Abstract
Our recent experience of the COVID-19 pandemic has highlighted the importance of easy-to-use, quick, cheap, sensitive and selective detection of virus pathogens for the efficient monitoring and treatment of virus diseases. Early detection of viruses provides essential information about possible efficient and targeted treatments, prolongs the therapeutic window and hence reduces morbidity. Graphene is a lightweight, chemically stable and conductive material that can be successfully utilized for the detection of various virus strains. The sensitivity and selectivity of graphene can be enhanced by its functionalization or combination with other materials. Introducing suitable functional groups and/or counterparts in the hybrid structure enables tuning of the optical and electrical properties, which is particularly attractive for rapid and easy-to-use virus detection. In this review, we cover all the different types of graphene-based sensors available for virus detection, including, e.g., photoluminescence and colorimetric sensors, and surface plasmon resonance biosensors. Various strategies of electrochemical detection of viruses based on, e.g., DNA hybridization or antigen-antibody interactions, are also discussed. We summarize the current state-of-the-art applications of graphene-based systems for sensing a variety of viruses, e.g., SARS-CoV-2, influenza, dengue fever, hepatitis C virus, HIV, rotavirus and Zika virus. General principles, mechanisms of action, advantages and drawbacks are presented to provide useful information for the further development and construction of advanced virus biosensors. We highlight that the unique and tunable physicochemical properties of graphene-based nanomaterials make them ideal candidates for engineering and miniaturization of biosensors.
Keywords: Graphene, Virus, Biosensor, Nanocomposite, SARS-CoV-2
Graphical abstract
Highlights
-
•
Methods for virus detection are summarized, need for rapid diagnosis highlighted.
-
•
The properties of graphene, its derivatives and composites are overviewed.
-
•
Principles of graphene-containing biosensor engineering for virus detection are presented.
-
•
Many examples for virus detection of graphene-based biosensors are shown.
-
•
Current trends and perspectives are discussed.
1. Methods of detection of viral pathogens of humans
1.1. Origin and discovery of viruses
Viruses are small microorganisms (of diameter ~20–400 nm), observable only via an electron microscope, that are now recognized as ancient structures that possibly preceded the divergence of life forms into the Bacteria, Archaea, and Eukarya (Prangishvili et al., 2006). Viruses lack metabolic activities and merely act as information carriers that need to hijack host mechanisms for replication, transcription, and protein synthesis to multiply and spread their particles, often causing the host cell to die. The existence of small pathogens that pass through bacterial filters was known from the end of 19th century as causative agents/germs for rabies, foot-and-mouth, and tobacco mosaic disease. The first full image of a virus was obtained only in 1955 by Rosalind Franklin, who crystallized and determined the structure of tobacco mosaic virus (Franklin, 1955). Since then, more than 6000 virus species have been described in detail. Viruses use different mechanisms to generate protein-coding mRNAs and replicate themselves from the genetic information they carry, which can be double-stranded DNA (dsDNA), single-stranded DNA (ssDNA), dsRNA, (+)ssRNA, or (−)ssRNA (Baltimore, 1971).
1.2. Viruses as human pathogens
Viruses cause many common human diseases, such as the common cold (Jacobs et al., 2013), influenza (Hutchinson, 2018), cold sores (Petti and Lodi, 2019), and chickenpox (Zerboni et al., 2014), but also some more harmful and even life-threatening ones, including rabies (Davis et al., 2015), hepatitis (Purcell, 1994), Ebola (Holmes et al., 2016), AIDS (Barré-Sinoussi et al., 1983), avian influenza (Tanner et al., 2015), SARS, MERS, and the most recently discovered COVID-19 (Cui et al., 2019). The severity of the disease depends on the type and quantity of infected and disrupted cells, which is described in terms of virulence. In addition to acute diseases, some viruses may remain dormant within the human body, e.g., herpesviruses (Petti and Lodi, 2019), whereas others, e.g., hepatitis B and C viruses (Purcell, 1994), cause chronic infections or even trigger cancer (Cao and Li, 2018). Infected people serve as vectors or carriers for spreading the virus in populations, with a high number of carriers causing an epidemic. Nowadays, with global travel and trade, good diagnostic tools are essential for gathering reliable data to aid decision-making on disease prevention, vaccination, and healthcare.
1.3. Classical diagnostics and detection methods
Clinical diagnostics of viral diseases and virus detection can be achieved by different methods. In recent years, methods in use have changed rapidly due to the fast development of molecular techniques that offer improved selectivity and increased sensitivity.
1.3.1. Methods based on culture
Many viruses can now be grown in cell cultures (Leland and Ginocchio, 2007), such as Vero cells (African monkey kidney cell line), human lung fibroblasts (MRC-5), and human epidermoid carcinoma cells (HEp-2) or in embryonated chicken eggs (avian influenza viruses) and other biological systems. Plaque-based assays are a standard technique conducted in Petri dishes or multi-well plates overlaid with a host cell culture (Dulbecco, 1952). Viral plaques formed of lysed infected host cells are counted to determine the viral dose as the number of plaque forming units (PFU). However, plaque formation can take days to more than a week depending on the virus. This technique has been improved to obtain results in a shorter time based on detection of infected host cells by using immunostaining with fluorescently labeled antibodies against a viral antigen (Yakimovich et al., 2015). For influenza, a fast assay based on virus surface protein hemagglutinin that agglutinates red blood cells is used (Hirst, 1942).
1.3.2. Antibody-based diagnostics
Other methods rely on the detection of specific IgM and IgA antibodies for acute infection and IgG for past infection present in the blood of an infected person that specifically recognize the virus. These methods are widely used with the enzyme-linked immunosorbent assay (ELISA) and can be automated for multiple screens at once (D. Wang et al., 2015a). An automated and multiplexed quantitative assay of several hepatitis B serology markers has been designed using giant magnetoresistive biosensor chips (Gani et al., 2019). A combined IgM and IgG antibody test has been developed for the rapid diagnosis of COVID-19 (Li et al., 2020b).
1.3.3. Electron microscopy
Transmission electron microscopy (TEM) allows visualization of the shape of the whole virus particle (virion), and therefore is an important tool for discovering and describing new virus isolates or explaining contradictory results obtained by other techniques (Goldsmith and Miller, 2009), but it is not routinely used for diagnostics due to the need for highly specialized sample preparation and expensive equipment requiring specific technical expertise.
1.3.4. Viral genome sequence detection
Techniques based on detection of the viral genome or a specific part of its sequence are the most sensitive diagnostic tests routinely used to diagnose viral infections. Real-time polymerase chain reaction (PCR), which measures fluorescence relative to amplified target DNA using a standard, has frequently been used to quantify the viral loads of a patient for several different viruses at once (Reijans et al., 2008; Santiago et al., 2018). For RNA viruses, this procedure should be preceded by a reverse transcriptase reaction step. PCR amplifies all targeted nucleic acids regardless of whether they originate from intact infectious viral particles, broken ones, or liberated nucleic acids. Therefore, higher totals are obtained than from other techniques that count virions (e.g., TEM) or determine active viral particles. The results are obtained in hours and owing to PCR amplification, the sensitivity is superior to other methods. A clear advantage is that the technique can be quickly adopted to newly emerging viruses, such as SARS-CoV-2 (Corman et al., 2020). Droplet digital PCR based on the absolute quantification of target DNA by partitioning samples into thousands of nanoliter-sized droplets each running an independent amplification has been used for human herpesviruses 6A and 6B (Vellucci et al., 2018).
1.3.5. Flow cytometry
In general, viral particles are too small for analysis by classical flow cytometry, but it is possible to use the technique for analyzing infected or damaged host cells (Mcsharry, 1994). Applications have been developed for the diagnosis of influenza (Badyda et al., 2013) and glandular fever (Crucian et al., 2001).
1.4. Emerging detection and quantification strategies
In recent years, novel techniques have been developed to detect directly, selectively, and with high sensitivity viral nucleic acids and proteins. Some of the techniques have already found practical applications in diagnostics or are getting close to commercial use.
1.4.1. Detection of viral proteins
Antigen-capture ELISA can be used for detecting viral proteins or particles of orthopoxvirus species (Stern et al., 2016). Direct detection of viral proteins has been demonstrated by using nanoparticles (NPs) and quantum dots (Agrawal et al., 2005) or biochips (Roh et al., 2010).
1.4.2. Strategies based on nucleic acids
The most complete information about a virus genome can be obtained by next-generation sequencing, which also provides information about tiny differences between viruses or mutated strains that perform similarly in other tests. This once expensive technique is becoming more affordable and faster and is likely to become the primary diagnostic tool in the future (Boonham et al., 2014). Simultaneous identification of known vertebrate RNA viruses, including their variants, and even novel viral sequences in complex samples, such as serum, blood, and various tissues, is now possible by virome capture sequencing using the VirCapSeq-VERT platform (Briese et al., 2015), which has a limit of detection (LOD) comparable to that of real-time PCR.
Loop-mediated isothermal amplification (LAMP) brings to virus diagnostics the advantage of isothermal amplification over PCR thermocycling but offers less flexible usage due to constrained primer design (Notomi et al., 2000). LAMP has also been designed for the detection of SARS-CoV-2 (Kashir and Yaqinuddin, 2020). The well-known CRISPR technology can also be adopted for fast and highly sensitive detection of a specific nucleic acid sequence. All-in-One Dual CRISPR-Cas12a (AIOD-CRISPR) has recently been designed for SARS-CoV-2 diagnostics (Ding et al., 2020).
1.4.3. Viral particles detection
Flow cytometry has evolved into flow virometry (Zamora and Aguilar, 2018) to allow the direct detection, analysis, and characterization of fluorescently labeled single viral particles, such as HIV (Bonar and Tilton, 2017). A novel technique of resistive pulse sensing based on driving particles through a nanopore allows high-throughput measurements of the size and concentration of virus particles (Yang and Yamamoto, 2016).
1.5. Biosensing for fast diagnosis
Viruses are a major cause of human diseases and mortality, especially in populated and developing countries, and economic burdens on human populations. Fast and early diagnosis helps to select appropriate treatments and could prevent and monitor the spread of infection and manage its elimination. Thus, there is a demand for biosensor development that can produce a quantitative signal selective for specific viral entities (Chowdhury et al., 2019; Grubaugh et al., 2019; Jacob et al., 2020). The conventional diagnostic tools for antigen detection include chemiluminescence (Meng et al., 2020; Wang et al., 2019; Yang et al., 2020; Zhang et al., 2020), ELISA (Arce et al., 2019; Shrivastava et al., 2019), and PCR (Maartens et al., 2020; Mishra et al., 2019; Oliveira et al., 2019a; Pathirana et al., 2019). However, the latter methods require highly skilled personnel and suitable laboratory environment. Furthermore, the cost of reagents and time required could be considered drawbacks of these methods. Also, although quantification of a disease can be accomplished, the selectivity may be a challenge in cases of cross-reactivity with other infectious diseases (Chowdhury et al., 2018; Xiang et al., 2018). These facts call for the development of new, fast, and accurate diagnostic tools for the detection of pathogenic viruses. The very recent lesson given by COVID-19 pandemic outbreak taught us that biosensing can help to get the pandemic under control (Morales-Narváez and Dincer, 2020).
In this review, we present an overview of biosensing methods that utilize emerging two-dimensional carbon-based materials based on graphene, its derivatives and composites for the detection of human viruses. The unique physicochemical properties of these carbon-based nanomaterials make them ideal candidates for engineering biosensors with high stability, sensitivity, and selectivity, all properties highly desirable in the field of human virus detection.
2. Carbon-based nanomaterials
2.1. Graphene as unique carbon-based nanomaterial
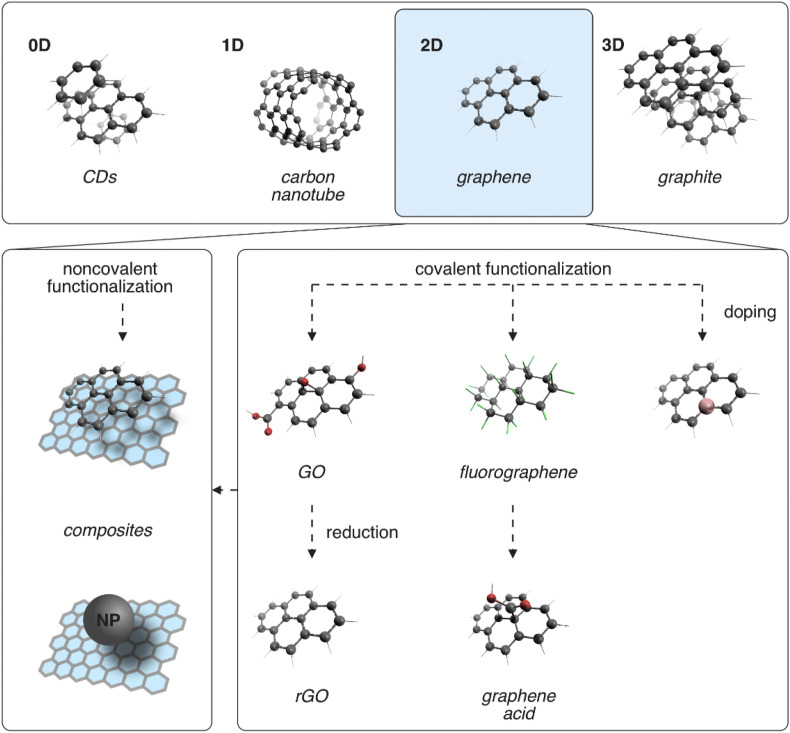
Carbon-based nanomaterials represent a large, diverse and important family of nanomaterials composed of the lightweight and abundant element carbon and possessing a wide range of physicochemical properties that can be exploited in many different applications. Carbon dots (CDs), carbon nanotubes (CNTs), and graphene are prototypical members (Fig. 1 ) which can be classified as formally zero- (0D), one- (1D), and two- (2D) dimensional carbon nanomaterials (Georgakilas et al., 2015). CDs were discovered in 2004 (Xu et al., 2004) and currently encompass a large family of small (<10 nm) objects, which typically share high stability, intensive photoluminescence, and biocompatibility, predisposing them for various applications, including (bio)imaging and sensing (Hola et al., 2014; Lim et al., 2015; Liu et al., 2019). The 1D CNTs share interesting electronic and mechanical properties that can be modulated by their structure, making them suitable for a wide variety of applications, including sensing and biosensing (Kong et al., 2000; Wang, 2005). A boom in research devoted to the 2D carbon allotrope graphene was triggered by its mechanical exfoliation from graphite in 2004 (Novoselov et al., 2004). Graphene is an atomically thin layer of sp2 carbon atoms covalently connected into a honeycomb lattice with a unique electronic structure because its valence and conductive bands touch at Dirac points, making it a semimetal/zero-gap semiconductor (Castro Neto et al., 2009).
Fig. 1.
The upper panel shows structures of selected carbon-based nanomaterials with their formal dimensionalities and the lower panels provide an overview of noncovalent and covalent graphene functionalization. CDs stands for carbon dots, GO graphene oxide, rGO reduced GO, and NP nanoparticle. Carbon atoms are shown as black and oxygen as red balls, hydrogen and fluorine atoms as white and green sticks, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.1.1. Graphene properties
Graphene is extremely electronically and thermally conductive, chemically stable, strong, flexible, highly optically transparent but impenetrable to any molecule. It is a lightweight material with an extraordinarily large surface area of 2630 m2/g. All the mentioned properties make graphene suitable for an enormous range of applications, ranging from mechanical reinforcement up to sensing (Georgakilas et al., 2012; Novoselov et al., 2012; “Graphene calling”, 2007). Methods of graphene preparation based on graphite exfoliation and chemical vapor deposition (CVD) have advanced so that both graphene colloidal solutions and up to square-meter-sized graphene sheets, respectively, are currently readily available on the market (Bae et al., 2010; Hernandez et al., 2008; Obraztsov, 2009). The ability of graphene to effectively quench photoluminescence makes it useful for the construction of optical sensors (He et al., 2010; Liu et al., 2012; Swathi and Sebastian, 2008). The sensitivity of the electrical resistance of graphene to the adsorption of even single molecules on its surface makes it highly advantageous for high-sensitivity sensing applications (Schedin et al., 2007). The graphene surface readily adsorbs guest molecules, which on the one hand is useful for high sensitivity graphene-based sensors, and on the other hand is challenging because the surface can become easily contaminated, e.g., by air-borne contaminants. Use of graphene in various sensing applications based on an electrochemical readout has been demonstrated in many practical sensors operating as gas sensors, chemical sensors, and biosensors (Ambrosi et al., 2016; Chen et al., 2010; Fowler et al., 2009; Kuila et al., 2011; Pumera et al., 2010; Shao et al., 2010). Graphene sensors can also be based on its electrocatalytic properties (Chen et al., 2010; Shang et al., 2008). A concept of field effect transistors (FETs) having graphene as an active zone is particularly attractive owing to the advantageous chemical and physical properties of graphene. However, the graphene zero band gap limits its straightforward application in FET technology based on the utilization of semiconductors (Burghard et al., 2009; Schwierz, 2010). Therefore, a large research effort has been devoted to graphene band gap opening.
2.2. Graphene functionalization
Opening of the band gap is one of the strategies for improving graphene's properties in order to enhance its application potential in biosensing. The hydrophobicity of graphene complicates its handing and use in water polar media and can be an obstacle for its broad application in biosensing. On the other hand, graphene offers a high surface area for the adsorption of proteins and nucleobases via hydrophobic interactions. Graphene itself lacks any chemically reactive functional groups that could be utilized for the covalent or noncovalent immobilization of molecules. These issues have triggered a great effort into modifying the properties of graphene by increasing the number of graphene layers, introducing defects, reducing the size (to nanoflakes and nanoribbons), doping graphene with other elements, and covalent and noncovalent graphene functionalization (Boukhvalov and Katsnelson, 2008; Georgakilas et al., 2016, 2012; Li et al., 2008; Narita et al., 2015). All these strategies have been successfully applied to make graphene a promising candidate for biosensing applications.
2.2.1. Covalent graphene derivatives
The covalent functionalization of graphene (Fig. 1) leads to significant changes in its physicochemical properties (Georgakilas et al., 2012). Graphene oxygen derivatives, i.e., graphene oxide (GO) and its reduced version reduced graphene oxide (rGO), contain functional groups, e.g., epoxy, carboxyl, and hydroxyl, allowing the development of a multitude of potential biosensors (Dreyer et al., 2010; Kim et al., 2017, 2010; Kris Erickson et al., 2010). Both GO and rGO exhibit similar physicochemical properties to those of graphene, including flexibility, transparency, and low cytotoxicity, but offer greater biosensing capabilities owing to their hydrophilic nature and availability of reactive functional groups (Chung et al., 2013; Compton and Nguyen, 2010). The hydrophilicity is also advantageous for biosensor assembly since it allows the preparation of films by dropcasting, spin coating, ink-jet printing, and processing for electrode materials. GO is usually prepared by the vigorous oxidation of graphite/graphene colloidal solutions, and therefore is a chemically complex material containing various functional groups, whose nature and amount depend on the preparation method. The functional groups of GO make it susceptible to surface modification by anchoring redox species or bio-recognition molecules, such as enzymes, antibodies, peptides, or DNA and RNA oligonucleotides (Draz et al., 2014). In addition, GO (and generally all carbon-based nanomaterials) may also contain metal impurities originating from the reagents used, which can affect their properties. This may present an obstacle for the construction of robust sensors. However, the electrochemical activity of these impurities can be utilized in electroanalysis (Chee and Pumera, 2012). The introduction of oxygen-containing functional groups onto graphene (and increase in sp3 carbon atoms in the sp2 graphene lattice) leads to a significant drop in conductivity and GO is considered a nonconductive material. The conductivity of GO can be restored by its chemical or thermal reduction to rGO (Becerril et al., 2008; Gilje et al., 2007; Gómez-Navarro et al., 2007; Stankovich et al., 2007). The degree of reduction of GO affects the sensitivity of rGO-based sensors (Robinson et al., 2008). The conductive rGO is to some extent water dispersible and its chemical functionalities make it an excellent platform for the immobilization of enzymes, DNA, metallic/metal oxide NPs and quantum dots (Abd Muain et al., 2018; Singh et al., 2017). Their unique properties, including conductivity, fluorescence quenching, peroxide-like enzyme activity, intrinsic Raman activity, oxidation/reduction capabilities, and ability to anchor various NPs, enzymes, proteins, and nucleic acids together with their commercial availability, make GO and rGO suitable platforms for many biosensors based on electrochemical, fluorescence, colorimetric, surface plasmon resonance (SPR), and surface-enhanced Raman spectroscopy (SERS) readouts (Fang and Wang, 2013; Jung et al., 2010; Sinha et al., 2016; Wang et al., 2011; Yan et al., 2020; Yang et al., 2019).
