SUMMARY
Rett syndrome (RTT), mainly caused by mutations in the methyl-CpG binding protein 2 (MeCP2), is one of the most prevalent intellectual disorders without effective therapeutics. Here, we used two-dimensional and three-dimensional human brain cultures to investigate MeCP2 function. We found that MeCP2 mutations cause severe abnormalities in human interneurons (INs). Surprisingly, the treatment with a BET inhibitor, JQ1, rescued the molecular and functional phenotypes of MeCP2 mutant INs. We uncovered that abnormal increases in the chromatin binding of BRD4 and the enhancer-promoter interactions underlie the abnormal transcription in MeCP2 mutant INs, which were recovered to normal levels by JQ1. We revealed the cell type-specific transcriptome impairment in MeCP2 mutant region-specific human brain organoids, which were rescued by JQ1. Finally, JQ1 ameliorated RTT-like phenotypes in mice. These data demonstrate BRD4 dysregulation is a critical driver for RTT etiology, and suggest targeting BRD4 could be a potential therapeutic opportunity for RTT.
eTOC Blurb
Xiang et al. report that dysregulation of the BRD4 function is a critical driver for the abnormal transcriptome in human RTT cells, and targeting BRD4 rescues molecular and functional deficiencies of human RTT cells and ameliorates RTT progression in mice.
Graphical Abstract
INTRODUCTION
Rett syndrome (RTT) is a severe X-linked neurodevelopmental disorder. In the majority of cases, it is caused by the loss of function of the methyl-CpG binding protein 2 (MeCP2) gene (Amir et al., 1999). Over the past few decades, the function of MeCP2 and the etiology of RTT has been extensively investigated (Lyst and Bird, 2015). MeCP2 is known to bind to the genome to suppress or activate transcription (Gabel et al., 2015; Mellen et al., 2012). The interaction of MeCP2 with NCoR/SMRT co-repressor complex is one of the major routes of the transcription repressor function of MeCP2 (Ebert et al., 2013; Lyst et al., 2013). Other functions attributed to MeCP2 include chromatin remodeling, alternative splicing, and miRNA processing (Lyst and Bird, 2015), all of which exemplify MeCP2’s role in providing genome-wide transcription balance.
Glutamatergic cortical pyramidal neurons (CNs), GABAergic interneurons (INs), astrocytes (ASs), and oligodendrocytes (OLs) are the major cell types within the cortical circuit where MeCP2 is expressed. Cell-type-specific deletion of MeCP2 demonstrated that MeCP2 is critical in all cell types, and thus deletion in CNs, INs, or ASs leads to distinct but significant phenotypes in animal models (Adachi et al., 2009; Chao et al., 2010; Samaco et al., 2009; Zhang et al., 2016). Studies in the murine brain also highlighted cell-type-specific alteration in the transcriptome (Johnson et al., 2017). For human studies, human pluripotent stem cell (hPSC)-based models have been established using either RTT patient-derived induced pluripotent stem cells (hiPSCs) (Kim et al., 2011; Marchetto et al., 2010) or MeCP2-edited human embryonic stem cells (hESCs) (Li et al., 2013). These studies defined the cellular and molecular phenotypes in human MeCP2 mutant neurons, mainly CNs. Recent single-cell transcriptome analysis (scRNA-seq) in RTT patients demonstrated that the upregulation of genes in MeCP2 mutant neurons could be predicted by cell-type-specific methylation (Renthal et al., 2018).
Here, we found that human RTT INs are severely impaired, and the BET inhibitor JQ1 could reverse the abnormalities of RTT INs both molecularly and functionally. JQ1 treatment attenuated the transcription hyperactivation phenotype in MeCP2 mutant INs, through decreasing the BRD4 binding and enhancer-promoter interaction. We further demonstrated that JQ1 could reverse abnormal transcriptomes in a variety of cortical cell types in the region-specific human brain organoids with MeCP2 mutation. Finally, we demonstrated that JQ1 could ameliorate RTT phenotypes in vivo. Our studies uncovered the previously unknown MeCP2 function in regulating neuronal development by suppressing BRD4.
RESULTS
Human RTT INs Are Severely Impaired
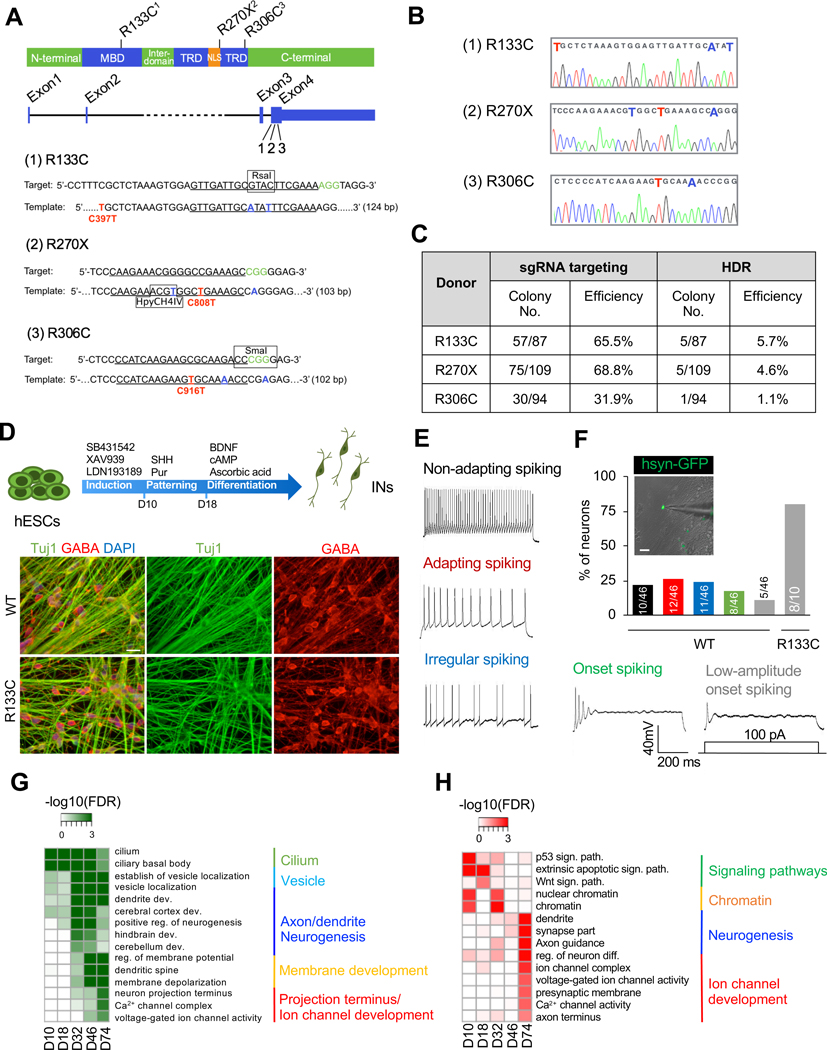
To exclude the unpredictable effect of unstable X-inactivation on MeCP2 expression (Kim et al., 2014; Mekhoubad et al., 2012), we generated a human RTT model using a male hESC line H1 by CRISPR/Cas9 system. The following changes, R133C (C397T), R270X (C808T), or R306C (C916T), representing the three most-frequent clinical MeCP2 mutations (Krishnaraj et al., 2017), were introduced using a one-time dissociation protocol specially optimized for hESC/hiPSC editing (Figure 1A–1C; Figure S1A–S1C; Table S1). Edited cell lines maintained pluripotency (Figure S1D and S1E), and no off-target mutations were found when assaying top potential off-targets (Table S2).
Figure 1. Severe Abnormalities in Human RTT INs.
(A) Design for CRISPR/Cas9-mediated MeCP2 editing. MBD: methyl-CpG binding domain; TRD: transcription repression domain; NLS: nuclear localization sequence.
(B) Sequences of targeted locus in selected MeCP2 mutant hESC clones. Red color: desired point mutation; blue color: synonymous nucleotide substitution.
(C) Summary of CRISPR/Cas9-mediated editing in MeCP2 locus. HDR: homology-directed repair.
(D) Schematic view of protocol for IN differentiation (top), and representative images of immunostainig for Tuj1 and GABA in day 78 MeCP2-WT and MeCP2-R133C INs (bottom). The scale bar represents 20 μm.
(E and F) Action potential patterns of MeCP2-WT and MeCP2-R133C INs (E) and the corresponding quantification (F). Colors in quantification bars in F correspond to the same colors for different spiking patterns in E. The numbers of cells recorded within each category are labeled. The scale bar represents 20 μm.
(G) GOs of genes up-regulated during WT IN development (at day 10, 18, 32, 46 and 74). Note genes related to projection terminus and ion channles showed the highest enrichment in mature INs (day 74).
(H) GOs of genes down-regulated in MeCP2-R133C progenitors and mature INs (at day 10, 18, 32, 46 and 74).
To decipher RTT etiology in a defined cell-type background, we focused on INs because IN malfunction is critically involved in RTT (Chao et al., 2010), yet how MeCP2 mutation affects IN development and function remain largely elusive, especially in human. To investigate MeCP2 function, we first focused on the R133C mutation (MeCP2-R133C) that is impaired in the DNA binding ability of MeCP2 protein (Mellen et al., 2012). INs were generated using a previously established protocol where medial ganglionic eminence (MGE) development was recapitulated and were cultured over ten weeks (Figure 1D, upper panel), which allowed functional maturation of INs (Maroof et al., 2013). Similar overall differentiation was achieved for MeCP2-WT and MeCP2-R133C hESCs (Figure 1D, bottom panel; Figure S2A–S2E). We found that MeCP2-R133C INs displayed a decrease in neurite growth, neurite coalescence, and soma size (Figure S2F and S2G). Whole-cell patch-clamp recordings were then performed to examine the activities of INs co-cultured with mouse astrocytes. MeCP2-WT INs action potential (AP) patterns were similar to those described by Ascoli and colleagues (Petilla Interneuron Nomenclature et al., 2008). However, MeCP2-R133C INs did not discharge APs or display small-amplitude onset spiking, which indicates a defect in functional maturation (Figure 1E and 1F; Figure S2H). Calcium imaging (Chen et al., 2013) further revealed that MeCP2-WT INs displayed robust individual firing and highly synchronized network activity (Figure S2I and S2J). In contrast, active calcium surges in MeCP2-R133C INs were rarely detectable (Figure S2I and S2J).
RNA-seq for developing INs (from day 10 to day 74) revealed that GABAergic INs were enriched in both MeCP2-WT and MeCP2-R133C IN cultures (Figure S2K). MeCP2-WT samples revealed a stepwise transcriptional transition of neuronal function during development (Figure 1G), whereas replication and cell cycle transition were significantly down-regulated at day 74 (Figure S3A). The most drastic difference appeared in mature INs (day 74), where deficits in multiple neuronal functions were observed, including axon/dendrite growth, synapse development, and ion channel activities (Figure 1H). This was further validated by comparing our IN transcriptome datasets against the BrainSpan Atlas of the Developing Human Brain (http://brainspan.org/) (Figure S3B). Differential expression analysis of well-defined biomarkers revealed 7 out of 10 highly correlated diseases were directly related to brain function (Figure S3C).
Proteomic profiling of mature INs (day 76) revealed the 8,250 quantified proteins and both up-and down-regulated protein subsets in MeCP2-R133C INs (Figure S3D). We found a significant correlation between the global transcriptome and proteome profiles for both MeCP2-WT and MeCP2-R133C INs (Figure S3E). Additionally, calcium-related cellular components were drastically affected by MeCP2 mutation (Figure S3F). In line with the importance of calcium signal transduction in development and the plasticity of neuronal networks (Zhou et al., 2006), MeCP2-R133C INs displayed less activity-dependent dendritic outgrowth when compared to MeCP2-WT INs (Figure S3G). Consistently, the depolarization-induced transcription of immediate early genes (IEGs; e.g., c-FOS, FOSB, NPTX2) was significantly reduced in MeCP2-R133C INs compared with MeCP2-WT INs (Figure S3H). Together, these results demonstrate the broad transcriptional dysregulation underlies the abnormalities in mature human RTT INs.
Reversing Transcriptome and Activity in RTT INs
Since we found a global dysregulation of transcription in RTT INs, we sought to identify small molecules that are capable of broadly altering transcription, rather than affecting a single gene target. JQ1, a thienotriazolodiazepine that affects the epigenetic pathways, is in our top list. To understand whether JQ1 alters transcription in RTT INs, we treated MeCP2-WT and MeCP2-R133C INs with low dose JQ1 (75 nM) (see methods) and performed RNA-seq in matured INs (day 74). We first confirmed JQ1-treated MeCP2-WT and MeCP2-R133C cultures were enriched with GABAergic INs (Figure S4A–S4D). Surprisingly, we found the transcriptome of MeCP2-R133C INs shifted to resemble MeCP2-WT INs after JQ1 treatment, while there was only a minimal effect of JQ1 on MeCP2-WT INs (Figure 2A). Specifically, a large number of genes showed up-regulated expression in MeCP2-R133C INs and became similar to MeCP2-WT INs upon JQ1 treatment, and these genes were linked to multiple protein and vesicle processing pathways (Figure 2B). Meanwhile, consistent with the previous gene profiling (Figure 1H), genes involved in the regulation of neuronal structure and function were repressed in MeCP2-R133C INs, but with JQ1 treatment their expressions were recovered (Figure 2B).
Figure 2. JQ1 Reverses Transcriptome and Activity in human RTT INs.
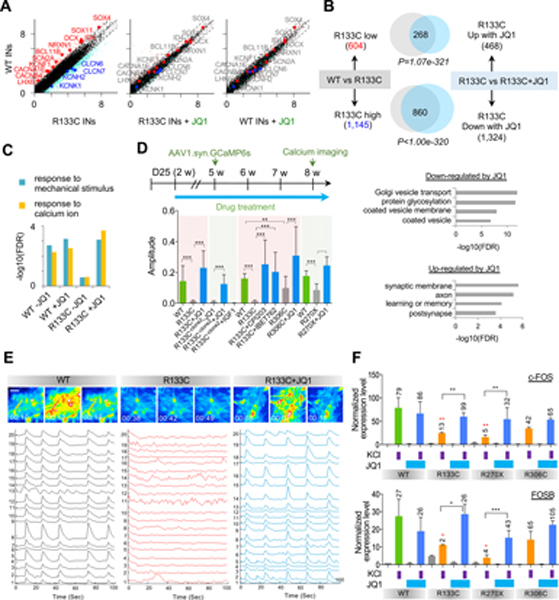
(A) Comparative analysis of gene expression profiles with MeCP2 mutation and JQ1 treatment (left: WT INs vs MeCP2-R133C INs, middle: WT INs vs MeCP2-R133C INs + JQ1, right: WT INs vs WT INs + JQ1). Differentially-expressed genes between WT and MeCP2-R133C INs are colored by pink and cyan colors. Representative differentially-expressed neurodevelopmental genes and ion channel genes are shown by red and blue colors.
