Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a positive-sense, single-stranded RNA virus that causes the potentially lethal Covid-19 respiratory tract infection. It does so by binding to host cell angiotensin converting enzyme 2 (ACE2) receptors, leading to endocytosis with the receptor, and subsequently using the host cell’s machinery to replicate copies of itself and invade new cells. The extent of the spread of infection in the body is dependent on the pattern of ACE2 expression and overreaction of the immune system. Additionally, by inducing an imbalance in the renin-angiotensin-aldosterone system (RAAS) and the loss of ACE2 would favour the progression of inflammatory and thrombotic processes in the lungs. No drug or vaccine has yet been approved to treat human coronaviruses. Hundreds of clinical trials on existing approved drugs from different classes acting on a multitude of targets in the virus life cycle are ongoing to examine potential effectiveness for the prevention and treatment of the infection. This review summarizes the SARS-CoV-2 virus life cycle in the host cell and provides a biological and pathological point of view for repurposed and experimental drugs for this novel coronavirus. The viral life cycle provides potential targets for drug therapy.
Keywords: Covid-19, SARS-CoV-2, ACE2, Ang (1-7), Remdesivir, Immunomodulators
Graphical abstract
Highlights
-
•
ACE2 is a cellular functional receptor for SARS-CoV-2.
-
•
Ang (1-7) counterbalances the adverse actions of Ang II on the heart and blood vessels.
-
•
The ss-positive sense RNA undergoes translation by using host cell machinery.
-
•
Cell attachment, translation, replication and inflammation are drug targets for Covid-19.
-
•
The RAAS inhibitors in the pathogenesis of Covid-19 is overly complex and controversial.
1. Introduction
The outbreak of severe acute respiratory syndrome (SARS) first occurred in 2003 and was largely contained within six months.[1] There was a large time gap before a second, closely-related coronavirus, Middle East respiratory syndrome coronavirus (MERS-CoV) appeared in 2012.[2] This frequently caused renal failure in addition to severe respiratory disease.[3] As of November 30, 2019, the World Health Organization (WHO) had registered 2494 MERS-CoV cases worldwide.[4] The first report of SARS-CoV-2, the seventh coronavirus known to infect humans, appeared in China in December 2019.[5] As of July 22, about 15 million SARS-CoV-2 related cases were reported worldwide with over 617,000 deaths (https://coronavirus.jhu.edu/map.html) surpassing the combined number of cases and deaths from two previous coronaviruses. The disease, Covid-19, is characterized by a range of symptoms, primarily including fever, cough, dyspnea, and myalgia.[6] In severe cases, the most common chest computed tomography finding is bilateral lung involvement with ground-glass opacity.[7] The primary cause of mortality from Covid-19 stems from compromised immune systems, leading to respiratory failure. Many more patients have exhibited cardiovascular related pathologies including congestive heart failure and brain medullary cardiorespiratory dysfunction and inflammatory thrombocytosis.[[8], [9], [10]]
Covid-19 killed more people on a weekly or monthly basis in early 2020 than any other cause of death except cardiovascular disease, which has still marginally maintained the number one spot.[11] The rapid spread of the virus causing Covid-19 has sparked fear worldwide and almost all countries are dealing with a surge in confirmed cases and deaths. The disease is increasingly affecting elderly people with underlying medical issues, although no one is invulnerable regardless of age, sex, or race.
Currently, there are no proven effective therapies for Covid-19. Within the first month of the Covid-19 global pandemic, there was a surge in clinical trials of repurposing of mostly approved drugs such as hydroxychloroquine (antimalarial drug), ritonavir and lopinavir (a combination of antiretroviral drugs), remdesivir (an experimental Ebola drug), ritonavir and lopinavir (antiretrovirals), interferon-beta, and several immunomodulators and anti-inflammatory drugs. In this context, the US Food and Drug Administration (FDA) issued an Emergency Use Authorization for the use of hydroxychloroquine (and chloroquine) and remdesivir on March 28 and May 1, 2020, respectively, for the treatment of hospitalized patients with Covid-19. Hundreds of clinical trials of potential Covid-19 related drugs have been launched globally, including the WHO’s large-scale SOLIDARITY trial. Additionally, several clinical trials to develop an effective vaccine to treat Covid-19 are underway.
The journey of SARS-CoV-2 begins as it enters the nasopharynx by attaching to angiotensin converting enzyme 2 (ACE2) receptor-enriched epithelial cells of the nasal and oral mucosa, precipitating a reduced sense of smell and taste as it passes down to the lungs, the primary site of infection. The subsequent events, including endocytosis, using the host cellular machinery for genome replication, transcription, assembly, and release of virus to other cells where it infiltrates organ systems from the brain to blood vessels to toes inflicting multiple organ damage [12] provide ample intervention sites to intercede in the progression of the disease.
This review presents the argument that drug targets should be identified at each of the multiple stages of the SARS-CoV-2 life cycle. Given the pathology of the disease, in which the virus attacks more than one organ and induces a dysregulated inflammatory response, an optimally successful strategy for identifying efficacious and safe drug therapy may require a concerted and integrated approach, matching drug mechanisms of action to the respective drug targets at each stage of the virus life cycle. Exploring the repurposing of existing drugs and developing new drugs are both of considerable interest. We discuss therapeutic strategies of drugs under active investigation based on the target site and mechanism of action, and the need for evaluation of promising approaches via multisite randomized, concurrently controlled clinical trials.
2. Structure of SARS-CoV-2
SARS-CoV-2, the spherical or pleomorphic Beta coronavirus,[13] possesses five distinct structures that include spike (S), membrane (M), envelope (E), nucleocapsid (N), and hemagglutinin-esterase dimer (HE) glycoproteins along with RNA as genetic material.[14,15]
Spike glycoprotein (S) projects in the form of specific spikes that hoop from the peripheral surface of the virus, resembling a crown (Latin meaning corona) under an electron microscope.[16] S proteins are known to assist in the entry of coronavirus into host cells. Further, for interaction with the host cell, S protein is cleaved by TMPRSS2 (cellular furin-like protease) into two distinct polypeptides, S1 and S2. S1 is known to attach to the cellular receptor ACE2, and S2 leads to the fusion of the viral and cellular membranes.
The Membrane protein (M) is the most abundant protein present in coronavirus. It possesses a triple-helical bilayer with a short-NH2 terminal domain at the extracellular region and a COOH terminal at the intracellular cytoplasmic region. The primary function of the protein is in the development of virus-specific humoral response. It acts as the most abundant structural protein that can neutralize antibodies developed in SARS patients. Furthermore, the transmembrane domain of the M protein may contain a T cell epitope cluster, which holds dominant cellular immunogenicity.[17]
The Envelope protein (E) is a transmembrane protein that functions as an ion channel, allowing human protease to release the viral genomic material to the host cell.[18] The protein, although not necessary for viral replication, allows pathogenesis via assembly and release of the virus. The Nucleocapsid protein (N) is a phosphorylated protein that enables the binding of viral RNA in vitro in a ‘bead on a string’ type conformation. The protein is also known to assist in encapsulation of genomic material into the virus particles by tethering the viral genome network of protein to replicase-transcriptase complex (RTC) machinery.[15,19]
Hemagglutinin-esterase dimer protein (HE) contains acetyl-esterase activity. It binds to sialic acids on the surface of the glycoprotein membrane, and assist coronavirus release from the infected cells after their hijack.[20,21]
Based on mutations, the virus is classified into S (~30%) and L lineage (~70%) types involving (8782C>T and 28144T>C) important co-mutations. During a study by Tang et al., it was revealed that only 3.7% of viral isolates were S type compared to 96.3 % of L type isolated from Wuhan, China. However, outside Wuhan, 61.3% was found to be L type and 38.4% S type.[22] Further, Nextstrain analysis on 2310 viral isolates disclosed that West Coast (United States) predominates with B1 clade, whereas A2a clade, was found to be predominant in East Coast including New York, Europe, and Italy. These clades further diverge in virulence. The non-synonymous mutation involving the spike protein of SARS-CoV-2 at codon 614 reveals presence of aspartic acid (D) at this residue is predominant on the West Coast, whereas glycine (G) is dominant on the East Coast thus involving D614G mutation in a highly glycosylated region of the viral spike protein.[23,24]
Genetically, coronaviruses share the largest genomes (26.4 - 31.7 kb) among all known RNA viruses with high guanine-cytosine content.[25] The nucleotide identification of SARS-CoV-2 revealed 80, 55, and 50% homology to SARS-CoV-1, MERS, and common cold CoV, respectively. SARS-CoV-2 contains a positive-sense (single-stranded) RNA genome of 30 kb and fourteen open reading frames (ORFs) underlying various conserved genes (ORF1ab, S, E, M, and N protein region)[13]. ORF1a and ORF1ab produce two essential polypeptides, pp1a and pp1ab, encoding for S, M, E, and N proteins (encoded at the 3’ end) and being vital for virus protein integrity. The amino acid sequences of the ORF1ab domain further reveal a 94.4% sequence identity with SARS-CoV.[26] The pp1ab polypeptide encodes for essential non-structural proteins required for the formation of the sophisticated replicase machinery Nsp1-16. The polypeptides are also processed by viral enzymes (chymotrypsin-like protease (3CLpro) and main protease (Mpro)) to yield non-structural proteins (Nsp1-16) which encode for endoribonuclease activity, hence playing a vital role in viral replication and transcription. [[27], [28], [29]] The presence of the receptor-binding domain (on the spike protein) and a polybasic cleavage site confer that the protein is naturally evolved and removes the possibility of the genome being constructed or manipulated in the laboratory.[30]
3. SARS-CoV-2 entry and initiation
The isolation of RaTG13, the closest source for SARS-CoV-2, indicates bats as the source point of origin of Covid-19. Additionally, research demonstrated the 80% genetic homology of SARS-CoV-2 with SARS-CoV. During the pandemic of SARS-CoV, it was demonstrated that this coronavirus consists of a receptor-binding domain in its S protein that allows receptor recognition by ACE2. Similar experiments conducted by Zhou et al. revealed that SARS-CoV-2 utilizes ACE2 receptors for its entry into host cells.[31] The group determined the virus’ infectivity using HeLa cells. The cells were obtained from humans, Chinese horseshoe bats, civets, pigs, and mice and differentiated into ACE2 (transfected) protein-expressing and non-expressing (non-transfected) types. The results revealed that the novel coronavirus strain invaded all the cells (except those from mice) expressing ACE2, whereas cells not expressing ACE2 were not affected. The study also revealed that aminopeptidase N (APN) and dipeptidyl peptidase 4 (DPP4) are not involved in virus interaction with host cells in the case of SARS-CoV-2.[31] Furthermore, the evidence of SARS-CoV-2 entry through host cells via ACE2 was independently provided by Walls et al.[15], Hoffmann et al.[32], and Verdecchia et al.[33] Further research indicated that the S proteins in all three coronaviruses belong to class 1 viral fusion protein, which indicates that the S protein needs to undergo cleavage by a fusion protein to activate its fusion potential. Proteases including furin, cathepsins, trypsin, human airway trypsin-like proteases, and transmembrane protease serine protease (2/4) (TMPRSS) have been identified that allow activation of S protein by proteolytic cleavage into subunits S1 and S2.[[34], [35], [36], [37]] Experiments conducted by Ou et al., tested various proteases that might be responsible for S protein cleavage. The experiments were performed using HEK293T cells expressing SARS-CoV-2 S protein and control HEK293 cells expressing hACE2 in the presence and absence of proteases. The results revealed that co-expression of TMPRSS on hACE2 expressing HEK293 cells enhanced cell-cell fusion mediated by S protein. The addition of trypsin as a protease leads to the formation of large syncytia in these cells (a fusion of the cellular surface without endocytosis), suggesting proteins could be triggered without exogenous protease priming upon the receptor binding. The study indicated that among all the serine proteases, TMPRSS2 is vital for S protein activation, and ACE2 acts as an entry receptor for SARS-CoV-2.[38] The study by Hoffmann et al. corroborated these findings and suggested a role of TMPRSS2 in proteolytic cleavage of S protein to S1 and S2 subunits.[32]
3.1. S protein cleavage
The role of S protein in CoV fusion has been well-documented in previous outbreaks.[39] The fusion of S protein with the human cell receptor allows the entry of viral genetic material to host cells, causing the invasive infection. In the previous outbreaks, it was found that CoV uses a furin activation site (FAS) to bind to the host receptor.[35] The furin and furin-like proteases are considered to be a group of proprotein convertases (type I transmembrane protein) that allow cleavage of S protein at two distinct sites, resulting in the formation of two subunits - S1 (globular domain) and S2 (biomembrane-anchored stalk domain).[31,40] Cleavage is essential for ensuring the acquaintance of the S2 site to host proteases. S1 segment allows attachment of CoV to ACE2, whereas S2 enables the fusion of CoV inside the host cell.[41,42] It is now known that apart from furin or furin-like proteases, SARS-CoV-2 uses TMPRSS2 for its interaction with the ACE2 receptor and entry into the host cell.[32,43] TMPRSS2 is a type II transmembrane serine protease expressed widely in epithelial cells forming respiratory, gastrointestinal, and urogenital tracts.[44] The S1/S2 region is known to contain multiple arginine or lysine residues (R/K), and, hence, activates the virus leading to the cleavage of S protein by TMPRSS2 at these monobasic cleavage sites.[45]
The sequence analysis studies suggest that in SARS-CoV-2, furin cleaves at (R-R-A-R685↓), which contain an insertion of four amino acids, including an alanine residue (non-basic) in position P2, recognized as a furin cleavage site.[45] Besides this, a paired dibasic motif with a single KR (Lysine-Arginine) segment (KR815↓) is recognized by trypsin-like serine proteases. This additional feature makes the new CoV much different in cell entry than SARS, and probably affects the stability of the virus and, consequently, the transmission process. Furthermore, it was demonstrated that both furin and TMPRSS2 do not compensate for each other and inhibiting either of them may halt virus binding to host cells and their invasion. The role of furin and TMPRSS2 has been elucidated recently by many experimental pieces of evidence signifying the importance of these two proteins in proteolytic activation and blocking by inhibiting these targets.[46] (Fig. 1 )
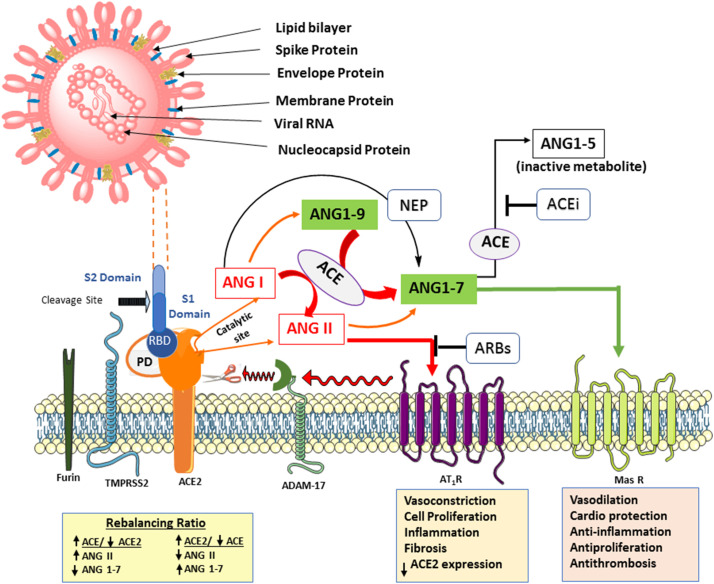
Fig. 1.
SARS-COV-2 virus binding ACE2 and the renin-angiotensin system axis.
Top left shows the virus structure identifying protruding surface spike proteins (enlarged to show S1 and S2 segments with the Receptor Binding Domain (RBD)), the most abundant Membrane protein, the Envelope protein that forms a simple multimeric ion channel, and the Nucleocapsid protein with the bound positive sense single strand (ss) RNA. Below: Furin pre-cleaves spike proteins at the S1/S2 site and promotes subsequent TMPRSS2-dependent entry into host cells. The RBD in the S1 segment binds with high affinity the protease domain (PD) in ACE2. Subsequent conformational changes in S2 facilitate fusion between the viral envelope and the host cell membrane, and internalization by endocytosis with ACE2. Ang II-AT-1 receptor activation of ADAM17 cleaves/sheds cell surface ACE2 which can no longer cleave Ang II or Ang I to protective peptides (Ang (1-9) and Ang (1-7)), resulting in myocardial dysfunction. This action is blocked by AT-1 receptor blockers upregulating ACE2 levels. The AT-1 receptor-mediated effects are opposite to that of Mas and MrgD (not shown) receptors. The catalytic subunit of ACE2 is independent of PD and may operate as long as ACE2 is not internalized. The protective axis comprising of recombinant ACE2, Ang (1–7), Mas receptor agonists, and RAS inhibitors enhance ACE2 action and serve as potential therapies in Covid-19. In addition, ACE inhibitors prevent the metabolism of Ang (1-7) by ACE to the inactive metabolite Ang (1-5) (see Table 1).
