Abstract
Receptor activity-modifying proteins (RAMPs) are a family of three single span transmembrane proteins in humans that interact with many GPCRs and can modulate their function. RAMPs were discovered as key components of the calcitonin gene-related peptide and adrenomedullin receptors. They are required for transport of this class B GPCR, calcitonin receptor-like receptor (CLR), to the cell surface and determine its peptide ligand binding preferences. Soon thereafter RAMPs were shown to modulate the binding of calcitonin and amylin peptides to the related calcitonin receptor (CTR) and in the years since an ever-growing number of RAMP-interacting receptors have been identified including most if not all of the fifteen class B GPCRs and several GPCRs from other families. Studies of CLR, CTR, and a handful of other GPCRs revealed that RAMPs are able to modulate various aspects of receptor function including trafficking, ligand binding, and signaling. Here, we review RAMP interactions and functions with an emphasis on class B receptors for which our understanding is most advanced. A key focus is to discuss recent evidence that RAMPs serve as endogenous allosteric modulators of CLR and CTR. We discuss structural studies of RAMP-CLR complexes and CTR and biochemical and pharmacological studies that collectively have significantly expanded our understanding of the mechanistic basis for RAMP modulation of these class B GPCRs. Last, we consider the implications of these findings for drug development targeting RAMP-CLR/CTR complexes.
Keywords: class B GPCR, RAMP, allostery, dynamics, peptide hormone
1. INTRODUCTION
The class B/Secretin family of G protein-coupled receptors (GPCRs) comprises 15 receptors in humans that mediate responses to a diverse collection of peptides that signal in an endocrine, paracrine, or autocrine manner (Hoare, 2005). This family includes two receptors for the calcitonin family peptides (CTR and CLR), two receptors for parathyroid hormone and related peptides (PTH1R and PTH2R), two receptors for the corticotropin releasing factor/urocortin peptide family (CRFR1 and CRFR2), four receptors for glucagon family peptides (GCGR, GLP1R, GLP2R, and GIPR), three receptors for pituitary adenylate cyclase-activating peptide and related peptides (PAC1R, VPAC1R, VPAC2R), and the receptors for secretin (SECR) and growth hormone-releasing hormone (GHRHR). Receptors in this family have clinical relevance for many diseases including migraine, diabetes, cardiovascular disorders, osteoporosis, depression, and cancer. Several peptide therapeutics targeting class B receptors are on the market; with the most successful example being the GLP1R agonists used for type II diabetes (Drucker, 2018). Enormous potential remains for developing novel therapeutics targeting this receptor family and it is expected that recent breakthroughs in the structural biology of class B receptors will facilitate progress in this area (Wootten & Miller, 2019). Most of these receptors, if not all (see section 3), can be modulated by receptor activity-modifying proteins (RAMPs) that are the focus of this chapter. These accessory membrane proteins can affect various aspects of receptor function, which adds considerable complexity to their pharmacology, but also significantly expands opportunities for drug discovery.
Class B receptors have a two-domain structure with an N-terminal extracellular domain (ECD) followed by a transmembrane domain (TMD). The ECD is ~120 amino acids and forms a conserved α/β fold with an N-terminal α-helix connected to a series of β-strands and loops, all stabilized by three conserved disulfide bonds (Pal, Melcher, & Xu, 2012). The TMD forms a 7-transmembrane helical bundle with a single conserved disulfide bond connecting ECL2 and the top of TM3 (Hollenstein et al., 2014). The bundle is similar to that of the class A GPCRs except that the top half is more open to accommodate the binding of large peptides (Hollenstein et al., 2013; Siu et al., 2013). The peptide ligands, which are generally ~30–40 amino acids in length, engage both receptor domains in what is known as the “two-domain” binding model and is thought to occur in two steps (Hoare, 2005). The C-terminal half of the peptide first binds the ECD in a somewhat high-affinity interaction and this then promotes binding of the N-terminal half of the peptide to the TMD, which is otherwise a low-affinity interaction. The latter interaction is responsible for receptor activation. Class B receptors predominantly couple to Gs, but they also couple to Gi and Gq and many signal through β-arrestins.
Structural studies of class B receptors have advanced our understanding of peptide binding and receptor activation, particularly with the recent determinations of full-length CTR, CLR, GCGR, GLP1R, and PTH1R structures by X-ray crystallography or cryo-EM methods (Ehrenmann et al., 2018; Jazayeri et al., 2017; Liang, Khoshouei, Deganutti, et al., 2018; Liang, Khoshouei, Glukhova, et al., 2018; Liang et al., 2017; H. Zhang et al., 2017; H. Zhang et al., 2018; Y. Zhang et al., 2017; Zhao et al., 2019). Most of the peptide ligands adopt continuous α-helical conformations spanning both receptor domains, except for the calcitonin family peptides for which the TMD-binding portions are α-helical and the ECD-binding portions are relatively unstructured (sections 5 and 6). Receptor activation involves TM helix movements similar to those observed in class A GPCRs (Weis & Kobilka, 2018), except that the changes in TM6 are more dramatic with a nearly 90° bend in the middle resulting in the outward movement of TM6 needed to accommodate Gs binding (Liang, Khoshouei, Deganutti, et al., 2018; Liang, Khoshouei, Glukhova, et al., 2018; Liang et al., 2017; Y. Zhang et al., 2017; Zhao et al., 2019). A consistent theme emerging from these structures is that there is inter-domain flexibility in the class B receptors and this can be considerable even in the agonist-bound active state as observed for CTR (Liang et al., 2017; Wootten & Miller, 2019).
The calcitonin and calcitonin-like receptors, CTR and CLR, are the best-studied class B receptors in terms of their modulation by RAMPs so much of this chapter focuses on them. The most notable effect of the RAMPs on CTR and CLR is modulation of their binding preferences for the six calcitonin family peptides: calcitonin (CT), amylin, calcitonin gene-related peptides α and β (αCGRP and βCGRP), adrenomedullin (AM) and adrenomedullin 2/intermedin (AM2/IMD). Here we introduce the functions and clinical applications of these peptides and refer the reader to excellent reviews for more information. The endocrine hormone CT inhibits osteoclast activity and high affinity forms such as salmon CT (sCT) are used to treat hypercalcemia, Paget’s disease, and osteoporosis (Naot, Musson, & Cornish, 2019). Amylin is a glucoregulatory endocrine hormone co-secreted with insulin that decreases food intake, gastric emptying, and glucagon secretion (Hay, Chen, Lutz, Parkes, & Roth, 2015). The amylin analog pramlintide is used as an insulin adjunct therapy for types I and II diabetes. α/βCGRP (hereafter CGRP) are neuropeptides with vasodilator and cardioprotective activities and roles in pain transmission and neurogenic inflammation (Kee, Kodji, & Brain, 2018; Russell, King, Smillie, Kodji, & Brain, 2014). CGRP contributes to migraine headache pathogenesis and monoclonal antibodies that either bind to CGRP or a CGRP receptor have recently obtained regulatory approval for this condition (Edvinsson, Haanes, Warfvinge, & Krause, 2018). AM is critical for lymphatic vasculature and heart development and its functions in the adult include vasodilation, cardioprotection, and promotion of embryo implantation (Kato & Kitamura, 2015; Klein & Caron, 2015; Lenhart & Caron, 2012; Tsuruda, Kato, Kuwasako, & Kitamura, 2019). AM holds promise for treating heart attack, heart failure, lymphedema, infertility, sepsis, and inflammatory bowel disease. AM2/IMD is a vasodilator and it has cardioprotective activity, promotes endothelial barrier integrity, and has roles in angiogenesis and metabolism (Holmes, Campbell, Harbinson, & Bell, 2013; Zhang, Xu, & Wang, 2018). AM2/IMD may be of value for cardiovascular disorders, sepsis, and metabolic syndrome.
In the sections below we begin with a brief history of the discovery of the RAMPs followed by discussion of their interacting GPCR partners identified to date. After this we discuss mechanisms by which the RAMPs modulate class B GPCRs with an emphasis on how they determine CTR/CLR ligand selectivity. We discuss recent studies that provide evidence that an important component of the mechanism involves allosteric modulation by the RAMPs. Last, we discuss the implications of these findings for drug development targeting RAMP-CLR/CTR complexes.
2. Discovery of RAMPs as accessory proteins for CLR and CTR
The RAMPs were discovered through an expression-cloning strategy seeking to identify the CGRP receptor (McLatchie et al., 1998). Using a cDNA library from a human neuroblastoma cell line a clone encoding RAMP1 was found that conferred enhanced CGRP responses in Xenopus oocytes. Rather than encoding a GPCR, RAMP1 encoded a single span transmembrane protein with a small ECD and a short cytoplasmic tail. It was realized that RAMP1 must potentiate the CGRP receptor and co-expression of RAMP1 and CLR in HEK293T cells was shown to yield a functional CGRP receptor at the cell surface. This explained earlier findings that CLR could act as a CGRP receptor in some cell lines, but not others, likely due to differential expression of RAMP1. RAMP2 and −3 were identified based on sequence similarity and co-expression of RAMP2 with CLR conferred preferential response to AM over CGRP. This original landmark report thus identified a novel family of accessory single span transmembrane proteins and showed that they can aid the transport of a class B GPCR to the cell surface and alter its ligand binding preferences.
Soon after the discovery of the RAMPs several groups reported that RAMP1 and −3 confer high-affinity amylin binding and signaling responses when co-expressed with CTR (Armour, Foord, Kenakin, & Chen, 1999; G. Christopoulos et al., 1999; Muff, Buhlmann, Fischer, & Born, 1999). RAMP1 also confers high affinity CGRP binding on CTR (Hay, Christopoulos, Christopoulos, Poyner, & Sexton, 2005; Tilakaratne, Christopoulos, Zumpe, Foord, & Sexton, 2000). RAMP2 could also elicit an amylin receptor when co-expressed with CTR, but this phenotype is harder to observe and was cell-type dependent indicating that cellular context can affect RAMP-GPCR interactions (Tilakaratne et al., 2000). In contrast to CLR, which is dependent on RAMPs for cell surface expression, CTR transports to the cell surface on its own and functions independently of RAMPs as the receptor for CT (Purdue, Tilakaratne, & Sexton, 2002).
