Abstract
MYC is a oncoprotein that coordinates the expression of genes involved in metabolism, cell differentiation and survival in various types of malignancies. However, the underlying oncogenic mechanisms and the clinical significance of MYC expression in the acute myeloid leukemia with myelodysplasia related changes (AML-MRC) remain to be answered. A total of 135 patients were retrospectively identified using Total Cancer Care (TCC) Moffitt Cancer Center (MCC) databases. Diagnosis of AML-MRC was based on the 2016 WHO classification utilizing bone marrow (BM) evaluation. MYC protein expression level was assessed by immunohistochemistry (IHC) staining using paraffin-embedded BM trephine biopsy samples obtained at the time of diagnosis or relapse. Concurrent somatic mutations were assessed using targeted next generation sequencing that include 54 genes. A total of 38% (n=51) and 62% (n=84) patients had high and low MYC expression, respectively. The most common somatic mutation in our cohort was TP53 followed by DNMT3A, and ASXL1. The median OS was significantly longer in low MYC patients (median OS 42.3 vs. 17.05 months, p=0.0109). Multivariate analysis including MYC expression level, transplantation status, gender and age demonstrated high MYC expression (HR 1.77, 95% CI 1.004–3.104, p=0.045) to be an independent poor prognostic factor. Further studies are needed to identify the underlying mechanism of MYC driven oncogenesis in AML-MRC. Additionally, the prognostic impact of MYC on the AML survival in a larger cohort that include diverse somatic mutations and chromosomal abnormalities requires further investigation.
Keywords: sAML, MYC
INTRODUCTION
Acute myeloid leukemia with myelodysplasia related changes (AML-MRC) is a hematopoietic clonal disorder that is characterized by dysplasia, increased myeloblasts and impaired hematopoiesis1. AML-MRC patients were shown to have aggressive clinical courses with 5 year overall survival (OS) less than 30%2,3. Recent studies demonstrated a complex composition of somatic mutations and cytogenetics that are associated with heterogeneous disease course in the de novo and secondary AML patients4–7. These somatic mutations and chromosomal abnormalities involve genes that regulate RNA splicing, metabolism, signaling cascades, and epigenetics4–6. Among these, acquired MYC somatic mutations and gene amplifications are frequently identified in both pediatric and adult AML patients8–11.
MYC is a well-known oncoprotein that coordinates the expression of genes involved in the metabolism, nutrient transport, and cell proliferation and growth12,13. Clinical significance of MYC translocation and amplification has been extensively studied in a variety of malignancies including lymphomas14–16, however, relatively less in AML. Recent preclinical studies have shown that MYC regulates down-stream genes that are important for the cell death and differentiation in the AML cells. Further, MYC was shown to be overexpressed and/or required for the myeloid leukemia triggered by FLT3-ITD and PML-RARα, RUNX1-RUNX1T1 and BCR-ABL fusion oncoproteins17–19.
Although the underlying oncogenic mechanism of MYC is unclear, a recent study showed that high levels of MYC expression is associated with inferior survival outcomes in de novo AML patients20,21. Compared to other subtypes of AML, AML-MRC patients were shown to have dynamic range of MYC protein expression20, yet the clinical significance of MYC expression in these unique patient population is unknown. Our study attempts to explore the prognostic impact of MYC protein levels on the survival outcomes in AML-MRC patients and assess somatic mutational landscapes in low vs. high MYC AML-MRC patients.
METHODS
Patients and Sample Acquisition
Using Total Cancer Care (TCC) Moffitt Cancer Center (MCC) databases, we retrospectively identified histologically confirmed AML-MRC patients from 2011 to 2018 (Figure S1). The research proposal was approved by institutional research board (IRB). Patient had provided written informed consent to be included in the database. Diagnosis of AML-MRC were based on bone marrow (BM) evaluation and classified based on the 2016 WHO classification of tumors of hematopoietic and lymphoid tissues22. We included both de novo AML-MRC cases and AML-MRC with preceding MDS or MDS/MPN. Therapy related AML (tAML) cases were not included in the study (Figure S1). The BM biopsy samples harvested at the time of AML-MRC diagnosis or at the time of relapse were collected to confirm morphologic and molecular diagnosis as well as for the retrospective immunohistochemistry study to assess expression of MYC protein. Detailed information regarding patient selection process is described in the Figure S1. The patients’ demographic data, diagnosis, laboratory results including complete blood counts (CBC), cytogenetics, NGS myeloid mutation profile, clinical treatment and overall survival were retrieved and summarized in Table 1.
Table 1.
