Abstract
Coronavirus disease 2019 (COVID-19) has caused thousands of deaths worldwide and has become an urgent public health concern. The extraordinary interhuman transmission of this disease has urged scientists to examine the various facets of its pathogenic agent, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Herein, based on publicly available genomic data, we analyzed the codon usage co-adaptation profiles of SARS-CoV-2 and other respiratory coronaviruses (CoVs) with their human host, identified CoV-responsive human genes and their functional roles on the basis of both the relative synonymous codon usage (RSCU)-based correlation of viral genes with human genes and differential gene expression analysis, and predicted potential drugs for COVID-19 treatment based on these genes. The relatively high codon adaptation index (CAI) values (>0.70) signposted the gene expressivity efficiency of CoVs in human. The ENc-GC3 plot indicated that SARS-CoV-2 genome was under strict selection pressure while SARS-CoV and MERS-CoV were under selection and mutational pressures. The RSCU-based correlation analysis indicated that the viral genomes shared similar codons with a panoply of human genes. The merging of RSCU-based correlation data and SARS-CoV-2-responsive differentially expressed genes allowed the identification of human genes potentially affected by SARS-CoV-2 infection. Functional enrichment analysis indicated that these genes were enriched in biological processes and pathways related to host response to viral infection and immune response. Using the drug-gene interaction database, we screened a list of drugs that could target these genes as potential COVID-19 therapeutics. Our findings not only will contribute in vaccine development but also provide a useful set of drugs that could guide practitioners in strategical monitoring of COVID-19. We recommend practitioners to scrupulously screen this list of predicted drugs in order to authenticate those qualified for treating COVID-19 symptoms.
Keywords: Coronavirus, COVID-19, Codon usage, Transcriptome, Virus-host co-adaptation, Treatment drugs
1. Introduction
The coronaviruses (CoVs) are a large family of RNA viruses responsible for illnesses ranging from the common cold to severe acute respiratory diseases both in wild animals and humans (Sutton and Subbarao, 2015; Tortorici and Veesler, 2019). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a CoV identified as the causal agent of the respiratory epidemic first detected in China (Wuhan) and subsequently declared by the World Health Organization (WHO) as the COVID-19 pandemics due to the spread of the outbreak to other countries worldwide (Lai et al., 2020; Zheng, 2020). The clinical symptoms of SARS-CoV-2 infection include fever, cough, headache, and shortness of breath (Lin et al., 2020; Xu et al., 2020; Yang et al., 2020; Zhang et al., 2020). COVID-19 has caused a significant loss in human lives and has resulted in a tremendous economic damage across the five continents (Ahn et al., 2020). At present, there is no accurate therapy for COVID-19 though some medications such as chloroquine/hydroxychloroquine (Wright et al., 2020), combinations of antiviral and anti-inflammatory treatments (Stebbing et al., 2020), traditional Chinese medicine (Ren et al., 2020) and other therapeutics (Ahn et al., 2020; Elfiky, 2020) have been proposed. The development of vaccines against COVID-9 is also at its infancy (Ahmed et al., 2020; Prompetchara et al., 2020; Robson, 2020). Due to the extent of the pandemic, experimental research on COVID-19 vaccine candidates and therapeutics as well as the identification of suitable experimental models are needed to limit the impact of SARS-CoV-2 infection. This requires the understanding of SARS-CoV-2 at molecular level.
The reference genome of SARS-CoV-2 has been made available and an in-depth genome annotation of this virus has been performed (Licastro et al., 2020; Paraskevis et al., 2020; Sah et al., 2020; Stefanelli et al., 2020; Yadav et al., 2020). Preliminary comparative genomics studies suggested the divergence between SARS-CoV-2 and other CoVs, namely SARS-CoV and MERS-CoV (Wu et al., 2020). Codon usage bias, the differential use of codons encoding a given amino acid, is vital for examining the expressivity and the adaptation of exogenous viral genes to the hosts (Deb et al., 2020; Oldfield et al., 2020; Yao et al., 2019). Codon usage can play a significant role in modulating the expression of viral genes via codon optimization (Morgunov and Babu, 2014; Victor et al., 2019; Zalucki et al., 2009). Codon usage is equally important for studying the evolution and ecological adaptation of diverse organisms (Liang et al., 2014; Ou et al., 2020). Studies on codon usage analysis of CoVs have been focused on the analysis of codon usage bias of some proteins among CoVs (Castells et al., 2017; Chen et al., 2017; Gu et al., 2004; Kandeel and Altaher, 2017; Sheikh et al., 2020). With the outbreak of COVID-19, scientists have put forward effort to analyze the codon usage bias of CoVs in order to improve our understanding on the emergence, adaptation, spread and evolution of SARS-CoV-2 (Tort et al., 2020). Apart from that, the phenotypic variation of genes encoded by SARS-CoV-2 has been also studied (Dilucca et al., 2020). Nonetheless, little is known about the molecular mechanism associated with the SARS-CoV-2 and its adaptation to its human host. Therefore, studies on the codon usage pattern of SARS-CoV-2 would be vital in elucidating the evolution of SARS-CoV-2, its adaptation to the host and the molecular mechanism involved in SARS-CoV-2 infection and pathological changes in the host. This will provide useful data on the virulence of SARS-CoV-2 and help understand the molecular mechanisms underlying the pathogenicity in human. Studying the codon usage of SARS-CoV-2 will pave the way for devising various approaches to engineer the SARS-CoV-2 genome to mitigate its virulence and uncover its replicative effectiveness in human hosts, which is relevant for safe and efficient vaccine and therapeutic drug development. Previous studies indicated that codon usage preferences of viruses and their animal and human hosts are strongly correlated, especially in tissues they infect (Miller et al., 2017). These correlations are the consequence of co-evolution between the virus and their host, which is necessary for the host evolution in the defense against the virus as well as the virus adaptation to the host physiological changes (Guo et al., 2019; Miller et al., 2017). Thus, the analysis of codon usage is important for uncovering the host genes which are affected by the viral infection. The knowledge on these genes is necessary for determining the mechanisms underlying the pathogenesis associated with viral infection in the host.
Systems biology is a multidisciplinary method that is systematically used for portraying the multifaceted aspects of biological processes and accurate prediction of the performance of biological systems (Kitano, 2002). This approach has been applied for examining the codons usage profile of various organism and their adaptation to their hosts. Systems biology is also useful for predicting drugs for a given disease. Systems biology has been recently applied for network-based drug repurposing for SARS-CoV-2 (Zhou et al., 2020), but this study was solely based on phylogenetic approaches, and thus present some limitations in the understanding of the interaction between the virus and its host; this could introduce some biases in drug prediction. As a systems biology approach, network-pharmacology has been also used for evaluating the potential of traditional Chinese medicines against COVID-19 (Yu et al., 2020). In silico prediction of drug candidates against COVID-19 has been also proposed on the basis of Angiotensin converting enzyme 2 (ACE2) regulatory network (Cava et al., 2020); however, this prediction had no direct connection with viral infection. To the best of our knowledge, systems biology approaches based on codon usage co-adaptation between SARS-CoV-2 and its human host as well as SARS-CoV-2-induced differential gene expression have not been applied for discovering potential therapeutics for COVID-19 and other respiratory CoVs. This is an important research avenue to deeply explore.
Hence, the primary purpose of the present study was to use systems biology approaches for analyzing the codon usage profile of SARS-CoV, SARS-CoV-2 and MERS-CoV in order to provide useful molecular data for the development of attenuated SARS-CoV-2 vaccine strains that may have vaccine potential against COVID-19 and a broader applicability for CoVs. Secondly, we aimed to exploit the codon usage co-adaptation of CoVs with their hosts to assess the impact of the interaction between these viruses and their human host on the expression of host genes in order to discover possible mechanisms of their pathogenicity. Finally, based on our knowledge of these genes, we also aimed to predict, in silico, the potential drugs for the treatment of the ongoing COVID-19 in particular, and CoV infection in general. Based on the model of CAI, ENc-GC3 and RSCU of the three CoVs, we uncovered the functional role of CoV-responsive human genes and how respiratory CoVs affect human genes, and revealed a useful set of drugs that could guide practitioners in strategical monitoring of COVID-19. By comparing the above aspects with previous studies (Dilucca et al., 2020; Tort et al., 2020), our findings may provide some new strategies to control or prevent SARS-CoV-2 and other CoV diseases.
2. Material and methods
2.1. Data collection
The full-length CDS sequences of SARS-CoV, SARS-CoV-2 and MERS-CoV were downloaded from the NCBI virus (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/) database. The data were downloaded on March 16, 2020. The reference genomes of SARS-CoV, SARS-CoV-2 and MERS-CoV and their hosts, including Camelus bactrianus, C. dromedarius, Canis lupus familiaris, Capra hircus, Chlorocebus sabeus, Equus asinus,Felis catus, Hipposideros armiger,Homo sapiens, Mus musculus, Panthera tigris, Manis javanica,Rhinolophus ferrumequinum and R. sinicus, were downloaded from RefSeq. Human CDS sequences (N = 20,091) of the Release 33 (GRCh38.p13) of human genome were downloaded from GENCODE (https://www.gencodegenes.org/human/). The gene expression dataset GSE147507 (Blanco-Melo et al., 2020) containing the gene expression of human cells transfected or untransfected with SARS-CoV-2 was downloaded from the GEO database(https://www.ncbi.nlm.nih.gov/geo/).
2.2. Sequence analysis in viruSITE
The viruSITE (http://www.virusite.org/), a database of viral genomes and genes (Stano et al., 2016), was used for analyzing the sequences of SARS-CoV, SARS-CoV-2 and MERS-CoV based on the reference sequences deposited in this database. Circoletto (Darzentas, 2010) was used for visualizing sequence similarity circos plot.
2.3. Determination of codon usage indices
The R package vhcub (Anwar et al., 2019) was used to compute codon usage bias indices including the effective number of codons (ENc), codon adaptation index (CAI), relative codon deoptimization index (RCDI), similarity index (SiD), and relative synonymous codon usage (RSCU). The vhcub package was also used for analyzing dinucleotide over- and under-representation based on base model, codon and syncodon models. Details on these indices are described below.
2.3.1. GC content analysis
The total GC content (percentage of G and C nucleotides relative to the total number of nucleotides), GC1 (percentage of G and C nucleotides at first position of codons), GC2 (percentage of G and C nucleotides at second position of codons) and GC3 (percentage of G and C nucleotides at second position of codons) were determined for each coding protein of SARS-CoV, SARS-CoV-2 or MERS-CoV and averaged.
2.3.2. Relative dinucleotide abundance analysis
Deviations in the ratio of dinucleotide pairs constitute an influential factor of codon usage (Kunec and Osterrieder, 2016; Sexton and Ebel, 2019). The ratio of dinucleotide pairs is useful for gaging the preferential use of various dinucleotide pairs in an organism (Kunec and Osterrieder, 2016; Sexton and Ebel, 2019). There are 16 dinucleotide combinations and the profiles of dinucleotide ratio can reflect mutational or selectional pressures. The ratio of dinucleotides was computed using the equation ρXY = fXY/fX*fY, wherein fX and fY stand for the frequencies of nucleotide X and nucleotide Y, respectively, whereas fXY is the frequency of XY dinucleotides in the same nucleotide sequence. The ratio of the factual to predicted dinucleotide frequency is the odds ratio. An odds ratio lower than 0.78 indicates underrepresented dinucleotide pairs while odds ratio greater than 1.25 indicates overrepresented dinucleotides (Kunec and Osterrieder, 2016).
