Abstract
Electrical events in neurons occur on the order of milliseconds, but the brain can process and reproduce intervals millions of times longer. We present what we believe to be the first neuronal mechanism for timing durations longer than a few seconds. An initial activation and gradual relaxation of calcium-independent CaMKII measures a 6-minute time interval to coordinate two male-specific events during Drosophila mating: sperm transfer and a simultaneous decrease in motivation. We localize these functions to four neurons whose electrical activity is only necessary to report the conclusion of the decline in CaMKII’s activity—not for the measurement of the interval. The computation of elapsed time is therefore largely invisible to standard methods of monitoring neuronal activity. Its broad conservation, ubiquitous expression, and tunable duration of activity suggest that CaMKII may time a wide variety of behavioral and cognitive processes.
INTRODUCTION
Many behaviors unfold over tens of seconds to several minutes, and animals are capable of learning intervals of time on this scale (often called interval time) (Buhusi et al., 2005; Lejeune and Wearden, 1991; Paton and Buonomano, 2018), but how neurons actually represent interval time remains almost entirely unknown (Mauk and Buonomano, 2004; Paton and Buonomano, 2018). These intervals pose a particularly interesting challenge because the electrical patterns of activity usually considered to represent information in the brain operate on far more rapid timescales (Goel and Buonomano, 2014; Johnston and Wu, 1995). The intracellular circadian clock is the best understood, and perhaps the only, example of a neuronal mechanism for measuring time on a scale longer than a few seconds—but its fixed period of oscillation limits its usefulness for timing shorter intervals that are independent of time of day. In principle, however, molecular representations of interval time would be attractive for several reasons. Biochemical processes often proceed over seconds to minutes, just as electrical means of sustaining activity begin to fail or become susceptible to noise (Seung et al., 2000). Molecular timers could also enable representations of temporal intervals that are independent of the electrical information a neuron is immediately conveying, allowing multiplexed information processing. We searched for molecular interval timing mechanisms within the compact sexually dimorphic circuitry of male Drosophila melanogaster, whose robust ~23 minute mating duration (Crickmore and Vosshall, 2013), together with automated scoring, enabled an analysis across tens of thousands of genetically manipulated animals.
RESULTS
CaMKII activity in four male-specific neurons sets the duration of copulation
We developed an automated scoring system to screen for the effects of genetic perturbations in male Drosophila that alter their average copulation duration (Supplementary Figure 1). The screen analyzed 36,557 flies across 2673 genetic manipulations (see Methods and Supplementary Table 1), ~99.1% of which had average mating durations that were consistent with variation by chance, attesting to the robustness of the underlying timing mechanism (Figure 1A, for p-values see Supplementary Table 2). The longest average duration, by nearly an order of magnitude, resulted from expression of a constitutively active version of calcium/calmodulin-dependent protein kinase II (CaMKII) (Park et al., 2002), a broadly-expressed enzyme that has remained remarkably unchanged over 1.2 billion years (Tombes et al., 2003). CaMKII’s hallmark is its function as a long-lasting molecular memory of calcium transients (Lisman et al., 2012; Miller and Kennedy, 1986): its activity initially requires calcium, but becomes calcium-independent through autophosphorylation of adjacent subunits of the dodecameric holoenzyme at the T286 or T287 site (Lai et al., 1986; Miller et al., 1988) (T287 in the single fly gene (Griffith, 2004)) (Figure 1B). Once initiated, calcium-independent kinase activity can last for at least several minutes (Bading et al., 1993; Chang et al., 2017; Michalski, 2013) and, through repeated bouts of autophosphorylation, may sometimes even outlast the lifetime of individual CaMKII subunits (Stratton et al., 2014). This property makes CaMKII well suited for a tunable interval timer, as the intracellular environment may be set to sustain autophosphorylation for a range of target intervals.
Figure 1. Expression of calcium-independent CaMKII in Crz neurons extends copulation duration by several hours.
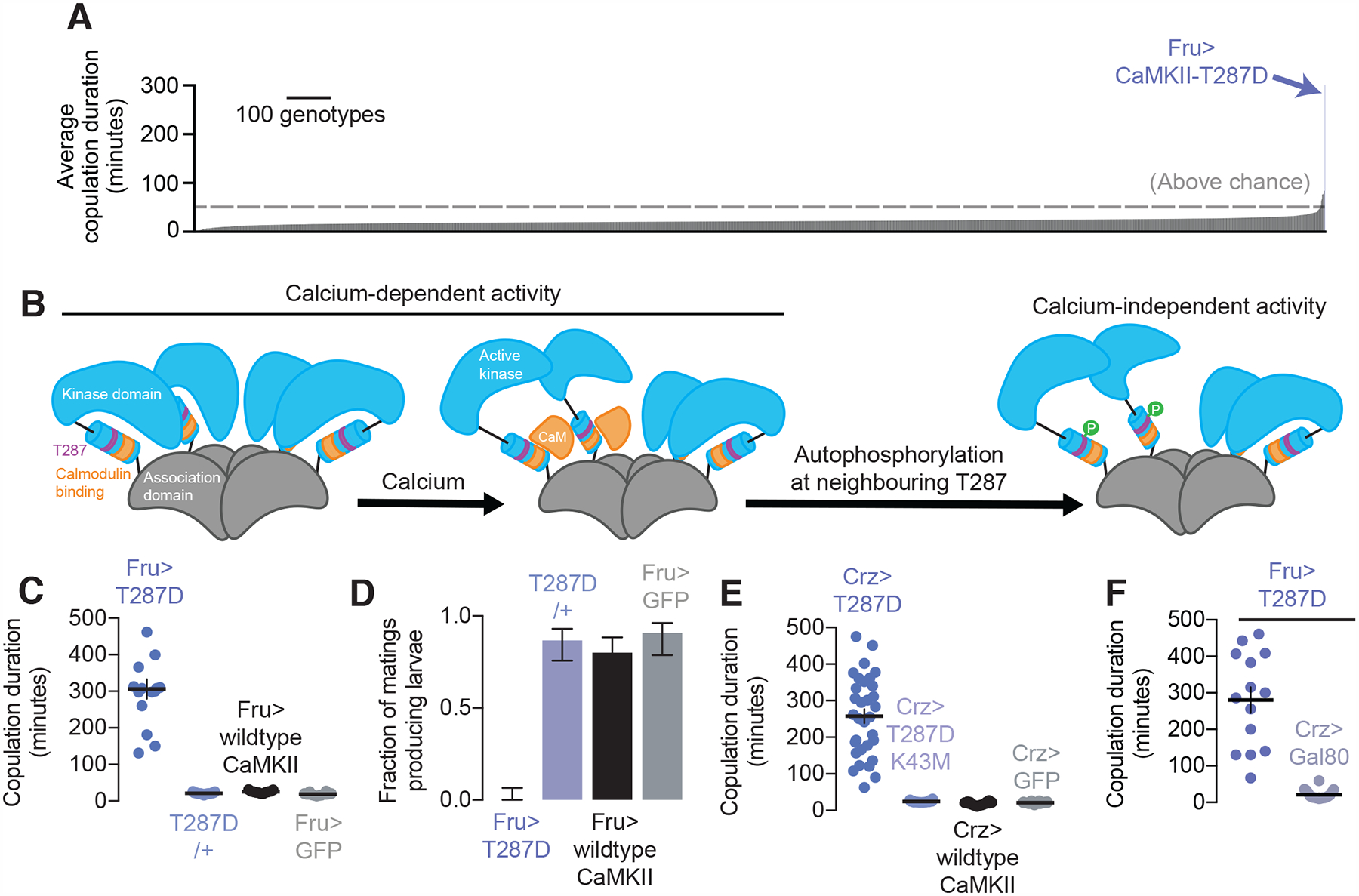
(A) Expression of calcium-independent CaMKII (T287D) in male Fruitless neurons causes the most dramatic lengthening of copulation duration out of ~3,000 genetic manipulations (statistics and n for all figures tabulated separately, see Methods).
(B) CaMKII activity is initially dependent on calcium/calmodulin (orange), but through autophosphorylation, can maintain its activity even after calcium levels decrease to baseline.
(C-D) Expressing T287D in Fruitless neurons extends copulation duration (C) and prevents sperm transfer (D) (horizontal lines indicate sample means, error bars in these and all other figures represent windows covering 68% of the density of the posterior distribution, see Methods).
(E) T287D expression in Crz neurons lengthens mating and this lengthening is prevented by further mutating CaMKII-T287D to render the catalytic domain nonfunctional (K43M).
(F) No extension of mating in Fru>T287D males is seen if expression of the transgene is excluded from the Crz neurons.
Autophosphorylation can be mimicked by replacing the T287 residue with an aspartic acid (T287D), resulting in constitutive activity (Fong et al., 1989). Expressing CaMKII-T287D in the ~2,500 neurons labeled by Fruitless-Gal4 (Stockinger et al., 2005) (Fru-Gal4), a sex-specific marker, dramatically increased mating duration (Figure 1C) and impaired fertility (Figure 1D). Matings were also lengthened when the manipulation was induced only in adult animals (Supplementary Figure 2A). Though we did not perform a quantitative analysis of behavior apart from mating duration, males expressing CaMKII-T287D in the Fruitless neurons did not have any obvious problems recognizing females, or courting, or deficits in the mechanics of initiating or terminating copulation (Supplementary Video 1), suggesting a specific role for CaMKII in measuring the passage of time during mating.
The four Corazonin-expressing (Crz) neurons of the male abdominal ganglion (Figure 2A, Supplementary Figure 2D–H) express Fruitless, are required for sperm transfer, and extend mating duration when silenced (Tayler et al., 2012). Driving CaMKII-T287D in the Crz neurons extended the mating to nearly the same extent as when expressed in the entire Fruitless population (Figure 1E), and extension of mating was precluded by additionally mutating the catalytic site of the kinase domain (K43M) (Figure 1E). Expression of CaMKII-T287D in the ~2,496 non-Crz Fruitless neurons did not extend mating duration, arguing that the effect is predominantly attributable to the Crz neurons (Figure 1F), though these neurons are not the sole determinants of overall copulation duration, even within the Fruitless population (Crickmore and Vosshall, 2013; Kim et al., 2013; Tayler et al., 2012). CaMKII-T287D did not dramatically affect the morphology of the Crz neurons (Supplementary Figure 2B), or their ability to induce sperm transfer upon stimulation (Supplementary Figure 2C). The phosphorylation status of this residue in four neurons can therefore control behavior over tens of minutes to hours.
Figure 2. Crz neurons coordinate sperm transfer and a motivational shift at 5–7 minutes into mating.
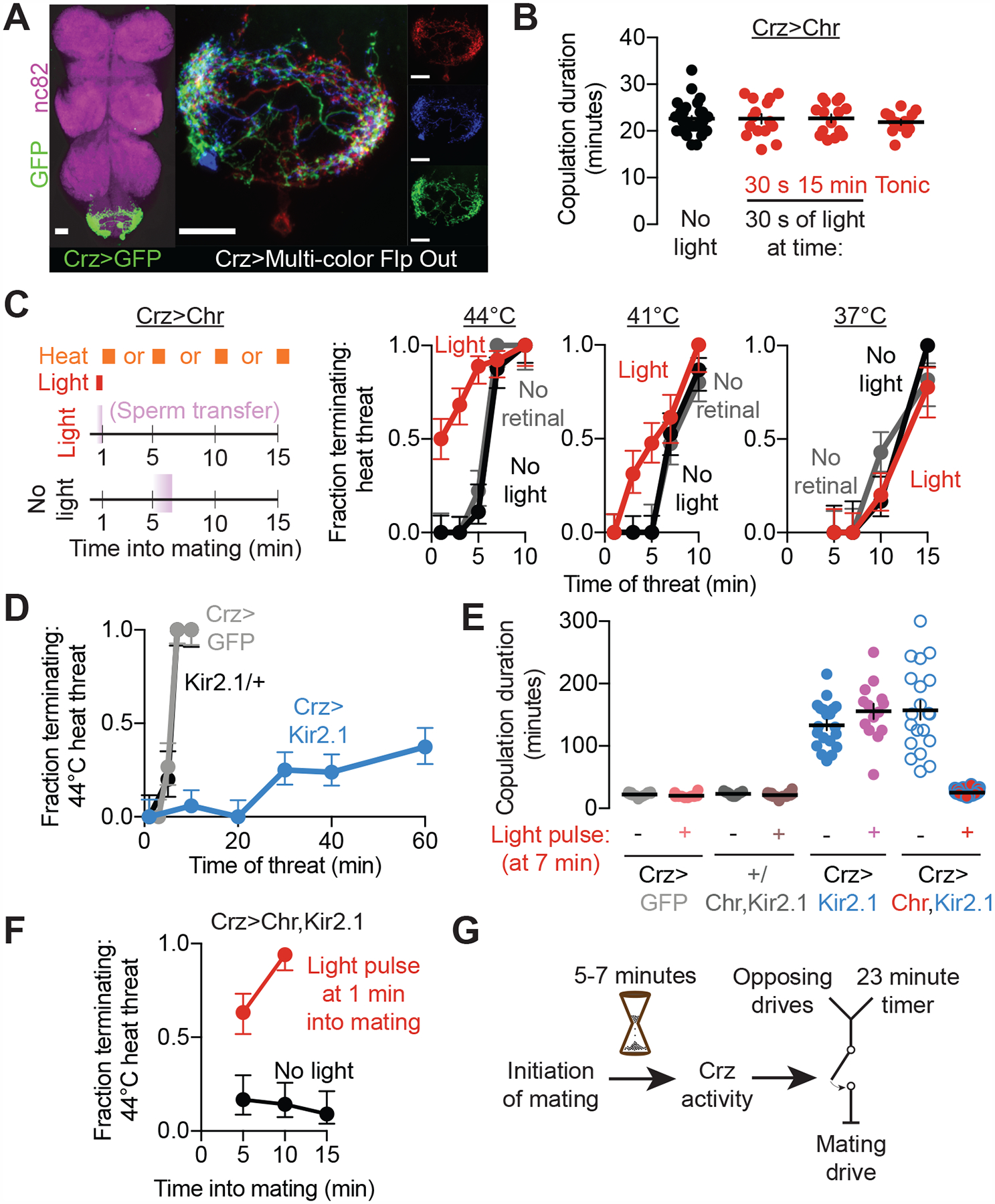
(A) The neurons in the abdominal ganglion labeled by Crz-Gal4 (left, nc82 labels neuropil, scale bar in this and all other figures is 20 μm). Individual neurons have similar morphologies (right, three of the four cells labeled using MCFO (Nern et al., 2015)). The soma are typically distributed near the abdominal nerve, at the posterior-most end of the abdominal ganglion. Each neuron sends a single projection that densely innervates both sides of the abdominal ganglion, with a few processes crossing the midline. All images are maximum-intensity projections.