(B) Venn diagrams of genes affected by JQ1 treatment in MeCP2-WT and MeCP2-R133C INs (upper panel) and the corresponding GOs (lower panel). Statistical significance of overlap is shown by p-value with hypergeometric test. In MeCP2-R133C INs, genes and GOs up-regulated in comparison to MeCP2-WT INs were significantly down-regulated by JQ1; vice versa, in MeCP2-R133C INs, genes and GOs down-regulated by MeCP2 mutation were significantly up-regulated by JQ1.
(C) GO analysis of global transcriptome revealed genes related to stimulation response were rescued by JQ1 in MeCP2-R133C INs.
(D) Schematic view of the strategy for JQ1 treatment (75 nM) and calcium imaging (top), and amplitude quantification for calcium spikes (bottom). Mean ± SD is shown (18~20 randomly recorded cells from 3 independently treated wells were quantified for each group). ***p < 0.001, **p < 0.01 (unpaired t-test). Green bars indicate parallel WT controls, gray bars indicate non-treated INs, and blue bars indicate JQ1 (75 nM)-treated INs.
(E) Representative images from calcium imaging in MeCP2-WT and MeCP2-R133C INs. The scale bar represents 20 μm.
(F) qPCR detection of depolarization-dependent c-FOS and FOSB inductions. Fold changes in comparison with non-KCl stimulated INs within each group are labeled with black numbers on top of each bar. Red stars represent comparison with non-JQ1 treated but KCl-stimulated MeCP2-WT INs. Mean ± SD is shown (n=3 biologically independent samples). ***p < 0.001, **p < 0.01, *p < 0.05 (unpaired t-test).
Also consistent with the above mentioned transcriptomic analysis (Figure 1H), MeCP2-R133C INs showed abnormal expression of ion channel genes (Figure S4E). After JQ1 treatment of MeCP2-R133C INs, expression levels of ion channel genes recovered to WT levels (Figure 2A). Importantly, JQ1 treatment also rescued the expression of IGF1, KCC2, and mGlu7 that were down-regulated in MeCP2-R133C INs in our study and are known to be down-regulated in other RTT models (Castro et al., 2014; Gogliotti et al., 2017; Tang et al., 2016) (Figure S4F).
While JQ1 treatment had minimal effect on the IEG induction in MeCP2-WT INs, it significantly recovered the activity-dependent IEG response in MeCP2-R133C INs (Figure S4G–S4I). Indeed, as shown by GO analysis, transcription of genes involved in pathways that respond to calcium changes and extracellular stimuli were improved in MeCP2-R133C INs by JQ1 treatment (Figure 2C). Together, these results indicate that JQ1 can reverse the abnormal IN development and the deficits in activity-dependent transcription caused by the MeCP2 mutation. Notably, we also found while 10 days of JQ1 treatment beginning at mature stage could rescue IEG induction defect in MeCP2-R133C INs, acute treatment of 24 hours failed to do so (Figure S4K and S4L), suggesting that cumulative effects during continuous JQ1 treatment may be required to achieve a functional rescue.
Upon treatment with JQ1, calcium surges of randomly recorded individual MeCP2-R133C INs were effectively rescued to a level comparable to MeCP2-WT INs (Figure 2D). More importantly, MeCP2-R133C INs displayed network synchronization of calcium surges under JQ1 treatment like MeCP2-WT INs (Figure 2E). INs from a different MeCP2-R133C clone (R133C-clone2) isolated from independent gene editing showed similar JQ1-mediated rescue of both single-cell calcium spiking and network synchronization (Figure 2D and Figure S5A). In contrast, IGF1 (20 ng/mL) did not improve INs activity (Figure 2D and Figure S5A). Other BET inhibitors, including CPI203 (50 nM) and IBET762 (100 nM) showed comparable levels of calcium spiking and network synchronization after drug treatment (Figure 2D and Figure S5B and S5C). Up to 300 nM of JQ1 treatment for 10 days was able to rescue IEG induction defect in MeCP2-R133C INs, suggesting an improvement in neuronal functions (Figure S4L); nevertheless, prolonged treatment of either high or too low dosage of JQ1 did not show efficient beneficiary improvement on INs activity (Figure S5D). These dosage-dependent rescue effects were also observed for other BET inhibitors CPI203 and IBET762 (Figure S5D).
We next assessed the effect of JQ1 (75 nM) on INs with other MeCP2 mutations, including TRD (R306C) or NLS (R270X) mutations (Figure S4J). MeCP2-R306C INs exhibited an increased frequency of calcium spiking and a decreased average spiking amplitude, whereas MeCP2-R270X INs displayed less frequent calcium spiking in addition to decreased spiking amplitude (Figure 2D and Figure S5B and S5C). These results suggest that MeCP2 R306C INs are functionally less impaired than R270X INs, in line with a previous study (Cuddapah et al., 2014). Regardless, the abnormal activity observed in MeCP2–306C and MeCP2-R270X INs was remarkably reversed by JQ1 treatment to mimic MeCP2-WT INs at both the single neuron level (increased amplitude) and the network level (recovered network synchronization) (Figure 2D; Figure S5B and S5C). In agreement with the rescued IN activity, we observed a significant improvement in the activity-dependent induction of IEGs (e.g., c-FOS and FOSB) in MeCP2-R133C and MeCP2-R270X INs with JQ1 treatment (Figure 2F). MeCP2-R306C INs did not display significant impairment in IEG induction and neuronal activity, and JQ1 had no significant effect on IEG induction (Figure 2D, F and S5B).
We then investigated the effects of JQ1 in a 3D context by generating human MGE organoids (hMGEOs) (Xiang et al., 2017). To facilitate real-time monitoring of calcium activity, the GCaMP6s reporter was knocked-into the AAVS1 locus of both MeCP2-WT and MeCP2-R133C hESCs (Figure S5E). We observed that MeCP2-WT hMGEOs, but not MeCP2-R133C hMGEOs, displayed area-scale synchronization of calcium surges (Figure S5F). This area-scale synchronization remained under JQ1 treatment. Importantly, JQ1 treatment rescued network activity in MeCP2-R133C hMGEOs (Figure S5F and S5G). Besides, KCl depolarization significantly induced the expression of c-FOS in MeCP2-WT hMGEOs without or with JQ1 treatment, whereas MeCP2-R133C hMGEOs displayed robust depolarization-dependent c-FOS induction only after JQ1 treatment (Figure S5H and S5I). These results further support that JQ1 can ameliorate the functional deficits of RTT INs in the brain.
Unlike IN cultures, MeCP2-WT and MeCP2-R133C CNs displayed similar activities without strong network synchronization, and the activity was not altered by JQ1 treatment (Figure S5J and S5K). We then generated human cortical organoids (hCOs) (Xiang et al., 2017) and found that both MeCP2-WT and MeCP2-R133C hCOs displayed area-scale synchronization of calcium surges with or without JQ1 treatment (Figure S5L). This finding indicates that brain organoids may offer a more favorable environment for functional neurodevelopment and that the cortical region may be less affected by the MeCP2 R133C mutation than the MGE region. Although CNs have this reduced sensitivity to MeCP2 dysfunction, MeCP2-R133C CNs displayed decreased depolarization-induced c-FOS induction, while the ability to induce FOSB expression was comparable to that of MeCP2-WT CNs (Figure S5M). Interestingly, JQ1 significantly improved the induction of c-FOS and FOSB in both MeCP2-WT and MeCP2-R133C CNs without altering neuronal identity (Figure S5M and S5N), suggesting that JQ1 may also benefit the activity-dependent transcription in CNs. Altogether, these results demonstrate that the abnormal transcriptome, activity, and activity-dependent transcriptional regulation in RTT INs can be reversed by JQ1 treatment.
Dysregulation of BRD4 Function in MeCP2 Mutant INs
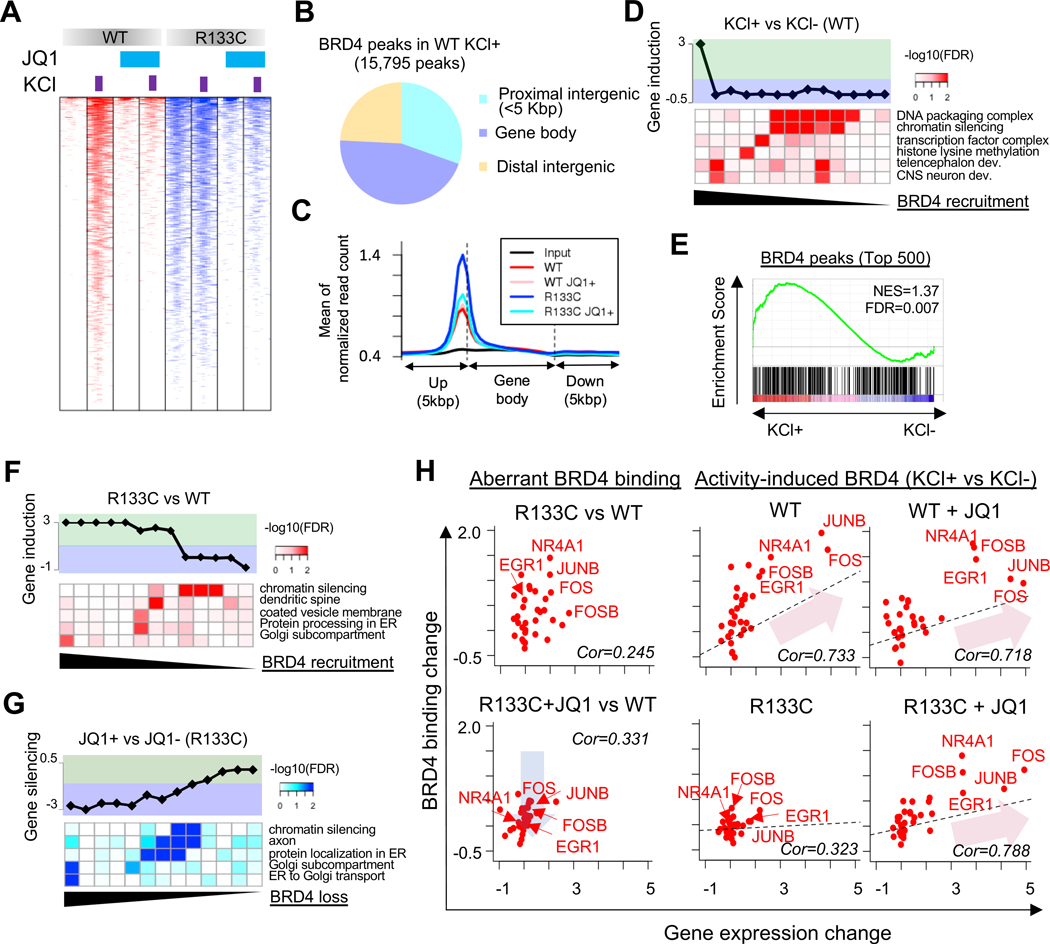
The epigenetic reader BRD4 is the primary target of JQ1 (Filippakopoulos et al., 2010). To understand how JQ1 reveres transcriptome in RTT INs, we then examined genome-wide occupancy of BRD4 in MeCP2-WT and MeCP2-R133C INs (day 74) with or without JQ1 treatment. To evaluate the changes at different status of neuronal activity, we also stimulated the samples by KCl depolarization. KCl stimulation significantly increased genome-wide binding of BRD4 in MeCP2-WT INs, which could be attenuated by JQ1 (Figure 3A and Figure S6A). Remarkably, we found a dramatic increase in genome-wide BRD4 binding in MeCP2-R133C INs even without KCl stimulation, which resembled the pattern observed in activated MeCP2-WT INs (Figure 3A). Besides, membrane depolarization with KCl barely induced further BRD4 binding in MeCP2-R133C INs, thus causing a defect in activity-dependent plasticity. Critically, JQ1 treatment robustly decreased the genome-wide occupancy of BRD4 in MeCP2-R133C INs, and more importantly, KCl exposure significantly increased the BRD4 binding in JQ1-treated MeCP2-R133C INs, a feature initially only observed in MeCP2-WT INs (Figure 3A and Figure S6A).
Figure 3. Excess Chromatin BRD4 Binding Dysregulates Transcriptome in human RTT INs.
(A) Global BRD4 chromatin binding pattern in MeCP2-WT and MeCP2-R133C INs.
(B) Distributions of BRD4 chromatin binding in MeCP2-WT INs under KCl depolarization.
(C) Distribution of BRD4 binding in gene body and 5kbp-flanking regions in MeCP2-WT and MeCP2-R133C INs with or without JQ1 (75 nM) treatment.
(D) Gene activation by BRD4 recruitment in MeCP2-WT INs induced by KCl depolarization. Lower panel shows GO terms with their significant over representation for BRD4 target genes, and top panel shows corresponding changes in gene transcription.
(E) GSEA of top 500 target genes bound by BRD4 and the corresponding transcriptional induction of target genes in MeCP2-WT INs. IEGs are significantly enriched in this category.
(F) Induction of gene transcription by gain of chromatin BRD4 compared between MeCP2-R133C and MeCP2-WT INs. BRD4 peaks are sorted by the gain level (log2(MeCP2-R133C/WT)).
(G) Silencing of gene transcription by loss of chromatin BRD4 compared between JQ1-treated and non-treated MeCP2-R133C INs. BRD4 peaks are sorted by log2(JQ1+/JQ1-) in MeCP2-R133C INs.
(H) Correlation between BRD4 binding and transcription for IEGs. Left panel: Relative BRD4 binding and transcription of potential IEGs (Tullai et al., 2007) in non- and JQ1-treated MeCP2-R133C INs to MeCP2-WT INs. Blue arrow represents the reduction of aberrant BRD4 binding in MeCP2-R133C INs by JQ1. Right panel: Relative BRD4 binding and transcription of potential IEGs in JQ1-treated INs to non-treated INs. Red arrows represent significant correlations between BRD4 recruitment and IEG induction in WT and JQ1-treated WT and MeCP2-R133C INs (Pearson correlation > 0.718 & p<1.83e-6). Linear regressions are shown by dashed lines. Five representative IEGs are also shown.
See also Figure S6.
