Abstract
SARS-CoV-2 incidence and mortality in Europe have shown wide variation. Northern Italy in particular the Lombardy region, north-eastern French regions, Switzerland and Belgium were amongst the hardest hit, while the central and southern Italian regions, all the Balkan countries from Slovenia to Greece and the Islands of Malta and Cyprus had much fewer cases and deaths per capita, and deaths per number of cases. Differences in public health measures, and health care delivery, in the author’s opinion, can only partly explain the difference. The geographical distribution of Phlebotomus sand-flies and the relative distribution of arthropod borne diseases Leishmaniasis and Phlebovirus infections especially the Sicilian Sandfly fever group corresponds to most areas of low prevalence of SARS-CoV-2. A hypothesis is proposed whereby repeated arthropod or sandfly vector infection of humans by novel viruses of zoonotic origins carrying bat or mammalian RNA/DNA, such as phleboviruses may have resulted in the development of an effective evolutionary immune response to most novel zoonotic viruses such as SARS-CoV-2 by means of survival of the fittest possibly over many generations. This process probably ran in parallel and concurrent with the progressive evolution of novel coronaviruses which spread from one mammalian species to another. Other possible, but less likely mechanisms for the role of sandfly meals within a much shorter time frame may have led to, (i) previous exposure and infection of humans with the SARS-Cov-2 virus itself, or a closely related corona virus in the previous decades, or (ii) exposure of human populations to parts coronavirus protein namely either S or more likely N protein carried mechanically by arthropods, but without clinical disease causing direct immunity or (iii) by causing infection with other arthropod borne viruses which could carry bat DNA/RNA and have similar functional proteins resulting in an immediate cross-reactive immune response rather than by natural selection. The Evidence possibly supporting or disputing this hypothesis is reviewed, however the major problem with the hypothesis is that to date no coronavirus has ever been isolated from arthropods. Such a hypothesis can only be supported by research investigating the possible biological relationship of arthropods and coronaviruses where paradoxically they may be promoting immunity rather than disease.
Keywords: SARS-CoV-2, Phlebomotus, Arthropod borne viruses, Immunity, Coronavirus, Phlebovirus
Introduction.
Novel Corona Virus SARS-CoV-2 has caused a global pandemic which is probably the worst to hit the world since the Spanish Flu in 1918 and HIV [1]. Phylogenetic analysis of the SARS-Cov-2 genome indicates that the virus is closely related (with 88% identity) to two bat-derived SARS-like coronaviruses collected in 2018 in eastern China (bat-SL-CoVZC45 and bat-SL-CoVZXC21) and genetically distinct from SARS-CoV-1(with about 79% similarity) and MERS-CoV [1]. How bat viruses actually cross species is not known for certain however Calisher et al questioned in their review of SARS-CoV-1 and other pathogenic viruses originating from bats speculate that ”Perhaps these emerging bat viruses are naturally transmitted by arthropods or by other potential vectors that have not been examined” and “Are insectivorous bats intermediate hosts between insects and vertebrates (or plants)?” [2]
Publicly available data for SARS-Cov-2 reported by European Union countries to ECDC, regional data for Italy was taken from the Dipartimento (department) di Protezione Civile website, Regional data for France was obtained from coronavirus.fr provided by the French health ministry was utilized.(French data was only limited to hospitals). Regional swabbing rates for French regions were calculated on the national average. ECDC data did not include the age groups of cases or deaths in their downloadable data even though it appears to have been provided by national authorities.
Fig. 1 and Table 1 show the death rates, testing rates and positive cases per million population and the case fatality rate (CFR) on the 1st June 2020. In the Alpine regions of northern Italy the epidemic had a major impact on mortality and morbidity to much higher levels when compared to central and southern Italy[3] and other southern European countries such as Malta, Cyprus and Greece, and the Balkan countries up to Slovenia and Bulgaria have also have data similar to southern Italy.[4] On the other hand alpine countries bordering with Italy had rates comparable to Lombardy, while north western European countries experienced even higher levels.
Fig. 1.
Total Mortality rate per million from SARS-COV-2 (1st June 2020).
Table 1.
Total positive cases, Total deaths, and Case fatality rate per million population. (1st June 2020).