Baseline characteristics of the study cohort
| Characteristic | Low MYC patients (n=84) |
High MYC patients (n=51) |
All patients (n=135) |
|---|---|---|---|
| Age at AML-MRC diagnosis (years) | 65.1 (22.3–85.6) | 68.5 (44.4–85.93) | 67.3 (22.3–85.9) |
| Gender (%) | Male 51 (61) | Male 33 (65) | Male 84 (62) |
| Female 33 (39) | Female 18 (35) | Female 51 (38) | |
| CBC1 | |||
| Hemoglobin (g/dL) | 9 (7.1–12.5) | 8.4 (6.5–10.7) | 8.7 (6.5–12.7)) |
| Platelet counts (/L) | 43 (3–697) | 40 (2–945) | 42 (2–945) |
| White blood counts (/L) | 2.53 (0.12–78.07) | 2.95 (0.1–216.8) | 2.8 (0.1–216.8) |
| ANC (/L) | 0.77 (0–42.94) | 0.33 (0–17.62) | 0.67 (0–42.94) |
| Blasts, % (range)1 | |||
| Peripheral blood | 25 (1–87) | 40 (5–93) | 28 (10–93) |
| Bone marrow | 25 (1–90) | 35 (10–95) | 29 (10–95) |
| Cytogenetics1 (%) | |||
| 5q deletion | 23 (27) | 24 (47) | 47 (35) |
| Trisomy 8 | 11 (13) | 15 (29) | 26 (19) |
| 7 deletion | 19 (23) | 13 (25) | 32 (24) |
| 12 deletion | 5 (6) | 5 (10) | 10 (7) |
| 20 deletion | 10 (12) | 2 (4) | 12 (9) |
| 17p deletion | 18 (21) | 17 (33) | 35 (26) |
| Complex karyotype2 | 27 (32) | 20 (39) | 47 (35) |
| NGS assessments (%)3 | |||
| TP53 | 23 (35) | 23 (51) | 46 (41) |
| DNMT3A | 8 (12) | 15 (33) | 23 (21) |
| ASXL1 | 15 (23) | 7 (16) | 22 (20) |
| SRSF2 | 9 (14) | 7 (16) | 16 (14) |
| IDH1 | 5 (8) | 8 (18) | 13 (12) |
| RUNX1 | 9 (14) | 4 (9) | 13 (12) |
| NRAS | 9 (14) | 3 (7) | 12 (11) |
| TET2 | 7 (11) | 5 (11) | 12 (11) |
| NPM1 | 5 (8) | 4 (9) | 9 (8) |
| BCOR | 5 (8) | 2 (4) | 7 (6) |
| KRAS | 3 (5) | 4 (9) | 7 (6) |
| SF3B1 | 6 (9) | 1 (2) | 7 (6) |
| Treatment (%) | |||
| Hypomethylating agents | 16 (19) | 8 (16) | 24 (18) |
| Intensive chemotherapy4 | 47 (56) | 27 (53) | 74 (55) |
| Low dose cytarabine | 0 (0) | 0 (0) | 0 (0) |
| Allo-SCT | 21 (25) | 10 (20) | 31 (23) |
| Median OS, months | 42.3 | 17.0 | 20.0 |
Abbreviation: complete blood count (CBC), next generation sequencing (NGS), allogeneic stem cell transplant (Allo-SCT), overall survival (OS).
Assessment of MYC protein expression
MYC protein expression was assessed by immunohistochemistry (IHC) staining using paraffin-embedded BM trephine biopsy samples. Blocks were sectioned 4.0 μm in thickness and unstained sides were deparaffinized using EZ Prep solution (Ventana Medical System, Tucson). Slides were stained with anti-MYC antibody (Ventana, Cat No. 790–4628; prediluted) using Ventana Discovery XT automated system. We used 5% as cut-off (calculated as MYC positive cells out of total counted blasts in the selected area with sheets of blasts) as previously reported20.
Assessment of Targeted Next Generation Sequencing and Cytogenetics
Somatic mutations were assessed by 54 myeloid targeted gene sequencing as described previously23. Genomic DNA was isolated from BM or peripheral blood (PB) mononuclear cells. DNA samples were subjected to targeted genome sequencing using Illumina HiSeq2000. For our pathogenic vs. non-pathogenic call algorithm, we used a modification of the ACMG classification scheme that was developed for germline variants for the classification of somatic sequence variants24. We established filters to determine clinically actionable pathogenic alterations and to filter out benign variants or polymorphism, which were clinically validated as described previously23. Conventional karyotyping or/and fluorescence in-situ hybridization (FISH) were performed on the patients’ BM specimens to assess any cytogenetic aberrations.