The pharmacology of the CT family peptides at the receptors arising from CLR/CTR and their complexes with RAMPs has been thoroughly reviewed (Hay, Garelja, Poyner, & Walker, 2018; Hong, Hay, Quirion, & Poyner, 2012; Poyner et al., 2002). Here we briefly summarize the receptors and their ligand selectivity profiles. Rank orders are for cAMP production, which tends to mirror affinity for radiologand binding experiments, where available. CLR-RAMP1 is the CGRP receptor that is activated with a potency rank order of CGRP > AM ≈ AM2/IMD. CLR-RAMP2 is the AM1 receptor that is activated with the rank order AM > AM2/IMD > CGRP. CLR-RAMP3 is the AM2 receptor that is activated with the rank order AM ≥ AM2/IMD > CGRP. CGRP has a clear preference for the CGRP receptor, whereas AM and AM2/IMD discriminate the three receptors to lesser extents. CTR alone is the CT receptor that is activated with the rank order CT > Amy. CTR-RAMP1 is the AMY1 receptor that is activated roughly equivalently by amylin and CGRP. CTR-RAMP2 and CTR-RAMP3 are the AMY2 and AMY3 receptors that are both activated by amylin.
3. RAMP modulation of additional class B GPCRs
Early efforts to identify additional RAMP interactions took advantage of the poor cell surface expression of the RAMPs when expressed on their own. Thus, cell surface expression of the RAMP when co-expressed with a receptor is indicative of an interaction between the two. Using this assay, RAMPs were reported to interact with the GCGR, the PTH1R and PTH2R, and the VPAC1R (A. Christopoulos et al., 2003). Subsequent studies demonstrated further RAMP interactions with the CRFR1, SCTR, and VPAC2R (Bailey et al., 2019; Harikumar, Simms, Christopoulos, Sexton, & Miller, 2009; Wootten et al., 2013). Notably, in contrast to CLR and CTR, some of the other class B receptors seemed to selectively interact with a subset of the RAMPs (Table 1). The effects of these interactions on the receptors’ functions, where known, are discussed in section 7.
Table 1.
RAMP interactions with human class B GPCRs.
These interactions have only been observed in a high-throughput multiplexed suspension bead immunoassay using dodecylmaltoside-solubilized cells transfected with epitope tagged receptor components.
Newer approaches to catalog putative RAMP-GPCR interactions appear to significantly expand the number of RAMP partners. A bioinformatics study analyzed phylogenetic trees in various organisms and expression correlations of RAMPs and GPCRs and found that RAMPs and GPCRs co-evolved, suggesting that RAMP-GPCR interactions are widespread (Barbash, Lorenzen, Persson, Huber, & Sakmar, 2017). A follow-up study used an experimental high-throughput approach to examine the interactions of the human RAMPs with 23 different GPCRs and identified RAMP interactions with all 15 class B receptors (Lorenzen et al., 2019) (Table 1). This study used a high throughput bead-based immunoassay with detergent-solubilized lysates from cells transfected with epitope-tagged receptors. As intriguing as these findings are, only a few interactions were further validated by an independent assay and the functional consequences of these interactions remains to be tested.
4. RAMP modulation beyond class B receptors
It is worth noting that RAMP effects are not limited to class B GPCRs (Hay & Pioszak, 2016). The first demonstration involved the class C calcium sensing receptor where RAMP1 and −3 facilitated its trafficking to the cell surface in model cell lines (Bouschet, Martin, & Henley, 2005). This finding was confirmed in human thyroid carcinoma cells (Desai, Roberts, Richards, & Skerry, 2014). The class A GPR30 estrogen receptor interacts with RAMP3 and this association may also be important for receptor trafficking. RAMP3 was required for GPR30-mediated sex-dependent cardioprotection in a mouse model demonstrating the relevance of this interaction (Lenhart, Broselid, Barrick, Leeb-Lundberg, & Caron, 2013). The bioinformatics study mentioned above suggested that RAMPs may interact with GPCRs on a widespread scale involving members from each of the GPCR families, but most of these putative interactions have not been validated (Barbash et al., 2017). Last, application of the high-throughput bead-based immunoassay mentioned above found RAMP interactions with several class A chemokine receptors, class A receptors GPR4 and GPR182, and the adhesion receptor ADGRF5 (Lorenzen et al., 2019). These findings await further confirmation.
5. Mechanisms of RAMP modulation of CLR ligand binding
How the RAMPs determine the peptide ligand binding preferences of CLR has been a fundamental question since their discovery. A priori one can imagine that they might perform this function by three possible mechanisms: 1) the RAMPs directly contribute some portion of the peptide binding site and thereby make direct contacts with the ligand, 2) the RAMPs allosterically modulate the conformation and/or dynamics of the receptor without directly contacting the ligand, or 3) the RAMPs provide both direct contacts to the ligand and allosteric modulation of the receptor. There is now good evidence that RAMP modulation of CLR peptide binding involves mechanism 3.
An obvious first question is what are the roles of the ECD and TMD. Chimeric RAMP experiments indicated that the RAMP ECD largely drives peptide preference at CLR (Fitzsimmons, Zhao, & Wank, 2003; Fraser et al., 1999). Bacterial expression and purification of the isolated RAMP1-CLR and RAMP2-CLR ECD complexes showed that the ECDs associate in the absence of the TMDs and the CGRP and AM binding preferences of these complexes generally reflected those of the intact receptors (Hill & Pioszak, 2013; Koth et al., 2010; Moad & Pioszak, 2013; Watkins et al., 2013). We characterized the binding of CGRP, AM, and AM2/IMD to purified, N-glycosylated RAMP1-, RAMP2-, and RAMP3-CLR ECD complexes produced in mammalian cells and found that the peptide selectivity profiles of these complexes were similar, although not identical to those of the intact CGRP, AM1, and AM2 receptors (Roehrkasse, Booe, Lee, Warner, & Pioszak, 2018). Thus, the RAMP ECD is a primary determinant of peptide selectivity, but other regions also contribute.
Structural studies considerably advanced our understanding of RAMP modulation of CLR peptide binding (Table 2). The RAMP1 and −2 ECDs form 3-helix bundles held together by conserved disulfide bonds and their α2-α3 face binds the α1 helix of the CLR ECD (Kusano et al., 2008; Kusano et al., 2012; ter Haar et al., 2010). We determined ECD complex structures with bound C-terminal antagonist peptide fragments including a high-affinity CGRP analog bound to RAMP1-CLR ECD, AM bound to RAMP2-CLR ECD, and AM2/IMD bound to RAMP1-CLR ECD (Booe et al., 2015; Roehrkasse et al., 2018) (Fig. 1A–C). The three peptides occupy a common peptide-binding site that is primarily on CLR and they adopt distinct, relatively unstructured conformations influenced by Pro residues. They share a type I β-turn prior to the C-terminal residue. CT family peptides are C-terminally amidated and this modification is important for activity. The C-terminal F37-amide of CGRP, Y52-amide of AM, and Y47-amide of AM2/IMD anchor the interactions by occupying a pocket in the CLR ECD over the W72 “trp shelf” (Fig. 1A). The amide group hydrogen bonds with the main chain of CLR T122 at the base of the “turret loop” and the side chain rests on the trp shelf. Small molecule CGRP receptor antagonists developed for migraine occupy this same pocket (ter Haar et al., 2010).
Table 2.
Experimentally determined structures of RAMPs, CLR, and CTR.
| Modulator/Receptor/Transducer protein(s) | Ligand | Resolution Å (method) | PDB ID | Reference |
|---|---|---|---|---|
| RAMP1 ECD | None | 2.4 (X-ray) | 2YX8 | (Kusano et al., 2008) |
| RAMP2 ECD | None | 2.1, 2.0 (X-ray) | 2XVT, 3AQE | Unpublished, (Kusano et al., 2012) |
| RAMP1 ECD:CLR ECD complex | None | 2.8 (X-ray) | 3N7P | (ter Haar et al., 2010) |
| RAMP1 ECD:CLR ECD complex | Telcagepant (small molecule antagonist) | 2.9 (X-ray) | 3N7R | (ter Haar et al., 2010) |
| RAMP1 ECD:CLR ECD complex | Olcegepant (small molecule antagonist) | 2.1 (X-ray) | 3N7S | (ter Haar et al., 2010) |
| RAMP2 ECD:CLR ECD complex | None | 2.6 (X-ray) | 3AQF | (Kusano et al., 2012) |
| 1MBP-RAMP1 ECD-CLR ECD fusion | CGRP(27–37) N31D/S34P/K35F (high-affinity antagonist) | 2.4 (X-ray) | 4RWG | (Booe et al., 2015) |
| 1MBP-RAMP2 ECD-CLR ECD fusion | AM(25–52) (peptide antagonist) | 1.8 (X-ray) | 4RWF | (Booe et al., 2015) |
| 1MBP-RAMP1 ECD-CLR ECD fusion | AM2/IMD(29–47) (peptide antagonist) | 2.1 (X-ray) | 6D1U | (Roehrkasse et al., 2018) |
| 1MBP-RAMP1 ECD-CLR ECD fusion | AM(37–52) S45W/K46L/Q50W/Y52F (high-affinity, altered selectivity peptide antagonist) | 2.8 (X-ray) | 5V6Y | (Booe et al., 2018) |
| CTR ECD | salmon CT(8–32) BrPhe22 (peptide antagonist) | 2.1 (X-ray) | 5II0 | (Johansson et al., 2016) |
| CTR:Gs heterotrimer:2Nb35 complex | salmon CT(1–32) (high-affinity peptide agonist) | 4.1, 3.3 (Cryo-EM) | 5UZ7, 6NIY | (Emma dal Maso et al., 2019; Liang et al., 2017) |
| CLR:RAMP1:3DNGs heterotrimer:2Nb35 complex | CGRP(1–37) (peptide agonist) | 3.3 (Cryo-EM) | 6E3Y | (Liang, Khoshouei, Deganutti, et al., 2018) |
Maltose binding protein (MBP) in the fusion was used to promote crystallization.
