Abstract
Objective(s):
Resistance to carbapenems as the last line for controlling resistant bacteria is increasing due to production of carbapenemase. The aim of this study was to detect the plasmid-encoded carbapenemases using phenotypic methods and multiplex PCR among the multi-drug resistant (MDR) isolates from patients with urinary tract infection (UTI) in northern Iran.
Materials and Methods:
Antimicrobial susceptibility testing and extended spectrum β-lactamase (ESBL) production test were performed for 91 MDR Escherichia coli strains by disc diffusion and double disk synergy tests (DDST), respectively. Carbapenemases production was confirmed using Hodge test, EDTA double disk synergy test (EDST) and combined disk test (CDT). The isolates were subjected to PCR targeting blaIMP, blaVIM, blaKPC and blaOXA-48 β-Lactamase genes.
Results:
Resistance of isolates to 1st, 2nd, 3rd, and 4th generations of cephalosporins, carbapenems, and penicillins were 73%, 84.5%, 62%, 37.5%, 17.5%, and 76%, respectively. Based on CDT and Hodge test, 1 (3%) and based on EDST, 2 (6%) of 33 ESBL producers synthesize a type of carbapenemase. The frequency of blaIMP, blaVIM, blaKPC, and blaOXA-48 genes was 8.7%, 9.8%, 2.1%, and 15.3%, respectively. Existence of blaIMP conferred more resistance to cephalotin, fosfomycin, and piperacillin (P≤0.01) and carrying blaVIM caused more resistance to cephalotin, cefepime, and ceftazidime (P≤0.01). The presence of blaKPC conferred more resistance to cephalotin and presence of blaOXA-48 caused more resistance to chloramphenicol and piperacillin (P≤0.05).
Conclusion:
Identification and controlling of this nearly low frequent ESBL and carbapenemase producing strains are important due to the presence of plasmid genes encoding carbapenemase.
Key Words: Carbapenemase, Cephalosporines, Multiplex PCR, Plasmid, Resistance
Introduction
Over the last decade, the emergence of resistance to carbapenems, has become a major public health crisis worldwide especially in developing countries, due to their rapid spread and the lack of development of new antimicrobial agents (1-3). Resistance to carbapenems was reported in 86% of Gram-negative bacterial strains in Iran in 2010 (4).
Throughout 2006–2018, incremental trend of resistance to carbapenems was evident in Iran (5). The least rate of resistance was reported in year 2010 at Milad Hospital (6). During years 2012–2015, a study evaluated the trend of antibiotic resistance in Acinetobacter baumannii. Based on their study results, resistance rate in A. baumannii increased from 83% in year 2012 to 96% in year 2015. Also 100% of A. baumannii isolates during these years were resistant to carbapenem (7). No specific trend was followed by the other microorganisms’ resistance patterns. Most of the carbapenem-resistant strains were isolated from burn patients, and many studies which were conducted in this group were from Motahari Hospital, Tehran, Iran (5).
Since 1993, wide varieties of carbapenemases have been recognized that belong to three molecular classes: the Ambler class A, B, and D β-lactamases (3).
In this investigation, four carbapenemases including IMP, VIM, KPC, and OXA-48 were studied. KPC stands for Klebsiella pneumoniae carbapenemase and is a class A β-lactamase that has the ability to hydrolyze penicillins, cephalosporins, and carbapenems. KPC was initially reported from a K. pneumoniae strain isolated in North Carolina in 1996 (8).
Class B metallo-β-lactamases (MBLs) are mostly of the Verona integron-encoded metallo-β-lactamase (VIM) and IMP types and, more recently, of the New Delhi metallo-β-lactamases-1 (NDM-1) type (3, 9). MBLs can hydrolyze all β-lactams except monobactam (e.g., aztreonam). Their activity is inhibited by EDTA but not by clavulanic acid (9).
The IMP-type enzymes, initially reported in 1991 in a Serratia marcescens clinical isolate from Japan (2), have now been reported all over the world in Enterobacteriaceae, Pseudomonas aeruginosa, and A. baumannii (2). The most commonly found class B carbapenemases are of the VIM type, which has been identified in all continents (2). The death rates associated with MBL producers are high (18% to 67%)(10, 11).
OXA stands for oxacillinase and is a diverse group of β-lactamases classified to class D. Some of OXA β-lactamases additionally have the capability to hydrolyze carbapenems. OXA-48 was first found in a K. pneumoniae strain isolated in Turkey in 2001 (12). Its production mediates resistance to penicillins and carbapenems (especially imipenem), but not to cephalosporins. In Iran, OXA-48 was first reported in 2017 in the Escherichia coli isolates (13).
Among the uropathogenic bacteria, E. coli is predominant in both community and nosocomial urinary tract infection (UTI) (14-16). These resistance patterns have shown large inter-regional variability. Understanding the spectrum and resistance patterns may help guide effective empirical antibiotic therapies and decrease treatment failure and costs.
Contact precautions and outbreak detections require reliable detection of carbapenemases. However, detection of carbapenemase in Enterobacteriaceae is challenging, because carbapenemase-producing K. pneumoniae with low carbapenem MICs have been described in the CLSI or EUCAST-susceptible range. Also, a difference between porin loss coupled with an ESBL or AmpC β-lactamase or carbapenemase is not feasible in carbapenem-resistant isolates alone on the basis of the antibiogram (17). Phenotypic tests such as the modified Hodge test are helpful to detect carbapenemases but have low sensitivity for NDM and low specificity (18). Phenotypic tests based on synergy with EDTA are available for detection of MBL but can produce false-positive outcomes with certain strains and cannot distinguish between kinds of MBL (19). Class A carbapenemases such as KPC can be identified through synergy with boronic acid, but if AmpC β-lactamases are coproduced, false-positive synergy test findings occur (20). Thus, confirmation using molecular analysis is essential.
Due to limited information on carbapenemase in Iran (21), identifying the resistant strains is a major challenge for diagnostic laboratories. The carbapenemases that were surveyed in this study were encoded by plasmids and due to their transfer to other isolates, the purpose of this study was to identify types of carbapenemases using phenotypic methods and to determine the frequency of plasmid genes encoding carbapenemases (IMP, VIM, KPC, and OXA-48) among the MDR isolates causing UTI in northern Iran.
