Summary
The mammalian heart contains multiple cell types that appear progressively during embryonic development. Advances in determining cardiac lineage diversification has often been limited by the unreliability of genetic tracers. Here we combine clonal analysis, genetic lineage tracing, tissue transplantation and mutant characterization to investigate the lineage relationships between epicardium, arterial mesothelial cells (AMCs) and the coronary vasculature. We report a contribution of the second heart field (SHF) to a vasculogenic niche composed of AMCs and sub-mesothelial cells at the base of the pulmonary artery. Sub-mesothelial cells from this niche differentiate into lymphatic endothelial cells and, in close association with AMC-derived cells, contribute to, and are essential for the development of ventral cardiac lymphatics. In addition, regionalized epicardial/mesothelial retinoic acid signaling regulates lymphangiogenesis, contributing to the niche properties. These results uncover a SHF vasculogenic contribution to coronary lymphatic development through a local niche at the base of the great arteries.
Graphical Abstract
eTOC Blurb
Lioux et al., identify a population of coronary lymphatics that originates from non-venous cells of the Second Heart Field and contributes to about 50% of lymphatic endothelial cells of the ventral heart. This population is nonetheless essential for ventral heart lymphatics development, highlighting the functional relevance of coronary lymphatics heterogeneity.
Introduction
The vertebrate heart develops from two pools of progenitor cells known as first and second heart fields (FHF, SHF) (Meilhac and Buckingham, 2018). The FHF differentiates first and contains a mix of unipotent progenitors that give rise to the endocardium, cardiomyocytes of the left ventricle (LV) and parts of the atria (Zaffran et al., 2004). The SHF is added to the heart later than the FHF and contains multipotent progenitors that contribute cardiomyocytes, endothelial cells, and smooth muscle cells of the right ventricle (RV), the base of the pulmonary artery and parts of the atria (Cai et al., 2003; Christoffels et al., 2006; Galli et al., 2008; Kelly et al., 2001; Mjaatvedt et al., 2001; Waldo et al., 2001).
Heart function depends critically on the coronary vasculature, which develops under strong influence of the epicardium, sub-epicardium and myocardium (Olivey and Svensson, 2010). The epicardium is the outer mesothelial layer of the heart and is added to the myocardial surface after the primitive heart tube has formed (Simoes and Riley, 2018). The rest of the pericardial cavity, including the base of the great arteries, is covered by a mesothelial cell layer that is continuous with, and abuts the epicardium at the myocardial-arterial interface (Gittenberger-de Groot et al., 2000; Perez-Pomares et al., 2003). Epicardial cells derive from the proepicardium, a group of proliferating cells that forms at the dorsal pericardial wall (Manner, 1992). The epicardium plays an important role in coronary formation (Tian et al., 2015) through contributing cells that undergo epithelial-mesenchymal transition (EMT), invade the myocardium and differentiate into endothelium, smooth muscle and fibroblasts. In addition, the epicardium produces signals, like retinoic acid and FGF, that help patterning the coronary vasculature.
Besides coronary arteries and veins, whose roles are well established in cardiac physiology, the coronary vasculature contains a lymphatic plexus whose misfunction leads to several cardiac problems (reviewed in (Brakenhielm and Alitalo, 2019)), including endocardial/myocardial edema (Miller et al., 1963), atherosclerosis (Xu et al., 2007), susceptibility to cardiac infection (Miller et al., 1964), fibrosis (Kline et al., 1994), and ventricular function impairment (Ludwig et al., 1997). Lymphangiogenesis plays an important role also in the response to cardiac injury. Following acute myocardial ischemic injury, lymphangiogenesis is naturally activated (Ishikawa et al., 2007; Klotz et al., 2015), whereas its impairment worsens the recovery from the injury (Vieira et al., 2018; Vuorio et al., 2018) and its activation improves it (Henri et al., 2016; Klotz et al., 2015; Taira et al., 1990).
During embryonic development, most lymphatic endothelial cells (LECs) are produced from the venous endothelium through a wave of specification along the cardinal veins (CVs) and associated inter-somitic vessels in the 10-day mouse embryo (Sabin, 1902; Srinivasan et al., 2007; Yang et al., 2012). A recent study indicates that most venous endothelium that produces LECs has a somitic origin (Stone and Stainier, 2019). Upon specification, LECs bud off from the vein to form bilateral antero-posterior chains of lymphatic sacs, which then evolve into the lymphatic vasculature by sprouting and further recruitment of LECs from the CVs and inter-somitic vessels (Escobedo and Oliver, 2016). While most embryonic lymphatics derive from this early specification wave, LEC generation from alternative cell populations has long been proposed and debated (Huntington and McClure, 1910). Such alternative sources include the mesenchyme (Eng et al., 2019; Ny et al., 2005; Wilting et al., 2006) and hematopoietic cell lineages (Maruyama et al., 2005; Sebzda et al., 2006; Stanczuk et al., 2015), however the characterization of the niches and mechanisms that mediate these alternative sources remain largely uncharacterized or controversial (Ma and Oliver, 2017).
Cardiac lymphatics start to form at mid-gestation, when LECs are first seen in the dorsal side of the heart around the venous pole and ventrally in the outflow region (Flaht et al., 2012). Although most cardiac lymphatics derive from the CV (Klotz et al., 2015), additional sources have also been proposed, including hematopoietic lineages (Klotz et al., 2015) and the second heart field (Klotz et al., 2015; Maruyama et al., 2019), the origin of these alternative cardiac LECs remains uncertain (Engleka et al., 2012; Stone and Stainier, 2019; Ulvmar et al., 2016). To a large extent, the controversies around the conclusions of these studies are due to the use of constitutive Cre lines, whose recombination pattern is often leaky and/or incompletely described.
Here, we use clonal analysis to identify a lineage relationship between non-venous SHF-derived precursors at the base of the pulmonary artery and the cardiac lymphatic vasculature. We present detailed insights into the origins and the molecular and cellular mechanisms involved in SHF-derived lymphatic vasculogenesis in the heart.
Results
Lineage relationships and contribution of arterial mesothelial cells
While analyzing a large-scale clonal analysis designed to interrogate the potency of single cardiac precursors at embryonic day 9 (E9) (Figure S1), we found characteristic clones that included arterial mesothelial cells (AMCs) at the base of the great arteries (Figure 1A) (see also (Gittenberger-de Groot et al., 2012) for definition AMCs). The clonal analysis strategy is based on the ubiquitous, low-frequency random recombination of two independent reporters. This approach allows the determination of clonal contributions from the frequency of color matching between labelled cells in different clusters or different cell types within single clusters ((Arques et al., 2007), Figure S1 and Methods).
Figure 1. Clonal analysis and lineage tracing identify arterial mesothelial cell-related lineages and their separation from epicardium.
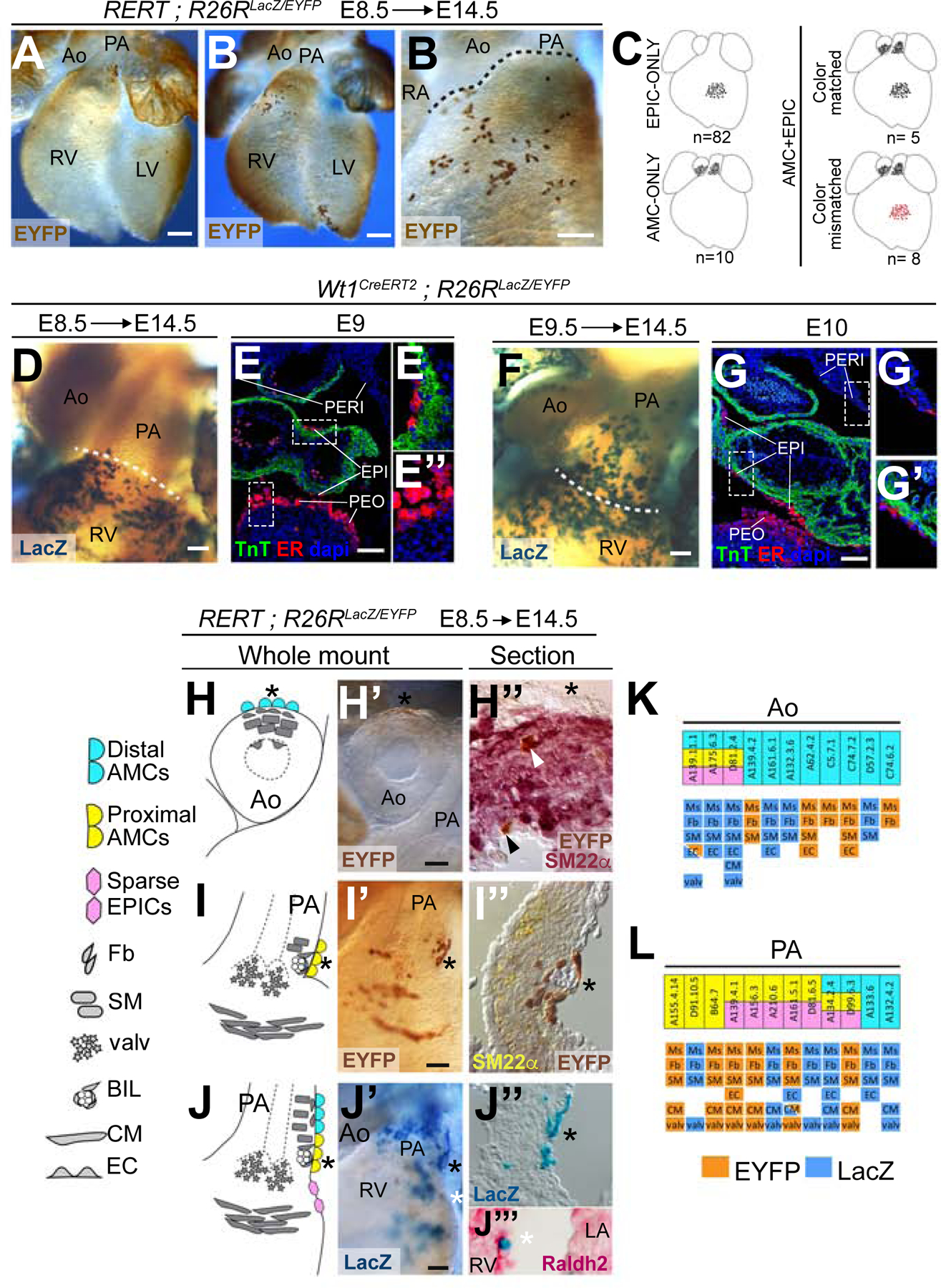
(A) Whole-mount ventral view of a specimen with a mesothelial clone in the PA. (B) Whole-mount ventral view of a specimen with an epicardial clone over the sub-pulmonary myocardium. (B’) high magnification of the OFT area of the specimen in B. (C) Distribution of Clonal Analysis specimens according to the presence AMCs and EPICs clones and their color. (D–G’’) Distribution and fate of CreERT2-expressing cells in Wt1CreERT2 embryos. Images show the expression of CreERT2 in the pericardial cavity of E9 (E-E’’) and E10 (G-G’’) embryos and the distribution at E14.5 of cells recombined in Wt1CreERT2 embryos upon TM injections at E8.5 (D) and E9.5 (F). (E’-E’’) and (G’–G’’) show magnifications of the boxed areas in E and G, respectively. Dotted lines in D and F indicate the approximate limit between the PA and the RV. (H–J’) Whole-mount examples of clonal contributions to the base of the great arteries and the sub-arterial regions of the ventricles, and schemes showing the lineages identified. (H’’-I’’’) Sections of specimens in (H’–I’) showing co-localization of clonally labelled cells with markers of smooth muscle (H’’, I’’) and epicardium (J’’’). Asterisks indicate equivalent positions at the mesothelial side of the whole-mounts, schemes and sections. White arrowhead in H’’ shows cells with co-localization of EYFP and SM22α. Black arrowhead in H’’ shows an arterial endothelial cell positive for EYFP and negative for SM22α. (K, L), tables scoring the results of clones containing AMCs in the Aorta (K) and PA (L) by specimen and position of the AMCs. The upper row indicates the contribution to AMCs and EPICs, according to the color code in H-J. The lower rows indicate the presence of additional cell lineages in the scored clones and their clonal label (LacZ or EYFP). All scale bars: 100µm. Ao: Aorta; PA: Pulmonary artery; RA: Right atrium; RV: Right ventricle; LV: Left ventricle; TnT: cardiac Troponin T; ER: estrogen receptor; Fb: Fibroblast; SM: Smooth muscle; Ms: Mesothelium; valv: valvular mesenchyme; BIL: Blood island-like coronary structure; CM: Cardiomyocyte; EPICs: epicardial cells; EC: Endothelial cell. See also Figures S1 and S2.
To study the AMC lineage, we first investigated the relationship between AMCs and epicardial cells (EPICs). Out of 737 hearts analyzed, we found 95 hearts with labelled cell clusters containing a substantial proportion of EPICs. None of these EPIC clusters extended across the border between the great arteries and the ventricles, which suggested that epicardial cells are restricted from colonizing the arterial mesothelium (Figure 1B, B’). In 13 of the 95 hearts that contained EPICs clusters, an AMC cluster was also found (Figure 1C and Figure S2). The frequency of color matching between AMC and EPIC clusters in these hearts indicated an independent origin of AMCs and EPICs (Figure 1C and Figure S2). To confirm these observations, we used time-controlled labeling of mesothelial/proepicardial cells using the Wt1CreERT2 driver. We first induced recombination by tamoxifen (TM) injection at E8.5, aiming for recombination around E9, a stage in which CreERT2 expression is only detected in the proepicardium and a few epicardial cells (Figure 1E–E’’). The cells that recombined at this stage colonized the ventricular surface but not the arterial mesothelial layer (Figure 1D). In contrast, induction at E9.5, aiming to produce recombination at around E10, when CreERT2 is not only expressed in the epicardium but also in the pericardium (Figure 1G–G’’), labelled both AMCs and EPICs (Figure 1F). These results show the proepicardial origin of the epicardium and the pericardial origin of AMCs, demonstrating a border of lineage restriction that prevents epicardial cells from colonizing the mesothelium of the great arteries. These results agree with previous transplantation and ablation studies in the chick (Gittenberger-de Groot et al., 2000; Perez-Pomares et al., 2003).
