Abstract
Macrophage metabolic pathways show changes in response to various external stimuli.
Especially, increased lipopolysaccharide, an important bacterial component and Toll-like receptor 4 agonist, can induce activity in various macrophage metabolic pathways, including energy production and biosynthesis, as well as high immune responses due to increase in differentiated M1 macrophages. In this study, we confirmed that Lactobacillus paracasei (L. paracasei) KBL382, KBL384 and KBL385, isolated from the feces of healthy Koreans, can modulate various enzymes and membrane transporters related to glycolysis or macrophage polarization including hypoxia-inducible factor 1-alpha (HIF1A), inducible nitric oxide synthase (iNOS) and arginase in stimulated macrophages at the mRNA level, using the in vitro rodent bone‐marrow‐derived macrophage (BMDM) model. All L. paracasei exhibited significant down-regulatory effects on mRNAs for glycolysis-related enzymes, including lactate dehydrogenase A, solute carrier family 2 member 1, and triosephosphate isomerase. Moreover, L. paracasei treatment could lead to significant reductions in HIF1A or iNOS mRNA, and induced arginase mRNA in the BMDM model. Therefore, further extensive studies should be performed to support the application of L. paracasei, such as in probiotics or therapeutics, in controlling abnormal immune responses related to macrophage.
Keywords: Lactobacillus paracasei, Macrophage, Immunomodulation, Macrophage polarization, Glycolysis, Probiotics
Highlights
-
•
Macrophage metabolic pathways can be altered by the various stimuli.
-
•
L. paracasei treatment reduces glycolysis-related mRNA in the BMDM model.
-
•
L. paracasei treatment modulates mRNA in association with macrophage polarization.
-
•
L. paracasei could be useful to control macrophage-mediated immune responses.
1. Introduction
Recently, important roles of metabolic reprograming in immune cells have been reported [1,2]. Especially, macrophages can alter their metabolic pathways in response to various environmental stimuli [3]. For example, increases in lipopolysaccharide (LPS), an important bacterial component and Toll-like receptor 4 agonist, can trigger various metabolic pathways related to energy production and biosynthesis in M1 macrophage. Previous studies have been reported that differentiated M1 macrophages are strongly related to various immune responses [[4], [5]].
During inflammation due to LPS, enzymes related to glycolysis and pentose phosphate pathway or inducible nitric oxide synthase (iNOS) can be induced in M1 macrophages to produce the energy sources adenosine triphosphates and nitric oxide (NO), respectively [[6], [7], [8]]. Moreover, induction of hypoxia-inducible factor 1-alpha (HIF1A) and arginase can affect macrophage polarization [[9], [10], [11]].
Probiotics such as Lactobacillus spp. have been reported to show strong immunomodulatory effects [12,13]. We previously confirmed the strong anti-inflammatory effects of Lactobacillus paracasei (L. paracasei) species, isolated from the feces of healthy Koreans, in the in vivo dextran sodium sulfate (DSS)-induced colitis animal model [12]. In this study, we investigated the modulatory effects of L. paracasei treatment on various enzymes related to glycolysis or macrophage polarization, including HIF1A, iNOS and arginase in stimulated macrophages at the mRNA level in vitro using the rodent bone‐marrow‐derived macrophage (BMDM) model.
2. Materials and methods
2.1. Preparation of L. paracasei strains
Previously, we isolated three L. paracasei strains (KBL382, KBL384 and KBL385) from the feces of healthy Koreans and confirmed their strong resistance to bile salts or low pH [12]. All L. paracasei strains were cultured in anaerobic conditions using Lactobacilli MRS agar (Becton, Dickinson and Company, Sparks, MD, USA) with 0.05% L-cysteine-hydrochloride and Anaeropack (Mitsubishi Gas Chemical Company Inc., Tokyo, Japan) at 37 °C for 24 h as described previously [12]. Bacteria were collected using centrifugation at 1200×g and washed twice using 1× phosphate-buffered saline (PBS). Bacterial concentrations were quantified as colony-forming units (CFUs) through the cultivation, and the bacteria were stored at −4 °C until further use.