3.2. Binding of S protein with ACE2 receptors and fusion with the host cell membrane
The interaction between the viral S protein and ACE2 on the host cell surface is a critical step soon after the virus is primed by TMPRSS2 as this initiates the infection process. ACE2 is a cellular functional receptor for coronavirus.[47] Different coronaviruses bind different receptors while ACE2 is highly specific (100%) for SARS-CoV-2 as it binds with high affinity (KD, 1 to 30 nM), about 20 times higher than does SARS-CoV.[48] Chan et al[18] hypothesized that such high specificity and affinity toward ACE2 may contribute to the apparent ease with which widespread transmission and contagiousness of SARS-CoV-2 occur from human to human. ACE2 is widely distributed throughout the body, starting with nasal epithelium, oral mucosa, brain, lungs, heart, vascular endothelium, kidney, small intestine, colon, and testis.[49,50] The resultant widespread infection that begins with acute respiratory distress syndrome, followed by high levels of circulating cytokines, is a plausible cause of myocardial injury, multiorgan failure, and mortality.[51] A study suggests that as many as 32 variants of ACE2 are characterized in different populations. There is a possibility that some people could be less susceptible to SARS-CoV-2 infection than others.[52] Additionally, it is reported that men have a higher ACE2 expression in the lungs than women and Asian people express ACE2 higher than Caucasian and African American populations.[53]
ACE2 is a non-specific (as it also hydrolyzes apelin, des-arginine bradykinin [to bradykinin], and dynorphin) metallopeptidase, carboxypeptidase enzyme, and a homolog of ACE (with 40% similarity) first discovered two decades ago in the heart.[54] Functionally, there are two forms of ACE2 - (a) the full-length ACE2 containing structural transmembrane domain which anchors its extracellular domain to the plasma membrane, and (b) the soluble form lacking the membrane anchor, which circulates in undetectable quantity in the blood.[55,56] Both forms of ACE2 comprise extracellularly the NH2-terminal peptide or protease domain and the catalytic site (Box 1 ).[57,58] The receptor-binding domain of the S1 segment from SARS-CoV-2 binds the extracellular N-terminus, the protease domain of the ACE2. Subsequent conformational changes in S2 (stalk domain) facilitate fusion between the viral envelope and the host cell membrane and internalization by endocytosis with ACE2.[32,39] This action results in the loss of ACE2 (downregulation as a result of the negative feedback loop) at the cell surface that removes an important pathway for the cell to degrade Ang I and Ang II to generate cardioprotective peptides (Fig. 1). An increase in the ratio of Ang II to Ang (1-7) following the removal of ACE2 contributes to the extent of pulmonary tissue damage, loss of its function, and generation of a cytokine storm contributing to widespread inflammation observed with Covid-19. The impact of the virus on the CVS, CNS, kidney, and other organs apart from the lungs is currently not known. Protease inhibitors block the protease domain, preventing binding to ACE2. Spike vaccine is purported to inhibit this initial step of cell entry.[59]
Box 1. ACE2, the functional receptor of SARS-CoV-2.
-
•
Angiotensin converting enzyme2 (ACE2), a monocarboxypeptidase enzyme, was discovered in 2000. It shares 40% homology and 61% similarity with the classic ACE.
-
•
ACE2 is widely expressed in most of the organs, beginning with nose and throat and from head to the gut and testicles. ACE2 converts decapeptide Ang I and octapeptide Ang II to nonapeptide, Ang (1-9), and heptapeptide, Ang (1-7), respectively.[58]
-
•
ACE2 actions are non-specific as it also cleaves and inactivates bioactive apelin peptides apelin-13 and apelin-36 through a negative feedback mechanism in the heart and vasculature[56] and also des-arginine bradykinin to bradykinin. Additionally, it is an amino acid transporter.
-
•
ACE2/Ang 1–7/MasR axis is a physiological antagonist that counter-regulates the activated RAAS. Thus, ACE2 is a critical protective pathway against heart failure, myocardial infarction and hypertension, and against lung disease and diabetes mellitus.[56]
-
•
There are two forms of ACE2, the full-length ACE2 containing structural domain anchors its extracellular domain to the cell membrane, and the soluble form lacking the membrane anchor circulates in undetectable quantity in the blood. [53,163] The both forms of ACE2 comprise the N-terminal protease domain with which the spike glycoprotein (S1) of SARS-CoV-2 binds. The catalytic site which is distinct from the protease domain catalyses Ang I and Ang II (Fig 1). [55,56]
-
•
Following binding by SARS-CoV-2, ACE2 is endocytosed. The loss of ACE2 function results in elevated levels of circulating Ang II.
-
•
ACE2 expression is increased by RAAS inhibitors and statins.
Alt-text: Box 1
The catalytic site of ACE2 removes the amino acid phenylalanine from octapeptide Ang II, the principal pressor hormone of the RAAS, to form Ang (1-7).[60] The latter also forms from Ang (1-9), a cleaved product of Ang I by ACE2, and by neutral endopeptidase (NEP). The pharmacologic effects of Ang (1-7) are opposite to that of Ang II, and counterbalance the adverse actions of Ang II on the heart and blood vessels. The spike protein from SARS-CoV-2 does not attenuate the hydrolysis of Ang II to Ang (1–7) by ACE2, suggesting that it may be unlikely that Ang II or other peptide substrates would directly interfere with SARS-CoV-2 binding and internalization.[61] This also suggests that Ang II might bind ACE2 as long as it is not internalized by the virus as their binding/catalyzing sites are different in recognizing their substrates (Fig. 1). However, it is not clear when the spike protein binds ACE2 whether it reduces the catalytic properties of ACE2.
The physiological actions of Ang II are mediated through AT-1 and AT-2 receptors while that of Ang (1-7) and its cleaved product, alamandine, are mediated through the Mas oncogene and Mas-related G-protein-coupled D (MrgD) receptors. Both are G protein-coupled receptors. Ang (1-7) is a biased agonist of the AT-1 receptor with a protective action in cardiac hypertrophy (Box 2 ).[62,63] Taken together, the RAAS system is comprised of two axes: ACE–Ang II–AT-1 receptor is the predominant axis that promotes vasoconstriction, proliferation, fibrosis, oxidative stress, thrombosis, and inflammation; and the ACE2–Ang (1–7)–Mas receptor axis counterbalances the effects of Ang II as Ang (1–7) has natriuretic, endothelial protective, cardioprotective, vasodilatory, anti-inflammatory, anti-thrombosis, and anti-proliferative properties (Fig. 1). [60]
Box 2. Cardioprotective Angiotensin (1-7).
-
•
Ang (1-7) is a major component of the protective arm of the Renin-Angiotensin-Aldosterone System, ACE2–Ang (1–7)–Mas R axis, considered as an ‘angel’ in the RAAS cascade.
-
•
Ang II has a high affinity for ACE2 to form Ang (1-7), which is also being formed by the action of ACE on Ang (1-9) and by the action of NEP on Ang I (see Fig. 1).
-
•
The physiological functions of Ang (1-7) are mediated through the Mas and MrgD receptors; both are G protein-coupled receptors distributed in numerous cardiovascular-related tissues. Ang (1-7) is also a biased agonist of the AT-1 receptor with a protective action in cardiac hypertrophy.[60]
-
•
Ang (1–7) is reported to produce effects opposite to that of Ang II, the major pressor hormone of the system, and it counterbalances the adverse actions of Ang II on the heart, kidney, and blood vessels.
-
•
The renal actions of Ang (1-7) are diuresis and natriuresis as it increases renal blood flow.[164]
-
•
AT-1 receptor blockade potentiates the vasodilatory effects of Ang (1-7) by increasing its availability at the target site through increased ACE2 expression.
-
•
Ang (1-7) promotes vasodilation (restores bioavailable nitric oxide), decreases inflammation, and fibrosis. It is shown to attenuate lung injury in an acute respiratory distress syndrome animal model. Lacking Ang (1-7) may be of negative effect on lung health.[165]
-
•
Nearly a dozen clinical trials are ongoing investigating cardiovascular effects and, more recently on ARDS in Covid-19 patients (see Table 1).
Alt-text: Box 2
ACE2 is the principal control component of the protective axis as it generates those key cardioprotective and hypotensive peptides.[64] It is upregulated in failing hearts. However, in hosting SARS-CoV-2, ACE2 is now playing a pathogenic role in easing virus infection, thus limiting its protective effect in lung and heart. ACE2 internalization worsens the pulmonary and cardiac damage initially triggered by SARS-CoV-2. On the other hand, drugs that can upregulate the expression of ACE2, such as ARBs and ACE inhibitors that are extensively used in hypertensive patients and statins in hyperlipidemic patients infected with SARS-CoV-2 have strengthened activity of the protective axis relative to the ACE–Ang II–AT-1 R axis, and these patients are unlikely to be at greater risk. [9,57,65,66]
3.3. Translation and RNA replication
Once the viral RNA is released into the host cell, the ss-positive sense RNA acts as messenger RNA (mRNA) and undergoes translation by using host cell machinery.[67] The series of events also leads to the formation of enzymes involved in viral replication, which generate and assemble new viral particles. The sequence of steps is illustrated in Fig 2 . The critical pathways utilized by SARS-CoV-2 include a) RNA Synthesis, b) proofreading of template, and c) capping.[68] The RNA machinery involves Nsp1-16 complex,[68,69] among which Nsp12 (RNA dependent RNA polymerase; RdRp)[70,71], Nsp13 (zinc-binding helicase; HEL)[72], Nsp14-16 complex (mRNA capping)[73], Nsp14 (RNA proofreading)[70,74], Nsp15 (uridylate-specific endoribonuclease activity (NendoU))[29], and Nsp7–Nsp10 (non-structural proteins) are critical for virus.[74] All the machinery system is present in host cell endoplasmic reticulum in complex with the transcription enzyme of SARS-CoV-2.[75] This leads to the generation of new genome molecules including sub-genomic (sg) messenger RNAs.[27]
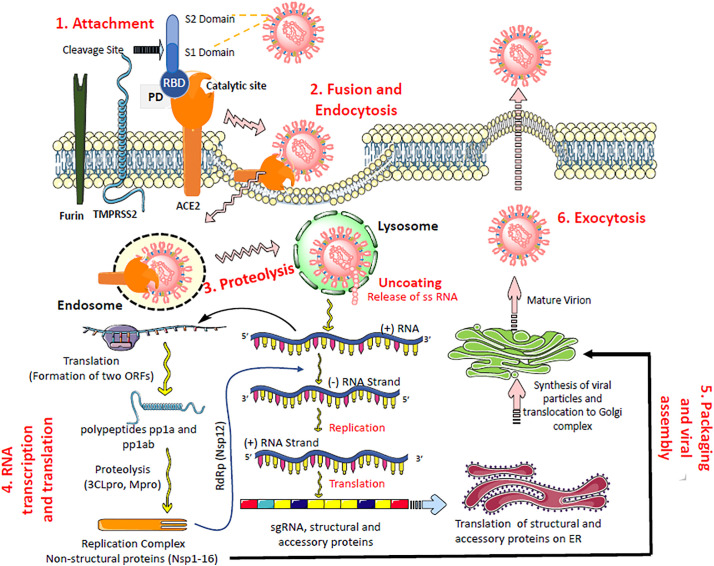
Fig. 2.
SARS-CoV-2 life cycle showing binding, membrane fusion, translation/replication, and virion release.
Following the receptor binding (1), conformational changes in S2 (stalk domain) facilitate fusion between the viral envelope and the host cell membrane, and internalization by endocytosis with ACE2 (2). The ss-positive sense RNA is released inside the cell. (3). Using the host cell protein translation machinery, the viral RNA is translated into viral polyproteins, proteases 3CLpro and PLpro, and the RNA-dependent RNA polymerase (RdRp) (4). Proteases process the polypeptide translation product from the gRNA into nonstructural proteins for the viral replication and packaging of a new generation of viruses. RdRp synthesizes a full-length negative-strand RNA template to be used by the RdRp to make more viral genomic RNA and structural proteins leading to completion of the assembly (5). The viruses that mature at the endoplasmic reticulum or Golgi apparatus get released via exocytosis (6) to infect other cells.
The RNA polymerization is catalyzed by Nsp12, in association with cofactors Nsp7-8. The N-terminal domain of Nsp12 (also referred to as Nidovirus RdRp-Associated Nucleotidyltransferase or NiRAN) is considered to be conserved in the majority of coronaviruses and possesses nucleotidylation activity.[76] The domain consists of α and β subunits (8 α helices and 5 stranded β-sheet). The β subunit possesses Serine/Threonine kinase PRP4 and Tyrosine-protein kinase JAK1 homologs that interact with the palm subdomain of the RdRp or Nsp12 and promotes viral growth. The C-terminal domain of Nsp12 is known to possess catalytic aspartic acid residues (Asp618, Asp623 Asp760, and Asp761) and are involved in recognition of nucleoside triphosphate and other divalent cation(s)(Mg2+) to perform the catalytic role (initiation and elongation) in viral growth.[71]
Furthermore, RNA proofreading and mRNA capping occur in conjunction with the bifunctional protein Nsp14. It contains two domains; the N terminal domain possesses exoribonuclease activity (DEDD exonucleases family) involved in proofreading, whereas the C terminal domain has (N7 guanine)-methyl transferase activity involved with viral mRNA cap synthesis. The cap consists of an N7-methylated GTP molecule linked 5′–5′ triphosphate bond transcribed by the first nucleotide.[70,74] The capping machinery apart from Nsp14 consists of Nsp13, 16, and cofactor Nsp10.[77,78] The capping is considered vital for mRNA stability and evading host immune response. The mRNA capping is initiated with multifunction Nsp13, which plays the role of helicase (nucleoside triphosphate enzymes (NTPs)) and plays a crucial role in virus replication. The Nsp13 is involved with the hydrolysis of NTPs, unwinding of RNA duplexes with 5'-3' directionality along with RNA 5'-triphosphatase activity.[72]
Finally, the termination of mRNA capping is done by Nsp16 in complex with Nsp10. Nsp16 is known to belong to a class of O-methyl Transferase characterized by reversed β hairpin at the carboxyl end. Finally, the processes catalyzed by Nsp13 (helicase), Nsp14 (GTPase), Nsp15 (N7-Methyl Transferase), and Nsp16 (2’-O- Methyl Transferase) activities leads to the synthesis of a nascent RNA strand possessing cap1 structure at its 5′ end terminal.[74,79] The host endoplasmic reticulum and Golgi apparatus further facilitate the assembly of viral RNAs and associated proteins into virions (SARS-CoV-2). [80] The M and E proteins interact together to form virus-like particles, whereas the N protein enhances their formation in the endoplasmic reticulum and Golgi apparatus. The S protein, however unimportant, is also incorporated into the new virions.