We found depolarization-induced BRD4 binding in MeCP2-WT INs occurred across enhancer, promoter, and transcribed regions (Figure 3B). A similar pattern was observed in MeCP2-R133C INs at a basal level and under depolarization (Figure S6B and S6C). Upon JQ1 treatment, the observed excessive genome-wide binding of BRD4 in MeCP2-R133C INs significantly decreased (Figure 3C). In MeCP2-WT INs, not all KCl-induced BRD4 recruitment led to transcriptional alteration. However, a subset of genes showed a significant correlation between increased BRD4 binding and induction of transcription (Figure 3D). Gene set enrichment analysis (GSEA) also revealed that genes (including IEGs) highly enriched with KCl-induced BRD4 binding in MeCP2-WT INs displayed a significant increase of transcription (Figure 3E).
Notably, the increased BRD4 binding in MeCP2-R133C INs corresponded to transcriptional up-regulation, including IEGs (Figure 3F and S6D). Supporting this, the reduction of BRD4 binding in MeCP2-R133C INs via JQ1 treatment led to the attenuation of transcription of these genes (Figure 3G). In both cases, affected genes were linked to pathways similar to those observed in transcriptome profiling (Figure 2B, down-regulated GOs), where large numbers of up-regulated genes in MeCP2-R133C INs were found to be rescued by JQ1. Similarly, JQ1-mediated BRD4 loss of binding from the genome led to transcriptional down-regulation of MeCP2-R133C INs under KCl depolarization (Figure S6E). These results strongly suggest that BRD4 is critically involved in the observed transcriptional dysregulation in MeCP2-R133C INs.
While JQ1 recovered the activity-dependent deposition of BRD4 in KCl-stimulated MeCP2-R133C INs, some BRD4 binding was still reduced, leading to the reduced-regulated transcription of the corresponding genes (Figure S6F). Nevertheless, the majority of the remaining BRD4 binding sites in KCl-stimulated MeCP2-R133C INs overlapped with those observed in KCl-stimulated MeCP2-WT INs (Figure S6G). Among them, genes linked to transcriptional regulation and vesicle or membrane-related processing were commonly targeted by BRD4 in MeCP2-WT INs and JQ1-treated MeCP2-R133C INs, indicating their essential roles in activity-dependent regulation (Figure S6G). As shown in Figure 3H, MeCP2-R133C INs showed increased chromatin binding of BRD4 to IEGs, which was attenuated by JQ1 treatment (blue arrow). While MeCP2-WT INs recruited BRD4 to IEGs in response to KCl depolarization with or without JQ1 treatment and showed induced transcription of target IEGs, MeCP2-R133C INs displayed similar level of response only when JQ1 treatment was performed (red arrows). This was further confirmed by examining individual gene tracks (Figure S6H), suggesting BRD4 is involved in mediating the activity-dependent expression of IEGs in MeCP2-R133C INs. Of note, some IEGs in MeCP2-R133C INs showed relatively normal BRD4 binding at basal level and improved activity-dependent BRD4 binding and gene expression upon JQ1 treatment (e.g., NPAS4 in Figure S6H); besides, MeCP2-WT INs may also show enhanced induction of IEGs after JQ1 treatment without apparent alterations in BRD4 binding (Figure S6H). These observations indicate besides BRD4-mediated transcriptional change, other indirect mechanisms may also play a role in regulating IEGs.
MeCP2-BRD4 Axis Controls Transcription in INs
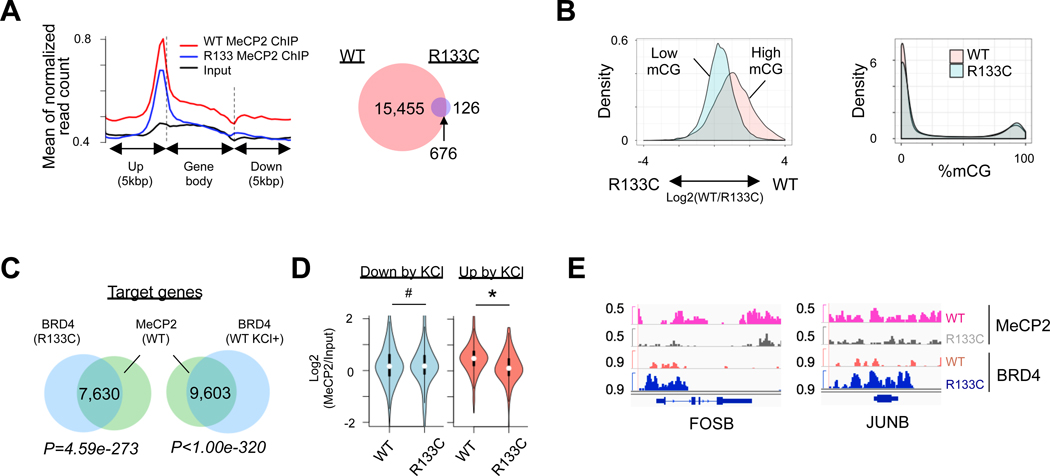
Although both KCl-stimulated MeCP2-WT INs and un-stimulated MeCP2-R133C INs exhibited similar BRD4 chromatin binding, MeCP2-R133C INs displayed a drastically different BRD4-dependent transcriptional upregulation (Figure 3D–3F). The mutation R133C within the methyl-CpG binding domain (MBD) of MeCP2 impairs its DNA binding ability (Mellen et al., 2012). We therefore hypothesized that loss of MeCP2 binding to methylated CpG leads to the de-repression of transcription in its targets. Genome-wide profiling of MeCP2 binding in mature MeCP2-WT and MeCP2-R133C INs (day 74) revealed a dramatic decrease in global MeCP2 binding in MeCP2-R133C INs (Figure 4A). Reduced representation bisulfite sequencing (RRBS) also revealed that while MeCP2-WT and MeCP2-R133C INs had similar methyl-CpG (mCpG) levels, only highly methylated CpG loci displayed decreased MeCP2 binding in MeCP2-R133C INs (Figure 4B). In order to examine the potential role of transcriptional activation through MeCP2 binding at 5hmC, we performed oxidative RRBS (oxRRBS) to distinguish 5mC and 5hmC (Mellen et al., 2012). However, MeCP2-WT and MeCP2-R133C INs exhibited similar levels of 5hmC, and we observed no significant alteration in MeCP2 binding at 5hmC in MeCP2-R133C INs (Figure S6I).
Figure 4. MeCP2 Mutation Leads to Increase of BRD4 Binding at mCpG Sites.
(A) Distribution of MeCP2 binding in gene body and flanking region (left), and Venn diagram showing overlaps of MeCP2 peaks between MeCP2-WT and MeCP2-R133C INs (right). Left: p=2.2e-16. Right: p=0.1307.
(B) Difference of MeCP2 binding to highly- or lowly-methylated CpG islands (CpGIs) between MeCP2-WT and MeCP2-R133C INs (left panel), and difference of their mCGs on CpGIs (right panel). CpGIs were classified by high (>80%) and low mCGs (<20%). Left: p=2.2e-16. Right: p=0.1307.
(C) Venn diagram showing common target genes between WT MeCP2 and BRD4 in MeCP2-R133C INs, and between WT MeCP2 and BRD4 in KCl-depolarized MeCP2-WT INs.
(D) Chromatin binding of MeCP2 in KCl-suppressed (<1.5 fold) or KCl-induced genes (>1,5 fold) in MeCP2-WT and MeCP2-R133C INs. # P=0.3621, * P=3.295e-9.
(E) Snapshots of MeCP2 and BRD4 binding peaks in representative IEG locus in MeCP2-WT and MeCP2-R133C INs.
See also Figure S6.
When examining the genome-wide binding of MeCP2 and BRD4, we found a significant overlap of chromatin targeting by MeCP2 and BRD4 (Figure S6H). Notably, genes with excessive BRD4 binding in unstimulated MeCP2-R133C INs showed a significant overlap with MeCP2 binding targets (Figure 4C). These results further suggest that MeCP2 and BRD4 co-regulate the transcription of common target genes. Especially in stimulated MeCP2-WT and unstimulated MeCP2-R133C INs, BRD4 targets similar subsets of genes, as revealed by GO analysis (e.g., ER-related, synapse, neuronal differentiation, histone-related, and transcriptional repression) (Figure S6K). Indeed, corresponding to the increased chromatin recruitment of BRD4 in depolarization-induced genes (Figure 3D and 3E), we found a significant loss of MeCP2 binding at these sites in MeCP2-R133C INs, but not in genes down-regulated by depolarization (Figure 4D). Examination of individual IEG tracks (e.g., FOSB and JUNB) also revealed a loss of MeCP2 and gain of BRD4 at these loci (Figure 4E), along with the up-regulation of target transcription (Figure 3H, see RPKM count). Altogether, these results reveal the importance of the MeCP2-BRD4 regulatory axis in controlling transcription.
Disrupted Chromatin Topology Underlies the Impaired Transcription in RTT INs
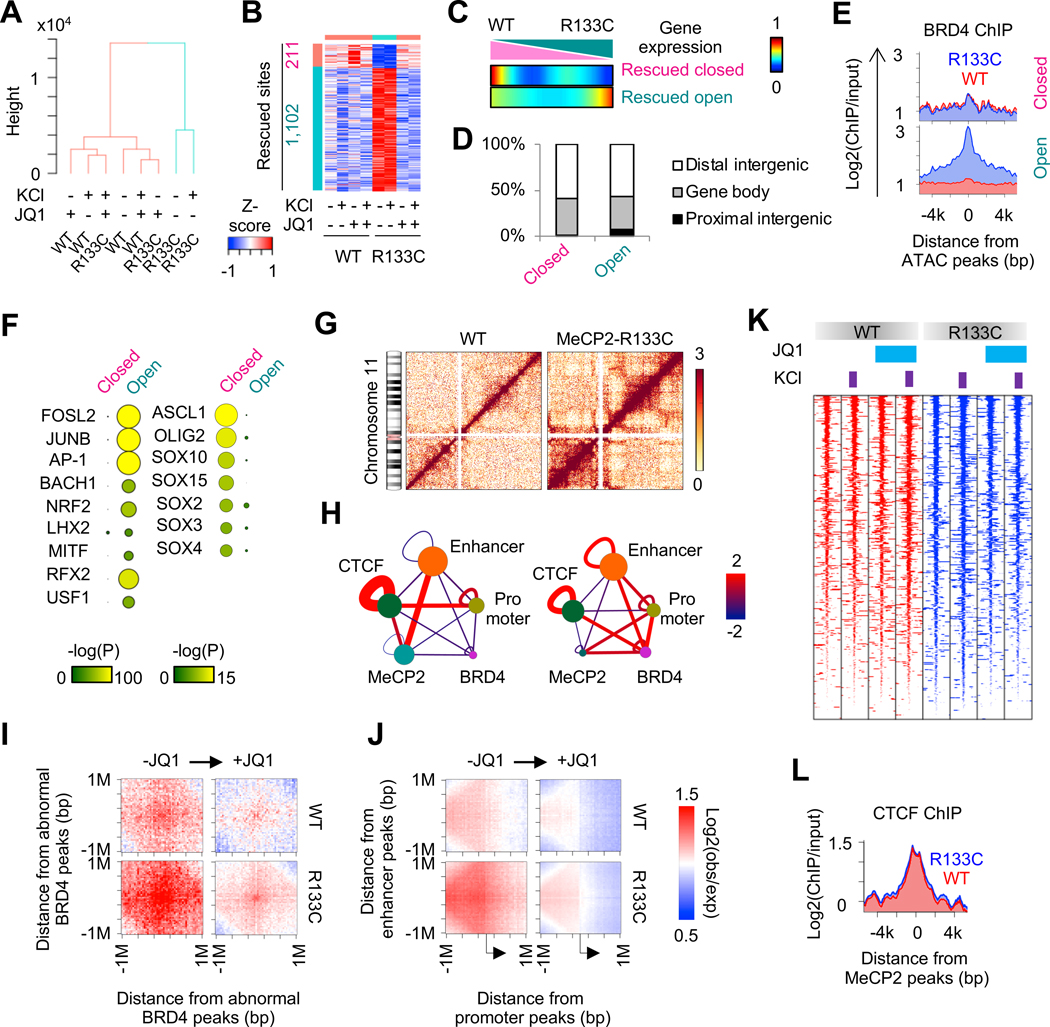
BRD4 mediates nucleosome remodeling and accelerates chromatin decompaction (Devaiah et al., 2017). We thus performed transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) in mature MeCP2-WT and MeCP2-R133C INs (day 74). Unsupervised clustering revealed that open chromatin profiles in MeCP2-R133C INs were distinct from those in MeCP2-WT INs. Notably, with JQ1 treatment, the global chromatin accessibilities in MeCP2-R133C INs became to resemble MeCP2-WT INs (Figure 5A). 1,102 out of 1313 JQ1-rescued differentially open chromatin regions (dOCRs) were at an abnormally open state in MeCP2-R133C INs. A smaller portion (211 out of 1313) of the recovered dOCRs were abnormally closed in MeCP2-R133C INs, but regained accessibility after JQ1 treatment (Figure 5B). Importantly, the dOCRs were preferentially observed in distal enhancer regions of genes that were differentially expressed between MeCP2-WT and MeCP2-R133C INs (Figure 5C and 5D). Furthermore, chromatin binding of BRD4 was significantly enriched in dOCRs that were abnormally open in MeCP2-R133C INs, suggesting that BRD4 binding leads to increased chromatin accessibility (Figure 5E). By scanning the 1,102 opened dOCRs for known transcription factor binding sequences, we detected significant enrichment of binding motifs for IEGs (e.g., FOSL2, JUNB, and AP-1) (Figure 5F). In contrast, motifs of transcription factors for neural development (e.g., ASCL1, OLIG2, and SOX family) were significantly enriched in dOCRs that were closed in MeCP2-R133C INs (Figure 5F), which is in line with the abnormal development and IEG regulation we found in MeCP2-R133C INs.
Figure 5. BRD4 Modulates 3D Genome Architecture.
(A) Hierarchical clustering of open chromatin profiles in MeCP2-WT and MeCP2-R133C INs.
(B) Heatmap representing dOCRs between MeCP2-WT and MeCP2-R133C INs.
(C) Comparison of dOCRs with gene expression change in MeCP2-WT and MeCP2-R133C INs. Genes were sorted by gene expression change and divided into 150 bins. The number of genes in the vicinity of dOCRs was counted into each bin. Heat color represents percentage of genes showing dOCRs.
(D) Distribution of abnormally closed or opened dOCRs in the genome of MeCP2-R133C INs.
(E) Enrichment of BRD4 chromatin binding around dOCRs in MeCP2-WT and MeCP2-R133C INs.
(F) Motif enrichment of dOCRs. Representative motifs uniquely enriched in open and closed dOCRs in MeCP2-R133C INs are shown.
(G) Hi-C contact matrices in chromosome 11 in MeCP2-WT and MeCP2-R133C INs. Heat color represents ICED value.
