Abstract
The subtelomeres, highly heterogeneous repeated sequences neighboring telomeres, are transcribed into coding and noncoding RNAs in a variety of organisms. Telomereproximal subtelomeric regions produce non-coding transcripts i.e., ARRET, αARRET, subTERRA, and TERRA, which function in telomere maintenance. The role and molecular mechanisms of the majority of subtelomeric transcripts remain unknown. This review depicts the current knowledge and puts into perspective the results obtained in different models from yeasts to humans.
Keywords: subtelomeres, noncoding RNA, repeated sequences, transcription
Graphical abstract
Highlights
-
•
The subtelomeres are highly heterogeneous repeated sequences next to telomeres.
-
•
They are transcribed into coding and noncoding (nc)RNAs in a variety of organisms.
-
•
The subtelomeric ncRNAs ARRET, αARRET, subTERRA and TERRA maintain telomere.
-
•
But the role and molecular mechanisms of most of subtelomeric RNA remain unknown.
-
•
This review puts into perspective different model organisms, from yeasts to humans.
General Overview
Subtelomeres are regions immediately adjacent to the telomeric repeats. They consist of repetitions of heterogeneous sequences, and have, therefore, been difficult to map and sequence. Only recently has the complete assembly of subtelomeres become possible, thanks to the progress in sequencing techniques, as well as the use of high throughput optical mapping of large single DNA molecules in nanochannel arrays [1], [[2], [3], [4], [5], [6]]. While our understanding of subtelomeric structure and activity is still developing, accumulating data support the idea that subtelomeres are chromosomal entities with specific functions. To date, subtelomeres have been involved in (1) Telomere maintenance and regulation of telomere length (in budding yeast [7,8], in fission yeast [9], Plasmodium falciparum and Tetrahymena [10,11]), (2) Proper chromosome segregation [12], (3) Chromosome recognition and pairing during meiosis [13], (4) Replicative senescence [14,15], (5) Nuclear positioning of telomeres [16], (6) Control of spreading of telomeric heterochromatin [14,17]), (7) Phenotypic polymorphism and genome plasticity [18,19], and finally, in (8) the regulation of TERRA transcription—a telomeric repeat-containing family of long noncoding RNAs (lncRNAs) transcribed into telomeric tracts from promoters embedded in subtelomeres [[20], [21], [22]].
Telomere and subtelomere features
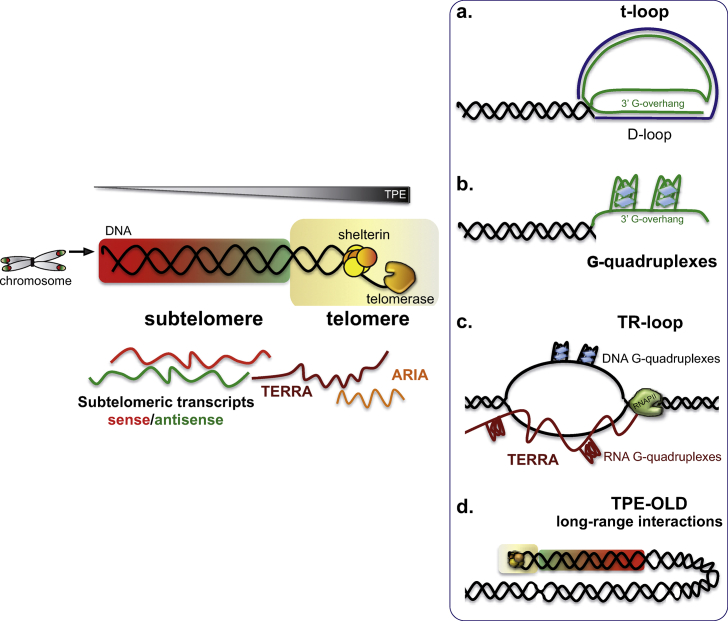
Telomeres are nucleoprotein caps protecting chromosome ends from erosion and from DNA repair machinery that could recognize them as DNA breaks. Indeed, this could trigger the formation of chromosomal aberrations, such as end-to-end fusions, which would destabilize the genome. Telomeres are composed of tandem arrays of short double-stranded repeats and a single-stranded 3′G-overhang [23,24]. Due to the so-called “end-replication problem,” telomeres shorten with each cell division [25]. Excluding those in D. melanogaster that are maintained thanks to a retrotransposon mechanism [26], telomeres in all Eukaryotes are elongated by telomerase [27,28]. Telomeric repeats are specifically bound by telomere-binding proteins, which in most organisms are called shelterin complex [29]. Moreover, telomeres can fold into different structures, such as the well-known telomeric loop (T-loop; Fig. 1a [30]) that protects the chromosome ends from unwanted fusions. To form this large loop, the single-stranded 3′G-overhang invades the homologous double-stranded region forming a displacement loop (D-loop [31,32]). The G-rich telomeric repeats can also generate G-quadruplex structures (Fig. 1b [[33], [34], [35]]). The most recently discovered telomeric R-loop (TRL) is a three-stranded structure that contains a DNA:RNA hybrid implicating TERRA (Fig. 1c). The TRL regulates telomere length and is instrumental in recombination [[36], [37], [38]]. Telomeres are enriched in heterochromatic marks (H3K9me3, H4K20me3, the PRC2-repressive mark H3K27me3, DNA methylation, and heterochromatin protein 1 – HP1; [[39], [40], [41], [42], [43]], which spread through subtelomeric regions by a mechanism called Telomere Position Effect (TPE). The proper organization of this heterochromatic domain is important for the maintenance of telomere length through regulation of telomerase activity and through recombination (ALT mechanism—alternative lengthening of telomeres).
Fig. 1.
Overview of telomeric structures and potential molecular mechanisms of action of telomeric and subtelomeric transcripts. Schematic representation of a chromosome highlighting the telomeres (rainbow squares) at its extremities. Telomeres (in yellow) are composed of tandem repeats bound by shelterin complex and telomerase, which elongates telomeric DNA. TPE intensity decreases toward the centromere. a. Schematic representation of a telomeric t-loop, including a displacement loop (D-loop). b. G-quadruplexes formed on a telomeric 3′G-overhang. c. Telomeric R-loop (TRL) constituted by RNAPII-transcribed TERRA and telomeric sequences. Additionally, G-quadruplexes can be formed by a single G-rich strand. d. Schematic DNA loop representing TPE-OLD and other telomere long-range interactions.
The size of subtelomeres varies greatly among organisms: ranging from 500 kb in each autosomal arm in humans, 100 kb in fission yeast to 10 kb in budding yeast. Furthermore, subtelomeres share little or no homology. Due to the presence of blocks of repeated homologous sequences, subtelomeres exhibit the highest instability in the genome [44,45]. Various types of large-scale rearrangements shape subtelomeres, such as segmental duplication amplification, the formation of extended tandem repeat arrays, and extended deletions (as described for humans and plants [2,3,46]). In consequence, subtelomeres are highly polymorphic [18,46,47]. These allelic variations affect the expression of coding and noncoding RNAs embedded in or located near subtelomeres.
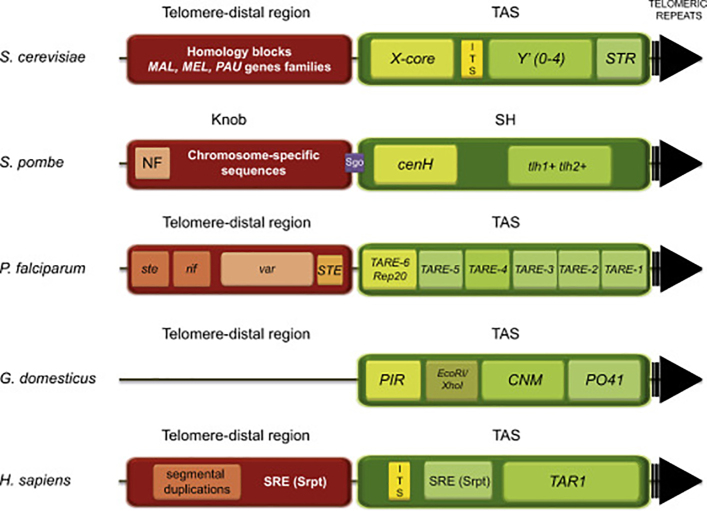
Although there is no sequence homology between subtelomeres of different organisms, their structures and features are similar [48,49]. In general, they can be divided into two domains. The telomere-proximal region found immediately adjacent to the telomere is called TAS (Telomere Associated Sequence). TAS is gene-poor and contains homologous blocks of sequences highly conserved across many chromosome ends [50,51]. The second domain, the telomere-distal region, which is centromere-proximal, contains sequences found at only a few chromosome ends, as well as sequences specific to a given subtelomeric region [52]. Devoid of essential genes, telomere-distal regions harbor nonessential genes and multiple gene families [[53], [54], [55]]. Tracts of degenerated telomeric repeats often separate these two domains (ITS, internal/interstitial telomeric sequences) (for general structure see Fig. 2). Furthermore, telomeres and subtelomeres form specialized heterochromatin structures, which are essential for chromosome stability [17,46,[56], [57], [58]]. Two main mechanisms influence subtelomeric transcription at chromosome ends, Copy Number Variation (CNV) and Telomeric Position Effect (TPE). CNV modifies the number of subtelomeric sequences through recombination [59]. TPE imposes transcriptional repression of nearby sequences [60] by changing their chromatin state. It operates by histone modifications, a quantity of telomere-associated proteins (cis-spreading), and higher-order structures (long-distance looping). A variant of TPE, called TPE-OLD (telomere position effect over long distance [61]; Fig. 1d), modulates the expression of target genes at any position in the genome by telomere looping [60]. An important factor regulating TPE is telomere length. Telomere shortening provokes changes in the protein content within telomeres and subtelomeric regions, which, in turn, become less compacted and loose heterochromatin marks. This reduces TPE and TPE-OLD and leads to the activation of subtelomeric genes [61].
Fig. 2.
Organization of subtelomeric regions in various organisms. In general subtelomeres are composed of two regions: a telomere-proximal region also called TAS (for Telomere-Associated Sequences; in green) and a telomere-distal region (in red). Telomeric repeats are represented as a black arrow. In yeast S. cerevisiae, TAS can be found in two flavors: TAS containing Y′ elements or not (X-only subtelomeres). The X-core element contains ARS (ACS) sequences binding the Abf1 protein and STR repeats with Tbf1 protein-binding sites. The telomere-distal region encloses subtelomeric genes families. ITS (Interspersed/Interstitial/Internal Telomeric Repeats; in yellow) are more or less degenerated telomeric repeats present in TAS between X and Y′ elements. In S. pombe, TAS are called SH regions and contain cenH (centromere-homologous sequence) and telomere-linked helicases (tlh) encoded at chromosomes I (tlh1+) and II (tlh2+). These putative helicases are members of the recQ family and show sequence homology with the dh and dg repeats found at centromeres [186]. The Sgo domain, shown in violet, represents a Sgo2-binding barrier, which controls the spreading of subtelomeric heterochromatin between proximal and distal regions. The telomere-distal region called “knob” contains chromosome-specific sequences and the nucleosome free region (NF) at its centromere-proximal end. In P. falciparum, six telomere-associated repeat elements (TARE1-6/Rap20) and several 12-base SPE sites (binding SPE2 interacting protein 2, PfSIP2) are located between var genes and TAS. 60 var genes are positioned in telomere-distal regions within subtelomeres; rif and ste genes are frequently found in their vicinity [187]. In G. domesticus, tandem repeats PO41, CNM, EcoRI/XhoI and PIR compose TAS. In H. sapiens, Subtelomeric Repeats (SRE/Srpt) comprise about 25% of the most distal 500 kb and 80% of the most distal 100 kb of the chromosome ends [50]. ITS repeats are found between different genetic elements. The TAR1 element, immediately adjacent to the telomeric repeats, is a variably sized (0–2 kb) sequence segment bearing similarity to the TAR1 repeat family.
Telomeric and subtelomeric regions can also form loops of various sizes with more or less distant genomic loci. Little is known about these structures as their existence has only recently been discovered thanks to chromosome capture methods. Described in yeast, as well as in human cells, they potentially change the transcriptional environment of the interacting regions [62]. These looping interactions depend on telomere length. Of note, the implication of subtelomeric and telomeric transcripts has still not been addressed. Telomeric and subtelomeric sequences (ITS, ACS) are bound by various specific proteins, offering platforms for potential interactions. Among these proteins are telomeric repeat-recognizing factors like Rap1 protein and other members of the shelterin and silencing complexes (i.e., SIR proteins). On-going subtelomeric and telomeric transcription modifies the state of these interactions by introducing newly-transcribed RNA into their structure, and by changing the chromatin state, as well as the content of interacting proteins [62].
