Abstract
Purpose:
Circulating tumor DNA in plasma may present a minimally invasive opportunity to identify tumor-derived mutations to inform selection of targeted therapies for individual patients, particularly in cases of oligometastatic disease where biopsy of multiple tumors is impractical. To assess the utility of plasma DNA as a “liquid biopsy” for precision oncology, we tested whether sequencing of plasma DNA is a reliable surrogate for sequencing of tumor DNA to identify targetable genetic alterations.
Methods:
Blood and biopsies of 1–3 tumors were obtained from 4 evaluable patients with advanced breast cancer. One patient provided samples from an additional 7 tumors post-mortem. DNA extracted from plasma, tumor tissues, and buffy coat of blood were used for probe-directed capture of all exons in 149 cancer-related genes and massively parallel sequencing. Somatic mutations in DNA from plasma and tumors were identified by comparison to buffy coat DNA.
Results:
Sequencing of plasma DNA identified 27.94 +/− 11.81% (mean +/− SD) of mutations detected in a tumor(s) from the same patient; such mutations tended to be present at high allelic frequency. The majority of mutations found in plasma DNA were not found in tumor samples. Mutations were also found in plasma that matched clinically undetectable tumors found post-mortem.
Conclusions:
The incomplete overlap of genetic alteration profiles of plasma and tumors warrants caution in the sole reliance of plasma DNA to identify therapeutically targetable alterations in patients, and indicates that analysis of plasma DNA complements, but does not replace, tumor DNA profiling.
Trial Registration:
Subjects were prospectively enrolled in trial NCT01836640 (registered April 22, 2013).
Keywords: Circulating tumor DNA, advanced breast cancer, DNA sequencing, cell-free DNA
Background
Tumor genetic alterations are increasingly being used to identify targeted therapies for individual patients. Conventional methods use sequencing of DNA extracted from tumor tissues, which may be archived or newly acquired by biopsy or surgical excision. Since tumors genetically evolve, particularly in response to treatment with targeted therapies [1–4], profiling of recently biopsied tumors is preferable over archived tumors. However, not all tumors are amenable to biopsy, and intra- and inter-tumoral heterogeneity raises concern over biopsy bias [5–8]. If a tumor biopsy contains genetic lesions present at different allelic frequencies (AF) compared to the bulk of the tumor, or if the biopsied tumor is not genetically reflective of most tumors within a patient, then the optimal targeted therapy may be overlooked. Identifying targetable genetic changes using minimally invasive procedures will be an increasing challenge for oncologists. One way to potentially overcome this issue is through analysis of cell-free circulating tumor DNA (ctDNA), which is detectable in the bloodstream [9–11]; this method provides the ability to identify targetable mutations through DNA sequencing, and the required blood draw is far less invasive than traditional biopsies.
Quantification and mutational profiling of ctDNA have shown promise for several clinical applications. In patients with early-stage breast cancer that has been surgically resected, detection of ctDNA in plasma is associated with higher likelihood of recurrence, suggesting that the presence of ctDNA reflects residual disease. In addition, the mutational profile of post-surgical ctDNA better reflects the mutational profile of the impending recurrent tumor than the primary breast tumor, suggesting that such ctDNA can be used to identify preemptive therapies to target micrometastatic disease [12,13]. Genetic profiling of plasma DNA may be useful to identify targeted therapies in patients with advanced disease [14,15], and such profiling is accepted in lieu of tumor genetic profiling in ongoing clinical trials (e.g., TAPUR study, clinicaltrials.gov identifier NCT02693535; NCI-MATCH trial, clinicaltrials.gov identifier NCT02465060). ctDNA levels also correlate with disease burden and can serve as an early marker of therapeutic response in cases of advanced disease [16].
Several technologies have been developed to analyze ctDNA from plasma [17–19], and service providers have begun offering FDA-approved genetic profiling of DNA from patient plasma. Although tumor-derived DNA is detectable in plasma, particularly in patients with heavy cancer burden [16], it remains unclear whether plasma DNA captures all of the clinically actionable mutations present in tumors in patients with advanced disease. Addressing this issue is critical to assignment of the weight of plasma DNA profiling in treatment decision-making.
Methods
Patients and study design
Tissue and blood specimens were obtained from patients under protocols approved by the Dartmouth College Institutional Review Board. All patients provided signed informed consent. This study was conducted in accordance with good clinical practice and all applicable regulatory requirements, including the 1996 version of the Declaration of Helsinki.
DNA sequencing
Blood was separated in plasma and buffy coat fractions. Tissues were obtained by biopsy or excision. Extracted DNA was used for library preparation, capture, and amplification per Illumina TruSight Tumor 170, which covers exons of 149 cancer-associated genes. DNA was sequenced using Illumina NextSeq 500. DNA sequencing data are available at NCBI Sequencing Read Archive (accession # PRJNA604239). Further details on DNA extraction, quantification, and bioinformatics procedures are described in Supplemental Methods and Fig. S1.
Results
Patient characteristics
Blood and tissue specimens from 4 patients were evaluated (Fig. S2). Patient #9 donated her body to research upon her death, which occurred 7 months after initial acquisition of blood and tumor specimens; she provided a second blood sample 3 months prior to death, and additional tumor specimens were obtained post-mortem (denoted as Patient #9A). Blood and tumor specimens were obtained from one patient (#2) recently diagnosed with oligometastatic disease who had not received anti-cancer therapy in the preceding 3 years, and from 3 patients who received systemic or tumor-directed radiation therapy within the preceding 4 months. Patient disease and treatment histories are detailed in Figs. S3 and S4.
DNA sequencing
DNA extracted from plasma gave the characteristic dominant ~167-base-pair fragments reported previously [20], as well as ~310-base-pair fragments in some samples (Fig. S5). DNA from plasma, tumor, and buffy coat specimens (normal liver sample was used for Patient #2) was used for capture-based sequencing of all exons in 149 cancer-associated genes of the Illumina TST170 panel. Plasma DNA samples were sequenced to an average depth of 719-fold (range of 129- to 1150-fold), tumor DNA samples were sequenced to an average depth of 1479-fold (range of 444- to 2411-fold), and buffy coat and liver DNA samples were sequenced to an average depth of 1498-fold (range of 224- to 1,506-fold) (Table S1).
Plasma DNA incompletely reflects mutational profiles of tumor DNA
Comparisons between mutations identified in plasma versus those found in tumors revealed that plasma DNA identified between 11.1% (ovary metastasis in Patient #9) and 100% (liver and omental metastases in Patient #9A) of the mutations found in tumors; Fig. 1 shows overlapping mutations identified in each patient. Overall, the ratio of mutations found in plasma to those found in tumors varied greatly: plasma from Patient #2 revealed only 6/36 (16.7%) total mutations detected in the 3 tumors sampled (1 in-breast recurrence and 2 liver metastases); plasma from Patient #10 revealed 8/17 (47.1%) total mutations found in the 3 liver metastases sampled among 5 lesions observed.
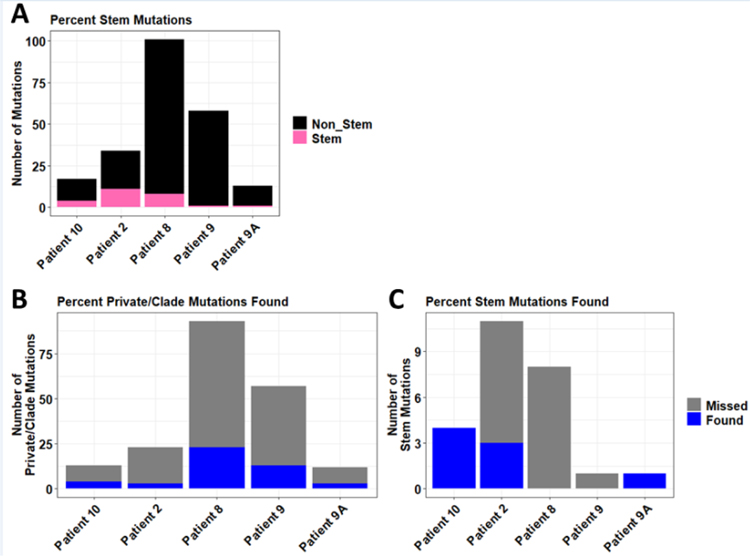
Fig. 1-. Sequencing of plasma DNA identifies tumor-derived mutations.