Statistical Analysis
Clinical variables and disease-related prognostic factors including age, gender, cytogenetics and somatic mutations were characterized at the time of AML-MRC diagnosis and were annotated using descriptive statistics. The overall survival (OS) outcomes were estimated with the Kaplan-Meier method and compared using the log-rank test. All statistical analyses were performed using SPSS v24.0 and GraphPad Prism 7.
RESULTS
Patient Characteristics
A total of 135 AML-MRC patients were included in this study. The median age at AML-MRC diagnosis was 67.3 (22.3–85.9) years and 62% of patients were male (n=84) (Table 1). A total of 55% (n=74) of patients were treated with intensive chemotherapy including 7+3 (cytarabine 100mg/m2/day continuous IV infusion for 7 days and daunorubicin 45–90mg/m2/day or idarubicin 12mg/m2/day for 3 days), CLAG (cladribine 5mg/m2/day and cytarabine 2g/m2/day for 5 days, G-CSF 300mcg for 6 days), CLAG-M (CLAG and mitoxantrone 10mg/m2/day for 3 days), or MEC (mitoxantrone 8mg/m2/day, etoposide 100mg/m2/day, and cytarabine 1g/m2/day for 5 days) and 18% (n=24) were treated with hypomethylating agents (decitabine or azacitidine). Allogeneic stem cell transplant (allo-SCT) was performed in 23% (n=31) patients (Table 1).
Spectrum of MYC Oncoprotein Expression and Associated Mutational Landscape and Cytogenetic Abnormalities
A total of 38% (n=51) patients had high MYC expression and 62% (n=84) patients had low MYC expression (Figure 1 and Table 1). Somatic mutations were assessed in 82% (n=111). In these patients, most common somatic mutation in all patients was TP53 (41%) followed by DNMT3A (21%), ASXL1 (20%), SRSF2 (14%), IDH1 (12%), RUNX1 (12%), NRAS (11%), and TET2 (8%). Among 51 patients with high MYC protein expression, the most common comutation was TP53 (51%) followed by DNMT3A (33%), IDH1 (18%), ASXL1 (16%), SRSF2 (16%), and TET2 (11%). In patients with low MYC protein expression, the most common comutation was TP53 (35%) followed by ASXL1 (23%), RUNX1 (14%), NRAS (14%), and DNMT3A (12%). Fisher’s Exact test revealed that the rates of TP53 (p=0.0255) and DNMT3A (p=0.0043) mutations were significant higher in high MYC patients (Table 1). AML-MRC patients with TP53 mutation had numerically higher expression of MYC oncoprotein (9.1% vs. 6.1%, p=0.2058), but this was not statistically significant (Figure 3). We also assessed the cytogenetics in our study cohort. A total of 26% (n=35) patients had chromosome 17p deletion [del(17p)] and 22% (n=30) had both del(17p) and TP53 mutation (Table 1). Additional chromosomal abnormalities including deletion 5q, trisomy 8, deletion 7q, deletion 20q, and complex karyotypes were observed in 35% (n=47), 19% (n=26), 24% (n=32), 9% (n=12), and 35% (n=47) of patients, respectively (Table 1). The most common chromosomal abnormality in low MYC patients was complex karyotype (32%, n=27) followed by 5q deletion (27%, n=23), deletion 7 (23%, n=19), and del(17p) (21%, n=18) (Table 1). In high MYC patients, the most common cytogenetics was 5q deletion (47%, n=24) that was followed by complex karyotypes (39%, n=20), del(17p) (33%, n=17), trisomy 8 (29%, n=15), and deletion 7 (25%, n=13) (Table 1).
Fig 1. MYC Immunohistochemistry Staining in AML-MRC Patients.
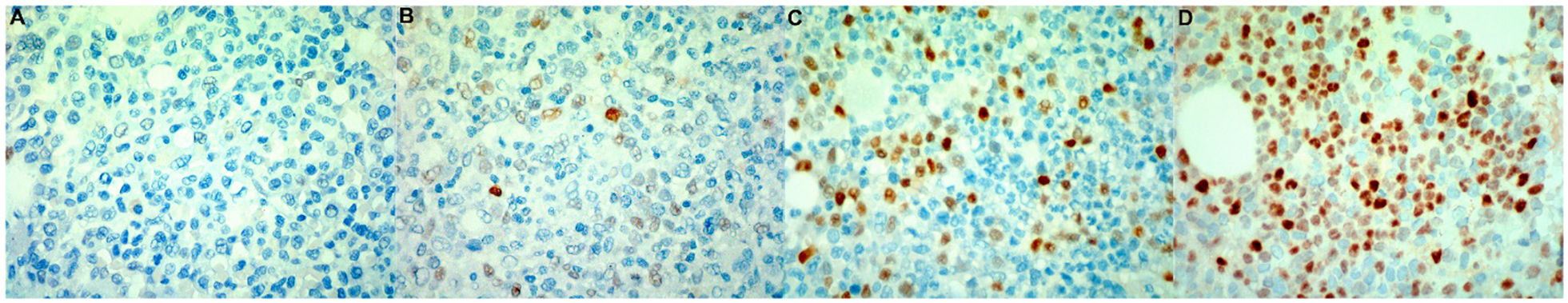
Examples of IHC staining results in low (A and B) and high (C and D) MYC protein expressing AM-MRC patients.