While the contribution of epicardial cells to the heart has been extensively studied, that of AMCs to the arteries has not been determined. We analyzed AMC-containing labelled cell clusters and found that all of them also contained non-mesothelial cells (Figure 1H–J’, K–L). Different cell compositions were found depending on whether the clusters contained AMCs in the proximity of the ventricle or in more distal regions (Figure 1H–J’). Eight clusters contained exclusively proximal AMCs, 10 exclusively distal AMCs and 5 contained both (Figure 1K, L), indicating that there is no lineage restriction between proximal and distal AMCs. Clusters containing exclusively proximal AMCs also included valve mesenchyme (8/8), cardiomyocytes (6/8), smooth muscle (8/8), fibroblasts (8/8) and arterial endothelium (2/8) (Figure 1K, L). Furthermore, these clusters contained sub-mesothelial blood island-like structures at the base of the PA that were surrounded by cells of endothelial appearance which, however, did not express endothelial markers (Figure 1I’’, J’’). Distal-only AMC clusters showed fibroblast-like cells (10/10), smooth muscle (10/10) and arterial endothelium (4/10), while cardiomyocytes were only found in one case (1/10). This single case does not necessarily indicate a lineage relationship, provided that one bicolor cluster of cardiomyocytes was also observed in the collection, and therefore the observation is within the range of expected random polyclonality (see Methods). In addition, 5 clusters contained both proximal and distal cells and, in this group, we observed a combination of the contributions of proximal-only and distal-only clusters (Figure 1L). Clusters containing proximal AMCs often showed a few sparse cells that colonized the epicardium over the RV near to the PA root (7/12) (Figure 1J’’’). This observation suggests that the border between the arterial mesothelium and the epicardium is not fully restrictive to arterial cells.
These results indicate that all AMCs share lineage with arterial endothelium, smooth muscle and fibroblasts, whereas proximal AMCs, in addition, share lineage with cardiomyocytes, valve mesenchyme, and cells in the vicinity of blood island-like structures. Given that there is a clonal relationship between proximal and distal AMCs, the fact that proximal AMCs, but not the distal ones, relate to ventricular lineages is due to their position in proximity to the ventricle and not to a lineage compartmentalization from the distal AMCs.
To understand the hierarchy of these lineage relationships, we recombined mesothelial cells at low frequency using the Wt1CreERT2 driver. We induced recombination at E10.5, a stage in which AMCs are already present, and studied their contribution to the lineages observed in AMC-containing clones (Figure S3). We found labelled cells in the arterial endothelium, smooth muscle and fibroblast layers of the arteries. In addition, we found contributions to peri-endothelial cells associated to coronary vascular structures but not to endothelial cells in these structures or to cardiomyocytes (Figure 2A–A’’). Furthermore, inductions at E11.5 and E12.5 produced similar results, indicating a continued contribution during several days of development (Figure S3). These results show that, similar to their epicardial counterparts, AMCs undergo EMT to colonize the underlying tissues. In agreement with this idea, AMCs express high levels of the EMT inductor snail1 (Figure 2B–B’’).
Figure 2. Analysis of the lineage relationships between the SHF, AMCs and arterial and cardiac cell lineages.
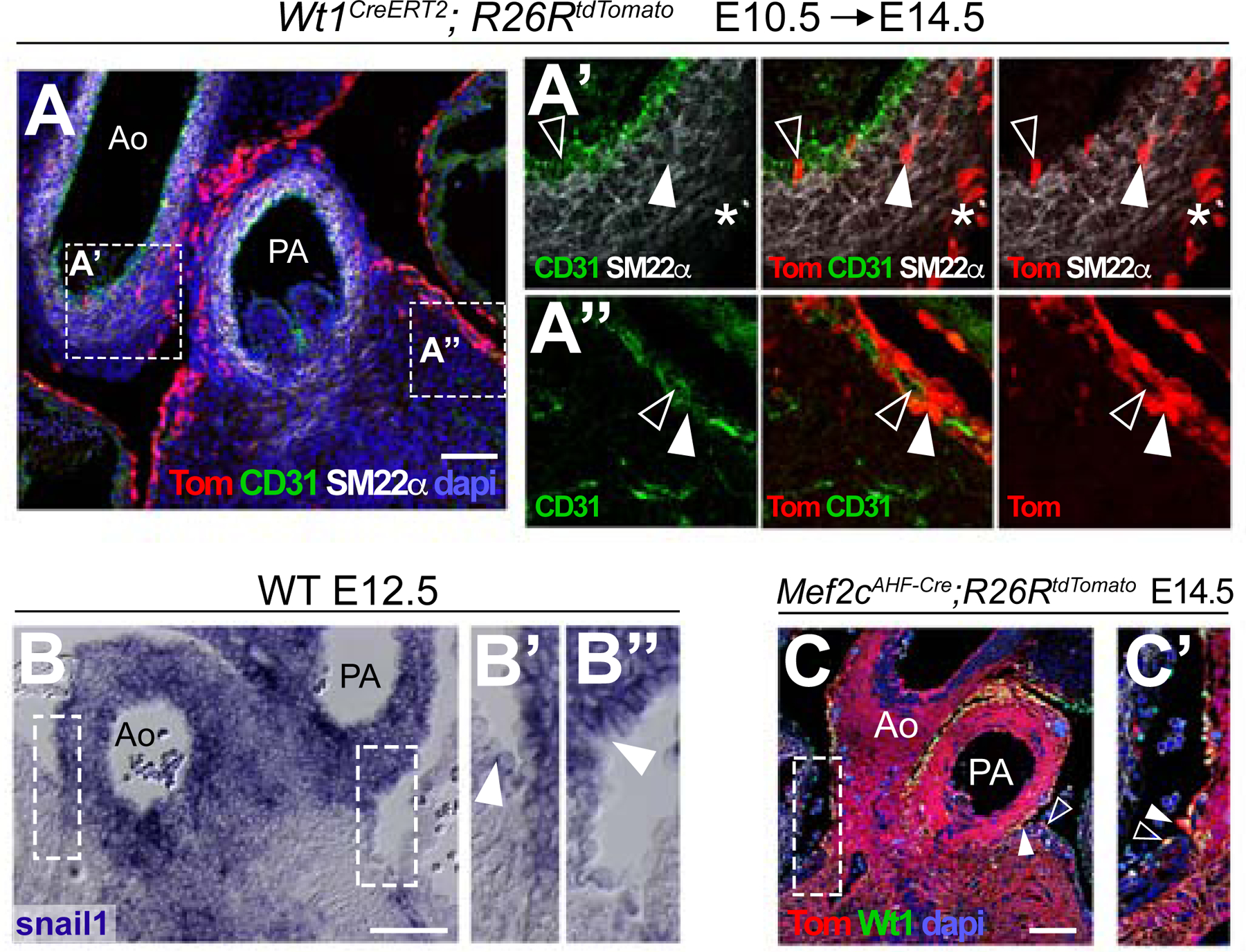
(A) Contribution of the Wt1CreERT2 lineage in co-detection with markers of endothelium (CD31) and smooth muscle (SM22α). (A’) and (A’’), magnification of the respective boxed areas indicated in (A) showing selected color channels. In A’, examples of lineage-positive cells are indicated by an empty arrowhead in the aortic endothelium, by a solid arrowhead in the media and by an asterisk in the adventitia. In A’’, empty arrowheads indicate endothelial cells and solid arrowheads endothelial-associated lineage-positive cells. (B-B’’) Expression of the EMT marker snail1 (mRNA) at the base of the great arteries. B’ and B’’ show a magnification of the boxed areas in B. Arrowheads indicate the border between the positive mesothelial layer and the negative epicardium. (C) Detection of the Mef2cAHF-Cre enhancer lineage, co-stained with the mesothelial/epicardial marker Wt1. The boundary between AMCs and epicardium is shown by a solid arrowhead. A Tom+ cell in the epicardium is indicated by an empty arrowhead. (C’) Magnification of the boxed area in E. All scale bars: 100 µm. Ao: aorta; PA: pulmonary artery; Tom: tomato. See also Figures S3 and S4.
Therefore, the relationship between AMCs, the coronary endothelium and the sub-pulmonary myocardium can only be explained by the existence of a common precursor between AMCs and these lineages. Given that the base of the great arteries, the valves and the RV receive contributions from the SHF (Waldo et al., 2005), we studied whether SHF precursors contribute to AMCs. We found that the SHF tracer lines Islet1Cre, Mef2cAHF-Cre, Hoxb1Cre and Islet1MER-Cre-MER (induced at low labelling density at E8.5), all consistently labelled AMCs and arterial sub-mesothelial cells at the base of the great arteries at E14.5 (Figure 2C–C’ and Figure S4). The epicardium was in general not labelled, with the exception of a few disperse cells close to the base of the PA (Figure 2C–C’ and Figure S4), which correlates with the distribution of cells in proximal AMC clones. These results indicate that the common precursor that contributes to AMCs and their clonally related lineages belongs to the SHF.
Arterial mesothelial cell-related clones contribute to ventral but not dorsal heart lymphatics
To further study the nature and contribution of the AMC-related coronary blood island-like structures, we generated new sets of clonal cell groups at E8.5, but analyzed them at progressively later stages. This series of clonal marks were generated with fluorescent reporters, which allowed using multiple fluorescent markers together with the clonal labeling. At E15, we obtained three labelled cell groups that contributed to AMCs (Figure 3A–B and S5G). These groups contained AMCs (Figure 3C), cardiomyocytes (Figure 3D), valve mesenchymal cells (not shown) and sub-epicardial/mesothelial endothelial cells forming vascular networks (Figure 3B, D–F). These vascular networks express the LEC markers Prox1 (Figure 3D, F) and Lyve1 (Figure 3B, C, E). The lymphatic cells in these clones were restricted to the base of the PA and the adjacent myocardium, which suggests that they are not related to a pre-existing lymphatic vascular network but originate in situ.
Figure 3. Clonal analysis identifies a lineage relationship between ventral coronary lymphatics and arterial mesothelial cells.
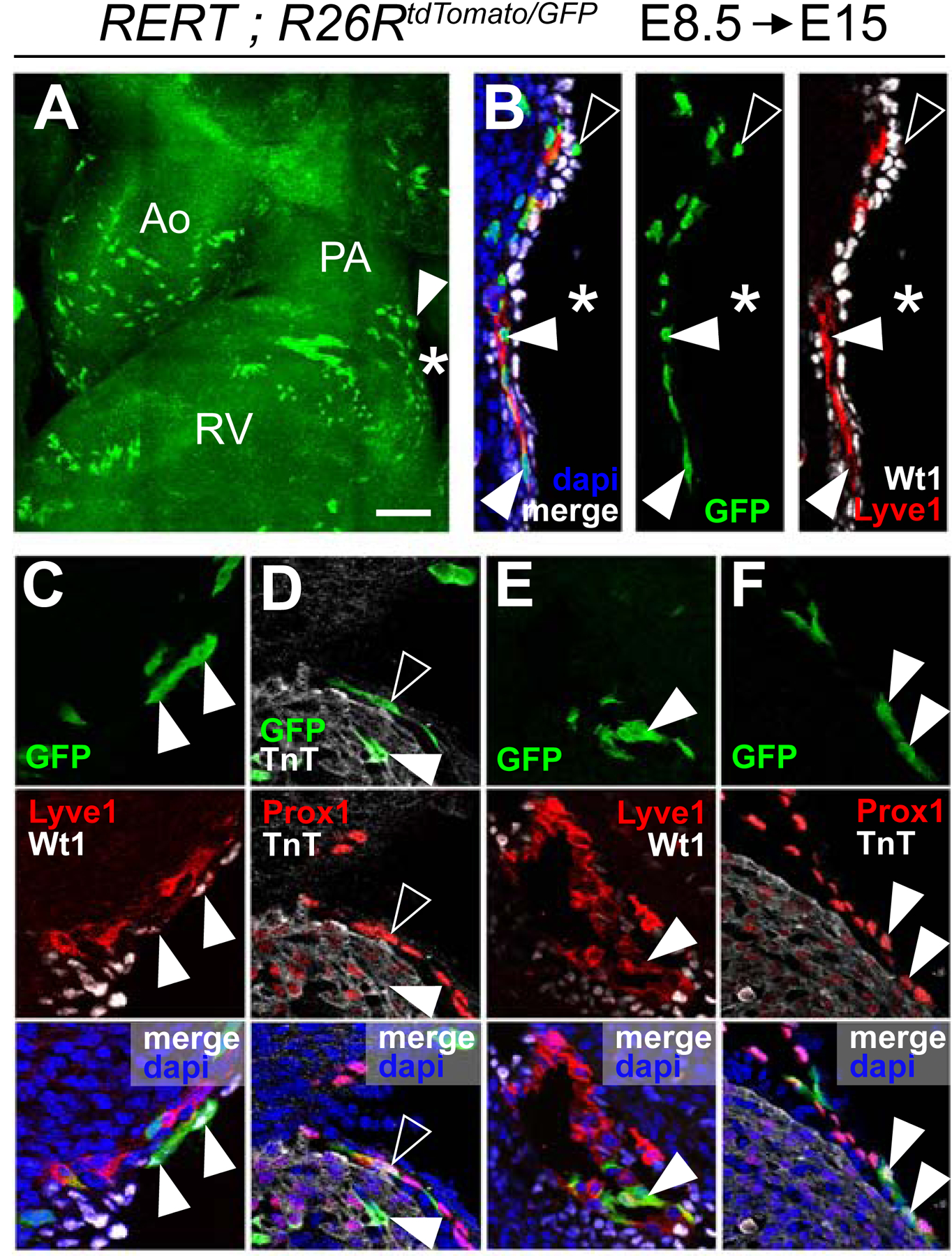
(A) Whole-mount view of the OFT region of a clone-containing heart. The arrowhead indicates proximal AMCs and the asterisk, cells extending towards the ventricle. (B) Section of the region indicated by an asterisk in A’ showing LECs (Lyve1+), positive for the GFP lineage (solid arrowheads), and EPICs (Wt1+), negative for the GFP lineage. Sparse cells over the epicardial surface are positive for the lineage but negative for the Wt1 (empty arrowhead). (C) shows AMCs (Wt1+, solid arrowhead) positive for the GFP lineage and sub-mesothelial cells positive for the GFP lineage in association with LECs (Lyve1+). (D) Sub-epicardial LECs (Prox1+, empty arrowhead) and Cardiomyocytes (TnT+, solid arrowhead) positive for the GFP lineage. (E) LECs over the ventricle (Lyve1+, arrowhead) are positive for the GFP lineage. (F) Sparse cells of the GFP-lineage found in the epicardial layer are Prox1+ (Arrowheads). All scale bars: 100µm. Ao: Aorta; PA: Pulmonary artery; RA: Right Atrium; LA: Left Atrium; SV: Sinus Venosus. See also Figures S5 and S6.