3.4. Virion release
The virus release from an infected cell primarily occurs via budding, exocytosis, or cell death. Mechanistically, for undeveloped virion particles, budding occurs. N protein interacts with the host cell membrane region where the viral E protein (glycosylated) has been inserted. Importantly, the N protein allows a proper orientation for budding of virion at the plasma membrane, endosomal, nuclear, or perinuclear membranes leading to the release of virus particles. The viruses that become matured at the endoplasmic reticulum or Golgi apparatus are released via exocytosis. In many cases, the virus attack destroys the host cell machinery leading to the release of lysosomes: this disruption of cell integrity results in cell death and consequent release of virus particles.[80,81] Finally, these progeny virions get released out of the host cell in vesicles by exocytosis. These newly released virions may either infect other healthy cells or shed into the environment via the opening of the mouth, breathing, or similar occurrences that may infect healthy persons via reported modes of transmission.[77,78]
4. Drug targets
At present, there are no drugs approved for the treatment of patients with Covid-19.[82] The pandemic that was set in the early months of 2020 worldwide has triggered an unprecedented stampede into hundreds of clinical trials evaluating both approved or licensed drugs and investigational agents.[83] All of these clinical trials are based on keeping in mind the viral life cycle, from the host cell attachment and entry to translation, replication and release, and the subsequent damage resulting from the compromised immune system.[84] Thus, there are multiple targets to consider for potential therapeutic intervention. Covid-19 is more than a lung infection as imaging and pathological investigations show that the syndrome is a thrombo-inflammatory process that initially affects lung perfusion, but consecutively affects all organs of the body.[85] Thus, a combination of drugs encompassing more than one target could potentially yield better results.[86]
The development of new antiviral drugs is time-consuming. To speed up the process, repurposing of existing drugs can potentially be an efficient strategy for drug development.[87] This has led to the quest for new antiviral molecules and, importantly, newer targets for therapeutic development.[88,89] Some successful examples of drug repurposing includes, Zidovudine (from cancer to HIV AIDS), Minoxidil (from hypertension to hair loss), Sildenafil (from angina to erectile dysfunction), Thalidomide (from morning sickness to leprosy and multiple myeloma), Ketoconazole (from fungal infections to Cushing syndrome), Aspirin (from analgesia to colorectal cancer), Paromomycin (from antibiotic in acute and chronic intestinal amebiasis to visceral and cutaneous leishmaniasis),[90] Doxycycline (from broad-spectrum bacteriostatic to antimalarial agent)[91], Miltefosine (from breast cancer to visceral leishmaniasis)[92], Allopurinol (antineoplastic to gout), and Bromocriptine (from initial use in Parkinson’s disease to Type 2 diabetes).[93]
The relevant repurposing data in SARS-CoV-2 is led by computational approaches and prior knowledge of previous pandemics. Computational methods, including protein-protein interaction models, enrich evaluation of the target profile of small molecules, allowing the possibility of already approved drugs being found useful.[88,89] In this context, Joshi and Poduri [94] have screened 8548 ligands (approved, investigational, and experimental drugs) on endoribonuclease NendoU (Nsp-15) via virtual screening and molecular modeling. The study showed eprosartan (AT-1 receptor blocker approved for the treatment of hypertension), investigational drugs DB15063 (Inarigivir soproxil), and DB12307 (Foretinib), and an experimental drug DB01813 are promising drugs for further evaluation in the treatment of Covid-19.[94]
Drugs under consideration that could have activity for Covid-19, such as antivirals, protease inhibitors, antibiotics, statins, RAAS inhibitors, corticosteroids, antirheumatics, and antineoplastic, are listed in Table 1 . Various classes of drugs including anti-inflammatory, immune modulators, and anesthetics are being evaluated preclinically or into clinical trials to assess their possible repurposing for Covid-19.[95,96] Therefore, drug repurposing is a potentially fast and reasonable strategy to discover new treatments from existing drugs, and may reduce the cost compared to de novo drug discovery.[97] However, for repurposed drugs, a benefit-risk profile in clinical trials may fail for any new indication. Other aspects such as selecting appropriate doses that affect the dose-response relationship may also be taken into consideration.[98][99] Therefore, newer development using repurposed drugs will depend not only on regulatory evidence of efficacy, safety, and quality but also on comparative cost-effectiveness and comparative clinical efficacy. [98,100,101]
Table 1.
Therapeutic strategies of drugs under active investigation based on the target site and mechanism of action for Covid-19.
| Drug | Class (Pharmacology/Chemical) | MOA | Clinical trial | Reference |
|---|---|---|---|---|
| 1. Drug interfering with the fusion of the virus with host cells | ||||
| Apilimod | Interleukin inhibitor | Inhibits lipid kinase PIKfyve (phosphoinositide kinase for position 5 containing a FYVE finger domain) in cellular regulation | Phase II | [165] |
| Camostat | Serine protease inhibitor | Blocks mechanism of SARS-CoV-2 which uses TMPRSS2 for docking to the ACE2. This hampers the binding of the spike protein of the virus to the host cell | Phase 1 | NCT:04321096[166,167] |
| Nafamostat | Serine protease inhibitor | Phase II | NCT:04321096[32,104] | |
| Tranexamic acid | Antifibrinolytic hemostatic | Inhibits activation of plasminogen thereby reducing the conversion of plasminogen to plasmin (fibrinolysin), an enzyme that degrades fibrin clots. Plasmin acts on coronavirus and cleaves the newly inserted furin site at the S protein portion. This results in increased virulence and infectivity. | Phase II | NCT:04338126[168] |
| Umifenovir | Antiviral | The antiviral properties are thought to occur due to interactions with aromatic residues within the viral glycoproteins involved in S protein /ACE2 membrane fusion and cellular recognition. | Phase IV | NCT:04260594[169] |
| 2. Drugs targeting RAAS-ACE2-AT-1 receptor | ||||
| rhACE2 (APN01) | Recombinant enzyme | The biologic is known to possess a dual mode of action. Firstly, it mimics human enzyme ACE2 misleading viruses to bind to a pseudo receptor; secondly, it reduces inflammatory reaction during infection, thus protecting against lung infection. | Phase II | NCT:04335136;04287686 (withdrawn) [[170], [171], [172], [173]], |
| Ang (1-7) | Cardioprotective angiotensin peptide | ACE2-Ang (1-7)-Mas receptor axis. It limits several detrimental effects of Ang II on AT-1 receptors by triggering counter-regulatory protective effects through binding to G protein-coupled Mas receptors. It is being explored clinically, for example, for stimulating hematopoietic recovery in patients with cancer after chemotherapy and for examining cardiovascular effects. |
Phase II/III | NCT:04332666[171,174,175] |
| Captopril Trandolapril Perindopril Fosinopril Benazepril Quinapril Ramipril |
ACE inhibitor |
Currently, these drugs are approved for the treatment of hypertension. By preventing the cleavage of Ang I to Ang II, Ang I levels are increased resulting in the increased formation of Ang (1-9) by ACE2 and subsequently Ang (1-7). Nonclinical studies have demonstrated that these drugs upregulate ACE2 expression by decreasing the availability of Ang II at the AT-1 receptor and subsequently masking the inhibitory activity of ADAM17 on ACE2 | Phase II | NCT:04355429[176] |
| Phase IV | NCT:04330300[177,178] | |||
| Phase II | NCT:04366050[179] | |||
| Losartan Valsartan Telmisartan |
AT-1 receptor blocker | All of the ARBs are approved for the treatment of hypertension. They reversibly and competitively inhibit Ang II binding to the AT-1 receptor. This action prevents Ang II-AT-1 receptor-mediated cytokine release. Preclinical studies have demonstrated that blockade of AT-1 receptors increases ACE2 expression by inhibiting ADAM17 that cleaves ACE2. Losartan is an inverse agonist. The PPAR partial agonist activity of telmisartan also contributes to increased ACE2 expression. | Phase II | NCT:04312009; NCT:04311177[[180], [181], [182]] |
| Phase IV | NCT:04335786[[180], [181], [182]] | |||
| Phase II | NCT:04355936[183] | |||
| Simvastatin (In combination with Ruxolitinib) |
Anti-hyperlipidemic | The drug is known to inhibit virus multiplication in early stages, decreases virus-induced cytotoxicity in affected host cells, along with inhibition of Rab/RhoA GTPase activity and LC3 membrane localization known to enhance the replication process in viruses. It also affects cytokine storm by decreasing pro-inflammatory cytokines (TNF-α, IL-6, INF-γ) released during the viral infection. | Phase II | NCT:04348695[184,185] |
| Atorvastatin | Anti-hyperlipidemic | The drug is known to inhibit virus multiplication in the early stages, along with decreasing the virus titer in infected cells, thus increasing host cell viability. | Phase II | NCT: 04380402 [184,186] |
| 3. Drug interfering with translocation of the virus within host cells | ||||
| Chloroquine phosphate HCQ |
Antimalarial | Approved or the treatment of rheumatoid arthritis and systemic lupus erythematosus. Targets endocytosis and endosomes of host cells. Disrupts intracellular trafficking and viral fusion events by increasing the pH in vacuoles. Additionally, the virus requires acidic pH for proper Golgi apparatus functioning and transport. It inhibits glycosylation of ACE2 receptors. Additionally, immunomodulatory effects through inhibition of cytokine production, autophagy, and lysosomal activity in host cells are reported. | Phase II | NCT:04303507[187] |
| Hydroxychloroquine sulfate and azithromycin | Antimalarial and antibiotic | HCQ Same mechanism as chloroquine; Azithromycin binds to 50s subunit of the ribosome and interferes with translation and prevents the expression of the replicase gene. | Phase II | NCT:04370782[188] |
| 4. Drugs interfering with proteolysis mechanism of the virus within host cells | ||||
| Danoprevir (in combination with ritonavir) | Antiviral | Interferes with the protease of virus and inhibits the release of genomic material from the virus to host cell. | Phase II | NCT: 04291729[38] |
| Ritonavir + Lopinavir | Antiretroviral | Ritonavir and Lopinavir decrease the viral load by inhibiting viral protease 3CLpro, target nucleocapsid during virion assembly, and also, target tumor necrosis factor receptor type 6. Ribavirin inhibits viral RNA-dependent RNA polymerase but is less potent than remdesivir. Interferon strengthens the innate immune system. | Phase II | NCT:04321174[131,189] |
| Ritonavir/Lopinavir and interferon-β1b | Antiviral | Phase II | NCT:02845843[131,189] | |
| Ribavirin + Ritonavir + Lopinavir | Antiviral | Phase II | NCT:04276688[190] | |
| Darunavir (with Cobicistat) | Antiretroviral | Prevents HIV replication through binding to the viral protease, inhibits cleavage of encoded viral protein Gag-Pol. Also, it targets nucleocapsid during virion assembly. Cobicistat (a CYP3A inhibitor) enhances the activity of darunavir. | Phase III | NCT:04252274[191] |
| Carmofur (1-hexylcarbamoyl-5-fluorouracil) | Anticancer agent | In vitro studies have shown carmofur inhibits potently the main protease Mpro activity. | None | [134] |
| 5. Drugs interfering with replication, transcription, and translation of virus genomic material | ||||
| Favipiravir | Antiviral | Inhibition of viral coded enzyme RNA-dependent RNA polymerase would block the formation of a negative-sense RNA strand during viral replication within the host cell. | Phase III | NCT:04336904[192] |
| Galidesivir (BCX4430) | Antiviral | Phase II | NCT:03891420[193] | |
| Remdesivir | Antiviral | Phase III | NCT:04292730[194] | |
| Levovir | Antiviral | Phase II | NCT:04347915[195] | |
| Emtricitabine + Tenofovir | Non-nucleoside reverse transcriptase inhibitor + Nucleotide reverse transcriptase inhibitor | This drug class is widely recommended as first-line treatment for HIV when co-administered with two nucleoside or nucleotide reverse transcriptase inhibitors. These drugs compete with deoxycytidine 5'-triphosphate for reverse transcriptase enzyme. | Phase III | NCT:00458393[196] |
| Ciclesonide | Glucocorticoid | It interacts with viral non-structural protein 15, either directly or indirectly leading to inhibition of viral replication of coronavirus. | Phase II | NCT:04330586[197,198] |
| Nitazoxanide | Broad-spectrum antiviral agent, anthelminthic | Approved for the treatment of diarrhea. It suppresses viral replication by inhibition of viral hemagglutinin maturation achieved by inhibition of viral transcription factor immediate-early 2 and activating eukaryotic translation initiation factor 2α (an antiviral intracellular protein). It is purported to induce the host immune response to produce interferons by the host’s fibroblasts. | Phase IV | NCT:04341493[170] |
| Selinexor | Anticancer | It is a selective inhibitor of nuclear export, blocks cellular protein XPO1. | Phase II | NCT:04349098[199] |
| Ivermectin | anti-parasite | Inhibit viral replication by inhibiting IMPα/β1-mediated nuclear import of viral proteins. | Alone: no; With Bicalutamide: Phase II | NCT:04373824; NCT:04374279[200] |
| 6. Drugs interfering with cytokine storm syndrome and compromised immune Function | ||||
| Tocilizumab | Anti-inflammatory | It is approved to treat rheumatoid arthritis. It alleviates inflammation of the lungs by suppressing cytokine storm (reduces IL-6 and TNFα). | Phase II | NCT: 04335071[196,197] |
| Sarilumab | Anti-inflammatory | Phase III | NCT:04327388[201] | |
| Siltuximab | Anti-inflammatory | NA | NCT: 04322188[202,203] | |
| Ruxolitinib | Anticancer | Inhibit the mediation of cytokine and growth factor signaling by inhibiting JAK 1 and 2, which otherwise affect immune function and hematopoiesis. | Phase II | NCT:04331665, 04334044[204] |
| Baricitinib | Anticancer | It selectively and reversibly inhibits tyrosine-protein kinase Janus associated kinases 1 and 2 to modulates their signalling pathways, thereby reduce the phosphorylation and activation of STATs leading to inhibition of cytokine and chemokine transcription and thereby modulate the immune response. Currently, it is approved for the treatment of rheumatoid arthritis. Also, a clinical trial in combination with remdesivir has been initiated. | Phase III | NCT: 04320277, NCT: 04340232, NCT:04321993[152] |
| Sirolimus (Rapamycin) | Immunosuppressant | It inhibits T lymphocyte activation and proliferation in response to antigenic and cytokine. It also inhibits the mammalian target of rapamycin pathway to block viral protein expression and virion release. | Phase II | NCT: 04341675[205] |
| L-ascorbic acid | Antioxidant | It affects the development and maturation of T-lymphocytes, in particular, Natural Killer cells involved in the immune response to viral agents. Also, it inhibits ROS production in cases of the systemic inflammatory syndrome. | Phase I | NCT: 04323514[206,207] |
| Piclidenoson | Anti-inflammatory, A3 adenosine receptor agonist | Targets A3 Adenosine receptor leading to deregulation of the Wnt/β-catenin pathway. Inhibits inflammatory cytokine release syndrome in cancer immunotherapy. | Phase II | NCT:04333472[208] |
| Methylprednisolone | Glucocorticoid, Anti-inflammatory, immunomodulator | It activates a specific type of nuclear receptors, leading to their reformed gene expression thus inhibiting cytokine production and decreases circulating lymphocytes, inhibits TNF-α expression and NF-κB activation. | Phase II, III | NCT:04273321, NCT: 04263402, NCT:04323592[58] |
| Thalidomide | Anti-angiogenic, anti-inflammatory, anti-fibrotic | It impairs the synthesis of TNF-alpha and reduces the infiltration of inflammatory cells, proinflammatory cytokine, and chemokine levels, and inhibits the activated p-NFκBp6. | Phase II | NCT: 04273581, NCT:04273529[209] |
| NORS (Nitric Oxide Releasing Solution) | Gasotransmitter, Endothelium-derived relaxing factor (EDRF) | The inhibitory effect is shown to be correlated with s-nitrosylation of viral proteins such as reductases and proteases, thus inhibiting viral protein and RNA synthesis. NO generated by inducible nitric oxide synthase inhibits the virus replication cycle. | Phase II | NCT:04337918[210] |
| Colchicine | Antigout, Anticancer | Colchicine inhibits tubulin polymerization and microtubule generation and, possibly, effects on cellular adhesion molecules, inflammatory chemokines, and the inflammasome. It reduces excessive inflammatory reaction by inhibiting the activation of NLRP3 inflammasome and inhibit the synthesis of TNF-α and IL-6. | Phase II | NCT:: 04328480, NCT: 04326790, NCT:04322682[154] |