Fig 3. MYC Protein Expression Levels in TP53 Wild Type vs. Mutant Patients and Peripheral Blast Counts in Low vs. High MYC Patients.
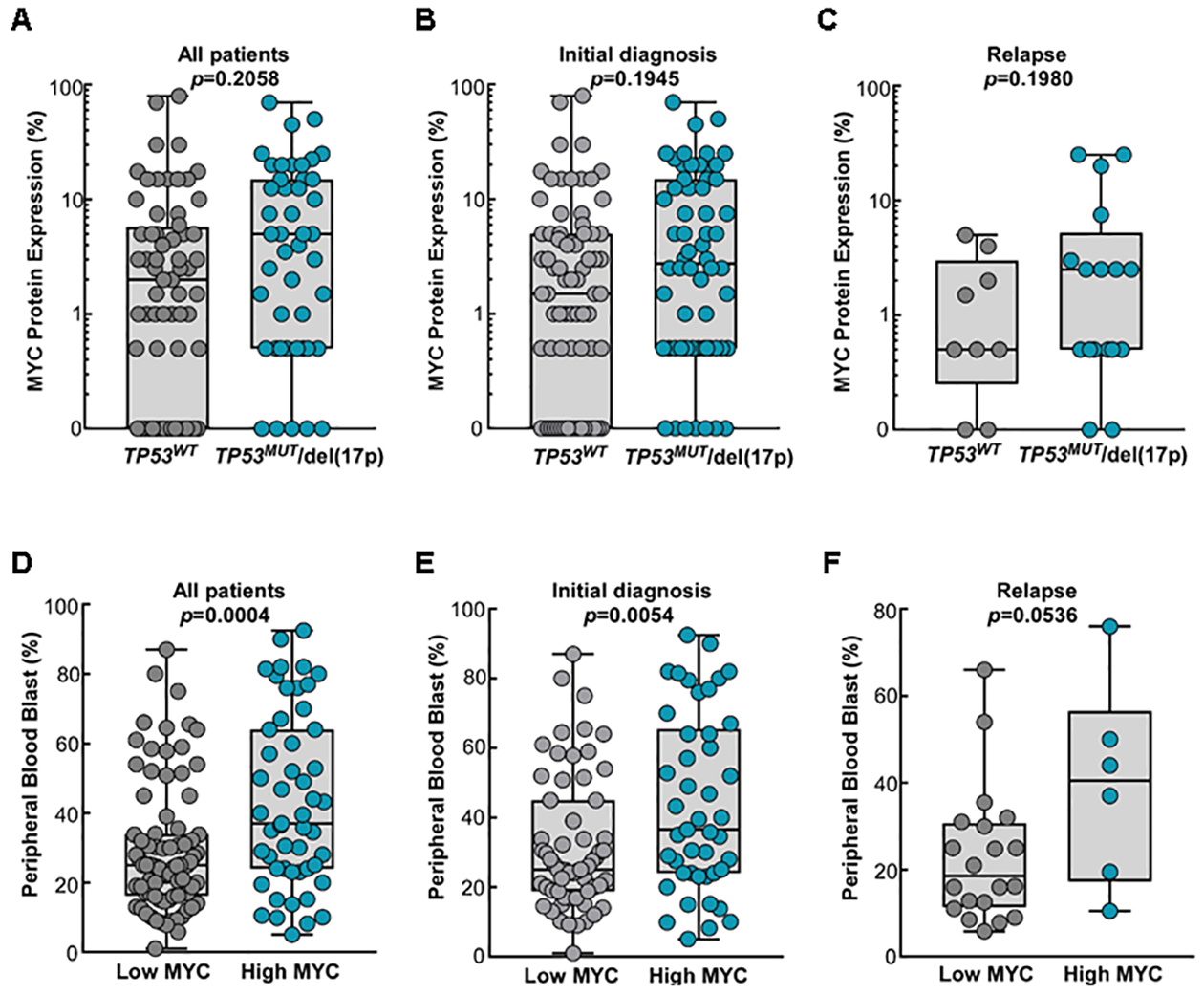
MYC protein expression levels were assessed at the time of AML-MRC in the majority of the patients (n=109) and some patients had MYC protein expression assessed at the time of relapse (n=26). MYC levels are plotted as the separate groups; all patients (A), patients with MYC assessment at the diagnosis (B), and patients with assessment at the time of relapse (C). Blast counts (%) in the peripheral blood are shown in three different patient groups: all patient (D), patients with MYC assessment at the time of diagnosis (E), and patients with assessment at the time of relapse (F).