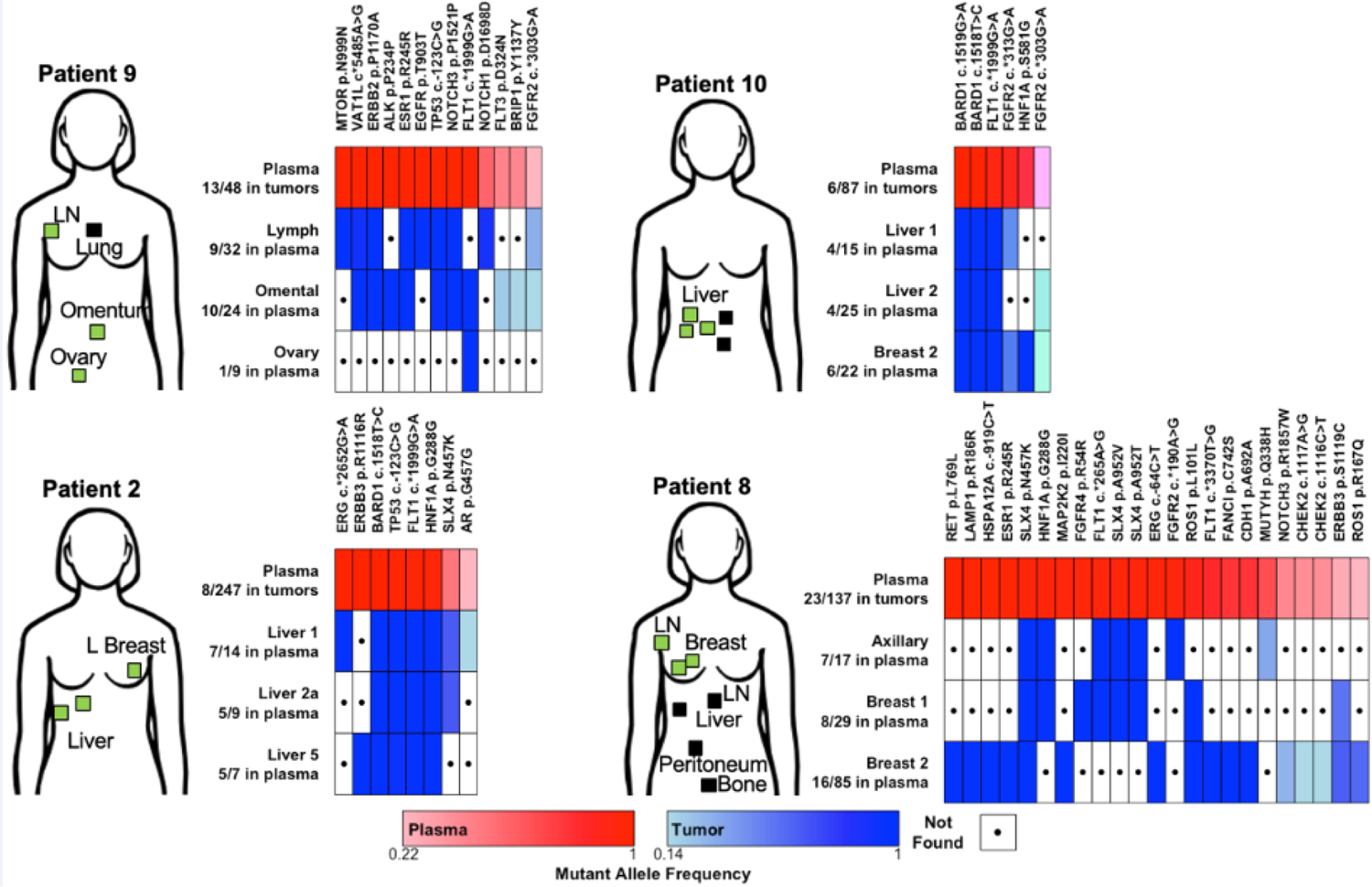
DNA from plasma, tumors, and buffy coat (or normal liver sample) from 4 patients with metastatic breast cancer was used for sequencing of all exons in 149 genes. Plasma and tumor DNA sequences were compared to buffy coat or normal liver to identify somatic mutations in each patient. Mutations present in both plasma (≥5% AF) and at least one tumor specimen (≥10% AF) are indicated in heatmaps. Samples are labeled along y-axes with number of mutations found in plasma out of number of mutations called in parentheses. Mutations are indicated along x-axes. Locations of known tumors in each patient at the time of biopsy are indicated with boxes: green indicates a sequenced lesion; black indicates a non-sequenced lesion.
Among mutations detected in both plasma and at least one tumor sample within a patient, we found significant positive correlations (p≤0.05) between plasma AF and tumor AF in 7/12 tumors across all 4 patients, but not in Patient #9A in which tumors were sampled post-mortem (likely due to the limited number of overlapping mutations found) (Table S2).
This analysis also revealed a large degree of intertumoral genetic heterogeneity (Fig. 1). For example, analysis of 3 metastases in Patient #9 revealed a total of 58 unique mutations (≥10% AF), only 1 of which was a “stem” mutation common to all 3 tumors sampled; the other 57 mutations were “private” (found in only 1 tumor) or “clade” (found in >1 but not all tumors) (Fig. 2A). Despite not occurring often, stem mutations were found at higher frequencies than private/clade mutations for most patients, with the exception of Patient #8 (Figs. 2B, C). In addition to not identifying the sole stem mutation in Patient #9, plasma samples identified 3/11 stem mutations in Patient #2, 0/8 stem mutations in Patient #8, 4/4 stem mutations in Patient #10, and 1/1 stem mutations in Patient #9A (Fig. 2C).
Fig. 2-. Plasma DNA sequencing identifies proportions of stem and non-stem mutations.
A) Proportions of stem mutations (found in all tumors) versus non-stem mutations (found in at least one but not all tumors) are shown for each patient. B/C) Proportions of non-stem (private/clade) (B) and stem (C) mutations identified in plasma DNA are shown.
Considering the low incidence of detection of tumor-derived mutations in plasma when plasma AF was ≥5%, we sought to determine if lowering the AF threshold in plasma to ≥1% captured more tumor-derived mutations (≥10% AF). The increase in numbers of mutations called in plasma samples at ≥1% AF can be seen in Table 1. Several samples saw minimal increases in the number of mutations called, with plasma from Patient #8 gaining only 2 mutations, and plasma from Patient #2 gained only 1 mutation. Lowering the threshold of plasma AF had drastic effects on the numbers of mutations called in plasma samples of Patients #9, #10, and #9A, which increased by as much as 14-fold. Despite these drastic increases in plasma mutation calls, lowering the threshold to ≥1% AF failed to identify any further mutations found in the sampled tumors, suggesting that the gaps in mutational overlap between tumors and plasma are likely not due to exclusion of rare (<1% AF) plasma mutations. Increasing the plasma DNA mutation threshold to ≥10% AF would further improve concordance with tumor mutations, as most concordant mutation calls can be characterized by greater AF in plasma (Fig. 3). Likewise, only 1 mutation <20% AF in any tumor sample was detected in a corresponding plasma sample (Fig. 4). We conclude that plasma DNA provides a snapshot of tumor mutational profiles, but the picture is incomplete and the majority of mutations identified in plasma are not attributable to sampled tumors.
Table 1-. Decreasing plasma AF threshold does not improve concordance with tumor samples.
Concordant mutations were assessed at plasma thresholds of ≥5% and ≥1% AF to determine how lower plasma AF thresholds affected concordance with tumor mutations.
| Patient | Plasma Calls at ≥5% AF | Plasma Calls at ≥1% AF | Concordant Mutations Gained |
|---|---|---|---|
| 2 | 87 | 88 | 0 |
| 8 | 137 | 139 | 0 |
| 9 | 48 | 486 | 0 |
| 9A | 122 | 1068 | 0 |
| 10 | 247 | 3477 | 0 |
Fig. 3-. Plasma DNA mutations with high allelic frequency tend to be detected in tumors.