(B) Optogenetic stimulation of the Crz neurons using Chrimson has no effect on copulation duration, whether provided acutely (30 second bursts near the start or end of mating) or constantly (with the light turned on at the initiation of mating).
(C) Early stimulation of the Crz neurons (schematized on the left) permits early responses to threats, without affecting motivational state after the usual time of sperm transfer.
(D) Hyperpolarization of the Crz neurons using Kir2.1 prolongs the period of high persistence.
(E-F) Brief optogenetic excitation (by stimulating CsChrimson) of otherwise-silenced (with Kir2.1) Crz neurons recovers naturalistic copulation duration (E) and a persistent reduction in motivation after stimulation (F). In F, 1 minute of optogenetic stimulation was provided at 1 minute into mating.
(G) The Crz neurons act as a permissive switch for the timely termination of mating. While many opposing drives originate in the brain, they must be conveyed to the VNS, which houses the 23-minute timer. The decision to terminate the mating results from a comparison between the motivation to continue mating and the severity of the competing drives, also within the VNS (Crickmore and Vosshall, 2013).
The Crz neurons coordinate sperm transfer and a switch in motivational state at 6 minutes into mating
Early in mating the male will persist through even life-threatening stimuli, but his persistence declines over time until even mild challenges cause him to flee by 15–20 minutes into the mating (Crickmore and Vosshall, 2013). The decision of when and whether to terminate the mating is made largely or entirely by the male, as it is not altered by inactivation of all female neurons (Supplementary Figures 1D and 1E). To better understand the motivational dynamics of mating, we designed mating chambers with individualized temperature control (Supplementary Figure 3). Using this more precise system for threat delivery, we noticed an abrupt shift in the male’s response occurring at 5–7 minutes: he will endure 1 minute of severe heat (44°C) when presented any time between 1 and 5 minutes into the mating (Figure 2C), whereas after 7 minutes he will truncate the mating in response to the same threat. The timing of this behavioral switch coincides with sperm transfer, which is induced by the activity of the Crz neurons: separation of the mating pair within 5 minutes of the start of mating results in infertility, while separation after ~7 minutes produces a normal number of offspring (Crickmore and Vosshall, 2013; Gilchrist, A. S.; Partridge, 2000; Tayler et al., 2012). The similarity in timing between both events suggests a coordination between sperm transfer and motivational state, such that once the most critical reproductive fluids have been transferred, the male will no longer risk his and his partner’s life to continue mating. We refer to the internal mechanisms underlying the abrupt change in responsiveness to competing stimuli as a motivational switch, following a definition of motivation given by the influential ethologist Nikolaas Tinbergen: “By confronting an animal that is striving towards the accomplishment of an instinctive act (feeding, mating, nursing young, etc.) with a measurable obstruction of varying strength, it is possible to determine, and to express in a quantitative way, the degree of obstruction required for the inhibition of the act. The greater the obstruction tolerated, the stronger the motivation must be” (Tinbergen, 1951).
The above results predict that, in addition to their previously described function in inducing sperm transfer (Tayler et al., 2012), activating the Crz neurons very early in mating should engage the motivational switch. Optogenetic stimulation of the Crz neurons using the red-light sensitive cation channel CsChrimson (Chr) did not alter copulation duration regardless of when it was applied (Figure 2B), so long as the mating was not challenged. However, transient stimulation of the Crz neurons at the beginning of mating caused the male to become immediately responsive to 44°C heat threats—an effect which endured long after the optogenetic stimulation had ceased. In the minutes following early Crz neuronal activation, the male’s persistence gradually declined to match that of control animals by the usual time of sperm transfer (Figure 2C). Rather than dictating the overall mating duration or the response to threats in real time, activation of the Crz neurons switches the male out of a period of insurmountably high motivation to sustain the mating and into an independently-controlled and steadily-diminishing tendency persist (Figure 2G) that is likely orchestrated by previously described motivational circuitry (Crickmore and Vosshall, 2013).
Hyperpolarizing the Crz neurons through expression of the leak potassium channel Kir2.1 (Baines et al., 2001) has been shown to prolong matings by over an hour (Tayler et al., 2012). We find that this manipulation also dramatically extends the epoch of insurmountably high persistence (Figure 2D), consistent with an inability to engage the motivational switch and allow termination in response to threats. Briefly stimulating the Crz neurons with CsChrimson while they were otherwise hyperpolarized restored a typical mating duration (Figure 2E), conferred responsiveness to threats at any time after stimulation (Figure 2F) (even minutes later), and induced sperm transfer (Supplementary Figure 4A). Importantly, a similar protocol reversed the effects of expressing CaMKII-T287D (Supplementary Figures 4B and 4C), demonstrating that CaMKII activity does not block the Crz neurons from communicating with downstream circuitry, so long as they receive sufficient excitation. CaMKII activity therefore prevents the Crz neurons from engaging a one-time switch in motivational state that irreversibly allows the male to respond to threats and eventually terminate the mating at a normal time. In wildtype flies, this switch is the output of a 6-minute timer, presumably to allow the initial mechanics of the copulation to play out in preparation for sperm transfer and a synchronous behavioral state change.
The duration of sustained CaMKII activity times the 6-minute behavioral switch
The above results led us to hypothesize that the duration of CaMKII’s activity within the Crz neurons is the 6-minute timer. We first confirmed that CaMKII-T287D increases levels of phosphorylated endogenous CaMKII in the Crz neurons using a phosphoT287 antibody (Figure 3A). This antibody does not recognize transgenic T287D mutant CaMKII, as expression of a kinase-dead CaMKII-K43M-T287D did not increase the signal. Like Kir2.1, sustaining CaMKII activity by driving CaMKII-T287D in the Crz neurons prevented the shift in motivation at 6 minutes and maintained high motivation for over an hour (Figure 3B), and simultaneously silencing the neurons while expressing CaMKII-T287D did not further extend copulation (Supplementary Figure 4D). Inversely, expression of a T287-unphosphorylatable, and therefore calcium-obligate, CaMKII (T287A) (Jin et al., 1998), which complexes with endogenous CaMKII to weaken its ability to sustain signaling (Supplementary Figure 5C), hastened the motivational shift (Figure 3C) and fertilization (Figure 3D) without impacting the overall duration of the mating (Figure 3E), similar to early stimulation of the Crz neurons. RNAi-mediated knockdown of CaMKII transcripts also induced a premature decrease in motivational state (Figure 3F) that was rescued by expression of mouse CaMKII (not targeted by the RNAi), but not calcium-obligate CaMKII (Figure 3F). As expected, these manipulations did not affect the durations of undisturbed matings (Figure 3G). Together these data suggest that the duration over which CaMKII remains active, and in particular the duration of its calcium-independent activity, determines when the Crz neurons induce sperm transfer and the motivational shift.
Figure 3. The duration of sustained CaMKII activity times the 6-minute behavioral switch.
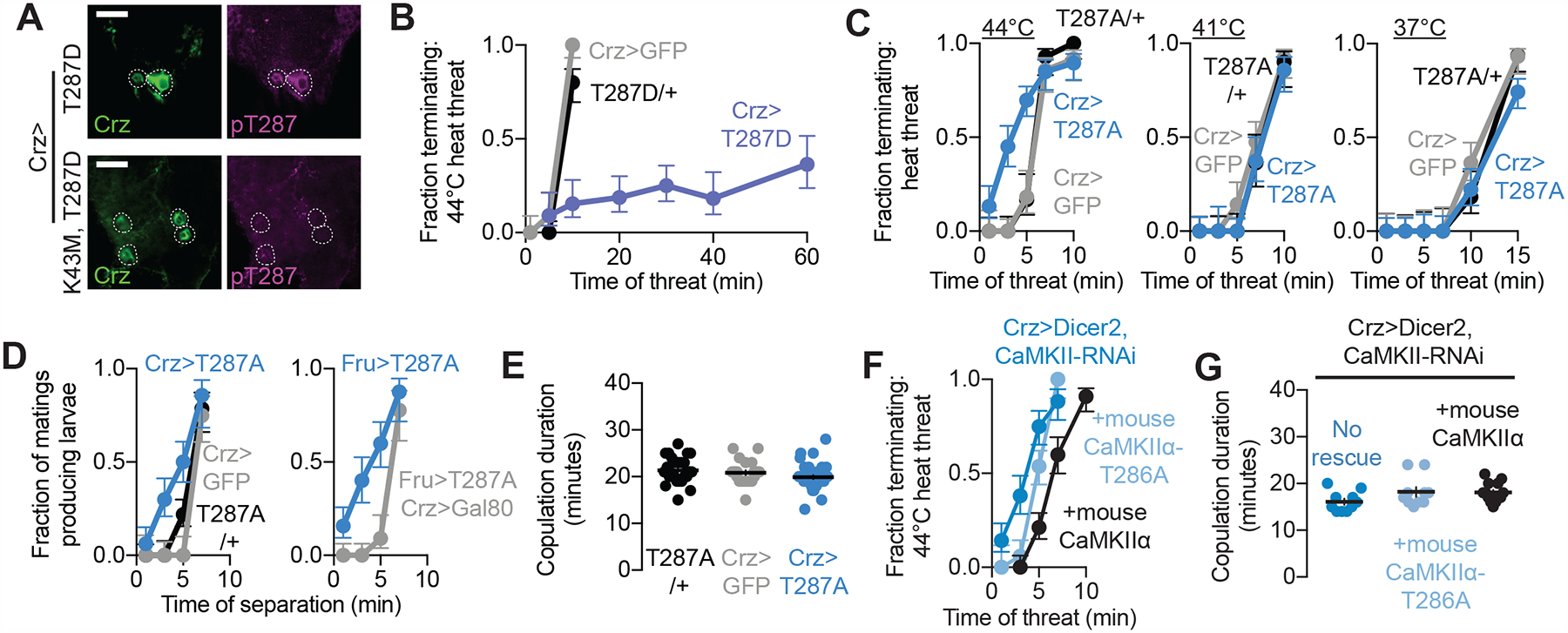
(A) T287-phosphorylated CaMKII levels increase when phosphomimic CaMKII is introduced to the Crz neurons. This increase is not seen in response to a kinase dead CaMKII (K43M), indicating that endogenous CaMKII is phosphorylated by T287D in the Crz neurons. These images are representative of 3 abdominal ganglia imaged for both conditions.
(B) Calcium-independent CaMKII (T287D) expression in the Crz neurons prolongs the epoch of high motivation.
(C-E) Expression of calcium-obligate CaMKII (T287A) in the Crz neurons hastens the motivational switch (C) and onset of sperm transfer (D) without affecting copulation duration (E).
(F,G) Knockdown of CaMKII in the Crz neurons (assisted by Dicer2) hastens the switch in motivational state (dark blue), and its timing is reverted to normal by additionally expressing wildtype mouse CaMKIIα (black). Expressing calcium-obligate mouse CaMKIIα (T286A) results in an incomplete rescue of the timing of the switch in motivational state (light blue) (F). None of these manipulations impacted overall copulation duration (G).
Our attempts to use the phospho-T287 antibody to monitor CaMKII at various times into mating gave inconsistent results, likely due to the challenge of reproducibly fixing the tissue rapidly enough to capture the active enzyme. We therefore generated flies capable of expressing the two-photon FRET-FLIM reporter of CaMKII activity, green-Camuiα (Lee et al., 2009). Green-Camuiα is a mouse CaMKIIα fused to an eGFP donor fluorophore and a darkened acceptor fluorophore (REACh), so that when the two are brought into close proximity, the excited eGFP donor can transfer energy to the acceptor through FRET (Förster resonant energy transfer). This results in a change in the measured fluorescence lifetime of the eGFP fluorophore, causing a longer fluoroscence lifetime when the two fluorophores are separated (i.e. CaMKII is active) and a shorter fluoroscence lifetime when they undergo FRET (i.e. CaMKII is inactive) (Figure 4A). The change in lifetime can be measured using fluorescence lifetime imaging microscopy (FLIM). We confirmed that this approach measures the activation of Drosophila CaMKII by showing that basal green-Camuiα fluorescence lifetime was increased by co-expression of constitutively active Drosophila CaMKII-T287D (Supplementary Figure 5A).