2.3.3. Relative synonymous codon usage (RSCU)
The RSCU value of a given codon is the ratio of the observed value of this codon over the expected value of this codon for a given amino acid (Sharp and Li, 1986). RSCU value is independent from sequence length of the content in amino acid. Codons with RSCU greater than 1.6 are considered as preferred codons while codons with RSCU values lower than 0.6 are considered as non-preferential codons. Unbiased codons are those with RSCU ranging from 0.6 to 1.6. RSCU values were generated based on the CDS sequences of SARS-CoV, SARS-CoV-2, MERS-CoV and their hosts.
2.3.4. Similarity index analysis
The similarity index (SiD) that assesses the impact of the codon usage of a given host on that of its pathogen. This approach is commonly used to gage the capacity of a host to give refuge to a virus (Zhou et al., 2013). SiD is the effect of the overall codon usage of the host on that of its pathogens. SiD is comprised between 0 and 1, and high SiD values indicates greater effect of the host on codon usage of the pathogens. SiD values of SARS-CoV, SARS-CoV-2 and MERS-CoV were examined relative to their human host.
2.3.5. Effective number of codons (ENc)
The effective number of codons (ENc), ranging between 20 and 61, is a codon usage index used for detecting the bias in the use of synonymous codons (Wright, 1990). ENc value = 20 when there is maximum bias and only one of the available synonymous codons is used for the encoding of a given amino acid. On the contrary, the maximum ENc value of 61 signposts no codon usage bias and indicates the same probability of the use of the available codons. ENc value <35, indicates high codon usage bias in the genome.
2.3.6. Neutrality and parity analysis
To analyze the effect of the translation selection and mutation bias on the codon usage, we generated the neutrality plots for SARS-CoV, SARS-CoV-2 and MERS-CoV, respectively. The regression line between GC12s and GC3s was generated and the slopes of the regression equations were used for evaluation the mutational force. The parity rule 2 (PR2)-bias plot was generated by plotting the three CoVs relative to human.
2.3.7. Codon adaptation index (CAI) analysis
Codon adaptation index (CAI) is a numerical measure portraying the frequency of the use of a preferred codon among genes with high expression (Carbone et al., 2003; Lee et al., 2010). CAI is used to assess the translation efficiency and finds application in the genetic engineering of nucleotide sequences optimal protein expression for vaccine development. Comprised between 0 and 1, elevated CAI values indicate greater gene expressivity and CAI values approximating the value of 1 signpost the use of codons with high RSCU values in the gene. We computed the CAI values of the CDS sequences of SARS-CoV, SARS-CoV-2 and MERS-CoV.
2.3.8. Relative codon deoptimization index (RCDI)
RCDI (Mueller et al., 2006) is a numerical variable used for comparing the similarity in codon usage in genes and given genomes. RCDI can depict the rate of translation of viral gene in a host genome. Similar codon usage between a pathogen and its host is characterized by RCDI values close to 1, which indicates high translation rate and high adaptive capacity to the host (Butt et al., 2016). RCDI values of the CDS sequences of SARS-CoV, SARS-CoV-2 and MERS-CoV for the host H. sapiens were determined.
2.4. Principal component analysis (PCA)
PCA is a multivariate analysis approach that is used for geometrical plotting of sets of columns and rows in a given dataset (Wold et al., 1987). We performed PCA based on the RSCU values of the CDS sequences of SARS-CoV, SARS-CoV-2, MERS-CoV and H. sapiens using the function dudi.pca in the ade4 R package.
2.5. Correspondence analysis
Correspondence analysis was performed based on the RSCU values of the three viruses using the R packages “FactoMineR” and “factoextra”.
2.6. Correlation analysis
The correlation between the RSCU values of human genes and those of viral genes was analyzed using the Hmisc library in R. The Pearson correlation coefficients and p values were generated for each pair of genes. The correlation significance was set for p < .05.
2.7. Differential gene expression analysis
The differential expression analysis of genes among uninfected and SARS-CoV-2-infected human cell lines A549, A549-ACE2, Calu3 and NHBE was performed based on the GSE147507 data. The differential expression among uninfected and infected groups was analyzed using the DESeq library in R. The screening parameters of differentially expressed genes (DEGs) were as follows: |log2FC| ≥ 1 and adjusted p value (p.adjust) > 0.05. To uncover the true DEGs in response to SARS-CoV-2, the DEGs screened from the four cell lines were merge while for broader applicability, these DEGs were combined.
2.8. Gene ontology and KEGG pathway enrichment analysis of potential human genes responding to viral infection
The R package clusterProfiler (Yu et al., 2012) was used for gene ontology (GO) and KEGG pathway enrichment analysis. The adjusted p values were also generated and the statistical significance was set for p < .05.
2.9. Prediction of drugs interacting with potential human genes responding to viral infection
The Drug-Gene Interaction Database (DGIdb at http://www.dgidb.org/search_interactions) (Cotto et al., 2018) was used for predicting the potential drugs that could target the potential human genes affected by viral infection. The prediction result TSV file was downloaded and imported in R for generating the summary of results using the summary function in R.
2.10. Statistical analysis and data visualization
One-way analysis of variance (ANOVA) followed by Bonferroni posttest was used to evaluate the differences between values in the genomes of SARS-CoV, SARS-CoV-2 and MERS-CoV in GraphPad Prism software (GraphPad Software, La Jolla California USA, www.graphpad.com). Graphs were plotted using the R packages ggplot2 and ggrepel. Various coloring schemes were employed for labelling sequences on the plots according to the different features being investigated.
3. Results
3.1. Sequence similarity analysis
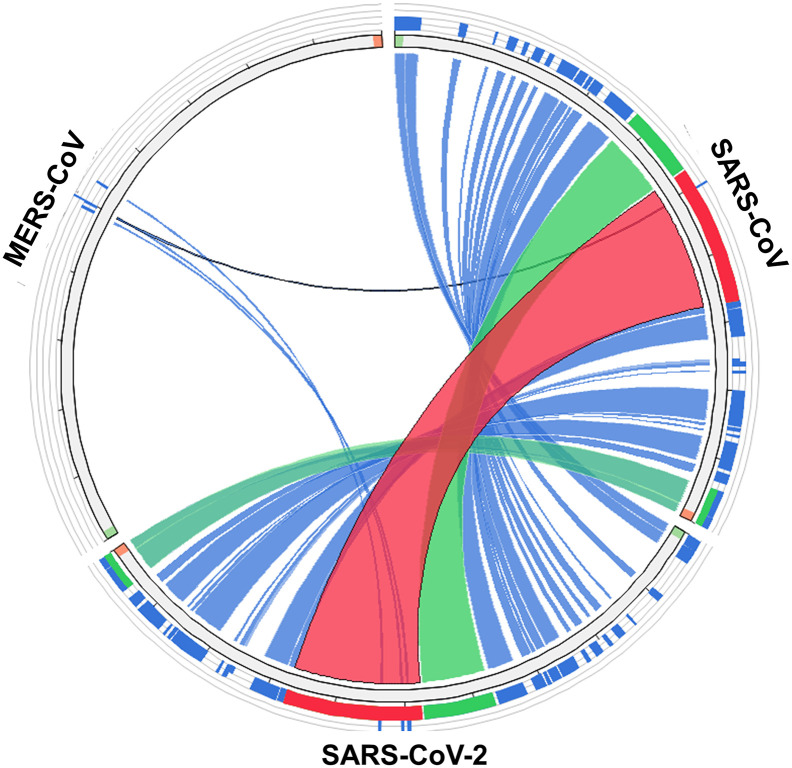
In order to examine the genomic similarity of SARS-CoV, SARS-CoV-2 and MERS-CoV, similarity analysis was performed using the viruSITE database. The circos plot depicted in Fig. 1 indicated high genetic divergence between the genome of MERS-CoV compared to SARS-CoV and SARS-CoV-2. On the contrary, high similarity of genomic features were observed between SARS-CoV and SARS-CoV-2. This suggested that clinical and experimental practices based on SARS-CoV may be applicable to SARS-CoV-2, especially in the search for vaccines and treatment approaches.
Fig. 1.
Similarity analysis of the genomes of human respiratory coronaviruses. The circos plot was generated using circolleto in viruSITE.
3.2. GC content in the genomes of coronaviruses
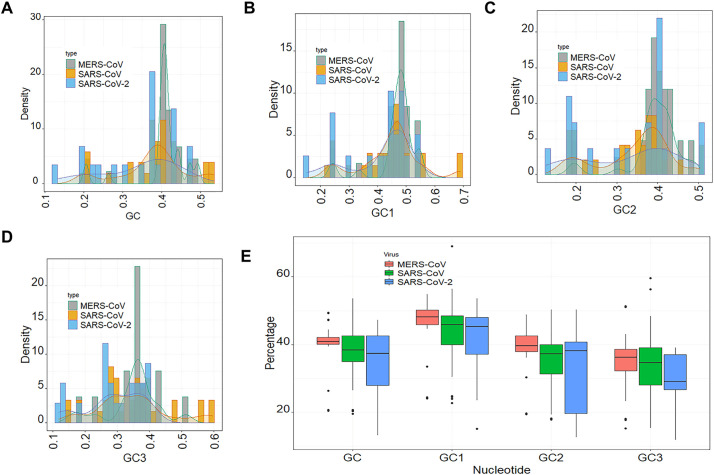
To examine the compositional constraints in the genome of CoVs, the nucleotide compositions of the CDSs from MERS-CoV, SARS-CoV and SARS-CoV-2 were determined. The GC% content of the CDS sequences of MERS-CoV, SARS-CoV and SARS-CoV-2 ranged from 13.16% to 53.52% (Fig. 2A). Most of the CDS sequences had GC content around 40% (Fig. 2A). The histogram showing the distribution of GC1 (Fig. 2B) indicated that most of the CDS sequences had GC1 content of 40–50% especially for MERS-CoV genome (Fig. 2B). The lowest GC1 content was 15% while the highest was 69.01% (Fig. 2B). Additionally, the CDS sequences with GC2 content of approximately 40% were the most abundant in the genomes of these CoVs with the lowest GC2 content of 12.56% and the highest GC2 content of 50.33% (Fig. 2C). The lowest GC3 content was 11.84% while the GC3 content was 59.59% (Fig. 2D). The CDS sequences with GC3 content between 20% and 40% were the most abundant (Fig. 2D). As shown in Fig. 2E, the average GC contents of CDS sequences were 0.34, 0.38 and 0.40 for SARS-CoV-2, SARS-CoV and MERS-CoV, respectively. The average GC1 contents were 0.41, 0.45 and 0.47 for SARS-CoV-2, SARS-CoV and MERS-CoV, separately (Fig. 2E). The average GC2 contents were 0.33, 0.34 and 0.39 for SARS-CoV-2, SARS-CoV and MERS-CoV, respectively (Fig. 2E). The average GC3 contents were 0.29, 0.35 and 0.35 for SARS-CoV-2, SARS-CoV and MERS-CoV, respectively (Fig. 2E). There were no significant differences in the mean of GC, GC1, GC2 and GC3 between the 3 viruses. These results indicated that the genomes of CoVs are rich in A/U nucleotides in comparison to G/C nucleotides.
Fig. 2.