2.2. Preparation of BMDM
BMDMs were isolated from 6-week-old female C57BL/6 mice (Central Lab Animals Inc., Seoul, Republic of Korea), as described previously [14,15]. BMDMs were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco, Thermo Fisher Scientific Inc., Waltham, MA, USA) containing 1% penicillin/streptomycin (Gibco), 1% gentamycin (Gibco), and 10% fetal bovine serum (Gibco) at 37 °C under a humidified atmosphere with 5% CO2. To measure the total cell concentration, BMDMs were stained with trypan blue and counted using a CKX31 inverted microscope (Olympus Corp., Tokyo, Japan).
2.3. Measurement of mRNA expression in the in vitro BMDM model
Approximately 3 × 105 cells of BMDMs were incubated with 100 ng/mL LPS (Sigma-Aldrich Corp., St. Louis, MO, USA) and 20 ng/mL interferon-γ (IFN-γ) or interleukin-4 (IL-4) (PeproTech Inc., Rocky Hill, NJ, USA) in each well of a 48-well plate (SPL Life Sciences Co., Ltd., Pocheon-si, Gyeonggi-do, Republic of Korea) at 37 °C for 24 h as previously decribed with some modification [[16], [17]]. Approximately 3 × 106 CFUs of each L. paracasei strain were co-incubated with the BMDMs. The cytotoxicity of Lactobacillus spp. for BMDM were confirmed using Lactate dehydrogenase (LDH) Assay Kit (Cytotoxicity) (Abcam Inc., Cambridge, MA, USA) following the manufacturer's protocol. To measure mRNA expression, total RNA was extracted using an easy-spin Total RNA Extraction Kit (iNtRON biotechnology Inc., Seongnam-si, Gyeonggi-do, Republic of Korea) following the manufacturer's protocol. Complementary DNA (cDNA) was synthesized using a High-Capacity RNA-to-cDNA Kit (Thermo Scientific). Then, real-time polymerase chain reaction (RT-PCR) was performed using a Power SYBR Green PCR Master Mix (Thermo Scientific) with 0.01 mM primers [[16], [17], [18]] (Table 1). PCR was carried out with an initial denaturation step at 95 °C for 5 min, followed by 40 cycles of 95 °C for 5 s and 60 °C for 10 s using a Step-One-Plus Real-Time PCR System (Applied Biosystems Inc., Förster City, CA USA). The expression levels of target mRNAs were normalized to hypoxanthine-guanine phosphoribosyl transferase (HPRT) [19].
Table 1.
The primers used in this study.
| Target | Sequence | Reference |
|---|---|---|
| Arginase | Fwa: 5′-CAG AAG AAT GGA AGA GTC AG-3′ | [18] |
| Rvb: 5′-CAG ATA TGC AGG GAG TCA CC-3′ | ||
| Enolase 1 | Fw: 5′-TGC GTC CAC TGG CAT CTA C-3′ | [16] |
| Rv: 5′-CAG AGC AGG CGC AAT AGT TTT A-3′ | ||
| HIF1A | Fw: 5′-AGC TTC TGT TAT GAG GCT CAC C-3′ | [16] |
| Rv: 5′-TGA CTT GAT GTT CAT CGT CCT C-3′ | ||