Nanobody 35 was used to stabilize the Gs heterotrimer for structural studies.
DN=dominant negative mutant of G alpha used to increase complex stability.
Figure 1.
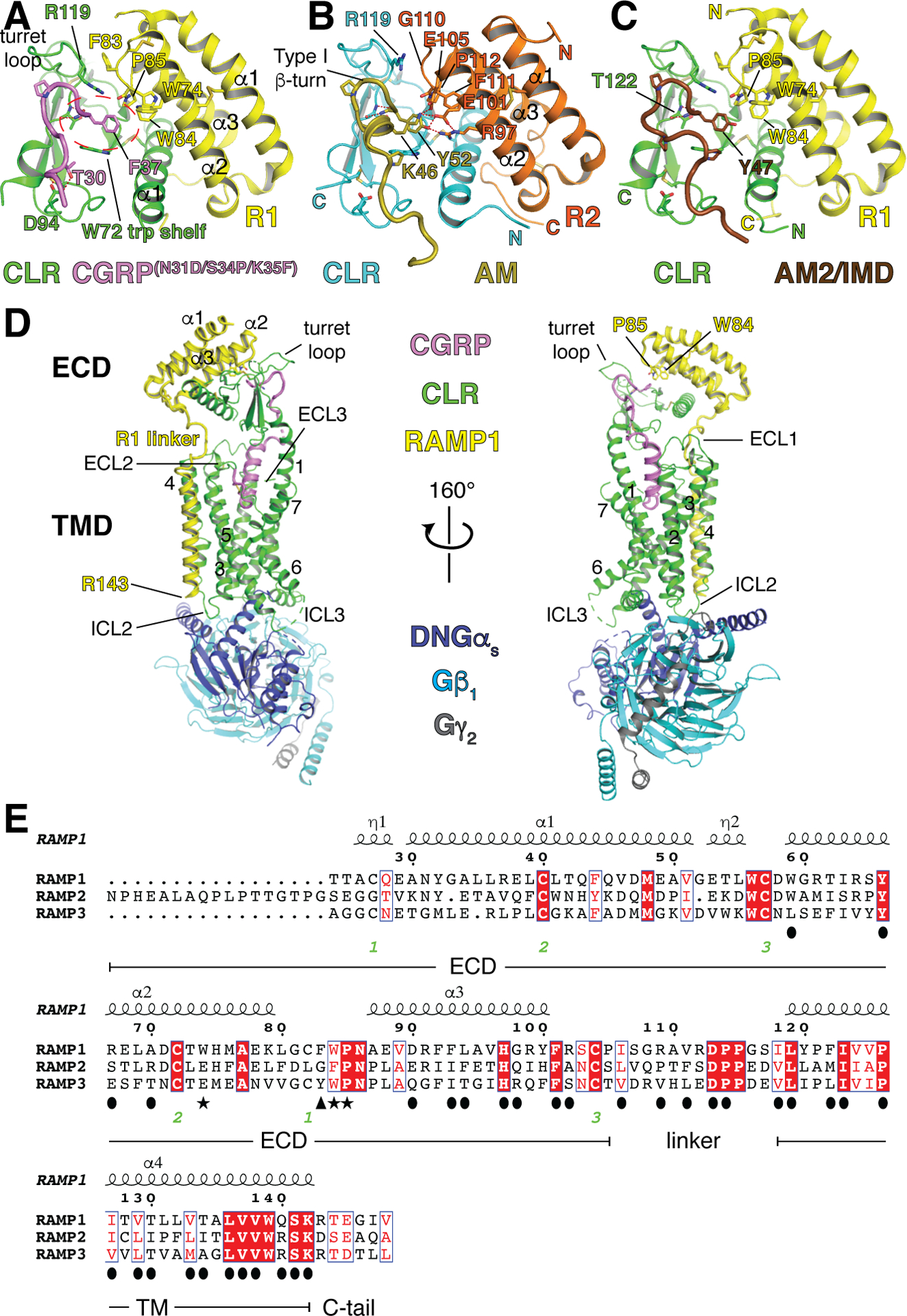
Structures of peptide-bound RAMP-CLR complexes. (A) RAMP1-CLR ECD in complex with a high-affinity variant CGRP C-terminal fragment (PDB 4RWG). The dotted ellipse highlights the pocket that is occupied by the peptide C-terminal residue. (B) RAMP2-CLR ECD in complex with an AM C-terminal fragment (PDB 4RWF). (C) RAMP1-CLR ECD in complex with an AM2/IMD C-terminal fragment (PDB 6D1U). (D) Full-length RAMP1-CLR complex (CGRP receptor) with full-length CGRP and Gs heterotrimer (PDB 6E3Y). (E) Amino acid sequence alignment of the three human RAMPs. The signal peptides are omitted for clarity. Residue numbering above the sequences corresponds to RAMP1 and numbers below indicate disulfide bond connectivity. Filled ovals mark residues at the interface with CLR, stars indicate key residues that augment the ECD binding pocket and in some cases directly contact the peptide ligands, and the filled triangle indicates the residue that appears to determine the position of R119 in the CLR turret loop. The alignment was performed with Clustal Omega (Madeira et al., 2019) and the depiction was made with ESpript 3 (Robert & Gouet, 2014).
The RAMP subunits augment the pocket with residues from their α2-α3 loop and α2 helix, with some of these shared and some unique among the RAMPs (Fig. 1E). A conserved cis-proline in the α2-α3 loop (P85 or P112) makes contact with the peptide C-terminal residue (Fig. 1A–C). Prior to the cis-proline RAMP1 and −3 share W84 and in RAMP1 this can contact CGRP F37 and AM2/IMD Y47 (Fig. 1A, C), whereas RAMP2 has F111 that does not reach the C-terminal residue (Fig. 1B). Three residues from RAMP2 α2, R97, E101, and E105, augment the pocket and are within hydrogen bonding distance of AM Y52 and/or K46 (Fig. 1B). Of these, only E101 significantly contributes to peptide binding because its mutation to Ala diminishes AM cAMP signaling whereas the R97A and E105A mutants had no effect (Booe et al., 2015). At the position equivalent to RAMP2 E101, RAMP1 has W74, which does not contact CGRP or AM2/IMD, and RAMP3 has E74, which presumably would hydrogen bond with AM Y52 and AM2/IMD Y47 (Fig. 1E). So RAMP1 W84 and RAMP2 E101 each provide unique contacts to the peptide that the other RAMP cannot and RAMP3 appears to be a hybrid with E74 and W84 able to provide either contact. This at least partially explains why peptides with a C-terminal Phe show reduced binding at RAMP2-CLR (Booe, Warner, Roehrkasse, Hay, & Pioszak, 2018; Roehrkasse et al., 2018).
Consistent with the structures, mutagenesis experiments indicated that the C-terminal residue of each of the peptides and RAMP1 W84, RAMP2 E101, and RAMP3 E74 are important for peptide-receptor interactions and that RAMP2 E101 and RAMP3 E74 promote AM binding (Moad & Pioszak, 2013; Moore, Gingell, Kane, Hay, & Salvatore, 2010; Qi et al., 2008; Qi et al., 2011; Roehrkasse et al., 2018; Watkins et al., 2013; Watkins et al., 2014). AM K46 mutagenesis also indicated its importance for receptor binding (Moad & Pioszak, 2013; Watkins et al., 2013), but its unclear if this is due to its packing against AM Y52 and CLR W72 or contact with RAMP2. Notably, the ECD-complex binding properties of peptides having reciprocal C-terminal residue exchange and the signaling activity of wild-type agonist peptides at receptors bearing reciprocal exchange of RAMP1/2 residues near the CLR ECD pocket confirmed the importance of AM Y52-RAMP2 E101 contact for binding the AM1 receptor, but the reciprocal peptide and RAMP exchanges were insufficient to swap selectivity (Booe et al., 2015). These data suggested that there must be an allosteric component to RAMP modulation of CLR.
Comparisons of the peptide-bound ECD complexes revealed conformational differences in CLR between the RAMP1- and RAMP2-bound states. The RAMP1/2 helical bundles are shifted in their position against CLR α1 and this shifts α1 (Fig. 2A). These changes likely result from differences in RAMP1/2 residues at the α2-α3 interface (Fig. 2B and 1E), and appear to propagate to the CLR pocket resulting in subtle shape changes that may affect the position of the peptide C-terminal residue (Fig. 2C). There is also a dramatic movement of CLR R119 that seems to be controlled by the RAMPs with RAMP1 F83 pushing it down towards the pocket and RAMP2 G110 allowing it to point up (Fig. 2C). Notably, the CLR R119A mutant is more deleterious in the RAMP1 complex than in the RAMP2 complex (Booe et al., 2015). The contour of the CLR pocket is different in the RAMP1- and RAMP-2 bound states (Fig. 2D, E) and this may impact selectivity because we were able to engineer an AM antagonist variant with altered preference for RAMP1-CLR over RAMP2-CLR via the K46L mutation designed to exploit the pocket shape changes and Y52F to remove RAMP2 E101 contact (Booe et al., 2018). A crystal structure of this variant bound to RAMP1-CLR ECD suggested that it filled out the pocket better than AM would and it may have been able to overcome the R119 down position because of additional affinity-enhancing mutations (Fig. 2F). We cannot rule out the possibility that the K46L effect on selectivity stemmed from loss of K46 contact with RAMP2, however, there is also evidence for an allosteric component to RAMP function from an engineered high-affinity CGRP antagonist variant with the F37Y substitution designed to enable contact with RAMP2 E101. This variant retained better binding to RAMP1-CLR than RAMP2-CLR, which was likely due to CLR ECD conformational differences because only the C-terminal residue of CGRP contacts the RAMP (Booe et al., 2018). The structural, biochemical, and pharmacological data thus support a mechanism in which direct RAMP-peptide contacts and allosteric modulation of CLR cooperate at the level of the ECDs to determine peptide-binding preferences.
Figure 2.