Besides the very popular and widely used graphene oxygen derivatives, other covalent graphene derivatives have been prepared and well characterized. Graphene hydrogenation leads to graphane (Elias et al., 2009) with accompanying band gap opening and decreased conductivity. Pumera et al. showed that this graphene derivative can be used for the electrochemical sensing of important biomarkers (Tan et al., 2013). Graphene halides (covalent graphene derivatives densely functionalized by halogen atoms) also exhibit an open band gap (Karlický et al., 2013), among them fluorographene (Fig. 1) is of particular interest. Each carbon atom is functionalized by the fluorine atom in this covalent graphene derivative. It can be prepared in a large scale from graphite fluoride, which is a commercially available solid lubricant, by chemical exfoliation (Zbořil et al., 2010). Owing to its wide band gap, it is considered one of the thinnest insulators (Nair et al., 2010). An interesting feature of this graphene derivative is that it can be chemically converted under rather mild conditions to a wide portfolio of graphene derivatives (Chronopoulos et al., 2017). The common feature of graphene derivatives prepared from fluorographene is that they contain well-defined chemical moieties (e.g., thiol, amine, hydroxyl, sulfonate, alkyl, aryl, alkenyl, etc.) homogeneously distributed over both sides of graphene surface (Bakandritsos et al., 2017). The well-defined chemistry with possibilities to tune the degree of functionalization make these materials very promising for many applications, including sensing (Chia et al., 2014; Heng Cheong et al., 2019; Lenarda et al., 2019; Urbanová et al., 2015).
2.2.2. Graphene doping
Besides the covalent modification of graphene, i.e., attachment of functional groups to sp2 carbons, doping of graphene, i.e., replacement of carbon atoms by other elements in the honeycomb graphene lattice (Fig. 1), is another strategy to tune graphene's properties. Particularly attractive are B and N elements because they are lightweight, well tolerated by the graphene lattice, and serve as electron withdrawing and donating groups (Panchakarla et al., 2009; Wang et al., 2012, 2009; Wei et al., 2009). N-doped graphene has been successfully utilized in electrochemical biosensing (Feng et al., 2015; Wang et al., 2010).
2.2.3. Noncovalent graphene composites
Graphene derivatives, namely GO and rGO, can be used as a component for the formation of a diverse range of graphene-based nanocomposites with enhanced sensitivity for biosensors by integration with metal and metal oxide NPs, quantum dots, nanoclusters, polymers and various biomolecules (C. Cheng et al., 2017; Chua and Pumera, 2014; Krishnan et al., 2019; Teymourian et al., 2013). Owing to the high specific surface area, good availability of functional chemical groups and unique interface properties, these graphene-based materials possess vast capacities for the adsorption of biomolecules (Karimi et al., 2015; Lawal, 2018; Suvarnaphaet and Pechprasarn, 2017). Noncovalent interactions of graphene derivatives with adsorbates may include electrostatics, polarization, London forces, and hydrophobic effects (Georgakilas et al., 2016; Lazar et al., 2013). They ensure contact of an adsorbate with the surface, leading to changes in the electronic properties, which can be utilized in sensing. They can also be exploited for the noncovalent functionalization of graphene and its derivatives in sensor construction. Pyrene derivatives are a prototypical example of molecules used for graphene functionalization (Fig. 1) via π-π stacking (Parviz et al., 2012). In addition, the noncovalent functionalization approach offers the possibility of binding functional groups without significant disturbance of the graphene electronic structure (Sreeprasad and Berry, 2013). The graphene surface can also be modified by polymers (Georgakilas et al., 2016) and such composites can be utilized in biosensing (Navakul et al., 2017). The loading ratio of GO for biomolecules can reach up to 200%, much higher than that of other nanocarriers, which is very beneficial for construction of sensitive electrochemical immunosensors (Liu et al., 2008; Yang et al., 2008). Metal and metal oxide NPs bind to graphene via noncovalent interactions (Granatier et al., 2012). Loading of metal/metal oxide NPs onto a graphenic surface can also lead to their stabilization and increased catalytic activity. More importantly, their attachment to a graphene surface can introduce new features that can be utilized in sensor construction. For instance, gold and silver NPs introduce surface plasmons, which can be exploited in SERS (Xu et al., 2013). The noncovalent functionalization of graphene and construction of 3D networks of graphene hybrids can introduce selectivity, e.g., via accommodation of specific guest molecules, and enhance the sensitivity by increasing the number of adsorbed biomolecules (W. Cheng et al., 2017).
It is worth noting that 2D functionalized graphene derivatives/composites may suffer from restacking, i.e., formation of multilayer van der Waals materials, because of strong noncovalent interlayer interactions. Constructing a solid 3D network is an important strategy for obtaining new and stable graphene nanocomposites that are free of restacking and have many advantageous properties, e.g., high surface area, accessible inner space, tunable electronic properties, better mechanical stability, and enhanced conductivity/electrocatalytic properties. These properties make such 3D architectures suitable for efficient and selective biosensing applications (Kim et al., 2017; Lu, 2018; Passaretti et al., 2019; Ramanathan et al., 2019; Vermisoglou et al., 2020).
2.2.4. Graphene quantum dots
Reducing the size of graphene can lead to formation of graphene quantum dots (GQDs). GQDs belong to CDs, which is a large family of 0D carbon nanostructures displaying significant photoluminescence (Burghard et al., 2009; Schwierz, 2010). GQDs have attracted a great deal of interest for both in vitro and in vivo biosensing applications owing to their stable and tunable photoluminescence, low photobleaching, favorable aqueous dispersibility, low toxicity and high biocompatibility (Shen et al., 2012; Sun et al., 2013; Xie et al., 2016; Zhou et al., 2016). Intensive research efforts have been devoted to designing and engineering GQDs. Ideal GQDs contain from 1 to 3 graphene layers with a lateral dimension of the order of 100 nm (Fan et al., 2015; Peng et al., 2012). GQDs can be prepared through various top-down approaches, e.g., solvothermal, electrochemical, and electrochemical exfoliation methods, and bottom-up methods, e.g., microwave or hydrothermal treatment of cheap carbohydrates (Fan et al., 2015). They may contain various oxygen functionalities, such as carboxyl, hydroxyl, carbonyl, and epoxide groups, at their edges that can act as reaction sites and endow the material with high water solubility and biocompatibility (Ghanbari et al., 2017). The properties of optical biosensors can be further tuned by heteroatomic doping by electron donor/acceptor atoms to alter the bandgap (HOMO-LUMO gap) and electronic properties compared to pristine GQDs (Ananthanarayanan et al., 2015; Chung et al., 2019; Iravani and Varma, 2020). Besides the electronic properties, doping by heteroatoms can also change the electrochemical characteristics and introduce anchoring sites for, e.g., metal NPs (Chowdhury et al., 2019; Hasanzadeh and Shadjou, 2017). GQDs can be coupled with specific sequence DNA molecules for use as probes that are complementary to target virus DNA molecules (Xiang et al., 2018). Besides π-π interaction with graphene surfaces, oxygen functionalities on GQD surfaces favor aptamer (short, single-stranded oligonucleotides) adsorption through electrostatic and hydrogen bonding interactions (Ghanbari et al., 2017). Moreover, thiol-functionalized GQDs (GQDs-SH) have been used for noble (silver) NP immobilization on SH groups via Ag–S bonding formation. This type of platform has been employed for antibody loading via interaction of antibody amino groups and AgNPs (Valipour and Roushani, 2017). Doping of GQDs with heteroatoms such as nitrogen and sulfur into the π-conjugated carbon system introduces useful functionalities. Nitrogen alters the electronic characteristics and amplifies the electrochemical activity, whereas sulfur creates numerous anchoring sites for noble nanoparticle attachment. In addition, the presence of edge carboxylic groups on GQDs allows antibody conjugation (Chowdhury et al., 2019, 2018). The tunable fluorescence properties, inherent redox activity, chemiluminescence, and catalytic effect of GQDs make them promising materials for optical, electrochemical, and electrochemiluminescence (ECL) biosensing applications with enhanced selectivity and sensitivity. The optical biosensor engineering is mainly based on fluorescence quenching (signal off) and enhancement (signal on) processes.
3. Graphene-based materials in virus sensing
Numerous approaches for sensing biomolecules, such as nucleic acids, peptides, enzymes, and antigen–antibody complexes, have been developed based on various physicochemical methods providing an electrical, electrochemical, nuclear magnetic resonance (NMR), mass spectrometry (MS), and optical signal. Methods based on optical and electrical signals are particularly attractive because of accessible and reasonably affordable instrumentation (compared to very expensive MS and NMR instruments). Owing to the electrical and optical properties of graphene, its derivatives and nanocomposites (discussed in the previous section), such materials are highly suitable for utilization in biosensing, including for the detection of viruses (Fig. 2 ). In the following text, we will focus mainly on optical and electrochemical sensing methods. Table 1 provides overview of so far published articles utilizing graphene-based materials in virus sensing.
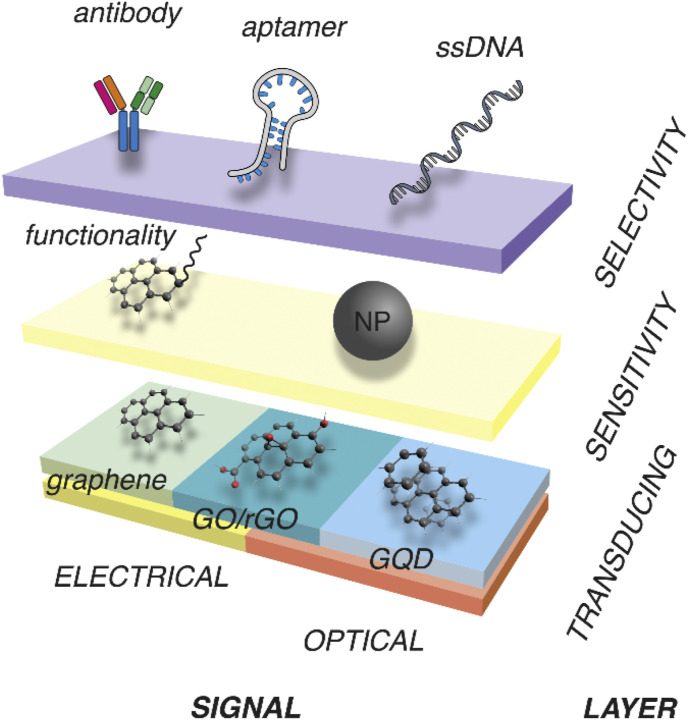
Fig. 2.
Materials commonly used for assembly of prototypical graphene-containing biosensors. Graphene and rGO are usually used in biosensors with an electrical readout, whereas GO and GQD are used in optical biosensors. Noncovalent functionalization by NPs can enhance the sensitivity, e.g., via enabling SPR and SERS signals. Biomolecules, such as antibodies, aptamers, and nucleic acids, are used to establish the biosensor selectivity.
Table 1.
The overview of so far reported biosensing works dealing with virus detection on graphene-based materials.
| Virus | Method of detection | Material | LoD | Ref |
|---|---|---|---|---|
| Influenza virus A (H1N1) | optical | G/Au-FeXOYNPs | 7.27 fg mL−1 | Lee et al. (2018) |
| Influenza virus A (H1N1) | electro | rGO/AuNPs | 10−8U mL−1 | Anik et al. (2018) |
| Influenza virus A (H1N1) | electro | rGO | 0.5 PFU mL−1 | Singh et al. (2017) |
| Influenza virus A (H1N1) | electro | rGO | 33 PFU mL−1 | Joshi et al. (2020) |
| Influenza virus A (H1N1) | electro | GO | 8 pM | Veerapandian et al. (2016) |
| Influenza virus A (H1N1) | optical | rGO | 3.8 pg mL−1 | Jeong et al. (2018) |
| Influenza virus A (H5N1) | electro | rGO | 5 pM | Chan et al. (2017) |
| Influenza virus H7 (AIV H7) | electro | G/AuNPs | 1.6 pg mL−1 | Huang et al. (2016) |
| Ebola virus | optical | GO | 1.4 pM | Wen et al. (2016) |
| Ebola virus | electro | rGO | 2.4 pg mL−1 | Jin et al. (2019) |
| Ebola virus | electro | rGO | 1 μg mL−1 | Maity et al. (2018) |
| Dengue virus | electro | GO | 0.12 PFU mL−1 | Navakul et al. (2017) |
| Dengue virus | optical | CdSQDs-NH2-GO | 1 pM | Omar et al. (2019) |
| Dengue virus | optical | rGO | 0.08 pM | Omar et al., 2020a, Omar et al., 2020b |
| Dengue virus | optical/electro | GO/Ru | 0.38 ng mL−1 | Kanagavalli and Veerapandian (2020) |
| Dengue virus | electro | GO/APTES | 1 fM | Jin et al. (2016) |
| Dengue virus | optical | rGO/PAMAM | 0.08 pM | Omar et al., 2020a, Omar et al., 2020b |
| Hepatitis C virus | electro | GQDs-SH/Ag | 3 fg mL−1 | Valipour and Roushani (2017) |
| Hepatitis C virus | optical | GO | NA | Kim et al. (2013) |
| Hepatitis C virus | optical | rGO | 10 fM | Fan et al. (2019) |
| Hepatitis C virus | electro | rGO/CuNPs | 0.4 nM | J. Li et al. (2020a) |
| Hepatitis C virus | electro | GO | 0.2 nM | Jiang et al. (2020) |
| SARS-COV-2 | electro | G | 1.6 PFU mL−1 | Seo et al. (2020) |
| ZIKA virus | electro | G/CVD | 0.5 nM | Afsahi et al. (2018) |
| HIV | electro | GO | 8.3 fM | Nehra et al. (2017) |
| HIV | electro | G/AuNPs | 30 aM | Y. Wang et al. (2015b) |
| HIV | electro | G/CVD | 0.1 ng mL−1 | Islam et al. (2019) |
| HIV | optical | GO | 0.4 nM | Wang et al. (2016) |
| HIV | optical | BN-SGQDs | 0.5 nM | Li et al. (2017b) |
| HIV | electro | GO/PANi | 100 aM | Q. Gong et al. (2019) |
| Rotavirus | optical | rGO | 102 PFU mL−1 | Liu et al. (2013) |
| Rotavirus | electro | GO | 103 PFU mL−1 | Liu et al. (2011) |
| Rotavirus | optical | GO | NA | Fan et al. (2014) |
| Hepatitis Bvirus | electro | GO | 0.1 ng mL−1 | Zhao et al. (2017) |
| Hepatitis B virus | electro | rGO/AuNPs | 3.8 ng mL−1 | Abd Muain et al. (2018) |
| Hepatitis B virus | electro | GQDs | 1 nM | Xiang et al. (2018) |
“NA” stands for not available.
3.1. Optical sensing
Optical biosensors offer many advantages by providing a real-time readout and exceptional qualities, such as high detection limits, specificity, biocompatibility, and sensitivity. They also enable portable, rapid, and economically affordable instrumentation, with respect to traditional culture or PCR-based assays. Such instrumentation can be further miniaturized into point-of-care (POC) devices. All these features can be effectively utilized in the detection of pathogenic microorganisms (Chen and Wang, 2020; Oshin et al. 2019; Yoo and Lee, 2016).
Among 2D materials, graphene and its nanocomposites have been widely exploited for optical biosensing applications (Fig. 3 ) owing to their remarkable physicochemical properties (discussed in detail in the previous sections) (Allen et al., 2010). During the last two decades, a wide variety of optical methods have been developed based on fluorescence, colorimetry, SPR and SERS, and utilizing graphene, graphene derivatives (namely rGO) and graphene-containing composites, including noble metals, metal oxides, quantum dots, polymers, and porous graphene networks.
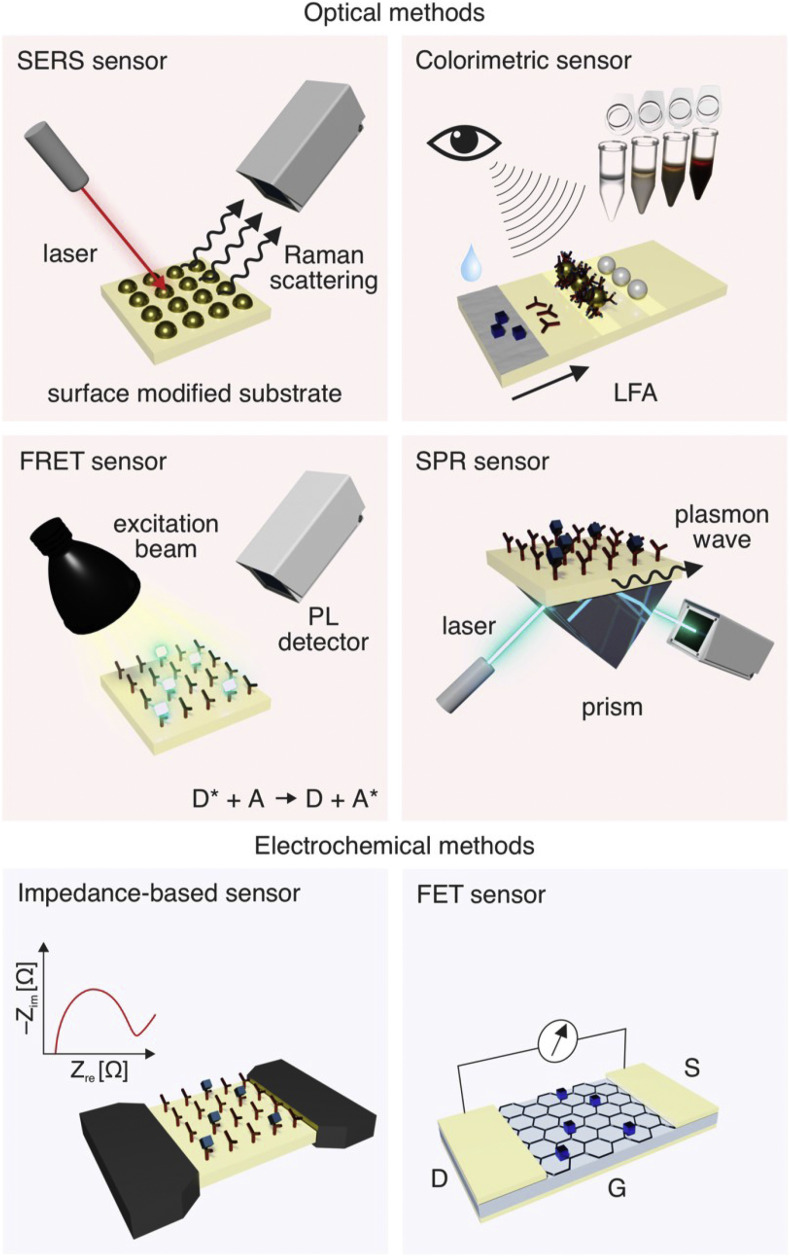
Fig. 3.
Overview of sensing methods utilizing graphene-based materials.
3.1.1. Photoluminescence-based biosensors
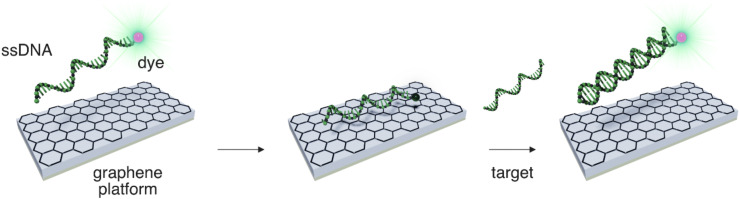
Fluorescence biosensors offer tremendous advantages, such as portability, rapid detection times, low detection limits and high sensitivity, making them ideal platforms for sensing various bioreceptors (Gao and Tang, 2017; Zhang et al., 2017). The fluorescence biosensors are usually based on registration of the wavelength (and intensity) of emitted light rather than other fluorescence features, such as lifetime, which can also provide useful information. They are also usually designed on the basis of the signal (fluorescence) being on/off, and therefore rely on the primary principle of Förster resonance energy transfer (FRET) (Tian et al., 2017). In FRET, the energy of an excited fluorophore (donor) is transferred in a nonradiative manner to another light-responsive molecule (acceptor). FRET can lead to changes in the fluorescence emission wavelength and intensity (i.e., by quenching or enhancement) and these features can be exploited in biosensor engineering. Strong acceptor properties of excitation energy over the entire visible region make GO and rGO suitable materials for the effective quenching of fluorescence, and they have therefore been exploited in fluorescence biosensors (Lu et al., 2009; Zhang et al., 2017). The fluorescence of fluorescent dyes covalently attached to nucleic acids as fluorescent probes can be quenched by GO upon the adsorption of single-stranded nucleic acids onto its surface. However, the fluorescence can be restored, e.g., by nucleic acid hybridization or binding to a specific protein (see Fig. 4 ). These features can be utilized in biosensor construction (Lu et al., 2009). The selectivity and detection limit in the FRET detection mode depend not only on the ability for Watson–Crick base-pairing but also the noncovalent binding of nucleobases to nanomaterials (Iliafar et al., 2014; Umadevi and Sastry, 2011; Waiwijit et al., 2015). It should be noted that the strength of interaction of biomacromolecules and their building blocks to 2D nanomaterials is still largely underexplored with respect to binding affinities among biomolecules. Taking advantage of this concept, other strategies have emerged, e.g., to replace organic fluorescent dyes by quantum dots to enhance the fluorescence signal and stability. For instance, boron and nitrogen-doped single layer GQDs display a high affinity for binding nucleic acids and appropriate noncovalent binding capability with respect to undoped individual layer GQDs (Li et al., 2019, 2017). The presence of boron has been shown to be beneficial for the absorption of ssDNA, whereas nitrogen promotes the release of ssDNA. These properties have been successfully applied for the detection of HIV DNA in living HeLa cells (Li et al., 2017b). Owing to their strong quenching capabilities, graphene-based materials and composites offer exciting opportunities for the development of fluorescent biosensing with improved sensitivity, selectivity, and low detection limits.