| HPRT | Fw: 5′-TTA TGG ACA GGA CTG AAA GAC-3′ | [19] |
| Rv: 5′-GCT TTA ATG TAA TCC AGC AGG T-3′ | ||
| iNOS | Fw: 5′-TTT GCT TCC ATG CTA ATG CGA AAG-3′ | [18] |
| Rv: 5′-GCT CTG TTG AGG TCT AAA GGC TCC G-3′ | ||
| LDHA | Fw: 5′-CAT TGT CAA GTA CAG TCC ACA CT-3′ | [16] |
| Rv: 5′-TTC CAA TTA CTC GGT TTT TGG GA-3′ | ||
| MCT4 | Fw: 5′-TCA CGG GTT TCT CCT ACG C-3′ | [16] |
| Rv: 5′-GCC AAA GCG GTT CAC ACA C-3′ | ||
| PFK1 | Fw: 5′-GGA GGC GAG AAC ATC AAG CC-3′ | [16] |
| Rv: 5′-CGG CCT TCC CTC GTA GTG A-3′ | ||
| PKM2 | Fw: 5′-GCC GCC TGG ACA TTG ACT C-3′ | [16] |
| Rv: 5′-CCA TGA GAG AAA TTC AGC CGA G-3′ | ||
| SLC2A1 | Fw: 5′-CAG TTC GGC TAT AAC ACT GGT G-3′ | [16] |
| Rv: 5′-GCC CCC GAC AGA GAA GAT G-3′ | ||
| TPI | Fw: 5′-CCA GGA AGT TCT TCG TTG GGG-3′ | [16] |
| Rv: 5′-CAA AGT CGA TGT AAG CGG TGG-3′ |
aFw represents sequences of forward primers.
bRv represents sequences of reverse primers.
2.4. Statistical analysis
Data are expressed as mean ± standard error of the mean (SEM) with at least three independent experiments. When appropriate, the Mann-Whitney U test was used, and P-values less than 0.05 were considered statistically significant. GraphPad Prism 5.04 (GraphPad Software, Inc., La Jolla, CA, USA) was used for all statistical analyses and visualizations.
3. Results
3.1. Modulation of mRNA expressions related to glycolysis with L. paracasei treatment in the in vitro BMDM model
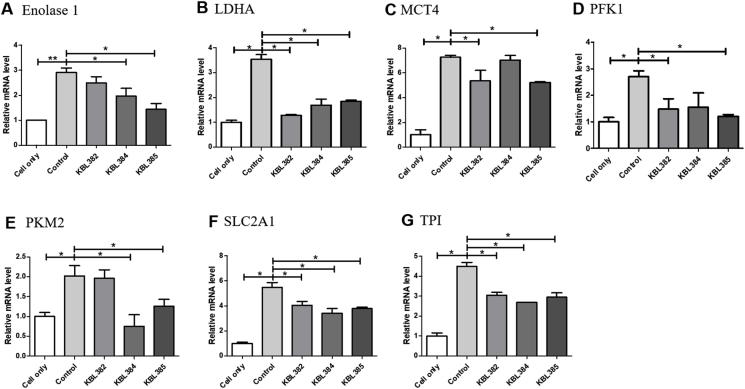
Fig. 1 shows the modulation of mRNA expressions related to glycolysis in BMDM with L. paracasei treatment. L. paracasei concentration had no cytotoxic effects in BMDM (data not shown). Overall, treatment with all L. paracasei strains had significant down-regulatory effects on the mRNAs of glycolysis-related enzymes or membrane transporters. mRNA levels of lactate dehydrogenase A (LDHA), solute carrier family 2 member 1 (SLC2A1), and triosephosphate isomerase (TPI) were significantly reduced after treatment of L. paracasei KBL382, KBL384 or KBL385 compared to control without L. paracasei treatment (P < 0.05) (Fig. 1B, F, G). Moreover, L. paracasei KBL385 exhibited strong down-regulatory effects on enolase 1, monocarboxylate transporter 4 (MCT4), phosphofructokinase 1 (PFK1), and pyruvate kinase M2 (PKM2) compared to the control (P < 0.05) (Fig. 1A, C, D, E). L. paracasei KBL382 showed significant down-regulatory effects at mRNA levels of MCT4 and PFK1 (Fig. 1C and D). L. paracasei KBL384 also modulated Enolase 1 and PKM2 significantly (Fig. 1A and E).
Fig. 1.