In addition, some of these clones contained a few cells on the epicardial surface close to the artery. Interestingly, the epicardial cells clonally related to cells of the PA and LECs are negative for Wt1, unlike their immediate epicardial neighbors (Figure 3B), suggesting they have a different identity than AMCs and EPICs. Instead, these cells were positive for Prox1 (Figure 3F). These observations indicate that SHF-derived cells at the base of the PA produce LECs, some of which transiently colonize the epicardial layer close to the base of the PA. Given their lymphatic nature and their epicardial location, we will refer to these cells as Lymphatic Epicardial Cells (LEPICs).
Cardiac lymphatics appear first around E14.5 as two growing vascular networks; a dorsal one that ramifies from the region of the sinus venosus, and a ventral one that is connected to a band of lymphatic cells that appears at the base of the PA (Figure S6). This band of lymphatic cells adopts a sinusoidal/sac like structure around E16.5 (Figure S6). Only occasionally similar lymphatic structures also appear at the base of the aorta (not shown).
We next studied a collection of 9 cell clusters, induced at E8.5 and retrieved at E16.5, that contained lymphatic endothelial cells (Figure S5A–B’’ and G). We observed that 3 of the clusters colonized only the dorsal part of the heart, 4 of them only the ventral side and 2 of them both sides. The clusters that contributed to the dorsal side showed a denser distribution of the labelled cells in the lymphatic endothelium than those exclusively present the ventral side (Figure S5A–B’’). In addition, just the 4 ventral-only clusters contained labelled cells at the PA root (Figure S5A–A’). Interestingly, all lymphatic clusters located at the dorsal heart also showed labelled extra-cardiac LECs in the jugular region cephalic to the heart, suggesting their relationship to the pre-existing lymphatic vasculature (Figure S5B, B’). In contrast, ventral lymphatics did not correlate with extracardiac lymphatic labeling other than that at the base of the great arteries (Figure S53A–A’). In agreement with these observations, two singular lymphatic clones induced at E8.5 and retrieved at P0 exclusively labeled either the dorsal or the ventral side of the heart, with the dorsal clone showing a much denser and widespread cellular distribution than the ventral one (Figure S5C–G).
These data suggest that the origin and dynamics of formation of the cardiac lymphatic vasculature are different in the ventral and dorsal parts of the heart. To test this, we traced the timing of specification of cardiac lymphatics using the Vegfr3CreERT2 driver (Martinez-Corral et al., 2016), which recombines lymphatic endothelial cell precursors (Figure 4A–L’). Following TM injections at E11.5 and E12.5 the frequency of ventral lymphatic colonization by Vegfr3CreERT2-labelled cells is below that observed for dorsal lymphatics (Figure 4M). The frequency of labeling in ventral and dorsal cardiac lymphatics only matches following inductions at E14.5 (Figure 4M). In addition, we found that the specification sequence of dorsal lymphatics matches that of extracardiac thoracic lymphatics; however, that of ventral lymphatics only matches following injections at E14.5 (Figure 4N).
Figure 4. Heterochronic specification of ventral and dorsal coronary lymphatics.
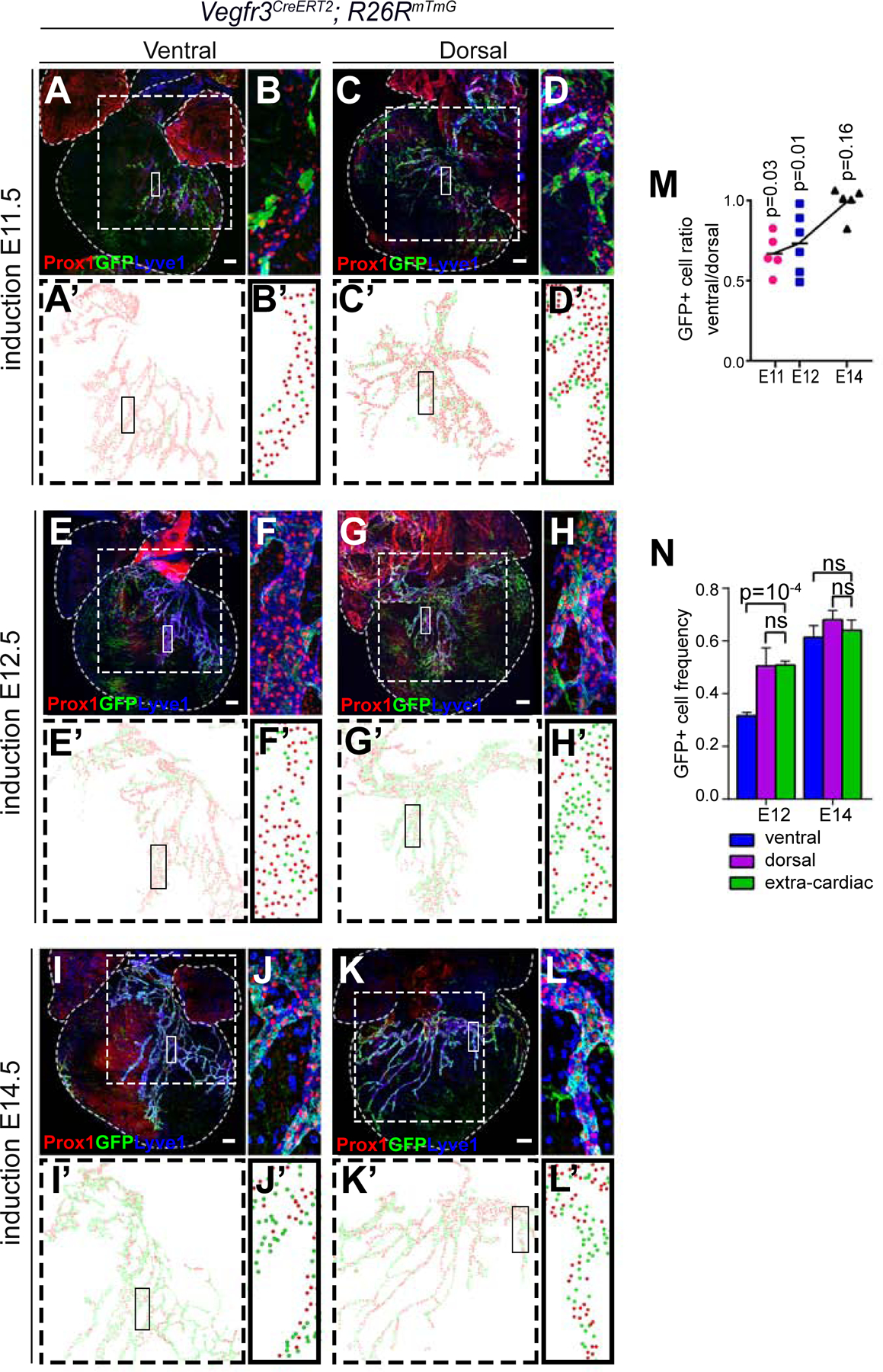
(A–L) Z-projections of confocal stacks showing the lymphatic vasculature and the lineage of Vegfr3-expressing cells at E17.5 following induction of recombination at E11 (A–D), E12 (E–H) and E14 (I–L). (B, D, F, H, J and L) show magnifications of the boxed areas in (A, C, E, G, I and K), respectively. (A, E and I) show ventral lymphatics and (C, G and K) show dorsal lymphatics. (A’-L’) show examples of the scoring of Vegfr3 lineage-positive (GFP+) and lineage-negative (GFP−) LECs. Green dots indicate positive LECs and red dots indicate negative LECs. (M) Graph showing the ratio of GFP+ LEC frequency between ventral and dorsal LECs following induction of recombination at different stages. p-values in each stage of induction show the significance of the difference in the proportion of positive cells between dorsal and ventral sides of each heart by paired T-test. (N) Graph showing the frequencies of GFP+ LECs in ventral, dorsal and extracardiac lymphatics following inductions at E12 and E14. N=4 hearts/stage in both M and N. Paired T-test was used for calculating the p values. Ns, p-value> 0.05. Error bars show standard error of the mean (SEM). All scale bars: 100µm.
These results show that SHF-derived, AMC-related lineages at the base of the pulmonary artery contribute to the ventral lymphatics of the heart through an in-situ specification process independent of the general wave of lymphatic specification from the CV.
Second heart field origin of ventral lymphatics
The results of the clonal analysis implied that part of the ventral lymphatics derive from SHF precursors. To directly test this, we determined the contribution of Islet1Cre and Mef2cAHF-Cre to cardiac lymphatics. In both cases, we found contribution of the labelled cell lineages to ventral endothelial lymphatic cells and LEPICs, while no contribution was found to dorsal lymphatics (Figure 5A–B’’ and Figure S7A–A’’’). The quantification of the contribution of the Islet1Cre labelled cells indicated that more than half of the lymphatic endothelium of the E17.5 ventral heart derives from the SHF (Figure 5C). These results were confirmed using the inducible Islet1MERCreMER driver (Figure S7B–B’’’).
Figure 5. Second heart field contribution to cardiac lymphatics.
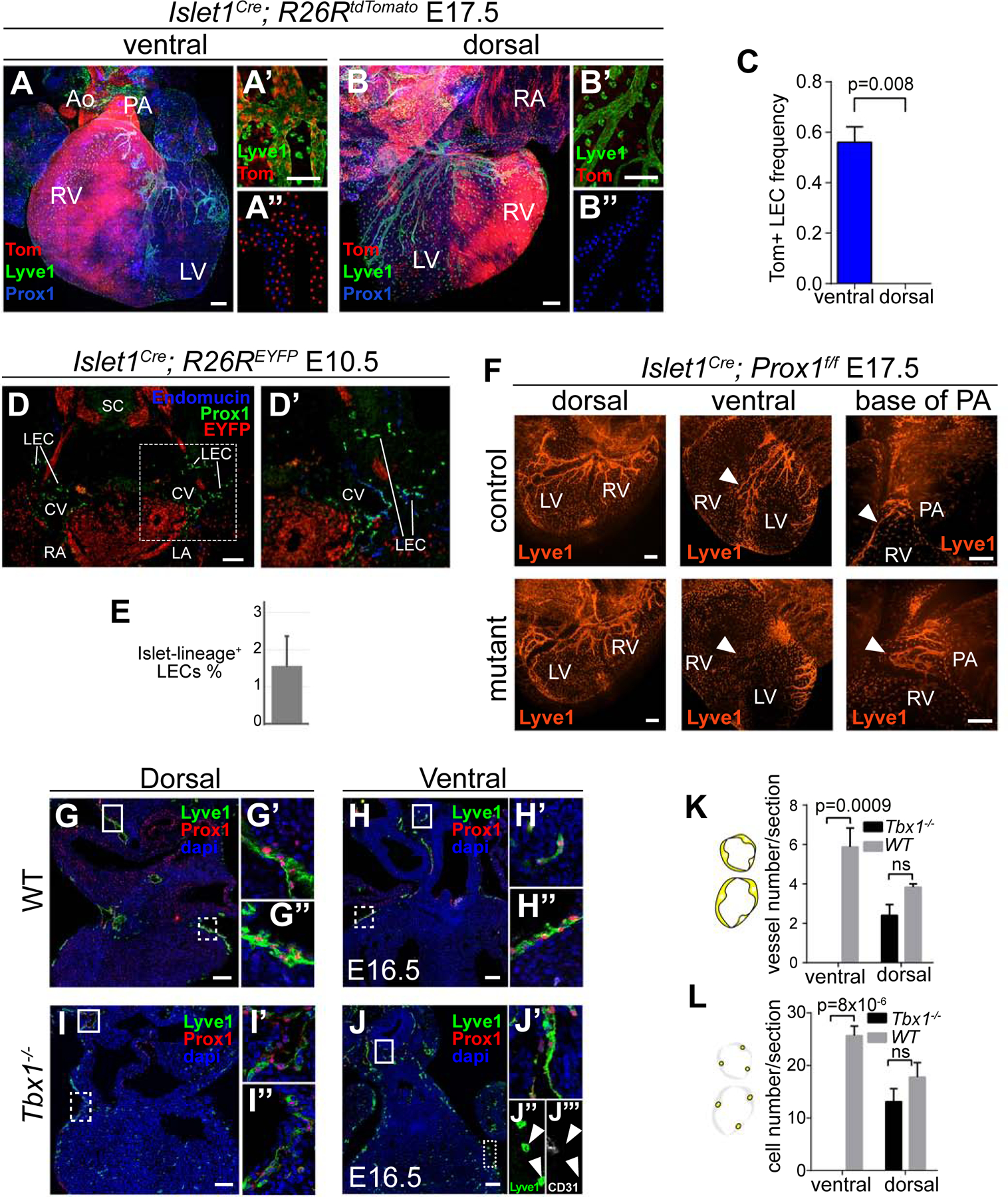
(A–C) Isl1+ lineage tracing. Confocal stack Z-projections of the ventral (A–A’’) and dorsal (B–B’’) sides of hearts at E17.5. (A’) Magnification of the ventral cardiac lymphatics. (A’’) example of the scoring of LECs positive (red dots) and negative (blue dots) for the Islet1 lineage. (B–B’’) similar analysis as in (A–A’’) for the dorsal cardiac lymphatics. (C) Graph showing the frequency of Isl1+ lineage LECs in the ventral and dorsal parts of the heart. N=5 and p-value calculated with a Mann-Whitney test. (D) Detection of the Islet1+ cell lineage in CV and CV-derived LECs. (D’) Magnification of boxed region in (D). (E) Graph showing the percentage of LECs positive for the Isl1+ lineage. N=2 embryos and 485 LECs scored. (F) Elimination of Prox1 function in the Islet1+ cell lineage. Dorsal and ventral views are provided showing coronary lymphatics (Lyve1+). Solid arrowheads show ventral lymphatic sprouts. (G–L) Cardiac lymphatic development in Tbx1 mutants. Frontal sections at the ventral or dorsal levels are shown for WT and Tbx1 null mutant hearts. Lymphatics were identified by simultaneous detection of Lyve1 and Prox1. (G’–J’) Magnification of the respective solid-line boxes in (G–J) showing extra-cardiac lymphatics. (G’’–J’’ and J’’’) Magnification of the respective dotted-line boxes in (G–J) showing the analysis of cardiac lymphatics. Lyve1+ cells in the ventral side of mutant hearts are negative for Prox1 (J) and CD31 (J’’’), indicating that they are not LECs but macrophages. (K) Graph showing the quantification of the number of lymphatic vessels scored/section. (L) Graph showing the quantification of the number of LECs/section. N=4 hearts for the ventral sides and N=3 hearts for the dorsal. p-values were calculated using a T-test. ns, p>0.05. Error bars show SEM in C, K and L and Standard Deviation in E. All scale bars: 100 µm. Ao: Aorta; PA: Pulmonary artery; RV: Right ventricle; LV: Left ventricle; RA: Right atrium; LA: left atrium. See also Figure S7.