(H) Enrichment of Hi-C interaction among promoters, enhancers, CTCF, BRD4 and MeCP2 binding sites. Node size represents the number of sites. Edge thickness and color represents statistical significance of interaction (−log(pvalue)). Left panel: MeCP2-WT, right panel: MeCP2-R133C.
(I) Aggregated Hi-C contact map around abnormal BRD4 peaks in MeCP2-WT and MeCP2-R133C INs, with or without JQ1 treatment.
(J) Aggregated Hi-C contact map between enhancer and promoter in MeCP2-WT and MeCP2-R133C INs, with or without JQ1 treatment.
(K) Comparison of CTCF chromatin binding profiles in MeCP2-WT and MeCP2-R133C INs.
(L) CTCF chromatin binding profile around MeCP2 chromatin binding sites in MeCP2-WT and MeCP2-R133C INs.
Given the dysregulation of distal enhancers in MeCP2-R133C INs (Figure 5C and 5D), we next examined chromosomal interactions between enhancers and promoters by performing Hi-C in MeCP2-WT and MeCP2-R133C INs (day 74) (Figure 5G). Enrichment analysis for Hi-C interactions revealed the limited chromosomal contacts between enhancers and promoters in WT INs. Interestingly, enhancers and promoters showed a preferential interaction with MeCP2 or CTCF binding sites, respectively (Left, Figure 5H). In MeCP2-R133C INs, the interaction between enhancers and promoters was highly increased (Right, Figure 5H). While the interaction of enhancers with MeCP2 binding sites and the interaction of promoters with CTCF binding sites were decreased, more interaction between BRD4 and enhancers or promoters were detected in MeCP2-R133C INs (Figure 5H). We found that the decrease in chromatin-bound BRD4 by JQ1 treatment consequently reduced the abnormal BRD4-mediated chromosomal interaction (Figure 5I). The abnormal enhancer-promoter interactions were also attenuated by JQ1 treatment (Figure 5J). We also noted that the chromatin recruitment of CTCF, an insulator for long-range DNA contact, was not affected by either MeCP2 mutation or JQ1 treatment (Figure 5K and 5L). Overall, our results underline the significance of BRD4-mediated disruption in chromatin accessibility and conformation in dysregulated transcription.
Single Cell Transcriptomic Impairment and Reversal in Human RTT Brain Organoids
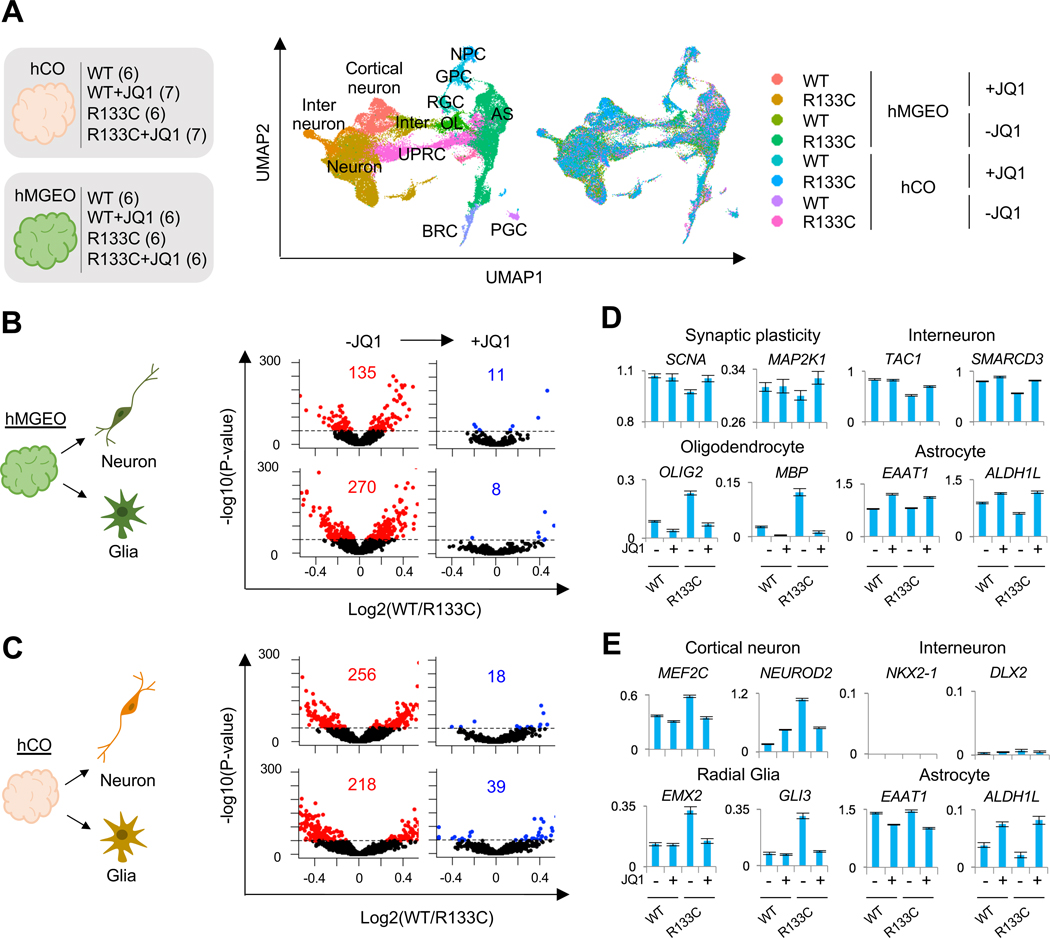
In order to investigate MeCP2 function in other cell types and in a region-specific manner, we generated hMGEOs and hCOs, representing the developing human MGE and cortex, respectively (Xiang et al., 2017) (Figure S7A–S7D). We treated hMGEOs and hCOs from MeCP2-WT or MeCP2-R133C background with JQ1, and profiled the transcriptome at single-cell level (Figure 6A and Figure S7E). MeCP2-WT and MeCP2-R133C brain organoids were cultured over 2.5 months before processing, which allowed cell maturation (Xiang et al., 2017). Analysis of genes that are exclusively expressed in distinct cell types verified a doublet rate of ~0.38%, demonstrating a presence of very limited doublets in our scRNA-seq (Figure S7F). We systematically labeled 30 clusters with major cell classes including radial glia cells (RGC), neuronal cells (cortical neuron [CN], interneuron [IN], and non-committed neurons [NC-neuron]), glial cells (immature oligodendrocyte [OL], astrocyte [AS]) as well as cells characterized with BMP signal response (BRC), unfolded protein response (UPRC) and proteoglycan (PGC) (Figure 6A and Figure S7G–S7I, See Experimental Procedures) (Xiang et al., 2019). The cell type assignment was validated with reference transcriptome profiles from in vivo human fetal brain (Figure S7J) (Darmanis et al., 2015).
Figure 6. JQ1 Rescues RTT cells in Human Brain Organoids.
(A) tSNE plot of single cells distinguished by cluster annotations. Numbers in brackets on the left indicate organoid quantities used for each condition. Within each condition, organoids (day 77 ~ day 86, see methods for details) were collected from 2 different culture dishes, pooled together, and dissociated for scRNA-seq. For drug treatment, 75 nM JQ1 was supplemented to the media starting from day 25 until the date of sample collection. CN: cortical neuron; IN: interneuron; RGC: radial glia cell; AS: astrocyte; MAS: mature astrocyte; NPC: neural progenitor cell; OPC: oligodendrocyte progenitor cell; iOL: immature oligodendrocyte; OB-like cell: osteoblast-like cell.
(B and C) Gene dysregulation in MeCP2-R133C neuron or glia clusters derived from hMGEOs (B) or hCOs (C) with or without JQ1 treatment. The dysregulation score (= −log10(p-value) by two-side T test comparing MeCP2-WT and MeCP2-R133C cells is plotted.
(D) Expression patterns of representative genes for cell type specification in hMGEO-derived cells. Cell types of each row correspond to the same row delineated in B.
(E) Expression patterns of representative genes for cell type specification in hCO-derived cells. Cell types of each row correspond to the same row delineated in C.
See also Figure S7.
To assess the effects of JQ1, we first classified cells from hCOs and hMGEOs into either neuron and glia groups, respectively (Figure 6B). We found that expression of between 100–500 genes was dysregulated in neurons and glial cells from hMGEOs or hCOs (Red in Figure 6B). Surprisingly, JQ1 treatment dramatically reduced the number of dysregulated genes in MeCP2-R133C hMGEO-derived neurons and glia, rescuing to MeCP2-WT levels (Figure 6B). In hCO-derived neurons and glial cells, JQ1 also robustly reduced the number of dysregulated genes, albeit to a lesser extent than hMGEO-derived cells (Figure 6C). GO analysis of JQ1-rescued genes revealed that genes of different functions were recovered in neurons and glia produced from hMGEOs or hCOs (Figure S7K). Deficits in forebrain development, GABAergic interneuon differentiation, and neuron fate commitment in hMGEO-derived MeCP2-R133C neurons were reversed by JQ1. Meanwhile, the reversal of abnormal transcription related to synaptic transmission suggests that synaptic function may be rescued in MeCP2-R133C hCO-derived neurons. Investigation of individual genes further revealed these cell type- and region-specific alterations (Figure 6D and 6E). TAC1 and SMARCD3 encoding an IN-specific neurotransmitter and transcriptional regulator were all significantly down-regulated and subsequently rescued by JQ1 (Figure 6D). However, in MeCP2-R133C hCO-derived neurons, transcription factors critical for CNs (e.g., MEF2C and NEUROD2) were aberrantly increased but recovered with JQ1 treatment (Figure 6E). We also found that JQ1 had a different effect on hMGEO-derived glial cells, compared to hCO-derived glial cells. Genes rescued by JQ1 in hMGEO-derived glia were predominantly involved in OL differentiation and OL-related functions (e.g., axon regeneration), while genes in hCO-derived glia were related to protein transport or radial-glia migration (Figure S7K). OLIG2 and MBP, which are transcription factors controlling OL development, were abnormally increased in MeCP2-R133C hMGEO-derived glia but returned to normal levels with JQ1 treatment (Figure 6D). Aberrant expression of a number of radial glia-specific transcription factors such as EMX2 and GLI3, were observed in MeCP2-R133C hCO-derived glia, which was again reversed by JQ1 (Figure 6E). However, no significant difference for AS-specific genes (e.g., EAAT1 and ALDH1L) was observed in either hMGEO- or hCO-derived glia. Overall, these results suggest that MeCP2 mutation causes varied transcriptional dysregulation in distinct cell types in the human brain and JQ1 rescues abnormal transcription in RTT cells.
Ameliorating RTT-like deficits in vivo
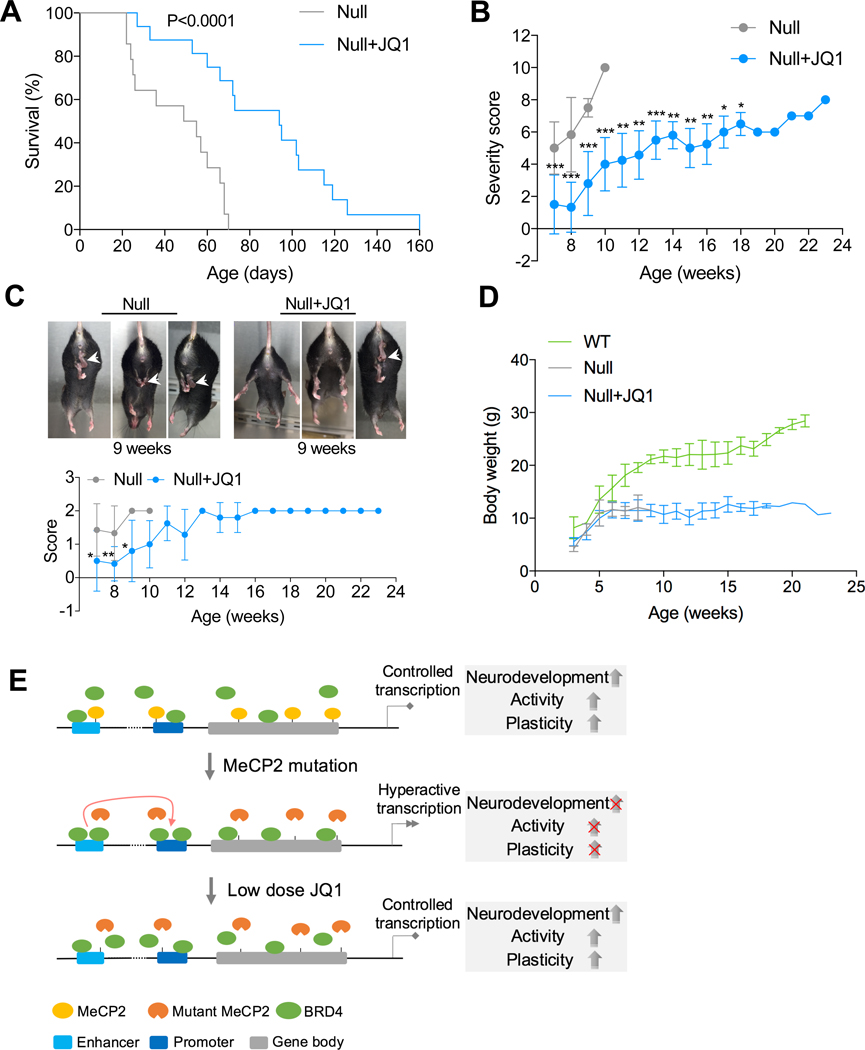
Finally, we examined whether JQ1 treatment could affect the development of RTT in vivo by utilizing the MeCP2-/Y murine model containing a deletion in exons 3 and 4 of MeCP2 (Guy et al., 2001). We performed a daily low-dose intraperitoneal injection of JQ1 (15 mg/kg) into MeCP2-/Y mice starting 2 weeks after birth. Death of MeCP2-/Y mice started to appear as early as 3 weeks after birth in the absence of JQ1 treatment (Figure 7A). With JQ1 treatment, MeCP2-/Y mice exhibited increased lifespan (approximately 81%), compared with non-treated MeCP2-/Y mice (Figure 7A, median survival of 94 days in JQ1 treated group vs. median survival of 52 days in non-treated group), which clearly demonstrates that JQ1 treatment enhances the survival of MeCP2-/Y mice. MeCP2-/Y mice showed typical progression of symptom scores and died during the progression. JQ1 treatment significantly decreased the progression of symptom scores in MeCP2-/Y mice (Figure 7B). The amelioration of RTT-like phenotypes by JQ1 was more prominent in earlier stages, such as in the prominent delay of the hind limb clasping phenotype (Figure 7C). However, JQ1 treatment did not have a significant impact on body weight throughout the lifespan of MeCP2-/Y mice (Figure 7D). Taken together, our results reveal the potential of JQ1 in ameliorating RTT progression in vivo.