Effects of L. paracasei treatment on mRNA expressions related to glycolysis. A) Enolase 1, B) lactate dehydrogenase A (LDHA), C) monocarboxylate transporter 4 (MCT4), D) phosphofructokinase 1 (PFK1), E) Pyruvate Kinase M2 (PFM2), F) solute carrier family 2 member 1 (SLC2A1), G) triosephosphate isomerase (TPI). Approximately 3 × 105 cells of rodent bone‐marrow‐derived macrophage cells (BMDMs) treated with 100 ng/mL lipopolysaccharide (LPS) and 20 ng/mL interferon-γ (IFN-γ) were co-incubated with 3 × 106 colony-forming units of each L. paracasei strain at 37 °C for 24 h. Total RNA was extracted using an easy-spin Total RNA Extraction Kit and cDNA was synthesized using a High-Capacity RNA-to-cDNA Kit. PCR reaction was performed and the expression levels were normalized to hypoxanthine-guanine phosphoribosyl transferase (HPRT). Samples without L. paracasei treatment were used as a control. Data are expressed as mean ± SEM of three independent experiments. Asterisks indicated a statistical significance (*, P < 0.05; **, P < 0.01).
3.2. Modulation of mRNA expressions related to macrophage polarization with L. paracasei treatment in the in vitro BMDM model
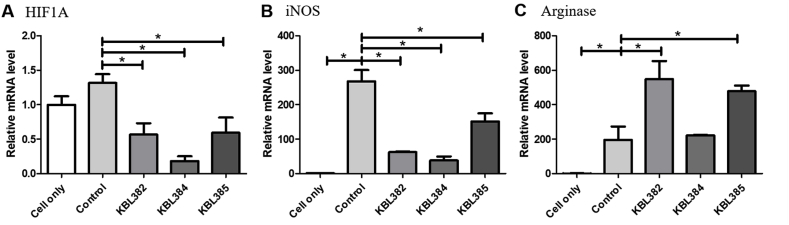
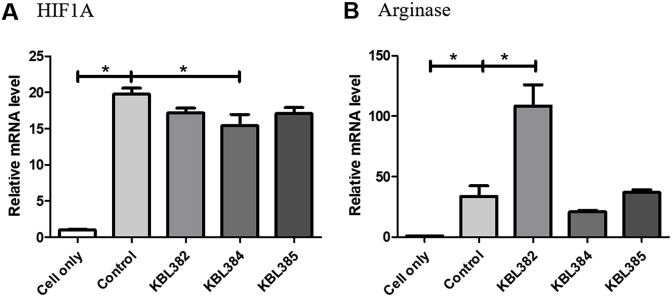
Treatment with L. paracasei KBL382, KBL384, or KBL385 had significant down-regulatory effects on the M1 macrophage-related marker HIF1A and iNOS (P < 0.05) (Fig. 2A and B). Moreover, L. paracasei KBL382 or KBL385 showed up-regulatory effects on the M2 macrophage–related marker arginase in the BMDMs stimulated with LPS and IFN-γ (P < 0.05) (Fig. 2C). However, mRNA levels of arginase did not change significantly with L. paracasei KBL384 treatment (Fig. 2C). Moreover, mRNA levels of HIF1A changed significantly with L. paracasei KBL384 treatment in BMDMs stimulated with LPS and IL-4 (P < 0.05) (Fig. 3A). The high arginase levels were observed in BMDMs stimulated with LPS and IL-4 after L. paracasei KBL382 treatment (P < 0.05) (Fig. 3B).
Fig. 2.
Effects of L. paracasei treatment on mRNA expression related to macrophage polarization in BMDM stimulated with LPS and IFN-γ. A) Hypoxia-inducible factor 1-alpha (HIF1A), B) inducible nitric oxide synthase (iNOS), C) arginase. PCR was performed using cDNA and the expression levels were normalized to HPRT. Samples without L. paracasei treatment were used as a control. Data are expressed as mean ± SEM of three independent experiments. Asterisks indicated a statistical significance (*, P < 0.05).