| Pyridostigmine Bromide | Acetylcholinesterase inhibitor | It inhibits acetylcholinesterase thereby improves acetylcholine (ACh) bioavailability. ACh then binds nicotinic-alpha receptors present in macrophages and T cells. This action reduces overactivated immune cells, which reduces inflammatory mediators and elevates CD4+ T cell counts. In experimental studies, it is shown to reduce inflammation and mortality. | Phase II | NCT:04343963[211,212] |
| Acalabrutinib | Anticancer | It is undergoing a Phase III trial for the treatment of mantle cell lymphoma and chronic lymphocytic leukemia. It selectively inhibits Bruton Tyrosine Kinase and thereby reduces the production of inflammatory cytokines associated with respiratory complications. | Phase II, Phase III | NCT:04346199[213] |
| Vazegepant (BHV-3500) | Calcitonin gene-related peptide (CGRP) receptor antagonist | Upon viral attack, Transient Receptor Potential channels on the plasma membrane are activated and release CGRP, which subsequently release interleukin 6 and pro-inflammatory mediators. This results in the development of cough, fever, migraine, and pain. Vazegepant blocks the CGRP receptor-mediated effects. | Phase II | NCT:04346615[214] |
| Tradipitant | Neurokinin1 receptor antagonist | It is indicated in the treatment for patients with atopic dermatitis. It prevents the activation of NK1 receptor by Substance P. | Phase III | NCT:04326426[215] |
| Etoposide, Teniposide (chemically related) | Anticancer | Both appear to act by causing breaks in DNA via interaction with DNA topoisomerase II or by the formation of free radicals. Teniposide is more potent as regards the production of DNA damage and cytotoxicity. The drugs are proposed to treat the cytokine storm in Covid-19 associated with the hyperinflammatory response to the virus. | Phase II | NCT:04356690[216] |
| 7. Drugs purported to prevent Covid-19 progression in patients with severe ARDS | ||||
| Nitric oxide Gas | Gasotransmitter | It reverses pulmonary hypertension and improves the condition of severe hypoxia thereby shortening the length of ventilatory support. | Phase II | NCT:04290871[217] |
| Sevoflurane | Anesthetic agent | It attenuates ARDS by vascular dilation, improves oxygenation in ventilator patients, thus decreases morbidity and mortality of patients. | Phase II | NCT:04355962[218] |
| Fingolimod | Immunomodulator | It is under clinical trial for treating multiple sclerosis. Structurally, it resembles lipid sphingosine-1-phosphate (S1P). It is a potent functional antagonist of S1P1 receptors in lymph node T cells. It suppresses the exit of lymphocytes from lymph node T cells, thus reducing the inflammation and attenuates immunopathogenesis. In Covid-19 patients, it is known to decrease pulmonary edema and hyaline membrane formation, thus preventing ARDS associated with the disease. | Phase II | NCT:04280588[219] |
| Fluvoxamine | Anti-depressant | It inhibits the uptake of 5-HT by blood platelets and brain synaptosomes. It is under trial to prevent more severe complications like shortness of breath in Covid-19 patients. | Phase II | NCT:04342663[101] |
| Tetrandrine | Anti-inflammatory and anti-tumor | It shows broad pharmacologic effects with antitumor and apoptotic activity. It decreases division and maturation of fibroblasts, that slows down lung damage in patients. | Phase IV | NCT:04308317[68] |
| Aviptadil (FDA Orphan Drug) |
Vasoactive intestinal polypeptide (VIP) analogue | Inhibits NMDA-induced caspase-3 activation, IL6 and TNF-α production in lungs. | Phase II | NCT:04311697[220] |
4.1. Spike protein
Viral attachment to the cellular receptor requires S protein priming by cellular proteases. The virus engages cellular protease TMPRSS2 for S protein priming for viral entry into target cells and viral spread in the infected host.[102] Furin or furin-like proteases (trypsin, cathepsin L) pre-cleavage S protein at the S1/S2 site,[103] which promotes subsequent TMPRSS2-dependent entry into host cells.[32] The blockade of these pathways might reduce the virus titer of SARS-CoV. Proprotein convertase inhibitor α1-PDX is demonstrated to inhibit cleavage activity.[103] Serine protease inhibitor camostat mesylate, which blocks TMPRSS2 activity, has been shown to significantly reduce MERS-S-, SARS-S-, and SARS-2-S-driven entry into the lung cells.[32] It has been approved in Japan for chronic pancreatitis and could be considered for clinical trials in the treatment of SARS-CoV-2-infected patients.[104] Much of the effort to develop vaccines and diagnostic tests has focused on a spike protein. However, other proteins might also be important determinants of immunity against SARS-CoV-2.[59]
4.2. Modulating SARS-CoV-2 receptor ACE2
As discussed earlier, SARS-CoV-2 exploits ACE2 for cellular entry with higher efficiency than SARS-CoV. This process induces the internalization of ACE2 that could cause loss of pulmonary function and increased tissue fibrosis as a result of elevated levels of circulating Ang II.[57,105] In the absence of ACE2, all available Ang I is converted to Ang II. Drugs that target various components of the RAAS such as ACE inhibitors, ARBs, aldosterone antagonists augmenting ACE2 activity or expression, and the product of ACE2 such as Ang (1-7), are subject to several clinical trials (Table 1).[106,107]
4.2.1. Drugs that bind to ACE2 receptor
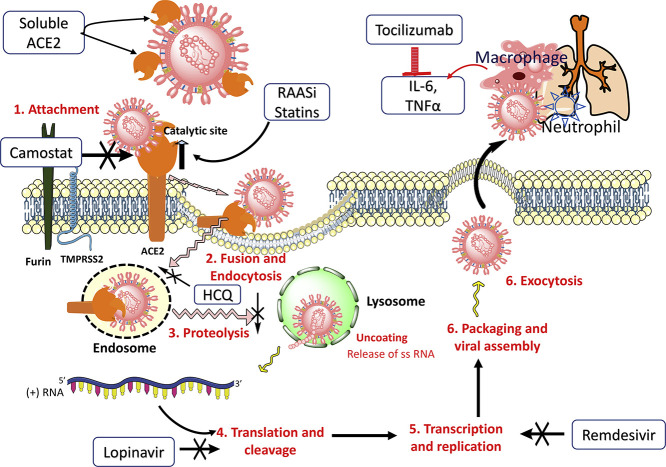
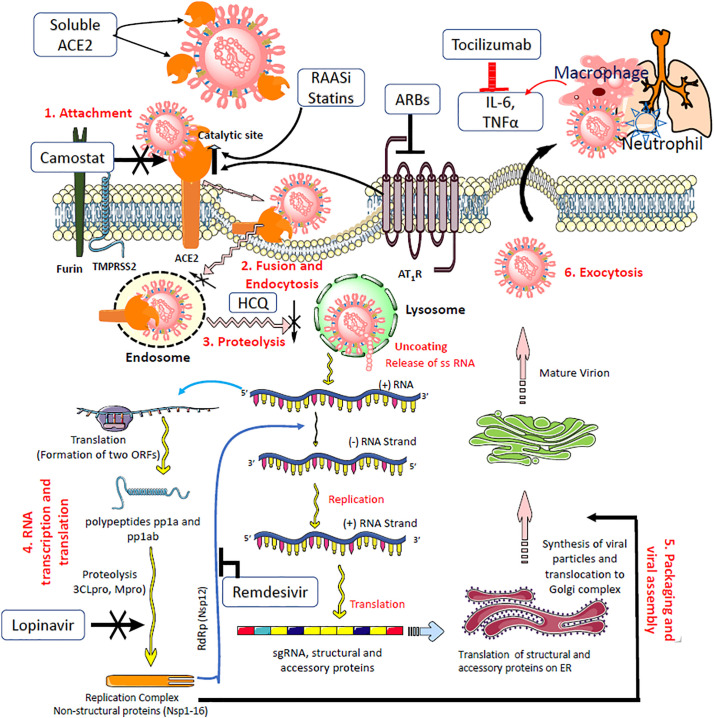
One of the measures that could successfully compete with endogenous ACE2 is soluble ACE2[56,108] or an Fc domain fused to ACE2 that may act as a decoy to direct SARS-CoV-2 away from endogenous ACE2 and itself bind the invading virus. The soluble form floats in the bloodstream and may act as a competitive interceptor of SARS-CoV-2 from binding to the full length ACE2 anchored in the cell membrane (Fig. 3 ). This prevents the virus from multiplying and damaging the cells. Endogenous ACE2 receptors are spared and may continue to function in counteracting the Ang II canonical pathway. However, endogenous circulating levels of soluble ACE2 are below the detection threshold and are unlikely to sequester the virus in circulation and disseminate it.[57,109] Experimental studies with a clinical grade human recombinant soluble ACE2 (hrsACE2) have been shown to inhibit the attachment of the virus to the cells and dose-dependently reduce the viral load by a factor of 1,000-5,000. These studies suggest that hrsACE2 can significantly block initial stages of SARS-CoV-2 infections.[108] It is thought that hrsACE2 may decrease Ang II levels while increasing ACE2 activity. The development of hrsACE2 has undergone two clinical trials for the treatment of acute respiratory distress syndrome.[110] Another strategy that is being investigated in clinical trials is the administration of an antibody or a single chain antibody fragment (scFv) that binds ACE2 and blocks the interaction of spike protein on the virion to ACE2.[111]
Fig. 3.
Potential pharmacological targets with select repurposed and investigational drugs in the life cycle of SARS-CoV-2.
To begin with, camostat inhibits cell entry of the virus spike protein by inhibiting TMPRSS2. The soluble form of rhACE2 that has no membrane anchoring collectrin-like domain binds the virus and sequesters it to prevent further interaction with cell-surface ACE2, thereby contributing to increased ACE2 levels. RAAS inhibitors and statins have been shown to increase endogenous ACE2 levels or expression. Chloroquine and HCQ inhibit viral entry and endocytosis by multiple mechanisms and host immunomodulatory effects. Lopinavir and darunavir inhibit viral proteinases 3CLpro and PLpro. Remdesivir inhibits RNA-dependent RNA polymerase (RdRp) and consequently halts the replication of the viral genome. The pathophysiology of organ damage in the lungs and other organs is caused by an immune response and cytokine release. Immunomodulatory agents such as tocilizumab and sarilumab target interleukin-6 and reduce cytokine storm.
The small intestine secretes two kinds of α defensins, HD5 and HD6.[112] Wang et al reported that HD5 with lectin-like ability interacts with the spike protein or ACE2 or both by targeting glycosylation sites as both are glycosylated proteins.[113] Besides, HD5 was shown to block the ligand-binding domain of intestinal epithelial ACE2 and thereby prevent the entry of the virus into host cells. Of note, people living in high altitude regions (above 2500 m sea level) are not prone to develop adverse effects to SARS-CoV-2 infection.[114] The reason could be hypoxia upregulates ACE and downregulates ACE2 expression in the heart[115] and pulmonary epithelial cells.[116] ARBs are known to decrease hypoxia-induced cell damage.[117]
4.2.2. RAAS inhibitors
Experimental studies have shown that Ang II overactivation of the AT-1 receptor activates metalloprotease A Disintegrin And Metalloprotease 17 (ADAM-17), also known as tumor necrosis factor-α converting enzyme (TACE), cleaves ACE2, which can no longer catalytically convert Ang II or Ang I to protective peptides (Ang (1-9) and Ang (1-7), respectively). This is one of the mechanisms that contribute to myocardial dysfunction and hypertension.[118] The cleaved ACE2 is endocytosed and degraded in lysosomes. With coronavirus infection, ACE2 is downregulated, causing increased levels of Ang II[105], the generator of a few inflammatory molecules. Limiting the synthesis of protective peptides may magnify the cytokine storm, resulting in acute lung injury, and overwhelming inflammatory response.[119] High cytokine levels may represent the leading cause of myocardial injury and endothelial dysfunction in patients with Covid-19 infection.[51] Although no evidence of histologically confirmed Covid-19 myocarditis has been published,[51] elevated plasma troponin and N-terminal pro-B-type natriuretic peptide levels with a significant (P <0.001) positive linear correlation with plasma high sensitivity C-reactive protein levels showing myocardial injury have been reported. [[119], [120], [121]] Rarely, the patient presents with electrocardiogram changes (acute ST-elevation myocardial infarction).[122] These results suggest that cardiac injury is a common condition among hospitalized patients with Covid-19 and it is associated with a higher risk of in-hospital mortality compared with lack of cardiac injury (P <0.001).[120]
In non-clinical studies, treatments with ARBs, ACE inhibitors, and aldosterone receptor antagonists strengthen the activity of the protective axis as these drugs upregulate the expression of ACE2 [57,106,123,124] (Fig. 3) and increase the production of Ang (1-7) by NEP on Ang I. Additionally, ACE inhibitors prevent the metabolism of Ang (1-7) to the inactive metabolite, Ang (1-5). However, it is not known whether RAAS inhibitor therapy increases ACE2 expression in patients. Based on preclinical studies, it is postulated that continuous therapy with RAAS inhibitors might cause an increased rate of cellular infection by a predicted compensatory induction of more ACE2 viral receptor sites and hence accelerate the spread of Covid-19 in an infected lung. However, it is unlikely to happen in the presence of AT-1 receptor blockade.[38] The compensatory increase of Ang II is a) unlikely to produce any inflammatory or oxidative responses; b) engaged by ACE2 to form Ang (1-7), and the operation might go in parallel with the virus binding ACE2 until it is internalized as their loci on ACE2 is different and do not compete with each other.[58]
There are no studies in humans demonstrating the effects of RAAS inhibitors on pulmonary or cardiac ACE2 expression and activity. Several reports coming from hospitals having acute inpatients with Covid-19 have observed that treatment with ARBs or ACE inhibitors was not associated with an increased risk of rapidly deteriorating severe disease.[125] A recent clinical study[9] observed an association of reduced all-cause mortality of Covid-19 patients with hypertension. Based on the analysis of 1128 hospitalized Covid-19 patients with hypertension, the investigators reported that the mortality rate was lower in the ACE inhibitors/ARBs group versus the non-ACE inhibitors/ARBs group (3.7% vs. 9.8%; P = 0.01). In another study, patients on ACE inhibitors/ARBs had statistically significantly lower levels of hypersensitive C-reactive protein and procalcitonin when compared with patients not receiving an ACE inhibitor/ARB, but with no effect on mortality[65], suggesting a potential anti-inflammatory function of RAAS inhibitors in Covid-19. This was substantiated in another study[126] wherein the absolute number of CD3+ and CD8+ T cells were statistically significantly higher in the ACE inhibitor/ARB group than in the non-ACE inhibitor/ARB group. Furthermore, there was a trend toward lower interleukin-6 levels in patients from the ACE inhibitor/ARB group. These findings support the fact that Ang II regulates the expression of inflammatory cytokines through the activation of AT-1 receptors.[127] RAAS inhibitors attenuate that inflammatory response and indirectly work to regulate immune function and inhibit inflammatory responses. A number of professional societies recommend that patients treated with ARBs/ACE inhibitors before the infection should not discontinue therapy if Covid-19 infection occurs (HFSA/ACC/AHA 2020). A possible trend towards a beneficial effect of RAAS inhibitors including aldosterone antagonists (spironolactone and eplerenone) will need to be explored as more data becomes available. The potential mechanisms in the complex interplay of the RAAS and SARS-CoV-2 need to be studied. To address this question directly in patients, a multicenter trial led by investigators at the University of Minnesota, USA, is planned (https://clinicaltrials.gov/ct2/show/NCT04312009). Nevertheless, the effect of RAAS inhibitors in the pathogenesis of Covid-19 is overly complex and controversial.
Similarly to hypertension, atherosclerosis [66,128], and hypoxia/high altitude[114] are also associated with reduced ACE2 expression. Statins have been shown to increase ACE2 expression in the heart and kidneys of rabbits fed a high cholesterol diet.[129] Additionally, PPARγ may also influence ACE2 expression as rosiglitazone and telmisartan (an ARB with a partial PPARγ agonist activity) have been shown to increase ACE2 expression in the aorta of hypertensive rats. Some warranted, clinical trials are already in progress to evaluate the impact of treatment with RAAS inhibitors, Ang (1-7), and statins (Table 1) given that these classes of drugs enhance ACE2, which in part contributes to the benefit of these regimens for Covid-19 related pathologies such as acute and chronic respiratory disease.
4.3. Drugs interfering with proteolysis mechanism of the virus within host cells
Upon entrance to the host cells, the viral genome is released as a single-stranded positive RNA. Subsequently, it is translated with a ribosomal frameshifting mechanism generating two polyproteins, pp1a and pp1ab. Encoded within the polyproteins are two proteinases, 3-chymotrypsin-like proteinase (3CLpro, also called main protease Mpro) and papain-like proteinase (PLpro). Both of these proteases are responsible for the cleavage of polyproteins translated from viral RNA into functional or effector proteins for virus replication and packaging within the host cells.[130] Inhibiting the activity of this enzyme would block viral replication. PLpro behaves as a deubiquitinase that may deubiquitinate certain host cell proteins such as interferon and NF-κB, resulting in suppression of the innate immune system.[65] Both are considered to be attractive drug targets as they play a central role in viral replication and transcription functions through extensive proteolysis of two replicase polyproteins, pp1a and pp1ab.
Drugs that target these proteases in other viruses, such as the HIV/AIDS drugs lopinavir and ritonavir (Fig. 3), have been explored in combination in patients with mild and moderate Covid-19 (Table 1). However, no benefit was observed with the treatment.[131] The efficacy of combination therapy of lopinavir/ritonavir and interferon β1b is being studied in a clinical trial.[132] A recently published X-ray crystal structure of the SARS-CoV-2 3CLpro provides a basis for structure-based drug discovery efforts.[133] A different class of drugs has been shown to inhibit viral replication in cells. Carmofur (1-hexylcarbamoyl-5-fluorouracil), an approved anticancer agent indicated for colorectal cancer since the 1980s, has been shown to inhibit the main protease Mpro activity. [134] Besides, a large number of inhibitors of these proteinases have been investigated in nonclinical studies, and are described in several publications.