Figure 4. CaMKII activity in the Crz neurons is sustained for minutes after super-threshold stimulation.
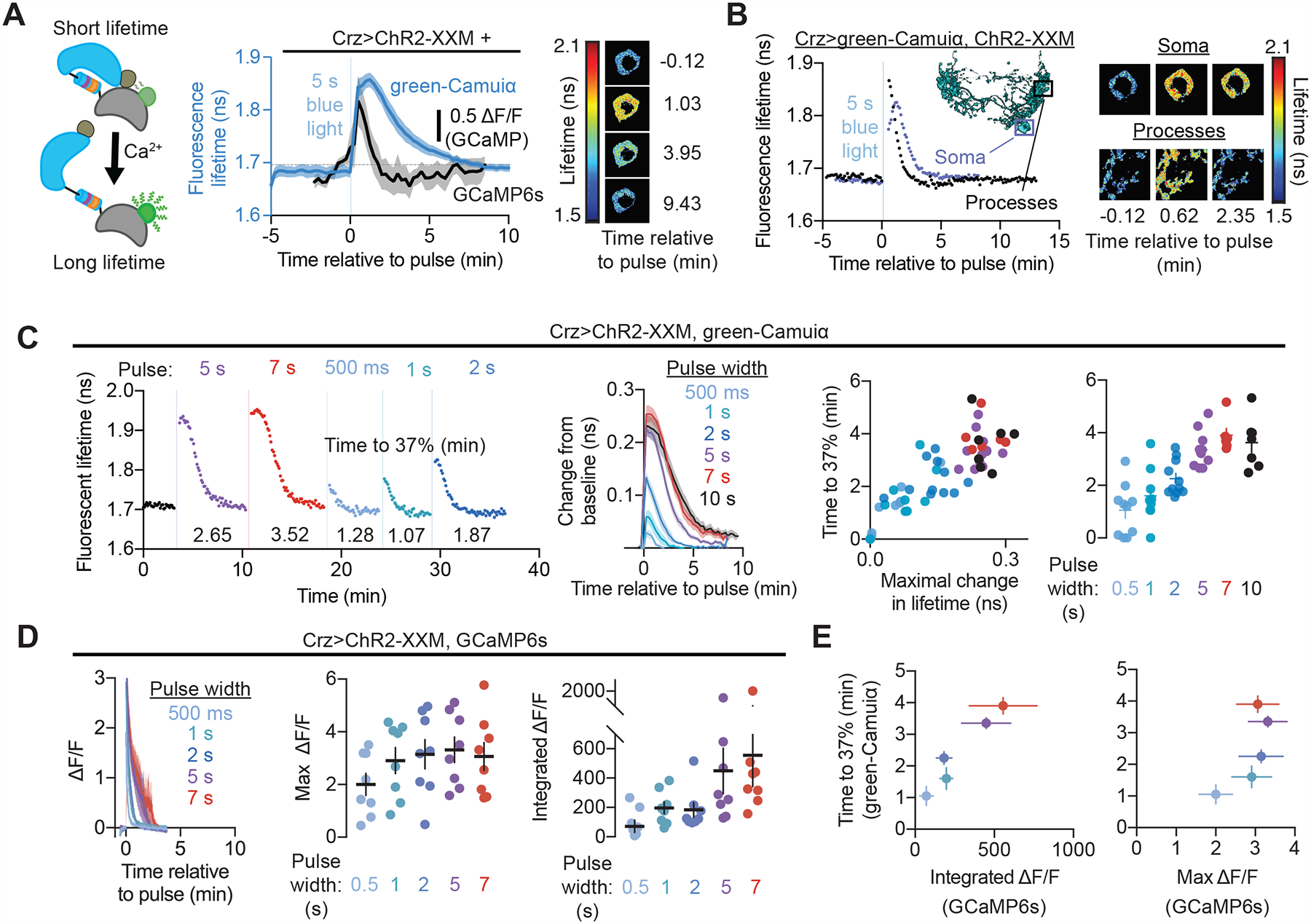
(A) CaMKII activity, as reported by the fluorescence lifetime of green-Camuiα (schematized on the left), decays over several minutes after brief stimulation (5 seconds) of the Crz neurons using a channelrhodopsin2 variant (ChR2-XXM) (blue). This decay lasts much longer than the increase in calcium induced by the same stimulation (black) Right: Fluorescent lifetime map of green-Camuiα signal of an example Crz neuron cell body before and after ChR2-XXM stimulation.
(B) Active green-Camuiα decays more rapidly in the processes of the Crz neurons than in the soma when stimulated for 5 seconds with ChR2-XXM. Data shown is from an example abdominal ganglion (also shown on the right) but held true for all flies examined (4/4). Inset: segmentation of a single Crz neuron using Multi-Color Flp Out illustrating the location of the soma and processes imaged.
(C) Sufficiently strong increases in CaMKII activity result in sustained activation for several minutes. Left: an individual experiment showing that five or more seconds of stimulation of ChR2-XXM elicits the sustained response. Compiled data on the right show that a long response with similar kinetics is seen after five or more seconds of stimulation. Weaker stimulations decay more rapidly with kinetics that scale with the amount of increased activity. We characterized the decay of the curves nonparametrically, to avoid making explicit assumptions about a parametric description of decay kinetics (e.g. by modeling them as a single or double exponential). We chose the time to reach 1/e of the maximal deflection from baseline (~37%), because this reflects the time constant of a single exponential.
(D) Optogenetic stimulation of the Crz neurons caused elevations in calcium that were much briefer than CaMKII activations evoked by the same stimulation.
(E) The longevity of CaMKII activation is better correlated with the time integral of calcium elevation than with peak calcium.
Stimulating the Crz neurons for five seconds using a calcium-permeable, high-photocurrent variant of the blue-light sensitive cation channel channelrhodopsin2 (ChR2-XXM (Scholz et al., 2017)) increased levels of active CaMKII in the soma (Figure 4A, Supplementary Figure 5B) and processes, where the signal was slightly shorter-lived (Figure 4B). We found that while five or more seconds of optogenetic stimulation resulted in CaMKII decay rates that approximated the behaviorally measured time interval, weaker stimulation produced reliable decay rates covering a range of time intervals (Figure 4C). These results are consistent with computational modeling predicting a slow decay following super-threshold CaMKII activation (Michalski, 2013) and suggest two possible timing paradigms for CaMKII: one in which the decay kinetics are fixed to generate a reproducible interval after super-threshold stimulation, and one that is adjustable and scaled to the intensity of sub-threshold input.
If CaMKII’s activity is the timer itself, as opposed to the output of some other timing mechanism, it should not rely on upstream input such as sustained intracellular calcium. When we measured the calcium response to the same optogenetic stimulation using GCaMP6s (Chen et al., 2013), we found that cellular calcium levels drop off much more rapidly than CaMKII activity (Figures 4A and 4D), indicating that CaMKII activity is sustained in a calcium-independent manner via its hallmark autophosphorylation. The extent to which CaMKII was activated covaried with how long levels of calcium were elevated at the onset of the timer (Figure 4E, Supplementary Figure 5I), but not necessarily peak calcium levels as registered by our reporter, suggesting that the dynamics of CaMKII are determined by more than just maximal neuronal activity. To test the necessity of autophosphorylation to maintain CaMKII activity, we performed the same experiments while expressing CaMKII-T287A to impair calcium-independent activity. Initial CaMKII activation was similar, but, as expected, returned to baseline more rapidly in the presence of non-autophosphorylatable subunits (Supplementary Figures 5F–H). In anatomically distinct Corazonin-producing neurons that do not affect mating behavior (Supplementary Figure 3) we observe a more rapid decay from a similar peak of CaMKII activity (Supplementary Figures 5D and 5E), and in hippocampal spines (Lee et al., 2009) active green-Camuiα decays within a minute. The duration of CaMKII’s calcium-independent state therefore appears to be tuned for different roles in different neurons, and in the Crz neurons is poised to measure out 6 minutes to delay sperm transfer and a switch in motivation.
To gain precise temporal control over CaMKII activity during behavior, we generated flies capable of expressing a blue-light-inducible peptide inhibitor of CaMKII kinase activity (UAS-paAIP2) (Murakoshi et al., 2017) (Figure 5A). Using paAIP2 to abruptly block CaMKII activity in the Crz neurons, either shortly after the initiation of mating or just before a threat, induced the motivational shift and allowed the male to terminate the mating in response to heat (Figure 5A). Activation of paAIP2 at the onset of mating induced a response to heat threats at any subsequent point into the mating (Figure 5B), and also reversed the extension of mating duration caused by CaMKII-T287D (Figure 5C) restoring copulation durations to more typical values (always above 13 and generally less than 30 minutes), further evidence that the failure of the Crz neurons to signal in the presence of constitutively high CaMKII activity does not result from developmental or structural defects. These results demonstrate that instantaneous CaMKII activity during the first minutes of mating is the sole impediment to the switch in motivational state. While inhibition of CaMKII with paAIP2 induced sperm transfer during mating (Supplementary Figure 4F) without affecting copulation duration (Supplementary Figure 4E), it did not cause ejaculation in non-mating males (Supplementary Figure 4G), suggesting that CaMKII activity impedes an excitatory process that only occurs during mating.
Figure 5. CaMKII actively prevents the output of the Crz neurons.
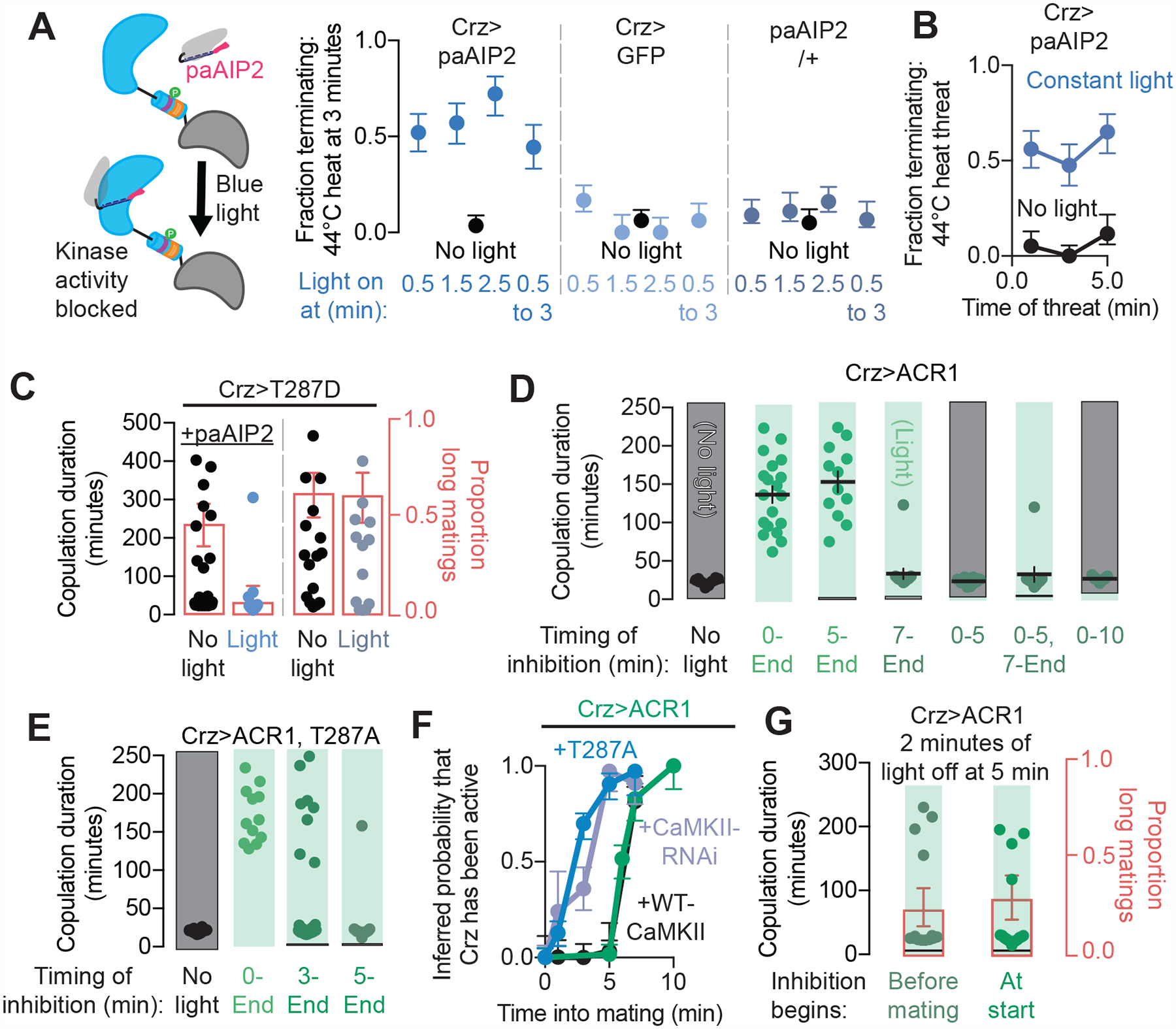
(A) Inhibiting CaMKII-mediated phosphorylation in the Crz neurons with blue light using paAIP2 (schematized on the left) induces the shift in motivation (right). The shift was maintained even after the removal of CaMKII inhibition (light from 1.5 to 3 minutes), showing that transiently impairing CaMKII activity suffices to cause the motivational switch.