High throughput transcriptional analyses have provided evidence regarding the transcriptional activity of telomeres and subtelomeres and suggested that the production of sense and antisense lncRNAs is their common feature. We propose here a comprehensive characterization of subtelomeric transcripts recently discovered in different organisms: ARRET, αARRET [63,64], subTERRA [65], lncRNA-TARE/var [66,67], PO41 [68,69], WASH, DDX11L, RPL23AP7, D4Z4 [2,55,70] and L. infantum’s subtelomeric transcripts [71]), as well as telomeric repeats containing transcripts: TERRA, ARIA, and PAR-TERRA, whose transcription depends on or starts at subtelomeres. We suggest dividing them into two groups, transcripts with functions related or nonrelated to telomeres.
Transcripts with Telomere-Related Functions
Here we present telomeric repeat-containing transcripts and subtelomeric transcripts whose transcription or binding impact telomere maintenance and function.
TERRA and other telomeric repeat-containing transcripts
TERRA
TERRA is a conserved evolutionary lncRNA containing mostly G-rich telomeric repeats. A tremendous quantity of data has been accumulated on TERRA expression in different cell-types or in specific conditions (e.g., stress, cancer, different developmental, or cell cycle states; reviewed in [[72], [73], [74], [75]]). TERRA is an integral component of a functional telomere. Deregulation of its expression or localization causes telomere dysfunction and genome instability [20,76]. From a molecular point of view TERRA can (i) bind specific proteins, (ii) form secondary structures like G-quadruplexes [77], and (iii) assemble in ternary DNA:RNA: protein complexes or DNA:RNA hybrids like in TRLs (Fig. 1c). Concerning telomere maintenance, TERRA acts through direct binding to telomerase components and through TRL formation in homologous recombination (ALT). In yeast and mammalian cells, higher TERRA levels correlate with increased numbers of TRLs, suggesting the importance of TERRA in regulating TRL formation. Interestingly, the ectopic expression of RNase H1, which specifically degrades RNA from DNA-RNA hybrids and resolves R-loops, reduced the DNA damage at telomeres [78], indicating that the TRL level is crucial for telomere instability. The regulation and functions of R-loops have been extensively reviewed in Ref. [38]. TERRA also impacts telomeric chromatin through interactions with shelterin and other telomere-interacting proteins like the Origin of Replication Complex (ORCs [76]), RNA helicase ATRX [79] or the RNA-binding protein hnRNPA1 [80]. Those proteins may assist TERRA in binding to telomeres or to other factors in order to regulate its stability or accessibility. In this way, TERRA could facilitate telomere replication directly through the formation of RNA:DNA:protein ternary complexes [81] or by binding to the ORCs [76]. As mentioned, TERRA has been shown to directly bind to telomerase components in mice, humans, and yeast [73,79,82]. In mammalian cells, TERRA inhibits telomerase in vitro [57,82]. In the same line, ASO-TERRA (antisense oligonucleotides), depleting the in vivo level of TERRA, provoked an increase of telomerase activity in murine ES cells [79], confirming TERRA’s negative function in telomere lengthening. In contrast, in fission and budding yeast, TERRA transcription is induced at short telomeres [83,84], which rather suggests a positive role for TERRA in regulating telomere length. Thus, the molecular mechanism of TERRA’s activity may not be conserved across species. Accordingly, we chose to present the telomeric and subtelomeric RNA features in different organisms.
Mammalian TERRA
Human and mouse telomeres are composed of TTAGGGn repeats conserved in all vertebrates. Human telomeres are quite short (5–15 kb) compared to mouse telomeres, which are 20–100 kb-long. This size variation could be explained by the differences in telomerase activity, which is active in embryonic nondifferentiated stem cells in both organisms but inactivated in differentiated human cells [85]. TERRA is transcribed from promoters embedded in subtelomeric regions toward the telomere. Its 5′-end contains subtelomeric sequences but is rather considered as a telomeric transcript since it consists of mostly tandem arrays of G-rich telomeric repeats. TERRA length is heterogeneous. It depends on the length of telomeric repeat tracts, the number of transcribed repeats, and the location of its Transcription Start Site (TSS) in subtelomeres. Using (TG) repeat probing by Northern blot experiments, TERRA appears as a smear of 0.2–10 kb in human and mouse cells. TERRA is transcribed by RNA Polymerase II (RNAPII [86]) and is (m7G)-capped [20]. There are two major TERRA fractions: long-lived polyadenylated, and short-lived nonpolyadenylated (in humans cancer cells, half-lives of 8 to less than 3 h, respectively; [[20], [21], [22]]. The polyadenylation status also impacts TERRA localization; polyadenylated molecules are found within the nucleoplasm while nonpolyadenylated localize to telomeres [87]. This suggests that different fractions have different fates and functions.
Human subtelomere regions are a patchwork of segmental duplications and structural variations, which vary from telomere to telomere [48,55,88]. Numerous studies have identified subtelomeres as hotspots of DNA breakage and repair. These hotspots allow interchromosomal recombination, and in consequence, the rapid evolution of subtelomeric regions, as well as the prevalence of large CNVs [3]. For simplifying, the subtelomeric region can be divided into two parts, depending on its general structure (Fig. 2): the telomere-proximal TAS sequences containing the TAR1 repeats and the telomere-distal part containing the subtelomere repeats (Srpt or SRE). These elements can be interspersed by internal (TTAGGG)n-like sequences (ITS [88]). The TAR1 region is composed of sequences variable in size (0–2 kb), similar to the TAR1 repeat family [89] and found at nearly all telomeres (Fig. 2). The subtelomeric repeat part encompasses the first 500 kb adjacent to each telomere and contains a high degree of segmental duplication. These duplicated sequences are also termed subtelomere duplicon blocks. Patterns of duplicated blocks in the first 25 kb of the subtelomere are shared between related subtelomeres, defining six duplicon families and seven unique subtelomeres (7q, 8q, 11q, 12q, 14q, 18q, XpYp [55]). The most recent and detailed subtelomeric sequence maps have been established by high-throughput single-molecule mapping [5], and a public browser of subtelomeric regions is available at www.vader.wistar.upenn.edu/humansubtel [4]. The subtelomeric region exhibits heterochromatic features; it is enriched with HP1, histone methylations H3K9me3, and H4K20me3 [43]. Furthermore, subtelomeric histones are hypoacetylated, and subtelomeric DNA is heavily methylated by DNA methyltransferases DNMT1, DNMT3a, DNMT3b [40].
Both genetic and epigenetic subtelomeric elements regulate TERRA levels [76,90,91], but still, little is known about TERRA promoters and transcriptional regulators. To date, they have been characterized only in humans and mice. Initially, it was proposed that each chromosome produced TERRA. Putative promoters have been identified at almost all subtelomeres in humans [90]. Then, further analysis discriminated two different types of promoters: those close to the telomeric repeats (at 8 chromosome ends) and those more distal (at 10 chromosomes), located at 5–10 kb from the telomeres [87]. More recently, it has been shown that chromosome 20q is the main TERRA locus in human cells [42]. The deletion of the 20q subtelomere region by the CRISPR-Cas9 method caused a significant decrease in TERRA levels, which led to the loss of telomere sequences, telomere de-protection and the induction of a massive DNA damage response [42]. In the same line, only two mouse chromosome ends have been shown to be responsible for TERRA production (chromosome 18 and 9 [92]). TERRA proximal promoters in mice and humans contain CpG-island sequences, but only in mouse cells are these sequences sensitive to DNA methylation by DNMT1 and 3b to repress TERRA transcription [93]. The chromatin at TERRA TSSs is enriched for active promoter histone-dedicated marks (such as H3K4me3) even though the predominant histone marks at the subtelomeres remain those associated with transcriptional repression and condensed chromatin, H3K9me3, and H4K20me3, respectively. CTCF, the transcriptional repressor, and insulator, form a barrier against the spreading of heterochromatin from telomeric repeats, and at the same time, control islands of open chromatin encompassing TERRA’s TSSs. Most of the human subtelomeres contain CTCF- and cohesin-binding sites within 1–2 kb of the telomeric repeats, which are proximal to CpG-islands controlling TERRA transcription [94]. It was proposed that CTCF recruits RNAPII and a cohesin ring at TERRA’s TSSs to orientate RNAPII transcription toward the telomere [4,94]. Interestingly, subtelomeric CTCF and cohesin (Rad21) also impact the binding of TRF1/2 proteins, two members of the shelterin complex, at telomeres and subtelomeres. In the same line, TRF2 depletion was shown to up-regulate TERRA [22]. This suggests that subtelomeres and telomeres communicate and that secondary loop-like structures could form and impact chromatin composition and TERRA transcription. Thus, it is likely that the transcriptional status of telomeres and the long-range epigenetic regulation interplay, which may indicate extra-telomeric functions for TERRA.
In contrast to these telomere-related functions, TERRA has recently been shown to be a part of genome-wide mechanisms regulating polycomb recruitment to pluripotency and cell-fate genes in mouse embryonic stem cells [95]. Earlier observations already suggested the role of the TRF1 shelterin protein in the regulation of pluripotency and tissue reprogramming. TRF1 is overexpressed in ES and iPS cells (induced pluripotent cells), in de-differentiated cells, as well as in the blastocyst inner membrane [96,97]. On the other hand, TRF1 depletion led to an important increase in TERRA levels [87,98]. Moreover, TRF1 has been previously shown to bind to the H3K27me3-rich regions in the genome and to interact with PRC2 in human models [79]. Inspired by these facts, Marion and colleagues (2019) assessed the genome-wide impact of TRF1 down-regulation in a murine model. They showed that upon TRF1 depletion, TERRA levels were greatly increased, correlating with enhanced association with PRC2-controlled (Polycomb Repressive Complex) genes [95]. Consequently, the lack of TRF1 decreased the expression of pluripotency genes and increased the expression of differentiation genes. This study indicates that TERRA could act as a factor binding specific chromatin domains to mediate the recruitment of Polycomb proteins (SUZ12 [95]).
TERRA in S. cerevisiae
In budding yeast S. cerevisiae, chromosome ends are classified in one of two types: X or Y’. X elements, composed of X core and associated variable sequences, are found in various forms at all chromosome ends. Most of the core X elements are approximately 475 bp long and contain an X-ACS region (ARS consensus sequence, which binds the Origin of Replication Complex/ORC [99]) and the conserved Abf1, a general regulatory factor binding site. The Y′ end (long or short, 6.7 kb or 5.2 kb, respectively) is found in about 70% of telomeres. It appears in 1–4 tandem copies separated by short tracks of (TG1-3)n telomeric repeats (ITS, internal telomeric sequences), which also border subtelomeres at the centromere-proximal end. Moreover, the telomeric repeats, X and Y’ elements are separated by small subtelomeric repeat sequences (STRs) that can vary in copy number. STRs contain potential binding sites for Tbf1p (DNA binding general regulatory factor). ITS and ARS sequences act as proto-silencers. X, as well as Y′ sequences, differ among telomeres by short, multiple insertions and/or deletions that correlate with a high frequency of recombination among subtelomeric regions [99,100]. Consequently, yeast subtelomeric regions are mosaic structures, which show a broad diversity not only among different strains but also within a population of the same strain. This heterogeneity is targeted by a variety of protein factors [101], leading to a tremendous diversity in chromatin structure and transcriptional output among individual chromosomal extremities [102]. Sequences of the last 30 kb of each chromosomal end, along with alignments of relevant subtelomeric elements, can be found at https://www2.le.ac.uk/colleges/medbiopsych/research/gact/resources/yeast-telomeres/telomere-data-sets [103]. Telomeric repeats in yeast are not organized in nucleosomes but are folded into multimers of the DNA-binding protein Rap1 (Repressor Activator Protein 1 [104]), which is recognized by different complexes. The Rap1-C-terminal domain binds Rif1 and Rif2, inhibiting telomerase activity and telomere lengthening [105], and tethers the silencing factors Sir3 and Sir4 [106], recruiting the NAD-dependent histone deacetylase Sir2p [107]. Sir2-mediated hypoacetylation of histones H3 and H4 creates a favorable environment for binding of Sir3 protein. Repetitive cycles of deacetylation and binding of Sir3 lead to the spreading of the SIR complex and transcriptional repression of the subtelomeric regions [108]. Sir2, Sir4, and Sir3 genomic occupancy are circumscribed to heterochromatin by Set1 and Dot1-mediated histone methylation [109,110], regulating both subtelomeric repression and the activity of a subset of promoters throughout the genome [106,[111], [112], [113]].
Yeast telomeres impose transcriptional repression on subtelomeres through TPE [[114], [115], [116]]. Contrary to telomeric repeats, subtelomeres contain histones. The X core is transcriptionally inactive, with large histone-free regions and low acetylation of histone H4 (H4K16). Y′ elements, especially the distal ones, are mostly euchromatic. X and Y′ elements underlie the maintenance of telomeric DNA through recombination [117] when cells enter into crisis and early senescence. Upon telomerase loss, telomeres can still be lengthened through ALTs [118]. The Type I (ALT1) results in multiple tandem Y’ elements that arise predominantly through replication-dependent recombination [117]; The type II (ALT2), probably lengthens TG1-3 tracks via rolling circle-mediated replication [119] and exhibits an increase of telomeric repeats. Extensive and comprehensive insights into yeast transient subtelomere associations and telomere biology are described in Ref. [120].