Materials and Methods
Bacterial isolates
Urine samples of the patients (138 samples including 31 male and 107 female specimens with mean age of 43 for male and 41 for female) were collected from appropriate patients in early morning mid-stream using sterile, wide mouthed glass bottles with screw cap tops between May and July 2017. Samples were maintained in an icebox until laboratory analysis. Sample collection and its analysis were no more than one hour apart. The usual bacteriological methods were applied for cultivation, isolation and identification of the strains. Urine samples were cultured on Nutrient Agar, Blood Agar, Eosin Methylene Blue Agar (EMB), and MacConkey agar plates and incubated at 37 °C for 18–24 hr. Urine culture was considered positive, if it contained ≥105 cfu/ml. E. coli from positive urine cultures identified by their characteristic appearance on the media, Gram staining reaction, by the pattern of biochemical tests such as catalase, oxidase, ONPG, IMViC tests, lactose fermentation, H2S and CO2 production, urea hydrolysis, and lysine decarboxylase (22). The isolates were stored at -70 °C in a Tryptic Soy Broth containing 15% glycerol until processing.
Antibacterial susceptibility testing
To identify the susceptibility of the isolates to antibiotics, the disc diffusion test was used according to Clinical and Laboratory Standards Institute (CLSI)(23) guidelines; the following antibiotics were utilized: ampicillin (AMP)(10 μg), amoxicillin (AMX)(25 μg), oxacillin (OXA)(5 μg), fosfomycin (FOF)(200 μg), piperacillin (PIP)(100 μg), streptomycin (STR)(10 μg), tetracycline (TET)(30 μg), chloramphenicol (CHL)(30 μg), cefepime (FEP)(30 μg), ceftriaxone (CRO)(30 μg), ceftazidime (CAZ)(30 μg), cephalothin (CF)(30 μg), cefazolin (CFZ)(30 μg), cefotaxime (CTX)(30 μg), cefixime (CFM)(5 μg), cefuroxime (CXM)(30 μg), imipenem (IMP)(10 μg), meropenem (MEM)(10 μg), amoxicillin-clavulanic acid (AMC)(20/10 μg), and ciprofloxacin (CIP)(5 μg). E. coli ATCC 25922 and ATCC 35218 were used as the standard strains to control the quality of the applied antimicrobial agents. MDR is defined as resistance to three or more antibiotics.
Detection of ESBL
In order to identify ESBL, double disk synergy test (DDST), which depends on comparing the inhibition zone given by CAZ (30 µg) and CAZ-plus-clavulanate (30 µg/10 µg) was used. A difference of ≥5 mm between the zone of CAZ-plus-clavulanate and CAZ alone was taken to indicate ESBLs production as advocated by CLSI (23).
Hodge test
Briefly, a 0.5 McFarland bacterial suspension of E. coli ATCC 25922 as control or susceptible strain was inoculated on the surface of a Mueller-Hinton agar (MHA) as a lawn culture. After brief drying, a 10 µg imipenem disk was placed at the center, and the test isolate was streaked from the center to the periphery of the plate and the plate was incubated overnight. Isolates which produced a cloverleaf-shaped inhibition zone were recognized as producers of carbapenemase (24).
Imipenem-EDTA combined disk test (CDT)
As recommended by CLSI, the control strain was cultured as a lawn on the MHA plate along with test isolates (turbidity of 0.5 McFarland). Then, two 10-μg meropenem discs were located on the lawn culture with 20 mm distance from center to center of the discs. In one of the meropenem disks, a 10 μl 0.5 M EDTA was added and incubated overnight. Isolates indicating a rise of ≥7 mm in the meropenem+EDTA disc’s inhibition zone size compared to the meropenem disc alone were known MBL producers (24).
EDTA-disk synergy test (EDST)
An overnight broth culture of the test isolate was suspended to the turbidity of a 0.5 McFarland and used to swab inoculate a MHA. A 10-μg meropenem disc and a blank disk (Whatmann filter paper no. 2, 6 mm in diameter) were located 10 mm apart from edge to edge, 10 μl EDTA solution 0.5 M was then used as the blank disc. The plates were incubated at 37 °C overnight and an expanded inhibition zone was interpreted as positive EDS (24).
Multiplex PCR technique
DNA extraction was performed with suspending one colony in 100 µl of distilled water (95 °C for 10 min) followed by centrifugation of the cell suspension. The DNA concentration and purity were determined by spectrophotometric measurement of absorbance at 260 and 280 nm by a UV spectrophotometer. All DNA samples were dissolved in water and stored at -20 °C. The PCR reactions were carried out using a 96-well mini PCR System Thermal Cycler (BioRad, USA) in a final volume of 25 µl containing 200 ng of each primer, 50 ng genomic DNA, 1.5 mM MgCl2, 200 µM dNTPs, and 1.0 U of Taq DNA Polymerase in the buffer provided by the manufacturer (25). The sequences of specific primers were designed based on relevant DNA sequences available in the NCBI GenBank database (http://www.ncbi.nlm.nih.gov/genbank) using Oligo-primer analysis software (Version 7.54, Molecular Biology Insights, USA). Primers sequences were listed in Table 1.
Table 1.