One possibility to explain the observed contribution of SHF tracer lines to lymphatics would be the contribution of this lineage to the venous endothelial cells of the CVs and their subsequent recruitment to form LECs. To test this possibility, we characterized the presence of Islet1Cre-labelled cells in the CVs and derived lymphatic vascular structures at E10.5 (Figure 5D–D’). We found that the vast majority of LECs in the CV and surrounding tissues was negative for the Islet1Cre lineage (Figure 5E). As Islet1Cre also contributes to the neural crest, we determined whether cardiac lymphatics were labelled by the neural crest tracer line Wnt1Cre and did not find any contribution to the regions where lymphatics are formed at E14.5 (Figure S7C).
These results show the contribution of SHF lineages to the lymphatic endothelium of the ventral heart and suggest a direct contribution from SHF to lymphatics, independent of the CV-derived LECs.
SHF-derived LECs are essential for the formation of ventral cardiac lymphatics
To study the functional relevance of SHF-derived LECs, we blocked SHF lymphangiogenesis by conditionally eliminating Prox1 in the SHF (Figure 5F). Despite the fact that only about half of ventral LECs in the heart derive from SHF, we observed complete agenesis of ventral lymphatics in fetuses developed to term (Figure 5F). In contrast, dorsal lymphatics were completely normal (Figure 5F). In SHF-specific Prox1 mutants, we found that the lymphatic plexus at the base of the pulmonary artery is present, however it does not sprout towards the ventral side of the heart as it normally does in control specimens (Figure 5F).
Next, to study whether the deployment of SHF lineages is relevant for ventral lymphatic development, we studied cardiac lymphatics in Tbx1−/− mutants, which are deficient in SHF incorporation to the developing heart (Rana et al., 2014; Xu et al., 2004) (Figure 5G–L). In these mutants, lymphatic vessels were normally observed in regions distal to the heart (Figure 5G’, H’, I’, J’) and dorsal cardiac lymphatics were only slightly affected (Figure 5G’’, I’’, K, L), in accordance with the generalized mild reduction of the lymphatic vasculature in Tbx1 mutants (Chen et al., 2010). In contrast, ventral heart lymphatics were completely absent (Figure 5H’’, J–J’’’, K, L). These results show the absolute requirement of SHF-derived lineages and SHF deployment for ventral cardiac lymphatics development.
A sub-mesothelial vasculogenic niche at the base of the PA generates coronary lymphatics
The data so far presented suggest that SHF-derived precursors related to the PA mesothelium contribute to lymphatics. To directly test this hypothesis, we performed mesothelial/epicardial grafting experiments in cultured whole hearts. As tissue donors, we used E14.5 Wt1Cre; R26RmTmG hearts, in which the epicardium/mesothelium is labelled with mGFP, while the sub-epicardium/sub-mesothelium is labelled with mTomato (Figure 6A). The donor tissue was obtained by peeling off the mesothelial/epicardial layers, together with their associated sub-epicardial/sub-mesothelial tissues (Figure 6A). The donor tissues were transplanted onto the ventricular surface of wild type hearts, in the region where the first ventral lymphatic vessels are observed at E15.5 (Figure 6B). Transplanted hearts were then placed in culture for three days, after which, they were still beating (see Methods). We performed heterotopic experiments transplanting the E14.5 PA mesothelium and orthotopic experiments, transplanting the epicardium. Mesothelial cells from heterotopic transplants showed a stronger tendency to contribute to the inner myocardium than epicardial cells from orthotopic transplants (Figure 6C–D’, E). In fact, the orthotopic transplants conserved a continuous layer of labelled epicardial cells, while the heterotopic ones kept only a few sparse cells in the epicardial layer (Figure 6C–D’). Similarly, the relative contribution of the sub-mesothelial cells to the inner myocardium was higher than that of the sub-epicardial layer of orthotopic transplants (Figure 6C–D’, E). The study of the fate of transplanted tissues indicated that sub-mesothelial cells from all heterotopic transplants formed LECs at considerable rates, while sub-epicardial cells showed contribution in only 1 out of 5 transplants (Figure 6F). Interestingly, the PA mesothelium did not contribute to the lymphatic endothelium, but profusely contributed to LEC-associated cells (Figure 6D, D’, F). As further evidence for this, we found that lymphatic vessels in Wt1Cre;R26RmTmG hearts are composed of Tomato+ LECs and GFP+ LEC-associated cells (Figure 6G–G’).
Figure 6. Transplantation experiments demonstrate specific Lymphangiogenetic activity of the mesothelium/sub-mesothelium at the base of the pulmonary artery.
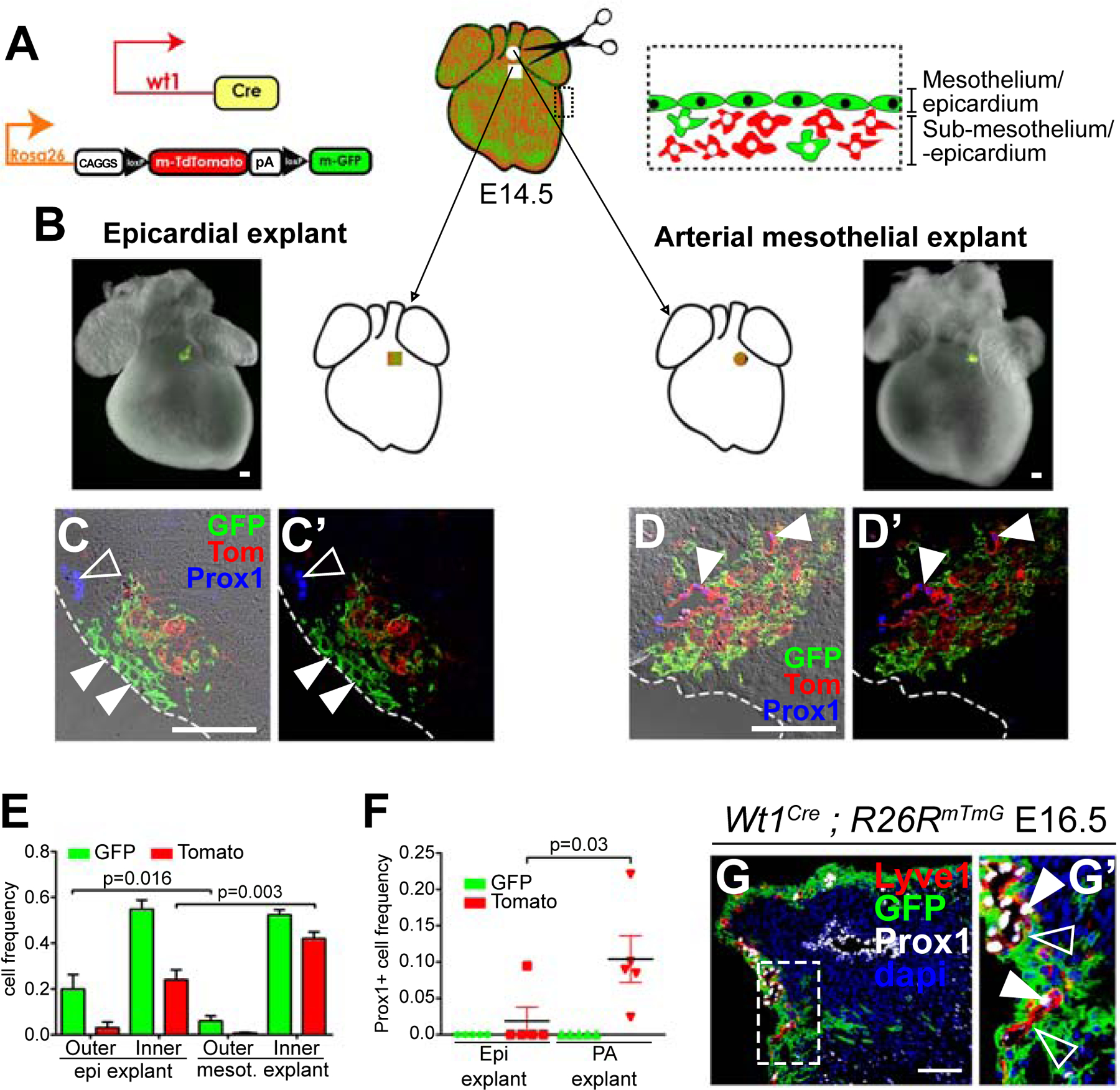
(A) scheme of the experimental design. (B) Scheme and compound fluorescent/bright field images of whole-mount hearts showing the transplantation strategy. (C, D) and (C’, D’) show sections with and without the brightfield channel of hearts with homotopic (C, C’) and heterotopic (D, D’) transplants. The dashed line represents the border of the epicardium. (C, C’) Transplanted epicardial cells are conserved in the epicardial layer (solid arrowheads). Prox1 was expressed in the host tissue (empty arrowhead) but not in the explant-derived cells. (D, D’) Most transplanted mesothelial cells invade the underlying myocardium. Prox1 was expressed in vascular structures derived from sub-mesothelial cells (solid arrowheads), which were surrounded by mesothelial-derived cells. (E) Graph showing the relative contributions of transplanted epicardial/sub-epicardial cells and mesothelial/sub-mesothelial cells. (F) Graph showing the proportion of Prox1+ cells in the transplanted GFP+ and Tomato+ populations. N=10 arterial mesothelial transplants and N=9 epicardial transplants. Mann-Whitney test was used to calculate the p-values. Error bars show SEM. (G) Section showing the distribution of the Wt1+ cell lineage (GFP+) in relationship to the lymphatic vasculature (Lyve1+ and Prox1+) at the base of the pulmonary artery. Tomato detection is not shown. (G’) Magnification of the boxed area in G. All scale bars: 100µm.
These results show that the mesothelial/sub-mesothelial layers at the base of the PA contain LECs and a microenvironment that promotes lymphangiogenesis. In addition, they show that the lymphatic endothelium derives exclusively from the sub-mesothelium, whereas the mesothelium undergoes EMT to produce lymphatic endothelium-associated cells.
Regionalized control of cardiac lymphangiogenesis by the retinoic acid pathway
To better understand the molecular cues that drive lymphatic specification at the base of the PA, we identified genes differentially expressed between the arterial mesothelium/sub-mesothelium (MsM) and the epicardium/sub-epicardium (EsE) at E16.5 by RNAseq. This analysis identified genes encoding regulators of the retinoic acid (RA) pathway (Figure 7A), including the gene coding for the main RA-synthesizing enzyme Raldh2 and the RA target Wt1, which showed lower expression levels in the MsM than in the EsE (Figure 7A). Conversely, markers of sub-regions of the SHF, like Fgf10, Hox genes and Foxc1 and Foxc2, were expressed at higher levels in the MsM than in the EsE. In agreement with the observations above, several markers of lymphatic development were also found expressed at higher levels in the MsM than in the EsE (Figure 7A).
Figure 7. Regionalized retinoic acid signaling contributes to the lymphangiongenic niche at the base of the great arteries.
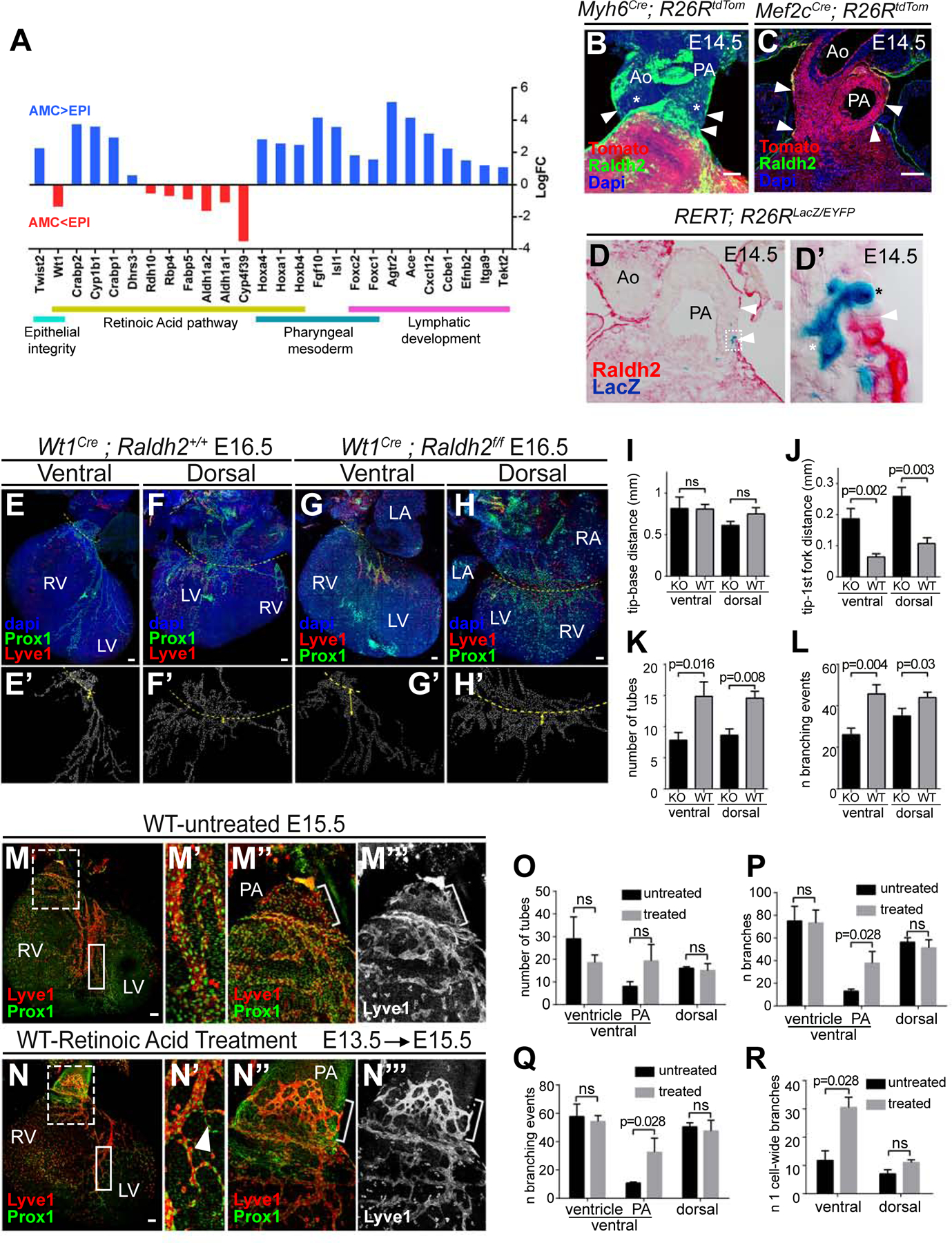
(A) Graph showing the expression changes detected by RNAseq between the arterial proximal MsM and the EsE. (B–D’) analysis of Raldh2 expression at the base of the great arteries shows a gap in mesothelial Raldh2 expression (arrowheads) in whole mount confocal analysis (B) and histological sections (C–D’). (C) AHF cell lineage (Tom+) was used as a reference to identify AMCs together with low Raldh2 levels in this region (arrowheads). (D) Histological section of the base of the great arteries showing Raldh2 expression and a LacZ clone containing AMCs. (D’) Magnification of the boxed area in (D) showing LacZ+ AMCs (black asterisk) and clonally related cells (white asterisk). (E-H) Confocal stack Z-projections of the lymphatic vasculature (Lyve1+ and Prox1+) of control hearts (E, F) and Wt1Cre-recombined (G, H) Raldh2 mutant hearts. (E’–H’) examples of the scoring of single LECs in E-H. Dashed lines indicate the reference used for length measurements. (I–L) Graphs showing different measurements of the lymphatic vasculature in E–H. Examples of measurements in J are shown in E’–H’. N=7 wild type hearts and N=5 mutant hearts. The Mann-Whitney test was used to calculate the p values. Ns, p-value>0.05. (M-N’’’) Confocal stack Z-projections showing the ventral lymphatic vasculature (Lyve1+, Prox1+) of hearts untreated (M) and treated with retinoic acid for 2 days (N). (M’–N’’’) Magnified views of the boxed areas in (M, N). Graph showing the quantification of the number of lymphatic vascular tubes in treated and untreated hearts. (O-R) Graphs showing different measurements of the lymphatic vasculature in M–N. For graphs in (O–R), N=4 wild type hearts and N=4 mutant hearts. The Mann-Whitney test was used to calculate the p values. Ns, p-value>0.05. Error bars show SEM. All scale bars: 100 µm. Ao: Aorta; PA: Pulmonary artery RV; Right ventricle; LA: Left atrium; RA: Right atrium; LV: Left ventricle.