Fig. 4.
Principle of ssDNA@graphene-based photoluminescence (PL) sensor. ssDNA labeled by PL dye adsorbs on graphene/GO via stacking of nucleobases, which leads to PL quenching. After hybridization with its complementary strand, it forms dsDNA, which does not effectively bind to graphene surface, desorbs, and enhances PL.
3.1.2. Colorimetric biosensors
Among optical biosensors, colorimetric biosensors are usually straightforward, fast, accurate, and cost-effective detection tools that generate signals visible to the naked eye (Hu et al., 2013; Yan et al., 2020). An important strategy used in colorimetric biosensor construction utilizes enzymes or enzyme-like materials that can catalyze chemical reactions accompanied by color changes. Another approach is based on the SPR of noble metal nanomaterials and corresponding change in color. Among graphene materials, GO shows inherent peroxide-based enzyme activity but with high stability compared to natural enzymes (Song et al., 2019; Yan et al., 2020). Recently, Huang et al. prepared a GO hybrid with AuNPs mimicking a peroxidase and its catalytic activity which was enhanced even by the chelation of mercury. They reported a successful colorimetric immunoassay based on the Hg2+-stimulated peroxidase activity to detect human respiratory syncytial virus (RSV) with a low LOD of 0.04 pg mL−1 (Zhan et al., 2014). Without doubt, graphene-based (and metal-ion enhanced) immunoassays show great potential for use in optical colorimetric biosensors.
3.1.3. Graphene-based surface plasmon resonance biosensors
SPR is an important tool for biosensing because of its real-time readout, nanomolar sensitivity, and label-free detection of bio-interactions (Malmqvist, 1993; Singh, 2016). The basic principle of an SPR biosensor is to monitor a change in the surface refractive index upon biomolecule binding as measured by a shift in the SPR angle (Jorgenson and Yee, 1993; Singh, 2016). Noble metals such as gold and silver amplify SPR signals and are therefore widely used in sensors and biosensors (Jorgenson and Yee, 1993; Malmqvist, 1993). AgNPs exhibit better performance than AuNPs in the visible light region. An SPR sensor can also incorporate graphene-based materials, such as GO, for the detection of various forms of nucleic acids. Such devices rely on monitoring the change in plasmonic signal upon an antibody–antigen affinity reaction in real-time (Omar et al., 2020a, Omar et al., 2020b; Wong et al., 2016). Mahdi et al. deposited a thin film on glass slides containing a layer of gold, a layer of a composite of CdS quantum dots with amino-functionalized GO, and a layer of IgM immobilized via EDC-NHS (Au/CdS–NH2GO/EDC-NHS/IgM). The resulting device was successfully employed to sense dengue virus (DENV) based on antibody–antigen affinity reaction (Omar et al., 2019). This strategy based on covalently bonding antibodies on the graphene surface provided a very low LOD of 1 pM. Other approaches have been reported to improve the sensitivity of SPR for DNA hybridization analysis (W. Gong et al., 2019).
3.1.4. Surface-enhanced Raman spectroscopy
SERS offers a tremendous application potential owing to the ability to enhance a Raman signal up to 106–1014 orders of magnitude with single-molecule/biomolecule detection featuring high sensitivity, label-free detection capability, low sample volumes, and real-time detection (Nie and Emory, 1997). SERS is an important technique for detecting not only chemical species but also biological analytes (Kim et al., 2017). Among graphene materials, GO has been extensively studied as a Raman active platform to sense biomolecules using SERS and GERS (Kim et al., 2017; Yu et al., 2011). Various strategies have been developed using GO as both a Raman active material and scaffold for the immobilization of SERS active (i.e., Au and Ag) NPs. GO exhibits its own SERS ability, but it is of the order of ~104 signal enhancement (Jabłońska et al., 2018), which is significantly lower than SERS of plasmonic metal NPs (106–1014) (Sujai et al., 2020; Yun and Koh, 2020). Thus, GO is used mainly as a SERS platform for anchoring SERS active NPs. This approach enables high sensitivity. However, the sensitivity can be enhanced further by using separation technologies, e.g., magnetic separation, which can be achieved by nanocomposite formation with the inclusion of magnetically active NPs. The high selectivity of SERS-based sensors is achieved by anchoring selectors, such as nucleic acids and antibodies, that selectively bind their counterparts. Ray et al. reported the selective detection and complete removal of rotavirus from water by using an antibody attached three-dimensional graphene network of plasmonic-magnetic architecture using SERS (Fan et al., 2014). Thus, antibody functionalized graphene-based materials and composites bearing SERS active NPs provide good selective, rapid, and label-free materials for the identification of pathogen.
3.2. Electrochemical sensing
Electrochemical biosensors, which combine electrochemical techniques with biosensors, meet requirements for high selectivity, high sensitivity (high signal-to-noise ratio), simplicity, and low cost (Anik et al., 2018; Lawal, 2018). Their portability (e.g., for use in POC systems), fast detection, real-time diagnostics, small analyte volume requirement, minimal manipulation, compatibility with microfabrication technology, and simple instrumentation are among their many advantages (Abd Muain et al., 2018; Huang et al., 2016). However, shortcomings of electrochemical sensing methods include sample damage during analysis, time-consuming preparation of electrodes, stability/aging of the biosensor, and the high price of instruments with low detection limits (Chekin et al., 2018; Joshi et al., 2020).
3.2.1. Graphene-based materials used in electrochemical biosensors
Several materials, including carbon nanomaterials, conducting polymers, metal NPs, and magnetic NPs or composites of these materials, have been used to amplify the signal of electrochemical biosensors to produce pioneering ultrasensitive nano-biosensors (Chowdhury et al., 2019; Zribi et al., 2016). Owing to its unique properties, graphene can serve as a highly suitable 2D nanoplatform for biosensing and its interaction with bio-interfaces can be desirably tuned through functionalization (as discussed in Section 2.2).
3.2.2. General strategies in electrochemical biosensor design
Electrochemical biosensors can detect the presence of virus-specific molecules, e.g., nucleic acids, antigens, and antibodies. Electrochemical graphene-based immunosensor design can be based on the selective affinity of ssDNA over dsDNA on graphene. Nucleobases interact with the graphene surface by noncovalent π-stacking interactions, which facilitate immobilization of single-stranded nucleic acids with exposed nucleobases on graphene surface (Li et al., 2018). Conversely, dsDNA has a lower affinity toward graphene due to its rigid structure and hydrophilic negatively charged backbone. The superior binding of graphene/GO to ssDNA over dsDNA has been used for the fabrication of biosensors that detect nucleic acids, proteins, and small molecules (Ahour and Shamsi, 2017; Li et al., 2016). RNA, having a similar structure to DNA, exhibits similar interactions with graphene. Nevertheless, there are fewer sensors using RNA due to its higher susceptibility to enzyme degradation than DNA. It is worth noting that GO has been reported to effectively protect ssRNA from this enzymatic cleavage (Cui et al., 2013; Li et al., 2018; Reina et al., 2018). Nucleic acid probes used in the electrochemical sensing based on DNA hybridization are designed to be highly selective and specific so that their binding to complementary target nucleic acid strains enables identification of target even in presence of a mixture of nucleic acids. Despite the hybridization depending on the temperature, pH, ionic strength, and DNA concentration, the DNA-based analytical techniques allow very specific, sensitive, qualitative, quantitative, and accurate target detection.
A strategy based on aptamer proximity binding has been used to produce an electrochemical aptasensor for the ultrasensitive detection of viruses, such as the hepatitis C virus (HCV) core antigen (Ghanbari et al., 2017). The design of an aptasensor is based on recognition between an aptamer and a target. Aptamers are short, single-stranded oligonucleotides or peptides that can act as important molecular tools for diagnostics and therapeutics. They are analogous to antibodies but offer many advantages over them as therapeutic reagents, including low cost, no immunogenicity, greater stability, easy synthesis and modification, and smaller molecular weight (Song et al., 2014; Wu et al., 2019). An example is an aptasensor based on porous rGO/MoS2 electrode covalently functionalized with an RNA aptamer targeted against human papillomavirus protein (HPV-16 L1 protein) (Chekin et al., 2018). Another biosensor was constructed by using graphene-stabilized gold nanoclusters as a platform and an aptamer with a cytosine-rich base set as a capture probe for HIV virus detection (Wang et al., 2015a, Wang et al., 2015b).
Enzymes and proteins can be immobilized onto graphene both covalently and noncovalently. Among the advantages of immobilized enzymes are their thermal and storage stability and easy separation from reaction mixtures. Enzyme immobilization on graphene or GO can be driven by noncovalent interactions (Zhang et al., 2013), which vary in various classes of enzymes/proteins (Duinhoven et al., 1995). The flexibility of GO assists protein binding by adopting a shape that better fits a protein surface (Alwarappan et al., 2012). In the noncovalent adsorption of an enzyme/protein onto a GO surface, the interactions depend greatly on the environmental conditions, such as pH and ionic strength of buffer. Thus, different enzymes exhibit different enzyme loadings and stabilities on GO. More specifically, if electrostatic repulsion occurs between the enzyme and GO, a sufficient enzyme loading will not be achieved. Since proteins have amine and carboxyl groups on their surfaces, chemical reactions with the functional groups of GO can take place in addition to noncovalent interactions, leading to the covalent binding of protein on the GO surface. For this purpose, free amine groups on the surface of the enzyme/protein and carboxylic groups on GO are needed. Besides direct covalent binding between the oxygen-containing functional groups of GO and enzyme/protein free amines, cross-linkers can also be used. These linking molecules act as spacers, minimizing the direct adsorption of enzyme onto GO sheets and preserving the enzyme activity (Xu et al., 2012). Graphene acid is a conductive graphene derivative with a well-defined chemistry, as both sides of graphene are densely (with ~15% degree of functionalization) and homogenously decorated by carboxyl groups (Bakandritsos et al., 2017). The carboxyl groups of graphene acid enable efficient covalent enzyme immobilization (Seelajaroen et al., 2020), which can be exploited in electrochemical sensing in the future.
Electrochemical immunosensor designs can also be based on the highly biospecific recognition interaction between an antigen and antibody for fast and sensitive target antigen detection. Antibody-based sensors can provide robust, sensitive, and rapid analysis. The key element is the quality of the antibody used and its ability for selective and sensitive antigen recognition. Potential issues related to antibody sensitivity may arise due to alterations in pH or temperature fluctuations or other external factors. Sandwich-type immunosensors offer improved sensitivity owing to different signal amplification strategies. The detectable signal of a conventional sandwich-type immunosensor usually originates from a labeled electroactive substance, such as metal NPs or redox-active small molecules. Nevertheless, there is a risk that the NPs in these immunosensors are not homogeneously distributed on the graphene surface and could leak from the electrode surface to the solution, resulting in an unstable output signal and poor reproducibility (Huang et al., 2016; Yang et al., 2015).
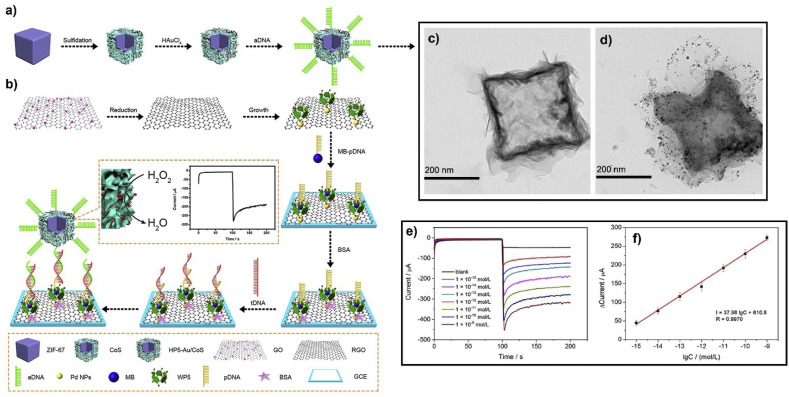
Nanomaterial-based signal amplification is crucial for achieving high sensitivity analysis. For sandwich-type DNA sensors, the detection sensitivity can be enhanced by using enzymes as electrocatalysts or metal NPs due to their intrinsic catalytic activity, high chemical stability, and electrical conductivity. An example of a sandwich-type DNA sensor used for electrode modification is presented in Fig. 5 . It comprises a water-soluble pillar[5]arene (WP5), where Pd NPs are deposited onto a rGO surface (WP5–Pd/rGO). WP5 serves the role of host, where guest-probe DNA labeled with methylene blue (MB-pDNA) is immobilized. The probe DNA (MB-pDNA) is appropriate for hybridization with target DNA (tDNA) related to a virus. The other half of the sandwich enables signal acquisition via hydrogen peroxide (H2O2) reduction. It comprises a ZIF-67-derived cobalt sulfide, where another host, i.e., hydroxylatopillar[5]arene (HP5) bearing AuNPs, accommodates auxiliary DNA (aDNA) that is hybridized with the part of tDNA not hybridized by MB-pDNA (Qian et al., 2020). The biosensor developed by Qian and coworkers addresses a common hurdle faced in electrochemical sensors based on DNA hybridization, which is related to the arrangement of adjacent DNA probes. A short distance between them could be in expense of hybridization efficiency due to a steric hindrance effect. By host-guest recognition the appropriate distance between adjacent DNA probes is reassured by utilizing the certain size of pillar[n]arenes. Furthermore, the strong host-guest interaction prevents DNA probe leakage from the electrode surface.
Fig. 5.
Construction of a sandwich DNA sensor. (a) Preparation of the hybrid. The synthesis is based on preparation of cobalt metal–organic framework nanocubes (ZIF-67) and their sulfudation (CoS). Subsequently, the nanocubes are functionalized by gold nanoparticles (HP5–Au/CoS) and an auxiliary DNA (aDNA). (b) Electrochemical procedure of the sensor. Probe DNA (pDNA) is loaded onto a graphene surface with methylene blue (MB) and the electrode is then coated with bovine serum albumin (BSA). The whole graphene composite is framed in a glassy carbon electrode (GCE). (c,d) TEM images of hollow CoS nanobox and HP5–Au/CoS. (e) The amperometric i-t responses. The quantitative curve of tDNA detection is obtained by amperometric i-t technique in phosphate buffer (pH = 7.0) solution including KCl and [Fe(CN)6]3-/4- as a redox active marker under a constant voltage of −0.4 V. After the background remains constant, H2O2 is mixed with phosphate buffer solution. The current responses are derived from the electrocatalytic activity of HP5–Au/CoS-aDNA label towards H2O2. The current values are related to the concentration of tDNA and more specifically current values are improved with the concentration of tDNA increasing from 1 × 10−15 mol/L to 1 × 10−9 mol/L. (f) The logarithmic calibration curve of the proposed DNA sensor towards different concentrations of HBV DNA. Reprinted by permission of (Qian et al., 2020). Copyright © 2020, Elsevier. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2.3. Electrochemical sensing based on DNA hybridization
Very sensitive and selective electrochemical sensing was achieved by utilizing DNA hybridization on graphene-based material modified electrodes. Nagar et al. designed an impedimetric sensing platform that allowed one-step sensing of a target ssDNA specific of Coxcakie B3 virus by modifying the electrode with a graphene/AuNPs composite (Nagar et al., 2019). The composite with the biorecognition element, ssDNA, was stamped onto a PET substrate. Reduced graphene oxide was employed as excellent conductive substrate for the deposition of AuNPs to assist the immobilization of probe ssDNA appropriate for hybridization with target ssDNA. Target ssDNA of Coxcakie B3 virus was selected as a model analyte. The change in electron transfer resistance after hybridization with the target ssDNA was monitored by electrochemical impedance spectroscopy (EIS), with a linear response ranging from 0.01 to 20 μM and LOD of 0.18 nM. This represents an example of an affinity biosensor which in combination with screen printing technology can be used to produce compact devices (Veerapandian et al., 2016). The analytical signals generated upon hybridization can be recorded using a redox active marker, e.g., [Fe(CN)6]3-/4- (Malecka et al., 2012; Nagar et al., 2019). This constitutes an example of POC device development, which employs a wax stamping technique in order to create the working electrode pattern without including any post printing step for keeping the biomaterial unharmed. Such technique allows the fabrication of a ready-to-use one-step electrochemical biosensor.
The electroactive indicator K3[Fe(CN)6] was used to detect and monitor changes on a glassy carbon electrode (GCE) surface modified with GQDs coupled with probe DNA designed to be complementary to hepatitis B virus DNA (HBV-DNA). When the probe DNA, was strongly bound to the surface of the GQD modified electrode, electron transfer from the electrode to the electrochemically active species [Fe(CN)6]3- was not facilitated. In contrast, when the target HBV-DNA was present in the test solution, the probe DNA bound to it instead of GQDs, enabling electron transfer. The peak current due to [Fe(CN)6]3- reduction increased with increasing concentration of the target HBV-DNA and the variation was directly monitored by differential pulse voltammetry (DPV). The sensor exhibited a linear detection range from 10 to 500 nM and LOD of 1 nM (Xiang et al., 2018). An advantage of this biosensor is that it does not need any fluorophore labelling or enzyme amplification step. So this ultrasensitive label-free electrochemical biosensor can be rather easily cost effectively fabricated.
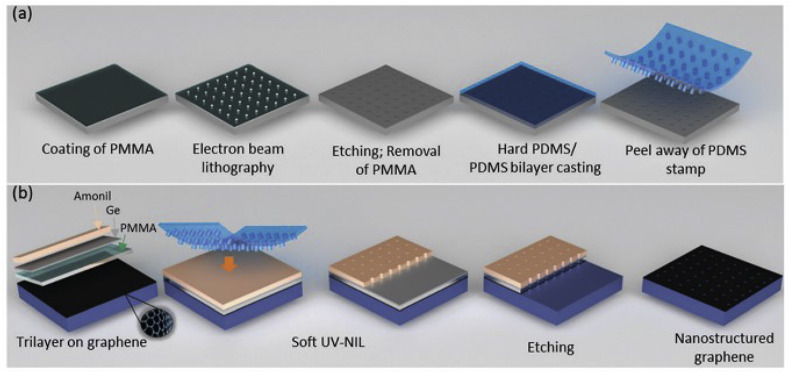
Technologies such as nanoimprint lithography (NIL) have been employed for the fabrication of large surface graphene nanomesh bearing controlled artificial edges (Fig. 6 ). In these artificial edges more than 90% of the target molecules were located allowing the detection of viruses such as hepatitis C. More specifically, low-cost poly(dimethylsiloxane) (PDMS) stamps were prepared from one master mold made by electron beam lithography (EBL). Soft UV-NIL based on a soft hard-PDMS/PDMS bilayer stamp obtained from EBL master mold was used to pattern high-quality single-layer graphene with lateral sizes of several centimeters square. The bioreceptor employed was an ssDNA covalently immobilized on graphene which was complementary to the target DNA related to hepatitis C. After hybridization reaction a decrease in the redox signal was observed which was attributed to dsDNA that had a more rigid conformation. This conformation led to a decrease of electron transfer from the ferrocenyl group (redox marker) to the surface of graphene (Zribi et al., 2016). Such a nanomesh platform exhibits an outstanding ultra-high sensitivity that allows direct detection at the sub-attomolar level of hepatitis C virus DNA.
Fig. 6.
Schematic illustration of the fabrication process (a) of the soft PDMS stamp based on electron beam lithography in a PMMA resist and casting of a hard PDMS/PDMS bilayer, (b) of the graphene nanomesh based on soft UV nanoimprint lithography in an Amonil/Ge/PMMA trilayer transferred by reactive ionic etching. Republished with permission of Royal Society of Chemistry, from Large area graphene nanomesh: an artificial platform for edge-electrochemical biosensing at the sub-attomolar level (Zribi et al., 2016); permission conveyed through Copyright Clearance Center, Inc.