4.4. Drugs interfering with replication, transcription, and translation of virus genomic material
The life cycle of the virus is dependent on enzymes that are excellent antiviral drug targets. One of the major viral enzymes that assist in translation and replication of the virus is RNA-dependent RNA polymerase. It is the major target of many existing nucleotide drugs such as remdesivir, favipiravir, ribavirin, galidesivir, and EIDD-2801 (Table 1). All of these are likely to inhibit RNA-dependent RNA polymerase (Fig. 3) through non-obligate RNA chain termination, a mechanism that requires the conversion of the parent compound to the active triphosphate form.[135]
Remdesivir, a nucleoside analog prodrug, is much in the news since the US FDA issued emergency use authorization for the treatment of Covid-19 to Gilead Sciences on May 1, 2020, based on Phase III clinical trial (FDA EAU-remdesivir). The National Institutes of Health’s clinical trial (Adaptive COVID-19 Treatment Trial, NCT04280705) was an adaptive, randomized, double-blind, placebo-controlled trial that evaluated 1,063 hospitalized adult patients with a broad mix of Covid-19 symptoms. In this trial, remdesivir outperformed placebo by showing a median time to recovery of 11 days relative to 15 days for those who received a placebo, thus demonstrating a 31% faster time to recovery. [9] The second trial was Gilead’s global SIMPLE trial (NCT04292899) evaluating 5-day and 10-day dosing durations in up to 6,000 Covid-19 patients. Nevertheless, some trials failed to show any benefit of remdesivir in patients with severe Covid-19.[136]
On May 1, 2020, the U.S. FDA issued an emergency use authorization for the investigational antiviral drug remdesivir for the treatment of suspected or laboratory-confirmed COVID-19 in adults and children hospitalized with severe disease. While there is limited information known about the safety and effectiveness of using remdesivir to treat people in the hospital with COVID-19, the investigational drug was shown in a clinical trial to shorten the time to recovery in some patients. Furthermore, according to the US FDA, remdesivir is administered intravenously by healthcare providers to treat suspected or confirmed severe Covid-19 cases such as patients with low blood oxygen levels needing oxygen therapy or more intensive breathing support (FDA EAU-Remdesivir). At present, worldwide, more than 15 clinical trials in several phases are investigating the efficacy and safety of intravenous remdesivir for Covid-19. It is also one of the four arms of WHO’s SOLIDARITY trial evaluating the potential benefit and safety of remdesivir for Covid-19.
4.5. Anti-inflammatory and immunomodulators in the control of cytokine storm
The invasion of the ACE2 receptor rich alveolar epithelial cells in the lung by the virus leads to the exuberant stimulation of the immune system, triggering a cytokine storm. Cytokines are chemical signaling molecules that essentially guard a healthy immune system. Thus, it should beat back the virus attack at the initial stages before the virus runs through the windpipe and invades the lungs and setting in motion the cytokine storm. This event causes a plethora of effects such as lung and cardiovascular damage, metabolic acidosis, coagulation dysfunction, septic shock, and multiple organ failure. [137,138]
The inflammatory response to the virus is excessive, resulting in the onset of a cytokine storm[12] with an increase in inflammation-inducing cytokines such as IL-2, IL-6, IL-7, granulocyte colony stimulating factor, and tumour necrosis factor-α (TNF-α).[65] The cytokine storm also leads to secondary hemophagocytic lymphohistiocytosis (sHLH) and tissue damage.[139] A recent retrospective multicentre analysis of circulating increases in IL- 6 levels can be a predictor of morbidity and mortality in Covid-19 patients.[140] Clinical pathology investigations in Covid-19 patients have shown a decrease in the number of lymphocytes, neutrophils, CD4+ and CD8+ T cells counts.[141]
Some studies have shown either enhanced helper T cells count or decline in the memory helper T cells and CD28+ cytotoxic suppressor T cells count in SARS-CoV-2 infected patients.[140] Reduction of B lymphocytes and natural killer cells were also observed in these patients. Immune dysregulation, abnormal chemokine response, and change in the composition of the subgroup of lymphocytes may cause a cytokine storm and subsequent tissue damage. The upregulation of ACE2 stimulates the RPS3 and SRC (hub genes), facilitating viral replication and the stimulation of neutrophils, natural killer cells, other T lymphocytes, and TNF-α, leading to an inflammatory response.[142] A large number of approved drugs for other diseases with anti-inflammatory and/or immunomodulatory activities targeting cytokines have a rationale for providing benefit, and therefore are reasonable to study in Covid-19. These drugs are listed in Table 1 with their possible mechanism of action and are candidates in several clinical trials worldwide. A selected few are discussed below.
Chloroquine (CQ) and hydroxychloroquine (HCQ) are approved for the treatment of malaria, lupus erythematosus, and rheumatoid arthritis. Both have antiviral activity including hepatitis B, HIV, H1N1, and Zika virus.[143] CQ and HCQ interfere with glycosylation and proteolytic maturation of proteins and terminal glycosylation of ACE2, and increase the pH of endosomes, Golgi vesicles, and lysosomes, thereby inhibiting viral release into the host cell. By altering membrane stability, signaling pathways, and transcriptional activity, CQ and HCQ inhibit cytokine production and modulation of certain costimulatory molecules. Studies have shown that CQ and HCQ inhibit antigen presentation in dendritic cells and cytokine production by leukocytes, Toll-like receptors, and immune cells.[144] The immunomodulatory mechanism of action of CQ and HCQ is mediated via multiple mechanisms involving inhibition of i) cytokine and its release by T-cells, ii) cytotoxic T cell and self-reactive CD4+ lymphocyte activities, iii) miR expression, iv) decreasing the levels of chemokines (CCL2 and CXCL10), and v) upregulating IFN-α and Treg activity.[144] There are no convincingly robust studies to support the use of CQ and HCQ for pre- or post-exposure prophylaxis of Covid-19. Some clinical trials evaluating the efficacy of CQ or HCQ in high-risk individuals are underway (Table 1). Meanwhile, the US FDA issued emergency use authorization for the use of hydroxychloroquine (and chloroquine) on March 28 for the treatment of hospitalized patients with Covid-19.
4.5.1. Tocilizumab (TCZ)
In the pathogenesis of SARS, there is a considerable release of proinflammatory cytokines (including IL-6) possibly by stimulation of immune cells.[145] IL-6 is also a biomarker of severe betacoronavirus infection. Its receptors are omnipresent in hematopoietic cells, including T-cells, activated B-cells, monocytes, and neutrophils. IL-6 confers numerous biological activities via IL-6Rα (gp-126; membrane bound) and IL-6Rβ (gp-130; soluble form). Upon binding to IL-6, both receptors undergo homodimerization and initiate a cascade of intracellular signaling pathways. Two types of signaling are reported: classical signaling conferred upon binding of IL-6 to IL-6Rα; and trans-signaling, activated upon binding of IL-6 to IL-6Rβ. Classical signaling plays a vital role in anti-inflammatory and regenerative mechanisms, whereas trans-signaling is associated with pro-inflammatory response upon activation of JAK-MAPK responses and is considered important in a cytokine storm.[146] Elevated production of IL-6 contributes to the pathogenesis of various autoimmune and inflammatory diseases. In immune cells, IL-6 induces proliferation and differentiation of cells.[147] and interfering at IL-6 might provide a new therapeutic strategy for Covid-19. TCZ, sarilumab, and siltuximab are classified as IL-6 antagonists. TCZ ameliorates hypoxemia, lymphopenia, and lung infiltration in Covid-19.[148] However, IL-6 antagonists increase the risk of infections[149] and might suppress the immune response that the body needs to fight off the virus.[12] Several clinical trials are in progress to determine the beneficial effects of tocilizumab, sarilumab, and siltixumab (Table 1).
4.5.2. Janus kinase (JAK)-signal transducer and activator of transcription (STAT) inhibitors
JAK-STAT inhibitors is IL-6 post-receptor downstream signaling pathway. The pathway mediates the effect of interleukins, IFN-(α, β, γ) and growth factors (GM-CSF, TGF-β, erythropoietin, and thrombopoietin).[150] JAK inhibitors are approved for rheumatoid arthritis, psoriatic arthritis, and inflammatory disorders.[151] Many proinflammatory cytokines involved in cytokine storms are antagonised by JAK inhibitors. A notable drug in this class is baricitinib, which inhibits viral entry and assembly.[152] Currently, JAK inhibitors baricitinib, ruxolitinib, and tofacitinib are undergoing clinical trials for the treatment of Covid-19.
4.5.3. Nod-like receptor family pyrin domain-containing-3 (NLRP3) inflammasome
NLRP3 has a major role in regulating innate immunity. Its activation occurs in response to invading pathogens including RNA viruses. It activates caspase-1 to produce the pro-inflammatory cytokines IL-1β and IL-18. Both fuel a cytokine storm and are inhibited by anakinra. It is a recombinant human IL-1 receptor antagonist approved for the treatment of rheumatoid arthritis. Anakinra improved the survival of patients with disseminated intravascular coagulation (DIC).[153] There are several anakinra studies registered for Covid-19 (NCT04339712, NCT04330638). Another drug that is reported to antagonise the NLRP3 inflammasome and the synthesis of TNF-α and IL-6 is colchicine.[154] Colchicine inhibits tubulin microtubule formation, cellular adhesion molecules, and inflammatory chemokines. It is approved for the treatment of gout and familial Mediterranean fever. Clinical trials are in progress to evaluate the efficacy of colchicine for the treatment of Covid-19.
4.6. Vaccines and biologics (Biopharmaceuticals)
The SARS-CoV-2 pandemic created havoc in the world and everyone is eagerly awaiting the arrival of a vaccine for Covid-19. The critical step for the successful development of a vaccine is the identification of the complete genome sequence of the pathogen. This information was made available on 25 January 2020 at NCBI (Gen. Bank: MN 908947.3). A detailed account of the difference between traditional vaccine development and development using the pandemic paradigm is reported.[155]
The pandemic paradigm requires multiple activities to be conducted at financial risk to developers and manufacturers and without knowing whether the vaccine candidate will be safe and effective, including a very early manufacturing scale-up to commercial scale before the establishment of clinical proof of concept.
The Coalition for Epidemic Preparedness Innovation (CEPI) is coordinating with health authorities throughout the world and developers of vaccines.
Owing to the high similarity of SARS-COV-2 with SARS-COV and the binding pattern to ACE2 receptors in the host cell, the pandemic has unlocked the scope for the development of vaccines and biopharmaceuticals at a more rapid pace than ever before.[156] These biologics are expected to broaden the spectrum of the current treatment options available.[65] As per the landscape of Covid-19 candidate vaccines published by WHO on 20 April 2020, approximately 7 vaccines have begun clinical trials and 75 vaccines are in preclinical investigations.[157] The first doses of Covid-19 trial vaccines were injected into patients as early as March 2020. Importantly, the vaccines are developed based upon a) live/attenuated status of the virus (virus vaccines); b) vector-based vaccines including Adenovirus Type 5 and Modified Vaccinia Virus encoding spike or membrane proteins; c) Nucleic acid-based vaccines based on DNA, plasmid, mRNA, SiRNA, and others; d) Protein-based vaccines which include vaccines derived from various Capsid-like particles, peptide antigen, recombinant protein developed using spike, membrane, nucleocapsid elements of SARS-CoV-2, etc.; and e) microneedle array delivered recombinant coronavirus vaccines.[158] These vaccines are expected to train the host immune system to recognize the pathogenic virus strains and combat them providing herd immunity.[159] Based on previous experience of virus pandemics, due consideration is being given to safety evaluations that could otherwise result in immunopotentiation marked by eosinophilic infiltration and increased infectivity.[156] A brief status of vaccines for SARS-CoV-2 is curated in Supplementary Table 1.
Biopharmaceuticals (biologics), which are engineered macromolecular products drugs comprising nucleic acids and proteins, are also being evaluated to combat the Covid-19. Important classes of biopharmaceuticals that are being explored include: a) protein-based monoclonal/polyclonal antibody; b) recombinant enzymes; c) interferon; d) oligonucleotides; e) hematopoietic growth factors; and f) interleukin-based products. The detailed classification, the mechanism of action of each biologic, and their clinical status are compiled in Supplementary Table 2. These are chiefly intended to target the most fragile and vulnerable sites/receptors expressed on viral surfaces (monoclonal antibodies),[160] provide antiviral immunity along with possessing immunomodulatory properties (interferons),[161] potentially interfere with the spike protein or replicative machinery of the virus (oligonucleotides),[162] enhance the production of hematopoietic cells (hematopoietic growth factors)[163] or via blocking key immune players (interleukin based products).[164]
4.7. Conclusion
At the time of writing this paper, Covid-19 is considered an important global public health crisis. Although most of the drugs under evaluation have demonstrated good in vitro activity or possible benefits in limited observational or non-randomized clinical studies against SARS-CoV-2, clinical benefits are awaiting confirmation in large scale randomized clinical trials such as WHO’s SOLIDARITY trial. Observational studies with hypertensive patients on ARB/ACE inhibitor therapy have shown no increased risk of Covid-19 acquisition, although the pattern of multiorgan dysfunction seen in Covid-19 patients is likely related to ACE2 expression in these organs. On the contrary, a few studies have reported benefits with ARB and ACE inhibitor therapy on parameters such as decreased mortality rate and lower levels of hypersensitive C-reactive protein and procalcitonin. The repurposing of existing drugs designed for other virus infections and pathologies may speed up the process of fighting the pandemic, as noted with the success of remdesivir gaining emergency authorization use. Based on the pathology of the disease, in which the virus attacks more than one organ and makes each of them worse by inducing a dysregulated inflammatory response, any one drug or mechanism of action may not combat the disease. It should be a multipronged approach to directly target the virus replication cycle and boost innate antiviral immune responses.
During the reviewing process of this manuscript, many related papers have been published. The following significant findings were reported during this period. The addendum is confined to the scope of the current manuscript.
-
1
Cardiovascular complications (JACC, 2020; doi.org/10.1016/j.jacc.2020.06.007; Heart, 2020;106:1154–1159. doi:10.1136/heartjnl-2020-317007) are upcoming as a significant threat in Covid-19 patients along with blood clotting incidences (Lancet, 2020;395,1417-1418, doi: 10.1016/S0140-6736(20)30937-5; Thrombosis Research, 2020;190,62, doi: 10.1016/j.thromres.2020.04.014). There are reports of abnormal coagulation with elevated D-dimer levels in hospitalized patients with severe Covid-19 (J Thromb Haemost, 2020;18:844; JACC 2020, doi.org/10.1016/j.jacc.2020.03.031)
-
2
Many publications have now shown that ACE2 upregulation with ACE inhibitors/ARBs therapy is not associated with increased susceptibility to SARS-CoV-2 infection (Hypertension,2020;76. DOI: 10.1161/HYPERTENSIONAHA.120.15289). Additionally, they have beneficial effects on pulmonary inflammation improving clinical outcomes (JAMA, 2020;doi:10.1001/jama.2020.11301). Statins besides increasing ACE2 expression reduce inflammation and thrombosis (Br J Pharmacol, 2020; doi:10.1111/bph.15166), and also shown to inhibit SARS-CoV-2’s main protease (Arch Med Sci, 2020;16:490–496, DOI: doi.org/10.5114/aoms.2020.94655).
-
3
The US FDA has revoked authorized use of HCQ/CQ on June 15, 2020 (https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-revokes-emergency-use-authorization-chloroquine-and).
-
4
Dexamethasone was found to reduce mortality and the recovery time in critically ill patients (RECOVERY TRIAL, June 29, 2020; Lancet Resp Med, 2020;8,267-276.doi: 10.1016/S2213-2600(19)30417-5.)
-
5
Favipiravir, a drug initially approved in Japan for treating novel epidemic influenza strains, has received regulatory clearance in Russia, followed by India for the treatment of Covid-19. The drug remains a subject of clinical trials across the world, including India and Russia (NCT04359615) (Engineering, 2020; Mar 18.: doi.org/10.1016/j.eng.2020.03.007; Clin Pharmacol Therap, 2020; doi/pdf/10.1002/cpt.1877).
-
6
The World Health Organisation has published a draft landscape on Covid-19 vaccines. There are currently 133 vaccines in development with 10 in clinical trials (The Lancet, 2020;395(10239),1751-1752. Doi: 10.1016/S0140-6736(20)31252-6).