Whole-mount detection of Raldh2 showed strong Raldh2 expression in the epicardium and pericardial mesothelium, with the exception of the mesothelium at the base of the aorta and PA (Figure 7B), which is consistent with previous data in the chick embryo (Pérez-Pomares et al., 2003). This region of low or no Raldh2 expression approximately coincides with the region in which the lymphatic vasculature appears as poorly remodeled sinusoids/sacs (Figure S6). Detection of Raldh2 expression in sections of Mef2cAHF-Cre; R26RtdTom embryos confirmed that SHF-derived proximal AMCs exhibit lower levels of Raldh2 and that a sharp expression boundary forms between EPICs and AMCs at the arterial/ventricular boundary (Figure 7C). To determine the relationship between this boundary and the proximal AMC clones described in Figure 1, we stained for Raldh2 expression in sections containing RERT-induced clones. We found that AMCs in the Raldh2-negative region are clonally related to cells in the sub-mesothelial space of the PA and in the ventricular sub-epicardial space beneath the Raldh2-expressing epicardial cells (Figure 7D–D’).
To functionally assess the role of RA in coronary lymphatic development, we studied a conditional deletion of Raldh2 using the Wt1Cre driver (Figure 7E–L). In mutant hearts, the lymphatic vessels growing on both the dorsal and ventral ventricular surfaces showed less ramifications, shorter extensions and thicker branches than in controls (Figure 7E–L). Overall, the observations suggested normal LEC proliferation but impaired maturation of the cardiac lymphatic plexus.
Finally, to study the relevance of the low-RA environment at the base of the PA, we exposed fetuses to excess RA by treating gestating females at E13.5 and analyzing them at E15.5 (Figure 7M–R). We did not observe evident alterations of ventricular lymphatics (Figure 7M–N’), which is consistent with the fact that this region naturally contains high RA levels. In contrast, we observed increased angiogenic maturation at the base of the PA (Figure 7N’’–M’’’), with increased branching and narrower vessel-like structures, instead of the normally un-patterned sac-like lymphatic structures found in this area at E15.5 (Figure 7N–R).
These results show the contribution of regionalized RA signaling to the vasculogenic niche at the base of the PA and indicate a role of epicardial RA signaling in ventricular lymphangiogenesis.
Discussion
The description of the cellular genealogy of the mammalian heart has been extensively explored by permanently labeling cell groups and their descendants using DNA recombinases under the control of regulatory elements of regionally expressed genes (Meilhac and Buckingham, 2018). While this approach has been extremely useful for the definition of heart tissue origins, it has also generated extensive controversy on the reliability of the conclusions, for example on the origin of cardiomyocytes (Cai and Molkentin, 2017; Christoffels et al., 2009; Rudat and Kispert, 2012) and the coronary vasculature (He and Zhou, 2018). The underlying causes of these controversies are the poor description of recombinase expression patterns. This limitation is aggravated by the use of constitutively active recombinases able to permanently label cell lineages despite very transient and/or low expression levels. An alternative to this approach is retrospective clonal analysis, whereby single cell precursors are interrogated for developmental potency (Buckingham and Meilhac, 2011). Previous clonal approaches to heart development have used lineage-restricted clone induction/detection to study the early segregation of heart lineages and fields (Lescroart et al., 2014; Meilhac et al., 2004; Miquerol et al., 2013). Here, we performed the first random, lineage-unrestricted clonal analysis of the developing heart, which allowed to explore all lineage relationships in an unbiased manner. While this approach resolves to a large extent the problems associated to the use of constitutive recombinases, the retrospective nature of the approach bears as well some limitations. On one side, the lineage relationships identified do not inform about the hierarchy of lineage diversification and, on the other, the identity and position of the cell that originated the clone are undetermined. To overcome these limitations, we combined clonal analysis with the use of several lines expressing constitutive and inducible recombinases tracer lines. The combination of the two approaches provided a detailed and solid knowledge on the lineage relationships analyzed.
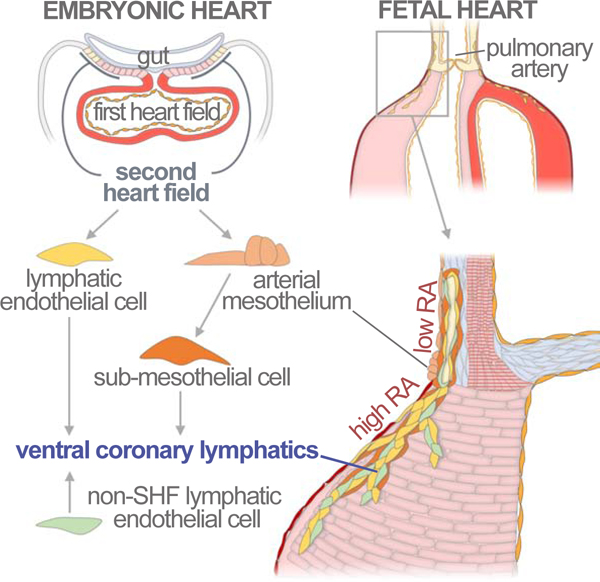
We characterized the fate of SHF precursors at the base of the great arteries and found a contribution of SHF to the coronary lymphatic vasculature. The clonal analysis identified a shared lineage for arterial mesothelial and sub-mesothelial cells and cell types known to derive from the SHF, like RV cardiomyocytes, valve mesenchyme and coronary endothelial cells. Interestingly, a population of ventral coronary lymphatics extending from the sub-mesothelium at the base of the PA into the ventricular sub-epicardium was also present in this type of clones. Several observations indicate that LECs at the base of the PA originate in situ from splanchnic mesoderm of the SHF. An alternative possible origin is the cardiac neural crest, which is labelled by Islet1Cre
(Engleka et al., 2012) and colonizes the arterial pole of the heart, however, Mef2cAHF-Cre, which does not contribute to neural crest (Engleka et al., 2012), also contributes to cardiac lymphatics, as shown here and in a recent independent study (Stone and Stainier, 2019). Furthermore, as previously reported (Klotz et al., 2015), we did not find any contribution from the neural crest to the lymphatic coronary endothelium using the Wnt1Cre tracer line. Another alternative to the direct specification of LECs from SHF mesenchymal precursors is an indirect contribution through transient colonization of the CV. Here, we showed that indeed some CV LECs are also positive for the Islet1Cre lineage, however, the contribution is below 2%, which does not explain the abundant contribution to coronary lymphatics (>50%) nor its restriction to the ventral heart. Finally, the observation that the first ventral lymphatics are disconnected from any preformed extracardiac lymphatics and the local distribution of the clonal cell groups containing SHF-related lymphatics are good evidence for the in situ specification of LECs at the base of the PA.
We characterized the niche in which vasculogenesis takes place at the base of the PA. AMC lineage tracing showed that, similar to their epicardial counterparts, these cells undergo active EMT and contribute to the underlying layers of the great arteries. Interestingly, this contribution included mesenchymal cells strongly associated to the coronary lymphatic vasculature but not LECs themselves. This observation, together with the common origin of AMCs and lymphatics deduced from the clonal analysis, suggested that a common SHF precursor contributes to both AMCs and the sub-mesothelial cell populations that originate LECs. To test this idea, we established an organotypic culture and transplantation approach for the E14.5 heart and used it to demonstrate the lymph-vasculogenic activity of the MsM at the base of the PA. In agreement with the lineage tracing results, the sub-mesothelium contributed to LECs and the mesothelium to LEC-associated cells. Importantly, similar experiments transplanting epicardium did not show significant lymphatic vessel formation, highlighting the relevance of the mesothelial niche at the base of the PA for the lymph-vasculogenic activity.
Several conclusions from our study are striking, like the restriction of the contribution of the SHF to the ventral lymphatics of the heart. This restriction agrees with the preferential detection of dorsal- and ventral-specific LEC clones and with the heterochronic specification of the ventral and dorsal LECs. Dorsal LEC clones include extra-cardiac lymphatics in the jugular region and their specification dynamics follows that of extra-cardiac lymphatics, suggesting that they develop by angiogenesis from CV-derived lymphatic vessels. SHF-derived lymphatics, in contrast, show delayed specification and differentiation, characteristics shared with other SHF-derived lineages. Another striking observation is that SHF contribution to the ventral side is mixed with LECs from a different source. While these additional ventral LECs may derive from the CV, we cannot exclude that they derive from yet a third source of cardiac lymphatics. SHF-derived LECs appear completely intermingled with this additional population of ventral LECs, which could imply their functional equivalence, however, the functional analysis of SHF-derived LECs suggests otherwise. Elimination of SHF-derived Lymphatics by mutating Prox1 in this population or elimination of SHF incorporation to the heart in Tbx1 mutants completely abolishes ventral Lymphatic development, but does not affect dorsal lymphatics. Our observation of selective involvement of Tbx1 in ventral lymphatics is consistent with recent findings of enhanced Tbx1 and Vegfr3 interactions during establishment of ventral over dorsal cardiac lymphatic vessels (Martucciello et al., 2019). These results again show the modularity and independence of dorsal and ventral lymphatic development. At the same time, it shows that ventral lymphatics critically depend on SHF LECs for their development. The fact that in SHF-Prox1 mutants lymphatics are still produced at the base of the PA, but cannot sprout into the ventricle, suggests that the specific roles of SHF-derived LECs involve angiogenic abilities that elicit the colonization of the ventricles. We anticipate that SHF-derived LECs bear lineage-specific properties that are not present in the non-SHF LECs and enable them to lead the angiogenic process.
Finally, we have explored the molecular cues that characterize the vasculogenic niche at the base of the PA. We found that restricted retinoic acid signaling from the epicardium influences the lymphangiogenic process by promoting vascular maturation in the ventricles, while a low-retinoic acid region at the base of the great arteries maintains the lymphatic vasculature in an immature state. Interestingly, alterations in jugular lymphatics relate to a fetal condition known as nuchal translucency (Haak et al., 2002) that is frequently associated with fetal cardiac defects (Hyett et al., 1999) and can be promoted by defective RA signaling (Burger et al., 2014). We also found that cells derived from AMCs undergo extensive EMT and intimately associate with nascent lymphatic vessels at the arterial pole. These observations suggest that interactions between AMCs and AMC-derived cells might contribute to the niche properties required for LEC specification in the PA and lymphatic angiogenic growth into the ventricles.
Our study identifies and characterizes a new vasculogenic niche that contributes to coronary lymphatic development and shows that cardiac lymphatics are heterogeneous in origin and function. This work opens avenues for future studies on the mechanism involved in lymphatic vasculogenesis in this new niche and on the functional diversity of cardiac lymphatics.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Miguel Torres (mtorres@cnic.es).
This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse strains were handled in accordance with CNIC Ethics Committee, Spanish laws and the EU Directive 2010/63/EU for the use of animals in research. All mouse experiments were approved by the CNIC and Universidad Autónoma de Madrid Committees for “Ética y Bienestar Animal” and the area of “Protección Animal” of the Community of Madrid with reference PROEX 220/15. Mouse experiments performed at Northwestern University were performed in accordance with protocols approved by Northwestern University Institutional Animal Care and Use Committee. Animal procedures performed at CNIO were approved by the Instituto de Salud Carlos III Ethics Committee for Research and Animal Welfare and by the competent authority in the Madrid Local Government (Proex 234/15). For this study, Mice were maintained on mixed C57Bl/6 and CD1 or NMRI backgrounds. Mice were genotyped by PCR as described in the original reports. Male and female mice of more than 8 weeks of age were used for mating. The mouse lines used are detailed in the Key resources table. Experimental specimens were retrieved during gestation and their sex was not determined.
METHOD DETAILS
Tamoxifen administration
For induction of the RERT mouse line, 10mg of 4-Hydroxy Tamoxifen (Sigma) was dissolved in 1ml absolute Ethanol and 9ml corn oil (Sigma) for a final concentration of 1mg/ml. The stock solution was then sonicated for 40 minutes on ice to prevent overheating. The 4-OHT solution was aliquoted and stored at 4°C for up to 4 weeks, and re-sonicated before being administered to female mice by intra-peritoneal injection. For induction of all the other CreERT lines, 200mg of Tamoxifen (Sigma) was dissolved in 10ml of corn oil (Sigma) for a final concentration of 20 mg/ml, excepting Vegfr3CreERT2, for which tamoxifen was prepared at 10 mg/ml in sunflower oil (Sigma). The stock solution was then sonicated for 20 minutes on ice. The solution was aliquoted and stored at RT for up to 3 months. Tamoxifen was administered by oral gavage using a 1ml syringe (Beckton Dickinson) via a feeding needle (BD Microlance) in a range of 1 to 4 mg/female, depending on the specific tracer line used.