Figure 7. JQ1 reverses RTT progression in vivo.
(A) Kaplan-Meier survival plot for JQ1 treated (n=16) and non treated (n=14) MeCP2-/y mice.
(B) Weekly progression of phenotypic severity scores for JQ1 treated (n=12) and non treated (n=7) MeCP2-/y mice. Mean ± SD is shown. ***p < 0.001, **p < 0.01, *p < 0.05 (unpaired t-test).
Comparison was performed between JQ1 treated and non treated mice at the same stage. After 10 weeks, comparison was performed between JQ1 treated mice and 10 week old non treated mice.
(C) Representative images of hind limb clasping (arrows) (top) and progressive scores in JQ1 treated (n=12) and non treated (n=7) MeCP2-/y mice (bottom). Mean ± SD is shown. **p < 0.01, *p <0.05 (unpaired t-test). Comparison was performed between JQ1 treated and non treated mice at the same stage.
(D) Weekly body weight of MeCP2+/y (n=7), JQ1-treated (n=14) and non treated (n=8) MeCP2-/y mice. Mean ± SD is shown.
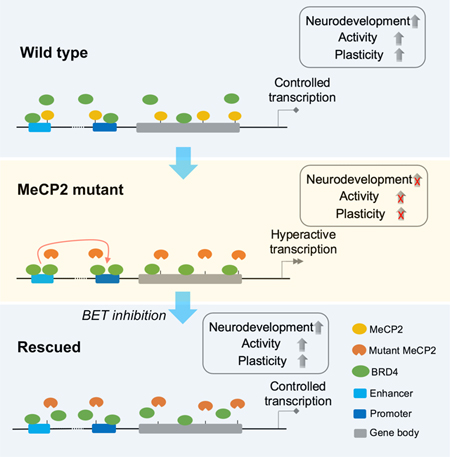
(E) Model of BRD4 and MeCP2 functions in transcriptional regulation. BRD4 and MeCP2 target common genes in INs, representing a balancing of machinery between gene activation and suppression, which lead to controlled transcription and development. RTT INs show loss of MeCP2 targeting and excessive BRD4 targeting, causing increased enhancer-promoter interaction and hyperactive transcription, and consequently impaired neurodevelopment, neuronal activity, and plasticity. Low dose JQ1 can partially block BRD4 targeting, which returns the enhancer-promoter interaction and hyperactive transcription under control, reverses the deficient development, neuronal activity, and plasticity of RTT INs.
DISCUSSION
Our study suggests that the loss of function in MeCP2 results in gain of BRD4 chromatin binding, which forces RTT INs to maintain a hyperactive transcription state. The persistent hyperactive transcription is detrimental to neural development, resulting in dysregulation of neural development genes, and impaired neuronal activity and plasticity. Low-dose JQ1 treatment could impede chromatin binding of BRD4, recover 3D genome architecture and transcription, and consequently reverse the developmental defect of RTT INs (Figure 7E).
Suppression of BET proteins has been shown to modulate brain plasticity in WT mice (Benito et al., 2017; Korb et al., 2015). A recent study reported that the phenotypes in fragile X syndrome (FXS) mice were alleviated with JQ1 treatment (Korb et al., 2017). Our results further underscore the potential of BET inhibition for the recovery of neurological phenotypes in patients. It was found while low dose JQ1 treatment was able to alleviate phenotypes in FXS (Korb et al., 2017), high dose JQ1 treatment could impair memory formation (Korb et al., 2015). Also, we found that while up to 300 nM JQ1 could rescue activity-dependent IEG induction in RTT INs after a period of up to 10 days treatment (Figure S4L), only low doses of JQ1 (50 nM~75 nM) were beneficial in achieving efficient functional rescue after prolonged treatment (Figure S5C), indicating that identification of an appropriate dosage for BET inhibitors will be crucial when rescuing RTT cells. Titrating the specific dosage could be especially critical for samples with different MeCP2 mutations. Although JQ1 effectively crosses blood-brain barrier (Matzuk et al., 2012), non-CNS related effects of JQ1 in MeCP2-/Y mice could not be excluded at this stage. The short plasma half-life of JQ1 (Filippakopoulos et al., 2010) may detriment its beneficial effects in the brain, which also urges the needs for identifying more stable and effective analogues and more optimized drug delivery approach for the purpose of in vivo treatment.
Overall, our results reveal the importance of BRD4-mediated mechanism in RTT and indicate the potential of targeting BRD4 in disease intervention.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for reagents may be directed to, and will be fulfilled by the lead contact, Dr. In-Hyun Park (inhyun.park@yale.edu).
Materials availability
Plasmids generated in this study is stored in the lab and will be shared upon request.
Data and code availability.
Single cell RNA-seq, bulk RNA-seq, ChIP-seq, ATAC-seq, and HiC data are available at GEO with accession number (GSE117513).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
hPSCs culture
H1 and HES-3 NKX2–1GFP/w human ES cells, human iPSC (RTT4–16M), and all derivative clones from genome manipulation were cultured on Matrigel coated tissue culture dish with mTeSR1 media. Cells were passaged every 7 days by Dispase (0.83 U/ml) treatment. All experiments involving hESCs and hiPSCs were approved by the Yale Embryonic Stem Cell Research Oversight Committee (ESCRO).
MeCP2-/Y mice.
B6.129P2(C)-Mecp2tm1.1Bird/J female mice and C57BL male mice, both of which were obtained from the Jackson Laboratory, were used to generate MeCP2-/Y mice. All animal experiments were approved by the Institutional Animal Care & Use Committee (IACUC) of the Yale University.
METHOD DETAILS
CRISPR/Cas9-based genome editing.
sgRNA was selected using the CRISPR Design Tool (http://crispr.mit.edu/), and corresponding coding sequence was cloned into pSpCas9(BB)-2A-Puro (PX459) construct. Single-stranded oligo donor were designed to introduce desired point mutation, which contained at least 45 bp homologous arm at each side. 2×106 of single cells dissociated from hESCs or hiPSCs culture were electroporated with 2 μg of PX459-sgRNA and 2 μg of ssODN donor using the Human Stem Cell Nucleofector Solution 1 (Lonza VPH-5012). After electroporation, cells were plated onto Matrigel coated 10-cm dishes with mTeSR1 media containing 10 μM Y27632 (Stem Cell Technologies). Two days of puromycin selection was then performed starting from 24 h after electroporation. After another 5~7 days of recovery, single clones were picked and cultured in Matrigel coated 96-well plate. After ~7 days of culture, half of each clone was collected for gDNA isolation and PCR amplification of target region. PCR products were purified and subjected to sequencing also at a scale of 96-well plate. Successfully targeted clones, including indels and HDR, were identified according to their target sequences. Representative clones from MECP2 editing are listed in Table S1. Off-target sequence analysis of selected clones is listed in Table S2. Selected target clones were then expanded for downstream use.
TALEN-based genome editing.
The hrGFP sequence in AAVS1-CAG-hrGFP plasmid was replaced with GCaMP6s sequence. 2×106 of single cells dissociated from hESCs were electroporated with 1 μg of AAVS1-TALEN-L plasmid, 1 μg of AAVS1-TALEN-R plasmid, and 8 μg of AAVS1-CAG-GCaMP6s donor, and then plated onto Matrigel coated 10-cm dish. Three days after electroporation, puromycin selection was performed, which lasted for 1 week. After another 5~7 days of recovery, single clones were picked. Functional GCaMP6s insertion was confirmed by the fluorescence responses under 55 mM KCl stimulation.
Cortical interneuron differentiation.
hESCs were differentiated into cortical interneurons as previously described(Maroof et al., 2013). Briefly, cells were dissociated by Accutase (Stem Cell Technologies AT104) treatment and cultured on Matrigel coated 24-well plate with mTeSR1 media until 95% confluence to start neural induction. Neural induction media contained DMEM-F12 (Life Technologies), 15% (v/v) knockout serum replacement (KSR) (Life Technologies 10828–028), 1% (v/v) MEM-NEAA (Life Technologies 11140050), 1% (v/v) Glutamax (Life Technologies 35050), 100 μM β-Mercaptoethanol (Sigma M7522), 100 nM LDN-193189 (Sigma SML0559), 10 μM SB-431542 (Abcam ab120163) and 2 μM XAV-939 (Sigma X3004). Media was replenished every day for the first 4 days. On day 5, day 7, and day 9, cells were fed with neural induction media mixed with N2 media at various ratios (3:1 ratio on day 5, 1:1 ratio on day 7, and 1:3 ratio on day 9). Neuroepithelial cells were dissociated with Accutase treatment and replated on Matrigel coated 24-well plate at 1:1 ratio on day 10 using the same media used on day 9, with the exception that 10 μM Y27632 was supplemented. N2 media contained DMEM-F12, 0.15% (w/v) Dextrose (Sigma G7021), 100 μM β-Mercaptoethanol, and 1% (v/v) N2 supplement (Life Technologies 17502–048). Ventral patterning was performed from day 10 to day 18, with the media replenished every other day. To prepare ventral patterning media, 2% (v/v) B27 supplement (Life Technologies 17504–044), 100 ng/ml recombinant SHH (R&D Systems 464-SH-200), and 1 μM purmorphamine (Stem Cell Biotech 72204) were added to N2 media. From day 19, neural maturation media was replenished every other day to promote further neuronal differentiation and maturation. Neural maturation media contained Neurobasal media (Life Technologies), 2% (v/v) B27 supplement, 20 ng/ml BDNF (Prepotech 450–02), 200 μM ascorbic acid (Sigma A92902), 200 μM cAMP (Millipore 28745), and 1% (v/v) Penicillin/Streptomycin (Life Technologies). Neuronal progenitor cells (NPCs) were dissociated by Accutase and replated on Matrigel coated plate at lower density (0.5×105 cells/cm2) at desired scale after 1 week of neuronal maturation. After 2 weeks of maturation, media was replenished every four days.
Cortical neuron differentiation.
Cortical neuron differentiation was performed based on a previously published protocol with minor modifications(Shi et al., 2012). Briefly, hESCs were dissociated to single cells and plated onto Matrigel coated 24-well plate with mTeSR1 media. After cells achieved over 95% confluence, media was changed to neural induction media, which contained 1:1 mixture of DMEM-F12 media and Neurobasal media, supplemented with 1% (v/v) N2 supplement, 2% (v/v) B27 supplement, 20 μg/ml Insulin (Sigma I9278), 1% (v/v) Glutamax, 1% (v/v) MEM-NEAA, 100 μM β-Mercaptoethanol, 100 nM LDN-193189, 10 μM SB-431542 and 2 μM XAV-939. Neural induction was performed for 10 days, with daily media replenishment. On day 11, neuroepithelial cells were dissociated by Accutase and plated onto Matrigel coated 24-well plate using neural expansion media at a density of 1.5×105 cells/cm2. 10 μM Y27632 was added to the media when replating the cells. The components of neural expansion media were the same with neural induction media, except that LDN-193189, SB-431542, and XAV-939 were not added. Neural expansion media was replenished every other day. From day 19, media was changed to maturation media, which contained Neurobasal media, 1% (v/v) N2 supplement, 2% (v/v) B27 supplement, 1% (v/v) Glutamax, 1% (v/v) Penicillin/Streptomycin, 20 ng/ml BDNF, 200 μM ascorbic acid, and 200 μM cAMP. For the first week of maturation, media was changed every other day. NPCs were disscociated by Accutase and replated on Matrigel coated plate at a density of 0.5×105 cells/cm2 after 1 week of maturation. After 2 weeks of maturation, media was replenished every four days.
Brain organoids generation.
hMGEOs and hCOs were generated as previously described(Xiang et al., 2017). hESCs were dissociated by Accutase and plated into U-bottom ultra-low-attachment 96-well plate in neural induction media (DMEM-F12, 15% (v/v) KSR, 1% (v/v) MEM-NEAA, 1% (v/v) Glutamax, 100 μM β-Mercaptoethanol, 100 nM LDN-193189, 10 μM SB-431542 and 2 μM XAV-939) supplemented with 50 μM Y27632 and 5% (v/v) heat-inactivated FBS (Life Technologies). 9,000 cells were plated into each well. Neural induction media was replenished every other day until day 10. FBS was removed from the media from day 2, and Y27632 was removed from day 4. On day 10, organoids were transferred to ultra-low-attachment 6-well plate for spinning culture (80 rpm/min). For hMGEO generation, ventral patterning media (DMEM-F12, 0.15% (w/v) Dextrose, 100 μM β-Mercaptoethanol, 1% (v/v) N2 supplement, 2% (v/v) B27 supplement without vitamin A, 100 ng/ml recombinant SHH, and 1 μM purmorphamine) was used from day 10 to day 18. For hCO generation, neural differentiation media (1:1 mixture of DMEM-F12 and Neurobasal media, 0.5% (v/v) N2 supplement, 1% (v/v) B27 supplement without vitamin A, 1% (v/v) Glutamax, 0.5% (v/v) MEM-NEAA, 0.025% (v/v) Insulin, 50 μM β-Mercaptoethanol and 1% (v/v) Penicillin/Streptomycin) was used from day 10 to day 18. For both hMGEO and hCO cultures, media was replenished every other day before day 18. After day 18, media was changed to neural differentiation media as used for hCO culture, except that B27 supplement with vitamin A was used, and 20 ng/ml BDNF, 200 μM ascorbic acid, as well as 200 μM cAMP were added. Media was replenished every other day before day 25, and every four days thereafter. To enhance the growth of hCOs, 20 ng/ml of FGF2 was added between day 18 and day 22 for hCO culture. We noticed that adding FGF2 is cell line dependent, and H1 hESCs require FGF2 supplement for better hCO development.
Neuronal stimulation.
Monolayer neurons and brain organoids were membrane depolarized with KCl as described previously(Ebert et al., 2013). KCl buffer (170 mM KCl, 2 mM CaCl2, 1 mM MgCl2 and 10 mM HEPES, pH7.5) was directly added to culture to achieve final concentration of 55 mM KCl. For stimulation of brain organoids, plate was returned to spinning culture after adding KCl buffer. Samples were immediately collected for down-stream assays 1 hour after membrane depolarization. For stimulation and IN morphology assay, cells were treated with 25 mM KCl for 48 h before sample collection.
Drug treatment.