Impact of MYC Expression on Blast Counts and Overall Survival
The PB blast counts were significantly higher in high MYC patients compared to low MYC patients (40% vs. 25%, p=0.0004) (Figure 3). Notably, the median OS was significantly longer in low MYC patients compared to high MYC patients (median OS 42.3 vs. 17.05 months, p=0.0109) (Figure 4A). Further, when considering only TP53 wild type patients without del(17p), low MYC patients had even longer median OS (median OS 58.6 vs. 17.0 months, p=0.0338) (Figure 4B). In AML-MRC patients with either TP53 mutation and/or del(17p), there was no statistical OS difference between low and high MYC groups (median OS 33.1 vs. 15.2 months, p=0.17) (Figure 4C). In an additional analysis including high MYC patient only, there was no OS difference between TP53 wild type patients vs. mutant patients (p=0.7995) (Figure S2).
Fig 4. Overall Survival Based on MYC Expression in AML-MRC Patients.
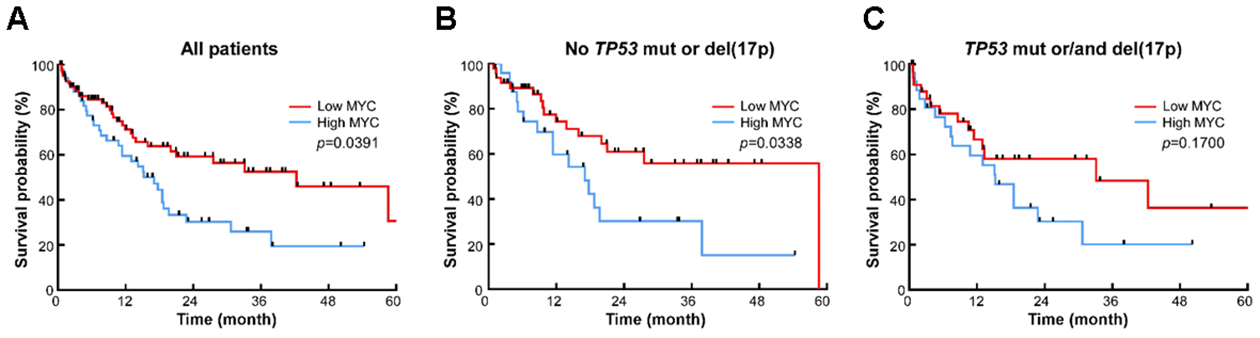
(A) Overall survival in all patients, (B) in patients with no TP53 mutation or del(17p), and (C) in patients with TP53 mutations or/and del(17p).
We performed additional survival analysis including newly diagnosed AML-MRC patients only (n=109) and the results were similar (Figure S3). In the univariate analysis with newly diagnosed AML-MRC patients (n=109), high MYC protein expression was associated with inferior OS (HR 1.817, 95% CI 1.042–3.169, p=0.034). Multivariate analysis including MYC expression level, transplantation status, gender and age demonstrated high MYC expression (HR 1.77, 95% CI 1.004–3.104, p=0.045) and no allogeneic transplant (HR 3.23, 95% CI 1.396–7.486, p=0.006) are poor prognostic factors for the OS outcome (Table 2).
Table 2.
Prognostic Impact of MYC oncoprotein expression in the univariate (log-rank) and multivariate (cox-regression) analyses*.
| Overall Survival | ||||||
|---|---|---|---|---|---|---|
| Univariate | Multivariable* | |||||
| Variable | HR | 95% CI | P | HR | 95% CI | P |
| High MYC | 1.817 | 1.042 to 3.169 | 0.034 | 1.765 | 1.004 to 3.104 | 0.048 |
| Age | 1.019 | 0.992 to 1.047 | 0.992 | 1.000 | 0.972 to 1.028 | 0.989 |
| Gender (female) | 1.557 | 0.860 to 2.819 | 0.144 | 1.444 | 0.785 to 2.656 | 0.238 |
| No allogeneic transplant | 3.445 | 1.526 to 7.779 | 0.003 | 3.232 | 1.396 to 7.486 | 0.006 |
Abbreviation: HR (hazard ratio), CI (confidence interval).
In the univariate and multivariate analyses, newly diagnosed AML-MRC patients whose MYC proteins expression levels were assessed at the time of diagnosis.