Retinoic Acid administration
RA was administered to pregnant CD1 females by oral gavage using a 1ml syringe and a feeding needle. 6.75 mg of crystalline all-trans RA (302–79-4 Sigma) was diluted in 0.8 ml of Ethanol (EtOH) in 9.2 ml sesame oil. The solution was sonicated, aliquoted and stored in the dark at 4C. Samples were sonicated before being administered to pregnant females. Pregnant mice were administered at E13.5 and E14.5 days of gestation with a 6.75mg RA dose. Control mice were administered with equivalent volumes of the vehicle (i.e., EtOH in sesame oil) at the corresponding times.
Embryo retrieval
Embryos were staged establishing 12 AM on the morning of the vaginal plug as embryonic day (E) 0.5. Females were sacrificed by CO2 inhalation followed by cervical dislocation. Mice embryos were extracted at different developmental stages as follow. The abdominal cavity of sacrificed females was opened to expose the uterus. The uterus was then placed in ice-cold PBS in a petri dish in order to open the muscle layer, decidual layer and amnion. Once extracted, embryos were decapitated and their thorax opened at the level of the sternum. If needed, the heart was collected by pulling it out with forceps. Unless stated differently, all tissues were dissected in ice-cold PBS and fixed in PFA (Merck) 2% in PBS overnight at 4°C.
Retrospective clonal analysis setting
For retrospective clonal analysis, we used mouse embryos with the inducer RERT and the reporters R26RlacZ; R26REYFP or R26RtdTomato; R26REYFP in trans-heterozygosis. These genotypes were generated upon breeding mice that have the inducer and one of the reporter alleles in double homozygosis with mice that have the second reporter allele in homozygosis. Random Cre-mediated recombination was provoked with 4-hydroxy-tamoxifen (4OHT) dissolved in corn oil. A single dose of 4OHT (0.1mg, 0.0675mg or 0.05mg) was injected intraperitoneally into pregnant females at E8.5 days of gestation. Hearts were dissected and analyzed at E14.5, E15, E16.5 and in neonates (P0).
Histology
For paraffin embedding, following overnight (ON) fixation in 4% PFA at 4°C, embryonic tissues were rinsed several times in PBS, and dehydrated through gradual concentrations of EtOH:H2O (30%, 50%, 70%, 85%, 90%, 96%, 100%) at room temperature. Samples were washed twice in xylol and twice in paraffin wax at 65°C. Embryos were oriented and embedded in warm paraffin and left at 4°C on to solidify. For cryosectioning, after overnight fixation, tissues were washed several times in PBS and cryopreserved with a solution of 15% sucrose in PBS overnight at 4°C. To build gelatin blocks tissues were incubated in a solution of sucrose 15% and gelatin 7.5% (Sigma Aldrich) in PBS at 37°C for 4 hours. The samples were then embedded in the same solution at RT. Gelatin blocks were left to cool down at 4°C and snap-frozen in a solution of isopentane at −70°C for 1 minute. Blocks were then stored at −80°C until cryo-sectioning. 8 µm-thick cryo-sections were obtained using a Leica CM1950 automated Cryostat. Sections were collected on Superfrost+ slides (Fisherbrand) and stored at −20°C on short term or −80°C for the long term.
RNA detection in sections
In situ mRNA hybridisation was performed on 7 µm-thick paraffin sections according to previously described protocols (Kanzler et al., 1998). After sectioning, samples were rehydrated and digested with proteinase K (10 µg/ml) at 37°C for 10 minutes. Riboprobe hybridisation was performed at 65°C ON. Section were washed on the next day, incubated with anti-DIG antibody and developed with BM-purple (Roche, ref. 11442074001). Images were acquired with a Nikon Eclipse 90i microscope.
Co-detection of GFP and LacZ in whole-mount
To visualize LacZ+ clones in whole mount, β-galactosidase (β-gal) activity staining was performed as previously described (Torres, 1998). Embryos were post-fixed in 4% paraformaldehyde (PFA).
To visualize EYFP+ clones, hearts went through a two-step process as follow:
-Whole mount immuno-staining (common protocol for immuno-histochemistry and immuno-fluorescence): Hearts were permeabilized using 0.5% Triton X-100 in PBS ON at 4°C. Hearts went through a process of endogenous peroxidase quenching using 0.03% hydrogen peroxide (H2O2) in PBS at room temperature (RT). Hearts were then blocked in PBST (PBS triton 0.1%) with 10% goat serum for 3 hours at RT. The samples were incubated for 3 days with the primary antibody (anti-GFP, living colors) at 4°C. Samples were washed in PBST for 3 hours and incubated in a biotinylated secondary antibody (Jackson ImmunoResearch) for 2 days at 4°C. Samples were finally washed for 3 additional hours at RT.
YFP signal was amplified with the Vectastain ABC Peroxidase kit and detected using the −3,3’-diaminobenzidine (DAB) Vectastain Elite ABC Kit (see the Key Resources Table). All procedures were performed as advised by the manufacturer. As a result, YFP+ cells and LacZ+ cells could be observed as brown and blue labels, respectively. Hearts were then stored in 80% glycerol at 4 °C.
Immunohistochemistry in sections
Stored frozen slides were allowed to defreeze at 4°C before being placed at Room temperature. Immuno-staining was usually performed on freshly cut gelatin sections. Slides were incubated at 37°C in PBS for 15 minutes to remove the gelatin and washed several times with PBS.
Immuno-histochemistry on sections followed the same steps as the whole mount immuno-staining, with the only difference that times of incubation were shorter. Permeabilization was performed between 10 and 30 min depending on the targeted antigen. Peroxidase block as well as protein blocks were performed for 30min each. All procedures were spaced by 3 times washes in PBST. After a final wash with PBS, sections were mounted with aquatex (108562 Merck) or dako fluorescence mounting medium (S3023).
To identify clones by immuno-histochemistry commercial kits were used as detailed in the Key Resources Table.
Immunofluorescence in sections and whole-mounts
Tissues were fixed overnight at 4 °C in 2%–4% paraformaldehyde in phosphate-buffered saline (PBS). For immunofluorescence on sections, tissues were gelatin-embedded and cryo-sectioned at 8–16µm. All the steps to perform immuno-fluorescence in whole mount or on sections were the same as in the immunohistochemistry except for the following points:
Peroxidase quenching was performed only in the case of using HRP-conjugated secondary antibodies followed by a peroxidase detection-based tyramide fluorescent amplification kit. A list of Tyramide amplification kits and reagents is provided in the Key Resources Table.
The universal TNB blocking reagent (FP1012) Perkin Elmer was used instead of goat serum to block unspecific proteins.
For biotin-streptavidin based amplification, streptavidin proteins conjugated with a fluorophore were used after incubation with a biotinylated secondary antibody. The streptavidin conjugated fluorophores used are detailed in the Key Resources Table.
Embryonic and heart tissues were mounted in Vectashield mounting medium (Vector Laboratories, USA).
Tissue processing for whole mount cardiac lymphatic imaging
Hearts stained for Prox1 and Lyve1 double immunofluorescence were embedded in low-melting-point agarose at 50 °C and were frontally sectioned with a blade at the plane between the great arteries and the atria. The agarose was then removed, and the hemi-hearts cleared in increasing concentrations of glycerol in PBS (see the tissue clearing and microscopy section). Each hemi-heart was then mounted in 80% glycerol on a MalTek glass bottom dish (the epicardial surface facing the objective).
Epicardial / arterial mesothelium dissection and RNA extraction
We will describe the common and specific steps used for mesothelial dissection of samples used for RNAseq and embryonic heart culture.
To preserve mRNA for RNAseq, all solutions were ice-cold, sterile and RNAse free. For tissue culture, we used a dissection medium at 37 °C. Samples were dissected under a Leica MZ12F stereoscope. The hearts were placed in the appropriate solution and sectioned at the border between the great arteries and myocardium. Each piece of tissue was isolated in different petri dishes. The samples for RNA analysis were pinned down on a bed of 2% agarose/PBS by thin metal threads. In the latter case, only the forceps were used to delicately handle the embryonic hearts. In both cases, fine forceps (Dumont #55 No11255–20) were used to grasp the mesothelium of the great arteries or the epicardium. The forceps were then used to maintain the embryonic tissues in place while carefully peeling the mesothelial/sub-mesothelial or epicardial/subepicardial layers. For RNA analysis, the ventricular epicardial and arterial mesothelial samples were frozen into different eppendorf tubes on dry ice, pooling together samples from each litter. A total of 6 litters were independently sequenced. RNA was then purified from each pool using the RNeasy Micro kit (Qiagen). For future culture, only small pieces of mesothelium/submesothelium or epicardium/subepicardium were harvested.
Ex vivo cardiac culture and transplantation
Hearts were dissected in 10% Calf Serum in DMEM/F12 1% Penicillin-Streptomycin kept in a water bath at 37 °C when not in use. E14.5 Wt1Cre; R26mTmG fluorescent hearts and their WT littermates, observed with a Leica MZ12F stereoscope (fitted with a fluorescent lamp), were placed in different petri dishes. The medium was replaced every 5 min to prevent cooling of the embryonic tissue. One heart was dissected at a time under the stereoscope. Meanwhile, the rest of hearts were placed in Petri dishes containing dissection medium on a histology flat table set at 37 °C. In WT hearts, a small piece of epicardium was peeled off from the ventricle under the PA (as described previously). A piece of Pulmonary mesothelium was then placed on the naked myocardium of the recipient WT heart. As a control, a homotopic piece of epicardium from a fluorescent donor was placed on the naked myocardium of WT recipients instead.
Immediately after dissection, the E14.5 transplanted hearts were placed in a solution of 49.5% modified Krebs Henseleit Buffer (25 mM NaHCO3, 118 mM NaCl, 4.7 mM KCl, 1.2 mM MgSO4, 2.5 mM CaCl2,11 mM glucose, 0.5 mM EDTA, 1.5 mM Na-pyruvate in DMEM/F12 instead of water), 49.5% Mouse serum (supplemented with N2 1:200 and B10 1:50) and 1% Penicillin-Streptomycin. For mouse serum preparation, mouse blood was collected by cardiac puncture from sacrificed adult WT mice. After clotting for 30min at RT, blood samples were submitted to two series of centrifugations at 4100 rpm for 10 min at RT. The serum was retrieved collecting the supernatant. Samples were then placed on a water bath at 52°C to be inactivated and stored at −20°C until use.
Hearts were cultured in rolling-bottles in a 5%CO2/ 95%O2 gas environment for three days. Three hearts were cultured per bottle separating AMC-transplanted hearts from epicardium-transplanted hearts. Hearts were finally washed in PBS and fixed in PFA 2% PBS ON at 4 °C.
Tissue clearing and microscopy
To detect superficial signal, tissues were dehydrated in gradual concentrations of glycerol (20%, 40%, 60% 80%). For thick samples or deep signal, tissues were cleared with cubic 1 solution as described in the literature (Susaki et al., 2014).
Depending on the specific samples, experiments were performed in the following microscopes:
Leica SP5 multiline inverted confocal microscope for E16.5 and P0 fluorescent hearts in whole-mount.
Leica SP8 inverted confocal equipped with white light laser for all embryonic fluorescent sections.
Leica TCS SP8 coupled to a DMi8 inverted confocal microscope Navigator module equipped with white light laser. We used this microscope to acquire several embryonic hearts from a same experiment at the same time and ensure comparable settings for further quantifications (E15.5 RA-induced hearts, E16.5 Raldh2f/f; Wt1Cre mutant hearts and E17.5 isl1Cre; R26RtdTomato hearts).
Zeiss LSM 780 multiphoton upright microscope. A MaiTai laser line at 1000 nm was used for two-channel two-photon imaging of td-Tomato and GFP fluorescence of E15.5 and E16.5 hearts in whole mount.
Maximum intensity Z-projections of whole hearts were acquired using both the tiling and z-stack functions, generating a single composite image.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical test and N
Statistical details of experiments, including the statistical test used, n numbers and specification of what n rand error bars represent have been included in the legends to the relevant Figures (4–7 and S1), except for the RNAseq analysis, which is described below. Samples were considered significantly different when p≤0.05. The specific p-values for significantly different data sets are shown for each specific comparison on the graphs in Figures 4–7 and Figure S1. All statistical analyses were performed using Prism (Graphpad) software, except for RNAseq analysis.
For RNAseq statistical analysis, data were then normalized and differential expression tested using the Bioconductor package EdgeR (Robinson et al., 2010). Genes were considered as differentially expressed in case of a Benjamini-Hochberg adjusted P-Value ≤0.05. Analysis of the data was performed using Gene set enrichment analysis and Ingenuity pathways analysis software (Biobase International).
Clonal Analysis
For estimation of the polyclonality in the collection of labelled cell clusters, we studied the frequency of bicolor clusters, which represent cases of randomly occurring polyclonality. Bicolor clusters represent only part of all polyclonal clusters, because independent progenitors can also recombine the same reporter by chance. The frequency of mono-color polyclonal clusters can then be estimated as a function of the frequencies of bicolor clusters and of single-color clusters, which is biased towards the production of LacZ+ clusters, with a frequency of 0.69 of clusters containing LacZ+ cells and 0.31 of clusters containing GFP+ cells (Figure S1F). Dismissing polyclonality levels above biclonality and assuming a stochastic distribution of clusters, the following functions apply for estimations:
Frequency of bicolor clusters= frequency of lacZ (A) × frequency of GFP (B) × 2 Frequency of polyclonal monocolor clusters= A2 + B2
Then,
Then, for a proportion of frequencies A/B of 2.22 (0,69/0.31), the following estimation applies for the frequency of polyclonality in monocolor clusters: 0.038 × 1.35 = 0.05
Quantification of the lymphatic vasculature in whole mount immunofluorescence
Cell counting
For the quantification of lineage-traced LECs following IsletCre or VEGFR3CreERT2 recombination, the number of cells expressing the recombination reporter and the LEC markers were counted manually. First, a mask was generated in adobe Photoshop using the paint tool to draw spots of two different colors. One color was used to draw spots identifying nuclei of the double positive Prox1+/ Lyve1+ cells while the other color was used to identify the nuclei of the triple positive Prox1+/ Lyve1+/ reporter+ cells. The mask was then loaded to imageJ and the spots counted with the “analyse particles” tool. The data of the ventral and dorsal parts of a same heart were systematically paired together for statistical analysis.
The same manual method was used to draw maps of lymphatic cells in the experiments of gain and loss of function of the RA pathway. For the analysis of the structure of the lymphatic plexus analysis in the RA pathway gain- and loss-of-function experiments, hemi-hearts were mounted in MatTek glass bottom dishes, as previously described, and forced flat using a sealed coverslip to allow the measurements to be performed in 2D.
Measurements
Branching events: A branching event is the branch point at the intersection of two or more branches. All the branching events were counted manually using the image J “multi-point” and “measure” tools.