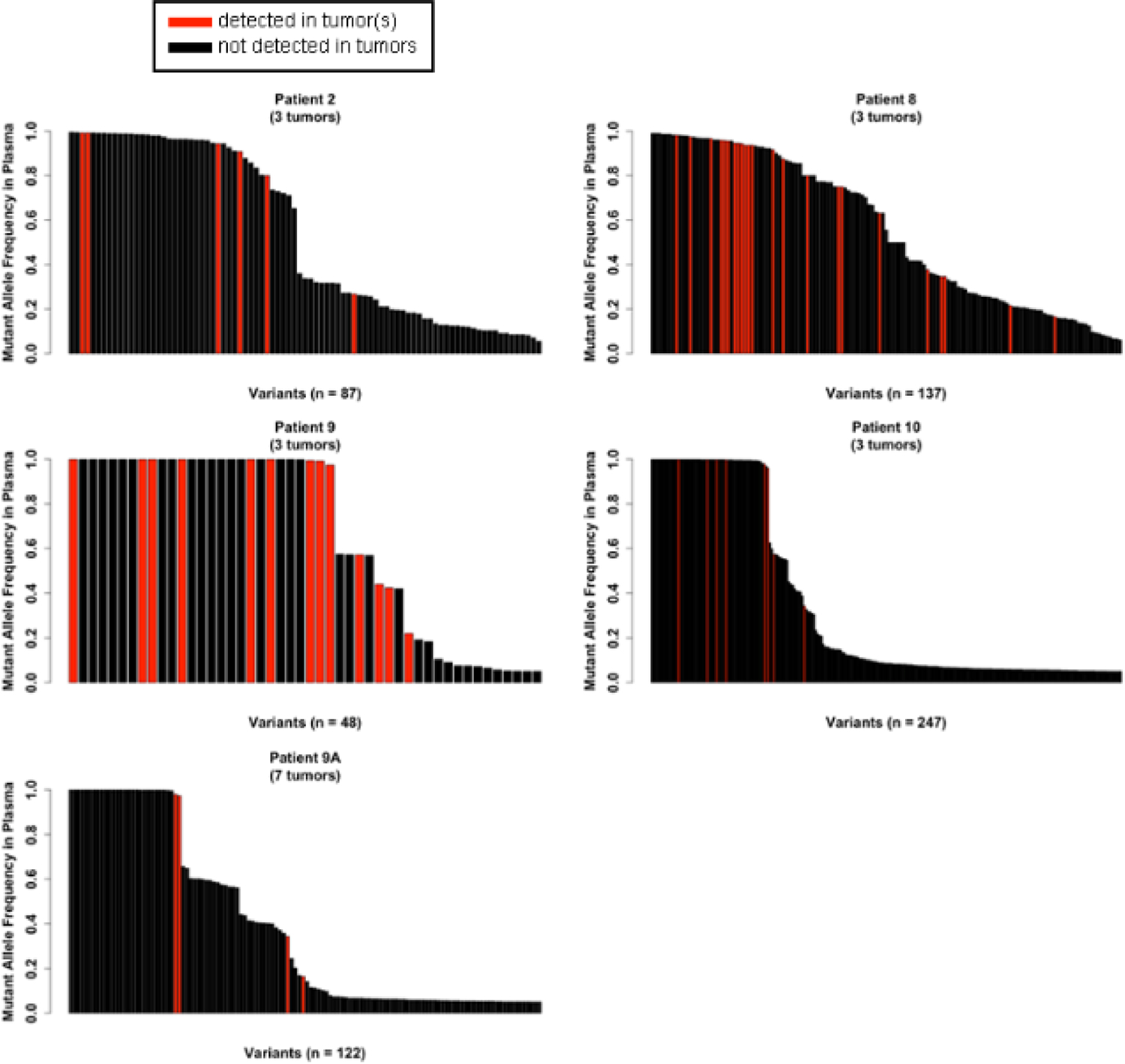
Each vertical bar indicates one mutation. Plasma AFs of mutations detected in plasma at ≥5% AF are noted for each patient along y-axes. Red bars indicate variants present in both plasma and ≥1 tumor (≥10% AF). Black bars indicate variants present in plasma only. Numbers of tumors evaluated from each patient are noted.
Fig. 4-. Tumor DNA mutations at high allelic frequency are often detected in plasma DNA.
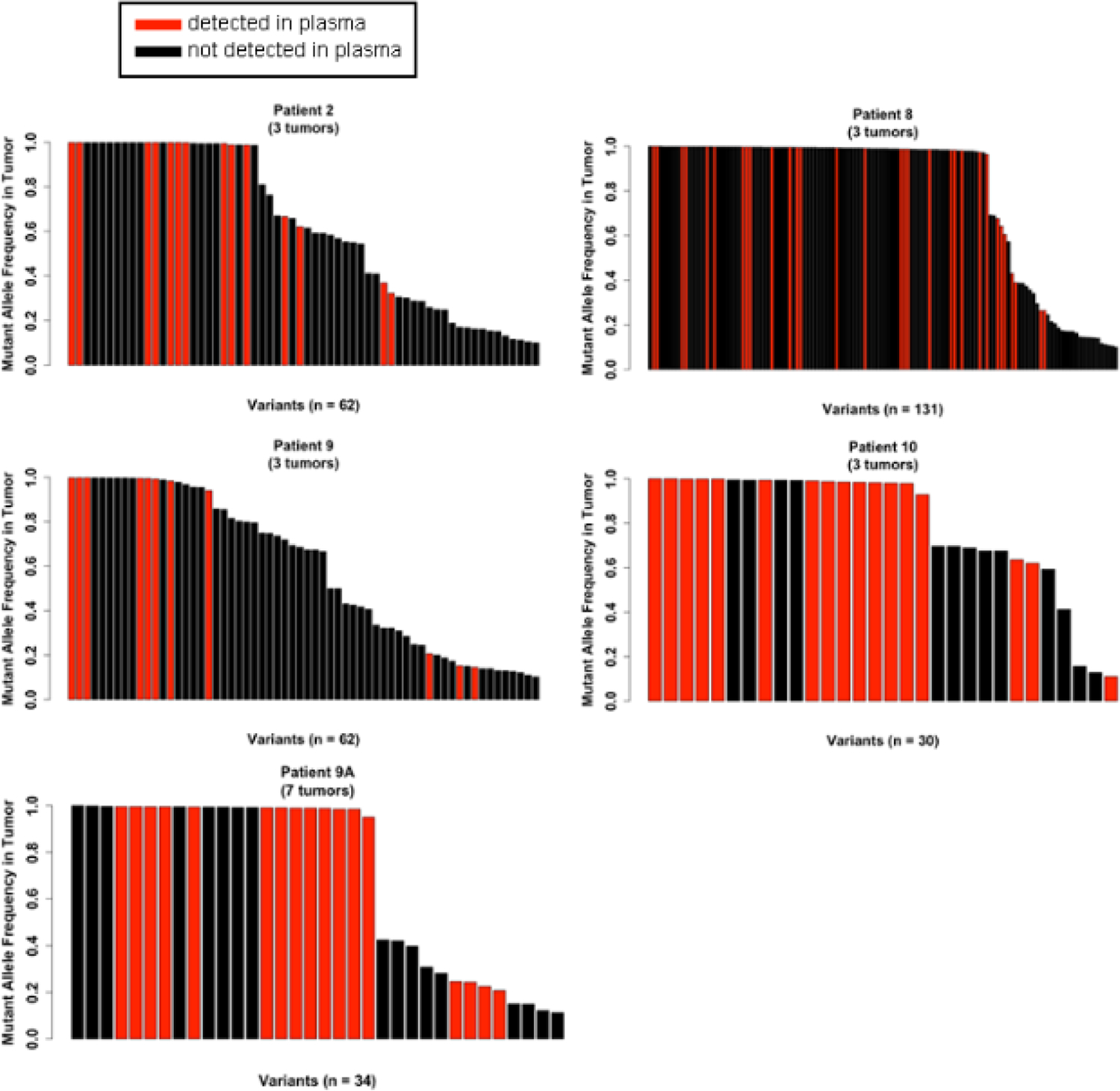
Each vertical bar indicates one mutation. Tumor AFs of mutations detected in a tumor(s) at ≥10% AF are noted for each tumor along y-axes. Red bars indicate variants present in both a tumor and plasma (≥5% AF). Black bars indicate variants present only in a tumor(s).