Group-specific primers used for multiplex PCR (23)
| Gene | Primer name | Sequence (5ʹ –3 ʹ) | Length (bases) | Annealing position | Product size (bp) | Primer concentration | |||
|---|---|---|---|---|---|---|---|---|---|
| bla OXA-48 like | MultiOXA-48_for MultiOXA-48_rev |
GCTTGATCGCCCTCGATT GATTTGCTCCGTGGCCGAAA |
18 20 |
230-247 490-510 |
281 | 0.4 0.4 |
|||
|
bla
IMP variants except blaIMP-9, blaIMP-16, blaIMP-18, blaIMP-22 and blaIMP-25 |
MultiIMP_for MultiIMP-48_rev |
TTGACACTCCATTTACDGb GATYGAGAATTAAGCCACYCTb |
18 21 |
194-211 332-313 |
139 | 0.5 0.5 |
|||
| bla VIM variants including blaVIM-1 and blaVIM-2 | MultiVIM_for MultiVIM_rev |
GATGGTGTTTGGTCGCATA CGAATGCGCAGCACCAG |
19 17 |
151-169 540-524 |
390 | 0.5 0.5 |
|||
| bla KPC-1 to blaKPC-5 | MultiKPC_for MultiKPC_rev |
CATTCAAGGGCTTTCTTGCTGC ACGACGGCATAGTCATTTGC |
22 20 |
209-230 746-727 |
538 | 0.2 0.2 |
|||
bY=T or C; R= A or G; S= G or C; D= A or G or T
Amplification was carried out with the following thermal cycling conditions: 10 min at 94 °C and 30 cycles of amplification consisting of 94 °C for 40 sec, 60 °C for 40 sec, and at 72 °C for 1 min, with 7 min at 72 °C for the final extension. For the multiplex PCR analysis, the annealing temperature was at 55 °C for amplification of blaVIM, blaIMP, and blaKPC genes, and 57 °C for amplification of blaOXA-48 gene. The PCR products were subjected to electrophoresis on a 2% agarose gel and observed under UV after staining with ethidium bromide (25).
Statistical analysis
The data were statistically analyzed using One-way analysis of variance (ANOVA) and differences among the means were determined at P≤0.01 using Duncan’s multiple range tests (by SAS, 9.1).
Results
Isolates and their resistance patterns
From 138 patients that enrolled in this study, 112 urine specimens were infected with bacteria, from which 91 were positive for E. coli. The remaining 21 strains included 15 Enterobacter spp. isolates, 5 Proteus mirabilis isolates and 1 of Group B Streptococcus isolates. E. coli isolates characterized with distinctive metallic green sheen on EMB and pink colored colonies on MacConkey’s agar, while white or creamy-colored colonies appeared on the nutrient agar. All 91 isolates of E. coli had given positive test for catalase, ONPG, lysine decarboxylase, indole, methyl-red test, CO2 production, and for lactose fermenting, and negative biochemical test for oxidase, Voges Proskuaer, citrate, urease, and H2S production; there we confirmed that these isolates belonged to E. coli. It should be noted that the patterns of resistance of these E. coli isolates to 20 antibiotics were completely different and therefore all isolates were distinctive. The results of antibiotic susceptibility tests were depicted in Figure 1. The sensitivity of isolates to different antibiotics was different, with meropenem being effective on 91.90%±1.89, piperacillin on 78.94%±3.41, cephalotin on 67.77%±2.20, imipenem and chloramphenicol on 65.34%±1.39 and 64.07%±3.15, respectively, and cefepime on 58.82%±2.37 of the tested isolates. All E. coli isolates were identified as MDR bacteria.
Figure 1.
The Susceptibility patterns of various antibiotics against 91 uropathogenic Escherichia coli strains isolated from urine samples
ESBL and carbapenemase detection
Of total 91 E. coli isolates, the synthesis of ESBL was detected in 33 isolates. In this study, phenotypic and genotypic tests were carried out for detection of different types of carbapenemases including IMP, VIM, KPC, and OXA-48. Based on the Hodge test, 3 (3%) of the 33 ESBL-producing E. coli isolates produced imipenemase. Using CDT and EDST methods, 1 (3%) and 2 (6%) of the 33 ESBL-producing E. coli isolates produced MBL (IMP, VIM, or both and/or other MBLs), respectively. Therefore, these two methods have somewhat different performances in MBL detection. It was interesting that by applying the phenotypic methods in this study, it was proven that none of the non-ESBL-producing E. coli isolates were able to produce different types of carbapenemases.
Multiplex PCR
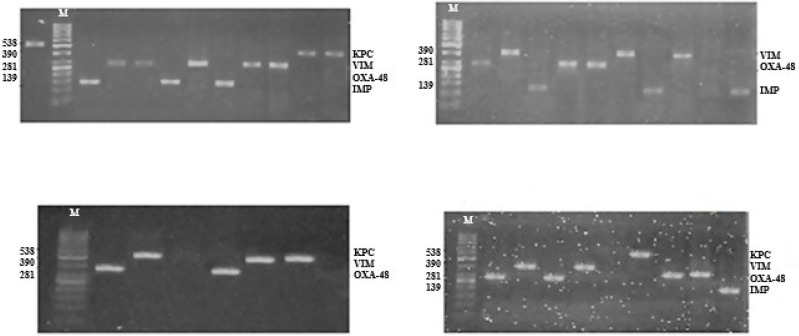
After optimizing the amplification conditions, amplicons with the desired sizes were obtained from the studied isolates and confirmed the specificity of the primers (Figure 2). The results of multiplex PCR analysis showed that frequency of carbapenemaeses genes including IMP, VIM, KPC, and OXA-48 in ESBL-producing E. coli isolates were 24% (8/33), 27% (9/33), 6% (2/33), and 42% (14/33) and in all isolates 8.7% (8/91), 9.8% (9/91), 2.1% (2/91), and 15.3% (14/91), respectively. Therefore, blaOXA-48 and blaKPC genes had the highest and the lowest abundance among the E. coli isolates, and none of the carbapenemase genes were detected in non-ESBL-producing E. coli isolates. Also, the results of phenotypic and multiplex PCR tests were consistent in non-ESBL-producing E. coli isolates.
Figure 2.