Yeast TERRA, which ranges from a few hundred to around 1.2 kb in length is rapidly degraded by the Rat1 exonuclease [86]. The impact of the subtelomeric region on TERRA transcription has been well studied in yeast, as has the importance of subtelomere structure on TERRA transcription and degradation [121]. Several TERRA TSSs have been identified on different chromosome ends. At Y′ telomeres, TERRA is repressed by the Rap1p-binding proteins Rif1/2, while the Sir2/3/4 complex has only a minor negative role. At X-only telomeres, both the Sir2/3/4 complex, as well as the Rif1/2 proteins, are important for promoting TERRA repression [121]. Moreover, TERRA transcription depends on telomere length and is probably affected by shortening-induced changes in subtelomeric chromatin structure. Experiments designed to monitor TERRA transcripts in single yeast cells by RNA fluorescence in situ hybridization (FISH), showed that telomere shortening induced TERRA expression. Overexpressed TERRA accumulated in a single perinuclear focus [83]. In the S phase, these foci form telomerase clusters, called T-Recs, suggesting that TERRA may act as a scaffold for the recruitment of telomerase to shorten telomeres [83]. Using cells lacking telomerase activity has further refined the characterization of this mechanism. Telomerase negative cells enter senescence due to critically short telomeres [122] that can be repaired by homology-directed repair (HDR). Graf and colleagues [123] have recently shown that TERRA accumulates at these very short telomeres, similarly to HDR-promoting RNA-DNA hybrid R-loops (for comprehensive review see Ref. [38]). TERRA downregulation reduced the level of telomeric R-loops, and in consequence, decreased levels of telomeric recombination in cis [124]. On the other hand, the upregulation of TERRA expression promoted recombination [125]. Thus telomeric R-loops mediate recombination at telomeres and induction of ALT type II survivors. Since telomeric R-loops form preferentially at short telomeres, they could prevent replicative senescence in telomerase-negative cells when the TERRA level is sufficient. Interestingly, the Rat1 nuclease, responsible for TERRA degradation, is locally depleted at short telomeres and preferentially recruited by the Rif2 protein at long telomeres [123].
Fission yeast’s TERRA
Fission yeast possesses three chromosomes with subtelomeres of approximately 100 kb. Chromosomes II and I contain telomeric repeats followed by Subtelomeric Heterochromatin (SH) sequences and a variety of genes. Chromosome III subtelomeres contain arrays of ribosomal DNA repeats [14,56]. The sequence of S. pombe subtelomeres can be found at http://www.pombase.org/status/sequencing-status. The telomere-proximal TAS region (Fig. 2) spans 50 kb and is composed of multiple segments of homologous sequences. The TAS heterochromatin is established at the centromere-homologous (cenH) sequence located in the SH region via the telomere-associated shelterin proteins and the RNAi machinery [56,126,127]. The telomere-distal region composed of unique sequences spans an additional 50 kb and forms a highly condensed particular chromatin structure called knob [128]. The levels of histone modifications at the knob region are particularly low when compared to adjacent chromosome euchromatin and to telomere-proximal subtelomeric heterochromatin [129]. The telomere-proximal region is characterized by high levels of H3K9me2 and the telomere-distal by low-levels of H3K9me2 [56]. The spreading of subtelomeric heterochromatin is controlled by a Shugoshin family protein, Sgo2, which serves as a barrier between telomere-proximal and -distal subtelomeres ([130], Fig. 2). Moreover, to prevent the spreading of heterochromatin into the chromosome, the telomere-distal knob is bordered by a nucleosome-free region. These important security mechanisms suggest that limiting the spread of telomeric heterochromatin is an important role of subtelomeric regions. Since S. pombe harbors only three chromosomes, the subtelomeric sequences can be relatively easily deleted, making possible the analysis of the lack of subtelomeres.
Surprisingly, experiments determining the role of subtelomere by complete deletion showed that the absence of the SH regions had no effect on mitotic cell growth, meiotic program, cellular stress responses, and telomere length [14]. However, recent experiments showed that epigenetic stability of subtelomeric chromatin is specifically controlled by TOR2, a major regulator of eukaryotic cellular growth [131], indicating that the subtelomere’s state responds to cellular growth conditions. Upon telomerase loss, subtelomeric deletions triggered inter- or intra-chromosomal circularized chromosomes. Furthermore, fusion around subtelomeres had occurred within homologous genomic regions—paralogous genes and LTRs—evidencing the importance of subtelomeres for telomere maintenance [14].
The role of SH regions in protecting gene expression was demonstrated by measuring the spreading of H3K9me2 using ChIP in strains lacking individual SH regions. In these strains, SH-adjacent subtelomeric regions were invaded by heterochromatin, which provoked gene repression in these regions [14]. Recent analyses demonstrated that TAS regions are highly recombinogenic and comprise a unique chromatin structure that is characterized by low levels of nucleosomes [9]. These characteristics are independent of TAS chromosomal position but are an intrinsic feature of these AT-rich sequences, as shown by their ectopic localization. TAS particular chromatin state is maintained by the interaction between shelterin protein Ccq1 and two heterochromatic repressor complexes, CLRC and SHREC [9]. All these elements suggest a role of subtelomeres in organizing chromatin and protecting gene expression homeostasis. This nucleosome-poor region marked by histone inactive marks serves as a buffering zone to protect the expression of subtelomeric genes from TPE. The telomere-distal subtelomere element organized into knob comprises chromatin boundaries that further block heterochromatin spreading [14].
The family of the fission yeast telomeric RNAs comprises the telomeric G-rich TERRA and the complementary C-rich transcripts, named ARIA (described below [63,64]). TERRA in S. pombe is transcribed by RNAPII, and only 10% of transcripts are polyadenylated. Polyadenylation has an impact on TERRA steady-state levels [63] and properties. For example, Trt1, the telomerase catalytic subunit, interacts only with polyadenylated molecules that lack large telomeric repeats. These TERRAs are spread in the nucleoplasm [84]. As in budding yeast, S. pombe TERRA levels increase when telomeres shorten. This polyadenylated fraction was suggested to tether telomerase present in the nucleus to the short telomeres. Recently, the generation of “transcriptionally inducible telomeres” evidenced that TERRA transcription stimulated telomere elongation in cis through telomerase action [84]. A detailed description of TERRA transcriptional regulation in S. pombe is presented below, together with subtelomeric transcripts. TERRA transcriptional regulation is mainly controlled by the Rap1 protein and its regulator Cactin [84,132].
ARIA
ARIA is a class of transcripts containing C-rich telomeric repeats, in which the telomere itself acts as a promoter [63]. ARIA is negatively regulated by Rap1 and Taz1 proteins, homologues of mammalian RAP1 and TRF1/2 respectively. Its transcription is neither affected by heterochromatin assembly factors (Clr4, Swi6) nor by mutations in the RNAi machinery (Dcr1, Rdp1, and Ago1-lacking cells). The role of ARIA is unknown, but its transcription regulation seems to directly depend on telomeric proteins, which suggests telomere-mediated function.
The existence of complementary telomeric transcripts (TERRA-ARIA) suggests that they may form double-stranded (ds)RNA intermediates, which could participate in gene silencing and the formation of heterochromatin. This could occur by at least two mechanisms, co-transcriptional gene silencing (CTGS) or post-transcriptional gene silencing (PTGS). CTGC is achieved by the degradation of nascent RNA and depends on both active transcription and the chromatin state of the transcribed locus. The siRNAs that guide chromatin-modifying enzymes in order to establish heterochromatin are generated from the “target” locus. PTGS comes in two flavors, (i) RNAi-mediated, which depends on chromatin modifications and involves siRNA and (ii) RNA-decay-mediated implicating recognition and modification of heterochromatic RNA by the Cid14/Trf4 complex and its degradation by the exosome. To date, no siRNA processed from telomeric and subtelomeric RNA pairs have been detected in fission yeast [64], suggesting that telomeric transcripts are not controlled via RNAi. However, the equivalent of ARIA-TERRA processing intermediates has recently been found in Arabidopsis thaliana [133].
PAR-TERRA a novel variant of TERRA
PAR-TERRA is a novel telomeric repeat-containing RNA. As shown by FISH experiments, only a fraction of total TERRA localizes to the telomeres (about 40% in humans; only a small amount in mouse cells). TERRA localization changes during differentiation; it co-localizes with the inactive X-chromosome (Xi) in differentiated cells, whereas in embryonic stem cells (murine and human), a significant TERRA fraction was found at the proximity of both sex chromosomes [21,134]. X-chromosome inactivation in early development has been intensively studied. It involves the X-inactivation center (XiC), which is composed of three noncoding genes Xist, Tsix, and Xite (reviewed in Ref. [135]). Analysis of the link between TERRA and noncoding transcription of sex chromosomes in mESCs showed that subtelomeric pseudoautosomal regions (PAR) from both sex chromosomes are transcribed [136]. Produced lncRNA was called PAR-TERRA [97]. As shown by Northern blot and FISH analyses, PAR-TERRA co-localizes with TERRA and has the same size distribution. Mechanistically, PAR-TERRA could tether PAR regions that initiate XiC pairing and lead to the inactivation of X-chromosome [136]. This suggests a nontelomeric function for this novel class of transcripts.
subTERRA in Saccharomyces cerevisiae
The yeast S. cerevisiae expresses subtelomeric lncRNAs called subTERRA [65]. SubTERRA is a family of heterogeneous and mostly unstable ncRNAs produced from Y′ subtelomeres. SubTERRA transcripts can clearly be distinguished from TERRA since they do not cover the terminal telomeric repeats. The size of yeast subTERRA transcripts ranges from 0.5 to 9 kb. They are RNAPII-dependent and are mainly regulated at a post-transcriptional level. Y′ antisense (reverse) transcription has previously been detected in mutants defective for RNA degradation pathways; reverse Y′ transcript accumulated in trf4 [137] and in rat1-1 mutants (as for the fission yeast’s ARRET [86]), already suggesting that these silenced regions are transcribed into unstable ncRNAs. While present at low levels in wild type cells, subTERRA strongly accumulates in mutants for both cytoplasmic (Xrn1p) and nuclear RNA decay pathways (Trf4p), indicating that these two RNA degradation pathways are implicated in subTERRA degradation. Each degradation pathway is specific to the different set of transcripts: subTERRA-CUTs (Cryptic Unstable Transcripts [138,139]), sensitive to nuclear degradation, are transcribed toward the centromere, and subTERRA-XUTs (Xrn1-dependent Unstable Transcripts [140]) are preferentially degraded in the cytoplasm by Xrn1p (Fig. 3) and transcribed toward the telomere.
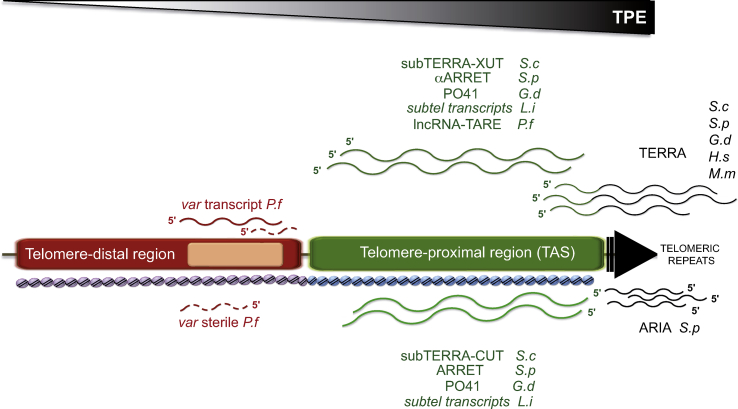
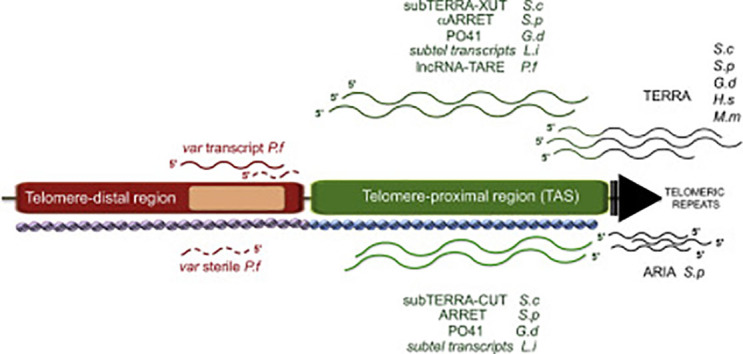
Fig. 3.
lncRNA expressed at subtelomeric regions. The telomere-proximal region is shown in green and the telomere-distal region in red. In various species, TERRA (G-rich) is transcribed from subtelomere-embedded promoters through telomeric repeats (black arrow) and contains subtelomeric sequences at its 5’. Telomeric repeats produce also ARIA (C-rich) transcripts composed exclusively of telomeric sequences. lncRNA species encoded within TAS regions are depicted in green. Upper transcripts are transcribed toward the telomere (subTERRA-XUT in S. cerevisiae (S.c), αARRET in S. pombe (S.p), PO41 in G. domesticus (G.d), subtelomeric transcripts in L. infantum (L.i) and lncRNA-TARE in P. falciparum (P.f)). lncRNAs transcribed toward the centromere (subTERRA-CUT in S. cerevisiae, ARRET in S. pombe, PO41 in G. domesticus, subtelomeric transcripts in L. infantum) are represented below. In P. falciparum different var transcripts are produced from var genes (pink rectangle); var coding full-length transcript (red line), and var sterile noncoding transcripts (dashed red lines) transcribed in both direction from the intron. TAS and telomere-distal regions harbor specific chromatin states represented here as arrays of violet and blue nucleosomes. Decreasing TPE direction is shown. H.s stands for Homo sapiens, M.m for Mus musculus.