GC composition of CDS sequences of human respiratory coronaviruses. A- Distribution of the overall GC content of CDS sequences of MERS-CoV, SARS-CoV and SARS-CoV-2. B- Distribution of the GC1 content of CDS sequences of MERS-CoV, SARS-CoV and SARS-CoV-2. C- Distribution of the GC2 content of CDS sequences of MERS-CoV, SARS-CoV and SARS-CoV-2. D- GC3 content of CDS sequences of MERS-CoV, SARS-CoV and SARS-CoV-2. E- Boxplot of GC, GC1, GC2 and GC3 contents in CDS sequences of MERS-CoV, SARS-CoV and SARS-CoV-2.
3.3. Dinucleotide over- and underrepresentation in coronaviruses
To determine codon pair preferences in CoV, we used three models for determining the underrepresented and the overrepresented dinucleotides. In the “base model” (Fig. 3A), CG, TA, TC, GA, AT and CC were identified as underrepresented dinucleotides whereas the other dinucleotide pairs were found as the over represented dinucleotide with GC, CT, TG and CA being the most overrepresented. In the “codon model” (Fig. 3B), AC, GT, AG, CT, GC, TG and CA were found as the overrepresented dinucleotide pairs in the genomes of SARS-CoV-2, SARS-CoV and MERS-CoV while the other dinucleotide pairs were underrepresented with CG being the most underrepresented. In the “syncodon model” (Fig. 3A), GC, TG and CA were found as the overrepresented dinucleotide pairs in the genomes of SARS-CoV-2, SARS-CoV and MERS-CoV while the other dinucleotide pairs were underrepresented with CG being the most underrepresented. The patterns of dinucleotide pair preferences were similar among SARS-CoV-2, SARS-CoV and MERS-CoV.
Fig. 3.
Dinucleotide over-representation and under-representation in CDS sequences of human respiratory coronaviruses. A- Heatmap of dinucleotide content of CDS sequences of MERS-CoV, SARS-CoV and SARS-CoV-2 based on the base model. B- Heatmap of dinucleotide content of CDS sequences of MERS-CoV, SARS-CoV and SARS-CoV-2 based on the codon model. C- Heatmap of dinucleotide content of CDS sequences of MERS-CoV, SARS-CoV and SARS-CoV-2 based on the syncodon model.
3.4. Codon usage bias in CoV CDS sequences and ENc-GC3 plot analysis
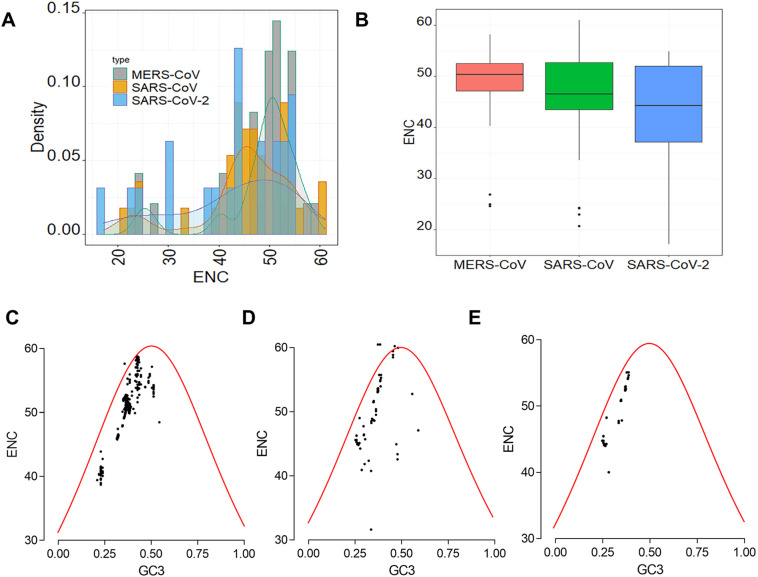
ENc values were computed to evaluate the intra-virus codon bias. Within SARS-CoV-2, the ENc values varied from 17.15 to 54.87, with a mean value of 42.47 ± 11.57 (Fig. 4A, B). For SARS-CoV, the ENc values varied from 20.71 to 61, with a mean value of 45.54 ± 9.72 (Fig. 4A, B). For MERS-CoV, the ENc values varied from 24.98 to 58.27, with a mean value of 50.40 ± 4.58 (Fig. 4A, B). The One-way ANOVA analysis followed by Bonferoni posttest at a 95% confidence level indicated no statistically significant difference in the mean ENc values among MERS, SARS-CoV and SARS-CoV-2 (Fig. 4B). The percentages of CDS sequences with ENc >35 were 76.19, 90.63 and 86.49% for SARS-CoV-2, MERS-CoV and SARS-CoV, respectively. Given the high number of sequences with ENc values >35, we inferred that there was little bias in codon usage in the CoVs.
Fig. 4.
ENc values and ENc-GC3 plots of CDS sequences of human respiratory coronaviruses. A- Distribution of the ENc of CDS sequences of MERS-CoV, SARS-CoV and SARS-CoV-2. B- Boxplot of ENC values of CDS sequences of MERS-CoV, SARS-CoV and SARS-CoV-2. C- ENc-GC3 plot of CDS sequences of MERS-CoV. D- ENc-GC3 plot of CDS sequences of SARS-CoV. E- ENc-GC3 plot of CDS sequences of SARS-CoV-2.
The ENc-GC3 plot was generated in order to assess whether mutational pressure or selection pressure has an impact on codon usage of viral genes. Points over the expected ENc curve suggest mutational pressure while points under the expected ENc curve indicate selection pressure. Fig. 4C and D shows that for MERS-CoV and SARS-CoV, a huge number of CDS sequences were under the expected ENc curve while a lesser number of CDS sequences were represented over this curve. This suggested that selectional pressure, in larger extent, and mutational pression, in lesser extent are the major forces influencing the codon usage in MERS-CoV (Fig. 4C) and SARS-CoV (Fig. 4D). For SARS-CoV-2, all the points were under the expected ENc curve, which suggested that selectional pressure is the exclusive factor influencing codon usage in SARS-CoV-2 (Fig. 4E).
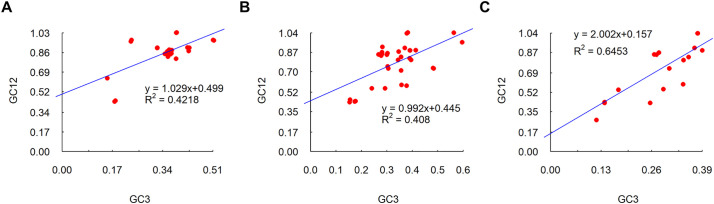
3.5. Neutrality plot
Neutrality plots of GC12 in function of GC3 was generated to assess the extent of mutational pressure on the usage of codons in CoVs. A correlation among GC12 and GC3 indicates mutational forces (Jenkins and Holmes, 2003). For MERS-CoV, SARS-CoV and SARS-CoV-2, GC12 was positively correlated with GC3 (R2 = 0.4218, P < .001 for MERS-CoV (Fig. 5A); R2 = 0.4218, P < .001 for SARS-CoV (Fig. 5B); R2 = 0.4218, P < .001 for SARS-CoV-2 (Fig. 5C)), which indicated the occurrence of mutational pressure. The slopes of the regression equations were 1.029, 0.992 and 2.002 for MERS-CoV, SARS-CoV and SARS-CoV-2, respectively, suggesting that the relative neutrality (mutation pressure) was 10.29%, 9.92% and 20.02% for MERS-CoV, SARS-CoV and SARS-CoV-2 compared to relative natural selection constraints on GC3. The neutrality plot suggested that natural selection forces were stronger than mutational pressure.
Fig. 5.
Neutrality plots of CDS sequences of human respiratory coronaviruses. A- Neutrality plot of CDS sequences of MERS-CoV. B- Neutrality plot of CDS sequences of SARS-CoV. C- Neutrality plot of CDS sequences of SARS-CoV-2.
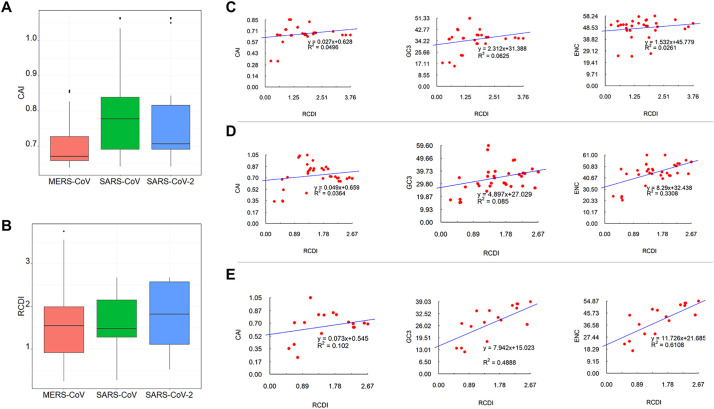
3.6. Measures of virus adaptation
CAI is a hallmark of the expressivity of viral proteins in the host and a defining marker of virus adaptation to a host. Genes with elevated CAI values are those which are adapted to the corresponding host. On the contrary low CAI values are indicative of weak adaptation to the host. The average CAI values were 0.71 ± 0.07, 0.79 ± 0.12, 0.77 ± 0.12 for MERS-CoV, SARS-CoV and SARS-CoV-2 (Fig. 6A), indicating that SARS-CoV was more adapted to human, followed by SARS-CoV-2 and MERS-CoV in this decreasing order.
Fig. 6.
CAI and RCDI of CDS sequences of human respiratory coronaviruses. A- Boxplot of CAI values of CDS sequences of MERS-CoV, SARS-CoV and SARS-CoV-2. B- Boxplot of RCDI values of CDS sequences of MERS-CoV, SARS-CoV and SARS-CoV-2. C- Correlation between RCDI and CAI, GC3 and ENC in MERS-CoV genome. D- Correlation between RCDI and CAI, GC3 and ENC in SARS-CoV genome. E- Correlation between RCDI and CAI, GC3 and ENC in SARS-CoV-2 genome.
RCDI values indicate the cumulative effects of codon biases on the expression of a gene. It is measured by comparing the codon usage of a virus with that of its host. These values also provide insight into the possible co-evolution of virus and host genomes. A lower RCDI value indicates higher adaptation of a virus to its host. Here, we found that the mean RCDI value of SARS-CoV-2 was higher than that of SARS-CoV and MERS-CoV (Fig. 6B). No significant difference was recorded among the three viruses, indicating that these viruses might have the same adaptive capacity for their human host.
The correlation analysis in each virus indicated a marked positive correlation of CAI values with RCDI values (p < .01; R2 = 0.049 for MERS-CoV; p < .01; R2 = 0.0364 for SARS-CoV; p < .01; R2 = 0.102 for SARS-CoV-2) (Fig. 6C-6E). Similarly, positive correlations were observed between RCDI and GC3 on one hand, and RCDI and ENC on the other end (Fig. 6C-6E). Designing virus genome with lower CAI and higher RCDI via modifying codon usage will result in low level viral protein expression and decreased viral infectivity, which important for the development attenuated vaccine candidates. A comparative analysis of RCDI values of the viruses in human showed that SARS-CoV is more adapted than MERS-CoV and SARS-CoV-2.
The analysis of the whole sequence after adaptation to human indicated that the CAI values of the genomic sequences after adaption were 0.957, 0.9511 and 0.9556, respectively (Additional Fig. S1).