Plasma DNA reflects clinically occult and evident disease
Patient #9 donated her body (post-mortem) to research: a second plasma sample was acquired 3 months before her death due to widespread metastatic disease; samples from 7 metastatic tumors were acquired at autopsy; these specimens were labeled “Patient #9A” (Fig. S4). More mutations (≥5% AF) were detected in the second plasma sample compared to the plasma sample acquired 4 months earlier (Table 1), with very little overlap between the two plasma samples; increasing the threshold to ≥10% AF or decreasing to ≥1% AF had little to no effect on the number of overlapping mutation calls (Fig. S6). Contrary to other patient cases (Fig. 1), we found increased overlap between mutational profiles of plasma and tumors in Patient #9A: the 1 stem mutation found in Patient #9 was the only stem mutation found in Patient #9A, and tumor mutations were identified in the plasma of Patient #9A at a rate of concordance as high as 100% (Fig. 5A). Despite the increased frequency of tumor-associated mutations identified in plasma, there were no significant correlations between plasma AF and tumor AF in samples from Patient #9A (Table S2).
Fig. 5-. Plasma DNA reflects clinically occult and evident tumors in Patient #9A.
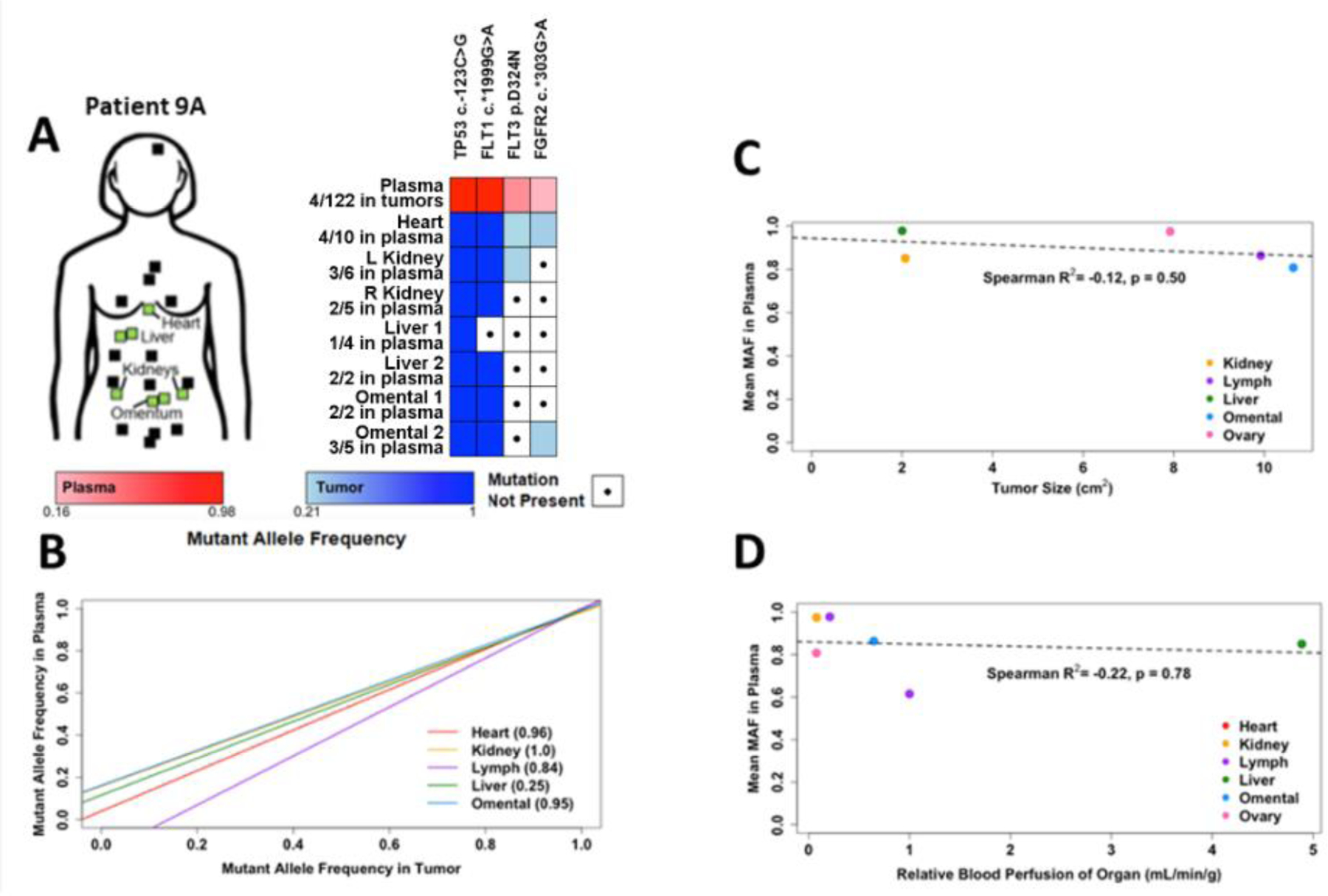
A) DNA from plasma acquired 3 months before death, buffy coat, and samples from 7 tumors acquired post-mortem from Patient #9 was used for DNA sequencing as in Fig. 1. Tumor sites sampled for DNA sequencing are indicated by green boxes. Additional tumor sites noted at autopsy (but not sampled) are indicated by black boxes. B) Mutant AFs were plotted according to organ site of tumor. For organ sites with multiple tumors, data were pooled. Data were analyzed by linear regression. C-D) Mutant AFs in plasma were compared to tumor size (mean from each organ site) determined from CT scans (C) and relative blood perfusion (mL/min) of tumor-containing organs (D) using Spearman correlation and linear regression analyses.
Patient #9A provided tissue samples from metastases found in the heart, omentum (n=2), kidneys (one from each), and liver (n=2). In patients with advanced breast cancer, metastases to the heart and peritoneum (including omentum) occur in 15.5% (evaluated at autopsy) and 7.6% (evaluated by imaging) of cases, respectively [21,22]. The heart is typically not evaluated during routine follow-up due to variability in cardiac uptake of [18F]FDG in PET imaging [23], and heart motion precluding measurement by CT. Omentum is often not evaluable by CT because it contains primarily fat that blends with subperitoneal fat upon anatomical imaging, but CT remains the best routine imaging modality to detect peritoneal metastasis [24,25]. Thus, heart and omentum are two sites of disease that are difficult to follow clinically, but metastases in these organs were potentially represented (FGFR2 c.*303G>A and FLT3 p.D324N) in the pool of ctDNA in plasma in Patient #9A (Fig. 5A).
We then combined data from all time points of sample acquisition from Patient #9 and #9A. Since some tumors sampled at autopsy were not detected or measurable in radiographs, mutational profiles of multiple tumors acquired from the same organ site (n=6) were combined for this analysis. Comparison of AF of mutations in plasma DNA vs. organ-specific tumor DNA revealed that all tumor samples aside from the lymph node metastasis exhibited high correlation with plasma (Spearman R2 > 0.8, Fig. 5B), indicating that most tumor samples are well-represented in the plasma. As the AF of mutations in tumor and plasma increased, concordance of mutually detected mutations also increased, lending confidence to higher-AF mutation calls in plasma. We considered whether tumor size (Fig. S7) was associated with contribution of DNA to plasma, and thus represented by mutation AF in plasma; however, we found a non-significant slightly negative correlation between tumor size (using mean of tumor sizes from a given organ site) and median AF of mutations in plasma (Fig. 5C).