Certain similarities can be observed between TERRA and subTERRA. Like TERRA, subTERRA localizes in the nucleus and the expression of both transcripts is cell cycle-regulated. However, they are anticorrelated and can hardly be found in the cell at the same time, at least at detectable levels. TERRA is mainly degraded by Rat1 exonuclease prior to telomere replication. This prevents the generation of new telomere R-loops, which in excess could be harmful to the cell [123]. subTERRA accumulates before cells enter the S phase i.e. before cells start to replicate their DNA. This suggests that an increased transcription level or accumulation of subTERRA could be necessary for the establishment of open DNA replication-prone structures in subtelomeric regions. Alternatively, increasing subTERRA could control the formation of S-phase-specific RNP-molecules required for telomere replication and elongation by telomerase. The role of subTERRA is not clear yet. Direct experiments aiming at its overexpression using GAL1-induced promoters have not enabled conclusions regarding an RNA-mediated function. Furthermore, cells have shown no or heterogeneous phenotypes regarding their growth and effects on silencing or replicative senescence ([65] and our unpublished data). However, indirect experiments using RNA decay mutants have shown specific phenotypes in mutants accumulating different sets of subTERRA transcripts and suggested a subTERRA action in cis. The effect of subTERRA transcript accumulation on TPE was tested using the native reporter system with a URA3 reporter gene integrated into nonmodified subtelomere Y’ sequences at telomere IXL [50]. These experiments showed that subTERRA-CUTs enhanced TPE. On the other hand, the accumulation of subTERRA-XUTs counteracted telomeric clustering and strongly affected the number of telomeric foci, suggesting a role of this subset of transcripts in telomere clustering. Interestingly, Rap1 protein was shown to be a negative regulator of subTERRA, acting rather post-transcriptionally by promoting subTERRA degradation. Altogether, those experiments suggest that subTERRA could be produced to facilitate the establishment of specific heterochromatic states of subtelomeric chromatin and should be taken into account in the complex picture of telomere metabolism regulation. The mechanism of action is not known, but an interesting idea is the possibility of complementary transcript pairing and the formation of dsRNAs. This could only be possible if all, sense and antisense, subTERRA transcripts are present in the same cell. Since S. cerevisiae is devoid of an RNAi pathway, the role of helicases [141] and RNases should be more carefully evaluated in single-cell dedicated experiments.
ARRET, αARRET – Schizosaccharomyces pombe
Extensive characterization of sub- and telomeric transcripts by two groups [63,64] allowed the discovery of ARRET and its antisense called αARRET. These subtelomeric RNAs are complementary to the centromere-proximal subtelomeric region of TERRA but lack telomere repeats (see Fig. 3). Both transcripts seem to be RNAPII-dependent as Chromatin Immunoprecipitation (ChIP) experiments show the localization of RNAPII at their TSSs. In addition, the αARRET cellular levels decrease upon functional inactivation of the RNAPII subunit Rpb7 [63]. The large majority of ARRET and αARRET are polyadenylated, and the differential polyadenylation by Cid12 and Cid14 impacts their steady-state levels [63].
The expression of telomeric and subtelomeric transcripts is differentially regulated in S. pombe. Telomere-associated proteins control telomeric and subtelomeric lncRNAs. Heterochromatin assembly factors (Clr4, Swi6) are only important for subtelomeric transcription [64] as clr4 and swi6 mutants exhibited increased levels of subtelomeric transcripts while the level of telomere repeat-containing transcripts remained unchanged [64]. Rap1, telomere-associated protein, and a member of the shelterin complex restricts RNAPII binding to TERRA TSS [64]. When there was a lack of Rap1 or Taz1 (another member of shelterin), both subtelomeric and telomeric RNAs accumulated. This suggests that Rap1 and Taz1 telomere-associated proteins are implicated in the regulation of subtelomeric and telomeric RNA expression at the transcriptional level. Access of RNAPII to the subtelomeric promoter/s may depend on the telomere state, i.e., closed at long telomeres or open at short ones [84].
The transcriptional control of the telomere and subtelomere lncRNAs remains poorly characterized, and transcriptional activators/repressors and RNA factors binding the telomere-associated transcripts have still not been extensively studied. Recent works addressing this question showed that Translin (Tsn1) and Trax (Tfx1) proteins, two highly conserved nucleic acid-binding proteins implicated in RNA regulation, both reciprocally regulate sub- and telomeric transcripts [142]. Trax represses subtelomeric ARRET while Translin induces its transcription. Vice versa, Translin represses TERRA transcripts whereas Trax maintains high levels of TERRA. This reveals a novel regulation mechanism of telomere-associated transcription and potential functional implications for TERRA and subTERRA in physiological contexts [142].
The comparison of the fission and budding yeast telomere-associated transcriptomes shows obvious analogies, suggesting functional conservation between these two yeasts for telomere metabolism. The budding yeast’s subTERRA family [65] share properties with ARRET and its complementary αARRET species and suggests that the only, still nonidentified transcript, ARIA, should be carefully looked at in budding yeast. Both organisms use noncanonical poly(A) polymerases for polyadenylation of telomere-associated transcripts; Cid14 and Trf4, component of the TRAMP complex in S. cerevisiae, are functional homologues. This suggests that polyadenylation is a mark directing these RNAs for decay by the exosome. Differential cellular localization and stability of polyA+ and polyA- TERRA fractions supports this idea [22,87]. Finally, analogous to subTERRA transcripts in budding yeast, the Rap1 telomeric protein impacts the expression of subtelomeric and telomeric transcripts in S. pombe.
As for telomeric transcripts, the existence of two subsets of subtelomeric transcripts suggests that they form dsRNA intermediates, which could participate in gene silencing and the formation of heterochromatin. Although the implication of an RNAi machinery has not been evidenced in the regulation of subtelomeric transcripts, one should bear in mind that the analyses of subtelomeric regions (DNA sequences and the transcriptome) are technically challenging. Therefore analyses of these regions are still not exhaustive. Indeed, subtelomeric siRNAs located several kilobases upstream of ARRET 3’ ends have only been detected recently in S. pombe [56,63].
Transcripts With NonTelomeric Functions
This chapter describes transcripts with different functions, mostly sensu stricto subtelomeric transcripts that do not contain telomeric repeats.
var and lncRNA-TARE – fascinating chromosome ends in Plasmodium falciparum
The subtelomeric regions of the protozoan malaria parasite Plasmodium falciparum follow the general organization scheme and consist of two regions: telomere-associated sequences (TAS) neighboring telomeric repeats and the telomere-distal region harboring members of gene families coding for virulence factors, including var genes responsible for antigenic variation and cytoadhesion [143,144]. TAS region (20–40 kb) is composed of six different telomere-associated repetitive elements (TARE 1 to 6) and has a highly conserved organization. Six TAREs are positioned in the same relative order within all chromosomes, while their sizes and DNA sequences are polymorphic [145]. The closest to the telomere, TARE-1, is built of tandem repeats, and its size varies between 0.9 and 1.9 kb. TARE-2 is 1.6 kb long and consists of a 135-bp long, degenerated sequence repeated 12 times, interspersed by two distinct 21-bp sequences [145]. TARE-3 is composed of three to four repeated elements of 0.7-kb. TARE-4 ranges from 0.7 to 2 kb. It is composed of highly degenerated, short repeats, and an interspersed nonrepetitive sequence of 230 bp. TARE-5 varies from 1.4 to 2 kb and is composed of moderately degenerated tandem repeats of 12 bp (5′ ACTAACA(T/A)(C/G)A(T/C)(T/C)). TARE-6 corresponds to the Rep20 element, which contains a degenerated 21-bp sequence ([146]; Fig. 2). Its length varies from 8.4 to up to 21 kb.
P. falciparum chromosome ends form clusters at the nuclear periphery (four to seven telomeres per cluster), facilitating ectopic recombination among heterologous subtelomeric chromosome regions, including genes coding for virulence factors [47]. In experiments analyzing deletions of TAS, the localization of chromosome ends to the nuclear periphery was not affected while cluster formation was. This suggests that chromosome ends lacking subtelomeric regions can dissociate from clusters [10]. Like in fission and budding yeast, TAS deletions do not detectably affect the parasite’s life. However, as already described for S. pombe, TAS regions serve as spacers, separating different chromatin states and protecting subtelomeric genes, like var genes, from TPE. Moreover, deletions of regions containing TAS provoked elongation of telomere repeats, suggesting that TAS elements are responsible for telomere length homeostasis [10]. The mechanism responsible for this effect is still not known, but it has been suggested that chromosome end folding may play a role in the process.
The var gene cluster constitutes a particular part of the subtelomeric region in P. falciparum. var genes are located downstream of TARE-6 (Rep20 [147]). The var gene family codes for PfEMP1 (erythrocyte membrane protein 1), which is expressed at the surface of infected erythrocytes. For avoiding clearance by the host’s anti-PfEMP1 antibodies, P. falciparum can change the variant form of expressed PfEMP1. Thus, P. falciparum evades the host’s immune response by switching expression of the var gene family coding for variant surface proteins. However, only one var gene expresses full-length transcripts in any one parasite, while other members of this family are kept silent [148]. Antigenic variation of PfEMP1 is generated through in situ activation of a silenced var gene and does not involve recombination. Duraisingh and colleagues [149] showed that PfSir2 protein, a homolog of yeast Sir2 histone deacetylase, is implicated in var gene silencing. Moreover, experiments with reporter genes inserted in subtelomeric locations showed that (i) local chromatin structure determines whether the transcriptional machinery has access to the var genes and (ii) var gene transcriptional activation depends on their nuclear location [149]. The importance of histone acetylation, histone methylation, and the propagation of heterochromatin in the control of var genes has been well documented [[150], [151], [152], [153]]. Recent data has shown that var silencing implicates lncRNA sense and antisense var transcripts produced from intron-containing bi-directional promoters [154] that participate in chromatin structure regulation. Two promoters regulate each var gene [[155], [156], [157]]; the upstream promoter from the only expressed gene produces var mRNA, while the intron promoter produces a noncoding RNA [158]. Surprisingly, the transcriptional activity of the intron promoter is required for the silencing of the upstream var promoter [155,157,159]. The first detected noncoding transcript transcribed from a var intron promoter was named “sterile var transcript” since it was truncated and could not produce a full-length protein (only part of exon2 was detected [160]). Recent investigations have identified an antisense RNA transcript of var exon1 produced by the intron promoters. This nonpolyadenylated RNA localizes to the chromatin of the var gene loci and potentially participates in the structural organization and epigenetic regulation of this gene family [154]. Furthermore, transcription of sense and antisense sterile var transcripts was shown to peak simultaneously with lncRNA-TARE during parasite invasion [67]. This suggests the unique functional coordination of lncRNA-TARE and sterile var transcripts upon parasite invasion and the importance of subtelomeric regions and their chromatin organization in this vital process.
Global transcriptional analyses of P. falciparum revealed a novel family of telomere-associated transcripts [66,67] termed lncRNA-TARE, from the telomere-associated repetitive elements present at subtelomeres. Homologous lncRNA-TARE loci were mapped on 22 of 28 chromosome ends, and 23 lncRNA-TARE transcripts have been detected and characterized. lncRNA-TARE is long (about 4 kb), with high-GC content, and are transcribed toward the telomere (see Fig. 3). All of them showed coordinated and stage-specific expression after parasite DNA replication during an invasion, with two major transcripts ranging from 1.5 kb to 3.1 kb, starting at the extremity of TARE-3 in the direction of the telomere. Moreover, identified lncRNA-TARE sequences were systematically enriched with transcription factor binding sites (SPE2 recognized by PfSip2) found only at upsB-type var gene promoters. PfSip2 specifically binds subtelomeric SPE2 sites to participate in the silencing of var genes [161]. Since PfSip2 is expressed and binds SPE2 sites at the stage of maximal lncRNA-TARE transcription, this suggests its implication in the regulation of the lncRNA-TARE locus. In turn, lncRNA-TARE locus could participate in regulating upsB-type var genes by directly or indirectly interacting with and/or recruiting PfSip2 [66].