DISCUSSION
Among many of the common somatic mutations and abnormal cytogenetics associated with AML, MYC gene rearrangement, copy number gain, and somatic mutations have been frequently reported in the pediatric and adult AML patients8–11. Previous studies using in vivo mice models have demonstrated that MYC is sufficient to induce AML and MYC was shown to be overexpressed or/and required for AML provoked by various fusion oncogenes such as PML-RARα, RUNX1-RUNX1T1 and BCR-ABL117–19. Although MYC was shown to play a pivotal role in regulating myeloid differentiation and cell death, the exact mechanisms underlying MYC-driven oncogenesis remains to be unanswered in AML. As the first step to understand the oncogenic role of MYC in AML patients, we attempted to analyze the prognostic impact of MYC oncoprotein expression in AML-MRC patients with and without preceding MDS, MPN, and MDS/MPN and the landscapes of co-mutations in high vs. low MYC patients.
Two previous studies have assessed the prognostic significance of MYC oncoprotein in AML patients20,21. In the first study performed by Mughal et al. that included a total of 199 AML patients, high MYC level (IHC staining at or above median score and more than 1+ staining intensity) was associated with poor OS in patients with favorable and intermediate cytogenetic groups although it did not reach the statistical significance in the multivariate analysis adjusted by age and cytogenetic risk group21. In an independent study performed by Ohanian et al. that included a total of 265 untreated AML patients, high MYC protein expression (>6%) was associated with inferior complete remission duration when compared to the low expression (12 vs. 23 months, p=0.028)20. Importantly, among 241 patients with higher risk for relapse (age ≥55 years, intermediate and high risk groups), high MYC expression was associated with inferior median OS (24 vs. 13 months, p=0.042), event free survival (14 vs. 6 months, p=0.048), and relapse free survival (25 vs. 12 months, p=0.024)20. In consistent with these reports20,21, we observed a dynamic range of MYC oncoprotein expression in AML-MRC patients (0–100%) and inferior OS in the high MYC patients. In patients without TP53 somatic mutations, high MYC level remains to be an independent poor prognostic factor although there was no OS difference between high and low MYC groups in patients with TP53 mutations or/and del(17p). Supporting these observations, concurrent TP53 mutation rate was significantly higher in high MYC patients and MYC levels were maintained higher in patients with TP53 mutation or/and del(17p) although the difference was not significant. In an additional analysis in high MYC patients only, there was no OS difference between patients with TP53 mutation or/and del(17p) vs. patients with no TP53/del(17p). Collectively, these observations suggest that TP53 mutation/del(17p) associated poor prognosis may result from high MYC accumulation driven by TP53 mutation/del(17p). In previous studies, wild type p53 was shown to transcriptionally repress MYC expression and induce miR-145 suppressing MYC expression25,26. Further studies are warranted to investigate the detail mechanisms of mutant p53 dependent MYC upregulation in AML patients.
The most common somatic mutation in de novo AML patients from TCGA database was FLT3 (28%) which is followed by NPM1 (27%), DNMT3A (24.5%), RUNX1 (13%), IDH2 (10%), IDH1 (9.5%), and TET2 (8.5%)27. In contrast, ASXL1 (32%) was the most common somatic mutation in the secondary AML patients and this was followed by RUNX1 (31%), NRAS (23%), TET2 (20%), SRSF2 (20%), DNMT3A (19%), FLT3 (19%), U2AF1 (16%), and TP53 (15%)7. Similar to this study, TP53 (41%), DNMT3A (21%), ASXL1 (20%), SRSF2 (14%), IDH1 (12%), RUNX1 (12%), NRAS (11%), and TET2 (8%) mutations were frequently identified in our AML-MRC patient cohort. Of note, we observed more frequent DNMT3A mutations and higher blasts counts at the time of diagnosis in high MYC patients. These observations suggest that MYC oncoprotein may provide proliferative potential to MDS cells harboring DNMT3A mutations, leading to MDS to AML progression. The initially BM biopsy specimens at the time of MDS diagnosis were not available in many of the referred patients in our study. Therefore, we were unable to compare the MYC levels between MDS vs. AML-MRC in the individual patient and to determine whether increase of MYC expression contributes to MDS to AML progression. However, it is worthy of exploration in the future study. Additionally, on-going study to investigate the underlying molecular contribution of MYC oncoprotein in AML cell differentiation and survival will answer this question (data not published).
In conclusion, AML-MRC patients with high MYC expression have inferior OS outcome compared to low MYC patients. Further, multivariate analysis established that high MYC level is a poor prognostic factor in AML-MRC patients. These findings warrant further study of the prognostic impact of MYC expression in addition to MYC gene amplification or/and somatic mutations in AML patients, with larger numbers of patients having other somatic mutations or chromosomal abnormalities that have adverse outcomes.