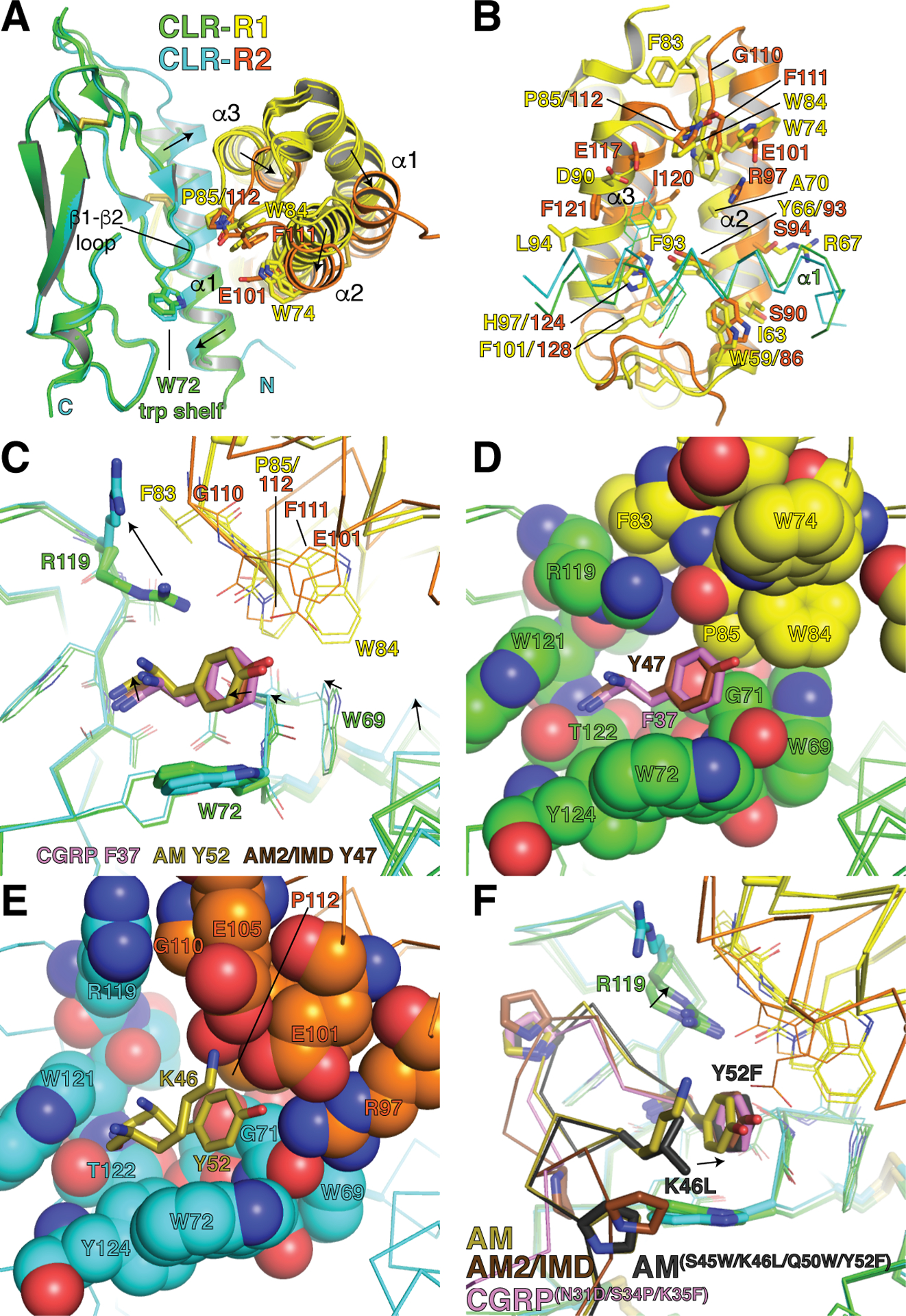
Allosteric modulation of CLR ECD conformation by RAMP1 and RAMP2. (A) Superimpositions of the high-affinity CGRP variant-bound RAMP1-CLR ECD (PDB 4RWG), AM-bound RAMP2-CLR ECD (PDB 4RWF), and AM2/IMD-bound RAMP1-CLR ECD (PDB 6D1U) complexes. For clarity the CLR turret loop is omitted. Arrows indicate shifts occurring from the RAMP1 complexes to the RAMP2 complex. (B) View of the RAMP1/2 α2-α3 interface with the CLR α1 helix (shown as α-carbon trace). (C) Close-up view of the pocket occupied by the peptides’ C-terminal residues, which are shown as sticks. Arrows indicate shifts occurring from the RAMP1 complexes to the RAMP2 complex. (D) View of the pocket in the RAMP1-CLR ECD complexes with residues in space-filling representation. The CGRP F37 and AM2/IMD Y47 C-terminal residues are shown as sticks. (E) View of the pocket in the RAMP2-CLR ECD complex with pocket residues in space-filling representation and the AM K46 and Y52 residues as sticks. (F) Superimpositions of the three structures in panels A and C with RAMP1-CLR ECD bound to a rationally-designed high-affinity, altered selectivity AM variant (PDB 5V6Y). Arrows indicate shifts in the positions of the AM variant C-terminal residue and CLR R119. In all panels the structures were aligned based on the CLR ECD.
The cryo-EM structure of the full-length CGRP receptor with bound CGRP and Gs heterotrimer showed that other than CGRP F37 being near RAMP1 W84 and P85 in the ECD, there were no direct contacts between RAMP1 and the CGRP agonist (Liang, Khoshouei, Deganutti, et al., 2018) (Fig. 1D). Additional modulation of peptide binding arising beyond the level of the ECD complex must therefore be allosteric. The RAMP1 TMD packs against CLR TM3, 4, and 5 and the RAMP linker connecting the ECD and TMD contacts CLR ECL2, which in turn contacts CGRP. RAMP2 and −3 likely occupy the same TM3/4/5 interface given the sequence similarity in the RAMP TMD (Fig. 1E). Differences among the RAMPs in the linker might differentially affect ECL2 to alter peptide binding. Pharmacological studies indicated that some mutations in the CLR TMD, including several in ECL2, have RAMP-dependent effects consistent with allosteric modulation of the CLR TMD and ECL2 by the RAMPs (Watkins et al., 2016; Woolley et al., 2017). Mutagenesis and modeling studies also support RAMP-dependent effects on ECL3, which is distant from the RAMP TMD (Watkins et al., 2016). Notably, allosteric modulation of a GPCR via the extra-helical bundle TM3/4/5 site is not without precedent. A small molecule agoPAM of the class A free fatty acid receptor GPR40 binds in this region (Lu et al., 2017). Last, given the inter-domain flexibility of the class B GPCRs it seems reasonable to speculate that the RAMPs might differentially modulate CLR inter-domain dynamics and this could also affect peptide selectivity. MD simulations indicated that RAMP1 restricts the flexibility of the CLR ECD relative to the TMD (Liang, Khoshouei, Deganutti, et al., 2018), so the other RAMPs probably have similar, although not identical effects. Additional cryo-EM structures of the AM1 and AM2 receptors expected in the near future should provide further insights into how the RAMPs allosterically modulate the CLR TMD and ECD-TMD dynamics.
6. Mechanisms of RAMP modulation of CTR ligand binding
Our understanding of the mechanisms of RAMP modulation of CTR peptide binding is less advanced than for CLR. Structures of CTR alone are available, but we lack structures of CTR-RAMP complexes (Table 2). Biochemical and pharmacological studies revealed similarities with CLR including evidence for an allosteric role of the RAMPs, but there are also differences and unresolved puzzles particularly regarding the role of the peptide C-terminus and its possible contact with RAMPs. The high-affinity ligands for CTR and the CTR-RAMP complexes, CT, amylin, and CGRP, are more similar to each other than the adrenomedullins, but those that require RAMPs (amylin, CGRP) and those that do not (CT) differ at their C-terminus. CT ends with a Pro-amide that is critical for activity, whereas amylin ends with a Tyr-amide as in the adrenomedullins and similar to the Phe-amide of CGRP.
Chimeric RAMP1/2 studies indicated that the RAMP ECD dictated CTR peptide selectivity (Udawela, Christopoulos, Tilakaratne, et al., 2006; Zumpe et al., 2000) and binding studies with purified CTR ECD and RAMP1-CTR and RAMP2-CTR ECD complexes showed that the RAMPs enhanced affinity for amylin and CGRP (Lee et al., 2017; Lee, Hay, & Pioszak, 2016). A crystal structure of the CTR ECD with a C-terminal antagonist fragment of sCT revealed that it occupied a binding site very similar to that in CLR and adopted a CGRP-like conformation except with a type II turn (Fig. 3A) (Johansson et al., 2016). The C-terminal Pro-amide occupied the pocket over the W79 trp shelf with the side chain packing against the shelf and the amide hydrogen bonding with the S129 backbone. Modeling suggested that the RAMPs augment the pocket with the conserved cis-Proline and the α2-α3 loop/α2 helix residues described in section 5 (Fig. 1E), and peptide mutagenesis supported a CGRP-like ECD-bound conformation for amylin (Lee et al., 2016).
Figure 3.