Detection of different carbapenemase genes in some of 33 ESBL-producing Escherichia coli isolates by multiplex PCR in 2% agarose gel. In all figures, lane M for molecular marker (50-1000 bp DNA ladder) and other lanes show the amplified fragments of carbapenemase genes including IMP, OXA-48, VIM and KPC
Carbapenemaeses genes and resistance patterns
In relation with the simultaneous presence of two or more carbapenemase genes in one isolate, only two strains (1%) included blaVIM and blaIMP genes and one strain (0.5%) included blaOXA-48 and blaVIM genes. There was no simultaneous presence of blaIMP and blaKPC, blaIMP and blaOXA-48, blaKPC and blaOXA-48, or blaKPC and blaVIM genes in any of the isolates. Only one isolate (0.5%) carried all four genes studied (Table 2). Existence of IMP conferred more resistance to cefuroxime (100%), cefazolin (87.5%), ampicillin (87.5%), amoxicillin (87.5%), and oxacillin (87.5%)(P<0.01), and carrying VIM caused more resistance to ampicillin (100%), amoxicillin (100%), oxacillin (100%), and amoxicillin-clavulanic acid (88.88%)(P<0.01). The existence of KPC conferred more resistance to cephalotin (100%) and existence of OXA-48 caused more resistance to ampicillin (100%), amoxicillin (100%), oxacillin (100%), and streptomycin (88.88%)(P<0.05). The results of association of two or more carbapenemases genes with the resistance pattern are presented in Table 3.
Table 2.
Comparison of the resistance rates between ESBL, non-ESBL-producing, and total Escherichia coli isolates against 20 antibiotics and association between carbapenemase genes with resistance pattern among ESBL-producing isolates a,b
| Antibiotic | ESBL (n=33) | non-ESBL (n=58) | Total (n=91) | IMP (n=8) | OXA-48 (n=14) | KPC (n=2) | VIM (n=9) |
|---|---|---|---|---|---|---|---|
| CF | 20 (60.60) | 13 (22.41) | 27 (30) | 6 (75) | 1 (7.14) | 2 (100) | 7 (77.77) |
| AMC | 21 (63.63) | 50 (86.20) | 75 (83) | 6 (75) | 1 (7.14) | 0 (0.00) | 8 (88.88) |
| FEP | 8 (24.24) | 24 (41.37) | 35 (38.5) | 4 (50) | 4 (28.5) | 0 (0.00) | 5 (55.55) |
| FOF | 12 (36.36) | 26 (44.82) | 40 (43.5) | 6 (75) | 3 (21.42) | 1 (50) | 4 (44.44) |
| TET | 19 (57.57) | 41 (70.68) | 62 (68.5) | 6 (75) | 9 (64.28) | 1 (50) | 2 (22.22) |
| CIP | 10 (30.30) | 33 (56.89) | 47 (51.64) | 5 (62.5) | 6 (42.85) | 1 (50) | 5 (55.55) |
| STR | 22 (66.66) | 41 (69.87) | 63 (69.23) | 3 (37.5) | 12 (85.71) | 0 (0.00) | 7 (77.77) |
| CHL | 3 (9.09) | 22 (37.93) | 30 (32.96) | 2 (25) | 7 (50) | 0 (0.00) | 3 (33.33) |
| PIP | 1 (3.03) | 13 (22.41) | 17 (18.68) | 3 (37.5) | 4 (28.5) | 0 (0.00) | 3 (33.33) |
| AMP | 30 (90.90) | 53 (91.37) | 89 (97.80) | 7 (87.5) | 14 (100) | 1 (50) | 9 (100) |
| AMX | 30 (90.90) | 53 (91.37) | 84 (92.30) | 7 (87.5) | 14 (100) | 1 (50) | 9 (100) |
| OXA | 31 (93.93) | 56 (96.55) | 88 (96.70) | 7 (87.5) | 14 (100) | 1 (50) | 9 (100) |
| CRO | 18 (54.54) | 38 (65.51) | 58 (63.73) | 5 (62.5) | 11 (78.57) | 1 (50) | 5 (55.55) |
| CTX | 12 (36.36) | 37 (63.79) | 54 (59.34) | 6 (75) | 12 (85.71) | 1 (50) | 6 (66.66) |
| CFM | 19 (57.57) | 43 (74.13) | 65 (71.42) | 5 (62.5) | 11 (78.57) | 1 (50) | 4 (44.44) |
| CAZ | 16 (48.48) | 32 (55.17) | 49 (53.84) | 4 (50) | 7 (50) | 1 (50) | 7 (77.77) |
| IMP | 7 (21.21) | 20 (34.48) | 30 (32.96) | 3 (37.5) | 2 (14.28) | 0 (0.00) | 3 (33.33) |
| MEN | 5 (15.15) | 2 (34.48) | 6 (65.93) | 0 (0.00) | 1 (7.14) | 0 (0.00) | 0 (0.00) |
| CFZ | 18 (54.54) | 45 (77.58) | 68 (74.72) | 7 (87.5) | 41.42 | 1 (50) | 6 (66.66) |
| CXM | 29 (87.87) | 51 (87.93) | 77 (84.61) | 8 (100) | 78.57 | 1 (50) | 7 (77.77) |
a Data are presented No. (%), n= number of isolates. b ESBL= extended spectrum β-lactamase
IMP: imipenem; OXA: oxacillin; KPC: Klebsiella pneumoniae carbapenemase; VIM: Verona integron-encoded metallo-β-lactamase
Table 3.