Supplementary Material
Fig 2. Landscape of Concurrent Somatic Mutations and Cytogenetic Abnormalities in High vs. Low MYC Patients.
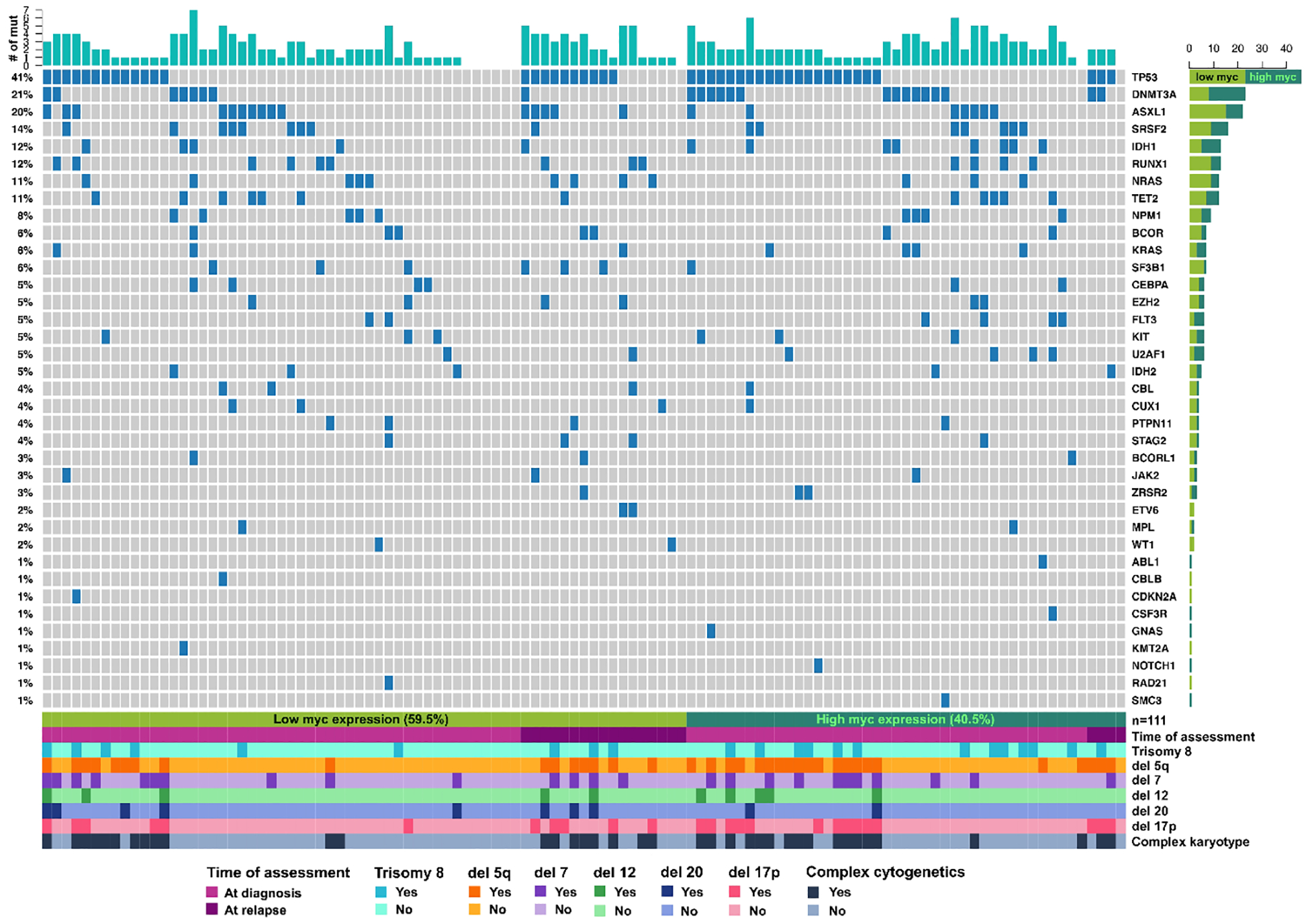
Individual column is somatic mutation data from individual patient. Each row is showing the presence of individual mutation in the patient cohort (name of mutation is described on the right side and rate of mutations are described on the left side). The presence of concurrent abnormal cytogenetics including trisomy 8, deletion 5q, monosomy 7, monosomy 12, deletion 20, and deletion 17p, and complex karyotypes are shown at the bottom of the plot.
HIGHLIGHTS.
AML-MRC patients express dynamic ranges of MYC oncoprotein.