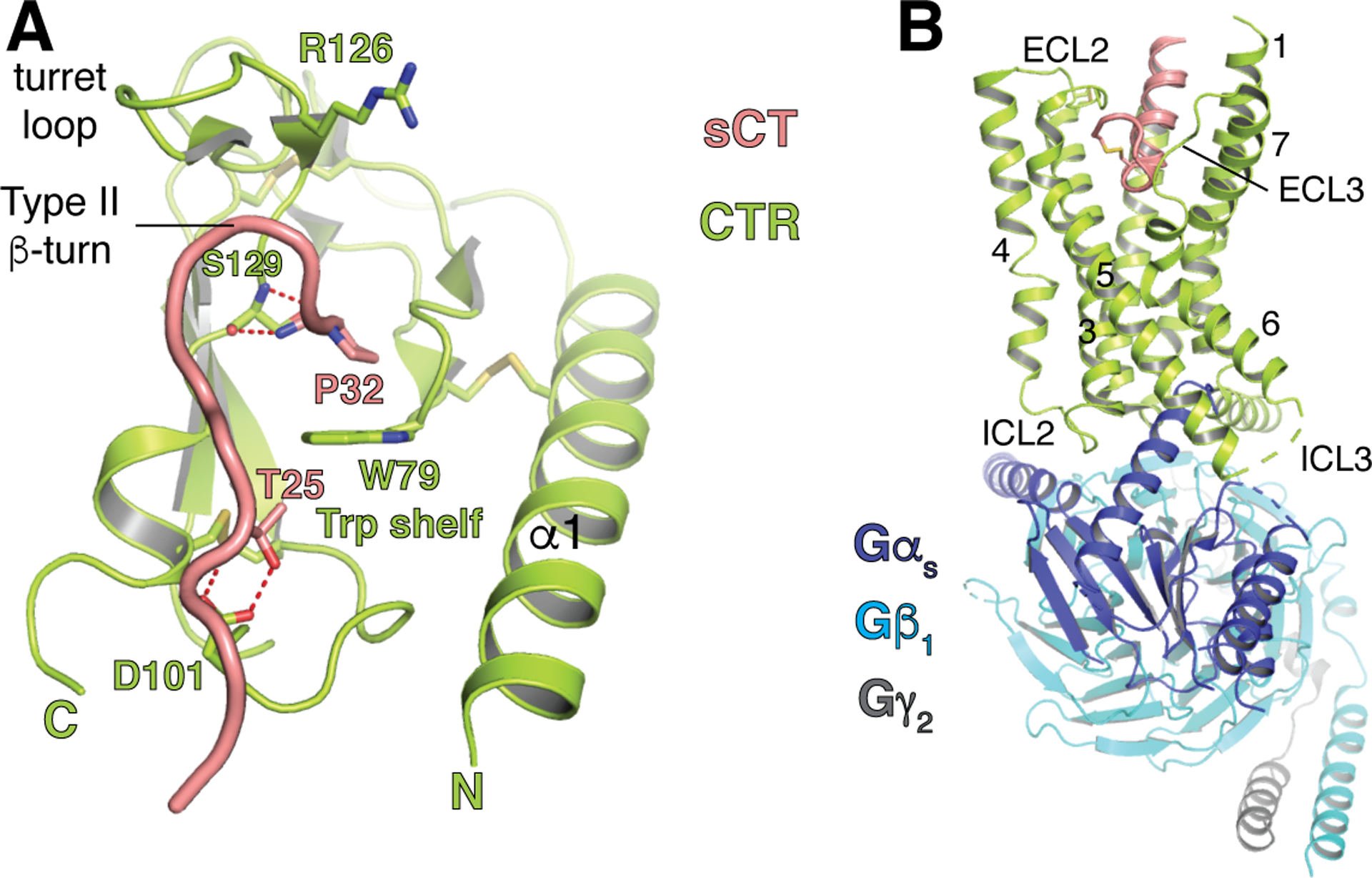
Structures of sCT-bound CTR. (A) CTR ECD in complex with a sCT C-terminal fragment (PDB 5II0). (B) Full-length CTR with full-length sCT and Gs heterotrimer (PDB 6NIY). The ECD and C-terminal half of sCT were not modeled because of weak cryo-EM density for this region, apparently due to significant inter-domain flexibility.
One might predict that the RAMPs enhance amylin affinity in part through contact of RAMP1 W84, RAMP2 E101, or RAMP3 W84/E74 with the amylin C-terminal Y37, however, mutation of RAMP1 W84 or RAMP2 E101 in the purified ECD complexes had no effect on amylin analog binding and the amylin Y37A mutation only modestly diminished binding and signaling at intact AMY1/3 receptors (Bower et al., 2018; Lee et al., 2016). In contrast, loss of the amylin C-terminal amide dramatically diminished binding and signaling at AMY1/3 (Bower et al., 2018). A C-terminal Tyr may contribute to AMY receptor selectivity as in the traditional antagonists AC187 and AC413 (Hay et al., 2005; Young et al., 1994), but substitution of the C-terminal Tyr with Pro in amylin or amylin analogs enhanced RAMP-CTR ECD complex binding and signaling at AMY1/3 (Bower et al., 2018; Lee et al., 2016). Overall, the amylin C-terminal residue side chain appears to be less important to receptor binding than those of CT or CGRP, AM, and AM2/IMD at the CLR complexes and its role in selectivity and RAMP contact remains unresolved. A crystal structure of an amylin-bound RAMP-CTR ECD complex would clarify these issues.
The diminished role of the amylin C-terminal Tyr may reflect more importance of allostery as a mechanism for RAMP modulation of CTR peptide binding. In the CTR ECD structure R126 in the turret loop was in the up position (Fig. 3A) similar to CLR R119 in the RAMP2 complex structure, so RAMP1 would be expected to alter R126 position to reshape the pocket. Consistent with this, mutation of R126 had little effect on signaling at CTR while significantly diminishing signaling at AMY1 (Gingell et al., 2016). In this study RAMP-dependent effects on ligand pharmacology were also observed with mutation of predicted CTR-RAMP interface residues, which may reflect disruption of an allosteric pathway. Moreover, modeling and MD simulations provided support for RAMP1 altering the dynamics of the CTR ECD such that two loops and the C-terminus of the CTR ECD near the peptide-binding site became more flexible in the presence of the RAMP. These results were interpreted in light of the concept of “dynamic” allostery whereby entropically driven changes in flexibility alter global domain dynamics to regulate function (Gingell et al., 2016).
The cryo-EM structure of full-length CTR with bound sCT agonist and Gs heterotrimer showed how the agonist binds the TMD and revealed substantial inter-domain flexibility such that the ECD was not modeled in this structure (Fig. 3B) (Emma dal Maso et al., 2019; Liang et al., 2017). Sequence similarity of CTR and CLR supports the RAMP TMD occupying the same TM3/4/5 site observed in the CGRP receptor so the RAMPs likely contact ECL2 and have no direct contact with the N-terminal half of the peptide. The RAMPs probably restrict the flexibility of the CTR ECD relative to the TMD and this modulation of inter-domain dynamics along with direct modulation of the CTR TMD may contribute to peptide selectivity. Support for the latter comes from extensive pharmacological studies assessing the impacts of mutagenesis of CTR ECL2 and ECL3 in the context of the CTR and the AMY3 receptors (Emma dal Maso et al., 2019; E. Dal Maso et al., 2018; Pham et al., 2019). These provided evidence for RAMP3-mediated changes in ECL2 and ECL3, the latter of which is likely distant from the RAMP. Cryo-EM structures of full-length CTR-RAMP complexes are needed to further advance our understanding of RAMP allosteric modulation of the CTR TMD and ECD-TMD dynamics.
7. RAMP modulation of class B GPCR signaling
RAMPs modulate the signaling of several class B GPCRs, but the mechanistic bases for these effects are poorly understood. Early studies on CTR indicated that RAMP1 and −3 alter G protein coupling efficiency in the AMY receptors and at least part of this is likely due to direct interaction of the RAMP C-tail with the transducer proteins (Morfis et al., 2008; Udawela, Christopoulos, Morfis, et al., 2006; Udawela, Christopoulos, Tilakaratne, et al., 2006). A more recent study provided support for an allosteric role of RAMP3 in altering the CTR signaling profile in the AMY3 receptor (Pham et al., 2019). RAMPs were also reported to bias coupling of CLR to Gs, Gi, and Gq (Weston et al., 2016). RAMP2 alters the G protein coupling efficiency of both VPAC receptors and CRFR1 while having no effect on ligand binding (A. Christopoulos et al., 2003; Wootten et al., 2013). Mixed results have been reported for RAMP2 modulation of GCGR signaling and ligand binding. In one study RAMP2 increased potency and efficacy of cAMP signaling, decreased Gi coupling, and decreased binding of a subset of peptide ligands (Weston et al., 2015). In another study RAMP2 decreased potency while enhancing efficacy of cAMP signaling, decreased efficacy of Ca2+ signaling, abolished β-arrestin recruitment, and had no effect on the binding of several peptide ligands (Cegla et al., 2017). Differences in these studies may be attributable in part to the different cell lines used and/or the use of stable vs. transient expression.
In most cases other than CLR/CTR it is unclear how the RAMPs interact with GPCRs. However, the interaction interface was mapped for the RAMP3-SECR pair (Harikumar et al., 2009). RAMP3 had no effect on the ligand binding and signaling of the SECR, but it did alter trafficking of a mutant receptor. Chimeric and truncated receptor and peptide TMD competition experiments were consistent with the interaction being mediated solely by the TMDs with the RAMP3 TMD contacting SECR TM6/7. This study raises the interesting possibility that the RAMPs may utilize various faces of the GPCR helical bundle for interaction depending on the receptor. It is unclear if any other class B GPCR-RAMP associations involve ECD-ECD interactions.
8. RAMPs in lower organisms
The CT family peptides and their receptors have been mostly studied in vertebrates, but recent work identified their existence in invertebrates (Sekiguchi, 2018). This has implications for understanding the mechanistic basis for RAMP modulation of GPCRs throughout evolution. The amphioxus Branchiostoma floridae (Bf) has a family of three CT-like peptides, a CTR/CLR-like receptor, and three RAMP-like proteins (Sekiguchi et al., 2016). Heterologous expression of the Bf receptor in mammalian cells enabled pharmacological characterization of this system. The three CT-like peptides stimulated cAMP accumulation when the Bf receptor was co-expressed with any one of the three Bf RAMPs, but not in their absence, and the RAMPs differentially modulated the responses to the peptides. Notably, the three Bf peptides all end with a Pro-amide like CT, rather than the Phe- or Tyr-amide residues in CGRP, the adrenomedullins, and amylin. As noted in a recent review by Poyner and colleagues (Simms, Routledge, Uddin, & Poyner, 2019), if the Bf receptors and their peptides are structurally similar to those in humans, then it would appear that the Bf RAMPs do not directly contact the peptides and instead probably function through an allosteric mechanism. The RAMPs may have started as trafficking chaperones and allosteric modulators and the utilization of direct RAMP-ligand contacts in the modulation mechanism may have emerged only as the peptide family expanded in complexity in higher organisms.