3.2.4. Electrochemical sensing based on antigen–antibody biospecific recognition interaction
Electrochemical biosensors utilizing antigen–antibody specific recognition enable high sensitivity and selectivity. Such an influenza virus sensor was fabricated by indium tin oxide (ITO)/glass electrodes modified with rGO derived from shellac flakes after thermal annealing. The resulting TrGO/ITO/glass electrodes were functionalized with a 1-pyrenebutyric acid N-hydroxysuccinimide ester (PBSE) linker. PBSE makes stable noncovalent interactions via π-electron donating and accepting interactions and also enables covalent attachment of biomolecules through amine groups. These electrodes were incubated with H1N1 antibodies which were bound to the amine groups. Electrochemical measurements were performed in PBS buffer solution containing [Fe(CN)6]3-/4- as a redox mediator. The deposition of TrGO led initially to an increase in the peak current due to its high electron transfer ability. However, after immobilization of H1N1 antibodies, there was a decrease in the peak current due to their more insulating behavior. The resistance of the sensor after H1N1 virus immobilization reached a value that was characteristic for the specific virus, demonstrating selectivity over other viruses. The LODs for the target virus in PBS and saliva samples were estimated to be 26 and 33 PFU mL−1, respectively (Joshi et al., 2020). It is worth noting that graphene-like sheets formation by thermal annealing of shellac, which is a natural biopolymer, is beneficial since it does not include chemical oxidation and chemical reduction processes which require toxic reagents. Moreover, the cyclization of shellac long aliphatic carbon chains takes place at lower temperatures than that of synthetic polymers.
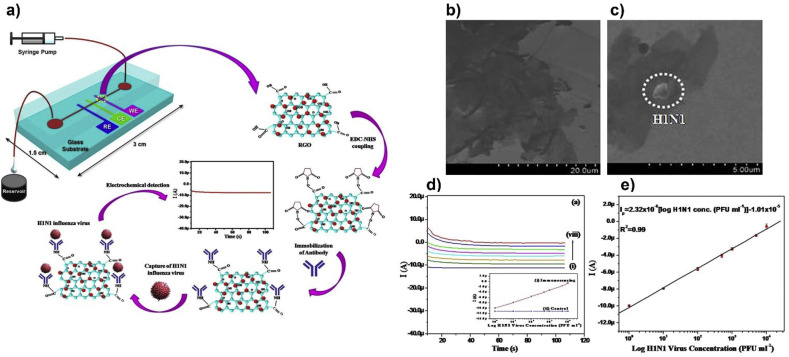
An electrochemical sensor can also be effectively incorporated into a microfluidic platform, which can constitute effective label-free virus detection system. Singh et al. constructed a rGO-coated electrochemical immunosensor integrated with a microfluidic platform for detection of an influenza A H1N1 virus (Fig. 7 ). The carboxyl groups of rGO were activated by using N-ethyl-N-(3-dimethylaminopropyl)carbodiimide (EDC) as a coupling agent and N-hydroxysuccinimide (NHS) as an activator. Thus, improved immobilization of biomolecules through direct linkage of carboxyl groups on the rGO surface and amino groups on an antibody (Ab) was accomplished without the need for a linker or spacer. The immunosensor was shown to detect whole viruses rather than their nucleic acids, facilitating sample preparation. Working, counter, and reference electrodes were fabricated on a glass substrate functionalized with biomolecules and encapsulated with PDMS microchannel. Electrochemical detection of H1N1 viruses was performed at different virus concentrations from 1 to 104 PFU mL−1 in the presence of [Fe(CN)6]3-/4-. An enhanced LOD of 0.5 PFU mL−1 was found (Singh et al., 2017). The integration of an electrochemical immunosensor with a microfluidic platform combines all the benefits of electrochemistry along with those of microfluidics such as low manufacturing cost, small sample volume per analysis, increased efficiency, analysis speed, and portability. It opens door for further biosensor miniaturization.
Fig. 7.
(a) Schematic illustration of an rGO-coated electrochemical immunosensor (chip) which is integrated with a microfluidic platform. EDC-NHS coupling improves the immobilization of antibodies capturing H1N1 influenza virus. The redox active marker [Fe(CN)6]3-/4- is present in the sample solution. Electrochemical response of BSA/Ab/rGO/CA/Au (BSA: Bovine serum albumin, CA: cysteamine) electrode as a function of H1N1 virus concentration was obtained using the chronoamperometric technique. The chronoamperometric current increases proportionally with the H1N1 virus concentration in the range of 1 to 104 PFU mL−1. (b) SEM of rGO/CA/Au and (c) Ab/rGO/CA/Au after capture of H1N1 virus. (d) Chronoamperometric response of the BSA/Ab/rGO/CA/Au-based immunochip as a function of H1N1 virus concentrations. The experiment was controlled using a syringe pump attached to the inlet of the microsystem (inset: response current of BSA/Ab/rGO/CA/Au immunosensing chip with (i) or without (control) H1N1 virus concentration). (e) Calibration plot showing H1N1 virus concentrations (PFU mL−1) and the amperometric current of the immunochip during sensing. Adapted from (Singh et al., 2017). Copyright © 2017 Springer Nature is licensed under CC BY 4.0 (http://creativecommons.org/licenses/by/4.0/).
Another possibility could be the simultaneous detection of more than one virus antigens in the same sample by a multi electrochemical immunosensor platform. An example of this type of sensors is a dual electrochemical immunosensor enabling simultaneous detection of two subtypes of influenza A virus antigens in one sample. The electrode surface composed of GO covered by electrochemically adsorbed MB was successively modified with chitosan and protein-A molecules and then with monoclonal antibodies adequate to detect influenza virus antigens (HA proteins of H5N1 and H1N1). MB is a positively charged organic molecule exhibiting electrocatalytic properties. GO has negatively charged oxygenated functional groups on its basal plane and edges (carboxylic acid, epoxy, carbonyl, and hydroxyl), which enable electrostatic interactions. Chitosan and protein-A molecules were used to enhance the bio-activity of the immobilized antibodies synergistically. These antibodies interacted with H5N1 or H1N1 antigens. Linearity of DPV signals was observed over a wide range of 25–500 pM, and a sensitivity of picomolar levels and rapid response time <1 min were achieved (Veerapandian et al., 2016). This dual screen-printed electrode (SPE) chip is advantageous compared to single working electrodes since it offers simultaneous detection of two different subtypes of influenza-A virus HA protein in one sample. The measurement of electrochemical signals and data processing can be performed sequentially. This method opens prospects for multiplexed detection and discrimination of various pathogenic virus strains.
Magnetic NPs in immunosensor fabrication enable magnetic deposition on the electrode surface. The magnetic deposition is more efficient and stable than a classical deposition and offers better stacking on the electrode surface reducing a contact resistance between the NPs and the electrode material. This results in improved sensing performance compared to a non-magnetic deposition. The aforementioned are evidenced in the case of binary metal NP (Au) and magnetic nanoparticle-decorated graphenes (MNP-GRPs) which were employed as a norovirus-sensing platform. The Au/MNP-GRPs composite was magnetically deposited onto a Pt-interdigitated electrode (Pt-IDE), which was then functionalized by binding GII type norovirus-specific antibody (Ab)onto the surface to produce a norovirus-like particle (NoV-LP)-sensing platform. This system monitors the target biomolecules according to the change in electrical resistance. The electrical conductivity decreased after Ab modification on the Au/MNP-GRPs. When the target NoV-LPs were added, the conductivity of the Ab-conjugated Au/MNP-GRPs decreased even more after the NoV-LPs bound Ab-conjugated Au/MNP-GRPs in the sensing system. High sensitivity and specificity over a concentration range 0.01–1 ng mL−1 was achieved and the LOD was calculated as ~1.16 pg mL−1. Influenza virus (H7N7) antigen and bovine serum albumin (BSA), were used in the specificity test and a high response was obtained only in the case of NoV-LP and this fact constitutes a major advantage of this method along with the benefits of excellent electrical conductivity and magnetic property (Lee et al., 2017).
An electrochemical aptasensor can offer good precision and accuracy in virus detection in real samples offering a simple, rapid, and economical approach towards ultrasensitive and selective measurement of antigens. Their simple fabrication renders them advantageous over other immunosensors. An aptasensor based on a GQD nanocomposite for ultrasensitive detection of HCV core antigen was developed. The aptamer (bearing amine groups) was immobilized on the surface of GQDs (bearing carboxyl groups) through non-covalent interactions such as electrostatic, hydrogen bonding, and π-stacking interactions. GCE was modified with the aforementioned GQDs. GQD was introduced as a suitable substrate due to the richness of hydrophilic edges as well as hydrophobic plane in GQD which enhances the aptamer adsorption on the electrode surface. Ferricyanide/Ferrocyanide has been introduced as a redox probe in detection of HCV core antigen. In the presence of antigen, the strong interaction between aptamer and antigen resulted in the formation of aptamer/antigen complex. Consequently, the charge transfer resistance increased with increasing concentration of HCV core antigen due to hindrance of electron transfer process of [Fe(CN)6]3-/4- at electrode surface. The resulted surface changes were confirmed by EIS, CV, and DPV (Ghanbari et al., 2017). The sensitivity of aptamer in environmental conditions is an issue of great importance. Parameters such as pH should be properly adjusted so that the aptamer will not be destroyed. These conditions influence not only the stability of the aptamer but also the analytical performance. Complex formation between aptamer and antigen also requires estimation of the optimum incubation time.
An alternative method that has the advantage of entire virions detection includes virus deposition on the electrode and observation of the signal changes when it interacts with the antibody compared to the signal in absence of the antibody. It represents an inverse approach to traditional methods, where an antibody is bound on an electrode surface and interacts with an antigen. Such a method was successfully applied for dengue virus detection and involves antibody screening of DENV. It is based on EIS and was developed by using a gold electrode coated with GO and reinforced by polymer matrix composites (PMCs). This electrode had a charge transfer resistance (Rct) that was influenced by the type and quantity of the virus exposed on the surface. Specifically, DENV was bound onto this surface. The addition of antibody changed the surface properties of DENV in terms of shape, size, and surface charge. Under these conditions the signal changed compared to the signal in absence of the antibody. This signal was related to the antibody affinity. [Fe(CN)6]3-/4- was used as a redox indicator. The linear dependence of Rct versus virus concentrations ranged from 1 to 2 × 103 pfu mL−1 DENV with a 0.12 pfu mL−1 detection limit. This method allows the detection of entire dengue virions and antibody recognition via a self-assembly process to PMC. (Navakul et al., 2017). The method benefits from a straightforward experimental protocol utilizing relatively low-cost reagents and conditions and high specificity, enabling, e.g., to discriminate between DENV and influenza A H5N1.
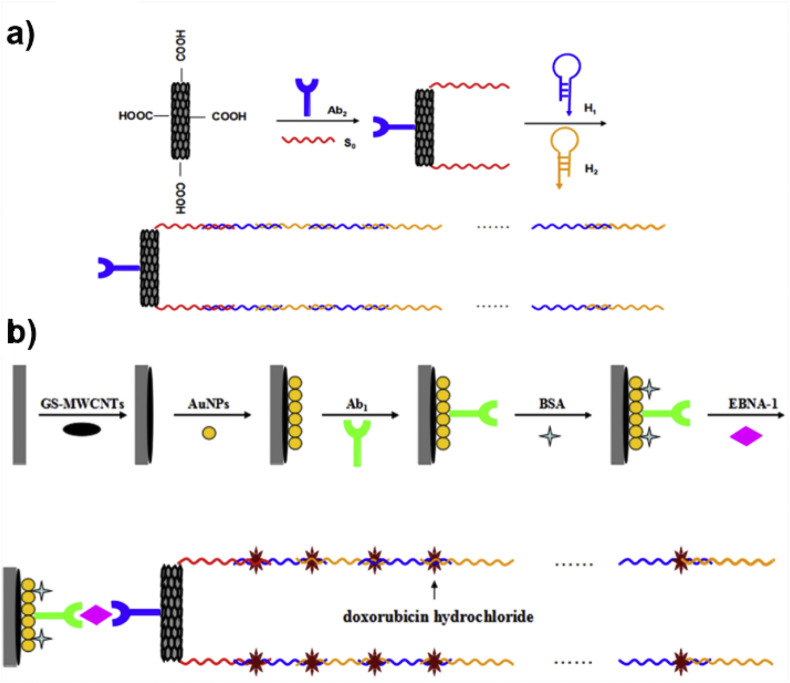
An immunosensor based on antigen–antibody recognition interactions may include also DNA hybridization for its fabrication. DNA hybridization serves the intercalation of the electroactive indicator. An example of the case is an ultrasensitive and selective sandwich electrochemical immunosensor for the detection of Epstein–Barr virus nuclear antigen 1 (EBNA-1) is shown in Fig. 8 . AuNPs were electrodeposited onto the surface of a graphene sheet-multi-walled carbon nanotube (GS-MWCNT) film deposited on GCE. Antibodies were immobilized on the composite and then specific sandwich immunoreactions were allowed to occur between the captured antibodies, EBNA-1 and a secondary antibody on MWCNTs conjugated with DNA (DNA–MWCNTs–Ab2). DNA initiator strands (S0) and secondary antibodies linked to the MWCNTs and double-helix DNA polymers were obtained by a hybridization chain reaction (HCR). S0 on the MWCNTs propagated a chain reaction of hybridization events between two alternating hairpins to form a nicked double-helix. Finally, the electroactive indicator doxorubicin hydrochloride was intercalated between the HCR products to produce an electrochemical signal that was monitored by DPV. Under optimum conditions, the amperometric signal increased linearly with increasing target concentration (0.05–6.4 ng mL−1), and the immunosensor exhibited a LOD as low as 0.7 pg mL−1. This immunosensor exhibits significant binding specificity, acceptable precision, and fabrication reproducibility. Regarding the regeneration, which is an important parameter for practice, it should be noted that after five cycles of regeneration, the immunosensor retained 88.2% of its origin current (C. Song et al., 2014).
Fig. 8.
(a) The first half part of the sandwich immunosensor is constructed by the attachment of antibodies (Ab2) and DNA initiator strands (S0) on carboxyl groups of MWCNTs activated with EDC/NHS. Two hairpins (H1 and H2) were hybridized with S0 DNA strands resulting in DNA–MWCNTs–Ab2 bioconjugates. Double helix DNA polymers were obtained through a hybridization chain reaction where S0 propagates this chain reaction between two alternated hairpins in order to form double helix. These long double helix DNA polymers provide a lot of sites for intercalation of the electroactive indicator, i.e., doxorubicin which significantly improves the sensitivity of the sensor and enhances the signal. The secondary antibody Ab2 is the one of the two antibodies that will capture the antigen in a sandwich-like structure (b) The second half of the sandwich is comprised of graphene sheets (GS) in a composite with MWCNTs, i.e., GS-MWCNTs which coated GCE. GS-MWCNTs was decorated with electrodeposited AuNPs forming AuNPs/GS–MWCNTs composite. Antibodies Ab1 were attached on the latter composite in BSA buffer (pH ~7.0) solution. EBNA-1 antigen was entraped in a ‘sandwich’ structure between DNA-MWCNTs-Ab2 and AuNPs/GS–MWCNTs carrying Ab1. Doxorubicin hydrochloride was used as the electroactive indicator. Reprinted by permission of (C. Song et al., 2014) Copyright © 2014, Elsevier.
3.2.5. Electrochemical sensing based on other approaches
Besides the above-discussed methods molecular amplification technologies can be included in the diagnostic tools for virus detection. These platforms offer portability, automation, speed, cost, and high efficiency. A sensing interface based on reduced graphene oxide-polyaniline (rGO-PANi) film was employed for the in situ detection of LAMP products by open circuit potential measurements. PANi, a pH sensitive conducting polymer, was electro-deposited onto rGO-coated screen-printed electrodes. Specifically, the electrode was electrochemically modified by sweeping in 0.5 M H2SO4 solution containing 0.3 M aniline at scan rate of 50 mV s−1 for ten scans. The final scan was stopped at 0 V to obtain emeraldine which is the only conductive form of PANi. This way electro-polymerization of aniline took place. Protonation and deprotonation of PANi are reversible and the transition between emeraldine salt and emeraldine base can be tuned by shifting the pH of the solution. By combining PANi and rGO, the pH sensitivity of the system was modulated up to about −64 mV per pH unit. This enabled the number of amplified amplicons resulting from the isothermal amplification process to be monitored. The sensor was examined by monitoring LAMP reactions using HBV as a model. The detection mechanism is based on the pH decrease caused by hydronium ions released during the elongation of DNA in LAMP reaction. Two linear regions were obtained at pH ranging from 6.0 to 7.5 and pH ranging from 7.5 to 9.0 (Thu et al., 2018). LAMP is considered as an advanced genetic technique that can provide extremely fast sequential progression (less than 1 h) and relatively high selectivity (100-fold greater than conventional PCR), because the gene amplification is performed under isothermal conditions within a short diagnostic time by using six gene regions, and one type of enzyme (Bst DNA polymerase).
Another approach based on molecular recognition between an RNA aptamer and a virus protein could lead to sensitive and selective sensing of a virus. This sensing approach appears to be an alternative to the currently prevailing nucleic hybridization assays. A composite matrix modified with aptamer electrodes were used for the detection of the L1-major capsid protein of HPV. Specifically, GCEs were modified first with porous reduced graphene oxide (prGO) and then molybdenum sulfide (MoS2) to generate a large surface area. The electrode was covalently functionalized with an aptamer Sc5-c3, i.e., an RNA aptamer targeted against the HPV-16 L1 protein. Using DPV and an optimized sensor interface, a linear relationship between the peak current density of the redox couple [Fe(CN)6]4− and the concentration of HPV-16 L1 proteins was found in the range 0.2–2 ng mL−1 (3.5–35.3 pM) and an LOD of 0.1 ng mL−1 (1.75 pM) was achieved. The sensor demonstrated high selectivity over potential interfering species, such as HPV-16 E6 (Chekin et al., 2018). A high stability of the composite based on prGO and another 2D layered material such as the semiconducting MoS2 benefits from a structural compatibility of both van der Waals materials. Such composites can be effectively modified by aptamers towards sensitive and selective electrochemical detection of viruses like HPV-16.
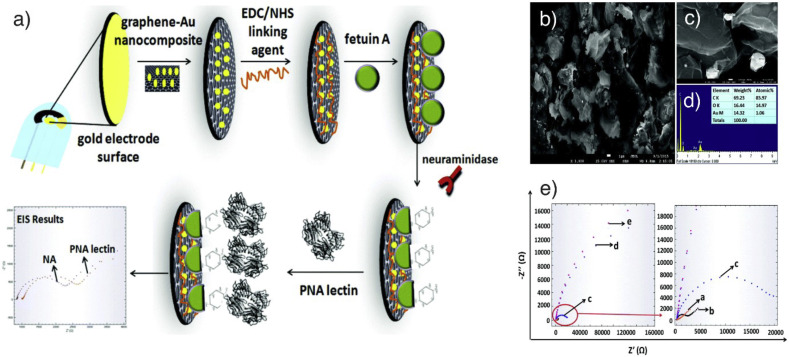
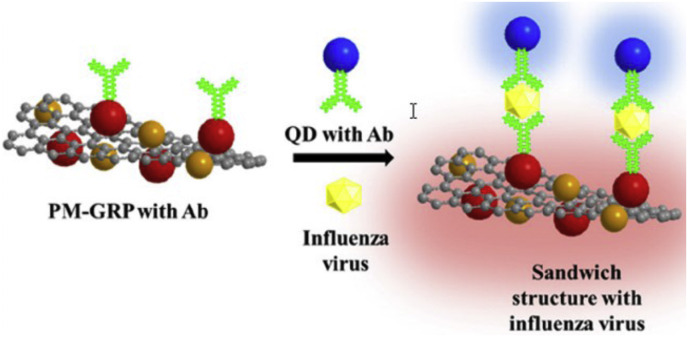
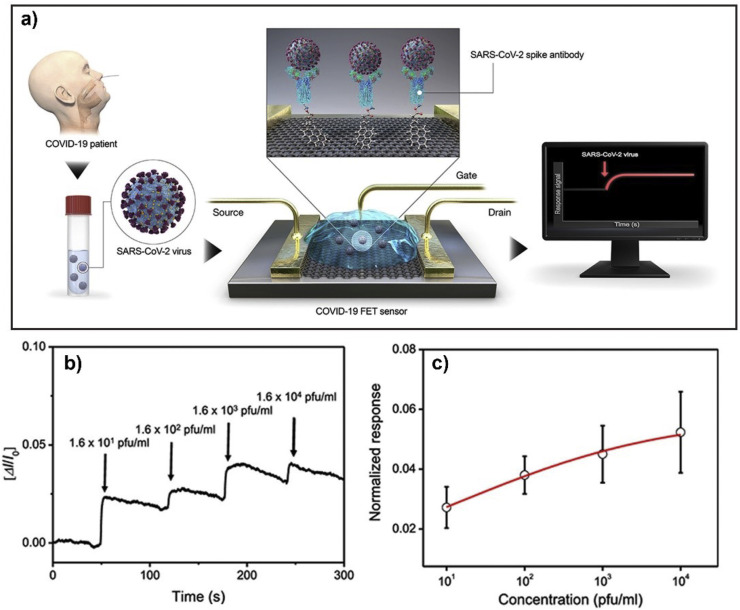
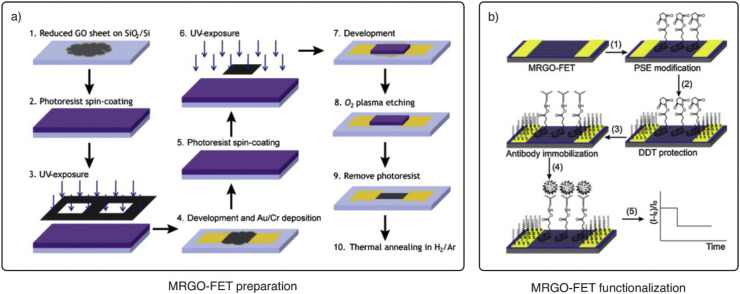
3.3. Virus detection examples
3.3.1. The influenza virus
Is a ssRNA virus belonging to the Orthomyxoviridae family which causes a serious disease (Park and Taubenberger, 2016), with millions of infections and approximately 500 000 deaths every year (according World Health Organization). Therefore, developing a sensor for rapid and sensitive early detection is needed. Influenza A virus has two surface glycoproteins, hemagglutinin and neuraminidase, which exhibit opposite functions. Hemagglutinin binds virions to cells through binding to terminal sialic acid residues on glycoproteins to initiate the infectious cycle. Neuraminidase cleaves terminal sialic acids and releases virions to end the infectious cycle (Kosik and Yewdell, 2019). Anik et al. have investigated an electrochemical diagnostic device based on GO modified by AuNPs for a screen-printed biosensor. The working principle of the sensor involved observing neuraminidase activity. GO functionalized by AuNPs was used to prepare a gold screen-printed electrode. When the electrode surface was covered by the glycoprotein fetuin-A, the resistance of the electrode surface increased because the active electrode surface area was blocked. In the next step, neuraminidase was immobilized on fetuin-A via sialic acid residues, again leading to a drop in electrode conductivity. In the last step, peanut agglutinin lectin was immobilized onto the electrode surface to monitor cleavage of fetuin-A by neuraminidase to form galactose molecules (Fig. 9 ). Thus, detection of the influenza virus was based on the observation of the specific interaction between the lectin and galactose molecules. Increasing the concentration of neuraminidase increased the concentration of galactose molecules, and hence lectin linked to the galactose ends, causing changes in the electrode resistance, which were monitored by EIS. Despite the sophisticated construction of the biosensor, a very low LOD of 10−8 U mL−1 was achieved (Anik et al., 2018).