High MYC expression is associated with inferior survival in the AML-MRC patients.
High MYC level is associated with higher rates of TP53 and DNMT3A mutations.
ACKNOWLEDGEMENTS
This work was supported by the American Society of Hematology (ASH) Research Training Award for Fellow (RTAF) grant (S.Y.), University of South Florida (USF) Graduate Medical Education (GME) grant (S.Y.), K08 CA237627 grant (S.Y.), Departmental Research Fund (L.Z.) and MDS TMA grant (R.S).
Footnotes
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
REFERENCES
- 1.Tefferi A, Vardiman JW. Myelodysplastic syndromes. N Engl J Med 2009; 361(19): 1872–85. [DOI] [PubMed] [Google Scholar]
- 2.Arana Yi CY, Kantarjian HM, Garcia-Manero G, et al. Comparing Outcomes of Patients with Secondary AML: Treatment-Related MDS/AML, AML Secondary to Myeloproliferative Neoplasms (t-MPN), and AML with Prior Malignancies. 2012; 120(21): 3557–. [Google Scholar]
- 3.Grimwade D, Hills RK. Independent prognostic factors for AML outcome. 2009; 2009(1): 385–95. [DOI] [PubMed] [Google Scholar]
- 4.Bejar R, Stevenson K, Abdel-Wahab O, et al. Clinical Effect of Point Mutations in Myelodysplastic Syndromes. New England Journal of Medicine 2011; 364(26): 2496–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papaemmanuil E, Gerstung M, Malcovati L, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013; 122(22): 3616–27; quiz 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med 2016; 374(23): 2209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindsley RC, Mar BG, Mazzola E, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 2015; 125(9): 1367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. New England Journal of Medicine 2016; 374(23): 2209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolouri H, Farrar JE, Triche T Jr, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nature Medicine 2017; 24: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tyner JW, Tognon CE, Bottomly D, et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018; 562(7728): 526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genomic and Epigenomic Landscapes of Adult De Novo Acute Myeloid Leukemia. 2013; 368(22): 2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol 2006; 16(4): 253–64. [DOI] [PubMed] [Google Scholar]
- 13.Dang CV. MYC on the path to cancer. Cell 2012; 149(1): 22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ott G, Rosenwald A, Campo E. Understanding MYC-driven aggressive B-cell lymphomas: pathogenesis and classification. 2013; 122(24): 3884–91. [DOI] [PubMed] [Google Scholar]
- 15.Johnson NA, Savage KJ, Ludkovski O, et al. Lymphomas with concurrent BCL2 and MYC translocations: the critical factors associated with survival. Blood 2009; 114(11): 2273–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sabò A, Kress TR, Pelizzola M, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 2014; 511: 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J, Woo AJ, Chu J, et al. A Myc Network Accounts for Similarities between Embryonic Stem and Cancer Cell Transcription Programs. Cell; 143(2): 313–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, Osdal T, Ho Y, et al. SIRT1 Activation by a c-MYC Oncogenic Network Promotes the Maintenance and Drug Resistance of Human FLT3-ITD Acute Myeloid Leukemia Stem Cells. Cell Stem Cell; 15(4): 431–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie S, Lin H, Sun T, Arlinghaus RB. Jak2 is involved in c-Myc induction by Bcr-Abl. Oncogene 2002; 21(47): 7137–46. [DOI] [PubMed] [Google Scholar]
- 20.Ohanian M, Rozovski U, Kanagal-Shamanna R, et al. MYC protein expression is an important prognostic factor in acute myeloid leukemia. Leukemia & Lymphoma 2018; 60(1): 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mughal MK, Akhter A, Street L, Pournazari P, Shabani-Rad M-T, Mansoor A. Acute myeloid leukaemia: expression of MYC protein and its association with cytogenetic risk profile and overall survival. 2017; 35(3): 350–6. [DOI] [PubMed] [Google Scholar]
- 22.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016; 127(20): 2391–405. [DOI] [PubMed] [Google Scholar]
- 23.Sallman DA, Komrokji R, Vaupel C, et al. Impact of TP53 mutation variant allele frequency on phenotype and outcomes in myelodysplastic syndromes. Leukemia 2016; 30(3): 666–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17(5): 405–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho JSL, Ma W, Mao DYL, Benchimol S. p53-Dependent Transcriptional Repression of c-myc Is Required for G1 Cell Cycle Arrest. 2005; 25(17): 7423–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sachdeva M, Zhu S, Wu F, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. 2009; 106(9): 3207–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.TCGA. Genomic and Epigenomic Landscapes of Adult De Novo Acute Myeloid Leukemia. 368(22): 2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