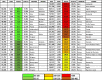 |
Fig. 2 shows the time trends of positive cases per million expressed as the moving 7 day average for Cyprus, Greece, Croatia, and Sicily and shows remarkable similarity in the time trend and the extent of the peak number of cases. There was only a statistically lower difference for daily cases per million in Greece. (p < 0.001, using unpaired t test, and kruskall wallis) when compared to Sicily.
Fig. 2.
Daily new positive cases per million population (7 days average) 4 Southern European countries/regions.
The geographical distribution of low prevalence appears to be remarkably similar to the geographical distribution of phlebotomus sandflies in Europe (Fig. 3, Fig. 4 ). The sandfly species is the vector of two diseases namely leishmaniaisis and phlebovirus infections of the Sicilian sandfly fever virus group which are prevalent in southern Europe. Notably sandflies feed on many animals including bats, cats, dogs, hares, rodents, cattle besides humans [5], [6].
Fig. 3.
Geographical distribution of Phlebotomus Pappatasi (European Centre for Disease control).
Fig. 4.
Geographical distribution of Phlebotomus Perfiliewi (European Centre for Disease control).
Proposed hypothesis
The geographical distribution of the SARS-Cov-2 in southern Europe is highly suggestive of herd immunity, or perhaps a more effective immune response to this novel virus. These areas of low prevalence and mortality are very similar to the distribution of Phlebotomus sandflies in Europe and the associated arthropod-borne disease caused by phleboviruses of the Sicilian Sandfly fever group. A hypothesis is proposed whereby repeated arthropod or sandfly vector infection of humans by novel viruses of zoonotic origins carrying bat or mammalian DNA or RNA possibly since many generations, may have resulted in the development of an effective evolutionary immune response to most novel zoonotic viruses such as SARS-CoV-2 by means of survival of the fittest. This process probably ran in parallel and concurrent with the progressive evolution of novel coronaviruses which spread from one mammalian species to another. Phylogenetic studies on RNA-dependent RNA polymerase (RdRp) sequences, suggest that a common bat ancestor for CoVs infecting most mammals and humans, appeared about 7000–8000 years ago. Other possible, but less likely mechanisms for the role of sandfly meals within a much shorter time frame may include, (i) previous exposure and infection of humans to the SARS-Cov-2 virus itself , or a closely related corona virus in the previous decades, (ii) exposure of human populations to parts of SARS-Cov-2 RNA or coronavirus protein namely either S or more likely N protein, but without clinical disease causing direct immunity or (iii) by causing infection with other arthropod borne viruses which could carry bat DNA/RNA and have similar functional proteins resulting in an immediate cross-reactive immune response rather than by natural selection.
Discussion
Limitations of the SARS-CoV-2 incidence and prevalence data
The swabbing testing rates showed great variation and complete data on the swabbing was unavailable for some regions. It is also unclear if the reported mortality data in the individual countries included deaths in hospitals only or in the community or both, and if deaths in the community were tested ante or post mortem for SARS-CoV-2. Another problem with the data is that it is not age standardized, so that mortality data is not directly comparable.
Is the epidemiological data indicative of herd immunity to SARS-Cov-2 in southern Europe?
As reviewed by Randolph et al [7] where the infrastructure and swabbing rates was high, one can assume that tracking and tracing, and subsequent isolation of cases would be more effective as for example Malta and Cyprus.
The level of preparedness, the organization of health services, and whether demand exceeded capacity would have significant impact on mortality. This was probably one component for the high death rates in the Lombardy and other northern Italian regions, or bordering states Switzerland and French regions. However all of the alpine countries and regions had high swabbing rates indicating a high level of organizational co-ordination. In contrast despite the fact that swabbing rates in Greece and the Balkan countries were comparatively low, the mortality per population and the CFR were still much lower than northern Italy, but very similar to central and southern Italian regions.
Where elderly patients in nursing homes, living independently in the community, and vulnerable groups such as the immunosuppressed were effectively isolated and quarantined early, the mortality would be kept low. There is no way to quantify this factor in the individual countries and regions. The problems of the highly regionalized Italian health care system were highlighted by Nardini et al.[8] Supporting evidence is provided by the CFR which was highest in Lombardy and neighbouring Italian regions. French data on CFR is actually higher, but must be interpreted with caution because it does not include community cases.