Author contributions
RP and Jagadeesh discussed the objectives and wrote the outline of the article. All three authors wrote and revised the manuscript before submission. Gaurav and Jagadeesh developed Figures. Gaurav created supplementary Tables.
Disclaimer
This article has been reviewed by FDA and determined not to be consistent with the Agency’s views or policies. It reflects only the views and opinions of the author (G. Jagadeesh).
Funding source
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare that they have no competing financial interests.
Acknowledgments
We thank Drs. J. Rick Turner and Monika Deshpande of the US FDA for a review of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cellsig.2020.109721.
Appendix A. Supplementary data
Supplementary material
References
- 1.Hilgenfeld R., Peiris M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antivir. Res. 2013;100(1):286–295. doi: 10.1016/j.antiviral.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 3.Eckerle I., Müller M.A., Kallies S., Gotthardt D.N., Drosten C. In-vitro renal epithelial cell infection reveals a viral kidney tropism as a potential mechanism for acute renal failure during Middle East Respiratory Syndrome (MERS) Coronavirus infection. Virol. J. 2013;10(1):359. doi: 10.1186/1743-422X-10-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. WHO; Geneva: 2020. Coronavirus Disease 2019 (COVID-19): Situation Report, 92. [Google Scholar]
- 5.Oberfeld B., Achanta A., Carpenter K., Chen P., Gilette N.M., Langat P., Said J.T., Schiff A.E., Zhou A.S., Barczak A.K., Pillai S. SnapShot: COVID-19. Cell. 2020;181(4) doi: 10.1016/j.cell.2020.04.013. 954-954 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., Fan Y., Zheng C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect. Dis. 2020;20(4):425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y., Yang Y., Zhang C., Huang F., Wang F., Yuan J., Wang Z., Li J., Li J., Feng C., Zhang Z., Wang L., Peng L., Chen L., Qin Y., Zhao D., Tan S., Yin L., Xu J., Zhou C., Jiang C., Liu L. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci. China Life Sci. 2020;63(3):364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng Y.Y., Ma Y.T., Zhang J.Y., Xie X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 2020;17(5):259–260. doi: 10.1038/s41569-020-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He F., Deng Y., Li W. Coronavirus disease 2019: what we know? J. Med. Virol. 2020 doi: 10.1002/jmv.25766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balakumar P., Maung U.K., Jagadeesh G. Prevalence and prevention of cardiovascular disease and diabetes mellitus. Pharmacol. Res. 2016;113(Pt A):600–609. doi: 10.1016/j.phrs.2016.09.040. [DOI] [PubMed] [Google Scholar]
- 12.Wadman M., Couzin-Frankel J., Kaiser J., Matacic C. A rampage through the body. 2020;368(6489):356–360. doi: 10.1126/science.368.6489.356. [DOI] [PubMed] [Google Scholar]
- 13.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fehr A.R., Perlman S. Springer; 2015. Coronaviruses: An Overview of their Replication and Pathogenesis, Coronaviruses; pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2) doi: 10.1016/j.cell.2020.02.058. 281-292 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robson B. COVID-19 Coronavirus spike protein analysis for synthetic vaccines, a peptidomimetic antagonist, and therapeutic drugs, and analysis of a proposed achilles’ heel conserved region to minimize probability of escape mutations and drug resistance. Comput. Biol. Med. 2020;103749 doi: 10.1016/j.compbiomed.2020.103749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J., Sun Y., Qi J., Chu F., Wu H., Gao F., Li T., Yan J., Gao G.F. The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes. J. Infect. Dis. 2010;202(8):1171–1180. doi: 10.1086/656315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong R., Yang G., Xue R., Liu M., Wang F., Hu J., Guo X., Chang S. 2020. COVID-19 Docking Server: An Interactive Server for Docking Small Molecules, Peptides and Antibodies Against Potential Targets of COVID-19. arXiv preprint. doi: arXiv:2003.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mu J., Xu J., Zhang L., Shu T., Wu D., Huang M., Ren Y., Li X., Geng Q., Xu Y. SARS-CoV-2-encoded nucleocapsid protein acts as a viral suppressor of RNA interference in cells. Sci. China Life Sci. 2020:1–4. doi: 10.1007/s11427-020-1692-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng Q., Langereis M.A., van Vliet A.L., Huizinga E.G., de Groot R.J. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc. Natl. Acad. Sci. 2008;105(26):9065–9069. doi: 10.1073/pnas.0800502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lang Y., Li W., Li Z., Koerhuis D., van den Burg A.C., Rozemuller E., Bosch B.-J., van Kuppeveld F.J., Boons G.-J.P., Huizinga E.G. Coronavirus hemagglutinin-esterase and spike proteins co-evolve for functional balance and optimal virion avidity. bioRxiv. 2020 doi: 10.1101/2020.04.03.003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang X., Wu C., Li X., Song Y., Yao X., Wu X., Duan Y., Zhang H., Wang Y., Qian Z. On the origin and continuing evolution of SARS-CoV-2. Nat. Sci. Rev. 2020 doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brufsky A. Distinct viral clades of SARS-CoV-2: implications for modeling of viral spread. J. Med. Virol. 2020 doi: 10.1002/jmv.25902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dilucca M., Pavlopoulou A. Analysis of codon usage and evolutionary rates of the 2019-nCoV genes. bioRxiv. 2020 doi: 10.1101/2020.03.25.006569. [DOI] [Google Scholar]
- 26.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mousavizadeh L., Ghasemi S. Genotype and phenotype of COVID-19: their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng Y., Ma Y., Zhang J., Xie X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 2020;17(5):259–260. doi: 10.1038/s41569-020-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim Y., Jedrzejczak R., Maltseva N.I., Wilamowski M., Endres M., Godzik A., Michalska K., Joachimiak A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020 doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2) doi: 10.1016/j.cell.2020.02.052. (271-280.e8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 2020;S0953-6205(20):30151–30155. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. 2009;106(14):5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U. S. A. 2014;111(42):15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gierer S., Bertram S., Kaup F., Wrensch F., Heurich A., Krämer-Kühl A., Welsch K., Winkler M., Meyer B., Drosten C. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J. Virol. 2013;87(10):5502–5511. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park J.E., Li K., Barlan A., Fehr A.R., Perlman S., McCray P.B., Jr., Gallagher T. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc. Natl. Acad. Sci. U. S. A. 2016;113(43):12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du L., He Y., Zhou Y., Liu S., Zheng B.-J., Jiang S. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7(3):226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan R., Zhang Y., Guo Y., Xia L., Zhou Q. Structural basis for the recognition of the 2019-nCoV by human ACE2. bioRxiv. 2020 doi: 10.1101/2020.02.19.956946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brielle E.S., Schneidman-Duhovny D., Linial M. The SARS-CoV-2 exerts a distinctive strategy for interacting with the ACE2 human receptor. Viruses. 2020;12(5):497. doi: 10.3390/v12050497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., Winter H., Meister M., Veith C., Boots A.W., Hennig B.P., Kreuter M., Conrad C., Eils R. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39(10):e105114. doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hussain M., Jabeen N., Amanullah A., Baig A.A., Aziz B., Shabbir S., Raza F. Structural basis of SARS-CoV-2 spike protein priming by TMPRSS2. bioRxiv. 2020 doi: 10.1101/2020.04.21.052639. [DOI] [Google Scholar]
- 45.Hofmann H., Pohlmann S. Cellular entry of the SARS coronavirus. Trends Microbiol. 2004;12(10):466–472. doi: 10.1016/j.tim.2004.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khadim Sheikh H., Arshad T., Mohammad Z.S., Arshad I., Hassan M. Repurposed single inhibitor for serine protease and spike glycoproteins of SAR-CoV-2. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12192660.v1. Preprint. [DOI] [Google Scholar]
- 47.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bar-On Y.M., Flamholz A., Phillips R., Milo R. SARS-CoV-2 (COVID-19) by the numbers. Elife. 2020;9 doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hikmet F., Mear L., Uhlen M., Lindskog C. 2020. The protein expression profile of ACE2 in human tissues, bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venkatakrishnan A., Puranik A., Anand A., Zemmour D., Yao X., Wu X., Chilaka R., Murakowski D.K., Standish K., Raghunathan B. Knowledge synthesis from 100 million biomedical documents augments the deep expression profiling of coronavirus receptors. arXiv. 2020 doi: 10.1101/2020.03.24.005702. preprint arXiv:2003.12773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tersalvi G., Vicenzi M., Calabretta D., Biasco L., Pedrazzini G., Winterton D. Elevated troponin in patients with Coronavirus Disease 2019 (COVID-19): possible mechanisms. J. Card. Fail. 2020;S1071-9164(20) doi: 10.1016/j.cardfail.2020.04.009. 30357-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Devaux C.A., Rolain J.M., Raoult D. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun P., Lu X., Xu C., Sun W., Pan B. Understanding of COVID-19 based on current evidence. J. Med. Virol. 2020;92(6):548–551. doi: 10.1002/jmv.25722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R. A novel angiotensin-converting enzyme–related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 2000;87(5):e1–e9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 55.Wysocki J., Ye M., Rodriguez E., González-Pacheco F.R., Barrios C., Evora K., Schuster M., Loibner H., Brosnihan K.B., Ferrario C.M. Targeting the degradation of angiotensin II with recombinant angiotensin-converting enzyme 2: prevention of angiotensin II–dependent hypertension. Hypertension. 2010;55(1):90–98. doi: 10.1161/HYPERTENSIONAHA.109.138420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Batlle D., Wysocki J., Satchell K. Soluble angiotensin-converting enzyme 2: a potential approach for coronavirus infection therapy? Clin. Sci. (Lond.) 2020;134(5):543–545. doi: 10.1042/CS20200163. [DOI] [PubMed] [Google Scholar]
- 57.South A.M., Diz D.I., Chappell M.C. COVID-19, ACE2, and the cardiovascular consequences. Am. J. Physiol. Heart Circ. Physiol. 2020;318(5):H1084–H1090. doi: 10.1152/ajpheart.00217.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.C., Turner A.J., Raizada M.K., Grant M.B., Oudit G.Y. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ. Res. 2020;126(10):1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang F., Kream R.M., Stefano G.B. An evidence based perspective on mRNA-SARS-CoV-2 vaccine development. Med. Sci. Monit. 2020;26:e924700. doi: 10.12659/MSM.924700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Balakumar P., Jagadeesh G. A century old renin–angiotensin system still grows with endless possibilities: AT1 receptor signaling cascades in cardiovascular physiopathology. Cell. Signal. 2014;26(10):2147–2160. doi: 10.1016/j.cellsig.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 61.Jia H.P., Look D.C., Tan P., Shi L., Hickey M., Gakhar L., Chappell M.C., Wohlford-Lenane C., McCray P.B., Jr. Ectodomain shedding of angiotensin converting enzyme 2 in human airway epithelia. Am. J. Phys. Lung Cell. Mol. Phys. 2009;297(1):L84–L96. doi: 10.1152/ajplung.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teixeira L.B., Parreiras-e-Silva L.T., Bruder-Nascimento T., Duarte D.A., Simões S.C., Costa R.M., Rodríguez D.Y., Ferreira P.A., Silva C.A., Abrao E.P. Ang-(1-7) is an endogenous β-arrestin-biased agonist of the AT 1 receptor with protective action in cardiac hypertrophy. Sci. Rep. 2017;7(1):1–10. doi: 10.1038/s41598-017-12074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Galandrin S., Denis C., Boularan C., Marie J., M’Kadmi C., Pilette C., Dubroca C., Nicaise Y., Seguelas M.-H., N’Guyen D. Cardioprotective angiotensin-(1–7) peptide acts as a natural-biased ligand at the angiotensin II type 1 receptor. Hypertension. 2016;68(6):1365–1374. doi: 10.1161/HYPERTENSIONAHA.116.08118. [DOI] [PubMed] [Google Scholar]
- 64.Balakumar P., Anand-Srivastava M.B., Jagadeesh G. Renin-angiotensin-aldosterone: an inclusive, an invigorative, an interactive and an interminable system. Pharmacol. Res. 2017;125(Pt A):1–3. doi: 10.1016/j.phrs.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 65.Yang G., Tan Z., Zhou L., Yang M., Peng L., Liu J., Cai J., Yang R., Han J., Huang Y., He S. 2020. Effects Of ARBs And ACEIs On Virus Infection, Inflammatory Status And Clinical Outcomes In COVID-19 Patients With Hypertension: A Single Center Retrospective Study, Hypertension. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Y.H., Hao Q.Q., Wang X.Y., Chen X., Wang N., Zhu L., Li S.Y., Yu Q.T., Dong B. ACE2 activity was increased in atherosclerotic plaque by losartan: possible relation to anti-atherosclerosis. J. Renin-Angiotensin-Aldosterone Syst. 2015;16(2):292–300. doi: 10.1177/1470320314542829. [DOI] [PubMed] [Google Scholar]
- 67.Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4) doi: 10.1016/j.cell.2020.04.011. 914-921 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117(1):17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shannon A., Le N.T.T., Selisko B., Eydoux C., Alvarez K., Guillemot J.-C., Decroly E., Peersen O., Ferron F., Canard B. Remdesivir and SARS-CoV-2: structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antivir. Res. 2020;104793 doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shu T., Huang M., Wu D., Ren Y., Zhang X., Han Y., Mu J., Wang R., Qiu Y., Zhang D.-Y. 2020. SARS-Coronavirus-2 nsp13 Possesses NTPase and RNA Helicase Activities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kachaev Z.M., Lebedeva L.A., Kozlov E.N., Shidlovskii Y.V. Interplay of mRNA capping and transcription machineries. Biosci. Rep. 2020;40(1) doi: 10.1042/BSR20192825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells. 2020;9(5):1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Astuti I., Ysrafil Severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab. Syndr. 2020;14(4):407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L. 2020. Structure of RNA-Dependent RNA Polymerase from 2019-nCoV, A Major Antiviral drug Target, BioRxiv. [DOI] [Google Scholar]
- 77.Ibrahim O.O. Coronavirus SARS-CoV-2 is the newly emerged zoonotic virus causing pandemic death and economic loss. EC Pulmonol. Respir. Med. 2020;9:65–75. [Google Scholar]
- 78.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Viswanathan T., Arya S., Chan S.-H., Qi S., Dai N., Hromas R.A., Park J.-G., Oladunni F., Martinez-Sobrido L., Gupta Y.K. 2020. Structural Basis of RNA Cap Modification by SARS-CoV-2 Coronavirus, bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Malik Y.A. Properties of coronavirus and SARS-CoV-2. Malays. J. Pathol. 2020;42(1):3–11. [PubMed] [Google Scholar]
- 81.Buratta S., Tancini B., Sagini K., Delo F., Chiaradia E., Urbanelli L., Emiliani C. Lysosomal exocytosis, exosome release and secretory autophagy: the autophagic-and endo-lysosomal systems go extracellular. Int. J. Mol. Sci. 2020;21(7):2576. doi: 10.3390/ijms21072576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dong L., Hu S., Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discov. Ther. 2020;14(1):58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- 83.Kalil A.C. Treating COVID-19—off-label drug use, compassionate use, and randomized clinical trials during pandemics. JAMA. 2020;323(19):1897–1898. doi: 10.1001/jama.2020.4742. [DOI] [PubMed] [Google Scholar]