Number of branches: A branch is considered as a unique lymphatic segment. The number of branches was counted manually, using the image J “multi-point” and “measure” tools, as the sum of segments originating at branching events. To count 1 cell-wide branches, only branches with an average diameter of one cell were counted.
Tip-base distance: The base of the lymphatic plexus was established at the base of the pulmonary artery for the ventral side and at the atrioventricular groove for the dorsal side. A line was drawn in ImageJ at these origins as a reference. A straight line was drawn from the tip of each main branch to the base of the plexus passing through the branch. The length of each line was measured, and the average length calculated using ImageJ.
Distance of the first fork: At the base of the heart the lymphatics are organised as sinusoids or lymph sac-like structures and start branching in the ventricles. The first forks were considered as the first observed points where branches diverge as distance increases from the base of the plexus. Straight lines were drawn connecting the base of the heart to all the first forks observed (as shown by the arrows in Figure 7 E’–H’) and their length was measured. The distance to the first fork represented in Figure 7 shows the average of all the measurements made on a given heart surface.
Number of tubes: Tubes are regions surrounded by a continuous network of branches. The number of tubes was counted manually using the image J “multi-point” and “measure” tool.
Transcriptomic analyses
Stranded RNA-seq libraries were sequenced on an Illumina HiSeq 2500 system. Sequencing reads were pre-processed by means of a pipeline that used FastQC, to asses read quality, and Cutadapt to trim sequencing reads, eliminating Illumina adaptor remains, and to discard reads that were shorter than 30 bp. The resulting reads were mapped against the mouse transcriptome (mm10) and quantified using RSEM v1.17. Data were then processed with a differential expression analysis pipeline that used Bioconductor package limma for normalization and differential expression testing, defining blocks of samples obtained from the same tissue source. Six biological replicates for each population were obtained and analyzed by RNA-sequencing.
The raw data analysis of RNA-seq data were generated by CNIC Bioinformatic unit. Contaminants were eliminated from reads using the Cutadapt software (Martin, 2011). The resulting reads were mapped and quantified on the transcriptome (Ensembl gene-build 70) using RSEM v1.2.3 (Li and Dewey, 2011). Only genes with at least 2 counts per million in at least five samples were considered for statistical analysis.
DATA AND CODE AVAILABILITY
RNAseq data have been deposited in GEO (GSE136750). All raw data have been deposited in Mendeley under DOIs: 10.17632/b98p6kg7yw.1 and 10.17632/p38b33x627.1
New code was not produced in this study.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti GFP @1:200 | Living Colors | 632460 |
| Chicken anti GFP @1:100/1000 | abcam | ab13970 |
| Chicken anti GFP @1:400 | Aves labs | GFP-1010 |
| Mouse anti c-TnT @1:200 | Thermo Scientific | MS-295 |
| Rat anti CD31 @1:100/500 | Pharmingen™ | 553370 |
| Rabbit anti Wt1 @1:200 | abcam | ab89901 |
| Rabbit anti Raldh2 @1:200 | abcam | ab96060 |
| Rat anti PDGFRα @1:100 | BD Pharmingen | 558774 |
| Mouse anti SMAcy3 @1:400 | Sigma | C6198 |
| Rabbit anti SM22α @1:200 | abcam | ab14106 |
| Goat anti Prox1 @1:300 | R&D systems | AF2727 |
| Rabbit anti Prox1 @1:500 | Angiobio | 11002 |
| Rabbit anti Lyve1 @1:200 | ReliaTech | 103-PA50S |
| Rat anti Lyve1 @1:100 | ReliaTech | 103-M130 |
| Goat anti Lyve1 @1:200 | R&D Systems | AF2125 |
| Rabbit anti ER @undiluted | abcam | ab27595 |
| Rabbit anti β-galactosidase @1:500 | Cappel | 56031 |
| Rabbit anti Periostin @1:200 | novusbio | NBP1–30042 |
| Hamster anti Podoplanin @1:200 | Fitzgerald | 10R-P155a |
| Rabbit anti Calponin @1:200 | abcam | Ab46794 |
| Rat anti Endomucin @1:500 | eBiosciences | 14–5851-82 |
| Anti-Mouse Alexa Fluor 488 @1:500 | A11029 life technologies ™ | A11029 life technologies ™ |
| Anti-Rabbit Alexa Fluor 488 @1:500 | A11034 life technologies ™ | A11034 life technologies ™ |
| Anti-Rabbit Alexa Fluor 488 @1:500 | A21206 Molecular Probes | A21206 Molecular Probes |
| Anti-chicken Alexa Fluor 488 @1:500 | A11039 life technologies ™ | A11039 life technologies ™ |
| Anti-Mouse Alexa Fluor 568 @1:500 | A11004 life technologies ™ | A11004 life technologies ™ |
| Anti-Rabbit Alexa Fluor 568 @1:500 | A11036 life technologies ™ | A11036 life technologies ™ |
| Anti-Rabbit Alexa Fluor 594 @1:500 | A11012 life technologies ™ | A11012 life technologies ™ |
| Anti-Mouse Alexa Fluor 594 @1:500 | A11005 life technologies ™ | A11005 life technologies ™ |
| Anti-Rabbit Alexa Fluor 633 @1:500 | A21071 life technologies ™ | A21071 life technologies ™ |
| Anti-Mouse Alexa Fluor 633 @1:500 | A21052 life technologies ™ | A21052 life technologies ™ |
| Anti-Mouse HRP @1:200 | P 0447 Dako | P 0447 Dako |
| Anti-Rabbit HRP @1:200 | P 0448 Dako | P 0448 Dako |
| Anti-Rabbit Biotin @1:500 | 111–066-003 Jackson ImmunoResearch | 111–066-003 Jackson ImmunoResearch |
| Anti-Goat Cy3 @1:300 | 705–165-147 Jackson ImmunoResearch | 705–165-147 Jackson ImmunoResearch |
| Anti-Rat Cy5 @1:200 | 712–175-150 Jackson ImmunoResearch | 712–175-150 Jackson ImmunoResearch |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Streptavidin-Cy3 @1:300 | Jackson ImmunoResearch | 016–160-084 |
| Streptavidin-647 @1:300 | ThermoFisher | S21374 |
| Streptavidin-633 @1:300 | ThermoFisher | S21375 |
| Streptavidin-405 @1:300 | ThermoFisher | S-32351 |
| Critical Commercial Assays | ||
| TSA plus Cyanine 3. @1:100 | Perkin Elmer | NEL744001KT |
| TSA plus Cyanine 5. @1:100 | Perkin Elmer | NEL745001KT |
| TSA plus Cyanine 5.5. @1:100 | Perkin Elmer | NEL766001KT |
| TSA plus Fluorescein. @1:100 | Perkin Elmer | NEL741001KT |
| Vector Red alkaline phosphatase kit | Vector Laboratories | SK-5100 |
| Vector VIP peroxidase substrate | Vector Laboratories | SK-4600 |
| Vectastain ABC Alkaline phosphatase | Vector Laboratories | AK-500 |
| Dako liquid DAB + substrate | Dako | K3468 |
| Vector DAB peroxidase substrate | Vector Laboratories | SK-4100KI01 |
| Vectastain Elite ABC HRP Kit | Vector Laboratories | PK-6100 |
| Deposited Data | ||
| RNAseq data | Gene Expression Omnibus | GSE136750 |
| Raw data | Mendeley | DOI: 10.17632/b98p6kg7yw.1 |
| Mendeley | 10.17632/p38b33x627.1 | |
| Experimental Models: Organisms/Strains | ||
| RERT | (Guerra et al., 2003) | Polr2atm1(cre/ERT2)Bbd |
| Islet1Cre | (Cai et al., 2003) | Isl1tm1(cre)Sev |
| Islet1MERCreMER | (Laugwitz et al., 2005) | Isl1tm1(cre/Esr1*)Krc |
| Mef2cAHF-Cre | (Verzi et al., 2005) | Tg(Mef2c-cre)#Blk |
| Wt1CreERT2 | (Zhou et al., 2008) | Wt1tm2(cre/ERT2)Wtp |
| Wt1Cre | (Wessels et al., 2012) | Tg(Wt1-cre)#Jbeb |
| Hoxb1Cre | (Arenkiel et al., 2003) | Hoxb1tm6(cre)Mrc |
| Vegfr3CreERT2 | (Martinez-Corral et al., 2016) | Flt4tm2.1(cre/ERT2)Sgo |
| Wnt1Cre | (Lewis et al., 2013) | E2f1Tg(Wnt1-cre)2Sor |
| Raldh2flox | (Vermot et al., 2003) | Aldh1a2tm1Dll |
| Tbx1- | (Jerome and Papaioannou, 2001) | Tbx1tm1Pa |
| Prox1flox | (Harvey et al., 2005) | Prox1tm2Gco |
| R26RLacZ | (Soriano, 1999) | Gt(ROSA)26Sortm1Sor |
| R26REYFP | (Srinivas et al., 2001) | Gt(ROSA)26Sortm1.1(EYFP)Cos |
| R26RtdTomato | (Madisen et al., 2010) | Gt(ROSA)26Sortm14(CAG-tdTomato)Hze |
| R26RmTmG | (Muzumdar et al., 2007) | Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo |
Highlights.
A Non-venous cell source partially contributes to ventral heart lymphatic endothelium
Non-venous heart lymphatics originate from SHF-derived arterial sub-mesothelial cells
Elimination of SHF-derived lymphatics abrogates ventral heart lymphatic development
Regionalized epicardial retinoic acid production modulates cardiac lymphangiogenesis
Acknowledgements
We thank Cristina Villa del Campo, Ester de la Cruz and other members of the Torres group for stimulating discussions and suggestions. We thank members of the Microscopy, Genomics, Bioinformatics, Transgenesis and Animal Facility CNIC units for excellent support. This work was supported by grants PGC2018-096486-B-I00 and RD16/0011/0019 (ISCIII) from the Spanish Ministry of Science, Innovation and Universities and grant S2017/BMD-3875 from the Comunidad de Madrid to M.T., grant ITN-CARDIONET-ref.289600 from EU to G.L. and by grant BFU2015-71376-R to S.O. The CNIC is supported by the Ministerio de Ciencia, Innovación y Universidades and the Pro CNIC Foundation, and is a Severo Ochoa Center of Excellence (SEV-2015-0505).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no conflict of interests.
