 2015;18:1119–1133. doi: 10.1111/ele.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C.K., Hitchens P.L., Pandit P.S., Rushmore J., Evans T.S., Young C.C.W., Doyle M.M. Global shifts in mammalian population trends reveal key predictors of virus spillover risk. Proc. R. Soc. Lond. B. 2020;287:20192736. doi: 10.1098/rspb.2019.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L., Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–994. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keesing F., Holt R.D., Ostfeld R.S. Effects of species diversity on disease risk. Ecol. Lett. 2006;9:485–498. doi: 10.1111/j.1461-0248.2006.00885.x. [DOI] [PubMed] [Google Scholar]
- Keesing F., Belden L.K., Daszak P., Dobson A., Harvell C.D., Holt R.D., Hudson P., Jolles A., Jones K.E., Mitchell C.E., Myers S.S. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature. 2010;468:647–652. doi: 10.1038/nature09575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keesing F., Belden L.K., Daszak P., Dobson A., Harvell C.D., Holt R.D., Hudson P., Jolles A., Jones K.E., Mitchell C.E. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature. 2010;468:647–652. doi: 10.1038/nature09575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefcheck J.S. piecewiseSEM: piecewise structural equation modeling in R for ecology, evolution, and systematics. Methods Ecol. Evol. 2016;7:573–579. [Google Scholar]
- Lloyd-Smith J.O., George D., Pepin K.M., Pitzer V.E., Pulliam J.R.C., Dobson A.P., Hudson P.J., Grenfell B.T. Epidemic dynamics at the human-animal interface. Science. 2009;326:1362–1367. doi: 10.1126/science.1177345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson M., Fischhoff I.R., Ecke F., Hornfeldt B., Ostfeld R.S. Effect of spatial scale and latitude on diversity–disease relationships. Ecology. 2020;101 doi: 10.1002/ecy.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowski D., Lüdecke D. R package version 0.1.0. 2020. Correlation: easy Peasy correlations.https://CRAN.R-project.org/package=correlation [Google Scholar]
- McNeill W.H. Anchor Press; New York: 1976. Plagues and People. [Google Scholar]
- Morand S. Editions Fayard; Paris: 2016. La Prochaine Peste. Une histoire globale des sociétés et de leurs épidémies. [Google Scholar]
- Morand S., Lajaunie C. Linking Life, Ecosystems and Societies. Elsevier; London: 2017. Biodiversity and health. [Google Scholar]
- Morand S., Walther B. Individualistic values are related to an increase in the outbreaks of infectious diseases and zoonotic diseases. Sci. Rep. 2018;8:3866. doi: 10.1038/s41598-018-22014-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morand S., Waret-Szkuta A. Determinants of human infectious diseases in Europe: biodiversity and climate variability influences. Bull Epidémiologique Hebdomadaire. 2012;12–13:156–159. [Google Scholar]
- Morand S., Owers K., Waret-Szkuta A., McIntyre K., Baylis M. Climate variability and outbreaks of infectious diseases in Europe. Sci. Rep. 2013;3:1774. doi: 10.1038/srep01774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morand S., McIntyre K.M., Baylis M. Domesticated animals and human infectious diseases of zoonotic origins: domestication time matters. Infection Genetics Evolution. 2014;24:76–87. doi: 10.1016/j.meegid.2014.02.013. [DOI] [PubMed] [Google Scholar]
- Morand S., Owers K., Bordes F. Biodiversity and emerging zoonoses. In: Yamada A., Kahn L.H., Kaplan B., Monath T.P., Woodall J., Conti L.A., editors. Confronting Emerging Zoonoses: The One Health Paradigm. Springer; 2014. pp. 27–41. [Google Scholar]
- Ostfeld R.S., Keesing F. Biodiversity and disease risk: the case of Lyme disease. Conserv. Biol. 2000;14:722–728. [Google Scholar]
- Poisot T., Nunn C., Morand S. Ongoing worldwide homogenization of human pathogens. BioRxiv. 2015 doi: 10.1101/009977. [DOI] [Google Scholar]
- R Development Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2019. R: A Language and Environment for Statistical Computing.http://www.R-project.org [Google Scholar]