Fig. 9.
(a) Preparation of the electrochemical biosensor based on GO functionalized by AuNPs (yellow balls). The gold nanoparticles were used as an anchor for the loading of EDC/NHS linker (orange line) via Au–N bond. The fetuin-A (green balls) was immobilized onto electrode surface through the linker and used as a holder for neuraminidase which is a surface glycoprotein of the influenza virus. The PNA (peanut agglutinin) lectin (shadow ball) was used as a monitor for galactose molecules that appear after the cleavage of fetuin-A by neuraminidase. Electrochemical impedance spectroscopy was used for a virus detection. (b), (c) SEM images of a graphene–Au nanocomposite and (d) EDS results of the graphene–Au nanocomposite. (e) Nyquist plots of the biosensor for influenza A virus. a. Plain AuSPE, b. AuSPE/graphene-AuNp, c. AuSPE/graphene-AuNp/fetuin A, d. AuSPE/graphene-AuNp/fetuin A/N, and e. AuSPE/graphene-AuNp/fetuin A/N/PNA lectin. The EIS procedure was set to measure the electron transfer resistance in the frequency range of 0.1 Hz–10 kHz at a potential of 0.1 V. Republished with permission of Royal Society of Chemistry, from Towards the electrochemical diagnostic of influenza virus: development of a graphene-Au–hybrid nanocomposite modified influenza virus biosensor based on neuraminidase activity (Anik et al., 2018); permission conveyed through Copyright Clearance Center, Inc. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
A binary combination of Au and iron oxide NPs (Au-FexOyNPs) decorated on a GO surface could function as an efficient platform for bio-detection. Lee et al. developed a new type of hybrid based on the plasmonic properties of AuNPs and magnetic features of iron oxide NPs immobilized on a GO surface for influenza virus detection. The material benefited from dual functionalization, where the magnetic properties of iron oxide NPs were used for magnetic separation of the hybrid from solution and the plasmonic properties of AuNPs were utilized for more sensitive optical detection. The target virus detection was based on formation of a sandwich structure consisting of two components. One component comprised GO decorated by Au-FexOyNPs, where the AuNPs were covalently functionalized by hemagglutinin antibody via a Au–N bond, and the other component consisted of quantum dots functionalized with the same hemagglutinin antibody. In the presence of the virus, the sandwich complex GO-AuNP-antibody-virus-antibody-quantum dots was formed (Fig. 10 ), leading to changes in the photoluminescence. The photoluminescence of the quantum dots in the absence of the virus was quenched by GO decorated by AuNPs via energy transfer, whereas in the presence of the virus, the distance between the quantum dots and graphene increased and photoluminescence was recovered. Thus, with increasing concentration of analyte, the photoluminescence intensity was increased by plasmon resonance energy transfer. This very sensitive method was able to reach an ultra-low LOD of 7 fg mL−1 (Lee et al., 2018).
Fig. 10.
Illustration of the mechanism of the detection of influenza virus by biosensor forming a sandwich structure. The red balls represent AuNPs, orange balls iron oxide NPs, and blue/green structures represent quantum dots with antibody (Reprinted by permission of (Lee et al., 2018), Copyright © 2018, Elsevier.). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
An electrochemical immunosensor based on a graphene oxide-H5-polychonal antibody-bovine serum albumin (GO-PAb-BSA) nanocomposite was reported by Xie et al. This sensor demonstrated selective detection of avian influenza virus H5 subtype (AIV H5). GO bearing H5-polychonal antibody (PAb) was used as a signal amplification material. The resulting sensor exhibited a 256-fold increase in detection sensitivity compared to an immunosensor without GO-PAb-BSA. The ultrasensitive and selective detection of AIV H5 was attributed to the PAb labeling GO strategy and signal amplification procedure. The established method responded to 2−15 HA unit/50 mL H5, with a linear calibration range from 2−15 to 2−8 HA unit/50 mL (Xie et al., 2014).
An amplified electrochemical immunosensor based on in situ generation of 1-naphthol as an electroactive substance and Pt/CeO2/GO composites as a catalytic amplifier was developed by Yang et al. toward sensitive influenza detection. Antibody-functionalized Pt/CeO2/GO bio-conjugates were captured on an electrode surface through a “sandwich” reaction. The rest (supporting) part of the sandwich comprised another antibody immobilized on AuPd/GCE. Between the two parts of the sandwich, the antigen of influenza was entrapped. 1-naphthol was produced in situ through the hydrolysis of 1-naphthyl phosphate catalyzed by an enzyme, i.e., alkaline phosphatase, giving rise to an electrochemical signal. Pt/CeO2/GO composites assisted the catalytic oxidation of 1-naphthol, resulting in detection signal amplification. The proposed immunosensor had linear ranges of 1.0 × 10−3–1.0 ng mL−1 and 5.0–1.0 × 102 ng mL−1 with LOD of 0.43 pg mL−1 (Yang et al., 2015).
3.3.2. Dengue fever
Is a type of disease caused by the DENV, which infects about 100 million people per annum with a death rate of 2.5%. Tropical and subtropical areas in South America, Africa, and South-East Asia are especially affected by DENV, which is spread by mosquitoes. Nevertheless, owing to global warming and extensive international travel, DENV is becoming a global problem (Liu-Helmersson et al., 2016). Generally, patients who are infected with the virus will develop immunity against it. However, subsequent infection by other serotypes of DENV can cause more serious problems. Early detection of DENV can help to ensure proper treatment, reducing fatalities (Halstead, 1988; Kurane, 2007). Kanagavalli et al. synthesized GO covalently functionalized by electrochemically active ruthenium bipyridine complex. The material was utilized as an immunoprobe for the detection of non-structural 1 protein. The research showed that Ru(II) formed intermolecular bonding with functional groups of GO. Bio-affinity layers of protein-G at the GO-Ru(II) electrode interface supported the localization of monoclonal antibodies selective for binding the epitopes of non-structural 1 protein antigen. Chronoamperometric or fluorescence quenching based immunoassays allowed detection of DENV with LOD of 0.5 ng mL−1 (Kanagavalli and Veerapandian, 2020). Jin et al. prepared an impedimetric biosensor based on 3-aminopropyltriethoxysilane (APTES) functionalized GO for DENV detection via primer hybridization using different oligonucleotide sequences. First, they functionalized the GO surface by APTES to obtain a positive surface charge. The positive amine functional groups in the hybrid were then utilized for immobilization of negatively charged oligonucleotide primers and their incorporation into the electrode. Subsequently, the specific primer was used for binding the DNA target and its selective detection. By impedimetric measurement and observing changes in resistance values, they achieved a very low LOD of 1 fM (Jin et al., 2016).
A dual mode DNA detection method based on nitrogen and sulfur doped GQDs with AuNPs nano assembly (N,S-GQDs@AuNP) was developed by Chowdhury et al. to monitor the serotype of DENV both qualitatively and quantitatively. This biosensor had the benefit that it required only a low volume of the analyte. Qualitative identification of the serotype was defined by the fluorometric signal from the sensor, whereas quantitative determination of the identified serotype was obtained from the DPV signal. Multiple serotypes of DNA could be detected by this method with a linear range of 10−14 to 10−6 M and LOD of 9.4 fM. MB was used as a redox indicator for the DPV measurements. This technique is also an example of DNA hybridization detection (Chowdhury et al., 2018).
3.3.3. Hepatitis C virus
Is a RNA virus that poses another serious human pathogen. HCV strikes the liver and causes inflammation. The World Health Organization (WHO) has estimated that about 71 million people in 2013 suffer from chronic HCV infection, which can develop into cirrhosis or liver cancer. WHO estimated that in 2016, approximately 399000 people died from hepatitis C, mostly from cirrhosis and liver cancer. Early and sensitive detection of the hepatitis virus in blood at the beginning of infection is very important for treatment. AgNPs and thiol graphene quantum dots (GQD-SH) have been employed for the ultrasensitive and selective detection of the HCV core antigen. AgNPs/GQD-SH was utilized as a substrate to load antibody for detection of the HCV core antigen. AgNPs were immobilized on SH groups of GQDs via bond formation of Ag–S. Anti-HCV was then loaded onto the electrode surface via interaction between –NH2 group of the antibodies and AgNPs. A decrease in the electrochemical signal was observed after specific recognition between the antibodies and antigens. Riboflavin was used as a redox probe for development of a HCV core antigen electrochemical immunosensor. The proposed immunosensor showed a wide linear range from 0.05 pg mL−1 to 60 ng mL−1 with LOD of 3 fg mL−1 (Valipour and Roushani, 2017). Kim et al. presented a different way to detect HCV genes in liver cells. They combined GO with deoxyribozyme (DNAzyme), which is an RNA-cleaving antisense oligo-DNAzyme mediating catalytic hydrolysis of a target mRNA. The graphene hybrid with loaded DNAzyme was utilized to monitor HCV mRNA in living cells. Initially, nucleobases of DNAzyme were immobilized on the graphene surface. Next, they were conjugated to fluorescence dye at the 5′ end of the DNAzyme. The fluorescence was quenched due to π−π stacking between the dye and graphene. The mechanism of detection was based on the change in distance between the graphene and DNAzyme caused by double helix formation in the presence of the complementary target sequence of HCV NS3 mRNA. This detachment resulted in recovery of fluorescence of the dye due to the loss of proximity with graphene. Using this method based on the on-off fluorescence effect, a LOD of 10 nM was achieved (Kim et al., 2013).
Li et al. proposed a DNA-assisted synthesis of a magnetic reduced graphene oxide-copper nanocomposite (mrGO-CuNCs) and its application for HCV DNA detection. A special DNA template was designed based on the different interactions of mrGO and Cu NPs with ssDNA or dsDNA. For Cu NPs, dsDNA acted as a template, whereas ssDNA helped to immobilize Cu NPs onto the surface of mrGO. In the presence of target DNA (HCV), a DNA composite was formed containing both ssDNA and dsDNA. After magnetic separation, the CuNPs were dissolved into Cu ions, which acted as catalysts for the oxidation of o-phenylenediamine. The oxidized product, i.e., 2,3-diaminobenazine, was detected via electrochemical assay. This strategy was able to sensitively detect HCV-DNA over a linear range of 0.5–10 nM and had LOD of 405.0 pM (J. Li et al., 2020a).
A rapid and sensitive diagnostic method for HCV has been developed Ping et al. based on a host-and-guest interaction between cucurbit[7]uril (CB7) and MB (Jiang et al., 2020). The CB7 molecules captured the MB molecules in the solution, promoting electron transport between MB and an electrode surface. GO was used for the embedment of CB7. CB7-N3-GO was generated after photo-crosslinking CB7 with azide-functionalized GO. Probe MB DNA was hybridized with the target HCV DNA, producing dsDNA. Afterwards, the MB DNA was degraded by DNA exonuclease III, releasing MB molecules. The HCV DNA was then free to hybridize with the next MB DNA and the process continued in cycles. After several cycles, the amount of released MB correlated to the amount of HCV DNA. The method was shown to linearly detect the HCV nucleic acid in the range 0.2–10 nmol L−1 with LOD as low as 160.4 pmol L−1 (Jiang et al., 2020).
Development of an electrochemical genosensor for the diagnosis of HCV in real samples was reported by Oliveira et al. based on a gold electrode modified with GO and ethylenediamine (GO-ETD) (Oliveira et al., 2019b). The modification was useful to probe DNA immobilization following interaction with the target DNA HCV through hybridization. DPV and EIS were employed for the evaluation of HCV infected sera and viral genomic RNA. An inverse linear relationship between the values of the peak current of ferrocyanide ion [Fe(CN)6]4- oxidation and the concentration of the samples was observed. The LOD was 1:483 (v/v) or 1.36 nmol L−1 of RNA. Thus, the selective and specific diagnosis of hepatitis C could be achieved by this genosensor (Oliveira et al., 2019b).
Ahour et al. constructed a label-free electrochemical biosensor for rapid and effective detection of short sequence oligonucleotides (ssDNA) related to HCV. A pencil graphite electrode was modified with GO. Probe ssDNA was attached on the surface of GO due to π-interactions of purine/pyrimidine rings with aromatic domains of GO. After hybridization of probe ssDNA with the target DNA HCV, dsDNA was formed, which easily desorbed from the GO surface. This had a direct effect on the guanine oxidation signal recorded through DPV. The oxidation peak current was proportional to the concentration of the complementary strand related to HCV in the range 0.1 nM–0.5 μM with LOD of 4.3 × 10−11 M (Ahour and Shamsi, 2017). The hepatitis B virus core antigen (HBcAg) was immobilized onto a AuNPs-decorated rGO (rGO-en-AuNPs) nanocomposite for use as an antigen-functionalized surface to sense the presence of anti-HBcAg. The modified rGO-en-AuNPs/HBcAg was used to impedimetrically detect anti-HBcAg in the presence of anti-estradiol antibody and bovine serum albumin as interferences. Successful detection of anti-HBcAg in spiked buffer samples and spiked human serum samples was carried out. The electrochemical response showed a linear relationship between the electron transfer resistance and the concentration of anti-HBcAg ranging from 3.91 ng mL−1 to 125.00 ng mL−1 with the lowest LOD of 3.80 ng mL−1 (Abd Muain et al., 2018).
A sensitive sandwich-type electrochemical immunoassay A was reported by Alizadeh et al. for the detection of hepatitis B virus surface antigen (HBsAg). Construction of this biosensor was based on the covalent conjugation of a monoclonal antibody anti-HBsAg (Ab1) on chitosan functionalized GO using glutaraldeyde as a linker molecule between the primary antibody and GO. Incubation with the target antigen HBsAg was then performed. The electrochemical signal due to the catalytic reduction of hydrogen peroxide in presence of MB was recorded. Two nanocomposites were found to promote the catalytic reduction: Ab2-Fe3O4-AuNPs-hemin/G-quadruplex and Ab2-hemin-amino-rGO-AuNPs-hemin/G-quadruplex. G-quadruplex DNAzyme composed of hemin and guanine-rich nucleic acid was used as an amplifier owing to its excellent peroxidase activity. The role of the Fe3O4-AuNPs or (H-amino-rGO-Au) nanocomposite was to provide an appropriate support for the immobilization of the hemin/G-quadruplex. The primary antibody immobilized on the GO and the secondary antibody immobilized on the Fe3O4-AuNPs or (H-amino-rGO-Au) nanocomposite entrapped the target protein in a sandwich-like structure. The synergetic action of the Fe3O4-AuNPs or (H-amino-rGO-Au) nanocomposite with the hemin/G-quadruplex toward the electrocatalytic reduction of H2O2 in the presence of MB as a mediator resulted in an immunosensor with a wide linear dynamic range: for Fe3O4-AuNPs, the range was 0.1–300 pg mL−1 with LOD of 60 fg mL−1, whereas for H-amino-rGO-Au, the range was 0.1–1000 pg mL−1 with LOD of 10 fg mL−1 (Alizadeh et al., 2018).
A composite comprising GO, ferrocene and chitosan was synthesized by Zhao et al. and used to develop an electrochemical immunosensor for hepatitis B surface antigen HBsAg detection. The composite was combined with AuNPs and applied as a film layer on a gold electrode. The presence of AuNPs provided binding sites for proteins and enhanced electron conductivity, which are both important for antibody immobilization used for antigen HBsAg detection. EIS was performed in a solution containing K3[Fe(CN)6]/K4[Fe(CN)6] and a strong reversible redox signal related to current changes was produced. The immunosensor showed a wide linear range from 0.05 to 150 ng mL−1 and LOD for HBsAg of 0.01 ng mL−1 (Zhao et al., 2018).
A pulse-induced highly sensitive electrochemical sensor has been fabricated by Chowdhury et al. for hepatitis E virus (HEV) detection. This immunosensor used a self-assembly approach and interfacial polymerization to combine GQDs and gold-embedded PANi nanowires in a composite. An antibody conjugated to the composite deposited on a GCE was used for HEV recognition. During this process, the induced electrical pulse increased the sensitivity toward HEV, which was attributed to the expanded surface of the virus particle. The best results were observed at +0.8 V (positive with respect to Ag/AgCl reference electrode). A good linear relationship between Rct and the logarithmic values of HEV-LP (LP: like-particles) concentration was recorded in the range 1 fg mL−1 to 100 pg mL−1. The LOD was found to be 0.8 fg mL−1, which can be considered exceptionally low (Chowdhury et al., 2019).
3.3.4. SARS-CoV-2
Is a new type virus that causes human disease (COVID-19) associated with severe health problems, e.g., respiratory distress. This type of virus belongs to the ssRNA virus family with a positive sense. The WHO classified the recent COVID-19 outbreak as a pandemic on March 12, 2020. As of July 23, 2020, more than 15 million cases of COVID-19 have been confirmed around the world, resulting in 630 000 deaths. Thus, a rapid and sensitive sensor for COVID-19 detection is urgently needed. Seo et al. have developed a very sophisticated method for virus detection by utilizing a graphene matrix for a FET as is shown in Fig. 11 . First, they immobilized a spike antibody (anti-SARS-CoV-2) onto a graphene surface via 1-pyrenebutyric acid N-hydroxysuccinimide ester (PBSE) as a probe linker. Electrochemical measurements were then used to evaluate the presence of the virus in a sample. The current-voltage of the graphene biosensor platform was measured before and after attachment of the virus through the spike antibody. In the presence of the virus, the slope of the current-voltage curve was decreased, indicating successful detection of the COVID-19 virus by immobilization on the antibody. This system can provide a very low LOD of 1.6 PFU mL−1 (Seo et al., 2020).
Fig. 11.
Schematic diagram of ‘‘COVID-19 FET sensor’’ operation procedure. (a) FET biosensor contains graphene layer noncovalently functionalized via PBSE with SARS-CoV-2 spike antibody. (b) Real-time response of COVID-19 FET toward SARS-CoV-2 cultured virus. In the presence of the virus, the slope of the current-voltage curve decreased. (c) Related dose-dependent response curve. Reprinted by permission of (Seo et al., 2020). Copyright © 2020, American Chemical Society.
3.3.5. Zika virus
Is a flavivirus that is spread by Aedes mosquitoes. It was first identified in 1947 and took its name after the Zika Forest in Uganda. This virus is associated with congenital malformations and microcephaly during pregnancy. Even though many infected people appear to be asymptomatic, the infection can still cause serious neurologic complications, including neuropathy and Guillain-Barré syndrome. In 2016, the WHO declared an emergency in public health due to the transmission of Zika virus (ZIKV) to a large of number of people in Central and South America. Apart from transmission through mosquitoes, blood transfusion, prenatal, and sexual transmission constitute possible infection paths.
An electrochemical genosensor for detection of the genomic RNA of Zika virus has been reported by Moço et al. and tested on real samples from infected patients. In this biosensor, graphite electrodes were modified with electrochemical rGO and polytyramine, which is a conducting polymer. An oligonucleotide ZIKV-probe was immobilized on the modified electrode, which could then be hybridized with complementary target ZIKV-RNA. Electrochemical measurements were conducted by DPV in KCl solution containing K4[Fe(CN)6/K3[Fe(CN)6]. The LOD reached 0.1 fg mL−1 (1.72 copies mL−1). The advantages of this method were rapid analysis (20 min), a reliable response even after 60 days of storage and the possibility of miniaturization (Moço et al., 2019).