Since a heart metastasis was potentially represented in plasma DNA (Fig. 5A/B), and a source of ctDNA is leakage of tumor DNA into the bloodstream, we considered whether tumor location and access to the bloodstream was associated with contribution of tumor DNA to plasma. As a surrogate for tumor access to the bloodstream, we used blood perfusion (mL/min) of an organ site [26–28] for each tumor-containing organ. Organ blood perfusion rate, relative to the heart, was not significantly correlated with AF of mutations in plasma DNA (Fig. 5D). These data suggest that neither tumor location and access to the bloodstream nor tumor size are strongly associated with contribution of DNA to plasma.
Timing or sampling bias may affect mutation detection in plasma
As previously shown in Fig. S6, mutational profiles of plasma samples can differ between time points within a patient; this difference may be due to disease evolution and/or plasma sampling bias. To examine the effect that this could have on mutation calls of tumor samples for Patient #9/9A, we tested whether the near-autopsy plasma sample (#9A) revealed mutations found in the first set of tumor biopsy samples (#9; acquired 4 months earlier) that the first plasma sample (#9) did not; conversely, we tested whether the first plasma sample (#9) revealed mutations found in the tumor samples taken at autopsy (#9A; acquired 7 months later) that the near-autopsy plasma sample (#9A) did not (Table 2).
Table 2-. Timing of plasma sample acquisition identifies new mutations.
To determine if multiple plasma samples over time could identify previously missed tumor mutations, tumor samples in Patient #9 collected at autopsy were compared to near autopsy mutations, and vice versa. Nucleotide changes are shown, with notations for synonymous (s), missense (m), or UTR variants (u).
| Patient | Sample | Concordant Mutations Newly Identified |
|---|---|---|
| 9 | Lymph Node Met | AKT3 c.*461A>G (u), ERBB3 c.3348G>A (s), |
| HNF1A c.1741A>G (m), FGFR4 c.1162G>A (m) | ||
| Omental Met | RET c.135A>G (s), ALK c.702T>A (s), | |
| FGFR4 c.1162G>A (m), SDK1 c.*5301G>A (u) | ||
| Ovarian Met | BRCA1 c.4900A>G (m), FGFR4 c.1162G>A (m), NOTCH1 | |
| c.6555C>T (s) | ||
| 9A | Heart Met | TSC2 c.5202T>C (s) |
| Left Kidney Met | None | |
| Right Kidney Met | SRC c.1218G>C (s) | |
| Liver 1 Met | None | |
| Liver 2 Met | None | |
| Omental 1 Met | None | |
In the tumor samples acquired at autopsy, using the first plasma sample would have identified 1 additional mutation found in the heart metastasis, and 1 additional mutation found in the right kidney metastasis. In the earlier set of tumor biopsies, using the near-autopsy plasma sample identified several new mutations in each of the tumors. This might have proved useful in the case of identifying mutations in the ovarian metastasis, where the first plasma sample only identified one mutation. The near-autopsy plasma sample identifies 4 additional mutations, including missense variants BRCA1 p.S1634G and FGFR4 p.G388R, the latter of which has been associated with progression and treatment resistance in breast cancer [29–31]. This suggests that identifying variants in plasma samples in a time-course manner may provide a more complete picture of the mutational profile of the patient and allow for better identification of therapeutic targets, as has also been suggested previously [32].
Discussion
Studies have shown that tumor-derived DNA is detectable in plasma, prompting the development of diagnostic tests using plasma as a “liquid biopsy.” Although liquid biopsy is undoubtedly useful for detection of residual disease and cancer progression [12,16], it remains unclear whether plasma DNA is suitable for genetic profiling to inform selection of targeted therapies. The results of our study were varied, as we found that sequencing of plasma DNA captured as many as 100% (Patient #9A) to as few as 11.1% (Patient #9) of mutations (≥5% AF in plasma) detected in ≥1 tumor at ≥10% AF in patients with oligometastatic breast cancer. The majority (mean 79.3% +/− SD 13.8%) of mutations found in plasma DNA at ≥5% AF were not found in corresponding sampled tumors; this result may have been affected by the sequencing depth of the plasma samples. Chae et al. similarly reported 31.7% of all tumor mutations were found in plasma samples of 45 patients with metastatic breast cancer using the Guardant360 sequencing panel [33].
There was a negative correlation (Spearman R2 = −0.53) between the number of plasma samples also found in corresponding tumors and sequencing depth of the plasma sample, suggesting that the proportion of plasma mutations found in tumor samples decreases as sequencing depth of plasma increases; this is likely due to increased sequencing depth leading to more low-AF mutation calls, which have already been shown to have no impact on concordance with tumor mutation calls (Table 1). The majority (68.5%) of mutations found in both plasma and tumors had a plasma AF ≥ 0.8 (Fig. 3), giving confidence in the tumor origin of high-AF mutations found in plasma
Studies have used several methodologies to evaluate the utility of plasma DNA sequencing as a surrogate for tumor DNA sequencing. Several studies used the amplicon-based Ion Torrent AmpliSeq platform that covers 2855 catalogued mutations across 50 genes. Rothé et al. found 0 to 2 mutations in 5 genes (TP53, PIK3CA, PTEN, AKT1, and IDH2) in plasma or tumor specimens among 17 patients with metastatic breast cancer [34]. Their study found that plasma samples identified mutations in tumors in 13/17 (76%) patients, yet overall correlation of AF reported was lower than our study; the relatively good concordance may have been a result of the low number of mutations in detected in the few genes reported. Frenel et al. evaluated specimens from patients with late-stage metastatic solid tumors, and identified 1 to 5 (median=2) mutations in each tumor [35]. Sequencing of plasma DNA identified ≥1 tumor-derived mutation in 23/39 (59%) patients. In patients with advanced solid tumors, Jovelet et al. found 0 to 4 (median=1) mutations per tumor, and plasma DNA provided mutational concordance with tumors in 145/283 (51%) patients [36]. A major limitation of such amplicon-based platforms is the short amplicon length (mean=154 bp for AmpliSeq), where most plasma DNA fragments (~167 bp) would not provide template for amplification. More akin to the capture-based method that we used herein (which is not subject to bias due to DNA fragment size), Dietz et al. performed capture-based whole-exome sequencing (Agilent High Sensitivity Kit, sequenced on Illumina HiSeq 2000) of DNA from serum and primary tumor specimens from 6 patients with early-stage lung cancer, achieving 68.5-fold mean depth of coverage for serum DNA [37]. Their analysis identified averages of 2557 (range: 1892–3322) and 1416 (range: 589–4105) mutations among tumor specimens (20–80% AF) and serum specimens (>0% AF and ≥10 mutant reads), respectively. Comparing patient-matched specimens, Dietz et al. detected 5–57% (mean=23%) of tumor mutations in serum, and 22–41% (mean=31%) of serum mutations in tumors. A limitation of their study was a lack of normal DNA (e.g., buffy coat) controls, which may have resulted in the inclusion of many false positives, including germline mutations and clustered mutation events [38,39]. More recently, Wyatt et al. reported an impressive 93.6% concordance among metastatic prostate cancer patients using targeted sequencing of 72 prostate cancer-related genes, finding 109 unique somatic mutations within those 72 genes [40]. In comparison, we found a total of 272 (range: 2–85) unique mutations within ≥1 tumor (≥10% AF) when considering all 4 patients among 149 cancer-related genes. These ranges in mutational burden are similar to those reported by Shah et al. [41], who reported that in cases of triple-negative breast cancer, numbers of somatic mutations ranged from as few as 5 to as many as 100 using whole exome sequencing and RNA-seq. The mutational overlap between tumors and plasma was lower in the current study, possibly due to the advanced stage of disease (advanced breast cancer vs. early-stage lung cancer), and increased genetic intra-and inter-tumor heterogeneity in patients with advanced disease [42]; however, in the case of Patient #2, all 3 clinically evident tumors were sampled, helping to rule out the variable of inter-tumor heterogeneity. We found that decreasing the plasma DNA mutation threshold to ≥1% AF had no effect on overlap with groups of tumor mutations, suggesting that low-AF mutations found in plasma were mostly A) below the threshold used for calling mutations in tumors, and/or B) sequencing errors. Differences in DNA sequencing platforms (Illumina and Ion Torrent have error rates of 1.78% vs. 0.4% [43]) and variant callers [37,44–48] may have also contributed to disparate results.