- Reid R.S., Bedelian C., Said M.Y., Kruska R.L., Mauricio R.M., Castel V., Olson J., Thornton P.K. Global livestock impacts on biodiversity. In: Steinfeld H., Mooney H.A., Schneider F., Neville L.E., editors. Livestock in a Changing Landscape. Drivers, Consequences, and Responses. Vol 1. Island Press; Washington, DC: 2010. pp. 111–138. [Google Scholar]
- Rohr J.R., Barrett C.B., Civitello D.J., Craft M.E., Delius B., DeLeo G.A., Hudson P.J., Jouanard N., Nguyen K.H., Ostfeld R.S. Emerging human infectious diseases and the links to global food production. Nature Sustainability. 2019;2:445–456. doi: 10.1038/s41893-019-0293-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr J.R., Civitello D.J., Halliday F.W., Hudson P.J., Lafferty K.D., Wood C.L., Mordecai E.A. Towards common ground in the biodiversity–disease debate. Nature Ecology. 2020;4:24–33. doi: 10.1038/s41559-019-1060-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salkeld D.J., Padgett K., Jones J.H. A meta-analysis suggesting that the relationship between biodiversity and risk of zoonotic pathogen transmission is idiosyncratic. Ecol. Lett. 2013;16:679–686. doi: 10.1111/ele.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schipper J., Chanson J.S., Chiozza F., Cox N.A., Hoffmann M., Katariya V., Lamoreux J., Rodrigues A.S., Stuart S.N., Temple H.J. The status of the world’s land and marine mammals: diversity, threat, and knowledge. Science. 2008;322:225–230. doi: 10.1126/science.1165115. [DOI] [PubMed] [Google Scholar]
- Simpson G.L. gratia: Graceful 'ggplot'-Based Graphics and Other Functions for GAMs Fitted Using 'mgcv'. R package version 0.2-8. 2019. https://gavinsimpson.github.io/gratia
- Smith K.F., Goldberg M., Rosenthal S., Carlson L., Chen J., Chen C., Ramachandran S. Global rise in human infectious disease outbreaks. Journal Royal Society Interface. 2014;11:20140950. doi: 10.1098/rsif.2014.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South A. Rworldmap: a new R package for mapping global data. The R Journal. 2011;3(1):35–43. [Google Scholar]
- Steinfeld H., Gerber P., Wassenaar T.D., Castel C., Rosales M., de Haan C. Food and Agriculture Organization of the United Nations; Roma: 2006. Livestock’s Long Shadow: Environmental Issues and Options. [Google Scholar]
- Thornton P.K. Livestock production: recent trends, future prospects. Philosophical Transactions of the Royal Society B: Biological Sciences. 2010;365:2853–2867. doi: 10.1098/rstb.2010.0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells K., Morand S., Wardeh M., Baylis M. Distinct spread of DNA and RNA viruses among mammals amid prominent role of domestic species. Glob. Ecol. Biogeogr. 2020;29:470–481. doi: 10.1111/geb.13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmee S., Haines A., Beyrer C., Boltz F., Capon A.G., de Souza Dias B.F., Ezeh A., Frumkin H., Gong P., Head P. Safeguarding human health in the Anthropocene epoch: report of the Rockefeller Foundation–Lancet Commission on planetary health. Lancet. 2015;386:1973–2028. doi: 10.1016/S0140-6736(15)60901-1. [DOI] [PubMed] [Google Scholar]
- Wickham H. httr: Tools for Working with URLs and HTTP. R package version 1.4.1. 2019. https://CRAN.R-project.org/package=httr
- Wickham H. rvest: Easily Harvest (Scrape) Web Pages. R package version 0.3.5. 2019. https://CRAN.R-project.org/package=rvest
- Wilcox B.A., Gubler D.J. Disease ecology and the global emergence of zoonotic pathogens. Environmental Health Preventive Medicine. 2005;10:263–272. doi: 10.1007/BF02897701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe N.D., Dunavan C.P., Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood S.N. 2nd edition. Chapman and Hall/CRC; New York: 2017. Generalized Additive Models: An Introduction with R. [Google Scholar]
- Young H.S., Wood C.L., Kilpatrick A.M., Lafferty K.D., Nunn C.L., Vincent J.R. Conservation, biodiversity and infectious disease: scientific evidence and policy implications. Philosophical Transactions Royal Society, B. 2017;372:20160124. doi: 10.1098/rstb.2016.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wu Q., Zhigang Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020 doi: 10.1016/j.cub.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]