Fig. 3.
Effects of L. paracasei treatment on mRNA expression related to macrophage polarization in BMDM stimulated with LPS and interleulin-4 (IL-4). A) HIF1A, B) arginase. Approximately 3 × 105 cells of rodent BMDMs treated with 100 ng/mL lipopolysaccharide (LPS) and 20 ng/mL IL-4 were co-incubated with 3 × 106 colony-forming units of each L. paracasei strain at 37 °C for 24 h. Total RNA was extracted using an easy-spin Total RNA Extraction Kit and cDNA was synthesized using a High-Capacity RNA-to-cDNA Kit. PCR was performed using cDNA and the expression levels were normalized to HPRT. Samples without L. paracasei treatment were used as a control. Data are expressed as mean ± SEM of three independent experiments. Asterisks indicated a statistical significance (*, P < 0.05).
4. Discussion
In this study, we demonstrated that treatment with L. paracasei KBL382, KBL384, or KBL385 clearly modulated the expression levels of various mRNA related to glycolysis and macrophage polarization in the in vitro BMDM model. Especially, L. paracasei treatment significantly reduced the mRNA levels of glycolysis-related enzymes or membrane transporters in BMDMs stimulated with LPS and IFN-γ (Fig. 1.). Increased enolase 1 can contribute to the production of pro-inflammatory cytokines and prostaglandins [20]. Moreover, previous study has been reported that LDHA in macrophages can boost the T Helper 17 (Th17) cell-dependent immune responses and exacerbate autoimmune diseases [21]. When glycolysis is suppressed, pyruvate production and oxidative phosphorylation also decrease [22]. Then, chain reduction of MCT4, which is involved in the conversion of pyruvate to lactate, and PFK1 can occur [22,23]. Decreases in the mRNA levels of PKM2, SLC2A1 and TPI, which are important enzyme or membrane transporter in the glycolytic pathway, were also exhibited in BMDM with L. paracasei treatment (Fig. 1.) [16].
Previous study has been reported that PKM2 is important for the activation of M1 macrophages and regulation of HIF1A activities, such as the production of M1 macrophages [24]. Moreover, increases in HIF1A can promote the glycolysis pathway and pro-inflammatory activities of M1 macrophages [16]. Therefore, L. paracasei treatment could be used to modulate the immune response by controlling both PKM2 and HIF1A levels in macrophages (Fig. 1E and 2A). M1 macrophages can induce iNOS, which mediates arginine breakdown and produces NO [16]. Therefore, increases in M1 macrophage differentiation are closely associated with the increased iNOS expression and down-regulation of arginase [25]. Our study confirmed that L. paracasei treatment can modulate the reduction of iNOS mRNA and induction of arginase mRNA, especially with KBL382 or KBL385 treatment (Fig. 2B and C). Moreover, L. paracasei KBL382 can induce arginase mRNA significantly in M2 polarization model using BMDM (Fig. 3B). However, to elucidate the mechanisms including metabolites underlying the effects of L. paracasei treatment on the expression patterns and differentiation of macrophages, further longitudinal studies with multi-omics approaches in in vivo models need to be performed.
Over-polarization of M1 macrophages, which is caused by the environmental stimuli, can be related to various autoimmune diseases [26]. Our data suggested the important effects of L. paracasei treatment to mRNA levels of macrophages in the in vitro conditions. Therefore, in conclusion, further extensive studies should be performed to support the application of L. paracasei, such as in probiotics or therapeutics, in controlling abnormal immune responses related to macrophage.
CRediT authorship contribution statement
Dae Hee Han: Data curation, Formal analysis, Writing - original draft. Woon-Ki Kim: Conceptualization, Project administration, Writing - original draft. SungJun Park: Conceptualization, Writing - original draft, Writing - review & editing. You Jin Jang: Formal analysis, Writing - review & editing. GwangPyo Ko: Supervision, Funding acquisition.