9. Drug development outlook for RAMP-CLR/CTR complexes
Novel opportunities for drug development provided by RAMP association with GPCRs has been reviewed (Sexton, Poyner, Simms, Christopoulos, & Hay, 2012; Wootten & Miller, 2019), but it is worth emphasizing a couple of aspects in light of our increasing understanding of the mechanisms of RAMP modulation of CLR and CTR. First, the RAMP-GPCR interface provides expanded opportunities for targeting with small molecules, peptides, and antibodies. This was first demonstrated with the development of small molecule CGRP receptor antagonists for migraine that occupy the pocket in the CLR ECD and make contacts with RAMP1 that enable selectivity (ter Haar et al., 2010). More recently this concept was confirmed and expanded with the development and FDA approval of a monoclonal antibody that binds the CGRP receptor ECD complex to antagonize CGRP signaling for migraine (Edvinsson et al., 2018; Shi et al., 2016). Going forward it should be possible to develop allosteric modulators that bind to various areas at the RAMP-CLR/CTR interface to modulate peptide binding and signaling. The additional protein, RAMP, in RAMP-GPCR complexes presumably creates many more allosteric ligand pockets that could be exploited with different types of molecules.
Second, the structural advances are enabling structure-guided drug design approaches to be applied to these receptors. Our work on this front has yielded short ECD complex binding CGRP, AM, and AM2/IMD antagonist variants with enhanced affinity for the CGRP and AM receptors (Booe et al., 2018; Roehrkasse et al., 2018). In a few cases we have been able to engineer receptor selectivity into these variants, but in general this is very challenging because the endogenous peptides make so few contacts with the RAMP subunits that determine receptor phenotype. Nonetheless, future developments have the potential to lead to agonist peptide variants with enhanced potencies or perhaps long-duration signaling capabilities. It might even become possible to engineer kinetic selectivity into these variants as we obtain a better understanding of the kinetics of their receptor binding. We anticipate significant advances in the coming years for the development of novel therapeutics targeting RAMP-CLR/CTR complexes.
10. CONCLUSION
Recent advances in our understanding of the biochemistry, pharmacology, and structural biology of RAMP-CLR/CTR complexes has revealed that RAMPs act as endogenous allosteric modulators of these class B GPCRs. Allosteric modulation by the RAMPs involves conformational effects on both the ECD and TMD of the receptors and alteration of receptor dynamics is likely also important. RAMP modulation of CLR and CTR determines their peptide binding phenotype and also has the capacity to alter their signaling. With the expanding list of GPCRs that appear to interact with RAMPs it is possible that allosteric modulation of GPCRs by RAMPs is widespread, but more studies are needed in this area. Although RAMP interactions complicate GPCR pharmacology, they provide new opportunities for drug development.
ACKNOWLEDGMENTS
Research in the Pioszak laboratory is supported by National Institutes of Health grant R01GM104251.
Abbreviations
- RAMP
receptor activity-modifying protein
- CLR
calcitonin receptor-like receptor
- CTR
calcitonin receptor
- CGRP
calcitonin gene-related peptide
- AM
adrenomedullin
- AM2/IMD
adrenomedullin 2/intermedin
- CT
calcitonin
- ECD
extracellular domain
- TMD
transmembrane domain
Footnotes
CONFLICT OF INTEREST
A. A. P. is inventor on a patent describing enhanced affinity and altered selectivity AM and CGRP variant peptides.
REFERENECES
- Armour SL, Foord S, Kenakin T, & Chen WJ (1999). Pharmacological characterization of receptor-activity-modifying proteins (RAMPs) and the human calcitonin receptor. J Pharmacol Toxicol Methods, 42(4), 217–224. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/11033437 [DOI] [PubMed] [Google Scholar]
- Bailey S, Harris M, Barkan K, Winfield I, Harper MT, Simms J, … Poyner D (2019). Interactions between RAMP2 and CRF receptors: The effect of receptor subtypes, splice variants and cell context. Biochim Biophys Acta Biomembr, 1861(5), 997–1003. doi: 10.1016/j.bbamem.2019.02.008 [DOI] [PubMed] [Google Scholar]
- Barbash S, Lorenzen E, Persson T, Huber T, & Sakmar TP (2017). GPCRs globally coevolved with receptor activity-modifying proteins, RAMPs. Proc Natl Acad Sci U S A, 114(45), 12015–12020. doi: 10.1073/pnas.1713074114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booe JM, Walker CS, Barwell J, Kuteyi G, Simms J, Jamaluddin MA, … Pioszak AA (2015). Structural Basis for Receptor Activity-Modifying Protein-Dependent Selective Peptide Recognition by a G Protein-Coupled Receptor. Mol Cell, 58(6), 1040–1052. doi: 10.1016/j.molcel.2015.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booe JM, Warner ML, Roehrkasse AM, Hay DL, & Pioszak AA (2018). Probing the Mechanism of Receptor Activity-Modifying Protein Modulation of GPCR Ligand Selectivity through Rational Design of Potent Adrenomedullin and Calcitonin Gene-Related Peptide Antagonists. Mol Pharmacol, 93(4), 355–367. doi: 10.1124/mol.117.110916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouschet T, Martin S, & Henley JM (2005). Receptor-activity-modifying proteins are required for forward trafficking of the calcium-sensing receptor to the plasma membrane. J Cell Sci, 118(Pt 20), 4709–4720. doi: 10.1242/jcs.02598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower RL, Yule L, Rees TA, Deganutti G, Hendrikse ER, Harris PWR, … Hay DL (2018). Molecular Signature for Receptor Engagement in the Metabolic Peptide Hormone Amylin. ACS Pharmacology & Translational Science, 1(1), 32–49. doi: 10.1021/acsptsci.8b00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegla J, Jones BJ, Gardiner JV, Hodson DJ, Marjot T, McGlone ER, … Bloom SR (2017). RAMP2 Influences Glucagon Receptor Pharmacology via Trafficking and Signaling. Endocrinology, 158(8), 2680–2693. doi: 10.1210/en.2016-1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopoulos A, Christopoulos G, Morfis M, Udawela M, Laburthe M, Couvineau A, … Sexton PM (2003). Novel receptor partners and function of receptor activity-modifying proteins. J Biol Chem, 278(5), 3293–3297. doi: 10.1074/jbc.C200629200 [DOI] [PubMed] [Google Scholar]
- Christopoulos G, Perry KJ, Morfis M, Tilakaratne N, Gao Y, Fraser NJ, … Sexton PM (1999). Multiple amylin receptors arise from receptor activity-modifying protein interaction with the calcitonin receptor gene product. Mol Pharmacol, 56(1), 235–242. doi: 10.1124/mol.56.1.235 [DOI] [PubMed] [Google Scholar]
- dal Maso E, Glukhova A, Zhu Y, Garcia-Nafria J, Tate CG, Atanasio S, … Sexton PM (2019). The Molecular Control of Calcitonin Receptor Signaling. ACS Pharmacology & Translational Science, 2(1), 31–51. doi: 10.1021/acsptsci.8b00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Maso E, Zhu Y, Pham V, Reynolds CA, Deganutti G, Hick CA, … Wootten D (2018). Extracellular loops 2 and 3 of the calcitonin receptor selectively modify agonist binding and efficacy. Biochem Pharmacol, 150, 214–244. doi: 10.1016/j.bcp.2018.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai AJ, Roberts DJ, Richards GO, & Skerry TM (2014). Role of receptor activity modifying protein 1 in function of the calcium sensing receptor in the human TT thyroid carcinoma cell line. PLoS One, 9(1), e85237. doi: 10.1371/journal.pone.0085237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drucker DJ (2018). Mechanisms of Action and Therapeutic Application of Glucagon-like Peptide-1. Cell Metab, 27(4), 740–756. doi: 10.1016/j.cmet.2018.03.001 [DOI] [PubMed] [Google Scholar]
- Edvinsson L, Haanes KA, Warfvinge K, & Krause DN (2018). CGRP as the target of new migraine therapies - successful translation from bench to clinic. Nat Rev Neurol, 14(6), 338–350. doi: 10.1038/s41582-018-0003-1 [DOI] [PubMed] [Google Scholar]
- Ehrenmann J, Schoppe J, Klenk C, Rappas M, Kummer L, Dore AS, & Pluckthun A (2018). High-resolution crystal structure of parathyroid hormone 1 receptor in complex with a peptide agonist. Nat Struct Mol Biol, 25(12), 1086–1092. doi: 10.1038/s41594-018-0151-4 [DOI] [PubMed] [Google Scholar]
- Fitzsimmons TJ, Zhao X, & Wank SA (2003). The extracellular domain of receptor activity-modifying protein 1 is sufficient for calcitonin receptor-like receptor function. J Biol Chem, 278(16), 14313–14320. doi: 10.1074/jbc.M211946200 [DOI] [PubMed] [Google Scholar]
- Fraser NJ, Wise A, Brown J, McLatchie LM, Main MJ, & Foord SM (1999). The amino terminus of receptor activity modifying proteins is a critical determinant of glycosylation state and ligand binding of calcitonin receptor-like receptor. Mol Pharmacol, 55(6), 1054–1059. doi: 10.1124/mol.55.6.1054 [DOI] [PubMed] [Google Scholar]
- Gingell JJ, Simms J, Barwell J, Poyner DR, Watkins HA, Pioszak AA, … Hay DL (2016). An allosteric role for receptor activity-modifying proteins in defining GPCR pharmacology. Cell Discov, 2, 16012. doi: 10.1038/celldisc.2016.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikumar KG, Simms J, Christopoulos G, Sexton PM, & Miller LJ (2009). Molecular basis of association of receptor activity-modifying protein 3 with the family B G protein-coupled secretin receptor. Biochemistry, 48(49), 11773–11785. doi: 10.1021/bi901326k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay DL, Chen S, Lutz TA, Parkes DG, & Roth JD (2015). Amylin: Pharmacology, Physiology, and Clinical Potential. Pharmacol Rev, 67(3), 564–600. doi: 10.1124/pr.115.010629 [DOI] [PubMed] [Google Scholar]