References
- Arenkiel BR, Gaufo GO, and Capecchi MR (2003). Hoxb1 neural crest preferentially form glia of the PNS. Dev Dyn 227, 379–386. [DOI] [PubMed] [Google Scholar]
- Arques CG, Doohan R, Sharpe J, and Torres M (2007). Cell tracing reveals a dorsoventral lineage restriction plane in the mouse limb bud mesenchyme. Development (Cambridge, England) 134, 3713–3722. [DOI] [PubMed] [Google Scholar]
- Brakenhielm E, and Alitalo K (2019). Cardiac lymphatics in health and disease. Nature reviews Cardiology 16, 56–68. [DOI] [PubMed] [Google Scholar]
- Buckingham ME, and Meilhac SM (2011). Tracing cells for tracking cell lineage and clonal behavior. Developmental cell 21, 394–409. [DOI] [PubMed] [Google Scholar]
- Burger NB, Stuurman KE, Kok E, Konijn T, Schooneman D, Niederreither K, Coles M, Agace WW, Christoffels VM, Mebius RE, et al. (2014). Involvement of neurons and retinoic acid in lymphatic development: new insights in increased nuchal translucency. Prenat Diagn 34, 1312–1319. [DOI] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, and Evans S (2003). Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Developmental cell 5, 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CL, and Molkentin JD (2017). The Elusive Progenitor Cell in Cardiac Regeneration: Slip Slidin’ Away. Circ Res 120, 400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Mupo A, Huynh T, Cioffi S, Woods M, Jin C, McKeehan W, Thompson-Snipes L, Baldini A, and Illingworth E (2010). Tbx1 regulates Vegfr3 and is required for lymphatic vessel development. The Journal of cell biology 189, 417–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels VM, Grieskamp T, Norden J, Mommersteeg MT, Rudat C, and Kispert A (2009). Tbx18 and the fate of epicardial progenitors. Nature 458, E8–9; discussion E9–10. [DOI] [PubMed] [Google Scholar]
- Christoffels VM, Mommersteeg MT, Trowe MO, Prall OW, de Gier-de Vries C, Soufan AT, Bussen M, Schuster-Gossler K, Harvey RP, Moorman AF, et al. (2006). Formation of the venous pole of the heart from an Nkx2-5-negative precursor population requires Tbx18. Circ Res 98, 1555–1563. [DOI] [PubMed] [Google Scholar]
- Eng TC, Chen W, Okuda KS, Misa JP, Padberg Y, Crosier KE, Crosier PS, Hall CJ, Schulte-Merker S, Hogan BM, et al. (2019). Zebrafish facial lymphatics develop through sequential addition of venous and non-venous progenitors. EMBO Rep 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engleka KA, Manderfield LJ, Brust RD, Li L, Cohen A, Dymecki SM, and Epstein JA (2012). Islet1 derivatives in the heart are of both neural crest and second heart field origin. Circ Res 110, 922–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobedo N, and Oliver G (2016). Lymphangiogenesis: Origin, Specification, and Cell Fate Determination. Annual review of cell and developmental biology 32, 677–691. [DOI] [PubMed] [Google Scholar]
- Flaht A, Jankowska-Steifer E, Radomska DM, Madej M, Gula G, Kujawa M, and Ratajska A (2012). Cellular phenotypes and spatio-temporal patterns of lymphatic vessel development in embryonic mouse hearts. Dev Dyn 241, 1473–1486. [DOI] [PubMed] [Google Scholar]
- Galli D, Dominguez JN, Zaffran S, Munk A, Brown NA, and Buckingham ME (2008). Atrial myocardium derives from the posterior region of the second heart field, which acquires left-right identity as Pitx2c is expressed. Development (Cambridge, England) 135, 1157–1167. [DOI] [PubMed] [Google Scholar]
- Gittenberger-de Groot AC, Vrancken Peeters MP, Bergwerff M, Mentink MM, and Poelmann RE (2000). Epicardial outgrowth inhibition leads to compensatory mesothelial outflow tract collar and abnormal cardiac septation and coronary formation. Circ Res 87, 969–971. [DOI] [PubMed] [Google Scholar]
- Gittenberger-de Groot AC, Winter EM, Bartelings MM, Goumans MJ, DeRuiter MC, and Poelmann RE (2012). The arterial and cardiac epicardium in development, disease and repair. Differentiation; research in biological diversity 84, 41–53. [DOI] [PubMed] [Google Scholar]
- Guerra C, Mijimolle N, Dhawahir A, Dubus P, Barradas M, Serrano M, Campuzano V, and Barbacid M (2003). Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell 4, 111–120. [DOI] [PubMed] [Google Scholar]
- Haak MC, Bartelings MM, Jackson DG, Webb S, van Vugt JM, and Gittenberger-de Groot AC (2002). Increased nuchal translucency is associated with jugular lymphatic distension. Hum Reprod 17, 1086–1092. [DOI] [PubMed] [Google Scholar]
- Harvey NL, Srinivasan RS, Dillard ME, Johnson NC, Witte MH, Boyd K, Sleeman MW, and Oliver G (2005). Lymphatic vascular defects promoted by Prox1 haploinsufficiency cause adult-onset obesity. Nature genetics 37, 1072–1081. [DOI] [PubMed] [Google Scholar]
- He L, and Zhou B (2018). The Development and Regeneration of Coronary Arteries. Curr Cardiol Rep 20, 54. [DOI] [PubMed] [Google Scholar]
- Henri O, Pouehe C, Houssari M, Galas L, Nicol L, Edwards-Levy F, Henry JP, Dumesnil A, Boukhalfa I, Banquet S, et al. (2016). Selective Stimulation of Cardiac Lymphangiogenesis Reduces Myocardial Edema and Fibrosis Leading to Improved Cardiac Function Following Myocardial Infarction. Circulation 133, 1484–1497; discussion 1497. [DOI] [PubMed] [Google Scholar]
- Huntington GS, and McClure CFW (1910). The anatomy and development of the jugular lymph sacs in the domestic cat (Felis domestica). American Journal of Anatomy 10, 177–312. [Google Scholar]
- Hyett J, Perdu M, Sharland G, Snijders R, and Nicolaides KH (1999). Using fetal nuchal translucency to screen for major congenital cardiac defects at 10–14 weeks of gestation: population based cohort study. BMJ 318, 81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa Y, Akishima-Fukasawa Y, Ito K, Akasaka Y, Tanaka M, Shimokawa R, Kimura-Matsumoto M, Morita H, Sato S, Kamata I, et al. (2007). Lymphangiogenesis in myocardial remodelling after infarction. Histopathology 51, 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerome LA, and Papaioannou VE (2001). DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nature genetics 27, 286–291. [DOI] [PubMed] [Google Scholar]
- Kanzler B, Kuschert SJ, Liu YH, and Mallo M (1998). Hoxa-2 restricts the chondrogenic domain and inhibits bone formation during development of the branchial area. Development (Cambridge, England) 125, 2587–2597. [DOI] [PubMed] [Google Scholar]
- Kelly RG, Brown NA, and Buckingham ME (2001). The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Developmental cell 1, 435–440. [DOI] [PubMed] [Google Scholar]
- Kline GM, Shen Z, Mohiuddin M, Ruggiero V, Rostami S, and DiSesa VJ (1994). Development of tolerance to experimental cardiac allografts in utero. Ann Thorac Surg 57, 72–74; discussion 75. [DOI] [PubMed] [Google Scholar]
- Klotz L, Norman S, Vieira JM, Masters M, Rohling M, Dube KN, Bollini S, Matsuzaki F, Carr CA, and Riley PR (2015). Cardiac lymphatics are heterogeneous in origin and respond to injury. Nature 522, 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M, et al. (2005). Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature 433, 647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescroart F, Chabab S, Lin X, Rulands S, Paulissen C, Rodolosse A, Auer H, Achouri Y, Dubois C, Bondue A, et al. (2014). Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nature cell biology 16, 829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AE, Vasudevan HN, O’Neill AK, Soriano P, and Bush JO (2013). The widely used Wnt1-Cre transgene causes developmental phenotypes by ectopic activation of Wnt signaling. Developmental biology 379, 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig LL, Schertel ER, Pratt JW, McClure DE, Ying AJ, Heck CF, and Myerowitz PD (1997). Impairment of left ventricular function by acute cardiac lymphatic obstruction. Cardiovasc Res 33, 164–171. [DOI] [PubMed] [Google Scholar]
- Ma W, and Oliver G (2017). Lymphatic Endothelial Cell Plasticity in Development and Disease. Physiology (Bethesda) 32, 444–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manner J (1992). The development of pericardial villi in the chick embryo. Anatomy and embryology 186, 379–385. [DOI] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal, North America 17, 10–12. [Google Scholar]
- Martinez-Corral I, Stanczuk L, Frye M, Ulvmar MH, Dieguez-Hurtado R, Olmeda D, Makinen T, and Ortega S (2016). Vegfr3-CreER (T2) mouse, a new genetic tool for targeting the lymphatic system. Angiogenesis 19, 433–445. [DOI] [PubMed] [Google Scholar]
- Maruyama K, Li M, Cursiefen C, Jackson DG, Keino H, Tomita M, Van Rooijen N, Takenaka H, D’Amore PA, Stein-Streilein J, et al. (2005). Inflammation-induced lymphangiogenesis in the cornea arises from CD11b-positive macrophages. The Journal of clinical investigation 115, 2363–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martucciello S, Turturo MG, Cioffi S, Chen L, Baldini A, Illingworth EA (2019) Tbx1 interacts genetically with Vegfr3 to regulate cardiac lymphangiogenesis in mice. bioRxiv. 10.1101/553578 [DOI]
- Maruyama K, Miyagawa-Tomita S, Mizukami K, Matsuzaki F, and Kurihara H (2019). Isl1-expressing non-venous cell lineage contributes to cardiac lymphatic vessel development. Developmental biology 452, 134–143. [DOI] [PubMed] [Google Scholar]
- Meilhac SM, and Buckingham ME (2018). The deployment of cell lineages that form the mammalian heart. Nature reviews Cardiology 15, 705–724. [DOI] [PubMed] [Google Scholar]
- Meilhac SM, Esner M, Kelly RG, Nicolas JF, and Buckingham ME (2004). The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Developmental cell 6, 685–698. [DOI] [PubMed] [Google Scholar]
- Miller AJ, Pick R, Katz LN, Jones C, and Rodgers J (1963). Ventricular Endomyocardial Changes after Impairment of Cardiac Lymph Flow in Dogs. British heart journal 25, 182–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AJ, Pick R, Kline IK, and Katz LN (1964). The Susceptibility of Dogs with Chronic Impairment of Cardiac Lymph Flow to Staphylococcal Vagular Endocarditis. Circulation 30, 417–424. [DOI] [PubMed] [Google Scholar]
- Miquerol L, Bellon A, Moreno N, Beyer S, Meilhac SM, Buckingham M, Franco D, and Kelly RG (2013). Resolving cell lineage contributions to the ventricular conduction system with a Cx40-GFP allele: a dual contribution of the first and second heart fields. Dev Dyn 242, 665–677. [DOI] [PubMed] [Google Scholar]
- Mjaatvedt CH, Nakaoka T, Moreno-Rodriguez R, Norris RA, Kern MJ, Eisenberg CA, Turner D, and Markwald RR (2001). The outflow tract of the heart is recruited from a novel heart-forming field. Developmental biology 238, 97–109. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605. [DOI] [PubMed] [Google Scholar]
- Ny A, Koch M, Schneider M, Neven E, Tong RT, Maity S, Fischer C, Plaisance S, Lambrechts D, Heligon C, et al. (2005). A genetic Xenopus laevis tadpole model to study lymphangiogenesis. Nature medicine 11, 998–1004. [DOI] [PubMed] [Google Scholar]
- Olivey HE, and Svensson EC (2010). Epicardial-myocardial signaling directing coronary vasculogenesis. Circ Res 106, 818–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pomares JM, Phelps A, Sedmerova M, and Wessels A (2003). Epicardial-like cells on the distal arterial end of the cardiac outflow tract do not derive from the proepicardium but are derivatives of the cephalic pericardium. Dev Dyn 227, 56–68. [DOI] [PubMed] [Google Scholar]
- Rana MS, Theveniau-Ruissy M, De Bono C, Mesbah K, Francou A, Rammah M, Dominguez JN, Roux M, Laforest B, Anderson RH, et al. (2014). Tbx1 coordinates addition of posterior second heart field progenitor cells to the arterial and venous poles of the heart. Circ Res 115, 790–799. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England) 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudat C, and Kispert A (2012). Wt1 and epicardial fate mapping. Circ Res 111, 165–169. [DOI] [PubMed] [Google Scholar]
- Sabin FR (1902). On the origin of the lymphatic system from the veins and the development of the lymph hearts and thoracic duct in the pig. American Journal of Anatomy 1, 367–389. [Google Scholar]
- Sebzda E, Hibbard C, Sweeney S, Abtahian F, Bezman N, Clemens G, Maltzman JS, Cheng L, Liu F, Turner M, et al. (2006). Syk and Slp-76 mutant mice reveal a cell-autonomous hematopoietic cell contribution to vascular development. Developmental cell 11, 349–361. [DOI] [PubMed] [Google Scholar]
- Simoes FC, and Riley PR (2018). The ontogeny, activation and function of the epicardium during heart development and regeneration. Development (Cambridge, England) 145. [DOI] [PubMed] [Google Scholar]
- Soriano P (1999). Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature genetics 21, 70–71. [DOI] [PubMed] [Google Scholar]
- Sousa VH, Miyoshi G, Hjerling-Leffler J, Karayannis T, and Fishell G (2009). Characterization of Nkx6–2-derived neocortical interneuron lineages. Cereb Cortex 19 Suppl 1, i1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, and Costantini F (2001). Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan RS, Dillard ME, Lagutin OV, Lin FJ, Tsai S, Tsai MJ, Samokhvalov IM, and Oliver G (2007). Lineage tracing demonstrates the venous origin of the mammalian lymphatic vasculature. Genes & development 21, 2422–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanczuk L, Martinez-Corral I, Ulvmar MH, Zhang Y, Lavina B, Fruttiger M, Adams RH, Saur D, Betsholtz C, Ortega S, et al. (2015). cKit Lineage Hemogenic Endothelium-Derived Cells Contribute to Mesenteric Lymphatic Vessels. Cell reports 10, 1708–1721. [DOI] [PubMed] [Google Scholar]
- Stone OA, and Stainier DYR (2019). Paraxial Mesoderm Is the Major Source of Lymphatic Endothelium. Developmental cell 50, 247–255 e243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, Yokoyama C, Onoe H, Eguchi M, Yamaguchi S, et al. (2014). Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157, 726–739. [DOI] [PubMed] [Google Scholar]
- Taira A, Uehara K, Fukuda S, Takenada K, and Koga M (1990). Active drainage of cardiac lymph in relation to reduction in size of myocardial infarction: an experimental study. Angiology 41, 1029–1036. [DOI] [PubMed] [Google Scholar]
- Tian X, Pu WT, and Zhou B (2015). Cellular origin and developmental program of coronary angiogenesis. Circ Res 116, 515–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M (1998). The use of embryonic stem cells for the genetic manipulation of the mouse. Current topics in developmental biology 36, 99–114. [DOI] [PubMed] [Google Scholar]
- Ulvmar MH, Martinez-Corral I, Stanczuk L, and Makinen T (2016). Pdgfrb-Cre targets lymphatic endothelial cells of both venous and non-venous origins. Genesis 54, 350–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermot J, Niederreither K, Garnier JM, Chambon P, and Dolle P (2003). Decreased embryonic retinoic acid synthesis results in a DiGeorge syndrome phenotype in newborn mice. Proceedings of the National Academy of Sciences of the United States of America 100, 1763–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi MP, McCulley DJ, De Val S, Dodou E, and Black BL (2005). The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Developmental biology 287, 134–145. [DOI] [PubMed] [Google Scholar]
- Vieira JM, Norman S, Villa Del Campo C, Cahill TJ, Barnette DN, Gunadasa-Rohling M, Johnson LA, Greaves DR, Carr CA, Jackson DG, et al. (2018). The cardiac lymphatic system stimulates resolution of inflammation following myocardial infarction. The Journal of clinical investigation 128, 3402–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuorio T, Yla-Herttuala E, Laakkonen JP, Laidinen S, Liimatainen T, and Yla-Herttuala S (2018). Downregulation of VEGFR3 signaling alters cardiac lymphatic vessel organization and leads to a higher mortality after acute myocardial infarction. Scientific reports 8, 16709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldo KL, Hutson MR, Ward CC, Zdanowicz M, Stadt HA, Kumiski D, Abu-Issa R, and Kirby ML (2005). Secondary heart field contributes myocardium and smooth muscle to the arterial pole of the developing heart. Developmental biology 281, 78–90. [DOI] [PubMed] [Google Scholar]
- Waldo KL, Kumiski DH, Wallis KT, Stadt HA, Hutson MR, Platt DH, and Kirby ML (2001). Conotruncal myocardium arises from a secondary heart field. Development (Cambridge, England) 128, 3179–3188. [DOI] [PubMed] [Google Scholar]
- Wessels A, van den Hoff MJ, Adamo RF, Phelps AL, Lockhart MM, Sauls K, Briggs LE, Norris RA, van Wijk B, Perez-Pomares JM, et al. (2012). Epicardially derived fibroblasts preferentially contribute to the parietal leaflets of the atrioventricular valves in the murine heart. Developmental biology 366, 111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilting J, Aref Y, Huang R, Tomarev SI, Schweigerer L, Christ B, Valasek P, and Papoutsi M (2006). Dual origin of avian lymphatics. Developmental biology 292, 165–173. [DOI] [PubMed] [Google Scholar]
- Xu H, Morishima M, Wylie JN, Schwartz RJ, Bruneau BG, Lindsay EA, and Baldini A (2004). Tbx1 has a dual role in the morphogenesis of the cardiac outflow tract. Development (Cambridge, England) 131, 3217–3227. [DOI] [PubMed] [Google Scholar]
- Xu X, Lin H, Lv H, Zhang M, and Zhang Y (2007). Adventitial lymphatic vessels -- an important role in atherosclerosis. Medical hypotheses 69, 1238–1241. [DOI] [PubMed] [Google Scholar]
- Yang Y, Garcia-Verdugo JM, Soriano-Navarro M, Srinivasan RS, Scallan JP, Singh MK, Epstein JA, and Oliver G (2012). Lymphatic endothelial progenitors bud from the cardinal vein and intersomitic vessels in mammalian embryos. Blood 120, 2340–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffran S, Kelly RG, Meilhac SM, Buckingham ME, and Brown NA (2004). Right ventricular myocardium derives from the anterior heart field. Circ Res 95, 261–268. [DOI] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, et al. (2008). Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 454, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAseq data have been deposited in GEO (GSE136750). All raw data have been deposited in Mendeley under DOIs: 10.17632/b98p6kg7yw.1 and 10.17632/p38b33x627.1
New code was not produced in this study.