Declaration of competing interest
GK is the founder of KoBioLabs, Inc. and SP is employee by KoBioLabs, Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgements
This work was supported by a grant of National Research Foundation of Korea (NRF) (NRF-2018R1A2A1A05078258) funded by the Korean government (MSIP), Republic of Korea.
References
- 1.O'Neill L.A.J., Grahame Hardie D. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 2013;493:346–355. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- 2.Krawczyk C.M., Holowka T., Sun J., Blagih J., Amiel E., DeBerardinis R.J., Cross J.R., Jung E., Thompson C.B., Jones R.G. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood. 2010;115:4742–4749. doi: 10.1182/blood-2009-10-249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ginhoux F., Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 2014;14:392–404. doi: 10.1038/nri3671. [DOI] [PubMed] [Google Scholar]
- 4.Mosser D.M., Edwards J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murray P.J., Wynn T.A. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vadiveloo P.K., Keramidaris E., Morrison W.A., Stewart A.G. Lipopolysaccharide-induced cell cycle arrest in macrophages occurs independently of nitric oxide synthase II induction. Biochim. Biophys. Acta Mol. Cell Res. 2001;1539:140–146. doi: 10.1016/S0167-4889(01)00102-1. [DOI] [PubMed] [Google Scholar]
- 7.Vadiveloo P.K. Macrophages - proliferation, activation, and cell cycle proteins. J. Leukoc. Biol. 1999;66:579–582. doi: 10.1002/jlb.66.4.579. [DOI] [PubMed] [Google Scholar]
- 8.Newsholme P., Costa Rosa L.F.B.P., Newsholme E.A., Curi R. The importance of fuel metabolism to macrophage function. Cell Biochem. Funct. 1996;14:1–10. doi: 10.1002/cbf.644. [DOI] [PubMed] [Google Scholar]
- 9.Wang T., Liu H., Lian G., Zhang S., Wang X., Jiang C. HIF1α -induced glycolysis metabolism is essential to the activation of inflammatory macrophages. Mediat. Inflamm. 2017 doi: 10.1155/2017/9029327. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Z., Ming X. Functions of arginase isoforms in macrophage inflammatory responses : impact on cardiovascular diseases and metabolic disorders. Front. Immunol. 2014;5:1–10. doi: 10.3389/fimmu.2014.00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Werno C., Menrad H., Weigert A., Dehne N., Goerdt S., Schledzewski K. Knockout of HIF-1α in tumor-associated macrophages enhances M2 polarization and attenuates their pro-angiogenic responses. Carcinogenesis. 2010;31:1863–1872. doi: 10.1093/carcin/bgq088. [DOI] [PubMed] [Google Scholar]
- 12.Kim W.K., Jang Y.J., Seo B., Han D.H., Park S.J., Ko G.P. Administration of Lactobacillus paracasei strains improves immunomodulation and changes the composition of gut microbiota leading to improvement of colitis in mice. Journal of Functional Foods. 2019;52:565–575. doi: 10.1016/j.jff.2018.11.035. [DOI] [Google Scholar]
- 13.Jang Y.J., Kim W.K., Han D.H., Lee K., Ko G. Lactobacillus fermentum species ameliorate dextran sulfate sodium-induced colitis by regulating the immune response and altering gut microbiota. Gut Microbes. 2019;10:696–711. doi: 10.3389/fmolb.2019.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xaus J., Cardó M., Valledor A.F., Soler C., Lloberas J., Celada A. Interferon γ induces the expression of p21(waf-1) and arrests macrophage cell cycle, preventing induction of apoptosis. Immunity. 1999;11:103–113. doi: 10.1016/S1074-7613(00)80085-0. [DOI] [PubMed] [Google Scholar]