Association between two carbapenemase genes with resistance pattern in 33 ESBL-producing Escherichia coli isolatesa,b
| Antibiotic | IMP and KPC (n=0) | IMP and OXA-48 (n=0) | IMP and VIM (n=2) | KPC and OXA-48 (n=0) | KPC and VIM (n=0) | OXA-48 and VIM (n=1) |
|---|---|---|---|---|---|---|
| CF | 0 (0.00) | 0 (0.00) | 2 (100) | 0 (0.00) | 0 (0.00) | 1 (100) |
| AMC | 0 (0.00) | 0 (0.00) | 2 (100) | 0 (0.00) | 0 (0.00) | 1 (100) |
| FEP | 0 (0.00) | 0 (0.00) | 1 (50) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| FOF | 0 (0.00) | 0 (0.00) | 1 (50) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| TET | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| CIP | 0 (0.00) | 0 (0.00) | 1 (50) | 0 (0.00) | 0 (0.00) | 1 (100) |
| STR | 0 (0.00) | 0 (0.00) | 1 (50) | 0 (0.00) | 0 (0.00) | 1 (100) |
| CHL | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| PIP | 0 (0.00) | 0 (0.00) | 1 (50) | 0 (0.00) | 0 (0.00) | 1 (100) |
| AMP | 0 (0.00) | 0 (0.00) | 2 (100) | 0 (0.00) | 0 (0.00) | 1 (100) |
| AMX | 0 (0.00) | 0 (0.00) | 2 (100) | 0 (0.00) | 0 (0.00) | 1 (100) |
| OXA | 0 (0.00) | 0 (0.00) | 2 (100) | 0 (0.00) | 0 (0.00) | 1 (100) |
| CRO | 0 (0.00) | 0 (0.00) | 1 (50) | 0 (0.00) | 0 (0.00) | 1 (100) |
| CTX | 0 (0.00) | 0 (0.00) | 2 (100) | 0 (0.00) | 0 (0.00) | 1 (100) |
| CFM | 0 (0.00) | 0 (0.00) | 2 (100) | 0 (0.00) | 0 (0.00) | 1 (100) |
| CAZ | 0 (0.00) | 0 (0.00) | 1 (50) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| IMP | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| MEN | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| CFZ | 0 (0.00) | 0 (0.00) | 2 (100) | 0 (0.00) | 0 (0.00) | 1 (100) |
| CXM | 0 (0.00) | 0 (0.00) | 2 (100) | 0 (0.00) | 0 (0.00) | 1 (100) |
a Data are presented No.(%), n= number of isolates
b ESBL= extended spectrum β-lactamases
IMP: imipenem; OXA: oxacillin; KPC: Klebsiella pneumoniae carbapenemase; VIM: Verona integron-encoded metallo-β-lactamase
Discussion
Resistance to carbapenems is due to carbapenemase and other resistance mechanisms, such as ESBLs, efflux pumps, and/or porin mutations (10). The current emergence of carbapenemase-producing bacteria especially Enterobacteriaceae is of concern because it is often associated with the occurrence of multidrug-resistant isolates, where there are very few drug options available for them, if any (10). Therefore, detection and initial identification of carbapenemase-producing bacteria are important. In some cases, due to the low sensitivity or specificity of phenotypic methods, molecular approaches may also be used (26). Reliable identification of carbapenemases is essential for the implementation of contact precautions and the detection of the outbreak.
In the current study, from the 112 urine specimens from UTI patients infected with bacteria, 91 were positive for E. coli. Carbapenems and pipiracillin were the most effective antibiotics and all cephalosporins other than 4th cephalosporin affected more than 50% of isolates. Compared to the ESBL-producing isolates, resistance of non-ESBL-producing isolates was higher than different antibiotics. This is due to another mechanism other than ESBL in resistance to antibiotics. Generally, the frequency of ESBL-producing isolates and the types of carbapenemaese genes among them were low and also in the detection of carbapenemases, there was no correlation between the results of phenotypic and molecular analyses. Due to the similarity of the results of the Hodge and CDT tests, it seems that in the Hodge test, the addition of EDTA did not have any effect on the improvement of the test. Also, the EDST test is comparison with CDT could detect more MBLs and therefore, EDST probably is more reliable. The results of current study in the detection of cabapenemases types in ESBL-producing E. coli showed that multiplex PCR is both more sensitive and also more reliable than phenotypic methods due to detection of different carbapenemases and more positive samples.
Given that the resistance of the isolates to imipenem and moropenem was 29% and 6%, respectively, the multiplex analysis identified fewer resistance genes to carbapenems (frequency of OXA-48, KPC, IMP, and VIM genes was 7% (10/33), 1% (2/33), 4% (6/33), and 4% (6/33), respectively), which was similar to that of Gheitani and Fazeli (21). The possible reason for this may be the presence of other types of resistance genes to carbapenems that have not been investigated in the current study or other mechanisms of resistance to carbapenem other than carbapenemase production.
Some previous studies in Iran indicated different outbreaks of ESBLs-producing E. coli. Contrary to our research, in another study, 115 (89.8%) E. coli strains were recognized as ESBL producers (27). Zaniani et al. (2012) reported that 43.9% of E. coli isolates were ESBL producers (28). Another study identified ESBL-producing E. coli in 44.1% and 21.2% of inpatients and outpatients isolates, respectively (29).
Most of the studies have used EDST, CDT, MHT, and E-test for detection of MBL. According to their findings, MBL production varied from 7% to 65%. Some studies recorded the use of EDST as one of the suitable methods to detect Ambler Class B MBL production and the positivity ranged from 14.8% to 72% (16-18). Their results demonstrate that EDST is more reliable and reproducible with elevated positivity rates. In our research like studies of Arakawa, Jesudasan, and John, EDST has shown the highest positivity (30-32).
Similar to previously published data, in the current study low positivity of Hodge test compared with other tests was shown, which varied from 14.8%–56.16% (30-34). Contrary to our study, in most studies, CDT is more robust than EDST and MHT (35).
Given that phenotypic tests may be false positive and/or low sensitive or specific, confirmation by molecular methods is required. In one study, 183 Enterobacteriaceae were identified from 442 patients with UTI and of them 160 (87.4%) were MDR. The most common isolates were K. pneumoniae and E. coli. Similar to our study, in their study the prevalence of carbapenemase was a low 2.73% (5/160) and all carbapenemase-producing Enterobacteriaceae (CPE) produced ESBL. In their study, the most effective antibiotic was ciprofloxacin. Significant drug resistances were detected among CPE compared to other MDR Enterobacteriaceae (36). In another study, detection of IMP carbapenemase in 600 Enterobacteriaceae clinical isolates was determined by the PCR method. The most common isolate was E. coli 52% (315/600) and the highest rates of resistance were towards ertapenem and imipenem. In combined disk tests by using of the ertapenem or imipenem, 25 isolates were screened positive. Unlike our study, the blaIMP gene was not detected in any of the 25 isolates (37).