3.3.6. Human immunodeficiency virus (HIV)
A member of the Retroviridae family, still causes significant worldwide public health issues. In fact, it has been considered one of the most concerning viruses of the last few decades. Advancements in the treatment of AIDS have improved the situation greatly, but it is still important to develop detection methods for the HIV gene that can be made readily available around the world. HIV contains two identical strands of RNA that can be transcribed into DNA for additional gene expression through reverse transcription. This type of RNA virus attacks cells of the immune system, such as CD4 or T cells. Due to the destruction of cells and subsequent decrease of cell numbers below the critical level, the immune system is weakened, and thus is not able to defend the body against common diseases that are not usually serious for a healthy person. Thus, detection of HIV biomarkers or genes is very important for early diagnosis and prevention of the virus (Horiya et al., 2014; Martínez et al., 1999; Parker et al., 2009; Sapsford et al., 2010). Islam et al. functionalized graphene with amino groups for covalent conjugation of antibodies (anti-p2) on the graphene surface via carbodiimide activation. Electrochemical measurements were conducted to monitor changes in the resistance of the electrode surface due to interaction of the antigen with the specific antibody (Islam et al., 2019). An electrochemical biosensor based on graphene for the detection of HIV DNA was also developed by Wang et al. (Y. Wang et al., 2015b) First, gold nanoclusters (AuNCs) were immobilized on graphene via a one-step ultrasonic method. The resulting composite, labeled graphene/AuNCs, was then used for GCE modification and to a capture probe aptamer with cytosine-rich base by conjugation. This probe aptamer was labeled with MB. The electrode was immersed in a target solution containing the enzyme exonuclease III (Exo III). When the capture probe and target DNA folded into a double DNA helix, the enzyme digested the capture probe and a MB molecule was released, triggering an electrochemical signal. The role of the enzyme was to assist the target recycling, resulting in signal amplification and increased sensitivity of the biosensor toward HIV DNA. The biosensor exhibited a low LOD of 30 aM (Y. Wang et al., 2015b).
Another approach based on an optical method was developed by Li et al. (2017b) They presented a simple and rapid method for analysis by utilizing boron and nitrogen co-doped GQDs as a fluorescent probe for high-affinity binding to ssDNA and dsDNA for visualizing the dynamic invasion of HIV DNA into living cells. The excellent absorption and photoluminescence properties of the quantum dot hybrid provided FRET processes for nucleic acid detection and by observation of changes in photoluminescence intensity achieved a very low LOD of 0.5 nM (Li et al., 2017b).
3.3.7. Rotavirus
Is a genus of dsRNA viruses in the family Reoviridae. There are ten species of the genus: A, B, C, D, E, F, G, H, I, and J. Type A, the most common species, causes more than 90% of rotavirus infections in humans (Dennehy, 2015). According to the WHO, even in the 21st century, more than one million children die each year due to rotavirus contamination of drinking water. Liu et al. reported a rapid and sensitive biosensor for pathogenic rotavirus detection in real time based on the micropatterned reduced graphene oxide field-effect transistor (MrGO-FET). In the first step, they prepared suitable matrix for antibody immobilization based on rGO as shown in Fig. 12 a. The second step was adsorbing of PBSE on the graphene surface, then used this functionalization to covalently bind the specific rotavirus antibodies as is shown in Fig. 12b. Injection of a rotavirus sample caused a decrease in the source-drain current and then gradual saturation. The lowest LOD for rotavirus was determined as 102 PFU, which is better than commonly used methods, such as ELISA (Liu et al., 2013).
Fig. 12.
(a) Preparation scheme of the micropatterned reduced graphene oxide field-effect transistor (MrGO-FET). (b) Schematic of the biosensor for rotavirus detection in real time. (1) PSE modification of rGO matrix, (2) blocking of the gold electrode by 1-dodecanethiol (DDT), (3) antibody immobilization and blocking the MrGO surface by ethanolamine, (4) rotavirus capture is detected by (5) monitoring the relative current changes (I) in real time. Adapted by permission of (Liu et al., 2013). Copyright © 2013, Elsevier. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3.8. Human papillomavirus (HPV)
Is a group of non-enveloped, dsDNA viruses composed of more than 100 genotypes (Piro et al., 2011). About 93% of worldwide invasive carcinomas have been shown to be associated with a limited spectrum of HPV types, mostly HPV 16 and 18 (Zari et al., 2009). Thus, the sensitive detection of HPV DNA sequences is crucial for the rapid and early diagnosis of diseases. Huang et al. constructed an ultrasensitive DNA electrochemical sensor for the detection of HPV based on modification of a GCE with a composite of graphene, Au nanorods (AuNRs) and polythionine (graphene/Au NR/PT). Specifically, a graphene/Au NRs film was deposited onto a GCE and then thionine was electropolymerized on this film. Graphene was used to increase the electrode surface area and electrical conductivity, whereas Au NRs were used to enhance the immobilization of the probe DNA for hybridization with the target DNA. Two auxiliary probes were designed and used for the long-range self-assembly of a DNA nanostructure. Thionine has good transfer ability and the nitrogen atom of the NH2 moieties of thionine strongly bound to the AuNRs surface. A redox mediator, 1,10-phenanthroline ruthenium dichloride [Ru(phen)3]2+, which could bind to the DNA long-range hybridized structure, was used to amplify the electrochemical signal. The DNA biosensor displayed excellent performance for HPV DNA detection over the range 1.0 × 10−13 to 1.0 × 10−10 mol L−1 with LOD of 4.03 × 10−14 mol L−1 (Huang et al., 2015).
3.3.9. Epstein–Barr virus (EBV)
Is a type of virus that infects over 90% of adults. EBV can contribute to oncogenesis and is often found in Burkitt's lymphoma, Hodgkin's lymphoma, nasopharyngeal carcinoma, and lymphoproliferative diseases in immunosuppressed individuals (Young and Murray, 2003). EBV damages human B lymphocytes in vitro, and expression of the latent membrane protein 1 (LMP-1) gene of EBV affects the morphology and growth of human epithelial cells (Banchereau and Rousset, 1991; Kaye et al., 1993) and contributes to oncogenesis. An electrochemical sandwich immunosensor with increased sensitivity for the determination of the tumor marker LMP-1 was designed by Zhang et al. (2016). The capture antibody (Ab1) was immobilized on a composite consisting of graphene sheets and carboxylated multi-walled carbon nanotubes (GS-MWCNTs) in a chitosan matrix. This assembly was placed on the surface of a GCE. The detection antibody (Ab2) marked with thionine was fixed on a composite of HRP/Pd@Pt-MWCNTs (HRP: enzyme horseradish peroxidase). Upon exposure to the antigen (LMP-1), an amplified signal was observed in DPV due to synergistic effects between the Pd@Pt NPs and HRP in the presence of added H2O2. The DPV signal was best acquired at a working potential of −250 mV (vs. Ag/AgCl) in the presence of H2O2. Under optimized conditions, the calibration range extended from 0.01 to 40 ng mL−1 of LMP-1 with a LOD of 0.62 pg mL−1 (Zhang et al., 2016).
4. Summary and conclusions
Rapid, easy-to-use and cheap virus detection methods could significantly help in pandemic situations as thought us by the very recent experience with COVID-19. On one hand, the rapid virus detection enables rapid tracking of primary contacts, prolongs therapeutic window and support targeted treatment, on the other hand it facilitates spread of a disease, which helps to get pandemic under control. Graphene, its derivatives and composites offer many benefits in biosensing applications for virus detection. The high electrical and thermal conductivity, flexibility, light weight, and biocompatibility of these materials together with their large surface areas make them ideal platforms for electrochemical biosensors. Graphene, GO, and rGO effectively quench photoluminescence, whereas GQD displays significant photoluminescence that can be tuned by doping and functionalization. These features also make graphene-based materials suitable for use in optical biosensors. Combining these materials with suitable nanomaterials can enhance the portfolio of available detection methods, e.g., Au/Ag NPs impart a significant SERS signal. The sensor sensitivity is usually connected with changing electronic properties in the presence of virus adsorbates. In addition, the large surface area of graphene-based materials facilitates the adsorption of analytes from the environment. Selectivity is achieved by combining graphene-based materials with antibodies, aptamers, or nucleic acids, which selectively bind counterparts specific for a particular virus. All these features make graphene-based biosensors a versatile but sensitive and selective platform for detection of pathogenic viruses. The developed strategies can be readily adopted for detection of new emerging pathogenic viruses, which can help to efficiently tackle future diseases caused by such pathogens.
5. Future perspectives
Both optical and electrochemical methods are relatively sensitive and selective, and highly suitable for rapid, real time, on-site, easy-to-use, and rather low-cost detection of viruses. It should be noted that the stability of modified electrodes may be a limiting factor for electrochemical methods, but properly stored electrodes (e.g., in the form of microchips) can provide sufficient stability. Among graphene derivatives, the emerging covalently modified graphene such as cyanographene and graphene acid, which are both conductive and display a well-defined chemical nature, open new doors toward graphene-based platforms that can be utilized for the precise construction of biosensors. For even broader and robust application of graphene derivatives in (bio)sensorics a high batch-to-batch reproducibility and standardization must be achieved. In addition, a commercial availability of such materials may boost further development in this field. The dispersibility of graphene-based nanocomposites can also be exploited as an ink for ink-printed electrodes and paper-based biosensors, broadening the application potential of these materials in biosensors and enabling miniaturization. A significant attention should be paid to miniaturization, standardization and multi-readout biosensors, which can lower rate of false negative/positive results. Further miniaturization, combination with fluid systems and integration into multi-chip systems represent important challenges for the next generation of graphene-based biosensors. These features combined with high sensitivity, selectivity and large-scale production are of primary importance for achieving the broad applicability of graphene sensors for the detection of various viral species. Particularly great potential promises a combination of graphene-based biosensors with smartphones enabling low-cost, reliable and versatile platform for (bio)sensing, which can be widely applied on-site, e.g., as POC devices. Without any doubt graphene and its derivatives are highly suitable materials for engineering of efficient biosensors for virus detection, which can be readily adopted for detection of newly emerging viruses. There is also a great potential for commercialization of emerging graphene-based biosensors.
CRediT author statement
Eleni Vermisoglou: Writing - review & editing. David Panáček: Writing - review & editing. Kolleboyina Jayaramulu: Writing - review & editing. Martin Pykal: Writing - review & editing. Ivo Frébort: Writing - review & editing. Milan Kolář: Writing - review & editing. Marián Hajdúch: Writing - review & editing. Radek Zbořil: Writing - review & editing. Michal Otyepka: Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We acknowledge the support from the Operational Programme Research, Development and Education – European Regional Development Fund, projects no. CZ.02.1.01/0.0/0.0/16_019/0000754 and CZ.1.05/2.1.00/19.0377 of the Ministry of Education, Youth and Sports of the Czech Republic, from the H2020 ERC project (683024) and from the Internal Student Grant Agency of the Palacký University Olomouc, Czech Republic (IGA_PrF_2020_022).
References
- Abd Muain M.F., Cheo K.H., Omar M.N., Amir Hamzah A.S., Lim H.N., Salleh A.B., Tan W.S., Ahmad Tajudin A. Bioelectrochemistry. 2018;122:199–205. doi: 10.1016/j.bioelechem.2018.04.004. [DOI] [PubMed] [Google Scholar]
- Afsahi S., Lerner M.B., Goldstein J.M., Lee J., Tang X., Bagarozzi D.A., Pan D., Locascio L., Walker A., Barron F., Goldsmith B.R. Biosens. Bioelectron. 2018;100:85–88. doi: 10.1016/j.bios.2017.08.051. [DOI] [PubMed] [Google Scholar]
- Agrawal A., Tripp R.A., Anderson L.J., Nie S. J. Virol. 2005;79:8625–8628. doi: 10.1128/JVI.79.13.8625-8628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahour F., Shamsi A. Anal. Biochem. 2017;532:64–71. doi: 10.1016/j.ab.2017.06.004. [DOI] [PubMed] [Google Scholar]
- Alizadeh N., Hallaj R., Salimi A. Electroanalysis. 2018;30:402–414. [Google Scholar]
- Allen M.J., Tung V.C., Kaner R.B. Chem. Rev. 2010;110:132–145. doi: 10.1021/cr900070d. [DOI] [PubMed] [Google Scholar]
- Alwarappan S., Boyapalle S., Kumar A., Li C.Z., Mohapatra S. J. Phys. Chem. C. 2012;116:6556–6559. [Google Scholar]
- Ambrosi A., Chua C.K., Latiff N.M., Loo A.H., Wong C.H.A., Eng A.Y.S., Bonanni A., Pumera M. Chem. Soc. Rev. 2016;9:2458–2493. doi: 10.1039/c6cs00136j. [DOI] [PubMed] [Google Scholar]
- Ananthanarayanan A., Wang Y., Routh P., Sk M.A., Than A., Lin M., Zhang J., Chen J., Sun H., Chen P. Nanoscale. 2015;7:8159–8165. doi: 10.1039/c5nr01519g. [DOI] [PubMed] [Google Scholar]
- Anik Ü., Tepeli Y., Sayhi M., Nsiri J., Diouani M.F. Analyst. 2018;143:150–156. doi: 10.1039/c7an01537b. [DOI] [PubMed] [Google Scholar]
- Arce L.P., Müller M.F., Martinez A., Baiker A., Marranzino G., Agote F., Vizoso-Pinto M.G. Front. Microbiol. 2019;10:2481. doi: 10.3389/fmicb.2019.02481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badyda A.J., Dabrowiecki P., Lubinski W., Czechowski P.O., Majewski G., Chcialowski A., Kraszewski A. Springer; Dordrecht: 2013. Neurobiology of Respiration; pp. 229–235. [DOI] [PubMed] [Google Scholar]
- Bae S., Kim H., Lee Y., Xu X., Park J.S., Zheng Y., Balakrishnan J., Lei T., Ri Kim H., Song Y. Il, Kim Y.J., Kim K.S., Özyilmaz B., Ahn J.H., Hong B.H., Iijima S. Nat. Nanotechnol. 2010;5:574–578. doi: 10.1038/nnano.2010.132. [DOI] [PubMed] [Google Scholar]
- Bakandritsos A., Pykal M., Boński P., Jakubec P., Chronopoulos D.D., Poláková K., Georgakilas V., Čépe K., Tomanec O., Ranc V., Bourlinos A.B., Zbořil R., Otyepka M. ACS Nano. 2017;11:2982–2991. doi: 10.1021/acsnano.6b08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D. Bacteriol. Rev. 1971;35:235–241. doi: 10.1128/br.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau J., Rousset F. Nature. 1991;353:678–679. doi: 10.1038/353678a0. [DOI] [PubMed] [Google Scholar]
- Barré-Sinoussi F., Chermann J.C., Rey F., Nugeyre M.T., Chamaret S., Gruest J., Dauguet C., Axler-Blin C., Vézinet-Brun F., Rouzioux C., Rozenbaum W., Montagnier L. Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- Becerril H.A., Mao J., Liu Z., Stoltenberg R.M., Bao Z., Chen Y. ACS Nano. 2008;2:463–470. doi: 10.1021/nn700375n. [DOI] [PubMed] [Google Scholar]
- Bonar M.M., Tilton J.C. Virology. 2017;505:80–90. doi: 10.1016/j.virol.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonham N., Kreuze J., Winter S., van der Vlugt R., Bergervoet J., Tomlinson J., Mumford R. Virus Res. 2014;186:20–31. doi: 10.1016/j.virusres.2013.12.007. [DOI] [PubMed] [Google Scholar]
- Boukhvalov D.W., Katsnelson M.I. Nano Lett. 2008;8:4374–4379. doi: 10.1021/nl802098g. [DOI] [PubMed] [Google Scholar]
- Briese T., Kapoor A., Mishra N., Jain K., Kumar A., Jabado O.J., Ian Lipkin W. 2015. mBio 6; p. e01491. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghard M., Klauk H., Kern K. Adv. Mater. 2009;21:2586–2600. doi: 10.1002/adma.200803582. [DOI] [PubMed] [Google Scholar]
- Cao J., Li D. J. Cell. Biochem. 2018;119:4897–4906. doi: 10.1002/jcb.26717. [DOI] [PubMed] [Google Scholar]
- Castro Neto A.H., Guinea F., Peres N.M.R., Novoselov K.S., Geim A.K. Rev. Mod. Phys. 2009;81:109–162. [Google Scholar]
- Chan C., Shi J., Fan Y., Yang M. Sensor. Actuator. B Chem. 2017;251:927–933. [Google Scholar]
- Chee S.Y., Pumera M. Analyst. 2012;137:2039–2041. doi: 10.1039/c2an00022a. [DOI] [PubMed] [Google Scholar]
- Chekin F., Bagga K., Subramanian P., Jijie R., Singh S.K., Kurungot S., Boukherroub R., Szunerits S. Sensor. Actuator. B Chem. 2018;262:991–1000. [Google Scholar]