Countries with small population have the least logistical organizational problems as evidenced by the high testing rates. Adequate and timely public health responses may partially explain the very low case fatality rates. The similarity of data from Croatia, Sicily, Cyprus and Greece, which have very different health care systems weighs in favour of the argument that these areas had some form of herd immunity together with a health care system working within its capacity.
Northern Italian regions, Belgium, and north-eastern French regions had high mortality and high case fatality rates, while Austria and Switzerland had lower mortality rates
Within the limitations stated above, the argument for herd immunity in southern Europe is strongest from mortality data. Notwithstanding this, the author acknowledges that high incidence of disease that overwhelmed the health care system in the alpine regions amplified the difference in outcomes.
Geographical distribution of phlebotomous and Sicilian sand-fly fever. Can phlebotomus sandflies spread viral disease across species?
An association between disease and vector is usually made after the analysis of epidemiological data. Temporal and geographical patterns of disease occurrence and spread are then associated with vector distributions. [9]. Many viruses may be transmitted mechanically by arthropods, however it is notoriously difficult to implicate a vector by laboratory data alone. The mechanically transmitted virus must be resistant to inactivation and survive exposure on the insect's mouth parts until it next feeds. Both DNA and RNA viruses can fulfil these conditions, and there seems to be little vector specificity [9].
To date a medical literature search did not result in any study on whether phlebotomous species can carry or spread corona virus, or else contribute to exposing humans to corona virus RNA or any related proteins present in humans or other mammals such as bats, cats, dogs, sheep or cattle.
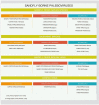
However based on the epidemiology of Leishmaniasis amongst mammals [10], the possibility of mechanical transmission of coronavirus RNA or proteins amongst different mammals is theoretically possible. Phleboviruses of the Sicilian sandfly fever group (Table 2 ) have been cultured from sandflies, but not from other mammals. Verani et al have isolated Toscana virus from bat brains, however no other authors have reported this finding again in the medical literature [11].
Table 2.
 |
Using large genomic and ecological datasets, based on RNA from known viruses and hosts Babayan et al predicted that the phlebovirus Sicilian sandfly fever Naples virus, has RNA from Pteropodiformes bats and even hooved mammals. Furthermore the bunyavirus group which includes phleboviruses predicted RNA from a wide variety of mammals including rodents and both Pteropodiformes and Vespertilioniformes bats [12]. This data also confirms that the sandfly was a possible for Flavivirus and Rhabdovirus.
No vector for the coronavirus group however was detected by this technique [12]. This may simply be the reflection that are no studies on coronaviruses in vectors in the database.
Sandflies in southern Europe are notorious because of transmission of visceral and cutaneous leishmaniasis a diseases caused by a protozoan [13]. However Phlebotomus sandflies are implicated in the transmission of several zoonotic viral agents, amongst which the most important are grouped into the Phlebovirus genus (family Bunyaviridae), which includes the sandfly fever Sicilian and Toscana and Naples viruses, and the Vesiculovirus genus (family Rhabdoviridae), which includes vesicular stomatitis, as well as the Chandipura and Isfahan viruses [13]. Table 2. Shows the currently known phleboviruses in the Mediterranean area.[14], [15]. It is likely that viral transmission by the sandflies is mechanical and not part of the lifecycle of either the vector or the virus itself.
Specifically, viruses belonging to the Sandfly fever Naples species were detected and isolated from P. perfiliewi, P. perniciosus, P. longicuspis, P. papataci, P. sergenti and Sergentomyia minuta. Viruses belonging to the tentative Sandfly fever Sicilian species and Corfu species were detected and isolated from P. ariasi, P. papatasi, P. neglectus, P. perniciosus and P. longicuspis. Viruses belonging to the Salehabad species were detected and isolated from P. perniciosus and P. perfiliewi.[6] Furthermore studies on bat guano (excrement) reveals that most bats are voracious insect eaters,[16]. To date, it has not been firmly established whether viruses are acquired by bats in this way.