- Hay DL, Christopoulos G, Christopoulos A, Poyner DR, & Sexton PM (2005). Pharmacological discrimination of calcitonin receptor: receptor activity-modifying protein complexes. Mol Pharmacol, 67(5), 1655–1665. doi: 10.1124/mol.104.008615 [DOI] [PubMed] [Google Scholar]
- Hay DL, Garelja ML, Poyner DR, & Walker CS (2018). Update on the pharmacology of calcitonin/CGRP family of peptides: IUPHAR Review 25. Br J Pharmacol, 175(1), 3–17. doi: 10.1111/bph.14075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay DL, & Pioszak AA (2016). Receptor Activity-Modifying Proteins (RAMPs): New Insights and Roles. Annu Rev Pharmacol Toxicol, 56, 469–487. doi: 10.1146/annurev-pharmtox-010715-103120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill HE, & Pioszak AA (2013). Bacterial expression and purification of a heterodimeric adrenomedullin receptor extracellular domain complex using DsbC-assisted disulfide shuffling. Protein Expr Purif, 88(1), 107–113. doi: 10.1016/j.pep.2012.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoare SR (2005). Mechanisms of peptide and nonpeptide ligand binding to Class B G-protein-coupled receptors. Drug Discov Today, 10(6), 417–427. doi: 10.1016/S1359-6446(05)03370-2 [DOI] [PubMed] [Google Scholar]
- Hollenstein K, de Graaf C, Bortolato A, Wang MW, Marshall FH, & Stevens RC (2014). Insights into the structure of class B GPCRs. Trends Pharmacol Sci, 35(1), 12–22. doi: 10.1016/j.tips.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenstein K, Kean J, Bortolato A, Cheng RK, Dore AS, Jazayeri A, … Marshall FH (2013). Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature, 499(7459), 438–443. doi: 10.1038/nature12357 [DOI] [PubMed] [Google Scholar]
- Holmes D, Campbell M, Harbinson M, & Bell D (2013). Protective effects of intermedin on cardiovascular, pulmonary and renal diseases: comparison with adrenomedullin and CGRP. Curr Protein Pept Sci, 14(4), 294–329. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/23745696 [DOI] [PubMed] [Google Scholar]
- Hong Y, Hay DL, Quirion R, & Poyner DR (2012). The pharmacology of adrenomedullin 2/intermedin. Br J Pharmacol, 166(1), 110–120. doi: 10.1111/j.1476-5381.2011.01530.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Rappas M, Brown AJH, Kean J, Errey JC, Robertson NJ, … Marshall FH (2017). Crystal structure of the GLP-1 receptor bound to a peptide agonist. Nature, 546(7657), 254–258. doi: 10.1038/nature22800 [DOI] [PubMed] [Google Scholar]
- Johansson E, Hansen JL, Hansen AM, Shaw AC, Becker P, Schaffer L, & Reedtz-Runge S (2016). Type II Turn of Receptor-bound Salmon Calcitonin Revealed by X-ray Crystallography. J Biol Chem, 291(26), 13689–13698. doi: 10.1074/jbc.M116.726034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J, & Kitamura K (2015). Bench-to-bedside pharmacology of adrenomedullin. Eur J Pharmacol, 764, 140–148. doi: 10.1016/j.ejphar.2015.06.061 [DOI] [PubMed] [Google Scholar]
- Kee Z, Kodji X, & Brain SD (2018). The Role of Calcitonin Gene Related Peptide (CGRP) in Neurogenic Vasodilation and Its Cardioprotective Effects. Front Physiol, 9, 1249. doi: 10.3389/fphys.2018.01249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein KR, & Caron KM (2015). Adrenomedullin in lymphangiogenesis: from development to disease. Cell Mol Life Sci, 72(16), 3115–3126. doi: 10.1007/s00018-015-1921-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koth CM, Abdul-Manan N, Lepre CA, Connolly PJ, Yoo S, Mohanty AK, … Moore JM (2010). Refolding and characterization of a soluble ectodomain complex of the calcitonin gene-related peptide receptor. Biochemistry, 49(9), 1862–1872. doi: 10.1021/bi901848m [DOI] [PubMed] [Google Scholar]
- Kusano S, Kukimoto-Niino M, Akasaka R, Toyama M, Terada T, Shirouzu M, … Yokoyama S (2008). Crystal structure of the human receptor activity-modifying protein 1 extracellular domain. Protein Sci, 17(11), 1907–1914. doi: 10.1110/ps.036012.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano S, Kukimoto-Niino M, Hino N, Ohsawa N, Okuda K, Sakamoto K, … Yokoyama S (2012). Structural basis for extracellular interactions between calcitonin receptor-like receptor and receptor activity-modifying protein 2 for adrenomedullin-specific binding. Protein Sci, 21(2), 199–210. doi: 10.1002/pro.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Booe JM, Gingell JJ, Sjoelund V, Hay DL, & Pioszak AA (2017). N-Glycosylation of Asparagine 130 in the Extracellular Domain of the Human Calcitonin Receptor Significantly Increases Peptide Hormone Affinity. Biochemistry, 56(26), 3380–3393. doi: 10.1021/acs.biochem.7b00256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Hay DL, & Pioszak AA (2016). Calcitonin and Amylin Receptor Peptide Interaction Mechanisms: INSIGHTS INTO PEPTIDE-BINDING MODES AND ALLOSTERIC MODULATION OF THE CALCITONIN RECEPTOR BY RECEPTOR ACTIVITY-MODIFYING PROTEINS. J Biol Chem, 291(16), 8686–8700. doi: 10.1074/jbc.M115.713628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhart PM, Broselid S, Barrick CJ, Leeb-Lundberg LM, & Caron KM (2013). G-protein-coupled receptor 30 interacts with receptor activity-modifying protein 3 and confers sex-dependent cardioprotection. J Mol Endocrinol, 51(1), 191–202. doi: 10.1530/JME-13-0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhart PM, & Caron KM (2012). Adrenomedullin and pregnancy: perspectives from animal models to humans. Trends Endocrinol Metab, 23(10), 524–532. doi: 10.1016/j.tem.2012.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang YL, Khoshouei M, Deganutti G, Glukhova A, Koole C, Peat TS, … Sexton PM (2018). Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor. Nature, 561(7724), 492–497. doi: 10.1038/s41586-018-0535-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang YL, Khoshouei M, Glukhova A, Furness SGB, Zhao P, Clydesdale L, … Wootten D (2018). Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex. Nature, 555(7694), 121–125. doi: 10.1038/nature25773 [DOI] [PubMed] [Google Scholar]
- Liang YL, Khoshouei M, Radjainia M, Zhang Y, Glukhova A, Tarrasch J, … Sexton PM (2017). Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature, 546(7656), 118–123. doi: 10.1038/nature22327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen E, Dodig-Crnkovic T, Kotliar IB, Pin E, Ceraudo E, Vaughan RD, … Sakmar TP (2019). Multiplexed analysis of the secretin-like GPCR-RAMP interactome. Sci Adv, 5(9), eaaw2778. doi: 10.1126/sciadv.aaw2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Byrne N, Wang J, Bricogne G, Brown FK, Chobanian HR, … Soisson SM (2017). Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40. Nat Struct Mol Biol, 24(7), 570–577. doi: 10.1038/nsmb.3417 [DOI] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, … Lopez R (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res, 47(W1), W636–W641. doi: 10.1093/nar/gkz268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLatchie LM, Fraser NJ, Main MJ, Wise A, Brown J, Thompson N, … Foord SM (1998). RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature, 393(6683), 333–339. doi: 10.1038/30666 [DOI] [PubMed] [Google Scholar]
- Moad HE, & Pioszak AA (2013). Selective CGRP and adrenomedullin peptide binding by tethered RAMP-calcitonin receptor-like receptor extracellular domain fusion proteins. Protein Sci, 22(12), 1775–1785. doi: 10.1002/pro.2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore EL, Gingell JJ, Kane SA, Hay DL, & Salvatore CA (2010). Mapping the CGRP receptor ligand binding domain: tryptophan-84 of RAMP1 is critical for agonist and antagonist binding. Biochem Biophys Res Commun, 394(1), 141–145. doi: 10.1016/j.bbrc.2010.02.131 [DOI] [PubMed] [Google Scholar]
- Morfis M, Tilakaratne N, Furness SG, Christopoulos G, Werry TD, Christopoulos A, & Sexton PM (2008). Receptor activity-modifying proteins differentially modulate the G protein-coupling efficiency of amylin receptors. Endocrinology, 149(11), 5423–5431. doi: 10.1210/en.2007-1735 [DOI] [PubMed] [Google Scholar]
- Muff R, Buhlmann N, Fischer JA, & Born W (1999). An amylin receptor is revealed following co-transfection of a calcitonin receptor with receptor activity modifying proteins-1 or −3. Endocrinology, 140(6), 2924–2927. doi: 10.1210/endo.140.6.6930 [DOI] [PubMed] [Google Scholar]
- Naot D, Musson DS, & Cornish J (2019). The Activity of Peptides of the Calcitonin Family in Bone. Physiol Rev, 99(1), 781–805. doi: 10.1152/physrev.00066.2017 [DOI] [PubMed] [Google Scholar]
- Pal K, Melcher K, & Xu HE (2012). Structure and mechanism for recognition of peptide hormones by Class B G-protein-coupled receptors. Acta Pharmacol Sin, 33(3), 300–311. doi: 10.1038/aps.2011.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham V, Zhu Y, Dal Maso E, Reynolds CA, Deganutti G, Atanasio S, … Sexton PM (2019). Deconvoluting the Molecular Control of Binding and Signaling at the Amylin 3 Receptor: RAMP3 Alters Signal Propagation through Extracellular Loops of the Calcitonin Receptor. ACS Pharmacology & Translational Science, 2(3), 183–197. doi: 10.1021/acsptsci.9b00010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyner DR, Sexton PM, Marshall I, Smith DM, Quirion R, Born W, … Foord SM (2002). International Union of Pharmacology. XXXII. The mammalian calcitonin gene-related peptides, adrenomedullin, amylin, and calcitonin receptors. Pharmacol Rev, 54(2), 233–246. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/12037140 [DOI] [PubMed] [Google Scholar]