PO41
Chicken subtelomeric regions contain multiple classes of tandemly repeated units comprising the same family and sharing a common ancestor. All subtelomeric repeats show structure and sequence similarities; their core repeat unit is 21 bp-long and contains (A)3–5 and (T)3–5 clusters separated by 5–7-bp sequences. Four major classes have been characterized to date: CNM, PIR, EcoRI/XhoI and PO41 [162]. The most abundant repeat type is CNM (chicken nuclear-membrane-associated [163]), a tandem repeat with a unit size of 40 bp. CNM is dispersed within a large number of microchromosomes but is not present in macrochromosomes 1–5, ZW, and in some of the intermediate chromosomes 6–10. PIR (partially inverted repeat [164]) represents a family of multiple types of partially inverted repeat units of about 1.2, 1.4, and 1.6 kb. The PIR sequence has been located only on chicken chromosome 8 and shown to spread over about 3.8 Mb. EcoRI/XhoI family comprises about 70% of the W sex chromosome [165]. PO41 that stands for “Pattern Of 41 bp” is a short subtelomeric sequence and is locus-conserved in all galliform species.
The transcription of these conserved subtelomeric repeats was firstly shown during the lampbrush stage of oogenesis [166], and recently in somatic tissues (in brain, muscles, oviduct, intestine, and eye of adult chicken and Japanese quail females), during chicken embryogenesis, as well as in the chicken MDCC-MSB1 malignant cell line [68,69]. The PO41 transcripts are transcribed from both strands, but in the cell, they predominantly exist in a single-stranded form with short double-stranded regions. In interphase, these long-lived ncRNAs seem to be retained in the nucleus and form one or two major foci with some PO41 transcripts dispersed in euchromatin. During mitosis, PO41 covers chromosomes. Arrays of the conserved PO41 repeat lie adjacent to the CNM repeat, which is transcribed during the lampbrush stage from both strands forming loop-specific patterns of G-and C-rich transcripts [68,69].
The role of these transcripts remains elusive, but their features suggest the existence of dsRNA generation and RNAi-dependent mechanisms involved in the assembly and maintenance of heterochromatin at subtelomeres [167]. Moreover, an interesting scenario concerning the role of these transcripts in embryo development has been proposed [167]: single-stranded G and C transcripts could form long dsRNAs, which could then be processed into short dsRNAs and accumulate in oocytes. After fertilization, the dsRNAs would be transferred into the embryo and play a role in the regulation of the early stages of embryogenesis before activation of the embryonic genome.
Subtelomeric transcripts in Leishmania infantum
L. infantum is a protozoan parasite, a prevalent pathogen in humans. The ability of these parasites to survive is dependent on tightly regulated gene expression. It was recently evidenced that a novel class of sense and antisense transcripts are produced from subtelomeric, head-to-tail tandem repeats in a developmentally dependent manner. These RNAPII-dependent transcripts are spliced and polyadenylated. Localization experiments (FISH and sedimentation) indicate their presence in the cytoplasm and potential association with a small RNP complex [71]. Overexpression of these subtelomeric transcripts at different developmental stages failed, proving their efficient degradation.
The role of these subtelomeric transcripts is still unknown, but their discovery confirms that our knowledge on subtelomeres is still incomplete and that more investigations need to be performed.
Human subtelomeric genes
While the functions of some human coding and noncoding transcripts at subtelomere regions have been described in the literature, most remain incompletely characterized [19,[168], [169], [170]]. The abundance of these transcripts and the regulation of their expression depend on their sequence and local chromatin status at the subtelomere encoding them, which potentially varies from one chromosome to another and within a population (reviewed in Ref. [59]). The most important events shaping their transcription are (i) the CNV, which results in changes in the number and form of subtelomere gene alleles, and (ii) location of the genes at specific subtelomeres, which, due to the important variations in sequence and GC skew, impacts the binding of various factors and the formation of secondary structures. Thus, interindividual and even allelic variations in the expression of subtelomeric genes, coding and noncoding, could be a determinant for phenotypic differences in humans. Except for TERRA, described above, little is known about the majority of these telomere-proximal lncRNAs.
WASH gene family
The WASH proteins participate in cytoskeletal organization and signal transduction and regulate the polymerization of actin filaments in response to external stimuli. They correspond to a third identified subclass of the highly conserved and widely expressed Wiskott-Aldrich Syndrome Protein (WASP) family [19]. The WASH genes duplicated during evolution to multiple subtelomeric locations. The WASH protein family is single-copy in lower organisms and in early primates, but multicopy in great apes, with the highest copy number in humans [59]. The WASH genes encode for short and long forms of the protein. The 3’ exons encoding the short form are embedded in subterminal duplicon block C, the subtelomere part nearest to telomeric repeats [55], whereas the full-length forms start near a CpG island, 20 kb from telomeres within 16 chromosome ends [171].
Inter-individual and even allelic variations in the subtelomeric organization (sequence, copy number and/or telomere length) could influence WASH gene expression. Accordingly, differences in the sequences and number of intact copies, as well as their location within the subtelomere, were observed [19]. Since WASH genes are widely expressed, mainly in hematopoietic tissues and some brain zones, those changes could lead to phenotypic differences and pathologies. In the same line, it was suggested that the overexpression of WASH genes observed in a breast cancer cell line could contribute to metastasis, as shown for other WASP proteins (N-WASP and the SCARs [172,173]).
RPL23AP7 gene family
The second major family of subtelomeric transcripts consists of genes that are related to a ribosomal protein pseudogene RPL23AP7. Similarly to the WASH gene family, RPL23AP7 exists in two forms, interrupted and full-length open reading frames [55]. Intriguingly, the RPL23AP7 pseudogene and WASH genes are transcribed in the same orientation, and their 3’ end positions correspond to TERRA TSSs. This raises the question of whether these transcripts are coregulated or how they impact each other [59].
DDX11L
This is a novel transcript family, which maps to 15 subtelomeric locations in humans [70]. The DDV11L gene originated as a rearranged portion of the primate DDX11 gene, which propagated in multiple subtelomeric locations in human chromosomes. Initially described as a pseudogene of the CHLR1-related helicase gene (CHLR1, homologous to CHL helicase of S. cerevisiae), the DDX11L family seems to be transcribed in many human tissues and subjected to canonical splicing. Although their biological function remains unknown, transcription of DDX1L regions suggests that this gene has emerged from an inactive pseudogene and is still undergoing a neofunctionalization process [70]. Interestingly, each DDX11L gene is neighbored by telomeric repeats and WASH genes at its centromere-proximal end and is transcribed in the opposite direction to WASH genes, away from the telomere. It is unknown if and how transcription of DDX11L impacts the transcription of WASH genes.
D4Z4
Analysis of the subtelomeric regions revealed that interchromosomal exchanges took place in primate subtelomeric regions in recent evolution [174,175]. By analogy with segmental duplications in general, subtelomeric repeats are important for chromosome rearrangement and instability. Subtelomeric loci have been proposed to increase gene diversification by subtelomere-to-subtelomere interchromosomal exchanges (ectopic recombination), which lead to sequence rearrangements (intels, duplications, SNP). Due to their repetitive character, subtelomeres are unstable and provide all the elements for the quickest genetic adaptation to occur by promoting sequence recombination and duplication in an allele independent manner [176].
Subtelomere-associated diseases are the direct consequences of these features. Long-range interactions of telomeres with subtelomeric genes can regulate the expression of specific subtelomeric genes, and in some contexts, the de novo deletion of duplicated subtelomeric genes can cause diseases. For example, the type I immunodeficiency centromeric instability and facial anomalies syndrome (ICF1), the fascioscapulohumeral muscular dystrophy (FSHD), and some mental retardations are due to subtelomeric dysfunction. ICF1 involves limited DNA hypomethylation caused by mutations in the DNMT3B gene [176]. In ICF1 patients, subtelomeres are hypomethylated, and abnormally elevated TERRA levels and short telomeres are observed. In consequence, some chromosome rearrangements occur. The FSHD is caused by deletions of the subtelomeric D4Z4 locus [177] due to the exchange of the tandemly repeated subtelomeric D4Z4 sequence tract between 4q and 10q telomere regions [178,179]. The size of the 4q and 10q’ D4Z4 repeats is highly variable in individuals. The contraction of D4Z4 to the critical size associated with local hypomethylation and changes in chromatin relaxation at subtelomeres increases the likelihood of toxic DUX4 (4q35.2) gene expression in skeletal muscle. It is thought that this disease results from CNV and TPE modifications at subtelomeres. Additionally, it has been proposed that cryptic subtelomeric rearrangements, known to occur in humans, could account for 7.4% of patients with unexplained mental disorders [180].
Conclusions
The subtelomeres are regions bearing various functions. They determine telomere maintenance, the formation of telomeric clusters, chromatin state regulation, chromosome stability, and production of a variety of transcripts. Telomere-proximal repeats, composing TAS elements, produce mainly noncoding transcripts (i.e. ARRET, αARRET, subTERRA, lncRNA-TARE, ARIA and TERRA) while telomere-distal regions contain mainly the subtelomeric gene families. Subtelomeric genes evolve faster than nonsubtelomeric gene families and support rapid clonal phenotypic switches. These are important for survival and proliferation in hosts, as shown for parasite antigenic variation genes (in Plasmodium, Leishmania, and Trypanosoma) or rapid adaptation to novel niches (for yeasts). In humans, well-known subtelomeric genes important for phenotypic variations are the olfactory receptor and immunoglobulin heavy chain genes [18,181].
The regulation of subtelomeric gene transcription also involves chromatin. One of the particularities of subtelomeric regions is reversible silencing mediated by TPE through binding of proteins to the telomeres and interacting with subtelomeric proteins and/or sequences. TPE and TAS noncoding transcription contributes to gene switching and mutually exclusive expression. Further investigations would be necessary to explore these novel types of regulation. Interestingly, coregulated expression of lncRNA-TARE and sterile var transcripts, as well as regulation of var genes by sterile var transcripts suggest that the expression of one noncoding RNA would impact the transcription of another one. This could be achieved by changes in chromatin state by ongoing transcription or the recruitment of specific factors by the produced RNA (in cis). It has not yet been addressed whether these transcripts could also act on loci located at different subtelomeres (in trans). Some inputs coming from TERRA studies show that TERRA acts in trans and in cis. TERRA forms telomeric R-loops impacting replicative senescence and parallel-stranded G-quadruplexes, which are important in higher-order structure formation potentially implicating RNA:DNA hybrids [182].
The presence of complementary subtelomeric noncoding RNAs, as detected in chicken, P. falciparum, and yeast cells, suggests their implication in chromatin formation through a siRNA mechanism. It is easily imaginable, but still not proved that long dsRNAs are processed to form siRNAs and nucleate heterochromatin. This mechanism would not be possible in the S. cerevisiae budding yeasts since they lack all components of the siRNA machinery, suggesting the implication of a different mechanism. One can imagine that dsRNA regulates the population of ssRNAs, which have been shown to regulate TPE and telomere clustering when accumulated [65]. dsRNAs, since potentially more stable, could serve as a reservoir of ss-forms. In this case, helicases and RNA decay systems would be implicated in their degradation and regulation [141]. Recent results have implicated TERRA in the regulation of gene silencing throughout the genome. These have given a new direction in the field of telomeric and subtelomeric transcription.
Perspectives
Subtelomeric transcription has been discovered quite recently; thus, the field is still in its early days. The role of subtelomeres and emerging domains of subtelomere-associated transcription will benefit from progress in the analysis of subtelomere structures and sequences in various organisms. This step is necessary for the creation of tools to answer more specific questions. The important input into subtelomeric transcript characterization could come from the identification of subtelomeric transcript interacting factors and binding through the genome. Using RIP [183], ChIRP [184], and CHART [185] experiments would be informatory for their role and mechanism of action. Nonetheless, the most important step forward, in our opinion, will be the improvement of methods allowing subtelomeric transcript gain and loss of function experiments (over-expression, as well as complete depletion), keeping in mind cell-cycle or cell development specificities. This may be achieved by specific and fine regulation of transcription (manipulation of promoter sequences and transcriptional activators). For example, the use of sequence-specific antisense oligonucleotides (ASO) already allows specific degradation of targeted transcripts, which permits the analysis of the impact of their depletion. In addition, single-cell experiments will be a precious source of information to determine the causal link between subtelomere genetic engineering and cell fate.
Acknowledgements
MK is supported by CNRS (UMR 5099); AM is supported by the European Research Council (DARK consolidator grant) and by the Agence Nationale de la Recherche (DNA-Life). We thank Melanie Gettings and Athula Murti for English proof reading.
Edited by Brian Luke.