Lessons on herd immunity in Military history
Military history indicates that, starting with Napoleonic forces in Egypt, Austrian troops in the Balkans, as was later shown by Alois Pict in 1887, that sandfly fever presented considerable risk to visitors in the Balkans while the local population was unaffected. In 1937 a small epidemic occurred in Greece and other outbreaks were described in German and British troops in the Mediterranean.[17] Sandfly fever Sicilian virus (SFSV) was first isolated, characterized and named Sicilian virus, from the serum of a US soldier by Sabin in 1943.[17] Since then Toscana TOSV and Naples and Sicilian-like groups have been described.[17]. Phleboviruses were either isolated or detected in humans by molecular techniques in France, Italy, Portugal, Greece, Albania, Croatia, Bosnia Herzegovina, Turkey, Iran, Tunisia, Algeria and Morocco in the last two decades.[15] One case of Toscana virus from Malta was reported in the international literature.[18]
Sero-epidemiological studies showed the presence of neutralizing antibodies against Toscana virus in several Mediterranean countries and middle east countries [19]. However, the rates vary depending on the region Mediterranean basin considered as endemic region of Toscana virus.
Bats and viral zoonosis
In recent years, bats have been increasingly recognized as important reservoir hosts for viruses that can cross species barriers to infect humans and other domestic and wild mammals [2]. Their role is important for hosts for alpahviruses, flavirviruses, rhabdoviruses and arenaviruses. Besides SARS-CoV-1, Nipah virus caused a major outbreak of encephalitis in humans and in pigs [16]. While Hendravirus caused another epidemic amongst horses [16], and Ebola led to another epidemic amongst humans.[16]
Coronaviruses (COVs)
The CoV family (Coronaviridae) has been described as a model in virology, because it infects more than 200 different hosts [20]. CoVs encode membrane-associated proteins that are incorporated into virions: spike (S), envelope (E), membrane (M), and nucleoprotein (N) [20]. Fig. 5 . Shows that Alphacoronavirus and Betacoronavirus include viruses that principally infect mammals, and are derived from the bat gene pool. Gammacoronavirus and Deltacoronavirus group viruses that infect birds and mammals and are derived from the avian and pig gene pool [21].
Fig. 5.
Corona virus Evolution. Is the sandfly the missing link? (Adapted from Woo et al [24]).
Phylogenetic studies, as shown of Fig. 5, on RNA-dependent RNA polymerase (RdRp) sequences, , suggested that the common bat ancestor, the most recent of the CoVs infecting mammals, appeared about 7000–8000 years ago, while the most recent common ancestor of avian CoVs dates back 10,000 years [20], [22]
Interspecies transmission bats cats and dogs and other mammals
According to Secondy “Host specificity is conditioned by the cell susceptibility to the virus, given by the host cell receptors, and by the cell permissivity that relies on the availability of cellular factors required for viral replication.” [23] Coronaviridae are the exception, as several viruses that are detected in humans have phylogenetic and genetic similarity to those isolated from other animal hosts [24], [25] These nonspecific properties that CoVs possess, may be due to accessory CoV genes, which are already thought to play a role in host tropism and adaptation to a new host. S-Glycoprotein (spike) appears to be the main determinant for the success of initial events of infection between species.
Canine Respiratory corona virus (CRCov) causes respiratory illness in dogs, Feline Coronavirus FeCOV causes mild diarrhoea and a carrier state in Cats. CRCov is widely prevalent according to serological studies.[26] in Italy (31.9%), Greece(38.5%), Spain (63.5%), and France (72.2%). [26]
The prevalence of antibodies to FCoV in the healthy field cats was found to vary between 14.6% in Japan to greater than 70% in Austria,50% in Switzerland[27]. Both viruses are thought to spread via the faeco-oral route.
While there is no evidence of spread to humans, more recent research points to a common ancestor between FCoV and CRCoV. An S protein from a yet-unknown virus was passed into the ancestor and gave rise to CCoV, whose S protein was again recombined into FCoV I to form FCoV II. [28]
S and N protein antibody cross reactivity is SARS-Cov-2.