- Purdue BW, Tilakaratne N, & Sexton PM (2002). Molecular pharmacology of the calcitonin receptor. Receptors Channels, 8(3–4), 243–255. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/12529940 [PubMed] [Google Scholar]
- Qi T, Christopoulos G, Bailey RJ, Christopoulos A, Sexton PM, & Hay DL (2008). Identification of N-terminal receptor activity-modifying protein residues important for calcitonin gene-related peptide, adrenomedullin, and amylin receptor function. Mol Pharmacol, 74(4), 1059–1071. doi: 10.1124/mol.108.047142 [DOI] [PubMed] [Google Scholar]
- Qi T, Ly K, Poyner DR, Christopoulos G, Sexton PM, & Hay DL (2011). Structure-function analysis of amino acid 74 of human RAMP1 and RAMP3 and its role in peptide interactions with adrenomedullin and calcitonin gene-related peptide receptors. Peptides, 32(5), 1060–1067. doi: 10.1016/j.peptides.2011.03.004 [DOI] [PubMed] [Google Scholar]
- Robert X, & Gouet P (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res, 42(Web Server issue), W320–324. doi: 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehrkasse AM, Booe JM, Lee SM, Warner ML, & Pioszak AA (2018). Structure-function analyses reveal a triple beta-turn receptor-bound conformation of adrenomedullin 2/intermedin and enable peptide antagonist design. J Biol Chem, 293(41), 15840–15854. doi: 10.1074/jbc.RA118.005062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell FA, King R, Smillie SJ, Kodji X, & Brain SD (2014). Calcitonin gene-related peptide: physiology and pathophysiology. Physiol Rev, 94(4), 1099–1142. doi: 10.1152/physrev.00034.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi T (2018). The Calcitonin/Calcitonin Gene-Related Peptide Family in Invertebrate Deuterostomes. Front Endocrinol (Lausanne), 9, 695. doi: 10.3389/fendo.2018.00695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi T, Kuwasako K, Ogasawara M, Takahashi H, Matsubara S, Osugi T, … Satake H (2016). Evidence for Conservation of the Calcitonin Superfamily and Activity-regulating Mechanisms in the Basal Chordate Branchiostoma floridae: INSIGHTS INTO THE MOLECULAR AND FUNCTIONAL EVOLUTION IN CHORDATES. J Biol Chem, 291(5), 2345–2356. doi: 10.1074/jbc.M115.664003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton PM, Poyner DR, Simms J, Christopoulos A, & Hay DL (2012). RAMPs as drug targets. Adv Exp Med Biol, 744, 61–74. doi: 10.1007/978-1-4614-2364-5_6 [DOI] [PubMed] [Google Scholar]
- Shi L, Lehto SG, Zhu DX, Sun H, Zhang J, Smith BP, … Xu C (2016). Pharmacologic Characterization of AMG 334, a Potent and Selective Human Monoclonal Antibody against the Calcitonin Gene-Related Peptide Receptor. J Pharmacol Exp Ther, 356(1), 223–231. doi: 10.1124/jpet.115.227793 [DOI] [PubMed] [Google Scholar]
- Simms J, Routledge S, Uddin R, & Poyner D (2019). The Structure of the CGRP and Related Receptors. Handb Exp Pharmacol, 255, 23–36. doi: 10.1007/164_2018_132 [DOI] [PubMed] [Google Scholar]
- Siu FY, He M, de Graaf C, Han GW, Yang D, Zhang Z, … Stevens RC (2013). Structure of the human glucagon class B G-protein-coupled receptor. Nature, 499(7459), 444–449. doi: 10.1038/nature12393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Haar E, Koth CM, Abdul-Manan N, Swenson L, Coll JT, Lippke JA, … Moore JM (2010). Crystal structure of the ectodomain complex of the CGRP receptor, a class-B GPCR, reveals the site of drug antagonism. Structure, 18(9), 1083–1093. doi: 10.1016/j.str.2010.05.014 [DOI] [PubMed] [Google Scholar]
- Tilakaratne N, Christopoulos G, Zumpe ET, Foord SM, & Sexton PM (2000). Amylin receptor phenotypes derived from human calcitonin receptor/RAMP coexpression exhibit pharmacological differences dependent on receptor isoform and host cell environment. J Pharmacol Exp Ther, 294(1), 61–72. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/10871296 [PubMed] [Google Scholar]
- Tsuruda T, Kato J, Kuwasako K, & Kitamura K (2019). Adrenomedullin: Continuing to explore cardioprotection. Peptides, 111, 47–54. doi: 10.1016/j.peptides.2018.03.012 [DOI] [PubMed] [Google Scholar]
- Udawela M, Christopoulos G, Morfis M, Christopoulos A, Ye S, Tilakaratne N, & Sexton PM (2006). A critical role for the short intracellular C terminus in receptor activity-modifying protein function. Mol Pharmacol, 70(5), 1750–1760. doi: 10.1124/mol.106.024257 [DOI] [PubMed] [Google Scholar]
- Udawela M, Christopoulos G, Tilakaratne N, Christopoulos A, Albiston A, & Sexton PM (2006). Distinct receptor activity-modifying protein domains differentially modulate interaction with calcitonin receptors. Mol Pharmacol, 69(6), 1984–1989. doi: 10.1124/mol.105.021915 [DOI] [PubMed] [Google Scholar]
- Watkins HA, Au M, Bobby R, Archbold JK, Abdul-Manan N, Moore JM, … Hay DL (2013). Identification of key residues involved in adrenomedullin binding to the AM1 receptor. Br J Pharmacol, 169(1), 143–155. doi: 10.1111/bph.12118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins HA, Chakravarthy M, Abhayawardana RS, Gingell JJ, Garelja M, Pardamwar M, … Hay DL (2016). Receptor Activity-modifying Proteins 2 and 3 Generate Adrenomedullin Receptor Subtypes with Distinct Molecular Properties. J Biol Chem, 291(22), 11657–11675. doi: 10.1074/jbc.M115.688218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins HA, Walker CS, Ly KN, Bailey RJ, Barwell J, Poyner DR, & Hay DL (2014). Receptor activity-modifying protein-dependent effects of mutations in the calcitonin receptor-like receptor: implications for adrenomedullin and calcitonin gene-related peptide pharmacology. Br J Pharmacol, 171(3), 772–788. doi: 10.1111/bph.12508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis WI, & Kobilka BK (2018). The Molecular Basis of G Protein-Coupled Receptor Activation. Annu Rev Biochem, 87, 897–919. doi: 10.1146/annurev-biochem-060614-033910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston C, Lu J, Li N, Barkan K, Richards GO, Roberts DJ, … Ladds G (2015). Modulation of Glucagon Receptor Pharmacology by Receptor Activity-modifying Protein-2 (RAMP2). J Biol Chem, 290(38), 23009–23022. doi: 10.1074/jbc.M114.624601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston C, Winfield I, Harris M, Hodgson R, Shah A, Dowell SJ, … Ladds G (2016). Receptor Activity-modifying Protein-directed G Protein Signaling Specificity for the Calcitonin Gene-related Peptide Family of Receptors. J Biol Chem, 291(42), 21925–21944. doi: 10.1074/jbc.M116.751362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley MJ, Reynolds CA, Simms J, Walker CS, Mobarec JC, Garelja ML, … Hay DL (2017). Receptor activity-modifying protein dependent and independent activation mechanisms in the coupling of calcitonin gene-related peptide and adrenomedullin receptors to Gs. Biochem Pharmacol, 142, 96–110. doi: 10.1016/j.bcp.2017.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootten D, Lindmark H, Kadmiel M, Willcockson H, Caron KM, Barwell J, … Poyner DR (2013). Receptor activity modifying proteins (RAMPs) interact with the VPAC2 receptor and CRF1 receptors and modulate their function. Br J Pharmacol, 168(4), 822–834. doi: 10.1111/j.1476-5381.2012.02202.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootten D, & Miller LJ (2019). Structural Basis for Allosteric Modulation of Class B G Protein-Coupled Receptors. Annu Rev Pharmacol Toxicol. doi: 10.1146/annurev-pharmtox-010919-023301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AA, Gedulin B, Gaeta LS, Prickett KS, Beaumont K, Larson E, & Rink TJ (1994). Selective amylin antagonist suppresses rise in plasma lactate after intravenous glucose in the rat. Evidence for a metabolic role of endogenous amylin. FEBS Lett, 343(3), 237–241. doi: 10.1016/0014-5793(94)80563-6 [DOI] [PubMed] [Google Scholar]
- Zhang H, Qiao A, Yang D, Yang L, Dai A, de Graaf C, … Wu B (2017). Structure of the full-length glucagon class B G-protein-coupled receptor. Nature, 546(7657), 259–264. doi: 10.1038/nature22363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Qiao A, Yang L, Van Eps N, Frederiksen KS, Yang D, … Wu B (2018). Structure of the glucagon receptor in complex with a glucagon analogue. Nature, 553(7686), 106–110. doi: 10.1038/nature25153 [DOI] [PubMed] [Google Scholar]
- Zhang SY, Xu MJ, & Wang X (2018). Adrenomedullin 2/intermedin: a putative drug candidate for treatment of cardiometabolic diseases. Br J Pharmacol, 175(8), 1230–1240. doi: 10.1111/bph.13814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sun B, Feng D, Hu H, Chu M, Qu Q, … Skiniotis G (2017). Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature, 546(7657), 248–253. doi: 10.1038/nature22394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LH, Ma S, Sutkeviciute I, Shen DD, Zhou XE, de Waal PW, … Zhang Y (2019). Structure and dynamics of the active human parathyroid hormone receptor-1. Science, 364(6436), 148–153. doi: 10.1126/science.aav7942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumpe ET, Tilakaratne N, Fraser NJ, Christopoulos G, Foord SM, & Sexton PM (2000). Multiple ramp domains are required for generation of amylin receptor phenotype from the calcitonin receptor gene product. Biochem Biophys Res Commun, 267(1), 368–372. doi: 10.1006/bbrc.1999.1943 [DOI] [PubMed] [Google Scholar]