- Chen C., Wang J. Analyst. 2020;145:1605–1628. doi: 10.1039/c9an01998g. [DOI] [PubMed] [Google Scholar]
- Chen D., Tang L., Li J. Chem. Soc. Rev. 2010;39:3157–3180. doi: 10.1039/b923596e. [DOI] [PubMed] [Google Scholar]
- Cheng C., Li S., Thomas A., Kotov N.A., Haag R. Chem. Rev. 2017;117:1826–1914. doi: 10.1021/acs.chemrev.6b00520. [DOI] [PubMed] [Google Scholar]
- Cheng W., Ding C., Nie X., Duan T., Ding R. ACS Sustain. Chem. Eng. 2017;5:5503–5511. [Google Scholar]
- Chia X., Ambrosi A., Otyepka M., Zbořil R., Pumera M. Chem. Eur J. 2014;20:6665–6671. doi: 10.1002/chem.201402132. [DOI] [PubMed] [Google Scholar]
- Chowdhury A.D., Ganganboina A.B., Nasrin F., Takemura K., Doong R.A., Utomo D.I.S., Lee J., Khoris I.M., Park E.Y. Anal. Chem. 2018;90:12464–12474. doi: 10.1021/acs.analchem.8b01802. [DOI] [PubMed] [Google Scholar]
- Chowdhury A.D., Takemura K., Li T.C., Suzuki T., Park E.Y. Nat. Commun. 2019;10:1–12. doi: 10.1038/s41467-019-11644-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronopoulos D.D., Bakandritsos A., Pykal M., Zbořil R., Otyepka M. Appl. Mater. Today. 2017;9:60–70. doi: 10.1016/j.apmt.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua C.K., Pumera M. Chem. Soc. Rev. 2014;324:762–772. doi: 10.1039/c3cs60303b. [DOI] [PubMed] [Google Scholar]
- Chung C., Kim Y.K., Shin D., Ryoo S.R., Hong B.H., Min D.H. Acc. Chem. Res. 2013;46:2211–2224. doi: 10.1021/ar300159f. [DOI] [PubMed] [Google Scholar]
- Chung S., Revia R.A., Zhang M. Adv. Mater. 2019;69:1426–1544. [Google Scholar]
- Compton O.C., Nguyen S.T. Small. 2010;6:711–723. doi: 10.1002/smll.200901934. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Euro Surveill. 2020;25:2000045. [Google Scholar]
- Crucian B.E., Stowe R.P., Pierson D.L., Sams C.F. J. Immunol. Methods. 2001;247:35–47. doi: 10.1016/s0022-1759(00)00326-4. [DOI] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L., Chen Z., Zhu Z., Lin X., Chen X., Yang C.J. Anal. Chem. 2013;85:2269–2275. doi: 10.1021/ac303179z. [DOI] [PubMed] [Google Scholar]
- Davis B.M., Rall G.F., Schnell M.J. Annu. Rev. Virol. 2015;2:451–471. doi: 10.1146/annurev-virology-100114-055157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennehy P.H. Infect. Dis. Clin. 2015;29:617–635. doi: 10.1016/j.idc.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Ding X., Yin K., Li Z., Liu C. BioRxiv. 2020 doi: 10.1101/2020.03.19.998724. [DOI] [Google Scholar]
- Draz M.S., Fang B.A., Zhang P., Hu Z., Gu S., Weng K.C., Gray J.W., Chen F.F. Theranostics. 2014;29:617–635. doi: 10.7150/thno.9404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer D.R., Park S., Bielawski C.W., Ruoff R.S. Chem. Soc. Rev. 2010;39:228–240. doi: 10.1039/b917103g. [DOI] [PubMed] [Google Scholar]
- Duinhoven S., Poort R., Van der Voet G., Agterof W.G.M., Norde W., Lyklema J. J. Colloid Interface Sci. 1995;170:340–350. [Google Scholar]
- Dulbecco R. Proc. Natl. Acad. Sci. Unit. States Am. 1952;38:747–752. doi: 10.1073/pnas.38.8.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias D.C., Nair R.R., Mohiuddin T.M.G., Morozov S.V., Blake P., Halsall M.P., Ferrari A.C., Boukhvalov D.W., Katsnelson M.I., Geim A.K., Novoselov K.S. Science. 2009;323:610–613. doi: 10.1126/science.1167130. [DOI] [PubMed] [Google Scholar]
- Fan J., Yuan L., Liu Q., Tong C., Wang W., Xiao F., Liu B., Liu X. Analyst. 2019;144:3972–3979. doi: 10.1039/c9an00179d. [DOI] [PubMed] [Google Scholar]
- Fan Z., Li S., Yuan F., Fan L. RSC Adv. 2015;5:19773–19789. [Google Scholar]
- Fan Z., Yust B., Nellore B.P.V., Sinha S.S., Kanchanapally R., Crouch R.A., Pramanik A., Chavva S.R., Sardar D., Ray P.C. J. Phys. Chem. Lett. 2014;5:3216–3221. doi: 10.1021/jz501402b. [DOI] [PubMed] [Google Scholar]
- Fang Y., Wang E. Chem. Commun. 2013;49:9526–9539. doi: 10.1039/c3cc44735a. [DOI] [PubMed] [Google Scholar]
- Feng X., Zhang Y., Zhou J., Li Y., Chen S., Zhang L., Ma Y., Wang L., Yan X. Nanoscale. 2015;7:2427–2432. doi: 10.1039/c4nr06623e. [DOI] [PubMed] [Google Scholar]
- Fowler J.D., Allen M.J., Tung V.C., Yang Y., Kaner R.B., Weiller B.H. ACS Nano. 2009;3:301–306. doi: 10.1021/nn800593m. [DOI] [PubMed] [Google Scholar]
- Franklin R.E. Nature. 1955;175:379–381. doi: 10.1038/175379a0. [DOI] [PubMed] [Google Scholar]
- Gani A.W., Wei W., Shi R.Z., Ng E., Nguyen M., Chua M.S., So S., Wang S.X. Sci. Rep. 2019;9:15615. doi: 10.1038/s41598-019-52147-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Tang B.Z. ACS Sens. 2017;2:1382–1399. doi: 10.1021/acssensors.7b00551. [DOI] [PubMed] [Google Scholar]
- Georgakilas V., Otyepka M., Bourlinos A.B., Chandra V., Kim N., Kemp K.C., Hobza P., Zboril R., Kim K.S. Chem. Rev. 2012;112:6156–6214. doi: 10.1021/cr3000412. [DOI] [PubMed] [Google Scholar]
- Georgakilas V., Perman J.A., Tucek J., Zboril R. Chem. Rev. 2015;115:4744–4822. doi: 10.1021/cr500304f. [DOI] [PubMed] [Google Scholar]
- Georgakilas V., Tiwari J.N., Kemp K.C., Perman J.A., Bourlinos A.B., Kim K.S., Zboril R. Chem. Rev. 2016;116:5464–5519. doi: 10.1021/acs.chemrev.5b00620. [DOI] [PubMed] [Google Scholar]
- Ghanbari K., Roushani M., Azadbakht A. Anal. Biochem. 2017;534:64–69. doi: 10.1016/j.ab.2017.07.016. [DOI] [PubMed] [Google Scholar]
- Gilje S., Han S., Wang M., Wang K.L., Kaner R.B. Nano Lett. 2007;7:3394–3398. doi: 10.1021/nl0717715. [DOI] [PubMed] [Google Scholar]
- Goldsmith C.S., Miller S.E. Clin. Microbiol. Rev. 2009;22:552–563. doi: 10.1128/CMR.00027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Navarro C., Weitz R.T., Bittner A.M., Scolari M., Mews A., Burghard M., Kern K. Nano Lett. 2007;7:3499–3503. doi: 10.1021/nl072090c. [DOI] [PubMed] [Google Scholar]
- Gong Q., Han H., Yang H., Zhang M., Sun X., Liang Y., Liu Z., Zhang W., Qiao J. J. Mater. 2019;5:313–319. [Google Scholar]
- Gong W., Jiang S., Li Z., Li C., Xu J., Pan J., Huo Y., Man B., Liu A., Zhang C. Optic Express. 2019;27:3483. doi: 10.1364/OE.27.003483. [DOI] [PubMed] [Google Scholar]
- Granatier J., Lazar P., Prucek R., Šafářová K., Zbořil R., Otyepka M., Hobza P. J. Phys. Chem. C. 2012;116:14151–14162. [Google Scholar]
- Grubaugh N.D., Ladner J.T., Lemey P., Pybus O.G., Rambaut A., Holmes E.C., Andersen K.G. Nat. Microbiol. 2019;4:10–19. doi: 10.1038/s41564-018-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead S.B. Science. 1988;239:476–481. doi: 10.1126/science.3277268. [DOI] [PubMed] [Google Scholar]
- Hasanzadeh M., Shadjou N. Mater. Sci. Eng. C. 2017;71:1313–1326. doi: 10.1016/j.msec.2016.11.068. [DOI] [PubMed] [Google Scholar]
- He S., Song B., Li D., Zhu C., Qi W., Wen Y., Wang L., Song S., Fang H., Fan C. Adv. Funct. Mater. 2010;20:453–459. [Google Scholar]
- Heng Cheong Y., Nasir M.Z.M., Bakandritsos A., Pykal M., Jakubec P., Zbořil R., Otyepka M., Pumera M. ChemElectroChem. 2019;6:229–234. [Google Scholar]
- Hernandez Y., Nicolosi V., Lotya M., Blighe F.M., Sun Z., De S., McGovern I.T., Holland B., Byrne M., Gun’ko Y.K., Boland J.J., Niraj P., Duesberg G., Krishnamurthy S., Goodhue R., Hutchison J., Scardaci V., Ferrari A.C., Coleman J.N. Nat. Nanotechnol. 2008;3:563–568. doi: 10.1038/nnano.2008.215. [DOI] [PubMed] [Google Scholar]
- Hirst G.K. J. Exp. Med. 1942;75:49–64. doi: 10.1084/jem.75.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hola K., Zhang Y., Wang Y., Giannelis E.P., Zboril R., Rogach A.L. Nano Today. 2014;9:590–603. [Google Scholar]
- Holmes E.C., Dudas G., Rambaut A., Andersen K.G. Nature. 2016;53:193–200. doi: 10.1038/nature19790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiya S., Bailey J.K., Temme J.S., Guillen Schlippe Y.V., Krauss I.J. J. Am. Chem. Soc. 2014;136:5407–5415. doi: 10.1021/ja500678v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., Zhang Y., Nie L., Xie C., Yan Z. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013;104:87–91. doi: 10.1016/j.saa.2012.11.026. [DOI] [PubMed] [Google Scholar]
- Huang H., Bai W., Dong C., Guo R., Liu Z. Biosens. Bioelectron. 2015;68:442–446. doi: 10.1016/j.bios.2015.01.039. [DOI] [PubMed] [Google Scholar]
- Huang J., Xie Zhixun, Xie Zhiqin, Luo S., Xie L., Huang L., Fan Q., Zhang Y., Wang S., Zeng T. Anal. Chim. Acta. 2016;913:121–127. doi: 10.1016/j.aca.2016.01.050. [DOI] [PubMed] [Google Scholar]
- Hutchinson E.C. Trends Microbiol. 2018;26:809–810. doi: 10.1016/j.tim.2018.05.013. [DOI] [PubMed] [Google Scholar]
- Iliafar S., Mittal J., Vezenov D., Jagota A. J. Am. Chem. Soc. 2014;136:12947–12957. doi: 10.1021/ja5055498. [DOI] [PubMed] [Google Scholar]
- Iravani S., Varma R.S. Environ. Chem. Lett. 2020;18:703–727. doi: 10.1007/s10311-020-00984-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S., Shukla S., Bajpai V.K., Han Y.K., Huh Y.S., Kumar A., Ghosh A., Gandhi S. Biosens. Bioelectron. 2019;126:792–799. doi: 10.1016/j.bios.2018.11.041. [DOI] [PubMed] [Google Scholar]
- Jabłońska A., Jaworska A., Kasztelan M., Berbeć S., Pałys B. Curr. Med. Chem. 2018;26:6878–6895. doi: 10.2174/0929867325666181004152247. [DOI] [PubMed] [Google Scholar]
- Jacob S.T., Crozier I., Fischer W.A., Hewlett A., Kraft C.S., Vega M.A. de La, Soka M.J., Wahl V., Griffiths A., Bollinger L., Kuhn J.H. Nat. Rev. Dis. Prim. 2020;6:1–31. doi: 10.1038/s41572-020-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S.E., Lamson D.M., Kirsten S., Walsh T.J. Clin. Microbiol. Rev. 2013;26:135–162. doi: 10.1128/CMR.00077-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S., Kim D.M., An S.Y., Kim D.H., Kim D.E. Anal. Biochem. 2018;561–562:66–69. doi: 10.1016/j.ab.2018.09.015. [DOI] [PubMed] [Google Scholar]
- Jiang P., Li Y., Ju T., Cheng W., Xu J., Han K. Chem. Res. Chin. Univ. 2020;36:323–337. [Google Scholar]
- Jin S.A., Poudyal S., Marinero E.E., Kuhn R.J., Stanciu L.A. Electrochim. Acta. 2016;194:422–430. [Google Scholar]
- Jin X., Zhang H., Li Y.T., Xiao M.M., Zhang Z.L., Pang D.W., Wong G., Zhang Z.Y., Zhang G.J. Microchim. Acta. 2019;186:435–453. doi: 10.1007/s00604-019-3256-5. [DOI] [PubMed] [Google Scholar]
- Jorgenson R.C., Yee S.S. Sensors and Acruofors. 1993;8:213–220. [Google Scholar]
- Joshi S.R., Sharma A., Kim G.H., Jang J. Mater. Sci. Eng. C. 2020;108:110–465. doi: 10.1016/j.msec.2019.110465. [DOI] [PubMed] [Google Scholar]
- Jung J.H., Cheon D.S., Liu F., Lee K.B., Seo T.S. Angew. Chem. 2010;122:5844–5847. doi: 10.1002/anie.201001428. [DOI] [PubMed] [Google Scholar]
- Kanagavalli P., Veerapandian M. Biosens. Bioelectron. 2020;150:456–524. doi: 10.1016/j.bios.2019.111878. [DOI] [PubMed] [Google Scholar]
- Karimi A., Othman A., Uzunoglu A., Stanciu L., Andreescu S. Nanoscale. 2015;150:435–471. doi: 10.1039/c4nr07586b. [DOI] [PubMed] [Google Scholar]
- Karlický F., Kumara Ramanatha Datta K., Otyepka M., Zbořil R. ACS Nano. 2013;7:6434–6464. doi: 10.1021/nn4024027. [DOI] [PubMed] [Google Scholar]
- Kashir J., Yaqinuddin A. Med. Hypotheses. 2020;141:109786. doi: 10.1016/j.mehy.2020.109786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye K.M., Izumi K.M., Kieff E. Proc. Natl. Acad. Sci. U.S.A. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Cote L.J., Kim F., Yuan W., Shull K.R., Huang J. J. Am. Chem. Soc. 2010;132:8180–8186. doi: 10.1021/ja102777p. [DOI] [PubMed] [Google Scholar]
- Kim J., Park S.J., Min D.H. Anal. Chem. 2017;89:232–248. doi: 10.1021/acs.analchem.6b04248. [DOI] [PubMed] [Google Scholar]
- Kim S., Ryoo S.R., Na H.K., Kim Y.K., Choi B.S., Lee Y., Kim D.E., Min D.H. Chem. Commun. 2013;49:8241–8243. doi: 10.1039/c3cc43368d. [DOI] [PubMed] [Google Scholar]
- Kong J., Franklin N.R., Zhou C., Chapline M.G., Peng S., Cho K., Dai H. Science. 2000;287:622–625. doi: 10.1126/science.287.5453.622. [DOI] [PubMed] [Google Scholar]
- Kosik I., Yewdell J.W. Viruses. 2019;11:34–49. doi: 10.3390/v11040346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kris Erickson B., Erni Rolf, Lee Zonghoon, Alem Nasim, Gannett Will, Zettl Alex, Erickson K., Alem N., Gannett W., Zettl A., Erni R., Lee Z. Adv. Mater. 2010;11:234–256. [Google Scholar]
- Krishnan S.K., Singh E., Singh P., Meyyappan M., Singh Nalwa H. RSC Adv. 2019;16:123–130. doi: 10.1039/c8ra09577a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuila T., Bose S., Khanra P., Mishra A.K., Kim N.H., Lee J.H. Biosens. Bioelectron. 2011;26:4637–4648. doi: 10.1016/j.bios.2011.05.039. [DOI] [PubMed] [Google Scholar]
- Kurane I. Comp. Immunol. Microbiol. Infect. Dis. 2007;30:329–340. doi: 10.1016/j.cimid.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Lawal A.T. Biosens. Bioelectron. 2018;106:149–178. doi: 10.1016/j.bios.2018.01.030. [DOI] [PubMed] [Google Scholar]
- Lazar P., Karlický F., Jurecka P., Kocman M., Otyepková E., Šafářová K., Otyepka M. J. Am. Chem. Soc. 2013;135:6372–6377. doi: 10.1021/ja403162r. [DOI] [PubMed] [Google Scholar]
- Lee J., Takemura K., Kato C.N., Suzuki T., Park E.Y. ACS Appl. Mater. Interfaces. 2017;9:27298–27304. doi: 10.1021/acsami.7b07012. [DOI] [PubMed] [Google Scholar]
- Lee J., Takemura K., Park E.Y. Sensor. Actuator. B Chem. 2018;276:254–261. [Google Scholar]
- Leland D.S., Ginocchio C.C. Clin. Microbiol. Rev. 2007;20:49–78. doi: 10.1128/CMR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenarda A., Bakandritsos A., Bevilacqua M., Tavagnacco C., Melchionna M., Naldoni A., Steklý T., Otyepka M., Zbořil R., Fornasiero P. ACS Omega. 2019;4:19944–19952. doi: 10.1021/acsomega.9b02881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Zhang W., Yu X., Wang Z., Su Z., Wei G. Nanoscale. 2016;19:115–132. doi: 10.1039/c6nr07249f. [DOI] [PubMed] [Google Scholar]
- Li J., Li Y., Zhai X., Cao Y., Zhao J., Tang Y., Han K. Electrochem. Commun. 2020;110:345–358. [Google Scholar]
- Li M., Wang C., Chen L., Liu D. Anal. Chim. Acta. 2019;1090:57–63. doi: 10.1016/j.aca.2019.09.018. [DOI] [PubMed] [Google Scholar]
- Li Q., Froning J.P., Pykal M., Zhang S., Wang Z., Vondrák M., Banáš P., Čépe K., Jurečka P., Šponer J., Zbořil R., Dong M., Otyepka M. 2D Mater. 2018;5 [Google Scholar]
- Li R.S., Yuan B., Liu J.H., Liu M.L., Gao P.F., Li Y.F., Li M., Huang C.Z. J. Mater. Chem. B. 2017;5:8719–8724. doi: 10.1039/c7tb02356a. [DOI] [PubMed] [Google Scholar]
- Li X., Wang X., Zhang L., Lee S., Dai H. Science. 2008;319:1229–1232. doi: 10.1126/science.1150878. [DOI] [PubMed] [Google Scholar]
- Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., Zhang Y., Wang J., Huang B., Lin Y., Yang J., Cai W., Wang X., Cheng J., Chen Z., Sun K., Pan W., Zhan Z., Chen L., Ye F. J. Med. Virol. 2020;16:25727. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.Y., Shen W., Gao Z. Chem. Soc. Rev. 2015;44:362–381. doi: 10.1039/c4cs00269e. [DOI] [PubMed] [Google Scholar]
- Liu-Helmersson J., Quam M., Wilder-Smith A., Stenlund H., Ebi K., Massad E., Rocklöv J. EBioMedicine. 2016;7:267–277. doi: 10.1016/j.ebiom.2016.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Choi K.S., Park T.J., Lee S.Y., Seo T.S. Biochip J. 2011;5:123–128. [Google Scholar]
- Liu F., Kim Y.H., Cheon D.S., Seo T.S. Sensor. Actuator. B Chem. 2013;186:252–257. [Google Scholar]
- Liu M.L., Chen B. Bin, Li C.M., Huang C.Z. Green Chem. 2019;32:567–581. [Google Scholar]
- Liu Y., Dong X., Chen P. Chem. Soc. Rev. 2012;41:2283–2307. doi: 10.1039/c1cs15270j. [DOI] [PubMed] [Google Scholar]
- Liu Z., Robinson J.T., Sun X., Dai H. J. Am. Chem. Soc. 2008;130:10876–10877. doi: 10.1021/ja803688x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C.H., Yang H.H., Zhu C.L., Chen X., Chen G.N. Angew. Chem. Int. Ed. 2009;48:4785–4787. doi: 10.1002/anie.200901479. [DOI] [PubMed] [Google Scholar]
- Lu L. Biosens. Bioelectron. 2018;110:180–192. doi: 10.1016/j.bios.2018.03.060. [DOI] [PubMed] [Google Scholar]
- Maartens G., Griesel R., Dube F., Nicol M., Mendelson M. Clin. Infect. Dis. 2020;70:1147–1152. doi: 10.1093/cid/ciz332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maity A., Sui X., Jin B., Pu H., Bottum K.J., Huang X., Chang J., Zhou G., Lu G., Chen J. Anal. Chem. 2018;90:14230–14238. doi: 10.1021/acs.analchem.8b03226. [DOI] [PubMed] [Google Scholar]
- Malecka K., Grabowska I., Radecki J., Stachyra A., Góra-Sochacka A., Sirko A., Radecka H. Electroanalysis. 2012;24:439–446. [Google Scholar]
- Malmqvist M. Nature. 1993;361:186–187. doi: 10.1038/361186a0. [DOI] [PubMed] [Google Scholar]
- Martínez P.M., Torres A.R., Lejarazu R.O. de, Montoya A., Martín J.F., Eiros J.M. J. Clin. Microbiol. 1999;37:1100–1106. doi: 10.1128/jcm.37.4.1100-1106.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcsharry J.J. Clin. Microbiol. Rev. 1994;7:576–604. doi: 10.1128/cmr.7.4.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y., Bai W., Zhang Y., Sun H., Li Y. Talanta. 2020;210:120597. doi: 10.1016/j.talanta.2019.120597. [DOI] [PubMed] [Google Scholar]
- Mishra N., Ng J., Rakeman J.L., Perry M.J., Centurioni D.A., Dean A.B., Price A., Thakkar R., Angus A.G., Williamson P., Delwart E., Carrington C., Sahadeo N., Che X., Briese T., Tokarz R., Lipkin W.I. J. Clin. Virol. 2019;120:44–50. doi: 10.1016/j.jcv.2019.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moço A.C.R., Guedes P.H., Flauzino J.M.R., da Silva H.S., Vieira J.G., Castro A.C.H., Gomes É.V.R., Tolentino F.M., Soares M.M.C.N., Madurro J.M., Brito‐Madurro A.G. Electroanalysis. 2019;31:1580–1587. [Google Scholar]