References
- 1.Young E., Abid H.Z., Kwok P.-Y., Riethman H., Xiao M. Comprehensive analysis of human subtelomeres by whole genome mapping. BioRxiv. 2019 doi: 10.1101/728519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riethman H., Ambrosini A., Paul S. Human subtelomere structure and variation. Chromosome Res. 2005;13:505–515. doi: 10.1007/s10577-005-0998-1. [DOI] [PubMed] [Google Scholar]
- 3.Riethman H. Human telomere structure and biology. Annu. Rev. Genom. Hum. Genet. 2008;9:1–19. doi: 10.1146/annurev.genom.8.021506.172017. [DOI] [PubMed] [Google Scholar]
- 4.Stong N., Deng Z., Gupta R., Hu S., Paul S., Weiner A.K. Subtelomeric CTCF and cohesin binding site organization using improved subtelomere assemblies and a novel annotation pipeline. Genome Res. 2014;24:1039–1050. doi: 10.1101/gr.166983.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young E., Pastor S., Rajagopalan R., McCaffrey J., Sibert J., Mak A.C.Y. High-throughput single-molecule mapping links subtelomeric variants and long-range haplotypes with specific telomeres. Nucleic Acids Res. 2017;45:e73. doi: 10.1093/nar/gkx017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Islam T., Ranjan D., Zubair M., Young E., Xiao M., Riethman H. Analysis of subtelomeric REXTAL assemblies using QUAST. IEEE ACM Trans. Comput. Biol. Bioinf. 2019 doi: 10.1109/TCBB.2019.2913845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craven R.J., Petes T.D. Dependence of the regulation of telomere length on the type of subtelomeric repeat in the yeast Saccharomyces cerevisiae. Genetics. 1999;152:1531–1541. doi: 10.1093/genetics/152.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berthiau A.-S., Yankulov K., Bah A., Revardel E., Luciano P., Wellinger R.J. Subtelomeric proteins negatively regulate telomere elongation in budding yeast. EMBO J. 2006;25:846–856. doi: 10.1038/sj.emboj.7600975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Emden T.S., Forn M., Forné I., Sarkadi Z., Capella M., Martín Caballero L. Shelterin and subtelomeric DNA sequences control nucleosome maintenance and genome stability. EMBO Rep. 2019;20 doi: 10.15252/embr.201847181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figueiredo L.M., Freitas-Junior L.H., Bottius E., Olivo-Marin J.-C., Scherf A. A central role for Plasmodium falciparum subtelomeric regions in spatial positioning and telomere length regulation. EMBO J. 2002;21:815–824. doi: 10.1093/emboj/21.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacob N.K., Stout A.R., Price C.M. Modulation of telomere length dynamics by the subtelomeric region of tetrahymena telomeres. Mol. Biol. Cell. 2004;15:3719–3728. doi: 10.1091/mbc.e04-03-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sadaie M., Naito T., Ishikawa F. Stable inheritance of telomere chromatin structure and function in the absence of telomeric repeats. Genes Dev. 2003;17:2271–2282. doi: 10.1101/gad.1112103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calderón M. del C., Rey M.-D., Cabrera A., Prieto P. The subtelomeric region is important for chromosome recognition and pairing during meiosis. Sci. Rep. 2014;4:6488. doi: 10.1038/srep06488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tashiro S., Nishihara Y., Kugou K., Ohta K., Kanoh J. Subtelomeres constitute a safeguard for gene expression and chromosome homeostasis. Nucleic Acids Res. 2017;45:10333–10349. doi: 10.1093/nar/gkx780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jolivet P., Serhal K., Graf M., Eberhard S., Xu Z., Luke B. A subtelomeric region affects telomerase-negative replicative senescence in Saccharomyces cerevisiae. Sci. Rep. 2019;9:1845. doi: 10.1038/s41598-018-38000-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ottaviani A., Schluth-Bolard C., Rival-Gervier S., Boussouar A., Rondier D., Foerster A.M. Identification of a perinuclear positioning element in human subtelomeres that requires A-type lamins and CTCF. EMBO J. 2009;28:2428–2436. doi: 10.1038/emboj.2009.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hocher A., Ruault M., Kaferle P., Garnier M., Descrimes M., Morillon A. Expanding heterochromatin reveals discrete subtelomeric domains delimited by chromatin landscape transitions. BioRxiv. 2017 doi: 10.1101/187138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linardopoulou E.V., Williams E.M., Fan Y., Friedman C., Young J.M., Trask B.J. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature. 2005;437:94–100. doi: 10.1038/nature04029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linardopoulou E.V., Parghi S.S., Friedman C., Osborn G.E., Parkhurst S.M., Trask B.J. Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS Genet. 2007;3:e237. doi: 10.1371/journal.pgen.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azzalin C.M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 21.Schoeftner S., Blasco M.A. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 22.Porro A., Feuerhahn S., Reichenbach P., Lingner J. Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol. Cell Biol. 2010;30:4808–4817. doi: 10.1128/MCB.00460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blackburn E.H. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett. 2005;579:859–862. doi: 10.1016/j.febslet.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 24.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 25.Hug N., Lingner J. Telomere length homeostasis. Chromosoma. 2006;115:413–425. doi: 10.1007/s00412-006-0067-3. [DOI] [PubMed] [Google Scholar]
- 26.Mason J.M., Frydrychova R.C., Biessmann H. Drosophila telomeres: an exception providing new insights. Bioessays. 2008;30:25–37. doi: 10.1002/bies.20688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greider C.W., Blackburn E.H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 28.Meyne J., Ratliff R.L., Moyzis R.K. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc. Natl. Acad. Sci. U.S.A. 1989;86:7049–7053. doi: 10.1073/pnas.86.18.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Lange T. Shelterin-mediated telomere protection. Annu. Rev. Genet. 2018;52:223–247. doi: 10.1146/annurev-genet-032918-021921. [DOI] [PubMed] [Google Scholar]
- 30.de Lange T. T-loops and the origin of telomeres. Nat. Rev. Mol. Cell Biol. 2004;5:323–329. doi: 10.1038/nrm1359. [DOI] [PubMed] [Google Scholar]
- 31.Doksani Y., Wu J.Y., de Lange T., Zhuang X. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013;155:345–356. doi: 10.1016/j.cell.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 33.Sundquist W.I., Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 34.Lin C., Yang D. Human telomeric G-quadruplex structures and G-quadruplex-interactive compounds. Methods Mol. Biol. 2017;1587:171–196. doi: 10.1007/978-1-4939-6892-3_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paeschke K., Simonsson T., Postberg J., Rhodes D., Lipps H.J. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 36.Arora R., Lee Y., Wischnewski H., Brun C.M., Schwarz T., Azzalin C.M. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014;5:5220. doi: 10.1038/ncomms6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Y., Bennett H.W., Liu N., Moravec M., Williams J.F., Azzalin C.M. RNA-DNA hybrids support recombination-based telomere maintenance in fission yeast. Genetics. 2019;213:431–447. doi: 10.1534/genetics.119.302606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toubiana S., Selig S. DNA:RNA hybrids at telomeres - when it is better to be out of the (R) loop. FEBS J. 2018;285:2552–2566. doi: 10.1111/febs.14464. [DOI] [PubMed] [Google Scholar]
- 39.Blasco M.A. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- 40.Gonzalo S., Jaco I., Fraga M.F., Chen T., Li E., Esteller M. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat. Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- 41.Benetti R., García-Cao M., Blasco M.A. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat. Genet. 2007;39:243–250. doi: 10.1038/ng1952. [DOI] [PubMed] [Google Scholar]
- 42.Montero J.J., López de Silanes I., Graña O., Blasco M.A. Telomeric RNAs are essential to maintain telomeres. Nat. Commun. 2016;7:12534. doi: 10.1038/ncomms12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.García-Cao M., O'Sullivan R., Peters A.H.F.M., Jenuwein T., Blasco M.A. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- 44.Rudd M.K., Friedman C., Parghi S.S., Linardopoulou E.V., Hsu L., Trask B.J. Elevated rates of sister chromatid exchange at chromosome ends. PLoS Genet. 2007;3:e32. doi: 10.1371/journal.pgen.0030032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen N.W.G., Thareau V., Ribeiro T., Magdelenat G., Ashfield T., Innes R.W. Common bean subtelomeres are hot spots of recombination and favor resistance gene evolution. Front. Plant Sci. 2018;9:1185. doi: 10.3389/fpls.2018.01185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Louis E.J., Becker M.M., editors. Subtelomeres. Illustrated. Springer Science & Business Media; 2013. [Google Scholar]
- 47.Freitas-Junior L.H., Bottius E., Pirrit L.A., Deitsch K.W., Scheidig C., Guinet F. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- 48.Mefford H.C., Trask B.J. The complex structure and dynamic evolution of human subtelomeres. Nat. Rev. Genet. 2002;3:91–102. doi: 10.1038/nrg727. [DOI] [PubMed] [Google Scholar]
- 49.Fabre E., Muller H., Therizols P., Lafontaine I., Dujon B., Fairhead C. Comparative genomics in hemiascomycete yeasts: evolution of sex, silencing, and subtelomeres. Mol. Biol. Evol. 2005;22:856–873. doi: 10.1093/molbev/msi070. [DOI] [PubMed] [Google Scholar]
- 50.Pryde F.E., Louis E.J. Saccharomyces cerevisiae telomeres. A review. Biochemistry (Mosc.) 1997;62:1232–1241. [PubMed] [Google Scholar]
- 51.McEachern M.J., Krauskopf A., Blackburn E.H. Telomeres and their control. Annu. Rev. Genet. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- 52.Flint J., Thomas K., Micklem G., Raynham H., Clark K., Doggett N.A. The relationship between chromosome structure and function at a human telomeric region. Nat. Genet. 1997;15:252–257. doi: 10.1038/ng0397-252. [DOI] [PubMed] [Google Scholar]
- 53.Brown C.A., Murray A.W., Verstrepen K.J. Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr. Biol. 2010;20:895–903. doi: 10.1016/j.cub.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bergström A., Simpson J.T., Salinas F., Barré B., Parts L., Zia A. A high-definition view of functional genetic variation from natural yeast genomes. Mol. Biol. Evol. 2014;31:872–888. doi: 10.1093/molbev/msu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ambrosini A., Paul S., Hu S., Riethman H. Human subtelomeric duplicon structure and organization. Genome Biol. 2007;8:R151. doi: 10.1186/gb-2007-8-7-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cam H.P., Sugiyama T., Chen E.S., Chen X., FitzGerald P.C., Grewal S.I.S. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 57.Schoeftner S., Blasco M.A. A “higher order” of telomere regulation: telomere heterochromatin and telomeric RNAs. EMBO J. 2009;28:2323–2336. doi: 10.1038/emboj.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zakian V.A. Telomeres: the beginnings and ends of eukaryotic chromosomes. Exp. Cell Res. 2012;318:1456–1460. doi: 10.1016/j.yexcr.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Riethman H. Human subtelomeric copy number variations. Cytogenet. Genome Res. 2008;123:244–252. doi: 10.1159/000184714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ottaviani A., Gilson E., Magdinier F. Telomeric position effect: from the yeast paradigm to human pathologies? Biochimie. 2008;90:93–107. doi: 10.1016/j.biochi.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 61.Kim W., Shay J.W. Long-range telomere regulation of gene expression: telomere looping and telomere position effect over long distances (TPE-OLD) Differentiation. 2018;99:1–9. doi: 10.1016/j.diff.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robin J.D., Ludlow A.T., Batten K., Magdinier F., Stadler G., Wagner K.R. Telomere position effect: regulation of gene expression with progressive telomere shortening over long distances. Genes Dev. 2014;28:2464–2476. doi: 10.1101/gad.251041.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bah A., Wischnewski H., Shchepachev V., Azzalin C.M. The telomeric transcriptome of Schizosaccharomyces pombe. Nucleic Acids Res. 2012;40:2995–3005. doi: 10.1093/nar/gkr1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Greenwood J., Cooper J.P. Non-coding telomeric and subtelomeric transcripts are differentially regulated by telomeric and heterochromatin assembly factors in fission yeast. Nucleic Acids Res. 2012;40:2956–2963. doi: 10.1093/nar/gkr1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kwapisz M., Ruault M., van Dijk E., Gourvennec S., Descrimes M., Taddei A. Expression of subtelomeric lncRNAs links telomeres dynamics to RNA decay in S. cerevisiae. Noncoding RNA. 2015;1:94–126. doi: 10.3390/ncrna1020094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Broadbent K.M., Park D., Wolf A.R., Van Tyne D., Sims J.S., Ribacke U. A global transcriptional analysis of Plasmodium falciparum malaria reveals a novel family of telomere-associated lncRNAs. Genome Biol. 2011;12:R56. doi: 10.1186/gb-2011-12-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Broadbent K.M., Broadbent J.C., Ribacke U., Wirth D., Rinn J.L., Sabeti P.C. Strand-specific RNA sequencing in Plasmodium falciparum malaria identifies developmentally regulated long non-coding RNA and circular RNA. BMC Genom. 2015;16:454. doi: 10.1186/s12864-015-1603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trofimova I., Popova D., Vasilevskaya E., Krasikova A. Non-coding RNA derived from a conservative subtelomeric tandem repeat in chicken and Japanese quail somatic cells. Mol. Cytogenet. 2014;7:102. doi: 10.1186/s13039-014-0102-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trofimova I., Chervyakova D., Krasikova A. Transcription of subtelomere tandemly repetitive DNA in chicken embryogenesis. Chromosome Res. 2015;23:495–503. doi: 10.1007/s10577-015-9487-3. [DOI] [PubMed] [Google Scholar]