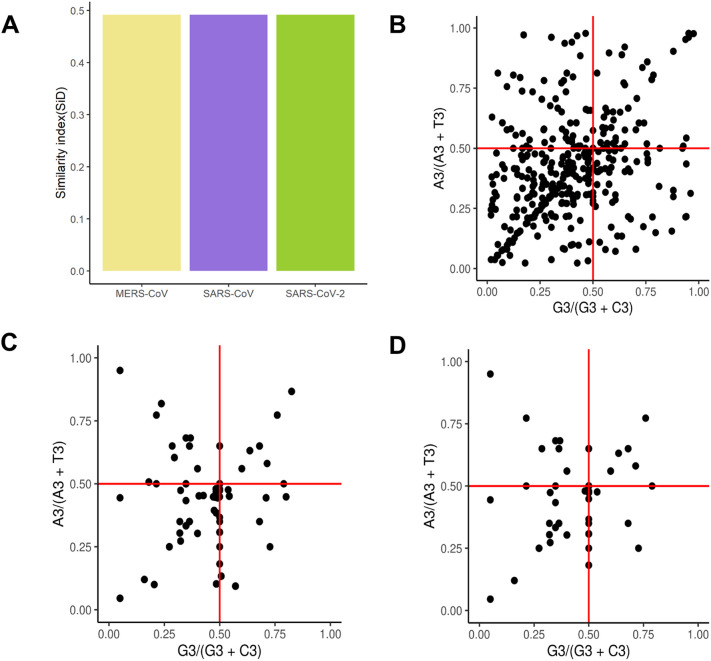
3.7. Similarity index and PR2-bias plot
To assess the influence of the host (human) on codon usage by MERS-CoV, SARS-CoV and SARS-CoV-2, similarity index (SiD) was computed. SiD, ranging from 0 to 1, is an indicator of the similarities in codon usage among the virus and its host. The similarity indexes for MERS-CoV, SARS-CoV and SARS-CoV-2 were 0.4921, 0.4919 and 0.4919, respectively (Fig. 7A). This indicated that the human genome had the highest effect on the MERS-CoV codon bias, followed by SARS-CoV and SARS-CoV-2, though there was no significant difference between the three viruses (Fig. 7A). The parity plot was used to examine natural selection and mutation pressure in viral genes. Variation among A and T and among C and G contents was recorded for MERS-CoV (Fig. 7B), SARS-CoV (Fig. 7C) and SARS-CoV-2 (Fig. 7D), signifying that not only natural selection but also mutation pressure cooperatively affect the codon usage bias of viral genes.
Fig. 7.
Similarity index and PR2-bias plot of CDS sequences of the human respiratory coronaviruses. A-Similarity index of human respiratory coronaviruses relative to their host. B- PR2-bias plot of MERS-CoV CDS sequences. C- PR2-bias plot of SARS-CoV CDS sequences. D- PR2-bias plot of SARS-CoV-2 CDS sequences. The PR2-bias plot was based on GC bias [G3/(G3 + C3)] and AT bias [A3/(A3 + T3)] in the third codon position. The two solid red lines represent both coordinates (ordinate and abscissa) equal to 0.5, where A = T and G = C. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.8. Relative synonymous codon usage (RSCU) analysis
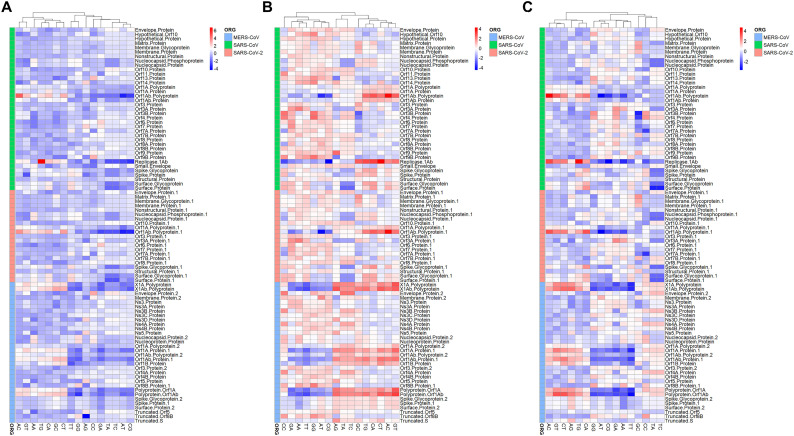
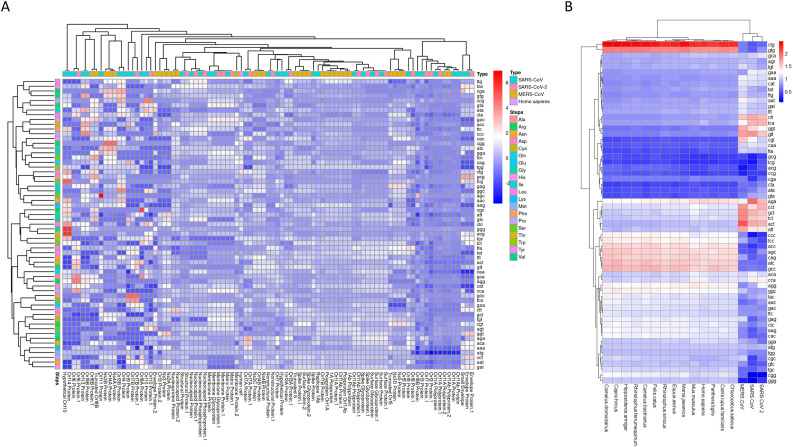
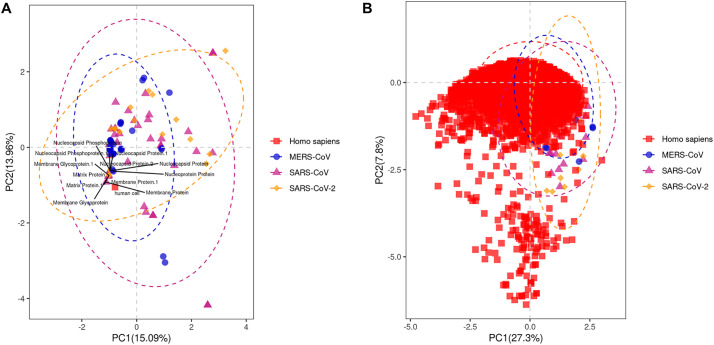
To explore the profiles of synonymous codon usage, we computed the RSCU values for each gene sequence from SARS-CoV-2, SARS-CoV and MERS-CoV. The average RSCU values of human CDS sequences were also calculated. A heatmap showing the profile of the codons in each viral gene and human cell is depicted in Fig. 8A. It can be seen that numerous viral gene shared similar RSCU profile with human cell. In addition, after averaging the RSCU values of each viral genomes, we found that C-and G-ending codons were those generally preferred in human cell and other hosts compared to viral genes (Fig. 8B). In addition, A- and T-ending codons were preferentially used in the viral genomes compared to the hosts (Fig. 8B). In addition, as shown in Fig. 8B, the three CoVs were clustered together on the basis of their RSCU values while their hosts were also clustered together. Moreover, a high similarity of RSCU values between the hosts was recorded (Fig. 8B). The PCA plots of the A-ended, C-ended, G-ended and U-ended for MERS-CoV, SARS-CoV and SARS-CoV-2 based on the RSCU values was depicted in Additional Fig. S2. In general, C-ended and G-ended codon were clustered together for each viral genome. The correlation between codon on the basis of RSCU values were as depicted in Additional Fig. S3. To identify proteins which were shared similar RSCU pattern with the human cells, The PCA plot of genes was generated and indicated that membrane protein, matrix protein, membrane glycoprotein nucleocapsid protein and nucleocapsid phosphoprotein shared the most similar RSCU patterns with human cells (Fig. 9A). To uncover the human genes that could be affected by the virus, we also generated the PCA plot based on the RSCU values of human genes and viral genes. This PCA plot indicated that viral genes of MERS-CoV, SARS-CoV and MERS-CoV-2 shared the similarities with a huge number of human genes (Fig. 9B).
Fig. 8.
Heatmaps representing the RSCU values of respiratory coronaviruses and that of human cell. A- Heatmap based on the RSCU values of each viral gene and the average RSCU of human gene. B- Heatmap based on the average RSCU values of genes in each virus and the average RSCU values of the original and intermediate hosts of the three coronaviruses.
Fig. 9.
PCA analysis of the CDS sequences of human coronaviruses based on RSCU values. A- PCA based on MERS-CoV, SARS-CoV and SARS-CoV-2 CDS sequences and human cell; genes showing high similarity with human cell were visualized on the plot. B- PCA based on MERS-CoV, SARS-CoV and SARS-CoV-2 CDS sequences and human cell CDS sequences.
3.9. Correspondence analysis
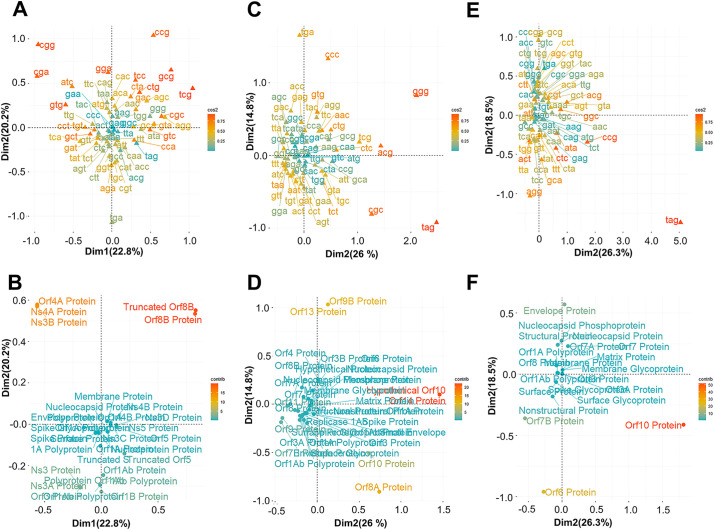
To identify the other factors that shape codon usage bias in CoVs, we performed correspondence analysis which is a statistical method used to study the variation of codon usage bias in genomes. Correspondence analysis reflects the distribution of genes and their corresponding codons, which helps identify potential factors influencing codon usage bias. The correspondence analysis of the variation in the use of codons in the genome of the MERS-CoV, SARS-CoV and SARS-CoV-2 viruses was performed on the basis of the RSCU values. For MERS-CoV, the result revealed that the first axis represented 22.8% of the total variation, while the second axis represented 20.2%, confirming that the first axis is the main factor governing the use of codons in the genes of this virus (Fig. 10A and B). For SARS-CoV, we found that the first axis represented 26% of the total variation, while the second axis represented 14.8% (Fig. 10C and D). For SARS-CoV-2, the first axis represented 26.3% of the total variation, while the second axis represented 18.5% (Fig. 10E and F). The distribution of synonymous codons (Fig. 10A, C and E) showed that codons with C/G endings were the most important factors contributing to variation along both axes, indicating that variations in the use of synonymous codons within the CoVs genes were based on the nucleotide content. For MERS-CoV, we found that the contributions of the truncated Orf8B protein, the Orf8B protein, the Orf4A protein, the NS4A protein and the NS3B protein to the variation on the first and second axes were the most representative (Fig. 10B). For SARS-CoV, the contributions of the hypothetical Orf10 protein, the Orf14 protein, the Orf9B protein, the Orf13 protein, the Orf8A protein and the Orf10 protein to the variation on the first and the second axis were higher than those of other proteins (Fig. 10D). For SARS-CoV-2, the contributions of the Orf10 protein, the Orf6 protein, the Orf7B protein and the envelope protein to the variation on the first and the second axis were more important than those of other proteins (Fig. 10F).