A preliminary study from the United States of human sera collected prior to the SARS-CoV-2 pandemic demonstrates overall high IgG reactivity to common human coronaviruses and low IgG cross reactivity to epidemic coronaviruses including SARS-CoV-2. Cross-reactivity of conserved antigenic domains including S2 domain of spike protein and nucleocapsid N protein was noted. The S1 domain is strain specific and had a low antibody response. [29] This re-enforces the concept that while part of the virus was novel, other parts like S2 and N were not.
In another study by Amanat et al on human antibody response to SARS-Cov-2 , the majority of control subjects had strong reactivity to the spike protein of common human coronavirus NL63 and 229E, but showed no cross-reactivity to SARS-CoV-2 RBD and spike.[30] Cross-reactivity by SARS-CoV Receptor binding-specific antibodies with SARS-CoV-2 RBD protein was also noted, but not to other coronaviruses mainly due to its ability to bind with ACE 2 receptors.[31]. No serological cross-reactivity was detected between the SARS-CoV-2 and type I or II feline infectious peritonitis virus (FIPV) while it was noted that SARS-CoV-2 infected the cat population in Wuhan during the outbreak.[32]
Problems with the hypothesis
There is no published research showing that coronaviruses can be isolated from sandflies. Furthermore neither has corona virus RNA, or spike molecules or N proteins from any coronavirus been isolated from sandflies. No arthropod vector has been predicted to date in corona viruses by Babayan et al, [12].
SARS-CoV-2 attaches to the ACE-2 receptor which only occurs in two Chinese bat viruses, SARS-CoV-1 and 2 and HCoV‐NL63 in humans which causes bronchiolitis in children. [1] SARS-Cov-2 is presumably spread by respiratory droplets and not by blood thus excluding arthropod spread. However Young et al detected SARS-CoV-2 in the blood of one out 18 patients,[33] while Huang at el detected Viral RNA in the serum of 6 out of 41 SARS-CoV-2 patients [34]. 2 out of 10 civets inoculated intranasally with SARS-CoV-1 had virus isolated from their serum. This would make arthropod spread theoretically possible.
Exposures to infected urine and aerosols generated during defecation have been suggested as possible routes of intraspecies and interspecies transmission of viruses from bats. [35] Alphaviruses, flaviviruses, and bunyaviruses, may infect bats via arthropods, but it is not clear whether bats are important reservoir hosts for these viruses [2].
Research to test hypothesis
Case control serological studies comparing antibody titres to local phleboviruses, in patients with SARS-Cov-2, to individuals exposed to SARS-Cov-2 who remained asymptomatic, and healthy controls could give an indication of cross immunity, can be performed.
Research is necessary to establish the presence of coronaviruses, coronavirus RNA, or functional proteins S and N, within sandflies and to extend the use molecular techniques looking for arthropod genes within coronavirus and vice versa, and widen the search performed by Babayan et al.[12].
Immunological techniques focusing on the human immune response in patients from different geographical areas, in particular looking for antibody response to S and N proteins in SARS-Cov-2, looking for cross immunity between other species of arthropod borne viruses and coronaviruses [29].
A much wider view and consequent research in novel arthropod borne diseases particularly those arising from bats may be necessary.
Conclusion
SARS-CoV, SARS-CoV-2 are a distant relative of the group alpha and beta coronaviruses that infect rodents, cattle, dogs, pigs, and humans. All coronaviruses affecting mammals have evolved in the last 10,000 years and carry common bat ancestory, probably coinciding with the start of human civilization and farming [20], [22] . The sandfly is able to inoculate large numbers of individual mammals including bats, cats, dogs, hares causing leishmaniasis [36]. The sandfly is also able to transmit phlebovirus effectively amongst humans and possibly to bats [10], [37]. To date sandflies have not been shown to carry corona viruses, however repeated exposure to novel arthropod borne viruses can lead to a more reactive immune system by natural selection.
There seems to be cross reactivity to corona virus spike or N protein present in the different species of coronavirus which may enhance immunity to novel corona [30], [31], however cross reactivity with other arthropod borne viruses such as bunyaviruses or other arthropod borne viruses has not been demonstrated.
Such a hypothesis can only be supported by research investigating the possible biological relationship of arthropods and coronaviruses where paradoxically they may be promoting immunity rather than disease.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mehy.2020.110121.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19(3):531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dipartimento di Protezione Civile COVID-19 Italia - . Monitoraggio della situazione.