For treatment of monolayer neuronal cultures (e.g. cortical INs and general cortical neurons), 75 nM JQ1 (ApexBio A1910) was added to the media starting from 2 weeks of neuronal maturation (day 32). Media containing JQ1 was replenished every four days. For treatment with other BET bromodomain inhibitors, 50 nM CPI203 (Cayman 15479) or 100 nM IBET762 (Cayman 10676) was added to the media from day 32, and drug-containing media was replenished every four days. For treatment at later stages (Figure S4K and S4L), day 65 INs were treated with 75 nM JQ1 for 24 hours, or day 61 INs were treated with 75 nM, 150 nM or 300 nM JQ1 for 10 days, followed by depolarization and gene expression assays. For treatment of brain organoids, 75 nM JQ1 was added to the media starting from day 25, and drug-containing media was replenished every four days. For in vivo treatment, JQ1 was administered to mice starting from 2 weeks of age via intraperitoneal injections. Each mouse was injected daily with JQ1 (15 mg/kg) dissolved in DMSO or DMSO along, which was diluted into cyclodextrin (Sigma C0926). Mice were subjected to weekly phenotypic scoring starting from 7 weeks of age as previously described (Guy et al., 2007; Tillotson et al., 2017). Body weights of mice were weekly measured. Mice survival was graphed using the Kaplan-Meier survival plot.
Real time quantitative PCR.
Total RNA was extracted using the RNeasy Mini Kit (Qiagen), and cDNA was generated using the iScript Select cDNA Synthesis Kit (Biorad). Real time quantitative PCR was performed using the SsoFast EvaGreen Supermix (Biorad) in the CFX96 Real-Time PCR System (Biorad). The PCR cycling conditions were: 95°C for 15 min, followed by 40 two-step cycles at 94°C for 10 s and 60°C for 45 s. Primers used were as follows: c-FOS forward: 5’-TCTGTGCGTGAAACACACCA-3’, reverse: 5’-GCCTGGCTCAACATGCTACT-3’; FOSB forward: 5’-GGTTCGTTGGGGACATGAGA-3’, reverse: 5’-AATTGTGGTTGGCAGGAGCA-3’; NPTX2 forward: 5’-GTCCTTCGCGCACAAGAAAT-3’, reverse: 5’-GAGAAGGCGGCTACAAGT CA-3’.
Immunostaining and fluorescence quantification.
For immunostaining of monolayer neuron cultures, samples were fixed in 4% paraformaldehyde (PFA) at RT for 15 min, incubated with 0.1% Triton-100 at RT for 15 min, blocked with 3% BSA/PBS at RT for 1h, incubated with primary antibody diluted in 3% BSA/PBS at RT for 1h, incubated with secondary antibody diluted in 3% BSA/PBS at RT for 1 h, and finally stained for nuclei by DAPI. Samples were washed with PBS 3 times in between each step, except that no wash was performed after BSA blocking. For immunostaining of brain organoids, samples were fixed in 4% PFA at 4°C for 4 days, washed 3 times with PBS (10 min incubation for each wash), and incubated in 30% sucrose solution at 4°C for 3 days. Organoids were transferred to O.C.T compound (Tissue-Tek 4583) for incubation at RT for 15 min and then transferred to tissue base molds for embedding with O.C.T compound on dry ice. Embedded samples were either stored at −80°C or immediately used for cryosectioning to obtain 40 mm slices. Slices were then washed with PBS, incubated with 0.1% Triton-100 at RT for 15 min, blocked with 3% BSA/PBS at RT for 2 h, and incubated with primary antibody diluted in 3% BSA/PBS at 4°C overnight. After PBS wash, slices were incubated with secondary antibody diluted in 3% BSA/PBS at RT for 1 h, and nuclei were stained by DAPI. Finally, slices were mounted with ProLong Gold Antifade Reagent (ThermoFisher P36930). Corrected total nuclei fluorescence (CTNF) of c-FOS was quantified using the ImageJ software as reported before(Gavet and Pines, 2010). Briefly, CTNF=Integrated density of selected nucleus - (Area of selected nucleus x Mean fluorescence of background reading). Antibodies used for staining included MAP2 (Millipore MAB3418), GFP (Sigma SAB4600051), β-Tubulin III (Sigma T8578), SOX2 (Cell Signaling 3579), FAM107A (Sigma HPA055888), S100 (Sigma S2644), GABA (Sigma A0310), vGAT (Synaptic Systems 131003), vGLUT1 (Synaptic Systems 135311), c-FOS (Abcam ab134122), SSEA3 (Millipore MAB4303), SSEA4 (BD Biosciences BD560218), Tra-1–60 (BD Biosciences BD560173), and OCT4 (Abcam ab19857).
Organoids dissociation.
Human brain organoid samples were dissociated using Papain Dissociation System (Worthington Biochemical Corporation) as previously described(Xiang et al., 2017).
Calcium imaging and data analysis.
For experiments in Figure 3 and Figure S6, neurons at 5 weeks of maturation (day 54) were replated onto Matrigel coated plate, and transduced with AAV1.syn.GCaMP6s.WPRE.SV40 vector (Penn Vector Core) 3 days after plating. Calcium imaging was performed ~3 weeks after transduction. For experiments in Figure S3, INs were at 7 weeks of maturation (day 67) were replated onto Matrigel coated plate, transduced with AAV1.syn.GCaMP6s.WPRE.SV40 vector 3 days after plating, and subjected to calcium imaging at ~4 weeks after transduction. Calcium imaging was performed using the Nikon inverted microscope (Eclipse TS100) with 20x objective (single cell scale) at 488 nm excitation. Time lapse images were captured using a Digital CCD camera (QICAM: FAST 1394) and Qcapture Pro7 software (QICAM) at a speed of 1 frame/sec (FPS). For organoid cultures, organoids were produced using the H1-MeCP2-WT/AAVS1-GCaMP6s or H1-MeCP2-R133C/AAVS1-GCaMP6s reporter hESCs, and calcium imaging was performed at ~70 days of in vitro culture. Microscope, camera, software, and other parameters were the same with monolayer calcium imaging, except that 10x objective (area scale) was used. Tracing of single cell calcium activities were analyzed using the MATLAB software with FluoroSNNAP code, and area-scale image data was analyzed using the Fiji software.
Global proteome profiling.
Cell pellets were collected at 8 weeks of neuronal maturation (day 76) and processed for proteomic profiling in Thermo Fisher Scientific Center for Multiplexed Proteomics at Harvard Medical School according to standardized workflow. Briefly, cells were lysed and proteins were quantified. Protein precipitation was performed using methanol/chloroform, and digestion was performed by using LysC and trypsin, with all of the peptides being labeled with TMT reagent. Samples were then processed for liquid chromatography–mass spectrometry (LC-MS), with peptides being detected (MS1) and quantified (MS3) in the orbitrap, and sequenced (MS2) in the ion trap. MS2 spectra were searched using the SEQUEST algorithm against a Uniprot composite database derived from the human proteome containing its reversed complement and known contaminants. Peptide spectral matches were filtered to a 1% false discovery rate (FDR) using the target-decoy strategy combined with linear discriminant analysis. The proteins from fractions were filtered to a <1% FDR. Proteins were quantified only from peptides with a summed SN threshold of >=100 and isolation specificity of 0.5.
Co-cultures of neurons and astrocytes.
Mouse astrocytes were isolated as previously described(Schildge et al., 2013). To facilitate patch-clamp recording, neurons were transduced with AAV1.hSyn.eGFP vectors (Penn Vector Core) at 2 weeks of neuronal maturation (day 32). 1 week after transduction, neurons were dissociated using Accutase, and re-plated onto astrocytes. Patch-clamp recordings were performed after 8 weeks of neuronal maturation (>day 74).
Dendrite complexity assay.
Cultured INs at 7 weeks of maturation were transduced with AAV1-hsyn-eGFP vectors. 10 days after transduction, INs were replated onto confluent mouse astrocytes for co-culture. 4 days after replating, cells were fixed with 4% PFA at RT for 15 min and processed to immunostaining against GFP antibody. For dendrite complexity in response to KCl stimulation, cells were treated with 25 mM KCl for 48 h, then fixed for immunostaining. Sholl analysis was performed using NeuronJ plugin in Fiji software.
Electrophysiological recordings.
Whole cell patches were established at room temperature using Multiclamp 700B amplifier (Molecular Devices) and Digidata 1440 (Axon Instruments), and the acquired data were analyzed using the pCLAMP version 10.2 (Axon instrument). Action potentials (APs) were triggered by a step current injection (100 pA) in current clamp mode for 800 ms. To block APs, 1 μM tetrodotoxin (TTX) was used. For sodium and potassium current measurement cells were clamped at −70 mV and depolarizing voltage steps were delivered for 250 ms in 10 mV increments. The pipette solution for current-clamp experiments contained (in mM): 122.5 K-gluconate, 12.5 KCl, 0.2 EGTA, 8 NaCl, 2 MgATP, 0.3 Na3GTP and 10 HEPES, pH adjusted to 7.3 with KOH. The bath solution contained (in mM): 119 NaCl, 2.5 KCl, 26 NaHCO3, 2 CaCl2, 1.3 MgSO4, and 25 glucose, pH7.4. Data were acquired with pClamp 10.2 (Molecular Devices); current was filtered at 0.1 kHz and digitized at 2 kHz. Blind test was performed for MeCP2-WT and MeCP2-R133C INs during recording. For both groups, neurons with bright and intact soma were randomly recorded. Data were excluded if cells sustain incomplete processing with gigaseal formation and whole-cell configuration.
High-throughput sequencing.
For bulk RNA-seq, total RNAs were isolated from cell pellets at specific time point (see detailed results) using the RNeasy Mini kit. Libraries were constructed using Tru-Seq RNA library preparation kits, followed by sequencing in HiSeq 2000 with manufacture’s protocol.
For scRNA-seq, for each condition brain organoids (MeCP2-WT hMGEOs: 6 organoids at day 79; MeCP2-R133C hMGEOs: 6 organoids at day 79; MeCP2-WT hMGEOs with JQ1 treatment: 6 organoids at day 77; MeCP2-R133C hMGEOs with JQ1 treatment: 6 organoids at day 77; MeCP2-WT hCOs: 6 organoids at day 84; MeCP2-R133C hCOs: 7 organoids at day 84; MeCP2-WT hCOs with JQ1 treatment: 6 organoids at day 86; MeCP2-R133C hCO with JQ1 treatment: 7 organoids at day 86) were collected from 2 different culture dishes, pooled together, and dissociated into single cells. cDNA libraries preparation and sequencing were performed as previously described(Xiang et al., 2017).
For ChIP-seq, monolayer IN cultures after ~8 weeks of neuronal maturation (day 74 in vitro) were collected. BRD4 and MeCP2 ChIP experiment were performed using anti-BRD4 (A301–985A50, Bethyl lab.) and anti-MeCP2 antibody (ab195393, Abcam) with iDeal ChIP-seq kit for Transcription Factors with manufacture protocol (C01010170, Diagenode).
RRBS was performed as described previously(Gu et al., 2011). Briefly, genomic DNA from day 74 MeCP2-WT and MeCP2-R133C INs was digested by MspI (NEB R0106S) overnight. Gap filling and A-tailing were conducted by adding Klenow fragment (NEB 3’ -> 5’ exo-M0212L). Illumina TruSeq adapter was ligated to the end-repaired DNA with T4 ligase (NEB M0202). Bisulfite reaction was then performed to the adapter-ligated DNA by EpiTech Bisulfite kit (Qiagen 59104). For oxRRBS, we performed oxidation step before MspI digestion. Genomic DNA was denatured with 0.05 M sodium hydroxide (NaOH) for 30 min at 37C. Then, oxidation reaction was performed with 15 mM potassium perruthenate solution (in 0.05M NaOH) for 1hr on ice (Booth et al., 2013).
For ATAC-seq, 50,000 cells were lysed in cold lysis buffer (10mM Tris-HCl, pH7.4, 10mM NaCl, 3mM MgCl2, 0.1% IGEPAL CA-630)(Buenrostro et al., 2013). After spinning at 500xg for 5min, the pellet was treated by Tn5 transposase by Nextra® DNA Library Prep Kit. After isolating digested DNA by Qiagen EinElute kit, the library was constructed and amplified by NEB High Fidelity 2xPCR master mix and purified by AxyPrepTM Mag PCR Clean-up kit.
For Hi-C, we performed the Dekkar Hi-C 2.0 protocol as previously reported (Belaghzal et al., 2017). Cells were fixed in 1% formaldehyde in HBSS for 10 minutes, followed by scraping, washing, and aliquoting into ~5×106 cell aliquots. One 5×106 cell aliquot per condition was then used. Cells had their plasma membranes lysed and were homogenized before overnight digestion with DpnII. The in-situ digested ends were then biotinylated and ligated together before the crosslinking was reversed with ProK overnight. The DNA was then sheared, size selected, and pulled down via streptavidin beads before Illumina adaptor ligation, production PCR, and 2×100 sequencing.
Data processing of bulk RNA-seq.
Human genomic sequences, CpGI and RefSeq gene coordinates (version hg19) were downloaded from UCSC genome browser. Mapping to human genome and calculation of gene expression genome were processed by Tophat2 (v2.1.0) and Cufflinks with Refseq genes as reference transcripts (v1.2.1)(Trapnell et al., 2009; Trapnell et al., 2010). Differentially-expressed genes in each time point to day 0 hESC, between WT and mutant and between treatment and control were defined by 2 fold cutoff. GO analysis was performed to the differentially-expressed genes using GOstats (v2.24.0) Bioconductor package (Falcon and Gentleman, 2007). Multiple tests were adjusted by Benjamini-Hochberg method with p.adjust function in R. GO terms with FDR < 0.05 were used as statistical significance. The list of immediate early genes was obtained from (Tullai et al., 2007). The enrichment of IEGs in BRD4 binding loci or KCl-stimulated INs was evaluated by Gene Set Enrichment Analysis (GSEA) software (v2.2.2) with 1,000 permutations for gene set, and weighted enrichment statistic without collapsing dataset (Subramanian et al., 2005).
Data processing of RRBS.
RRBS reads were mapped to human genome by bsmap software (v2.7) with “-s 12 -g 3 -w 3 -D C-CGG -r 0” options(Xi and Li, 2009). Methylation ratio was calculated by methratio.py python script in bsmap package with “-q –z –u” options. Methylation level of each CpGI was estimated by calculating average methylation ratio of CpGs, which were covered by more than three reads. oxRRBS reads were also processed with the same way. 5hmC level were calculated by subtracting methylation ratio in oxRRBS from that in RRBS. 5hmC-enriched CpGI was defined with more than 0.05 average 5hmC level.
Data processing of ChIP-seq.