- 15.Schreiber R.D., Jolla L., Hicks L.J., Celada A. Evidence for a gamma-interferon receptor that regulates macrophage tumoricidal activity. J. Exp. Med. 1984;160:55–74. doi: 10.1084/jem.160.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L., Luc Y., Martinez J., Bi Y., Lian G., Wang T., Milasta S., Wang J., Yang M., Liu G. Proinflammatory signal suppresses proliferation and shifts macrophage metabolism from Myc-dependent to HIF1α-dependent. Proc. Natl. Acad. Sci. U.S.A. 2016;113:1564–1569. doi: 10.1073/pnas.1518000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palsson-McDermott E.M., Curtis A.M., Goel G., Lauterbach M.A.R., Sheedy F.J., Gleeson L.E., van den Bosch M.W.M., Quinn S.R., Domingo-Fernandez R., Johnston D.G.W., Jiang J., Israelsen W.J., Keane J., Thomas C., Clish C., Heiden M.V., Xavier R.J., O’Neill L.A.J. Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metab. 2015;21:65–80. doi: 10.1016/j.cmet.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao Z., Zhang J., Zhang B., Liang G., Chen X., Yao F. Imatinib prevents lung cancer metastasis by inhibiting M2-like polarization of macrophages. Pharmacol. Res. 2018;133:121–131. doi: 10.1016/j.phrs.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Kwon H.-K., Lee C.-G., So J.-S., Chae C.-S., Hwang J.-S., Sahoo A., Nam J.H., Rhee J.H., Hwang K.-C., Im S.-H. Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc. Natl. Acad. Sci. U.S.A. 2010;107:2159–2164. doi: 10.1073/pnas.0904055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bae S., Kim H., Lee N., Won C., Kim H.-R., Hwang Y., Song Y.W., Kang J.S., Lee W.J. α-Enolase expressed on the surfaces of monocytes and macrophages induces robust synovial inflammation in rheumatoid arthritis. J. Immunol. 2012;189:365–372. doi: 10.4049/jimmunol.1102073. [DOI] [PubMed] [Google Scholar]
- 21.Seth P., Csizmadia E., Hedblom A., Vuerich M., Xie H., Li M., Longhi M.S., Wegiel B. Deletion of lactate dehydrogenase-A in myeloid cells triggers antitumor immunity. Cancer Res. 2017;77:3632–3643. doi: 10.1158/0008-5472.CAN-16-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan Z., Xie N., Banerjee S., Cui H., Fu M., Thannickal V.J., Liu G. The monocarboxylate transporter 4 is required for glycolytic reprogramming and inflammatory response in macrophages. J. Biol. Chem. 2015;290:46–55. doi: 10.1074/jbc.M114.603589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamin D., Robay D., Hindupur S.K., Pohlmann J., Colombi M., El-Shemerly M.Y., Maira S.M., Moroni C., Lane H.A., Hall M.N. Dual inhibition of the lactate transporters MCT1 and MCT4 is synthetic lethal with metformin due to NAD+ depletion in cancer cells. Cell Rep. 2018;25:3047–3058. doi: 10.1016/j.celrep.2018.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palsson-McDermott E.M., Palsson-McDermott E.M., Curtis A.M., Curtis A.M., Goel G., Goel G., Lauterbach M.A.R., Lauterbach M.A.R., Sheedy F.J., Sheedy F.J. Pyruvate kinase M2 regulates hif-1α activity and IL-1β induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metabol. 2015;21:65–80. doi: 10.1016/j.cmet.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rath M., Müller I., Kropf P., Closs E.I., Munder M. Metabolism via arginase or nitric oxide synthase: two competing arginine pathways in macrophages. Front. Immunol. 2014;5:1–10. doi: 10.3389/fimmu.2014.00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Funes S.C., Rios M., Escobar-Vera J., Kalergis A.M. Implications of macrophage polarization in autoimmunity. Immunology. 2018;154:186–195. doi: 10.1111/imm.12910. [DOI] [PMC free article] [PubMed] [Google Scholar]