(B) Inhibition of CaMKII activity with paAIP2 throughout the mating is capable of inducing a behavioral response to threats at any time after inhibition has begun.
(C) Inhibition of CaMKII activity using paAIP2 is sufficient to prevent the lengthening of mating duration resulting from expression of CaMKII-T287D. The proportion of long matings for each condition is shown in red when there is significant mixing of long and normal durations. No manipulations of the Crz neurons in this paper lead to matings that are shorter than we observe in control genotypes (~13 min).
(D) Optogenetic inhibition of the Crz neurons (light green shading) shows that electrical activity is not required for the first five minutes of mating as the CaMKII timer is winding down, but is required after 5 minutes to report the output of the timer and induce the motivational switch. The x-axis indicates the times into mating when constant inhibition of the Crz neurons was imposed.
(E-F) Expression of calcium-obligate CaMKII (T287A) (dark blue) or knockdown of CaMKII levels (purple), but not expression of wildtype CaMKII (black) in the Crz neurons decreases the time interval between the onset of mating and the time of transient Crz neuronal activation, as measured by whether beginning inhibition of the Crz neurons at the indicated time can extend copulation (curves fit to experimental data of the same type of experiments as in D and E with a Gaussian mixture model, see Methods).
(G) The initiation of the CaMKII timer is voltage-independent: similar proportions of normal-duration matings were obtained when inhibition was relieved from the 5th to 7th minute after mating began regardless of whether inhibition commenced before mating onset (13/17) or at mating onset (9/13), indicating that the Crz neurons do not require a voltage-dependent signal at the onset of mating to activate the CaMKII timer.
Electrical activity is only required for the output of the timing system
The imaging experiments above show that active CaMKII can be sustained for several minutes longer than elevated calcium (Figure 4A), suggesting that the timer does not rely on either calcium or electrical activity to track time during mating. To further explore the requirements for electrical activity in this system we conditionally inhibited the Crz neurons using the green-light gated chloride channel GtACR1 (Mohammad et al., 2017) (ACR1). Tonic silencing for the entire mating resulted in a long copulation duration, as did silencing that was initiated at 5 minutes into mating (Figure 5D), suggesting that electrical activity is required beginning around the time of the motivational switch. Inhibition that began at 7 minutes (soon after the switch) had no effect on mating duration (Figure 5D). If the Crz neurons were inhibited throughout the mating except during the normal time of the switch—from the 5th through 7th minutes—mating duration was normal, showing that the neurons successfully measure the 6 minute interval even in the absence of electrical activity, but require membrane voltage dynamics to report its conclusion (Figure 5D). Finally, we asked whether the switch could be induced any time after the CaMKII timer has run down by relieving inhibition 10 minutes after copulation began. This protocol always produced matings of normal duration (Figure 5D). Together with the ability of acute CaMKII inhibition to change the behavioral state (Figure 5A), this result argues that the Crz neurons are poised to throw the switch at all times during mating, and that CaMKII activity delays this voltage-dependent signaling.
Optogenetic silencing at specific times into mating provided another means of validating the role of CaMKII in timing the motivational switch. When the Crz neurons expressed the CaMKII-T287A transgene as well as ACR1, nearly half of the matings were of normal duration if the inhibition commenced at 3 minutes (Figure 5D), consistent with the idea that decreased calcium-independent CaMKII activity shortens the interval between mating initiation and the motivational switch. We used a statistical analysis (See Methods) on these inhibition-onset data to estimate the probability that the motivational switch has occurred within the allowed time window (Figure 5F). The switch occurred several minutes earlier in flies expressing CaMKII-T287A or in which CaMKII expression was reduced by RNAi (~3 minutes into mating) as compared to those overexpressing wildtype CaMKII (~6 minutes) or with no overexpression (~6 minutes) (Figure 5F).
Voltage-independent initiation and voltage-dependent output of the CaMKII timing system
Our results suggest that the CaMKII timer is initiated at the onset of mating, presumably by an increase in intracellular calcium, and reports the output of the timer 6 minutes later, likely using a second increase in intracellular calcium. How, then, do the Crz neurons distinguish the initiating cue from the output signal? In our ex vivo studies, we start the CaMKII timer by optogenetic depolarization and calcium influx. However, in vivo inhibition of the Crz neurons that commenced before the onset of copulation (i.e. before and during courtship) and was relaxed only from 5 to 7 minutes into mating most often resulted in a normal motivational switch and mating duration (Figure 5G), strong evidence that depolarization is not required to initiate the timer. A minority of flies still showed long copulation durations in this paradigm, likely those with the slowest CaMKII timers. The opposing requirements for membrane voltage at the start and end of the timer suggest mechanistic differences between the signals used to trigger its onset and report its conclusion.
To further explore the electrical requirements at the end of the timer, we inhibited the Crz neurons until 10 minutes into mating to ensure that the CaMKII timer had elapsed. We then relaxed inhibition for various durations before re-imposing it for the remainder of the mating. These experiments revealed a surprisingly long, and surprisingly robust, period of sustained electrical activity that is required to cause the motivational switch: relaxation windows of 60 seconds or less almost always resulted in long matings, while windows of 90 seconds or longer almost always threw the switch (Figure 6A, Supplementary Figure 6A–C). This 60–90 second window is much longer than the 1–2 seconds required for membrane voltage to return to baseline after GtACR1-mediated inhibition (Supplementary Figure 6C). If the relaxation window was applied early in mating while CaMKII is still active, the switch did not occur (Figure 6A), consistent with a requirement for depolarization only during the output phase of the timing system. The duration of relieved inhibition required to induce the switch was the same no matter when the relief was applied, so long as it was after the end of the CaMKII timer: a 90 second relaxation window applied even 45 minutes into mating caused termination 15 minutes later (Figure 6B), demonstrating that the Crz neurons are capable of retaining the information that the timer has run down for an extremely long time and suggesting that CaMKII activity counteracts an excitatory input onto the Crz neurons that builds over ~75 seconds.
Figure 6. The CaMKII timer relies on a voltage-independent initiation phase and voltage-dependent output phase.
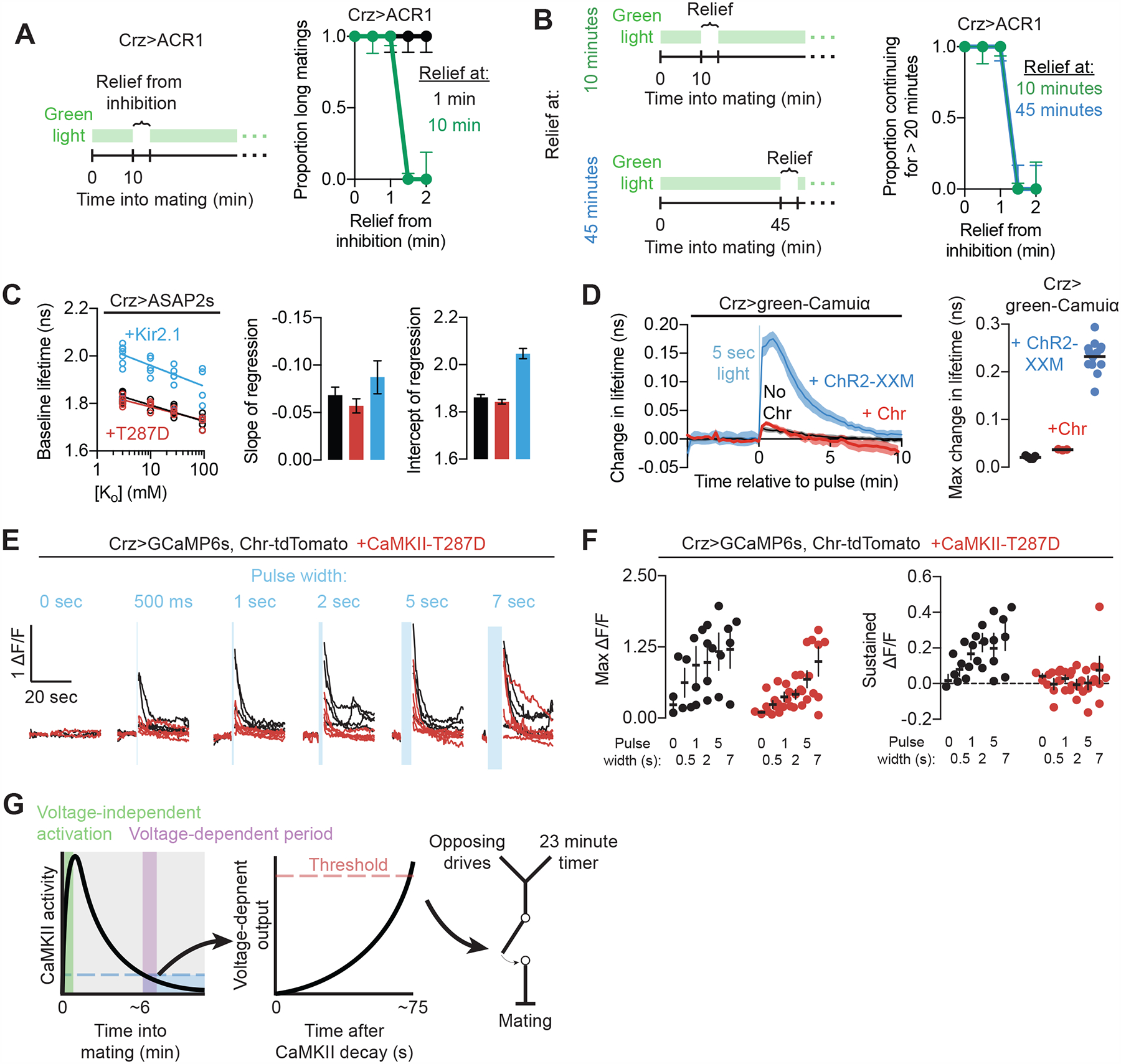
(A) The Crz neurons require ~75 seconds of voltage-dependent activity after the conclusion of the CaMKII timer (here, 10 minutes into mating) to induce the motivational shift (green trace), but cannot do so while CaMKII is active at the beginning of mating (black trace).
(B) The duration of the window of relief from optogenetic inhibition required to induce the shift in motivation is invariant to time into mating (lines are perfectly overlapping), so long as the CaMKII timer has finished. In this experiment, inhibition is removed from the neurons either after 10 minutes of mating or 45 minutes of mating. The voltage-dependent period is the same length, arguing that the decay of CaMKII does not itself trigger the activity of the Crz neurons, but instead that CaMKII prevents a voltage-dependent signal that is constant throughout the mating.
(C) Expression of constitutively active CaMKII does not modify baseline fluorescence lifetime of ASAP2s (a reflection of membrane voltage) or the slope of the relationship between the fluorescent lifetime and extracellular potassium (a reflection of potassium conductance). Expressing the leak potassium channel Kir2.1 greatly increases fluorescent lifetime (consistent with a more hyperpolarized cell at rest), as well as increasing the intercept and (to a lesser extent) the slope of the fit to external potassium, reflecting the increase in potassium conductance.
(D) Excitation of the Crz neurons using the calcium-impermeable channel CsChrimson (Chr) does not elicit CaMKII activity.
(E) Excitation of the Crz neurons with CsChrimson elicits transient increases in intracellular calcium with sustained tails (black). Constitutively active CaMKII (CaMKII-T287D) blocks the persistent elevation in calcium (red).
(F) Quantification of data in (E). “Sustained” ΔF/F refers to mean ΔF/F levels from 10 to 25 seconds after the optogenetic stimulation.
(G) Model for CaMKII timing of Crz neuronal output: the decay of CaMKII after a voltage-independent increase times the activity of the Crz neurons by permitting a voltage-dependent process that requires ~75 seconds to induce the change in motivational state.
Since increasing the activity of CaMKII has a similar behavioral effect as electrical inhibition of the Crz neurons, we examined the effects of CaMKII manipulations on membrane voltage. Absolute measures of membrane voltage using 2p-FLIM (Brinks et al., 2015) (Supplementary Figure 6D, E) did not show a change in resting voltage or baseline potassium conductance when constitutively active CaMKII-T287D was expressed in the Crz neurons (Figure 6C). Though these measurements were made outside of the mating context, they argue that activated CaMKII prevents the output of the Crz neurons without modifying baseline electrical excitability.
In our in vitro stimulation experiments above, we used ChR2-XXM to stimulate the Crz neurons and activate CaMKII. We used this Channelrhodopsin variant for its ability to directly pass calcium (Scholz et al., 2017), as we found that CsChrimson (which does not pass calcium (Vierock et al., 2017)) stimulated the Crz neurons without activating the CaMKII sensor (Figures 6D–F). In the absence of CaMKII activation, CsChrimson-mediated stimulation of the Crz neurons led to sustained elevation of intracellular calcium that persisted for tens of seconds (Figure 6E and 6F). Introduction of CaMKII-T287D completely abolished this sustained calcium signal, while imposing at most a modest reduction of peak calcium levels, especially when the Crz neurons were provided with strong excitation (Figure 6F). These results provide two potentially useful insights into the initiation and output phases of the timer: i) calcium influx through voltage-gated calcium channels alone is incapable of activating CaMKII in this system (since calcium increases from CsChrimson-mediated depolarization do not activate green-Camuiα), pointing to calcium release from intracellular stores as the timer-initiating signal; and ii) active CaMKII blocks sustained calcium elevations, which may play a role in meeting the 60–90 second requirement to induce the motivational switch at the conclusion of the timer. These results suggest that differences in point-of-origin and timescale (rapid vs. sustained) distinguish between the calcium-mediated signals that trigger the activation and conclusion of the timer (Figure 6G).