The clinical importance of the discrepancy between tumor and plasma mutational profiles is exemplified by the lack of detection of a TP53 p.P72R mutation found in all tumor samples in Patient #2, which was not detected in the corresponding plasma sample. This mutation has previously been reported as a strong indicator of toxicity due to cyclophosphamide-based neoadjuvant chemotherapy in breast cancer patients [49,50]. Patient #2 did not receive cyclophosphamide as part of their treatment regimen, but an accurate plasma sample denoting this mutation could have been invaluable in making treatment decisions for this patient.
As liquid biopsy profiling assays have been developed, a recurring concern has been whether plasma DNA reflects the clinically important (i.e., life-threatening) tumor(s) in a patient. Results of our analysis of tumor specimens acquired at autopsy from Patient #9 highlight an inherent disconnect between tumors in vital organs and plasma DNA. Among the 7 tumors evaluated, the mutational profile of a heart metastasis was potentially represented in plasma DNA; this was confounded by the detection of the same mutations in kidney and/or omental metastases (Fig. 2). If Patient #9 had diagnostic plasma-only DNA sequencing, a genetically informed therapy could have been selected based on a private or clade mutation(s) found in the heart tumor. In this scenario, the heart tumor (which was not detected by routine imaging) may have responded to treatment, while other (detectable) tumors that did not harbor the same mutation(s) could have progressed. As a result, the patient would have been taken off treatment due to disease progression, which would be a failure of genetically informed therapy.
Conclusions
Plasma contains a mixture of DNA from tumors in patients with oligometastatic disease, so liquid biopsy creates a gap between tumor identity and tumor genetic profile: mutations identified in plasma DNA cannot be attributed to a specific tumor, so response of a given tumor to therapy may not be attributable to mutations identified in plasma DNA. In the case of Patient #9, only 13/48 (27%) mutations found in plasma were also found in tumor samples. This low rate of concordance could be due to several factors, most notably the presence of other tumors in the body that were occult or not biopsied.
The oncology community must ultimately determine the value of plasma DNA profiling for selecting targeted therapies. Our findings indicate that plasma DNA sequencing is a weak surrogate for tumor DNA sequencing, sporadically detecting a minority of tumor mutations. We conclude that liquid biopsy should not yet replace, but may complement, tumor biopsy for genetic profiling.
Supplementary Material
Acknowledgments
We are grateful to the patients who participated in this study and their family members who supported them. We thank the Norris Cotton Cancer Center Genomics and Pathology Shared Resources for assistance.
Funding: This study was funded by Norris Cotton Cancer Center (Hopeman Foundation Pilot Grant to MDC and TWM), and the NIH (F30CA216966 to KS; R01CA200994 and R01CA211869 to TWM; Dartmouth College Norris Cotton Cancer Center Support Grant P30CA023108).
Abbreviations Used
- AF
allelic frequency
- ctDNA
cell-free circulating tumor DNA
- FFPE
formalin-fixed and paraffin-embedded
- GATK
Genome Analysis Toolkit
- [18F]FDG
fluorodeoxyglucose
- PET
positron emission tomography
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Conflict of Interest: The authors declare that they have no conflict of interest.
Ethical approval: All procedures performed involving human subjects were approved by the Dartmouth College Institutional Review Board and were consistent with the 1996 version of the Declaration of Helsinki.
Informed consent: Informed consent was obtained from all individual participants included in the study.
References
- 1.Engelman JA, Janne PA, Mermel C, Pearlberg J, Mukohara T, Fleet C, Cichowski K, Johnson BE, Cantley LC (2005) ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proceedings of the National Academy of Sciences of the United States of America 102 (10):3788–3793. 10.1073/pnas.0409773102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin LA, Pancholi S, Farmer I, Guest S, Ribas R, Weigel MT, Thornhill AM, Ghazoui Z, A’Hern R, Evans DB, Lane HA, Johnston SR, Dowsett M (2012) Effectiveness and molecular interactions of the clinically active mTORC1 inhibitor everolimus in combination with tamoxifen or letrozole in vitro and in vivo. Breast cancer research : BCR 14 (5):R132 10.1186/bcr3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Juric D, Castel P, Griffith M, Griffith OL, Won HH, Ellis H, Ebbesen SH, Ainscough BJ, Ramu A, Iyer G, Shah RH, Huynh T, Mino-Kenudson M, Sgroi D, Isakoff S, Thabet A, Elamine L, Solit DB, Lowe SW, Quadt C, Peters M, Derti A, Schegel R, Huang A, Mardis ER, Berger MF, Baselga J, Scaltriti M (2015) Convergent loss of PTEN leads to clinical resistance to a PI(3)Kalpha inhibitor. Nature 518 (7538):240–244. 10.1038/nature13948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu ZY, Xie N, Tian C, Yang X, Liu L, Li J, Xiao H, Wu H, Lu J, Gao J, Hu X, Cao M, Shui Z, Xiao M, Tang Y, He Q, Chang L, Xia X, Yi X, Liao Q, Ouyang Q (2018) Identifying Circulating Tumor DNA Mutation Profiles in Metastatic Breast Cancer Patients with Multiline Resistance. EBioMedicine 32:111–118. 10.1016/j.ebiom.2018.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinberg AL, Carter D, Ahonen M, Alarid ET, Murdoch FE, Fritsch MK (2007) The DNA binding domain of estrogen receptor alpha is required for high-affinity nuclear interaction induced by estradiol. Biochemistry-Us 46 (31):8933–8942. 10.1021/bi700018w [DOI] [PubMed] [Google Scholar]
- 6.Dunn CA, Clark W, Black EJ, Gillespie DAF (2003) Estrogen receptor activation function 2 (AF-2) is essential for hormone-dependent transactivation and cell transformation induced by a v-Jun DNA binding domain-estrogen receptor chimera. Bba-Gene Struct Expr 1628 (3):147–155. 10.1016/S0167-4781(03)00136-2 [DOI] [PubMed] [Google Scholar]
- 7.von Lindern M, Boer L, Wessely O, Parker M, Beug H (1998) The transactivation domain AF-2 but not the DNA-binding domain of the estrogen receptor is required to inhibit differentiation of avian erythroid progenitors. Mol Endocrinol 12 (2):263–277. 10.1210/Me.12.2.263 [DOI] [PubMed] [Google Scholar]
- 8.Klinge CM, Brolly CL, Bambara RA, Hilf R (1997) hsp70 is not required for high affinity binding of purified calf uterine estrogen receptor to estrogen response element DNA in vitro. J Steroid Biochem 63 (4–6):283–301. 10.1016/S0960-0760(97)00091-5 [DOI] [PubMed] [Google Scholar]