- 4.ECDC. Data on the geographic distribution of COVID-19 cases worldwide; 2020.
- 5.Stahn B., Sudeck H., Frickmann H., Kruger A., Burchard H.G., Wiemer D. Sandfly fever-a “neglected” disease. Hautarzt. 2018;69(11):928–937. doi: 10.1007/s00105-018-4251-1. [DOI] [PubMed] [Google Scholar]
- 6.Ayhan N., Charrel R.N. Of phlebotomines (sandflies) and viruses: a comprehensive perspective on a complex situation. Curr Opin Insect Sci. 2017;22:117–124. doi: 10.1016/j.cois.2017.05.019. [DOI] [PubMed] [Google Scholar]
- 7.Randolph H.E., Barreiro L.B. Herd Immunity: Understanding COVID-19. Immunity. 2020;52(5):737–741. doi: 10.1016/j.immuni.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nardini S.G., Sanguinetti C.M., De Benedetto F., Baccarani C., Del Donno M., Polverino M. SARS-CoV-2 pandemic in Italy: ethical and organizational considerations. Multidisciplinary. Respir Med. 2020;15 doi: 10.4081/mrm.2020.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carn V.M. The role of dipterous insects in the mechanical transmission of animal viruses. Br Vet J. 1996;152(4):377–393. doi: 10.1016/s0007-1935(96)80033-9. [DOI] [PubMed] [Google Scholar]
- 10.T.D. Serafim E. Iniguez F. Oliveira Leishmania infantum Trends in Parasitol 2020;36(1):80-. [DOI] [PubMed]
- 11.Verani P., Ciufolini M.G., Caciolli S., Renzi A., Nicoletti L., Sabatinelli G. Ecology of Viruses Isolated from Sand Flies in Italy and Characterization of a New Phlebovirus (Arbia Virus) Am J Trop Med Hygiene. 1988;38(2):433–439. doi: 10.4269/ajtmh.1988.38.433. [DOI] [PubMed] [Google Scholar]
- 12.Babayan S.A., Orton R.J., Streicker D.G. Predicting reservoir hosts and arthropod vectors from evolutionary signatures in RNA virus genomes. Science. 2018;362(6414):577–580. doi: 10.1126/science.aap9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maroli M., Feliciangeli M.D., Bichaud L., Charrel R.N., Gradoni L. Phlebotomine sandflies and the spreading of leishmaniases and other diseases of public health concern. Med Vet Entomol. 2013;27(2):123–147. doi: 10.1111/j.1365-2915.2012.01034.x. [DOI] [PubMed] [Google Scholar]
- 14.Ayhan N. Emerging sandfly-borne Phleboviruses in Balkan countries : virus isolation, characterization, evolution and seroepidemiology 2017.
- 15.Ayhan N., Charrel R.N. Emergent Sand Fly-Borne Phleboviruses in the Balkan Region. Emerg Infect Dis. 2018;24(12):2324–2330. [Google Scholar]
- 16.Scott H., Newman H.F., Epstein J., de Jong C. Investigating the role of Bats in Emerging Zoonosis. FAO Animal Product Health. 2011;12 [Google Scholar]
- 17.Alkan C., Bichaud L., de Lamballerie X., Alten B., Gould E.A., Charrel R.N. Sandfly-borne phleboviruses of Eurasia and Africa: Epidemiology, genetic diversity, geographic range, control measures. Antiviral Res. 2013;100(1):54–74. doi: 10.1016/j.antiviral.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Schultze D., Korte W., Rafeiner P., Niedrig M. First report of sandfly fever virus infection imported from Malta into Switzerland, October 2011. Eurosurveillance. 2012;17(27):20209. doi: 10.2807/ese.17.27.20209-en. [DOI] [PubMed] [Google Scholar]
- 19.Tesh R.B., Saidi S., Gajdamovic S.J., Rodhain F., Vesenjak-Hirjan J. Serological studies on the epidemiology of sandfly fever in the Old World. Bull World Health Organ. 1976;54(6):663–674. [PMC free article] [PubMed] [Google Scholar]
- 20.Kasmi Y., Khataby K., Souiri A., Ennaji M.M. Chapter 7 - Coronaviridae: 100,000 Years of Emergence and Reemergence. In: Ennaji M.M., editor. Emerging and Reemerging Viral Pathogens. Academic Press; 2020. pp. 127–149. [Google Scholar]