- 70.Costa V., Casamassimi A., Roberto R., Gianfrancesco F., Matarazzo M.R., D'Urso M. DDX11L: a novel transcript family emerging from human subtelomeric regions. BMC Genom. 2009;10:250. doi: 10.1186/1471-2164-10-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dumas C., Chow C., Müller M., Papadopoulou B. A novel class of developmentally regulated noncoding RNAs in Leishmania. Eukaryot. Cell. 2006;5:2033–2046. doi: 10.1128/EC.00147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Azzalin C.M., Lingner J. Telomere functions grounding on TERRA firma. Trends Cell Biol. 2015;25:29–36. doi: 10.1016/j.tcb.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 73.Cusanelli E., Chartrand P. Telomeric repeat-containing RNA TERRA: a noncoding RNA connecting telomere biology to genome integrity. Front. Genet. 2015;6:143. doi: 10.3389/fgene.2015.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oliva-Rico D., Herrera L.A. Regulated expression of the lncRNA TERRA and its impact on telomere biology. Mech. Ageing Dev. 2017;167:16–23. doi: 10.1016/j.mad.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 75.Bettin N., Oss Pegorar C., Cusanelli E. The emerging roles of TERRA in telomere maintenance and genome stability. Cells. 2019;8 doi: 10.3390/cells8030246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Deng Z., Norseen J., Wiedmer A., Riethman H., Lieberman P.M. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martadinata H., Phan A.T. Structure of human telomeric RNA (TERRA): stacking of two G-quadruplex blocks in K(+) solution. Biochemistry. 2013;52:2176–2183. doi: 10.1021/bi301606u. [DOI] [PubMed] [Google Scholar]
- 78.Sagie S., Toubiana S., Hartono S.R., Katzir H., Tzur-Gilat A., Havazelet S. Telomeres in ICF syndrome cells are vulnerable to DNA damage due to elevated DNA:RNA hybrids. Nat. Commun. 2017;8:14015. doi: 10.1038/ncomms14015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chu H.-P., Cifuentes-Rojas C., Kesner B., Aeby E., Lee H.-G., Wei C. TERRA RNA antagonizes ATRX and protects telomeres. Cell. 2017;170:86–101. doi: 10.1016/j.cell.2017.06.017. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Redon S., Zemp I., Lingner J. A three-state model for the regulation of telomerase by TERRA and hnRNPA1. Nucleic Acids Res. 2013;41:9117–9128. doi: 10.1093/nar/gkt695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Beishline K., Vladimirova O., Tutton S., Wang Z., Deng Z., Lieberman P.M. CTCF driven TERRA transcription facilitates completion of telomere DNA replication. Nat. Commun. 2017;8:2114. doi: 10.1038/s41467-017-02212-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Redon S., Reichenbach P., Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;38:5797–5806. doi: 10.1093/nar/gkq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cusanelli E., Romero C.A.P., Chartrand P. Telomeric noncoding RNA TERRA is induced by telomere shortening to nucleate telomerase molecules at short telomeres. Mol. Cell. 2013;51:780–791. doi: 10.1016/j.molcel.2013.08.029. [DOI] [PubMed] [Google Scholar]
- 84.Moravec M., Wischnewski H., Bah A., Hu Y., Liu N., Lafranchi L. TERRA promotes telomerase-mediated telomere elongation in Schizosaccharomyces pombe. EMBO Rep. 2016;17:999–1012. doi: 10.15252/embr.201541708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shay J.W., Wright W.E. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis. 2005;26:867–874. doi: 10.1093/carcin/bgh296. [DOI] [PubMed] [Google Scholar]
- 86.Luke B., Panza A., Redon S., Iglesias N., Li Z., Lingner J. The Rat1p 5’ to 3’ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 87.Porro A., Feuerhahn S., Delafontaine J., Riethman H., Rougemont J., Lingner J. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat. Commun. 2014;5:5379. doi: 10.1038/ncomms6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Riethman H., Ambrosini A., Castaneda C., Finklestein J., Hu X.-L., Mudunuri U. Mapping and initial analysis of human subtelomeric sequence assemblies. Genome Res. 2004;14:18–28. doi: 10.1101/gr.1245004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brown W.R., MacKinnon P.J., Villasanté A., Spurr N., Buckle V.J., Dobson M.J. Structure and polymorphism of human telomere-associated DNA. Cell. 1990;63:119–132. doi: 10.1016/0092-8674(90)90293-n. [DOI] [PubMed] [Google Scholar]
- 90.Nergadze S.G., Farnung B.O., Wischnewski H., Khoriauli L., Vitelli V., Chawla R. CpG-island promoters drive transcription of human telomeres. RNA. 2009;15:2186–2194. doi: 10.1261/rna.1748309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Britt-Compton B., Rowson J., Locke M., Mackenzie I., Kipling D., Baird D.M. Structural stability and chromosome-specific telomere length is governed by cis-acting determinants in humans. Hum. Mol. Genet. 2006;15:725–733. doi: 10.1093/hmg/ddi486. [DOI] [PubMed] [Google Scholar]
- 92.López de Silanes I., Graña O., De Bonis M.L., Dominguez O., Pisano D.G., Blasco M.A. Identification of TERRA locus unveils a telomere protection role through association to nearly all chromosomes. Nat. Commun. 2014;5:4723. doi: 10.1038/ncomms5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Feretzaki M., Renck Nunes P., Lingner J. Expression and differential regulation of human TERRA at several chromosome ends. RNA. 2019 doi: 10.1261/rna.072322.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Deng Z., Wang Z., Stong N., Plasschaert R., Moczan A., Chen H.-S. A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. EMBO J. 2012;31:4165–4178. doi: 10.1038/emboj.2012.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marión R.M., Montero J.J., López de Silanes I., Graña-Castro O., Martínez P., Schoeftner S. TERRA regulate the transcriptional landscape of pluripotent cells through TRF1-dependent recruitment of PRC2. Elife. 2019;8 doi: 10.7554/eLife.44656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boué S., Paramonov I., Barrero M.J., Izpisúa Belmonte J.C. Analysis of human and mouse reprogramming of somatic cells to induced pluripotent stem cells. What is in the plate? PloS One. 2010;5 doi: 10.1371/journal.pone.0012664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marión R.M., López de Silanes I., Mosteiro L., Gamache B., Abad M., Guerra C. Common telomere changes during in vivo reprogramming and early stages of tumorigenesis. Stem Cell Rep. 2017;8:460–475. doi: 10.1016/j.stemcr.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zeng S., Liu L., Sun Y., Lu G., Lin G. Role of telomeric repeat-containing RNA in telomeric chromatin remodeling during the early expansion of human embryonic stem cells. Faseb. J. 2017;31:4783–4795. doi: 10.1096/fj.201600939RR. [DOI] [PubMed] [Google Scholar]
- 99.Chan C.S., Tye B.K. A family of Saccharomyces cerevisiae repetitive autonomously replicating sequences that have very similar genomic environments. J. Mol. Biol. 1983;168:505–523. doi: 10.1016/s0022-2836(83)80299-x. [DOI] [PubMed] [Google Scholar]
- 100.Louis E.J., Haber J.E. The structure and evolution of subtelomeric Y’ repeats in Saccharomyces cerevisiae. Genetics. 1992;131:559–574. doi: 10.1093/genetics/131.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mak H.C., Pillus L., Ideker T. Dynamic reprogramming of transcription factors to and from the subtelomere. Genome Res. 2009;19:1014–1025. doi: 10.1101/gr.084178.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Londoño-Vallejo J.A., Wellinger R.J. Telomeres and telomerase dance to the rhythm of the cell cycle. Trends Biochem. Sci. 2012;37:391–399. doi: 10.1016/j.tibs.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 103.Louis E.J. The chromosome ends of Saccharomyces cerevisiae. Yeast. 1995;11:1553–1573. doi: 10.1002/yea.320111604. [DOI] [PubMed] [Google Scholar]
- 104.Shore D., Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987;51:721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- 105.Marcand S., Gilson E., Shore D. A protein-counting mechanism for telomere length regulation in yeast. Science. 1997;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- 106.Moretti P., Freeman K., Coodly L., Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 107.Imai S., Armstrong C.M., Kaeberlein M., Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 108.Hecht A., Strahl-Bolsinger S., Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 109.Santos-Rosa H., Bannister A.J., Dehe P.M., Géli V., Kouzarides T. Methylation of H3 lysine 4 at euchromatin promotes Sir3p association with heterochromatin. J. Biol. Chem. 2004;279:47506–47512. doi: 10.1074/jbc.M407949200. [DOI] [PubMed] [Google Scholar]
- 110.van Leeuwen F., Gafken P.R., Gottschling D.E. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/S0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 111.Palladino F., Laroche T., Gilson E., Axelrod A., Pillus L., Gasser S.M. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 1993;75:543–555. doi: 10.1016/0092-8674(93)90388-7. [DOI] [PubMed] [Google Scholar]
- 112.Cockell M., Gasser S.M. Nuclear compartments and gene regulation. Curr. Opin. Genet. Dev. 1999;9:199–205. doi: 10.1016/S0959-437X(99)80030-6. [DOI] [PubMed] [Google Scholar]
- 113.Taddei A., Schober H., Gasser S.M. The budding yeast nucleus. Cold Spring Harb. Perspect. Biol. 2010;2:a000612. doi: 10.1101/cshperspect.a000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gottschling D.E., Aparicio O.M., Billington B.L., Zakian V.A. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-Z. [DOI] [PubMed] [Google Scholar]
- 115.de Bruin D., Kantrow S.M., Liberatore R.A., Zakian V.A. Telomere folding is required for the stable maintenance of telomere position effects in yeast. Mol. Cell Biol. 2000;20:7991–8000. doi: 10.1128/mcb.20.21.7991-8000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Guillemette B., Gaudreau L. Reuniting the contrasting functions of H2A.Z. Biochem. Cell. Biol. 2006;84:528–535. doi: 10.1139/o06-077. [DOI] [PubMed] [Google Scholar]
- 117.Lundblad V., Blackburn E.H. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 118.Teng S.C., Zakian V.A. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Teng S.C., Chang J., McCowan B., Zakian V.A. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell. 2000;6:947–952. doi: 10.1016/s1097-2765(05)00094-8. [DOI] [PubMed] [Google Scholar]
- 120.Kupiec M. Biology of telomeres: lessons from budding yeast. FEMS Microbiol. Rev. 2014;38:144–171. doi: 10.1111/1574-6976.12054. [DOI] [PubMed] [Google Scholar]
- 121.Iglesias N., Redon S., Pfeiffer V., Dees M., Lingner J., Luke B. Subtelomeric repetitive elements determine TERRA regulation by Rap1/Rif and Rap1/Sir complexes in yeast. EMBO Rep. 2011;12:587–593. doi: 10.1038/embor.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fallet E., Jolivet P., Soudet J., Lisby M., Gilson E., Teixeira M.T. Length-dependent processing of telomeres in the absence of telomerase. Nucleic Acids Res. 2014;42:3648–3665. doi: 10.1093/nar/gkt1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Graf M., Bonetti D., Lockhart A., Serhal K., Kellner V., Maicher A. Telomere length determines TERRA and R-loop regulation through the cell cycle. Cell. 2017;170:72–85. doi: 10.1016/j.cell.2017.06.006. e14. [DOI] [PubMed] [Google Scholar]
- 124.Balk B., Maicher A., Dees M., Klermund J., Luke-Glaser S., Bender K. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat. Struct. Mol. Biol. 2013;20:1199–1205. doi: 10.1038/nsmb.2662. [DOI] [PubMed] [Google Scholar]
- 125.Pfeiffer V., Crittin J., Grolimund L., Lingner J. The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. EMBO J. 2013;32:2861–2871. doi: 10.1038/emboj.2013.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kanoh J., Sadaie M., Urano T., Ishikawa F. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr. Biol. 2005;15:1808–1819. doi: 10.1016/j.cub.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 127.Wang J., Cohen A.L., Letian A., Tadeo X., Moresco J.J., Liu J. The proper connection between shelterin components is required for telomeric heterochromatin assembly. Genes Dev. 2016;30:827–839. doi: 10.1101/gad.266718.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Matsuda A., Chikashige Y., Ding D.-Q., Ohtsuki C., Mori C., Asakawa H. Highly condensed chromatins are formed adjacent to subtelomeric and decondensed silent chromatin in fission yeast. Nat. Commun. 2015;6:7753. doi: 10.1038/ncomms8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Buchanan L., Durand-Dubief M., Roguev A., Sakalar C., Wilhelm B., Strålfors A. The Schizosaccharomyces pombe JmjC-protein, Msc1, prevents H2A.Z localization in centromeric and subtelomeric chromatin domains. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tashiro S., Handa T., Matsuda A., Ban T., Takigawa T., Miyasato K. Shugoshin forms a specialized chromatin domain at subtelomeres that regulates transcription and replication timing. Nat. Commun. 2016;7:10393. doi: 10.1038/ncomms10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cohen A., Habib A., Laor D., Yadav S., Kupiec M., Weisman R. TOR complex 2 in fission yeast is required for chromatin-mediated gene silencing and assembly of heterochromatic domains at subtelomeres. J. Biol. Chem. 2018;293:8138–8150. doi: 10.1074/jbc.RA118.002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lorenzi L.E., Bah A., Wischnewski H., Shchepachev V., Soneson C., Santagostino M. Fission yeast Cactin restricts telomere transcription and elongation by controlling Rap1 levels. EMBO J. 2015;34:115–129. doi: 10.15252/embj.201489559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vrbsky J., Akimcheva S., Watson J.M., Turner T.L., Daxinger L., Vyskot B. siRNA-mediated methylation of Arabidopsis telomeres. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang L.-F., Ogawa Y., Ahn J.Y., Namekawa S.H., Silva S.S., Lee J.T. Telomeric RNAs mark sex chromosomes in stem cells. Genetics. 2009;182:685–698. doi: 10.1534/genetics.109.103093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Galupa R., Heard E. X-chromosome inactivation: a crossroads between chromosome architecture and gene regulation. Annu. Rev. Genet. 2018;52:535–566. doi: 10.1146/annurev-genet-120116-024611. [DOI] [PubMed] [Google Scholar]