- 84.Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. JAMA. 2020 doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- 85.Pan F., Ye T., Sun P., Gui S., Liang B., Li L., Zheng D., Wang J., Hesketh R.L., Yang L., Zheng C. Time course of lung Changes at chest CT during recovery from coronavirus disease 2019 (COVID-19) Radiology. 2020;295(3):715–721. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rosa S.G.V., Santos W.C. Clinical trials on drug repositioning for COVID-19 treatment. Rev. Panam. Salud Publica. 2020;44:e40. doi: 10.26633/RPSP.2020.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A., Doig A., Guilliams T., Latimer J., McNamee C., Norris A., Sanseau P., Cavalla D., Pirmohamed M. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019;18(1):41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 88.Mani D., Wadhwani A., Krishnamurthy P.T. Drug repurposing in antiviral research: a current scenario. J. Young Pharm. 2019;11(2):117. [Google Scholar]
- 89.Aubé J. Drug Repurposing and the Medicinal Chemist. CS Med. Chem. Lett. 2012;3(6):442–444. doi: 10.1021/ml300114c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ben Salah A., Ben Messaoud N., Guedri E., Zaatour A., Ben Alaya N., Bettaieb J., Gharbi A., Belhadj Hamida N., Boukthir A., Chlif S. Topical paromomycin with or without gentamicin for cutaneous leishmaniasis. N. Engl. J. Med. 2013;368(6):524–532. doi: 10.1056/NEJMoa1202657. [DOI] [PubMed] [Google Scholar]
- 91.Tan K.R., Magill A.J., Parise M.E., Arguin P.M., C. Centers for Disease, Prevention Doxycycline for malaria chemoprophylaxis and treatment: report from the CDC expert meeting on malaria chemoprophylaxis. Am. J. Trop. Med. Hyg. 2011;84(4):517–531. doi: 10.4269/ajtmh.2011.10-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dorlo T.P., Balasegaram M., Beijnen J.H., de Vries P.J. Miltefosine: a review of its pharmacology and therapeutic efficacy in the treatment of leishmaniasis. J. Antimicrob. Chemother. 2012;67(11):2576–2597. doi: 10.1093/jac/dks275. [DOI] [PubMed] [Google Scholar]
- 93.Shaughnessy A.F. Old drugs, new tricks. BMJ. 2011;342:d741. doi: 10.1136/bmj.d741. [DOI] [PubMed] [Google Scholar]
- 94.Joshi G., Poduri R. Virtual screening enabled selection of antiviral agents against Covid-19 disease targeting coronavirus endoribonuclease NendoU: plausible mechanistic interventions in the treatment of new virus strain. Chemrxiv. 2020 doi: 10.26434/chemrxiv.12198966. [DOI] [Google Scholar]
- 95.Khan R.J., Jha R.K., Amera G., Jain M., Singh E., Pathak A., Singh R.P., Muthukumaran J., Singh A.K. Targeting SARS-Cov-2: a systematic drug repurposing approach to identify promising inhibitors against 3C-like Proteinase and 2’-O-RiboseMethyltransferase. J. Biomol. Struct. Dyn. 2020:1–40. doi: 10.1080/07391102.2020.1753577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu S., Zheng Q., Wang Z. Potential covalent drugs targeting the main protease of the SARS-CoV-2 coronavirus. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6(1):14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Breckenridge A., Jacob R. Overcoming the legal and regulatory barriers to drug repurposing. Nat. Rev. Drug Discov. 2019;18(1):1–2. doi: 10.1038/nrd.2018.92. [DOI] [PubMed] [Google Scholar]
- 99.Baker N.C., Ekins S., Williams A.J., Tropsha A. A bibliometric review of drug repurposing. Drug Discov. Today. 2018;23(3):661–672. doi: 10.1016/j.drudis.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cavalla D. Predictive methods in drug repurposing: gold mine or just a bigger haystack? Drug Discov. Today. 2013;18(11-12):523–532. doi: 10.1016/j.drudis.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 101.Glebov O. 2020. Understanding the cell Biology of SARS-CoV-2 Endocytosis for COVID-19 drug Repurposing: Looking Beyond Chloroquine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011;85(2):873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hasan A., Paray B.A., Hussain A., Qadir F.A., Attar F., Aziz F.M., Sharifi M., Derakhshankhah H., Rasti B., Mehrabi M., Shahpasand K., Saboury A.A., Falahati M. A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1754293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yamamoto M., Matsuyama S., Li X., Takeda M., Kawaguchi Y., Inoue J.-i., Matsuda Z. Identification of nafamostat as a potent inhibitor of Middle East respiratory syndrome coronavirus S protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob. Agents Chemother. 2016;60(11):6532–6539. doi: 10.1128/AAC.01043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu Y., Yang Y., Zhang C., Huang F., Wang F., Yuan J., Wang Z., Li J., Li J., Feng C. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci. China Life Sci. 2020;63(3):364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Keidar S., Gamliel-Lazarovich A., Kaplan M., Pavlotzky E., Hamoud S., Hayek T., Karry R., Abassi Z. Mineralocorticoid receptor blocker increases angiotensin-converting enzyme 2 activity in congestive heart failure patients. Circ. Res. 2005;97(9):946–953. doi: 10.1161/01.RES.0000187500.24964.7A. [DOI] [PubMed] [Google Scholar]
- 107.Medina D., Arnold A.C. Angiotensin-(1-7): translational avenues in cardiovascular control. Am. J. Hypertens. 2019;32(12):1133–1142. doi: 10.1093/ajh/hpz146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Del Pozo C.H., Prosper F. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181(4):905–913. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chappell M.C. Biochemical evaluation of the renin-angiotensin system: the good, bad, and absolute? Am. J. Physiol. Heart Circ. Physiol. 2016;310(2):H137–H152. doi: 10.1152/ajpheart.00618.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Khan A., Benthin C., Zeno B., Albertson T.E., Boyd J., Christie J.D., Hall R., Poirier G., Ronco J.J., Tidswell M., Hardes K., Powley W.M., Wright T.J., Siederer S.K., Fairman D.A., Lipson D.A., Bayliffe A.I., Lazaar A.L. A pilot clinical trial of recombinant human angiotensin-converting enzyme 2 in acute respiratory distress syndrome. Crit. Care. 2017;21(1):234. doi: 10.1186/s13054-017-1823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kruse R.L. Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China. F1000Res. 2020;9:72. doi: 10.12688/f1000research.22211.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wehkamp J., Chu H., Shen B., Feathers R.W., Kays R.J., Lee S.K., Bevins C.L. Paneth cell antimicrobial peptides: topographical distribution and quantification in human gastrointestinal tissues. FEBS Lett. 2006;580(22):5344–5350. doi: 10.1016/j.febslet.2006.08.083. [DOI] [PubMed] [Google Scholar]
- 113.Wang Y., Jiang W., He Q., Wang C., Wang B., Zhou P., Dong N., Tong Q. Early, low-dose and short-term application of corticosteroid treatment in patients with severe COVID-19 pneumonia: single-center experience from Wuhan, China. medRxiv. 2020 doi: 10.1101/2020.03.06.20032342. [DOI] [Google Scholar]
- 114.Arias-Reyes C., Zubieta-DeUrioste N., Poma-Machicao L., Aliaga-Raudan F., Carvajal-Rodriguez F., Dutschmann M., Schneider-Gasser E., Zubieta-Calleja G., Soliz J. Does the pathogenesis of SAR-CoV-2 virus decrease at high-altitude? Respir. Physiol. Neurobiol. 2020;103443 doi: 10.1016/j.resp.2020.103443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dang Z., Su S., Jin G., Nan X., Ma L., Li Z., Lu D., Ge R. Tsantan Sumtang attenuated chronic hypoxia-induced right ventricular structure remodeling and fibrosis by equilibrating local ACE-AngII-AT1R/ACE2-Ang1-7-Mas axis in rat. J. Ethnopharmacol. 2020;250:112470. doi: 10.1016/j.jep.2019.112470. [DOI] [PubMed] [Google Scholar]
- 116.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Villapol S., Saavedra J.M. Neuroprotective effects of angiotensin receptor blockers. Am. J. Hypertens. 2015;28(3):289–299. doi: 10.1093/ajh/hpu197. [DOI] [PubMed] [Google Scholar]
- 118.Balakumar P., Jagadeesh G. EDITORIAL [Hot Topic-II: PPAR ligands and cardiovascular disorders: friend or Foe (Guest Editors: Pitchai Balakumar and Gowraganahalli Jagadeesh)] Curr. Mol. Pharmacol. 2012;5(2):219–223. doi: 10.2174/1874467211205020219. [DOI] [PubMed] [Google Scholar]
- 119.Guo T., Fan Y., Chen M., Wu X., Zhang L., He T., Wang H., Wan J., Wang X., Lu Z. Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19) JAMA Cardiol. 2020 doi: 10.1001/jamacardio.2020.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shi S., Qin M., Shen B., Cai Y., Liu T., Yang F., Gong W., Liu X., Liang J., Zhao Q., Huang H., Yang B., Huang C. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiol. 2020 doi: 10.1001/jamacardio.2020.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lippi G., Plebani M. Laboratory abnormalities in patients with COVID-2019 infection. Clin. Chem. Lab. Med. 2020 doi: 10.1515/cclm-2020-0198. [DOI] [PubMed] [Google Scholar]
- 122.Siddamreddy S., Thotakura R., Dandu V., Kanuru S., Meegada S. Corona virus disease 2019 (COVID-19) presenting as acute ST elevation myocardial infarction. Cureus. 2020;12(4) doi: 10.7759/cureus.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ferrario C.M., Jessup J., Chappell M.C., Averill D.B., Brosnihan K.B., Tallant E.A., Diz D.I., Gallagher P.E. Effect of angiotensin-converting enzyme inhibition and angiotensin II receptor blockers on cardiac angiotensin-converting enzyme 2. Circulation. 2005;111(20):2605–2610. doi: 10.1161/CIRCULATIONAHA.104.510461. [DOI] [PubMed] [Google Scholar]
- 124.Furuhashi M., Moniwa N., Mita T., Fuseya T., Ishimura S., Ohno K., Shibata S., Tanaka M., Watanabe Y., Akasaka H. Urinary angiotensin-converting enzyme 2 in hypertensive patients may be increased by olmesartan, an angiotensin II receptor blocker. Am. J. Hypertens. 2015;28(1):15–21. doi: 10.1093/ajh/hpu086. [DOI] [PubMed] [Google Scholar]
- 125.Li J., Wang X., Chen J., Zhang H., Deng A. Association of Renin-angiotensin system inhibitors with severity or risk of death in Patients with hypertension hospitalized for coronavirus disease 2019 (COVID-19) Infection in Wuhan, China. JAMA Cardiol. 2020 doi: 10.1001/jamacardio.2020.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Meng J., Xiao G., Zhang J., He X., Ou M., Bi J., Yang R., Di W., Wang Z., Li Z., Gao H., Liu L., Zhang G. Renin-angiotensin system inhibitors improve the clinical outcomes of COVID-19 patients with hypertension. Emerg. Microbes Infect. 2020;9(1):757–760. doi: 10.1080/22221751.2020.1746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xianwei W., Magomed K., Ding Z., Sona M., Jingjun L., Shijie L., Mehta J.L. Cross-talk between inflammation and angiotensin II: studies based on direct transfection of cardiomyocytes with AT1R and AT2R cDNA. Exp. Biol. Med. 2012;237(12):1394–1401. doi: 10.1258/ebm.2012.012212. [DOI] [PubMed] [Google Scholar]
- 128.Lovren F., Pan Y., Quan A., Teoh H., Wang G., Shukla P.C., Levitt K.S., Oudit G.Y., Al-Omran M., Stewart D.J., Slutsky A.S., Peterson M.D., Backx P.H., Penninger J.M., Verma S. Angiotensin converting enzyme-2 confers endothelial protection and attenuates atherosclerosis. Am. J. Physiol. Heart Circ. Physiol. 2008;295(4):H1377–H1384. doi: 10.1152/ajpheart.00331.2008. [DOI] [PubMed] [Google Scholar]
- 129.Tikoo K., Patel G., Kumar S., Karpe P.A., Sanghavi M., Malek V., Srinivasan K. Tissue specific up regulation of ACE2 in rabbit model of atherosclerosis by atorvastatin: role of epigenetic histone modifications. Biochem. Pharmacol. 2015;93(3):343–351. doi: 10.1016/j.bcp.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 130.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281(18):4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Arabi Y.M., Alothman A., Balkhy H.H., Al-Dawood A., AlJohani S., Al Harbi S., Kojan S., Al Jeraisy M., Deeb A.M., Assiri A.M. Treatment of Middle East Respiratory Syndrome with a combination of lopinavir-ritonavir and interferon-β1b (MIRACLE trial): study protocol for a randomized controlled trial. Trials. 2018;19(1):81. doi: 10.1186/s13063-017-2427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jin Z., Zhao Y., Sun Y., Zhang B., Wang H., Wu Y., Zhu Y., Zhu C., Hu T., Du X., Duan Y., Yu J., Yang X., Yang X., Yang K., Liu X., Guddat L.W., Xiao G., Zhang L., Yang H., Rao Z. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat. Struct. Mol. Biol. 2020:1–4. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 135.Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020 doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., Hu Y., Luo G., Wang K., Lu Y., Li H., Wang S., Ruan S., Yang C., Mei C., Wang Y., Ding D., Wu F., Tang X., Ye X., Ye Y., Liu B., Yang J., Yin W., Wang A., Fan G., Zhou F., Liu Z., Gu X., Xu J., Shang L., Zhang Y., Cao L., Guo T., Wan Y., Qin H., Jiang Y., Jaki T., Hayden F.G., Horby P.W., Cao B., Wang C. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395(10236):1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xie P., Ma W., Tang H., Liu D. Severe COVID-19: a review of recent progress with a look toward the future. Front. Public Health. 2020;8:189. doi: 10.3389/fpubh.2020.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Guzik T.J., Mohiddin S.A., Dimarco A., Patel V., Savvatis K., Marelli-Berg F.M., Madhur M.S., Tomaszewski M., Maffia P., D'Acquisto F., Nicklin S.A., Marian A.J., Nosalski R., Murray E.C., Guzik B., Berry C., Touyz R.M., Kreutz R., Wang D.W., Bhella D., Sagliocco O., Crea F., Thomson E.C., McInnes I.B. COVID-19 and the cardiovascular system: implications for risk assessment, diagnosis, and treatment options. Cardiovasc. Res. 2020 doi: 10.1093/cvr/cvaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.McGonagle D., O'Donnell J.S., Sharif K., Emery P., Bridgewood C. Immune mechanisms of pulmonary intravascular coagulopathy in COVID-19 pneumonia. Lancet Rheumatol. 2020 doi: 10.1016/S2665-9913(20)30121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020:1–3. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.C.N.H. Commission . 2020. Chinese Clinical Guidance For COVID-19 Pneumonia Diagnosis and Treatment. 4 March 2020. [Google Scholar]
- 142.Li G., He X., Zhang L., Ran Q., Wang J., Xiong A., Wu D., Chen F., Sun J., Chang C. Assessing ACE2 expression patterns in lung tissues in the pathogenesis of COVID-19. J. Autoimmun. 2020;102463 doi: 10.1016/j.jaut.2020.102463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.D’Alessandro S., Scaccabarozzi D., Signorini L., Perego F., Ilboudo D.P., Ferrante P., Delbue S. The use of antimalarial drugs against viral infection. Microorganisms. 2020;8(1):85. doi: 10.3390/microorganisms8010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schrezenmeier E., Dorner T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat. Rev. Rheumatol. 2020;16(3):155–166. doi: 10.1038/s41584-020-0372-x. [DOI] [PubMed] [Google Scholar]
- 145.Zhou Y., Fu B., Zheng X., Wang D., Zhao C., Qi Y., Sun R., Tian Z., Xu X., Wei H. Aberrant pathogenic GM-CSF+ T cells and inflammatory CD14+ CD16+ monocytes in severe pulmonary syndrome patients of a new coronavirus. BioRxiv. 2020 doi: 10.1101/2020.02.12.945576. [DOI] [Google Scholar]
- 146.Mihara M., Hashizume M., Yoshida H., Suzuki M., Shiina M. IL-6/IL-6 receptor system and its role in physiological and pathological conditions. Clin. Sci. (Lond.) 2012;122(4):143–159. doi: 10.1042/CS20110340. [DOI] [PubMed] [Google Scholar]
- 147.Uciechowski P., Dempke W.C.M. Interleukin-6: A Masterplayer in the Cytokine Network. Oncology. 2020;98(3):131–137. doi: 10.1159/000505099. [DOI] [PubMed] [Google Scholar]
- 148.Mihai C., Dobrota R., Schroder M., Garaiman A., Jordan S., Becker M.O., Maurer B., Distler O. COVID-19 in a patient with systemic sclerosis treated with tocilizumab for SSc-ILD. Ann. Rheum. Dis. 2020;79(5):668–669. doi: 10.1136/annrheumdis-2020-217442. [DOI] [PubMed] [Google Scholar]
- 149.Ferro F., Elefante E., Baldini C., Bartoloni E., Puxeddu I., Talarico R., Mosca M., Bombardieri S. COVID-19: the new challenge for rheumatologists. Clin. Exp. Rheumatol. 2020;38(2):175–180. [PubMed] [Google Scholar]