Fig. 10.
Correspondence analysis (CA) of the CDS sequences of human coronaviruses based on RSCU values. A- Plot showing the distribution of codons based on the first and second axes obtained from CA of RSCU values in MERS-CoV. B- Plot showing the distribution of genes based on the first and second axes obtained from CA of RSCU values in MERS-CoV. C- Plot showing the distribution of codons based on the first and second axes obtained from CA of RSCU values in SARS-CoV. D- Plot showing the distribution of genes based on the first and second axes obtained from CA of RSCU values in SARS-CoV. E- Plot showing the distribution of codons based on the first and second axes obtained from CA of RSCU values in SARS-CoV-2. F- Plot showing the distribution of genes based on the first and second axes obtained from CA of RSCU values in SARS-CoV-2.
3.10. Functional roles of coronavirus-responsive human genes
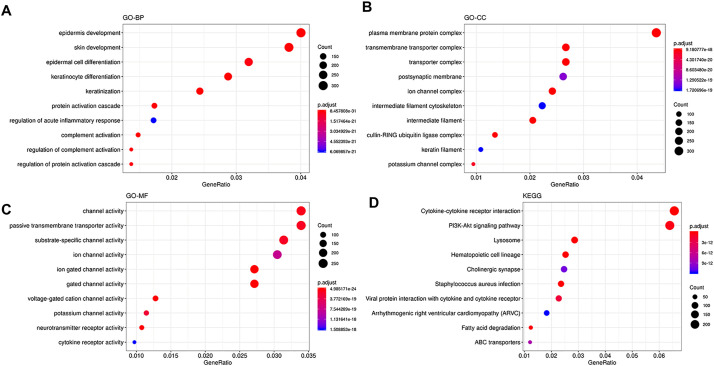
Further, we performed Pearson correlation between human genes and viral genes. Human genes showing correlation r with p < .05 were retained for subsequent analysis. In total, 14,091 human genes were correlated with viral genes (Additional File 1) with 8603 of them showing positive correlations whereas 5488 showed negative correlations (Additional File 1). To uncover the functional role of these genes, KEGG and GO enrichment analysis was performed. As shown in Fig. 11A and Additional File 2, the results showed that all of the human genes correlated with viral genes were enriched in the biological processes (GO-BP) of epidermis development, skin development, epidermal cell differentiation, keratinocyte differentiation, keratinization, protein activation cascade, regulation of acute inflammatory response, complement activation, regulation of complement activation, and regulation of protein activation cascade. In the category of cellular component (GO-CC), plasma membrane protein complex, and transmembrane transporter complex were the most enriched terms (Fig. 11B) while in the category of molecular function (GO-MF), the channel activity, passive transmembrane transporter activity, and substrate-specific channel activity were the most enriched terms (Fig. 11C). In KEGG pathway enrichment analysis, we found that cytokine-cytokine receptor interaction, PI3K-Akt signaling pathway, lysosome, and hematopoietic cell lineage were the most enriched pathways (Fig. 11D). It is worth noting that viral protein interaction with cytokine and cytokine receptor pathway was among the top enriched pathways (Fig. 11D).
Fig. 11.
Functional enrichment analysis results of human genes correlated with viral genes based on the RSCU values. A- Terms enriched in the category of GO-BP (Gene Ontology- Biological processes). B-Terms enriched in the category of GO-CC (Gene Ontology- Cellular component). C- Terms enriched in the category of GO-MF (Gene Ontology-Molecular Function). D- Pathways enriched in the KEGG (Kyoto Encyclopedia of Genes and Genomes) database.
In order to identify the terms directly linked to viral infection, we searched the results for the corresponding terms. We found that 54 terms were related to virus in the category of GO-BP (Additional File 2, sheet “Virus-related terms”). Among these terms, response to virus, regulation of defense response to virus by virus, defense response to virus, viral entry into host cell, and modulation by virus of host morphology or physiology were the most significantly enriched GO-terms. In the category of molecular function, virus receptor activity was the only term related to virus. In KEGG pathway enrichment results, we found that viral protein interaction with cytokine and cytokine receptor, human papillomavirus infection, human cytomegalovirus infection, Kaposi sarcoma-associated herpesvirus infection, viral myocarditis, human immunodeficiency virus 1 infection, human T-cell leukemia virus 1 infection, and Epstein-Barr virus infection pathways were the significant pathways related to viral infection. Our results demonstrated that the codon usage of MERS-CoV, SARS-CoV and SARS-CoV-2 have significant impact on human and gave insight in the related mechanisms of corresponding diseases. In total, 879 genes were involved in these virus-related enrichment terms.
3.11. Screening and functional enrichment analysis of SARS-CoV-2-responsive human genes
In order to further confirm the potential genes affected by SARS-CoV-2, we performed differential expression analysis between uninfected and SARS-CoV-2-infected cells. For the A549, A549-ACE2, Calu3 and NHBE cells, we obtained 460, 1109, 1967 and 117 DEGs, respectively (Additional File 3). After merging of DEGs from the four cell lines, we obtained LGALS9, ICAM1, CCL20 and IL6 as common genes. In addition, the combination of the results from the four cell lines led to 3067 DEGs in total. Further, we merged the results of differential expression with those of the RSCU-based correlation analysis described above. The merging of DEGs obtained by combination of DEGs from the four cell lines with the RSCU-based correlation data allowed the identification of common 1348 DEGs as SARS-CoV-2 responsive genes while merging of DEGs obtained by merging of differential expression analysis results and those of RSCU-based correlation analysis indicated that LGALS9, ICAM1, CCL20 and IL6 were the common genes found in the differential expression analysis from the four cell lines and RSCU-based correlation analysis.
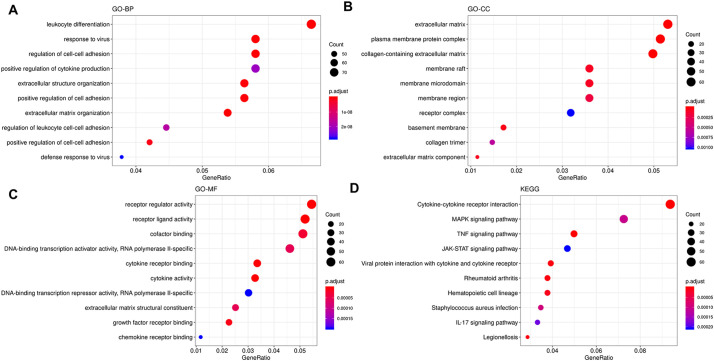
Functional analysis of the 1348 genes affected by both codon usage bias and differential expression (Fig. 12, Additional File 4) indicated that these genes were majorly enriched in biological processes (GO-BP, Fig. 12A) of leukocyte differentiation, response to virus, regulation of cell-cell adhesion, positive regulation of cytokine production. The most enriched terms in the category of cellular component (GO-CC, Fig. 12B) were extracellular matrix and plasma membrane protein complex while the representative molecular functions (GO-MF, Fig. 12C) were receptor regulator activity, receptor ligand activity, and cofactor binding. The major KEGG pathways (Fig. 12D) were cytokine-cytokine receptor interaction, MAPK signaling pathway, TNF signaling pathway, JAK-STAT signaling pathway, and viral protein interaction with cytokine and cytokine receptor.
Fig. 12.
Functional enrichment analysis results of human genes correlated with viral genes based on the RSCU values and that are differentially expressed in human cells infected by SARS-CoV-2. A- Terms enriched in the category of GO-BP (Gene Ontology- Biological processes). B-Terms enriched in the category of GO-CC (Gene Ontology- Cellular component). C- Terms enriched in the category of GO-MF (Gene Ontology-Molecular Function). D- Pathways enriched in the KEGG (Kyoto Encyclopedia of Genes and Genomes) database.
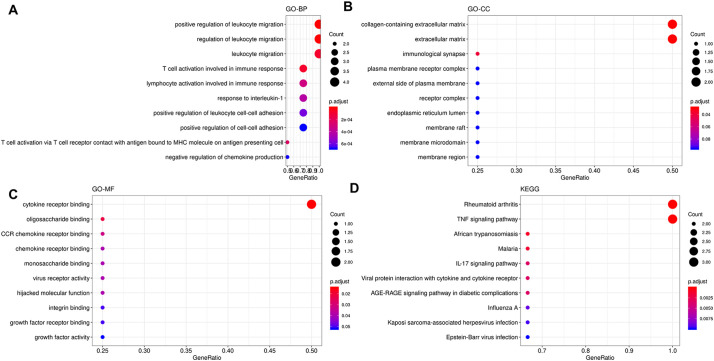
Functional analysis of the 4 common genes obtained from merging of the DEGs obtained from differential expression analysis of each cell line and the RSCU-based correlation analysis indicated that these genes were enriched in biological processes of positive regulation of leukocyte migration, regulation of leukocyte migration, leukocyte migration, T cell activation involved in immune response and other immune-related processes (GO-BP, Fig. 13A). The most enriched cellular components were collagen-containing extracellular matrix, extracellular matrix, immunological synapse and plasma membrane receptor complex (GO-CC, Fig. 13B). The most representative molecular functions were cytokine receptor binding, oligosaccharide binding, and CCR chemokine receptor binding (GO-MF, Fig. 13C). The most representative pathways were rheumatoid arthritis, TNF signaling pathway, African trypanosomiasis, malaria, IL-17 signaling pathway, viral protein interaction with cytokine and cytokine receptor, AGE-RAGE signaling pathway in diabetic complications, and influenza related pathways (Fig. 13D).
Fig. 13.
Functional enrichment analysis results of the 4 common human genes considering all the comparative instances. A- Terms enriched in the category of GO-BP (Gene Ontology- Biological processes). B-Terms enriched in the category of GO-CC (Gene Ontology- Cellular component). C- Terms enriched in the category of GO-MF (Gene Ontology-Molecular Function). D- Pathways enriched in the KEGG (Kyoto Encyclopedia of Genes and Genomes) database.
3.12. Prediction of drug candidates for treating viral infection
Using the Drug-Gene Interaction database (DGIdb), we identified drug candidates targeting the potential human genes affected by viral infection. All the human genes showing significant correlations with the viral (MERS-CoV, SARS-CoV and SARS-CoV-2) genes were first used for drug prediction. The results were summarized in Additional File 5. A total of 5650 drugs were predicted. The drug CHEMBL1161866 had the highest number (78) of target genes in the list of retrieved genes, followed by L-glutamate (54 target genes), staurosporine (49 target genes), celecoxib (47 target genes), clozapine (45 target genes), dalfampridine (41 target genes), fluorouracil (41 target genes), olanzapine (41 target genes) and nerispirdine (40 target genes). Among the remaining genes we found hydroxychloroquine (3 target genes) and chloroquine (2 target genes) which have been recently proposed as therapeutics for COVID-19.
Further, based on genes screened by the intersection of the 3067 DEGs between SARS-CoV-2-infected cells and uninfected cells with the RSCU-based correlation genes, we predicted a total of 2135 drugs (Additional File 6). The drugs with the highest number of target genes were paclitaxel (15 targets), CHEMBL1161866 (14 targets), cyclosporine (14 targets), staurosporine (14 targets), alcohol (13 targets), phorbol myristate acetate (13 targets), dexamethasone (12 targets), fluorouracil (12 targets), tretinoin (12 targets) and ocriplasmin (11 targets). Hydroxychloroquine (2 target genes) was also among the retrieved drugs. Based on the four common genes that were found in all of the comparison analyses, we predicted 30 drugs, with 24 of them targeting IL6 and six of them targeting ICAM1.