- 21.Woo P.C.Y., Lau S.K.P., Lam C.S.F., Lau C.C.Y., Tsang A.K.L., Lau J.H.N. Discovery of Seven Novel Mammalian and Avian Coronaviruses in the Genus Deltacoronavirus Supports Bat Coronaviruses as the Gene Source of Alphacoronavirus and Betacoronavirus and Avian Coronaviruses as the Gene Source of Gammacoronavirus and Deltacoronavirus. J Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong S., Lau S., Woo P., Kwok-Yung Y. Bats as a continuing source of emerging infections in humans. Rev Med Virol. 2007;17(2):67–91. doi: 10.1002/rmv.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segondy M. Spécificité d’hôte des virus et passages inter-espèces. Revue Francophone des Laboratoires. 2010;2010(423):37-42. [DOI] [PMC free article] [PubMed]
- 24.Woo P.C.Y., Lau S.K.P., Huang Y., Yuen K.-Y. Coronavirus Diversity, Phylogeny and Interspecies Jumping. Exp Biol Med. 2009;234(10):1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- 25.J.-F.-W. Chan K.-K.-W. To H. Tse D.-Y. Jin K.-Y. Yuen Interspecies transmission and emergence of novel viruses: lessons from bats and birds Trend Microbiol 2013;21(10):544. [DOI] [PMC free article] [PubMed]
- 26.Mitchell J.A., Cardwell J.M., Leach H., Walker C.A., Le Poder S., Decaro N. European surveillance of emerging pathogens associated with canine infectious respiratory disease. Vet Microbiol. 2017;212:31–38. doi: 10.1016/j.vetmic.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kummrow M., Meli M.L., Haessig M., Goenczi E., Poland A., Pedersen N.C. Feline Coronavirus Serotypes 1 and 2: Seroprevalence and Association with Disease in Switzerland. Clin Diagn Lab Immunol. 2005;12(10):1209. doi: 10.1128/CDLI.12.10.1209-1215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gary R.W., Nicole M.A., Jean Kaoru M., Ana F.-S., Bart L.H., Benjamin N. Improving Virus Taxonomy by Recontextualizing Sequence-Based Classification with Biologically Relevant Data: the Case of the Alphacoronavirus 1 Species. mSphere. 2018;3(1):e00463–e517. doi: 10.1128/mSphereDirect.00463-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan S, Nakajima R, Jain A, de Assis RR, Jasinskas A, Obiero JM, et al. Analysis of Serologic Cross-Reactivity Between Common Human Coronaviruses and SARS-CoV-2 Using Coronavirus Antigen Microarray. bioRxiv : the preprint server for biology. 2020:2020.03.24.006544.
- 30.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020 doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17(6):613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q., Zhang H., Huang K., Yang Y., Hui X., Gao J. SARS-CoV-2 neutralizing serum antibodies in cats: a serological investigation. BioRxiv. 2020 [Google Scholar]
- 33.Young B.E., Ong S.W.X., Kalimuddin S., Low J.G., Tan S.Y., Loh J. Epidemiologic Features and Clinical Course of Patients Infected With SARS-CoV-2 in Singapore. JAMA. 2020;323(15) doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan J.-F.-W., To K.-K.-W., Tse H., Jin D.-Y., Yuen K.-Y. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21(10):544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spada E., Canzi I., Baggiani L., Perego R., Vitale F., Migliazzo A. Prevalence of Leishmania infantum and co-infections in stray cats in northern Italy. Comp Immunol Microbiol Infect Dis. 2016;45:53–58. doi: 10.1016/j.cimid.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kassahun A., Sadlova J., Benda P., Kostalova T., Warburg A., Hailu A. Natural infection of bats with Leishmania in Ethiopia. Acta Trop. 2015;150:166–170. doi: 10.1016/j.actatropica.2015.07.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.