DISCUSSION
We present what we believe to be the first mechanistic description of a neuronal interval timer longer than a few seconds. Several lines of evidence argue that CaMKII is the timer itself, rather than an effector of some other sustained signal: i) the core timing mechanism does not require electrical activity; ii) selectively impairing calcium-dependent CaMKII activity (via the T287A mutation) shortens the timer, indicating that it relies on CaMKII autophosphorylation rather than sustained calcium; iii) elevating calcium-independent CaMKII activity (via the T287D mutation) prevents the timer from running down; iv) acute inhibition of CaMKII concludes the time interval; v) the dynamics of CaMKII in the Crz neurons match the timed interval. The gradual decline in CaMKII’s sustained activity therefore appears to be the central timing mechanism in this system, but our work provides less specific information about the processes that initiate and report the conclusion of the timer, or the factors that control the rate of CaMKII’s decline over time.
Since the Crz neurons do not extend projections outside of the abdominal ganglion, there must exist upstream neurons that detect the onset of mating and relay this information to start the timer. Voltage changes in the Crz neurons are not required for initiation of the timer, suggesting that the start signal may be relayed through GPCR signaling, and perhaps through liberation of internal calcium stores, though other, non-calcium-mediated forms of CaMKII activation are also possible (Bayer and Schulman, 2019). Activated CaMKII then delays the output of the Crz neurons for a duration that depends on the ability of autophosphorylated subunits to sustain activity against inactivating factors (e.g. phosphatases). When CaMKII activity eventually returns to baseline, the Crz neurons use a voltage-dependent process to signal the conclusion of the timer to downstream neurons. Inhibiting CaMKII outside of the mating context does not induce sperm transfer, one of several lines of evidence suggesting a constant excitatory force acting on the Crz neurons during mating. CaMKII activity opposes this excitation through a mechanism that remains obscure, but which likely involves the suppression of sustained calcium elevation. A sustained response may amplify the input signal over time, explaining the ~75 seconds of membrane voltage required for the Crz neurons to signal after the CaMKII timer has run down. In this model, a high and sustained level of electrical activity may be necessary to gate the release of the neurotransmitter(s) that trigger(s) sperm transfer and the motivational switch. The localization of the timer to four neurons, the identification of the core timekeeping mechanism, the ability to infer the activity of the timer through a robust behavioral readout, and a system for automated scoring should allow a rapid resolution of these molecular- and circuit-level details.
Downstream of the Crz neurons, the sperm transfer signal is likely processed by serotonergic neurons that innervate the ejaculatory bulb (Tayler et al., 2012), while the motivational output may adjust the activity of previously described dopaminergic and/or GABAergic neurons in the abdominal ganglion that regulate the changing motivational state during mating (Crickmore and Vosshall, 2013). The Crz, dopaminergic, and GABAergic neurons clearly impact motivation, as opposed to motor function, as experimentally altering their activity biases the tendency to terminate the mating in response to competing stimuli, as opposed to simply inducing termination, and silencing the neurons does not obviously impact motor functions. We were initially surprised to find these neurons in a region of the nervous system most often called the ventral nerve cord (recently updated to the ventral nervous system, VNS (Court et al., 2017)). Like the spinal cord, the VNS is situated in the thorax and contains motor neurons. But unlike the spinal cord, the VNS does not physically resemble a cord and is comparable in size to the central brain (if the optic lobes are not considered). The VNS is far more interconnected with the fly brain than its vertebrate counterparts (e.g. there are ~105 descending neurons in mice out of ~108 brain neurons (0.1%), while there are ~103 descending neurons in flies of their ~105 brain neurons (1%) (Namiki et al., 2017)). While there are many anatomical similarities between invertebrates and vertebrates, there are important differences; for example, insects enclose their entire bodies, not only their CNS, in a hard shell. This, together with their generally much smaller body size, may lead to different spatial constraints in the positioning of neurons with various functions in the CNS, including the location of a motivational control center in the abdominal ganglion of the VNS.
How extensively is this type of fixed time measurement used in the nervous system? Perhaps more so than is currently appreciated. We, for example, did not anticipate a 6-minute timer nested within the 23-minute mating duration when we began this study. The passage of time is unique among information about the external world in that it passes inexorably and ubiquitously. Unlike traditional sensory modalities, any neuron can, in principle, detect the passage of time directly and use that information to organize or synchronize circuit functions. The duration of CaMKII’s sustained activity is differentially tuned in different contexts; our sub-threshold activation experiments show that, even within the same neurons, CaMKII can produce a range of time intervals that are adjusted to the level of input. Though not apparent in our system, in principle a CaMKII-based timer could be adjusted in real-time by modulatory inputs that increase or decrease phosphorylation at T286/7. The mechanism described here may therefore be used beyond the measurement of fixed time intervals, generating a range of solutions for translating neuronal timescales into behavioral ones. Temporal intervals are implicit in almost all functions of the nervous system, especially those organizing behavior, necessitating some form of representation of these intervals for each of these contexts. There are hints that CaMKII is involved in timing across animals and behaviors, for example in the precisely timed circalunar eclosion of midges (Kaiser et al., 2016) and the intestine-controlled rhythmic defecation of C. elegans every 45 seconds (Reiner et al., 1999).
CaMKII is most often studied in the nervous system for its role in memory formation and storage, with a particular focus on the potentiation of synaptic transmission in the hippocampus (Bayer and Schulman, 2019; Lisman et al., 2012). But CaMKII evolutionarily predates the ionotropic glutamate receptors that underlie this synaptic plasticity by hundreds of millions of years (Alié and Manuel, 2010). Here we show that CaMKII delays the output of the Crz neurons in a memory-independent interval timing system. Though the idea that biochemical reactions progress over reliable timescales of many seconds or minutes is certainly not new, theoretical work on interval timing has focused largely on electrical network mechanisms (Goel and Buonomano, 2014). Similarly, early hypotheses about circadian timing (e.g. Enright 1980) were developed around the production of oscillations via electrical and synaptic mechanisms, before Drosophila genetics revealed its molecular nature (reviewed in Song & Rogulja 2017). Our work points to timing mechanisms that may not even be detectable by standard electrical recording and 0calcium imaging techniques.
Single-celled organisms use biochemical computations to respond to their environment in sophisticated ways: for example, the cyanobacterial circadian clock keeps time using autophosphorylation in a process strikingly similar to CaMKII (Snijder et al., 2017). It is unlikely that neurons have abandoned these evolutionarily ancient strategies, given that they enable processing of much more information than immediate electrical input provides (Thomson and Gunawardena, 2009). While in principle many biochemical processes might function to measure intervals of time, the long-recognized function of CaMKII as a molecular memory of earlier events makes it especially suitable for timekeeping. Its broad expression, strict evolutionary conservation, and tunable duration of its activity make CaMKII a candidate for timing functions and slowly evolving dynamics underlying a wide variety of behavioral states and other emergent neuronal network properties.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michael Crickmore (michael.crickmore@childrens.harvard.edu).
DATA AND SOFTWARE AVAILABILITY
Raw data for the screen in Figure 1A is available upon request. All code used in analyses is available at https://github.com/CrickmoreRoguljaLabs/CaMKIICode.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly stocks
Flies were maintained on conventional cornmeal-agar-molasses medium under a 12 hour light/12 hour dark cycle at 25 °C. Unless otherwise stated, males were collected 0–6 days after eclosion and group-housed away from females for 3–6 days before testing. Flies expressing Chrimson, and all experimental controls for experiments using Chrimson, were housed with rehydrated potato food (Carolina Bio Supply Formula 4–24 Instant Drosophila Medium, Blue) coated with all-trans-retinal (Sigma Aldrich R2500) diluted to 50 mM in ethanol for at least 3 days, unless marked as “no retinal.” These vials were kept inside aluminum foil sheaths to prevent degradation of the retinal due to light exposure. Virgin females used as partners for copulation assays were generated by heat-shocking a CsChrimson-mVenus stock with a hs-hid transgene integrated on the Y-chromosome (from Bloomington stock #55135) in a 37 °C water bath for 90 minutes. This stock was selected because we found the females to be highly receptive to courtship, resulting in a large number of mating pairs shortly after the initiation of assays; and because copulation duration is robust to variations in the female’s genetic composition, as well as independent of the behavior of the female, as paralyzing the female by total inactivation of her nervous system does not affect copulation duration or the response to heat threats (Supplementary Figure 1D and E). Virgins were group-housed for 3–13 days before use. Experiments with Chrimson-expressing males were not performed at specific times relative to the light-dark cycle of the incubator because these animals were housed in constant dark conditions to preserve all-trans-retinal integrity. We did not observe any dependency of time of day on any of the behaviors described here, but all flies experiencing light-dark cycles were tested between ZT (Zeitgeber Time) 2 and ZT 14 (lights are turned on at ZT 0 and off at ZT 12). Detailed genotypes of all strains used in the paper are listed in the supplement. Oregon R (OR) and Canton S (CS) were used as wildtype stocks to which single transgene stocks were crossed in controls. The 20x-UAS-CaMKII-T287D stock in Figure 1E was produced near the end of this study, and so was not used in any other experiments detailed in this manuscript. The previously existing UAS-CaMKII-T287D (Park et al., 2002) stock used in other experiments typically only produced long matings ~60% of the time when expressed with Crz-Gal4 (though it was 100% effective when expressed together with CsChrimson), and so we attempt to address this with quantification of the proportions of matings that are long when relevant, as in Figure 5B.
METHOD DETAILS
Copulation duration screen
Initially, UAS-RNAi lines were selected randomly from the collections at Bloomington, Kyoto, and Vienna, and were crossed to flies expressing Gal4 either pan-neuronally (elav-Gal4) or in sexually dimorphic neurons (Fru-Gal4) along with UAS-Dicer2. If the cross produced progeny, we generally screened 8–10 male flies in the automated system described below and in Supplementary Figure 1. If a line caused unusually long or short matings, we ordered lines targeting related genes suggested by the literature. These were typically other RNAi lines or UAS-driven expression of mutants of that gene or related genes. We performed the experiment using CaMKII-T287D because of an initial and relatively weak hit from knockdown of calcineurin, a known regulator of CaMKII activity. All data and code is available upon request.
Evaluation of mating
Automated:
Females and males were loaded into 32-well arenas (schematics posted at https://github.com/CrickmoreRoguljaLabs) and recorded from a height of ~9” using a Canon camera (VIXIA HF R600) and saved on an SD card. These files were then processed using custom MATLAB code (also at https://github.com/CrickmoreRoguljaLabs). In brief, the analysis pipeline identified individual behavioral arenas by thresholding and then using a flood fill algorithm. Within each arena, and within a single frame, it identified flies by thresholding, applying a morphological erosion, and then using MATLAB’s built-in segmentation function bwlabel. If this method failed to find either of the two flies, we used a watershedding algorithm to find the missing animals. The centroid of each segmented fly was calculated, and then the distance between those centroids was compared. If the distance between the centroids was calculated to be less than a pre-determined value (we used 0.17 cm) for longer than a pre-determined value (we used 1 minute), the pair was scored as “mating.”
Manual:
Experiments involving heat threats or delivery of light stimuli at a set time into mating were evaluated manually in real time. Video was streamed from an infrared camera connected to Raspberry Pi (see below). A pair of flies was scored as “mating” when they adopted a stereotyped mating posture for at least 30 seconds. This posture consists of the male mounting the female and propping himself up on her abdomen using his forelegs, while curling his own abdomen and keeping the genitalia in contact. The posture is starkly different from anything exhibited during other naturalistic behaviors, and the pose is not sustained for 30 seconds during unsuccessful attempts to initiate a mating. It is also maintained even in the presence of threats, unless the male elects to terminate the mating. If the flies are physically pulled apart without disengaging the genitalia (such as if the female falls or if they collide with an obstacle), the male is able to climb back into place. Thus persistence in the face of a threat is clearly distinguished from any defects of musculature. When actually “stuck”, the male dismounts the female, orients himself away from her, and attempts to walk away, but cannot decouple their genitalia. Occasionally, the flies become stuck after mating for too long after ejaculation, perhaps because the seminal fluids harden and adhere the flies together. Stuck behavior is not scored as mating.
Termination in response to threats:
Matings were scored as ended in response to a heat threat if they were terminated any time between the initiation of the threat and 30 seconds after the end of the threat. The extra 30-second period was used to accommodate the sometimes-slow process of dismounting and decoupling the genitalia, as well as the several seconds required for the temperature inside the behavioral arena to return to ambient levels.