When considering the 879 genes involved in virus-related GO and KEGG enrichment terms, we predicted 2014 drugs (Additional File 7). Among these drugs, ocriplasmin (26 target genes), staurosporine (18 target genes), paclitaxel (16 target genes), cyclosporine (15 target genes), tretinoin (15 target genes), collagenase clostridium histolyticum (14 target genes), CHEMBL1213492 (13 target genes), dasatinib (13 target genes), CUDC-101 (12 target genes), fluorouracil (12 target genes), panobinostat (12 target genes), romidepsin (12 target genes), valproic acid (12 target genes) and vorinostat (12 target genes) were those with the highest number of target genes. We inferred that the list of drugs found in this study could be useful for the treatment of COVID-19.
4. Discussion
SARS-CoV-2 is currently a virus that has caused serious public health problems worldwide. There is an urgent need for viable treatments and the development of effective vaccines to contain it. Information on the interaction between this virus and similar viruses with their human host can facilitate the identification of prophylactic and therapeutic measures. Here, based on the use of codons, we tried to identify the susceptibility of humans to carry the three respiratory viruses. Based on the gene analysis, there are remarkable genomic differences between the CDS sequences of SARS-CoV-2 and SARS-CoV and MERS-CoV with respect to the use of codons. Our results showed that the GC content and the GC3 content of MERS-CoV, SARS-CoV and SARS-CoV-2 were relatively low, indicating that the U/A-ending codons are preferentially used compared to the GC-ending codons. We also noted that selection pressure plays a major role in the codon usage of the three viruses and that in addition to this selection pressure, some SARS-CoV and MERS-CoV genes are under the influence of mutation forces. The CAI index of the different viruses showed that SARS-CoV and SARS-CoV-2 are highly adapted to human sequences than MERS-CoV and that human cells were very favorable to virus replication, given the high CAI values. Based on the virus-interaction analysis, we discovered potential human genes involved in the pathogenesis of CoV infections in human. Drug-gene interaction analysis allowed the discovery of available drugs for potential treatment of the symptoms of CoV infections, particularly COVID-19.
The occurrence of dinucleotide codons is a defining variable for evaluating mutational and codon usage biases as well as the effect of compositional constraints and selective pressure in a given genome (Ellis et al., 1993; Smutzer and Chamberlin, 1994; Yamagishi et al., 2002). Studies have shown that the relative abundance of dinucleotides has a strong influence on the use of viral codons (Tan et al., 2004). GC dinucleotide depletion is a selective force that has a direct impact on the frequency of GC dinucleotide codons (Johnson, 1990; Tan et al., 2004). In the present study, we found an under-representation of GC dinucleotides. This low GC dinucleotide frequency is a strategy of the virus to avoid host immunization that identifies any sequence containing unmethylated GC dinucleotides as an antigen to be degraded (Vetsigian and Goldenfeld, 2009; Woo et al., 2007). In addition, the low frequency of GC dinucleotides may be a direct cause of methylation of cytosine residues induced by genetic events such as X chromosome inactivation, genetic silencing and genetic fingerprinting (Li and Zhang, 2014; Zhang et al., 2014). Spontaneous deamination of methylated cytosines into thymine may therefore lead to low levels of GC dinucleotides. An under-representation of GC dinucleotides has been similarly observed in the genome of various viruses (Castells et al., 2017; Rothberg and Wimmer, 1981; Tan et al., 2004). Studies indicate that artificially increasing the frequency of GC dinucleotides attenuates the virus and inhibits its replication (Takata et al., 2017). This approach could be adopted in the search for vaccines against respiratory viruses such as the SARS-CoV2 responsible for COVID-19.
The mean GC3 contents were 0.29, 0.35 and 0.35 for SARS-CoV-2, SARS-CoV and MERS-CoV, respectively. If the use of the codon is only affected by the GC3 content, there is mutation pressure and, in this case, the ENc value is above the predicted ENc curve (Gu et al., 2004; Zhong et al., 2007). In all three genomes of MERS-CoV, SARS-CoV and SARS-CoV-2, ENc values for the vast majority of CDS sequences were below the predicted ENc curve, suggesting that selection pressure is the main force impacting codon utilization in these viruses. For SARS-CoV-2, ENc values for some CDS sequences were above the expected ENc curve, suggesting that these sequences are under the force of mutational pressure. Subsequently, we also examined the role of selection pressure using neutrality plots. A significant correlation between GC12 and GC3 with a slope of the regression line around 1 indicates that the mutation pressure is the key coefficient that affects codon use. On the other hand, a slope of zero or close to the horizontal line suggests that selection pressure is the major factor influencing codon use. The slopes of the regression lines were 1.029, 0.992 and 2.002 for MERS-CoV, SARS-CoV and SARS-CoV-2, respectively, indicating that the force of natural selection takes precedence over the force of mutation.
CAI, RCDI and similarity indicators were evaluated and verified between MERS-CoV, SARS-CoV and SARS-CoV-2 for their host. The mean CAI values were found to be 0.71 ± 0.07, 0.79 ± 0.12, 0.77 ± 0.12 for MERS-CoV, SARS-CoV and SARS-CoV-2, respectively. This result shows that SARS-CoV is more adapted to humans compared to SARS-CoV-2 and MERS-CoV, with MERS-CoV being the least adapted to humans among the three viruses. In addition, the ENc is a parameter correlated with a shift caused by mutation or selection pressure. The correlation between CAI and ENc indicates a relative balance between selection and mutation pressure (Vicario et al., 2007). A positively significant correlation between CAI and ENc observed in our study indicates that high expressivity is partly associated with ENc and confirms the primacy of selection pressure over mutation strength in the genomes of SARS-CoV-2 and MERS-CoV.
The similarity index (SiD) analysis showed that the human genome has the greatest impact on the use of codons by SARS-CoV-2 and MERS-CoV. Similarity indicators have been reported for other viruses, including the Zika virus (Butt et al., 2016), Influenza D virus (Yan et al., 2019) and the Henipaviruses (Kumar et al., 2018). In this study, the mean SiD values indicated that SARS-CoV, SARS-CoV-2 and MERS-CoV can replicate efficiently in humans without significant effect on host codon utilization.
Currently, the mechanism underlying the pathogenesis of COVID-19 is unknown. Making use of the results obtained in the present study, we found human genes that could be affected by viral (SARS-CoV-2, SARS-CoV and MERS-CoV) genes. Similar to previous studies (Miller et al., 2017), we found that the codon usage of the SARS-CoV, SARS-CoV-2, and MERS-CoV were significantly correlated with a huge number of human genes. The GO and KEGG enrichment analysis of these genes allowed us to uncover their functional roles. Biological processes related to epidermis development, keratinization, protein activation cascade, regulation of acute inflammatory response and regulation of complement activation were the most represented. The enrichment of genes in skin-related biological processes can be explained by the fact that the skin may play a vital role in the protection of the host against the virus (Friedman, 2006). Keratinization may also help the virus to skip the host immune system (Friedman, 2006). Previous studies indicated the activation of protein cascades such as coagulation protease cascade in viral infections (Antoniak and Mackman, 2014). Our study indicated that human genes correlated with viral genes on the basis of RSCU values were involved in protein activation cascade. In addition, the regulation of complement activation found in the present study corroborated with previous studies indicating that complement plays a significant role in the fight against viral infections (Stoermer and Morrison, 2011) and viral evasion to the host immune-system (Agrawal et al., 2017). Also, we found that 879 genes were involved in “virus”-related GO terms such as host response to virus infection and virus entry in the host. Cytokine-cytokine receptor interaction, hematopoietic cell lineage, lysosome, fatty acid degradation, PI3K-Akt signaling pathway and viral protein interaction with cytokine and cytokine receptor were the pathways affected by the dysregulated human genes. These enrichment in virus-related terms, especially terms associated with the interaction between the virus and the host, indicated the reliability of the approach adopted in this study. In addition, recent studies have reported that segmental pulmonary emboli (hemoptysis) occurs in COVID-19 infection (Casey et al., 2020), which corroborated with our finding that the pathway of hematopoietic cell lineage is potentially dysregulated in CoV infections. Thus, our findings shed light on the mechanisms of CoV pathogenesis in the host.
Currently, there is no precise therapy for treating the ongoing COVID-19. Here, we predicted a set of drugs that could be efficient in the treatment of COVID-19 and other CoV infections. In the early search of treatment for SARS-CoV-2 infection, researchers have proposed hydroxycholoroquine and chloroquine as treatment for COVID-19 (Singh et al., 2020). Our study also predicted these drugs as a treatment for COVID-19 and other CoV infections. However, the limited number of genes targeted by these drugs indicated that their efficiencies may be limited. Nevertheless, these results ascertain the credibility of the present study. In our study, we found that some drugs had huge numbers of target genes. Thus, we suggested that these drugs could be the most potentially efficient drugs for counteracting the deleterious pathogenic effect of the viral infection. As a model, CHEMBL1161866, also known as dihydronicotinamide adenine dinucleotide or NADH had the highest number of target genes among the potential CoV-responsive human genes. Nicotinamide adenine dinucleotide is a cofactor molecule allowing the action of certain enzymes and the proper functioning of our cells. It exists in reduced form (NADH) or in oxidized form (NAD+). NAD+ plays a key role in the production of energy by the mitochondria and its metabolism is considered a drug target for many human diseases such as tumors, inflammatory, cardiovascular and neurodegenerative diseases. NAD+ also plays a first-order role in the regulation of the innate or adaptive immune response, which gives it a very important place in the mechanisms of interaction between the host and pathogenic germs (Mesquita et al., 2016) and, therefore, in the treatment of infectious diseases. Nicotinamide and its derivatives have remarkable therapeutic implications for viral infections due to the role played by NAD+ metabolism in host infections (Singhal and Cheng, 2019). Studies have shown that NAD+ plays an important regulatory role in depressing genes involved in the response mechanisms to persistent adenovirus infection of lymphocytes (Dickherber and Garnett-Benson, 2019). Nicotinamide has also been shown to be an antimicrobial agent with activity against the tuberculosis pathogen (Mycobacterium tuberculosis) and the human immunodeficiency virus (HIV) (Murray, 2003). Reports of other scientific research indicate that ethanol, through its inducing action on lipid metabolism and NADH/NAD+, is effective in inhibiting the replication of the hepatitis C virus (Seronello et al., 2010). Our study also found alcohol as a potential drug against CoV infections. However, this should be dealt with caution as studies have shown that alcohol regulates the inflammatory and anti-viral activities of monocytes but that prolonged alcohol consumption reduces the anti-viral activity of interferon type 1 (IFN) while inducing inflammation through modulation of the pro-inflammatory cytokine TNFα (Pang et al., 2011). Our study uncovered L-GLUTAMATE as a potential therapeutic drug for CoV infections. Our results reaffirm those of previous studies stating that glutamate plays an important role against viral infections such as HIV-1 (Erdmann et al., 2007) and HIV-1 associated neuro-invasive dementia (Zhao et al., 2004). Thus, we believe that its administration to CoV-infected patients will probably be salutary for patients especially due to the fact that studies have demonstrated that the neuroinvasive ability of SARS-CoV2 is partially associated with the respiratory failure of COVID-19 patients (Li et al., 2020).