From a total of 50 carbapenem-resistant E. coli isolates in a study, the highest resistance rate was detected to ceftazidime (100%), tetracycline (88%), and amoxicillin (100%). Contrary to our study, the frequency of the blaIMP gene was greater (16%), and the blaVIM gene was not detected in any of their studied isolates. The prevalence of the blaOXA-48 gene was almost the same as our study (8%) (38). The disk diffusion test in research on 160 E. coli isolates showed that the highest resistance rates were against cefotaxime (20%) and ceftazidime (17%) and the lowest was to tetracycline (1%). Unlike our study, only five isolates (3%) were detected as resistant to imipenem and all five imipenem-resistant strains were confirmed positive for MBL enzymes based on the combination disk diffusion test (CDDT), but PCR did not detect any blaIMP or blaVIM genes in MBL-producing strains (39).
According to the study by Gheitani and Fazeli (2018) on 183 K. pneumoniae isolates, the highest and lowest rates of resistance were detected against cefotaxime (98.2%) and gentamicin (43.6%), respectively. Among the 183 isolates, 134 (73.2 %) were positive based on MHT. Also, in accordance with our study, the prevalence of blaVIM, blaIMP, and blaKPC genes were low (21).
In a different study, 111 CPE were isolated from different clinical samples. Fifty isolates (55%) were resistant to imipenem and/or meropenem. All the study isolated exhibited a positive MHT. MBL screen test using EDTA and KPC screen test using phenylboronic acid were positive in 54 and 36 isolates, respectively. By using multiplex PCR, carbapenemase-encoding genes were detected in 63 isolates including 58 NDM, 1 VIM, 2 OXA-181, and 6 both NDM and VIM (40). In this study, similar to our study, the prevalence of OXA-48 and VIM carbapenemas was low.
Research showed out of the 100 carbapenem resistant isolates (E. coli (25), K. pneumoniae (35), P. aeruginosa (18), and A. baumannii (22)), 70 isolates were MHT positive, while 65 isolates were CDT positive. In five isolates which were MHT positive but CDT negative, none of the 4 genes including blaNDM-1, blaVIM, blaIMP, and blaKPC were detected. The results of the multiplex PCR for four target genes showed only 5 strains with the blaVIM gene, 1 strain with the blaKPC gene, and none of the strains produced blaIMP. Out of 100 carbapenem resistant isolates, 65 isolates were harboring one or more than one genes, while in 35 isolates none of the genes was detected. The most common resistance gene was blaNDM-1 (59/100) followed by blaKPC (15/100) while the blaVIM gene was least frequent (6/100). Contrary to our research, the blaIMP gene did not detect in any of the isolates. Correlation of multiplex PCR with MHT and CDT among carbapenemase-producing isolates is observed (41).
In the study by Pavelkovich et al. in the Baltic States and St. Petersburg, Russia on CPE, of all 9757 strains including 1983 K. pneumoniae and 7774 E. coli isolated from intensive care patients and different clinical samples, 77 isolates (73 K. pneumoniae and 4 E. coli)(0.7%) were resistant to carbapenem. In this study unlike our study, of 77 strains, in 15 strains the blaNDM gene was detected and in the other 62 strains blaIMP, blaVIM, blaGIM, blaOXA-48, blaNDM, or blaKPC genes were not identified (42).
In a recent study, 210 MDR Gram-negative bacilli were obtained from different specimens such as urine (n=108) screened for carbapenemase resistance. They used uniplex PCR for detection of blaNDM-1 and blaKPC genes in E. coli and Klebsiella and applied multiplex PCR for detection of blaIMP and blaVIM genes in P. aeruginosa and A. baumannii isolates. Twenty three (11%) isolates (E. coli (6), K. pneumoniae (3), P. aeruginosa (5), and A. baumannii (9)) were found resistant to meropenem and imipenem by disc diffusion. The results of MHT showed that out of the 23 carbapenem-resistant isolates, 17 (74%) produced MBL. These were further confirmed by the E-test. MHT was negative for all isolates. All 17 isolates were subjected to PCR and found to contain at least one carbapenemase gene. Unlike our study, none of blaKPC, blaIMP, or blaVIM genes were detected in Enterobacteriaceae isolates (43).
In a study on frequency of carbapenemase genes including VIM, IMP, NDM, KPC, and OXA- 48 in 227 MDR Gram-negative bacteria, similar to our study, the most effective antibiotic was meropenem, and 80 strains (35%) were positive for one or more carbapenemase genes. Contrary to our study, IMP-types were the most predominant gene followed by VIM, in 49 (21.59%) and 28 (12%) isolates, respectively. Carbapenemase genes were most detected in K. pneumoniae (24, 11%), followed by P. aeruginosa (23, 10%), and E. coli with 19 isolates (8%)(44).
In one study, 60 bacteria were isolated from urine specimen including 26 strains of E. coli and 34 strains of K. pneumoniae. It was determined that meropenem and amikacin were the most effective antibiotics on E. coli, and imipenem the most effective antibiotic on K. pneumoniae. Four E. coli and 23 K. pneumoniae isolates were positive for carbapenemase production by using the MHT test. Although some results of phenotypic assays matched with the definite PCR identification, some results were misleading. Out of the 29 positive PCR samples, three samples of K. pneumoniae were negative for MHT and one E. coli sample was MHT positive but negative for PCR. Nine samples were positive for PCR but were determined as carbapenem sensitive by MicroScan (45).
In a study to detect MBL among 100 A. baumannii strains, 30 (30%) of the strains were positive for the blaVIM gene, but the blaIMP gene was not detected in any of the strains (46).
Conclusion
Although the prevalence of the ESBL-producing strains and the simultaneous presence of several carbapenemase genes in the studied population were nearly low, these low prevalent strains and genes are responsible for resistance to some antibiotics. Thus, identification and controlling of these strains is important due to the presence of plasmid genes encoding carbapenemases and their easy transferability to other clinical isolates.
Acknowledgment
The results presented in this paper were part of a student thesis. The authors would like to extend their thanks to the staff from Razi Pathobiology Laboratory, Rasht, Iran, for providing samples.
Conflicts of Interest
The authors declare that they have no conflicts of interest related to the subject matter or materials discussed in this article.