Assessing fertility
Mating pairs were manually separated by forcefully and repeatedly aspirating the flies through a narrow opening until mechanically separated, and then the female fly was collected and placed in isolation in the above-described cornmeal food vials. One week later, the vial was visually inspected for the presence of larvae as an indicator of successful fertilization. This approach was used to measure sperm transfer in lieu of methods of labeling the sperm with fluorescent markers because i) it is easy to do at scale, ii) it did not require any additional genetic tools (which would have made several experiments considerably more challenging in terms of genetics), iii) fluorescent approaches for measuring sperm transfer in the female are complicated by autofluorescent seminal fluids whose transfer is not controlled by the Crz neurons (data not shown), and iv) it reflects the supposed physiological goal of copulation: transfer of sufficient amounts of sperm to produce progeny.
Scoring ejaculation induced by light
Males were glued to a glass slide by their wings and illuminated by a 405 nm laser pointer while examining the flies under a dissecting microscope. The fly was scored as “ejaculating” if it began thrusting its aedeagus in response to light and the aedeagus became coated in seminal fluid, which fluoresced green under the UV light. For paAIP2 experiments, we used a blue LED as described below in addition to the UV laser pointer.
Optogenetic stimulation and heat threats during behavior
For Chrimson experiments: One male and one virgin female fly were placed in each of 0.86” diameter 1/8” thick acrylic wells sitting 4” above 655 nm LEDs (Luxeon Rebel, Deep Red, LXM3-PD01–0350) driven using 700 mA constant current drivers (LuxDrive BuckPuck, 03021-D-E-700) and passed through frosted collimating optics (Carclo #10124). This spot of light was scattered using a thin diffuser film (Inventables, 23114–01) under the wells to ensure a uniform light intensity of ~0.1 mW/mm2. The LEDs were controlled using an Arduino Mega2560 (Adafruit) running a custom script, which itself was controlled by a Raspberry Pi (either 2 or 3, running Raspbian, a Debian variant). Flies were observed by recording from above using the Raspberry Pi with a Raspberry Pi NoIR camera (Adafruit) and infrared illumination from below using IR LED arrays (Crazy Cart 48-LED CCTV Ir Infrared Night Vision Illuminator reflected off the bottom of the box) while streaming the video to a computer for observation.
For GtACR1 experiments: The set-up was as above except using the green (530 nm) Luxeon Rebel, LXML-PM01–0100, and a pulse-width modulated signal to set the time-average intensity of the light to ~5 μW/mm2 (approximately six times brighter than the ambient light).
For paAIP2 experiments: The set-up was as above, but additionally included a blue (470 nm) Luxeon Rebel (LXML-PB01–0040) with an average intensity within the well of ~0.25 mW/mm2. This intensity certainly disturbs flies to some extent, but is likely necessary for a sufficient amount of light to activate paAIP2 to pass through the cuticle. For these experiments, controls with the light but no paAIP2 are very important. The LEDs were arranged in a “3-UP” configuration (LEDSupply) and light was passed through a frosted collimating lens (Carclo 10511).
For threats: A similar device to the one described above was constructed, with the addition of a ¼” thick water bath underneath each well. Room temperature water was continually passed through this bath, except when heat threat manipulations occurred, when water of the temperature described in each experiment was used (controlled by a separate stopcock for each well). The LEDs above were driven with 1A BuckPucks controlled by a pulse-width modulated signal selected to ensure the average intensity of illumination is the same as in the other behavioral experiments (~0.1 mW/mm2 for red light, ~ 5 μW/mm2 for green, 0.25 mW/mm2 for blue) despite having to pass through the water (see Supplementary Figure 3).
Additional notes about behavior
All reported termination probabilities are conditional: they are only the subset of flies that persisted in mating until the noted time of the stimulus. For the 15-minute and earlier time points, this accounts for 100% of experimental flies, but data at the 20-minute time point or later should be considered in this light, rather than as a cumulative termination probability that includes the flies that terminated without intervention.
Generation of transgenic flies
Crz-LexA was generated using the 391 nucleotide sequence upstream of the putative transcription start site of Drosophila corazonin (CG3302) on chromosome 3R spanning nucleotides 14314545–14314935 using forward primer TTCGGATTTTGCCCGGG and reverse primer GTTTCGGCAGAAGTTAGTCC. The resulting LexA construct was inserted into the attP2 and attP40 landing sites by Rainbow Transgenics.
UAS-paAIP2 was created by inserting paAIP2 (a gift from Ryohei Yasuda) into the 20x UAS-IVS-mCD8GFP vector (Addgene 26220) after digestion with XhoI and XbaI, using the InFusion assembly kit (Clontech 638909). paAIP2 was amplified using the forward primer TTCTTATCCTTTACTTCAGGCGGCCGCGGCTCGAGATGCTGGCGACCACCC and reverse primer ACAGAAGTAAGGTTCCTTCACAAAGATCCTCTAGATTACAGCGCATCAAACGCTTCC. The construct was inserted into the attP2 landing site by BestGene, Inc.
UAS-CaMKII-K43M,T287D and 20x-UAS-CaMKII-T287D were made by assembling fragments amplified from the CaMKII ORF (DGRC FI03620) with digested 20x-UAS-IVS-mCD8GFP as above. The 5’ and 3’ primers, respectively, were TTCTTATCCTTTACTTCAGGCGGCCGCGGCTCGAGATGGCTGCACCAGCAGCCTGTAC and ACAGAAGTAAGGTTCCTTCACAAAGATCCTCTAGACTATTTTTGGGGTATAAAATCGAATGTAGTTG. The K43M mutation was introduced using the primer pair TTTGCTGCAATGATTATCAATACAAAAAAATTAACTGCCAGAGACTTTC and TTGATAATCATTGCAGCAAATTCAAAGCCAGTTGACTTTTG, and the T287D mutation was introduced with the primer pair GCATCGCCAAGAAGACGTAGACTGTCTCAAGA and TCTTGAACAGTCTACGTCTTTCTTGGCGATGC. Both constructs were inserted into the attP2 landing site by BestGene, Inc.
UAS-green-Camuiα was created by inserting green-Camuiα (from Addgene 26933) into the 20x UAS-IVS-mCD8GFP vector as above. Because of the sequence similarity of the 5’ and 3’ ends of the eGFP and darkened mVenus in green-Camuiα, the coding sequence was amplified in two halves split in the middle of the CaMKII segment, and the fragments were assembled together with the 20x UAS construct at once using InFusion. The forward primer and reverse primers for the first half were ttacttcaggcggccgcggcATGGTGAGCAAGGGCGAG and ggacgggttgATGGTCAGCATCTTATTGATCAG while those for the second half were ATGCTGACCATCAACCCGTCCAAACGCATC and ggttccttcacaaagatcctTTACTTGTACAGCTCGTCCATGC. The construct was inserted into the attP2 landing site by BestGene, Inc.
All lines are available upon request.
Antibodies and immunohistochemistry
All samples were fixed in PBS with Triton X-100 and 4% paraformaldehyde for 20 minutes, then washed three times with PBS and Triton X-100 for 20 minutes each before application of antibodies. All samples were incubated with the primary antibody for two days, washed three times with PBS and Triton X-100 for 20 minutes each, incubated with the secondary antibody for two days, then washed three times as before and mounted on coverslips using VectaShield (Vector Labs). The exception is for MCFO staining, in which we followed the protocol of Nern et al. (Nern et al., 2015)
Antibodies used are as in the Key Resources Table (except those in MCFO, which are as described in Nern et al.(Nern et al., 2015)):
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-GFP | Invitrogen | A-11122 |
| Chicken anti-GFP | Aves Labs | GFP-1010 |
| Mouse anti-GFP | Invitrogen | A11120 |
| Rabbit anti-DsRed | Clontech | 632496 |
| Mouse anti-nc82 | Developmental Studies Hybridoma Bank | Nc82 |
| Donkey anti-chicken 488 | Jackson ImmunoResearch | 703-545-155 |
| Donkey anti-rabbit 488 | Invitrogen | A11008 |
| Donkey anti-mouse 488 | Invitrogen | A21202 |
| Donkey anti-rabbit 555 | Invitrogen | A-31572 |
| Donkey anti-mouse Cy3 | Jackson ImmunoResearch | 715-166-150 |
| Donkey anti-rabbit 647 | Jackson ImmunoResearch | 711-605-152 |
| Donkey anti-mouse 647 | Jackson ImmunoResearch | 711-605-151 |
| Mouse anti-phosphoT286 CaMKII alpha | U.S. Biological | C1035–25F5 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| All-trans-retinal | Sigma Aldrich | R2500 |
| Experimental Models: Organisms/Strains | ||
| Elav-Gal4 | Bloomington Stock Center | BSC 458 |
| Fru-Gal4 | Bloomington Stock Center | BSC 66696 |
| Dsx-Gal4 | Stephen Goodwin lab | |
| Crz-Gal4 | Bloomington Stock Center | BSC 51976 |
| UAS-T287D | Bloomington Stock Center | BSC 29665 |
| MCFO | Bloomington Stock Center | BSC 64086 |
| UAS-Kir2.1 | Baines, R.A., J. Neurosci, 2001 | |
| LexAop-Gal80 | Bloomington Stock Center | BSC 32213 |
| UAS-CsChrimson-tdTomato | David Anderson lab | |
| UAS-T287A | Bloomington Stock Center | BSC 29663 |
| Crz-LexA | Michael Crickmore lab | |
| UAS-paAIP2 | Michael Crickmore lab | |
| UAS-CaMKII-K43M,T287D | Michael Crickmore lab | |
| 20x-UAS-CaMKII-T287D | Michael Crickmore lab | |
| UAS-green-Camuiα | Michael Crickmore lab | |
| UAS-green-Camuiα-T286A | Michael Crickmore lab | |
| UAS-ChR2-XXM | Robert Kittel lab | |
| UAS-CsChrimson-mVenus | Bloomington Stock Center | BSC 55135 |
| CS (Canton S) | Barry Dickson lab | |
| UAS-Shibire-ts | Bloomington Stock Center | BSC 5822 |
| UAS-GtACR1-eYFP | Adam Claridge-Chang lab | |
| tub-Gal80ts | Bloomington Stock Center | BSC 7071 |
| UAS-myr-tdTomato | Bloomington Stock Center | BSC 32222 |
| UAS-myr-GFP | Bloomington Stock Center | BSC 32197 |
| UAS-OPGCaMP6s | David Anderson lab | |
| LexAop-OPGCaMP6s | David Anderson lab | |
| Crz#8 (CrzNoAG) | Jae H. Park lab | |
| Software and Algorithms | ||
| Custom Python 2.7 and MATLAB code | This paper | https://github.com/CrickmoreRoguljaLabs/CaMKIICode |
| Analysis of copulation duration videos (for the screen) | This paper | https://github.com/CrickmoreRoguljaLabs/FlyKnight |
| Other | ||
| Potato food | Carolina Bio Supply | 173200 |
| Phosphate buffered saline (10x) | MediaTech | 46–013-CM |
| Raspberry Pi 3 Model B+ | Raspberry Pi Foundation | |
| Arduino ATMEGA2560 | Arduino | MEGA2560 REV 3 |
| LEDs | Luxeon Rebels | LXM3-PD01–0350, LXML-PM01–0100, LXML-PD01–040 |
Confocal microscopy
Confocal images were collected using a Zeiss LSM 710 through a 20x air objective (Olympus PLAN-APOCHROMAT) controlled by Zen software, and analyzed using ImageJ or custom Python code.
Two-photon FRET FLIM
FLIM was performed as described in Chen et al., 2014 (Chen et al., 2014). Two-photon imaging was achieved by a custom-built microscope with a mode-locked Ti-sapphire laser source (Chameleon Vision II, 80 MHz, Coherent). Photons were collected with fast photomultipler tubes (PMTs) (H10770PA-40MOD, Hamamatsu) using a 60X (NA 1.1) objective (Olympus). Image acquisition was performed using the custom-written software ScanImage running in Matlab. Fluorescence lifetime was quantified using time-domain single photon counting, with 256 time channels of width ~48.6 picoseconds, using a SPC-150 (Becker and Hickl GmbH) FLIM board. The donor fluorophore was excited using a 920 nm laser (with time-average light power ~1.5 mW under the objective), and ChR2 was excited by an arc lamp passed through a GFP filter. We acquired 128 pixel by 128 pixel FLIM images at ~0.143 Hz (one frame every 7 seconds) as an average of 20 consecutive frames of imaging at ~3 Hz. Experiments and dissections were performed in the same saline as used for calcium imaging. The ventral nervous system was removed in chilled saline and transferred to a room temperature stage for FRET-FLIM experiments. Samples were only used if lifetime measurements were stable for at least 10 minutes before manipulation to minimize the effects of bleaching or rundown on the baseline signal.
Some data in Figure 4C. all of Figure 4D, and Supplementary Figures S6G–I were collected on a modified Thorlabs Bergamo microscope. Samples were excited using a Coherent Chameleon Vision II Ti:Sapphire laser emitting a 920 nm beam and emission was detected using cooled Hamamatsu H7422P-40 GaAsP photomultiplier tubes, with light collected through a 16x water immersion objective (Olympus). The PMT signal was amplified using Becker-Hickl fast PMT amplifiers (HFAC-26) and passed to a PicoQuant TimeHarp 260 photon counting board, which was synchronized to the laser emission by a photodiode (Thorlabs DET110A2) inverted using a fast inverter (Becker-Hickl A-PPI-D). The TimeHarp signal was acquired by custom software (FLIMage, Florida Lifetime Imaging) which was also used to control the microscope. For intensity imaging (GCaMP), all detected photons within a pixel were summed together, regardless of arrival time relative to the excitation pulse. Images were acquired at a resolution of 256×256 and a rate of 4 Hz and, for FLIM, averaged to 0.66 Hz. Optogenetic excitation was provided by illumination with a liquid light guide coupled to a 488 nm LED, placed ~ 1 centimeter from the sample.