- 9.McBride DJ, Orpana AK, Sotiriou C, Joensuu H, Stephens PJ, Mudie LJ, Hamalainen E, Stebbings LA, Andersson LC, Flanagan AM, Durbecq V, Ignatiadis M, Kallioniemi O, Heckman CA, Alitalo K, Edgren H, Futreal PA, Stratton MR, Campbell PJ (2010) Use of cancer-specific genomic rearrangements to quantify disease burden in plasma from patients with solid tumors. Genes Chromosomes Cancer 49 (11):1062–1069. 10.1002/gcc.20815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, Antipova A, Lee C, McKernan K, De La Vega FM, Kinzler KW, Vogelstein B, Diaz LA Jr., Velculescu VE (2010) Development of personalized tumor biomarkers using massively parallel sequencing. Science translational medicine 2 (20):20ra14 10.1126/scitranslmed.3000702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorenson GD, Pribish DM, Valone FH, Memoli VA, Bzik DJ, Yao SL (1994) Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 3 (1):67–71 [PubMed] [Google Scholar]
- 12.Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, Cheang M, Osin P, Nerurkar A, Kozarewa I, Garrido JA, Dowsett M, Reis-Filho JS, Smith IE, Turner NC (2015) Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Science translational medicine 7 (302):302ra133 10.1126/scitranslmed.aab00217/302/302ra133 [pii] [DOI] [PubMed] [Google Scholar]
- 13.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, Abbott RM, Hoog J, Dooling DJ, Koboldt DC, Schmidt H, Kalicki J, Zhang Q, Chen L, Lin L, Wendl MC, McMichael JF, Magrini VJ, Cook L, McGrath SD, Vickery TL, Appelbaum E, Deschryver K, Davies S, Guintoli T, Lin L, Crowder R, Tao Y, Snider JE, Smith SM, Dukes AF, Sanderson GE, Pohl CS, Delehaunty KD, Fronick CC, Pape KA, Reed JS, Robinson JS, Hodges JS, Schierding W, Dees ND, Shen D, Locke DP, Wiechert ME, Eldred JM, Peck JB, Oberkfell BJ, Lolofie JT, Du F, Hawkins AE, O’Laughlin MD, Bernard KE, Cunningham M, Elliott G, Mason MD, Thompson DM Jr., Ivanovich JL, Goodfellow PJ, Perou CM, Weinstock GM, Aft R, Watson M, Ley TJ, Wilson RK, Mardis ER (2010) Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464 (7291):999–1005. 10.1038/nature08989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taniguchi K, Uchida J, Nishino K, Kumagai T, Okuyama T, Okami J, Higashiyama M, Kodama K, Imamura F, Kato K (2011) Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clinical cancer research : an official journal of the American Association for Cancer Research 17 (24):7808–7815. 10.1158/1078-0432.CCR-11-1712 [DOI] [PubMed] [Google Scholar]
- 15.Baselga J, Im S-A, Iwata H, Clemons M, Ito Y, Awada A, Chia S, Jagiello-Gruszfeld A, Pistilli B, Tseng L-M, Hurvitz S, Masuda N, Cortés J, De Laurentiis M, Arteaga CL, Jiang Z, Jonat W, Hachemi S, Le Mouhaër S, Di Tomaso E, Urban P, Massacesi C, Campone M (2015) PIK3CA status in circulating tumor DNA (ctDNA) predicts efficacy of buparlisib (BUP) plus fulvestrant (FULV) in postmenopausal women with endocrine-resistant HR+/HER2– advanced breast cancer (BC): First results from the randomized, phase III BELLE-2 trial. Presented at the San Antonio Breast Cancer Symposium (Dec 11, 2015) Abstract S6–01 [Google Scholar]
- 16.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, Rajan S, Humphray S, Becq J, Halsall D, Wallis M, Bentley D, Caldas C, Rosenfeld N (2013) Analysis of circulating tumor DNA to monitor metastatic breast cancer. The New England journal of medicine 368 (13):1199–1209. 10.1056/NEJMoa1213261 [DOI] [PubMed] [Google Scholar]
- 17.Haber DA, Velculescu VE (2014) Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov 4 (6):650–661. 10.1158/2159-8290.CD-13-10142159-8290.CD-13-1014 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ignatiadis M, Dawson SJ (2014) Circulating tumor cells and circulating tumor DNA for precision medicine: dream or reality? Annals of oncology : official journal of the European Society for Medical Oncology 25 (12):2304–2313. 10.1093/annonc/mdu480 [DOI] [PubMed] [Google Scholar]
- 19.Smith NG, Gyanchandani R, Shah OS, Gurda GT, Lucas PC, Hartmaier RJ, Brufsky AM, Puhalla S, Bahreini A, Kota K, Wald AI, Nikiforov YE, Nikiforova MN, Oesterreich S, Lee AV (2019) Targeted mutation detection in breast cancer using MammaSeq. Breast Cancer Res 21 (1):22 10.1186/s13058-019-1102-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo YM, Chan KC, Sun H, Chen EZ, Jiang P, Lun FM, Zheng YW, Leung TY, Lau TK, Cantor CR, Chiu RW (2010) Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Science translational medicine 2 (61):61ra91 10.1126/scitranslmed.30017202/61/61ra91 [pii] [DOI] [PubMed] [Google Scholar]
- 21.Bussani R, De-Giorgio F, Abbate A, Silvestri F (2007) Cardiac metastases. Journal of clinical pathology 60 (1):27–34. 10.1136/jcp.2005.035105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertozzi S, Londero AP, Cedolini C, Uzzau A, Seriau L, Bernardi S, Bacchetti S, Pasqual EM, Risaliti A (2015) Prevalence, risk factors, and prognosis of peritoneal metastasis from breast cancer. SpringerPlus 4:688 10.1186/s40064-015-1449-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maurer AH, Burshteyn M, Adler LP, Steiner RM (2011) How to Differentiate Benign versus Malignant Cardiac and Paracardiac F-18 FDG Uptake at Oncologic PET/CT. Radiographics 31 (5):1287–1305. 10.1148/rg.315115003 [DOI] [PubMed] [Google Scholar]
- 24.Raptopoulos V, Gourtsoyiannis N (2001) Peritoneal carcinomatosis. European radiology 11 (11):2195–2206. 10.1007/s003300100998 [DOI] [PubMed] [Google Scholar]
- 25.Laghi A, Bellini D, Rengo M, Accarpio F, Caruso D, Biacchi D, Di Giorgio A, Sammartino P (2017) Diagnostic performance of computed tomography and magnetic resonance imaging for detecting peritoneal metastases: systematic review and meta-analysis. La Radiologia medica 122 (1):1–15. 10.1007/s11547-016-0682-x [DOI] [PubMed] [Google Scholar]
- 26.Edginton AN, Ritter L (2009) Predicting plasma concentrations of bisphenol A in children younger than 2 years of age after typical feeding schedules, using a physiologically based toxicokinetic model. Environ Health Perspect 117 (4):645–653. 10.1289/ehp.0800073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hay JB, Hobbs BB (1977) The flow of blood to lymph nodes and its relation to lymphocyte traffic and the immune response. The Journal of experimental medicine 145 (1):31–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Virtanen KA, Lonnroth P, Parkkola R, Peltoniemi P, Asola M, Viljanen T, Tolvanen T, Knuuti J, Ronnemaa T, Huupponen R, Nuutila P (2002) Glucose uptake and perfusion in subcutaneous and visceral adipose tissue during insulin stimulation in nonobese and obese humans. J Clin Endocrinol Metab 87 (8):3902–3910. 10.1210/jcem.87.8.8761 [DOI] [PubMed] [Google Scholar]
- 29.Bange J, Prechtl D, Cheburkin Y, Specht K, Harbeck N, Schmitt M, Knyazeva T, Muller S, Gartner S, Sures I, Wang H, Imyanitov E, Haring HU, Knayzev P, Iacobelli S, Hofler H, Ullrich A (2002) Cancer progression and tumor cell motility are associated with the FGFR4 Arg(388) allele. Cancer Res 62 (3):840–847 [PubMed] [Google Scholar]
- 30.Thussbas C, Nahrig J, Streit S, Bange J, Kriner M, Kates R, Ulm K, Kiechle M, Hoefler H, Ullrich A, Harbeck N (2006) FGFR4 Arg388 allele is associated with resistance to adjuvant therapy in primary breast cancer. J Clin Oncol 24 (23):3747–3755. 10.1200/JCO.2005.04.8587 [DOI] [PubMed] [Google Scholar]