References
- 1.Carmeli Y, Akova M, Cornaglia G, Daikos GL, Garau J, Harbarth S, et al. Controlling the spread of carbapenemase-producing Gram-negatives: therapeutic approach and infection control. Clin Microbiol Infect. 2010;16:102–111. doi: 10.1111/j.1469-0691.2009.03115.x. [DOI] [PubMed] [Google Scholar]
- 2.Cornaglia G, Giamarellou H, Rossolini GM. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect Dis. 2011;11:381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- 3.Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yousefi S, Farajnia S, Nahaei MR, Ghojazadeh M, Akhi M, Sharifi Y, et al. Class 1 integron and imipenem resistance in clinical isolates of Pseudomonas aeruginosa: prevalence and antibiotic susceptibility. Iran J Microbiol. 2010;2:115–121. [PMC free article] [PubMed] [Google Scholar]
- 5.Davoudi-Monfared E, Khalili H. The threat of carbapenem-resistant Gram-negative bacteria in a Middle East region. Infect Drug Resist. 2018;11:1831–1880. doi: 10.2147/IDR.S176049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahbar M, Mehragan H, Haji Ali Akbari N. Prevalence of drug resistance in nonfermenter Gram-negative bacilli. Iran J Pathol. 2010;5:90–96. [Google Scholar]
- 7.Tarashi S, Goudarzi H, Erfanimanesh S, Pormohammad A, Hashemi A. Phenotypic and molecular detection of metallo-beta-lactamase genes among imipenem resistant Pseudomonas aeruginosa and Acinetobacter baumannii strains isolated from patients with burn injuries. Arch Clin Infect Dis. 2016;11:e39036. [Google Scholar]
- 8.Ahn C, Syed A, Hu F, O’Hara JA, Rivera JI, Doi Y. Microbiological features of KPC-producing Enterobacter isolates identified in a US hospital system. Diagn Microbiol Infect Dis. 2014;80:154–158. doi: 10.1016/j.diagmicrobio.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-beta-lactamases: the quiet before the storm? Clin Microbiol Rev. 2005;18:306–325. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daikos GL, Petrikkos P, Psichogiou M, Kosmidis C, Vryonis E, Skoutelis A, et al. Prospective observational study of the impact of VIM-1 metallo-β-lactamase on the outcome of patients with Klebsiella pneumoniae bloodstream infections. Antimicrob Agents Chemother. 2009;53:1868–1873. doi: 10.1128/AAC.00782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poirel L, Héritier C, Tolün V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Solgia H, Giskeb CG, Badmastia F, Aghamohammada S, Havaeic SA, Sabetid S. Emergence of carbapenem resistant Escherichia coli isolates producing blaNDM and blaOXA-48-like carried on IncA/C and IncL/M plasmids at two Iranian university hospitals. Infect Genet Evol. 2017;55:318–323. doi: 10.1016/j.meegid.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 14.Schito G, Naber K, Botto H, Palou J, Mazzei T, Gualco L, et al. The ARESC Study: an international survey on the antimicrobial resistance of pathogens involved in uncomplicated urinary tract infections. Int J Antimicrob Agents. 2009;34:407–413. doi: 10.1016/j.ijantimicag.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 15.El Bouamri M, Arsalane L, El Kamouni Y, Zouhair S. Antimicrobial susceptibility of urinary Klebsiella pneumonia and the emergence of carbapenem-resistance strain. Afr J Urol. 2015;21:36–40. [Google Scholar]
- 16.Ngwai Y, Akpotu M, Okidake R, Sounyo AA, Onanuga A, Origbo SO. Antimicrobial susceptibility of Escherichia coli and other coliforms isolated from urine of asymptomatic students in Bayelsa State, Nigeria. Afr J Microbiol Res. 2011;5:184–191. [Google Scholar]
- 17.Nordmann P, Gniadkowski M, Giske CG, Poirel L, Woodford N, Miriagou V, et al. Identification and screening of carbapenemase-producing Enterobacteriaceae. Clin Microbiol Infect. 2012;18:432–438. doi: 10.1111/j.1469-0691.2012.03815.x. [DOI] [PubMed] [Google Scholar]
- 18.Hrabak J, Studentov V, Walkov´ R, Žemličkováb H, Jakubůb V, Chudáčkováa E, et al. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapenemases by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol. 2012;50:2441–2443. doi: 10.1128/JCM.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burckhardt I, Zimmermann S. Using matrix-assisted laser desorption ionization-time of flight mass spectrometry to detect carbapenem resistance within 1 to 25 hours. J Clin Microbiol. 2011;49:3321–3324. doi: 10.1128/JCM.00287-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barraud O, Baclet MC, Denis F, Ploy MC. Quantitative multiplex real-time PCR for detecting class 1, 2 and 3 integrons. J Antimicrob Chemother. 2010;65:1642–1645. doi: 10.1093/jac/dkq167. [DOI] [PubMed] [Google Scholar]
- 21.Gheitani L, Fazeli H. Prevalence of blaVIM, blaIMP and blaKPC genes among carbapenem-resistant Klebsiella pneumoniae (CRKP) isolated from Kurdistan and Isfahan hospitals, Iran. Res Mol M. 2018;6:12–20. doi: 10.14715/cmb/2018.64.1.13. [DOI] [PubMed] [Google Scholar]
- 22.Cheesbrough M. Manual of medical microbiology. Low price ed. Britain: Oxford Press; 2000. pp. 251–260. [Google Scholar]
- 23.CLSI . CLSI document M100eS24. Wayne, PA: CLSI; 2014. Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplements. [Google Scholar]
- 24.Lee K, Chong Y, Shin HB, Kim YA, Yong D, Yum JH. Modified Hodge test and EDTA-disk synergy tests to screen metallo-β-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin Microbiol Infect. 2001;7:88–91. doi: 10.1046/j.1469-0691.2001.00204.x. [DOI] [PubMed] [Google Scholar]