The fluorescence lifetime was computed using the empirical method in Lee et al. (Lee et al., 2009). Briefly, the empirical lifetime τ was computed by numerically approximating the integral where F(t) represents the sum of all photons collected across all pixels in the image in time channel t. This value can be interpreted as representing the average latency to emit a photon across all fluorescing molecules in the image, which for a single exponential would be the time constant of that exponential. The offset t0 was estimated in the period before the experiment began by fitting the decay to a double exponential model plus an instrument response function (which was also fit to a Gaussian). Then the empirical estimate would give , and so our estimate of t0 (we will call it ) becomes the difference between the analytical value for the empirical lifetime and the measured value:
After producing our initial estimate , for the remainder of the experiment we computed and report Pixel-wise lifetime maps were generated using the empirical method (Lee et al., 2009). A matrix τi,j was computed corresponding to the empirical lifetime estimate of pixel as follows:
Where is the number of collected photons in time channel , tlower is the estimated time channel in which the decay begins, and tbin is the width of a time channel (in our experiments, ~48.86 picoseconds). Pixels whose total number of collected photons fell below a threshold (typically ~40 photons) were set to 0 as a proxy for “background.” A median filter (in which each pixel was assigned the value of the median of itself and the 8 pixels surrounding it) was applied to produce example images.
Analysis of FLIM data was performed using MATLAB. Code for analyzing FLIM data is available online at www.github.com/CrickmoreRoguljaLabs/
Stimulation using ChR2-XXM, as in Figure 4 and Supplementary Figure 5, would often result in a short period (about 30 seconds) in which fluorescence lifetime would continue to increase, even after the blue light stimulation had been shut off, plausibly due to the slow closing kinetics of ChR2-XXM (Scholz et al., 2017) or due to recurrent excitation of the Crz neurons.
By “brain” neurons, we mean a particular pair of neurons expressing Crz in the brain. These two cells are very close together, are small and very round, are some of the most lateral, and are directly ventral to a more elongated cell body. We were careful to choose the same cell bodies each time because the neurons do not all project to the same place and so are presumably different in an unknown number of ways. In fact, we noticed that the larger Crz neuron in the brain showed much longer decay kinetics than those in the abdominal ganglion (around 10–12 minutes), and the cells in the optic lobe showed even shorter responses than those we are labeling “brain” neurons. Different cells also showed different baselines, varying from 1.4–1.5 ns (in the optic lobe) to 1.8 ns (the large cell).
We noticed a weak response to blue light in the green-Camuiα signal even without ChR2. We suspect this artifact is due to bleaching of the darkened YFP acceptor protein (sREACh) so that the donor eGFP is unable to transfer energy via FRET, resulting in a longer fluorescent lifetime independent of the configuration of green-Camuiα. It may also abolish short-lifetime contamination from the low levels of sREACh fluorescence (Murakoshi et al., 2015) or reflect a light-sensitive process capable of causing calcium to enter the cytoplasm, though our behavioral experiments with paAIP2 suggest that the Crz neurons are not intrinsically activated or silenced by blue light (though they may be sensitive to the higher intensities delivered ex vivo). We also noted that the size of this response increased with the duration of the blue light pulse, with a 30 second stimulation creating as large as a 0.1 ns shift in measured lifetime. The effect in the 5 second pulse experiments is much weaker than that induced by the presence of ChR2-XXM, and so we are confident that the signal we measure is dominated by calcium influx resultant from light-sensitive currents through ChR2-XXM.
Flies expressing ChR2-XXM typically showed longer green-Camuiα lifetimes at the beginning of an imaging session that would decay to a baseline level over ~10 minutes and remain there indefinitely unless the neurons were stimulated. We interpret this initial activation to reflect the sensitivity of ChR2-XXM to the two-photon laser. We then think some portion of the channels are eventually (but reversibly) bleached, so that the laser no longer causes additional excitation as it bleaches at the same rate at which channels recover. This basal stimulation also likely accounts for the small increase in baseline lifetime measurements (and variability of baseline) in flies expressing ChR2-XXM, which was not seen when expressing the much less sensitive original ChR2.
The responses to pulses of varying width were collected in random order, generally collecting 3 pulse widths from the same cell, but sometimes collecting data for all five pulse widths.
Two-photon calcium imaging and voltage imaging
GCaMP6s and ASAP2s fluorescence was measured using the microscope described above for FRET-FLIM. Images were analyzed in a manner intended to emulate how lifetime was estimated during FLIM experiments: fluorescence intensity was simply summed across the image after a background subtraction. Almost all fluorescence came from the Crz neurons, so no additional processing was necessary. Baseline fluorescence F was computed using the average of ~ 1 minute before optogenetic stimulation.
Experiments involving Chrimson and GCaMP6s used excitation with a blue LED (Thorlabs M470F3) through a fiber optic cable (M28L02) placed ~500 μm from the abdominal ganglion, shielding the microscope’s detectors during stimulation using a shutter. The light power measured at the end of the fiber was ~3–5 mW. These experiments were done this way because they shortened the time between the end of optogenetic stimulation and the ability to resume imaging. Images were acquired at ~3.6 Hz.
Two-photon absolute voltage imaging with FLIM
Fluorescence lifetime was measured as described above for FRET-FLIM but using the voltage sensor ASAP2s excited with a 920 nm laser. Extracellular potassium was varied by gently pipetting 1M KCl into the edge of the dish. Baseline lifetime measurements were made by averaging lifetime measurements 2–5 minutes after introduction of KCl to allow the bath to equilibrate. It has been reported that the fluorescence lifetime of ASAP1 varies approximately linearly with voltage; if so, we can use baseline fluorescence lifetime as a reliable proxy for time-average membrane voltage. The previous experiments (Brinks et al., 2015) confirmed the linear relationship between ASAP1 fluorescence lifetime and membrane voltage using simultaneous electrophysiology, but this approach was not available to us. We attempted to confirm the same of ASAP2s by testing whether fluorescence lifetime varies approximately log-linearly with extracellular potassium, which would be a consequence of varying linearly with voltage by the following logic (schematized in Supplementary Figure 6E):
We suppose the cell is permitted to equilibrate and only consider passive conductances. This assumption is almost certainly wrong in all cases, but enables a simple analysis that still seems to reasonably accurately describe our data. In this case, the membrane voltage V is
Where I is the set of ions, gi is the membrane conductance of ion i, G = Σigi, and where φi depends on the ionic charge as per the Nernst equation, [i]out is the concentration of i outside of the cell, and [i]in is the concentration of i inside the cell. We then see that
If we then assume a linear relationship between the voltage V and fluorescence lifetime L so that L ≈ αV + β then
where we have grouped terms that depend on [K]out on the left side and those that do not in brackets on the right. We then see that lifetime L is linearly related to log([K]out) and the slope of this relationship is proportional to , the potassium conductance of the cell relative to the total conductances of the cell. If we assume all other conductances remain constant between two manipulations, then the ratio of the slopes of the linear fits between L and log([K]out) estimates the factor by which gK has changed between those manipulations. We also note that a change in gK will also affect the offset, both by adjusting G and through the term involving [K]in. This effect is visible in the manipulation involving Kir2.1, where we see a large increase in offset (reflection the hyperpolarization of the neuron), as well as a change in the slope. Here it is notable that the linear relationship between L and log([K]out) is a less good description, presumably because Kir2.1 is inwardly-rectifying and presumably contributes substantially to the overall potassium conductance so gK is no longer constant as we vary .
QUANTIFICATION AND STATISTICAL ANALYSIS
General framework
Throughout this manuscript, we take a Bayesian approach to estimating proportions because it more closely corresponds to the inference procedures we are performing, and as such all reported windows and intervals correspond to the mass of the posterior distribution for the inferred model parameter. This is because the Bayesian perspective corresponds to inference about the values of descriptors of our model (e.g. in our data, the probability of terminating the mating in response to some stimulus), rather than consistency of a data set with a particular value that the model might take (the “null hypothesis”). We take this approach for philosophical reasons: we can make statistical claims about our belief in the magnitude of effects, rather than simply reporting their deviation from that produced by a null hypothesis, or performing inference on a procedure that itself cannot make claims about true parameter values. We do, however, recognize that the frequentist approach is more commonplace, and so present our data in a manner that is as consistent as possible with typical frequentist reporting and hypothesis testing. We use noninformative priors (Jaynes, 1968), so this trivially corresponds to the usual Central Limit Theorem statistics in the case of estimating the variability of means, but a slightly different estimator on proportions. The extent of the Bayesian approach is limited to the graphical presentation of data; all statistics are performed in the traditional frequentist fashion (see hypothesis testing section below). Our results and their interpretation do not hinge, in any case, on precise statistical methodology, as our effects tend to be very large, and so this decision is more philosophical than effectual.
We use standard maximum likelihood estimates for fluorescence data, even though Bayesian approaches can be more robust, because these Bayesian analyses are considerably less commonplace than they are with proportions.
We report p-values in a table, rather than on the plots, to avoid the reduction of complex data into true-or-false hypothesis testing (Mcshane et al., 2017). We are careful to only make claims supported by standard null hypothesis analyses.
Credible intervals for proportions
All proportions are modeled as Bernoulli random processes with probability p and presented as the sample estimate for the proportion p ( with x the number of observed successes and n the total number of observations, the maximum likelihood estimate, rather than the maximum posterior estimate, for consistency with standard data presentation). This point is surrounded by a 68% credible interval (selected to be similar to the broadly familiar SEM metric, which is itself a 68% credible interval on the mean under a uniform prior), demarcated by error bars, generated by sampling from the posterior distribution using Markov Chain Monte Carlo (MCMC, Metropolis-Hastings algorithm) with the noninformative Jeffreys prior and selecting the 16–84% window of this empirical estimate of the posterior. The Jeffreys prior was selected because it gives a posterior that is in a sense invariant under reparameterizations (Jeffreys, 1945), and thus gives a consistent result between our posterior distributions even when we invert or transform the inference problem. The window generated by this method is also a numerical approximation of a 68% confidence interval with the corresponding frequentist properties. All estimation of the posterior was performed using Markov Chain Monte Carlo via the python package pymc.
For the experiments inferring the probability that the Crz neurons have been active at a certain time, we used a Bayesian Gaussian mixture model with two components: “long matings” and “short matings” whose parameters were common across experiments. For a given time t, we estimated the posterior distribution of the parameter p of a Bernoulli process, which was used to select the posterior distributions used to sample the mean and variance of the Gaussian from which an individual fly’s copulation duration was drawn. Explicitly:
with and the mean and variance of the “no light” condition, and and the mean and variance of the “light at start” condition, and likewise Nk the corresponding size of those samples. The prior on pt was a Beta distribution with α = β = 0.5, the Jeffrey’s prior, and we used MCMC to estimate the posterior distribution of pt. We then plotted the maximum a posteriori value for pt along with 68% credible intervals. A similar model was used for the “relief from inhibition” experiments in which inhibition was relieved after 45 minutes of mating. These flies would mate for ~15 minutes longer (in contrast to those flies in which inhibition was not relieved, that would typically mate for at least another hour). The flies in which inhibition was removed at 45 minutes were used instead of the “no light” condition for generating the priors on and .
Hypothesis testing on proportions
To test the hypothesis that two sample proportions were drawn from the same Bernoulli process, we used Fisher’s exact test. We then correct for multiple comparisons by using the Holm-Bonferroni correction on our criteria for statistical significance (with the number of hypotheses being the number of unique pairs of comparisons, n(n − 1)/2). In experiments involving heat threats at distinct times across distinct genotypes, the number of corrections was the number of hypotheses: n × t where n is the number of genotypes and t is the number of time points. This estimate is overly conservative (not all hypotheses are independent; in fact, almost all are likely to be highly correlated) but is the most principled given the challenge of estimating the correlations between motivation at different time points and how differences between genotypes are conserved across time points.
Credible intervals on means
We use the standard SEM estimator for variability of sample means with the unbiased estimator of sample variance and N the sample size, which corresponds to the 68% credible interval for the sample mean using the “improper” uniform prior.
Hypothesis testing on distributions
We use the nonparametric Mann-Whitney U test on rank sums for differences in distributions of copulation duration. We then correct for multiple comparisons by using the Holm-Bonferroni correction on our criteria for statistical significance (with the number of hypotheses being the number of unique pairs of comparisons, n(n − 1)/2). For experiments measuring copulation duration, this correction was done across all experimental conditions in the figure.
Power analyses
For proportions, we performed pilot experiments to estimate the effect size, and then used that estimated proportion to compute necessary sample size given a power of 0.8 and a significance criterion of 0.05, and then used approximately that many samples. The sample used to estimate the effect size was separate from the sample presented in the data (and analyzed using statistics), and so represented an independent estimate of the effect size. This approach is known to be imperfect, and results in an imprecise estimate of the effect, resulting in a suggested sample size that may not be accurately tuned to the true effect size. Thus some experiments may be slightly underpowered or overpowered, and indeed some observed effect sizes consistently trend towards statistical significance without achieving it (see, e.g., Figure 3D), a natural consequence of a slightly underpowered analysis analyzing an effect that is nonetheless present. We use this procedure only because we have no other pre hoc means of estimating the strength of an anticipated manipulation using traditional frequentist approaches (i.e. without priors).