- 136.Chu H.-P., Froberg J.E., Kesner B., Oh H.J., Ji F., Sadreyev R. PAR-TERRA directs homologous sex chromosome pairing. Nat. Struct. Mol. Biol. 2017;24:620–631. doi: 10.1038/nsmb.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Houseley J., Kotovic K., El Hage A., Tollervey D. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 2007;26:4996–5006. doi: 10.1038/sj.emboj.7601921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Neil H., Malabat C., d'Aubenton-Carafa Y., Xu Z., Steinmetz L.M., Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 139.Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Münster S., Camblong J. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.van Dijk E.L., Chen C.L., d'Aubenton-Carafa Y., Gourvennec S., Kwapisz M., Roche V. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475:114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- 141.Wery M., Descrimes M., Vogt N., Dallongeville A.-S., Gautheret D., Morillon A. Nonsense-mediated decay restricts LncRNA levels in yeast unless blocked by double-stranded RNA structure. Mol. Cell. 2016;61:379–392. doi: 10.1016/j.molcel.2015.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gomez-Escobar N., Almobadel N., Alzahrani O., Feichtinger J., Planells-Palop V., Alshehri Z. Translin and Trax differentially regulate telomere-associated transcript homeostasis. Oncotarget. 2016;7:33809–33820. doi: 10.18632/oncotarget.9278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rubio J.P., Thompson J.K., Cowman A.F. The var genes of Plasmodium falciparum are located in the subtelomeric region of most chromosomes. EMBO J. 1996;15:4069–4077. [PMC free article] [PubMed] [Google Scholar]
- 144.Hernandez-Rivas R., Mattei D., Sterkers Y., Peterson D.S., Wellems T.E., Scherf A. Expressed var genes are found in Plasmodium falciparum subtelomeric regions. Mol. Cell Biol. 1997;17:604–611. doi: 10.1128/mcb.17.2.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Figueiredo L.M., Pirrit L.A., Scherf A. Genomic organisation and chromatin structure of Plasmodium falciparum chromosome ends. Mol. Biochem. Parasitol. 2000;106:169–174. doi: 10.1016/S0166-6851(99)00199-1. [DOI] [PubMed] [Google Scholar]
- 146.Scherf A. Plasmodium telomeres and telomere proximal gene expression. Semin. Cell Dev. Biol. 1996;7:49–57. doi: 10.1006/scdb.1996.0008. [DOI] [Google Scholar]
- 147.Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Scherf A., Hernandez-Rivas R., Buffet P., Bottius E., Benatar C., Pouvelle B. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J. 1998;17:5418–5426. doi: 10.1093/emboj/17.18.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Duraisingh M.T., Voss T.S., Marty A.J., Duffy M.F., Good R.T., Thompson J.K. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 150.Freitas-Junior L.H., Hernandez-Rivas R., Ralph S.A., Montiel-Condado D., Ruvalcaba-Salazar O.K., Rojas-Meza A.P. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 151.Lopez-Rubio J.J., Gontijo A.M., Nunes M.C., Issar N., Hernandez Rivas R., Scherf A. 5’ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol. Microbiol. 2007;66:1296–1305. doi: 10.1111/j.1365-2958.2007.06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Malmquist N.A., Moss T.A., Mecheri S., Scherf A., Fuchter M.J. Small-molecule histone methyltransferase inhibitors display rapid antimalarial activity against all blood stage forms in Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16708–16713. doi: 10.1073/pnas.1205414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jiang L., Mu J., Zhang Q., Ni T., Srinivasan P., Rayavara K. PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature. 2013;499:223–227. doi: 10.1038/nature12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Epp C., Li F., Howitt C.A., Chookajorn T., Deitsch K.W. Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum. RNA. 2009;15:116–127. doi: 10.1261/rna.1080109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Calderwood M.S., Gannoun-Zaki L., Wellems T.E., Deitsch K.W. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J. Biol. Chem. 2003;278:34125–34132. doi: 10.1074/jbc.M213065200. [DOI] [PubMed] [Google Scholar]
- 156.Amit-Avraham I., Pozner G., Eshar S., Fastman Y., Kolevzon N., Yavin E. Antisense long noncoding RNAs regulate var gene activation in the malaria parasite Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E982–E991. doi: 10.1073/pnas.1420855112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dzikowski R., Li F., Amulic B., Eisberg A., Frank M., Patel S. Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Rep. 2007;8:959–965. doi: 10.1038/sj.embor.7401063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kyes S.A., Christodoulou Z., Raza A., Horrocks P., Pinches R., Rowe J.A. A well-conserved Plasmodium falciparum var gene shows an unusual stage-specific transcript pattern. Mol. Microbiol. 2003;48:1339–1348. doi: 10.1046/j.1365-2958.2003.03505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gannoun-Zaki L., Jost A., Mu J., Deitsch K.W., Wellems T.E. A silenced Plasmodium falciparum var promoter can be activated in vivo through spontaneous deletion of a silencing element in the intron. Eukaryot. Cell. 2005;4:490–492. doi: 10.1128/EC.4.2.490-492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Su X.Z., Heatwole V.M., Wertheimer S.P., Guinet F., Herrfeldt J.A., Peterson D.S. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 161.Flueck C., Bartfai R., Niederwieser I., Witmer K., Alako B.T.F., Moes S. A major role for the Plasmodium falciparum ApiAP2 protein PfSIP2 in chromosome end biology. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wicker T., Robertson J.S., Schulze S.R., Feltus F.A., Magrini V., Morrison J.A. The repetitive landscape of the chicken genome. Genome Res. 2005;15:126–136. doi: 10.1101/gr.2438004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Matzke M.A., Varga F., Berger H., Schernthaner J., Schweizer D., Mayr B. A 41–42 bp tandemly repeated sequence isolated from nuclear envelopes of chicken erythrocytes is located predominantly on microchromosomes. Chromosoma. 1990;99:131–137. doi: 10.1007/BF01735329. [DOI] [PubMed] [Google Scholar]
- 164.Wang X., Li J., Leung F.C. Partially inverted tandem repeat isolated from pericentric region of chicken chromosome 8. Chromosome Res. 2002 doi: 10.1023/a:1014226412339. [DOI] [PubMed] [Google Scholar]
- 165.Klein S., Ellendorff F. Localization of Xho1 repetitive sequences on autosomes in addition to the W chromosome in chickens and its relevance for sex diagnosis. Anim. Genet. 2000;31:104–109. doi: 10.1046/j.1365-2052.2000.00568.x. [DOI] [PubMed] [Google Scholar]
- 166.Deryusheva S., Krasikova A., Kulikova T., Gaginskaya E. Tandem 41-bp repeats in chicken and Japanese quail genomes: FISH mapping and transcription analysis on lampbrush chromosomes. Chromosoma. 2007;116:519–530. doi: 10.1007/s00412-007-0117-5. [DOI] [PubMed] [Google Scholar]
- 167.Trofimova I., Krasikova A. Transcription of highly repetitive tandemly organized DNA in amphibians and birds: a historical overview and modern concepts. RNA Biol. 2016;13:1246–1257. doi: 10.1080/15476286.2016.1240142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Linardopoulou E., Mefford H.C., Nguyen O., Friedman C., van den Engh G., Farwell D.G. Transcriptional activity of multiple copies of a subtelomerically located olfactory receptor gene that is polymorphic in number and location. Hum. Mol. Genet. 2001;10:2373–2383. doi: 10.1093/hmg/10.21.2373. [DOI] [PubMed] [Google Scholar]
- 169.Kermouni A., Van Roost E., Arden K.C., Vermeesch J.R., Weiss S., Godelaine D. The IL-9 receptor gene (IL9R): genomic structure, chromosomal localization in the pseudoautosomal region of the long arm of the sex chromosomes, and identification of IL9R pseudogenes at 9qter, 10pter, 16pter, and 18pter. Genomics. 1995;29:371–382. doi: 10.1006/geno.1995.9992. [DOI] [PubMed] [Google Scholar]
- 170.Mah N., Stoehr H., Schulz H.L., White K., Weber B.H. Identification of a novel retina-specific gene located in a subtelomeric region with polymorphic distribution among multiple human chromosomes. Biochim. Biophys. Acta Gene Struct. Expr. 2001;1522:167–174. doi: 10.1016/S0167-4781(01)00328-1. [DOI] [PubMed] [Google Scholar]
- 171.Martin-Gallardo A., Lamerdin J., Sopapan P., Friedman C., Fertitta A.L., Garcia E. Molecular analysis of a novel subtelomeric repeat with polymorphic chromosomal distribution. Cytogenet. Cell Genet. 1995;71:289–295. doi: 10.1159/000134129. [DOI] [PubMed] [Google Scholar]
- 172.Yamaguchi H., Condeelis J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim. Biophys. Acta. 2007;1773:642–652. doi: 10.1016/j.bbamcr.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Leirdal M., Shadidy M., Røsok Ø., Sioud M. Identification of genes differentially expressed in breast cancer cell line SKBR3: potential identification of new prognostic biomarkers. Int. J. Mol. Med. 2004 doi: 10.3892/ijmm.14.2.217. [DOI] [PubMed] [Google Scholar]
- 174.Martin C.L., Wong A., Gross A., Chung J., Fantes J.A., Ledbetter D.H. The evolutionary origin of human subtelomeric homologies--or where the ends begin. Am. J. Hum. Genet. 2002;70:972–984. doi: 10.1086/339768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.van Geel M., Eichler E.E., Beck A.F., Shan Z., Haaf T., van der Maarel S.M. A cascade of complex subtelomeric duplications during the evolution of the hominoid and Old World monkey genomes. Am. J. Hum. Genet. 2002;70:269–278. doi: 10.1086/338307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yehezkel S., Segev Y., Viegas-Péquignot E., Skorecki K., Selig S. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum. Mol. Genet. 2008;17:2776–2789. doi: 10.1093/hmg/ddn177. [DOI] [PubMed] [Google Scholar]
- 177.Sacconi S., Salviati L., Desnuelle C. Facioscapulohumeral muscular dystrophy. Biochim. Biophys. Acta. 2015;1852:607–614. doi: 10.1016/j.bbadis.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 178.van Deutekom J.C., Wijmenga C., van Tienhoven E.A., Gruter A.M., Hewitt J.E., Padberg G.W. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet. 1993;2:2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- 179.Lemmers R.J.L.F., Van Overveld P.G.M., Sandkuijl L.A., Vrieling H., Padberg G.W., Frants R.R. Mechanism and timing of mitotic rearrangements in the subtelomeric D4Z4 repeat involved in facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2004;75:44–53. doi: 10.1086/422175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Knight S.J., Regan R., Nicod A., Horsley S.W., Kearney L., Homfray T. Subtle chromosomal rearrangements in children with unexplained mental retardation. Lancet. 1999;354:1676–1681. doi: 10.1016/S0140-6736(99)03070-6. [DOI] [PubMed] [Google Scholar]
- 181.Cook G.P., Tomlinson I.M., Walter G., Riethman H., Carter N.P., Buluwela L. A map of the human immunoglobulin VH locus completed by analysis of the telomeric region of chromosome 14q. Nat. Genet. 1994;7:162–168. doi: 10.1038/ng0694-162. [DOI] [PubMed] [Google Scholar]
- 182.Song J., Perreault J.-P., Topisirovic I., Richard S. RNA G-quadruplexes and their potential regulatory roles in translation. Translation (Austin) 2016;4 doi: 10.1080/21690731.2016.1244031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Gagliardi M., Matarazzo M.R. RIP: RNA immunoprecipitation. Methods Mol. Biol. 2016;1480:73–86. doi: 10.1007/978-1-4939-6380-5_7. [DOI] [PubMed] [Google Scholar]
- 184.Chu C., Quinn J., Chang H.Y. Chromatin isolation by RNA purification (ChIRP) JoVE. 2012 doi: 10.3791/3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Simon M.D., Wang C.I., Kharchenko P.V., West J.A., Chapman B.A., Alekseyenko A.A. The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 2011;108:20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bühler M., Gasser S.M. Silent chromatin at the middle and ends: lessons from yeasts. EMBO J. 2009;28:2149–2161. doi: 10.1038/emboj.2009.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wyse B.A., Oshidari R., Jeffery D.C., Yankulov K.Y. Parasite epigenetics and immune evasion: lessons from budding yeast. Epigenet. Chromatin. 2013;6:40. doi: 10.1186/1756-8935-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]