Taken together, our study predicted a panoply of drugs with potential in the treatment of CoV infections. Some of the predicted drugs have shown antiviral potential in previous studies whereas the role of others in the treatments of viral infections is unknown. Further studies combining efforts from practitioners and scientific researchers are needed to validate these drugs as therapeutics for COVID-19 and other CoV diseases. Especially, in vitro, animal and clinical studies are needed to confirm the potential of the predicted drugs in treating CoV infections, especially the current COVID-19 pandemics.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
The following are the supplementary data related to this article.Additional Fig. S1. Adaptiveness of human respiratory coronaviruses to human host analyzed from JCAT.
Additional Fig. S2. PCA analysis of A-, C-, G, and U-ended codons in the CDS sequences of human corona viruses based on RSCU values. A- PCA based on MERS-CoV CDS sequences. B- PCA based on SARS-CoV CDS sequences. B- PCA based on SARS-CoV-2 CDS sequences.
Additional Fig. S3. Correlation analysis of codons in the CDS sequences of human coronaviruses based on RSCU values. A- Correlation based on MERS-CoV CDS sequences. B- Correlation based on SARS-CoV CDS sequences. B- Correlation based on SARS-CoV-2 CDS sequences.
Results of the correlation analysis between human genes and viral genes based on the RSCU values. Only potential human genes affected by viral infection and showing correlation significance of <0.05 with viral genes were selected.
Functional enrichment analysis of potential human genes affected by viral infection screened based on the results of RSCU-correlation between human genes and viral genes.
Results of differential gene expression analysis of SARS-CoV-2-infected and uninfected human cell lines.
Functional enrichment analysis of potential human genes obtained based on common genes obtained by the intersection of RSCU-based correlation results and differential gene expression analysis results.
Prediction of the potential therapeutic drugs based on all of the human genes correlated with viral genes on the basis of RSCU values.
Prediction of the potential therapeutic drugs based on common genes obtained by the intersection of RSCU-based correlation results and differential gene expression analysis results
Prediction of the potential therapeutic drugs based on genes directly enriched in virus functional terms.
Declaration of competing interest
The authors declared that they have no conflict of interest.
References
- Agrawal P., Nawadkar R., Ojha H., Kumar J., Sahu A. Complement evasion strategies of viruses: An overview. Front. Microbiol. 2017;8:1117. doi: 10.3389/fmicb.2017.01117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12 doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn D.G., Shin H.J., Kim M.H., Lee S., Kim H.S., Myoung J., Kim B.T., Kim S.J. Current status of epidemiology, diagnosis, therapeutics, and vaccines for novel coronavirus disease 2019 (COVID-19) J. Microbiol. Biotechnol. 2020;30:313–324. doi: 10.4014/jmb.2003.03011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniak S., Mackman N. Multiple roles of the coagulation protease cascade during virus infection. Blood. 2014;123:2605–2613. doi: 10.1182/blood-2013-09-526277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar A.M., Soudy M., Mohamed R. vhcub: Virus-host codon usage co-adaptation analysis. F1000Res. 2019;8:2137. doi: 10.12688/f1000research.21763.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., tenOever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. e1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt A.M., Nasrullah I., Qamar R., Tong Y. Evolution of codon usage in Zika virus genomes is host and vector specific. Emerg. Microbes Infect. 2016;5 doi: 10.1038/emi.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone A., Zinovyev A., Képès F. Codon adaptation index as a measure of dominating codon bias. Bioinformatics. 2003;19:2005–2015. doi: 10.1093/bioinformatics/btg272. [DOI] [PubMed] [Google Scholar]
- Casey K., Iteen A., Nicolini R., Auten J. COVID-19 pneumonia with hemoptysis: Acute segmental pulmonary emboli associated with novel coronavirus infection. Am. J. Emerg. Med. 2020;38:1544.e1541–1544.e1543. doi: 10.1016/j.ajem.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castells M., Victoria M., Colina R., Musto H., Cristina J. Genome-wide analysis of codon usage bias in Bovine Coronavirus. Virol. J. 2017;14:115. doi: 10.1186/s12985-017-0780-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cava C., Bertoli G., Castiglioni I. In silico discovery of candidate drugs against Covid-19. Viruses. 2020;12 doi: 10.3390/v12040404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Xu Q., Yuan X., Li X., Zhu T., Ma Y., Chen J.L. Analysis of the codon usage pattern in Middle East Respiratory Syndrome Coronavirus. Oncotarget. 2017;8:110337–110349. doi: 10.18632/oncotarget.22738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotto K.C., Wagner A.H., Feng Y.Y., Kiwala S., Coffman A.C., Spies G., Wollam A., Spies N.C., Griffith O.L., Griffith M. DGIdb 3.0: A redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018;46:D1068–d1073. doi: 10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzentas N. Circoletto: Visualizing sequence similarity with Circos. Bioinformatics. 2010;26:2620–2621. doi: 10.1093/bioinformatics/btq484. [DOI] [PubMed] [Google Scholar]
- Deb B., Uddin A., Chakraborty S. Codon usage pattern and its influencing factors in different genomes of hepadnaviruses. Arch. Virol. 2020;165:557–570. doi: 10.1007/s00705-020-04533-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickherber M.L., Garnett-Benson C. NAD-linked mechanisms of gene de-repression and a novel role for CtBP in persistent adenovirus infection of lymphocytes. Virol. J. 2019;16:161. doi: 10.1186/s12985-019-1265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilucca M., Forcelloni S., Georgakilas A.G., Giansanti A., Pavlopoulou A. Codon usage and phenotypic divergences of SARS-CoV-2 genes. Viruses. 2020;12 doi: 10.3390/v12050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248:117477. doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J., Griffin H., Morrison D., Johnson A.M. Analysis of dinucleotide frequency and codon usage in the phylum Apicomplexa. Gene. 1993;126:163–170. doi: 10.1016/0378-1119(93)90363-8. [DOI] [PubMed] [Google Scholar]
- Erdmann N., Zhao J., Lopez A.L., Herek S., Curthoys N., Hexum T.D., Tsukamoto T., Ferraris D., Zheng J. Glutamate production by HIV-1 infected human macrophage is blocked by the inhibition of glutaminase. J. Neurochem. 2007;102:539–549. doi: 10.1111/j.1471-4159.2007.04594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman H.M. Keratin, a dual role in herpes simplex virus pathogenesis. J. Clin. Virol. 2006;35:103–105. doi: 10.1016/j.jcv.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Gu W., Zhou T., Ma J., Sun X., Lu Z. Analysis of synonymous codon usage in SARS Coronavirus and other viruses in the Nidovirales. Virus Res. 2004;101:155–161. doi: 10.1016/j.virusres.2004.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Shen X., Irwin D.M., Shen Y. Avian influenza A viruses H5Nx (N1, N2, N6 and N8) show different adaptations of their codon usage patterns to their hosts. J. Inf. Secur. 2019;79:174–187. doi: 10.1016/j.jinf.2019.04.013. [DOI] [PubMed] [Google Scholar]
- Jenkins G.M., Holmes E.C. The extent of codon usage bias in human RNA viruses and its evolutionary origin. Virus Res. 2003;92:1–7. doi: 10.1016/s0168-1702(02)00309-x. [DOI] [PubMed] [Google Scholar]
- Johnson A.M. Comparison of dinucleotide frequency and codon usage in Toxoplasma and Plasmodium: Evolutionary implications. J. Mol. Evol. 1990;30:383–387. doi: 10.1007/BF02101892. [DOI] [PubMed] [Google Scholar]
- Kandeel M., Altaher A. Synonymous and biased codon usage by MERS CoV papain-like and 3CL-proteases. Biol. Pharm. Bull. 2017;40:1086–1091. doi: 10.1248/bpb.b17-00168. [DOI] [PubMed] [Google Scholar]
- Kitano H. Systems biology: A brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- Kumar N., Kulkarni D.D., Lee B., Kaushik R., Bhatia S., Sood R., Pateriya A.K., Bhat S., Singh V.P. Evolution of codon usage bias in henipaviruses is governed by natural selection and is host-specific. Viruses. 2018:10. doi: 10.3390/v10110604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunec D., Osterrieder N. Codon pair bias is a direct consequence of dinucleotide bias. Cell Rep. 2016;14:55–67. doi: 10.1016/j.celrep.2015.12.011. [DOI] [PubMed] [Google Scholar]
- Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Weon S., Lee S., Kang C. Relative codon adaptation index, a sensitive measure of codon usage bias. Evol. Bioinformatics Online. 2010;6:47–55. doi: 10.4137/ebo.s4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E., Zhang Y. DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 2014;6:a019133. doi: 10.1101/cshperspect.a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.C., Bai W.Z., Hashikawa T. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J. Med. Virol. 2020;92:552–555. doi: 10.1002/jmv.25728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., He M., Teng C.B. Evolution of the vesicular stomatitis viruses: Divergence and codon usage bias. Virus Res. 2014;192:46–51. doi: 10.1016/j.virusres.2014.08.013. [DOI] [PubMed] [Google Scholar]
- Licastro D., Rajasekharan S., Dal Monego S., Segat L., D’Agaro P., Marcello A. Isolation and full-length genome characterization of SARS-CoV-2 from COVID-19 cases in northern Italy. J. Virol. 2020;94:e00543–e00620. doi: 10.1128/JVI.00543-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Lu L., Cao W., Li T. Hypothesis for potential pathogenesis of SARS-CoV-2 infection-a review of immune changes in patients with viral pneumonia. Emerg. Microbes Infect. 2020;9:727–732. doi: 10.1080/22221751.2020.1746199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita I., Varela P., Belinha A., Gaifem J., Laforge M., Vergnes B., Estaquier J., Silvestre R. Exploring NAD+ metabolism in host-pathogen interactions. Cell. Mol. Life Sci. 2016;73:1225–1236. doi: 10.1007/s00018-015-2119-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.B., Hippen A.A., Wright S.M., Morris C., Ridge P.G. Human viruses have codon usage biases that match highly expressed proteins in the tissues they infect. Biomed. Genet. Genomics. 2017;2:1–5. [Google Scholar]
- Morgunov A.S., Babu M.M. Optimizing membrane-protein biogenesis through nonoptimal-codon usage. Nat. Struct. Mol. Biol. 2014;21:1023–1025. doi: 10.1038/nsmb.2926. [DOI] [PubMed] [Google Scholar]
- Mueller S., Papamichail D., Coleman J.R., Skiena S., Wimmer E. Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. J. Virol. 2006;80:9687–9696. doi: 10.1128/JVI.00738-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M.F. Nicotinamide: An oral antimicrobial agent with activity against both Mycobacterium tuberculosis and human immunodeficiency virus. Clin. Infect. Dis. 2003;36:453–460. doi: 10.1086/367544. [DOI] [PubMed] [Google Scholar]
- Oldfield C.J., Peng Z., Uversky V.N., Kurgan L. Codon selection reduces GC content bias in nucleic acids encoding for intrinsically disordered proteins. Cell. Mol. Life Sci. 2020;77:149–160. doi: 10.1007/s00018-019-03166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J., Chen R., Yan Z., Ou S., Dong N., Lu G., Li S. Codon usage bias of H3N8 equine influenza virus - An evolutionary perspective. J. Inf. Secur. 2020;80:671–693. doi: 10.1016/j.jinf.2020.01.004. [DOI] [PubMed] [Google Scholar]