- 25.Dallenne C, Costa AD, Decre´D , Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 26.Cornaglia G, Akova M, Amicosante G, Cantón R, Cauda R, Docquier JD, et al. Metallo-β-lactamases as emerging resistance determinants in Gram-negative pathogens: open issues. Int J Antimicrob Agents. 2007;29:380–388. doi: 10.1016/j.ijantimicag.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Soltan Dallal M, Sabbaghi A, Molla Aghamirzaeie H, Rastegar Lari A, Eshraghian MR, Fallah Mehrabad J, et al. Prevalence of AmpC and SHV β-lactamases in clinical isolates of Escherichia coli From Tehran jospitals. Jundishapur J Microbiol. 2013;6:176–180. [Google Scholar]
- 28.Zaniani FR, Meshkat Z, Naderi Nasab M, Khaje-Karamadini M, Ghazvini K, Rezaee A, et al. The prevalence of TEM and SHV genes among extended-spectrum beta-lactamases producing Escherichia coli and Klebsiella pneumoniae. Iran J Basic Med Sci. 2012;15:654–660. [PMC free article] [PubMed] [Google Scholar]
- 29.Mobasherizadeh S, Shokri D, Zargarzadeh A, Jalalpour S, Ebneshahidi S, Sajadi M. Antimicrobial resistance surveillance among hospitalized and non-hospitalized extend-spectrum beta-lactamase producing Escherichia coli from four tertiary care hospitals in Isfahan, Iran; 2008-2011. Afr J Microbiol Res. 2012;6:953–999. [Google Scholar]
- 30.Arakawa Y, Shibata N, Shibayama K, Kurokawa H, Yagi T, Fujiwara H, et al. Convenient test for screening metallo-β-lactamase-producing Gram-negative bacteria by using thiol compounds. J Clin Microbiol. 2000;38:40–43. doi: 10.1128/jcm.38.1.40-43.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jesudasan MV, Kandathil AJ, Balaji V. Comparison of two methods to detect carbapenemase and metallo-β-lactamase production in clinical isolates. Indian J med Res. 2005;121:780–783. [PubMed] [Google Scholar]
- 32.John S, Balagurunathan R. Metallo beta-lactamase producing Pseudomonas aeruginosa and Acinetobacter baumannii. Indian J Med Microbiol. 2011;29:302–304. doi: 10.4103/0255-0857.83918. [DOI] [PubMed] [Google Scholar]
- 33.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: Emergence of a successful pathogen. Clin Microbiol Res. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee K, Lim YS, Yong D, Yum JH, Chong Y. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-β-lactamase producing isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2003;41:4623–4629. doi: 10.1128/JCM.41.10.4623-4629.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Behra B, Mathur P, Das A, Kapil A, Sharma V. An evaluation of four different phenotypic techniques for detection of metallo-β-lactamase producing Pseudomonas aeruginosa. Indian J Med Microbiol. 2008;26:233–237. doi: 10.4103/0255-0857.39587. [DOI] [PubMed] [Google Scholar]
- 36.Eshetie S, Unakal C, Gelaw A, Ayelign B, Endris M, Moges F. Multidrug resistant and carbapenemase producing Enterobacteriaceae among patients with urinary tract infection at referral Hospital, Northwest Ethiopia. Antimicrob Resist Infect Control. 2015;4:12–20. doi: 10.1186/s13756-015-0054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mosavian M, Koraei D. Molecular detection of IMP carbapenemase-producing Gram-negative bacteria isolated from clinical specimens in Ahvaz, Iran. Jentashapir J Health Res. 2016;7:e36394. [Google Scholar]
- 38.Nojoomi F, Ghasemian A. Resistance and virulence factor determinants of carbapenem-resistant Escherichia coli clinical isolates in three hospitals in Tehran, IR Iran. Infect Epidemiol Microbiol. 2017;3:107–111. [Google Scholar]
- 39.Shams S, Hashemi A, Esmkhani M, Kermani S, Shams E, Piccirillo A. Imipenem resistance in clinical Escherichia coli from Qom, Iran. BMC Res Notes. 2018;11:314–318. doi: 10.1186/s13104-018-3406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mariappan S, Sekar U, Kamalanathan A. Carbapenemase-producing Enterobacteriaceae: risk factors for infection and impact of resistance on outcomes. Int J Appl Basic Med Res. 2017;7:32–39. doi: 10.4103/2229-516X.198520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solanki R, Vanjari L, Subramanian S, Aparna B, Nagapriyanka E, Lakshmi V. Comparative evaluation of multiplex PCR and routine laboratory phenotypic methods for detection of carbapenemases among Gram-negative bacilli. J Clin Diagn Res. 2014;8:DC23–26. doi: 10.7860/JCDR/2014/10794.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pavelkovich A, Balode A, Edquist P, Egorova S, Ivanova M, Kaftyreva L, et al. Detection of carbapenemase-producing Enterobacteriaceae in the Baltic countries and St. Petersburg area. BioMed Res Int. 2014:Article ID 548960. doi: 10.1155/2014/548960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swaminathan A, Ardra M, Manoharan A, Prithi Nair K, Girija KR. Characterization of carbapenemase-producing Gram-negative bacilli among clinical isolates in a tertiary care centre in Kerala, South India. J Acad Clin Microbiol. 2016;18:100–104. [Google Scholar]
- 44.Mushi MF, Mshana SE, Imirzalioglu C, Bwanga F. Carbapenemase genes among multidrug resistant Gram negative clinical isolates from a tertiary hospital in Mwanza, Tanzania. BioMed Res Int. 2014:Article ID:303104. doi: 10.1155/2014/303104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.AlTamimi M, AlSalamah A, AlKhulaifi M, AlAjlan H. Comparison of phenotypic and PCR methods for detection of carbapenemases production by Enterobacteriaceae. Saudi J Biol Sci. 2017;24:155–161. doi: 10.1016/j.sjbs.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Safari M, Mozaffari Nejad AS, Bahador A, Jafari R, Alikhani MY. Prevalence of ESBL and MBL encoding genes in Acinetobacter baumannii strains isolated from patients of intensive care units (ICU) Saudi J Biol Sci. 2015;22:424–429. doi: 10.1016/j.sjbs.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]