ChIP-seq reads were mapped to human genome by Bowtie2 software (v2.3.0) under SAMtools (v1.3.1) with “--local -D 15 -R 3 -N 1 -L 20 -i S,1,0.50 -k 1” options (Langmead and Salzberg, 2012; Li et al., 2009). BRD4 and CTCF peaks were identified by MACS2 software with “-g hs –q 0.05” option. MeCP2 peaks were detected with “-g hs –q 0.05 -- broad” option. Genes with ChIP-seq peaks within 10kbp upstream or gene body were selected as target genes. When we compared BRD4 peaks between two distinct conditions, peaks within 100-bp distance were merged. To measure differential binding level, ChIP-seq read from two distinct samples was counted within ±250bp of peak center and then normalized by total number of mapped reads. Then, peaks were classified into 13 categories according to fold change between two samples. The induction and silencing of target genes by KCl/JQ1 treatment or MeCP2 mutation were analyzed by GSEA software as described above. If target genes were induced (NES > 0), −log10(FDR) was used. In contrast, log10(FDR) was used for gene silencing (NES < 0).
Data processing of ATAC-seq.
ATAC-seq reads were mapped by Bowtie2 as described above. Open chromatin regions were then identified by findPeaks script with “-localSize 50000 -size 100 - minDist 50 -fragLength 0 -o auto” in HOMER package (v4.10.1). After merging ATAC peaks within 100-bp distance, ATAC-seq reads were counted and normalized by total number of mapped reads in each peak. Hierarchical clustering was then performed to log2-transformed normalized count matrix with hclust function in R. dOCRs were obtained by 2 fold difference and less than 0.05 T test p-value. Motif enrichment in dOCRs were analyzed by findMotifsGenome.pl scripts in HOMER with default parameters. ATAC-seq peaks within TSS±5kbp were classified into promoters and the others as enhancers.
Data processing of HiC.
Mapping, quality assessment, isolation of valid interaction pairs of HiC reads were processed by HiC-Pro software (v2.9.0) with DpnII restriction sites in hg19 genome and “GATCGATC” ligation site(Servant et al., 2015). Intrachromosomal HiC contact matrix was visualized by HiCPlotter software(Akdemir and Chin, 2015). Significant interactions were then identified by analyzeHiC script with 5kbp resolution and annotated by annotateInteractions.pl script with MeCP2, CTCF, BRD4 and ATAC promoter and enhancer peaks. Symmetric and asymmetric HiC contact matrices were drawn by hicAggregateContacts in HiCExplorer library(Ramírez et al., 2018).
Data processing of single-cell RNA-seq.
Mapping to hg19 human genome, quality control, UMI counting of Ensembl genes and k-mean clustering were performed by cellranger software with default parameter (v2.1.0). Secondary analyses and data visualization were conducted using Seurat (v3.0.1) (Heintzman et al., 2007; Stuart et al., 2019). Briefly, we first filtered out cells less than 500 detected genes and genes expressed in less than five cells from subsequent analyses. After normalizing UMI count to the total count per run, highly-variable genes were investigated by variance stabilizing transformation. Top 5,000 variable genes were used to identify cell pairs anchoring different single-cell transcriptome runs with canonical component (CC) analysis. Using 1st to 30th CCs, we integrated all single-cell transcriptome runs into a shared space. After scaling gene expression values across all integrated cells, principal compoenent analysis (PCA) was performed to reduce the dimension of the expression profile into 30 PCs. Subsequently, uniform manifold approximation and projection (UMAP) was implemented to visualize all cells into two-dimensional space. Cell clusters were identified with shared nearest neighbor graph method with 1st to 30th PCs and 0.8 resolution value (Figure S7E). Cluster specific genes were obtained by comparing one cluster with the rest of clusters by >1.5 fold change and p-value < 0.05 with two-side T test. Doublet frequency of our scRNA-seq was estimated by the number of cells expressing TBR1 or GFAP, which were exclusively expressed in cortical excitatory neuron and astrocyte cells (Figure S7F).
Cell clusters were then annotated according to the expression of unique markers and significant GO terms for cluster-specific genes (Figure S7G and S7H). First, cell clusters were classified into two groups by genes related to early neurogenesis (VIM, HES1 and NES) and neuronal growth cone (STMN2, GAP43 and DCX), which were almost exclusively expressed in neuron and non-neuronal cells, respectively (Figure S7I). 13 clusters were categorized into “neuron” and then further separated into cortical and interneuron clusters according to the expression of their unique markers (NEUROD2 and vGLUT1 (SLC17A7) for cortical neuron, vGAT and GAD1 (SLC32A1) for interneuron). Neuron clusters with a limited expression of cortical and interneuron markers were annotated as non-committed neuron. One cluster was mixtures of VIM/HES1/NES high and STMN2/GAP43/DCX high cells and annotated as “intermediate”.
Among 16 non-neuronal clusters, 11 clusters showed statistical significance of “Gliogenesis (GO:0042063)” or “Glia cell differentiation (GO:0010001)” and categorized into “glia” clusters. Unique expression of oligodendrocyte markers (OLIG1/2 and SOX10) and weak expression of its maturation markers (MAG and MOG) were detected in one glia cluster (labeled as immature oligodendrocyte). Another cluster displayed the highest expression of radial glia cell markers (i.e. LHX2, GLI3) (Figure S2H). Astrocyte (AS) clusters were separated from the other glia cell clusters with the significant enrichment of genes related to “astrocyte differentiation (GO:0048708)”. Two out of five astrocyte clusters highly expressed GFAP, which is enriched at late developmental stage of fibrous astrocyte (Molofsky et al., 2012), and separated as mature astrocytes. One out of the rest three glia clusters displayed high expression of genes involved in “response to unfolded protein (GO:0006986)” and separated as unfolded protein response cell (UPRC). The other two glia clusters were categorized as glia progenitor cell (GPC)
One non-glia cluster uniquely expressed biglycan and decorin (BRN and DCN) that are proteoglycans. Despite previous reports about their biosynthesis in astrocytes (Barkho et al., 2006; Koops et al., 1996), the BRN/DCN high cluster lacked astrocyte markers. Thus, we distinguished this cluster as the other astrocytes and tentatively named as proteoglycan-expressing cell (PGC). “BMP signaling pathway (GO:0030509)” was significantly overrepresented in one non-glia cluster and labeled as BMP responsible cell (BRC). As observed in the glia cluster, we identified the significant enrichement of “response to unfolded protein (GO:0006986)” in one non-glia cluster. Because of higher expression of cell cycle-related genes, one glia cluster was annotated as proliferative neuronal precursor cells (NPCs). The other non-glia cluster did not show any significant GO terms related to cell development and cell type specific markers and was not assigned to specific cell type (called unknown (UN)).
Public single-cell RNA-seq data from human fetal and adult cortex were downloaded from NCBI Short Read Archive (SRP057196) (Darmanis et al., 2015). Gene signatures for neuron, NPC, astrocyte and oligodendrocyte of each cell type were obtained as described previously (Xiang et al., 2017). GSEA was performed to gene expression profile of each single cell, which were pre-ranked by relative expression to average of all cells. A python wrapper, GSEAPY (v0.9.3), with options “-- max-size 50000 --min-size 0 -n 1000” was used for GSEA to each single cell (Figure S7J).
To measure the gene dyregulation by MeCP2 mutation, we compared transcriptional profiles of neuron and glia clusters between MeCP2-WT and MeCP2-R133C under JQ1 or no treatment. Top 200 genes were used for GO analysis. Network construction and grouping of significant GO terms were performed ClueGO plug-in (v1.8) in Cytospace software (v2.8) with default parameters (Bindea et al., 2009).
QUANTIFICATION AND STATISTICAL ANALYSIS.
For statistical analysis of immunostaining and qPCR, unpaired t test was used. Statistical significance was accepted for p < 0.05. For processing of high-throughput sequencing data, please refer to the method description for details of statistical analysis. The statistical details of experiments are also present in the figure legends and result description.
Supplementary Material
Table S1. Representative clones from MECP2 editing. (Related to Figure 1)
Table S2. Off target evaluation of selected clones. (Related to Figure 1)
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| MAP2 | Millipore | Cat# MAB3418 |
| GFP | Sigma | Cat# SAB4600051 |
| β-Tubulin III | Sigma | Cat# T8578 |
| GABA | Sigma | Cat# A0310 |
| vGAT | Synaptic Systems | Cat# 131003 |
| VGLUT 1 | Synaptic Systems | Cat# 135311 |
| c-FOS | Abcam | Cat# ab134122 |
| BRD4 | Bethyl | Cat# A301-985A100 |
| MeCP2 | Abcam | Cat# ab195393 |
| SSEA3 | Milipore | Cat# MAB4303 |
| SSEA4 | BD Biosciences | Cat# BD560218 |
| Tra-1–60 | BD Biosciences | Cat# BD560173 |
| OCT4 | Abcam | Cat# Ab19857 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| mTeSR1 | Stem Cell Technologies | Cat# 05875 |
| DMEM-F12 | Life Technologies | Cat# 11330057 |
| DMEM | Life Technologies | Cat# 11965092 |
| Neurobasal Media | Life Technologies | Cat# 2110349 |
| HBSS | Life Technologies | Cat# 14170112 |
| FBS | Life Technologies | Cat# 10437028 |
| Amino acids, non-essential | Life Technologies | Cat# 11140050 |
| Penicillin/Streptomycin | Life Technologies | Cat# 15140-122 |
| Glutamax | Life Technologies | Ca# 35050 |
| Insulin | Sigma | Ca# I9278 |
| β-Mercaptoethanol | Sigma | Ca# M7522 |
| N2 | Life Technologies | Cat# 17502-048 |
| B27 | Life Technologies | Cat# 17504–044 |
| B27 supplement without vitamin A | Life Technologies | Cat# 12587010 |
| bFGF | Millipore | Cat# GF003AF |
| KnockOut Serum Replacement | Life Technologies | Cat# 10828–028 |
| Matrigel | BD | Cat# 354230 |
| poly-L-ornithine | Sigma | Cat# P4957 |
| Y-27632 | Stem Cell Technologies | Cat# 72304 |
| Dispase (100ml) | Stem Cell Technologies | Cat# 07913 |
| Accutase (100ml) | Stem Cell Technologies | Cat# AT104 |
| LDN-193189 | Sigma | Cat# SML0559 |
| SB431542 | Abcam | Cat# ab120163 |
| XAV939 | Sigma | Cat# X3004 |
| JQ1 | ApexBio | Cat# A1910 |
| CPI203 | Cayman | Cat# 15479 |
| IBET762 | Cayman | Cat# 10676 |
| (2-Hydroxypropyl)-β-cyclodextrin | Sigma | Cat# C0926–10G |
| Nefiracetam | Tocris | Cat# 2851 |
| Human IGF1 | Peprotech | Cat# 100–11 |
| SHH | R&D Systems | Cat# 464-SH-200 |
| Purmorphamine | Stem Cell Biotech | Cat# 72204 |
| Puromycin | Sigma | Cat# P8833 |
| BDNF | Prepotech | Cat# 450–02 |
| cAMP | Millipore | Cat# 28745 |
| Ascorbic acid | Sigma | Cat# A92902 |
| O.C.T compound | Tissue-Tek | Cat# 4583 |
| Bovine serum albumin | American Bioanalytical | Cat# AB01088 |
| ProLong Gold Antifade Reagent | ThermoFisher | Cat# P36930 |
| Biocytin | Sigma | Cat #B4261 |
| Alexa Fluor 594 Streptavidin | Jackson Immunoresearch | Cat # 016–580-084 |
| Critical Commercial Assays | ||
| Papain Dissociation System | Worthington Biochemical Corporation | Cat# LK003150 |
| RNeasy mini kit | QIAGEN | Cat# 74104 |
| RNeasy MinElute Cleanup Kit | QIAGEN | Cat# 74204 |
| DNeasy Blood & Tissue Kits | QIAGEN | Cat# 69506 |
| Plasmid Mini Kit | QIAGEN | Cat# 12123 |
| Plasmid Midi Kit | QIAGEN | Cat# 12143 |
| Human Stem Cell Nucleofector Kit 1 | Lonza | Cat# VPH-5012 |
| RNase-Free DNase Set | QIAGEN | Cat# 79254 |
| iScript cDNA synthesis kit | Biorad | Cat# 1708891 |
| SsoFast EvaGreen Supermix | Biorad | Cat# 1725201 |
| Single Cell 3’ Reagent Kits | 10x GENOMICS | |
| Nextera® DNA Library Prep Kit | Illumina | Cat# FC-121–1030 |
| EinElute kit | QIAGEN | Cat# 28004 |
| High Fidelity 2xPCR master mix | NEB | Cat# M0541S |
| AxyPrepTM Mag PCR Clean-up kit | AXYGEN | Cat# 14–223-153 |
| Deposited Data | ||
| Raw and proposed RNA-seq and ChIP-seq | This paper | GEO:GSE117513 |
| Human genome assembly hg19 | N/A | https://genome.ucsc.edu/ |
| scRNA-seq for fetal and adult brains | Darmanis et al., 2015 | SRA:SRP057196 |
| Experimental Models: Cell Lines | ||
| H1 hESC line | WiCell | https://www.ncbi.nlm.nih.gov/pubmed/9804556/ |
| H1-MeCP2-R133C | This paper | |
| H1-MeCP2-R270X | This paper | |
| H1-MeCP2-R306C | This paper | |
| H1-AAVS1-CAG-GCaMP6s | This paper | |
| H1-MeCP2-R133C/AAVS1-CAG-GCaMP6s | This paper | |
| RTT4–16M | Kim et al., 2010 | https://www.ncbi.nlm.nih.gov/pubmed/21807996 |
| RTT4-MeCP2-R306C | This paper | |
| RTT4-MeCP2-WT | This paper | |
| HES-3 NKX2–1GFP/w | Elefanty lab | https://www.ncbi.nlm.nih.gov/pubmed/21425409 |
| Clones from MECP2 editing are listed in Table S1 | This paper | This paper |
| Off-target analysis of MECP2 edited clones is listed in Table S2 | This paper | This paper |
| Recombinant DNA | ||
| AAV1.syn.GCaMP6s.WPRE.SV40 | Penn Vector Core | AV-1-PV2824 |
| AAV1.hSyn.eGFP.WPRE.bGH | Penn Vector Core | AV-1-PV1696 |
| pSpCas9(BB)-2A-Puro (PX459) | Addgene | 48139 |
| pSpCas9(BB)-2A-Puro (PX459)_MeCP2 R133 sgRNA | This paper | |
| pSpCas9(BB)-2A-Puro (PX459)_MeCP2 R270 sgRNA | This paper | |
| pSpCas9(BB)-2A-Puro (PX459)_MeCP2 R306 sgRNA | This paper | |
| AAVS1-TALEN-L | Addgene | 59025 |
| AAVS1-TALEN-R | Addgene | 59026 |
| AAVS1-CAG-hrGFP | Addgene | 52344 |
| pGP-CMV-GCaMP6s-CAAX | Addgene | 52228 |
| AAVS1-CAG-GCaMP6s | This paper | |
| Software and Algorithms | ||
| Tophat (v2.1.0) | Trapnell et al., 2009 | https://ccb.jhu.edu/software/tophat/index.shtml |
| Samtools (v1.3.1) | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Seurat (v3.0.1) | Stuart et al., 2019 | https://satijalab.org/seurat/ |
| Bowtie (v2.3.0) | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Cufflinks (v1.2.1) | Trapnell et al., 2010 | http://cole-trapnell-lab.github.io/cufflinks/ |
| BSMAP (v2.7) | Xi and Li, 2009 | https://code.google.com/archive/p/bsmap/ |
| GSEA (v2.2.2) | Subramanian et al., 2005 | http://software.broadinstitute.org/gsea/index.jsp |
| GSEAPY (v0.9.3) | N/A | https://pypi.python.org/pypi/gseapy |
| GOstats (v2.24.0) | Falcon and Gentleman, 2007 | https://www.bioconductor.org/packages/release/bioc/html/GOstats.html |
| Cellranger (v2.1.0) | N/A | https://support.10xgenomics.com/ |
| CellrangerRkit (v1.1.0) | N/A | https://support.10xgenomics.com/ |
| R (v3.5) | N/A | https://www.r-project.org/ |
| Cytospace (v2.8) | N/A | http://www.cytoscape.org/ |
| ClueGO (v1.8) | Bindea et al., 2009 | http://apps.cytoscape.org/apps/cluego |
| HiC-Pro (v2.9.0) | (Servant et al., 2015) | https://github.com/nservant/HiC-Pro |
| HiCPlotter (v0.8.1) | (Akdemir and Chin, 2015) | https://github.com/kcakdemir/HiCPlotter |
| HiCExplorer (v2.1.4) | (Ramírez et al., 2018) | https://hicexplorer.readthedocs.io/en/latest/ |
| Qcapture Pro7 software | QICAM | https://www.qimaging.com/resources/pdfs/QCaptureProManual.pdf |
| Matlab | MathWorks | https://www.mathworks.com/products/matlab.html |
| FluoroSNNAP | N/A | http://www.seas.upenn.edu/~molneuro/fluorosnnap.html |
| Fiji | N/A | https://fiji.sc |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Clampfit 10 | Molecular Devices | |
| Other | ||
| U-bottom ultra-low-attachment 96-well plate | Corning | CLS7007–24EA |
| Ultra-low-attachment 6-well plate | Corning | CLS3471–24EA |
| Orbital shaker | IKA | KS260 |
| Nucleofector | Lonza | AAB-1001 |
Highlights.