- Pang M., Bala S., Kodys K., Catalano D., Szabo G. Inhibition of TLR8- and TLR4-induced Type I IFN induction by alcohol is different from its effects on inflammatory cytokine production in monocytes. BMC Immunol. 2011;12:55. doi: 10.1186/1471-2172-12-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect. Genet. Evol. 2020;79:104212. doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020;38:1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- Ren J.L., Zhang A.H., Wang X.J. Pharmacol. Res. 2020;55:104743. doi: 10.1016/j.phrs.2020.104743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson B. Computers and viral diseases. Preliminary bioinformatics studies on the design of a synthetic vaccine and a preventative peptidomimetic antagonist against the SARS-CoV-2 (2019-nCoV, COVID-19) coronavirus. Comput. Biol. Med. 2020;119:103670. doi: 10.1016/j.compbiomed.2020.103670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg P.G., Wimmer E. Mononucleotide and dinucleotide frequencies, and codon usage in poliovirion RNA. Nucleic Acids Res. 1981;9:6221–6229. doi: 10.1093/nar/9.23.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah R., Rodriguez-Morales A.J., Jha R., Chu D.K.W., Gu H., Peiris M., Bastola A., Lal B.K., Ojha H.C., Rabaan A.A., Zambrano L.I., Costello A., Morita K., Pandey B.D., Poon L.L.M. Complete genome sequence of a 2019 novel coronavirus (SARS-CoV-2) strain isolated in Nepal. Microbiol. Resour. Announc. 2020;9 doi: 10.1128/MRA.00169-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seronello S., Ito C., Wakita T., Choi J. Ethanol enhances hepatitis C virus replication through lipid metabolism and elevated NADH/NAD+ J. Biol. Chem. 2010;285:845–854. doi: 10.1074/jbc.M109.045740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton N.R., Ebel G.D. Effects of arbovirus multi-host life cycles on dinucleotide and codon usage patterns. Viruses. 2019;11 doi: 10.3390/v11070643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.M., Li W.H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol. 1986;24:28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- Sheikh A., Al-Taher A., Al-Nazawi M., Al-Mubarak A.I., Kandeel M. Analysis of preferred codon usage in the coronavirus N genes and their implications for genome evolution and vaccine design. J. Virol. Methods. 2020;277:113806. doi: 10.1016/j.jviromet.2019.113806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.K., Singh A., Shaikh A., Singh R., Misra A. Chloroquine and hydroxychloroquine in the treatment of COVID-19 with or without diabetes: A systematic search and a narrative review with a special reference to India and other developing countries. Diabetes Metab. Syndr. 2020;14:241–246. doi: 10.1016/j.dsx.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal A., Cheng C.Y. Host NAD+ metabolism and infections: Therapeutic implications. Int. Immunol. 2019;31:59–67. doi: 10.1093/intimm/dxy068. [DOI] [PubMed] [Google Scholar]
- Smutzer G., Chamberlin L.L. Dinucleotide frequencies and codon usage in jawless and cartilaginous fishes. Mol. Mar. Biol. Biotechnol. 1994;3:112–119. [PubMed] [Google Scholar]
- Stano M., Beke G., Klucar L. viruSITE-integrated database for viral genomics. Database (Oxford) 2016;2016 doi: 10.1093/database/baw162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbing J., Phelan A., Griffin I., Tucker C., Oechsle O., Smith D., Richardson P. COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infect. Dis. 2020;20:400–402. doi: 10.1016/S1473-3099(20)30132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanelli P., Faggioni G., Lo Presti A., Fiore S., Marchi A., Benedetti E., Fabiani C., Anselmo A., Ciammaruconi A., Fortunato A., De Santis R., Fillo S., Capobianchi M.R., Gismondo M.R., Ciervo A., Rezza G., Castrucci M.R., Lista F., On Behalf Of Iss Covid-Study, G. Whole genome and phylogenetic analysis of two SARS-CoV-2 strains isolated in Italy in January and February 2020: Additional clues on multiple introductions and further circulation in Europe. Euro. Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.13.2000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoermer K.A., Morrison T.E. Complement and viral pathogenesis. Virology. 2011;411:362–373. doi: 10.1016/j.virol.2010.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton T.C., Subbarao K. Development of animal models against emerging coronaviruses: From SARS to MERS coronavirus. Virology. 2015;479-480:247–258. doi: 10.1016/j.virol.2015.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M.A., Gonçalves-Carneiro D., Zang T.M., Soll S.J., York A., Blanco-Melo D., Bieniasz P.D. CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature. 2017;550:124–127. doi: 10.1038/nature24039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D.Y., Hair Bejo M., Aini I., Omar A.R., Goh Y.M. Base usage and dinucleotide frequency of infectious bursal disease virus. Virus Genes. 2004;28:41–53. doi: 10.1023/B:VIRU.0000012262.89898.c7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tort F.L., Castells M., Cristina J. A comprehensive analysis of genome composition and codon usage patterns of emerging coronaviruses. Virus Res. 2020;283:197976. doi: 10.1016/j.virusres.2020.197976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Veesler D. Structural insights into coronavirus entry. Adv. Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetsigian K., Goldenfeld N. Genome rhetoric and the emergence of compositional bias. Proc. Natl. Acad. Sci. U. S. A. 2009;106:215–220. doi: 10.1073/pnas.0810122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicario S., Moriyama E.N., Powell J.R. Codon usage in twelve species of Drosophila. BMC Evol. Biol. 2007;7:226. doi: 10.1186/1471-2148-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor M.P., Acharya D., Begum T., Ghosh T.C. The optimization of mRNA expression level by its intrinsic properties-insights from codon usage pattern and structural stability of mRNA. Genomics. 2019;111:1292–1297. doi: 10.1016/j.ygeno.2018.08.009. [DOI] [PubMed] [Google Scholar]
- Wold S., Esbensen K., Geladi P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987;2:37–52. [Google Scholar]
- Woo P.C., Wong B.H., Huang Y., Lau S.K., Yuen K.-Y. Cytosine deamination and selection of CpG suppressed clones are the two major independent biological forces that shape codon usage bias in coronaviruses. Virology. 2007;369:431–442. doi: 10.1016/j.virol.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright F. The ‘effective number of codons’ used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- Wright C., Ross C., Mc Goldrick N. Are hydroxychloroquine and chloroquine effective in the treatment of SARS-COV-2 (COVID-19)? Evid. Based Dent. 2020;21:64–65. doi: 10.1038/s41432-020-0098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.W., Wu X.X., Jiang X.G., Xu K.J., Ying L.J., Ma C.L., Li S.B., Wang H.Y., Zhang S., Gao H.N., Sheng J.F., Cai H.L., Qiu Y.Q., Li L.J. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: Retrospective case series. BMJ. 2020;368:m606. doi: 10.1136/bmj.m606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav P.D., Potdar V.A., Choudhary M.L., Nyayanit D.A., Agrawal M., Jadhav S.M., Majumdar T.D., Shete-Aich A., Basu A., Abraham P., Cherian S.S. Full-genome sequences of the first two SARS-CoV-2 viruses from India. Indian J. Med. Res. 2020;151:200–209. doi: 10.4103/ijmr.IJMR_663_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi K., Oshima T., Masuda Y., Ara T., Kanaya S., Mori H. Conservation of translation initiation sites based on dinucleotide frequency and codon usage in Escherichia coli K-12 (W3110): non-random distribution of A/T-rich sequences immediately upstream of the translation initiation codon. DNA Res. 2002;9:19–24. doi: 10.1093/dnares/9.1.19. [DOI] [PubMed] [Google Scholar]
- Yan Z., Wang R., Zhang L., Shen B., Wang N., Xu Q., He W., He W., Li G., Su S. Evolutionary changes of the novel influenza D virus hemagglutinin-esterase fusion gene revealed by the codon usage pattern. Virulence. 2019;10:1–9. doi: 10.1080/21505594.2018.1551708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Chen M., Tang Z. Analysis of synonymous codon usage bias in Flaviviridae virus. Biomed. Res. Int. 2019;2019:5857285. doi: 10.1155/2019/5857285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Wang J., Shen H. Network pharmacology-based analysis of the role of traditional Chinese herbal medicines in the treatment of COVID-19. Ann. Palliat. Med. 2020;9:437–446. doi: 10.21037/apm.2020.03.27. [DOI] [PubMed] [Google Scholar]
- Zalucki Y.M., Beacham I.R., Jennings M.P. Biased codon usage in signal peptides: a role in protein export. Trends Microbiol. 2009;17:146–150. doi: 10.1016/j.tim.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Mao R., Yan R., Cai D., Zhang Y., Zhu H., Kang Y., Liu H., Wang J., Qin Y. Transcription of hepatitis B virus covalently closed circular DNA is regulated by CpG methylation during chronic infection. PLoS One. 2014;9 doi: 10.1371/journal.pone.0110442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.J., Dong X., Cao Y.Y., Yuan Y.D., Yang Y.B., Yan Y.Q., Akdis C.A., Gao Y.D. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020;00:1–12. doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- Zhao J., Lopez A.L., Erichsen D., Herek S., Cotter R.L., Curthoys N.P., Zheng J. Mitochondrial glutaminase enhances extracellular glutamate production in HIV-1-infected macrophages: linkage to HIV-1 associated dementia. J. Neurochem. 2004;88:169–180. doi: 10.1046/j.1471-4159.2003.02146.x. [DOI] [PubMed] [Google Scholar]
- Zheng J. SARS-CoV-2: An emerging coronavirus that causes a global threat. Int. J. Biol. Sci. 2020;16:1678–1685. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J., Li Y., Zhao S., Liu S., Zhang Z. Mutation pressure shapes codon usage in the GC-rich genome of foot-and-mouth disease virus. Virus Genes. 2007;35:767–776. doi: 10.1007/s11262-007-0159-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.H., Zhang J., Sun D.J., Ma Q., Chen H.T., Ma L.N., Ding Y.Z., Liu Y.S. The distribution of synonymous codon choice in the translation initiation region of dengue virus. PLoS One. 2013;8 doi: 10.1371/journal.pone.0077239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article.Additional Fig. S1. Adaptiveness of human respiratory coronaviruses to human host analyzed from JCAT.
Additional Fig. S2. PCA analysis of A-, C-, G, and U-ended codons in the CDS sequences of human corona viruses based on RSCU values. A- PCA based on MERS-CoV CDS sequences. B- PCA based on SARS-CoV CDS sequences. B- PCA based on SARS-CoV-2 CDS sequences.
Additional Fig. S3. Correlation analysis of codons in the CDS sequences of human coronaviruses based on RSCU values. A- Correlation based on MERS-CoV CDS sequences. B- Correlation based on SARS-CoV CDS sequences. B- Correlation based on SARS-CoV-2 CDS sequences.
Results of the correlation analysis between human genes and viral genes based on the RSCU values. Only potential human genes affected by viral infection and showing correlation significance of <0.05 with viral genes were selected.
Functional enrichment analysis of potential human genes affected by viral infection screened based on the results of RSCU-correlation between human genes and viral genes.
Results of differential gene expression analysis of SARS-CoV-2-infected and uninfected human cell lines.
Functional enrichment analysis of potential human genes obtained based on common genes obtained by the intersection of RSCU-based correlation results and differential gene expression analysis results.
Prediction of the potential therapeutic drugs based on all of the human genes correlated with viral genes on the basis of RSCU values.
Prediction of the potential therapeutic drugs based on common genes obtained by the intersection of RSCU-based correlation results and differential gene expression analysis results
Prediction of the potential therapeutic drugs based on genes directly enriched in virus functional terms.