- Morales-Narváez E., Dincer C. Biosens. Bioelectron. 2020;13:112274. doi: 10.1016/j.bios.2020.112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagar B., Balsells M., de la Escosura-Muñiz A., Gomez-Romero P., Merkoçi A. Biosens. Bioelectron. 2019;129:238–244. doi: 10.1016/j.bios.2018.09.073. [DOI] [PubMed] [Google Scholar]
- Nair R.R., Ren W., Jalil R., Riaz I., Kravets V.G., Britnell L., Blake P., Schedin F., Mayorov A.S., Yuan S., Katsnelson M.I., Cheng H.M., Strupinski W., Bulusheva L.G., Okotrub A.V., Grigorieva I.V., Grigorenko A.N., Novoselov K.S., Geim A.K. Small. 2010;6:2877–2884. doi: 10.1002/smll.201001555. [DOI] [PubMed] [Google Scholar]
- Narita A., Wang X.Y., Feng X., Müllen K. Chem. Soc. Rev. 2015;44:6616–6643. doi: 10.1039/c5cs00183h. [DOI] [PubMed] [Google Scholar]
- Navakul K., Warakulwit C., Yenchitsomanus P. thai, Panya A., Lieberzeit P.A., Sangma C. Nanomed. Nanotechnol. Biol. Med. 2017;13:549–557. doi: 10.1016/j.nano.2016.08.009. [DOI] [PubMed] [Google Scholar]
- Nehra A., Chen W., Dimitrov D.S., Puri A., Singh K.P. ACS Appl. Mater. Interfaces. 2017;9:32621–32634. doi: 10.1021/acsami.7b12103. [DOI] [PubMed] [Google Scholar]
- Nie S., Emory S.R. Science. 1997;275:1102–1106. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoselov K.S., Fal’Ko V.I., Colombo L., Gellert P.R., Schwab M.G., Kim K. Nature. 2012;490:192–200. doi: 10.1038/nature11458. [DOI] [PubMed] [Google Scholar]
- Novoselov K.S., Geim A.K., Morozov S.V., Jiang D., Zhang Y., Dubonos S.V., Grigorieva I.V., Firsov A.A. Science. 2004;306:666–669. doi: 10.1126/science.1102896. [DOI] [PubMed] [Google Scholar]
- Obraztsov A.N. Nat. Nanotechnol. 2009;4:212–213. doi: 10.1038/nnano.2009.67. [DOI] [PubMed] [Google Scholar]
- Oliveira D.A., Silva J.V., Flauzino J.M.R., Sousa H.S., Castro A.C.H., Moço A.C.R., Soares M.M.C.N., Madurro J.M., Brito-Madurro A.G. J. Electroanal. Chem. 2019;844:6–13. [Google Scholar]
- Oliveira D.A., Silva J.V., Flauzino J.M.R., Sousa H.S., Castro A.C.H., Moço A.C.R., Soares M.M.C.N., Madurro J.M., Brito-Madurro A.G. J. Electroanal. Chem. 2019;844:6–13. [Google Scholar]
- Omar N.A.S., Fen Y.W., Abdullah J., Mustapha Kamil Y., Daniyal W.M.E.M.M., Sadrolhosseini A.R., Mahdi M.A. Sci. Rep. 2020;10:1–15. doi: 10.1038/s41598-020-59388-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omar N.A.S., Fen Y.W., Abdullah J., Sadrolhosseini A.R., Kamil Y.M., Fauzi N.I.M., Hashim H.S., Mahdi M.A. 2020. Nanomaterials 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omar N.A.S., Fen Y.W., Abdullah J., Zaid M.H.M., Daniyal W.M.E.M.M., Mahdi M.A. Optic Laser. Technol. 2019;114:204–208. [Google Scholar]
- Oshin O., Kireev D., Akinwande D., Adetiba E., Idachaba F., Atayero A. J. Phys. Conf. Ser. 2019;1378 [Google Scholar]
- Panchakarla L.S., Subrahmanyam K.S., Saha S.K., Govindaraj A., Krishnamurthy H.R., Waghmare U.V., Rao C.N.R. Adv. Mater. 2009;21:4726–4730. [Google Scholar]
- Park J.K., Taubenberger J.K. ACS Infect. Dis. 2016;2:5–7. doi: 10.1021/acsinfecdis.5b00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C.G., Domaoal R.A., Anderson K.S., Spiegel D.A. J. Am. Chem. Soc. 2009;131:16392–16394. doi: 10.1021/ja9057647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parviz D., Das S., Ahmed H.S.T., Irin F., Bhattacharia S., Green M.J. ACS Nano. 2012;6:8857–8867. doi: 10.1021/nn302784m. [DOI] [PubMed] [Google Scholar]
- Passaretti P., Sun Y., Khan I., Chan K., Sabo R., White H., Dafforn T.R., Goldberg Oppenheimer P. Nanoscale. 2019;11:458–469. doi: 10.1039/c9nr03670a. [DOI] [PubMed] [Google Scholar]
- Pathirana J., Groome M., Dorfman J., Kwatra G., Boppana S., Cutland C., Jones S., Madhi S.A. Clin. Infect. Dis. 2019;69:1789–1796. doi: 10.1093/cid/ciz019. [DOI] [PubMed] [Google Scholar]
- Peng J., Gao W., Gupta B.K., Liu Z., Romero-Aburto R., Ge L., Song L., Alemany L.B., Zhan X., Gao G., Vithayathil S.A., Kaipparettu B.A., Marti A.A., Hayashi T., Zhu J.J., Ajayan P.M. Nano Lett. 2012;12:844–849. doi: 10.1021/nl2038979. [DOI] [PubMed] [Google Scholar]
- Petti S., Lodi G. Oral Dis. 2019;25:1850–1865. doi: 10.1111/odi.13234. [DOI] [PubMed] [Google Scholar]
- Piro B., Kapella A., Le V.H., Anquetin G., Zhang Q.D., Reisberg S., Noel V., Tran L.D., Duc H.T., Pham M.C. Elsevier Ltd; 2011. Electrochimica Acta; pp. 10688–10693. [Google Scholar]
- Prangishvili D., Forterre P., Garrett R.A. Nat. Rev. Microbiol. 2006;4:837–848. doi: 10.1038/nrmicro1527. [DOI] [PubMed] [Google Scholar]
- Pumera M., Ambrosi A., Bonanni A., Chng E.L.K., Poh H.L. TrAC Trends Anal. Chem. (Reference Ed.) 2010;14 [Google Scholar]
- Purcell R.H. Proc. Natl. Acad. Sci. U.S.A. 1994;91:2401–2406. doi: 10.1073/pnas.91.7.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Tan S., Li Z., Qu Q., Li L., Yang L. Biosens. Bioelectron. 2020;153:112051. doi: 10.1016/j.bios.2020.112051. [DOI] [PubMed] [Google Scholar]
- Ramanathan S., Gopinath S.C.B., Md Arshad M.K., Poopalan P. Biosens. Bioelectron. 2019;141:478–503. doi: 10.1016/j.bios.2019.111434. [DOI] [PubMed] [Google Scholar]
- Reijans M., Dingemans G., Klaassen C.H., Meis J.F., Keijdener J., Mulders B., Eadie K., Van Leeuwen W., Van Belkum A., Horrevorts A.M., Simons G. J. Clin. Microbiol. 2008;46:1232–1240. doi: 10.1128/JCM.02294-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina G., Chau N.D.Q., Nishina Y., Bianco A. Nanoscale. 2018;10:5965–5974. doi: 10.1039/c8nr00333e. [DOI] [PubMed] [Google Scholar]
- Robinson J.T., Perkins F.K., Snow E.S., Wei Z., Sheehan P.E. Nano Lett. 2008;8:3137–3140. doi: 10.1021/nl8013007. [DOI] [PubMed] [Google Scholar]
- Roh C., Lee H.Y., Kim S.E., Jo S.K. Int. J. Nanomed. 2010;5:323–329. doi: 10.2147/ijn.s10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago G.A., Vázquez J., Courtney S., Matías K.Y., Andersen L.E., Colón C., Butler A.E., Roulo R., Bowzard J., Villanueva J.M., Muñoz-Jordan J.L. Nat. Commun. 2018;9:1–10. doi: 10.1038/s41467-018-03772-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapsford K.E., Blanco-Canosa J.B., Dawson P.E., Medintz I.L. Bioconjugate Chem. 2010;21:393–398. doi: 10.1021/bc9003712. [DOI] [PubMed] [Google Scholar]
- Schedin F., Geim A.K., Morozov S.V., Hill E.W., Blake P., Katsnelson M.I., Novoselov K.S. Nat. Mater. 2007;6:652–655. doi: 10.1038/nmat1967. [DOI] [PubMed] [Google Scholar]
- Schwierz F. Nat. Nanotechnol. 2010;5:487–496/. doi: 10.1038/nnano.2010.89. [DOI] [PubMed] [Google Scholar]
- Seelajaroen H., Bakandritsos A., Otyepka M., Zbořil R., Sariciftci N.S. ACS Appl. Mater. Interfaces. 2020;12:250–259. doi: 10.1021/acsami.9b17777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim T., Changkyun Park E., Kim S., Il ACS Nano. 2020;14:5142. [Google Scholar]
- Shang N.G., Papakonstantinou P., McMullan M., Chu M., Stamboulis A., Potenza A., Dhesi S.S., Marchetto H. Adv. Funct. Mater. 2008;18:3506–3514. [Google Scholar]
- Shao Y., Wang J., Wu H., Liu J., Aksay I.A., Lin Y. Electroanalysis. 2010;22:1027–1036. [Google Scholar]
- Shen J., Zhu Y., Yang X., Li C. Chem. Commun. 2012;48:3686–3699. doi: 10.1039/c2cc00110a. [DOI] [PubMed] [Google Scholar]
- Shrivastava N., Shrivastava A., Ninawe S.M., Sharma S., Kumar J.S., Alam S.I., Kanani A., Sharma S.K., Dash P.K. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.01822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P. Sensor. Actuator. B Chem. 2016;229:110–130. [Google Scholar]
- Singh R., Hong S., Jang J. Sci. Rep. 2017;7:42–771. doi: 10.1038/srep42771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S.S., Jones S., Pramanik A., Ray P.C. Acc. Chem. Res. 2016;49:2725–2735. doi: 10.1021/acs.accounts.6b00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Xie G., Wang L., Liu L., Tian G., Xiang H. Biosens. Bioelectron. 2014;58:68–74. doi: 10.1016/j.bios.2014.02.031. [DOI] [PubMed] [Google Scholar]
- Song J., Lau P.S., Liu M., Shuang S., Dong C., Li Y. ACS Appl. Mater. Interfaces. 2014;6:21806–21812. doi: 10.1021/am502138n. [DOI] [PubMed] [Google Scholar]
- Song W., Zhao B., Wang C., Ozaki Y., Lu X. J. Mater. Chem. B. 2019;7:850–875. doi: 10.1039/c8tb02878h. [DOI] [PubMed] [Google Scholar]
- Sreeprasad T.S., Berry V. Small. 2013;9:341–350. doi: 10.1002/smll.201202196. [DOI] [PubMed] [Google Scholar]
- Stankovich S., Dikin D.A., Piner R.D., Kohlhaas K.A., Kleinhammes A., Jia Y., Wu Y., Nguyen S.B.T., Ruoff R.S. Carbon N. Y. 2007;45:1558–1565. [Google Scholar]
- Stern D., Pauly D., Zydek M., Miller L., Piesker J., Laue M., Lisdat F., Dorner M.B., Dorner B.G., Nitsche A. PloS One. 2016;11:545–562. doi: 10.1371/journal.pone.0150110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujai P.T., Joseph M.M., Saranya G., Nair J.B., Murali V.P., Maiti K.K. Nanoscale. 2020;12:6971–6975. doi: 10.1039/d0nr00809e. [DOI] [PubMed] [Google Scholar]
- Sun H., Wu L., Wei W., Qu X. Mater. Today. 2013;19:434–465. [Google Scholar]
- Suvarnaphaet P., Pechprasarn S. Sensors (Switzerland) 2017;17:34–50. doi: 10.3390/s17102161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swathi R.S., Sebastian K.L. J. Chem. Phys. 2008;129 doi: 10.1063/1.2956498. [DOI] [PubMed] [Google Scholar]
- Tan S.M., Sofer Z., Pumera M. Electroanalysis. 2013;25:703–705. [Google Scholar]
- Tanner W.D., Toth D.J.A., Gundlapalli A.V. Epidemiol. Infect. 2015;143:3359–3374. doi: 10.1017/S0950268815001570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teymourian H., Salimi A., Khezrian S. Biosens. Bioelectron. 2013;49:1–8. doi: 10.1016/j.bios.2013.04.034. [DOI] [PubMed] [Google Scholar]
- Thu V.T., Tien B.Q., Ngoc Nga D.T., Thanh L.C., Sinh L.H., Le T.C., Lam T.D. RSC Adv. 2018;8:25361–25367. doi: 10.1039/c8ra04050h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F., Lyu J., Shi J., Yang M. Biosens. Bioelectron. 2017;89:123–135. doi: 10.1016/j.bios.2016.06.046. [DOI] [PubMed] [Google Scholar]
- Umadevi D., Sastry G.N. J. Phys. Chem. Lett. 2011;2:1572–1576. [Google Scholar]
- Urbanová V., Holá K., Bourlinos A.B., Čépe K., Ambrosi A., Loo A.H., Pumera M., Karlický F., Otyepka M., Zbořil R. Adv. Mater. 2015;27:2305–2310. doi: 10.1002/adma.201500094. [DOI] [PubMed] [Google Scholar]
- Valipour A., Roushani M. Biosens. Bioelectron. 2017;89:946–951. doi: 10.1016/j.bios.2016.09.086. [DOI] [PubMed] [Google Scholar]
- Veerapandian M., Hunter R., Neethirajan S. Talanta. 2016;155:250–257. doi: 10.1016/j.talanta.2016.04.047. [DOI] [PubMed] [Google Scholar]
- Vellucci A., Leibovitch E.C., Jacobson S. Humana Press Inc.; 2018. Methods in Molecular Biology; pp. 99–109. [DOI] [PubMed] [Google Scholar]
- Vermisoglou E.C., Jakubec P., Malina O., Kupka V., Schneemann A., Fischer R.A., Zbořil R., Jayaramulu K., Otyepka M. Front. Chem. 2020;8:544. doi: 10.3389/fchem.2020.00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waiwijit U., Phokaratkul D., Kampeera J., Lomas T., Wisitsoraat A., Kiatpathomchai W., Tuantranont A. J. Biotechnol. 2015;212:44–49. doi: 10.1016/j.jbiotec.2015.08.003. [DOI] [PubMed] [Google Scholar]
- Wang D., Zheng Y., Kang X., Zhang X., Hao H., Chen W., Liu L., Li X., Li L., Yuan Q., Chen F., Yang Y., Jiang Y., Jiang H. Eur. J. Clin. Microbiol. Infect. Dis. 2015;34:1327–1336. doi: 10.1007/s10096-015-2353-6. [DOI] [PubMed] [Google Scholar]
- Wang H., Maiyalagan T., Wang X. ACS Catal. 2012;2:781–794. [Google Scholar]
- Wang J. Electroanalysis. 2005;17:7–14. [Google Scholar]
- Wang L., Tian J., Yang W., Zhao Y., Zhao S. Luminescence. 2016;31:573–579. doi: 10.1002/bio.2997. [DOI] [PubMed] [Google Scholar]
- Wang X., Li X., Zhang L., Yoon Y., Weber P.K., Wang H., Guo J., Dai H. Science. 2009;324:768–771. doi: 10.1126/science.1170335. [DOI] [PubMed] [Google Scholar]
- Wang X., Shang L., Zhang W., Jia L.P., Ma R.N., Jia W.L., Wang H.S. Biosens. Bioelectron. 2019;141:234–250. doi: 10.1016/j.bios.2019.111436. [DOI] [PubMed] [Google Scholar]
- Wang Y., Bai X., Wen W., Zhang X., Wang S. ACS Appl. Mater. Interfaces. 2015;7:18872–18879. doi: 10.1021/acsami.5b05857. [DOI] [PubMed] [Google Scholar]
- Wang Y., Li Z., Wang J., Li J., Lin Y. 2011. Trends Biotechnol. [Google Scholar]
- Wang Y., Shao Y., Matson D.W., Li J., Lin Y. ACS Nano. 2010;4:1790–1798. doi: 10.1021/nn100315s. [DOI] [PubMed] [Google Scholar]
- Wei D., Liu Y., Wang Y., Zhang H., Huang L., Yu G. Nano Lett. 2009;9:1752–1758. doi: 10.1021/nl803279t. [DOI] [PubMed] [Google Scholar]
- Wen J., Li W., Li J., Tao B., Xu Y., Li H., Lu A., Sun S. Sensor. Actuator. B Chem. 2016;227:655–659. [Google Scholar]
- Wong W.R., Sekaran S.D., Mahamd Adikan F.R., Berini P. Biosens. Bioelectron. 2016;78:132–139. doi: 10.1016/j.bios.2015.11.030. [DOI] [PubMed] [Google Scholar]
- Wu J.F., Gao X., Ge L., Zhao G.C., Wang G.F. RSC Adv. 2019;9:19813–19818. doi: 10.1039/c9ra02475a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Q., Huang J., Huang H., Mao W., Ye Z. RSC Adv. 2018;8:1820–1825. doi: 10.1039/c7ra11945c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R., Wang Z., Zhou W., Liu Y., Fan L., Li Y., Li X. Anal. Methods. 2016;36:568–591. [Google Scholar]
- Xie Zhixun, Huang J., Luo S., Xie Zhiqin, Xie L., Liu J., Pang Y., Deng X., Fan Q. PloS One. 2014;9 doi: 10.1371/journal.pone.0094685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Chen X., Hu J., Yang P., Yang D., Wei L. Analyst. 2012;137:2757–2761. doi: 10.1039/c2an35093a. [DOI] [PubMed] [Google Scholar]
- Xu W., Mao N., Zhang J. Small. 2013;6:123–136. [Google Scholar]
- Xu X., Ray R., Gu Y., Ploehn H.J., Gearheart L., Raker K., Scrivens W.A. J. Am. Chem. Soc. 2004;126:12736–12737. doi: 10.1021/ja040082h. [DOI] [PubMed] [Google Scholar]
- Yakimovich A., Andriasyan V., Witte R., Wang I.H., Prasad V., Suomalainen M., Greber U.F. PloS One. 2015;10:3535. doi: 10.1371/journal.pone.0138760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Yuan H., Zhao Q., Xing L., Zheng X., Wang W., Zhao Y., Yu Y., Hu L., Yao W. Analyst. 2020;35:1323–1333. doi: 10.1039/d0an00339e. [DOI] [PubMed] [Google Scholar]
- Yang C., Huang Y., Cheng H., Jiang L., Qu L. Adv. Mater. 2019;31:3454–3468. doi: 10.1002/adma.201805705. [DOI] [PubMed] [Google Scholar]
- Yang L., Yamamoto T. Front. Microbiol. 2016;7:1500. doi: 10.3389/fmicb.2016.01500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Zhang X., Liu Z., Ma Y., Huang Y., Chen Y. J. Phys. Chem. C. 2008;112:17554–17558. [Google Scholar]
- Yang Y., Hu G.B., Liang W. Bin, Yao L.Y., Huang W., Zhang Y.J., Zhang J.L., Wang J.M., Yuan R., Xiao D.R. Nanoscale. 2020;12:5932–5941. doi: 10.1039/c9nr10712f. [DOI] [PubMed] [Google Scholar]
- Yang Z.H., Zhuo Y., Yuan R., Chai Y.Q. Biosens. Bioelectron. 2015;69:321–327. doi: 10.1016/j.bios.2015.01.035. [DOI] [PubMed] [Google Scholar]
- Yoo S.M., Lee S.Y. Trends Biotechnol. 2016;372:229–234. doi: 10.1016/j.tibtech.2015.09.012. [DOI] [PubMed] [Google Scholar]
- Young L.S., Murray P.G. Oncogene. 2003;22:5108–5121. doi: 10.1038/sj.onc.1206556. [DOI] [PubMed] [Google Scholar]
- Yu X., Cai H., Zhang W., Li X., Pan N., Luo Y., Wang X., Hou J.G. ACS Nano. 2011;5:952–958. doi: 10.1021/nn102291j. [DOI] [PubMed] [Google Scholar]
- Yun B.J., Koh W.G. J. Ind. Eng. Chem. 2020;82:341–348. [Google Scholar]
- Zamora J.L.R., Aguilar H.C. Methods. 2018;134:87–97. doi: 10.1016/j.ymeth.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zari N., Amine A., Ennaji M.M. Anal. Lett. 2009;42:519–535. [Google Scholar]
- Zbořil R., Karlický F., Bourlinos A.B., Steriotis T.A., Stubos A.K., Georgakilas V., Šafářová K., Jančík D., Trapalis C., Otyepka M. Small. 2010;6:2885–2891. doi: 10.1002/smll.201001401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerboni L., Sen N., Oliver S.L., Arvin A.M. Nat. Rev. Microbiol. 2014;12:197–210. doi: 10.1038/nrmicro3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan L., Li C.M., Wu W.B., Huang C.Z. Chem. Commun. 2014;50:11526–11528. doi: 10.1039/c4cc05155f. [DOI] [PubMed] [Google Scholar]
- Zhang Huan, Zhang Honglu, Aldalbahi A., Zuo X., Fan C., Mi X. Biosens. Bioelectron. 2017;89:96–106. doi: 10.1016/j.bios.2016.07.030. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhou D., Sheng S., Yang J., Chen X., Xie G., Xiang H. Microchim. Acta. 2016;183:2055–2062. [Google Scholar]
- Zhang Y., Wu C., Zhang J. Nanotechnol. Rev. 2013;2:27–45. [Google Scholar]
- Zhang Y., Zhang Z., Rong S., Yu H., Gao H., Sha Q., Ding P., Pan H., Chang D. Anal. Chim. Acta. 2020;1109:98–106. doi: 10.1016/j.aca.2020.02.056. [DOI] [PubMed] [Google Scholar]
- Zhao F., Bai Y., Zeng R., Cao L., Zhu J., Han G., Chen Z. Electroanalysis. 2018;30:2774–2780. [Google Scholar]
- Zhao F., Cao L., Liang Y., Wu Z., Chen Z., Zeng R. J. Biomed. Nanotechnol. 2017;13:1300–1308. [Google Scholar]
- Zhou S., Xu H., Gan W., Yuan Q. RSC Adv. 2016;16:454–469. [Google Scholar]
- Zribi B., Castro-Arias J.M., Decanini D., Gogneau N., Dragoe D., Cattoni A., Ouerghi A., Korri-Youssoufi H., Haghiri-Gosnet A.M. Nanoscale. 2016;8:15479–15485. doi: 10.1039/c6nr04289a. [DOI] [PubMed] [Google Scholar]