- 31.Xu B, Tong N, Chen SQ, Hua LX, Wang ZJ, Zhang ZD, Chen M (2011) FGFR4 Gly388Arg polymorphism contributes to prostate cancer development and progression: a meta-analysis of 2618 cases and 2305 controls. BMC Cancer 11:84 10.1186/1471-2407-11-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hojbjerg JA, Madsen AT, Schmidt HH, Sorensen SF, Stougaard M, Meldgaard P, Sorensen BS (2019) Intra-individual variation of circulating tumour DNA in lung cancer patients. Mol Oncol 13 (10):2098–2106. 10.1002/1878-0261.12546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chae YK, Davis AA, Jain S, Santa-Maria C, Flaum L, Beaubier N, Platanias LC, Gradishar W, Giles FJ, Cristofanilli M (2017) Concordance of Genomic Alterations by Next-Generation Sequencing in Tumor Tissue versus Circulating Tumor DNA in Breast Cancer. Mol Cancer Ther 16 (7):1412–1420. 10.1158/1535-7163.MCT-17-0061 [DOI] [PubMed] [Google Scholar]
- 34.Rothe F, Laes JF, Lambrechts D, Smeets D, Vincent D, Maetens M, Fumagalli D, Michiels S, Drisis S, Moerman C, Detiffe JP, Larsimont D, Awada A, Piccart M, Sotiriou C, Ignatiadis M (2014) Plasma circulating tumor DNA as an alternative to metastatic biopsies for mutational analysis in breast cancer. Annals of oncology : official journal of the European Society for Medical Oncology 25 (10):1959–1965. 10.1093/annonc/mdu288 [DOI] [PubMed] [Google Scholar]
- 35.Frenel JS, Carreira S, Goodall J, Roda D, Perez-Lopez R, Tunariu N, Riisnaes R, Miranda S, Figueiredo I, Nava-Rodrigues D, Smith A, Leux C, Garcia-Murillas I, Ferraldeschi R, Lorente D, Mateo J, Ong M, Yap TA, Banerji U, Tandefelt DG, Turner N, Attard G, de Bono JS (2015) Serial Next-Generation Sequencing of Circulating Cell-Free DNA Evaluating Tumor Clone Response To Molecularly Targeted Drug Administration. Clinical cancer research : an official journal of the American Association for Cancer Research 21 (20):4586–4596. 10.1158/1078-0432.CCR-15-0584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jovelet C, Ileana E, Le Deley MC, Motte N, Rosellini S, Romero A, Lefebvre C, Pedrero M, Pata-Merci N, Droin N, Deloger M, Massard C, Hollebecque A, Ferte C, Boichard A, Postel-Vinay S, Ngo-Camus M, De Baere T, Vielh P, Scoazec JY, Vassal G, Eggermont A, Andre F, Soria JC, Lacroix L (2016) Circulating Cell-Free Tumor DNA Analysis of 50 Genes by Next-Generation Sequencing in the Prospective MOSCATO Trial. Clinical cancer research : an official journal of the American Association for Cancer Research 22 (12):2960–2968. 10.1158/1078-0432.CCR-15-2470 [DOI] [PubMed] [Google Scholar]
- 37.Dietz S, Schirmer U, Merce C, von Bubnoff N, Dahl E, Meister M, Muley T, Thomas M, Sultmann H (2016) Low Input Whole-Exome Sequencing to Determine the Representation of the Tumor Exome in Circulating DNA of Non-Small Cell Lung Cancer Patients. PloS one 11 (8):e0161012 10.1371/journal.pone.0161012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Fritz MH, Konkel MK, Malhotra A, Stutz AM, Shi X, Casale FP, Chen J, Hormozdiari F, Dayama G, Chen K, Malig M, Chaisson MJP, Walter K, Meiers S, Kashin S, Garrison E, Auton A, Lam HYK, Mu XJ, Alkan C, Antaki D, Bae T, Cerveira E, Chines P, Chong Z, Clarke L, Dal E, Ding L, Emery S, Fan X, Gujral M, Kahveci F, Kidd JM, Kong Y, Lameijer EW, McCarthy S, Flicek P, Gibbs RA, Marth G, Mason CE, Menelaou A, Muzny DM, Nelson BJ, Noor A, Parrish NF, Pendleton M, Quitadamo A, Raeder B, Schadt EE, Romanovitch M, Schlattl A, Sebra R, Shabalin AA, Untergasser A, Walker JA, Wang M, Yu F, Zhang C, Zhang J, Zheng-Bradley X, Zhou W, Zichner T, Sebat J, Batzer MA, McCarroll SA, Genomes Project C, Mills RE, Gerstein MB, Bashir A, Stegle O, Devine SE, Lee C, Eichler EE, Korbel JO (2015) An integrated map of structural variation in 2,504 human genomes. Nature 526 (7571):75–81. 10.1038/nature15394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whelan S, Goldman N (2004) Estimating the frequency of events that cause multiple-nucleotide changes. Genetics 167 (4):2027–2043. 10.1534/genetics.103.023226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wyatt AW, Annala M, Aggarwal R, Beja K, Feng F, Youngren J, Foye A, Lloyd P, Nykter M, Beer TM, Alumkal JJ, Thomas GV, Reiter RE, Rettig MB, Evans CP, Gao AC, Chi KN, Small EJ, Gleave ME (2017) Concordance of Circulating Tumor DNA and Matched Metastatic Tissue Biopsy in Prostate Cancer. J Natl Cancer Inst 109 (12). 10.1093/jnci/djx118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, Bashashati A, Prentice LM, Khattra J, Burleigh A, Yap D, Bernard V, McPherson A, Shumansky K, Crisan A, Giuliany R, Heravi-Moussavi A, Rosner J, Lai D, Birol I, Varhol R, Tam A, Dhalla N, Zeng T, Ma K, Chan SK, Griffith M, Moradian A, Cheng SW, Morin GB, Watson P, Gelmon K, Chia S, Chin SF, Curtis C, Rueda OM, Pharoah PD, Damaraju S, Mackey J, Hoon K, Harkins T, Tadigotla V, Sigaroudinia M, Gascard P, Tlsty T, Costello JF, Meyer IM, Eaves CJ, Wasserman WW, Jones S, Huntsman D, Hirst M, Caldas C, Marra MA, Aparicio S (2012) The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486 (7403):395–399. 10.1038/nature10933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia-Saenz JA, Ayllon P, Laig M, Acosta-Eyzaguirre D, Garcia-Esquinas M, Montes M, Sanz J, Barquin M, Moreno F, Garcia-Barberan V, Diaz-Rubio E, Caldes T, Romero A (2017) Tumor burden monitoring using cell-free tumor DNA could be limited by tumor heterogeneity in advanced breast cancer and should be evaluated together with radiographic imaging. BMC Cancer 17 (1):210 10.1186/s12885-017-3185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y (2012) A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC genomics 13:341 10.1186/1471-2164-13-341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hwang S, Kim E, Lee I, Marcotte EM (2015) Systematic comparison of variant calling pipelines using gold standard personal exome variants. Sci Rep-Uk 5 10.1038/Srep17875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kroigard AB, Thomassen M, Laenkholm AV, Kruse TA, Larsen MJ (2016) Evaluation of Nine Somatic Variant Callers for Detection of Somatic Mutations in Exome and Targeted Deep Sequencing Data. PloS one 11 (3). 10.1371/journal.pone.0151664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G (2013) Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature biotechnology 31 (3):213–219. 10.1038/nbt.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK (2012) VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome research 22 (3):568–576. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK (2012) Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28 (14):1811–1817. 10.1093/bioinformatics/bts271 [DOI] [PubMed] [Google Scholar]
- 49.Henriquez-Hernandez LA, Murias-Rosales A, Gonzalez-Hernandez A, de Leon AC, Diaz-Chico N, Fernandez-Perez L (2010) Distribution of TYMS, MTHFR, p53 and MDR1 gene polymorphisms in patients with breast cancer treated with neoadjuvant chemotherapy. Cancer Epidemiol 34 (5):634–638. 10.1016/j.canep.2010.06.013 [DOI] [PubMed] [Google Scholar]
- 50.Henriquez-Hernandez LA, Perez LF, Hernandez AG, de Leon AC, Diaz-Chico B, Rosales AM (2010) TYMS, MTHFR, p53 and MDR1 gene polymorphisms in breast cancer patients treated with adjuvant therapy. Cancer Epidemiol 34 (4):490–493. 10.1016/j.canep.2010.03.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.