- 150.Virtanen A.T., Haikarainen T., Raivola J., Silvennoinen O. Selective JAKinibs: prospects in inflammatory and autoimmune diseases. Biodrugs. 2019;33(1):15–32. doi: 10.1007/s40259-019-00333-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fragoulis G.E., McInnes I.B., Siebert S. JAK-inhibitors. new players in the field of immune-mediated diseases, beyond rheumatoid arthritis. Rheumatology. 2019;58(Supplement_1):i43–i54. doi: 10.1093/rheumatology/key276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet (London, England) 2020;395(10223) doi: 10.1016/S0140-6736(20)30304-4. e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shakoory B., Carcillo J.A., Chatham W.W., Amdur R.L., Zhao H., Dinarello C.A., Cron R.Q., Opal S.M. Interleukin-1 receptor blockade is associated with reduced mortality in sepsis patients with features of the macrophage activation syndrome: re-analysis of a prior Phase III trial. Crit. Care Med. 2016;44(2):275. doi: 10.1097/CCM.0000000000001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Deftereos S.G., Siasos G., Giannopoulos G., Vrachatis D.A., Angelidis C., Giotaki S.G., Gargalianos P., Giamarellou H., Gogos C., Daikos G., Lazanas M., Lagiou P., Saroglou G., Sipsas N., Tsiodras S., Chatzigeorgiou D., Moussas N., Kotanidou A., Koulouris N., Oikonomou E., Kaoukis A., Kossyvakis C., Raisakis K., Fountoulaki K., Comis M., Tsiachris D., Sarri E., Theodorakis A., Martinez-Dolz L., Sanz-Sanchez J., Reimers B., Stefanini G.G., Cleman M., Filippou D., Olympios C.D., Pyrgakis V.N., Goudevenos J., Hahalis G., Kolettis T.M., Iliodromitis E., Tousoulis D., Stefanadis C. The Greek study in the effects of colchicine in Covid-19 complications prevention (GRECCO-19 study): Rationale and study design. Hell. J. Cardiol. 2020 doi: 10.1016/j.hjc.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lurie N., Saville M., Hatchett R., Halton J. Developing Covid-19 vaccines at pandemic speed. N. Engl. J. Med. 2020;382:1969–1973. doi: 10.1056/NEJMp2005630. [DOI] [PubMed] [Google Scholar]
- 156.Chen W.H., Strych U., Hotez P.J., Bottazzi M.E. The SARS-CoV-2 vaccine pipeline: an overview. Curr. Trop. Med. Rep. 2020:1–4. doi: 10.1007/s40475-020-00201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.T.S.A. Board The Science Advisory Board. 2020. https://www.scienceboard.net/index.aspx?sec=sup&sub=bioproc&pag=dis&ItemID=677
- 158.Amanat F., Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52(4):583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pulendran B., Ahmed R. Immunological mechanisms of vaccination. Nat. Immunol. 2011;12(6):509–517. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang C., Li W., Drabek D., Okba N.M., van Haperen R., Osterhaus A.D., van Kuppeveld F.J., Haagmans B.L., Grosveld F., Bosch B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11(1):1–6. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Deng X., Yu X., Pei J. 2020. Regulation of Interferon Production as a Potential Strategy for COVID-19 Treatment. arXiv preprint arXiv:2003.00751. [Google Scholar]
- 162.Rossi J.J., Rossi D. Oligonucleotides and the COVID-19 pandemic: a perspective. Nucl. Acid Ther. 2020 doi: 10.1089/nat.2020.0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Terpos E., Ntanasis-Stathopoulos I., Elalamy I., Kastritis E., Sergentanis T.N., Politou M., Psaltopoulou T., Gerotziafas G., Dimopoulos M.A. Hematological findings and complications of COVID-19. Am. J. Hematol. 2020 doi: 10.1002/ajh.25829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Liu B., Li M., Zhou Z., Guan X., Xiang Y. Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)? J. Autoimmun. 2020;102452 doi: 10.1016/j.jaut.2020.102452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Burgstaller S., Pache L., De Jesus P., Hull M.V., Chang M. 2020. A Large-scale Drug Repositioning Survey for SARS-CoV-2 Antivirals, bioRxiv. [DOI] [Google Scholar]
- 166.Bittmann S., Luchter E., Moschüring-Alieva E., Villalon G., Weissenstein A.C. 19: Camostat and The Role of Serine Protease Entry Inhibitor TMPRSS2. J. Regen Biol. Med. 2020;2(2):1–2. doi: 10.14740/jocmr4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hoffmann M., Kleine-Weber H., Krüger N., Mueller M.A., Drosten C., Pöhlmann S. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. BioRxiv. 2020 doi: 10.1101/2020.01.31.929042. [DOI] [Google Scholar]
- 168.Rosa S.G.V., Santos W.C. Clinical trials on drug repositioning for COVID-19 treatment. Rev. Panam. Salud Publica. 2020;44 doi: 10.26633/RPSP.2020.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lai C.-C., Shih T.-P., Ko W.-C., Tang H.-J., Hsueh P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55(3) doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Guo J., Huang Z., Lin L., Lv J. Coronavirus disease 2019 (COVID-19) and cardiovascular disease: a viewpoint on the potential influence of Angiotensin-Converting enzyme inhibitors/angiotensin receptor blockers on onset and Severity of severe acute respiratory syndrome coronavirus 2 infection. J. Am. Heart Assoc. 2020;9(7) doi: 10.1161/JAHA.120.016219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Medina D., Arnold A.C. Angiotensin-(1-7): Translational Avenues in Cardiovascular Control. Am. J. Hypertens. 2019;32(12):1133–1142. doi: 10.1093/ajh/hpz146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Alhenc-Gelas F., Drueke T.B. Blockade of SARS-CoV-2 infection by recombinant soluble ACE2. Kidney Int. 2020;97(6):1091–1093. doi: 10.1016/j.kint.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Batlle D., Wysocki J., Satchell K. Soluble angiotensin-converting enzyme 2: a potential approach for coronavirus infection therapy? Clin. Sci. 2020;134(5):543–545. doi: 10.1042/CS20200163. [DOI] [PubMed] [Google Scholar]
- 174.Santos S.H.S., Andrade J.M.O. Angiotensin 1–7: A peptide for preventing and treating metabolic syndrome. Peptides. 2014;59:34–41. doi: 10.1016/j.peptides.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 175.Peiró C., Moncada S. Substituting angiotensin-(1-7) to prevent lung damage in SARSCoV2 infection? Circulation. 2020 doi: 10.1161/CIRCULATIONAHA.120.047297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Vaduganathan M., Vardeny O., Michel T., McMurray J.J., Pfeffer M.A., Solomon S.D. Renin–angiotensin–aldosterone system inhibitors in patients with Covid-19. N. Engl. J. Med. 2020;382:1653–1659. doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Verdecchia P., Angeli F., Reboldi G. Angiotensin-converting enzyme inhibitors, angiotensin II receptor blockers and coronavirus. J. Hypertens. 2020;38(6):1190–1191. doi: 10.1097/HJH.0000000000002469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Talreja H., Tan J., Dawes M., Supershad S., Rabindranath K., Fisher J., Valappil S., van der Merwe V., Wong L., van der Merwe W. A consensus statement on the use of angiotensin receptor blockers and angiotensin converting enzyme inhibitors in relation to COVID-19 (corona virus disease 2019) The New Zealand Med. J. 2020;133(1512):85–87. [PubMed] [Google Scholar]
- 179.Kai H., Kai M. Interactions of coronaviruses with ACE2, angiotensin II, and RAS inhibitors—lessons from available evidence and insights into COVID-19. Hypertens. Res. 2020:1–7. doi: 10.1038/s41440-020-0455-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Brown J.D. Antihypertensive drugs and risk of COVID-19? Lancet Respir. Med. 2020;8(5) doi: 10.1016/S2213-2600(20)30158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mahnam K., Hejazi H., Damavandi M., Sadeghi P., Zeinalian M., Tabesh F., Mirbod S., Khanahmad H. 2020. Inhibition of Viral Macrodomain of COVID-19 and Human TRPM2 by Losartan. [Google Scholar]
- 182.Zeinalian M., Salari-Jazi A., Jannesari A., Khanahmad H. A potential protective role of losartan against coronavirus-induced lung damage. Infect. Control Hosp. Epidemiol. 2020;41(6):752–753. doi: 10.1017/ice.2020.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rothlin R.P., Vetulli H.M., Duarte M., Pelorosso F.G. Telmisartan as tentative angiotensin receptor blocker therapeutic for COVID-19. Drug Dev. Res. 2020 doi: 10.1002/ddr.21679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Bifulco M., Gazzerro P. Statins in coronavirus outbreak: it’s time for experimental and clinical studies. Pharmacol. Res. 2020;156:104803. doi: 10.1016/j.phrs.2020.104803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Katsiki N., Banach M., Mikhailidis D.P. Lipid-lowering therapy and renin-angiotensin-aldosterone system inhibitors in the era of the COVID-19 pandemic. Arch. Med. Sci. 2020;16(3):485–489. doi: 10.5114/aoms.2020.94503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Castiglione V., Chiriacò M., Emdin M., Taddei S., Vergaro G. Statin therapy in COVID-19 infection. Eur. Heart J. Cardiovasc. Pharmacother. 2020;6(4):258–259. doi: 10.1093/ehjcvp/pvaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;14(1):72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 188.Andreania J., Le Bideaua M., Duflota I., Jardota P., Rollanda C., Boxbergera M., Khalila J.Y.B., Baudouinb J.-P., Wurtza N., Rolaina J.-M. In vitro testing of hydroxychloroquine and azithromycin on SARS-CoV-2 shows 1 synergistic effect 2. Lung. 2020;21:22. [Google Scholar]
- 189.Baldelli S., Corbellino M., Clementi E., Cattaneo D., Gervasoni C. Lopinavir/ritonavir in COVID-19 patients: maybe yes, but at what dose? J. Antimicrob. Chemother. 2020 doi: 10.1093/jac/dkaa190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hung I.F.-N., Lung K.-C., Tso E.Y.-K., Liu R., Chung T.W.-H., Chu M.-Y., Ng Y.-Y., Lo J., Chan J., Tam A.R. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020 doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Davis D.A., Soule E.E., Davidoff K.S., Daniels S.I., Naiman N.E., Yarchoan R. Activity of human immunodeficiency virus type 1 protease inhibitors against the initial autocleavage in Gag-Pol polyprotein processing. Antimicrob. Agents Chemother. 2012;56(7):3620–3628. doi: 10.1128/AAC.00055-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Du Y.X., Chen X.P. Favipiravir: pharmacokinetics and concerns about clinical trials for 2019-nCoV infection. Clin. Pharmacol. Ther. 2020;108:242–247. doi: 10.1002/cpt.1844. [DOI] [PubMed] [Google Scholar]
- 193.Elfiky A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.-X. Compassionate use of remdesivir for patients with severe COVID-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Rouviere C.P., Dousson C.B., Tavis J.E. HBV replication inhibitors. Antivir. Res. 2020;179 doi: 10.1016/j.antiviral.2020.104815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Jockusch S., Tao C., Li X., Anderson T.K., Chien M., Kumar S., Russo J.J., Kirchdoerfer R., Ju J. 2020. Triphosphates of the Two Components in DESCOVY and TRUVADA are Inhibitors of the SARS-CoV-2 Polymerase, bioRxiv. [DOI] [Google Scholar]
- 197.Matsuyama S., Kawase M., Nao N., Shirato K., Ujike M., Kamitani W., Shimojima M., Fukushi S. 2020. The Inhaled Corticosteroid Ciclesonide blocks Coronavirus RNA Replication by Targeting Viral NSP15, bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Nakajima K., Ogawa F., Sakai K., Uchiyama M., Oyama Y., Kato H., Takeuchi I., Nakajima K. A Case of Coronavirus Disease 2019 Treated With Ciclesonide. Mayo Clin. Proc. 2020;95(6):1295–1297. doi: 10.1016/j.mayocp.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ferreira B.I., Cautain B., Grenho I., Link W. Small molecule inhibitors of CRM1. Front. Pharmacol. 2020;11:625. doi: 10.3389/fphar.2020.00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved Drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Misra D.P., Agarwal V., Gasparyan A.Y., Zimba O. Rheumatologists perspective on coronavirus disease 19 (COVID-19) and potential therapeutic targets. Clin. Rheumatol. 2020:1–8. doi: 10.1007/s10067-020-05073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Gritti G., Raimondi F., Ripamonti D., Riva I., Landi F., Alborghetti L., Frigeni M., Damiani M., Micò C., Fagiuoli S. Use of siltuximab in patients with COVID-19 pneumonia requiring ventilatory support. MedRxiv. 2020 doi: 10.1101/2020.04.01.20048561. [DOI] [Google Scholar]
- 203.Roumier M., Paule R., Valle A., Ackermann F. medrxiv; 2020. Interleukin-6 Blockade for Severe COVID-19. [DOI] [Google Scholar]
- 204.Stebbing J., Phelan A., Griffin I., Tucker C., Oechsle O., Smith D., Richardson P. COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infect. Dis. 2020;20(4):400–402. doi: 10.1016/S1473-3099(20)30132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zheng Y.-F., Li R., Liu S. Immunoregulation with mTOR inhibitors to prevent COVID‐19 severity: A novel intervention strategy beyond vaccines and specific antiviral medicines. J. Med. Virol. 2020 doi: 10.20944/preprints202004.0060.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Carr A.C. A new clinical trial to test high-dose vitamin C in patients with COVID-19. Crit. Care. 2020;24(1):133. doi: 10.1186/s13054-020-02851-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Boretti A., Banik B.K. Intravenous vitamin C for reduction of cytokines storm in acute respiratory distress syndrome. PharmaNutrition. 2020;12:100190. doi: 10.1016/j.phanu.2020.100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Cohen S., Fishman P. Targeting the A3 adenosine receptor to treat cytokine release syndrome in cancer immunotherapy. Drug Des. Devel. Ther. 2019;13:491–497. doi: 10.2147/DDDT.S195294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Dastan F., Tabarsi P., Marjani M., Moniri A., Hashemian S.M., Tavakoli-Ardakani M., Saffaei A. Thalidomide against coronavirus disease 2019 (COVID-19): A medicine with a thousand faces. Iran. J. Pharm. Res. 2020;19(1):1–2. doi: 10.22037/ijpr.2020.113369.14259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Alvarez R.A., Berra L., Gladwin M.T. Home NO therapy for COVID-19. Am. J. Respir. Crit. Care Med. 2020;ja doi: 10.1164/rccm.202005-1906ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Osman A.H. COVID-19: Targeting the cytokine storm via cholinergic anti-inflammatory (Pyridostigmine) Int. J. Clin. Virol. 2020;4:041–046. doi: 10.29328/journal.ijcv.1001014. [DOI] [Google Scholar]
- 212.Annemarie H., Lascano A.M., Lalive P.H. Management of patients with generalised myasthenia gravis and COVID-19: four case reports. J. Neurol. Neurosurgery Psychiatry. 2020 doi: 10.1136/jnnp-2020-323565. [DOI] [PubMed] [Google Scholar]
- 213.Chong E.A., Roeker L.E., Shadman M., Davids M.S., Schuster S.J., Mato A.R. BTK inhibitors in cancer patients with COVID19:" The winner will be the one who controls that chaos"(Napoleon Bonaparte) Clin. Cancer Res. 2020 doi: 10.1158/1078-0432.CCR-20-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Robertson C.E. Could CGRP antagonists be helpful in the fight against COVID-19? Headache J. Head Face Pain. 2020;10(1111):13853. doi: 10.1111/head.13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Farag A., Wang P., Ahmed M., Sadek H. 2020. Identification of FDA Approved Drugs Targeting COVID-19 Virus by Structure-Based Drug Repositioning. [DOI] [Google Scholar]
- 217.Ogen Y. Assessing nitrogen dioxide (NO2) levels as a contributing factor to the coronavirus (COVID-19) fatality rate. Sci. Total Environ. 2020;726:138605. doi: 10.1016/j.scitotenv.2020.138605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Chen R., Zhang Y., Huang L., Cheng B.H., Xia Z.Y., Meng Q.T. Safety and efficacy of different anesthetic regimens for parturients with COVID-19 undergoing Cesarean delivery: a case series of 17 patients. Can. J. Anaesth. 2020;67(6):655–663. doi: 10.1007/s12630-020-01630-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Willis M.D., Robertson N.P. Multiple sclerosis and the risk of infection: considerations in the threat of the novel coronavirus, COVID-19/SARS-CoV-2. J. Neurol. 2020;267(5):1567–1569. doi: 10.1007/s00415-020-09822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Lythgoe M.P., Middleton P. Ongoing clinical trials for the management of the COVID-19 pandemic. Trends Pharmacol. Sci. 2020;41(6):363–382. doi: 10.1016/j.tips.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material