For other distributions, no pilot studies were performed since these distributions are nonparametric and thus proper power analysis is not technically possible. Instead, if the effect sizes were large (as are almost all effects in the paper), we generally used ~10 flies. If we thought differences were small or unlikely, we used ~20 flies.
Randomization
Animals were usually alternatingly assigned to conditions, when applicable (e.g. animal 1 was subjected to condition A, animal 2 was subjected to condition B, animal 3 was subjected to condition C (when applicable, else condition A)), animal n was subjected to condition n modulo m where m is the number of conditions. Because not every animal mates, this often resulted in uneven sample sizes, and thus in some conditions more animals used than power analysis required (because experiments were performed in large batches, e.g. of 30 animals in parallel).
Blinding
Experimenters were not blinded in any experiments.
Supplementary Material
Supplemental Video 1, Related to Figure 1. Expression of CaMKII-T287D in the Crz neurons lengthens mating duration without obvious deficits in courtship or the mechanics of copulation
Acknowledgements
We thank Whitney Wallace, Alexa Soares, Henna Morani, Gabriel Verderame, Mary Dello Russo, Megan Hoffman, Fatih Toy, and Xavier du Maine for their assistance performing the screen in Figure 1A; Ofer Mazor and Pavel Gorelik at the Harvard Neuroinstrumentation Core for advice and facilities for creating our experimental apparatuses; Rachel Wilson’s lab for saline and advice on calcium imaging; Yao Chen and Bernardo Sabatini for providing the 2p-FRET-FLIM equipment and for substantial advice and assistance on green-Camuiα experiments; Gary Yellen for advice on FLIM and the code off of which our FLIM analysis code was based; Georg Nagel and Robert Kittel for the ChR2-XXM stock; Barret Pfeiffer, David Anderson, and Gerry Rubin for UAS and LexAop2 CsChrimson-tdTomato and OPGCaMP6s stocks; Ryohei Yasuda for the paAIP2 plasmid; Michael Rust for the insightful comparison to the KaiABC cyanobacterial oscillator; Jesse Goldberg, Jonathan Lipton, and the Rogulja and Crickmore labs for comments on the manuscript.
Funding: S.C.T. was supported by the National Science Foundation Graduate Research Fellowship (NSF Grant No. DGE1144152). S.X.Z. is a Stuart H.Q. & Victoria Quan Fellow. D.R. is a New York Stem Cell Foundation-Robertson Investigator. This work was supported by The New York Stem Cell Foundation
Footnotes
Competing interests: The authors declare no competing interests.
Data and material availability: All data and materials are available upon request.
REFERENCES
- Alié A, and Manuel M (2010). The backbone of the post-synaptic density originated in a unicellular ancestor of choanoflagellates and metazoans. BMC Evol. Biol 10, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bading H, Ginty DD, and Greenberg ME (1993). Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science (80-.) 260, 181–186. [DOI] [PubMed] [Google Scholar]
- Baines R. a, Uhler JP, Thompson a, Sweeney ST, and Bate M (2001). Altered electrical properties in Drosophila neurons developing without synaptic transmission. J. Neurosci 21, 1523–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer KU, and Schulman H (2019). CaM Kinase: Still Inspiring at 40. Neuron 103, 380–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinks D, Klein AJ, and Cohen AE (2015). Two-Photon Lifetime Imaging of Voltage Indicating Proteins as a Probe of Absolute Membrane Voltage. Biophys. J 109, 914–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhusi CV, Meck WH, and Carolina N (2005). What makes us tick? Functional and neural mechanisms of interval timing. Nat. Rev. Neurosci 6, 755–765. [DOI] [PubMed] [Google Scholar]
- Chamberland S, Yang HH, Pan MM, Evans SW, Guan S, Chavarha M, Yang Y, Salesse C, Wu H, Wu JC, et al. (2017). Fast two-photon imaging of subcellular voltage dynamics in neuronal tissue with genetically encoded indicators. Elife. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J-Y, Parra-Bueno P, Laviv T, Szatmari EM, Lee S-JR, and Yasuda R (2017). CaMKII Autophosphorylation Is Necessary for Optimal Integration of Ca 2+ Signals during LTP Induction, but Not Maintenance. Neuron 94, 800–808.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr R. a, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Saulnier JL, Yellen G, and Sabatini BL (2014). A PKA activity sensor for quantitative analysis of endogenous GPCR signaling via 2-photon FRET-FLIM imaging. Front. Pharmacol 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court R, Armstrong D, Börner J, Card G, Dickinson M, Duch C, Korff W, Mann R, Merritt D, Murphey R, et al. (2017). A Systematic Nomenclature for the Drosophila Ventral Nervous System. BioRxiv 1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crickmore MA, and Vosshall LB (2013). Opposing dopaminergic and GABAergic neurons control the duration and persistence of copulation in Drosophila. Cell 155, 881–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright JT (1980). Temporal Precision in Circadian Systems : A Reliable Neuronal Clock from Unreliable Components? Science (80-.) 209, 1542–1545. [DOI] [PubMed] [Google Scholar]
- Fong YL, Taylor WL, Means AR, and Soderling TR (1989). Studies of the regulatory mechanism of Ca2+/calmodulin-dependent protein kinase II. Mutation of threonine 286 to alanine and aspartate. J. Biol. Chem 264, 16759–16763. [PubMed] [Google Scholar]
- Gilchrist AS; Partridge L (2000). Why it is difficult to model sperm displacement in Drosophila melanogaster: the relation between sperm transfer and copulation duration. Evolution (N. Y) 54, 534–542. [DOI] [PubMed] [Google Scholar]
- Goel A, and Buonomano DV (2014). Timing as an intrinsic property of neural networks : evidence from in vivo and in vitro experiments Timing as an intrinsic property of neural networks : evidence from in vivo and in vitro experiments. Philos. Trans. R. Soc. B 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith LC (2004). Regulation of calcium/calmodulin-dependent protein kinase II activation by intramolecular and intermolecular interactions. J. Neurosci 24, 8394–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaynes ET (1968). Prior Probabilities. IEEE Trans. Syst. Sci. Cybern 4, 227–241. [Google Scholar]
- Jeffreys H (1945). An invariant form for the prior probability in estimation problems. Proc. R. Soc. Lond. A. Math. Phys. Sci 453–461. [DOI] [PubMed] [Google Scholar]
- Jin P, Griffith LC, and Murphey RK (1998). Presynaptic Calcium / Calmodulin-Dependent Protein Kinase II Regulates Habituation of a Simple Reflex in Adult Drosophila. J. Neurosci 18, 8955–8964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston D, and Wu S (1995). Foundations of Cellular Neurophysiology.
- Kaiser TS, Poehn B, Szkiba D, Preussner M, Sedlazeck FJ, Zrim A, Neumann T, Nguyen L, Betancourt AJ, Hummel T, et al. (2016). The genomic basis of circadian and circalunar timing adaptations in a midge. Nature 540, 69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WJ, Jan LY, and Jan YN (2013). A PDF/NPF neuropeptide signaling circuitry of male Drosophila melanogaster controls rival-induced prolonged mating. Neuron 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Nairn AC, and Greengard P (1986). Autophosphorylation reversibly regulates the Ca2 + / calmodulin-dependence of Ca2 + / calmodulin-dependent protein kinase II. PNAS 83, 4253–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-JR, Escobedo-Lozoya Y, Szatmari EM, and Yasuda R (2009). Activation of CaMKII in single dendritic spines during long-term potentiation. Nature 458, 299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune H, and Wearden JH (1991). The comparative psychology of fixed-interval responding: Some quantitative analyses. Learn. Motiv 22, 84–111. [Google Scholar]
- Lisman J, Yasuda R, and Raghavachari S (2012). Mechanisms of CaMKII action in long-term potentiation. Nat. Rev. Neurosci 13, 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauk MD, and Buonomano DV (2004). The neural basis of temporal processing. Annu. Rev. Neurosci 307–340. [DOI] [PubMed] [Google Scholar]
- Mcshane BB, Gal D, Gelman A, Robert C, and Tackett JL (2017). Abandon Statistical Significance. ArXiv 1–12. [Google Scholar]
- Michalski PJ (2013). The delicate bistability of CaMKII. Biophys. J 105, 794–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SG, and Kennedy MB (1986). Regulation of Brain Type II Ca2+/Calmodulin-Dependent Protein Kinase by Autophosphorylation: A Ca2+-Triggered Molecular Switch. Cell 44, 861–870. [DOI] [PubMed] [Google Scholar]
- Miller SG, Patton BL, and Kennedy MB (1988). Sequences of Autophosphorylation Sites in Neuronal Type II CaM Kinase That Control Ca2+-Independent Activity. Neuron 1, 593–604. [DOI] [PubMed] [Google Scholar]
- Mohammad F, Stewart JC, Ott S, Chlebikova K, Chua JY, Koh T, Ho J, and Claridge-chang A (2017). Optogenetic inhibition of behavior with anion channelrhodopsins. Nat. Methods [DOI] [PubMed] [Google Scholar]
- Murakoshi H, Shibata ACE, Nakahata Y, and Nabekura J (2015). A dark green fluorescent protein as an acceptor for measurement of Förster resonance energy transfer. Sci. Rep 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakoshi H, Shin ME, Parra-Bueno P, Szatmari EM, Shibata ACE, and Yasuda R (2017). Kinetics of Endogenous CaMKII Required for Synaptic Plasticity Revealed by Optogenetic Kinase Inhibitor. Neuron 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namiki S, Dickinson MH, Wong AM, Korff W, and Card G (2017). The functional organization of descending sensory-motor pathways in Drosophila. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Pfeiffer BD, and Rubin GM (2015). Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc. Natl. Acad. Sci. U. S. A 112, E2967–E2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Coleman MJ, Hodge JJL, Budnik V, and Griffith LC (2002). Regulation of Neuronal Excitability in Drosophila by Constitutively Active CaMKII. J. Neurobiol 52, 24–42. [DOI] [PubMed] [Google Scholar]
- Paton JJ, and Buonomano DV (2018). The Neural Basis of Timing : Distributed Mechanisms for Diverse Functions. Neuron 98, 687–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner DJ, Newton EM, Tian H, and Thomas JH (1999). Diverse behavioural defects caused by mutations in Caenorhabditis elegans unc-43 CaM Kinase II. Nature 402. [DOI] [PubMed] [Google Scholar]
- Scholz N, Guan C, Nieberler M, Grotemeyer A, Maiellaro I, Gao S, Beck S, Pawlak M, Sauer M, Asan E, et al. (2017). Mechano-dependent signaling by Latrophilin / CIRL quenches cAMP in proprioceptive neurons. Elife 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung HS, Lee DD, Reis BY, and Tank DW (2000). Stability of the memory of eye position in a recurrent network of conductance-based model neurons. Neuron 26, 259–271. [DOI] [PubMed] [Google Scholar]
- Snijder J, Schuller JM, Wiegard A, Lössl P, Schmelling N, Axmann IM, Plitzko JM, Förster F, and Heck AJR (2017). Structures of the cyanobacterial circadian oscillator frozen in a fully assembled state. Science (80-.) 1184, 1181–1184. [DOI] [PubMed] [Google Scholar]
- Song BJ, and Rogulja D (2017). SnapShot: Circadian Clock. Cell 171, 1224–1225. [DOI] [PubMed] [Google Scholar]
- Stockinger P, Kvitsiani D, Rotkopf S, Tirián L, and Dickson BJ (2005). Neural circuitry that governs Drosophila male courtship behavior. Cell 121, 795–807. [DOI] [PubMed] [Google Scholar]
- Stratton M, Lee IH, Bhattacharyya M, Christensen SM, Chao LH, Schulman H, Groves JT, and Kuriyan J (2014). Activation-triggered subunit exchange between CaMKII holoenzymes facilitates the spread of kinase activity. Elife 3, e01610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayler TD, Pacheco D. a, Hergarden AC, Murthy M, and Anderson DJ (2012). A neuropeptide circuit that coordinates sperm transfer and copulation duration in Drosophila. Proc. Natl. Acad. Sci. U. S. A 109, 20697–20702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson M, and Gunawardena J (2009). Unlimited multistability in multisite phosphorylation systems. Nature 460, 274–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinbergen N (1951). The study of instinct.
- Tombes RM, Faison MO, and Turbeville JM (2003). Organization and evolution of multifunctional Ca(2+)/CaM-dependent protein kinase genes. Gene 322, 17–31. [DOI] [PubMed] [Google Scholar]
- Vierock J, Grimm C, Nitzan N, and Hegemann P (2017). Molecular determinants of proton selectivity and gating in the red- light activated channelrhodopsin Chrimson. Sci. Rep 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Video 1, Related to Figure 1. Expression of CaMKII-T287D in the Crz neurons lengthens mating duration without obvious deficits in courtship or the mechanics of copulation
Data Availability Statement
Raw data for the screen in Figure 1A is available upon request. All code used in analyses is available at https://github.com/CrickmoreRoguljaLabs/CaMKIICode.