MeCP2 mutation severely impairs human cortical interneuron
BRD4 dysregulation contributes to abnormal transcriptome in RTT interneurons
MeCP2 mutation causes cell-type-specific impairments in human brain organoids
BET inhibition rescues RTT-like phenotypes
Acknowledgements
We thank Dr. Andrew G. Elefanty for sharing HES-3 NKX2-1GFP/w human ES cell line, Dr. Stewart A. Anderson for sharing cortical interneuron differentiation protocol, and Dr. Guilin Wang for Chromium service. I.-H. P. was partly supported by NIH (GM111667-01, R01MH118344-01A1, R01MH118554-01A1, R01AA025080-01, R01CA203011-2), CSCRF (14-SCC-YALE-01, 16-RMB-YALE-04), Kavli Foundation, Simons Foundation, Nomis Foundation, and KRIBB/KRCF research initiative program (NAP-09-3). This work was supported by the College of Medicine, University of Arkansas for Medical Sciences to S-HL and Core Facilities of the Center for Translational Neuroscience, Award P30 GM110702 from the IDeA program at NIGMS. Computation time was provided by Yale University Biomedical High Performance Computing Center.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi M, Autry AE, Covington HE, and Monteggia LM (2009). MeCP2-Mediated Transcription Repression in the Basolateral Amygdala May Underlie Heightened Anxiety in a Mouse Model of Rett Syndrome. J Neurosci 29, 4218–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akdemir KC, and Chin L (2015). HiCPlotter integrates genomic data with interaction matrices. Genome Biol 16, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, and Zoghbi HY (1999). Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 23, 185–188. [DOI] [PubMed] [Google Scholar]
- Barkho BZ, Song H, Aimone JB, Smrt RD, Kuwabara T, Nakashima K, Gage FH, and Zhao X (2006). Identification of astrocyte-expressed factors that modulate neural stem/progenitor cell differentiation. Stem Cells Dev 15, 407–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaghzal H, Dekker J, and Gibcus JH (2017). Hi-C 2.0: An optimized Hi-C procedure for high-resolution genome-wide mapping of chromosome conformation. Methods 123, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito E, Ramachandran B, Schroeder H, Schmidt G, Urbanke H, Burkhardt S, Capece V, Dean C, and Fischer A (2017). The BET/BRD inhibitor JQ1 improves brain plasticity in WT and APP mice. Transl Psychiatry 7, e1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, and Galon J (2009). ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth MJ, Ost TW, Beraldi D, Bell NM, Branco MR, Reik W, and Balasubramanian S (2013). Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat Protoc 8, 1841–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro J, Garcia RI, Kwok S, Banerjee A, Petravicz J, Woodson J, Mellios N, Tropea D, and Sur M (2014). Functional recovery with recombinant human IGF1 treatment in a mouse model of Rett Syndrome. Proc Natl Acad Sci U S A 111, 9941–9946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao HT, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, Neul JL, Gong S, Lu HC, Heintz N, et al. (2010). Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature 468, 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah VA, Pillai RB, Shekar KV, Lane JB, Motil KJ, Skinner SA, Tarquinio DC, Glaze DG, McGwin G, Kaufmann WE, et al. (2014). Methyl-CpG-binding protein 2 (MECP2) mutation type is associated with disease severity in Rett syndrome. J Med Genet 51, 152–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Hayden Gephart MG, Barres BA, and Quake SR (2015). A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A 112, 7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Case-Borden C, Gegonne A, Hsu CH, Chen QR, Meerzaman D, Dey A, Ozato K, and Singer DS (2017). BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin (vol 23, pg 540, 2016). Nat Struct Mol Biol 24, 194–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert DH, Gabel HW, Robinson ND, Kastan NR, Hu LS, Cohen S, Navarro AJ, Lyst MJ, Ekiert R, Bird AP, et al. (2013). Activity-dependent phosphorylation of MeCP2 threonine 308 regulates interaction with NCoR. Nature 499, 341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon S, and Gentleman R (2007). Using GOstats to test gene lists for GO term association. Bioinformatics 23, 257–258. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. (2010). Selective inhibition of BET bromodomains. Nature 468, 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel HW, Kinde B, Stroud H, Gilbert CS, Harmin DA, Kastan NR, Hemberg M, Ebert DH, and Greenberg ME (2015). Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 522, 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavet O, and Pines J (2010). Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev Cell 18, 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogliotti RG, Senter RK, Fisher NM, Adams J, Zamorano R, Walker AG, Blobaum AL, Engers DW, Hopkins CR, Daniels JS, et al. (2017). mGlu7 potentiation rescues cognitive, social, and respiratory phenotypes in a mouse model of Rett syndrome. Sci Transl Med 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, and Meissner A (2011). Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc 6, 468–481. [DOI] [PubMed] [Google Scholar]
- Guy J, Gan J, Selfridge J, Cobb S, and Bird A (2007). Reversal of neurological defects in a mouse model of Rett syndrome. Science 315, 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin JE, and Bird A (2001). A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet 27, 322–326. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. (2007). Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39, 311–318. [DOI] [PubMed] [Google Scholar]
- Johnson BS, Zhao YT, Fasolino M, Lamonica JM, Kim YJ, Georgakilas G, Wood KH, Bu D, Cui Y, Goffin D, et al. (2017). Biotin tagging of MeCP2 in mice reveals contextual insights into the Rett syndrome transcriptome. Nat Med 23, 1203-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Hysolli E, and Park IH (2011). Neuronal maturation defect in induced pluripotent stem cells from patients with Rett syndrome. Proc Natl Acad Sci U S A 108, 14169–14174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Hysolli E, Tanaka Y, Wang B, Jung YW, Pan X, Weissman SM, and Park IH (2014). X Chromosome of female cells shows dynamic changes in status during human somatic cell reprogramming. Stem Cell Reports 2, 896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koops A, Kappler J, Junghans U, Kuhn G, Kresse H, and Müller HW (1996). Cultured astrocytes express biglycan, a chondroitin/dermatan sulfate proteoglycan supporting the survival of neocortical neurons. Brain Res Mol Brain Res 41, 65–73. [DOI] [PubMed] [Google Scholar]
- Korb E, Herre M, Zucker-Scharff I, Darnell RB, and Allis CD (2015). BET protein Brd4 activates transcription in neurons and BET inhibitor Jq1 blocks memory in mice. Nat Neurosci 18, 1464–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korb E, Herre M, Zucker-Scharff I, Gresack J, Allis CD, and Darnell RB (2017). Excess Translation of Epigenetic Regulators Contributes to Fragile X Syndrome and Is Alleviated by Brd4 Inhibition. Cell 170, 1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaraj R, Ho G, and Christodoulou J (2017). RettBASE: Rett syndrome database update. Hum Mutat 38, 922–931. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Subgroup GPDP (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang HY, Muffat J, Cheng AW, Orlando DA, Loven J, Kwok SM, Feldman DA, Bateup HS, Gao Q, et al. (2013). Global Transcriptional and Translational Repression in Human-Embryonic-Stem-Cell-Derived Rett Syndrome Neurons. Cell Stem Cell 13, 446–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyst MJ, and Bird A (2015). Rett syndrome: a complex disorder with simple roots. Nat Rev Genet 16, 261–275. [DOI] [PubMed] [Google Scholar]
- Lyst MJ, Ekiert R, Ebert DH, Merusi C, Nowak J, Selfridge J, Guy J, Kastan NR, Robinson ND, de Lima Alves F, et al. (2013). Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nat Neurosci 16, 898–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MCN, Carromeu C, Acab A, Yu D, Yeo GW, Mu YL, Chen G, Gage FH, and Muotri AR (2010). A Model for Neural Development and Treatment of Rett Syndrome Using Human Induced Pluripotent Stem Cells. Cell 143, 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroof AM, Keros S, Tyson JA, Ying SW, Ganat YM, Merkle FT, Liu B, Goulburn A, Stanley EG, Elefanty AG, et al. (2013). Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell 12, 559–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk MM, McKeown MR, Filippakopoulos P, Li Q, Ma L, Agno JE, Lemieux ME, Picaud S, Yu RN, Qi J, et al. (2012). Small-molecule inhibition of BRDT for male contraception. Cell 150, 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhoubad S, Bock C, de Boer AS, Kiskinis E, Meissner A, and Eggan K (2012). Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell 10, 595–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, and Heintz N (2012). MeCP2 Binds to 5hmC Enriched within Active Genes and Accessible Chromatin in the Nervous System. Cell 151, 1417–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Krencik R, Krenick R, Ullian EM, Ullian E, Tsai HH, Deneen B, Richardson WD, Barres BA, and Rowitch DH (2012). Astrocytes and disease: a neurodevelopmental perspective. Genes Dev 26, 891–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petilla Interneuron Nomenclature G, Ascoli GA, Alonso-Nanclares L, Anderson SA, Barrionuevo G, Benavides-Piccione R, Burkhalter A, Buzsaki G, Cauli B, Defelipe J, et al. (2008). Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat Rev Neurosci 9, 557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Bhardwaj V, Arrigoni L, Lam KC, Grüning BA, Villaveces J, Habermann B, Akhtar A, and Manke T (2018). High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun 9, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Boxer LD, Hrvatin S, Li E, Silberfeld A, Nagy MA, Griffith EC, Vierbuchen T, and Greenberg ME (2018). Characterization of human mosaic Rett syndrome brain tissue by single-nucleus RNA sequencing. Nat Neurosci 21, 1670–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaco RC, Mandel-Brehm C, Chao HT, Ward CS, Fyffe-Maricich SL, Ren J, Hyland K, Thaller C, Maricich SM, Humphreys P, et al. (2009). Loss of MeCP2 in aminergic neurons causes cell-autonomous defects in neurotransmitter synthesis and specific behavioral abnormalities. P Natl Acad Sci USA 106, 21966–21971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildge S, Bohrer C, Beck K, and Schachtrup C (2013). Isolation and culture of mouse cortical astrocytes. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant N, Varoquaux N, Lajoie BR, Viara E, Chen CJ, Vert JP, Heard E, Dekker J, and Barillot E (2015). HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol 16, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Kirwan P, and Livesey FJ (2012). Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc 7, 1836–1846. [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Kim J, Zhou L, Wengert E, Zhang L, Wu Z, Carromeu C, Muotri AR, Marchetto MC, Gage FH, et al. (2016). KCC2 rescues functional deficits in human neurons derived from patients with Rett syndrome. Proc Natl Acad Sci U S A 113, 751–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillotson R, Selfridge J, Koerner MV, Gadalla KKE, Guy J, De Sousa D, Hector RD, Cobb SR, and Bird A (2017). Radically truncated MeCP2 rescues Rett syndrome-like neurological defects. Nature 550, 398–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, and Salzberg SL (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, and Pachter L (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullai JW, Schaffer ME, Mullenbrock S, Sholder G, Kasif S, and Cooper GM (2007). Immediate-early and delayed primary response genes are distinct in function and genomic architecture. J Biol Chem 282, 23981–23995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, and Li W (2009). BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Tanaka Y, Cakir B, Patterson B, Kim KY, Sun P, Kang YJ, Zhong M, Liu X, Patra P, et al. (2019). hESC-Derived Thalamic Organoids Form Reciprocal Projections When Fused with Cortical Organoids. Cell Stem Cell 24, 487–497.e487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Tanaka Y, Patterson B, Kang YJ, Govindaiah G, Roselaar N, Cakir B, Kim KY, Lombroso AP, Hwang SM, et al. (2017). Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain Development and Interneuron Migration. Cell Stem Cell 21, 383–398 e387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cao SX, Sun P, He HY, Yang CH, Chen XJ, Shen CJ, Wang XD, Chen Z, Berg DK, et al. (2016). Loss of MeCP2 in cholinergic neurons causes part of RTT-like phenotypes via alpha7 receptor in hippocampus. Cell Res 26, 728–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, et al. (2006). Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron 52, 255–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Representative clones from MECP2 editing. (Related to Figure 1)
Table S2. Off target evaluation of selected clones. (Related to Figure 1)
Data Availability Statement
Single cell RNA-seq, bulk RNA-seq, ChIP-seq, ATAC-seq, and HiC data are available at GEO with accession number (GSE117513).